Abstract
Type 2 diabetes mellitus is a major risk factor for cardiovascular disease and mortality. Uncontrolled type 2 diabetes mellitus results in a systemic milieu of increased circulating glucose and fatty acids. The development of insulin resistance in cardiac tissue decreases cellular glucose import and enhances mitochondrial fatty acid uptake. While triacylglycerol and cytotoxic lipid species begin to accumulate in the cardiomyocyte, the energy substrate utilization ratio of free fatty acids to glucose changes to almost entirely free fatty acids. Accumulating evidence suggests a role of miRNA in mediating this metabolic transition. Energy substrate metabolism, apoptosis, and the production and response to excess reactive oxygen species are regulated by miRNA expression. The current momentum for understanding the dynamics of miRNA expression is limited by a lack of understanding of how miRNA expression is controlled. While miRNAs are important regulators in both normal and pathological states, an additional layer of complexity is added when regulation of miRNA regulators is considered. miRNA expression is known to be regulated through a number of mechanisms, which include, but are not limited to, epigenetics, exosomal transport, processing, and posttranscriptional sequestration. The purpose of this review is to outline how mitochondrial processes are regulated by miRNAs in the diabetic heart. Furthermore, we will highlight the regulatory mechanisms, such as epigenetics, exosomal transport, miRNA processing, and posttranslational sequestration, that participate as regulators of miRNA expression. Additionally, current and future treatment strategies targeting dysfunctional mitochondrial processes in the diseased myocardium, as well as emerging miRNA-based therapies, will be summarized.
Keywords: metabolism, exosomes, epigenetics, miRNA processing, long noncoding RNA, diabetes
INTRODUCTION
Diabetes mellitus affects 29.1 million Americans and 25.9% of Americans age 65 yr and older. Adults with diabetes mellitus have two to four times the rate of mortality from heart disease compared with those without diabetes mellitus, and 68% of diabetic patients aged 65 yr and older die of heart disease (3, 4). The diabetic heart is metabolically characterized by insulin resistance, reduced cellular glucose import and oxidation, and increased mitochondrial fatty acid import and oxidation (131). This chronic alteration in energy substrate utilization and loss of the dynamic ability of the myocardium to metabolically adapt to its environment, such as upregulating anaerobic glycolysis during cardiac ischemia, initiates a pathological state termed diabetic cardiomyopathy (5, 92). Increased cardiac hypertrophy and fibrosis are structural changes observed in diabetic cardiomyopathy that are accompanied molecularly by increased oxidative stress, mitochondrial dysfunction, and cardiomyocyte apoptosis (28, 46, 69, 111, 173). Diabetic heart failure is the culmination of these pathologic insults that is characterized by severe contractile dysfunction of the myocardium (43).
The proteomic alterations of the heart in the diabetic state have been well characterized. Metabolically, the decrease in glucose import and oxidation in the diabetic cardiomyocyte is associated with decreased expression of insulin-stimulated glucose transporter 4 (GLUT4) and decreased activity of pyruvate dehydrogenase (PDH) (131). The increase in mitochondrial fatty acid import and oxidation is found in conjunction with decreased acetyl-CoA carboxylase activity, decreased malonyl-CoA concentration, increased carnitine palmitoyl transferase I activity, and increased β-hydroxyacyl-CoA dehydrogenase activity (131). Increased reactive oxygen species (ROS) generation in diabetic cardiomyopathy results from contributors such as excess fatty acid oxidation, increased NADPH oxidase activity, and uncoupled nitric oxide synthases (NOS) (69). What contributes to increased cardiomyocyte apoptosis in the diabetic setting is the decreased expression of the antiapoptotic protein Bcl-2 and increased expression of the proapoptotic protein p53 (108, 171). At the heart of energy substrate metabolism, ROS generation, and apoptosis lies the mitochondrion.
Although transcription factor regulation of gene expression, posttranslational modification, and substrate inhibition via negative feedback have shown to be foundational in regulating protein expression and activity, miRNA and epigenetic mechanisms of regulation have more recently emerged (6). miRNAs are single-stranded noncoding RNA molecules, ~22 nt long, that associate into a multiprotein RNA-induced silencing complex (RISC), which inhibits its target mRNA species from being translated into a functional protein (13). Differential expression of miRNAs have been characterized in a variety of cardiovascular conditions, including atherosclerosis, myocardial infarction, and heart failure to modulate processes, such as energy substrate metabolism, fibrosis, and cardiac remodeling (104, 126, 168). Nuclear- and, more recently, mitochondrial genome-encoded proteins, which function in the mitochondria, have been shown to be regulated by miRNA (12, 15, 36, 72). Because cardiomyocyte mitochondria are central to the pathogenesis of diabetic heart disease, the first section of this review will focus on miRNA regulation of key pathological pathways intersecting at the mitochondrion. The following section will focus on the role of regulation on the miRNA regulator, including epigenetic, exosomal, processing, and posttranscriptional control (Fig. 1). Finally, treatment strategies for cardiomyocyte energy substrate metabolism, pharmacological manipulation, and miRNA-based therapies for the treatment of diabetic heart disease will be discussed.
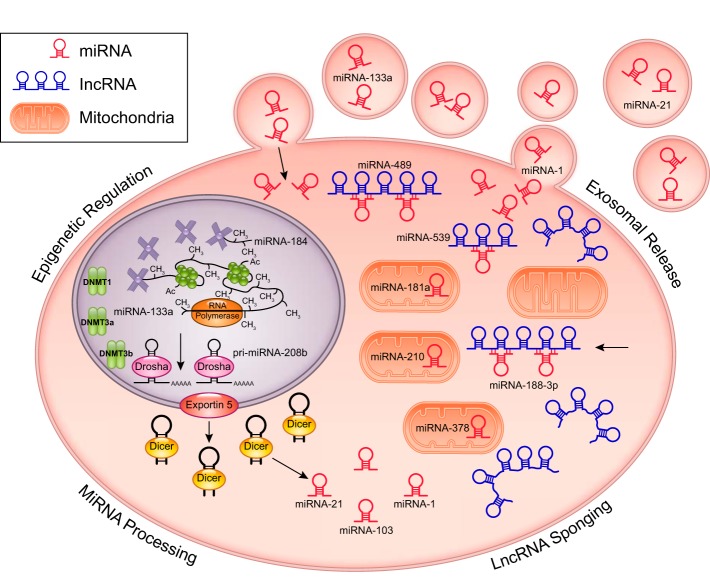
Fig. 1.
Regulation of miRNA regulators. The expression of miRNAs is controlled through multiple factors. Epigenetic regulation includes hypermethylation of the miRNA-184 CpG loci, pri-miRNA-208b tethering to increase enhancer of zeste homolog 2 binding, and miRNA interactions with DNA methyl transferase (DNMT)1, DNMT3a, and DNMT3b. Regulation of miRNA expression through miRNA processing is dependent on Dicer expression (miRNA-103) and protein-protein interactions with Dicer (miRNA-21). Other proteins, such as GTPase-activating protein-binding protein 1, can affect processing (miRNA-1). Long noncoding RNA (lncRNA) can regulate miRNA expression by acting as a “sponge,” sequestering native and exogenous miRNAs, such as miRNA-188-3p, miRNA-539, and miRNA-489. Also, exosomal release/transport of miRNA-1, miRNA-21, and miRNA-133a can alter miRNA expression.
miRNA REGULATION OF THE MITOCHONDRION IN THE DIABETIC HEART
miRNAs can be transcribed through two primary pathways, either a canonical or noncanonical pathway (61, 97, 156). In the canonical pathway, the pri-miRNA is transcribed from its existing exon and intron, containing one to several hairpin loop structures. After transcription, DiGeorge Syndrome critical region 8 (DGCR8) and Drosha, known as the microprocessing complex, cut the pri-miRNA in the nucleus, resulting in a shorter (~65 nt) pre-miRNA (57, 63). The pre-miRNA is then exported through exportin-5 and RanGTP (161). Finally, in the cytoplasm, Dicer edits the pre-miRNA, resulting in the mature miRNA (19–22 nt), which can then associate with the RISC. In the noncanonical pathway, hairpin structures within introns can be spliced out, resulting in pre-miRNAs that are referred to as mirtrons (156). After splicing, the mirtrons proceed through the same pathway as canonical miRNA to be exported from the nucleus.
After export from the nucleus, miRNAs have been implicated in regulating the expression of proteins essential to mitochondrial health. Recently, miRNAs have been found to affect mitochondrial function through targeting pathways, including energy substrate metabolism, ROS generation, and apoptosis. This section will focus on miRNA regulation of these processes within the cardiomyocyte in the context of the diabetic heart (Table 1).
Table 1.
miRNA regulation of mitochondrial pathways in the diabetic heart
| miRNA | Environmental Regulation | Expression in the Diabetic Heart | miRNA Target | miRNA Target Function | Diabetic Model | Source |
|---|---|---|---|---|---|---|
| let-7 | Pro | Upregulated | Insulin recptor, insulin receptor substrate 2 | Insulin signaling | STZ, ischemia-reperfusion | 9, 15, 77 |
| miRNA-195 | Upregulated | Sirtuin 1 | Insulin signaling | STZ; db/db | 1, 6, 156 | |
| miRNA-216a | Upregulated | Caveolin 2 | Insulin signaling | Human diabetic; ischemic heart failure | 50, 74 | |
| miRNA-199a-3p | Upregulated | Caveolin 2 | Insulin signaling | STZ | 3, 11, 39 | |
| miRNA-133a | Epi, Exo | Downregulated | Testicular receptor 4 | Fatty acid transport | STZ | 27, 95 |
| miRNA-210 | Upregulated | Iron-sulfur cluster assembly protein 1/2 | ETC Complex I | Human diabetic; ischemic heart failure | 20, 50 | |
| miRNA-141 | Upregulated | Slc25a3 | Mitochondrial phosphate carrier | STZ | 12 | |
| miRNA-378 | Upregulated | ATP6 | ATP production | STZ | 66 | |
| miRNA-29a | Downregulated | Peroxisome proliferator-activated receptor-γ coactivator-1α | Fatty acid oxidation | STZ | 39, 81 | |
| miRNA-30c | Downregulated | p53 | Apoptosis | Human diabetic; rat diabetic cardiomyopathy; high glucose-treated H9C2 | 100 | |
| miRNA-181a | Downregulated | p53 | Apoptosis | Human diabetic; rat diabetic cardiomyopathy; high glucose-treated H9C2 | 100 | |
| miRNA-30d | Upregulated | FOXO3a | Survival | STZ; high glucose-treated H9C2 | 8, 14, 78 | |
| miRNA-34a | Upregulated | Bcl-2 | Survival | High glucose-treated H9C2 | 154 | |
| miRNA-1 | Exo, Pro | Upregulated | Bcl-2 | Survival | STZ; high glucose-treated H9C2 | 9, 14, 70 |
The differential expression of each miRNA species in diabetes is indicated along with its protein target. The process or pathway the protein participates is also shown. The models in which the reported observations have been made are noted. Environmental regulation refers to the participation of the miRNA in either epigenetic (Epi), exosomal transport (Exo), or miRNA processing (Pro) pathways known to influence the diabetic cardiovascular system. STZ, streptozotocin.
miRNA REGULATION OF ENERGY SUBSTRATE METABOLISM IN THE DIABETIC HEART
miRNA regulation of insulin signaling and glycolysis.
The downregulation of insulin signaling components in the diabetic heart is influenced by miRNA regulation. Li et al. (84) showed that let-7 miRNA was overexpressed in the myocardium of streptozotocin (STZ)-induced diabetic rats, whereas the protein expression of insulin-like growth factor 1 receptor (IGF-1R), insulin receptor (IR), and GLUT4 were significantly lower. Interestingly, they found that inhibition of let-7 via the administration of let-7 antimiR conferred cardioprotection against ischemia-reperfusion injury through increased expression of phosphorylated Akt and phosphorylated mammalian target of rapamycin (84). IR and IR substrate 2 (IRS2) have been validated as direct targets of let-7, and the observation of a normalization of IGF-1R, IR, and GLUT4 expression upon administration of let-7 antimiR supports the assertion that let-7 targets insulin signaling and the glucose transport pathway (84, 175). Furthermore, Greco et al. (56) found that miRNA-216a was overexpressed in failing human hearts of patients with and without diabetes mellitus and that its expression was negatively correlated with left ventricular ejection fraction. miRNA-216a targets and represses caveolin 2, a scaffolding protein and substrate of the IR that helps to recruit IRS-1 to the IR and propagate insulin signaling (56, 81). The role of miRNA-216a overexpression in regulating intermediary metabolism in heart failure patients with and without diabetes is unanswered and of much interest to the field. miRNA-199a-3p has been observed to be overexpressed in STZ-induced diabetic mice, and it also has a validated target of caveolin 2 (44, 121). Zheng et al. (172) showed that miRNA-195 is overexpressed in STZ-induced and db/db mouse hearts as well as in cardiomyocytes isolated from db/db mice, whereas sirtuin 1 (Sirt1) protein levels were significantly decreased. Inhibition of miRNA-195 increased Sirt1 expression in diabetic mice (172). Sirt1 is a downstream effector of the insulin signaling pathway, indicating that miRNAs not only serve to fine tune IR and IRS2 but also regulate the expression of proteins further along the insulin signaling cascade (41). Altogether, miRNA targeting of insulin signaling components of the diabetic heart may help explain the proteomic alterations of this pathway observed in the diabetic condition.
miRNA REGULATION OF FATTY ACID TRANSPORT, FATTY ACID OXIDATION, TRICARBOXYLIC ACID CYCLE, ELECTRON TRANSPORT CHAIN, AND ATP PRODUCTION
The upregulation of fatty acid import and oxidation in the diabetic heart is impacted by miRNA modulation. Chen et al. (30) demonstrated that miRNA-133a was significantly decreased in the hearts of STZ-induced diabetic mice and that miRNA-133a was a direct regulator of testicular receptor 4 (TR4), which induces the expression of lipid importer CD36 and promoter of lipid accumulation through solute carrier family 27 member 1 (Slc27a1/FATP1) (102). This could help to explain the increase in CD36 expression in cardiac tissue of STZ-induced diabetic rats that has been reported (101). Peroxisome proliferator-activated receptor (PPAR)-α is the primary transcription factor responsible for the fatty acid oxidation gene expression program (117). PPAR-γ coactivator (PGC)-1α, an essential coactivator of PPAR-α and inducer of medium-chain acyl-CoA dehydrogenase, has been found to be directly regulated by the miRNA-29 family (miRNA-29a-c) (86, 88). Interestingly, miRNA-29a has been found to be decreased in STZ-induced diabetic hearts, which may help explain the induction of PPAR-α and increased fatty acid oxidation (44). Greco et al. (56) observed that miRNA-210 is upregulated in failing diabetic human hearts compared with failing nondiabetic human hearts. miRNA-210 is a known negative regulator of iron-sulfur cluster assembly scaffold protein ISCU1/2, which play essential roles in the function of aconitase and complex I of the electron transport chain (23). Baseler et al. (15) observed a significant upregulation of miRNA-141 in STZ-induced diabetic mice and found that miRNA-141 is a direct negative regulator of solute carrier family 25 member 3 (Slc25a3), which is essential for inorganic phosphate import into the mitochondrial matrix and thus ATP synthesis. It has also been shown that miRNA-378 was overexpressed in interfibrillar mitochondria (IFM), mitochondria located between the myofibrils, of STZ-induced diabetic mice and that miRNA-378 is a direct negative regulator of ATP synthase F0 component ATP6 (72). These observations provide compelling evidence that miRNAs play a critical role in the regulation of energy metabolism in the diabetic heart.
RECIPROCAL REGULATION OF miRNA AND ROS IN THE DIABETIC HEART
Oxidative stress has been shown to be a result of, and cause of, miRNA dysregulation. Saito et al. (113) demonstrated that glucose fluctuations in STZ-induced diabetic rats increased cardiomyocyte miRNA-200c and miRNA-141 levels, with an accompanying increase in ROS generation and upregulation of NADPH oxidase and thioredoxin-interacting protein, while decreasing catalase and superoxide dismutase activities. As glycemic control has been shown to attenuate some of the peripheral diabetic symptoms, diabetic cardiomyopathy progresses even after blood glucose normalization. One possible explanation for this finding comes from a study showing that even after glycemic control in STZ-induced diabetic mice, miRNA dysregulation of many myocardial damage pathways, including oxidative stress (dysregulated miRNA-221, miRNA-146a, miRNA-34a, miRNA-210, miRNA-19b, miRNA-27a, miRNA-155) persisted (33). Because of its upregulation in cardiac fibrosis, hypertrophy, and oxidative stress in both diabetic and insulin-treated groups compared with control, miRNA-125b seems to have broad-ranging effects in mediating diabetic cardiac dysfunction (33). Excess ROS production in STZ-induced diabetic cardiomyopathy has been linked to decreased expression of miRNA-499, miRNA-1, miRNA-133a, and miRNA-133b, as treatment with the antioxidant N-acetylcysteine restored the levels of these miRNA species to normal (162). The authors further demonstrated that Junctin, a key component of cardiomyocyte Ca2+ handling, is a direct target of miRNA-1 and is consequently upregulated in the diabetic heart, which has previously been shown to impair cardiac relaxation and induce cardiac hypertrophy and arrhythmia (162). The mechanisms by which miRNAs regulate ROS production and by which oxidative stress influences differential expression of miRNAs to regulate a variety of pathophysiological pathways in the diabetic heart remains a relatively unexplored area of research.
miRNA REGULATION OF APOPTOSIS IN THE DIABETIC HEART
The diabetic heart, in the setting of diabetic cardiomyopathy and diabetic heart failure, experiences an increased rate of cardiomyocyte apoptosis. This increase in apoptosis is determined by a variety of factors, including lipotoxicity, glucotoxicity, and increased oxidative stress. miRNA-34b is upregulated in diabetic heart failure, and it has been shown to promote apoptosis by acting as an important downstream effector of p53 (16, 56, 66). miRNA-30c and miRNA-181a have been shown to be downregulated in diabetic patients, a diabetic cardiomyopathy rat model, and high-glucose-treated cardiomyocytes (108). p53 is a validated target of miRNA-30c and miRNA-181a, and decreased levels of these miRNA species have been correlated with increased p53 pathway activation of hypertrophy and apoptosis (108). Upregulation of miRNA-30d in high glucose-treated cardiomyocytes and diabetic rats has also been shown to play an important role in cardiac mitochondria-implicated pyroptosis, where it has been verified to directly target and inhibit forkhead box O3 (FOXO3a), with downstream effects including decreased expression of apoptosis repressor with caspase recruitment domain (ARC) and upregulation of caspase-1 (85, 163). Zhao et al. (170) showed that high glucose-treated rat cardiomyocyte H9C2 cells have enhanced expression of miRNA-34a, decreased expression of the miRNA-34a target antiapoptotic protein Bcl-2, and increased apoptosis. Bcl-2 has been found to be targeted and downregulated by overexpression of miRNA-195 in a mouse model of STZ-induced type 1 diabetes, and inhibition of miRNA-195 reduced ROS production and inhibited apoptosis (172). Furthermore, Yu et al. (164) showed that rat cardiomyocyte H9C2 cells treated with high glucose had increased miRNA-1 expression, downregulation of the miRNA-1 target IGF-1, increased cytochrome c release, and increased apoptosis. To add validity to the importance of miRNA-1 in high glucose-mediated cardiomyocyte apoptosis, it has been found that high glucose in vitro and in vivo increases miRNA-1/miRNA-206 expression precipitating posttranslational modification of heat shock protein 60, a protein involved in protection against diabetic myocardial injury (119). Furthermore, Katare et al. (76) found upregulation of miRNA-1, downregulation of the miRNA-1 targets protooncogene serine/threonine-protein kinase (Pim-1) and Bcl-2 (antiapoptotic proteins), and increased proapoptotic caspase-3 activity in the hearts of STZ-induced type 1 diabetic mice.
miRNA impact on energy substrate metabolism, ROS interactions within the cell, and apoptotic pathways all show the significance of the interaction between miRNAs, mitochondria, and the diabetic heart (Fig. 2). With the significant impact to cellular and physiological health from miRNAs in the diabetic heart, a more fundamental question arises to address the driving mechanisms behind changing miRNA expression.
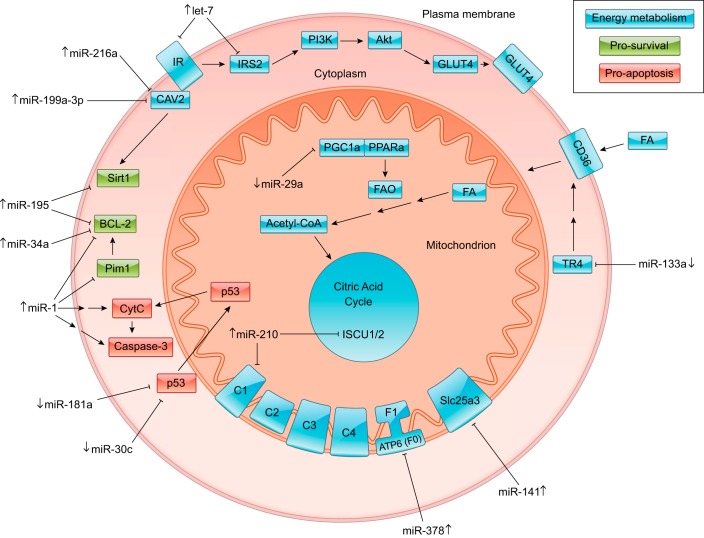
Fig. 2.
miRNA influence on signaling pathways in the diabetic heart. miRNA regulation of cardiomyocyte energy metabolism (blue), survival (green), and apoptosis (red) in the diabetic heart is shown. The up or down arrows beside each miRNA species indicate its upregulation or downregulation in the diabetic heart, respectively (Table 1). The marked line connecting each miRNA with its target demonstrates that the miRNA inhibits its target. Subcellular components have been labeled as follows: plasma membrane (yellow), cytoplasm (green), and mitochondrion (blue). CAV2, caveolin 2; IR, insulin receptor; IRS2, insulin receptor substrate 2; PI3K, phosphatidylinositol 4,5-bisphosphate 3-kinase; GLUT4, glucose transporter type 4; FA, fatty acid; TR4, testicular receptor 4; FAO, fatty acid oxidation; PPARa, peroxisome proliferator-activated receptor-α; PGC1a, peroxisome proliferator-activated receptor-γ coactivator-1α; ISCU1/2, iron-sulfur cluster assembly proteins; C1−C4, electron transport chain complexes 1–4; F0, ATP synthase subunit F0; F1, ATP synthase subunit F1; Slc25a3, mitochondrial phosphate carrier protein; CytC, cytochrome c; Sirt1, sirtuin 1.
miRNA IMPORT INTO THE MITOCHONDRION
What still remains a highly relevant, and hotly debated, topic is the process involved in miRNA import into the mitochondrion. Although it has been hypothesized that some miRNAs could be transcribed through the mitochondrial genome (130), miRNA import is predominantly favored as the mechanism for miRNA accumulation in the mitochondrion. Research has demonstrated the presence of miRNA within the mitochondrion (9, 37, 78, 124, 169) and even how fluctuating concentrations within the organelle contributes toward the development of pathologies (35, 36, 45, 122, 129). Two currently proposed mechanisms of miRNA import into the mitochondria include the direct movement across the mitochondrial membrane through a chaperone, Argonaut 2 (AGO2) (36, 169), and diffusion through small RNA import machinery, polynucleotide phosphorylase (PNPase) (122).
Both Das et al. (36) and Zhang et al. (169) have shown that AGO2 is present in the mitochondria and that cross-linking immunoprecipitation revealed associations with miRNAs. Although it has been suggested that AGO2 could have a localization sequence on its NH2 terminus for mitochondrial targeting (10), no definitive research has validated the precise mechanisms involved in AGO2/miRNA mitochondrial import. A new mechanism proposed for mitochondrial miRNA import includes the mitochondrial inner membrane protein PNPase and potential associations with AGO2. Wang et al. (150, 151) demonstrated the importance of PNPase in facilitating the import of RNase P, 5S rRNA, and mitochondrial RNA processing (MRP) RNAs, suggested through stem-loop recognition. Recently, our group has shown that the regulation of PNPase can alter miRNA-378 levels within the mitochondrion (122), indicating that the function of PNPase may extend to other small RNA species. The expression of PNPase is also decreased during diabetes mellitus. Although this novel mechanism of miRNA import has not been fully characterized mechanistically, it provides a basis for future research.
REGULATION OF miRNAS AFFECTING THE DIABETIC AND NONDIABETIC CARDIOVASCULAR SYSTEM
As described above, many cardiovascular diseases, including diabetic cardiomyopathy and diabetic heart failure, have unique miRNA expression signatures. These regulatory molecules have an impact in modulating cellular pathways in health and disease. Although miRNAs are highly active in posttranscriptional repression and degradation with complex downstream effects, the miRNA expression landscape is just as complexly regulated by environmental stimuli, including mechanisms of epigenetic modulation, exosomal transport between cells and tissues, processing of pri-miRNA and pre-miRNA pathways, long noncoding RNA (lncRNA) “sponging,” and many others (Table 2).
Table 2.
Regulation of miRNA in the heart and circulatory system
| miRNA | Type of Regulation | Pathway | Expression in the Heart or Circulatory System | Disease Model | Source |
|---|---|---|---|---|---|
| miRNA-133a | Epigenetic | Class I and IIb histone deacetylases decrease the expression of miRNA-133a | Downregulated | Mouse heart: transverse aortic constriction model | 101 |
| miRNA-184 | Epigenetic | DNA hypermethylation through DNMT1 | Downregulated | HL-1 cardiomyocytes: arrhythmogenic cardiomyopathy model | 54 |
| pri-miR-208b | Epigenetic | Pri-miRNA tether to enhancer of zeste homolog 2 | Upregulated | Mouse heart: transverse aortic constriction model | 88 |
| *miRNA-375 | Epigenetic | CpG island methylation | Upregulated | Human plasma | 21, 120 |
| *miRNA-145 | Epigenetic | Promoter hypomethylation | Upregulated | Human saphenous vein smooth muscle | 102 |
| *miRNA-125b | Epigenetic | Suppressor of variegation 3-9 homolog 1 3′-untranslated region binding, decreasing histone 3 lysine 9 trimethylation expression | Upregulated | Mouse microvascular smooth muscle cells (db/db model) | 133 |
| *miRNA-101 | Epigenetic | Binding to enhancer of zeste homolog 2, decreasing histone 3 lysine 27 trimethylation expression | Upregulated | Human fetal embryonic stem cells of the umbilical cord vein | 45 |
| *miRNA-133a | Epigenetic | Binding to DNMT1, DNMT3A, and DNMT3B | Downregulated | Ins2+/− Akita diabetic mice and HL-1 cardiomyocytes | 24 |
| miRNA-1 and miRNA-133a | Exosomal transport | Potential biomarker for cardiomyocyte death, released from the injured myocardium | Upregulated release into serum | Human serum from acute myocardial infarction patients | 73 |
| miRNA-142–3p | Exosomal transport | Endothelial barrier integrity breakdown through RAB11 family interacting protein 2 interaction | Upregulated release into serum | Human serum from heart transplant surgery patients | 119 |
| miRNA-21 | Exosomal transport | Binds programmed cell death 4, offering cardioprotection | Upregulated release into media | Mouse cardiac progenitor cells under oxidative stress | 142 |
| miRNA-30a | Exosomal transport | Hypoxia-inducible factor-1α upregulates miRNA-30a, potentially offering cardioprotection from autophagy | Upregulated release into serum | Human serum from acute myocardial infarction patients | 144 |
| miRNA-143 | Exosomal transport | Upregulation of miRNA-143-3p compared with miRNA-143-5p | Upregulated release into media | Human pulmonary artery smooth muscle cells from pulmonary arterial hypertension patients | 37 |
| miRNA-194 | Exosomal transport | Contribution to the p53 response and potential biomarker for ischemic heart failure | Upregulated release into serum | Human serum from acute myocardial infarction patients | 89 |
| *miRNA-1 and miRNA-133a | Exosomal transport | Levels positively correlated with predicting myocardial steatosis in type 2 diabetic patients | Upregulated release into media | HL-1 cardiomyocyte: lipid-loaded preconditioning | 35 |
| *miRNA-7 | Exosomal transport | Changes in serum levels were not significantly contributed by exosomes | Upregulated in serum, not changing in exosomes | Human serum from patients with microvascular complications | 135 |
| *miRNA-455, miRNA-29b, miRNA-323-5p and miRNA466 | Exosomal transport | Downregulation of matrix metalloproteinase-9 | Upregulated release into serum | Mouse: db/db model with exercise | 22 |
| *miRNA-320 | Exosomal transport | Targets insulin-like growth factor-1, heat shock protein 20, and Ets2, producing an antiangiogenic effect in cardiac endothelial cells | Upregulated release from cardiomyocytes | Rat cardiac endothelial cell and cardiomyocyte coculture: Goto-Kakizaki model | 140 |
| miRNA-21 | miRNA processing | DICER1 and phosphorylated SMAD-2 interactions increase mature miRNA-21 expression | Upregulated | Mouse: transverse aortic constriction model | 48 |
| miRNA-103 | miRNA processing | Downregulation of Dicer leads to decreased NF-κB activation through decreased miRNA-103/Krüppel-like factor 4 binding | Downregulated | Mouse: high-fat diet apolipoprotein E knockout model | 58 |
| miRNA-1 | miRNA processing | GTPase-activating protein (SH3 domain)-binding protein 1 binds the stem loop of miRNA-1, decreasing its expression | Downregulated | Rat cardiomyocyte: transverse aortic constriction model | 61 |
| *let-7a, miRNA-130, miRNA-142–3p, miRNA-148, miRNA-338, miRNA-345–3p, miRNA-384–3p, miRNA-433, miRNA-450, miRNA-451, miRNA-455, miRNA-494, miRNA-499, miRNA-500, miRNA-542–3p, miRNA-744, and miRNA-872 | miRNA processing | Dicer was shown to be downregulated, although some miRNAs showed increased expression | Upregulated | Ins2+/− Akita diabetic mice | 23 |
| miRNA-188-3p | lncRNA sponging | Increased expression of autophagy-promoting factor lncRNA decreases miRNA-188-3p levels while increasing autophagy-related protein 7 | Downregulated | Mouse cardiomyocytes: anoxia/reoxygenation model | 137 |
| miRNA-539 | lncRNA sponging | Increased expression of cardiac apoptosis-related lncRNA decreases miRNA-539 levels while increasing prohibitin 22 | Downregulated | Mouse cardiomyocytes: anoxia/reoxygenation model | 139 |
| miRNA-489 | lncRNA sponging | Increased expression of cardiac hypertrophy-related factor lncRNA decreases miRNA-489 levels while increasing myeloid differentiation primary response gene 88 | Downregulated | Mouse: ANG II treatment | 138 |
Each miRNA that is regulated or involved in mechanisms altering miRNA expression is listed for the heart and/or circulatory system. The type of regulation, the specific pathway involved, and the expression change of the miRNA are included. *Use of a diabetic model for the relevant miRNA. DMNT, DNA methyltransferase; lncRNA, long noncoding RNA.
REGULATION OF miRNAS ALTERING THE EPIGENOME OF THE CARDIOVASCULAR SYSTEM
What are epigenetics?
The most described forms of epigenetic regulation consist primarily of two transient alterations: DNA methylation and histone modification (21). DNA methylation proceeds through the addition of a 5′-methyl group, which occurs on cytosine nucleotides, where it is added by DNA methyltransferases [DNMTs (DNMT1, DNMT3a, and DNMT3b)] (17). Most often, this methyl group addition will occur at CpG islands (paired cytosine and guanine nucleotides within a DNA sequence). The addition of methyl groups to a gene sequence attenuates transcription by decreasing the propensity of transcription factors and other transcription initiation proteins to bind to the sequence (74).
Histone modifications, such as acetylation, methylation, and phosphorylation, change the confirmation of chromatin by either relaxing or condensing the DNA/protein structure (14). The processes involved in histone modifications include histone methyltransferases (HMTs), which generally condense chromatin, and histone acetyl transferases (HATs), which primarily relax chromatin (174). The interplay of DNA methylation and histone modifications can significantly alter gene expression, specifically in diseases such as type 2 diabetes mellitus, such that the epigenome can be completely reshaped (147).
Role of epigenetics in altering miRNA expression.
Of the variety of mechanisms known to regulate miRNA expression, the beginning of the regulatory chain, starts at transcription. miRNAs can either be intergenic, found within the introns of genes, or intragenic, found within the coding region of a gene, and can assume multidirectional or overlapping features with the gene (32, 94). At each gene locus containing a miRNA, the expression of the miRNA can be governed both by the DNA methylation of a promoter/intergenic/intragenic region or through histone confirmations around the reading frame (115). Researchers have begun to investigate how miRNAs can be controlled epigenetically in a variety of pathological conditions. Conversely, miRNAs can be implemented as a direct mechanism for altering cellular epigenetics. The epigenetic machinery of the cell contains multiple forms of epigenetic regulation: polycomb repressor complex 1 (PRC1), PRC2, histone deacetylases (HDACs), HATs, HMTs, and DNMTs (70, 115).
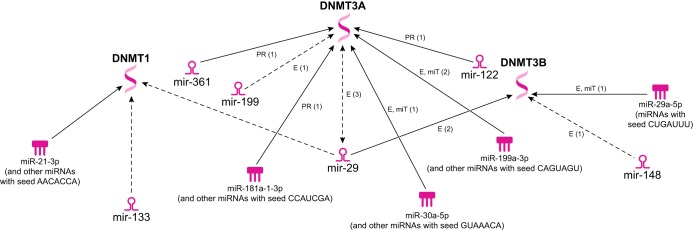
miRNAs have been found to directly affect major constituents of the epigenetic machinery, and many miRNAs have been identified with changed expression levels through epigenetic regulation. Ingenuity Pathway Analysis can be implemented as a protein/miRNA ontology system through the use of known molecular association data combined with predictive software analyzing sequence homology. Figure 3 shows how mature miRNAs are known to interact with the DNMT machinery and also reveals miRNAs associated with the mitochondrion of the diabetic heart that theoretically could interact with DNMTs through their seed sequence regions.
Fig. 3.
Predicted interactions of miRNA with DNA methyltransferase (DNMT)1. Ingenuity Pathway Analysis provides potential areas for future epigenetic/miRNA analyses. The predicted connections include mature miRNAs and miRNA seed sequences, which have been suggested to interact with DNMT1, DNMT3a, or DNMT3b. miRNAs with listed seed sequences can account for one or many miRNAs that contain similar regions suggested to promote protein/miRNA or mRNA/miRNA interactions with DNMTs.
In human femoral artery atherosclerotic plaques, Aavik et al. (1) measured global DNA methylation levels and found hypomethylation of the genome and that two-thirds of the genes exhibited a significant, differential methylation pattern compared with normal mammary artery. One of the major findings was the 14q32 locus being significantly hypomethylated, which contains miRNA-127, miRNA-136, miRNA-410, miRNA-431, miRNA-432, and miRNA-433 (1). The authors suggest that this epigenetic paradigm in atherosclerotic plaques is a potential mechanism for initiating the progression of the lesions. In the heart, during overload-induced fibrosis through transverse aortic constriction, class I and IIb HDACs were shown to negatively impact the heart through the regulation of miRNA-133a (109). HDACs are known to increase in a pressure overload-induced model, and by introducing HDAC inhibitors to overload-induced mice, miRNA-133a expression was restored and functional deficits were ameliorated.
Arrhythmogenic cardiomyopathy has been linked to mutations in specific genes, including plakophilin 2 (PKP2) (7, 144). A PKP2 knockdown model in HL-1 cardiomyocyte cells was used to evaluate miRNA expression, revealing 59 differentially regulated miRNAs (60). Of these differentially regulated miRNAs, miRNA-184 showed the most significant downregulation. The authors suggested that miRNA-184 expression is controlled through DNA hypermethylation of its gene loci through DNMT1. A newly proposed mechanism of miRNA/epigenetic interaction suggests that pri-miRNAs can be used as tethers to recruit histone modification proteins to specific DNA sites (95). Enhancer of zester homolog-2 (EZH2) was shown to bind to the bidirectional promoter at the myosin heavy chain (MHC) genes in the heart and promote a switch from α-MHC to antisense β-MHC. Through RNA-chromatin immunoprecipitation experiments, the authors demonstrated the capacity for pri-miR-208b to tether specific genomic sites and recruit chromatin modification machinery.
Impact of epigenetics and miRNA during diabetes mellitus.
With the progression toward type 2 diabetes mellitus (i.e., insulin resistance, increased adiposity, increased body mass index), changes in the epigenome can be observed. A patient cohort (n = 10,261) revealed that genome-wide changes in DNA methylation in blood were linked to an increased body mass index (147). The resulting differential methylation was found to encompass genes involved in lipid metabolism, inflammatory signaling, and substrate transport. The group also found that changes in genome-wide DNA methylation can be used as a predictive measure for the development of type 2 diabetes mellitus. Methylation of genes involved in the progression of type 2 diabetes mellitus also varies between ethnicities (52, 79, 99), with Indian Asians showing increased levels of DNA methylation at ATP-binding cassette subfamily G member 1, phosphoethanolamine/phosphocholine phosphatase, suppressor of cytokine signaling 3, sterol regulatory element binding transcription factor 1, and thioredoxin-interacting protein. The increased methylation of these genetic loci in the Indian Asian population correlates to the increased risk of developing type 2 diabetes mellitus (22).
Pheiffer et al. (103) found that miRNA expression in type 2 diabetes mellitus can be controlled through DNA methylation. Specifically, this group established the importance of intergenic regions in controlling miRNA expression and showed that most differentially regulated peaks between diabetic and nondiabetic groups were found in the intergenic regions, as opposed to the promoter or intragenic regions. These findings raise a number of new questions concerning miRNAs and epigenetics, such as how does epigenetic regulation occur in a specific pathology, such as diabetes mellitus, or tissue, such as the heart, and how can therapeutics take advantage of these processes? In plasma from type 2 diabetic patients, miRNA-375 was increased (133). Furthermore, miRNA-375 has 17 different CpG islands within or surrounding the DNA loci, in which some of these regions were differentially regulated between diabetic and control populations. Other studies focusing on two different populations in Chinese, Han, and Kazak people revealed differences in the methylation pattern of miRNA-375 (24). The authors postulated that this may be a potential mechanism for the increased prevalence of type 2 diabetes mellitus in the Han population.
The role that miRNAs and epigenetics play in defining pathological states, such as diabetes mellitus, and how this interaction occurs in a variety of tissue types has been well studied. What remains to be fully understood is the impact of the epigenetic/miRNA interplay within the diabetic heart and cardiovascular system. In the saphenous vein of human diabetic and nondiabetic patients, smooth muscle cells were cultured, and miRNA-145 promoter methylation was measured (110). Expression of miRNA-145 was increased in the diabetic phenotype, but highly variable CpG island methylation between groups did not allow for any overall significant changes between groups. However, specific CpG loci were shown to exhibit hypomethylation, irrespective of group. Microvascular smooth muscle cells (MVSMCs) from db/db and control mice were used to assess the effects of miRNA-125b, which is overexpressed in the diabetic phenotype (146). miRNA-125b was validated to bind to the suppressor of variegation 3-9 homolog 1 3′-untranslated region controlling the expression of histone 3 lysine 9 trimethylation (H3K9me3). Increased miRNA-125b expression resulted in less restrictive H3K9me3 marks, ultimately increasing the expression of inflammatory genes.
Gestational diabetes mellitus can affect the health and future function of the growing progeny. In human fetal embryonic stem cells of the umbilical cord vein (HUVECs), EZH2 was decreased in mothers with gestational diabetes mellitus, whereas miRNA-101 was increased (51). EZH2 is responsible for the trimethylation of histone 3 lysine 27 (H3K27me3). EZH2 is also directly targeted by miRNA-101, resulting in decreased H3K27me3 in HUVECs from gestational diabetic mothers and subsequent altered gene transcriptional patterns. Through the use of Ins2+/− Akita diabetic mice and HL-1 cardiomyocytes, Chavali et al. (27) defined the role of miRNA-133a in regulating DNMTs. The group found that DNMT1, DNMT3a, and DNMT3b were inversely correlated to miRNA-133a (i.e., silencing of miRNA-133a induced overexpression of the DNMTs, whereas expression of miRNA-133a inhibited DNMT expression). In the Akita model, miRNA-133a is decreased, with DNMT1 and DNMT3b being induced. Through involvement in both the processes of DNA methylation and histone modifications, the unique miRNA/epigenetic signature involved in the progression of a pathology, such as diabetes mellitus, could be an invaluable predictor of cardiac function and cardiovascular health.
EXOSOMAL RELEASE, TRANSPORT, AND UPTAKE OF miRNA IN THE CARDIOVASCULAR SYSTEM
Exosomes and exosomal cargo.
Exosomes are cell-derived vesicular carriers used for cell signaling and communication and have been implicated in numerous disease states, including diabetic cardiomyopathy (112) and various cancers (8). Generally, exosomes range in size from 50 to 100 nm (107, 127, 137), but small discrepancies in size are common in the literature. The central role of exosomes is the delivery of various cargo types, including lipids, nucleic acids, proteins, RNAs, and miRNAs to different cells (18, 137, 140, 142). Exosomes transport cargo, transmitting a unique biochemical signal from the host cell to a specific target cell or tissue or, more systemically, by the bloodstream. Exosomal cargo is of importance due to the specificity of the cargo loaded into each vesicle by the host cell. This cargo is then delivered to targeted acceptor cells, and the exosomes release their protein and lipid cargo to activate downstream signaling events or simply deliver genetic material, including DNA and various RNAs (128). Secretion of exosomes occurs via multiple mechanisms, such as release upon exocytic fusion of multivesicular bodies with the plasma membrane and miRNA release through a ceramide-dependent mechanism (140). The different pathways available for exosome secretion depend on the cell types (137) and, currently, a lack of classification of the mechanisms of exosome biogenesis exists.
Exosomal transport of miRNA.
The transport of miRNAs via exosomes can directly impact cellular dynamics. Exosomal transport in the immune system through macrophages has also proven to be an important mechanism for changing cellular dynamics (2, 38, 71, 136), specifically through the regulation of inflammation during cardiac injury (149). This section will focus explicitly on the regulation of exosomal transport of miRNA within the heart and circulatory system.
miRNA-1 and miRNA-133a are increased in the serum of patients who have suffered from an acute myocardial infarction (80). In a mouse model of myocardial infarction, the infarcted and peri-infarcted tissue displays a significant reduction of miRNA-1 and miRNA-133a. Through measuring the exosomal release of miRNA-133a in dead, H9c2 cardiomyoblasts, the authors suggested that the injured myocardium releases miRNA-133a and that this marker may be used in the serum to account for cardiomyocyte death. Sukma Dewi et al. (132) measured the miRNAs of exosomes taken from the serum of patients who underwent heart transplant surgery, both with and without experiencing acute cardiac allograft rejection. These authors found miRNA-142-3p to be significantly increased in the exosomes. When the exosomes from the serum were used to treat endothelial cells, miRNA-142-3p also increased in the endothelial cells. After acute cardiac allograft rejection, the authors concluded that exosomes in the serum can affect endothelial barrier integrity through the interaction of miRNA-142-3p with RAB11 family interacting protein 2. The capacity for exosomes released from the heart to affect the systemic circulation and peripheral tissues has been explored (48). When serum was taken from patients before and after undergoing coronary artery bypass grafting, the authors found an increase in the total plasma concentration of exosomes as well as increased miRNA cargo. They observed that high-sensitivity cardiac troponin I was positively correlated with the plasma expression of total exosomes.
Cardiac progenitor cells (CPCs) were used to understand how exosomal release from cells can contribute toward alleviation of oxidative stress (157). During oxidative stress, it has been shown that CPCs released more miRNA-21, which targets the protein programmed cell death 4, ultimately reducing the cleaved version of caspase-3. These authors suggested that CPCs may offer cardioprotection during oxidative stress through release of exosomal miRNA-21 to surrounding tissue. After acute myocardial infarction, exosomes isolated from the serum of patients contained increased levels of miRNA-30a (159). miRNA-30a is also released in exosomes from cardiomyocytes after hypoxic conditioning. Hypoxia inducible factor-1α regulates the expression of miRNA-30a in exosomes released by the cell, which directly target autophagy machinery. In this way, cardiomyocytes may protect surrounding tissue in a hypoxic environment by releasing miRNA-30a and decreasing autophagy. During pulmonary arterial hypertension, the selective upregulation of miRNA-143-3p compared with miRNA-143-5p was shown in pulmonary artery smooth muscle cells (42). Pulmonary artery smooth muscle cells also excreted more miRNA-143-3p through exosomes to influence vascular endothelial cells. Blockade of the actions of miRNA-143-3p resulted in decreased pulmonary hypertension in mouse models.
After acute myocardial infarction, Matsumoto et al. (96) examined the role of circulating miRNAs as a contributing factor to the incidence of heart failure. Eighteen days postinfarction, serum was collected and miRNA-192 was shown to be significantly increased, which plays a role in the p53 response. In the p53 pathway, miRNA-194 was shown to be significantly elevated in exosomes, and the authors correlated serum levels of miRNA-192, miRNA-194, and miRNA-34a in patients who experience acute myocardial infarction with the development of heart failure.
Exosomal transport of miRNA in diabetes mellitus.
Through the progression of diabetes mellitus, it has been well documented that the miRNA profile changes throughout the body (11, 49, 59, 77, 118, 166). What is not entirely known is how the miRNA profile changes within exosomes. Questions concerning the transport of miRNAs in diabetes mellitus, the protective or detrimental effects of exosomes during the pathology from a temporal standpoint, and how exosomal transport of miRNAs may differ mechanistically between tissues have begun to be explored.
In uncomplicated type 2 diabetes mellitus, the role of serum miRNAs in predicting myocardial steatosis was evaluated (40). These authors found that miRNA-1 and miRNA-133a were positively correlated with myocardial steatosis and that miRNA-133a was elevated in Type 2 diabetic patients. Using HL-1 cardiomyocytes, miRNA-1 and miRNA-133a were found to be higher in exosomes released from lipid-loaded preconditioning. Patients with type 2 diabetes mellitus may exhibit microvascular complications (148). miRNA-7 was shown to be overexpressed in the serum of patients with type 2 diabetes mellitus or type 2 diabetes mellitus with microvascular complications compared with nondiabetic patients. These authors found that miRNA-7 was not primarily packaged in exosomes but free floating in the serum. The authors concluded that miRNA-7 serum levels may have the potential to predict microvascular complications in type 2 diabetes mellitus. Examining the generational effects of diabetes mellitus and exosome delivery, Shi et al. (123) found that mice made type 1 diabetic through STZ injections before mating had serum containing increased levels of exosomes with miRNAs involved in cardiac developmental regulation. They also showed that these exosomes can be directly transferred to the fetal pups, which may account for the observed increased in congenital heart defects. By injecting the exosomes of the diabetic mothers into the control pups, they also observed an increase in congenital heart defects in the F1 generation.
The effects of exercise on the diabetic condition and how complications of the disease can be mitigated were explored through matrix metalloprotease (MMP)-9 regulation (25). MMP-9 has been shown to be significantly decreased in a db/db model after exercise. The authors demonstrated that exosomal transport of miRNAs decreasing the expression of MMP-9 (i.e., miRNA-455, miRNA-29b, miRNA-323-5p, and miRNA466) were increased after exercise. In the Goto-Kakizaki rat, a model of type 2 diabetes mellitus, cardiac endothelial cells (MCECs) cocultured with cardiomyocytes from diabetic rats resulted in a decrease in proliferation and migration compared with control Wistar rats (155). Inhibition of exosome formation from diabetic mice restored function of MCECs, which was correlated with a significant decrease in miRNA-320 within MCECs. The importance of miRNA-320 is that it binds to targets that decrease angiogenesis. The authors concluded that in diabetes mellitus, cardiomyocytes may be involved in producing an antiangiogenic effect on the surrounding environment.
miRNA PROCESSING, lncRNA, AND OTHER REGULATORS OF miRNA IN THE CARDIOVASCULAR SYSTEM
Mechanisms of miRNA processing and post-transcriptional control.
Indirect mechanisms of miRNA regulation, such as epigenomic changes and alterations in exosomal transport, are not the only ways that miRNA levels can be affected in the diabetic heart. More direct means of miRNA alteration within the cell can include miRNA processing as well as regulation by small biomolecules or other RNA species. The milieu of the pathology, such as changes in glucose and other metabolite concentrations within the cell, can directly alter the efficiency of miRNA processing and modify the dynamics of cellular processes.
After transcription of miRNA, posttranscriptional regulation through lncRNAs can occur. It has been shown that lncRNAs have binding sites for miRNAs (73). One mechanism, referred to as sponging, involves the expression of lncRNA to sequester miRNAs (65). This lncRNA/miRNA is considered part of a larger network of competing endogenous RNA (ceRNA), in which the cell uses both coding and noncoding RNA to expand the functional genetic information that a cell can produce (114). At the current time, this field is underexplored, with few lncRNAs and even fewer lncRNA/miRNA interactions characterized. Ultimately, the potential for information regarding the control, sequestering, and further posttranscriptional regulation of miRNAs is immense.
miRNA REGULATION
miRNA processing.
The most well-known and studied role of Drosha and Dicer is to regulate the transcription of miRNAs in developed organisms. Studies have shown that a complete depletion of Dicer from an embryonic time point can significantly alter the development of cardiac tissue (116, 125), likely through a depletion of most, if not all, miRNAs. Additionally, miRNA processing through Dicer has been shown to have epigenetic control (143). During hypoxic conditions, the demethylases KDM6A/B have decreased expression, resulting in increased H3K27me3. Other epigenetic mechanisms, such as DNA methylation or further histone modifications, could also likely control the expression and processing of Dicer or Drosha.
García et al. (54) demonstrated that myocardial remodeling by miRNA-21 may be mechanistically controlled by the presence of SMAD2/3 and DICER1. In mice undergoing pressure overload, SMAD2/3 and DICER1 expression was increased in the left ventricle, which was also upregulated in patients with aortic stenosis. miRNA-21 was isolated bound to DICER1, whereas DICER1 and phosphorylated SMAD2/3 were shown to have protein-protein interactions. The authors concluded that this tranforming growth factor-β-dependent mechanism of phosphorylated SMAD2/3 and DICER1 regulating the increased, mature miRNA-21 processing could be important in myocardial remodeling. In some tissues, the downregulation of Dicer may be beneficial. It was shown that in endothelial cells where Dicer is downregulated, it decreased lesional macrophage content and monocyte adhesion in a high-fat diet apolipoprotein E knockout mice (64). Mechanistically, downregulation of Dicer also decreases miRNAs, such as miRNA-103, which binds Krüppel-like factor 4. Krüppel-like factor 4, through NF-κB, leads to an exacerbated atherosclerotic pathology, suggesting that increased Dicer expression is a maladaptation in endothelial cells.
Other than the canonical Drosha and Dicer miRNA editing pathway, other mechanisms have been proposed for editing miRNA. GTPase-activating protein (SH3 domain) binding protein 1 (G3bp1) is upregulated in cardiac hypertrophy and binds to the stem loop of miRNA-1 (67). miRNA-1 is known to regulate important transcription and translational machinery in the cell. Overexpression of G3bp1 can lead to decreased miRNA-1 levels and, therefore, increased transcriptional and translational capacity of the cell. Although G3bp1 does not directly induce cardiac hypertrophy, this novel posttranscriptional regulation of miRNA by an endoribonuclease may be mimicked by other proteins as well.
miRNA posttranscriptional regulation.
Gene regulation through lncRNA is becoming recognized as a mechanism for the control of pathological development. A lncRNA named autophagy promoting factor promotes autophagy in the heart through targeting miRNA-188-3p (152). miRNA-188-3p targets autophagy-related protein 7, suppressing autophagy. Thus, regulation of autophagy promoting factor lncRNA in the heart could be beneficial in reducing autophagy-related heart conditions, such as myocardial infarction and heart failure. Wang et al. (154) also investigated the role of cardiac apoptosis-related lncRNA in regulating mitochondrial fission and apoptosis. Cardiac apoptosis-related lncRNA targets miRNA-539, acting as a sponge, to increase the expression of the miRNA-539 target prohibitin-2. Prohibitin-2 has been suggested to play a role in suppressing apoptosis and increasing mitochondrial fission. In mice, after ANG II treatment, miRNA-489 expression was found to be reduced (153). By overexpressing miRNA-489 in mouse cardiomyocytes, the hypertrophic response was decreased and miRNA-489 was shown to target myeloid differentiation primary response gene 88 as the mechanism. miRNA-489 expression was further found to be regulated by a lncRNA named cardiac hypertrophy-related factor, which can act as an endogenous sponge and increase the hypertrophic response.
Acute myocardial infarction was experimentally induced in swine (75). Using global run-on sequencing, RNA was taken from both the infarcted area and the peripheral, healthy cardiac tissue. In the infarcted tissue, 450 lncRNAs were differentially regulated. Some of the differentially regulated lncRNAs were novel, conserved sequences found within myocardial transcription factors in an antisense orientation. The sequencing experiment demonstrates the breadth of lncRNAs that could potentially impact miRNA and mitochondrial function. The regulation of miRNAs and the dynamic of cellular homeostasis in the heart and circulatory system surpass the scope of this review, with research teams reporting miRNA regulation through transcription factors (20, 68, 105), gene expression (29, 60), other noncoding RNAs (55, 160), and even the microbiome (145).
miRNA processing and lncRNA regulation in the diabetic heart.
Limited research has been conducted in exploring the diabetic heart and the mechanisms of miRNA processing and posttranscriptional regulation. During gestational diabetes mellitus and pregnancy, it has been demonstrated that the content of DGCR8, Drosha, and Dicer were increased compared with nonpregnant controls (106). Furthermore, DGCR8, Drosha, and Dicer were more highly expressed in gestational diabetes mellitus than in those with normal pregnancies, suggesting a role for increased miRNA processing in diabetes mellitus. An increase in Dicer expression is also observed in mutant Ins2+/− Akita mice (26). In Akita hearts, even though Dicer expression is increased, let-7a, miRNA-130, miRNA-142-3p, miRNA-148, miRNA-338, miRNA-345-3p, miRNA-384-3p, miRNA-433, miRNA-450, miRNA-451, miRNA-455, miRNA-494, miRNA-499, miRNA-500, miRNA-542-3p, miRNA-744, and miRNA-872 expression is decreased.
lncRNAs have been connected to the diabetic heart, being independent predictors of grade I diastolic dysfunction and the left ventricular mass-to-left ventricular end-diastolic volume ratio in human blood (39). Also, blockade of the function of lncRNA NONRATT021972 could serve as a therapeutic for diabetic cardiac autonomic neuropathy, decreasing TNF-α and increasing IRS1 expression (158). Although no studies at the current time have implicated lncRNAs in controlling miRNA expression in the diabetic heart, the impact of lncRNAs on the pathogenesis of the diabetic heart is noted, and miRNA targets are likely present.
THERAPEUTIC APPLICATIONS OF miRNA IN DIABETES MELLITUS
Because of the significant cardiovascular complications that arise from diabetes mellitus, investigating the therapeutic applications that are available, or could be feasible options in the future, is important. The purpose of this section is to evaluate both current and potential future therapeutic applications in the diabetic heart. We will evaluate how direct targeting of miRNAs, exosomal transport of miRNAs, epigenetic manipulation of/by miRNAs, and other therapeutics involving miRNAs could impact patient health.
Targeting dysregulated energy substrate metabolism in the viable, but diseased, myocardium has been extensively investigated as a potential therapy for heart disease patients. Diabetic cardiomyopathy and diabetic heart failure are characterized by decreased glycolysis, decreased glucose oxidation, and increased fatty acid metabolism. Previous strategies to address these metabolic alterations have been to pharmacologically improve myocardial glucose import, increase pyruvate dehydrogenase activity to allow for greater glucose oxidation, and decrease fatty acid oxidation (135). These drugs include glucose transport activators (glucagon-like peptide-1 mimetics/agonists), pyruvate dehydrogenase activators (dichloroacetate and l-carnitine), carnitine palmitoyltransferase I inhibitors (perhexiline and etomoxir), and fatty acid oxidation inhibitors (ranolazine and trimetazine) (62, 90, 91, 93). Because of extensive regulation of fatty acid metabolism by the PPAR family of transcription factors, PPAR agonists are being considered for improving mitochondrial function in diabetic cardiomyopathy (82). Interestingly, metoprolol, a β-blocker, has been shown to inhibit fatty acid oxidation and improve cardiac function in STZ-induced diabetic rats (120). Recent studies of antianginal drugs have shown effects in improving myocardial O2 utilization and efficiency of energy substrate metabolism (31).
miRNA-directed therapies.
As differential miRNA expression in cardiovascular disease has been proven to modulate the expression of components of energy substrate metabolism, one may ask whether normalizing the expression of miRNAs that target the most important metabolic players through the administration of antimiRs/antagomiRs or miRNA mimics would recalibrate energy substrate metabolism back to healthy flux. This strategy of pharmacologically regulating the regulators (miRNAs) has been successfully deployed preclinically to improve pathological cardiac abnormalities found in diabetic cardiomyopathy or diabetic heart failure. As an example, Thum et al. (138) showed that administering an antagomiR specific for miRNA-21 reduced cardiac fibrosis in a mouse pressure overload-induced model. Others have reported that antagomiR-mediated inhibition of miRNA-199b inhibited and reversed cardiac hypertrophy and fibrosis in mouse models of heart failure (34). Therapeutic inhibition of miRNA-208a via antimiR action has shown much promise in preventing pathological cardiac remodeling in hypertension-induced heart failure in Dahl hypertensive rats and in improving systemic insulin sensitivity and glucose tolerance in a mouse model of high-fat diet-induced obesity (58, 98). To our knowledge, there have been no preclinical miRNA-based studies to investigate the potential of normalizing the differential expression of miRNAs targeting components of energy substrate metabolism in the diseased diabetic heart. Because metabolic dysregulation is foundational to the etiology of diabetic heart disease and miRNA-based therapeutics may offer precise regulatory potential, we believe the field of miRNA-based therapies targeting energy substrate metabolism in diabetic cardiomyopathy and diabetic heart failure has a very bright future.
Epigenetics in miRNA regulation during therapeutic intervention.
In ob/ob mice, unacylated ghrelin was used as a therapeutic to increase resistance against oxidative stress (139). Unacylated ghrelin increased the expression of miRNA-126, leading to indirect increases in SIRT1 and decreased histone 3 lysate 56 deacetylation. The results of the study showed that unacylated ghrelin can reduce cell senescence by protecting endothelial cells from ROS damage. Curcumin (diferuloylmethane) has been implicated as a potential drug for modifying both epigenetic and miRNA expression. Curcumin supplementation has been suggested to inhibit DNA methylation through inhibition of the catalytic thiolate of DNMT1 (89). Restriction of DNA methylation may be a contributing factor toward modified miRNA expression after curcumin supplementation (134, 167). EGCG, which is an ester of epigallocatechin and gallic acid, has shown very similar results as curcumin. EGCG is a direct inhibitor of DNMT1 and also has been shown to alter as many as 61 miRNAs and their expression (50, 83, 141).
Exosomes in miRNA regulation during therapeutic intervention.
Exosome-based therapeutics currently revolve around the idea of collecting exosomes from healthy cell populations to directly transfer to those in a pathological setting. Gallet et al. (53) showed that in pigs, cardiosphere-derived cell (CDC) exosomes, which were delivered through open-chest intramyocardial injection, improved the condition of acute and convalescent myocardial infarction subjects. Alleviation of conditions included decreased scarring and improved left ventricular ejection fraction. The content of CDCs has also been profiled, revealing that, aside from traditional miRNAs, exosomes derived from CDCs also contain noncoding RNA species, such as Y RNA, which are understudied (19). Future applications for the heart could even include the use of other modified exosomes. One such study examined the transport of a cargo protein through injected macrophage exosomes across the blood-brain barrier (165). With the capacity to alter exosomal cargo, such as introducing specific miRNA or miRNA sponges, the potential therapeutic applications in the heart are numerous.
miRNA PROCESSING AND OTHER MECHANISMS OF REGULATION DURING THERAPEUTIC INTERVENTION
In both diabetic human patients and mice, DICER1 protein levels were found to be increased in peripheral blood mononuclear cells after treatment with metformin (100). Metformin was shown to posttranscriptionally regulate the expression of RNA-binding protein AU-binding factor 1, disrupting binding with DICER1 mRNA and leading to an increased expression of DICER1 protein. Metformin also resulted in an increase in miRNA-20a, miRNA-34a, miRNA-130a, miRNA-106b, miRNA-125, and let-7c as well as a decrease in genes associated with senescence. Elgheznawy et al. (47) demonstrated that platelets from diabetic human patients and mice had a lower expression of miRNA-142, miRNA-143, miRNA-155, and miRNA-223. In diabetic platelets, DICER1 expression was decreased, which the authors hypothesized was due to calpain cleavage of the enzyme. When diabetic mice were treated with a calpain inhibitor, miRNA-223 expression was restored.
Although lncRNAs have not yet been implemented in diabetic cardiovascular studies, their application could be effective. For cancer therapy, an interfering lncRNA was designed to contain specific complementary miRNA sequences, which impacted the disease progression (87). The results included decreased abilities for proliferation, migration, and invasion of hepatocellular carcinoma cells.
CONCLUSIONS
miRNAs are key players in the regulation, maintenance, and function of the cardiovascular system. The impact of miRNA on mitochondrial function in the diabetic heart is evidence that managing the expression of these small endogenous RNAs is important and that the onset of pathologies may largely be controlled, and sustained, by miRNA. This review has examined how the miRNA profile is altered in the diabetic heart and how the changes in miRNA expression specifically target mitochondrial function, through energy substrate utilization, apoptosis, and ROS production. Furthermore, we examined how miRNA may be controlled in the cardiovascular system and diabetes mellitus through epigenetics, exosomes, processing, and posttranscriptional sequestration. While research concerning miRNA expression is maturing, studies focusing on the regulation of the regulating miRNAs are in its infancy. Much information can be gained mechanistically of disease and pathologies, specifically in the heart and diabetes mellitus, through additional research in this area. Therapeutically, regulating the regulators provides a method with the potential for improved treatment strategies and health effects with greater longevity.
GRANTS
This work was supported by National Heart, Lung, and Blood Institute Grant R01-HL-128485 (to J. M. Hollander), by National Science Foundation IGERT: Research and Education in Nanotoxicology at West Virginia University Fellowship 1144676 (to Q. A. Hathaway), and by American Heart Association Predoctoral Fellowship AHA 17PRE33660333 (to Q. A. Hathaway). Ingenuity Pathway Analyses were supported by WV-INBRE Grant P20-GM-10343412.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
Q.A.H., M.V.P., A.J.D., S.W., D.L.S., and J.M.H. drafted manuscript; Q.A.H., M.V.P., A.J.D., S.W., D.L.S., and J.M.H. approved final version of manuscript; Q.A.H., and M.V.P. edited and revised manuscript; Q.A.H. and M.V.P. prepared figures.
ACKNOWLEDGMENTS
We acknowledge Sarah A. Roberts for contributions to the figure artwork.
REFERENCES
- 1.Aavik E, Lumivuori H, Leppänen O, Wirth T, Häkkinen SK, Bräsen JH, Beschorner U, Zeller T, Braspenning M, van Criekinge W, Mäkinen K, Ylä-Herttuala S. Global DNA methylation analysis of human atherosclerotic plaques reveals extensive genomic hypomethylation and reactivation at imprinted locus 14q32 involving induction of a miRNA cluster. Eur Heart J 36: 993–1000, 2015. doi: 10.1093/eurheartj/ehu437. [DOI] [PubMed] [Google Scholar]
- 2.Alipoor SD, Mortaz E, Tabarsi P, Farnia P, Mirsaeidi M, Garssen J, Movassaghi M, Adcock IM. Bovis Bacillus Calmette-Guerin (BCG) infection induces exosomal miRNA release by human macrophages. J Transl Med 15: 105, 2017. doi: 10.1186/s12967-017-1205-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.American Diabetes Association Statistics about Diabetes. http://www.diabetes.org/diabetes-basics/statistics/ [7 December 2017].
- 4.American Heart Association Cardiovascular Disease & Diabetes. http://www.heart.org/HEARTORG/Conditions/More/Diabetes/WhyDiabetesMatters/Cardiovascular-Disease-Diabetes_UCM_313865_Article.jsp/#.WXtO7ITyupp [7 December 2017].
- 5.An D, Rodrigues B. Role of changes in cardiac metabolism in development of diabetic cardiomyopathy. Am J Physiol Heart Circ Physiol 291: H1489–H1506, 2006. doi: 10.1152/ajpheart.00278.2006. [DOI] [PubMed] [Google Scholar]
- 6.Asrih M, Steffens S. Emerging role of epigenetics and miRNA in diabetic cardiomyopathy. Cardiovasc Pathol 22: 117–125, 2013. doi: 10.1016/j.carpath.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 7.Awad MM, Calkins H, Judge DP. Mechanisms of disease: molecular genetics of arrhythmogenic right ventricular dysplasia/cardiomyopathy. Nat Clin Pract Cardiovasc Med 5: 258–267, 2008. doi: 10.1038/ncpcardio1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Azmi AS, Bao B, Sarkar FH. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Metastasis Rev 32: 623–642, 2013. doi: 10.1007/s10555-013-9441-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bandiera S, Matégot R, Girard M, Demongeot J, Henrion-Caude A. MitomiRs delineating the intracellular localization of microRNAs at mitochondria. Free Radic Biol Med 64: 12–19, 2013. doi: 10.1016/j.freeradbiomed.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 10.Bandiera S, Rüberg S, Girard M, Cagnard N, Hanein S, Chrétien D, Munnich A, Lyonnet S, Henrion-Caude A. Nuclear outsourcing of RNA interference components to human mitochondria. PLoS One 6: e20746, 2011. doi: 10.1371/journal.pone.0020746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banerjee J, Nema V, Dhas Y, Mishra N. Role of microRNAs in type 2 diabetes and associated vascular complications. Biochimie 139: 9–19, 2017. doi: 10.1016/j.biochi.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 12.Baradan R, Hollander JM, Das S. Mitochondrial miRNAs in diabetes: just the tip of the iceberg. Can J Physiol Pharmacol 95: 1156–1162, 2017. doi: 10.1139/cjpp-2016-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233, 2009. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bártová E, Krejcí J, Harnicarová A, Galiová G, Kozubek S. Histone modifications and nuclear architecture: a review. J Histochem Cytochem 56: 711–721, 2008. doi: 10.1369/jhc.2008.951251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baseler WA, Thapa D, Jagannathan R, Dabkowski ER, Croston TL, Hollander JM. miR-141 as a regulator of the mitochondrial phosphate carrier (Slc25a3) in the type 1 diabetic heart. Am J Physiol Cell Physiol 303: C1244–C1251, 2012. doi: 10.1152/ajpcell.00137.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benchimol S. p53-dependent pathways of apoptosis. Cell Death Differ 8: 1049–1051, 2001. doi: 10.1038/sj.cdd.4400918. [DOI] [PubMed] [Google Scholar]
- 17.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev 16: 6–21, 2002. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 18.Buschow SI, Liefhebber JM, Wubbolts R, Stoorvogel W. Exosomes contain ubiquitinated proteins. Blood Cells Mol Dis 35: 398–403, 2005. doi: 10.1016/j.bcmd.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 19.Cambier L, de Couto G, Ibrahim A, Echavez AK, Valle J, Liu W, Kreke M, Smith RR, Marbán L, Marbán E. Y RNA fragment in extracellular vesicles confers cardioprotection via modulation of IL-10 expression and secretion. EMBO Mol Med 9: 337–352, 2017. doi: 10.15252/emmm.201606924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caporali A, Meloni M, Nailor A, Mitić T, Shantikumar S, Riu F, Sala-Newby GB, Rose L, Besnier M, Katare R, Voellenkle C, Verkade P, Martelli F, Madeddu P, Emanueli C. p75(NTR)-dependent activation of NF-κB regulates microRNA-503 transcription and pericyte-endothelial crosstalk in diabetes after limb ischaemia. Nat Commun 6: 8024, 2015. doi: 10.1038/ncomms9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet 10: 295–304, 2009. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 22.Chambers JC, Loh M, Lehne B, Drong A, Kriebel J, Motta V, Wahl S, Elliott HR, Rota F, Scott WR, Zhang W, Tan ST, Campanella G, Chadeau-Hyam M, Yengo L, Richmond RC, Adamowicz-Brice M, Afzal U, Bozaoglu K, Mok ZY, Ng HK, Pattou F, Prokisch H, Rozario MA, Tarantini L, Abbott J, Ala-Korpela M, Albetti B, Ammerpohl O, Bertazzi PA, Blancher C, Caiazzo R, Danesh J, Gaunt TR, de Lusignan S, Gieger C, Illig T, Jha S, Jones S, Jowett J, Kangas AJ, Kasturiratne A, Kato N, Kotea N, Kowlessur S, Pitkäniemi J, Punjabi P, Saleheen D, Schafmayer C, Soininen P, Tai ES, Thorand B, Tuomilehto J, Wickremasinghe AR, Kyrtopoulos SA, Aitman TJ, Herder C, Hampe J, Cauchi S, Relton CL, Froguel P, Soong R, Vineis P, Jarvelin MR, Scott J, Grallert H, Bollati V, Elliott P, McCarthy MI, Kooner JS. Epigenome-wide association of DNA methylation markers in peripheral blood from Indian Asians and Europeans with incident type 2 diabetes: a nested case-control study. Lancet Diabetes Endocrinol 3: 526–534, 2015. doi: 10.1016/S2213-8587(15)00127-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan SY, Zhang YY, Hemann C, Mahoney CE, Zweier JL, Loscalzo J. MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2. Cell Metab 10: 273–284, 2009. doi: 10.1016/j.cmet.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang X, Li S, Li J, Yin L, Zhou T, Zhang C, Chen X, Sun K. Ethnic differences in microRNA-375 expression level and DNA methylation status in type 2 diabetes of Han and Kazak populations. J Diabetes Res 2014: 761938, 2014. doi: 10.1155/2014/761938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chaturvedi P, Kalani A, Medina I, Familtseva A, Tyagi SC. Cardiosome-mediated regulation of MMP9 in diabetic heart: role of mir29b and mir455 in exercise. J Cell Mol Med 19: 2153–2161, 2015. doi: 10.1111/jcmm.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chavali V, Tyagi SC, Mishra PK. Differential expression of Dicer, miRNAs, and inflammatory markers in diabetic Ins2+/− Akita hearts. Cell Biochem Biophys 68: 25–35, 2014. doi: 10.1007/s12013-013-9679-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chavali V, Tyagi SC, Mishra PK. MicroRNA-133a regulates DNA methylation in diabetic cardiomyocytes. Biochem Biophys Res Commun 425: 668–672, 2012. doi: 10.1016/j.bbrc.2012.07.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chavali V, Tyagi SC, Mishra PK. Predictors and prevention of diabetic cardiomyopathy. Diabetes Metab Syndr Obes 6: 151–160, 2013. doi: 10.2147/DMSO.S30968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J, Young ME, Chatham JC, Crossman DK, Dell’Italia LJ, Shalev A. TXNIP regulates myocardial fatty acid oxidation via miR-33a signaling. Am J Physiol Heart Circ Physiol 311: H64–H75, 2016. doi: 10.1152/ajpheart.00151.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen S, Puthanveetil P, Feng B, Matkovich SJ, Dorn GW II, Chakrabarti S. Cardiac miR-133a overexpression prevents early cardiac fibrosis in diabetes. J Cell Mol Med 18: 415–421, 2014. doi: 10.1111/jcmm.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chong CR, Sallustio B, Horowitz JD. Drugs that affect cardiac metabolism: focus on perhexiline. Cardiovasc Drugs Ther 30: 399–405, 2016. doi: 10.1007/s10557-016-6664-3. [DOI] [PubMed] [Google Scholar]
- 32.Corcoran DL, Pandit KV, Gordon B, Bhattacharjee A, Kaminski N, Benos PV. Features of mammalian microRNA promoters emerge from polymerase II chromatin immunoprecipitation data. PLoS One 4: e5279, 2009. doi: 10.1371/journal.pone.0005279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Costantino S, Paneni F, Lüscher TF, Cosentino F. MicroRNA profiling unveils hyperglycaemic memory in the diabetic heart. Eur Heart J 37: 572–576, 2016. doi: 10.1093/eurheartj/ehv599. [DOI] [PubMed] [Google Scholar]
- 34.da Costa Martins PA, Salic K, Gladka MM, Armand AS, Leptidis S, el Azzouzi H, Hansen A, Coenen-de Roo CJ, Bierhuizen MF, van der Nagel R, van Kuik J, de Weger R, de Bruin A, Condorelli G, Arbones ML, Eschenhagen T, De Windt LJ. MicroRNA-199b targets the nuclear kinase Dyrk1a in an auto-amplification loop promoting calcineurin/NFAT signalling. Nat Cell Biol 12: 1220–1227, 2010. doi: 10.1038/ncb2126. [DOI] [PubMed] [Google Scholar]
- 35.Das S, Bedja D, Campbell N, Dunkerly B, Chenna V, Maitra A, Steenbergen C. miR-181c regulates the mitochondrial genome, bioenergetics, and propensity for heart failure in vivo. PLoS One 9: e96820, 2014. doi: 10.1371/journal.pone.0096820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Das S, Ferlito M, Kent OA, Fox-Talbot K, Wang R, Liu D, Raghavachari N, Yang Y, Wheelan SJ, Murphy E, Steenbergen C. Nuclear miRNA regulates the mitochondrial genome in the heart. Circ Res 110: 1596–1603, 2012. doi: 10.1161/CIRCRESAHA.112.267732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dasgupta N, Peng Y, Tan Z, Ciraolo G, Wang D, Li R. miRNAs in mtDNA-less cell mitochondria. Cell Death Dis 1: 15004, 2015. doi: 10.1038/cddiscovery.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Couto G, Gallet R, Cambier L, Jaghatspanyan E, Makkar N, Dawkins JF, Berman BP, Marbán E. Exosomal MicroRNA transfer into macrophages mediates cellular postconditioning. Circulation 136: 200–214, 2017. doi: 10.1161/CIRCULATIONAHA.116.024590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Gonzalo-Calvo D, Kenneweg F, Bang C, Toro R, van der Meer RW, Rijzewijk LJ, Smit JW, Lamb HJ, Llorente-Cortes V, Thum T. Circulating long-non coding RNAs as biomarkers of left ventricular diastolic function and remodelling in patients with well-controlled type 2 diabetes. Sci Rep 6: 37354, 2016. doi: 10.1038/srep37354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Gonzalo-Calvo D, van der Meer RW, Rijzewijk LJ, Smit JW, Revuelta-Lopez E, Nasarre L, Escola-Gil JC, Lamb HJ, Llorente-Cortes V. Serum microRNA-1 and microRNA-133a levels reflect myocardial steatosis in uncomplicated type 2 diabetes. Sci Rep 7: 47, 2017. doi: 10.1038/s41598-017-00070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Kreutzenberg SV, Ceolotto G, Papparella I, Bortoluzzi A, Semplicini A, Dalla Man C, Cobelli C, Fadini GP, Avogaro A. Downregulation of the longevity-associated protein sirtuin 1 in insulin resistance and metabolic syndrome: potential biochemical mechanisms. Diabetes 59: 1006–1015, 2010. doi: 10.2337/db09-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deng L, Blanco FJ, Stevens H, Lu R, Caudrillier A, McBride M, McClure JD, Grant J, Thomas M, Frid M, Stenmark K, White K, Seto AG, Morrell NW, Bradshaw AC, MacLean MR, Baker AH. MicroRNA-143 activation regulates smooth muscle and endothelial cell crosstalk in pulmonary arterial hypertension. Circ Res 117: 870–883, 2015. [Erratum in Circ Res 120: e6, 2017] doi: 10.1161/CIRCRESAHA.115.306806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dhingra R, Vasan RS. Diabetes and the risk of heart failure. Heart Fail Clin 8: 125–133, 2012. doi: 10.1016/j.hfc.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Diao X, Shen E, Wang X, Hu B. Differentially expressed microRNAs and their target genes in the hearts of streptozotocin-induced diabetic mice. Mol Med Rep 4: 633–640, 2011. doi: 10.3892/mmr.2011.489. [DOI] [PubMed] [Google Scholar]
- 45.Duarte FV, Palmeira CM, Rolo AP. The emerging role of MitomiRs in the pathophysiology of human disease. Adv Exp Med Biol 888: 123–154, 2015. doi: 10.1007/978-3-319-22671-2_8. [DOI] [PubMed] [Google Scholar]
- 46.Duncan JG. Mitochondrial dysfunction in diabetic cardiomyopathy. Biochim Biophys Acta 1813: 1351–1359, 2011. doi: 10.1016/j.bbamcr.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elgheznawy A, Shi L, Hu J, Wittig I, Laban H, Pircher J, Mann A, Provost P, Randriamboavonjy V, Fleming I. Dicer cleavage by calpain determines platelet microRNA levels and function in diabetes. Circ Res 117: 157–165, 2015. doi: 10.1161/CIRCRESAHA.117.305784. [DOI] [PubMed] [Google Scholar]
- 48.Emanueli C, Shearn AI, Laftah A, Fiorentino F, Reeves BC, Beltrami C, Mumford A, Clayton A, Gurney M, Shantikumar S, Angelini GD. Coronary artery-bypass-graft surgery increases the plasma concentration of exosomes carrying a cargo of cardiac MicroRNAs: an example of exosome trafficking out of the human heart with potential for cardiac biomarker discovery. PLoS One 11: e0154274, 2016. doi: 10.1371/journal.pone.0154274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Erener S, Marwaha A, Tan R, Panagiotopoulos C, Kieffer TJ. Profiling of circulating microRNAs in children with recent onset of type 1 diabetes. JCI Insight 2: e89656, 2017. doi: 10.1172/jci.insight.89656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fang MZ, Wang Y, Ai N, Hou Z, Sun Y, Lu H, Welsh W, Yang CS. Tea polyphenol (-)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res 63: 7563–7570, 2003. [PubMed] [Google Scholar]
- 51.Floris I, Descamps B, Vardeu A, Mitić T, Posadino AM, Shantikumar S, Sala-Newby G, Capobianco G, Mangialardi G, Howard L, Dessole S, Urrutia R, Pintus G, Emanueli C. Gestational diabetes mellitus impairs fetal endothelial cell functions through a mechanism involving microRNA-101 and histone methyltransferase enhancer of zester homolog-2. Arterioscler Thromb Vasc Biol 35: 664–674, 2015. doi: 10.1161/ATVBAHA.114.304730. [DOI] [PubMed] [Google Scholar]
- 52.Galanter JM, Gignoux CR, Oh SS, Torgerson D, Pino-Yanes M, Thakur N, Eng C, Hu D, Huntsman S, Farber HJ, Avila PC, Brigino-Buenaventura E, LeNoir MA, Meade K, Serebrisky D, Rodríguez-Cintrón W, Kumar R, Rodríguez-Santana JR, Seibold MA, Borrell LN, Burchard EG, Zaitlen N. Differential methylation between ethnic sub-groups reflects the effect of genetic ancestry and environmental exposures (Abstract). eLife 6: e20532, 2017. doi: 10.7554/eLife.20532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gallet R, Dawkins J, Valle J, Simsolo E, de Couto G, Middleton R, Tseliou E, Luthringer D, Kreke M, Smith RR, Marbán L, Ghaleh B, Marbán E. Exosomes secreted by cardiosphere-derived cells reduce scarring, attenuate adverse remodelling, and improve function in acute and chronic porcine myocardial infarction. Eur Heart J 38: 201–211, 2017. 10.1093/eurheartj/ehw240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.García R, Nistal JF, Merino D, Price NL, Fernández-Hernando C, Beaumont J, González A, Hurlé MA, Villar AV. p-SMAD2/3 and DICER promote pre-miR-21 processing during pressure overload-associated myocardial remodeling. Biochim Biophys Acta 1852: 1520–1530, 2015. doi: 10.1016/j.bbadis.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 55.Geng HH, Li R, Su YM, Xiao J, Pan M, Cai XX, Ji XP. The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PLoS One 11: e0151753, 2016. doi: 10.1371/journal.pone.0151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Greco S, Fasanaro P, Castelvecchio S, D’Alessandra Y, Arcelli D, Di Donato M, Malavazos A, Capogrossi MC, Menicanti L, Martelli F. MicroRNA dysregulation in diabetic ischemic heart failure patients. Diabetes 61: 1633–1641, 2012. doi: 10.2337/db11-0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The microprocessor complex mediates the genesis of microRNAs. Nature 432: 235–240, 2004. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 58.Grueter CE, van Rooij E, Johnson BA, DeLeon SM, Sutherland LB, Qi X, Gautron L, Elmquist JK, Bassel-Duby R, Olson EN. A cardiac microRNA governs systemic energy homeostasis by regulation of MED13. Cell 149: 671–683, 2012. doi: 10.1016/j.cell.2012.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guay C, Regazzi R. Circulating microRNAs as novel biomarkers for diabetes mellitus. Nat Rev Endocrinol 9: 513–521, 2013. doi: 10.1038/nrendo.2013.86. [DOI] [PubMed] [Google Scholar]
- 60.Gurha P, Chen X, Lombardi R, Willerson JT, Marian AJ. Knockdown of plakophilin 2 downregulates miR-184 through CpG hypermethylation and suppression of the E2F1 pathway and leads to enhanced adipogenesis in vitro. Circ Res 119: 731–750, 2016. doi: 10.1161/CIRCRESAHA.116.308422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15: 509–524, 2014. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 62.Hamilton DJ. Metabolic recovery of the failing heart: emerging therapeutic options. Methodist DeBakey Cardiovasc J 13: 25–28, 2017. doi: 10.14797/mdcj-13-1-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev 18: 3016–3027, 2004. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hartmann P, Zhou Z, Natarelli L, Wei Y, Nazari-Jahantigh M, Zhu M, Grommes J, Steffens S, Weber C, Schober A. Endothelial Dicer promotes atherosclerosis and vascular inflammation by miRNA-103-mediated suppression of KLF4. Nat Commun 7: 10521, 2016. doi: 10.1038/ncomms10521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hausser J, Zavolan M. Identification and consequences of miRNA-target interactions−beyond repression of gene expression. Nat Rev Genet 15: 599–612, 2014. doi: 10.1038/nrg3765. [DOI] [PubMed] [Google Scholar]
- 66.He L, He X, Lowe SW, Hannon GJ. microRNAs join the p53 network–another piece in the tumour-suppression puzzle. Nat Rev Cancer 7: 819–822, 2007. doi: 10.1038/nrc2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.He M, Yang Z, Abdellatif M, Sayed D. GTPase-activating protein (Sh3 domain) binding protein 1 regulates the processing of MicroRNA-1 during cardiac hypertrophy. PLoS One 10: e0145112, 2015. doi: 10.1371/journal.pone.0145112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang Z, Chen XJ, Qian C, Dong Q, Ding D, Wu QF, Li J, Wang HF, Li WH, Xie Q, Cheng X, Zhao N, Du YM, Liao YH. Signal transducer and activator of transcription 3/microRNA-21 feedback loop contributes to atrial fibrillation by promoting atrial fibrosis in a rat sterile pericarditis model. Circ Arrhythm Electrophysiol 9: e003396, 2016. doi: 10.1161/CIRCEP.115.003396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huynh K, Bernardo BC, McMullen JR, Ritchie RH. Diabetic cardiomyopathy: mechanisms and new treatment strategies targeting antioxidant signaling pathways. Pharmacol Ther 142: 375–415, 2014. doi: 10.1016/j.pharmthera.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 70.Iorio MV, Piovan C, Croce CM. Interplay between microRNAs and the epigenetic machinery: an intricate network. Biochim Biophys Acta 1799: 694–701, 2010. doi: 10.1016/j.bbagrm.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 71.Ismail N, Wang Y, Dakhlallah D, Moldovan L, Agarwal K, Batte K, Shah P, Wisler J, Eubank TD, Tridandapani S, Paulaitis ME, Piper MG, Marsh CB. Macrophage microvesicles induce macrophage differentiation and miR-223 transfer. Blood 121: 984–995, 2013. doi: 10.1182/blood-2011-08-374793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jagannathan R, Thapa D, Nichols CE, Shepherd DL, Stricker JC, Croston TL, Baseler WA, Lewis SE, Martinez I, Hollander JM. Translational regulation of the mitochondrial genome following redistribution of mitochondrial MicroRNA in the diabetic heart. Circ Cardiovasc Genet 8: 785–802, 2015. doi: 10.1161/CIRCGENETICS.115.001067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jalali S, Bhartiya D, Lalwani MK, Sivasubbu S, Scaria V. Systematic transcriptome wide analysis of lncRNA-miRNA interactions. PLoS One 8: e53823, 2013. doi: 10.1371/journal.pone.0053823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13: 484–492, 2012. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 75.Kaikkonen MU, Halonen P, Liu OH, Turunen TA, Pajula J, Moreau P, Selvarajan I, Tuomainen T, Aavik E, Tavi P, Ylä-Herttuala S. Genome-wide dynamics of nascent noncoding RNA transcription in porcine heart after myocardial infarction. Circ Cardiovasc Genet 10: e001702, 2017. doi: 10.1161/CIRCGENETICS.117.001702. [DOI] [PubMed] [Google Scholar]
- 76.Katare R, Caporali A, Zentilin L, Avolio E, Sala-Newby G, Oikawa A, Cesselli D, Beltrami AP, Giacca M, Emanueli C, Madeddu P. Intravenous gene therapy with PIM-1 via a cardiotropic viral vector halts the progression of diabetic cardiomyopathy through promotion of prosurvival signaling. Circ Res 108: 1238–1251, 2011. doi: 10.1161/CIRCRESAHA.110.239111. [DOI] [PubMed] [Google Scholar]
- 77.Kong L, Zhu J, Han W, Jiang X, Xu M, Zhao Y, Dong Q, Pang Z, Guan Q, Gao L, Zhao J, Zhao L. Significance of serum microRNAs in pre-diabetes and newly diagnosed type 2 diabetes: a clinical study. Acta Diabetol 48: 61–69, 2011. doi: 10.1007/s00592-010-0226-0. [DOI] [PubMed] [Google Scholar]
- 78.Kren BT, Wong PY, Sarver A, Zhang X, Zeng Y, Steer CJ. MicroRNAs identified in highly purified liver-derived mitochondria may play a role in apoptosis. RNA Biol 6: 65–72, 2009. doi: 10.4161/rna.6.1.7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kulkarni H, Kos MZ, Neary J, Dyer TD, Kent JW Jr, Göring HH, Cole SA, Comuzzie AG, Almasy L, Mahaney MC, Curran JE, Blangero J, Carless MA. Novel epigenetic determinants of type 2 diabetes in Mexican-American families. Hum Mol Genet 24: 5330–5344, 2015. doi: 10.1093/hmg/ddv232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kuwabara Y, Ono K, Horie T, Nishi H, Nagao K, Kinoshita M, Watanabe S, Baba O, Kojima Y, Shizuta S, Imai M, Tamura T, Kita T, Kimura T. Increased microRNA-1 and microRNA-133a levels in serum of patients with cardiovascular disease indicate myocardial damage. Circ Cardiovasc Genet 4: 446–454, 2011. doi: 10.1161/CIRCGENETICS.110.958975. [DOI] [PubMed] [Google Scholar]
- 81.Kwon H, Lee J, Jeong K, Jang D, Pak Y. Fatty acylated caveolin-2 is a substrate of insulin receptor tyrosine kinase for insulin receptor substrate-1-directed signaling activation. Biochim Biophys Acta 1853: 1022–1034, 2015. doi: 10.1016/j.bbamcr.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 82.Lee TW, Bai KJ, Lee TI, Chao TF, Kao YH, Chen YJ. PPARs modulate cardiac metabolism and mitochondrial function in diabetes. J Biomed Sci 24: 5, 2017. doi: 10.1186/s12929-016-0309-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee WJ, Shim JY, Zhu BT. Mechanisms for the inhibition of DNA methyltransferases by tea catechins and bioflavonoids. Mol Pharmacol 68: 1018–1030, 2005. doi: 10.1124/mol.104.008367. [DOI] [PubMed] [Google Scholar]
- 84.Li J, Ren Y, Shi E, Tan Z, Xiong J, Yan L, Jiang X. Inhibition of the Let-7 family MicroRNAs induces cardioprotection against ischemia-reperfusion injury in diabetic rats. Ann Thorac Surg 102: 829–835, 2016. doi: 10.1016/j.athoracsur.2016.02.016. [DOI] [PubMed] [Google Scholar]
- 85.Li X, Du N, Zhang Q, Li J, Chen X, Liu X, Hu Y, Qin W, Shen N, Xu C, Fang Z, Wei Y, Wang R, Du Z, Zhang Y, Lu Y. MicroRNA-30d regulates cardiomyocyte pyroptosis by directly targeting foxo3a in diabetic cardiomyopathy. Cell Death Dis 5: e1479, 2014. doi: 10.1038/cddis.2014.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li X, Monks B, Ge Q, Birnbaum MJ. Akt/PKB regulates hepatic metabolism by directly inhibiting PGC-1α transcription coactivator. Nature 447: 1012–1016, 2007. doi: 10.1038/nature05861. [DOI] [PubMed] [Google Scholar]
- 87.Li X, Su Y, Sun B, Ji W, Peng Z, Xu Y, Wu M, Su C. An artificially designed interfering lncRNA expressed by oncolytic adenovirus competitively consumes OncomiRs to exert antitumor efficacy in hepatocellular carcinoma. Mol Cancer Ther 15: 1436–1451, 2016. doi: 10.1158/1535-7163.MCT-16-0096. [DOI] [PubMed] [Google Scholar]
- 88.Liang J, Liu C, Qiao A, Cui Y, Zhang H, Cui A, Zhang S, Yang Y, Xiao X, Chen Y, Fang F, Chang Y. MicroRNA-29a-c decrease fasting blood glucose levels by negatively regulating hepatic gluconeogenesis. J Hepatol 58: 535–542, 2013. doi: 10.1016/j.jhep.2012.10.024. [DOI] [PubMed] [Google Scholar]
- 89.Liu Z, Xie Z, Jones W, Pavlovicz RE, Liu S, Yu J, Li PK, Lin J, Fuchs JR, Marcucci G, Li C, Chan KK. Curcumin is a potent DNA hypomethylation agent. Bioorg Med Chem Lett 19: 706–709, 2009. doi: 10.1016/j.bmcl.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 90.Lopaschuk GD. Treating ischemic heart disease by pharmacologically improving cardiac energy metabolism. Am J Cardiol 82, 5A: 14K–17K, 1998. doi: 10.1016/S0002-9149(98)00532-3. [DOI] [PubMed] [Google Scholar]
- 91.Lopatin YM, Rosano GM, Fragasso G, Lopaschuk GD, Seferovic PM, Gowdak LH, Vinereanu D, Hamid MA, Jourdain P, Ponikowski P. Rationale and benefits of trimetazidine by acting on cardiac metabolism in heart failure. Int J Cardiol 203: 909–915, 2016. doi: 10.1016/j.ijcard.2015.11.060. [DOI] [PubMed] [Google Scholar]
- 92.Lorenzo O, Ramírez E, Picatoste B, Egido J, Tuñón J. Alteration of energy substrates and ROS production in diabetic cardiomyopathy. Mediators Inflamm 2013: 461967, 2013. doi: 10.1155/2013/461967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Magoon R, Choudhury A, Malik V, Sharma R, Kapoor PM. Pharmacological update: New drugs in cardiac practice: A critical appraisal. Ann Card Anaesth 20, Suppl: S49–S56, 2017. doi: 10.4103/0971-9784.197798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Marsico A, Huska MR, Lasserre J, Hu H, Vucicevic D, Musahl A, Orom U, Vingron M. PROmiRNA: a new miRNA promoter recognition method uncovers the complex regulation of intronic miRNAs. Genome Biol 14: R84, 2013. doi: 10.1186/gb-2013-14-8-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mathiyalagan P, Okabe J, Chang L, Su Y, Du XJ, El-Osta A. The primary microRNA-208b interacts with Polycomb-group protein, Ezh2, to regulate gene expression in the heart. Nucleic Acids Res 42: 790–803, 2014. doi: 10.1093/nar/gkt896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Matsumoto S, Sakata Y, Suna S, Nakatani D, Usami M, Hara M, Kitamura T, Hamasaki T, Nanto S, Kawahara Y, Komuro I. Circulating p53-responsive microRNAs are predictive indicators of heart failure after acute myocardial infarction. Circ Res 113: 322–326, 2013. doi: 10.1161/CIRCRESAHA.113.301209. [DOI] [PubMed] [Google Scholar]
- 97.Miyoshi K, Miyoshi T, Siomi H. Many ways to generate microRNA-like small RNAs: non-canonical pathways for microRNA production. Mol Genet Genomics 284: 95–103, 2010. doi: 10.1007/s00438-010-0556-1. [DOI] [PubMed] [Google Scholar]
- 98.Montgomery RL, Hullinger TG, Semus HM, Dickinson BA, Seto AG, Lynch JM, Stack C, Latimer PA, Olson EN, van Rooij E. Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation 124: 1537–1547, 2011. doi: 10.1161/CIRCULATIONAHA.111.030932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mosher MJ, Melton PE, Stapleton P, Schanfield MS, Crawford MH. Patterns of DNA methylation across the leptin core promoter in four diverse Asian and North American populations. Hum Biol 88: 121–135, 2016. doi: 10.13110/humanbiology.88.2.0121. [DOI] [PubMed] [Google Scholar]
- 100.Noren Hooten N, Martin-Montalvo A, Dluzen DF, Zhang Y, Bernier M, Zonderman AB, Becker KG, Gorospe M, de Cabo R, Evans MK. Metformin-mediated increase in DICER1 regulates microRNA expression and cellular senescence. Aging Cell 15: 572–581, 2016. doi: 10.1111/acel.12469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pelsers MM, Lutgerink JT, Nieuwenhoven FA, Tandon NN, van der Vusse GJ, Arends JW, Hoogenboom HR, Glatz JF. A sensitive immunoassay for rat fatty acid translocase (CD36) using phage antibodies selected on cell transfectants: abundant presence of fatty acid translocase/CD36 in cardiac and red skeletal muscle and up-regulation in diabetes. Biochem J 337: 407–414, 1999. doi: 10.1042/bj3370407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Peng XP, Huang L, Liu ZH. miRNA-133a attenuates lipid accumulation via TR4-CD36 pathway in macrophages. Biochimie 127: 79–85, 2016. doi: 10.1016/j.biochi.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 103.Pheiffer C, Erasmus RT, Kengne AP, Matsha TE. Differential DNA methylation of microRNAs within promoters, intergenic and intragenic regions of type 2 diabetic, pre-diabetic and non-diabetic individuals. Clin Biochem 49: 433–438, 2016. doi: 10.1016/j.clinbiochem.2015.11.021. [DOI] [PubMed] [Google Scholar]
- 104.Pinti MV, Hathaway QA, Hollander JM. Role of microRNA in metabolic shift during heart failure. Am J Physiol Heart Circ Physiol 312: H33–H45, 2017. doi: 10.1152/ajpheart.00341.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Qiu Y, Yang J, Bian S, Chen G, Yu J. PPARγ suppresses the proliferation of cardiac myxoma cells through downregulation of MEF2D in a miR-122-dependent manner. Biochem Biophys Res Commun 474: 560–565, 2016. doi: 10.1016/j.bbrc.2016.04.112. [DOI] [PubMed] [Google Scholar]
- 106.Rahimi G, Jafari N, Khodabakhsh M, Shirzad Z, Dogaheh HP. Upregulation of microRNA processing enzymes Drosha and Dicer in gestational diabetes mellitus. Gynecol Endocrinol 31: 156–159, 2015. doi: 10.3109/09513590.2014.969700. [DOI] [PubMed] [Google Scholar]
- 107.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 200: 373–383, 2013. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Raut SK, Singh GB, Rastogi B, Saikia UN, Mittal A, Dogra N, Singh S, Prasad R, Khullar M. miR-30c and miR-181a synergistically modulate p53-p21 pathway in diabetes-induced cardiac hypertrophy. Mol Cell Biochem 417: 191–203, 2016. doi: 10.1007/s11010-016-2729-7. [DOI] [PubMed] [Google Scholar]
- 109.Renaud L, Harris LG, Mani SK, Kasiganesan H, Chou JC, Baicu CF, Van Laer A, Akerman AW, Stroud RE, Jones JA, Zile MR, Menick DR. HDACs regulate miR-133a expression in pressure overload-induced cardiac fibrosis. Circ Heart Fail 8: 1094–1104, 2015. doi: 10.1161/CIRCHEARTFAILURE.114.001781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Riches K, Huntriss J, Keeble C, Wood IC, O’Regan DJ, Turner NA, Porter KE. Mapping the methylation status of the miR-145 promoter in saphenous vein smooth muscle cells from individuals with type 2 diabetes. Diab Vasc Dis Res 14: 122–129, 2017. doi: 10.1177/1479164116677968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Russo I, Frangogiannis NG. Diabetes-associated cardiac fibrosis: cellular effectors, molecular mechanisms and therapeutic opportunities. J Mol Cell Cardiol 90: 84–93, 2016. doi: 10.1016/j.yjmcc.2015.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sahoo S, Emanueli C. Exosomes in diabetic cardiomyopathy: the next-generation therapeutic targets? Diabetes 65: 2829–2831, 2016. doi: 10.2337/dbi16-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Saito S, Thuc LC, Teshima Y, Nakada C, Nishio S, Kondo H, Fukui A, Abe I, Ebata Y, Saikawa T, Moriyama M, Takahashi N. Glucose fluctuations aggravate cardiac susceptibility to ischemia/reperfusion injury by modulating microRNAs expression. Circ J 80: 186–195, 2016. doi: 10.1253/circj.CJ-14-1218. [DOI] [PubMed] [Google Scholar]
- 114.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146: 353–358, 2011. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sato F, Tsuchiya S, Meltzer SJ, Shimizu K. MicroRNAs and epigenetics. FEBS J 278: 1598–1609, 2011. doi: 10.1111/j.1742-4658.2011.08089.x. [DOI] [PubMed] [Google Scholar]
- 116.Saxena A, Tabin CJ. miRNA-processing enzyme Dicer is necessary for cardiac outflow tract alignment and chamber septation. Proc Natl Acad Sci USA 107: 87–91, 2010. doi: 10.1073/pnas.0912870107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schilling JD. The mitochondria in diabetic heart failure: from pathogenesis to therapeutic promise. Antioxid Redox Signal 22: 1515–1526, 2015. doi: 10.1089/ars.2015.6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sebastiani G, Nigi L, Grieco GE, Mancarella F, Ventriglia G, Dotta F. Circulating microRNAs and diabetes mellitus: a novel tool for disease prediction, diagnosis, and staging? J Endocrinol Invest 40: 591–610, 2017. doi: 10.1007/s40618-017-0611-4. [DOI] [PubMed] [Google Scholar]
- 119.Shan ZX, Lin QX, Deng CY, Zhu JN, Mai LP, Liu JL, Fu YH, Liu XY, Li YX, Zhang YY, Lin SG, Yu XY. miR-1/miR-206 regulate Hsp60 expression contributing to glucose-mediated apoptosis in cardiomyocytes. FEBS Lett 584: 3592–3600, 2010. doi: 10.1016/j.febslet.2010.07.027. [DOI] [PubMed] [Google Scholar]
- 120.Sharma V, Dhillon P, Wambolt R, Parsons H, Brownsey R, Allard MF, McNeill JH. Metoprolol improves cardiac function and modulates cardiac metabolism in the streptozotocin-diabetic rat. Am J Physiol Heart Circ Physiol 294: H1609–H1620, 2008. doi: 10.1152/ajpheart.00949.2007. [DOI] [PubMed] [Google Scholar]
- 121.Shatseva T, Lee DY, Deng Z, Yang BB. MicroRNA miR-199a-3p regulates cell proliferation and survival by targeting caveolin-2. J Cell Sci 124: 2826–2836, 2011. doi: 10.1242/jcs.077529. [DOI] [PubMed] [Google Scholar]
- 122.Shepherd DL, Hathaway QA, Pinti MV, Nichols CE, Durr AJ, Sreekumar S, Hughes KM, Stine SM, Martinez I, Hollander JM. Exploring the mitochondrial microRNA import pathway through polynucleotide phosphorylase (PNPase). J Mol Cell Cardiol 110: 15–25, 2017. doi: 10.1016/j.yjmcc.2017.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shi R, Zhao L, Cai W, Wei M, Zhou X, Yang G, Yuan L. Maternal exosomes in diabetes contribute to the cardiac development deficiency. Biochem Biophys Res Commun 483: 602–608, 2017. doi: 10.1016/j.bbrc.2016.12.097. [DOI] [PubMed] [Google Scholar]
- 124.Shinde S, Bhadra U. A complex genome-microRNA interplay in human mitochondria. BioMed Res Int 2015: 206382, 2015. doi: 10.1155/2015/206382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Singh MK, Lu MM, Massera D, Epstein JA. MicroRNA-processing enzyme Dicer is required in epicardium for coronary vasculature development. J Biol Chem 286: 41036–41045, 2011. doi: 10.1074/jbc.M111.268573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature 469: 336–342, 2011. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sokolova V, Ludwig AK, Hornung S, Rotan O, Horn PA, Epple M, Giebel B. Characterisation of exosomes derived from human cells by nanoparticle tracking analysis and scanning electron microscopy. Colloids Surf B Biointerfaces 87: 146–150, 2011. doi: 10.1016/j.colsurfb.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 128.Squadrito ML, Baer C, Burdet F, Maderna C, Gilfillan GD, Lyle R, Ibberson M, De Palma M. Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Reports 8: 1432–1446, 2014. doi: 10.1016/j.celrep.2014.07.035. [DOI] [PubMed] [Google Scholar]
- 129.Sripada L, Singh K, Lipatova AV, Singh A, Prajapati P, Tomar D, Bhatelia K, Roy M, Singh R, Godbole MM, Chumakov PM, Singh R. hsa-miR-4485 regulates mitochondrial functions and inhibits the tumorigenicity of breast cancer cells. J Mol Med (Berl) 95: 641–651, 2017. doi: 10.1007/s00109-017-1517-5. [DOI] [PubMed] [Google Scholar]
- 130.Sripada L, Tomar D, Singh R. Mitochondria: one of the destinations of miRNAs. Mitochondrion 12: 593–599, 2012. doi: 10.1016/j.mito.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 131.Stanley WC, Lopaschuk GD, McCormack JG. Regulation of energy substrate metabolism in the diabetic heart. Cardiovasc Res 34: 25–33, 1997. doi: 10.1016/S0008-6363(97)00047-3. [DOI] [PubMed] [Google Scholar]
- 132.Sukma Dewi I, Celik S, Karlsson A, Hollander Z, Lam K, McManus JW, Tebbutt S, Ng R, Keown P, McMaster R, McManus B, Öhman J, Gidlöf O. Exosomal miR-142-3p is increased during cardiac allograft rejection and augments vascular permeability through down-regulation of endothelial RAB11FIP2 expression. Cardiovasc Res 113: 440–452, 2017. doi: 10.1093/cvr/cvw244. [DOI] [PubMed] [Google Scholar]
- 133.Sun K, Chang X, Yin L, Li J, Zhou T, Zhang C, Chen X. Expression and DNA methylation status of microRNA-375 in patients with type 2 diabetes mellitus. Mol Med Rep 9: 967–972, 2014. doi: 10.3892/mmr.2013.1872. [DOI] [PubMed] [Google Scholar]
- 134.Sun M, Estrov Z, Ji Y, Coombes KR, Harris DH, Kurzrock R. Curcumin (diferuloylmethane) alters the expression profiles of microRNAs in human pancreatic cancer cells. Mol Cancer Ther 7: 464–473, 2008. doi: 10.1158/1535-7163.MCT-07-2272. [DOI] [PubMed] [Google Scholar]
- 135.Taegtmeyer H. Cardiac metabolism as a target for the treatment of heart failure. Circulation 110: 894–896, 2004. doi: 10.1161/01.CIR.0000139340.88769.D5. [DOI] [PubMed] [Google Scholar]
- 136.Tang N, Sun B, Gupta A, Rempel H, Pulliam L. Monocyte exosomes induce adhesion molecules and cytokines via activation of NF-κB in endothelial cells. FASEB J 30: 3097–3106, 2016. doi: 10.1096/fj.201600368RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol 2: 569–579, 2002. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 138.Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, Castoldi M, Soutschek J, Koteliansky V, Rosenwald A, Basson MA, Licht JD, Pena JT, Rouhanifard SH, Muckenthaler MU, Tuschl T, Martin GR, Bauersachs J, Engelhardt S. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 456: 980–984, 2008. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 139.Togliatto G, Trombetta A, Dentelli P, Gallo S, Rosso A, Cotogni P, Granata R, Falcioni R, Delale T, Ghigo E, Brizzi MF. Unacylated ghrelin induces oxidative stress resistance in a glucose intolerance and peripheral artery disease mouse model by restoring endothelial cell miR-126 expression. Diabetes 64: 1370–1382, 2015. doi: 10.2337/db14-0991. [DOI] [PubMed] [Google Scholar]
- 140.Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brügger B, Simons M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319: 1244–1247, 2008. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 141.Tsang WP, Kwok TT. Epigallocatechin gallate up-regulation of miR-16 and induction of apoptosis in human cancer cells. J Nutr Biochem 21: 140–146, 2010. doi: 10.1016/j.jnutbio.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 142.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9: 654–659, 2007. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 143.van den Beucken T, Koch E, Chu K, Rupaimoole R, Prickaerts P, Adriaens M, Voncken JW, Harris AL, Buffa FM, Haider S, Starmans MHW, Yao CQ, Ivan M, Ivan C, Pecot CV, Boutros PC, Sood AK, Koritzinsky M, Wouters BG. Hypoxia promotes stem cell phenotypes and poor prognosis through epigenetic regulation of DICER. Nat Commun 5: 5203, 2014. doi: 10.1038/ncomms6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.van der Zwaag PA, Jongbloed JD, van den Berg MP, van der Smagt JJ, Jongbloed R, Bikker H, Hofstra RM, van Tintelen JP. A genetic variants database for arrhythmogenic right ventricular dysplasia/cardiomyopathy. Hum Mutat 30: 1278–1283, 2009. doi: 10.1002/humu.21064. [DOI] [PubMed] [Google Scholar]
- 145.Vikram A, Kim YR, Kumar S, Li Q, Kassan M, Jacobs JS, Irani K. Vascular microRNA-204 is remotely governed by the microbiome and impairs endothelium-dependent vasorelaxation by downregulating Sirtuin1. Nat Commun 7: 12565, 2016. doi: 10.1038/ncomms12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Villeneuve LM, Kato M, Reddy MA, Wang M, Lanting L, Natarajan R. Enhanced levels of microRNA-125b in vascular smooth muscle cells of diabetic db/db mice lead to increased inflammatory gene expression by targeting the histone methyltransferase Suv39h1. Diabetes 59: 2904–2915, 2010. doi: 10.2337/db10-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wahl S, Drong A, Lehne B, Loh M, Scott WR, Kunze S, Tsai PC, Ried JS, Zhang W, Yang Y, Tan S, Fiorito G, Franke L, Guarrera S, Kasela S, Kriebel J, Richmond RC, Adamo M, Afzal U, Ala-Korpela M, Albetti B, Ammerpohl O, Apperley JF, Beekman M, Bertazzi PA, Black SL, Blancher C, Bonder MJ, Brosch M, Carstensen-Kirberg M, de Craen AJ, de Lusignan S, Dehghan A, Elkalaawy M, Fischer K, Franco OH, Gaunt TR, Hampe J, Hashemi M, Isaacs A, Jenkinson A, Jha S, Kato N, Krogh V, Laffan M, Meisinger C, Meitinger T, Mok ZY, Motta V, Ng HK, Nikolakopoulou Z, Nteliopoulos G, Panico S, Pervjakova N, Prokisch H, Rathmann W, Roden M, Rota F, Rozario MA, Sandling JK, Schafmayer C, Schramm K, Siebert R, Slagboom PE, Soininen P, Stolk L, Strauch K, Tai ES, Tarantini L, Thorand B, Tigchelaar EF, Tumino R, Uitterlinden AG, van Duijn C, van Meurs JB, Vineis P, Wickremasinghe AR, Wijmenga C, Yang TP, Yuan W, Zhernakova A, Batterham RL, Smith GD, Deloukas P, Heijmans BT, Herder C, Hofman A, Lindgren CM, Milani L, van der Harst P, Peters A, Illig T, Relton CL, Waldenberger M, Järvelin MR, Bollati V, Soong R, Spector TD, Scott J, McCarthy MI, Elliott P, Bell JT, Matullo G, Gieger C, Kooner JS, Grallert H, Chambers JC. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature 541: 81–86, 2017. doi: 10.1038/nature20784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wan S, Wang J, Wang J, Wu J, Song J, Zhang CY, Zhang C, Wang C, Wang JJ. Increased serum miR-7 is a promising biomarker for type 2 diabetes mellitus and its microvascular complications. Diabetes Res Clin Pract 130: 171–179, 2017. doi: 10.1016/j.diabres.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 149.Wang C, Zhang C, Liu L, A X, Chen B, Li Y, Du J. Macrophage-derived mir-155-containing exosomes suppress fibroblast proliferation and promote fibroblast inflammation during cardiac injury. Mol Ther 25: 192–204, 2017. doi: 10.1016/j.ymthe.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wang G, Chen HW, Oktay Y, Zhang J, Allen EL, Smith GM, Fan KC, Hong JS, French SW, McCaffery JM, Lightowlers RN, Morse HC III, Koehler CM, Teitell MA. PNPASE regulates RNA import into mitochondria. Cell 142: 456–467, 2010. doi: 10.1016/j.cell.2010.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wang G, Shimada E, Koehler CM, Teitell MA. PNPASE and RNA trafficking into mitochondria. Biochim Biophys Acta 1819: 998–1007, 2012. doi: 10.1016/j.bbagrm.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wang K, Liu CY, Zhou LY, Wang JX, Wang M, Zhao B, Zhao WK, Xu SJ, Fan LH, Zhang XJ, Feng C, Wang CQ, Zhao YF, Li PF. APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nat Commun 6: 6779, 2015. doi: 10.1038/ncomms7779. [DOI] [PubMed] [Google Scholar]
- 153.Wang K, Liu F, Zhou LY, Long B, Yuan SM, Wang Y, Liu CY, Sun T, Zhang XJ, Li PF. The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circ Res 114: 1377–1388, 2014. doi: 10.1161/CIRCRESAHA.114.302476. [DOI] [PubMed] [Google Scholar]
- 154.Wang K, Long B, Zhou LY, Liu F, Zhou QY, Liu CY, Fan YY, Li PF. CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat Commun 5: 3596, 2014. doi: 10.1038/ncomms4596. [DOI] [PubMed] [Google Scholar]
- 155.Wang X, Huang W, Liu G, Cai W, Millard RW, Wang Y, Chang J, Peng T, Fan GC. Cardiomyocytes mediate anti-angiogenesis in type 2 diabetic rats through the exosomal transfer of miR-320 into endothelial cells. J Mol Cell Cardiol 74: 139–150, 2014. doi: 10.1016/j.yjmcc.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol 11: 228–234, 2009. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 157.Xiao J, Pan Y, Li XH, Yang XY, Feng YL, Tan HH, Jiang L, Feng J, Yu XY. Cardiac progenitor cell-derived exosomes prevent cardiomyocytes apoptosis through exosomal miR-21 by targeting PDCD4. Cell Death Dis 7: e2277, 2016. doi: 10.1038/cddis.2016.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Xu H, Liu C, Rao S, He L, Zhang T, Sun S, Wu B, Zou L, Wang S, Xue Y, Jia T, Zhao S, Li G, Liu S, Li G, Liang S. LncRNA NONRATT021972 siRNA rescued decreased heart rate variability in diabetic rats in superior cervical ganglia. Auton Neurosci 201: 1–7, 2016. doi: 10.1016/j.autneu.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 159.Yang Y, Li Y, Chen X, Cheng X, Liao Y, Yu X. Exosomal transfer of miR-30a between cardiomyocytes regulates autophagy after hypoxia. J Mol Med (Berl) 94: 711–724, 2016. doi: 10.1007/s00109-016-1387-2. [DOI] [PubMed] [Google Scholar]
- 160.Yang ZG, Awan FM, Du WW, Zeng Y, Lyu J, Wu D, Gupta S, Yang W, Yang BB. The circular RNA interacts with STAT3, increasing its nuclear translocation and wound repair by modulating Dnmt3a and miR-17 function. Mol Ther 25: 2062–2074, 2017. doi: 10.1016/j.ymthe.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 17: 3011–3016, 2003. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Yildirim SS, Akman D, Catalucci D, Turan B. Relationship between downregulation of miRNAs and increase of oxidative stress in the development of diabetic cardiac dysfunction: junctin as a target protein of miR-1. Cell Biochem Biophys 67: 1397–1408, 2013. doi: 10.1007/s12013-013-9672-y. [DOI] [PubMed] [Google Scholar]
- 163.Yu J, Nagasu H, Murakami T, Hoang H, Broderick L, Hoffman HM, Horng T. Inflammasome activation leads to caspase-1-dependent mitochondrial damage and block of mitophagy. Proc Natl Acad Sci USA 111: 15514–15519, 2014. doi: 10.1073/pnas.1414859111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Yu XY, Song YH, Geng YJ, Lin QX, Shan ZX, Lin SG, Li Y. Glucose induces apoptosis of cardiomyocytes via microRNA-1 and IGF-1. Biochem Biophys Res Commun 376: 548–552, 2008. doi: 10.1016/j.bbrc.2008.09.025. [DOI] [PubMed] [Google Scholar]
- 165.Yuan D, Zhao Y, Banks WA, Bullock KM, Haney M, Batrakova E, Kabanov AV. Macrophage exosomes as natural nanocarriers for protein delivery to inflamed brain. Biomaterials 142: 1–12, 2017. doi: 10.1016/j.biomaterials.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zampetaki A, Kiechl S, Drozdov I, Willeit P, Mayr U, Prokopi M, Mayr A, Weger S, Oberhollenzer F, Bonora E, Shah A, Willeit J, Mayr M. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res 107: 810–817, 2010. doi: 10.1161/CIRCRESAHA.110.226357. [DOI] [PubMed] [Google Scholar]
- 167.Zhang J, Zhang T, Ti X, Shi J, Wu C, Ren X, Yin H. Curcumin promotes apoptosis in A549/DDP multidrug-resistant human lung adenocarcinoma cells through an miRNA signaling pathway. Biochem Biophys Res Commun 399: 1–6, 2010. doi: 10.1016/j.bbrc.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 168.Zhang X, Schulze PC. MicroRNAs in heart failure: non-coding regulators of metabolic function. Biochim Biophys Acta 1862: 2276–2287, 2016. doi: 10.1016/j.bbadis.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhang X, Zuo X, Yang B, Li Z, Xue Y, Zhou Y, Huang J, Zhao X, Zhou J, Yan Y, Zhang H, Guo P, Sun H, Guo L, Zhang Y, Fu XD. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell 158: 607–619, 2014. doi: 10.1016/j.cell.2014.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhao F, Li B, Wei YZ, Zhou B, Wang H, Chen M, Gan XD, Wang ZH, Xiong SX. MicroRNA-34a regulates high glucose-induced apoptosis in H9c2 cardiomyocytes. J Huazhong Univ Sci Technolog Med Sci 33: 834–839, 2013. doi: 10.1007/s11596-013-1207-7. [DOI] [PubMed] [Google Scholar]
- 171.Zhao Y, Ponnusamy M, Dong Y, Zhang L, Wang K, Li P. Effects of miRNAs on myocardial apoptosis by modulating mitochondria related proteins. Clin Exp Pharmacol Physiol 44: 431–440, 2017. doi: 10.1111/1440-1681.12720. [DOI] [PubMed] [Google Scholar]
- 172.Zheng D, Ma J, Yu Y, Li M, Ni R, Wang G, Chen R, Li J, Fan GC, Lacefield JC, Peng T. Silencing of miR-195 reduces diabetic cardiomyopathy in C57BL/6 mice. Diabetologia 58: 1949–1958, 2015. doi: 10.1007/s00125-015-3622-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zhou Q, Lv D, Chen P, Xu T, Fu S, Li J, Bei Y. MicroRNAs in diabetic cardiomyopathy and clinical perspectives. Front Genet 5: 185, 2014. doi: 10.3389/fgene.2014.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet 12: 7–18, 2011. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 175.Zhu H, Shyh-Chang N, Segrè AV, Shinoda G, Shah SP, Einhorn WS, Takeuchi A, Engreitz JM, Hagan JP, Kharas MG, Urbach A, Thornton JE, Triboulet R, Gregory RI, Altshuler D, Daley GQ, DIAGRAM Consortium; MAGIC Investigators . The Lin28/let-7 axis regulates glucose metabolism. Cell 147: 81–94, 2011. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]