Abstract
Plasticity is a remarkable feature of the brain, allowing neuronal structure and function to accommodate to patterns of electrical activity. One component of these long-term changes is the activity-driven induction of new gene expression, which is required for both the long-lasting long-term potentiation of synaptic transmission associated with learning and memory, and the activitydependent survival events that help to shape and wire the brain during development. We have characterized molecular mechanisms by which neuronal membrane depolarization and subsequent calcium influx into the cytoplasm lead to the induction of new gene transcription. We have identified three points within this cascade of events where the specificity of genes induced by different types of stimuli can be regulated. By using the induction of the gene that encodes brain-derived neurotrophic factor (BDNF) as a model, we have found that the ability of a calcium influx to induce transcription of this gene is regulated by the route of calcium entry into the cell, by the pattern of phosphorylation induced on the transcription factor cAMP-response element (CRE) binding protein (CREB), and by the complement of active transcription factors recruited to the BDNF promoter. These results refine and expand the working model of activity-induced gene induction in the brain, and help to explain how different types of neuronal stimuli can activate distinct transcriptional responses.
Electrical activity within the brain rapidly encodes information about the world, but for these fleeting perceptions to have a lasting impact, long-term changes in the structure and function of neurons must follow. At the synapse, neurotransmitter reception initiates a number of biochemical signaling cascades in the postsynaptic cell, one of the most important of which is the elevation of intracellular calcium (1). This calcium rise is a critical component of signaling pathways whose functional consequences include activity-dependent survival (2) and the synaptic plasticity of long-term potentiation (LTP; ref. 3).
Early events induced by the rise of calcium in dendrites are likely to be local, resulting from posttranslational modifications of the synaptic machinery. However, for long-term structural and functional changes in the neuron, the calcium signal must regulate the expression of new gene products. There are several potential calcium-dependent steps in the process of new gene expression: elements of mRNA transcription, elongation, splicing, stability, and translation have all been suggested to be regulated by calcium in neurons. Especially exciting evidence has been accumulating in favor of the idea that some calcium-regulated mRNA translation occurs locally at postsynaptic sites, providing a means for rapid and accurate expression of activity-induced gene products at activated synapses (4, 5). The relative importance of dendritic mRNA translation in calcium-dependent biological processes will become clearer when the specific mRNAs that are regulated in this manner are identified.
Our laboratory has focused on the mechanisms by which calcium regulates new gene transcription. This line of investigation began with the observation that application of acetylcholine receptor agonists to the PC12 pheochromocytoma cell line induces rapid transcriptional initiation of the c-fos proto-oncogene in a manner that depends on calcium influx (6). c-Fos is a transcription factor whose mRNA expression had been shown to be rapidly induced when quiescent fibroblasts are stimulated by growth factors to reenter the cell cycle (7). That c-fos could also be induced in neurons suggested that an equally intricate pattern of regulated transcription might be responsible for cellular responses to neuronal activity. Indeed it has subsequently been shown that transcription of a large number of genes, encoding both transcription factors and molecules that function at synapses, is induced by synaptic activity and subsequent calcium influx (8, 9). These activity-dependent changes in gene transcription have been shown to be required for both neuronal survival (10) and for the maintenance of the late phase of LTP (11).
One of the best studied of the activity-induced genes is brain-derived neurotrophic factor (BDNF). BDNF is a small secreted protein that acts by binding to its receptors, the tyrosine kinase TrkB and the low-affinity neurotrophin receptor p75 (12). Ligation of TrkB receptors by BDNF promotes the activation of signaling pathways that both rapidly modify the function of local synaptic targets and also have long-term effects on gene transcription. BDNF was originally identified as an important neuronal survival factor in the central nervous system (13, 14). BDNF promotes survival through inactivation of components of the cell death machinery and also through activation of the transcription factor cAMP-response element binding protein (CREB), which drives expression of the pro-survival gene Bcl-2 (15, 16). In addition to its role in neuronal survival, BDNF also modulates synaptic activity (17). Mice lacking the BDNF gene show impaired hippocampal LTP, and overexpression of BDNF rescues this defect (18, 19). Both presynaptic and postsynaptic mechanisms may underlie this effect on synaptic plasticity. BDNF application to presynaptic terminals of Xenopus spinal neurons enhances both spontaneous and evoked release of neurotransmitter via a mechanism that may involve the phosphorylation and functional regulation of synaptic vesicle proteins (17, 20). At the postsynaptic membrane, BDNF may act as a neurotransmitter itself, as rapid pulsatile application of BDNF to neurons can cause TrkB-dependent membrane depolarization within milliseconds (21).
Perhaps because BDNF plays so many important roles in neuronal development and function, the expression and action of this protein is tightly regulated in time and space. In particular, expression of BDNF mRNA is highly responsive to electrical activity in the brain, being induced by a wide range of stimuli including seizures (22), electrical stimulation that produces LTP (23), and patterned visual input (24). BDNF is categorized as an immediate-early gene, meaning that the stimulus-induced transcriptional initiation of new BDNF mRNA occurs rapidly and without the need for new protein synthesis (25); stimuli induce the expression of new BDNF mRNA through the posttranslational modification of preexisting transcription factors. The function of BDNF protein expressed as a result of this activity-induced transcription can be assessed by acutely blocking the action of BDNF, either through the addition of function-blocking anti-BDNF Abs or soluble TrkB receptor extracellular domains that sequester newly synthesized and secreted BDNF. This approach has been used to study the role of BDNF in calcium-dependent neuronal survival. Depolarization and subsequent calcium influx promote the survival of cortical neurons in culture and also drive an increase in the transcription of BDNF mRNA (26). To determine whether stimulus-induced expression of BDNF is required for calcium-dependent neuronal survival, function-blocking BDNF Abs were added to the culture medium when the cells were depolarized, leading to a blockade of the ability of BDNF to activate TrkB receptors. Under these conditions, neuronal survival is reduced, suggesting that activity-induced BDNF expression is required for calcium-dependent neuronal survival. Similar experiments have implicated acutely synthesized BDNF in LTP. Sequestration of BDNF up to 1 h after the induction of hippocampal LTP causes previously potentiated synaptic transmission to return to baseline, suggesting that BDNF expression driven by stimuli that produce LTP may be required for the maintenance of synaptic potentiation (27).
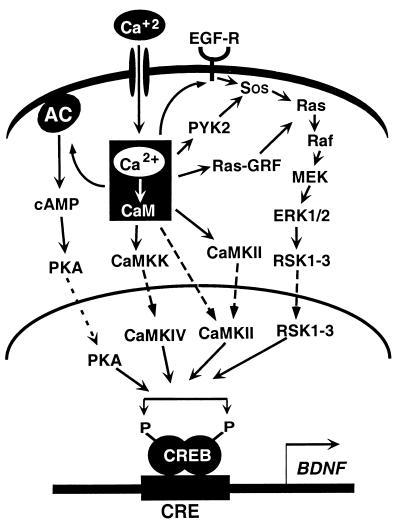
By using the induction of BDNF mRNA as an assay, we have explored in detail the mechanism of calcium-induced neuronal gene expression. Our studies, in combination with the work of numerous other labs, have revealed pathways that lead from the outer membrane of the cell, where calcium first enters the cell to the nucleus, where transcription is initiated. A summary of these pathways is presented in Fig. 1. Neurotransmitter reception and membrane depolarization open ligand- and voltage-gated calcium channels in the cell membrane, allowing the influx of extracellular calcium into the cell. This calcium is quickly bound by proteins that sit at the top of calcium-activated signaling cascades. A large number of pathways can respond to the elevation of intracellular calcium, and activation of these signal-transduction cascades amplifies the calcium signal while carrying it to the nucleus. Within the nucleus, the transcription factor CREB seems to be prebound to the BDNF promoter in an inactive form. Phosphorylation of CREB by calcium-regulated kinase cascades stimulates the recruitment of components of the basal transcription machinery to the BDNF promoter, and then new BDNF mRNA is synthesized.
Figure 1.
Calcium-activated signaling pathways that regulate gene transcription. In neurons, neurotransmitter reception and membrane depolarization lead to the opening of ligand- and voltage-gated calcium channels. Subsequent calcium influx across the plasma membrane drives the activation of a number of signaling molecules, including the calcium-sensitive adenylate cyclase, calcium/calmodulin-activated kinases, and Ras. Each of these molecules activates a cascade of signaling proteins that amplifies the calcium signal and carries it to the nucleus. Dashed lines represent the components of each pathway that are proposed to translocate into the nucleus. Nuclear kinases including protein kinase A, CaMK-IV, and members of the Rsk family phosphorylate CREB at Ser-133, rendering it competent to mediate transcription of genes such as BDNF. [Reproduced with permission from ref. 60 (Copyright 1999, Annual Reviews, http://AnnualReviews.org).]
Despite the elucidation of this general mechanism by which calcium elevation induces neuronal gene expression, a number of questions remain to be addressed. For example, there are many routes of calcium entry in neurons, and it has been widely observed that not all paths of entry are equally efficient at inducing the expression of activity-induced genes including BDNF. This observation raises the question, how is it possible that some calcium channels are tightly coupled to gene expression whereas others are not? Downstream of calcium entry, calcium-activated signaling pathways couple to transcription through the phosphorylation of CREB, but certain phosphorylation events render CREB competent to drive transcription whereas others do not. We have studied the molecular mechanisms of calcium-activated CREB-dependent transcription and investigated how CREB phosphorylation affects the formation of active transcriptional complexes. Finally, although CREB is one calcium-responsive transcription factor that contributes to BDNF transcription, mutational analysis of the BDNF promoter has revealed that there are additional non-CREB binding elements required for calcium-dependent BDNF transcription. We have used these elements to identify other calcium-regulated transcription factors that drive BDNF transcription and have explored how this complex of transcriptional activators gives specificity to transcriptional initiation at the BDNF promoter. In this article, we present our past and current work addressing these issues and describe how these findings have contributed to our working model of neuronal calcium-induced gene expression.
The Route of Calcium Entry Matters: Channel-Associated Signaling Complexes
The first step in calcium regulation of neuronal gene expression is the influx of calcium into the cytoplasm. There are four primary routes of calcium entry into the cytoplasm of the postsynaptic neuron: extracellular calcium can enter through the ligand-gated ion channels of the N-methyl-d-aspartate-type (NMDA) and α-amino-3-hydroxy-5-methyl-4-isoxazolepropionate-type (AMPA) (28) glutamate receptors, through voltage-gated calcium channels, or calcium can be released from intracellular stores (29).
The NMDA subtype of glutamate receptor is responsible for a substantial fraction of the total calcium influx in response to synaptic activity, especially early in development (30). The NMDA receptor (NMDA-R) is uniquely suited to play a role in Hebbian synaptic plasticity, because maximal ion flow through this channel depends on the coincidence of presynaptic activity, which releases glutamate to bind the receptor, and postsynaptic depolarization, which is required to relieve blockade of the NMDA-R channel by extracellular magnesium (31). Calcium entry through the NMDA-R has been shown to play a critical role in the induction of LTP (32), and it also induces the transcription of many immediate-early genes (33, 34). The other major route of calcium influx across the plasma membrane is through the voltage-sensitive calcium channels. In neurons, L-type, dihydropyridine-sensitive, alpha 1C- or 1D-containing voltage-sensitive calcium channels (L-VSCCs, designated Cav1.2 and Cav1.3) are especially concentrated in the basal dendrites and cell soma, where they are well positioned to transduce calcium-regulated signaling events to the nucleus (35, 36). Despite the fact that pharmacological inhibitors of L-VSCCs block a relatively minor fraction of the calcium influx that is induced by spontaneous synaptic activity in cultured cells, this blockade largely suppresses IEG transcription, suggesting that calcium flux through L-VSCCs is tightly linked to gene expression (37).
Although calcium is a ubiquitous second messenger, cells have found ways to endow distinct calcium signals with specific functions. Numerous studies have shown that not all routes of calcium entry are equivalent in their ability to induce specific gene-transcription events (38–40). The transcription of BDNF is preferentially driven by calcium influx through L-VSCCs, whereas it is poorly induced by calcium entering through NMDA-Rs (26). This channel specificity is despite the fact that there is little difference in the duration or magnitude of the somatic calcium rise mediated by direct stimulation of NMDA-Rs and L-VSCCs (41). Similarly, although calcium influx through L-VSCCs strongly activates CREB, calcium entering through non-L-VSCCs [including somatodendritically localized N-type channels (42)], generates a calcium rise in the cell soma that is equivalent in amplitude to that of L-type channels, but activation of these channels does not lead to CREB-dependent gene transcription (ref. 43 and R.E.D. and M.E.G., unpublished observations).
There are at least two possibilities to explain why the entry of calcium through different channels might lead to differences in the activation of gene transcription. By using the activation of CREB phosphorylation as an assay, one set of studies has led to the hypothesis that this specificity comes from the tethering of molecules required for the activation of calcium signaling pathways near the mouths of the channels (43, 44). The extremely high levels of calcium reached within the microdomain at the mouth of the channel would activate signaling pathways that travel to the nucleus to phosphorylate and activate CREB. However, another set of studies has suggested instead that the critical parameter for the activation of CREB-dependent transcription is the elevation of nuclear calcium (45, 46). In this case, the specificity of channel signaling for transcription would depend on the ability of each type of channel to generate a nuclear calcium rise leading to the activation of nuclear kinases that phosphorylate and activate CREB. To gain further insight into where and how calcium influxes gain their specificity toward transcription, we have examined the properties of calcium channels that enable them to drive CREB-dependent transcription. Our data support the hypothesis that the physical association of signaling molecules with calcium channels is an important means of regulating the coupling of calcium influxes to gene transcription.
During early neuronal development, calcium influx through the NMDA-R is thought to play a critical role in the regulation of a number of developmental processes including the plasticity of synapses. Recent studies have revealed that there are a large number of proteins in close association with the NMDA-R in the postsynaptic density (47, 48). Of particular interest is evidence that the NMDA-R directly associates with the EphB family of receptor tyrosine kinases (49). Both EphB receptors and their ephrin ligands are found concentrated at synapses, suggesting that they have important signaling functions at this site of cell–cell contact (50), and one of the roles of synaptic EphB receptors seems to be the regulation of synapse formation or function. Ligation of EphB receptors causes them to cluster within minutes, and within an hour promotes the coclustering of NMDA-Rs (49). In addition to these immediate events, activation of EphB receptors also has long-term effects on the number of synaptic sites. Stimulation of EphB receptors over a period of days increases the number of NMDA-R clusters as well as the number of presynaptic release sites. One way EphB receptor stimulation might mediate these long-term effects on synapse formation is through the regulation of genes required for synaptogenesis or synaptic plasticity. Our preliminary experiments suggest that activation of EphB receptors modulates the ability of calcium influx through NMDA-Rs to induce expression of immediate-early genes (M.A.T., M.B.D., and M.E.G., unpublished observations). These results suggest that in developing neurons, the association between the EphB receptor and the NMDA-R may play two roles, both immediately affecting channel clustering and later having a long-term effect on the regulation of gene products required for the development of new synapses.
The ability of NMDA-Rs to drive changes in CREB-dependent gene expression is a developmentally regulated event. Whereas in embryonic neurons cultured for 7 days in vitro (DIV), activation of calcium influx through NMDA-Rs leads to robust CREB-dependent transcription, by 14 DIV, a similar stimulus does not induce CREB-dependent transcription (51). The ability of calcium influx to induce CREB-dependent transcription is correlated with the ability of calcium to promote the sustained phosphorylation of CREB at a serine residue required for activation (Ser-133). In contrast to calcium influx through L-VSCCs that leads to sustained CREB Ser-133 phosphorylation, the entry of calcium through NMDA-Rs, except in very young neurons, triggers only transient phosphorylation of CREB at Ser-133 (41, 51). Recently it has been demonstrated that in embryonic cortical neurons cultured for 14 DIV, calcium entry through the NMDA-R induces a phosphatase that dephosphorylates CREB at Ser-133 (51). Because calcium entry through L-VSCCs does not activate this phosphatase, there is a source-specific ability of calcium to drive either transient or sustained CREB phosphorylation. Protein phosphatase 1 (52) has been shown to associate with the NMDA-R, however it remains to be determined whether the local tethering of this phosphatase near the receptor is required for the rapid dephosphorylation of CREB at Ser-133.
These results help to explain why NMDA-Rs are developmentally regulated activators of CREB-dependent transcription, but what allows L-VSCCs to efficiently drive CREB? One possible explanation is that specific signaling molecules that transduce the calcium signal into CREB phosphorylation are physically coupled to L-VSCCs. Several signaling molecules are known to bind L-VSCCs, including the protein kinase A anchoring protein AKAP (53), the nonreceptor tyrosine kinase Src (54), and the calcium transducer calmodulin (55). Studies to date have concentrated on how association of these signaling proteins with the L-VSCCs affects the function of the channel. However, our data suggest that binding of signaling molecules may also regulate the ability of L-VSCCs to mediate calcium-dependent cellular responses such as transcriptional activation. We propose that the physical association of signaling molecules with the L-VSCC is required for activation of signaling pathways such as the Ras/mitogen-activated protein kinase (MAPK) or calcium/calmodulin-dependent kinases (CaMKs) that are downstream of the channel itself (56).
Calcium-Regulated Transcription Factors: CREB
It is well established that changes in intracellular calcium levels are transduced into activation of gene transcription by signal transduction pathway-induced modifications of transcription factors. A number of transcription factors have been identified whose transcriptional activity is regulated by calcium-activated signaling pathways. The challenge has been to identify calcium-dependent sites of phosphorylation on these transcription factors, and to link these phosphorylation events mechanistically to the resulting change in transcriptional competence of the transcription factors. Recent studies have begun to provide an understanding of how calcium-signaling pathways regulate the transcription factor CREB.
CREB was first purified as a binding protein for the sequence 5′-TGACGTCA-3′ in the somatostatin promoter (57). This sequence (named the CRE for cAMP response element) had been identified as an element within the somatostatin promoter required for the transcriptional up-regulation of somatostatin expression in response to elevation of cellular cAMP levels and the activation of protein kinase A. Mutagenesis of the c-fos promoter resulted in the identification of a similar sequence element (5′-TGACGTTT-3′) that is required for the induction of c-fos transcription in response to membrane depolarization and calcium influx (58). CREB was subsequently identified as the c-fos promoter calcium-response element (CaRE) binding protein and shown to mediate both cAMP and calcium induction of c-fos expression through the CRE/CaRE sequence (59).
CREB has been implicated in a number of biological functions, all of which depend on its ability to act as a stimulus-induced transcription factor that can be activated by a variety of extracellular signals. In the brain, CREB mediates both activity-dependent synaptic plasticity and trophic factor-dependent neuronal survival (60, 61). Consistent with its role as an activity-dependent transcriptional factor, CREB is phosphorylated at a serine critical for its function (Ser-133) in physiologically active brain areas during a wide range of behaviors, including birdsong, cocaine reward, fear conditioning, and spatial learning. Genetic manipulations of CREB expression in Aplysia, Drosophila, and mice suggest that CREB is required for the potentiation of synaptic strength in paradigms of learning and memory (62). Neurotrophic factor stimulation also leads to the phosphorylation of CREB at Ser-133, and in two studies, the inhibition of CREB function was found to block BDNF-mediated survival of cerebellar granule neurons (15, 16). Further identification of the range of target genes induced by activated CREB in response to different stimuli will help to refine our understanding of the role played by CREB in cellular behaviors including synaptic plasticity and survival.
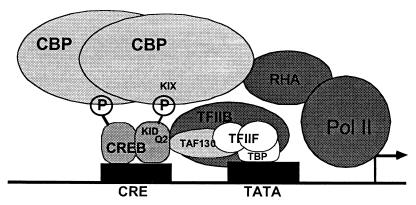
How do extracellular stimuli lead to the activation of CREB? The prevailing view is that in unstimulated cells, CREB is found in the nucleus bound to CREs within the promoters of CREB-regulated genes (Fig. 2; ref. 60). After cellular stimulation, signaling pathways are activated that transmit the signal to the nucleus and cause the phosphorylation of CREB at Ser-133. CREB is phosphorylated on this residue in response to cAMP elevation, membrane depolarization/calcium influx, and growth-factor reception, and phosphorylation of this site is critical for the function of CREB as a transcriptional activator in response to these stimuli (63–65). After phosphorylation at Ser-133, CREB recruits the CREB binding protein (CBP), which acts as a transcriptional coactivator (66, 67). CBP promotes transcription through its recruitment of components of the RNA polymerase II transcription machinery and through its function as a histone acetyl transferase. CBP-catalyzed acetylation of lysine residues within histones helps to remodel chromatin structure into a form that is accessible to active transcription. Recent experiments have tested the importance of the CREB–CBP association for transcriptional activation (68). We used an Escherichia coli two-hybrid binding system to screen for mutations of the CREB binding domain of CBP (called the KIX domain) that interact more or less strongly than wild-type CBP with CREB phosphorylated at Ser-133 in the presence of cotransfected protein kinase A (69). When these mutations were introduced into full-length CBP, and the mutant CBPs were transfected into mammalian cells, a CBP that bound with higher affinity to CREB-enhanced CREB reporter gene transcription. By contrast, expression of a mutant CBP that bound CREB with lower affinity than wild type was less effective at promoting CREB-dependent transcription. These data indicate that the strength of the CREB–CBP interaction determines the level of CREB-dependent transcription.
Figure 2.
The prevailing view for the mechanism of calcium-dependent CREB activation. CREB sits prebound as a dimer to the CRE in unstimulated cells. In response to neuronal stimulation and the activation of CREB kinases, CREB is phosphorylated at Ser-133 within the kinase-inducible domain (KID), allowing it to bind the KIX domain of CBP (or potentially a CBP dimer) recruiting this coactivator to the promoter. CBP promotes transcriptional activation in part through binding indirectly to the RNA polymerase II via an interaction with the RNA helicase A (RHA) protein, and thus CBP recruits the polymerase complex onto promoters bound by Ser-133-phosphorylated CREB. Other CREB-mediated interactions with the basal transcription machinery may also help to promote transcription. Through a glutamine-rich region (Q2), CREB binds to TAF130, a component of TFIID, and also to TFIIB. These complexes of proteins associate with the TATA binding protein (TBP) at the TATA box that is found just proximal to the initiation site of many genes. [Reproduced with permission from ref. 60 (Copyright 1999, Annual Reviews, http://AnnualReviews.org).]
CREB is phosphorylated at Ser-133 in response to a wide variety of stimuli, and a number of signaling pathways have been proposed to culminate in the nuclear phosphorylation of CREB at this residue. Among these pathways are the calcium/calmodulin-activated kinases and the Ras/MAPK pathway. Activation of plasma membrane voltage- and ligand-gated calcium channels leads to the elevation of cytoplasmic and nuclear calcium levels and the activation of the CaMKs. Although CaMKs I, II, and IV can each phosphorylate Ser-133 in vitro, CaMK-IV seems to be the most relevant CaMK in cells. CaMK-IV is localized to the nucleus, and the kinetics of its activation correlate in time with CREB phosphorylation and dephosphorylation on Ser-133 (44). Disruption of CaMK-IV activity by treatment of cells with CaMK-IV antisense oligonucleotides or by genetic homologous recombination reduces activity-dependent CREB Ser-133 phosphorylation, providing strong evidence that this kinase contributes a substantial fraction of the endogenous membrane depolarization-activated CREB kinase activity (44, 70). Calcium influx also triggers activation of the Ras/MAPK pathway, which culminates in the activation of the nuclear Rsk kinases, including Rsk2, a kinase first shown to phosphorylate CREB at Ser-133 in response to growth factor signaling (65, 71). Rsks are also activated by membrane depolarization (72), suggesting that they could mediate both growth factor- and calcium-dependent CREB phosphorylation. In response to many stimuli, the activation of the Ras/MAPK/Rsk and CaMK pathways occur in concert, suggesting that these pathways may play cooperative roles in CREB activation. Careful examination of the kinetics of CREB-dependent phosphorylation has revealed that CaMKs seem to dominate the rapid phase of CREB phosphorylation after membrane depolarization, whereas Ras/MAPK pathway activation is slower and becomes the predominant CREB kinase at later times after stimulation (56, 73). The timing and magnitude of activation of each of the kinase pathways could transmit information about the nature or extent of the stimulus that may subsequently be translated into the type or amount of transcription driven by CREB under different conditions.
Although phosphorylation of CREB at Ser-133 is required for the calcium induction of CREB-dependent transcription, there are many instances in which phosphorylation of CREB at Ser-133 is not sufficient for target gene activation. Depolarization of neurons leads to phosphorylation of CREB at Ser-133 within minutes, but CREB-dependent transcription takes longer to initiate. Ser-133 phosphorylation also persists past the time when transcription has ceased. In PC12 cells, depolarization, elevation of cAMP, and the neurotrophic factor nerve growth factor (NGF) are all able to induce the phosphorylation of CREB on Ser-133; however, NGF is unable to drive transcription from a CREB-dependent reporter gene (74). These results suggest that in addition to phosphorylation of CREB at Ser-133, other phosphorylation sites on either CREB, CBP, or some other transcriptional coactivator, may be regulated in concert with Ser-133 for calcium to effectively drive CREB-dependent transcription.
What is the nature of these additional signaling events? CREB is known to be phosphorylated on sites in addition to Ser-133, and our recent experiments suggest that phosphorylation at some of these sites is required for the activation of CREB in response to membrane depolarization. In vitro studies showed that CaMK-II could phosphorylate CREB at Ser-142, however phosphorylation at this site had been proposed to be inhibitory, as mutation of the serine at 142 to an alanine enhances the CREB-dependent activation of a reporter gene (75). In contrast to this prediction, Ser-142 phosphorylation is induced by membrane depolarization at times when CREB-dependent transcription is activated (J.M.K., A.J.S, and M.E.G., unpublished observations). Our preliminary evidence indicates that membrane depolarization-induced Ser-142 phosphorylation may be accompanied by the phosphorylation of CREB at additional sites. Given that phosphorylation of Ser-142 and/or Ser-143 has been shown to disrupt the association of CREB with the KIX domain of CBP (76), it may be that phosphorylation at these sites allows CREB to form a complex with a coactivator other than CBP or with a region of CBP other than the KIX domain. In either case, the formation of this alternative transcription complex in response to these specific signaling events may offer a way to modulate the range of target genes induced by CREB in response to different types of stimuli, and may help to explain the ability of CREB to function in a wide range of biological functions.
Calcium Regulation of BDNF Transcription
CREB can be activated by a wide variety of stimuli and signaling pathways in neurons and other cells, but at least one of its target genes, the gene encoding BDNF, is induced under a more restricted set of conditions. Although membrane depolarization of the PC12 neuroendocrine cell line leads to robust transcription of a CREB-dependent reporter gene at levels similar to those seen in neurons, BDNF expression is poorly induced in these cells by membrane depolarization (A.E.W., X.T., and M.E.G., unpublished observations). In neurons, BDNF expression is only strongly induced by membrane depolarization, whereas CREB-dependent transcription can also be driven by intracellular cAMP elevation or extracellular BDNF application.
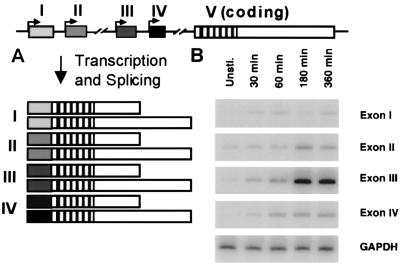
To understand the calcium- and neural-specific regulation of BDNF transcription, we have identified elements within the BDNF promoters that are required for induction of expression in response to membrane depolarization in neurons. The gene for BDNF has a complex organization with four initial exons, each of which can be spliced to a single 3′ exon containing the coding domain for the BDNF protein (Fig. 3A; ref. 77). Each of these four splice variants uses one of two alternative polyadenylation sites within the 3′-untranslated region. At present, the differential functions of these eight transcripts, all of which encode the same protein, are not known. One possibility is that these mRNA transcripts might be differentially targeted and translated within the cell, as some BDNF mRNA has been found to be transported into dendrites (78).
Figure 3.
Calcium-dependent induction of BDNF exon III expression. (A) The BDNF gene has four potential initial exons, each of which can be alternately spliced to a single 3′ exon containing the complete BDNF coding sequence. Each of these splice variants can use one of two alternative polyadenylation sites within the 3′ untranslated region, generating a total of eight distinct transcripts, all of which encode the same protein sequence. (B) Cultured embryonic cortical neurons were depolarized with 50 mM KCl for the times indicated, then RNA was harvested from the cells for quantitative reverse transcription (RT)-PCR analysis by using probes specific for the four initial exons of the BDNF transcripts or for glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Exon III is strongly induced within 180 min of depolarization. GAPDH is constant over the time course examined, serving as a control for RNA input and reverse transcription efficiency. [Reproduced with permission from ref. 79 (Copyright 1998, Elsevier Science).]
Regulatory elements that control transcription are most commonly found in the DNA sequences that flank the 5′ end of the initial exon of a transcript. Because there are four possible initial exons in the BDNF gene, it is important to determine which of these promoters are responsive to neuronal calcium-signaling pathways. Transcripts containing exons I, II, and III are expressed throughout the brain, whereas exon IV-containing transcripts are expressed primarily outside of the brain. Both exon I- and III-containing transcripts are induced by seizure activity in adult brain (77). Reverse transcription (RT)-PCR analysis demonstrates that transcripts containing exon III are the most highly induced BDNF transcripts in membrane-depolarized embryonic cortical neuron cultures (Fig. 3B; ref. 79). In these cells, induction of exon III-containing transcripts is not blocked when new protein synthesis is inhibited with cycloheximide, indicating that exon III transcription is driven by the posttranslational modification of preexisting transcription factors.
To identify sequences within promoter III of the BDNF gene that mediate membrane depolarization induction of BDNF exon III, we and others developed a reporter gene assay in cultured embryonic cortical neurons (79, 80). The 170 base pairs 5′ to the BDNF exon III initiation site when fused to a luciferase reporter gene were able to drive calcium-induced transcription of the reporter gene in response to membrane depolarization. We made a series of deletions of this promoter region to identify the sequences that are required for calcium induction of the reporter gene. The initial analysis revealed a requirement for a DNA sequence resembling a CREB binding site at -35 bp relative to the transcription initiation site, suggesting that CREB might drive BDNF transcription (79, 80). Point mutations within this DNA sequence element abolish membrane depolarization-driven induction of the BDNF promoter III-luciferase reporter gene. Although the BDNF CRE (B-CRE) is a variant of the canonical CRE sequence, recombinant CREB is capable of binding the B-CRE sequence. A protein present in neuronal nuclear extracts was identified that also binds the B-CRE in an electrophoretic mobility shift assay. This protein binds to the B-CRE in vitro with the same specificity as the protein that drives B-CRE-dependent transcription, because point mutations in the B-CRE that block calcium induction of transcription also block binding of the nuclear protein to the B-CRE in the electrophoretic mobility shift assay. Anti-CREB Abs were found to bind to the nuclear protein/B-CRE complex, implying that the endogenous B-CRE binding protein is indeed CREB or a closely related CREB family member. In further support of this idea, overexpression of a dominant interfering form of CREB in neurons blocks the induction of endogenous BDNF mRNA by membrane depolarization. In total, these data suggest that CREB itself or a CREB family member is an important regulator of BDNF transcription in cells.
In addition to the CREB binding site, further deletion analysis and detailed point mutagenesis of promoter III has revealed that there are two other DNA elements that are also required for calcium induction of the BDNF promoter III-luciferase reporter (A.E.W., W.G.C., X.T., and M.E.G., unpublished observations; refs. 79 and 80). The other two CaREs lie between -77 and -45 bp 5′ of the initiation site. The more proximal element contains an E-box sequence, and we have found that it binds a member of the basic helix–loop–helix family of transcription factors. The distal CaRE does not resemble any known transcription factor binding sites, so we used a yeast one-hybrid approach to identify proteins capable of binding to this DNA element. The yeast one-hybrid screen utilizes a selection strategy in which an auxotrophic survival gene is placed under the regulatory control of the DNA element we had identified from the deletion analysis. Then the yeast are transfected with a neuronal cDNA library in which each clone is expressed as a fusion protein with a yeast transcriptional activation domain. Any clones from the library that are capable of binding the DNA element should recruit the transcriptional activation domain onto the promoter of the survival gene, and then the yeast will grow on plates deficient in the amino acid used for the selection. From this screen we cloned a novel transcription factor that binds the distal CaRE from BDNF promoter III. Point mutations of the distal CaRE that block calcium induction in the context of the BDNF promoter III-luciferase reporter also eliminate binding of the novel transcription factor, consistent with a role for this factor in the calcium-regulated transcription of BDNF exon III.
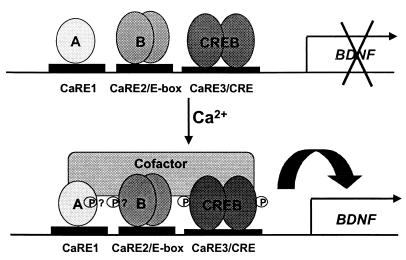
What is the purpose of having three distinct CaREs within BDNF promoter III? Mutation of any one of the three CaREs nearly eliminates calcium induction of the BDNF promoter III reporter gene, implying that coordinated activation of all three factors on the promoter is required for efficient transcriptional initiation. This requirement for cooperativity may serve to restrict activation of BDNF transcription until a number of signaling events can be integrated into the activation of the three transcription factors. Nuclear run-on assays support the idea that BDNF transcription may require several events, as transcriptional initiation of BDNF takes 60 min after membrane depolarization, whereas initiation of c-fos, another calcium-regulated gene, occurs in less than 30 min (79). In addition, requiring the activation of all three transcription factors to initiate transcription from the promoter may explain the preference of BDNF induction for membrane depolarization over other types of stimuli. If phosphorylation of CREB at Ser-133 were sufficient to drive BDNF transcription, elevation of intracellular cAMP should induce BDNF, but it does so poorly. One potential explanation is that when intracellular cAMP is elevated, although CREB may be found in an activated form on the BDNF promoter III, at least one of the other two transcription factors is not, and thus transcription at the promoter cannot be initiated. Why phosphorylated CREB is not sufficient to initiate transcription at BDNF promoter III is not yet clear, however, one possibility is that the three transcription factors may cooperatively recruit a common transcriptional coactivator (Fig. 4).
Figure 4.
Three transcription factors bind to the BDNF promoter III CaREs and coordinately regulate transcription. Three elements within BDNF promoter III are required for calcium-dependent induction of a luciferase reporter gene, and three distinct transcription factors bind to these elements. “A” represents the novel calcium-regulated transcription factor that binds to CaRE1, and “B” represents the basic helix–loop–helix family member that binds the CaRE2/E-box element. CREB binds the CaRE3/CRE. In response to depolarization and the activation of calcium-signaling pathways, all three factors are activated to promote transcription, potentially through the phosphorylation-dependent recruitment of a common transcriptional coactivator.
Conclusions/Future Directions
The specificity of signaling within the brain is a critical feature that allows neurons to have a wide dynamic range of responses to a myriad of stimuli. We have identified at least three points of specificity that influence the induction of gene expression in response to neuronal activity. First, we have shown that the physical association of signaling molecules with calcium channels influences the ability of these channels to promote gene expression. Next, we have demonstrated that different stimuli drive phosphorylation of the transcription factor CREB at unique sites, altering its transcriptional activity. Finally, we have seen that the neuron- and calcium-specific induction of BDNF promoter III is mediated in part by the coordinated action of three separate transcription factors, each of which seems to be activated by a distinct but overlapping set of intracellular signaling pathways. The challenge for the future is to link these steps together and to understand the molecular mechanisms that regulate them. For example, if calcium influx through L-VSCCs leads to specific sites of phosphorylation on CREB, is this the result of local tethering of signaling molecules to the L-VSCCs that then mediate these phosphorylation events? Could calcium-specific phosphorylation of CREB play a role in calcium-specific induction of BDNF promoter III? If so, how does altered CREB phosphorylation regulate its ability to interact with the other two transcription factors at the promoter? These and other similar questions will drive future efforts to obtain a more detailed understanding of long-term activity-dependent plasticity and gene expression in the brain.
Acknowledgments
M.E.G. acknowledges the generous support of the F. M. Kirby Foundation to the Division of Neuroscience. This work was supported by a Mental Retardation Research Center Grant (HD18655) and a National Institutes of Health Grant (NS28829-07) (to M.E.G.), an American Cancer Society Postdoctoral Fellowship (to A.E.W.), a Howard Hughes Medical Institute Predoctoral Fellowship (to W.G.C.), and by a Helen Hay Whitney Foundation Postdoctoral Fellowship (to R.E.D.). M.B.D. is a Chiron Life Science Research Foundation Fellow.
Abbreviations
- BDNF
brain-derived neurotrophic factor
- LTP
long-term potentiation
- NMDA
N-methyl-d-aspartate type glutamate receptor
- NMDA-R
NMDA receptor
- L-VSCC
L-type voltage-sensitive calcium channel
- CaRE
calcium-response element
- CRE
cAMP-response element
- CREB
CRE binding protein
- CBP
CREB binding protein
- MAPK
mitogen-activated protein kinase
- B-CRE
BDNF CRE
- CAMK
calcium/calmodulin-dependent kinase
Footnotes
This paper was presented at the Inaugural Arthur M. Sackler Colloquium of the National Academy of Sciences, “Neural Signaling,” held February 15–17, 2001, at the National Academy of Sciences in Washington, DC.
References
- 1.Ghosh A, Greenberg M E. Science. 1995;268:239–247. doi: 10.1126/science.7716515. [DOI] [PubMed] [Google Scholar]
- 2.Franklin J L, Johnson E M. Trends Neurosci. 1992;15:501–508. doi: 10.1016/0166-2236(92)90103-f. [DOI] [PubMed] [Google Scholar]
- 3.Lynch G, Larson J, Kelso S, Barrionuevo G, Schottler F. Nature (London) 1983;305:716–721. doi: 10.1038/305719a0. [DOI] [PubMed] [Google Scholar]
- 4.Wu L, Wells D, Tay J, Mendis D, Abbott M A, Barnitt A, Quinlan E, Heynen A, Fallon J R, Richter J D. Neuron. 1998;21:1129–3119. doi: 10.1016/s0896-6273(00)80630-3. [DOI] [PubMed] [Google Scholar]
- 5.Aakalu G, Smith W B, Nguyen N, Jiang C, Schuman E M. Neuron. 2001;30:489–502. doi: 10.1016/s0896-6273(01)00295-1. [DOI] [PubMed] [Google Scholar]
- 6.Greenberg M E, Ziff E B, Greene L A. Science. 1986;234:80–83. doi: 10.1126/science.3749894. [DOI] [PubMed] [Google Scholar]
- 7.Greenberg M E, Ziff E B. Nature (London) 1984;311:433–437. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- 8.Rosen L B, Ginty D D, Greenberg M E. Adv Second Messenger Phosphoprotein Res. 1995;30:225–253. doi: 10.1016/s1040-7952(05)80009-6. [DOI] [PubMed] [Google Scholar]
- 9.Lanahan A, Worley P. Neurobiol Learn Mem. 1998;70:37–43. doi: 10.1006/nlme.1998.3836. [DOI] [PubMed] [Google Scholar]
- 10.Mao Z, Bonni A, Xia F, Nadal-Vicens M, Greenberg M E. Science. 1999;286:785–790. doi: 10.1126/science.286.5440.785. [DOI] [PubMed] [Google Scholar]
- 11.Nguyen P V, Abel T, Kandel E R. Science. 1994;265:1104–1107. doi: 10.1126/science.8066450. [DOI] [PubMed] [Google Scholar]
- 12.Barbacid M. Ann NY Acad Sci. 1995;766:442–458. doi: 10.1111/j.1749-6632.1995.tb26693.x. [DOI] [PubMed] [Google Scholar]
- 13.Jones K R, Farinas I, Backus C, Reichardt L F. Cell. 1994;76:989–999. doi: 10.1016/0092-8674(94)90377-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwartz P M, Borghesani P R, Levy R L, Pomeroy S L, Segal R A. Neuron. 1997;19:269–281. doi: 10.1016/s0896-6273(00)80938-1. [DOI] [PubMed] [Google Scholar]
- 15.Bonni A, Brunet A, West A E, Datta S R, Takasu M A, Greenberg M E. Science. 1999;286:1358–1362. doi: 10.1126/science.286.5443.1358. [DOI] [PubMed] [Google Scholar]
- 16.Riccio A, Ahn S, Davenport C M, Blendy J A, Ginty D D. Science. 1999;286:1363–1367. doi: 10.1126/science.286.5448.2358. [DOI] [PubMed] [Google Scholar]
- 17.Poo M-m. Nat Rev Neurosci. 2001;2:1–9. doi: 10.1038/35049004. [DOI] [PubMed] [Google Scholar]
- 18.Korte M, Carroll P, Wolf E, Brem G, Thoenen H, Bonhoeffer T. Proc Natl Acad Sci USA. 1995;92:8856–8860. doi: 10.1073/pnas.92.19.8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patterson S L, Abel T, Deuel T A S, Martin K C, Rose J C, Kandel E R. Neuron. 1996;16:1137–1145. doi: 10.1016/s0896-6273(00)80140-3. [DOI] [PubMed] [Google Scholar]
- 20.Jovanovic J N, Czernik A J, Fienberg A A, Greengard P, Sihra T S. Nat Neurosci. 2000;3:323–329. doi: 10.1038/73888. [DOI] [PubMed] [Google Scholar]
- 21.Kafitz K W, Rose C R, Thoenen H, Konnerth A. Nature (London) 1999;401:918–921. doi: 10.1038/44847. [DOI] [PubMed] [Google Scholar]
- 22.Ernfors P, Bengzon J, Kokaia Z, Persson H, Lindvall O. Neuron. 1991;7:165–176. doi: 10.1016/0896-6273(91)90084-d. [DOI] [PubMed] [Google Scholar]
- 23.Patterson S L, Grover L M, Schwartzkroin P A, Bothwell M. Neuron. 1992;9:1081–1088. doi: 10.1016/0896-6273(92)90067-n. [DOI] [PubMed] [Google Scholar]
- 24.Tokuyama W, Okuno H, Hashimoto T, Li Y X, Miyashita Y. Nat Neurosci. 2000;3:1134–1142. doi: 10.1038/80655. [DOI] [PubMed] [Google Scholar]
- 25.Lauterborn J C, Rivera S, Stinis C T, Haynes V Y, Isackson P J, Gall C M. J Neurosci. 1996;16:7428–7436. doi: 10.1523/JNEUROSCI.16-23-07428.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghosh A, Carnahan J, Greenberg M E. Science. 1994;263:1618–1623. doi: 10.1126/science.7907431. [DOI] [PubMed] [Google Scholar]
- 27.Kang H, Welcher A A, Shelton D, Schuman E M. Neuron. 1997;19:653–664. doi: 10.1016/s0896-6273(00)80378-5. [DOI] [PubMed] [Google Scholar]
- 28.Jonas P, Burnashev N. Neuron. 1995;15:987–990. doi: 10.1016/0896-6273(95)90087-x. [DOI] [PubMed] [Google Scholar]
- 29.Berridge M J. Neuron. 1998;21:13–26. doi: 10.1016/s0896-6273(00)80510-3. [DOI] [PubMed] [Google Scholar]
- 30.Yuste R, Katz L C. Neuron. 1991;6:333–344. doi: 10.1016/0896-6273(91)90243-s. [DOI] [PubMed] [Google Scholar]
- 31.Bourne H R, Nicoll R. Cell. 1993;72,Suppl.:65–75. doi: 10.1016/s0092-8674(05)80029-7. [DOI] [PubMed] [Google Scholar]
- 32.Perkel D, Petrozzino J, Nicoll R, Connor J. Neuron. 1993;11:817–823. doi: 10.1016/0896-6273(93)90111-4. [DOI] [PubMed] [Google Scholar]
- 33.Cole A J, Saffen D W, Baraban J M, Worley P F. Nature (London) 1989;340:474–476. doi: 10.1038/340474a0. [DOI] [PubMed] [Google Scholar]
- 34.Bading H, Segal M M, Sucher N J, Dudek H, Lipton S A, Greenberg M E. Neuroscience. 1995;64:653–664. doi: 10.1016/0306-4522(94)00462-e. [DOI] [PubMed] [Google Scholar]
- 35.Catterall W A. Ann Rev Cell Dev Biol. 2000;16:521–555. doi: 10.1146/annurev.cellbio.16.1.521. [DOI] [PubMed] [Google Scholar]
- 36.Westenbroek R E, Ahlijanian M K, Catterall W A. Nature (London) 1990;347:281–284. doi: 10.1038/347281a0. [DOI] [PubMed] [Google Scholar]
- 37.Murphy T H, Worley P F, Baraban J M. Neuron. 1991;7:625–635. doi: 10.1016/0896-6273(91)90375-a. [DOI] [PubMed] [Google Scholar]
- 38.Gallin W J, Greenberg M E. Curr Opin Neurobiol. 1995;5:367–374. doi: 10.1016/0959-4388(95)80050-6. [DOI] [PubMed] [Google Scholar]
- 39.Ginty D D. Neuron. 1997;18:183–186. doi: 10.1016/s0896-6273(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 40.Bading H, Ginty D D, Greenberg M E. Science. 1993;260:181–186. doi: 10.1126/science.8097060. [DOI] [PubMed] [Google Scholar]
- 41.Hardingham G E, Chawla S, Cruzalegui F H, Bading H. Neuron. 1999;22:789–798. doi: 10.1016/s0896-6273(00)80737-0. [DOI] [PubMed] [Google Scholar]
- 42.Westenbroek R E, Hell J W, Warner C, Dubel S J, Snutch T P, Catterall W A. Neuron. 1992;9:1099–1115. doi: 10.1016/0896-6273(92)90069-p. [DOI] [PubMed] [Google Scholar]
- 43.Deisseroth K, Heist E K, Tsien R W. Nature (London) 1998;392:198–202. doi: 10.1038/32448. [DOI] [PubMed] [Google Scholar]
- 44.Bito H, Deisseroth K, Tsien R W. Cell. 1996;87:1202–1214. doi: 10.1016/s0092-8674(00)81816-4. [DOI] [PubMed] [Google Scholar]
- 45.Hardingham G E, Chawla S, Johnson C M, Bading H. Nature (London) 1997;385:260–265. doi: 10.1038/385260a0. [DOI] [PubMed] [Google Scholar]
- 46.Hardingham G E, Arnold F J L, Bading H. Nat Neurosci. 2001;4:261–267. doi: 10.1038/85109. [DOI] [PubMed] [Google Scholar]
- 47.Husi H, Ward M A, Choudhary J S, Blackstock W P, Grant S G N. Nat Neurosci. 2000;3:661–669. doi: 10.1038/76615. [DOI] [PubMed] [Google Scholar]
- 48.Walikonis R S, Jensen O N, Mann M, Provance D W, Jr, Mercer J A, Kennedy M B. J Neurosci. 2000;20:4069–4080. doi: 10.1523/JNEUROSCI.20-11-04069.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dalva M B, Takasu M A, Lin M Z, Shamah S M, Hu L, Gale N W, Greenberg M E. Cell. 2000;103:945–956. doi: 10.1016/s0092-8674(00)00197-5. [DOI] [PubMed] [Google Scholar]
- 50.Torres R, Firestein B L, Dong H, Staudinger J, Olsen E N, Huganir R L, Bredt D S, Gale N W, Yancopoulos G D. Neuron. 1998;21:1453–1463. doi: 10.1016/s0896-6273(00)80663-7. [DOI] [PubMed] [Google Scholar]
- 51.Sala C, Rudolph-Correia S, Sheng M. J Neurosci. 2000;20:3529–3536. doi: 10.1523/JNEUROSCI.20-10-03529.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Westphal R S, Tavalin S J, Lin J W, Alto N M, Fraser I D C, Langeberg L K, Sheng M, Scott J D. Science. 1999;285:93–96. doi: 10.1126/science.285.5424.93. [DOI] [PubMed] [Google Scholar]
- 53.Gray P C, Johnson B D, Westenbroek R E, Hays L G, Yates J R, Sheuer T, Catterall W A, Murphy B J. Neuron. 1998;20:1017–1026. doi: 10.1016/s0896-6273(00)80482-1. [DOI] [PubMed] [Google Scholar]
- 54.Bence-Hanulek K K, Marshall J, Blair L A C. Neuron. 2000;27:121–131. doi: 10.1016/s0896-6273(00)00014-3. [DOI] [PubMed] [Google Scholar]
- 55.Levitan I B. Neuron. 1999;22:645–648. doi: 10.1016/s0896-6273(00)80722-9. [DOI] [PubMed] [Google Scholar]
- 56.Dolmetsch, R. D., Pajvani, U., Fife, K., Spotts, J. M. & Greenberg, M. E. (2001) Science, in press. [DOI] [PubMed]
- 57.Montminy M R, Bilezikjian L M. Nature (London) 1987;328:175–178. doi: 10.1038/328175a0. [DOI] [PubMed] [Google Scholar]
- 58.Sheng M, Dougan S T, McFadden G, Greenberg M E. Mol Cell Biol. 1988;8:2787–2796. doi: 10.1128/mcb.8.7.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sheng M, McFadden G, Greenberg M E. Neuron. 1990;4:571–582. doi: 10.1016/0896-6273(90)90115-v. [DOI] [PubMed] [Google Scholar]
- 60.Shaywitz A J, Greenberg M E. Annu Rev Biochem. 1999;68:821–861. doi: 10.1146/annurev.biochem.68.1.821. [DOI] [PubMed] [Google Scholar]
- 61.Silva A J, Korgan J H, Frankland P W, Kida S. Annu Rev Neurosci. 1998;21:127–148. doi: 10.1146/annurev.neuro.21.1.127. [DOI] [PubMed] [Google Scholar]
- 62.Frank D A, Greenberg M E. Cell. 1994;79:5–8. doi: 10.1016/0092-8674(94)90394-8. [DOI] [PubMed] [Google Scholar]
- 63.Sheng M, Thompson M A, Greenberg M E. Science. 1991;252:1427–1430. doi: 10.1126/science.1646483. [DOI] [PubMed] [Google Scholar]
- 64.Yamamoto K K, Gonzalez G A, Biggs W H, Montminy M R. Nature (London) 1988;334:494–498. doi: 10.1038/334494a0. [DOI] [PubMed] [Google Scholar]
- 65.Ginty D D, Bonni A, Greenberg M E. Cell. 1994;77:713–725. doi: 10.1016/0092-8674(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 66.Chrivia J C, Kwok R P, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Nature (London) 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 67.Kwok R P S, Lundblad J R, Chrivia J C, Richards J P, Bächinger H P, Brennan R G, Roberts S G E, Green M R, Goodman R H. Nature (London) 1994;379:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 68.Cardinaux J-R, Notis J C, Zhang Q, Vo N, Craig J C, Fass D M, Brennan R G, Goodman R H. Mol Cell Biol. 2000;20:1546–1552. doi: 10.1128/mcb.20.5.1546-1552.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shaywitz A J, Dove S L, Kornhauser J M, Hochschild A, Greenberg M E. Mol Cell Biol. 2000;20:9409–9422. doi: 10.1128/mcb.20.24.9409-9422.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ho N, Liauw J A, Blaeser F, Wei F, Hanissian S, Muglia L M, Wozniak D F, Nardi A, Arvin K L, Holtzman D M, et al. J Neurosci. 2000;20:6459–6472. doi: 10.1523/JNEUROSCI.20-17-06459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xing J, Ginty D D, Greenberg M E. Science. 1996;273:959–963. doi: 10.1126/science.273.5277.959. [DOI] [PubMed] [Google Scholar]
- 72.Impey S, Obrietan K, Wong S T, Poser S, Yano S, Wayman G, Deloulme J C, Chan G, Storm D R. Neuron. 1998;21:869–883. doi: 10.1016/s0896-6273(00)80602-9. [DOI] [PubMed] [Google Scholar]
- 73.Wu G-Y, Deisseroth K, Tsien R W. Proc Natl Acad Sci USA. 2001;98:2808–2813. doi: 10.1073/pnas.051634198. . (First Published February 20, 2001; 10.1073/pnas.051634198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bonni A, Ginty D D, Dudek H, Greenberg M E. Mol Cell Neurosci. 1995;6:168–183. doi: 10.1006/mcne.1995.1015. [DOI] [PubMed] [Google Scholar]
- 75.Sun P, Enslen H, Myung P S, Maurer R A. Genes Dev. 1994;8:2527–2539. doi: 10.1101/gad.8.21.2527. [DOI] [PubMed] [Google Scholar]
- 76.Parker D, Jhala U S, Radhakrishnan I, Yaffe M B, Reyes C, Shulman A I, Cantley L C, Wright P E, Montminy M. Mol Cell. 1998;2:353–359. doi: 10.1016/s1097-2765(00)80279-8. [DOI] [PubMed] [Google Scholar]
- 77.Timmusk T, Palm K, Metsis M, Reintam T, Paalme V, Saarma M, Persson H. Neuron. 1993;10:475–489. doi: 10.1016/0896-6273(93)90335-o. [DOI] [PubMed] [Google Scholar]
- 78.Tongiorgi E, Righi M, Cattaneo A. J Neurosci. 1997;17:9492–9505. doi: 10.1523/JNEUROSCI.17-24-09492.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tao X, Finkbeiner S, Arnold D B, Shaywitz A J, Greenberg M E. Neuron. 1998;20:709–726. doi: 10.1016/s0896-6273(00)81010-7. [DOI] [PubMed] [Google Scholar]
- 80.Shieh P B, Hu S-C, Bobb K, Timmusk T, Ghosh A. Neuron. 1998;20:727–740. doi: 10.1016/s0896-6273(00)81011-9. [DOI] [PubMed] [Google Scholar]