Abstract
Pantoea ananatis LMG 2665T synthesizes and utilizes acyl homoserine lactones (AHLs) for signalling. The complete set of genes regulated by the EanI/R quorum sensing (QS) system in this strain is still not fully known. In this study, RNA-sequencing (RNA-seq) was used to identify the EanI/R regulon in LMG 2665T. Pairwise comparisons of LMG 2665T in the absence of AHLs (Optical density (OD)600 = 0.2) and in the presence of AHLs (OD600 = 0.5) were performed. Additionally, pairwise comparisons of LMG 2665T and its QS mutant at OD600 = 0.5 were undertaken. In total, 608 genes were differentially expressed between LMG 2665T at OD600 = 0.5 versus the same strain at OD600 = 0.2 and 701 genes were differentially expressed between LMG 2665T versus its QS mutant at OD600 = 0.5. A total of 196 genes were commonly differentially expressed between the two approaches. These constituted approximately 4.5% of the whole transcriptome under the experimental conditions used in this study. The RNA-seq data was validated by reverse transcriptase quantitative polymerase chain reaction (RT-qPCR). Genes found to be regulated by EanI/R QS were those coding for redox sensing, metabolism, flagella formation, flagella dependent motility, cell adhesion, biofilm formation, regulators, transport, chemotaxis, methyl accepting proteins, membrane proteins, cell wall synthesis, stress response and a large number of hypothetical proteins. The results of this study give insight into the genes that are regulated by the EanI/R system in LMG 2665T. Functional characterization of the QS regulated genes in LMG 2665T could assist in the formulation of control strategies for this plant pathogen.
Keywords: Pantoea ananatis, quorum sensing, acyl homoserine lactones, regulon, RNA-seq, LMG 2665T
1. Introduction
Pantoea ananatis is an ubiquitous, Gram-negative, yellow pigmented, rod shaped bacterium that has been described as an “emerging pathogen” of global importance [1]. This bacterium infects plants and is also capable of surviving in different environments [1]. This pathogen causes disease symptoms in a number of economically important plants such as Eucalyptus [1,2], staple food crops, namely maize, rice and sorghum [3] as well as cash crops, for example, honeydew melons, pineapples and onions [1]. Quorum sensing (QS) has been described as a pathogenicity determinant of P. ananatis [4,5], together with Type 6 Secretion System (T6SS) [6,7] and motility [8]. Unlike many other plant pathogens, P. ananatis does not possess Type 2 Secretion System (T2SS) and Type 3 Secretion System (T3SS) [9]. Thus, it is likely that other yet to be identified factors are involved in the pathogenicity of this bacterium.
Quorum sensing, a density dependent communication mechanism between bacterial cells in a population, is achieved by sensing the presence of accumulated QS signals called autoinducers [10]. In many Gram-negative bacteria, the acylated homoserine lactones (AHLs) are used as communication signals and expression of LuxI protein is required for the production of AHLs [11]. The LuxI/R QS regulon has been identified in a number of phytopathogens including P. stewartii subspecies (subsp.) stewartii [12], a close relative of P. ananatis. The draft genome sequence of P. ananatis LMG 2665T was reported recently and contains 4893 genes, including 4787 protein coding regions [13].
Previously, we generated a QS mutant 2665T eanΔI/R and showed that the EanI/R QS system is important for pathogenicity and biofilm formation [5]. To date, no transcriptome-wide studies have been conducted to investigate the QS regulons in any strain of P. ananatis. In the current study, we used transcriptome profiling through RNA-sequencing (RNA-seq) to identify the full set of genes under EanI/R QS regulation of the type strain of P. ananatis. This was done by comparing gene expression between optical density (OD)600 = 0.2 (before QS) and OD600 = 0.5 (during QS) in the wild-type, P. ananatis LMG 2665T. Additionally, pairwise comparison of the transcriptional changes in the wild-type strain and its QS mutant 2665TeanΔI/R during QS at OD600 = 0.5 was conducted. The genes that were differentially expressed between the wild-type and EanI/R mutant at OD600 = 0.5 and between the wild-type at the two points (OD600 = 0.2 (before QS) and OD600 = 0.5 (during QS)) were considered to be under the influence of QS. The results showed that the EanI/R QS system regulates, under the tested conditions, a variety of processes including redox sensing, metabolism, flagella formation and flagella dependent motility, cell adhesion and biofilm formation, regulators, transport, chemotaxis and methyl accepting proteins, membrane proteins, cell wall synthesis, stress response and a large number of hypothetical proteins.
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
The bacterial strains used in this study are P. ananatis LMG 2665T (virulent natural isolate from pineapple obtained from Belgian co-ordinated collections of microorganisms (BCCM)/LMG, University of Gent, Gent, Belgium), LMG 2665T ean∆I/R (P. ananatis LMG 2665T ∆ (eanI, eanR) resistant to kanamycin) [5], Chromobacterium violaceum 026 (mini-Tn5 mutant of C. violaceum [14]. Bacteria were routinely cultured in M9 minimal media [15] (supplemented with 2% casein hydrolysate, 1 mM MgCl2) at 30 °C with a shaker (Gallenkamp, Moriguchi, Japan) at 150 rev/min.
2.2. Identification of RNA Sampling Points
In a previous study, we generated a QS mutant and showed that EanI/R QS is responsible for pathogenicity and biofilm formation phenotypes [5]. The growth curves of LMG 2665T and LMG 2665T ean∆I/R under the studied conditions are shown in Figure S1. Here we wanted to identify the full set of genes that are under QS control using in vitro growth conditions. We first established the time at which AHLs are produced. To do this, we grew the P. ananatis wild-type strain in minimal media and determined the sampling points based on AHL production using the C.violaceum CV026 bio reporter strain [14]. The CV026 bioassay was conducted following a protocol that was previously described by [5]. Single colonies of LMG 2665T wild-type were inoculated into (150 mL) M9 salts minimal media and incubated at 30 °C with shaking at 150 rev/min. The OD600 of bacterial cultures was determined every hour. In addition, CV026 bioassays were performed on samples that were collected every hour. The RNA sampling points were OD600 = 0.2 (QS absent (absence of detectable AHLs)) and OD600 = 0.5 (QS present (presence of detectable AHLs)). The sampling points were at early exponential phase (OD600 = 0.2) and mid exponential phase (OD600 = 0.5) (Figure S1).
2.3. Total RNA Isolation
RNA samples were harvested from in vitro grown P. ananatis LMG 2665T and 2665TeanΔI/R and designated W2-1, W2-2, W2-3 for wild-type samples at OD600 = 0.2; W5-1, W5-2, W5-3 for wild-type samples in the presence of detectable AHLs at OD600 = 0.5 and M5-1, M5-2, M5-3 for mutant samples at OD600 = 0.5 (Table 1). Total RNA was isolated using the RNeasy Mini Kit (Qiagen, GmbH, Hilden, Germany) following instructions from the manufacturer. Three biological replicates of each bacterial culture were analysed (Table 1 shows those samples that were used for bioinformatics analysis; samples with poor mapped reads were excluded in bioinformatics analysis). To stabilize RNA, 10 mL of RNAlaterTM RNA stabilization reagent was added to 5 mL of culture in M9 salts minimal media followed by overnight incubation at 4 °C. The samples were then centrifuged for 10 min at 10,000 rev/min with Eppendorf centrifuge 5810R (Sigma-Aldrich, Hamburg, Germany) Cell pellets were re-suspended in a guanidine-thiocyanate-containing lysis buffer (Buffer RLT) (Qiagen). To remove residual genomic DNA (gDNA) contamination, the gDNA wipe out buffer (Qiagen) was used following the protocol from the manufacturer. Total RNA was tested for residual gDNA by polymerase chain reaction (PCR) and gel electrophoresis (results not shown). The quantity and quality of RNA were determined using the Nanodrop (Thermo Scientific, Sugarland, TX, USA) and 2100 Bioanalyser (Agilent Genomics, Santa Clara, CA, USA). RNA samples with RNA integrity number above eight were considered to be intact and not degraded.
Table 1.
Statistics of reads that mapped to Pantoea ananatis LMG 2665T genome per sample analysed.
| Sample | Total Mapped Reads (%) | Uniquely Mapped Reads (%) | RNA Integrity Number (RIN) |
|---|---|---|---|
| P. ananatis 2665TeanΔI/R samples | |||
| M5-1 | 19,318,790 | 16,835,822 | 9.5 |
| M5-2 | 20,712,376 | 18,833,927 | 9.2 |
| M5-3 | 20,108,558 | 17,421,192 | 9.5 |
| P. ananatis 2665T samples | |||
| W2-1 | 19,364,330 | 17,023,603 | 9.5 |
| W2-2 | 19,483,482 | 17,520,238 | 9.6 |
| W5-1 | 19,953,814 | 15,768,738 | 8.8 |
| W5-3 | 20,417,384 | 18,445,793 | 9.1 |
* Samples M5-1, M5-2 and M5-3 represent biological replicates for RNA isolated from 2665TeanΔI/R at OD600 = 0.5. Samples W2-1, W2-2 represent biological replicates for RNA samples isolated from 2665T at OD600 = 0.2, samples W5-1, W5-3 are biological replicates for RNA isolated at OD600 = 0.5 from 2665T. OD: Optical density; M: Mutant; W: Wild-Type.
2.4. Library Construction and Sequencing
Complementary DNA (cDNA) library construction and sequencing were conducted at the Beijing Genomics Institute (BGI-Shenzhen, Shenzhen, China; http://www.genomics.cn/en/index). In brief, ribosomal RNA (rRNA) was depleted using the RiboZero Magnetic Kit (Illumina Inc., San Diego, CA, USA) and messenger RNA (mRNA) was fragmented in fragmentation buffer. Synthesis of cDNA was done using mRNA fragments as templates. Purification and resolution of short fragments for end reparation and single nucleotide addition of adenine was done using the elution buffer followed by connection of adapters. Suitable cDNA fragments were selected following agarose gel electrophoresis and used as templates for PCR. The quality control of the constructed library was determined using the Agilent 2100 Bioanalyser followed by sequencing of the library using the Illumina HiSeq 2000 platform, generating 90 bp paired-end reads.
2.5. RNA Sequencing Data Analysis
Preliminary quality control was performed at BGI, including removal of adaptors and poor-quality reads. Prior to read mapping using Bowtie2 [16] against P. ananatis LMG 2665T draft genome sequence, the quality of the data set was assessed using FastQC, Version 0.11.5 [17]. P. ananatis LMG 2665T draft genome sequence data and annotation files were obtained from the NCBI Genome database (https://www.ncbi.nlm.nih.gov/genome/). A summary of mapping statistics was obtained using bam_stat.py implemented in the RNA sequencing quality control (RSeQC) package [18]. Mapped reads were filtered based on mapping quality and only uniquely mapped reads (mapping quality > 10) were used for further analysis. High-throughput sequencing (HTSeq)-count [19] was used to make the read counts, hierarchical clustering and principal component analysis (PCA) plot were used to assess the similarity and suitability of biological replicates prior to performing differential expression analysis. DESeq2 package [20] was used to perform differential expression analysis. In order to identify those genes influenced by EanI/R QS, pairwise comparisons of the wild-type strain LMG 2665T transcripts in the absence of AHLs (OD600 = 0.2) and in the presence of AHLs (OD600 = 0.5) were made. Additionally, pairwise comparisons of P. ananatis LMG 2665T and its QS mutant at OD600 = 0.5 were performed. Genes with a False Discovery Rate (FDR, q-value) < 0.05 were considered as significantly differentially expressed, for each comparison. InterProScan5 (v5.11-51) [21] was used to assign Gene Ontology (GO) IDs to the differentially expressed genes.
2.6. Complementary DNA Synthesis and Quantitative Reverse Transcription Polymerase Chain Reaction Validation of RNA-Sequencing Data
Synthesis of cDNA and quantitative reverse transcription (RT-qPCR) were conducted on randomly selected genes (Table 2). The selected genes included those encoding for methyl-accepting chemotaxis protein I (tsr), flagellar hook-associated protein 2 (fliD), carbamoyl 5 phosphate synthase (carB), galacturan 1,4-alpha 6 and shikimate 5-dehydrogenase (aroE) and one that codes for glycoside hydrolase and a hypothetical protein (WP_028715941.1). RNA samples from two sampling points, before QS (wild-type strain at OD600 = 0.2) and in the presence of QS (wild-type strain at OD600 = 0.5) were used. Synthesis of cDNA was done using the QuantiTect Reverse Transcription kit (Qiagen Inc., Germantown, MA, USA) as described by the manufacturer. Primers for the genes used for RT-qPCR were designed using OligoPerfect™ Designer (Thermo Fisher Scientific, Waltham, MA, USA) to amplify DNA fragments between 100 base pairs (bp) to 300 bp. The primers (Inqaba Biotech, Pretoria, South Africa) are listed in Table 2. The slope of a standard curve of cycle threshold (CT) values against concentration of serially diluted template cDNA determines the primer efficiency. The RT-qPCR reaction mix was made up of 5 µL of Applied Biosystems SYBR Green Master Mix, 1 µL of sample cDNA and each primer. The ffh gene that encodes the signal recognition particle protein [22,23] was used as an internal control to normalize the gene expression levels. The ffh gene was stable across all samples in the RNA-seq data.
Table 2.
Primers used for quantitative reverse transcription polymerase chain reaction (RT-qPCR).
| Gene Name | Protein ID | Primer Name and Sequence | Source |
|---|---|---|---|
| Methyl-accepting chemotaxis protein I (tsr) | WP_014605659.1 | TsRF CATGAATGAGATCGTCAGTGCG | This study |
| TsRR GTTGTGTTACGCGATCCATCTC | |||
| Flagellar hook-associated protein 2 (fliD) | WP_014593941.1 | FliDF CAAATGATGGCAGTCTGTCGC | This study |
| FliDR GTGATCGACACGCCGTTAATC | |||
| Carbamoyl-phosphate synthase (carB) | WP_014592889.1 | CarBF GATCCGAAAGTCCACCTTG | This study |
| CarBR GATTGAATACGCCGTCCAC | |||
| Hypothetical protein | WP_028715941.1 | HypF CAACTGGCGGACTACCAAC | This study |
| HypR GCCCTGACCAGTAATTGTCAG | |||
| Shikimate 5-dehydrogenase (aroE) | WP_014606596.1 | ShikiF CGACAGCGTTATTCTGACC | This study |
| ShikiR AATAAGCTCAGGACGCAGG | |||
| Glycoside hydrolase | WP_028715822.1 | GlyF GCGTTGCTACCGCAAATCAAG | This study |
| GlyR GTAACACCTTGCGTGTGACC |
To further validate ffh gene as a good housekeeping gene for data normalization in P. ananatis, RT-qPCR was conducted and the gene was found to be stable in both conditions, OD600 = 0.2 and OD600 = 0.5 in all samples (results not shown). The RT-qPCR reactions were performed using the QuantiStudio 12 K Flex Real-Time PCR machine (Life Technologies, Carlsbad, CA, USA). The PCR protocol was as follows: initial denaturation 50 °C for 2 min, 95 °C for 2 min, 45 cycles of 95 °C for 15 s and 60 °C for 1 min and a melt curve stage: 95 °C for 15 s, 60 °C for 1 min and 95 °C for 15 s. Three technical replicates were used per sample. The fold change in gene expression levels between sampling points, before QS (wild-type strain at OD600 = 0.2) and in the presence of QS (wild-type strain at OD600 = 0.5) were calculated using the comparative CT method [24] and data normalization was done using the ffh gene. The fold change in RNA-seq data were calculated using 2(log2FoldChange).
2.7. Accession Number(s)
The RNA-seq data from this study have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through the GEO accession number, GSE87207.
3. Results
3.1. RNA Sampling Points
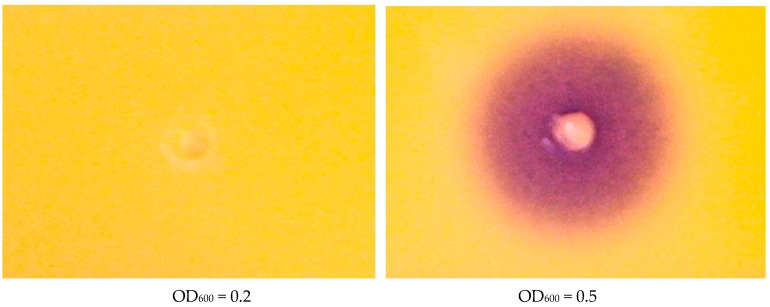
The sampling points were determined by detection of AHLs using the CV026 assay as described in Materials and Methods. At OD600 = 0.2 there were no detectable AHLs from the wild-type strain indicative of absence of QS. At OD600 = 0.5, a purple halo by the CV026 strain indicates presence of AHLs and thus presence of QS in LMG 2665T wild-type strain (Figure 1).
Figure 1.
CV026 bioassay results from samples of the wild-type collected at OD600 = 0.2 and OD600 = 0.5. The purple halo indicates presence of acyl homoserine lactones (AHLs) in culture supernatant at OD600 = 0.5. The purple colour is a result of violacein pigment production by the CV026 bio reporter strain. The absence of purple colour at OD600 = 0.2 shows absence of AHLs and absence of quorum sensing.
3.2. Illumina Sequencing and Read Mapping
To determine the transcriptome wide gene expression profiles of P. ananatis LMG 2665T, cDNA libraries from LMG 2665T were synthesized at OD600 = 0.2 (in the absence of detectable AHLs) and in the presence of AHLs (OD600 = 0.5). For 2665TeanΔI/R, the cDNA library was synthesized at OD600 = 0.5. The cDNA libraries were subjected to sequencing. Three independent biological replicates per strain were sequenced. Approximately, 20 million raw paired-end reads were produced per sample (Table 1). About 90% of these reads were successfully mapped to the P. ananatis LMG 2665T genome sequence (Table 1).
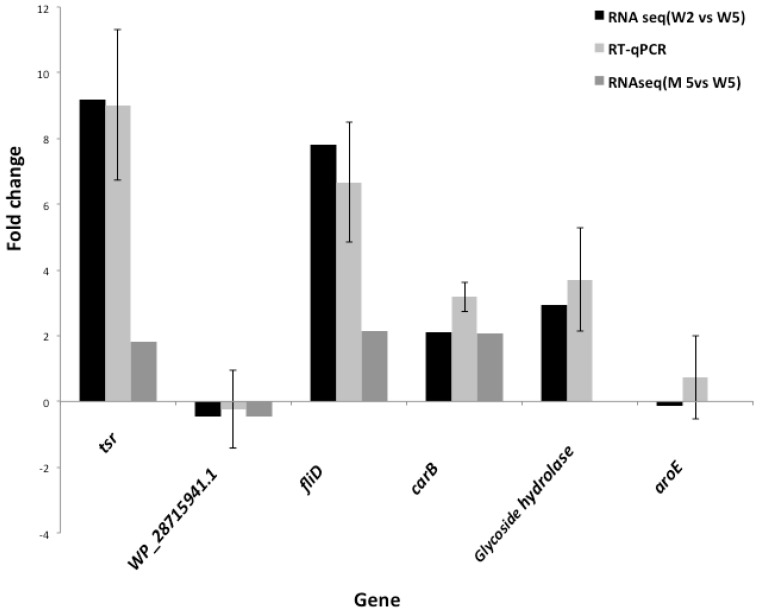
3.3. Quantitative Reverse Transcription Polymerase Chain Reaction Validation of RNA-Sequencing Data
The RT-qPCR results were in agreement with RNA-seq data, thus confirmed the reliability of the sequencing data (Figure 2). The following aroE and WP_028715941.1 were not differentially expressed genes (DEG) (Figure 2), they showed no change in expression under the tested conditions. These genes were simply included as control genes.
Figure 2.
RT-qPCR validation of RNA-sequencing (RNA-seq) data using six selected genes and wild-type, P. ananatis LMG 2665T RNA samples. The fold change in gene expression levels between sampling points, before Quorum Sensing (QS) (wild-type strain at OD600 = 0.2) and in the presence of QS (wild-type strain at OD600 = 0.5) were calculated using the comparative cycle threshold (CT) method [24] and data normalization was done using the ffh gene. The RT-qPCR results indicate fold change as cells shifted from before quorum sensing (OD600 = 0.2) to during quorum sensing (OD600 = 0.5) in LMG 2665T. The fold change in RNA-seq data were calculated using 2(log2FoldChange). Error bars represent the range of relative expression calculated using 2−(ΔΔCT ± StandardDeviation). Triplicates were used per biological sample. The genes aroE and the one encoding for glycoside hydrolase were not differentially expressed in RNA-seq data (M5 versus W5).
3.4. Identification of the EanI/R Quorum Sensing Regulon of Pantoea Ananatis LMG 2665T
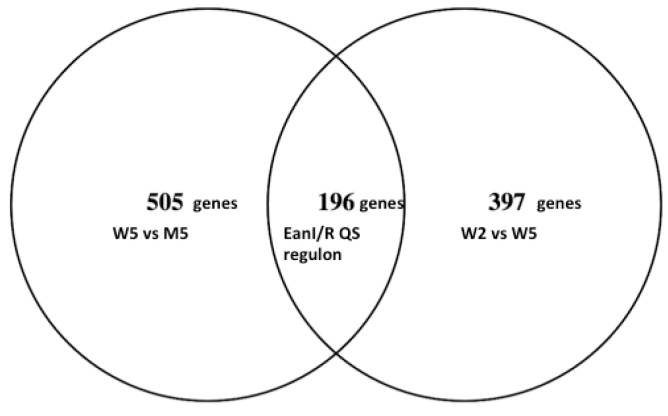
To unravel the QS regulon in P. ananatis LMG 2665T, two approaches were used. Firstly, genes that were differentially expressed in the wild-type LMG 2665T strain between OD600 = 0.2 (W2) and OD600 = 0.5 (W5) were determined. A total of 608 genes were differentially expressed between the wild-type strain before QS and after the onset of QS (Table S1; W2 versus W5). In the second approach, pairwise comparisons of transcriptional changes between the wild-type LMG 2665T (at OD600 = 0.5 indicating presence of QS) and the mutant 2665T eanΔI/R (at OD600 = 0.5, without QS) were conducted. A total of 701 genes were differentially expressed between the two strains at OD600 = 0.5 (Table S2; M5 versus W5). We hypothesized that the genes that are differentially expressed in the wild-type strain between the two points (before QS and during QS) as well as between the two strains (wild-type and mutant at OD = 0.5) are influenced by EanI/R QS (Figure 3). Using this approach, we identified a total of 194 genes (excluding two genes encoding for ribosomal proteins) that were commonly differentially expressed in both approaches (Figure 3). This set of genes was selected for further analysis and is here referred to as the EanI/R QS regulon. In this regard, genes found to be regulated by EanI/R QS constituted approximately 4.5% of the whole transcriptome. A rather large difference in the genes identified in the W2/W5 versus W5/M5 comparison was observed, we speculate that this could be due differential expression of those genes that are important for the mutant’s survival and ability to cope irrespective of its lack QS.
Figure 3.
Identification of the EanI/R QS regulon. The genes that were differentially expressed between the sampling points before QS and during QS (W2 versus W5) that were also differentially expressed between the wild type and its QS mutants (M5 versus W5) were considered to be under QS regulation. A total of 196 genes were found to be influenced by EanI/R QS system. The number of genes in W2 versus W5 in the Venn diagram is 593 since 15 genes in W2 versus W5 data set encode for proteins with unknown protein ID. The 15 genes have protein IDs indicated as not available (N/A) in Table S2.
3.5. Functional Annotation of the EanI/R Regulon in Pantoea ananatis
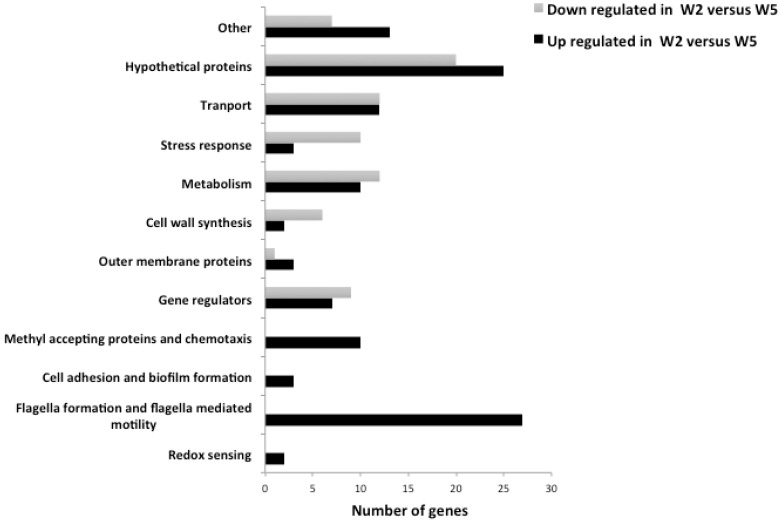
To understand the biological functions of the EanI/R QS regulon, the identified 194 DEGs were classified into functional categories. The DEGs within the EanI/R QS system were found to be those involved in redox sensing, metabolism, flagella formation and flagella dependent motility, cell adhesion and biofilm formation, regulators, transport, chemotaxis and methyl accepting proteins. In addition, other genes in the EanI/R QS regulon included those genes encoding for membrane proteins, cell wall synthesis, stress response and a large number of hypothetical proteins under the tested conditions. The list of genes in the EanI/R QS regulon and their respective log2 ratios in W2 versus W5 and in M5 versus W5 are shown in Table 3. The numbers of genes in each functional category are indicated in Figure 4 (the figure shows those genes differentially expressed in the wild-type strain P. ananatis LMG 2665T between OD600 = 0.5 (during QS) and OD600 = 0.2 (before QS); W2 versus W5).
Table 3.
Genes regulated by the EanI/R quorum sensing system in P. ananatis LMG 2665T. Genes were identified by pairwise comparison between wild-type at OD600 = 0.2 (no QS) and wild type at OD600 = 0.5 (during QS) (W2 versus W5) and pairwise comparison of the wild-type at OD600 = 0.5 and QS mutant at OD600 = 0.5 (M5 versus W5).
| Functional Category | Gene or Protein Encoded | Log2 Ratio | |
|---|---|---|---|
| W2 vs. W5 | M5 vs. W5 | ||
| Genes up regulated between the wild-type strain LMG 2665T in the presence of AHLs (at OD600 = 0.5) versus the same strain in the absence if AHLs (OD600 = 0.2) | |||
| Redox sensing | 3-oxoacyl-ACP reductase | 1.833 | 1.638 |
| Oxidoreductase | 2.115 | 1.93 | |
| Metabolism | pyruvate oxidase | 1.156 | 1.217 |
| malate:quinone oxidoreductase | 1.42 | 0.561 | |
| transketolase/WP_014593932.1 | 1.401 | 1.588 | |
| lysine 6-monooxygenase | 1.351 | 0.966 | |
| ketol-acid reductoisomerase | 1.187 | 0.888 | |
| alcA | 1.532 | 0.869 | |
| IucA | 2.106 | 1.444 | |
| siderophore-interacting protein (WP_014606889.1) | 1.686 | 1.014 | |
| pyridine nucleotide-disulphide oxidoreductase | 0.845 | −0.738 | |
| Wza | 0.971 | −0.984 | |
| Flagella formation and flagella mediated Motility | fliA | 2.837 | 0.848 |
| flgK | 2.447 | 0.795 | |
| flagellin/WP_014593940.1 | 3.053 | 0.952 | |
| fliZ | 2.607 | 1.012 | |
| fliL | 2.336 | 0.797 | |
| fligL | 2.2 | 0.881 | |
| fliM | 2.032 | 0.699 | |
| flhA | 2.086 | 1.488 | |
| flgG | 2.39 | 1.855 | |
| flagellar basal body P-ring protein | 2.104 | 1.542 | |
| fliS | 2.681 | 1.161 | |
| fliE | 2.293 | 1.921 | |
| flgN | 1.325 | 0.468 | |
| flgA | 2.279 | 1.357 | |
| flagellin/WP_013027859.1 | 1.6 | 4.051 | |
| flagellar basal body rod modification protein | 2.779 | 1.991 | |
| motB | 1.728 | 0.595 | |
| flit | 2.475 | 1.313 | |
| flgF | 2.508 | 1.697 | |
| flgJ | 1.934 | 1.267 | |
| flhB | 2.199 | 1.559 | |
| flgE | 2.392 | 1.892 | |
| flgC | 2.743 | 2.05 | |
| flgL | 2.201 | 0.881 | |
| flagellar basal body L-ring protein | 2.487 | 1.699 | |
| flgB | 2.875 | 1.96 | |
| flip | 1.89 | 0.595 | |
| Cell adhesion and biofilm formation | fliD/WP_014593941.1 | 2.93 | 1.104 |
| bcsO | 1.136 | 0.699 | |
| yhjQ | 0.942 | 0.717 | |
| Gene regulators | luxR | 0.816 | 1.276 |
| Fis family transcriptional regulator | 0.874 | 0.961 | |
| transcriptional regulator/WP_028714798.1 | 1.669 | 0.877 | |
| transcriptional regulator/WP_013027687.1 | 0.589 | 0.639 | |
| proQ | 2.497 | 0.534 | |
| luxI | 0.734 | 6.21 | |
| dgdR | 1.747 | −0.55 | |
| Methyl accepting proteins and Chemotaxis | WP_019105711.1 | 2.45 | 1.955 |
| WP_028714945.1 | 0.994 | 0.683 | |
| WP_050442519.1 | 1.888 | 0.734 | |
| WP_014605659.1 | 3.141 | 0.869 | |
| exbD | 2.259 | 1.512 | |
| chemotaxis protein-glutamate O-methyltransferase | 2.259 | 0.761 | |
| WP_014605660.1 | 2.716 | 0.882 | |
| chemotaxis response regulator (cheZ) | 2.194 | 0.717 | |
| Regulator of chemotaxis (cheA) | 1.888 | 0.743 | |
| chemotaxis response regulator (cheY) | 2.497 | 0.777 | |
| Outer membrane proteins | ligand-gated channel protein | 1.061 | 1.621 |
| lpoA | 1.102 | 0.431 | |
| Outer Membrane protein/ WP_026020991.1 | 1.806 | −0.929 | |
| Cell wall synthesis | endopeptidase | 2.145 | 0.907 |
| D-alanyl-D-alanine carboxypeptidase | 1.618 | 0.953 | |
| Stress response | hslV | 1.741 | 1.539 |
| carbamoyl phosphate synthase large subunit | 1.053 | 1.514 | |
| universal stress protein B | 1.457 | −0.751 | |
| Transport | magnesium-translocating P-type ATPase | 2.884 | 1.793 |
| MFS transporter/WP_028715176.1 | 2.149 | 1.176 | |
| pyridine nucleotide-disulphide oxidoreductase | 2.32 | 1.387 | |
| anion permease | 2.394 | 1.498 | |
| ligand-gated channel protein/WP_014595119.1 | 1.491 | 0.954 | |
| MFS transporter/WP_028715950.1 | 1.363 | 0.958 | |
| NCS2 family permease | 0.687 | 0.836 | |
| pyridine nucleotide-disulphide oxidoreductase | 2.32 | 1.387 | |
| glucose dehydrogenase | 0.732 | 0.544 | |
| WP_014593931.1/Transketolase | 1.267 | 1.129 | |
| sugar ABC transporter substrate-binding protein | 1.686 | 0.847 | |
| ferrous iron transporter A | 0.928 | −2.909 | |
| Hypothetical proteins | WP_013026151.1 | 2.51 | 2.214 |
| WP_019106310.1 | 1.013 | 0.601 | |
| WP_014593929.1 | 1.724 | 1.689 | |
| WP_014333251.1 | 1.197 | 0.612 | |
| WP_014604614.1 | 1.65 | 1.154 | |
| WP_028715174.1 | 1.039 | 0.632 | |
| WP_013027988.1 | 0.902 | 0.654 | |
| WP_013026097.1 | 1.84 | 1.293 | |
| WP_013024239.1 | 0.998 | 0.882 | |
| WP_014593930.1 | 0.902 | 0.847 | |
| WP_028715108.1 | 0.703 | 0.814 | |
| WP_050442523.1 | 0.955 | 1.876 | |
| WP_013025497.1 | 0.671 | 0.711 | |
| WP_014332946.1(putative motility protein) | 3.038 | 0.542 | |
| WP_013025421.1 | 3.227 | 0.923 | |
| WP_033765947.1 | 2.576 | 1.051 | |
| WP_028715248.1 | 1.979 | 1.442 | |
| WP_028715109.1 | 1.95 | 1.442 | |
| WP_014593989.1 | 1.376 | 0.594 | |
| WP_013024991.1 | 1.996 | −0.63 | |
| WP_014604946.1 | 0.597 | −0.777 | |
| WP_028715989.1 | 0.77 | −0.76 | |
| WP_050442541.1 | 1.443 | −2.11 | |
| WP_013025383.1 | 1.579 | −0.98 | |
| WP_028716027.1 | 1.033 | −0.715 | |
| Other | srfB (virulence factor) | 1.681 | 1.21 |
| Maa | 2.816 | 2.347 | |
| N6-hydroxylysine O-acetyltransferase | 1.74 | 0.936 | |
| Aminotransferase | 1.281 | 1.153 | |
| acyl—CoA ligase | 1.049 | 1.111 | |
| dTDP-4-dehydrorhamnose 3,5-epimerase | 1.467 | 1.13 | |
| traF | 1.88 | 0.524 | |
| peptidase C39 | 0.831 | 1.103 | |
| dusB | 0.983 | 0.713 | |
| cytochrome ubiquinol oxidase subunit II | 0.698 | 1.17 | |
| 2,5-didehydrogluconate reductase A | 1.438 | −0.598 | |
| aspartyl β-hydroxylase | 1.016 | −1.295 | |
| DNA polymerase III subunit ε | 1.574 | −0.854 | |
| Genes down regulated between the wild-type strain LMG 2665T in the presence of AHLs (at OD600 = 0.5) versus the same strain in the absence of AHLs (OD600 = 0.2). | |||
| Metabolism | erythrose-4-phosphate dehydrogenase | −0.977 | −0.762 |
| apbE | −0.62 | −1.113 | |
| nicotinate-nucleotide diphosphorylase | −1.559 | −0.78 | |
| dGTPase | −0.498 | −0.44 | |
| suhB | −0.649 | −0.472 | |
| L-aspartate oxidase | −1.706 | −0.601 | |
| Protease | −1.64 | −0.711 | |
| cystathionine β-lyase | −0.789 | −0.575 | |
| peptidylprolyl isomerase | −0.722 | 0.624 | |
| betaine-aldehyde dehydrogenase | −2.498 | 0.571 | |
| choline dehydrogenase | −2.154 | 0.674 | |
| 5-oxopent-3-ene-1,2,5-tricarboxylate decarboxylase | −0.837 | 2.096 | |
| Outer membrane proteins | ompC/WP_013026835.1 | −1.236 | −2.393 |
| Stress response | FAD/NAD(P) binding domain-containing protein | −0.93 | −1.039 |
| GNAT family N-acetyltransferase | −0.751 | −0.596 | |
| class C β-lactamase | −0.953 | −0.743 | |
| exodeoxyribonuclease I | −0.533 | −0.487 | |
| NUDIX hydrolase | −0.751 | −0.759 | |
| htpG | −1.09 | 0.877 | |
| groEL | −0.921 | 0.814 | |
| dnaJ | −0.84 | 0.742 | |
| sulfurtransferase | −0.733 | 0.971 | |
| hslU | −0.619 | 0.473 | |
| Transport | MFS transporter/WP_033765526.1 | −2.103 | −0.716 |
| sulfate transporter subunit | −1.613 | −0.567 | |
| C4-dicarboxylate ABC transporter | −1.445 | −0.993 | |
| MATE family efflux transporter | −1.266 | −0.806 | |
| tcyP | −0.908 | −1.025 | |
| MFS transporter/WP_028714804.1 | −0.957 | −0.716 | |
| ABC transporter permease | −0.712 | −0.522 | |
| flavocytochrome c | −0.712 | −0.934 | |
| microcin B17 transporter | −0.954 | 1.872 | |
| MFS transporter/WP_014598266.1 | −1.048 | 1.327 | |
| MFS transporter/WP_013024876.1 | −0.701 | 1.31 | |
| nickel transporter | −0.58 | 1.433 | |
| Gene regulators | DNA-binding response regulator | −0.6 | −0.603 |
| lysR/WP_028714815.1 | −1.675 | −0.718 | |
| draR | −0.974 | −1.295 | |
| lclR | −0.862 | −1.004 | |
| pbsX | −0.863 | −0.758 | |
| DNA-binding response regulator | −0.734 | 1.974 | |
| yqeI | −0.747 | 1.789 | |
| cdaR | −1.262 | 0.891 | |
| ompR | −0.734 | 1.974 | |
| Hypothetical proteins | WP_028715941.1 | −1.129 | −1.493 |
| WP_014593863.1 | −1.269 | −0.683 | |
| WP_028715707.1 | −1.481 | −1.226 | |
| WP_026021031.1 | −1.69 | −1.957 | |
| WP_028715464.1 | −0.864 | −1.032 | |
| WP_014594750.1 | −0.711 | −0.535 | |
| WP_028714922.1 | −1.067 | −1.138 | |
| WP_014605434.1 | −1.964 | −1.308 | |
| WP_014606741.1 | −1.173 | −0.939 | |
| WP_028715967.1 | −0.788 | −0.779 | |
| WP_028715704.1 | −0.939 | −0.922 | |
| WP_013024364.1 | −0.83 | −0.595 | |
| WP_028715521.1 | −0.742 | −0.578 | |
| WP_028715342.1 | −0.874 | −1.013 | |
| WP_050442548.1 | −1.663 | −1.265 | |
| WP_026021018.1 | −1.426 | −0.732 | |
| WP_014593149.1 | −1.931 | −1.472 | |
| WP_013026749.1 | −0.722 | 1.177 | |
| WP_022622675.1 | −0.63 | 1.053 | |
| WP_013024276.1 | −1.146 | 0.744 | |
| Cell wall synthesis | penicillin-binding protein 2 | −0.598 | 1.418 |
| ftsI | −0.726 | 1.406 | |
| murJ | −0.562 | 0.549 | |
| D-alanyl-D-alanine carboxypeptidase/WP_013026332.1 | −1.335 | 3.127 | |
| N-acetylmuramoyl-L-alanine amidase | −1.08 | 1.914 | |
| murein transglycosylase B | −1.155 | 0.608 | |
| Other | FUSC family protein | −0.918 | −0.704 |
| quinolinate synthetase | −1.122 | −0.863 | |
| dGTPase | −0.498 | −0.44 | |
| diaA | −0.658 | 0.527 | |
| U32 family peptidase | −0.641 | 0.884 | |
| peptidylprolyl isomerase | −0.722 | 0.624 | |
| K+/H+ antiporter | −0.727 | 0.879 | |
Figure 4.
Functional categories of genes in the EanI/R QS regulon. The figure represents those genes in the EanI/R regulon that were differentially expressed in the wild type strain, LMG 2665T between the two sampling points before QS and during QS. The 194 genes were grouped into different functional categories. The up regulated groups included those for flagella formation and flagella mediated motility, methyl accepting proteins and chemotaxis, redox sensing and cell adhesion. The groups with genes that were either up or down regulated included hypothetical proteins, transport, stress response, metabolism, cell wall synthesis, outer membrane proteins and regulators.
4. Discussion
The ability of LuxR proteins to fold, bind DNA and effect QS activity is dependent on the presence of AHLs [25]. The present study analysed the complete regulatory networks associated with the EanI/R QS system of P. ananatis in the presence of AHLs based on the CV026 bio reporter. The LuxI/R QS regulon of other plant pathogenic bacteria has been elucidated, for example, in P. stewartii subsp. stewartii, the LuxI/R regulon constituted about 8% of the transcriptome [12] whereas in Pectobacterium atrosepticum it constituted about 26% [26] representing the largest QS regulon in a plant pathogenic bacterium. In this study, transcriptome profiling showed that the EanI/R regulon in LMG 2665T constituted about 4.5% of the entire transcriptome. The differences in the percentage coverages of QS regulons in different bacteria could be due to differences in bacterial species and experimental conditions. For example, P. ananatis has a limited number of pathogenicity factors since it neither possesses a T3SS and T2SS nor does its genome encode genes of known phytotoxins [9,27]. The success of host infection and disease development lies on the effective regulation of pathogenicity factors throughout the infection process. Importantly, it has been shown that QS regulates pathogenicity in P. ananatis strain LMG 2665T [5] and in P. ananatis SK-1 [4], indicating that AHLs play a pivotal role in disease caused by P. ananatis.
Motility is a critical pathogenicity determinant in plant pathogenic bacteria including Dickeya dadantii [28], Ralstonia solanacearum [29] and Pectobacterium carotovorum subsp. carotovorum [30]. Moreover, a recent study showed that in P. ananatis motility contributes to pathogenesis and biofilm formation [8]. However, up to now there has been no information regarding QS regulation of motility in this bacterium. This study showed that the EanI/R QS positively regulates genes that encode for flagella formation and flagella mediated motility, methyl accepting proteins and chemotaxis (Table 3 and Figure 4) in LMG 2665T. Notably, most genes important for flagella formation and flagella mediated motility as well as methyl accepting and chemotaxis proteins were up regulated, they showed log2 ratios that are close to or above 2 in LMG 2665T as it shifted from before QS to presence of QS (Table 3). It is thus possible that, in P. ananatis, the genes for motility are essential for movement towards sites that are favourable for host infection and disease outbreak. Quorum sensing was found to positively regulate genes associated with flagella biosynthesis in other plant pathogenic bacteria such as Burkholderia glumae [31].
The ability of a bacterium to adapt to changing environment and take advantage of available nutrients is important for pathogen survival and success of infection of plant hosts [32]. Metabolism genes up regulated by EanI/R include those encoding for pyruvate oxidase (PoxB), malate: quinone oxidoreductase (Mqo) and transketolase/WP_014593932.1. The transketolase/WP_014593932.1 is an enzyme in glycolysis and pentose phosphate pathways. Pyruvate oxidase and malate:quinone oxidoreductase (Mqo) are key enzymes in pyruvate metabolism where the former is involved in controlling production of acetic acid from pyruvate. Notably, acidic environments do not favour growth and proliferation of plant pathogenic bacteria [33,34]. In some bacteria, for example, in P. syringae pv. tomato strain DC3000, Mqo was found to be required for pathogenicity and effective utilization of nutrients available in hosts [35]. It can be speculated that this enzyme could be important for pathogenicity and nutrient acquisition in LMG 2665T.
Amino acids serve as a carbon source in plant pathogenic bacteria [32]. The EanI/R up regulates genes encoding lysine 6-monooxygenase and ketol-acid reductoisomerase, an enzyme important for biosynthesis of valine, leucine and isoleucine, suggesting a possible utilization of these amino acids by LMG 2665T. Furthermore, the present study shows that transport of various metabolic products is regulated by EanI/R QS systems (Table 3). Transport systems in plant pathogenic bacteria are essential for acquisition of nutrients in nutrient poor niches [32]. The regulation of genes for metabolism and transport by EanI/R suggests that this system could be important for survival of the pathogen in nutrient poor niches. In addition, the present study shows that the gene wza important for polysaccharide biosynthesis is influenced by EanI/R QS in LMG 2665T. Importantly, in P. ananatis, AHLs were found to regulate exopolysaccharides production [36].
Genes associated with iron acquisition and siderophore biosynthesis including alcaA, lucA and siderophore-interacting protein/WP_014606889.1 are up regulated by EanI/R QS in LMG 2665T (Table 3). In P. stewartii subsp. stewartii, mutation of genes for siderophore biosynthesis, transport and iron acquisition resulted in impaired motility and pathogenicity [37]. In addition, iron has been shown to be important for pathogenicity in bacteria such as Pectobacterium spp. [38] and Erwinia spp. [39]. The ability to outcompete other micro-organism in iron acquisition is important for survival. Given the importance of genes encoding for iron acquisition and siderophore biosynthesis in other plant pathogenic bacteria, the role of those genes in LMG 2665T that are important for iron uptake and siderophore biosynthesis that were found to be QS regulated in this study deserves future investigation.
Bacterial attachment is the initial step in biofilm formation and cell adhesion in several bacterial species. It has been shown that AHL QS regulates biofilm formation [4,5]. The present study shows that the EanI/R QS system up regulates genes important for cell adhesion and biofilm formation (Table 3 and Figure 4). The fliD gene is important for mucus specific cell adhesion and is up regulated by EanI/R. The interaction of flagella with mucus is well documented in non-plant pathogenic bacteria, however not much research has been done on the interaction of mucilage (an analogous of mucus in plants) [40] with flagella in plant pathogens. The compelling up regulation of fliD gene and flagella genes observed in this study should be an incentive for investigation of a possible interaction of these proteins that are encoded by these genes in plant pathogens.
Quorum sensing regulates stress response genes in other plant pathogenic bacteria including P. stewartii subsp. stewartii [12], notably the universal stress proteins (UspA and UspB). The EanI/R QS system positively and negatively influences response to stress. Stress response genes that are upregulated by EanI/R include the gene encoding for carbamoyl phosphate synthase large subunit that is important for detoxification of ammonia and hslV important for response to heat shock (Table 3). Stress related genes that are down regulated by EanI/R include flavin adenine dinucleotide/nicotinamide adenine dinucleotide phosphate FAD/NAD(P) binding domain-containing protein important for response to oxidative stress, exodeoxyribonuclease I, class C beta-lactamase, GNAT family N-acetyltransferase NUDIX hydrolase (Table 3). The EanI/R QS system up regulates 3-oxoacyl-ACP reductase and oxidoreductase (Table 3), the two genes are involved in redox sensing that is important in sensing environmental changes [41], an important attribute for survival and adaptation.
Transcriptome profiling showed that the EanI/R up regulates expression of other gene regulators such as Fis family transcriptional regulator, transcriptional regulator/WP_028714798.1, transcriptional regulator/WP_013027687.1, proQ and luxI (Table 3). Most of the gene regulators that are influenced by EanI/R QS are those important for virulence, nutrients utilisation and response to changes in the environment. The EanI/R QS down regulates some regulators including the LysR type and transcriptional repressor (IclR) under the tested conditions. LysR transcriptional regulators are known for regulating pathogenicity related genes in other bacterial plant pathogens such as D. dadantii [42] whereas LcIR regulates pathogenicity in P. carotovorum subsp. carotovorum [43]. It is possible that EanI/R regulates some pathogenicity genes indirectly by regulating other transcriptional regulators in P. ananatis LMG 2665T. The role of these regulators in pathogenicity of LMG 2665T merits investigations.
Membrane proteins are upregulated by the EanI/R system, for example lpoA and the ligand gated channel protein that helps facilitate entry of iron into cells whilst ompC is down regulated (Table 3). OmpC is a non-selective membrane porin that allows entry of all substances, including antibiotics and other toxic substances, into cells [44]. It can be reasoned that EanI/R QS down regulates ompC in order to protect the bacterial cells from toxic substances. The EanI/R QS system up regulates genes important for cell wall synthesis and peptidoglycan, an important component of cell walls. These include endopeptidase and D-alanyl-D-alanine carboxypeptidase (Table 3). Furthermore, genes coding for penicillin-binding protein 2, FtsI, MurJ, D-alanyl-D-alanine carboxypeptidase, N-acetylmuramoyl-L-alanine amidase and murein transglycosylase B are down regulated in W2 vs. W5. The regulation of outer membrane proteins by QS was noted in other plant pathogenic bacteria including Xanthomonas citri subsp. citri [45]. It can be speculated that, in LMG 2665T, the EanI/R QS modulates the outer membrane proteins and genes associated with cell wall synthesis in order to prevent the entrance of antimicrobial compounds that could be fatal to the pathogen. A large number of hypothetical proteins, were up regulated by the EanI/R system. Furthermore, a substantial number of hypothetical proteins, were down regulated by the EanI/R system (Table 3). The hypothetical proteins that are influenced by QS form the basis for future work, where an understanding of their biological role could lead to better knowledge of the role of QS gene regulation in P. ananatis.
A recent transcriptome analysis study showed that QS regulates conjugative transfer in Agrobacterium tumefaciens [46]. Transcriptome profiling showed that the EanI/R up regulates a conjugal transfer gene traF, a gene important for plasmid transfer in P. ananatis LMG 2665T (Table 3). Some plasmids encode fitness traits required to colonize a given niche and bacteria can take up plasmids from neighbouring cells in a biofilm. Other genes upregulated by EanI/R include srfB, maa, dusB and those encoding for N6-hydroxylysine O-acetyltransferase, aminotransferase, acyl—CoA ligase, dTDP-4-dehydrorhamnose 3,5-epimerase, peptidase C39 and cytochrome ubiquinol oxidase subunit II. The cytochrome ubiquinol oxidase subunit II is important for survival in oxygen limited environments [47], possibly in a biofilm. Peptidase C39 is a bacteriocin processing enzyme, bacteriocin are antibiotics secreted by some bacteria that inhibit growth of other bacteria. This suggests the involvement of QS in regulation of a trait important for competitive advantage in LMG 2665T.
5. Conclusions
A small number of genes have been functionally characterized and found to play a role in pathogenicity of P. ananatis [4,5,6,8,36,48]. The present study provides information on EanI/R QS regulon in LMG 2665T and adds to the list of genes that could be essential for pathogenicity and survival of this pathogen. This study provided a broader picture of the role of the EanI/R QS in P. ananatis LMG 2665T. Future studies are aimed at functional characterization of those QS regulated genes that have not yet been studied in LMG 2665T as well as investigation of this QS system in planta or in the presence of a plant tissue. Such knowledge could help in the formulation of control strategies for this plant pathogen.
Acknowledgments
The authors would like to thank the National Research Foundation (NRF) of South Africa, the University of Pretoria, the Forestry and Agricultural Biotechnology Institute (FABI), the Tree Protection Cooperative Program (TPCP) and Centre of Excellence in Tree Health Biotechnology (CTHB) for supporting this research.
Supplementary Materials
The following are available online at http://www.mdpi.com/2073-4425/9/3/148/s1. Table S1: Genes that were DE between the wild type strain P. ananatis LMG 2665T before QS and the same strain after the onset of QS (W2 versus W5); Table S2: Genes that were DE between the two strains LMG 2665T ean∆I/R and P. ananatis LMG 2665T at OD600 = 0.5 (M5 versus W5); Figure S1: Growth of P. ananatis LMG 2665T and its QS mutant LMG 2665T ean∆I/R in vitro.
Author Contributions
S.S., D.Y.S., T.A.C. and L.N.M. conceived and designed the experiments; S.S. and C.K.T. performed the experiments; S.S., S.K. and L.N.M. analysed the data; T.A.C. contributed reagents/materials/analysis tools; S.S. wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Coutinho T.A., Venter S.N. Pantoea ananatis: An unconventional plant pathogen. Mol. Plant Pathol. 2009;10:325–335. doi: 10.1111/j.1364-3703.2009.00542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coutinho T., Preisig O., Mergaert J., Cnockaert M., Riedel K.-H., Swings J., Wingfield M. Bacterial blight and dieback of Eucalyptus species, hybrids, and clones in South Africa. Plant Dis. 2002;86:20–25. doi: 10.1094/PDIS.2002.86.1.20. [DOI] [PubMed] [Google Scholar]
- 3.Cota L., Costa R., Silva D., Parreira D., Lana U., Casela C. First report of pathogenicity of Pantoea ananatis in sorghum (Sorghum bicolor) in Brazil. Australas. Plant Dis. Notes. 2010;5:120–122. doi: 10.1071/DN10044. [DOI] [Google Scholar]
- 4.Morohoshi T., Nakamura Y., Yamazaki G., Ishida A., Kato N., Ikeda T. The plant pathogen Pantoea ananatis produces N-acylhomoserine lactone and causes center rot disease of onion by quorum sensing. J. Bacteriol. 2007;189:8333–8338. doi: 10.1128/JB.01054-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sibanda S., Theron J., Shyntum D.Y., Moleleki L.N., Coutinho T.A. Characterization of two LuxI/R homologs in Pantoea ananatis LMG 2665T. Can. J. Microbiol. 2016;62:893–903. doi: 10.1139/cjm-2016-0143. [DOI] [PubMed] [Google Scholar]
- 6.Shyntum D.Y., Theron J., Venter S.N., Moleleki L.N., Toth I.K., Coutinho T.A. Pantoea ananatis utilizes a type VI secretion system for pathogenesis and bacterial competition. Mol. Plant Microbe Interact. 2015;28:420–431. doi: 10.1094/MPMI-07-14-0219-R. [DOI] [PubMed] [Google Scholar]
- 7.Shyntum D.Y., Venter S.N., Moleleki L.N., Toth I., Coutinho T.A. Comparative genomics of type VI secretion systems in strains of Pantoea ananatis from different environments. BMC Genom. 2014;15:163. doi: 10.1186/1471-2164-15-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weller-Stuart T., Toth I., de Maayer P., Coutinho T. Swimming and twitching motility are essential for attachment and virulence of Pantoea ananatis in onion seedlings. Mol. Plant Pathol. 2017;18:734–745. doi: 10.1111/mpp.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Maayer P., Chan W.Y., Venter S.N., Toth I.K., Birch P.R., Joubert F., Coutinho T.A. Genome sequence of Pantoea ananatis LMG20103, the causative agent of Eucalyptus blight and dieback. J. Bacteriol. 2010;192:2936–2937. doi: 10.1128/JB.00060-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Kievit T.R., Iglewski B.H. Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 2000;68:4839–4849. doi: 10.1128/IAI.68.9.4839-4849.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuqua C., Parsek M.R., Greenberg E.P. Regulation of gene expression by cell-to-cell communication: Acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 2001;35:439–468. doi: 10.1146/annurev.genet.35.102401.090913. [DOI] [PubMed] [Google Scholar]
- 12.Ramachandran R., Burke A.K., Cormier G., Jensen R.V., Stevens A.M. Transcriptome-based analysis of the Pantoea stewartii quorum-sensing regulon and identification of EsaR direct targets. Appl. Environ. Microbiol. 2014;80:5790–5800. doi: 10.1128/AEM.01489-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adam Z., Tambong J.T., Lewis C.T., Lévesque C.A., Chen W., Bromfield E.S., Khan I.U., Xu R. Draft genome sequence of Pantoea ananatis strain LMG 2665T, a bacterial pathogen of pineapple fruitlets. Genome Announc. 2014;2:e00489-14. doi: 10.1128/genomeA.00489-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McClean K.H., Winson M.K., Fish L., Taylor A., Chhabra S.R., Camara M., Daykin M., Lamb J.H., Swift S., Bycroft B.W. Quorum sensing and Chromobacterium violaceum: Exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology. 1997;143:3703–3711. doi: 10.1099/00221287-143-12-3703. [DOI] [PubMed] [Google Scholar]
- 15.Minogue T.D., Trebra M.W.V., Bernhard F., Bodman S.B.V. The autoregulatory role of EsaR, a quorum-sensing regulator in Pantoea stewartii ssp. stewartii: Evidence for a repressor function. Mol. Microbiol. 2002;44:1625–1635. doi: 10.1046/j.1365-2958.2002.02987.x. [DOI] [PubMed] [Google Scholar]
- 16.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.FastQC. Simon Andrews; Babraham, UK: 2017. [(accessed on 30 January 2018)]. version 0.11.5. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Google Scholar]
- 18.Wang L., Wang S., Li W. RSeQC: Quality control of RNA-seq experiments. Bioinformatics. 2012;28:2184–2185. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- 19.Anders S., Pyl P.T., Huber W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones P., Binns D., Chang H.-Y., Fraser M., Li W., McAnulla C., McWilliam H., Maslen J., Mitchell A., Nuka G. InterProScan 5: Genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takle G.W., Toth I.K., Brurberg M.B. Evaluation of reference genes for real-time RT-PCR expression studies in the plant pathogen Pectobacterium atrosepticum. BMC Plant Biol. 2007;7:50. doi: 10.1186/1471-2229-7-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moleleki L.N., Pretorius R.G., Tanui C.K., Mosina G., Theron J. A quorum sensing-defective mutant of Pectobacterium carotovorum ssp. brasiliense 1692 is attenuated in virulence and unable to occlude xylem tissue of susceptible potato plant stems. Mol. Plant Pathol. 2017;18:32–44. doi: 10.1111/mpp.12372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 25.Zhu J., Winans S.C. The quorum-sensing transcriptional regulator TraR requires its cognate signaling ligand for protein folding, protease resistance, and dimerization. Proc. Natl. Acad. Sci. USA. 2001;98:1507–1512. doi: 10.1073/pnas.98.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu H., Coulthurst S.J., Pritchard L., Hedley P.E., Ravensdale M., Humphris S., Burr T., Takle G., Brurberg M.-B., Birch P.R. Quorum sensing coordinates brute force and stealth modes of infection in the plant pathogen Pectobacterium atrosepticum. PLoS Pathog. 2008;4:e1000093. doi: 10.1371/journal.ppat.1000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Maayer P., Chan W.Y., Rubagotti E., Venter S.N., Toth I.K., Birch P.R., Coutinho T.A. Analysis of the Pantoea ananatis pan-genome reveals factors underlying its ability to colonize and interact with plant, insect and vertebrate hosts. BMC Genom. 2014;15:404. doi: 10.1186/1471-2164-15-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Antúnez-Lamas M., Cabrera-Ordonez E., Lopez-Solanilla E., Raposo R., Trelles-Salazar O., Rodríguez-Moreno A., Rodríguez-Palenzuela P. Role of motility and chemotaxis in the pathogenesis of Dickeya dadantii 3937 (ex Erwinia chrysanthemi 3937) Microbiology. 2009;155:434–442. doi: 10.1099/mic.0.022244-0. [DOI] [PubMed] [Google Scholar]
- 29.Yao J., Allen C. The plant pathogen Ralstonia solanacearum needs aerotaxis for normal biofilm formation and interactions with its tomato host. J. Bacteriol. 2007;189:6415–6424. doi: 10.1128/JB.00398-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hossain M.M., Shibata S., Aizawa S.-I., Tsuyumu S. Motility is an important determinant for pathogenesis of Erwinia carotovora subsp. carotovora. Physiol. Mol. Plant Pathol. 2005;66:134–143. doi: 10.1016/j.pmpp.2005.06.001. [DOI] [Google Scholar]
- 31.Jang M.S., Goo E., An J.H., Kim J., Hwang I. Quorum sensing controls flagellar morphogenesis in Burkholderia glumae. PLoS ONE. 2014;9:e84831. doi: 10.1371/journal.pone.0084831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fatima U., Senthil-Kumar M. Plant and pathogen nutrient acquisition strategies. Front. Plant Sci. 2015;6 doi: 10.3389/fpls.2015.00750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee S.-W., Jeong K.-S., Han S.-W., Lee S.-E., Phee B.-K., Hahn T.-R., Ronald P. The Xanthomonas oryzae pv. oryzae PhoPQ two-component system is required for AvrXA21 activity, HRPG expression, and virulence. J. Bacteriol. 2008;190:2183–2197. doi: 10.1128/JB.01406-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakka S., Qi M., Zhao Y. The Erwinia amylovora PhoPQ system is involved in resistance to antimicrobial peptide and suppresses gene expression of two novel type III secretion systems. Microbiolo. Res. 2010;165:665–673. doi: 10.1016/j.micres.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 35.Mellgren E.M., Kloek A.P., Kunkel B.N. Mqo, a tricarboxylic acid cycle enzyme, is required for virulence of Pseudomonas syringae pv. tomato strain DC3000 on Arabidopsis thaliana. J. Bacteriol. 2009;191:3132–3141. doi: 10.1128/JB.01570-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morohoshi T., Oseki K., Ikeda T. Exopolysaccharide production is influenced by sugars, N-acylhomoserine lactone, and transcriptional regulators RcsA and RcsB, but does not affect pathogenicity in the plant pathogen Pantoea ananatis. Biosci. Biotechnol. Biochem. 2011;75:997–999. doi: 10.1271/bbb.100888. [DOI] [PubMed] [Google Scholar]
- 37.Burbank L., Mohammadi M., Roper M.C. Siderophore-mediated iron acquisition influences motility and is required for full virulence of the xylem-dwelling bacterial phytopathogen Pantoea stewartii subsp. stewartii. Appl. Environ. Microbiol. 2015;81:139–148. doi: 10.1128/AEM.02503-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanui C.K., Shyntum D.Y., Priem S.L., Theron J., Moleleki L.N. Influence of the ferric uptake regulator (Fur) protein on pathogenicity in Pectobacterium carotovorum subsp. brasiliense. PLoS ONE. 2017;12:e0177647. doi: 10.1371/journal.pone.0177647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Expert D. Withholding and exchanging iron: Interactions between Erwinia spp. and their plant hosts. Annu. Rev. Phytopathol. 1999;37:307–334. doi: 10.1146/annurev.phyto.37.1.307. [DOI] [PubMed] [Google Scholar]
- 40.Rossez Y., Wolfson E.B., Holmes A., Gally D.L., Holden N.J. Bacterial flagella: Twist and stick, or dodge across the kingdoms. PLoS Pathog. 2015;11:e1004483. doi: 10.1371/journal.ppat.1004483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Orué Lucana D.O. Redox Sensing: Novel Avenues and Paradigms. Mary Ann Liebert, Inc.; New Rochelle, NY, USA: 2012. [Google Scholar]
- 42.Hérault E., Reverchon S., Nasser W. Role of the LysR-type transcriptional regulator PecT and DNA supercoiling in the thermoregulation of PEL genes, the major virulence factors in Dickeya dadantii. Environ. Microbiol. 2014;16:734–745. doi: 10.1111/1462-2920.12198. [DOI] [PubMed] [Google Scholar]
- 43.Thomson N.R., Nasser W., McGowan S., Sebaihia M., Salmond G.P. Erwinia carotovora has two KdgR-like proteins belonging to the IciR family of transcriptional regulators: Identification and characterization of the RexZ activator and the KdgR repressor of pathogenesis. Microbiology. 1999;145:1531–1545. doi: 10.1099/13500872-145-7-1531. [DOI] [PubMed] [Google Scholar]
- 44.Blair J.M., Webber M.A., Baylay A.J., Ogbolu D.O., Piddock L.J. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015;13:42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- 45.Moreira L., Soares M., Facincani A., Ferreira C., Ferreira R., Ferro M., Gozzo F., Felestrino É., Assis R., Garcia C. Proteomics-based identification of differentially abundant proteins reveals adaptation mechanisms of Xanthomonas citri subsp. citri during Citrus sinensis infection. BMC Microbiol. 2017;17:155. doi: 10.1186/s12866-017-1063-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mhedbi-Hajri N., Yahiaoui N., Mondy S., Hue N., Pélissier F., Faure D., Dessaux Y. Transcriptome analysis revealed that a quorum sensing system regulates the transfer of the pAt megaplasmid in Agrobacterium tumefaciens. BMC Genom. 2016;17:661. doi: 10.1186/s12864-016-3007-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dalsing B.L., Truchon A.N., Gonzalez-Orta E.T., Milling A.S., Allen C. Ralstonia solanacearum uses inorganic nitrogen metabolism for virulence, ATP production, and detoxification in the oxygen-limited host xylem environment. mBio. 2015;6:e02471-14. doi: 10.1128/mBio.02471-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morohoshi T., Ogata Y., Ikeda T. Cell aggregation is negatively regulated by N-acylhomoserine lactone-mediated quorum sensing in Pantoea ananatis SK-1. J. Biosci. Bioeng. 2011;112:566–569. doi: 10.1016/j.jbiosc.2011.08.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.