Abstract
Listeria monocytogenes is a major human foodborne pathogen that is prevalent in the natural environment and has a high case fatality rate. Whole genome sequencing (WGS) analysis has emerged as a valuable methodology for the classification of L. monocytogenes isolates and the identification of virulence islands that may influence infectivity. In this study, WGS was used to provide an insight into 25 L. monocytogenes isolates from cases of clinical infection in Ireland between 2013 and 2015. Clinical strains were either lineage I (14 isolates) or lineage II (11 isolates), with 12 clonal complexes (CC) represented, of which CC1 (6) and CC101 (4) were the most common. Single nucleotide polymorphism (SNP) analysis demonstrated that clinical isolates from mother–infant pairs (one isolate from the mother and one from the infant) were highly related (3 SNP differences in each) and also identified close similarities between isolates from otherwise distinct cases (1 SNP difference). Clinical strains were positive for common virulence-associated loci and 13 isolates harbour the LIPI-3 locus. Pulsed-field gel electrophoresis (PFGE) was used to compare strains to a database of 1300 Irish food and food processing environment isolates and determined that 64% of clinical pulsotypes were previously encountered in the food or food processing environment. Five of the matching food and food processing environment isolates were sequenced and results demonstrated a correlation between pulsotype and genotype. Overall, the work provides insights into the nature of L. monocytogenes strains currently causing clinical disease in Ireland and indicates that similar isolates can be found in the food or food processing environment.
Keywords: Listeria monocytogenes, clinical, genome, genomics, sequence, single nucleotide polymorphism, SNP
1. Introduction
Listeria monocytogenes is the causative agent of listeriosis, a serious infection that can manifest as meningitis and/or septicaemia in adults, infection of the fetus and miscarriage in pregnant women, or neonatal infection [1,2]. Although the disease is relatively rare, listeriosis is severe, with high hospitalisation and mortality rates [1,3]. In 2014, there were 2161 confirmed cases of human listeriosis in the European Union (EU) [4]. The fatality rate among 1524 confirmed cases with known outcome was 17.7% (270 cases), indicating the potential for this pathogen to pose a significant public health concern. In Ireland, listeriosis is a notifiable disease and the number of reported cases has been subject to an increase recently from eight cases in 2013, 15 cases in 2014 to 19 cases in 2015. The case incidence rate for 2015 was approximately 0.41 per 100,000 population, which is below the EU average of 0.48 cases per 100,000 for the same year [4,5]. However, listeriosis in Ireland remains a significant hazard for immunocompromised persons and other vulnerable groups, especially the elderly. A recent report suggests a trend towards an increased percentage of cases in adults (in particular adults over 65 years of age), which present as blood stream infection or meningitis [5].
L. monocytogenes consists of four evolutionary lineages and 13 serotypes with serotypes 4b and 1/2b (in lineage I) and 1/2a and 1/2c (in lineage II) being the most common causes of human listeriosis [6]. Whole genome sequencing (WGS) and subsequent genomic analyses can differentiate isolates that are otherwise indistinguishable by other typing methodologies (including pulsed-field gel electrophoresis (PFGE)) and can also produce significant insights into loci associated with pathogenesis in virulent or hypervirulent strains [7,8]. WGS and multi-locus sequence typing (MLST) can subdivide isolates according to sequence type (ST) or clonal complex (CC) whilst analysis of single nucleotide polymorphisms (SNPs) provides even greater granularity for the purposes of strain comparisons in the context of molecular epidemiological investigations [9,10].
Crucial to L. monocytogenes pathogenesis is an ability to invade host cells using internalins [11]. In particular, the interaction between internalin A (InlA) and host E-cadherin is essential for oral infection [12,13]. However, a significant proportion of environmental and food isolates of L. monocytogenes produce a truncated form of the InlA protein due to premature stop codons in the inlA gene and are therefore significantly compromised in virulence potential [14,15]. L. monocytogenes also encode genomic islands, known as Listeria pathogenicity islands (LIPIs), which play important roles in the virulence of the pathogen. LIPI-1 is the major pathogenicity island (encoding factors that are essential for phagosomal escape (listeriolysin O (LLO)) and cell-to-cell spread (ActA)) and is well conserved across strains independent of lineage [16]. The presence of LIPI-3, which includes a second haemolysin known as listeriolysin S (LLS), is strongly associated with lineage I strains, including a number of serotype 4b strains that have caused epidemic listeriosis [17]. Recent work suggests that LLS functions as a bacteriocin and plays a significant role in the gastrointestinal phase of Listeria infection [18]. Finally, LIPI-4, which encodes a cellobiose family phosphotransferase (PTS) system, is strongly associated with certain lineage I strains that are associated with invasion of the central nervous system (CNS) [7]. In turn, other factors may play a role in the environmental survival of specific isolates [19]. Such loci include a five gene cluster known as the stress survival islet 1 (SSI-1), which may contribute to the survival of cells in suboptimal conditions including high salt and low pH [20].
The objectives of this study were to use WGS to characterise distribution of virulence determinants in Irish clinical L. monocytogenes isolates, to compare methods for determining relatedness of the isolates (PFGE, ST, CC, core genome and SNP analysis) and to relate clinical isolates and food-related isolates. The data reveal the predominant sequence types causing clinical disease in Ireland, highlight the presence or absence of particular virulence associated genes and indicate that similar strains are present in foods and food processing environments.
2. Materials and Methods
2.1. Bacterial Isolates
To determine the genetic diversity and to identify the genetic characteristics of L. monocytogenes, a total of 25 Irish L. monocytogenes isolates from clinical samples were obtained from the national Listeria monocytogenes collection database at the National Reference Laboratory Service, University Hospital Galway (Table 1). These strains have also been included in a pan EU study of similarity between L. monocytogenes isolates by cgMLST. In addition, the PFGE profiles of the 25 clinical isolates were compared to a database of about 1300 isolates obtained from about 70 small food processing companies from 2013 to 2015 [21]. Six strains of L. monocytogenes from food and the food processing environment (five with similar PFGE profiles to the clinical isolates and one unrelated control strain), were sequenced to investigate whether there is genomic relatedness between food/food environment isolates and the clinical cases (Table 1). A subset of suitable reference strains were also selected from publicly available complete genomes including EGD-e (NC_003210.1), F2365 (NC_002973.6) and CLIP 80459 (NC_012488.1).
Table 1.
Listeria monocytogenes strains sequenced in this study.
| Isolate | Genbank Accession Number | ST 1 | CC 2 | Lineage | Serotype | Year of Isolation | Sample Type | Pulsotype |
|---|---|---|---|---|---|---|---|---|
| MQ130026 | MUZG00000000 | ST-1 | CC1 | I | 4b | 2013 | Blood | P2 * |
| L970 | PJJD00000000 | ST-1 | CC1 | I | 4b | 2013 | Food production ** | P2 * |
| MQ130029 | MVED00000000 | ST-1 | CC1 | I | 4b | 2013 | CSF 3 | P2 * |
| MQ130032 | MVEE00000000 | ST-1 | CC1 | I | 4b | 2013 | Blood | P2 * |
| MQ130042 | MVEG00000000 | ST-1 | CC1 | I | 4b | 2013 | Pleural Swab | P1 * |
| MQ140025 | MVEK00000000 | ST-1 | CC1 | I | 4b | 2014 | Ear Swab | P68 |
| MQ140031 | MVEN00000000 | ST-1 | CC1 | I | 4b | 2014 | Blood | P2 * |
| MQ140033 | MVEP00000000 | ST-1 | CC1 | I | 4b | 2014 | Blood | P1 * |
| L2113 | PJJE00000000 | ST-1 | CC1 | I | 4b | 2015 | Food production | P2 * |
| MQ150012 | MVEY00000000 | ST-6 | CC6 | I | 4b | 2015 | Blood | P13 * |
| MQ150005 | MVEU00000000 | ST-6 | CC6 | I | 4b | 2015 | Blood | P13 * |
| MQ130058 | MVEH00000000 | ST-6 | CC6 | I | 4b | 2013 | Blood | P13 * |
| MQ140030 | MVEM00000000 | ST-4 | CC4 | I | 4b | 2014 | CSF 3 | NC 4 |
| MQ150004 | MVET00000000 | ST-54 | CC54 | I | 4b | 2015 | Placental Swab | P6 * |
| MQ130033 | MVEF00000000 | ST-54 | CC54 | I | 4b | 2013 | Blood | P12 |
| L2259 | PJJF00000000 | ST-54 | CC54 | I | 4b | 2015 | Food production | P6 * |
| MQ150013 | MVEZ00000000 | ST-2 | CC2 | I | 4b | 2015 | Blood | P16 * |
| MQ130037 | MVFA00000000 | ST-18 | CC18 | II | 1/2a | 2013 | Blood | P32 * |
| MQ150011 | MVEX00000000 | ST-20 | CC20 | II | 1/2a | 2015 | Nasal Swab | NC |
| MQ150001 | MVES00000000 | ST-37 | CC37 | II | 1/2a | 2015 | Blood | P32 * |
| MQ140029 | MVEL00000000 | ST-7 | CC7 | II | 1/2a | 2014 | Blood | P31 * |
| L1445 | PJJG00000000 | ST-7 | CC7 | II | 1/2a | 2014 | Food production | P31 * |
| L1976 | PJJI00000000 | ST-8 | CC8 | II | 1/2c | 2015 | Food production | P48 * |
| MQ140034 | MVEQ00000000 | ST-121 | CC121 | II | 1/2a | 2014 | Blood—Mother/Infant | P59 * |
| L2256 | PJJH00000000 | ST-121 | CC121 | II | 1/2c | 2015 | Food production | P59 * |
| MQ140035 | MVER00000000 | ST-121 | CC121 | II | 1/2a | 2014 | Ear Swab—Mother/Infant | P59 * |
| MQ140032 | MVEO00000000 | ST-425 | CC90 | II | 1/2a | 2014 | Blood | NC |
| MQ140012 | MVEJ00000000 | ST-101 | CC101 | II | 1/2a | 2014 | Blood—Mother/Infant | P30 |
| MQ140011 | MVEI00000000 | ST-101 | CC101 | II | 1/2a | 2014 | Placental Surface Swab—Mother/Infant | P30 |
| MQ150008 | MVEW00000000 | ST-431 | CC101 | II | 1/2a | 2015 | Blood | P30 |
| MQ150007 | MVEV00000000 | ST-431 | CC101 | II | 1/2a | 2015 | CSF 3 | P30 |
1 Sequence Type; 2 Clonal Complex; 3 Cerebrospinal fluid; 4 NC = not classified; * Indicates pulsotypes associated with persistence in the food processing environment in a 3 year study of Irish foods and food production facilities [21]; ** These samples are from food or food production environments (non-clinical) and were chosen for sequencing based upon pulsed-field gel electrophoresis (PFGE) similarities to clinical isolates (see text).
2.2. Pulsed Field Gel Electrophoresis
Pulsed field gel electrophoresis analysis was carried out using the International Standard PulseNet 2013 protocol. The DNA was digested with 10 U/µL of the restriction enzyme Sgs1 (Asc1) FastDigest (Fisher Scientific, Dublin, Ireland) and 50 U/µL of the restriction enzyme ApaI FastDigest (Fisher Scientific); the restricted DNA was run in a 1% SeaKem Gold agarose gel for 21 h as described in the PulseNet protocol, on a CHEF-DR III (Bio-Rad, Hercules, CA, USA). After staining with 1 µg/mL ethidium bromide solution, the gels were observed with the Alpha Imager (Alpha Innotech, Kasendorf, Germany). Analysis of the gels was performed with BioNumerics v7.0 software (Applied Maths, Sint-Martens-Latem, Belgium) using UPGMA (unweighted pair group method with averages) and the Pearson coefficient with 1% tolerance.
2.3. Genome Sequencing and Annotation
The 25 clinical isolates were sequenced previously by our group [22]. The same protocol and platform was utilised to sequence the 6 selected food/food environment isolates described herein. Briefly, DNA was prepared using the GenElute Bacterial Genomic DNA kit (Sigma Aldrich, St. Louis, MO, USA) as per the manufacturer’s instructions. Library preparation and 250-bp paired end sequencing were performed using the Illumina HiSeq 2500 platform (Microbes NG, University of Birmingham, Birmingham, UK). Raw reads were mapped to a reference genome using the Burrows-Wheeler aligner-maximum exact matches (BWA-mem) [23] and de novo assembly was performed using the SPAdes genome assembler [24]. Contigs were re-ordered using Mauve aligner (v2.4.0) [25]. Prediction of putative open reading frames (ORFs) was performed using PRODIGAL prediction software [26] and supported by BLASTX and BLASTP [27]. Artemis [28] was employed for visualisation and manual editing in order to verify, and, where necessary, redefine the start of predicted coding regions. Genomes have been submitted to GenBank and assigned the following accession numbers (in brackets); L970 (PJJD00000000), L1445 (PJJG00000000), L1976 (PJJI00000000), L2113 (PJJE00000000), L2256 (PJJH00000000), L2259 (PJJF00000000).
2.4. Multilocus Sequence Typing and Clonal Complex
In order to determine the level of phylogenetic diversity between isolates, STs were determined by MLST using seven housekeeping genes, including ABC transporter abcZ, beta-glucosidase bglA, catalase cat, succinyl diaminopimelate dessucinylase dapE, D-amino acid aminotransferase dat, L-lactate dehydrogenase ldh and histidine kinase IhkA. The contig files for each of the draft genomes were uploaded to the Center for Genomic Epidemiology MLST 1.8 with L. monocytogenes as the MLST scheme. The clonal complex (CC) was defined based on the MLST profile of the isolate having matching profiles at 6 out of 7 genes [8,29]. The evolutionary history was inferred by using the Maximum Likelihood method based on the Tamura 3-parameter model [30]. The tree with the highest log likelihood is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. A discrete γ distribution was used to model evolutionary rate differences among sites (5 categories (+G, parameter = 0.5970)). The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 77.4196% sites). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 31 nucleotide sequences. Codon positions included were 1st + 2nd + 3rd + Noncoding. All positions with less than 95% site coverage were eliminated. That is, fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position. There were a total of 3288 positions in the final dataset. Evolutionary analyses were conducted in MEGA7 [31].
2.5. Single Nucleotide Polymorphisms Analysis
Single nucleotide polymorphisms were identified using the CSI Phylogeny 1.4 pipeline available on the Center for Genomic Epidemiology [32]. Separate analyses were carried out on 4b serotype strains (with strain F2365 as a reference) and 1/2a serotype strains (with EGDe as a reference). Phylogenetic trees were generated using the CSI Phylogeny tool available at the Center of Genomic Epidemiology. Multi-FASTA files, containing the sequences for each contig of the draft genome, were uploaded to the CSI Phylogeny 1.4 pipeline. Default parameters were used [32]. A maximum likelihood tree was created using FastTree and the subsequent Newick file was visualised using Figtree v1.4.2 [33]. The evolutionary history was inferred by using the Maximum Likelihood method, based on the General Time Reversible model [34]. The tree with the highest log likelihood is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the MCL approach, and then selecting the topology with superior log likelihood value. The trees are drawn to scale, with branch lengths measured in the number of substitutions per site. Codon positions included were 1st + 2nd + 3rd + Noncoding. All positions with less than 95% site coverage were eliminated. That is, fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position. Evolutionary analyses were conducted in MEGA7 [31].
2.6. Public Data Sources
Several publicly available genome sequences of various MLST types of L. monocytogenes were included in the SNP analysis. These were complete or draft sequences of the L. monocytogenes genome (Supplementary Material File S1). The genome sequences were downloaded from the Genbank database.
2.7. Pan- and Core-Genomic Profiling of Protein-Coding Genes
A comparative analysis of the genome content using GView [35] generated a pangenome profile of the L. monocytogenes isolates. The pangenome was constructed by iteratively appending unique regions onto an initial seed genome. Genbank files for three reference strains (F2365 seed genome, Clip80459 and EGD-e) were compared with the clinical and food/food processing environment isolates.
PanCoreGen [36] was subsequently used to construct the pan-genomic database of orthogolous genes using the clinical and food associated isolates along with suitable references including F2365, CLIP80459 and EGD-e. This algorithm performs BLASTN against genes from each annotated genome to detect orthologs in the query genomes based on user-defined threshold values of nucleotide identity and length-coverage. Stringent cut-off values (80–95%) for both sequence-identity and gene length-coverage were used to determine the set of core genes. A complete profile of core, mosaic and strain-specific genes was created which was processed to generate a matrix of 1s and 0s to indicate the presence or absence of genes across each genome. The pan-genome distance-matrix was generated on the basis of distance between profiles using the binary distance measure.
3. Results and Discussion
3.1. Serotypes and Clonal Complexes of Clinical Isolates
The objectives of this study were to use WGS to characterise virulence determinants of Irish clinical L. monocytogenes isolates, in order to compare a variety of methods for determining relatedness of the isolates (PFGE, ST, CC, core genome and SNP analysis) and to relate clinical isolates to a selection of food/food processing environment isolates from a three year longitudinal study [21]. Twenty-five isolates were available from the National Reference Laboratory Service culture collection and as expected strains were either lineage 1 (serotype 4b) or lineage II (serotype 1/2a or 1/2c). Following WGS [22], the clinical isolates were also analysed for ST using the Center for Genomic Epidemiology MLST tool (Table 1). The isolates included 13 sequence types which corresponded to twelve CCs [8,29]. Clonal complex 1 (CC1) accounts for 7/25 isolates from sampled Irish clinical cases. Other predominant clones include CC101 (4/25), CC121 (3/25) and CC6 (3/25).
Data indicates that the number of reported listeriosis infections in the EU has increased by 17% since 2013 and that serogroup 4 has become the most prevalent serogroup (EFSA, 2015). Similarly, Ireland has experienced an increase in cases of listeriosis since 2013 (HPSC, 2015) with serotype 4b the most prevalent serotype followed by 1/2a. The majority of CC identified in our cohort of isolates were CC1, CC2, CC6 and CC101, which are predominantly associated with clinical sources [7], while CC121 is primarily considered a food-associated isolate [7,37]. A single isolate (from a cerebrospinal fluid (CSF) sample) was identified as belonging to CC4, a clonal complex that is generally prevalent in cases of human listeriosis and is neuroinvasive and hypervirulent [7]. Most CCs are stable over decades, however, CC101, which was common in the 1950s but decreased subsequently, has begun to re-emerge globally [37]. It is also worth noting that ST-1/CC1 accounts for approximately 30% of the isolates in this study. This clone is typically associated with clinical sources [7] and has the same ST as several outbreak strains including an isolate (F2365) that was responsible for epidemic listeriosis associated with a Mexican-style cheese in 1985 [38].
3.2. Relationship between Sequence Type and PFGE Profiles
Pulse-field gel electrophoresis analysis of the 25 clinical L. monocytogenes isolates was undertaken in order to allow comparisons with a database of L. monocytogenes strains isolated from ready-to-eat (RTE) foods and food production facilities in Ireland [21]. Pulsotype numbers were assigned using both enzymes, based upon an in-house classification system that was previously used to type food and food processing environment isolates [21,39]. The data indicate (Table 1) that certain clinical isolates dating from 2013–2015 closely matched strains that were isolated from contaminated foods and food processing environments in Ireland during the same time period. In particular, pulsotypes P2, P6, P31 and P59 were commonly associated with RTE foods or food processing environments across multiple food sectors and were considered as persistent strains within food production facilities and associated foods (isolated more than once from the same facility over 6 months apart) [21]. Interestingly, the clinical isolates MQ140030 (CC4), MQ150011 (CC20) and MQ140032 (CC90) represented pulsotypes that were not previously encountered in our previous analysis of Irish food and environmental strains [21] and most likely represent isolates that are very rare in foods and food environments in Ireland.
To allow further comparisons, six food-associated strains were sequenced, five of which (strains L2259, L970, L2113, L2256, L1445) have pulsotypes that match those of the clinical isolates (one strain, L1976, was used as a control outlier for phylogenetic comparisons). In this, admittedly low, sample set, pulsotype generally correlated with CC as defined by MLST, as expected. For instance, the pulsotype P2 isolates L2113 and L970 which were isolated from a food processing environment, were both CC1 as determined following WGS, and closely related to clinical CC1 strains.
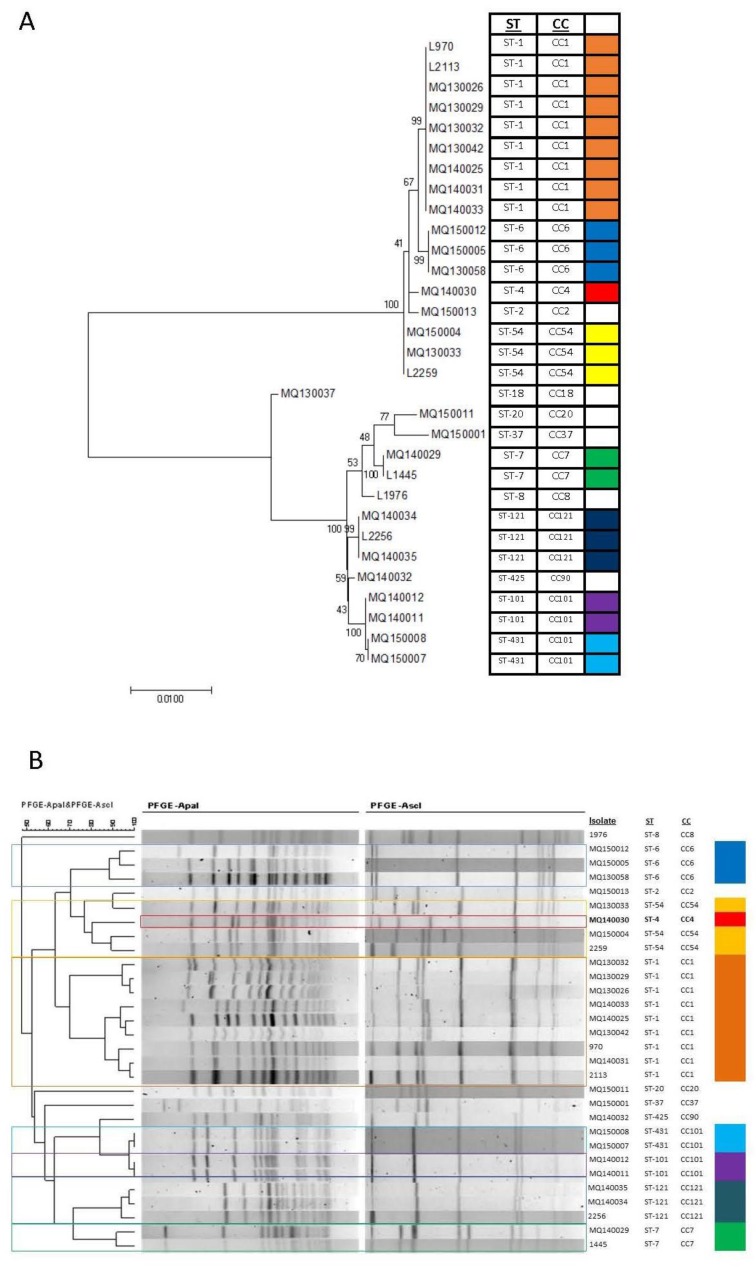
Figure 1A shows a maximum likelihood phylogeny constructed from the nucleotide sequences of the seven house-keeping genes used for sequence typing of L. monocytogenes by MLST while Figure 1B shows a dendrogram of the PFGE profiles. Both techniques are used to sub-type isolates, and can give results that are not comparable. Using MLST, the three CC54 isolates (two clinical and one food isolate) are shown as indistinguishable, while the ST-4 isolate MQ140030 is closely related. Using PFGE, the three CC54 isolates are more or less distinguishable, particularly MQ130033 and MQ150004, which show <80% similarity, while L2259 and MQ150004 show about 90% similarity. The ST-4 isolate shows about 80% similarity to the ST-54 isolates. On the other hand, strains MQ150013 and MQ130033 are indistinguishable (>90% similarity) by PFGE, but can be differentiated by ST.
Figure 1.
(A) Clustering of isolates based upon multi-locus sequence typing (MLST). The evolutionary history was inferred by using the Maximum Likelihood method based on the Tamura 3-parameter model [30] as outlined in detail in Materials and Methods. The tree with the highest log likelihood (−5971.3729) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Evolutionary analyses were conducted in MEGA7 [31]; (B) Clustering of isolates based upon pulsed field gel electrophoresis (PFGE). Dendograms were generated with BioNumerics v7.0 software (Applied Maths) using UPGMA (unweighted pair group method with averages) and the Pearson coefficient with 1% tolerance. Using either method (MLST or PFGE) strains cluster into two distinct groups dependent upon lineage. ST: sequence type; CC: clonal complex.
Although PFGE has greater discriminatory power than MLST, it does not have sufficient discriminatory power to consistently distinguish epidemiologically unrelated strains of L. monocytogenes [6]. In addition, while the methods of analysis have been standardised internationally, the naming of profiles unfortunately has not. This makes the comparison of strains amongst research groups and epidemiological monitoring bodies difficult. MLST based on the sequence of seven housekeeping genes overcomes some of these limitations and provides a highly standardised genotype that allows comparison with strains isolated internationally [7].
3.3. Single-Nucleotide Polymorphism Analysis of Isolates
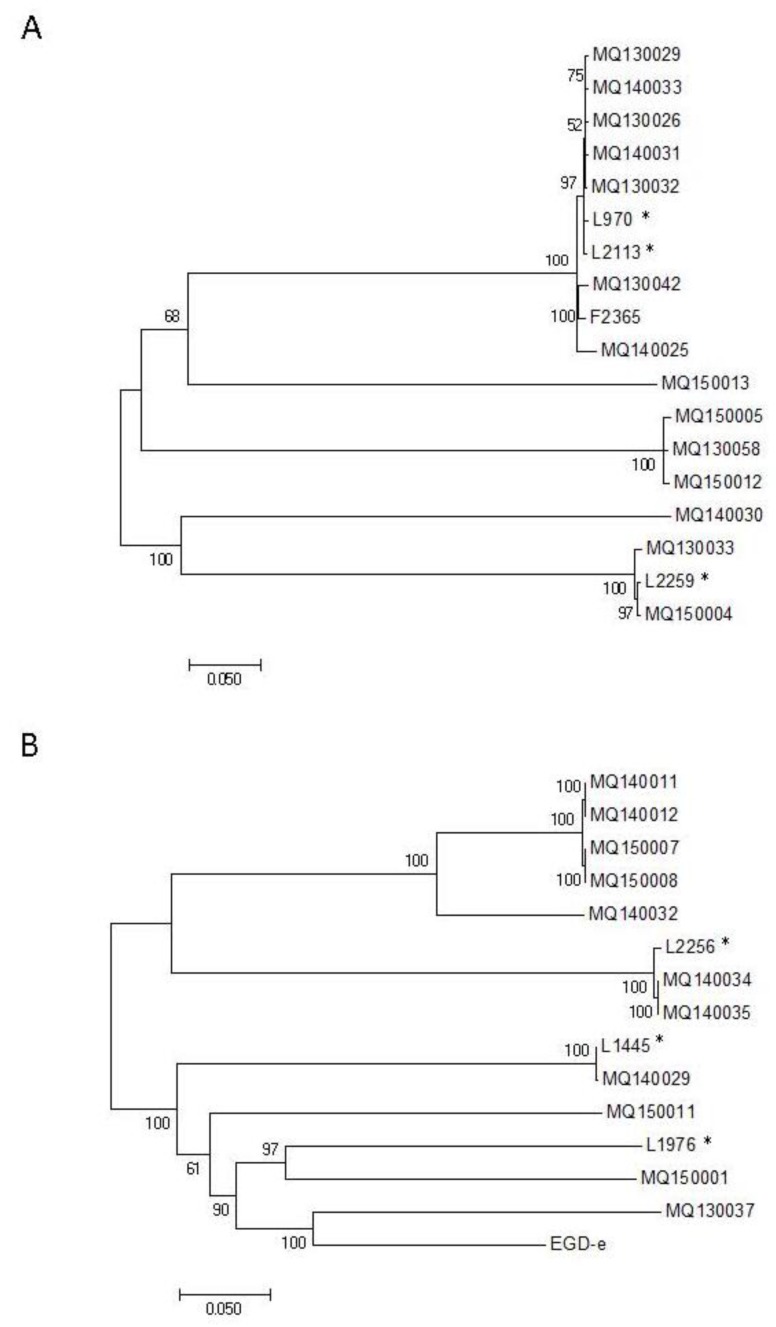
Due to the significant number of ST-1 isolates in both the clinical (28%) and food groups (33%), SNP analysis was carried out to assess whether any of the isolates may be closely linked. To obtain a broad overview of the SNP profiles, each isolate was initially compared to a reference strain of the same serotype. All serotype 4b isolates were initially compared to the reference F2365 (Supplementary Table S1) and all serotype 1/2a isolates were compared to the reference EGDe (Supplementary Table S2). The serotype 4b isolates differed by between a minimum of 41 and a maximum of 5862 SNPs while the serotype 1/2a isolates differed by a minimum of 3 and a maximum of 12,515 SNPs. A maximum likelihood tree was generated for each comparison showing the relationship between the isolates. The SNP tree clustered the L. monocytogenes isolates according to their ST/CC (Figure 2).
Figure 2.
Phylogeny of the isolates as determined by single nucleotide polymorphism (SNP) analysis. (A) All serotype 4b isolates were compared with strain F2365 as the reference genome. The evolutionary history was inferred by using the Maximum Likelihood method based on the General Time Reversible model [34]. The tree with the highest log likelihood (−67933.7374) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 18 nucleotide sequences. Codon positions included were 1st + 2nd + 3rd + Noncoding. All positions with less than 95% site coverage were eliminated. That is, fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position. There were a total of 12,069 positions in the final dataset. Evolutionary analyses were conducted in MEGA7 [31]. (B) All serotype 1/2a isolates were compared with strain EGDe as the reference genome. The evolutionary history was inferred by using the Maximum Likelihood method based on the General Time Reversible model [34]. The tree with the highest log likelihood (−214301.5165) is shown. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 15 nucleotide sequences. Codon positions included were 1st + 2nd + 3rd + Noncoding. All positions with less than 95% site coverage were eliminated. That is, fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position. There were a total of 30,102 positions in the final dataset. Evolutionary analyses were conducted in MEGA7 [31]. * Indicates strains from a food production source.
For reference-based methods of SNP analysis, the choice of reference genome can significantly influence the results [40]. Comparisons of genetically distant groups of isolates using a single reference may result in loss of resolution particularly if the chosen reference is genetically distant from the isolates under investigation [41]. Following the initial analyses, each cluster of closely related isolates (i.e., a single ST or CC) was analysed separately using a closely related reference genome to maximise the accuracy and resolution of SNP identification. The number of SNP differences between isolates within an outbreak of foodborne disease is considered to range from 0–12 SNPs [41,42,43,44]. However, there is some variation between studies regarding the classification of strains as closely linked. In the current study the criteria described by Kwong and coworkers [41] was used for estimating the possible genomic relatedness of L. monocytogenes isolates (Supplementary Material File S1).
The nine ST-1 isolates, including seven clinical isolates and two food processing environment isolates, were compared first using F2365 (ST-1) as the reference genome. Similar numbers of SNPs were revealed when using either of the food processing isolates, L970 or L2113, as the reference genome. Analysing the ST-1 isolates grouped by year of isolation, 2013 or 2014, revealed a relatively high minimum of 44 and 66 SNPs, respectively (Supplementary File S1). The ST-6 and ST-54 isolates also had a high minimum number of SNPs, 199 and 65, respectively (Supplementary File S1). The ST-7 isolates, MQ140029 (clinical) and L1445 (food) were shown to differ by only one SNP (Supplementary File S1) and therefore are phylogenetically highly similar strains [41]. The likelihood of uncovering epidemiological links in historic, sporadic cases of listeriosis is extremely low and indeed we could determine no clear links between this case and the food in question. However, the high phylogenetic similarity between these food and clinical strains indicates that L. monocytogenes strains causing cases of clinical infection can be found in Irish foods and highlights the importance of ongoing vigilance to ensure RTE foods remain free of the pathogen.
In our study, the mother–infant ST-121 isolates (one from the mother and one from the infant), MQ140034 and MQ140035, were expected to be highly related and indeed were found to differ by only three SNPs (Supplementary File S1). Similarly, the ST-101 mother–infant isolates (MQ140011 and MQ140012) were found to differ by just three SNPs (Supplementary File S1). The unbiased identification of phylogenetic similarities within each mother–infant pair indicate the validity of the SNP approach for identifying highly related strains.
Finally, the ST-431 isolates, MQ150007 and MQ150008, were found to differ by just two SNPs (Supplementary File S1), indicating that these isolates are likely to be phylogenetically linked [41]. These strains were isolated from separate patients but were isolated by the same local health authority within a one-month period. However, following further retrospective investigation, no epidemiological links between the cases could be determined (personal communication with relevant health authorities). However, the study indicates the ability of SNP analysis to reveal similarities between isolates and to identify the potential for common source outbreaks should they arise. Findings revealed through fine SNP analysis are summarized in Table 2.
Table 2.
Relatedness of strains as determined by fine single nucleotide polymorphism (SNP) analysis with appropriate reference strains.
| Sequence Type | Isolates | Reference Genome | Minimum SNPs | Maximum SNPs |
|---|---|---|---|---|
| ST1 | L970, L2113, MQ130026, MQ130029, MQ130032 *, MQ130042, MQ140025, MQ140031, MQ140033 | F2365 | 43 | 261 |
| F2365, L2113, MQ130026, MQ130029, MQ130032 *, MQ130042, MQ140025, MQ140031, MQ140033 | L970 | 42 | 256 | |
| F2365, L970, MQ130026, MQ130029, MQ130032 *, MQ130042, MQ140025, MQ140031, MQ140033 | L2113 | 42 | 254 | |
| F2365, L970, MQ130029, MQ130032 *, MQ130042 | MQ130026 | 44 | 190 | |
| F2365, L2113, MQ140031, MQ140033 * | MQ140025 | 66 | 259 | |
| ST6 | H7858, MQ130058, MQ150012 * | MQ150005 | 199 | 373 |
| ST54 | LM07-01337, MQ130033, MQ150004 * | L2259 | 65 | 115 |
| ST7 | J2692, L1846, L2676, MQ140029 * | L1445 | 1 | 409 |
| ST121 | 4423, 6179, L2256, La111, Lm1880, N53-1, MQ140035 * | MQ140034 | 3 | 461 |
| ST101 | 2012-L5240, 2012-L5323, Lm1840, MQ140012, MQ150007, MQ150008 * | MQ140011 | 1 | 145 |
| 2012-L5240, 2012-L5323, Lm1840, MQ150008 * | MQ150007 | 2 | 146 |
* Indicates strain with closest similarity to the reference genome (lowest number of SNPs).
3.4. Pan- and Core-Genome Analysis
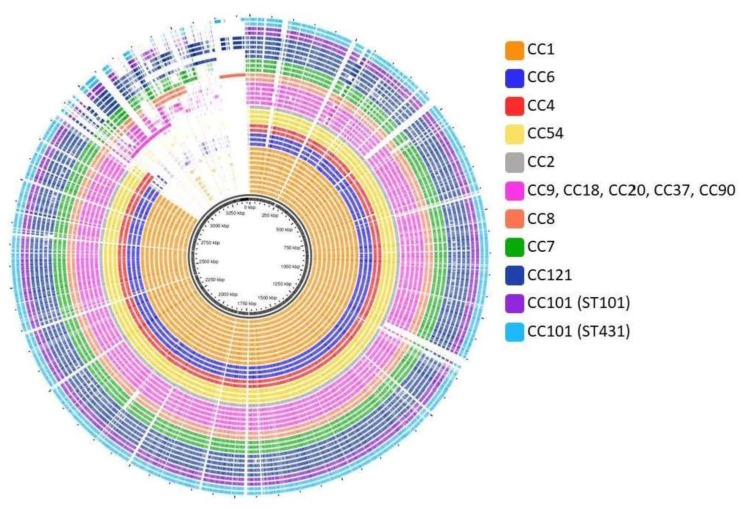
The pan-genome for 38 isolates, including 25 clinical isolates, six food/food processing environment isolates and seven reference genomes, was generated using GView [35]. Approximately 85% of the coding sequences consist of mutually conserved core genes as shown in Figure 3. This analysis demonstrates that a significant proportion of the L. monocytogenes genome is conserved between serotypes as well as STs. Analysis of the accessory genes that are common to a particular CC may yield candidate genes that contribute to successful adaption of isolates to a particular environment [7]. In particular, the significant variation in the accessory genome of ST-121 isolates, indicative of clonal diversity within this group, was noted (Figure 3).
Figure 3.
Pan-genome, constructed using GView [35], of 34 genomes including 25 clinical isolates, six food-associated isolates and three reference genomes. The pangenome is constructed by iteratively appending unique regions onto the initial seed genome in this case F2365. Gaps indicate that the region is missing in a particular gene but is found in others. Strains are grouped according to CC and colour coded as indicated.
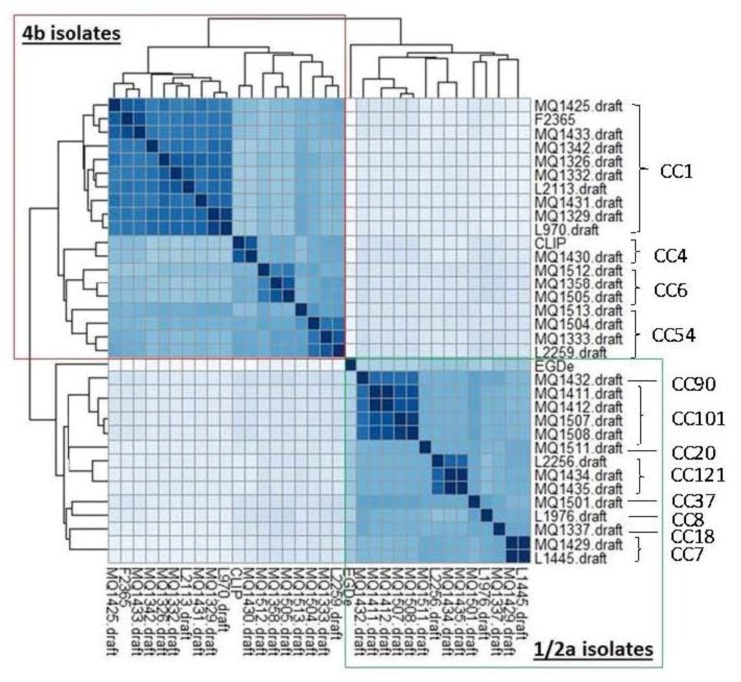
The pan-genome of a species is the sum of non-redundant genomic regions from its representative genomes [45]. It is composed of core genomic regions present in all genomes and accessory regions found in some but not all genomes. The L. monocytogenes pan-genome has been estimated in several independent studies [45,46,47,48] and has a repertoire of approximately 4000 unique genes. Those studies estimate that there are an average of 2900 genes per strain and approximately 2400 of these genes are part of the core genome (≈80%). A pan-genomic database of orthogolous genes was constructed using PanCoreGen [36]. A cut-off of 80% for sequence identity and gene length coverage resulted in a total of 3105 orthologous genes amongst the genome sequences analysed. The observed core genome shared by the three L. monocytogenes reference strains (F2365, CLIP 80459 and EGD-e) is 2431 genes, which is in agreement with the current estimates. These three reference strains cover a minimum of 91% of the coding sequences detected in the draft genomes of the isolates. A matrix of the genetic content of each isolate was generated to indicate the presence and absence of each core and accessory gene. Subsequently, the presence or absence of genes can be used to cluster the isolates into serotypes and CCs (Figure 4).
Figure 4.
The presence and absence of genes can be used to cluster the isolates into serotypes and sequence types.
When implemented as a health-care surveillance approach, the application of systematic WGS of all L. monocytogenes isolates, as applied in many different countries recently, can potentially link seemingly sporadic cases of listeriosis to common-source outbreaks [49]. However, although there may be similarities in WGS between strains, linking L. monocytogenes isolates to listeriosis outbreaks in the absence of epidemiological data is not feasible.
3.5. Listeria Pathogenicity Islands
As indicated, WGS can act as an epidemiological tool to identify highly-related isolates, but it can also be used to identify the presence of genes/pathogenicity islands associated with hypervirulence or particular modes of pathogenesis (such as ability to invade the CNS) [7]. In our study, each of the 31 genomes encodes LIPI-1 which is a Prf-A dependent virulence gene cluster consisting of six genes (prfA, plcA, hly, mpl, actA and plcB) that are key for the infection cycle of L. monocytogenes (Table 3). LIPI-3 is a gene cluster that encodes a potential haemolytic factor with homology to Streptolysin S (SLS) [17] and which has recently been shown to possess antimicrobial potential and to play a role in gastrointestinal colonization [18]. LIPI3 is strongly associated with lineage I strains and was found to be present in all Irish serotype 4b isolates with the exception of MQ150013. LIPI-4 was recently described as a gene cluster involved in neural and placental infection [7]. This pathogenicity island encodes six genes annotated as a cellobiose family PTS system and appears to be strongly associated with CC4 isolates. The presence of LIPI4 was confirmed in the only CC4 clinical isolate (MQ140030) in this dataset, a strain originally isolated from the CSF of a patient presenting with listeriosis in 2014. In support of a previous study [7], this island appears to be associated with CC4 and was not identified in any other strains in this study.
Table 3.
Presence or absence of key loci encoding L. monocytogenes virulence-associated elements or loci putatively involved in environmental survival.
| Isolate | ST 1 | CC 2 | LIPI1 | LIPI3 | LIPI4 | InlA | SSI-1 |
|---|---|---|---|---|---|---|---|
| MQ130026 | ST-1 | CC1 | + 3 | + | − 4 | + 5 | − |
| L970 | ST-1 | CC1 | + | + | − | + | − |
| MQ130029 | ST-1 | CC1 | + | + | − | + | − |
| MQ130032 | ST-1 | CC1 | + | + | − | + | − |
| MQ130042 | ST-1 | CC1 | + | + | − | + | − |
| MQ140025 | ST-1 | CC1 | + | + | − | + | − |
| MQ140031 | ST-1 | CC1 | + | + | − | + | − |
| MQ140033 | ST-1 | CC1 | + | + | − | + | − |
| L2113 | ST-1 | CC1 | + | + | − | + | − |
| MQ150012 | ST-6 | CC6 | + | + | − | + | − |
| MQ150005 | ST-6 | CC6 | + | + | − | + | − |
| MQ130058 | ST-6 | CC6 | + | + | − | + | − |
| MQ140030 | ST-4 | CC4 | + | + | + | + | − |
| MQ150004 | ST-54 | CC54 | + | + | − | + | − |
| MQ130033 | ST-54 | CC54 | + | + | − | + | − |
| L2259 | ST-54 | CC54 | + | + | − | + | − |
| MQ150013 | ST-2 | CC2 | + | − | − | + | − |
| MQ130037 | ST-18 | CC18 | + | − | − | + | + |
| MQ150011 | ST-20 | CC20 | + | − | − | + | − |
| MQ150001 | ST-37 | CC37 | + | − | − | + | − |
| MQ140029 | ST-7 | CC7 | + | − | − | + | + |
| L1445 | ST-7 | CC7 | + | − | − | + | + |
| L1976 | ST-8 | CC8 | + | − | − | + | + |
| MQ140034 | ST-121 | CC121 | + | − | − | + | − |
| L2256 | ST-121 | CC121 | + | − | − | − | − |
| MQ140035 | ST-121 | CC121 | + | − | − | + | − |
| MQ140032 | ST-425 | CC90 | + | − | − | + | − |
| MQ140012 | ST-101 | CC101 | + | − | − | + | − |
| MQ140011 | ST-101 | CC101 | + | − | − | + | − |
| MQ150008 | ST-431 | CC101 | + | − | − | + | − |
| MQ150007 | ST-431 | CC101 | + | − | − | + | − |
1 Sequence Type; 2 Clonal Complex; 3 Presence of genes; 4 Absence of genes; 5 Indicates encoding predicted full-length InlA.
3.6. Internalins
L. monocytogenes encode surface proteins known as internalins that are used to invade host cells [2]. The number of internalins encoded by the genomes ranged from 10 to 13. All isolates encoded full length internalin A with the exception of the food processing isolate L2256. The CC6 isolates (MQ130058, MQ150005 and MQ150012) all have a characteristic three amino acid deletion in the pre-anchor region of InlA [50]. Internalins B, C, C2 (inlH in EGDe), E, I and J are also present in all isolates. inlD is present in all isolates with the exception of EGDe while inlF is present in all isolates with the exception of MQ140034, MQ140035 and L2256.
3.7. Stress Survival Islet (SSI-1)
SSI-1 is a five gene islet that contributes to the growth of L. monocytogenes under suboptimal conditions [20]. This islet is present in two clinical isolates MQ130037 (CC18), MQ140029 (CC7), and two food-associated isolates L1445 (CC7) and L1976 (CC8). These isolates are all lineage II serotype 1/2. This islet has been recently shown to be a feature of ST-7 (CC7) and ST-8 (CC8) strains associated with persistence in a study of L. monocytogenes strains isolated over 20 years from food-processing plants but is also found in sporadic environmental strains [51]. Given that the islet is rare amongst clinical isolates, in this study, it is likely to be dispensable for virulence but may play a role in environmental survival and survival of some strains in foods [20].
4. Conclusions
Whole genome sequencing of L. monocytogenes isolates from cases of human listeriosis in Ireland between 2013 and 2015 has provided an overview of locally circulating clinical strains of the pathogen. The work identified particular CCs that were responsible for disease in Ireland and permitted comparison of clinical isolates to a comprehensive database of food and food production isolates from the same time period (based upon PFGE type) [21]. SNP analysis revealed that pairs of strains isolated from mother–infant cases were highly related. SNP analysis also identified phylogenetically identical isolates from a patient and a food source and another pair of isolates from distinct patients. Whilst retrospective follow up failed to prove any clear epidemiological links, the study highlights the potential for WGS to identify listeriosis disease patterns within a single national health authority.
Acknowledgments
This work was supported by the Irish Department of Agriculture and Food and the Marine under the Food Institutional Research Measure (FIRM) project number 11F008. We acknowledge funding received from Science Foundation Ireland in the form of a Center grant (APC Microbiome Ireland, SFI/12/RC/2273).
Supplementary Materials
The following are available online at www.mdpi.com/2073-4425/9/3/171/s1, File S1: Fine single nucleotide polymorphism analysis of clinical isolates, Table S1: Number of single nucleotide polymorphisms identified when serotype 4b strains were compared, using F2365 as a reference genome, Table S2: Number of single nucleotide polymorphisms identified when serotype 1/2a strains were compared, using EGDe as a reference genome.
Author Contributions
C.G.M.G., M.C. and K.J. conceived the project. A.H., C.G.M.G., M.C., C.H. and K.J. designed the experiments. A.H., D.L., A.O., E.P.C., C.A.M., N.D. performed the experimental work. A.H. and A.O. carried out the bioinformatic analysis. A.H., C.G.M.G., M.C., C.H. and K.J. analysed and interpreted the data. A.H. and C.G.M.G. wrote the paper with significant input from all authors.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.De Noordhout C.M., Devleesschauwer B., Angulo F.J., Verbeke G., Haagsma J., Kirk M., Havelaar A., Speybroeck N. The global burden of listeriosis: A systematic review and meta-analysis. Lancet Infect. Dis. 2014;14:1073–1082. doi: 10.1016/S1473-3099(14)70870-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vazquez-Boland J.A., Kuhn M., Berche P., Chakraborty T., Dominguez-Bernal G., Goebel W., Gonzalez-Zorn B., Wehland J., Kreft J. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 2001;14:584–640. doi: 10.1128/CMR.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scallan E., Hoekstra R.M., Angulo F.J., Tauxe R.V., Widdowson M.A., Roy S.L., Jones J.L., Griffin P.M. Foodborne illness acquired in the United States—Major pathogens. Emerg. Infect. Dis. 2011;17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.European Food Safety Authority The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2014. EFSA J. 2015;13:4329. doi: 10.2903/j.efsa.2018.5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Health Protection Surveillance Centre (HPSC) HPSC Annual Epidemiological Report 2015. Volume 75 Health Protection Surveillance Centre; Dublin, Ireland: 2016. [Google Scholar]
- 6.Lomonaco S., Nucera D. DNA Methods in Food Safety: Molecular Typing of Foodborne and Waterborne Bacterial Pathogens. Wiley; Hoboken, NJ, USA: 2014. Molecular subtyping methods for Listeria monocytogenes: Tools for tracking and control; pp. 303–336. [Google Scholar]
- 7.Maury M.M., Tsai Y.H., Charlier C., Touchon M., Chenal-Francisque V., Leclercq A., Criscuolo A., Gaultier C., Roussel S., Brisabois A., et al. Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat. Genet. 2016;48:308–313. doi: 10.1038/ng.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moura A., Criscuolo A., Pouseele H., Maury M.M., Leclercq A., Tarr C., Bjorkman J.T., Dallman T., Reimer A., Enouf V., et al. Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat. Microbiol. 2016;2:16185. doi: 10.1038/nmicrobiol.2016.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z., Perez-Osorio A., Wang Y., Eckmann K., Glover W.A., Allard M.W., Brown E.W., Chen Y. Whole genome sequencing analyses of Listeria monocytogenes that persisted in a milkshake machine for a year and caused illnesses in Washington state. BMC Microbiol. 2017;17:134. doi: 10.1186/s12866-017-1043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kvistholm Jensen A., Nielsen E.M., Bjorkman J.T., Jensen T., Muller L., Persson S., Bjerager G., Perge A., Krause T.G., Kiil K., et al. Whole-genome sequencing used to investigate a nationwide outbreak of listeriosis caused by ready-to-eat delicatessen meat, Denmark, 2014. Clin. Infect. Dis. 2016;63:64–70. doi: 10.1093/cid/ciw192. [DOI] [PubMed] [Google Scholar]
- 11.Gaillard J.L., Berche P., Frehel C., Gouin E., Cossart P. Entry of L. monocytogenes into cells is mediated by internalin, a repeat protein reminiscent of surface antigens from gram-positive cocci. Cell. 1991;65:1127–1141. doi: 10.1016/0092-8674(91)90009-N. [DOI] [PubMed] [Google Scholar]
- 12.Disson O., Grayo S., Huillet E., Nikitas G., Langa-Vives F., Dussurget O., Ragon M., Le Monnier A., Babinet C., Cossart P., et al. Conjugated action of two species-specific invasion proteins for fetoplacental listeriosis. Nature. 2008;455:1114–1118. doi: 10.1038/nature07303. [DOI] [PubMed] [Google Scholar]
- 13.Lecuit M., Vandormael-Pournin S., Lefort J., Huerre M., Gounon P., Dupuy C., Babinet C., Cossart P. A transgenic model for listeriosis: Role of internalin in crossing the intestinal barrier. Science. 2001;292:1722–1725. doi: 10.1126/science.1059852. [DOI] [PubMed] [Google Scholar]
- 14.Jacquet C., Doumith M., Gordon J.I., Martin P.M., Cossart P., Lecuit M. A molecular marker for evaluating the pathogenic potential of foodborne Listeria monocytogenes. J. Infect. Dis. 2004;189:2094–2100. doi: 10.1086/420853. [DOI] [PubMed] [Google Scholar]
- 15.Nightingale K.K., Ivy R.A., Ho A.J., Fortes E.D., Njaa B.L., Peters R.M., Wiedmann M. InlA premature stop codons are common among Listeria monocytogenes isolates from foods and yield virulence-attenuated strains that confer protection against fully virulent strains. Appl. Environ. Microbiol. 2008;74:6570–6583. doi: 10.1128/AEM.00997-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gouin E., Mengaud J., Cossart P. The virulence gene cluster of Listeria monocytogenes is also present in Listeria ivanovii, an animal pathogen, and Listeria seeligeri, a nonpathogenic species. Infect. Immun. 1994;62:3550–3553. doi: 10.1128/iai.62.8.3550-3553.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cotter P.D., Draper L.A., Lawton E.M., Daly K.M., Groeger D.S., Casey P.G., Ross R.P., Hill C. Listeriolysin S, a novel peptide haemolysin associated with a subset of lineage I Listeria monocytogenes. PLoS Pathog. 2008;4:e1000144. doi: 10.1371/journal.ppat.1000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quereda J.J., Dussurget O., Nahori M.A., Ghozlane A., Volant S., Dillies M.A., Regnault B., Kennedy S., Mondot S., Villoing B., et al. Bacteriocin from epidemic Listeria strains alters the host intestinal microbiota to favor infection. Proc. Natl. Acad. Sci. USA. 2016;113:5706–5711. doi: 10.1073/pnas.1523899113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gahan C.G., Hill C. Listeria monocytogenes: Survival and adaptation in the gastrointestinal tract. Front. Cell. Infect. Microbiol. 2014;4:9. doi: 10.3389/fcimb.2014.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryan S., Begley M., Hill C., Gahan C.G. A five-gene stress survival islet (SSI-1) that contributes to the growth of Listeria monocytogenes in suboptimal conditions. J. Appl. Microbiol. 2010;109:984–995. doi: 10.1111/j.1365-2672.2010.04726.x. [DOI] [PubMed] [Google Scholar]
- 21.Leong D., NicAogain K., Luque-Sastre L., McManamon O., Hunt K., Alvarez-Ordonez A., Scollard J., Schmalenberger A., Fanning S., O’Byrne C., et al. A 3-year multi-food study of the presence and persistence of Listeria monocytogenes in 54 small food businesses in Ireland. Int. J. Food Microbiol. 2017;249:18–26. doi: 10.1016/j.ijfoodmicro.2017.02.015. [DOI] [PubMed] [Google Scholar]
- 22.O’Callaghan A., Hilliard A., Morgan C.A., Culligan E.P., Leong D., DeLappe N., Hill C., Jordan K., Cormican M., Gahan C.G.M. Draft genome sequences of 25 Listeria monocytogenes isolates associated with human clinical listeriosis in Ireland. Genome Announc. 2017;5:e00184-17. doi: 10.1128/genomeA.00184-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-mem. arXiv. 2013. 1303.3997
- 24.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. Spades: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rissman A.I., Mau B., Biehl B.S., Darling A.E., Glasner J.D., Perna N.T. Reordering contigs of draft genomes using the Mauve aligner. Bioinformatics. 2009;25:2071–2073. doi: 10.1093/bioinformatics/btp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hyatt D., Chen G.L., Locascio P.F., Land M.L., Larimer F.W., Hauser L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 28.Rutherford K., Parkhill J., Crook J., Horsnell T., Rice P., Rajandream M.A., Barrell B. Artemis: Sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 29.Ragon M., Wirth T., Hollandt F., Lavenir R., Lecuit M., Le Monnier A., Brisse S. A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 2008;4:e1000146. doi: 10.1371/journal.ppat.1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamura K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol. Biol. Evol. 1992;9:678–687. doi: 10.1093/oxfordjournals.molbev.a040752. [DOI] [PubMed] [Google Scholar]
- 31.Kumar S., Stecher G., Tamura K. Mega7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaas R.S., Leekitcharoenphon P., Aarestrup F.M., Lund O. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS ONE. 2014;9:e104984. doi: 10.1371/journal.pone.0104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H., Wang J., Yu P., Ge P., Jiang Y., Xu R., Chen R., Liu X. Identification of antibiotic resistance genes in the multidrug-resistant Acinetobacter baumannii strain, MDR-SHH02, using whole-genome sequencing. Int. J. Mol. Med. 2017;39:364–372. doi: 10.3892/ijmm.2016.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nei M., Kumar S. Molecular Evolution and Phylogenetics. Oxford University Press; New York, NY, USA: 2000. [Google Scholar]
- 35.Petkau A., Stuart-Edwards M., Stothard P., Van Domselaar G. Interactive microbial genome visualization with GView. Bioinformatics. 2010;26:3125–3126. doi: 10.1093/bioinformatics/btq588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paul S., Bhardwaj A., Bag S.K., Sokurenko E.V., Chattopadhyay S. Pancoregen—Profiling, detecting, annotating protein-coding genes in microbial genomes. Genomics. 2015;106:367–372. doi: 10.1016/j.ygeno.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haase J.K., Didelot X., Lecuit M., Korkeala H., Achtman M. The ubiquitous nature of Listeria monocytogenes clones: A large-scale multilocus sequence typing study. Environ. Microbiol. 2014;16:405–416. doi: 10.1111/1462-2920.12342. [DOI] [PubMed] [Google Scholar]
- 38.Linnan M.J., Mascola L., Lou X.D., Goulet V., May S., Salminen C., Hird D.W., Yonekura M.L., Hayes P., Weaver R., et al. Epidemic listeriosis associated with mexican-style cheese. N. Engl. J. Med. 1988;319:823–828. doi: 10.1056/NEJM198809293191303. [DOI] [PubMed] [Google Scholar]
- 39.Leong D., Alvarez-Ordonez A., Zaouali S., Jordan K. Examination of Listeria monocytogenes in seafood processing facilities and smoked salmon in the Republic of Ireland. J. Food Prot. 2015;78:2184–2190. doi: 10.4315/0362-028X.JFP-15-233. [DOI] [PubMed] [Google Scholar]
- 40.Henri C., Leekitcharoenphon P., Carleton H.A., Radomski N., Kaas R.S., Mariet J.F., Felten A., Aarestrup F.M., Gerner Smidt P., Roussel S., et al. An assessment of different genomic approaches for inferring phylogeny of Listeria monocytogenes. Front. Microbiol. 2017;8:2351. doi: 10.3389/fmicb.2017.02351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwong J.C., Mercoulia K., Tomita T., Easton M., Li H.Y., Bulach D.M., Stinear T.P., Seemann T., Howden B.P. Prospective whole-genome sequencing enhances national surveillance of Listeria monocytogenes. J. Clin. Microbiol. 2016;54:333–342. doi: 10.1128/JCM.02344-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leekitcharoenphon P., Nielsen E.M., Kaas R.S., Lund O., Aarestrup F.M. Evaluation of whole genome sequencing for outbreak detection of Salmonella enterica. PLoS ONE. 2014;9:e87991. doi: 10.1371/journal.pone.0087991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor A.J., Lappi V., Wolfgang W.J., Lapierre P., Palumbo M.J., Medus C., Boxrud D. Characterization of foodborne outbreaks of Salmonella enterica serovar enteritidis with whole-genome sequencing single nucleotide polymorphism-based analysis for surveillance and outbreak detection. J. Clin. Microbiol. 2015;53:3334–3340. doi: 10.1128/JCM.01280-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wuyts V., Denayer S., Roosens N.H., Mattheus W., Bertrand S., Marchal K., Dierick K., De Keersmaecker S.C. Whole genome sequence analysis of Salmonella enteritidis PT4 outbreaks from a national reference laboratory's viewpoint. PLoS Curr. 2015;7:159. doi: 10.1371/currents.outbreaks.aa5372d90826e6cb0136ff66bb7a62fc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deng X., Phillippy A.M., Li Z., Salzberg S.L., Zhang W. Probing the pan-genome of Listeria monocytogenes: New insights into intraspecific niche expansion and genomic diversification. BMC Genomics. 2010;11:500. doi: 10.1186/1471-2164-11-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Den Bakker H.C., Cummings C.A., Ferreira V., Vatta P., Orsi R.H., Degoricija L., Barker M., Petrauskene O., Furtado M.R., Wiedmann M. Comparative genomics of the bacterial genus Listeria: Genome evolution is characterized by limited gene acquisition and limited gene loss. BMC Genomics. 2010;11:688. doi: 10.1186/1471-2164-11-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dunn K.A., Bielawski J.P., Ward T.J., Urquhart C., Gu H. Reconciling ecological and genomic divergence among lineages of Listeria under an “extended mosaic genome concept”. Mol. Biol. Evol. 2009;26:2605–2615. doi: 10.1093/molbev/msp176. [DOI] [PubMed] [Google Scholar]
- 48.Kuenne C., Billion A., Mraheil M.A., Strittmatter A., Daniel R., Goesmann A., Barbuddhe S., Hain T., Chakraborty T. Reassessment of the Listeria monocytogenes pan-genome reveals dynamic integration hotspots and mobile genetic elements as major components of the accessory genome. BMC Genomics. 2013;14:47. doi: 10.1186/1471-2164-14-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moura A., Tourdjman M., Leclercq A., Hamelin E., Laurent E., Fredriksen N., van Cauteren D., Bracq-Dieye H., Thouvenot P., Vales G., et al . Real-time whole-genome sequencing for surveillance of Listeria monocytogenes, France. Emerg. Infect. Dis. 2017;23:1462–1470. doi: 10.3201/eid2309.170336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cantinelli T., Chenal-Francisque V., Diancourt L., Frezal L., Leclercq A., Wirth T., Lecuit M., Brisse S. “Epidemic clones” of Listeria monocytogenes are widespread and ancient clonal groups. J. Clin. Microbiol. 2013;51:3770–3779. doi: 10.1128/JCM.01874-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Knudsen G.M., Nielsen J.B., Marvig R.L., Ng Y., Worning P., Westh H., Gram L. Genome-wide-analyses of Listeria monocytogenes from food-processing plants reveal clonal diversity and date the emergence of persisting sequence types. Environ. Microbiol. Rep. 2017;9:428–440. doi: 10.1111/1758-2229.12552. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.