SUMMARY
To identify novel genes associated with ALS, we undertook two lines of investigation. We carried out a genome-wide association study comparing 20,806 ALS cases and 59,804 controls. Independently, we performed a rare variant burden analysis comparing 1,138 index familial ALS cases and 19,494 controls. Through both approaches, we identified kinesin family member 5A (KIF5A) as a novel gene associated with ALS. Interestingly, mutations predominantly in the N-terminal motor domain of KIF5A are causative for two neurodegenerative diseases, hereditary spastic paraplegia (SPG10) and Charcot-Marie-Tooth Type 2 (CMT2). In contrast, ALS associated mutations are primarily located at the C-terminal cargo-binding tail domain and patients harboring loss of function mutations displayed an extended survival relative to typical ALS cases. Taken together, these results broaden the phenotype spectrum resulting from mutations in KIF5A and strengthen the role of cytoskeletal defects in the pathogenesis of ALS.
eTOC
Using large-scale genome-wide association study and exome sequencing, we identified KIF5A as a novel gene associated with ALS. Our data broaden the phenotype resulting from mutations in KIF5A and highlight the importance of cytoskeletal defects in the pathogenesis of ALS.
INTRODUCTION
Amyotrophic lateral sclerosis (ALS, OMIM #105400) is a neurodegenerative disorder clinically characterized by rapidly progressive muscle weakness and death due to respiratory failure, typically within two to four years of symptom onset (van Es et al., 2017). Although ALS is perceived as being rare, approximately 6,000 Americans die annually from the condition (Hirtz et al., 2007). Furthermore, the number of ALS cases across the globe will increase to nearly 400,000 in 2040, predominantly due to aging of the population (Arthur et al., 2016). This increase is anticipated to place an enormous socioeconomic burden on global healthcare systems, in particular because the annual healthcare cost per patient with ALS is among the highest for any neurological disease (Gladman and Zinman, 2015).
Approximately 10% of ALS display a family history (FALS) whereas the remaining 90% of ALS cases are sporadic (SALS) in nature. Driven in large part by advances in genotyping and sequencing technology, the genetic etiology of two-thirds of familial cases and about 10% of sporadic ALS cases is now known (Chia et al., 2018; Renton et al., 2014). Mutations in SOD1 were the first identified cause of ALS (Rosen et al., 1993) contributing to ~20% of FALS and ~2% of SALS. More recently, pathogenic hexanucleotide repeat expansions located within the first intron of the C9orf72 gene on chromosome 9p21 were identified as the most common cause of both FALS (~40%) and SALS (~7%) (DeJesus-Hernandez et al., 2011; Renton et al., 2011). Interestingly, this repeat expansion contributes to ~10% of all FTD cases thus genetically explaining much of the overlap between these clinical syndromes (Majounie et al., 2012). As a result of these major discoveries, there are several ongoing efforts towards directed silencing of these mutant genes which could result in a therapeutic treatment for up to 10% of all ALS cases and for a similar portion of FTD cases.
In addition to the insights provided by each novel ALS gene, the collective knowledge gained from genetic factors provides a more comprehensive understanding of the interacting pathways underlying motor neuron degeneration. For example, the identification of ALS genes has revealed at least three pathways believed to contribute to the development of ALS: (1) RNA metabolism (based on the observation of mutations in C9orf72, TDP-43, FUS, HNRNPA1, and MATR3); (2) protein homeostasis (UBQLN2, VCP, OPTN, VAPB); (3) cytoskeletal dynamics (PFN1, TUBA4A, DCTN1) (Chia et al., 2018; Robberecht and Eykens, 2015; Taylor et al., 2016). Understanding the mechanisms leading to disease pathogenesis again provides targets for therapeutic intervention that may be applicable to all forms of ALS.
Due to the decreased accessibility of multiple affected family members with unknown genetic etiology, there has been an increased focus on the identification of ALS associated genes with moderate to low impact. Despite their lower effect, such genes continue to provide valuable insight into ALS pathogenesis. For example, the product of the risk factor TBK1 is known to interact with the product of ALS associated gene OPTN, further solidifying the role of autophagy and protein homeostasis in disease development (Cirulli et al., 2015; Freischmidt et al., 2015; Maruyama et al., 2010; Morton et al., 2008). Similarly, the risk factor NEK1, identified through a rare variant burden analysis of index FALS (i.e., one affected sample per family), is a known binding partner of C21orf2, an ALS risk factor found through genome-wide association studies (GWAS) (Cirulli et al., 2015; Kenna et al., 2016; Malovannaya et al., 2011; van Rheenen et al., 2016). The interaction of these two proteins is required for efficient DNA damage repair (Fang et al., 2015), a pathway which is becoming increasingly implicated as a contributing factor in ALS and other neurodegenerative diseases (Coppedè and Migliore, 2015; Lopez-Gonzalez et al., 2016; Madabhushi et al., 2014; Wang et al., 2013).
RESULTS
Genome-wide Association Studies Identify KIF5A as a Novel ALS Associated Gene
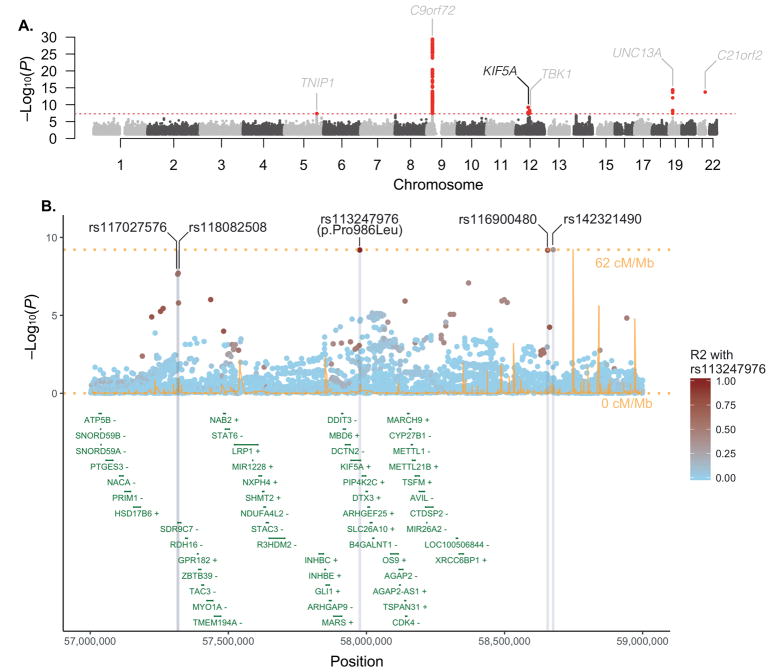
To identify new susceptibility loci operating in ALS, we undertook a large-scale GWAS involving 12,663 patients diagnosed with ALS and 53,439 control subjects (Table S1, S2). Our data were then incorporated into a meta-analysis with a recently published GWAS involving 12,577 ALS cases and 23,475 control subjects (van Rheenen et al., 2016). After imputation and quality-control measures (see Methods, Figure S1 for the workflow and Figure S2 for the multidimensional scaling plot), 10,031,630 genotyped and imputed variants from 20,806 ALS cases and 59,804 control samples were available for association analysis (Figure 1A). Quantile-quantile plots did not show evidence of significant population stratification (λ1000 = 1.001, Figure S3). Single nucleotide polymorphisms (SNPs) achieving genome-wide significance (P < 5.0×10−8) are listed in Table 1, Table S3 and suggestive loci with SNPs associated at P < 5.0×10−7 are listed in Table S4.
Figure 1. Identification of association between KIF5A locus and ALS risk through GWAS.
(A) Manhattan plot showing P values from the discovery set GWAS. Analysis of a combined set of 20,806 cases and 59,804 controls is shown. The dashed red line denotes the threshold for genome-wide significance after multiple test correction (P < 5.0×10−8). Five previously reported ALS associated loci are labeled in grey and one novel loci, containing the KIF5A gene, is labeled in black. (B) Regional association plot of the KIF5A locus. Recombination rates are from HapMap phase 2 European ancestry samples. The R2 pattern is based on the rs113247976 SNP using 85 European ancestry samples (CEU) from the November 2010 release of the 1000 Genomes Project dataset. R2 of the p.Pro986Leu (rs113247976) with additional SNPs achieving genome-wide significance was 0.544 (rs117027576), 0.544 (rs118082508), 0.741 (rs116900480), and 0.347 (rs142321490).
Table 1. SNPs achieving genome-wide significance in the discovery GWAS.
Position is based on Human Genome Assembly build 37. Nearest gene or previously published gene names are included. Chr, chromosome; MAF, minor allele frequency; OR, odds ratio; 95% CI, confidence interval;
| SNP Information | Present Study (8,229 Cases/36,329 Controls) | Van Rheenen et al. (12,577 Cases/23,475 Controls) | Combined Discovery Set (20,806 Cases/59,804 Controls) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||
| SNP | Chr | Position | Gene | Case MAF | Control MAF | OR [95% CI] | P | Case MAF | Control MAF | OR [95% CI] | P | Case MAF | Control MAF | OR [95% CI] | P |
| Novel Loci | |||||||||||||||
| rs117027576 | 12 | 57,316,603 | KIF5A | 1.55% | 1.27% | 1.45 [1.20–1.76] | 1.1×10−4 | 1.98% | 1.59% | 1.33 [1.16–1.53] | 4.3×10−5 | 1.81% | 1.40% | 1.37 [1.23–1.54] | 2.3×10−8 |
| rs118082508 | 12 | 57,318,819 | KIF5A | 1.56% | 1.28% | 1.45 [1.20–1.76] | 1.0×10−4 | 1.98% | 1.60% | 1.33 [1.16–1.53] | 3.8×10−5 | 1.81% | 1.41% | 1.37 [1.23–1.54] | 2.0×10−8 |
| rs113247976* | 12 | 57,975,700 | KIF5A | 1.83% | 1.42% | 1.46 [1.23–1.74] | 9.2×10−6 | 2.14% | 1.70% | 1.33 [1.17–1.52] | 1.1×10−5 | 2.02% | 1.53% | 1.38 [1.24–1.53] | 6.4×10−10 |
| rs116900480 | 12 | 58,656,105 | KIF5A | 1.75% | 1.46% | 1.42 [1.21–1.68] | 1.9×10−5 | 2.08% | 1.66% | 1.34 [1.18–1.53] | 7.1×10−6 | 1.95% | 1.54% | 1.37 [1.24–1.52] | 6.6×10−10 |
| rs142321490 | 12 | 58,676,132 | KIF5A | 1.79% | 1.48% | 1.43 [1.21–1.68] | 1.5×10−5 | 2.08% | 1.66% | 1.34 [1.18–1.53] | 8.0×10−6 | 1.97% | 1.55% | 1.37 [1.24–1.52] | 6.1×10−10 |
| Previously Published Loci | |||||||||||||||
| rs10463311 | 5 | 150,410,835 | TNIP1 | 73.19% | 74.84% | 0.94 [0.89–0.98] | 7.8×10−3 | 73.34% | 75.79% | 0.91 [0.87–0.94] | 8.5×10−7 | 73.28% | 75.21% | 0.92 [0.89–0.95] | 4.0×10−8 |
| rs3849943 | 9 | 27,543,382 | C9orf72 | 71.79% | 76.31% | 0.84 [0.80–0.88] | 1.4×10−12 | 72.78% | 76.5% | 0.83 [0.80–0.87] | 4.0×10−19 | 72.39% | 76.38% | 0.84 [0.81–0.86] | 3.8×10−30 |
| rs74654358 | 12 | 64,881,967 | TBK1 | 3.77% | 4.01% | 1.20 [1.07–1.34] | 1.6×10−3 | 5.12% | 4.61% | 1.23 [1.13–1.34] | 7.7×10−7 | 4.59% | 4.25% | 1.22 [1.14–1.30] | 4.7×10−9 |
| rs12973192 | 19 | 17,753,239 | UNC13A | 67.62% | 69.37% | 0.86 [0.82–0.91] | 1.3×10−8 | 64.52% | 66.00% | 0.9 [0.87–0.93] | 2.4×10−8 | 65.75% | 68.05% | 0.89 [0.86–0.91] | 3.9×10−15 |
| rs75087725 | 21 | 45,753,117 | C21orf2 | 0.70% | 0.46% | 1.99 [1.44–2.75] | 2.2×10−5 | 1.83% | 1.27% | 1.61 [1.39–1.87] | 8.7×10−11 | 1.38% | 0.78% | 1.67 [1.46–1.91] | 1.8×10−14 |
rs113247976 represents the p.Pro986Leu variant in KIF5A (NM_004984.2).
Our analysis revealed five previously identified loci that achieved genome-wide significance (loci including TNIP1, C9orf72, TBK1, UNC13A, C21orf2) (Benyamin et al., 2017; Laaksovirta et al., 2010; Shatunov et al., 2010; van Es et al., 2009; van Rheenen et al., 2016). In addition, we observed a strong association signal for five SNPs in linkage disequilibrium on chromosome 12q14.1 that reached genome-wide statistical significance (Table 1, Figure 1B) spanning a region of several hundred kilobases. Of the five SNPs, two of them resided in close proximity to each other within a large intergenic region and two in proximity to short-chain dehydrogenase/reductase family 9C member 7 (SDR9C7), a gene expressed primarily in skin. However, one SNP (rs113247976) results in a p.Pro986Leu coding change within the kinesin family member 5A (KIF5A) gene (P = 6.4×10−10, OR = 1.38, 95% CI = 1.24–1.53). The case:control allele frequencies for the combined discovery cohort were 2.07%:1.55% and genotype counts were 5: 529: 12,043 to 7: 786: 22,682 (homozygotes alternative allele: heterozygotes: homozygous reference allele, Figure 2). Calculations based on our cohort size as well as the OR and allele frequency of rs113247976 result in a ~99.5% power to detect this as an ALS associated SNP.
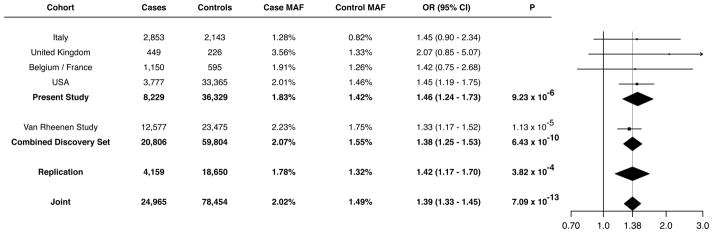
Figure 2. Discovery and replication for the association of the KIF5A p.Pro986Leu (rs113247976) variant with ALS.
Analysis of the p.Pro986Leu (rs113247976) variant within each of the described cohorts is shown. Allelic association for all subcohorts were analyzed by logistic regression followed by a fixed-effects meta-analysis. The Forest plot (right) displays the distribution of OR estimates across study cohorts with the vertical dotted line denoting the OR estimated under the meta-analysis.
Rare Variant Burden Analysis Identifies KIF5A as an ALS gene
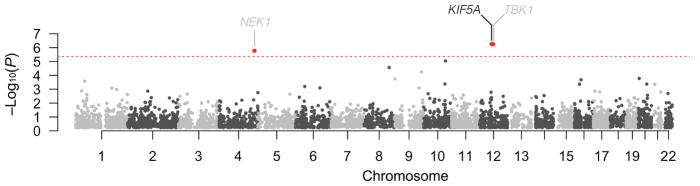
In an independent line of investigation, we attempted to identify novel ALS genes through exome-wide rare variant burden analysis (RVB). In brief, RVB compares the frequency of variants within each gene below a user defined frequency threshold in a case-control cohort. As the last two ALS associated genes identified by this methodology (TBK1, NEK1) displayed an increased frequency of loss of function (LOF) variants, we focused our initial analysis on such variants (consisting of nonsense and predicted splice-altering) (Cirulli et al., 2015; Freischmidt et al., 2015; Kenna et al., 2016).
Towards this end, we performed RVB testing for association of LOF variants in a cohort of 1,138 index FALS cases and 19,494 controls, after applying quality control filters (Experimental Methods, Figure S4, Table S5, S6). Genes displaying P < 5.0×10−4 are shown in Table 2. The previously identified ALS genes, TBK1 (P = 5.58×10−7, OR = 15.11, 95% CI = 5.81–38.69) and NEK1 (P = 1.68×10−6, OR = 6.64, 95% CI = 3.32–12.51), yielded strong associations with ALS reaching exome-wide significance (Figure 3). In addition, we observed a single novel gene reaching exome-wide significance, KIF5A (P = 5.55×10−7; OR = 32.07, 95% CI = 9.05–135.27). Within this gene, we observed 6 LOF variants in our 1,138 cases (0.53%) compared to 3 such variants in our comparison cohort of 19,494 controls (0.015%, Table 2). There was no evidence of genomic inflation (λ=0.93, Figure S5), sequencing center or other sub-cohort bias (Figure S6), or call rate bias (Figure S7) in our analysis. Of the index FALS cases carrying KIF5A LOF mutations, we obtained DNA from two siblings of the proband carrying a c.2993-3C>T, exon 27 - 5′ splice junction variant, and from a sibling of a different proband carrying a c.3020+2T>A, exon 27 - 3′ splice junction variant. These variants segregated with disease within each of these families. Sanger sequencing validated all identified LOF variant containing samples in the discovery set and affected relatives.
Table 2.
Top ALS associations identified through RVB of FALS and control exome sequencing results
| Gene | FALS | Control | OR | P |
|---|---|---|---|---|
| KIF5A | 6 (0.53%) | 3 (0.02%) | 32.07 (9.05–135.27) | 5.55×10−7 |
| TBK1 | 8 (0.70%) | 9 (0.05%) | 15.11 (5.81–38.69) | 5.58×10−7 |
| NEK1 | 12 (1.05%) | 32 (0.16%) | 6.64 (3.32–12.51) | 1.68×10−6 |
| CALHM2 | 7 (0.62%) | 9 (0.05%) | 12.13 (4.47–31.79) | 9.19×10−6 |
| COL14A1 | 8 (0.70%) | 16 (0.08%) | 8.04 (3.32–18.08) | 2.72×10−5 |
| AK1 | 10 (0.88%) | 34 (0.17%) | 5.37 (2.55–10.41) | 5.62×10−5 |
| ATRN | 5 (0.44%) | 9 (0.05%) | 11.06 (3.57–31.02) | 1.66×10−4 |
| VLDLR | 5 (0.44%) | 9 (0.05%) | 10.87 (3.51–30.43) | 1.79×10−4 |
| FUS | 4 (0.35%) | 4 (0.02%) | 16.53 (4.25–64.33) | 2.08×10−4 |
| ZMYND12 | 6 (0.53%) | 12 (0.06%) | 7.92 (2.86–19.96) | 2.61×10−4 |
Figure 3. Identification of association between KIF5A and ALS risk through rare variant burden analysis of exome sequencing.
Manhattan plot showing gene-level P values from an exome-wide rare variant burden analysis. Analyses of 1,138 index FALS cases versus 19,494 controls were restricted to rare LOF variants (splice altering/nonsense, MAF < 0.001). A minimum of 3 LOF gene variants were required for analysis. The dashed red line denotes the threshold for exome-wide significance after correction for 11,472 genes (4.36×10−6). Previously reported (grey) and novel (black) genes exhibiting a significant excess of rare LOF variants in patients are shown.
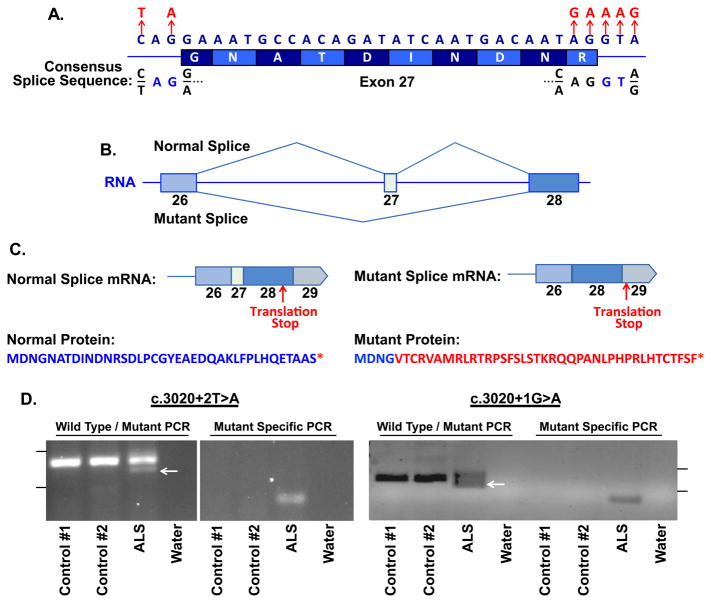
Interestingly, when we investigated the location of the six ALS associated variants present in KIF5A, all occurred within a 34 bp stretch of DNA and were predicted to effect splicing of exon 27 that encodes amino acids 998–1007 (Table 3, Figure 4A). Five of the six variants were located on sequential base pairs on the 3′ end of the exon, whereas one was located 5′ end of the exon. We used the application ASSEDA (Automated Splice Site and Exon Definition Analyses) to predict any mutant mRNA splice isoforms resulting from these variants (Tompson et al., 2007). This algorithm was chosen as it is known to have high performance in splice prediction (Caminsky et al., 2014). ASSEDA predicted a complete skipping of exon 27 for all variants, yielding a transcript with a frameshift at coding amino acid 998, the deletion of the normal C-terminal 34 amino acids of the cargo-binding domain, and the extension of an aberrant 39 amino acids to the C-terminus (Table 3, Figure 4B, 4C). The presence of transcripts with skipped exon 27 was demonstrated by performing RT-PCR in two patients carrying exon 27 - 3′ splice junction variants (c.3020+2T>A and c.3020+1G>A) using RNA from lymphoblasts and peripheral blood mononuclear cells, respectively. This splice form was not detected in four control lines (Figure 4D). Sequence analysis of the smaller RT-PCR products obtained from the patient cells confirmed the exon 26–28 splicing event. Material for RT-PCR was not available for any other patient carrying a KIF5A LOF variant.
Table 3. Loss of function variants within KIF5A identified in probands.
P, possible; Y, yes; N, no; M, male; F, female; L, limb onset; B, bulbar onset, n/a, not available or applicable. Note, ASSEDA does not predict exon skipping based on frameshifts or nonsense mutations (Tompson et al., 2007).
| Position | Variant | Exon | cDNA | Description | Predicted Exon Skipping | Gender | Age of Onset (years) | Site of Onset | Survival (months) | Alive (yes/no) |
|---|---|---|---|---|---|---|---|---|---|---|
| Control Variants | ||||||||||
| 57,963,470 | A>G | 11 | c.1117+4A>G | 3′ Splice Junction | P | M | n/a | n/a | n/a | n/a |
| 57,966,423 | C>T | 15 | c.1630C>T | p.Arg544* | - | F | n/a | n/a | n/a | n/a |
| 57,976,884 | G>C | 28 | c.3021G>C | 5′ Splice Junction | N | F | n/a | n/a | n/a | n/a |
| FALS Variants | ||||||||||
| 57,975,729 | GA>A | 26 | c.2987delA | p.Asp996fs | - | M | 45 | n/a | n/a | n/a |
| 57,976,382 | C>T | 27 | c.2993-3C>T | 5′ Splice Junction | Y | M | 29 | L | >264 | Y |
| 57,976,385 | GA>G | 27 | c.2996delA | p.Asn999fs | - | M | 42 | L | >12 | Y |
| 57,976,411 | A>G | 27 | c.3019A>G | p.Arg1007Gly | Y | F | 53 | L | 45 | N |
| 57,976,412 | G>A | 27 | c.3020G>A | p.Arg1007Lys | Y | M | 50 | L | >108 | Y |
| 57,976,412 | G>A | 27 | c.3020G>A | p.Arg1007Lys | Y | F | 50 | n/a | >240 | Y |
| 57,976,413 | G>A | 27 | c.3020+1G>A | 3′ Splice Junction | Y | M | 45 | B | >220 | Y |
| 57,976,414 | T>A | 27 | c.3020+2T>A | 3′ Splice Junction | Y | M | 46 | B | 124 | N |
| 57,976,415 | A>G | 27 | c.3020+3A>G | 3′ Splice Junction | Y | M | 50 | B | 54 | N |
| SALS Variants | ||||||||||
| 57,957,481 | G>A | 3 | c.291+5G>A | 3′ Splice Junction | N | n/a | n/a | n/a | n/a | n/a |
| 57,975,731 | CA>C | 26 | c.2989delA | p.Asn997fs | - | F | 50 | L | >96 | Y |
| 57,976,384 | G>A | 27 | c.2993-1G>A | 5′ Splice Junction | Y | n/a | 52 | B | n/a | n/a |
Figure 4. ALS associated loss of function variants of KIF5A disrupt C-terminal sequence by inducing skipping of exon 27.
(A) Single nucleotide variants (SNVs) within KIF5A identified in ALS patients are clustered at the 5′ and 3′ splice junctions of exon 27. The consensus splice sequence is shown. (B) ALS associated SNVs are predicted to induce skipping of exon 27 and result in an aberrant mRNA transcript. (C) The skipping of exon 27 of KIF5A yields an out-of-frame and extended disrupted C-terminal peptide sequence. The amino acids in red signify the divergence from the normal protein. (D) RT-PCR was performed using RNA derived from ALS patients with the indicated LOF variant or without (controls) using primers to either amplify both wild-type (155 bp) and mutant (127 bp) splice forms or specifically the mutant splice form (80 bp, right panel). The arrow represents the position of the mutant specific product. The tick marks represent 200 bp (upper) and 100 bp (lower) markers.
Our initial RVB was restricted to single nucleotide variants due to the limited sensitivity and comparatively high false positive rates associated with identifying small insertions and deletions (indels) within exome sequencing data (Fang et al., 2014). Based on our discovery of increased LOF variants within KIF5A, we re-evaluated this region for the presence of indels. Our analysis revealed two (0.026%) indels within our cohort of 1,138 FALS cases, compared to zero (0%) indels among 19,494 control samples. Both of these indels (p.Asp996fs, p.Asn999fs) resulted in a frameshift of the KIF5A protein coding sequence, and were located close to the splice junction variants that we previously observed to cause skipping of exon 27 resulting in a frameshift at amino acid 998 (Table 3). Sanger sequencing confirmed the presence of both indels. Combining the results of the single nucleotide and indel variant analysis yielded a highly significant P of 3.8×10−9 (OR = 41.16, 95% CI = 12.61–167.57). We failed to detect any signals of RVB association for rare missense variants across KIF5A or within any sub-domain of the gene (Table S8).
Replication Analysis of rs113247976 and LOF Variants in KIF5A
Given the strong signal of the missense variant identified by our GWAS (p.Pro986Leu, rs113247976) and its close proximity to the LOF variants identified by our RVB analysis (amino acids 996–999), we attempted to replicate its association with ALS by analyzing additional cohorts. To accomplish this, we evaluated this variant in a cohort of 4,159 ALS cases and 18,650 controls that were non-overlapping with our GWAS discovery analysis (Methods, Figure S8). This included non-overlapping samples from our RVB analysis (673 FALS, 17,696 controls). Analysis of the cohort revealed an allele frequency of 1.78% in cases and 1.32% in controls (rs113247976, P = 3.82×10−4, OR = 1.42, 95% CI = 1.17–1.70), thereby replicating the association of the original GWAS. A meta-analysis of the GWAS and replication cohort (n = 24,965 cases, 78,454 controls) yielded a highly significant P of 7.09×10−13 (OR = 1.39, 95% CI = 1.33–1.45, Figure 2). These results support the association of KIF5A p.Pro986Leu with ALS. However, at this point we cannot definitely state that the missense variant is the primary risk factor, as we cannot rule out other variants in linkage disequilibrium.
We next performed mutational screening of KIF5A in an additional cohort of 9,046 ALS cases that had not been included in our original RVB analysis. This revealed three additional carriers of C-terminal variants. One sporadic patient harbored an exon 26 frameshift mutation (p.Asn997fs) and a second sporadic patient harbored an exon 27 splice altering mutation (c.2993-1G>A, Table 3). The third patient carried a p.Arg1007Lys (c.3020G>A) mutation and had a familial history of ALS. This mutation was also observed in a FALS patients from our RVB analysis, however, a comparison of 240,715 common variant sites between the two patients failed to reveal a familial relationship (Experimental Methods). Additionally, one patient was observed to carry a predicted splice altering variant proximal to exon 3 (c.291+5G>A). However, this variant was not supported as creating an aberrant transcript by ASSEDA. The cohort used for this analysis was comprised mainly frequency of sporadic ALS cases. LOF variants were not observed in a follow up panel of 1,955 controls. Comparison of the LOF variants in sporadic patients (2/9,046 cases, 0.022%) with either the 1,955 replication controls or all controls analyzed in this study (21,449 controls) both yielded insignificant P values (0.868 and 0.423, respectively). Interestingly, the frequency of LOF variants in sporadic cases is lower than that observed in our original FALS cohort (0.703%), suggesting that KIF5A LOF variants display a high penetrance. Furthermore, the rate of LOF variants reported in the Exome Aggregation Consortium (ExAC) database is lower than we observed in the control samples used in our discovery cohort (0.007% versus 0.015%).
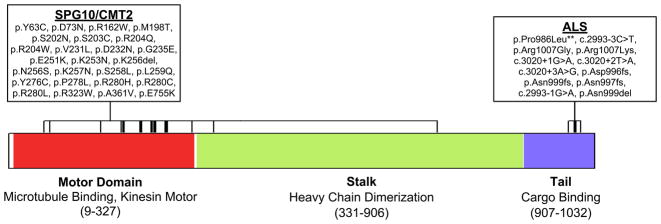
ALS-Associated Mutations in KIF5A are Distinct from SPG10/CMT2 Mutations
Missense mutations within KIF5A are a known cause of hereditary spastic paraparesis (spastic paraplegia type 10, autosomal dominant; OMIM #604187) and of Charcot-Marie-Tooth disease Type 2 (CMT2) (Crimella et al., 2011; Jennings et al., 2017; Liu et al., 2014; Reid et al., 2002). Although SPG10 and CMT2 share clinical features with ALS, a careful examination of the clinical records of the ALS cases with LOF mutations in KIF5A ruled out misdiagnosis. Furthermore, we detected no variants previously associated with SPG10 or CMT2 in our FALS cohort (Liu et al., 2014).
To further elucidate genotype-phenotype relationships, we evaluated the location of mutations within KIF5A. Interestingly, mutations contributing to SPG10 and to CMT2 are almost exclusively to be missense changes and are located in the N-terminal motor domain (amino acids 9–327) of KIF5A (Figure 5). In contrast, the mutations identified as contributing to ALS are found predominantly in the C-terminal cargo binding region of KIF5A (amino acids 907–1032) with the highly penetrant FALS mutations showing LOF. These results indicate that the functional domain mutated in KIF5A dictates the clinical phenotype, resulting in distinct yet overlapping neurodegenerative diseases.
Figure 5. KIF5A ALS mutations show distinct localization from missense mutations previously associated with SPG10 and CMT2.
Causative mutations for SPG10 and CMT2 described within the literature (Crimella et al., 2011; Jennings et al., 2017; Liu et al., 2014; Reid et al., 2002) and ALS associated mutations identified within this study are shown. As illustrated, mutations causative for SPG10/CMT2 are predominantly missense changes located in the N-terminal motor domain. In contrast, ALS mutations are primarily located at the C-terminal motor domain and are LOF.
Patients with KIF5A LOF Mutations Display Younger Age at Onset and Longer Survival
To establish the existence of any commonalities between patients with LOF mutations in the C-terminal region of KIF5A, we evaluated their clinical phenotype. Cases with LOF mutations exhibited a median age of onset at 46.5 years (n = 19, Table S7). This is lower than the age of onset reported for ALS in epidemiological studies (65.2 years, interquartile range 56.0 – 72.2) (ALSGEN Consortium et al., 2013). Interestingly, we also observed an increased disease duration (survival) in patients harboring these LOF mutations. The median survival time of ALS patients is 20 – 36 months (ALSGEN Consortium et al., 2013). In contrast, cases with LOF mutations exhibited a median survival of nearly 10 years (117 months, n = 17, Table S7). ALS patients with symptom onset before 40 years of age have been shown to have longer survival, often exceeding 10 years (Chio et al., 2009). In contrast, patients with uncomplicated types of hereditary spastic paraparesis and CMT2 display a normal life expectancy (Patzkó and Shy, 2011).
DISCUSSION
We previously identified KIF5A as a candidate gene for ALS in our prior study that lacked the power to draw a definitive conclusion (Kenna et al., 2016). KIF5A was also a candidate ALS gene in a previous GWAS, though it similarly failed to reach genome-wide significance (McLaughlin et al., 2017; van Rheenen et al., 2016) as well as a single gene study selected based on the a priori knowledge of its role in HSP/CMT2 and cytoskeletal function (Brenner et al., 2018). Here, we have confirmed KIF5A as an ALS-associated gene with genome-wide significance through two independent approaches. By performing a GWAS involving ~80,000 samples, in addition to replicating five previously published loci, as well as the previously reported locus SCFD1 using a linear mixed model analysis (data not shown), we identified a missense variant within the KIF5A gene that reached genome-wide significance for association with ALS risk. It should be stated though that, as with all GWAS, we cannot rule out that other variants in linkage disequilibrium represent the primary risk factor. In an independent line of investigation, we applied RVB analysis to exome sequencing of ~21,000 samples and identified an exome-wide significant association between FALS risk and rare KIF5A LOF variants. Analyses of KIF5A in independent replication cohorts confirmed our initial finding for both the p.Pro986Leu variant and revealed three additional carriers of LOF variants in 9,046 ALS cases. Taken together our results indicate that the p.Pro986Leu KIF5A variant may represent a relatively common, but low penetrance risk allele for ALS, while LOF variants constitute rare, but high penetrance risk factors.
Kinesins are microtubule-based motor proteins involved in intracellular transport of organelles within eukaryotic cells. In mammals, there are three heavy chain isoforms of KIF5: KIF5A, KIF5B and KIF5C (Miki et al., 2001). The three proteins homo- and heterodimerize through their coiled-coiled stalk domain, and create a complex with two kinesin light chains via binding to the tail domain (Hirokawa et al., 1989). All three KIF5 genes are expressed in neurons (Kanai et al., 2000) and function to transport many cargos by binding to distinct adaptor proteins.
The central role of kinesins in axonal transport leads us to speculate that mutations in KIF5A cause disease by disrupting axonal transport. Indeed, defects in axonal transport are a common observation in ALS patients and are already known to directly contribute to motor neuron degeneration pathogenesis (Chevalier-Larsen and Holzbaur, 2006; Hirokawa et al., 2010; Millecamps and Julien, 2013). KIF5 mediates the transport of granules containing both RNA and RNA binding proteins within neuronal dendrites and axons (Kanai et al., 2004). Among these cargos are the ALS-associated proteins FUS and hnRNPA1 (Guo et al., 2017; Kim et al., 2013; Kwiatkowski et al., 2009; Vance et al., 2009). Similarly, KIF5 mediates the transport of VAPB through the adaptor protein protrudin (Matsuzaki et al., 2011), and mutations in the VAPB gene have been identified in ALS and late-onset spinal muscular atrophy (Nishimura et al., 2005; 2004). KIF5 is responsible for the axonal transport of neurofilaments (Wang and Brown, 2010) and KIF5A knockout mice display abnormal transport of neurofilaments (Xia et al., 2003). Abnormal accumulation of neurofilaments are a pathological hallmark of ALS and rare mutations in neurofilament heavy polypeptide (NEFH) are associated with ALS (Al-Chalabi et al., 1999).
KIF5 also contributes to the transport of mitochondria (Kanai et al., 2000; Tanaka et al., 1998) and motor neurons derived from KIF5A−/− mice display transport deficits and reduced survival (Karle et al., 2012). Impaired transport and dysfunction of mitochondria represent another common hallmark observed in ALS patients (Chevalier-Larsen and Holzbaur, 2006; Guo et al., 2017; Palomo and Manfredi, 2015; Smith et al., 2017). KIF5 also contributes to the transport of AMPA-type (Heisler et al., 2014; Setou et al., 2002) and GABAA receptors (Nakajima et al., 2012). In keeping with reported ALS genes such as NEK1 (Thiel et al., 2011) and PFN1 (Wu et al., 2012), modulation of KIF5A expression has been shown to influence the formation of neurite like membrane protrusions (Matsuzaki et al., 2011). Given its critical interactions with the cytoskeleton, the identification of KIF5A mutations further extends the list of cytoskeletal related proteins implicated in ALS pathogenesis, such as PFN1, TUBA4A, NEFH and peripherin (Al-Chalabi et al., 1999; Gros-Louis, 2004; Smith et al., 2014; Wu et al., 2012).
An important question raised by the current study is why variation within the C-terminal cargo binding domain is associated with ALS, while missense variations of the N-terminal motor domain are associated with hereditary spastic paraparesis and Charcot-Marie-Tooth, type 2. Missense mutations within this latter domain have been shown to affect microtubule binding and/or ATP hydrolysis, resulting in a defective KIF5A-mediated anterograde transport of cargo along dendrites and axons. This, in turn, leads to the axonal retrograde degeneration observed both in hereditary spastic paraparesis and Charcot-Marie-Tooth, type 2, two length-dependent axonopathies (Ebbing et al., 2008). In contrast, the primary cellular lesion in ALS is believed to occur within motor neuron cell bodies, where cytoplasmic protein aggregates are consistently observed, and to propagate anterograde along neurites. We anticipate that LOF variants within the C-terminal domain of KIF5A will disrupt binding with specific cargo proteins. This is supported by a study in zebrafish in which truncation of the C-terminal resulted in a dramatic disruption of axonal localization of mitochondria (Campbell et al., 2014). One possible mechanism is that disruption of binding to cargo may possibly lead to their accumulation and seed aggregation within the cell body resulting in a deficiency at neurite terminals. Deficiency in KIF5A expression and cargo binding has been associated with accumulation of phosphorylated neurofilaments and amyloid precursor protein within neuronal cell bodies, and subsequent neurodegeneration, in patients with multiple sclerosis (Hares et al., 2016). While differences in KIF5A kinetics and KIF5A interactions constitute one possibility to explain the phenotypic heterogeneity, it is also possible C-terminal and N-terminal variants act through a common mechanism, but that a difference in the relative extent of loss or gain of function toxicities leads to milder (i.e. hereditary spastic paraplegia, Charcot-Marie-Tooth, type 2) or more severe (i.e. ALS) phenotypes.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, John Landers (john.landers@umassmed.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Study cohorts
GWAS cohort I
We undertook a GWAS of patients diagnosed with ALS (case cohort) and neurologically normal control individuals (control cohort). DNA was extracted from either whole blood or frozen brain tissue samples using standard procedures. All 12,663 patients included in the case cohort had been diagnosed with ALS according to the El Escorial criteria (Brooks, 1994) by a neurologist specializing in ALS, had onset of symptoms after age 18 years, and were of non-Hispanic white race/ethnicity. Both patients with familial ALS and patients with sporadic ALS were included in the analysis.
For the control cohort, we used genotype data obtained from (a) the database of Genotypes and Phenotypes (dbGaP) web repository (n = 44,017 US samples); (b) the HYPERGENES Project (n = 887 Italian samples) (Salvi et al., 2012); and (c) the Wellcome Trust Case Control Consortium (n = 5,663 British samples). An additional 2,112 US and Italian control samples were genotyped in the Laboratory of Neurogenetics, National Institute on Aging. The control cohort was matched to the case cohort for race and ethnicity, but not for age or sex. A detailed description of the cohorts is available in Table S1, S2.
Written consent was obtained from all individuals enrolled in this study, and the study was approved by the institutional review board approval of the National Institute on Aging (protocol number 03-AG-N329).
GWAS cohort II
Summary statistics from a recently published GWAS based on logistic regression analysis involving 12,577 cases and 23,475 controls were downloaded from the Project MinE Variant Browser. Additional details of the cohorts used in this study are available in van Rheenen et al (van Rheenen et al., 2016).
FALS discovery cohort
A total of 1,463 FALS patients were included in the initial cohort (pre-QC). Patients were recruited at specialist clinics in Australia (n = 92), Belgium (n = 13), Canada (n = 34), Germany (n = 228), Ireland (n = 18), Israel (n = 26), Italy (n = 230), Netherlands (n = 50), Spain (n = 60), Turkey (n = 72), UK (n = 223), and USA (n = 417). All samples were exome sequenced except those from the Netherlands which were whole genome sequenced. Familial history was considered positive for ALS if the proband had at least one affected relative within three degrees of relatedness.
Control discovery cohort
Read level sequencing data were obtained from dbGAP and the European Genome-Phenome Archive (EGA) and are listed in Table S5.
ALS WXS/WGS replication cohort
Replication analyses included sequencing data for a further 9,046 ALS cases and 1,955 non-ALS controls that were not also represented in the FALS discovery set. These samples included 2,742 cases subjected to WXS by the ALS Sequencing Consortium, as described previously (Cirulli et al., 2015); 719 cases subjected to WXS by the Laboratory of Neurogenetics, National Institute on Aging; 307 cases and 296 controls subjected to WGS by the Laboratory of Neurogenetics, National Institute on Aging; 155 cases subjected to WGS by the CReATe Consortium; 1,017 cases subjected to WGS by the NYGC ALS Consortium, Genomic Translation for ALS Care (GTAC) Consortium and Answer ALS Foundation; and 4,100 cases and 1,659 controls subjected to WGS by the Project MinE Sequencing Consortium.
All samples included in the case cohort had been diagnosed with ALS according to the El Escorial criteria (Brooks, 1994) by a neurologist specializing in ALS. We received approval for this study from the institutional review boards of the participating centers, and written informed consent was obtained from all patients (consent for research).
METHOD DETAILS
Data generation and pre-processing
Generation of SNP array callset
The case cohort (n = 12,663 samples) and part of the control cohort (n = 2,112) were genotyped in the Laboratory of Neurogenetics, National Institute on Aging, using HumanOmniExpress BeadChips (version 1.0, Illumina Inc., San Diego, CA) according to the manufacturer’s protocol. These SNP genotyping arrays assay 716,503 SNPs across the genome. Individual-level genotypes for these samples are available on the dbGaP web portal (accession number phs000101.v4.p1). The remainder of the control cohort had been previously genotyped on HumanOmni BeadChips (Illumina) as part of other GWAS efforts (see Table S2). Analyses were confined to the 595,692 autosomal SNPs that were common across the SNP genotyping arrays.
Generation of FALS case-control callset for exome-wide RVB discovery analysis
Exome sequencing of cases was performed as previously described (Kenna et al., 2016). Control exome sequences were generated as described under the relevant dbGAP and EGA project accessions. Sequence reads were aligned to human reference GRCh37 using BWA (Burrows-Wheeler Aligner) and processed according to recommended Genome Analysis Toolkit’s (GATK) best practices (https://software.broadinstitute.org/gatk/best-practices/). Joint variant detection and genotyping of all samples were performed using the GATK HaplotypeCaller. Variant quality control was performed using the GATK variant quality score recalibration method with default filters. A minimum variant quality by depth (QD) score of 2 was also imposed and all genotypes associated with genotype quality (GQ)< 20 were reset to missing. Variants were also excluded in the event of case or control call rates <70% (post genotype QC). Identified variants can be viewed through our web based ALS Variant Server (see link below).
Generation of ALS case-control callset for KIF5A replication analysis
Data for the KIF5A locus was extracted from all independently generated sequencing datasets and remapped to GRCh37. Variant calling was performed using the GATK haplotype caller as described above. In addition to the KIF5A locus, data was also extracted for a panel of 240,715 common variant sites and used to perform a single unified sample QC as described below.
Functional annotation of variants identified by WXS/WGS
Variant calls were assigned predicted functional consequences using snpEFF (Single Nucleotide Polymorphism Effect)(Cingolani et al., 2012), dbNSFP (A Database of Human Non-synonymous SNVs and Their Functional Predictions and Annotations)(Liu et al., 2013) and dbscSNV (database of splice site consequences of Single Nucleotide Variants)(Jian et al., 2014), which is incorporated into dbNSFP. Variants were classified as “loss of function” (LOF) where the sequence change was predicted to encode a premature stop codon, a frameshift causing insertion-deletion or a splice site disrupting SNV. Variants were classified as potentially splice altering if assigned an “ada” or “rf” score >0.7 by dbscSNV. Splice variants of potential interest were further assessed for putative effects on exon skipping using a secondary algorithm - automated splice site and exon definition server (ASSEDA)(Tompson et al., 2007).
RT-PCR Analysis
Total RNA was prepared from lymphoblast lines using Trizol reagent. Reverse transcription using Applied Biosystems RNA to cDNA kit (# 4368814) was performed with 0.5 ug with RNAse inhibitor in a 20 ul reaction according to the manufacturer’s protocol. PCR was carried out using New England Biolabs One Taq Hot Start DNA Polymerase (# M0481S), 2 ul RT reaction (representing 50 ng input RNA) and forward and reverse primer (0.15 uM each) in a 20 ul reaction volume. Amplification conditions were as follows: 94°C for 30 seconds, {94°C for 20 seconds, 58°C for 20 seconds, 68°C for 1 minute} x 35 cycles, followed by an extension stage of 68°C for 5 minutes and a 4°C hold. Amplification of both normal and mutant splice forms used primers F1 (CAGTGGAGCCACATCTTCTG) and R1 (TCTCTTGGTGGAGAGGGAAA). Primers used for the specific amplification of the mutant splice form were F2 (CCAACATGGACAATGGAGTGA), which spans exons 26 and 28, and R1.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses
Analysis of SNP array genotypes
Standard quality-control procedures were applied to our genotype data using PLINK software package (version 1.9)(Chang et al., 2015), and a summary of the workflow is shown in Figure S1. We excluded samples that demonstrated: call rates of less than 97.5%; non-European ancestry; abnormal F inbreeding coefficient; mismatch between phenotypic and genotypic gender; or, cryptic relatedness defined as identity-by-descent proportion of inheritance (pi_hat from PLINK) greater than 0.125. Samples in common between our study and van Rheenen’s study were identified using the checksum program id_geno_checksum and were removed from our analyses. We excluded palindromic SNPs, as well as SNPs with: call rates less than 95% in the US and Italian cohorts or less than 99% in the UK, French and Belgium cohorts; minor allele frequency less than 0.05 in the control cohorts; Hardy-Weinberg equilibrium P less than 10−7 in the US and Italian control cohorts and less than 10−5 in the UK, French and Belgium cohorts; missingness by case-control status P less than 10−5; or SNPs associated between the UK and French control cohorts with P less than 5.0×10−8. After quality control, 8,229 case and 36,329 control samples were included in the analysis, and 436,746 SNPs were available for imputation in the USA and Italy cohorts, and 420,131 SNPs were available in the UK, French and Belgium cohorts.
Estimation of the haplotypes was performed with SHAPEIT (version 2.r790)(Delaneau et al., 2013). Imputation was performed for individual batches based on ethnicity using the 1000 Genomes Project dataset (phase 3, version 5a, release 2013-05-02, 1000genomes.org) as reference and using Minimac3 software (version 1.0.11)(Das et al., 2016) with default settings. After imputation, principal components were calculated using PLINK software after removing known hypervariable regions and the 1 MB surrounding the C9orf72 region. After analysis of the Scree plots, 2 to 4 principal components were retained per cohort as covariates in the association analyses to compensate for any residual population stratification.
Logistic regression was performed per batch using mach2dat software (version 1.0.24)(Marchini and Howie, 2010) incorporating 2 to 4 principal components, age and gender as covariates, with dosage of imputed SNPs selected based on a Minimac3 R2 value of imputation accuracy greater than 0.3. SNPs with an absolute beta coefficient value above 5 or with a minor allele frequency less than 0.01 were excluded from meta-analysis. Meta-analysis was then performed combining the association results of the 13 batches of our individual-level studies with van Rheenen’s study summary statistics using METAL software (version 2011-03-25)(Willer et al., 2010) under an inverse-weighted, fixed effect model. A threshold P of 5.0×10−8 was set for genome-wide significance after Bonferroni correction for multiple testing in the GWAS (Pe’er et al., 2008).
The programming code used to analyze these data is freely available on GitHub (see link below), and GWAS summary statistics results for all tested SNPs are available from (link to be supplied at publication).
Analysis of WXS/WGS genotypes
For both the discovery and replication phases, samples were excluded from the study in the event of failing to meet standard genotype call rate, heterozygosity, duplication, relatedness or population stratification filters as summarized in Table S6. Each of these filters was performed using a set of autosomal markers meeting all of the following criteria: call rate > 0.95, minor allele frequency (MAF) > 0.01, P > 0.001 for deviation from Hardy-Weinberg equilibrium, linkage disequilibrium pruning (R2 < 0.5, window size = 50, step = 5). Filtering of autosomal markers, sample call rate assessments and sample heterozygosity assessments were performed using PLINK software. Study duplicates and sample relatedness within the WXS/WGS cohorts was identified using KING software (Manichaikul et al., 2010). Study duplicates between WXS/WGS cohorts and GWAS datasets were identified using the checksum program id_geno_checksum. LASER was used to generate PCA coordinates for samples from the Human Genome Diversity Panel (HGDP). Samples from the FALS discovery cohort were then mapped to this reference co-ordinate space. The discovery cohort was restricted to cases and controls occurring within 3 standard deviations of the mean for European HGDP samples along principal components 1–4.
RVB analyses were performed by penalized logistic regression of case-control status with respect to number of minor alleles observed per sample per gene with and MAF <0.001. Analyses were only performed where the dataset contained more than 3 variant allele occurrences. Replication analyses of rs113247976 were performed using the same logistic regression protocol as used for RVB analyses. All analyses were conditioned on the first 4 eigenvectors generated by principal components analysis of common variant profiles. Genomic inflation factors were calculated using genome-wide association analysis for quantitative, binary and time-till-event traits using GenABEL software. Candidate associations were tested for signs of call-rate or subcohort biases as outlined in Figures S6, S7. Meta-analysis of rs113247976 association results between sequencing and GWAS was performed using METAL. Unless otherwise indicated, all statistical analyses were performed using R (version 3.2.0).
Control-control analyses
To identify genes potentially subject to confounding biases in FALS RVB analyses and to assess the potential impact of batch effects with non-ALS-related data, population or phenotypic stratifiers, the control sample cohort was divided into 28 pseudo case-control groups based on the sequencing center or associated dbGaP/EGA project (Table S5). Genes shown in gray achieve for possible confounder association. Loci achieving a minimu P < 1 × 10–3 were deemed as displaying possible association with non-ALS related batch effects.
DATA AND SOFTWARE AVAILABILITY
Datasets
The programming code used to analyze the GWAS data including the imputation with SHAPEIT and Minimac3, individual-based association analysis using Mach2dat and a meta-analysis using METAL is freely available on GitHub: https://github.com/AudeDN/ALS_GWAS_1000G_mach2dat_2017). GWAS summary statistics results for all tested SNPs and identified SNVs from our 1,138 FALS cohort used for the RVB analysis can be viewed through our web based ALS Variant Server (http://als.umassmed.edu). For each variant, information on over 50 annotation fields and the results can be downloaded directly into Excel.
Data Resources and Databases
1000 Genomes Project dataset: http://www.internationalgenome.org
Database of Genotypes and Phenotypes (dbGaP): www.ncbi.nlm.nih.gov/gap
dbNSFP: https://sites.google.com/site/jpopgen/dbNSFP
dbscSNV: incorporated into dbNSFP (see previous link).
European Genome-phenome Archive (EGA): https://ega-archive.org
HapMap project: http://www.sanger.ac.uk/resources/downloads/human/hapmap3.html.
Human Genome Diversity Panel (HGDP): http://www.hagsc.org/hgdp/
HYPERGENES Project: http://www.hypergenes.eu
Project MinE Variant Browser: http://databrowser.projectmine.com
snpEFF: http://snpeff.sourceforge.net/SnpEff.html
Wellcome Trust Case Control Consortium: www.wtccc.org.uk
Software
ASSEDA: http://www.cytognomix.com/?post_type=duka&p=2670
BWA: http://bio-bwa.sourceforge.net
GenABEL: http://www.genabel.org
GATK: https://software.broadinstitute.org/gatk/
id_geno_checksum: https://personal.broadinstitute.org/sripke/share_links/checksums_download/
KING: http://people.virginia.edu/~wc9c/KING/
LASER: http://csg.sph.umich.edu/chaolong/LASER/
Mach2dat: https://genome.sph.umich.edu/wiki/Mach2dat:_Association_with_MACH_output
METAL: http://csg.sph.umich.edu/abecasis/metal/index.html
Minimac3: https://genome.sph.umich.edu/wiki/Minimac3
PLINK: http://zzz.bwh.harvard.edu/plink/
SHAPEIT: https://mathgen.stats.ox.ac.uk/genetics_software/shapeit/shapeit.html
Supplementary Material
Figure S1. Related to Figure 1; Workflow showing the quality control procedures applied to the present study.
Figure S2. Related to Figure 1; Multi-dimensional scaling plot of the 44,558 genotyped samples included in analysis compared to the HapMap populations.
Figure S3. Related to Figure 1; Quartile-Quartile plot of P-values from the meta-analysis based on logistic regression analysis.
Figure S4. Related to Figure 3; Principal components analysis of samples included in the RVB analysis compared to the Human Diversity Panel.
Figure S5. Related to Figure 3; Quartile-Quartile plot of P values from the gene-based rare variant burden analysis of exome data.
Figure S6. Related to Figure 3; Control-control analyses.
Figure S7. Related to Fgure 3; Plot of variant call rates across the KIF5A protein-coding region in FALS versus controls analyzed by RVB testing.
Figure S8. Related to Figure 2; Principal components analysis of samples included in KIF5A replication cohort.
Table S1. Related to Figure 1; Demographics and baseline characteristics of patients diagnosed with ALS and control individuals included in the GWAS analysis.
Table S2. Related to Figure 1; DbGaP studies contributing to the GWAS analysis.
Table S3. Related to Figure 1; SNPs achieving genome-wide significance in the GWAS analysis.
Table S4. Related to Figure 1; Suggestive SNPs with P values less than 5.0×10-7 in the GWAS analyses.
Table S5. Related to Figure 3; DbGaP/EGA studies contributing to the RVB analysis.
Table S6. Related to Figure 2, 3; Quality control filtering of the FALS discovery and KIF5A replication cohorts.
Highlights.
Loss of function mutations in KIF5A are a cause of amyotrophic lateral sclerosis
ALS-associated KIF5A mutations are distinct from HSP and CMT mutations in KIF5A
Identification of KIF5A highlights role of cytoskeleton in ALS pathogenesis
Acknowledgments
Funding/Support: The ALS Association (ALSA) provided funding support to Project MinE (15-LGCA-235), NYGC ALS Consortium (15-LGCA-234), the CReATe Consortium (17-LGCA-331), the GTAC Consortium (16-LGCA-310), the Target ALS Human Postmortem Tissue Core (16-LGCA-308) and NeuroLINCS, an NIH-funded collaborative effort. P.V.D. is a senior investigator of FWO-Vlaanderen. Project MinE Belgium has been supported by ALS liga België, Flanders Innovation & Enterpreneurship (IWT grant Project MinE), the Belgian National Lottery and a grant from Opening the Future Fund (KU Leuven). W.R. is supported through the E. von Behring Chair for Neuromuscular and Neurodegenerative Disorders and ERC (grant agreement n° 340429). Additional funding support includes NINDS R35 NS097261 (R.R.) and P01NS084974 (R.R. and K.B.B.). A.N.B. thanks Suna and Inan Kirac Foundation, Istanbul, TR for its generous support of the Neurodegeneration Research Laboratory throughout this study. Funding for this work was provided by the Heaton-Ellis Trust, The Middlemass Family, Motor Neurone Disease Association, Medical Research Council, Medical Research Foundation, The Psychiatry Research Trust of the Institute of Psychiatry, Guy’s and St. Thomas’ Charity, the Wellcome Trust, and the Noreen Murray Foundation (C.E.S). This work was also supported by the UK Dementia Research Institute which is funded by the Medical Research Council, Alzheimer’s Society and Alzheimer’s Research UK (C.E.S.). The salary for B.N.S was funded by the Medical Research Foundation (MRF) (MRF-060-0003-RG-SMITH). P.C.S. was supported through the auspices of Dr. H. Robert Horvitz (Massachusetts Institute of Technology), an Investigator of the Howard Hughes Institute. Support from the Department of Veterans Affairs and NIH (P30AG13846) to N.W.K. I.P.B. is supported by the Motor Neurone Disease Research Institute of Australia and the National Health and Medical Research Council of Australia (1107644, 1095215). P.F. is supported by an MRC/MNDA LEWF and by NIHR UCLH BRC. Research support from NIH/NIEHS (K23ES027221), the ALS Association, Target ALS, and Cytokinetics was provided to S.A.G. M.C. was awarded funding from ALS Finding a Cure. N.T., C.Ti., C.G., V.S. and J.E.L. received research support from AriSLA - Fondazione Italiana di Ricerca per la SLA (grants EXOMEFALS and NOVALS) and the Italian Ministry of Health (grant GR-2011-02347820 - IRisALS). R.L.M. was supported by Science Foundation Ireland and the MND Association of England, Wales and Northern Ireland. O.H. is funded by the Health Research Board Clinician Scientist Programme and Science Foundation Ireland. P.J.S. is supported as an NIHR Senior Investigator (NF-SI-0512-10082). P.J.S. and J.K. are supported by the Sheffield NIHR Biomedical Research Centre for Translational Neuroscience (IS-BRC-1215-20017). A.C. receives research support from the Italian Ministry of Health (Ricerca Finalizzata), Regione Piemonte (Ricerca Finalizzata), University of Turin, Fondazione Vialli e Mauro onlus, and the European Commission (Health Seventh Framework Programme). P.M.A. is supported by research grants from the Swedish Brain Foundation, the Swedish Science Council, the Knut and Alice Wallenberg Foundation, the Bertil Hållsten Foundation, the Ulla-Carin Lindquist Foundation, the Neuroförbundet Association, the Torsten and Ragnar Söderberg Foundation, the Stratneuro Initiative, Västerbotten County Council. R.B. received funding support from NINDS/NS061867 and Target ALS. R.H.B.J. received funding from the Angel Fund, Project ALS/P2ALS, and the ALS Therapy Alliance. E.R. received funding support from the Canadian Consortium on Neurodegeneration in Aging. L.M. received funding support from Helsinki University Hospital and the Academy of Finland (grant 294817). T.P. received funding support from Helsinki University Hospital and the Sigrid Juselius Foundation. J.D.G. received funding support from the ALS Association and Muscular Dystrophy Association. Additional funding was provided by the NIH/NINDS (R01NS073873, J.E.L.), the ALS Association (N.T., V.S. C.E.S., R.H.B.J., J.E.L.), and the MND Association (N.T., V.S., C.E.S., J.E.L.). J.Ka., S.F., S.W., A.L., E.F., C.N.S., L.M.T., J.E.V.E., and J.D.R. received funding through NeuroLINCS (NIH U54 NS091046).
The sequencing activities at NYGC were additionally supported by the TOW Foundation. The CReATe consortium (U54NS092091) is part of Rare Diseases Clinical Research Network (RDCRN), an initiative of the Office of Rare Diseases Research (ORDR), NCATS. This consortium is funded through collaboration between NCATS, and the NINDS. The Target ALS Human Postmortem Tissue Core received funding support from Target ALS (grant 90072272). The InCHIANTI study baseline (1998–2000) was supported as a “targeted project” (ICS110.1/RF97.71) by the Italian Ministry of Health and in part by the U.S. National Institute on Aging (Contracts: 263 MD 9164 and 263 MD 821336); the InCHIANTI Follow-up 1 (2001–2003) was funded by the U.S. National Institute on Aging (Contracts: N.1-AG-1-1 and N.1-AG-1-2111); the InCHIANTI Follow-ups 2 and 3 studies (2004–2010) were financed by the U.S. National Institute on Aging (Contract: N01-AG-5-0002). This work was supported in part by the Intramural Research Programs of the US National Institutes of Health (NIH), National Institute on Aging (Z01-AG000949-02); by the National Institute of Neurological Disorders and Stroke; and by Merck & Co., Inc. The work was also supported by the Center for Disease Control and Prevention, the Muscular Dystrophy Association, Microsoft Research, the Packard Center for ALS Research at Johns Hopkins, the ALS Association, UK MND Association, Medical Research Council (MRC) UK, Wellcome Trust/MRC Joint Call in Neurodegeneration Award, MRC Neuromuscular Centre, UK National Institute for Health Research Biomedical Research Unit, Italian Health Ministry (Ricerca Sanitaria Finalizzata 2007), Fondazione Vialli e Mauro Onlus, Compagnia di San Paolo, European Community’s Health Seventh Framework Programme (FP7/2007-2013) under grant agreements 259867.
Role of the Sponsors: The sponsors did not participate in the design and conduct of the study; collection, management, analysis or interpretation of the data; or preparation, review, or approval of the manuscript; or decision to submit the manuscript for publication.
Additional contributions: We thank the patients and research subjects who contributed samples for this study. This study used DNA samples and clinical data from the Target ALS Human Postmortem Tissue Core, the NINDS Repository at Coriell, the North East ALS (NEALS) Consortium Biorepository, the New York Brain Bank-The Taub Institute, Columbia University, Department of Veterans Affairs Biorepository Brain Bank (grant #BX002466; C.B.B.), The Baltimore Longitudinal Study of Aging (BLSA) and the Johns Hopkins University Alzheimer’s Disease Research Center, the NICHD Brain and Tissue Bank for Developmental Disorders at the University of Maryland and the Australian MND DNA Bank. We also thank Crystal Pacut, Blake Swihart, and Jayna Duell, RN for assistance in study coordination (S.A.G.). This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, Maryland (http://biowulf.nih.gov) and the Massachusetts Green High Performance Computing Center at the University of Massachusetts Medical School (https://www.mghpcc.org). This study also used genotype and clinical data from the Wellcome Trust Case Control Consortium.
CONSORTIA
ALS Sequencing Consortium
Andrew S. Allen, Stanley Appel, Robert H. Baloh, Richard S. Bedlack, Braden E. Boone, Robert Brown, John P. Carulli, Alessandra Chesi, Wendy K. Chung, Elizabeth T. Cirulli, Gregory M. Cooper, Julien Couthouis, Aaron G. Day-Williams, Patrick A. Dion, Summer Gibson, Aaron D. Gitler, Jonathan D. Glass, David B. Goldstein, Yujun Han, Matthew B. Harms, Tim Harris, Sebastian D. Hayes, Angela L. Jones, Jonathan Keebler, Brian J. Krueger, Brittany N. Lasseigne, Shawn E. Levy, Yi-Fan Lu, Tom Maniatis, Diane McKenna-Yasek, Timothy M. Miller, Richard M. Myers, Slavé Petrovski, Stefan M. Pulst, Alya R. Raphael, John M. Ravits, Zhong Ren, Guy A. Rouleau, Peter C. Sapp, Neil A. Shneider, Ericka Simpson, Katherine B. Sims, John F. Staropoli, Lindsay L. Waite, Quanli Wang, Jack R. Wimbish, Winnie W. Xin
Answer ALS Foundation
Julia Kaye, Steven Finkbeiner, Stacia Wyman, Alexander LeNail, Leandro Lima, Ernest Fraenkel, Jeffrey D. Rothstein, Clive N. Svendsen, Leslie M. Thompson, Jenny Van Eyk, Nicholas J. Maragakis, James D. Berry, Jonathan D. Glass, Timothy M. Miller, Stephen J. Kolb, Robert H Baloh, Merit Cudkowicz, Emily Baxi
Clinical Research in ALS and Related Disorders for Therapeutic Development (CReATe) Consortium
Michael Benatar, J. Paul Taylor, Gang Wu, Evadnie Rampersaud, Joanne Wuu, Rosa Rademakers, Stephan Züchner, Rebecca Schule, Jacob McCauley, Sumaira Hussain, Anne Cooley, Marielle Wallace, Christine Clayman, Richard Barohn, Jeffrey Statland, John Ravits, Andrea Swenson, Carlayne Jackson, Jaya Trivedi, Shaida Khan, Jonathan Katz, Liberty Jenkins, Ted Burns, Kelly Gwathmey, James Caress, Corey McMillan, Lauren Elman, Erik Pioro, Jeannine Heckmann, Yuen So, David Walk, Samuel Maiser, Jinghui Zhang
French ALS Consortium
William Camu, Kevin Mouzat, Serge Lumbroso, Philippe Corcia, Vincent Meininger, Gérard Besson, Emmeline Lagrange, Pierre Clavelou, Nathalie Guy, Philippe Couratier, Patrick Vourch, Véronique Danel, Emilien Bernard, Gwendal Lemasson
Genomic Translation for ALS Care (GTAC) Consortium
Matthew B. Harms, David B. Goldstein, Neil A. Shneider, Stephen Goutman, Zachary Simmons, Timothy M. Miller, Siddharthan Chandran, Suvankar Pal, George Manousakis, Stanley H. Appel, Ericka Simpson, Leo Wang, Robert H Baloh, Summer Gibson, Richard Bedlack, David Lacomis, Dhruv Sareen, Alexander Sherman, Lucie Bruijn, Michelle Penny
ITALSGEN Consortium
Francesco O. Logullo, Isabella Simone, Giancarlo Logroscino, Fabrizio Salvi, Ilaria Bartolomei, Giuseppe Borghero, Maria Rita Murru, Emanuela Costantino, Carla Pani, Roberta Puddu, Carla Caredda, Valeria Piras, Stefania Tranquilli, Stefania Cuccu, Daniela Corongiu, Maurizio Melis, Antonio Milia, Francesco Marrosu, Maria Giovanna Marrosu, Gianluca Floris, Antonino Cannas, Stefania Tranquilli, Margherita Capasso, Claudia Caponnetto, Gianluigi Mancardi, Paola Origone, Paola Mandich, Francesca L. Conforti, Sebastiano Cavallaro, Gabriele Mora, Kalliopi Marinou, Riccardo Sideri, Silvana Penco, Lorena Mosca, Christian Lunetta, Giuseppe Lauria Pinter, Massimo Corbo, Nilo Riva, Paola Carrera, Paolo Volanti, Jessica Mandrioli, Nicola Fini, Antonio Fasano, Lucio Tremolizzo, Alessandro Arosio, Carlo Ferrarese, Francesca Trojsi, Gioacchino Tedeschi, Maria Rosaria Monsurrò, Giovanni Piccirillo, Cinzia Femiano, Anna Ticca, Enzo Ortu, Vincenzo La Bella, Rossella Spataro, Tiziana Colletti, Mario Sabatelli, Marcella Zollino, Amelia Conte, Marco Luigetti, Serena Lattante, Giuseppe Marangi, Marialuisa Santarelli, Antonio Petrucci, Maura Pugliatti, Angelo Pirisi, Leslie D. Parish, Patrizia Occhineri, Fabio Giannini, Stefania Battistini, Claudia Ricci, Michele Benigni, Tea B. Cau, Daniela Loi, Andrea Calvo, Cristina Moglia, Maura Brunetti, Marco Barberis, Gabriella Restagno, Federico Casale, Giuseppe Marrali, Giuseppe Fuda, Irene Ossola, Stefania Cammarosano, Antonio Canosa, Antonio Ilardi, Umberto Manera, Maurizio Grassano, Raffaella Tanel, Fabrizio Pisano
NYGC ALS Consortium
Hemali Phatnani, Justin Kwan, Dhruv Sareen, James R. Broach, Zachary Simmons, Ximena Arcila-Londono, Edward B. Lee, Vivianna M. Van Deerlin, Neil A. Shneider, Ernest Fraenkel, Lyle W. Ostrow, Frank Baas, Noah Zaitlen, James D. Berry, Andrea Malaspina, Pietro Fratta, Gregory A. Cox, Leslie M. Thompson, Steve Finkbeiner, Efthimios Dardiotis, Timothy M. Miller, Siddharthan Chandran, Suvankar Pal, Eran Hornstein, Daniel J. MacGowan, Terry Heiman-Patterson, Molly G. Hammell, Nikolaos. A. Patsopoulos, Joshua Dubnau, Avindra Nath
Project MinE ALS Sequencing Consortium
Ahmad Al Kheifat, Ammar Al-Chalabi, Peter Andersen, A. Nazli Basak, Ian P Blair, Adriano Chio, Jonathan Cooper-Knock, Philippe Corcia, Philippe Couratier, Mamede de Carvalho, Annelot Dekker, Vivian Drory, Alberto Garcia Redondo, Marc Gotkine, Orla Hardiman, Winston Hide, Alfredo Iacoangeli, Jonathan Glass, Kevin Kenna, Matthew Kiernan, Maarten Kooyman, John Landers, Russell McLaughlin, Bas Middelkoop, Jonathan Mill, Miguel Mitne Neto, Mattieu Moisse, Jesus Mora Pardina, Karen Morrison, Stephen Newhouse, Susana Pinto, Sara Pulit, Wim Robberecht, Aleksey Shatunov, Pamela Shaw, Chris Shaw, Vincenzo Silani, William Sproviero, Gijs Tazelaar, Nicola Ticozzi, Philip van Damme, Leonard van den Berg, Rick van der Spek, Kristel van Eijk, Michael van Es, Wouter van Rheenen, Joke van Vugt, Jan Veldink, Markus Weber, Kelly L Williams, Mayana Zatz, Denis C. Bauer, Natalie A. Twine
SLAGEN Consortium
Vincenzo Silani, Nicola Ticozzi, Cinzia Gellera, Antonia Ratti, Franco Taroni, Giuseppe Lauria, Federico Verde, Isabella Fogh, Cinzia Tiloca, Giacomo P. Comi, Gianni Sorarù, Cristina Cereda, Sandra D’Alfonso, Lucia Corrado, Fabiola De Marchi, Stefania Corti, Mauro Ceroni, Letizia Mazzini, Gabriele Siciliano, Massimiliano Filosto, Maurizio Inghilleri, Silvia Peverelli, Claudia Colombrita, Barbara Poletti, Luca Maderna, Roberto Del Bo, Stella Gagliardi, Giorgia Querin, Cinzia Bertolin, Viviana Pensato, Barbara Castellotti
Footnotes
Author Contributions:
Sample Collection, Preparation and Clinical Evaluation: N.T., B.J.K., P.K., W.v.R., J.J.v.V., R.A.V.d.S., B.N.S., G.Ma., S.D.T., A.S.G., A.Ke., ITALSGEN Consortium, G.M., A.Ca., L.Ma., N.R., J.M., C.Ca., S.B., P.V., V.L.B., F.L.C., G.B., S.M., I.L.S., F.Tr., F.S., F.O.L., S.D., L.C., M.Ca., L.F., Genomic Translation for ALS Care (GTAC) Consortium, C.d.A.M.M., S.K., D.B.G., A.D.G., T.H., R.M.M., NYGC ALS Consortium, H.P., R.L.Mu., U.S.E., A.Ab., M.C.Z., J.Ka., S.F., S.W., A.L., L.L., E.F., C.N.S., L.M.T., J.E.V.E., J.D.Be., T.M.M., S.J.K., M.C., E.B., Clinical Research in ALS and Related Disorders for Therapeutic Development (CReATe) Consortium, M.B., J.W., SLAGEN Consortium, G.L., F.V., I.F., C.Ti., G.P.C., G.S., French ALS Consortium, P.C., H.L., L.M., L.J., M.V., J.Ea., H.H., S.R., S.P., R.W.O., K.C.S., A.M., J.H., P.C.S., D.M., M.P., A.K., C.T., C.V., J.d.B., F.B., A.L.t.A., J.L.M., S.W.S., M.K.F., F.L., R.B., S.M.P., J.M.R., D.J.M., J.K., E.P., R.P., J.B., G.G., T.L.D., C.B.B., N.W.K., J.C.T., I.L.B., K.M., S.L., T.D.H., F.K., L.V.D.B., R.H.B., T.M.S., T.M., A.S., K.R.V.E., M.d.C., M.K., B.M., M.M., R.L.M., M.A.V.E., M.W., K.B.B., M.V.B., R.R., K.E.M., A.N.B., J.S.M., V.E.D., P.J.S., M.R.T., K.T., O.H., K.L.W., J.A.F., G.A.N., I.P.B., G.A.R., J.E., A.G., A.A., Project MinE ALS Sequencing Consortium, E.R., L.Z., L.O., N.J.M., J.D.R., Z.S., J.C., A.B., S.A.G., E.L.F., S.B.G., F.T., A.R., C.G., P.V.D., W.R., P.F., M.S., C.L., A.C.L., P.M.A., J.H.W., W.C., J.Q.T., V.M.V.D., R.H.B.J., L.H.v.d.B., J.H.V., M.B.H., J.D.G., D.J.S., P.T., V.S., A.C., C.E.S., B.J.T., J.E.L.
Performed Experiments and Data Analysis: A.N., K.P.K., A.E.R., N.T., F.F., R.C., J.A.D., B.J.K., M.A.N., P.K., A.M.R., W.v.R., N.A.M., J.J.v.V., J.T.G., R.A.V.d.S., H.A.P., F.N.S., B.N.S., G.Ma., S.D.T., Y.A., A.S.G., J.D.E., A.Ke., Genomic Translation for ALS Care (GTAC) Consortium, C.d.A.M.M., S.K., D.B.G., A.D.G., T.H., R.M.M., NYGC ALS Consortium, H.P., R.L.Mu., U.S.E., A.Ab., M.C.Z., J.Ka., S.F., S.W., A.L., L.L., E.F., C.N.S., L.M.T., J.E.V.E., J.D.Be., T.M.M., S.J.K., M.C., E.B., Clinical Research in ALS and Related Disorders for Therapeutic Development (CReATe) Consortium, E.Ra., G.W., SLAGEN Consortium, G.L., F.V., I.F., C.Ti., G.P.C., G.S., A.B.S., J.O.J., S.Ar., P.C.S., D.M., M.P., C.T., C.V., J.d.B., F.B., A.L.t.A., J.L.M., D.G.H., J.D., J.R.G., S.W.S., R.H.C., T.D.H., L.V.D.B., R.H.B., T.M.S., T.M., A.S., K.R.V.E., M.d.C., M.K., B.M., M.M., R.L.M., M.A.V.E., M.W., K.B.B., M.V.B., R.R., K.E.M., A.N.B., J.S.M., V.E.D., P.J.S., M.R.T., K.T., O.H., K.L.W., J.A.F., G.A.N., I.P.B., G.A.R., J.E., A.G., A.A., Project MinE ALS Sequencing Consortium, E.R., L.O., N.J.M., J.D.R., Z.S., J.C., S.A.G., E.L.F., F.T., A.R., C.G., P.V.D., W.R., A.C.L., P.M.A., J.H.W., W.C., J.Q.T., V.M.V.D., R.H.B.J., L.H.v.d.B., J.H.V., M.B.H., J.D.G., D.J.S., P.T., V.S., A.C., C.E.S., B.J.T., J.E.L.
Scientific Planning and Direction: A.N., K.P.K., A.E.R., N.T., F.F., R.C., J.A.D., B.J.K., M.A.N., P.K., A.M.R., W.v.R., N.A.M., J.J.v.V., J.T.G., R.A.V.d.S., H.A.P., F.N.S., B.N.S., G.Ma., S.D.T., Y.A., J.D.E., ITALSGEN Consortium, G.M., A.Ca., L.Ma., N.R., J.M., C.Ca., S.B., P.V., V.L.B., F.L.C., G.B., S.M., I.L.S., F.Tr., F.S., F.O.L., S.D., L.C., M.Ca., L.F., Genomic Translation for ALS Care (GTAC) Consortium, D.B.G., A.D.G., T.H., R.M.M., NYGC ALS Consortium, H.P., J.Ka., S.F., S.W., A.L., L.L., E.F., C.N.S., L.M.T., J.E.V.E., J.D.Be., T.M.M., S.J.K., M.C., E.B., Clinical Research in ALS and Related Disorders for Therapeutic Development (CReATe) Consortium, M.B., J.P.T., J.W., French ALS Consortium, P.C., H.L., L.M., L.J., M.V., J.Ea., H.H., S.R., S.P., R.W.O., K.C.S., A.M., J.H., A.B.S., C.T., C.V., J.d.B., F.B., A.L.t.A., J.L.M., D.G.H., J.R.G., S.W.S., M.K.F., F.L., R.B., S.M.P., J.M.R., D.J.M., J.K., E.P., R.P., J.B., G.G., T.L.D., C.B.B., N.W.K., J.C.T., I.L.B., K.M., S.L., T.D.H., F.K., L.V.D.B., R.H.B., T.M.S., T.M., A.S., K.R.V.E., M.d.C., M.K., B.M., M.M., R.L.M., M.A.V.E., M.W., K.B.B., M.V.B., R.R., K.E.M., A.N.B., J.S.M., V.E.D., P.J.S., M.R.T., K.T., O.H., K.L.W., J.A.F., G.A.N., I.P.B., G.A.R., J.E., A.G., A.A., Project MinE ALS Sequencing Consortium, E.R., L.Z., L.O., N.J.M., J.D.R., Z.S., J.C., A.B., S.A.G., E.L.F., S.B.G., F.T., A.R., C.G., P.V.D., W.R., P.F., M.S., C.L., A.C.L., P.M.A., J.H.W., W.C., J.Q.T., V.M.V.D., R.H.B.J., L.H.v.d.B., J.H.V., M.B.H., J.D.G., D.J.S., P.T., V.S., A.C., C.E.S., B.J.T., J.E.L.
Initial Manuscript Preparation: A.N., K.P.K., A.E.R., N.T., F.F., R.C., J.A.D., M.A.N., A.B.S., S.W.S., J.H.V., D.J.S., P.T., V.S., A.C., C.E.S., B.J.T., J.E.L.
Declaration of Interests: J.D.Be. is a consultant to Neuraltus Pharmaceuticals and Denali Therapeutics, and held a research fellow position funded by Voyager Therapeutics. M.C. has been a consultant for Eli Lilly and Company, Mitsubishi Tanabe Pharma America (MT Pharma America), Denali Therapeutics, Karyopharm Therapeutics and Cytokinetics. S.A.G. has served as a consultant and received research support from Cytokinetics. O.H. has received speaking honoraria from Novarits, Biogen Idec, Sanofi Aventis and Merck-Serono and has been a member of advisory panels for Biogen Idec, Allergen, Ono Pharmaceuticals, Novartis, Cytokinetics and Sanofi Aventis. O.H. serves as Editor-in-Chief of Amyotrophic Lateral Sclerosis and Frontotemporal Dementia. L.H.v.d.B. serves on scientific advisory boards for the Prinses Beatrix Spierfonds, Thierry Latran Foundation, Biogen and Cytokinetics. Serves on the editorial board of Amyotrophic Lateral Sclerosis And Frontotemporal Degeneration and The Journal of Neurology, Neurosurgery, and Psychiatry. J.H.V. reports that his institute received consultancy fees from Vertex Pharmaceuticals. A.C. serves on scientific advisory boards for Biogen Idec, Cytokinetics, Italfarmaco, and Neuraltus. P.M.A. serve on advisory board panels for Biogen and Orphazyme. V.S. serves as a consultant for Cytokinetics.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Al-Chalabi A, Andersen PM, Nilsson P, Chioza B, Andersson JL, Russ C, Shaw CE, Powell JF, Leigh PN. Deletions of the heavy neurofilament subunit tail in amyotrophic lateral sclerosis. Human Molecular Genetics. 1999;8:157–164. doi: 10.1093/hmg/8.2.157. [DOI] [PubMed] [Google Scholar]
- ALSGEN Consortium. Ahmeti KB, Ajroud-Driss S, Al-Chalabi A, Andersen PM, Armstrong J, Birve A, Blauw HM, Brown RH, Bruijn L, et al. Age of onset of amyotrophic lateral sclerosis is modulated by a locus on 1p34.1. Neurobiol Aging. 2013;34:357.e7–.e19. doi: 10.1016/j.neurobiolaging.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur KC, Calvo A, Price TR, Geiger JT, Chio A, Traynor BJ. Projected increase in amyotrophic lateral sclerosis from 2015 to 2040. Nat Commun. 2016;7:12408. doi: 10.1038/ncomms12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benyamin B, He J, Zhao Q, Gratten J, Garton F, Leo PJ, Liu Z, Mangelsdorf M, Al-Chalabi A, Anderson L, et al. Cross-ethnic meta-analysis identifies association of the GPX3-TNIP1 locus with amyotrophic lateral sclerosis. Nat Commun. 2017;8:611. doi: 10.1038/s41467-017-00471-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner D, Yilmaz R, Müller K, Grehl T, Petri S, Meyer T, Grosskreutz J, Weydt P, Ruf W, Neuwirth C, et al. Hot-spot KIF5A mutations cause familial ALS. Brain. 2018 doi: 10.1093/brain/awx370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks BR. El Escorial World Federation of Neurology criteria for the diagnosis of amyotrophic lateral sclerosis. Subcommittee on Motor Neuron Diseases/Amyotrophic Lateral Sclerosis of the World Federation of Neurology Research Group on Neuromuscular Diseases and the El Escorial “Clinical limits of amyotrophic lateral sclerosis” workshop contributors. J Neurol Sci. 1994;124(Suppl):96–107. doi: 10.1016/0022-510x(94)90191-0. [DOI] [PubMed] [Google Scholar]
- Caminsky NG, Mucaki EJ, Rogan PK. Interpretation of mRNA splicing mutations in genetic disease: review of the literature and guidelines for information-theoretical analysis. F1000Res. 2014;3:282. doi: 10.12688/f1000research.5654.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell PD, Shen K, Sapio MR, Glenn TD, Talbot WS, Marlow FL. Unique function of Kinesin Kif5A in localization of mitochondria in axons. Journal of Neuroscience. 2014;34:14717–14732. doi: 10.1523/JNEUROSCI.2770-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:1–16. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier-Larsen E, Holzbaur ELF. Axonal transport and neurodegenerative disease. Biochim Biophys Acta. 2006;1762:1094–1108. doi: 10.1016/j.bbadis.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Chia R, Chio A, Traynor BJ. Novel genes associated with amyotrophic lateral sclerosis: diagnostic and clinical implications. Lancet Neurol. 2018;17:94–102. doi: 10.1016/S1474-4422(17)30401-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chio A, Logroscino G, Hardiman O, Swingler R, Mitchell D, Beghi E, Traynor BG On Behalf of the Eurals Consortium. Prognostic factors in ALS: A critical review. Amyotroph Lateral Scler. 2009;10:310–323. doi: 10.3109/17482960802566824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirulli ET, Lasseigne BN, Petrovski S, Sapp PC, Dion PA, Leblond CS, Couthouis J, Lu YF, Wang Q, Krueger BJ, et al. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science. 2015;347:1436–1441. doi: 10.1126/science.aaa3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppedè F, Migliore L. Mutation Research/Fundamental and MolecularMechanisms of Mutagenesis. Mutation Research - Fundamental and Molecular Mechanisms of Mutagenesis. 2015;776:84–97. doi: 10.1016/j.mrfmmm.2014.11.010. [DOI] [PubMed] [Google Scholar]
- Crimella C, Baschirotto C, Arnoldi A, Tonelli A, Tenderini E, Airoldi G, Martinuzzi A, Trabacca A, Losito L, Scarlato M, et al. Mutations in the motor and stalk domains of KIF5A in spastic paraplegia type 10 and in axonal Charcot-Marie-Tooth type 2. Clin Genet. 2011;82:157–164. doi: 10.1111/j.1399-0004.2011.01717.x. [DOI] [PubMed] [Google Scholar]
- Das S, Forer L, Schönherr S, Sidore C, Locke AE, Kwong A, Vrieze SI, Chew EY, Levy S, McGue M, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Meth. 2013;10:5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- Ebbing B, Mann K, Starosta A, Jaud J, Schols L, Schule R, Woehlke G. Effect of spastic paraplegia mutations in KIF5A kinesin on transport activity. Human Molecular Genetics. 2008;17:1245–1252. doi: 10.1093/hmg/ddn014. [DOI] [PubMed] [Google Scholar]
- Fang H, Wu Y, Narzisi G, O’Rawe JA, Barrón LTJ, Rosenbaum J, Ronemus M, Iossifov I, Schatz MC, Lyon GJ. Reducing INDEL calling errors in whole genome and exome sequencing data. Genome Med. 2014;6:89. doi: 10.1186/s13073-014-0089-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Lin H, Wang X, Zuo Q, Qin J, Zhang P. The NEK1 interactor, C21ORF2, is required for efficient DNA damage repair. Acta Biochim Biophys Sin. 2015;47:834–841. doi: 10.1093/abbs/gmv076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freischmidt A, Wieland T, Richter B, Ruf W, Schaeffer V, Müller K, Marroquin N, Nordin F, Hübers A, Weydt P, et al. Haploinsufficiency of TBK1 causes familial ALS and fronto-temporal dementia. Nat Neurosci. 2015;18:631–636. doi: 10.1038/nn.4000. [DOI] [PubMed] [Google Scholar]
- Gladman M, Zinman L. The economic impact of amyotrophic lateral sclerosis: a systematic review. Expert Rev Pharmacoecon Outcomes Res. 2015;15:439–450. doi: 10.1586/14737167.2015.1039941. [DOI] [PubMed] [Google Scholar]
- Gros-Louis F. A Frameshift Deletion in Peripherin Gene Associated with Amyotrophic Lateral Sclerosis. Journal of Biological Chemistry. 2004;279:45951–45956. doi: 10.1074/jbc.M408139200. [DOI] [PubMed] [Google Scholar]
- Guo W, Naujock M, Fumagalli L, Vandoorne T, Baatsen P, Boon R, Ordovás L, Patel A, Welters M, Vanwelden T, et al. HDAC6 inhibition reverses axonal transport defects in motor neurons derived from FUS-ALS patients. Nat Commun. 2017;8:861. doi: 10.1038/s41467-017-00911-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hares K, Redondo J, Kemp K, Rice C, Scolding N, Wilkins A. Axonal motor protein KIF5A and associated cargo deficits in multiple sclerosis lesional and normal-appearing white matter. Neuropathol Appl Neurobiol. 2016;43:227–241. doi: 10.1111/nan.12305. [DOI] [PubMed] [Google Scholar]
- Heisler FF, Lee HK, Gromova KV, Pechmann Y, Schurek B, Ruschkies L, Schroeder M, Schweizer M, Kneussel M. GRIP1 interlinks N-cadherin and AMPA receptors at vesicles to promote combined cargo transport into dendrites. Proceedings of the National Academy of Sciences. 2014;111:5030–5035. doi: 10.1073/pnas.1304301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Pfister KK, Yorifuji H, Wagner MC, Brady ST, Bloom GS. Submolecular domains of bovine brain kinesin identified by electron microscopy and monoclonal antibody decoration. Cell. 1989;56:867–878. doi: 10.1016/0092-8674(89)90691-0. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Niwa S, Tanaka Y. Molecular Motors in Neurons:Transport Mechanisms and Rolesin Brain Function, Development, and Disease. Neuron. 2010;68:610–638. doi: 10.1016/j.neuron.2010.09.039. [DOI] [PubMed] [Google Scholar]
- Hirtz D, Thurman DJ, Gwinn-Hardy K, Mohamed M, Chaudhuri AR, Zalutsky R. How common are the “common” neurologic disorders? Neurology. 2007;68:326–337. doi: 10.1212/01.wnl.0000252807.38124.a3. [DOI] [PubMed] [Google Scholar]
- Jennings S, Chenevert M, Liu L, Mottamal M, Wojcik EJ, Huckaba TM. Characterization of kinesin switch I mutations that cause hereditary spastic paraplegia. PLoS ONE. 2017;12:e0180353. doi: 10.1371/journal.pone.0180353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian X, Boerwinkle E, Liu X. In silico prediction of splice-altering single nucleotide variants in the human genome. Nucleic Acids Research. 2014;42:13534–13544. doi: 10.1093/nar/gku1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Okada Y, Tanaka Y, Harada A, Terada S, Hirokawa N. KIF5C, a novel neuronal kinesin enriched in motor neurons. J Neurosci. 2000;20:6374–6384. doi: 10.1523/JNEUROSCI.20-17-06374.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Karle KN, Möckel D, Reid E, Schols L. Axonal transport deficit in a KIF5A –/– mouse model. Neurogenetics. 2012;13:169–179. doi: 10.1007/s10048-012-0324-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenna KP, van Doormaal PTC, Dekker AM, Ticozzi N, Kenna BJ, Diekstra FP, van Rheenen W, van Eijk KR, Jones AR, Keagle P, et al. NEK1 variants confer susceptibility to amyotrophic lateral sclerosis. Nat Genet. 2016;48:1037–1042. doi: 10.1038/ng.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495:467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski TJ, Bosco DA, LeClerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, et al. Mutations in the FUS/TLS Gene on Chromosome 16 Cause Familial Amyotrophic Lateral Sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- Laaksovirta H, Peuralinna T, Schymick JC, Scholz SW, Lai SL, Myllykangas L, Sulkava R, Jansson L, Hernandez DG, Gibbs JR, et al. Chromosome 9p21 in amyotrophic lateral sclerosis in Finland: a genome-wide association study. Lancet Neurol. 2010;9:978–985. doi: 10.1016/S1474-4422(10)70184-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Jian X, Boerwinkle E. dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum Mutat. 2013;34:E2393–E2402. doi: 10.1002/humu.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YT, Laurá M, Hersheson J, Horga A, Jaunmuktane Z, Brandner S, Pittman A, Hughes D, Polke JM, Sweeney MG, et al. Extended phenotypic spectrum of KIF5A mutations: From spastic paraplegia to axonal neuropathy. Neurology. 2014;83:612–619. doi: 10.1212/WNL.0000000000000691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Gonzalez R, Lu Y, Gendron TF, Karydas A, Tran H, Yang D, Petrucelli L, Miller BL, Almeida S, Gao FB. Poly(GR) in C9ORF72-Related ALS/FTD Compromises Mitochondrial Function and Increases Oxidative Stress and DNA Damage in iPSC-Derived Motor Neurons. Neuron. 2016;92:383–391. doi: 10.1016/j.neuron.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madabhushi R, Pan L, Tsai LH. DNA Damage and Its Links to Neurodegeneration. Neuron. 2014;83:266–282. doi: 10.1016/j.neuron.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majounie E, Renton AE, Mok K, Dopper EGP, Waite A, Rollinson S, Chio A, Restagno G, Nicolaou N, Simón-Sánchez J, et al. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 2012;11:323–330. doi: 10.1016/S1474-4422(12)70043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malovannaya A, Lanz RB, Jung SY, Bulynko Y, Le NT, Chan DW, Ding C, Shi Y, Yucer N, Krenciute G, et al. Analysis of the human endogenous coregulator complexome. Cell. 2011;145:787–799. doi: 10.1016/j.cell.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- Maruyama H, Morino H, Ito H, Izumi Y, Kato H, Watanabe Y, Kinoshita Y, Kamada M, Nodera H, Suzuki H, et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465:223–226. doi: 10.1038/nature08971. [DOI] [PubMed] [Google Scholar]
- Matsuzaki F, Shirane M, Matsumoto M, Nakayama KI. Protrudin serves as an adaptor molecule that connects KIF5 and its cargoes in vesicular transport during process formation. Molecular Biology of the Cell. 2011;22:4602–4620. doi: 10.1091/mbc.E11-01-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin RL, Schijven D, van Rheenen W, van Eijk KR, O’Brien M, Kahn RS, Ophoff RA, Goris A, Bradley DG, Al-Chalabi A, et al. Genetic correlation between amyotrophic lateral sclerosis and schizophrenia. Nat Commun. 2017;8:14774. doi: 10.1038/ncomms14774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H, Setou M, Kaneshiro K, Hirokawa N. All kinesin superfamily protein, KIF, genes in mouse and human. Proc Natl Acad Sci USa. 2001;98:7004–7011. doi: 10.1073/pnas.111145398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millecamps S, Julien J-P. Axonal transport deficits and neurodegenerative diseases. Nat Rev Neurosci. 2013;14:161–176. doi: 10.1038/nrn3380. [DOI] [PubMed] [Google Scholar]
- Morton S, Hesson L, Peggie M, Cohen P. Enhanced binding of TBK1 by an optineurin mutant that causes a familial form of primary open angle glaucoma. FEBS Lett. 2008;582:997–1002. doi: 10.1016/j.febslet.2008.02.047. [DOI] [PubMed] [Google Scholar]
- Nakajima K, Yin X, Takei Y, Seog DH, Homma N, Hirokawa N. Molecular Motor KIF5A Is Essential for GABA. Neuron. 2012;76:945–961. doi: 10.1016/j.neuron.2012.10.012. [DOI] [PubMed] [Google Scholar]
- Nishimura AL, Al-Chalabi A, Zatz M. A common founder for amyotrophic lateral sclerosis type 8 (ALS8) in the Brazilian population. Hum Genet. 2005;118:499–500. doi: 10.1007/s00439-005-0031-y. [DOI] [PubMed] [Google Scholar]
- Nishimura AL, Mitne-Neto M, Silva HCA, Richieri-Costa A, Middleton S, Cascio D, Kok F, Oliveira JRM, Gillingwater T, Webb J, et al. A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. The American Journal of Human Genetics. 2004;75:822–831. doi: 10.1086/425287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomo GM, Manfredi G. Exploring new pathways of neurodegeneration in ALS: the role of mitochondria quality control. Brain Research. 2015;1607:36–46. doi: 10.1016/j.brainres.2014.09.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzkó A, Shy ME. Update on Charcot-Marie-Tooth disease. Curr Neurol Neurosci Rep. 2011;11:78–88. doi: 10.1007/s11910-010-0158-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pe’er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32:381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- Reid E, Kloos M, Ashley-Koch A, Hughes L, Bevan S, Svenson IK, Graham FL, Gaskell PC, Dearlove A, Pericak-Vance MA, et al. A kinesin heavy chain (KIF5A) mutation in hereditary spastic paraplegia (SPG10) The American Journal of Human Genetics. 2002;71:1189–1194. doi: 10.1086/344210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Chio A, Traynor BJ. State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci. 2014;17:17–23. doi: 10.1038/nn.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robberecht W, Eykens C. The genetic basis of amyotrophic lateral sclerosis. Agg. 2015:327–345. [Google Scholar]
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O’Regan JP, Deng HX. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- Salvi E, Kutalik Z, Glorioso N, Benaglio P, Frau F, Kuznetsova T, Arima H, Hoggart C, Tichet J, Nikitin YP, et al. Genomewide association study using a high-density single nucleotide polymorphism array and case-control design identifies a novel essential hypertension susceptibility locus in the promoter region of endothelial NO synthase. Hypertension. 2012;59:248–255. doi: 10.1161/HYPERTENSIONAHA.111.181990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setou M, Seog DH, Tanaka Y, Kanai Y, Takei Y, Kawagishi M, Hirokawa N. Glutamate-receptor-interacting protein GRIP1 directly steers kinesin to dendrites. Nature. 2002;417:83–87. doi: 10.1038/nature743. [DOI] [PubMed] [Google Scholar]
- Shatunov A, Mok K, Newhouse S, Weale ME, Smith B, Vance C, Johnson L, Veldink JH, van Es MA, van den Berg LH, et al. Chromosome 9p21 in sporadic amyotrophic lateral sclerosis in the UK and seven other countries: a genome-wide association study. Lancet Neurol. 2010;9:986–994. doi: 10.1016/S1474-4422(10)70197-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BN, Ticozzi N, Fallini C, Gkazi AS, Topp S, Kenna KP, Scotter EL, Kost J, Keagle P, Miller JW, et al. Exome-wide Rare Variant Analysis Identifies TUBA4A Mutations Associated with Familial ALS. Neuron. 2014;84:324–331. doi: 10.1016/j.neuron.2014.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EF, Shaw PJ, De Vos KJ. The role of mitochondria in amyotrophic lateral sclerosis. Neuroscience Letters. 2017 doi: 10.1016/j.neulet.2017.06.052. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Kanai Y, Okada Y, Nonaka S, Takeda S, Harada A, Hirokawa N. Targeted disruption of mouse conventional kinesin heavy chain, kif5B, results in abnormal perinuclear clustering of mitochondria. Cell. 1998;93:1147–1158. doi: 10.1016/s0092-8674(00)81459-2. [DOI] [PubMed] [Google Scholar]
- Taylor JP, Brown RH, Cleveland DW. Decoding ALS: from genes to mechanism. Nature. 2016;539:197–206. doi: 10.1038/nature20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel C, Kessler K, Giessl A, Dimmler A, Shalev SA, Haar von der S, Zenker M, Zahnleiter D, Stöss H, Beinder E, et al. REPOR TNEK1 Mutations CauseShort-Rib Polydactyly Syndrome Type Majewski. The American Journal of Human Genetics. 2011;88:106–114. doi: 10.1016/j.ajhg.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompson SWJ, Ruiz-Perez VL, Blair HJ, Barton S, Navarro V, Robson JL, Wright MJ, Goodship JA. Sequencing EVC and EVC2 identifies mutations in two-thirds of Ellis-van Creveld syndrome patients. Hum Genet. 2007;120:663–670. doi: 10.1007/s00439-006-0237-7. [DOI] [PubMed] [Google Scholar]
- van Es MA, Hardiman O, Chio A, Al-Chalabi A, Pasterkamp RJ, Veldink JH, van den Berg LH. Amyotrophic lateral sclerosis. Lancet. 2017;390:2084–2098. doi: 10.1016/S0140-6736(17)31287-4. [DOI] [PubMed] [Google Scholar]
- van Es MA, Veldink JH, Saris CGJ, Blauw HM, van Vught PWJ, Birve A, Lemmens R, Schelhaas HJ, Groen EJN, Huisman MHB, et al. Genome-wide association study identifies 19p13.3 (UNC13A) and 9p21.2 as susceptibility loci for sporadic amyotrophic lateral sclerosis. Nat Genet. 2009;41:1083–1087. doi: 10.1038/ng.442. [DOI] [PubMed] [Google Scholar]
- van Rheenen W, Shatunov A, Dekker AM, McLaughlin RL, Diekstra FP, Pulit SL, van der Spek RAA, Võsa U, de Jong S, Robinson MR, et al. Genome-wide association analyses identify new risk variants and the genetic architecture of amyotrophic lateral sclerosis. Nat Genet. 2016;48:1043–1048. doi: 10.1038/ng.3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, et al. Mutations in FUS, an RNA Processing Protein, Cause Familial Amyotrophic Lateral Sclerosis Type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Brown A. A hereditary spastic paraplegia mutation in kinesin-1A/KIF5A disrupts neurofilament transport. Mol Neurodegeneration. 2010;5:52–64. doi: 10.1186/1750-1326-5-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WY, Pan L, Su SC, Quinn EJ, Sasaki M, Jimenez JC, Mackenzie IRA, Huang EJ, Tsai LH. Interaction of FUS and HDAC1 regulates DNA damage response and repair in neurons. Nat Neurosci. 2013;16:1383–1391. doi: 10.1038/nn.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Fallini C, Ticozzi N, Keagle PJ, Sapp PC, Piotrowska K, Lowe P, Koppers M, McKenna-Yasek D, Baron DM, et al. Mutations in the profilin 1 gene cause familial amyotrophic lateral sclerosis. Nature. 2012;488:499–503. doi: 10.1038/nature11280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia CH, Roberts EA, Her LS, Liu X, Williams DS, Cleveland DW, Goldstein LSB. Abnormal neurofilament transport caused by targeted disruption of neuronal kinesin heavy chain KIF5A. The Journal of Cell Biology. 2003;161:55–66. doi: 10.1083/jcb.200301026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Related to Figure 1; Workflow showing the quality control procedures applied to the present study.
Figure S2. Related to Figure 1; Multi-dimensional scaling plot of the 44,558 genotyped samples included in analysis compared to the HapMap populations.
Figure S3. Related to Figure 1; Quartile-Quartile plot of P-values from the meta-analysis based on logistic regression analysis.
Figure S4. Related to Figure 3; Principal components analysis of samples included in the RVB analysis compared to the Human Diversity Panel.
Figure S5. Related to Figure 3; Quartile-Quartile plot of P values from the gene-based rare variant burden analysis of exome data.
Figure S6. Related to Figure 3; Control-control analyses.
Figure S7. Related to Fgure 3; Plot of variant call rates across the KIF5A protein-coding region in FALS versus controls analyzed by RVB testing.
Figure S8. Related to Figure 2; Principal components analysis of samples included in KIF5A replication cohort.
Table S1. Related to Figure 1; Demographics and baseline characteristics of patients diagnosed with ALS and control individuals included in the GWAS analysis.
Table S2. Related to Figure 1; DbGaP studies contributing to the GWAS analysis.
Table S3. Related to Figure 1; SNPs achieving genome-wide significance in the GWAS analysis.
Table S4. Related to Figure 1; Suggestive SNPs with P values less than 5.0×10-7 in the GWAS analyses.
Table S5. Related to Figure 3; DbGaP/EGA studies contributing to the RVB analysis.
Table S6. Related to Figure 2, 3; Quality control filtering of the FALS discovery and KIF5A replication cohorts.