Abstract
Introduction
Cigarette smoking is a physiologically harmful habit. Nicotinic acetylcholine receptors (nAChRs) are bound by nicotine and upregulated in response to chronic exposure to nicotine. It is known that upregulation of these receptors is not due to a change in mRNA of these genes, however, more precise details on the process are still uncertain, with several plausible hypotheses describing how nAChRs are upregulated. We have manually curated a set of genes believed to play a role in nicotine-induced nAChR upregulation. Here, we test the hypothesis that these genes are associated with and contribute risk for nicotine dependence (ND) and the number of cigarettes smoked per day (CPD).
Methods
Studies with genotypic data on European and African Americans (EAs and AAs, respectively) were collected and a gene-based test was run to test for an association between each gene and ND and CPD.
Results
Although several novel genes were associated with CPD and ND at P < 0.05 in EAs and AAs, these associations did not survive correction for multiple testing. Previous associations between CHRNA3, CHRNA5, CHRNB4 and CPD in EAs were replicated.
Conclusions
Our hypothesis-driven approach avoided many of the limitations inherent in pathway analyses and provided nominal evidence for association between cholinergic-related genes and nicotine behaviors.
Implications
We evaluated the evidence for association between a manually curated set of genes and nicotine behaviors in European and African Americans. Although no genes were associated after multiple testing correction, this study has several strengths: by manually curating a set of genes we circumvented the limitations inherent in many pathway analyses and tested several genes that had not yet been examined in a human genetic study; gene-based tests are a useful way to test for association with a set of genes; and these genes were collected based on literature review and conversations with experts, highlighting the importance of scientific collaboration.
Introduction
Cigarette smoking is a personally harmful and societally detrimental habit. Despite substantial progress since the first Surgeon General’s report was released 50 years ago, smoking is still the largest cause of preventable disease and death in the United States. An estimated half a million Americans die prematurely from smoking each year, and more than 16 million Americans suffer from smoking-related diseases. The economic costs from smoking and exposure to tobacco smoke are estimated at $300 billion annually, with productivity losses of $150 billion per year.1
It is widely accepted that nicotine is the major addictive component in tobacco smoke2,3 and exerts its effect by binding to nicotinic acetylcholine receptors (nAChRs, encoded by the CHRN genes) in the peripheral and central nervous systems.4 Accordingly, numerous studies have identified polymorphisms in several CHRN genes associated with nicotine dependence (ND) and cigarettes per day (CPD). The most well-replicated of these associations lies within a cluster of three CHRN genes on chromosome 15q25 (CHRNA3/A5/B4),5 although genes for other subunits have been associated with these phenotypes as well. Most notably, CHRNB3/A6 on chromosome 8p11 has been consistently associated with CPD6 and ND.7–13 Other CHRN genes associated with CPD and ND include CHRND/G, CHRNB1, CHRNA10, CHRNA4, and CHRNB2.10,11,14–19
Twin studies have estimated that ND and smoking quantity are roughly 56%–72% and 51%–61% heritable in men and women, respectively,20–24 yet associations between the CHRN genes and ND and CPD account for a very small proportion of the variance in smoking (roughly 1 CPD).5,11 Although this is small percentage, 1 additional CPD over a person’s lifetime will accumulate, often to greater than 22000 additional cigarettes in one’s lifetime in the case of the Genetic Epidemiology of Chronic Obstructive Pulmonary Disease25 study (COPDGene, described in the Samples section). In addition, studies have shown an association between higher levels of adolescent ND and heavier smoking trajectory patterns.26,27 Based on work providing evidence that common single nucleotide polymorphisms (SNPs) explain a large proportion of the heritability in height, Crohn’s disease, bipolar disorder, and type I diabetes,28,29 we hypothesize that common SNPs also explain a considerable proportion of the variation in smoking quantity and ND. Although it is highly likely rare variants play an equally important role in these behaviors, the purpose of this study was to examine common variants from available datasets. The goal of this study was to utilize set-based approaches to test for association between SNPs within selected genes and nicotine behaviors. We applied SNP-set based methods as a way to increase power to detect associations, and organize associations in a biologically meaningful way.30,31 Associations can be more easily detected by grouping SNPs into sets, and insights into their combined effects on biological function are more apparent.
Upon chronic exposure to nicotine, nAChRs undergo an upregulation of receptor number32–34 that is independent of upregulation of CHRN mRNA.35 Although there are many theories describing how this upregulation occurs, including increased receptor trafficking,36 decreased subunit degradation,37,38 increased nAChR subunit maturation and folding,39–41 pharmacological chaperoning by nicotine,42 and increased translation and second messenger signaling,43 researchers have yet to test several of the proteins suggested to impact upregulation in concert. Studies have demonstrated that nicotine-induced nAChR upregulation leads to nicotine sensitization in brain regions known to play a role in addiction and reward.44 Furthermore, nAChR upregulation has been well-documented in rodents exposed to nicotine during prenatal development45–51 and the long term and widespread consequences of prenatal nicotine exposure have been proposed to play a role in various neurobehavioral and physiological disorders.52 We posit that gene products are involved in nAChR upregulation through their interaction with nAChRs, and thus play a role in the development of ND.
We have assembled a list of genes that encode proteins known to interact with nAChRs or are known to play a role in their downstream signaling, by curating the literature, and conversing with experts (listed in the acknowledgements) in the field (Figure 1). The goal of this study was to assess the evidence for association with each gene and CPD and ND in African and European Americans (AAs and EAs, respectively). These analyses were carried out in several studies in which genome-wide genotyping was available and results were later combined as a meta-analysis. Although many studies use smoking quantity as a proxy for ND, research has demonstrated unique genetic effects on CPD, suggesting that a smoking quantity measure may not serve as a simple proxy for the genetic influences on ND.54 Thus, while these phenotypes may be correlated, they represent different aspects of smoking behavior and exposure. Based on these findings, both CPD and ND were used as phenotypes in the present study.
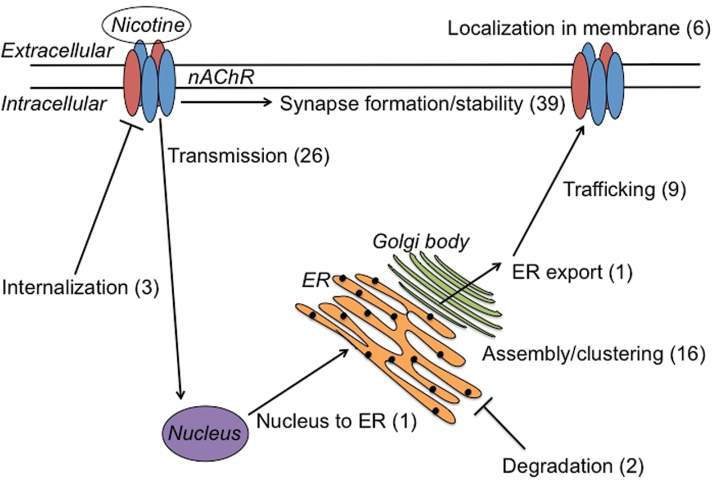
Figure 1.
Cellular processes proposed to play a role in nAChR upregulation; the number of genes from our curated list of 96 that play a role in each process is shown in parentheses (several genes were proposed to be involved in more than one cellular process). ER = endoplasmic reticulum. Modified from Melroy-Greif et al.53
Methods
Samples
Four independent samples with genome-wide genotype data and smoking phenotypes were included in the study (Table 1): (1) the Study of Addiction: Genetics and Environment (SAGE),55 (2) COPDGene,25 (3) the Multi-Ethnic Study of Atherosclerosis (MESA),56 and (4) an in-house sample comprised of unrelated Caucasian subjects from the Colorado Center on Antisocial Drug Dependence and the Genetics of Antisocial Drug Dependence (CADD-GADD).57 The SAGE sample is comprised of three substudies recruited via studies on cocaine, alcohol, or ND.55 The COPDGene study consists of smokers who have smoked at least 10 pack-years.25 MESA is a population-based study designed to investigate the characteristics of subclinical cardiovascular disease.56 Finally, the CADD-GADD sample is a mixture of various samples including probands targeted for drug use, their families and matched control families, as well as community twin samples. Additional details about the CADD-GADD sample recruitment and assessments are provided in Derringer et al.57 Study-level principal components (PCs) were supplied along with the SAGE, COPDGene, and CADD-GADD genotype data. PCs were computed in the MESA sample using shellfish (http://www.stats.ox.ac.uk/~davison/software/shellfish/shellfish.php). All genotype data were imputed to Phase 1 of the 1000 Genomes dataset58 and cleaned using standard quality control procedures: SNPs with low imputation accuracy (<0.9), low minor allele frequency (MAF) (<0.01), and out of Hardy–Weinberg equilibrium (HWE, P < .001) were excluded.
Table 1.
Study and Sample Characteristics
| Study | N CPD | Agea | Sex (% male) | CPDa | N FTND | Agea | Sex (% male) | FTNDa | |
|---|---|---|---|---|---|---|---|---|---|
| EA | COPDGene | 6670 | 62.09±8.84 | 52.37 | 25.84±11.44 | 2568 | 57.50±7.86 | 53.35 | 4.83±2.42 |
| SAGE | 1255 | 35.74±6.92 | 37.83 | 25.90±19.94 | 1673 | 37.53±8.65 | 43.93 | 2.99±3.29 | |
| CADD-GADD | 588 | 23.44±4.40 | 73.98 | 17.96±11.54 | NA | NA | NA | NA | |
| MESA | 628 | 63.03±9.95 | 56.53 | 20.59±21.66 | NA | NA | NA | NA | |
| AA | COPDGene | 3300 | 54.68±7.21 | 55.94 | 21.30±10.41 | 2567 | 53.34±6.04 | 57.78 | 4.93±2.34 |
| SAGE | 594 | 39.30±6.81 | 47.47 | 23.64±17.39 | 779 | 40.03±7.13 | 49.81 | 3.92±2.87 | |
| MESA | 53 | 63.64±9.91 | 49.06 | 12.77±11.10 | NA | NA | NA | NA |
NA = not applicable.
aMean ± standard deviation.
We analyzed two phenotypes when available from each study: quantitative ND symptoms as assessed by the Fagerström Test for Nicotine Dependence (FTND),59 and CPD, binned as follows: 0–10, 11–20, 21–30, 31–40, and 41 or more CPD. CPD was only assessed in two of the substudies from SAGE whereas FTND was collected in all substudies. Likewise, FTND was only assessed in current smokers in the COPDGene sample. FTND was not available in the MESA or CADD-GADD samples. Only subjects who had smoked 100 cigarettes or more in their lifetime were included in the analysis.
Analyses
SNPs were annotated to our genes of interest using the hg19 build. SNPs 20kb up- and down-stream of each gene were included, given reports that the majority of genetic variants that influence expression are located within 20kb of a gene.60 Only SNPs common across all datasets were evaluated. Among the EAs, this was limited to imputed SNPs from the SAGE, COPDGene, MESA, and CADD-GADD samples. Similarly, for the AA analyses, only SNPs common across the AA SAGE, MESA, and COPDGene samples were included. Age, sex, and the first five PCs from each study were included as covariates in each analysis. Additionally, substudy was used as a covariate in the SAGE analysis.
Joint Association of Genetic Variants
JAG61 (available at http://ctglab.nl/software/jag/) uses raw data and invokes PLINK (http://pngu.mgh.harvard.edu/~purcell/plink/index.shtml)62 to run a genome-wide association on each SNP included in the gene. A multivariate SNP test statistic is calculated by summing the −log10 of each SNP P-value and the empirical P-value for each gene is calculated by summing the −log10(P-value) for each permutation of the phenotype. The empirical P-value (Pemp) represents the number of times the Σ−log10(P-value) exceeds or equals the −log10(P-value) from the genome-wide association. The Pemp based on the Σ−log10(P-value) test statistic tests the hypothesis that, given the linkage disequilibrium (LD) structure of the included SNPs, the multivariate pattern of P-values of all SNPs in a gene is significantly different than what is expected under the null hypothesis of no association. Ten thousand permutations were performed for each test. This analysis was applied to our data in order to test the association between individual genes and CPD and FTND.
Meta-analysis
The Pemp for each gene was combined across studies using the weighted Z-score method.63 False discovery rate (FDR) corrected P-values were generated for each meta-analyzed P-value using the stats package in R.64
Replication
Summary P-values for CPD were used from the Tobacco and Genetics (TAG) Consortium65 as a replication. These data are freely available at http://www.med.unc.edu/pgc/downloads. MAGMA,66 a tool that can perform a gene-based test on summary SNP P-values, was used to test for association between genes trending toward association with CPD in EAs after meta-analysis. Briefly, SNPs in the 1000 Genomes CEU data were annotated to genes using the hg19 build, including SNPs 20kb up- and down-stream of each gene. A gene-based test was run on 17 genes: CHRNA3, CHRNA5, CHRNB4, NCAM1, CHRNE, DNAJA3, IQGAP1, MAPRE1, UNC50, CHRNB1, EPHA4, NRXN1, LRP4, CRELD2, CAMK2A, ITGA7, and APC. The 1000 Genomes CEU data was used as a reference panel for LD as evaluation of the gene test-statistic requires LD estimation between SNPs in the gene. Similar to the JAG analysis, MAGMA uses a SNP-wise model in which the SNP P-values are converted by taking the −log10 of the P-value and combined by computing their sum to obtain a gene-based P-value.
Results
Study and sample characteristics for the COPDGene, SAGE, MESA, and CADD-GADD samples are given in Table 1. Subjects in the COPDGene and MESA samples were the oldest and the CADD-GADD the youngest. EAs in the COPDGene and SAGE had similar smoking patterns, as did AAs in the COPDGene and SAGE samples. The AAs had somewhat lower CPD and higher FTND than their EA counterparts. CPD and FTND were correlated at 0.74 and 0.58 in the SAGE EA and AA samples, respectively. CPD and FTND were less correlated in the COPDGene EA and AA samples (0.37 and 0.35, respectively), likely due to FTND assessment in current smokers only.
Of the 112 total genes in our list (96 genes collected from the literature and 16 nAChR subunit genes), 100 genes had SNPs annotated and were used in the analysis in AAs, and 101 genes had SNPs annotated and were used in the EA analysis.
CPD and ND in EAs
Thirty-three thousand four hundred eighty-three SNPs from our gene set were tested for association with CPD in EAs from the COPDGene, SAGE, MESA, and CADD-GADD samples. The gene P-values were combined using the weighted Z-score method. Although 17 genes were trending at P < .1 (and used in the subsequent replication analysis), only CHRNA3, CHRNA5, and CHRNB4 were associated with CPD with controlling FDR at P < .05 (Table 2).
Table 2.
Results From the JAG Test in EAs
| Gene | No. of SNPs | No. of effective SNPsa | COPDGene Pemp | SAGE Pemp | MESA Pemp | CADD-GADD Pemp | Combined Pemp | FDR corrected P |
|---|---|---|---|---|---|---|---|---|
| CPD | ||||||||
| CHRNA3 | 102 | 2 | .0001 | .128 | .209 | .434 | .0001 | .013 |
| CHRNA5 | 29 | 1 | .0001 | .304 | .348 | .620 | .0001 | .013 |
| CHRNB4 | 17 | 3b | .0001 | .639 | .742 | .212 | .0001 | .013 |
| NCAM1 | 402 | 5c | .006 | .154 | .721 | .337 | .004 | .193 |
| CHRNE | 79 | 3 | .004 | .182 | .924 | .800 | .005 | .193 |
| DNAJA3 | 111 | 2 | .004 | .527 | .900 | .472 | .007 | .258 |
| IQGAP1 | 246 | 2 | .010 | .898 | .268 | .813 | .022 | .556 |
| MAPRE1 | 148 | 5 | .015 | .829 | .784 | .196 | .025 | .601 |
| UNC50 | 53 | 1 | .043 | .181 | .514 | .249 | .029 | .644 |
| CHRNB1 | 63 | 3 | .038 | .216 | .878 | .531 | .039 | .729 |
| EPHA4 | 212 | 5 | .133 | .073 | .238 | .105 | .064 | .782 |
| NRXN1 | 2433 | 22d | .022 | .992 | .516 | .493 | .064 | .782 |
| LRP4 | 106 | 3e | .089 | .178 | .599 | .441 | .070 | .782 |
| CRELD2 | 127 | 1f | .097 | .009 | .638 | .991 | .071 | .782 |
| CAMK2A | 89 | 5 | .178 | .059 | .034 | .174 | .075 | .782 |
| ITGA7 | 41 | 1 | .075 | .451 | .725 | .304 | .078 | .782 |
| APC | 231 | 2 | .061 | .245 | .876 | .920 | .079 | .782 |
| FTND | ||||||||
| CHRNA3 | 102 | 2 | .003 | .028 | NA | NA | .00042 | .042 |
| CHRNA5 | 29 | 1 | .012 | .060 | NA | NA | .003 | .193 |
| CHRNB4 | 17 | 2 | .012 | .080 | NA | NA | .004 | .193 |
| CHRNA9 | 239 | 4 | .009 | .221 | NA | NA | .008 | .280 |
| DNAJA3 | 111 | 2 | .008 | .338 | NA | NA | .012 | .381 |
| CHRNE | 79 | 3 | .218 | .004 | NA | NA | .017 | .482 |
| EPHA4 | 212 | 5 | .116 | .112 | NA | NA | .048 | .782 |
| CAMK2G | 61 | 2 | .008 | .794 | NA | NA | .056 | .782 |
| SVEP1 | 598 | 13 | .263 | .035 | NA | NA | .064 | .782 |
| CAMK2A | 89 | 5 | .368 | .013 | NA | NA | .067 | .782 |
| ERBB2 | 64 | 1 | .049 | .458 | NA | NA | .075 | .782 |
| RAPSN | 65 | 2 | .253 | .056 | NA | NA | .077 | .782 |
| CRELD2 | 127 | 1 | .237 | .073 | NA | NA | .081 | .782 |
| LRP4 | 106 | 3g | .106 | .269 | NA | NA | .084 | .782 |
The P-value is shown for each study as well as the combined P-value for genes associated with CPD or FTND at P < .1.
aThe number of signals tested in the gene in each study (unless otherwise noted) as calculated by JAG.
bOnly two signals calculated in the COPDGene sample.
cSix signals calculated in the COPDGene sample.
dTwenty-three signals calculated in the CADD-GADD and MESA samples.
eTwo signals calculated in the SAGE sample.
fTwo signals calculated in the MESA sample.
gTwo signals calculated in SAGE.
Thirty-three thousand four hundred eighty-three SNPs were tested for association with FTND in EAs from the COPDGene and SAGE samples. The gene P-values were combined using the weighted Z-score method. Similar to the results with CPD, while several genes were associated at P < .05, only the association with CHRNA3 survived correction for multiple testing at FDR P < .05 (Table 2).
CPD and ND in AAs
Thirty-eight thousand seven hundred eighty-seven SNPs from our set of genes were tested for association with CPD in AAs from the COPDGene, SAGE, and MESA samples and the gene P-values combined using the weighted Z-score method. Five genes were associated with CPD at P < .1 but none of these associations survived correction for multiple testing at FDR P < .05 (Table 3).
Table 3.
Results From the JAG Test in AAs
| Gene | No. of SNPs | No. of effective SNPsa | COPDGene Pemp | SAGE Pemp | MESA Pemp | Combined Pemp | FDR corrected P |
|---|---|---|---|---|---|---|---|
| CPD | |||||||
| CHRNA9 | 247 | 12b | .005 | .321 | .424 | .004 | .193 |
| PICK1 | 29 | 4c | .067 | .082 | .043 | .040 | .729 |
| NTRK2 | 443 | 19d | .031 | .824 | .667 | .048 | .782 |
| DLG4 | 7 | 2 | .080 | .458 | .841 | .083 | .782 |
| CAMK2B | 50 | 3e | .092 | .504 | .877 | .098 | .799 |
| FTND | |||||||
| IQGAP1 | 400 | 7 | .020 | .016 | NA | .005 | .193 |
| PDIA3 | 120 | 5 | .011 | .701 | NA | .021 | .556 |
| UNC50 | 56 | 2 | .019 | .649 | NA | .031 | .653 |
| UBXN2A | 271 | 5 | .012 | .918 | NA | .040 | .729 |
| EPHA4 | 221 | 10 | .028 | .791 | NA | .055 | .782 |
| NLGN1 | 1668 | 36 | .062 | .493 | NA | .070 | .782 |
| NCAM1 | 427 | 10f | .106 | .170 | NA | .071 | .782 |
| FYN | 635 | 15 | .085 | .406 | NA | .084 | .782 |
| PPP3CA | 408 | 12 | .068 | .607 | NA | .089 | .799 |
| LAMB1 | 187 | 10 | .156 | .111 | NA | .093 | .799 |
| CACNA2D1 | 1325 | 40g | .077 | .567 | NA | .094 | .799 |
The P-value for each gene is shown for each study as well as the combined P-value for genes associated with CPD or FTND at P < .1.
aThe number of signals tested in the gene in each study (unless otherwise noted) as calculated by JAG.
bEleven signals in the MESA data.
cThree signals in the MESA data.
dEighteen signals in the MESA data.
eTwo signals in the MESA data.
fNine signals in the SAGE data.
gThirty-nine signals in the SAGE data.
The same set of SNPs was tested for association with FTND and the gene P-values combined in AAs from the SAGE and COPDGene samples. More genes were nominally associated with FTND at P < .1 than CPD, but none survived correction for multiple testing at FDR P < .05 (Table 3).
Replication
Gene-based tests on summary P-values for CPD from the TAG data were run on 17 genes (CHRNA3, CHRNA5, CHRNB4, NCAM1, CHRNE, DNAJA3, IQGAP1, MAPRE1, UNC50, CHRNB1, EPHA4, NRXN1, LRP4, CRELD2, CAMK2A, ITGA7, and APC). No gene was trending toward association at P < .1, with the exception of CHRNA3, CHRNA5, and CHRNB4, which were highly associated at P < 5E−14 (results not shown).
Discussion
Given the highly replicated associations with CHRN genes and smoking behaviors, we examined 96 other genes, known to interact with nAChRs, to test for association in four existing samples for which genome-wide data and phenotypic measures were available. As expected, the CHRN gene cluster on chromosome 15q25 comprised of CHRNA3, CHRNB4, and CHRNA5 showed the strongest associations with nicotine behaviors in EAs in this study, replicating previous associations with these genes and CPD6,65,67–76 and ND.7,10,13,18,77–87 Although there was some overlap between the genes nominally associated with CPD and FTND (CHRNE, DNAJA3, CRELD2, CAMK2A, and EPHA4), no genes were associated after correction for multiple testing at FDR P < .05. Similarly, while primarily driven by the largest sample, COPDGene, two genes were trending toward association in more than one study: CRELD2 and CAMK2A.
The TAG results mirror our own study in that only the gene-wise P-values for CHRNA3/A5/B4 were highly significant. Although no other genes survived correction for multiple testing in our study, we would hypothesize that some of the genes would have been trending toward association with CPD in the TAG data. However, there are some notable differences between the original and replication analyses. First, we limited the analyses to SNPs annotated on all datasets used in our study, while additional signals were likely used in the replication sample, and this may have washed out associated SNPs. Second, there likely exist differences in sample ascertainment and phenotypic variables between the four samples used in our meta-analysis and those in TAG.
Unlike in EAs, no genes, particularly CHRNA3/A5/B4, were associated with CPD or FTND after controlling FDR at P < .05 in AAs. However, two genes showed consistent results across studies and bear future investigation: PICK1 and IQGAP1. None of the cluster genes were associated with CPD in AAs from the SAGE, MESA, or COPDGene samples. This result is perhaps not surprising given previous results with CPD in AAs in the COPDGene sample.76 However, a previous study using over 32000 AAs found an association between CHRNA3/A5/B4 with CPD. In that study, only one SNP, residing in the 5′-distal enhancer region of CHRNA5, exceeded genome-wide significance.88 It is possible that since our analyses sum individual SNP statistics to get an overall gene P-value this SNP was overshadowed by SNPs of lower significance. In our study, CHRNA5 was nominally associated with FTND in the SAGE sample using an alternative set-based test implemented in PLINK62 at P = .094, but this association was not replicated in the COPDGene sample. Although this suggests heterogeneity between the two samples, it aligns with what has been previously seen in the literature. Saccone et al82 reported an association between SNPs in CHRNA5 and ND in AAs alone in COGEND, yet no association between CHRNA3, CHRNB4, or CHRNA5 with ND was seen in the COPDGene sample.76
Although not significant after correction for multiple testing, several genes previously associated with substance use phenotypes in the literature were nominally associated with nicotine behaviors in the present study and will be discussed briefly here in an attempt to guide future replication efforts. EPHA4, nominally associated with CPD and FTND in EAs, has been associated with opioid dependence in subjects with a history of cocaine dependence.89CHRNA9 was nominally associated with FTND in EAs and CPD in AAs and has been previously associated with ND in female Israeli students,90 as well as with lung cancer in Caucasians.91 Recently, rare and common variation in CHRNA9 has been associated with smoking status in EAs and AAs from the Mid-South Tobacco Case-Control study.92
It is interesting there was very little overlap in the genes associated with CPD and FTND between EAs and AAs. These findings might indicate underlying genetic differences in the development of smoking behaviors in EAs and AAs. AAs tend to start smoking later in life93–96 and have a lower lifetime prevalence of ND compared to EAs.97 Additionally, AAs report higher cravings and more pleasurable sensations after smoking,96,98 lower rates of regular smoking,94–97,99–103 higher nicotine intake per cigarette,104 and slower metabolism rates of nicotine103–107 compared to EAs. Finally, EAs have higher smoking cessation rates than AAs.99 Together with our data, this suggests differences in smoking patterns between EAs and AAs may be partially genetically driven.
Although there was significant overlap in the genes implicated in ND and CPD in EAs, our results show little overlap in the genes associated with CPD and FTND in AAs. Given previous studies demonstrating both unique and common genetic influences on CPD and ND,54 we expected to see some overlap between the two phenotypes. This discrepancy in AAs may be due to the fact that FTND was only measured in current smokers in the COPDGene sample and our results could reflect differences in smoking cessation as well. The AA sample was also considerably smaller than the EA sample; thus these analyses had lower power to detect genetic effects and a higher likelihood of false negatives.
While not without limitations, several strengths of the present study help mitigate these limitations. JAG has higher power than several other pathway algorithms. Specifically, using 1694 cases and 2917 controls from the Wellcome Trust Case Control Consortium the gene-based test in JAG had better power than other algorithms to detect known genes associated with Crohn’s Disease.61 SNP-set based approaches can increase power to detect genetic loci with individually small effects by consolidating SNP associations and can help to prioritize associations based on biological relevance.30,31 A meta-analysis approach was used as opposed to a mega-analysis so associations with genes across studies could be compared, although differences in age and study ascertainment may have clouded these comparisons. By curating a unique list of genes to test, we avoided many potential limitations when examining already curated gene sets from the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO); for example, when examining gene sets from KEGG or GO, genes that have been well studied are more thoroughly annotated. As of 2011, only about 5000 human genes had been annotated to KEGG pathways.31 However, despite being able to curate a unique set of genes, this study was limited to testing SNPs both annotated on the hg19 build and imputed within each sample. In addition, trans effects were ignored by necessity. Similarly, while both JAG and MAGMA take LD within a gene into account when computing the test statistic,61,66 LD between genes was not taken into account. Finally, gene-based tests do not provide an effect size of the gene so the weighted Z-score method was used to meta-analyze the results. The direction of effect of each individual SNP is thus ignored and the resulting P-value differs from that of a traditional meta-analysis where small effects of opposite direction cancel each other out. To summarize, the direction of effect for each signal in an associated gene would be unknown as well as which signals are driving the association.
In conclusion, we tested several genes believed to be involved in nAChR upregulation with FTND and CPD and performed a meta-analysis over four independent samples. The list of genes tested is by no means an exhaustive list, and simply served to test the hypothesis that SNPs in the identified genes are associated with nicotine behaviors by altering nAChR function and/or expression through protein–protein interactions. Although no genes were associated after correction for multiple testing, except for the reliably replicated CHRNA3/A5/B4 association, it is possible these genes may nevertheless be affecting nAChR upregulation in a manner not directly driven by SNPs; for example, increased transcription factor binding, or enhancer effects. Future studies could assess trans effects in these genes, perform functional bioinformatics, or examine gene-by-gene interactions among this list. This work represents collaboration between neuroscience experts and statistical geneticists and although it did not bear fruit with respect to the current study, future collaborations will continue to be important to identify and characterize novel genetic associations.
Funding
This work was supported by grants from the National Institute on Alcohol Abuse and Alcoholism (R01 AA017889 and T32 AA013525); the National Institute on Drug Abuse (T32 DA017637); and the National Heart, Lung, and Blood Institute of the National Institutes of Health (NIH) (K01 HL125858). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The authors have no conflicts of interest to report. Funding sources for the datasets used in this study are listed as follows. CADD-GADD: Supported by NIH grants from the National Institute on Drug Abuse (DA011015 and DA012845). COPDGene: Supported by NIH grants (U01HL089897 and U01HL089856) from the National Heart, Lung, and Blood Institute. Use of this data was approved by the COPDGene Ancillary Studies and Publications Committee. MESA: Supported by contracts N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168 and N01-HC-95169, from the National Heart, Lung, and Blood Institute, and by grants (UL1-TR-000040, and UL1-TR-001079) from the National Center for Research Resources. These data were obtained through dbGaP. SAGE: Supported by NIH grants from the National Institute on Alcohol abuse and Alcoholism (AA008401); the National Institute on Drug Abuse (DA013423); and the National Cancer Institute (CA089392). These data were obtained through dbGaP.
Declaration of Interests
None declared.
Acknowledgments
The authors would like to thank Drs. Millet Treinin, Henry Lester, Marina Picciotto, Roger Papke, and Jon Lindstrom for their suggestions and expertise regarding our nAChR-associated gene set. Additionally, the authors would like to thank Dr Jaime Derringer for her work on the CADD-GADD dataset, as well as Dr Esther Lips for her invaluable assistance running JAG and performing the meta-analyses.
References
- 1. U.S. Department of Health and Human Services. The Health Consequences of Smoking—50 Years of Progress: A Report of the Surgeon General. Atlanta, GA: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Chronic Disease Prevention and Health Promotion, Office on Smoking and Health; 2014. [Google Scholar]
- 2. Gunby P. Surgeon General emphasizes nicotine addiction in annual report on tobacco use, consequences. JAMA. 1988;259(19):2811. [PubMed] [Google Scholar]
- 3. Stolerman IP, Jarvis MJ. The scientific case that nicotine is addictive. Psychopharmacology (Berl). 1995;117(1):2–10; discussion 14–20. [DOI] [PubMed] [Google Scholar]
- 4. Gotti C, Clementi F, Fornari A, et al. Structural and functional diversity of native brain neuronal nicotinic receptors. Biochem Pharmacol. 2009;78(7):703–711. [DOI] [PubMed] [Google Scholar]
- 5. Berrettini WH, Doyle GA. The CHRNA5-A3-B4 gene cluster in nicotine addiction. Mol Psychiatry. 2012;17(9):856–866. [DOI] [PubMed] [Google Scholar]
- 6. Thorgeirsson TE, Gudbjartsson DF, Surakka I, et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010;42(5):448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bierut LJ, Madden PA, Breslau N, et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet. 2007;16(1):24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Saccone SF, Hinrichs AL, Saccone NL, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16(1):36–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hoft NR, Corley RP, McQueen MB, Schlaepfer IR, Huizinga D, Ehringer MA. Genetic association of the CHRNA6 and CHRNB3 genes with tobacco dependence in a nationally representative sample. Neuropsychopharmacology. 2009;34(3):698–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Saccone NL, Saccone SF, Hinrichs AL, et al. Multiple distinct risk loci for nicotine dependence identified by dense coverage of the complete family of nicotinic receptor subunit (CHRN) genes. Am J Med Genet B Neuropsychiatr Genet. 2009;150B(4):453–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Saccone NL, Schwantes-An TH, Wang JC, et al. Multiple cholinergic nicotinic receptor genes affect nicotine dependence risk in African and European Americans. Genes Brain Behav. 2010;9(7):741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rice JP, Hartz SM, Agrawal A, et al. CHRNB3 is more strongly associated with Fagerstrom test for cigarette dependence-based nicotine dependence than cigarettes per day: phenotype definition changes genome-wide association studies results. Addiction. 2012;107(11):2019–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Haller G, Druley T, Vallania FL, et al. Rare missense variants in CHRNB4 are associated with reduced risk of nicotine dependence. Hum Mol Genet. 2012;21(3):647–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Han S, Yang BZ, Kranzler HR, Oslin D, Anton R, Gelernter J. Association of CHRNA4 polymorphisms with smoking behavior in two populations. Am J Med Genet B Neuropsychiatr Genet. 2011;156B(4):421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen HI, Shinkai T, Utsunomiya K, et al. Possible association of nicotinic acetylcholine receptor gene (CHRNA4 and CHRNB2) polymorphisms with nicotine dependence in Japanese males: an exploratory study. Pharmacopsychiatry. 2013;46(2):77–82. [DOI] [PubMed] [Google Scholar]
- 16. Kamens HM, Corley RP, McQueen MB, et al. Nominal association with CHRNA4 variants and nicotine dependence. Genes Brain Behav. 2013;12(3):297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li MD, Beuten J, Ma JZ, et al. Ethnic- and gender-specific association of the nicotinic acetylcholine receptor alpha4 subunit gene (CHRNA4) with nicotine dependence. Hum Mol Genet. 2005;14(9):1211–1219. [DOI] [PubMed] [Google Scholar]
- 18. Wessel J, McDonald SM, Hinds DA, et al. Resequencing of nicotinic acetylcholine receptor genes and association of common and rare variants with the Fagerström test for nicotine dependence. Neuropsychopharmacology. 2010;35(12):2392–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xie P, Kranzler HR, Krauthammer M, et al. Rare nonsynonymous variants in alpha-4 nicotinic acetylcholine receptor gene protect against nicotine dependence. Biol Psychiatry. 2011;70(6):528–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kendler KS, Neale MC, Sullivan P, Corey LA, Gardner CO, Prescott CA. A population-based twin study in women of smoking initiation and nicotine dependence. Psychol Med. 1999;29(2):299–308. [DOI] [PubMed] [Google Scholar]
- 21. True WR, Xian H, Scherrer JF, et al. Common genetic vulnerability for nicotine and alcohol dependence in men. Arch Gen Psychiatry. 1999;56(7):655–661. [DOI] [PubMed] [Google Scholar]
- 22. Lessov CN, Martin NG, Statham DJ, et al. Defining nicotine dependence for genetic research: evidence from Australian twins. Psychol Med. 2004;34(5):865–879. [DOI] [PubMed] [Google Scholar]
- 23. Vink JM, Beem AL, Posthuma D, et al. Linkage analysis of smoking initiation and quantity in Dutch sibling pairs. Pharmacogenomics J. 2004;4(4):274–282. [DOI] [PubMed] [Google Scholar]
- 24. Broms U, Silventoinen K, Madden PA, Heath AC, Kaprio J. Genetic architecture of smoking behavior: a study of Finnish adult twins. Twin Res Hum Genet. 2006;9(1):64–72. [DOI] [PubMed] [Google Scholar]
- 25. Regan EA, Hokanson JE, Murphy JR, et al. Genetic epidemiology of COPD (COPDGene) study design. COPD. 2010;7(1):32–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Karp I, O’Loughlin J, Paradis G, Hanley J, Difranza J. Smoking trajectories of adolescent novice smokers in a longitudinal study of tobacco use. Ann Epidemiol. 2005;15(6):445–452. [DOI] [PubMed] [Google Scholar]
- 27. Lessov-Schlaggar CN, Hops H, Brigham J, et al. Adolescent smoking trajectories and nicotine dependence. Nicotine Tob Res. 2008;10(2):341–351. [DOI] [PubMed] [Google Scholar]
- 28. Yang J, Benyamin B, McEvoy BP, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42(7):565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet. 2011;88(3):294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Purcell SM, Wray NR, Stone JL, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460(7256):748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang L, Jia P, Wolfinger RD, Chen X, Zhao Z. Gene set analysis of genome-wide association studies: methodological issues and perspectives. Genomics. 2011;98(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Marks MJ, Burch JB, Collins AC. Effects of chronic nicotine infusion on tolerance development and nicotinic receptors. J Pharmacol Exp Ther. 1983;226(3):817–825. [PubMed] [Google Scholar]
- 33. Schwartz RD, Kellar KJ. Nicotinic cholinergic receptor binding sites in the brain: regulation in vivo. Science. 1983;220(4593):214–216. [DOI] [PubMed] [Google Scholar]
- 34. Benwell ME, Balfour DJ, Anderson JM. Evidence that tobacco smoking increases the density of (−)-[3H]nicotine binding sites in human brain. J Neurochem. 1988;50(4):1243–1247. [DOI] [PubMed] [Google Scholar]
- 35. Marks MJ, Pauly JR, Gross SD, et al. Nicotine binding and nicotinic receptor subunit RNA after chronic nicotine treatment. J Neurosci. 1992;12(7):2765–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Darsow T, Booker TK, Piña-Crespo JC, Heinemann SF. Exocytic trafficking is required for nicotine-induced up-regulation of alpha 4 beta 2 nicotinic acetylcholine receptors. J Biol Chem. 2005;280(18):18311–18320. [DOI] [PubMed] [Google Scholar]
- 37. Rezvani K, Teng Y, Shim D, De Biasi M. Nicotine regulates multiple synaptic proteins by inhibiting proteasomal activity. J Neurosci. 2007;27(39):10508–10519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rezvani K, Teng Y, Pan Y, et al. UBXD4, a UBX-containing protein, regulates the cell surface number and stability of alpha3-containing nicotinic acetylcholine receptors. J Neurosci. 2009;29(21):6883–6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Harkness PC, Millar NS. Changes in conformation and subcellular distribution of alpha4beta2 nicotinic acetylcholine receptors revealed by chronic nicotine treatment and expression of subunit chimeras. J Neurosci. 2002;22(23):10172–10181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nashmi R, Dickinson ME, McKinney S, et al. Assembly of alpha4beta2 nicotinic acetylcholine receptors assessed with functional fluorescently labeled subunits: effects of localization, trafficking, and nicotine-induced upregulation in clonal mammalian cells and in cultured midbrain neurons. J Neurosci. 2003;23(37):11554–11567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sallette J, Pons S, Devillers-Thiery A, et al. Nicotine upregulates its own receptors through enhanced intracellular maturation. Neuron. 2005;46(4):595–607. [DOI] [PubMed] [Google Scholar]
- 42. Henderson BJ, Lester HA. Inside-out neuropharmacology of nicotinic drugs. Neuropharmacology. 2015;96(Pt B):178–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gopalakrishnan M, Molinari EJ, Sullivan JP. Regulation of human alpha4beta2 neuronal nicotinic acetylcholine receptors by cholinergic channel ligands and second messenger pathways. Mol Pharmacol. 1997;52(3):524–534. [PubMed] [Google Scholar]
- 44. Govind AP, Vezina P, Green WN. Nicotine-induced upregulation of nicotinic receptors: underlying mechanisms and relevance to nicotine addiction. Biochem Pharmacol. 2009;78(7):756–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hagino N, Lee JW. Effect of maternal nicotine on the development of sites for [(3)H]nicotine binding in the fetal brain. Int J Dev Neurosci. 1985;3(5):567–571. [DOI] [PubMed] [Google Scholar]
- 46. Navarro HA, Seidler FJ, Eylers JP, et al. Effects of prenatal nicotine exposure on development of central and peripheral cholinergic neurotransmitter systems. Evidence for cholinergic trophic influences in developing brain. J Pharmacol Exp Ther. 1989;251(3):894–900. [PubMed] [Google Scholar]
- 47. Popke EJ, Tizabi Y, Rahman MA, Nespor SM, Grunberg NE. Prenatal exposure to nicotine: effects on prepulse inhibition and central nicotinic receptors. Pharmacol Biochem Behav. 1997;58(4):843–849. [DOI] [PubMed] [Google Scholar]
- 48. Slotkin TA, Orband-Miller L, Queen KL. Development of [3H]nicotine binding sites in brain regions of rats exposed to nicotine prenatally via maternal injections or infusions. J Pharmacol Exp Ther. 1987;242(1):232–237. [PubMed] [Google Scholar]
- 49. Tizabi Y, Perry DC. Prenatal nicotine exposure is associated with an increase in [125I]epibatidine binding in discrete cortical regions in rats. Pharmacol Biochem Behav. 2000;67(2):319–323. [DOI] [PubMed] [Google Scholar]
- 50. Tizabi Y, Popke EJ, Rahman MA, Nespor SM, Grunberg NE. Hyperactivity induced by prenatal nicotine exposure is associated with an increase in cortical nicotinic receptors. Pharmacol Biochem Behav. 1997;58(1):141–146. [DOI] [PubMed] [Google Scholar]
- 51. van de Kamp JL, Collins AC. Prenatal nicotine alters nicotinic receptor development in the mouse brain. Pharmacol Biochem Behav. 1994;47(4):889–900. [DOI] [PubMed] [Google Scholar]
- 52. Abbott LC, Winzer-Serhan UH. Smoking during pregnancy: lessons learned from epidemiological studies and experimental studies using animal models. Crit Rev Toxicol. 2012;42(4):279–303. [DOI] [PubMed] [Google Scholar]
- 53. Melroy-Greif WE, Stitzel JA, Ehringer MA. Nicotinic acetylcholine receptors: upregulation, age-related effects and associations with drug use. Genes Brain Behav. 2016;15(1):89–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Haberstick BC, Timberlake D, Ehringer MA, et al. Genes, time to first cigarette and nicotine dependence in a general population sample of young adults. Addiction. 2007;102(4):655–665. [DOI] [PubMed] [Google Scholar]
- 55. Bierut LJ, Agrawal A, Bucholz KK, et al. A genome-wide association study of alcohol dependence. Proc Natl Acad Sci U S A. 2010;107(11):5082–5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bild DE, Bluemke DA, Burke GL, et al. Multi-Ethnic Study of Atherosclerosis: objectives and design. Am J Epidemiol. 2002;156(9):871–881. [DOI] [PubMed] [Google Scholar]
- 57. Derringer J, Corley RP, Haberstick BC, et al. Genome-wide association study of behavioral disinhibition in a selected adolescent sample. Behav Genet. 2015;45(4):375–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Abecasis GR, Auton A, Brooks LD, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Heatherton TF, Kozlowski LT, Frecker RC, Fagerström KO. The Fagerström Test for Nicotine Dependence: a revision of the Fagerström Tolerance Questionnaire. Br J Addict. 1991;86(9):1119–1127. [DOI] [PubMed] [Google Scholar]
- 60. Veyrieras JB, Kudaravalli S, Kim SY, et al. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 2008;4(10):e1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lips ES, Kooyman M, de Leeuw C, Posthuma D. JAG: a computational tool to evaluate the role of gene-sets in complex traits. Genes (Basel). 2015;6(2):238–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Whitlock MC. Combining probability from independent tests: the weighted Z-method is superior to Fisher’s approach. J Evol Biol. 2005;18(5):1368–1373. [DOI] [PubMed] [Google Scholar]
- 64. R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2012. http://www.R-project.org. [Google Scholar]
- 65. Tobacco and Genetics Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42(5):441–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11(4):e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Berrettini W, Yuan X, Tozzi F, et al. Alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol Psychiatry. 2008;13(4):368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Stevens VL, Bierut LJ, Talbot JT, et al. Nicotinic receptor gene variants influence susceptibility to heavy smoking. Cancer Epidemiol Biomarkers Prev. 2008;17(12):3517–3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Caporaso N, Gu F, Chatterjee N, et al. Genome-wide and candidate gene association study of cigarette smoking behaviors. PLoS One. 2009;4(2):e4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Liu JZ, Tozzi F, Waterworth DM, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet. 2010;42(5):436–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Saccone NL, Culverhouse RC, Schwantes-An TH, et al. Multiple independent loci at chromosome 15q25.1 affect smoking quantity: a meta-analysis and comparison with lung cancer and COPD. PLoS Genet. 2010;6(8):e1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sarginson JE, Killen JD, Lazzeroni LC, et al. Markers in the 15q24 nicotinic receptor subunit gene cluster (CHRNA5-A3-B4) predict severity of nicotine addiction and response to smoking cessation therapy. Am J Med Genet B Neuropsychiatr Genet. 2011;156B(3):275–284. [DOI] [PubMed] [Google Scholar]
- 73. Sorice R, Bione S, Sansanelli S, et al. Association of a variant in the CHRNA5-A3-B4 gene cluster region to heavy smoking in the Italian population. Eur J Hum Genet. 2011;19(5):593–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Cannon DS, Mermelstein RJ, Hedeker D, et al. Effect of neuronal nicotinic acetylcholine receptor genes (CHRN) on longitudinal cigarettes per day in adolescents and young adults. Nicotine Tob Res. 2014;16(2):137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gabrielsen ME, Romundstad P, Langhammer A, Krokan HE, Skorpen F. Association between a 15q25 gene variant, nicotine-related habits, lung cancer and COPD among 56,307 individuals from the HUNT study in Norway. Eur J Hum Genet. 2013;21(11):1293–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Frederiksen B, Lutz S, Cho MH, et al. , for the COPDGene Investigators. Genome-wide association study of nicotine dependence in COPDGene. In: Society for Research on Nicotine and Tobacco’s 19th Annual Meeting; March 13–16; Boston, MA. [Google Scholar]
- 77. Thorgeirsson TE, Geller F, Sulem P, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452(7187):638–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Bierut LJ, Stitzel JA, Wang JC, et al. Variants in nicotinic receptors and risk for nicotine dependence. Am J Psychiatry. 2008;165(9):1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Weiss RB, Baker TB, Cannon DS, et al. A candidate gene approach identifies the CHRNA5-A3-B4 region as a risk factor for age-dependent nicotine addiction. PLoS Genet. 2008;4(7):e1000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Spitz MR, Amos CI, Dong Q, Lin J, Wu X. The CHRNA5-A3 region on chromosome 15q24-25.1 is a risk factor both for nicotine dependence and for lung cancer. J Natl Cancer Inst. 2008;100(21):1552–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Baker TB, Weiss RB, Bolt D, et al. Human neuronal acetylcholine receptor A5-A3-B4 haplotypes are associated with multiple nicotine dependence phenotypes. Nicotine Tob Res. 2009;11(7):785–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Saccone NL, Wang JC, Breslau N, et al. The CHRNA5-CHRNA3-CHRNB4 nicotinic receptor subunit gene cluster affects risk for nicotine dependence in African-Americans and in European-Americans. Cancer Res. 2009;69(17):6848–6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Chen X, Chen J, Williamson VS, et al. Variants in nicotinic acetylcholine receptors alpha5 and alpha3 increase risks to nicotine dependence. Am J Med Genet B Neuropsychiatr Genet. 2009;150B(7):926–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Sherva R, Kranzler HR, Yu Y, et al. Variation in nicotinic acetylcholine receptor genes is associated with multiple substance dependence phenotypes. Neuropsychopharmacology. 2010;35(9):1921–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Erlich PM, Hoffman SN, Rukstalis M, et al. Nicotinic acetylcholine receptor genes on chromosome 15q25.1 are associated with nicotine and opioid dependence severity. Hum Genet. 2010;128(5):491–499. [DOI] [PubMed] [Google Scholar]
- 86. Maes HH, Neale MC, Chen X, Chen J, Prescott CA, Kendler KS. A twin association study of nicotine dependence with markers in the CHRNA3 and CHRNA5 genes. Behav Genet. 2011;41(5):680–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Broms U, Wedenoja J, Largeau MR, et al. Analysis of detailed phenotype profiles reveals CHRNA5-CHRNA3-CHRNB4 gene cluster association with several nicotine dependence traits. Nicotine Tob Res. 2012;14(6):720–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. David SP, Hamidovic A, Chen GK, et al. Genome-wide meta-analyses of smoking behaviors in African Americans. Transl Psychiatry. 2012;2:e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Levran O, Peles E, Randesi M, et al. Synaptic plasticity and signal transduction gene polymorphisms and vulnerability to drug addictions in populations of European or African ancestry. CNS Neurosci Ther. 2015;21(11):898–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Greenbaum L, Kanyas K, Karni O, et al. Why do young women smoke? I. Direct and interactive effects of environment, psychological characteristics and nicotinic cholinergic receptor genes. Mol Psychiatry. 2006;11(3):312–322, 223. [DOI] [PubMed] [Google Scholar]
- 91. Chikova A, Bernard HU, Shchepotin IB, Grando SA. New associations of the genetic polymorphisms in nicotinic receptor genes with the risk of lung cancer. Life Sci. 2012;91(21–22):1103–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Yang J, Wang S, Yang Z, et al. The contribution of rare and common variants in 30 genes to risk nicotine dependence. Mol Psychiatry. 2015;20(11):1467–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Duncan AE, Lessov-Schlaggar CN, Sartor CE, Bucholz KK. Differences in time to onset of smoking and nicotine dependence by race/ethnicity in a Midwestern sample of adolescents and young adults from a high risk family study. Drug Alcohol Depend. 2012;125(1–2):140–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Chatila WM, Wynkoop WA, Vance G, Criner GJ. Smoking patterns in African Americans and whites with advanced COPD. Chest. 2004;125(1):15–21. [DOI] [PubMed] [Google Scholar]
- 95. White HR, Nagin D, Replogle E, Stouthamer-Loeber M. Racial differences in trajectories of cigarette use. Drug Alcohol Depend. 2004;76(3):219–227. [DOI] [PubMed] [Google Scholar]
- 96. Finkenauer R, Pomerleau CS, Snedecor SM, Pomerleau OF. Race differences in factors relating to smoking initiation. Addict Behav. 2009;34(12):1056–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Edens EL, Glowinski AL, Pergadia ML, Lessov-Schlaggar CN, Bucholz KK. Nicotine addiction in light smoking African American mothers. J Addict Med. 2010;4(1):55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Carter BL, Paris MM, Lam CY, et al. Real-time craving differences between black and white smokers. Am J Addict. 2010;19(2):136–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Hahn LP, Folsom AR, Sprafka JM, Norsted SW. Cigarette smoking and cessation behaviors among urban blacks and whites. Public Health Rep. 1990;105(3):290–295. [PMC free article] [PubMed] [Google Scholar]
- 100. Kabat GC, Morabia A, Wynder EL. Comparison of smoking habits of blacks and whites in a case-control study. Am J Public Health. 1991;81(11):1483–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Ellickson PL, Orlando M, Tucker JS, Klein DJ. From adolescence to young adulthood: racial/ethnic disparities in smoking. Am J Public Health. 2004;94(2):293–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Moolchan ET, Franken FH, Jaszyna-Gasior M. Adolescent nicotine metabolism: ethnoracial differences among dependent smokers. Ethn Dis. 2006;16(1):239–243. [PubMed] [Google Scholar]
- 103. Signorello LB, Cai Q, Tarone RE, McLaughlin JK, Blot WJ. Racial differences in serum cotinine levels of smokers. Dis Markers. 2009;27(5):187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Pérez-Stable EJ, Herrera B, Jacob P, III, Benowitz NL. Nicotine metabolism and intake in black and white smokers. JAMA. 1998;280(2):152–156. [DOI] [PubMed] [Google Scholar]
- 105. St Helen G, Dempsey D, Wilson M, Jacob P, III, Benowitz NL. Racial differences in the relationship between tobacco dependence and nicotine and carcinogen exposure. Addiction. 2013;108(3):607–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Benowitz NL, Dains KM, Dempsey D, Wilson M, Jacob P. Racial differences in the relationship between number of cigarettes smoked and nicotine and carcinogen exposure. Nicotine Tob Res. 2011;13(9):772–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Rubinstein ML, Shiffman S, Rait MA, Benowitz NL. Race, gender, and nicotine metabolism in adolescent smokers. Nicotine Tob Res. 2013;15(7):1311–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]