Abstract
This study examined Sal-like protein (SALL)1 methylation profiles in head and neck squamous-cell carcinoma (HNSCC) patients at diagnosis and follow-up, and evaluated their prognostic significance and value as a biomarker. SALL1 expression was examined in a panel of cell lines by quantitative reverse transcription PCR (qRT-PCR). Promoter methylation was determined by quantitative methylation-specific polymerase chain reaction (qMSP) and was compared to the clinical characteristics of 205 samples. SALL1 promoter methylation was associated with transcriptional inhibition and was correlated with disease recurrence in 31.7% of cases, with an odds ratio of 1.694 (95% confidence interval: 1.093-2.626; P = 0.018) by multivariate Cox proportional hazard regression analysis. SALL1 promoter hypermethylation showed highly discriminatory receiver operator characteristic curve profiles that clearly distinguished HNSCC from adjacent normal mucosal tissue, and was correlated with reduced disease-free survival in early stage T1 and T2 patients (log-rank test, P < 0.001). SALL1 methylation was significantly correlated with the methylation status of both SALL3 and CDH1. This study suggests that CpG hypermethylation is a likely mechanism of SALL1 gene inactivation, supporting the hypothesis that SALL1 might play a role in HNSCC tumorigenesis and could serve as an important biomarker.
Keywords: SALL1, methylation, early tumor stage patients, biomarker, HNSCC
Introduction
Head and neck squamous-cell carcinoma (HNSCC) is the sixth most frequently occurring type of cancer in the world 1. It is a highly heterogeneous disease that develops through one of two primary mechanisms: chemical carcinogenesis through exposure to tobacco and alcohol or virus-induced tumorigenesis 2. Surgical intervention is challenging for HNSCC, as this disease is associated with a limited surgical margin, as tumors are located close to vital tissues such as the central nervous system, carotid artery, trachea, and esophagus. In addition, despite aggressive, site-specific multimodal therapy, a significant proportion of patients will develop disease recurrence, with a risk of local failure of up to a 60% and a risk of distant failure of up to a 30% 3, 4. To improve the survival rate of HNSCC patients, it is necessary to identify reliable biomarkers that provide sufficient prognostic power for the effective clinical management of this disease.
The spalt (Sal) genes were first identified in Drosophila. Mice and humans have four known Sal-related genes named sall1 to sall4 and SALL1 to SALL4, respectively 5. Heterozygous early-truncating SALL1 is known to cause autosomal dominant Townes-Brocks syndrome, a malformation syndrome, without severe central nervous system involvement 6. Recent studies have investigated the association between SALL1 expression and carcinogenesis. One group demonstrated that the inhibition of SALL1 correlates with reduced levels of CDH1, an important contributor to epithelial-to-mesenchymal transition 7. Another report showed that SALL1 hypermethylation resulted in reduced levels of SALL1 mRNA in breast carcinoma 8, and aberrant hypermethylation of SALL1 was found to contribute to carcinogenesis in chronic lymphocytic leukemia 9. Despite accumulating evidence implicating this event in adenocarcinomas, the hypermethylation of SALL1 in SCCs of tissues such as head and neck, esophagus, lung, and cervix, needs to be explored.
We demonstrate that loss of SALL1 expression is associated with hypermethylation of key CpG sites within transcription factor binding domains and that expression can be restored after treatment with the demethylating agent, 5-azacytidine. Furthermore, assessment of primary tumor specimens confirmed that hypermethylation is common in in-patient tumors and is directly associated with recurrence. This study suggests that hypermethylation of SALL1 in primary tumors is an independent predictor of survival for head and neck cancer.
Materials and Methods
Tumor samples and cell lines
A total of 205 primary HNSCC specimens were obtained during surgery at the Department of Otolaryngology of Hamamatsu University School of Medicine. Clinical information including age, gender, tumor location, smoking status, alcohol consumption, tumor size, lymph node status, and tumor stage was obtained from clinical records. The mean patient age was 64.8 years (range: 32-90 years), and the male:female ratio was 175:30. Primary tumors were located in the hypopharynx (n = 51), larynx (n = 40), oropharynx (n = 51), and oral cavity (n = 63). Patients provided written, informed consent for participation in the study and the protocol was approved by the Institutional Review Boards at the Hamamatsu University School of Medicine. Normal tonsillar specimens, Ton7, Ton8, and Ton10, were surgically obtained from patients with chronic tonsillitis. DNA and cDNA from eight UM-SCC cell lines, MJF, BDF, 81F, 99F fibroblasts cell lines were provided by Dr. Thomas E. Carey of the University of Michigan. Normal human keratinocytes were a gift from Dr. No Hee Park of the University of California at Los Angeles School of Dentistry 10. For reactivation of SALL1 expression, cultures were incubated for 48 h with 5-azacytidine (15 μg/mL, Sigma-Aldrich, St. Louis, MO, USA), a DNA methyltransferase inhibitor 11.
Bisulfite modification and quantitative methylation-specific polymerase chain reaction (qMSP)
Extraction and bisulfite conversion of genomic DNA was carried out as previously described 12,13. SALL1 promoter methylation was assessed by qMSP with the TP800 Thermal Cycler Dice Real-Time System (Takara Bio, Otsu, Japan) using the primer sequences shown in Supplementary Table S1. SALL1 primers amplified sequences upstream of, around, and downstream of the transcription start site. A standard curve was generated using serial dilutions of EpiScope Methylated HeLa genomic DNA (Takara Bio, Otsu, Japan), with fully methylated (FM) DNA used as a control. Normalized methylation values (NMV) was defined as follows: NMV = (SALL1-S/SALL1-FM)/(ACTB-S/ACTB-FM), where SALL1-S and SALL1-FM represent SALL1 methylation levels in the sample and universally methylated DNA, respectively, and ACTB-S and ACTB-FM correspond to β-actin in the sample and the universally methylated DNA, respectively 14. To analyze the methylation status of SALL3 15, CDH1 16, RASSF1A 16, p16 16, MGMT 17, DAPK 17, DCC 18, CDH13 19, and COL1A2 20 primers and conditions were used as previously described.
RNA extraction and quantitative reverse transcription-PCR (qRT-PCR)
Total RNA was isolated with the RNeasy Plus Mini kit (Qiagen, Valencia, CA, USA), and cDNA was synthesized with the ReverTra Ace qPCR RT kit (Toyobo, Osaka, Japan) 14. Primer sequences are shown in Supplementary Table S1. Target mRNA expression was compared between samples by normalization to glyceraldehyde 1-phosphate dehydrogenase (GAPDH) mRNA expression.
The Cancer Genome Atlas (TCGA) data
Aberrant DNA methylation data from the TCGA (available in June, 2017) was collected from MethHC, a database of DNA methylation and gene expression in human cancer (http://methhc.mbc.nctu.edu.tw/php/index.php) 21. The DNA methylation data were collected using the Infinium HumanMethylation450 platform (Illumina, Inc., San Diego, CA, USA) and are presented as β values.
Data analysis and statistics
Receiver-operator characteristic ROC curve analysis was performed using the NMVs for 36 HNSCC and 36 adjacent normal mucosal samples and the Stata/SE 13.0 system (Stata Corporation, TX, USA). The area under the ROC curve indicated the optimal sensitivity and specificity cut-off levels for distinguishing between methylation levels in normal and HNSCC tissue, and the NMV thresholds were calculated for each target gene. The cut-off values were used to determine the methylation frequencies of the SALL1 genes.
Associations between variables were assessed by performing a Student's t-test and Pearson's chi-square test. Disease-free survival (DFS) was measured from the date the initial treatment to the date of diagnosis of locoregional recurrence or distant metastasis. The Kaplan-Meier test was used to calculate survival probabilities and the log-rank test was used to compare survival rates. The prognostic value of methylation status was assessed by performing a multivariate Cox proportional hazards analysis adjusted for age (≥ 70 versus < 70 years), gender, alcohol intake, smoking status, and tumor stage (I-III versus IV). Differences with P < 0.05 were considered significant. All statistical analyses were performed using StatMate IV software (ATMS Co., Ltd., Tokyo, Japan).
Results
Initial Screening: Matched Pairs of Head and Neck Tumors and Adjacent Normal Mucosal Tissues
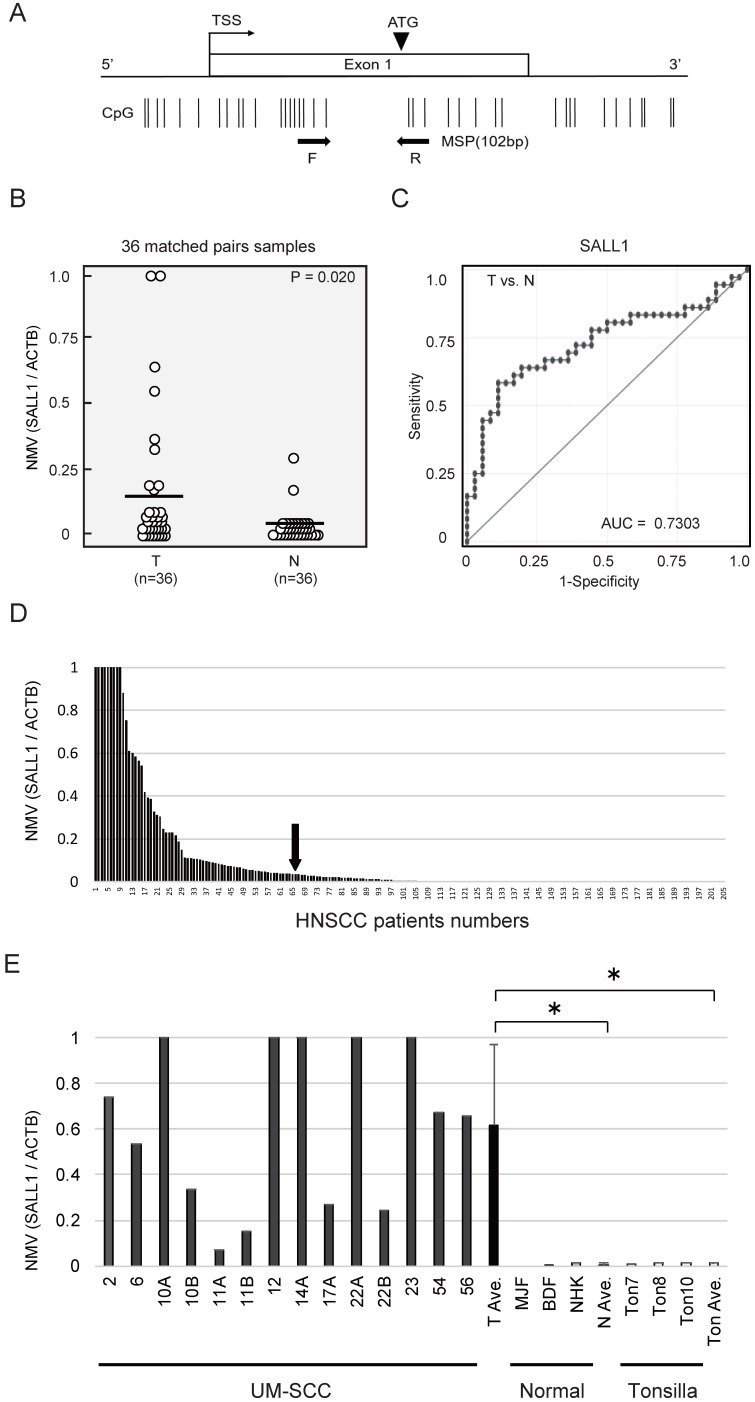
SALL1 CpG islands and regions, analyzed by qMSP, are shown in Fig. 1A. Initially, the methylation status of the SALL1 promoter was analyzed in 36 cancerous and paired noncancerous mucosae by qMSP. Promoter methylation levels were represented by NMVs, which is the ratio of methylated DNA at the target sequence in each specimen to a fully methylated control DNA. The SALL1 methylation level was significantly higher in primary HNSCCs than in noncancerous mucosal tissues (median NMV = 0.194 versus 0.025, P = 0.020; paired Student's t-test; Fig. 1B). SALL1 promoter hypermethylation exhibited highly discriminative ROC curve profiles, which clearly distinguished HNSCC from normal mucosal tissues (area under the ROC = 0.730). At the cutoff value of 0.036, sensitivity was 58.3% and specificity was 88.9% (Fig. 1C).
Figure 1.
Schematic representation of SALL1 gene methylation analysis by quantitative methylation-specific polymerase chain reaction (qMSP) and evaluation of SALL1 methylation in head and neck squamous-cell carcinoma (HNSCC) patients and UM-SCC cell lines. (A) SALL1 exon structure and CpG sites within expanded views of the promoter region relative to the transcript start site (TSS). Vertical lines, individual CpG sites. Straight arrows, relative location of the primers used for qMSP; bent arrow, TSS; arrowhead, translation start site (ATG). Straight arrows, relative location of the forward (F) and reverse (R) primers used for qMSP. (B) SALL1 normalized methylation values (NMVs) of head and neck tumors were higher than those of adjacent normal mucosal tissues (P = 0.020). (C) AUROC value for SALL1 was 0.7303. At the cutoff value of 0.036, sensitivity was 58.3% and specificity was 88.9%. (D) Two hundred and five DNA samples from untreated primary tumors were tested with qMSP. The cutoff NMVs (0.036) are indicated by straight arrows. (E) Representative examples of qMSP of SALL1 in eight UM-SCC cell lines, fibroblasts (MJF and BDF), keratinocytes (NHK), and normal tonsillar tissues (Ton7, Ton8, and Ton10). * P < 0.01.
SALL1 methylation status in tumor samples and cancer cell lines
Among the 205 DNA samples obtained from the examined untreated primary tumors, a specimen was classified as methylated when the NMV exceeded 0.036. The SALL1 promoter was methylated in 65/205 (31.7%) of cases and unmethylated in 140/205 (68.3%) of cases (Fig. 1D). Matched pairs of head and neck tumors and adjacent normal mucosal tissues were obtained from surgical specimens collected from 96 patients for methylation screening. These data are presented as Supplementary Table S2. qMSP indicated a significantly increased NMV for SALL1 promoter methylation in cancer cell lines versus that in normal cell lines and normal tonsillar tissues (P = 0.010 and P = 0.011, respectively; Fig. 1E).
SALL1 expression analysis by qRT-PCR
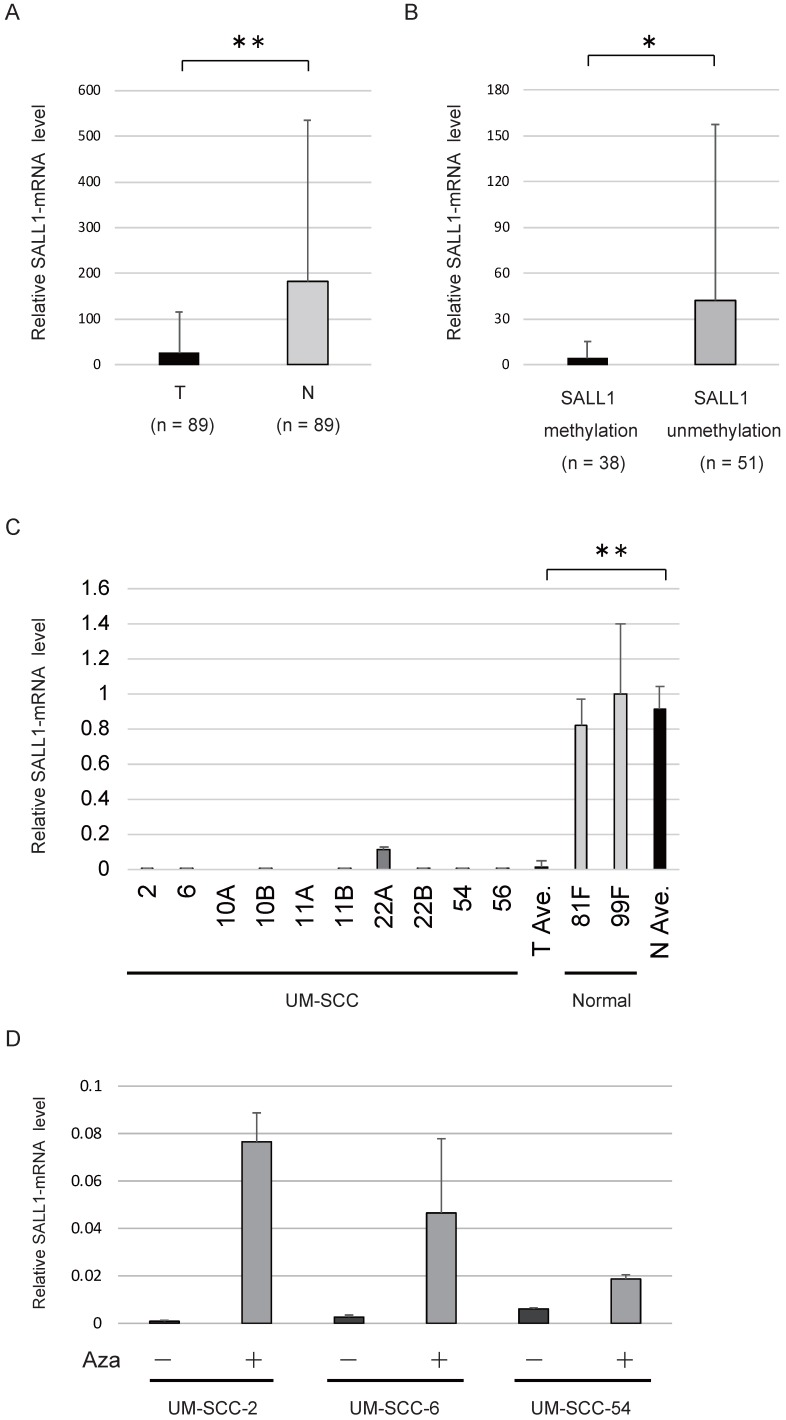
The levels of SALL1 mRNA were determined in 89 HNSCC tissues and adjacent normal mucosal tissues using qRT-PCR. The SALL1 mRNA level in HNSCC tissues was found to be 6.9-fold lower compared to that in the paired non-cancerous mucosa (P < 0.001; Fig. 2A). The levels of SALL1 mRNA were significantly decreased in cases in which the SALL1 promoter region was methylated (P = 0.045; Fig. 2B). qRT-PCR of SALL1 transcripts from 10 UM-SCC cell lines revealed lower expression in cancer cell lines than in normal fibroblast cell lines (P < 0.001; Fig. 2C). We treated the three silenced cell lines (UM-SCC-2, UM-SCC-6, and UM-SCC-54) with a DNA methyltransferase inhibitor (5-azacytidine), and all three cell lines exhibited restored SALL1 expression after inhibition of methyltransferase activity (Fig. 2D).
Figure 2.
SALL1 expression as assessed by quantitative reverse transcription PCR (qRT-PCR). (A) Relative mRNA expression level of SALL1 in 89 matched pairs of head and neck tumors and adjacent normal mucosal tissues. T, tumor samples; N, normal samples. ** P < 0.01. (B) SALL1 mRNA expression was inversely correlated with SALL1 hypermethylation. * P < 0.05. (C) Relative mRNA expression level of SALL1, as determined by qRT-PCR in 10 UM-SCC and two normal cell lines. ** P < 0.01. (D) Effect of 5-azacytidine (Aza) on SALL1 expression in three cell lines with densely methylated SALL1, as evaluated by qRT-PCR. Controls were cells that were similarly treated, but without Aza.
SALL1 promoter methylation correlates with clinicopathologic parameters and predicts poor patient outcome
Age, gender, smoking status, alcohol intake, tumor size, lymph node status, and clinical stage were not associated with SALL1 promoter methylation status in 205 patients. However, SALL1 methylation was significantly correlated with recurrence events, SALL3 methylation status, and CDH1 methylation status (P < 0.001, P = 0.003, and P < 0.001, respectively). No other molecular markers were significantly correlated with SALL1 methylation status (Table 1, Supplementary Table S3).
Table 1.
SALL1 Gene Methylation Status in Primary Samples of HNSCC in 205 patients
| Patient and tumorcharacteristics (n=205) | Methylation | P-value† | |
|---|---|---|---|
| Present (n=74) | Absent (n=131) | ||
| Age | |||
| 70 and older (67) | 24 | 43 | |
| Under 70 (138) | 50 | 88 | 1 |
| Gender | |||
| Male (175) | 62 | 113 | |
| Female (30) | 12 | 18 | 1 |
| Smoking status | |||
| Smoker (157) | 55 | 102 | |
| Nonsmoker (48) | 19 | 29 | 1 |
| Alcohol intake | |||
| Ever (149) | 55 | 94 | |
| Never (56) | 19 | 37 | 0.746 |
| Tumor size | |||
| T1, 2 (98) | 37 | 61 | |
| T3, 4 (107) | 37 | 70 | 1 |
| Lympho-node status | |||
| N0 (86) | 32 | 54 | |
| N+ (119) | 42 | 77 | 1 |
| Stage | |||
| I, II, III (89) | 32 | 57 | |
| IV (116) | 42 | 74 | 1 |
| Recurrence events | |||
| Positive (85) | 43 | 42 | |
| Negative (120) | 31 | 89 | < 0.001* |
†Fisher's exact probability test.
* P < 0.05.
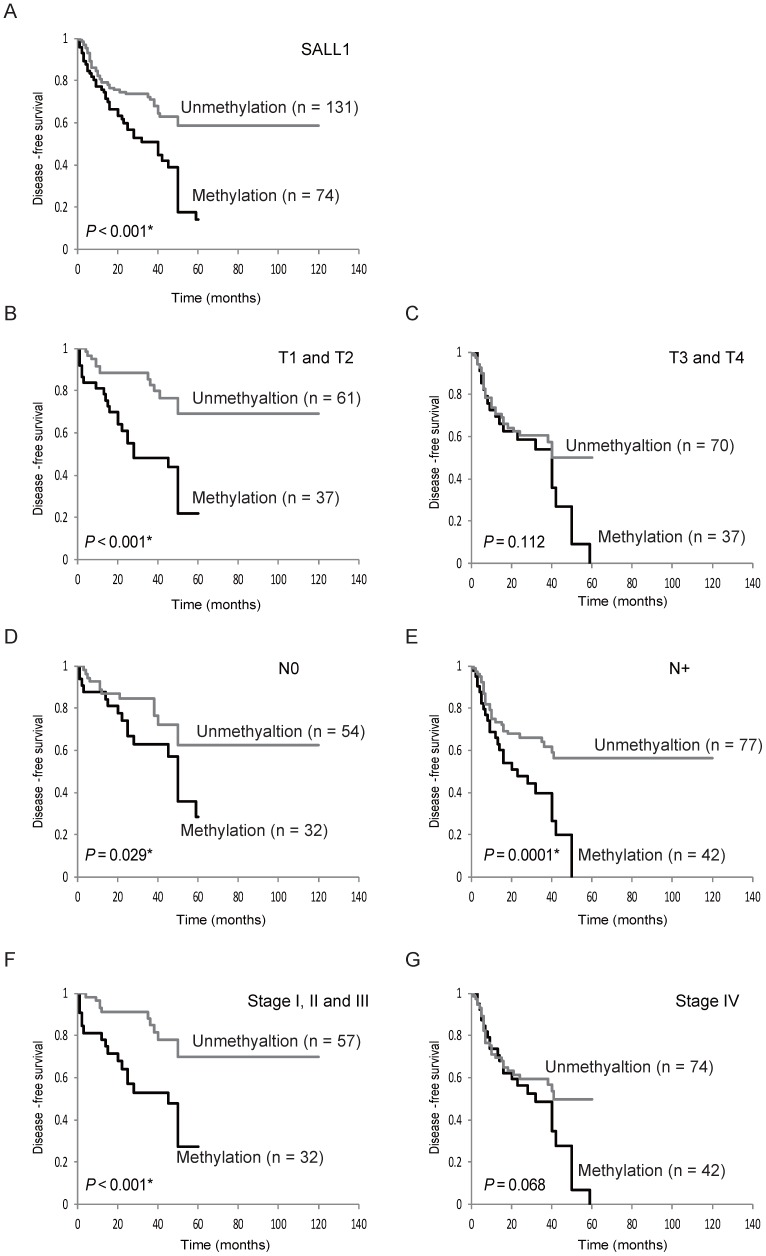
Kaplan-Meier plots revealed that methylation of the SALL1 gene in 205 tumors were related to DFS in some patients (log-rank test, P < 0.001; Fig. 3A). Patients with T1 and T2, negative lymph node metastasis, positive lymph node metastasis, and Stage I, II, and III disease had decreased DFS compared to those without SALL1 methylation (P < 0.001 P = 0.029, P < 0.001, and P < 0.001, respectively; Fig. 3B-F). SALL1 methylation status in patients with T3 and T4 and Stage IV tumors was not related to outcome (Fig. 3C, G). In addition, results based on multivariate Cox proportional hazard regression analysis, which included age, gender, alcohol consumption, smoking status, and tumor stage, indicated that survival rates were 1.694-fold (95% CI, 1.093-2.626) lower in patients with SALL1 gene methylation than in those without methylation (P = 0.018; Table 2). Moreover, based on 98 patients with T1 and T2 tumor stages, the group with methylated SALL1 had a 2.602-fold greater hazard (95% CI, 1.296-5.224) than the group without methylation (P = 0.007; Table 3). When both SALL1 and SALL3 were methylated in the tumor, there was a significantly lower rate of DFS than if SALL1 and SALL3 were unmethylated (log-rank test, P = 0.012) (Supplementary Fig. S1A). Concurrent hypermethylation of SALL1 and CDH1 promoters was also significantly correlated with reduced DFS (log-rank test, P = 0.005; Supplementary Fig. S1B).
Figure 3.
Kaplan-Meier survival curves for head and neck squamous-cell carcinoma (HNSCC) patients. Disease-free survival (DFS) for (A) all 205 HNSCC cases, (B) tumor size in T1 and T2 cases (n = 98), (C) tumor size in T3 and T4 cases (n = 107), (D) lymph node status in N0 cases (n = 86), (E) lymph node status in N+ cases (n = 119), (F) stage I, II, and III cases (n = 89), and (G) stage IV cases (n = 116). Gray and black lines indicate patients with tumors without and with methylation, respectively.
Table 2.
Multivariate analysis of factors affecting survival using Cox proportional hazards model in 205 patients
| Variables | Disease-free survival | |
|---|---|---|
| HR (95% CI) | P-value | |
| Age | ||
| 70 and older vs. <70 | 1.100 (0.685-1.752) | 0.702 |
| Sex | ||
| Male vs. Female | 1.164 (0.627-2.160) | 0.631 |
| Smoking status | ||
| Smoker vs. Non smoker | 1.990 (0.259-15.28) | 0.508 |
| Alcohol exposure | ||
| Ever vs. Never | 1.234 (0.738-2.065) | 0.423 |
| Stage | ||
| I, II, III vs. IV | 2.042 (1.284-3.250) | 0.003* |
| SALL1 methylation | ||
| Yes vs. No | 1.694 (1.093-2.626) | 0.018* |
HR: hazard ratio.
95% CI: 95% confidence interval.
* P < 0.05.
Table 3.
Multivariate analysis of factors affecting survival using Cox proportional hazards model in 98 patients with T1 and T2 stages
| Variables | Disease-free survival | |
|---|---|---|
| HR (95% CI) | P-value | |
| Age | ||
| 70 and older vs. <70 | 1.551 (0.776-3.098) | 0.214 |
| Sex | ||
| Male vs. Female | 0.711 (0.289-1.748) | 0.457 |
| Smoking status | ||
| Smoker vs. Non smoker | 0.769 (0.091-6.494) | 0.809 |
| Alcohol exposure | ||
| Ever vs. Never | 1.405 (0.635-3.107) | 0.402 |
| Stage | ||
| I, II, III vs. IV | 1.552 (0.749-3.218) | 0.237 |
| SALL1 methylation | ||
| Yes vs. No | 2.602 (1.296-5.224) | 0.007* |
HR: hazard ratio.
95% CI: 95% confidence interval.
* P < 0.05.
External validation of The Cancer Genome Atlas (TCGA) data
HNSCC data from TCGA were examined for SALL1 DNA methylation. SALL1 methylation showed an average β value of 0.381 in the HNSCC TCGA cohort as compared to 0.129 in normal samples (P < 0.001, Student's t-test; Supplementary Fig. S2).
Discussion
Clarifying the epigenetic regulation of SALL1 can provide insights into the mechanisms of tumorigenesis and the risk of disease recurrence for various tumor types. The present study investigated SALL1 promoter methylation profiles in 205 HNSCC patient tissues. We found that hypermethylation of CpG islands in the SALL1 promoter was independently associated with early T1 and T2 tumor behavior, suggesting that SALL1 acts as a tumor suppressor gene and can serve as a prognostic biomarker in HNSCC.
SALL1 (16q12.1) has been associated with Townes-Brocks Syndrome, a disorder characterized by multiorgan dysgenesis including renal, heart, and genital malformation 22, 23. Moreover, targeted deletion of mouse Sall1 results in kidney agenesis or severe dysgenesis 24, 25. Sall1 is a nuclear factor that is required for maintaining the stemness of renal progenitor cells by restricting their differentiation 26, 27. It is expressed not only in nephron progenitors but also in stromal progenitors 28. Furthermore, SALL1 expression is consistently high in Wilms tumor 29. Sall1 also affects the induction of neuronal and cardiogenic differentiation markers in pluripotent mouse embryonic stem cells (ESCs) 30. SALL1 binds NANOG directly and is required for open heterochromatin organization in ESCs 31. Further, transcriptional regulation by Sall1 maintains microglial identity and physiological properties in the central nervous system 32.
Three additional SALL genes have been identified to date: SALL2 on chromosome 14q11.2, SALL3 on chromosome 18q23, and SALL4 on chromosome 20q11.2 33. SALL2 binds the neurotrophin receptor and regulates neuronal development 34. SALL2 promoter methylation has been reported in ovarian cancer and esophageal cancer cells 35, 36. Aberrant hypermethylation of SALL3 along with HPV infection was found to be related to carcinogenesis in cervical cancer 9. Moreover, SALL3 was found to be silenced by DNA methylation and this protein interacts with DNA methyltransferases 3 alpha (DNMT3A) in hepatocellular carcinoma 37. Another report showed that SALL3 hypermethylation reduced the levels of SALL3 mRNA in hepatocellular carcinoma 38. Moreover, Sall3 is an epigenetic hotspot for aberrant DNA methylation associated with abnormal placental development of cloned mice 39. The last SALL gene, Sall4, is important for maintaining a pluripotent state in mouse embryonic stem cells, 40-42 and hypomethylation of its promoter is a common event in myelodysplastic syndrome 42. SALL4 is highly expressed in hepatocellular carcinoma with poor prognosis, and is often associated with chronic hepatitis B virus infection 43 and several cancers with worse prognosis 44. SALL4 can also act as a proto-oncogene by regulating the PTEN signaling pathway in glioma cells and myeloid leukemia 45, 46.
SALL1 acts as a transcriptional repressor in mammalian cells when fused to a heterologous DNA-binding domain 47. SALL1 expression is also correlated with the expression of CDH1, and this is consistent with its tumor suppressive function and suggests its potential involvement in epithelial-to-mesenchymal transition in breast cancer 7. SALL1 hypermethylation has been described in several malignancies including non-small cell lung cancer 8, prostate cancer 8, acute lymphocytic leukemia 48, chronic lymphocytic leukemia 49, and colon cancer 8. The frequency of SALL1 promoter methylation was determined to be 4/16 (25.0%) in non-small cell lung cancer, 5/14 (35.7%) in prostate cancer, 69/78 (88.5%) in chronic lymphocytic leukemia, and 44/53 (83.0%) in colon cancer 8, 49. Both the inactivation of SALL1 through methylation and the loss of its transcriptional repressor function in different cancers are supportive of its role in tumor progression and metastasis.
Early reports from our group noted that the expression of SALL3 mRNA is lost in HNSCC as a consequence of DNA methylation. SALL3 methylation was also found in 64.8% of primary tumor specimens 15. A concurrent analysis showed that the methylation of SALL1 is significantly correlated with SALL3 methylation. Furthermore, hypermethylation of both genes was associated with significantly shorter DFS, compared to that without methylation of both genes. Additionally, CDH1 promoter methylation has been detected in many tumor types 50. Low CDH1 expression is associated with an increased risk of late cervical lymph node metastasis in stage I and II oral cancer patients 51, and downregulation of CDH1 predicts delayed neck metastasis in stage I and II oral carcinoma after partial glossectomy 52. We found that aberrant patterns of promoter methylation in primary tumors are indicators of increased risk of recurrence in patients with early stage HNSCC.
Thus, detecting aberrant SALL1 methylation can serve as a means to identify patients at high risk for relapse. We demonstrated for the first time that SALL1 mRNA is downregulated in HNSCC due to DNA methylation; this might be a discriminative event for early T1/T2 HNSCC progression that is associated with DFS. Our findings suggest that these methylation markers can be used clinically to identify patients that could benefit from adjuvant therapy after initial surgical treatment; however, this must be confirmed in additional prospective studies using other HNSCC patient groups.
Supplementary Material
Supplementary figures and tables.
Acknowledgments
The authors would like to thank Ms. Yuko Mohri for her excellent technical support.
Author contributions
Conceptualization: KM. Methodology: KM. Software: YM and TK. Validation: AI, SE, HK, RI, and MM. Formal analysis: DM. Investigation: TK. Resources: RI and MM. Resources: MM and YM. Writing - original draft: KM. Writing - review and editing: TK and TY. Visualization: KM. Supervision: HK. Project administration: KM. Funding acquisition: KM. All authors read and approved the final manuscript.
References
- 1.Vokes EE, Weichselbaum RR, Lippman SM, Hong WK. Head and neck cancer. N Engl J Med. 1993;328:184–94. doi: 10.1056/NEJM199301213280306. [DOI] [PubMed] [Google Scholar]
- 2.Kang H, Kiess A, Chung CH. Emerging biomarkers in head and neck cancer in the era of genomics. Nat Rev Clin Oncol. 2015;12:11–26. doi: 10.1038/nrclinonc.2014.192. [DOI] [PubMed] [Google Scholar]
- 3.Argiris A, Karamouzis MV, Raben D, Ferris RL. Head and neck cancer. Lancet. 2008;371:1695–709. doi: 10.1016/S0140-6736(08)60728-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seiwert TY, Cohen EE. State-of-the-art management of locally advanced head and neck cancer. Br J Cancer. 2005;92:1341–8. doi: 10.1038/sj.bjc.6602510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karantzali E, Lekakis V, Ioannou M, Hadjimichael C, Papamatheakis J, Kretsovali A. Sall1 regulates embryonic stem cell differentiation in association with nanog. J Biol Chem. 2011;286:1037–45. doi: 10.1074/jbc.M110.170050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohlhase J, Wischermann A, Reichenbach H, Froster U, Engel W. Mutations in the SALL1 putative transcription factor gene cause Townes-Brocks syndrome. Nat Genet. 1998;18:81–3. doi: 10.1038/ng0198-81. [DOI] [PubMed] [Google Scholar]
- 7.Wolf J, Muller-Decker K, Flechtenmacher C, Zhang F, Shahmoradgoli M, Mills GB. et al. An in vivo RNAi screen identifies SALL1 as a tumor suppressor in human breast cancer with a role in CDH1 regulation. Oncogene. 2014;33:4273–8. doi: 10.1038/onc.2013.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hill VK, Hesson LB, Dansranjavin T, Dallol A, Bieche I, Vacher S. et al. Identification of 5 novel genes methylated in breast and other epithelial cancers. Molecular cancer. 2010;9:51. doi: 10.1186/1476-4598-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei X, Zhang S, Cao D, Zhao M, Zhang Q, Zhao J. et al. Aberrant Hypermethylation of SALL3 with HPV Involvement Contributes to the Carcinogenesis of Cervical Cancer. PLoS One. 2015;10:e0145700. doi: 10.1371/journal.pone.0145700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park NH, Min BM, Li SL, Huang MZ, Cherick HM, Doniger J. Immortalization of normal human oral keratinocytes with type 16 human papillomavirus. Carcinogenesis. 1991;12:1627–31. doi: 10.1093/carcin/12.9.1627. [DOI] [PubMed] [Google Scholar]
- 11.Misawa K, Ueda Y, Kanazawa T, Misawa Y, Jang I, Brenner JC. et al. Epigenetic inactivation of galanin receptor 1 in head and neck cancer. Clin Cancer Res. 2008;14:7604–13. doi: 10.1158/1078-0432.CCR-07-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Misawa Y, Misawa K, Kanazawa T, Uehara T, Endo S, Mochizuki D. et al. Tumor suppressor activity and inactivation of galanin receptor type 2 by aberrant promoter methylation in head and neck cancer. Cancer. 2014;120:205–13. doi: 10.1002/cncr.28411. [DOI] [PubMed] [Google Scholar]
- 13.Misawa K, Mochizuki D, Endo S, Mima M, Misawa Y, Imai A. et al. Site-specific methylation patterns of the GAL and GALR1/2 genes in head and neck cancer: Potential utility as biomarkers for prognosis. Mol Carcinog. 2017;56:1107–16. doi: 10.1002/mc.22577. [DOI] [PubMed] [Google Scholar]
- 14.Misawa K, Mochizuki D, Imai A, Endo S, Mima M, Misawa Y. et al. Prognostic value of aberrant promoter hypermethylation of tumor-related genes in early-stage head and neck cancer. Oncotarget. 2016;7:26087–98. doi: 10.18632/oncotarget.8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Misawa K, Mochizuki D, Imai A, Misawa Y, Endo S, Mima M. et al. Epigenetic silencing of SALL3 is an independent predictor of poor survival in head and neck cancer. Clin Epigenetics. 2017;9:64. doi: 10.1186/s13148-017-0363-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kristensen LS, Mikeska T, Krypuy M, Dobrovic A. Sensitive Melting Analysis after Real Time- Methylation Specific PCR (SMART-MSP): high-throughput and probe-free quantitative DNA methylation detection. Nucleic Acids Res. 2008;36:e42. doi: 10.1093/nar/gkn113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martone T, Gillio-Tos A, De Marco L, Fiano V, Maule M, Cavalot A. et al. Association between hypermethylated tumor and paired surgical margins in head and neck squamous cell carcinomas. Clin Cancer Res. 2007;13:5089–94. doi: 10.1158/1078-0432.CCR-07-0119. [DOI] [PubMed] [Google Scholar]
- 18.Park HL, Kim MS, Yamashita K, Westra W, Carvalho AL, Lee J. et al. DCC promoter hypermethylation in esophageal squamous cell carcinoma. Int J Cancer. 2008;122:2498–502. doi: 10.1002/ijc.23434. [DOI] [PubMed] [Google Scholar]
- 19.Jin Z, Cheng Y, Olaru A, Kan T, Yang J, Paun B. et al. Promoter hypermethylation of CDH13 is a common, early event in human esophageal adenocarcinogenesis and correlates with clinical risk factors. Int J Cancer. 2008;123:2331–6. doi: 10.1002/ijc.23804. [DOI] [PubMed] [Google Scholar]
- 20.Misawa K, Kanazawa T, Misawa Y, Imai A, Endo S, Hakamada K. et al. Hypermethylation of collagen alpha2 (I) gene (COL1A2) is an independent predictor of survival in head and neck cancer. Cancer Biomark. 2011;10:135–44. doi: 10.3233/CBM-2012-0242. [DOI] [PubMed] [Google Scholar]
- 21.Huang WY, Hsu SD, Huang HY, Sun YM, Chou CH, Weng SL. et al. MethHC: a database of DNA methylation and gene expression in human cancer. Nucleic Acids Res. 2015;43:D856–61. doi: 10.1093/nar/gku1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma Y, Chai L, Cortez SC, Stopa EG, Steinhoff MM, Ford D. et al. SALL1 expression in the human pituitary-adrenal/gonadal axis. J Endocrinol. 2002;173:437–48. doi: 10.1677/joe.0.1730437. [DOI] [PubMed] [Google Scholar]
- 23.Miller EM, Hopkin R, Bao L, Ware SM. Implications for genotype-phenotype predictions in Townes-Brocks syndrome: case report of a novel SALL1 deletion and review of the literature. Am J Med Genet A. 2012;158A:533–40. doi: 10.1002/ajmg.a.34426. [DOI] [PubMed] [Google Scholar]
- 24.Sato A, Kishida S, Tanaka T, Kikuchi A, Kodama T, Asashima M. et al. Sall1, a causative gene for Townes-Brocks syndrome, enhances the canonical Wnt signaling by localizing to heterochromatin. Biochem Biophys Res Commun. 2004;319:103–13. doi: 10.1016/j.bbrc.2004.04.156. [DOI] [PubMed] [Google Scholar]
- 25.Barry JS, Reddy MA. The association of an epibulbar dermoid and Duane syndrome in a patient with a SALL1 mutation (Townes-Brocks Syndrome) Ophthalmic Genet. 2008;29:177–80. doi: 10.1080/13816810802354224. [DOI] [PubMed] [Google Scholar]
- 26.Basta JM, Robbins L, Kiefer SM, Dorsett D, Rauchman M. Sall1 balances self-renewal and differentiation of renal progenitor cells. Development. 2014;141:1047–58. doi: 10.1242/dev.095851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanda S, Tanigawa S, Ohmori T, Taguchi A, Kudo K, Suzuki Y. et al. Sall1 maintains nephron progenitors and nascent nephrons by acting as both an activator and a repressor. J Am Soc Nephrol. 2014;25:2584–95. doi: 10.1681/ASN.2013080896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohmori T, Tanigawa S, Kaku Y, Fujimura S, Nishinakamura R. Sall1 in renal stromal progenitors non-cell autonomously restricts the excessive expansion of nephron progenitors. Sci Rep. 2015;5:15676. doi: 10.1038/srep15676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma Y, Singer DB, Gozman A, Ford D, Chai L, Steinhoff MM. et al. Hsal 1 is related to kidney and gonad development and is expressed in Wilms tumor. Pediatr Nephrol. 2001;16:701–9. doi: 10.1007/s004670100624. [DOI] [PubMed] [Google Scholar]
- 30.Buck A, Kispert A, Kohlhase J. Embryonic expression of the murine homologue of SALL1, the gene mutated in Townes-Brocks syndrome. Mech Dev. 2001;104:143–6. doi: 10.1016/s0925-4773(01)00364-1. [DOI] [PubMed] [Google Scholar]
- 31.Novo CL, Tang C, Ahmed K, Djuric U, Fussner E, Mullin NP. et al. The pluripotency factor Nanog regulates pericentromeric heterochromatin organization in mouse embryonic stem cells. Genes Dev. 2016;30:1101–15. doi: 10.1101/gad.275685.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buttgereit A, Lelios I, Yu X, Vrohlings M, Krakoski NR, Gautier EL. et al. Sall1 is a transcriptional regulator defining microglia identity and function. Nat Immunol. 2016;17:1397–406. doi: 10.1038/ni.3585. [DOI] [PubMed] [Google Scholar]
- 33.Parrish M, Ott T, Lance-Jones C, Schuetz G, Schwaeger-Nickolenko A, Monaghan AP. Loss of the Sall3 gene leads to palate deficiency, abnormalities in cranial nerves, and perinatal lethality. Mol Cell Biol. 2004;24:7102–12. doi: 10.1128/MCB.24.16.7102-7112.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pincheira R, Baerwald M, Dunbar JD, Donner DB. Sall2 is a novel p75NTR-interacting protein that links NGF signalling to cell cycle progression and neurite outgrowth. EMBO J. 2009;28:261–73. doi: 10.1038/emboj.2008.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sung CK, Li D, Andrews E, Drapkin R, Benjamin T. Promoter methylation of the SALL2 tumor suppressor gene in ovarian cancers. Mol Oncol. 2013;7:419–27. doi: 10.1016/j.molonc.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo J, Wang W, Tang Y, Zhou D, Gao Y, Zhang Q. et al. mRNA and methylation profiling of radioresistant esophageal cancer cells: the involvement of Sall2 in acquired aggressive phenotypes. J Cancer. 2017;8:646–56. doi: 10.7150/jca.15652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shikauchi Y, Saiura A, Kubo T, Niwa Y, Yamamoto J, Murase Y. et al. SALL3 interacts with DNMT3A and shows the ability to inhibit CpG island methylation in hepatocellular carcinoma. Mol Cell Biol. 2009;29:1944–58. doi: 10.1128/MCB.00840-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang XX, Sun JZ, Li FX, Wu YS, Du HY, Zhu W. et al. Aberrant methylation and downregulation of sall3 in human hepatocellular carcinoma. World J Gastroenterol. 2012;18:2719–26. doi: 10.3748/wjg.v18.i21.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohgane J, Wakayama T, Senda S, Yamazaki Y, Inoue K, Ogura A. et al. The Sall3 locus is an epigenetic hotspot of aberrant DNA methylation associated with placentomegaly of cloned mice. Genes Cells. 2004;9:253–60. doi: 10.1111/j.1356-9597.2004.00720.x. [DOI] [PubMed] [Google Scholar]
- 40.Yang J, Gao C, Chai L, Ma Y. A novel SALL4/OCT4 transcriptional feedback network for pluripotency of embryonic stem cells. PLoS One. 2010;5:e10766. doi: 10.1371/journal.pone.0010766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J, Tam WL, Tong GQ, Wu Q, Chan HY, Soh BS. et al. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat Cell Biol. 2006;8:1114–23. doi: 10.1038/ncb1481. [DOI] [PubMed] [Google Scholar]
- 42.Lin J, Qian J, Yao DM, Qian W, Yang J, Wang CZ. et al. Aberrant hypomethylation of SALL4 gene in patients with myelodysplastic syndrome. Leuk Res. 2013;37:71–5. doi: 10.1016/j.leukres.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 43.Fan H, Cui Z, Zhang H, Mani SK, Diab A, Lefrancois L. et al. DNA demethylation induces SALL4 gene re-expression in subgroups of hepatocellular carcinoma associated with Hepatitis B or C virus infection. Oncogene. 2017;36:2435–45. doi: 10.1038/onc.2016.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nicole L, Sanavia T, Veronese N, Cappellesso R, Luchini C, Dabrilli P. et al. Oncofetal gene SALL4 and prognosis in cancer: A systematic review with meta-analysis. Oncotarget. 2017;8:22968–79. doi: 10.18632/oncotarget.14952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu C, Wu H, Li Y, Shen L, Yu R, Yin H. et al. SALL4 suppresses PTEN expression to promote glioma cell proliferation via PI3K/AKT signaling pathway. J Neurooncol. 2017;135:263–72. doi: 10.1007/s11060-017-2589-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu J, Jeong HW, Kong N, Yang Y, Carroll J, Luo HR. et al. Stem cell factor SALL4 represses the transcriptions of PTEN and SALL1 through an epigenetic repressor complex. PLoS One. 2009;4:e5577. doi: 10.1371/journal.pone.0005577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Netzer C, Bohlander SK, Hinzke M, Chen Y, Kohlhase J. Defining the heterochromatin localization and repression domains of SALL1. Biochim Biophys Acta. 2006;1762:386–91. doi: 10.1016/j.bbadis.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 48.Kalari S, Pfeifer GP. Identification of driver and passenger DNA methylation in cancer by epigenomic analysis. Adv Genet. 2010;70:277–308. doi: 10.1016/B978-0-12-380866-0.60010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tong WG, Wierda WG, Lin E, Kuang SQ, Bekele BN, Estrov Z. et al. Genome-wide DNA methylation profiling of chronic lymphocytic leukemia allows identification of epigenetically repressed molecular pathways with clinical impact. Epigenetics. 2010;5:499–508. doi: 10.4161/epi.5.6.12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–53. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 51.Lim SC, Zhang S, Ishii G, Endoh Y, Kodama K, Miyamoto S. et al. Predictive markers for late cervical metastasis in stage I and II invasive squamous cell carcinoma of the oral tongue. Clin Cancer Res. 2004;10:166–72. doi: 10.1158/1078-0432.ccr-0533-3. [DOI] [PubMed] [Google Scholar]
- 52.Sakamoto K, Imanishi Y, Tomita T, Shimoda M, Kameyama K, Shibata K. et al. Overexpression of SIP1 and downregulation of E-cadherin predict delayed neck metastasis in stage I/II oral tongue squamous cell carcinoma after partial glossectomy. Ann Surg Oncol. 2012;19:612–9. doi: 10.1245/s10434-011-2052-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and tables.