Abstract
Background: The role of oncolytic viruses in cancer treatment is increasingly studied. The first oncolytic virus (Rigvir®, ECHO-7) was registered in Latvia over a decade ago. In a recent retrospective study Rigvir® decreased mortality 4.39-6.57-fold in stage IB-IIC melanoma patients. The aims of the present study are to test the effect of Rigvir® on cell line viability in vitro and to visualize the cellular presence of Rigvir® by immunocytochemistry.
Methods: The cytolytic effect of Rigvir® on the viability of FM-9, RD, AGS, A549, HDFa, HPAF‑II, MSC, MCF7, HaCaT, and Sk-Mel-28 cell lines was measured using live cell imaging. PBMC viability was measured using flow cytometry. The presence of ECHO-7 virus was visualized using immunocytochemistry. Statistical difference between treatment groups was calculated using two-way ANOVA.
Results: Rigvir® (10%, volume/volume) reduced cell viability in FM-9, RD, AGS, A549, HDFa, HPAF‑II and MSC cell lines by 67-100%. HaCaT cell viability was partly affected while Rigvir® had no effect on MCF7, Sk-Mel-28 and PBMC viability. Detection of ECHO-7 by immunocytochemistry in FM-9, RD, AGS, A549, HDFa, HPAF-II and Sk-Mel-28 cell lines suggests that the presence of Rigvir® in the cells preceded or coincided with the time of reduction of cell viability. Rigvir® (10%) had no effect on live PBMC count.
Conclusions: The results suggest that Rigvir® in vitro reduces the viability of cells of human melanoma, rhabdomyosarcoma, gastric adenocarcinoma, lung carcinoma, pancreas adenocarcinoma but not in PBMC. The presence of Rigvir® in the sensitive cells was confirmed using anti-ECHO-7 antibodies. The present results suggest that a mechanism of action for the clinical benefit of Rigvir® is its cytolytic properties. The present results suggest that the effect of Rigvir® could be tested in other cancers besides melanoma. Further studies of possible Rigvir® entry receptors are needed.
Keywords: ECHO-7 virus, immunocytochemistry, oncolytic virus, cell viability, Rigvir.
Background
Oncolytic viruses are emerging as a new class of cancer treatment although the connection between virus infections and remission of cancer has been observed for more than a century 1, 2. Starting from the middle of the 20th century the effect of viruses on cancers has been investigated. Today, an oncolytic virus is defined as a virus that selectively infects and lyses cancer cells while not affecting healthy cells of the organism. A few oncolytic viruses are already approved and registered for cancer therapy. Rigvir® was registered in 2004 in Latvia, Oncorine in 2005 in China, and Imlygic in 2015 in the USA and Europe 1, 3-5. It has been reported that oncolytic viruses possess natural preference (100-10000-fold) to cancer over healthy cells 6-8. The selectivity is mainly explained by impaired defence mechanisms in cancer cells 9.
Two groups of viruses are mainly being investigated for use in virotherapy. First, unmodified viruses that have natural selectivity to tumour cells and are non-pathogenic in humans. It has been suggested that human viruses with low pathogenicity may show natural specificity for tumour cells 10-12. Secondly, genetically modified viruses whether used as vaccine vectors (e.g., measles virus, poliovirus, vaccinia virus) or with those genes that are needed for replication in normal tissue modified or deleted, but with retained ability to replicate in cancer cells (e.g., adenovirus, herpes simplex virus, vesicular stomatitis virus) 13, 14. Oncolytic viruses have many appealing properties in comparison to current therapeutic approaches. Replication for most oncolytic viruses tested is tumour selective and does not damage healthy tissue. Usually more than one oncogenic pathway is targeted and cytolytic activity can be caused in several different ways. Resistance to oncolytic viruses has not been reported so far, and last, in the case of genetically modified viruses, safety as well as efficacy-enhancing properties can be inserted 9.
The ECHO-7 virus strain Rigvir® belongs to the Picornaviridae family, Enterovirus genus. Enteroviruses are positive sense single-stranded RNA, non-enveloped, icosahedric viruses approximately 25-30 nm in diameter. The host for ECHO viruses is human. Rigvir® is a non‑pathogenic oncolytic virus selected and adapted for melanoma that was originally isolated from the gastrointestinal tract of healthy children and has not been genetically modified; it was approved for treatment of melanoma in Latvia in 2004 4, 5.
The aim of the present study is to test the cytolytic effect of Rigvir® on human cell lines in vitro using an automated real-time cell imaging system and to determine if the effect is dose- and time-dependent. In one series of experiments virus entry into cells was determined by immunocytochemistry.
Methods
Cell culture conditions
Cell lines of malignant melanoma (FM-9; ECACC 13012416) were obtained from Public Health England, muscle rhabdomyosarcoma (RD; CCL-136), gastric adenocarcinoma (AGS; CRL-1739), lung carcinoma (A549; CCL-185), pancreas adenocarcinoma (HPAF‑II; CRL-1997), human bone marrow-derived mesenchymal stem cells (MSC; PCS-500-012), mammary gland adenocarcinoma (MCF7; HTB-22), and malignant melanoma (Sk‑Mel-28; HTB-72) were obtained from the American Type Culture Collection (ATCC), human normal dermal fibroblasts (HDFa; C0135C) were from Thermo Fisher Scientific, and immortalized human keratinocytes (HaCaT; cat. nr. 300493) were from Cell Lines Service. Negative control was peripheral blood mononuclear cells (PBMC) isolated from blood of three healthy volunteers.
Cells were cultivated in Dulbecco's Modified Eagle Medium (DMEM) with 10% foetal bovine serum (FBS) supplement, 100 U/ml penicillin, 100 µg/ml streptomycin and incubated at 37 °C in a humidified atmosphere of 5% CO2 in air, and sub-cultured after trypsinization (0.25% trypsin/EDTA). MSC cells were grown in mesenchymal stem cell basal medium for adipose, umbilical and bone marrow-derived MSCs (ATCC PCS-500-030) supplemented with mesenchymal stem cell growth kit for bone marrow-derived MSCs (ATCC PCS-500-041) and Penicillin-Streptomycin-Amphotericin B solution (ATCC PCS-999-002).
When the cell monolayer had reached approximately 10% confluency, Rigvir® (stock titre 106 -107 TCID50/ml) was added at final concentrations of 1% and 10% to the cell cultivation medium (volume/volume). An equal volume of medium (without Rigvir®) was added to control cells; the cells were subsequently observed for 96 h.
PBMC isolation and incubation with Rigvir®
Venous blood, 15 ml from each of three healthy volunteers, was collected in K2EDTA vacutainers. Blood samples were diluted with sterile 0.9% NaCl supplemented with 10 IU/ml heparin and slowly layered on Ficoll-Paque solution (GE Healthcare, Sweden) (blood:Ficoll-Paque; 2:1). Density gradient centrifugation was performed at 800xg for 20 min at room temperature with no brake. The buffy coat containing the mononuclear cells formed in the Ficoll-Paque solution was aspirated and transferred to new centrifugation tubes. The buffy coat was washed twice with 10 IU/ml heparin containing 0.9% NaCl and centrifuged at 600 x g for 20 min at room temperature. The cell pellet was suspended in 10% FBS/RPMI medium (Thermo Fisher Scientific); 1 million PBMCs/ml were incubated with Rigvir® (1% or 10% v/v) or PBS (10% v/v) for 24h, 48h and 96h at 37ºC, with agitation (130 rpm).
Viable cell count: live cell imaging
A live cell imaging system (Cell-IQ, now CellActivision, Yokogawa) was used to monitor changes in viable cell count. Phase contrast images were taken every hour.
The live cell imaging system was set to recognize differences in cell population and morphology of different cell growth phases.
Fluorescence immunocytochemistry
Cell lines AGS (ATCC® CRL-1739) and A549 (ATTC® CC1-185) were used for the detection of ECHO-7 virus by fluorescence immunocytochemistry. Cells were cultivated on 4-well cell culture slides. When a confluence of ≥70% had been reached, Rigvir® (10%) solution (diluted with 0.9% NaCl) was added to the cells and incubated for 24h or 96h at 37 °C. The cells were then fixed with 4% formaldehyde solution and washed with phosphate buffered saline (PBS). Then the cells were washed in a blocking buffer (1% BSA/PBS) and incubated in blocking solution (5% BSA / 0.1%TritonX-100/ PBS) for 45 min at room temperature, and rinsed with 3% hydrogen peroxide.
Samples were incubated with anti-ECHO-7 antibody (Centre for Infectious Diseases Control, National Institute for Public Health and the Environment, Bilthoven, NL), in 1% BSA/PBS (1:1000) for 2 h at room temperature in the dark. As a secondary antibody, donkey anti‑rabbit IgG H&L (Alexa Fluor® 555) (AbCam, UK) was used; the cells were incubated for 1 h at room temperature in the dark. Subsequently to visualize the cell nuclei, samples were stained with 4',6-diamidino-2-phenylindole (DAPI) (Applichem, Germany) in 1% BSA/PBS (1 μg/ml) for 10 min at room temperature in the dark.
Flow cytometry
Cells were washed with 2 ml PBS, centrifuged 300 x g, 5 min; the cell pellet was re-suspended in 100 µl PBS containing 0.1 µg propidium iodide (Thermo Fisher Scientific) and 4 µl ZombieGreen Viability Dye (Biolegend), incubated for 30 min at room temperature in the dark. Cells were washed with 2 ml PBS, centrifuged 300 x g, 5 min, re-suspended in 2% FBS/PBS and analysed on FACS Calibur flow cytometer. 50000 events per sample were recorded for analysis. Gating strategy involved singlet discrimination and ZombieGreen / propidium iodide negative cells were defined as live cells. Cell viability assay was performed on the minimum required number of cells (1x106 cells) according to the manufacturers protocol after 24, 48 and 96h incubation.
Calculation of the cytolytic effect of Rigvir® on cell viability
The inhibitory effect of Rigvir® on viable cell count was calculated using the formula: 100‑(100xA/B) = reduction (%), where A is cell count with Rigvir®, and B is the control cell count.
Statistics
The statistical difference between treatment groups was calculated using GraphPad Prism 7.02 software. Two-way ANOVA test was applied in the cell viability experiment.
Results
Cytolytic effect of Rigvir® on cell viability
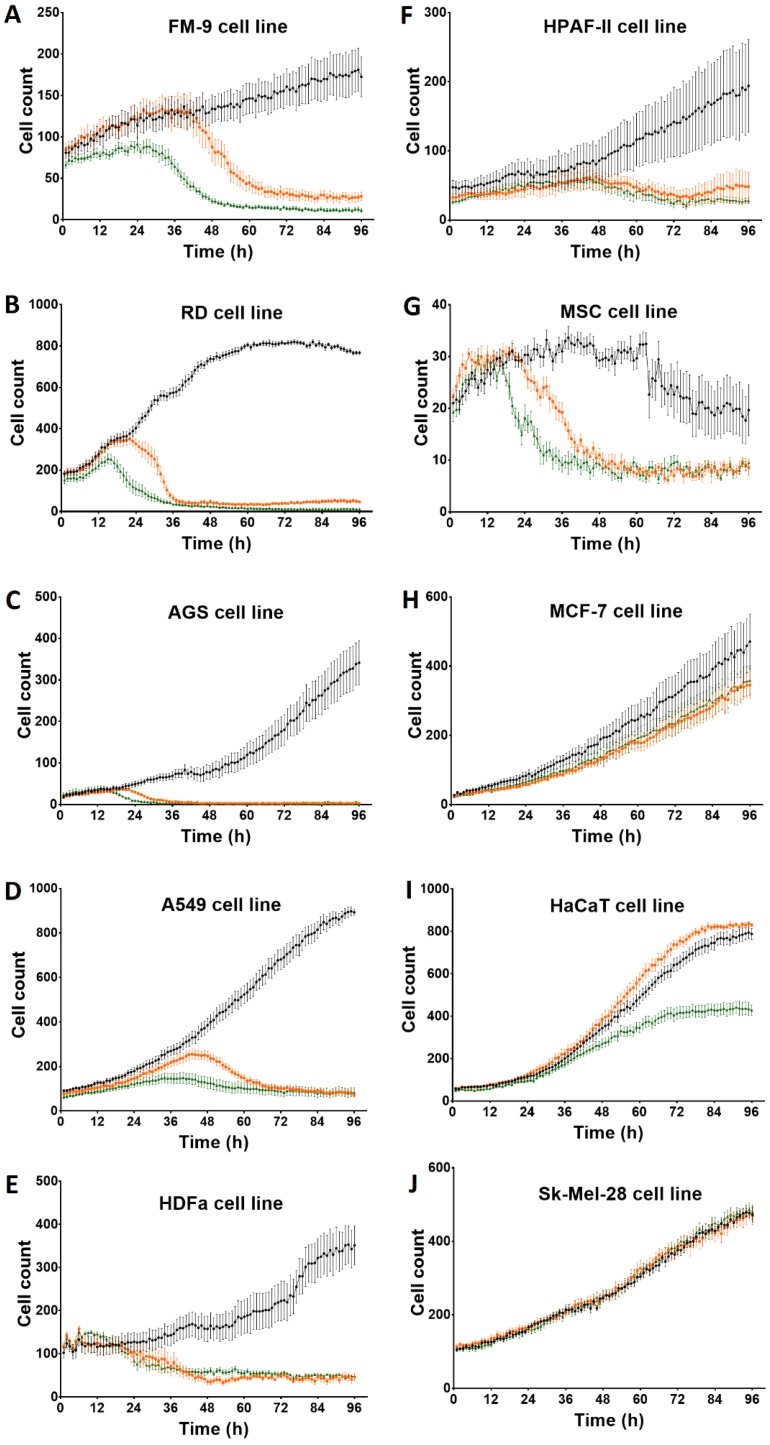
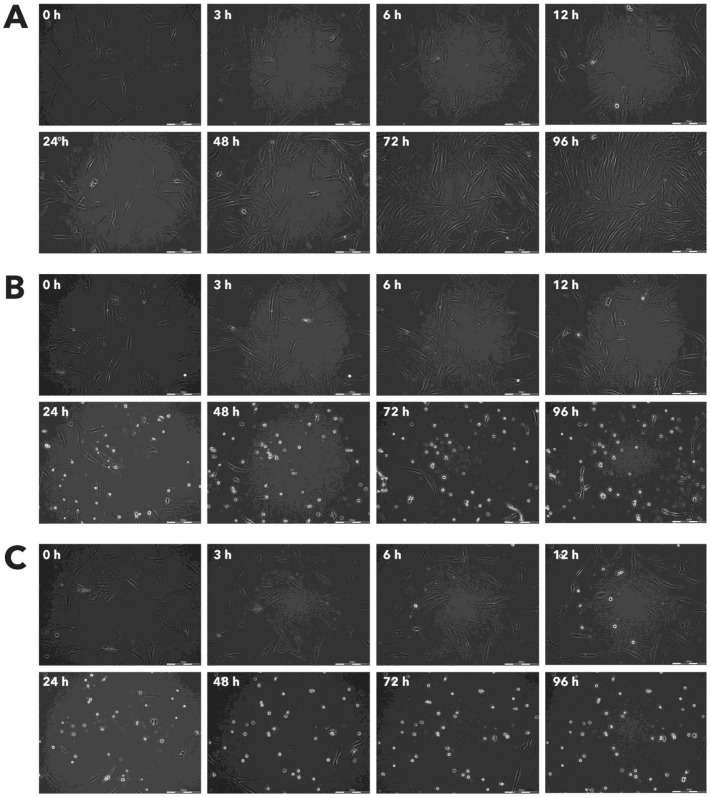
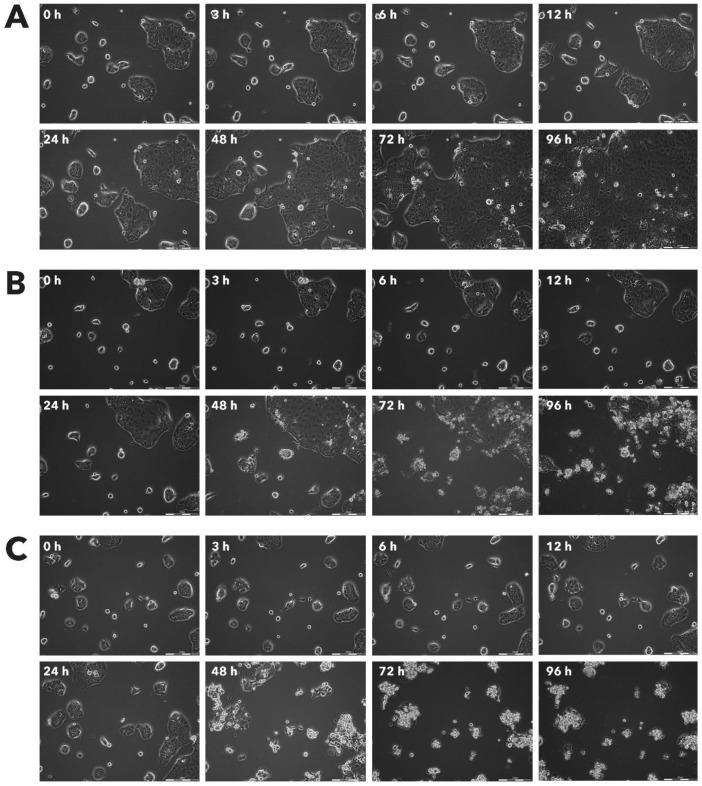
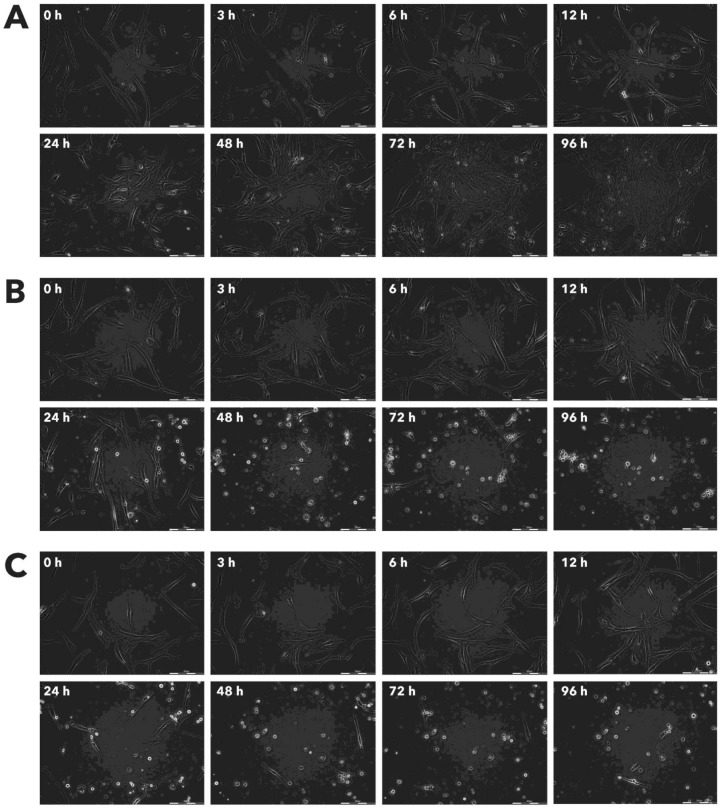
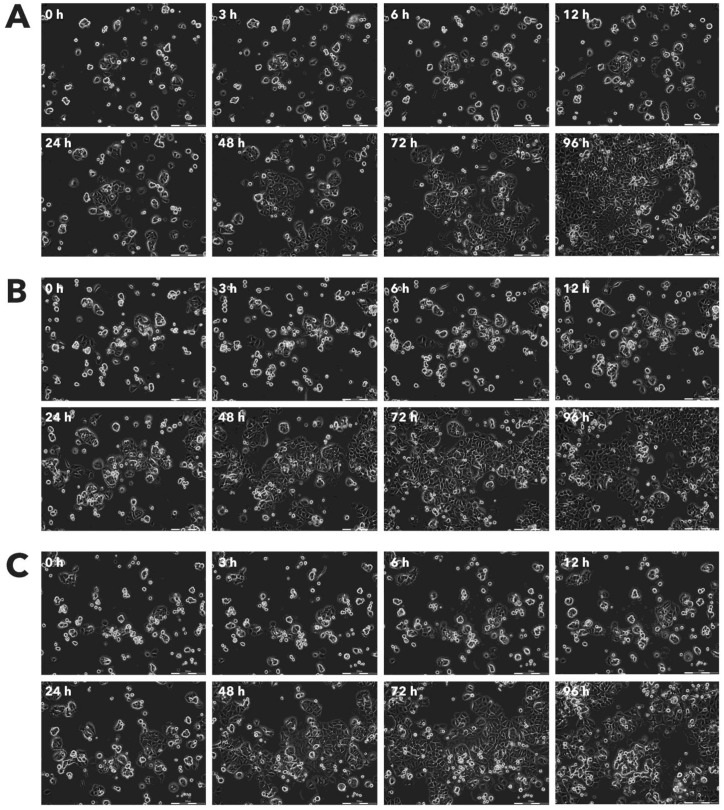
After incubation with Rigvir® (10%) of FM-9, RD, AGS, A549, HDFa, HPAF-II and MSC cell line cells almost no viable cells were observed by 96h. The effect of Rigvir® (1%) was slower in onset. In contrast, HaCaT cell viability was partly affected while Rigvir® appeared to have no effect on MCF7, and Sk-Mel-28 cell viability (Table 1, Fig. 1 - Fig. 11).
Table 1.
Effect of Rigvir® on cell viability observed at 96h and time of onset of the reduction.
| Cells | Rigvir® (1%) | Rigvir® (10%) | (N) | ||||
|---|---|---|---|---|---|---|---|
| Reduction of cell viability (%) | P | Time of onset of reduction (h) | Reduction of cell viability (%) | P | Time of onset of reduction (h) | ||
| FM-9 | 84 | * | 44 | 94 | * | 14 | 3 |
| RD | 94 | * | 23 | 98 | * | 11 | 3 |
| AGS | 100 | * | 27 | 100 | * | 23 | 3 |
| A549 | 92 | * | 21 | 91 | * | 10 | 3 |
| HDFa | 87 | * | 29 | 87 | * | 24 | 3 |
| HPAF-II | 75 | * | 51 | 100 | * | 49 | 3 |
| MSC | 71 | * | 27 | 67 | * | 19 | 3 |
| MCF-7 | 27 | * | 44 | 24 | * | 47 | 3 |
| HaCaT | 0 | n.s. | - | 46 | * | 42 | 3 |
| Sk-Mel-28 | 0 | n.s. | - | 0 | n.s. | - | 3 |
Statistically significantly difference vs. control P<0.05 (*), not statistically different (n.s.). The time of onset is defined as the first three consecutive time points when the reduction was statistically significantly difference vs. control (P<0.05).
Figure 1.
Effect of Rigvir® on viable cell count 0 to 96h. Statistical difference in cell viability between Rigvir®-treated and control was observed from the following time-points. Statistically significantly difference vs. control P<0.05 (*), not statistically different (n.s.). Data are expressed as means ± S.E.M. Control (●), Rigvir® (1%) (■) and Rigvir® (10%) (▲). A. FM-9: Rigvir® (1%) at 44h (P=0.0459) and Rigvir® (10%) at 14h (P=0.0036). B. RD: Rigvir® (1%) at 23h (P=0.0260) and Rigvir® (10%) at 11h (P=0.0083). C. AGS: Rigvir® (1%) at 27h (P=0.0362) and Rigvir® (10%) at 23h (P=0.0241). D. A549: Rigvir® (1%) at 21h (P=0.0360) and Rigvir® (10%) at 10h (P=0.0313). E. HDFa: Rigvir® (1%) at 29h (P=0.0147) and Rigvir® (10%) at 24h (P=0.0107). F. HPAF-II: Rigvir® (1%) at 51h (P=0.0191) and Rigvir® (10%) at 49h (P=0.0423). G. MSC: Rigvir® (1%) at 27h (P=0.0123) and Rigvir® (10%) at 19h (P=0.0188). H. MCF-7: Rigvir® (1%) at 44h (P=0.0154) and Rigvir® (10%) at 47h (P=0.0358). I. HaCaT: Rigvir® (1%) at 96h (n.s.) and Rigvir® (10%) at 42h (P=0.0171). J. Sk-Mel-28: Rigvir® (1% and 10%) 0-96h (n.s.).
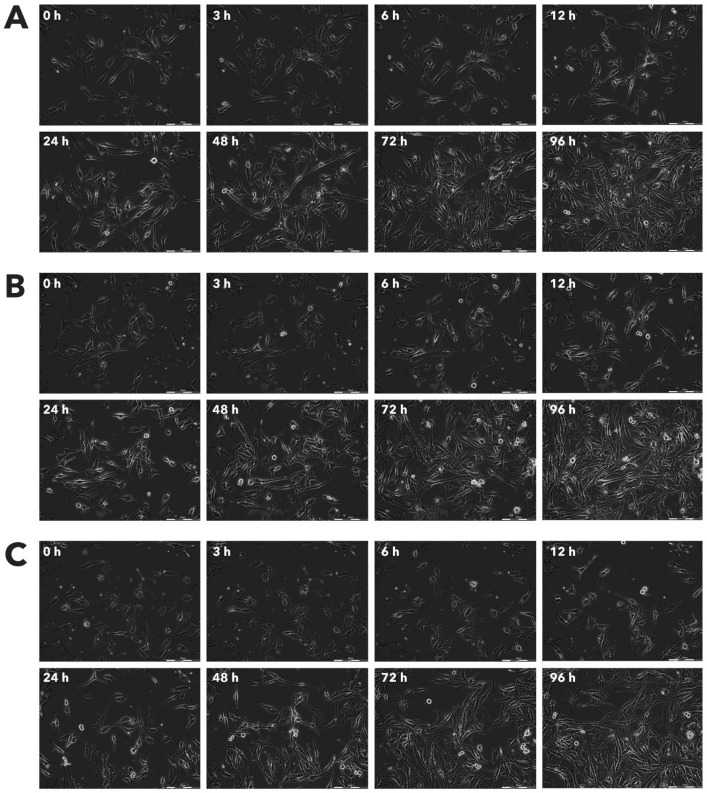
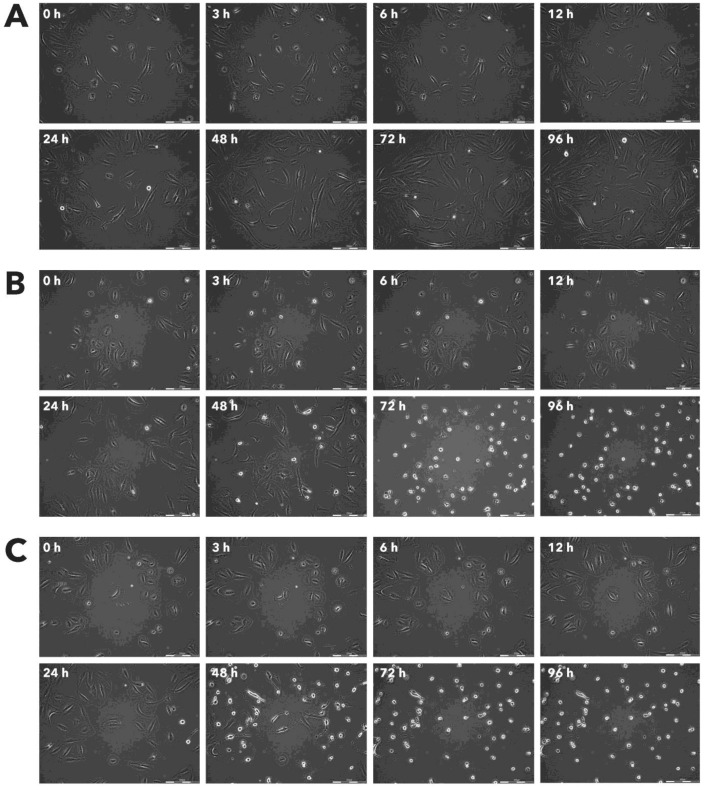
Figure 11.
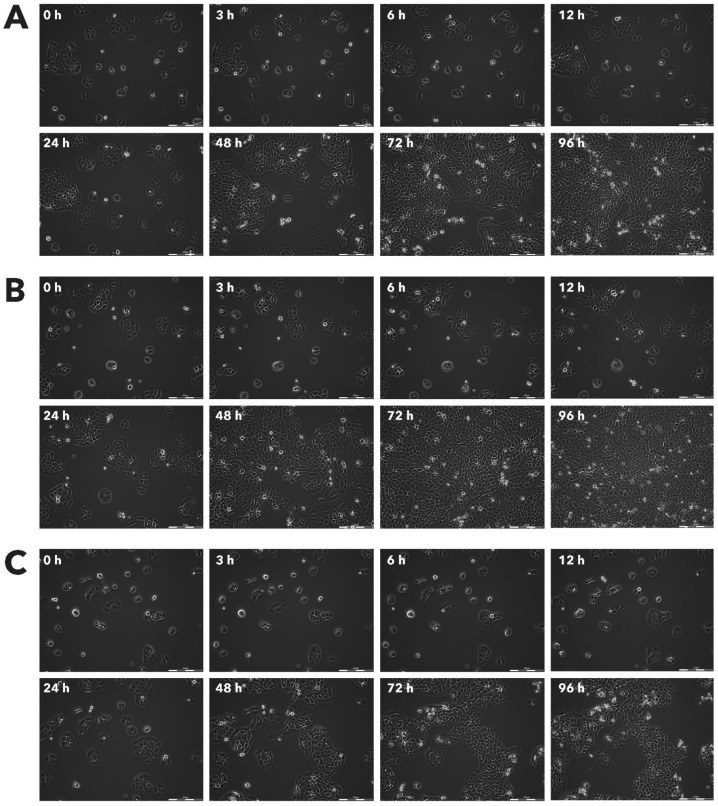
Effect of Rigvir® on Sk-Mel-28 viable cell count. Photographs taken at time points 0, 3, 6, 12, 24, 48, 72, and 96h of incubation. Control (A), Rigvir® (1%) (B) and Rigvir® (10%) (C). Scale bar is 200 µm.
Effect of Rigvir® on peripheral blood mononuclear cells (PBMC)
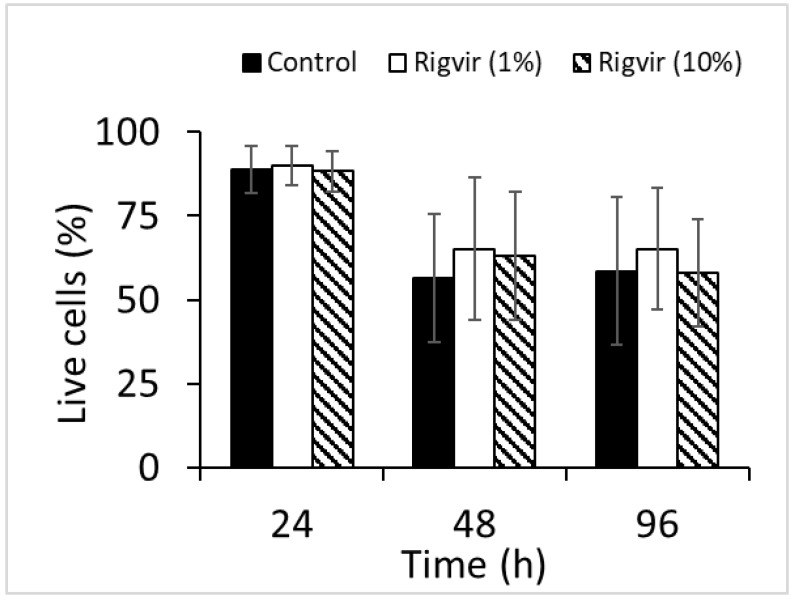
Flow cytometry analysis of cell viability monitored by ZombieGreen / PI staining of both 1% and 10% Rigvir® effect on cell viability after incubation for 24, 48, and 96h showed no significant changes in live cell count compared to control sample (Fig. 12).
Figure 12.
Effect of Rigvir on PBMC viability. Control, Rigvir® (1%), Rigvir® (10%) after 24h, 48h and 96h incubation (n=3). Differences in live cell count compared to control are not statistically significant (P>0.05). Data are expressed as means ± S.D.
Visualization of Rigvir® using anti-ECHO-7 antibody
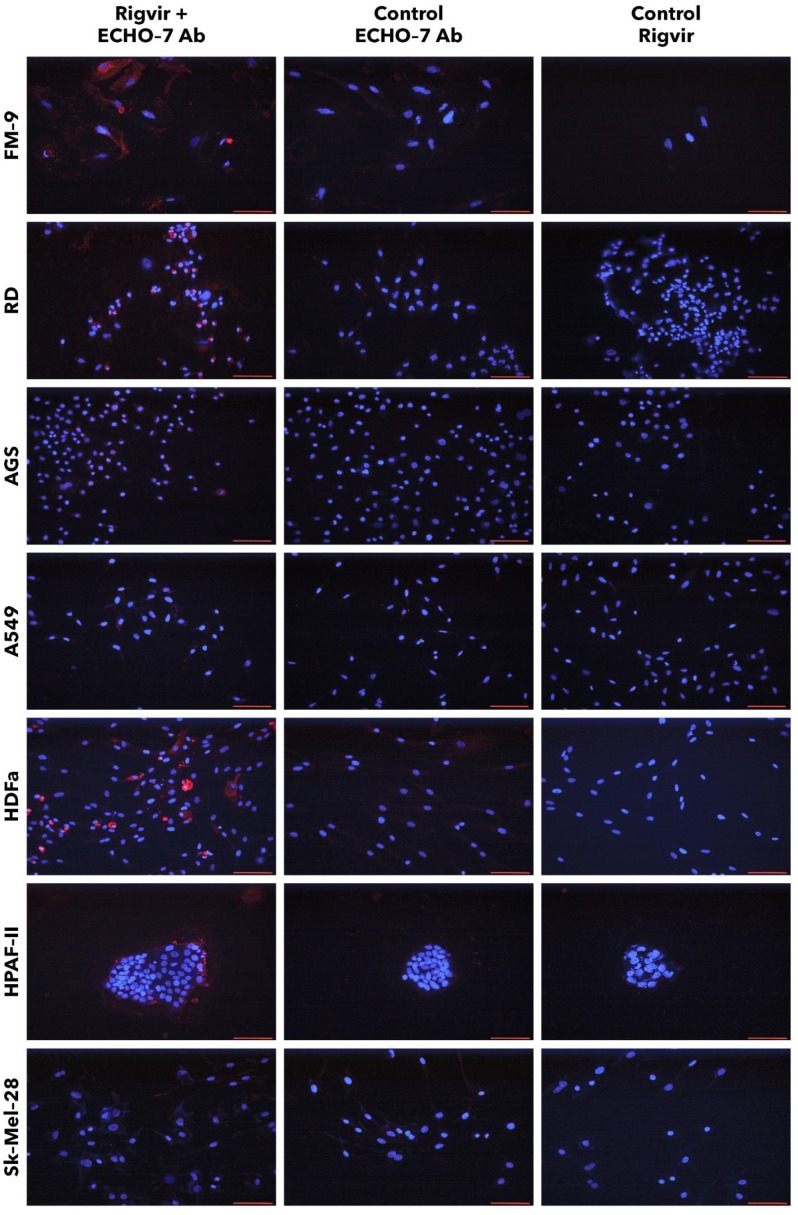
FM-9, RD, AGS, and A549 cells incubated with Rigvir® and anti-ECHO-7 antibody showed specific staining in (red) around nuclei (blue) in most cells (Fig. 13). Control cells incubated only with anti‑ECHO‑7 antibody (without Rigvir) or only with Rigvir® (10%, without antibody) did not show any staining (Fig. 13). In HPAF-II cells after incubation with Rigvir® even for 96 h the anti-ECHO‑7 antibody staining was differently expressed than in RD or FM-9 cells after 24h incubation; control incubations in HPAF-II cells showed no specific staining (Fig. 13).
Figure 13.
Presence of ECHO-7 antibody (red) around nuclei (blue). Incubation with Rigvir® (10%) was for 24h with FM-9, RD, AGS, and A549 cells, and for 96h with HDFa, HPAF-II and Sk-Mel-28 cells. The cells were incubated with ECHO-7 antibody (Ab) and donkey anti rabbit antibody and stained. Scale bar is 100 µm.
Staining of HPAF-II cells showed that anti-ECHO-7 antibody was localised mainly in the cells located on surface of the aggregates. These results agree with the live cell imaging results and suggests that in Rigvir® sensitive cells Rigvir® enters the cells and can be detected in the cytoplasm.
Sk-Mel-28 cell line cells did not show any significant staining in the presence of Rigvir® even after 96h cultivation with Rigvir® (10%). HDFa cells showed detectable anti-ECHO-7 antibody levels after the incubation of Rigvir® for 96 h, but not after 24h, in accordance with the timing of the effect on cell viability. Both control incubations in HDFa cells showed no relevant staining (Fig. 13). The results suggest that Rigvir® does not enter SK-Mel-28 cells, which is in accordance with the live cell imaging cell viability results.
Discussion
The aim of the present study was to test the cytolytic effect of Rigvir® in cancer cell lines of human origin in vitro. Rigvir® is shown to reduce the viability (by 75-100%) of FM-9, RD, AGS, A549 and HPAF-II cell lines in a time- and dose dependent manner (Table 1). The maximal reduction of MCF‑7 cell viability was below 35% at 96h. In contrast, Sk-Mel-28 cell viability was not affected (Table 1); the reason for this should be investigated in future experiments.
PBMC from healthy volunteers were not affected by Rigvir®, which corresponds to previous findings that enteroviruses such as Coxsackievirus B3, ECHO-1 and poliovirus 1 do not replicate in PBMC 15.
The immunocytochemistry results suggest that anti-ECHO-7 antibodies were present only in the cell line cells that showed reduced cell viability in the presence of Rigvir®. FM-9, RD, AGS, A549, HDFa, and HPAF-II cell lines showed ECHO-7 stain, while Sk-Mel-28 cells did not. This corresponds to the results of live cell imaging; the cells that showed antibody stain were the ones whose viability was reduced by Rigvir®, suggesting that in these cell lines Rigvir® entered the cell. In the case of HPAF‑II cells, it can be observed that anti-ECHO-7 antibodies are located around cell islets, suggesting that it might take longer for the virus to enter the cell aggregates.
Both HDFa and MSC cells were also susceptible to Rigvir® infection. Previous studies have shown that when Rigvir® is injected locally around the tumour, lymphocyte infiltration in tumour lesions can be observed after biopsy, suggesting specific infection of cancer cells 16. However, it must be noted that two-dimensional (2D) cell culture systems, such as the one used in the present study, although widely used, do not represent the exact in vivo environment in the body. Monolayer or suspension cell cultures lack properties such as cell-cell interaction, extracellular matrix, biochemical and mechanical cues, and heterogenous cell content 17 18. In several studies, it has been observed that cells grown in three-dimensional (3D) culture systems show reduced sensitivity to chemotherapy drugs 17. The structure of the culture system is also important in viral infection; cells in conventional cell cultures may often yield a phenotype that is different from that in an organized structure 19. Several ECHO viruses have been reported to show cytolytic effect in monolayer cell cultures, while investigation of cytolytic effect in spheroids have shown that not all cells, when grown in spheroids, are lysed by viruses 20.
Replication of enteroviruses occurs in the cytoplasm of infected cells 21 and is followed by cell lysis. Several receptors have been suggested to be involved in the infection mechanism of ECHO-7 virus. CD55 (also known as decay accelerating factor, DAF) has been reported as one of the main ECHO-7 receptors necessary for entry and infection 22-24, although it has been noted that other factors may also be involved such as β-2 microglobulin 25, 26. Studies carried out in polarized epithelial cells show that ECHO-7 virus first binds to the CD55 receptor, followed by internalization involving the clathrin-mediated endocytosis. After penetration of the cell, the ECHO-7 virus moves to early endosome before viral RNA is released into the cytoplasm, which is controlled by Rab5 and then moves to late endosome in a process where Rab7 GTPase is involved 27. Moreover, it has been reported that anti-CD59 antiserum also blocks ECHO-7 virus infection in RD cells 26.
One of the defence mechanisms of cancer cells to the immune system is to express membrane complement regulatory proteins. CD35, CD46, CD55, and CD59 expression is associated with blockade of the complement cascade. In normal tissues, these proteins protect from accidental injury by an activated complement system, however, in cancer cells the same proteins protect from the immune system 28. It is possible that cancer cells express larger amounts of the proteins and receptors that are responsible for viral entry (such as CD55) than normal cells, thus, creating apparent viral tropism for these cells.
The present experiments were limited by focusing only on the cell susceptibility to viral infection in vitro. Immune cell responses and interactions between immune cells and cancer cells might also play an important role in the in vivo / clinical response to Rigvir®.
In conclusion, the present in vitro study suggests that a mechanism of action for the clinical benefit of Rigvir® is its cytolytic properties. While Rigvir® is registered for melanoma, other cancer types such as lung, gastric, rhabdomyosarcoma, and pancreatic cancer might also be sensitive to Rigvir®. Further studies regarding how Rigvir® targets specific cancer cells in the body, entry mechanisms and immune response(s) are also needed. It is likely that observed effect on HDFa and MSC cell viability does not represent an in vivo action of Rigvir®.
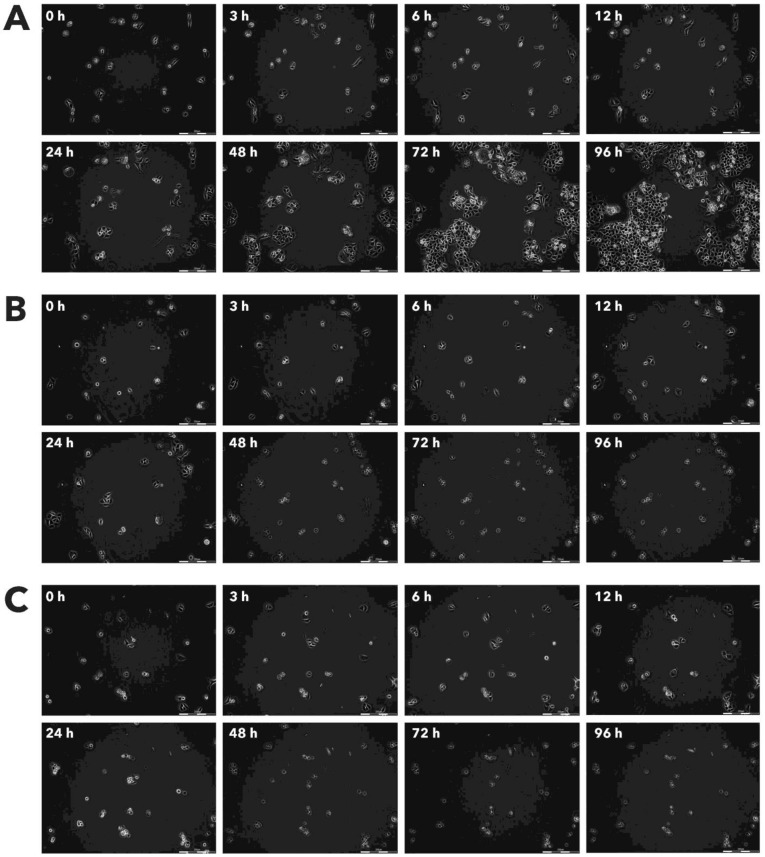
Figure 2.
Effect of Rigvir® on FM-9 viable cell count. Photographs taken at time points 0, 3, 6, 12, 24, 48, 72, and 96h of incubation. Control (A), Rigvir® (1%) (B) and Rigvir® (10%) (C). Scale bar is 200 µm.
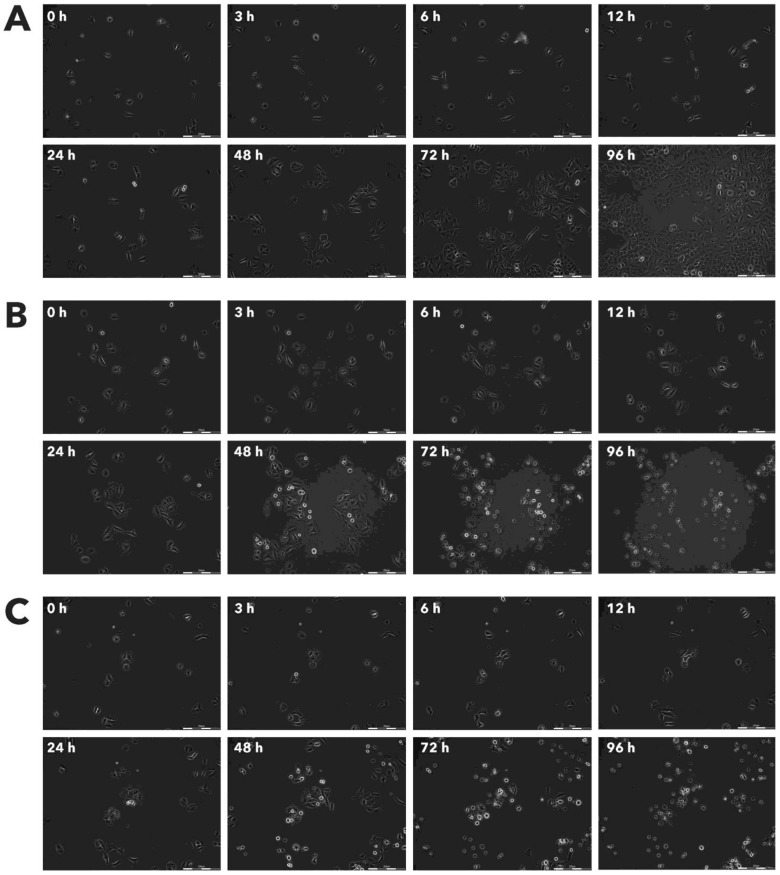
Figure 3.
Effect of Rigvir® on RD viable cell count. Photographs taken at time points 0, 3, 6, 12, 24, 48, 72, and 96h of incubation. Control (A), Rigvir® (1%) (B) and Rigvir® (10%) (C). Scale bar is 200 µm.
Figure 4.
Effect of Rigvir® on AGS viable cell count. Photographs taken at time points 0, 3, 6, 12, 24, 48, 72, and 96h of incubation. Control (A), Rigvir® (1%) (B) and Rigvir® (10%) (C). Scale bar is 200 µm.
Figure 5.
Effect of Rigvir® on A549 viable cell count. Photographs taken at time points 0, 3, 6, 12, 24, 48, 72, and 96h of incubation. Control (A), Rigvir® (1%) (B) and Rigvir® (10%) (C). Scale bar is 200 µm.
Figure 6.
Effect of Rigvir® on HDFa viable cell count. Photographs taken at time points 0, 3, 6, 12, 24, 48, 72, and 96h of incubation. Control (A), Rigvir® (1%) (B) and Rigvir® (10%) (C). Scale bar is 200 µm.
Figure 7.
Effect of Rigvir® on HPAF-II viable cell count. Photographs taken at time points 0, 3, 6, 12, 24, 48, 72, and 96h of incubation. Control (A), Rigvir® (1%) (B) and Rigvir® (10%) (C). Scale bar is 200 µm.
Figure 8.
Effect of Rigvir® on MSC viable cell count. Photographs taken at time points 0, 3, 6, 12, 24, 48, 72, and 96h of incubation. Control (A), Rigvir® (1%) (B) and Rigvir® (10%) (C). Scale bar is 200 µm.
Figure 9.
Effect of Rigvir® on MCF-7 viable cell count. Photographs taken at time points 0, 3, 6, 12, 24, 48, 72, and 96h of incubation. Control (A), Rigvir® (1%) (B) and Rigvir® (10%) (C). Scale bar is 200 µm.
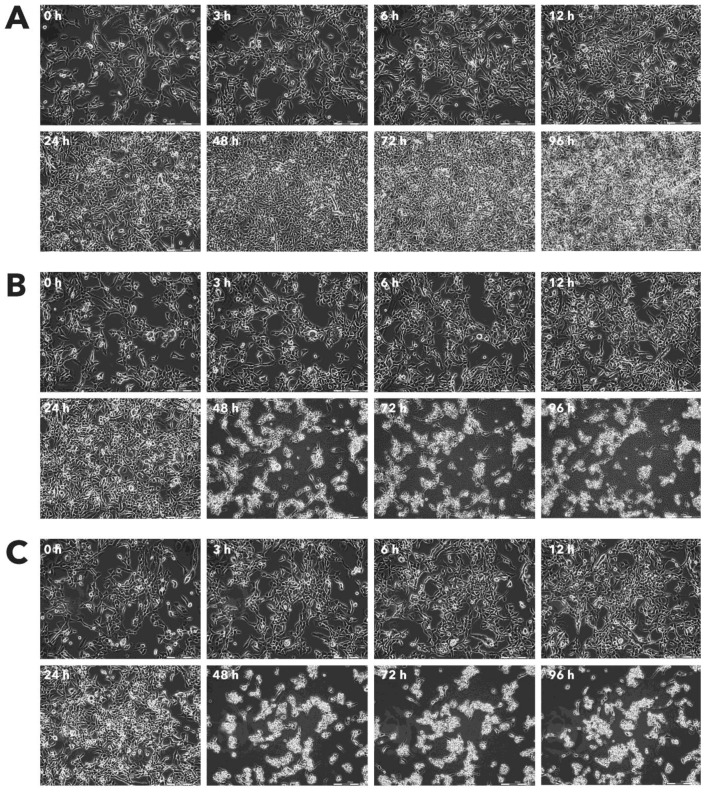
Figure 10.
Effect of Rigvir® on HaCaT viable cell count. Photographs taken at time points 0, 3, 6, 12, 24, 48, 72, and 96h of incubation. Control (A), Rigvir® (1%) (B) and Rigvir® (10%) (C). Scale bar is 200 µm.
Acknowledgments
The technical assistance of Ms. V. Telle is acknowledged.
Ethics approval and consent to participate
The study was approved by the local ethics committee.
Authors' contributions
ARS, MB, and PA made substantial contributions to conception and design, AT, LP, and IB made substantial contributions to acquisition of data, analysis, and interpretation of data. AT and PA drafted the manuscript. All authors have read and approved the final version for publication.
Availability of data and material
All relevant data and materials are included in the manuscript.
Abbreviations
- Ab
antibody
- BSA
Bovine serum albumin
- DMEM
Dulbecco's Modified Eagle Medium
- ECHO
enteric cytopathic human orphan
- FBS
foetal bovine serum
- PBMC
peripheral blood mononuclear cells
- PBS
phosphate buffered saline
- RPMI
Roswell Park Memorial Institute.
References
- 1.Kelly E, Russell SJ. History of oncolytic viruses: genesis to genetic engineering. Mol Ther. 2007;15:651–9. doi: 10.1038/sj.mt.6300108. [DOI] [PubMed] [Google Scholar]
- 2.Kaufman HL, Kohlhapp FJ, Zloza A. Oncolytic viruses: a new class of immunotherapy drugs. Nat Rev Drug Discov. 2015;14:642–62. doi: 10.1038/nrd4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andtbacka RH, Kaufman HL, Collichio F, Amatruda T, Senzer N, Chesney J. et al. Talimogene Laherparepvec improves durable response rate in patients with advanced melanoma. JClinOncol. 2015;33:2780–8. doi: 10.1200/JCO.2014.58.3377. [DOI] [PubMed] [Google Scholar]
- 4.Doniņa S, Strēle I, Proboka G, Auziņš J, Alberts P, Jonsson B. et al. Adapted ECHO-7 virus Rigvir immunotherapy (oncolytic virotherapy) prolongs survival in melanoma patients after surgical excision of the tumour in a retrospective study. Melanoma Research. 2015;25:421–6. doi: 10.1097/CMR.0000000000000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alberts P, Olmane E, Brokāne L, Krastiņa Z, Romanovska M, Kupčs K. et al. Long-term treatment with the oncolytic ECHO-7 virus Rigvir of a melanoma stage IV M1c patient, a small cell lung cancer stage IIIA patient, and a histiocytic sarcoma stage IV patient-three case reports. APMIS. 2016;124:896–904. doi: 10.1111/apm.12576. [DOI] [PubMed] [Google Scholar]
- 6.Fiola C, Peeters B, Fournier P, Arnold A, Bucur M, Schirrmacher V. Tumor selective replication of Newcastle disease virus: association with defects of tumor cells in antiviral defence. International journal of cancer. 2006;119:328–38. doi: 10.1002/ijc.21821. [DOI] [PubMed] [Google Scholar]
- 7.Lam HY, Yeap SK, Pirozyan MR, Omar AR, Yusoff K, Suraini AA. et al. Safety and clinical usage of newcastle disease virus in cancer therapy. Journal of biomedicine & biotechnology. 2011;2011:718710. doi: 10.1155/2011/718710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parato KA, Breitbach CJ, Le BF, Wang J, Storbeck C, Ilkow C. et al. The oncolytic poxvirus JX-594 selectively replicates in and destroys cancer cells driven by genetic pathways commonly activated in cancers. Mol Ther. 2012;20:749–58. doi: 10.1038/mt.2011.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bell J, McFadden G. Viruses for Tumor Therapy. Cell Host Microbe. 2014;15:260–5. doi: 10.1016/j.chom.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Atsumi S, Matsumine A, Toyoda H, Niimi R, Iino T, Nakamura T. et al. Oncolytic virotherapy for human bone and soft tissue sarcomas using live attenuated poliovirus. International journal of oncology. 2012;41:893–902. doi: 10.3892/ijo.2012.1514. [DOI] [PubMed] [Google Scholar]
- 11.Campbell SA, Gromeier M. Oncolytic viruses for cancer therapy II. Cell-internal factors for conditional growth in neoplastic cells. Onkologie. 2005;28:209–15. doi: 10.1159/000084010. [DOI] [PubMed] [Google Scholar]
- 12.Campbell SA, Gromeier M. Oncolytic viruses for cancer therapy I. Cell-external factors: virus entry and receptor interaction. Onkologie. 2005;28:144–9. doi: 10.1159/000083659. [DOI] [PubMed] [Google Scholar]
- 13.Chiocca EA, Rabkin SD. Oncolytic Viruses and Their Application to Cancer Immunotherapy. Cancer Immunol Res. 2014;2:295–300. doi: 10.1158/2326-6066.CIR-14-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pol J, Buque A, Aranda F, Bloy N, Cremer I, Eggermont A. et al. Trial Watch-Oncolytic viruses and cancer therapy. Oncoimmunology. 2016;5:e1117740. doi: 10.1080/2162402X.2015.1117740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vuorinen T, Vainionpaa R, Heino J, Hyypia T. Enterovirus receptors and virus replication in human leukocytes. J Gen Virol. 1999;80( Pt 4):921–7. doi: 10.1099/0022-1317-80-4-921. [DOI] [PubMed] [Google Scholar]
- 16.Glinkina LS, Bruvere RZ. [The reaction of the T-immunity system in patients with malignant skin melanoma and stomach cancer to active nonspecific immunotherapy] (In Russian) Vopr Onkol. 1992;38:659–66. [PubMed] [Google Scholar]
- 17.Lovitt CJ, Shelper TB, Avery VM. Advanced Cell Culture Techniques for Cancer Drug Discovery. Biology (Basel) 2014;3:345–67. doi: 10.3390/biology3020345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang Y, Eglen RM. Three-Dimensional Cell Cultures in Drug Discovery and Development. SLAS Discov. 2017;22:456–72. doi: 10.1177/1087057117696795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramanan V. New Methods in Tissue Engineering: Improved Models for Viral Infection. Annu Rev Virol. 2014;1:475–99. doi: 10.1146/annurev-virology-031413-085437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Israelsson S, Jonsson N, Gullberg M, Lindberg AM. Cytolytic replication of echoviruses in colon cancer cell lines. Virol J. 2011;8:473. doi: 10.1186/1743-422X-8-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Summers DF, Maizel JV. Evidence for large precursor proteins in poliovirus synthesis. Proc Natl Acad Sci U S A. 1968;59:966–71. doi: 10.1073/pnas.59.3.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ward T, Pipkin PA, Clarkson NA, Stone DM, Minor PD, Almond JW. Decay-accelerating factor CD55 is identified as the receptor for echovirus 7 using CELICS, a rapid immuno-focal cloning method. EMBO J. 1994;13:5070–4. doi: 10.1002/j.1460-2075.1994.tb06836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bergelson JM, Chan M, Solomon KR, St John NF, Lin H, Finberg RW. Decay-accelerating factor (CD55), a glycosylphosphatidylinositol-anchored complement regulatory protein, is a receptor for several echoviruses. Proc Natl Acad Sci USA. 1994;91:6245–8. doi: 10.1073/pnas.91.13.6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clarkson NA, Kaufman R, Lublin DM, Ward T, Pipkin PA, Minor PD. et al. Characterization of the echovirus 7 receptor: domains of CD55 critical for virus binding. J Virol. 1995;69:5497–501. doi: 10.1128/jvi.69.9.5497-5501.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ward T, Powell RM, Pipkin PA, Evans DJ, Minor PD, Almond JW. Role for beta2-microglobulin in echovirus infection of rhabdomyosarcoma cells. J Virol. 1998;72:5360–5. doi: 10.1128/jvi.72.7.5360-5365.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goodfellow IG, Powell RM, Ward T, Spiller OB, Almond JW, Evans DJ. Echovirus infection of rhabdomyosarcoma cells is inhibited by antiserum to the complement control protein CD59. J GenVirol. 2000;81:1393–401. doi: 10.1099/0022-1317-81-5-1393. [DOI] [PubMed] [Google Scholar]
- 27.Kim C, Bergelson JM. Echovirus 7 entry into polarized intestinal epithelial cells requires clathrin and Rab7. MBio. 2012;3:e00304–11. doi: 10.1128/mBio.00304-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fishelson Z, Donin N, Zell S, Schultz S, Kirschfink M. Obstacles to cancer immunotherapy: expression of membrane complement regulatory proteins (mCRPs) in tumors. Molecular Immunology. 2003;40:109–23. doi: 10.1016/s0161-5890(03)00112-3. [DOI] [PubMed] [Google Scholar]