Abstract
Endometrial cancer is the most common gynecological malignancy in the developed world. Although there is evidence of genetic predisposition to the disease, most of the genetic risk remains unexplained. We present the meta-analysis results of four genome-wide association studies (4907 cases and 11 945 controls total) in women of European ancestry. We describe one new locus reaching genome-wide significance (P < 5 × 10 −8) at 6p22.3 (rs1740828; P = 2.29 × 10 −8, OR = 1.20), providing evidence of an additional region of interest for genetic susceptibility to endometrial cancer.
Introduction
Endometrial carcinoma (EC), which arises from the epithelial lining of the uterus, is the sixth most common cancer among females worldwide and the most common gynecological malignancy in developed countries (1). According to SEER data (2), between 2005 and 2011, 18.3% of women with EC in the United States did not survive 5 or more years after diagnosis. Incidence rates of EC in developed countries are increasing over time (3,4), with most diagnoses made after age 55, making this a significant concern for older women in an aging population. A number of modifiable risk factors have been established, including obesity, estrogen-only post-menopausal hormone therapy and reproductive history. However, not much is known about the genetic etiology of EC.
Evidence suggests a component of genetic predisposition to EC. Multiple studies have seen a >2-fold risk in those with a family history of EC (5–7) and risk for women with first-degree female relatives with early onset disease increases nearly 3-fold (8). Additionally, women with Lynch Syndrome, a hereditary autosomal dominant genetic condition due to germline pathogenic variants in DNA mismatch repair genes, have an estimated lifetime risk of EC between 40% and 70% (9). Heritability estimates for EC are as high as 52% (10–12), though inconsistency in heritability estimates indicates the true value is likely lower.
Genome-wide association studies (GWAS) have discovered more than 1500 common variants associated with a variety of cancer types (13). However, the statistical power of GWAS may be limited by the modest effect sizes of common variants and by inadequate sample sizes (14,15). To date, three independent GWAS have been conducted to identify single nucleotide polymorphisms (SNPs) that contribute to EC risk. One GWAS found a significant association between rs4430796, in 17q12 near HNF1B, and EC risk (16). Fine-mapping of this region identified likely variants underlying this association in HNF1B intron 1 (17). Analysis including a more comprehensive validation phase of this GWAS has since identified an additional six loci associated with EC risk at genome-wide levels of significance ((18), Cheng et al. submitted for publication). However, no other novel genome-wide significant loci associated with EC risk were identified by the two other published GWAS (14,15).
Meta-analysis methods synthesize summary data from multiple independent studies, increasing power and reducing false-positive findings (19). We thus conducted a discovery meta-analysis of four GWAS datasets of women of European ancestry for a total of 4907 cases and 11 945 controls, comprising the largest discovery dataset for EC yet.
Results
Meta-analysis of GWAS results for risk of EC
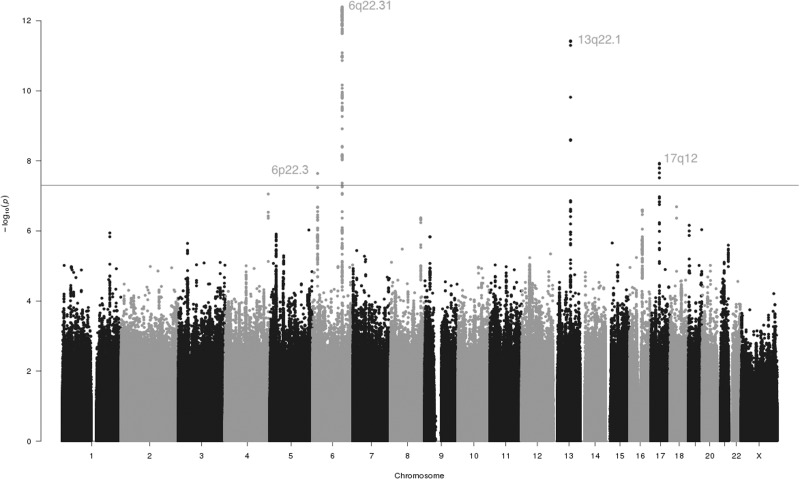
Meta-analysis of GWAS results from the Australian National Endometrial Cancer Study (ANECS), the US Epidemiology of Endometrial Cancer Consortium (E2C2), the UK National Study of Endometrial Cancer Genetics (NSECG) and the UK Studies of Epidemiology and Risk factors in Cancer Heredity (SEARCH) in 4907 cases and 11 945 controls of European ancestry examined 9 486 271 SNPs for association with risk of EC. No evidence of genomic inflation was observed in the meta-analysis (λGC = 1.013, Supplementary Material, Fig. S1). After implementing quality control, including removal of SNPs with P-values for heterogeneity <0.05 from further consideration, a total of 137 SNPs clustered in four chromosomal regions reached genome-wide significance at P < 5 × 10 −8 (Fig. 1, Supplementary Material, Table S1).
Figure 1.
Manhattan plot of meta-analysis results for EC in four cohorts. Association results between imputed and genotyped SNPs and risk of EC in women of European ancestry are depicted. Dashed line indicates the log of the threshold for genome-wide significance (P < 5.0 × 10 −8).
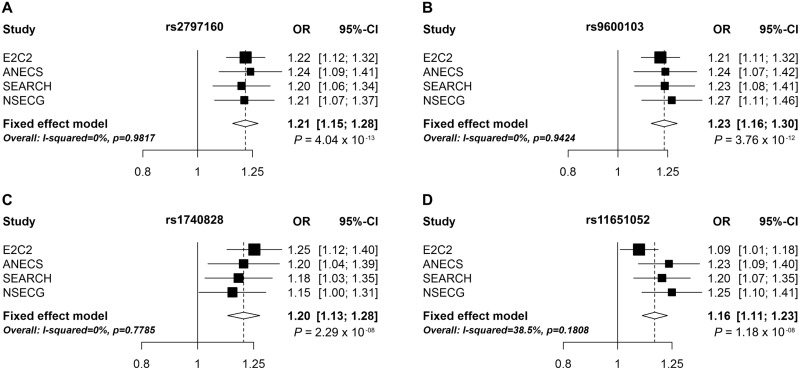
This meta-analysis of four independent EC GWAS datasets identified four loci with genome-wide levels of significance (Table 1). Three loci have been discovered previously by analyses that included the ANECS, SEARCH and NSECG GWAS datasets ((16,18), Cheng, TH et al. submitted for publication): 17q12 near HNF1B, 13q22.1 near KLF5 and 6q22.31 intronic to LOC643623. The direction of effect for all three previously identified loci in the E2C2 GWAS alone was consistent with that observed in the original studies (Fig. 2). In the E2C2 GWAS alone, P-values for the most significant SNPs in 13q22.1 (rs9600103, E2C2 P = 1.74 × 10−5) and 6q22.31 (rs2797160, E2C2 P = 1.18 × 10−6) exceeded the confirmation threshold of P = 0.017 based on a Bonferroni correction for three tests, representing an independent validation of these two previously reported EC GWAS hits.
Table 1.
Association results for loci reaching genome-wide significance with no evidence of significant study heterogeneity
| Lead SNP | Chromosome | Position (hg19) | Nearby gene | Description | Alleles | OR | P | RAFa |
|---|---|---|---|---|---|---|---|---|
| rs2797160 | 6q22.31 | 126010116 | LOC643623b | Intronic | A/G | 1.21 | 4.04E-13 | 0.578 |
| rs9600103 | 13q22.1 | 73811879 | KLF5 | Intergenic | A/T | 1.23 | 3.76E-12 | 0.722 |
| rs1740828 | 6p22.3 | 21649085 | SOX4 | Intergenic | G/A | 1.20 | 2.29E-08 | 0.516 |
| rs11651052 | 17q12 | 36102381 | HNF1B | Intronic | G/A | 1.16 | 1.18E-08 | 0.535 |
Risk allele frequency.
Uncharacterized gene region.
Figure 2.
Forest plots of the odds ratios for the association between rs2797160, rs1740828, rs9600103, rs11651052 and EC.
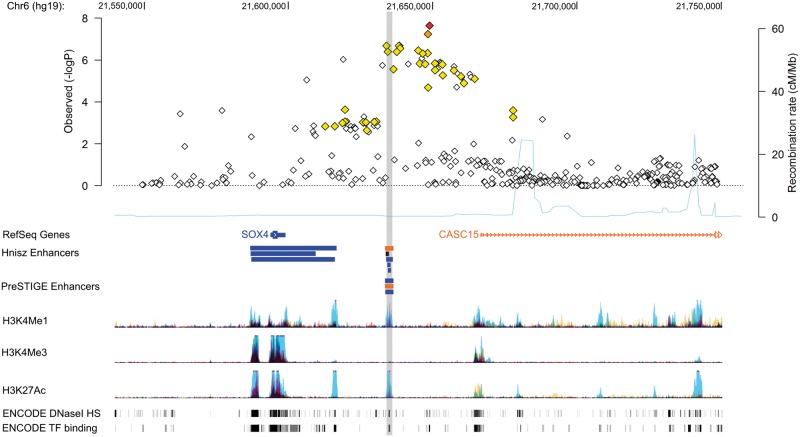
The fourth locus at 6p22.3 is a novel risk region for EC, represented by rs1740828 (OR = 1.20, P = 2.29 × 10−8) (Table 1). This locus at 6p22.3 falls in an intergenic region between SOX4 and CASC15 (Fig. 3). SOX4 encodes a transcription factor involved in the regulation of several aspects of development (20). CASC15 is a long intergenic non-coding RNA that has been identified as a neuroblastoma susceptibility locus (21,22).
Figure 3.
Regional association plot of 6p22.3 with annotation of genomic features, likely enhancers, and target genes. Association results for all SNPs in the 6p22.3 locus with EC risk from the meta-analysis are shown in the first panel. SNPs are plotted as the negative log of the P-value against relative position across the locus (base position [hg19] displayed across the top). The lead SNP, rs1740828, is shown as a red filled diamond. LD with surrounding SNPs are indicated by color (SNPs 0.5 ≤ r2 < 0.8 are orange, 0.2 ≤ r2 <0.5 are yellow, and r2 < 0.2 are unfilled). There were no SNPs with an r2 ≥ 0.8 to the lead SNP. The second panel displays genes as identified by RefSeq. Likely enhancers predicted by Hnisz et al. (23) and PreSTIGE (24) that overlap SNPs in LD (r2 > 0.2) with the lead SNP are depicted as colored bars, where the color matches the schematic of its predicted target gene (the black bar is predicted to target CDKAL1, not shown in this figure). Histone modification associated with promoters (H3K4Me1) and enhancers (H3K4Me1 and H3K27Ac) from seven ENCODE Project cell types and DNaseI hypersensitivity sites (DHS) and transcription factor (TF) binding sites identified in 125 and 91 ENCODE Project cell types, respectively, are also displayed.
Conditional and joint analyses of these four regions did not identify any secondary association signals, indicating no additional independently associated SNPs after conditioning on the region’s lead SNP.
Functional annotation
Though the most significant risk-associated SNP at 6p22.3 is located in an intergenic region, it may be a marker for an underlying variant that may modulate or regulate nearby or distant genes. To pursue a putative functional role that variants at 6p22.3 may have in risk of EC, we annotated SNPs in linkage disequilibrium (LD) (r2 > 0.2 in EU 1000 Genomes) with the region’s lead SNP, rs1740828, with publicly available data on relevant regulatory elements located near the susceptibility region. Candidate causal SNPs with log likelihood ratios of >1:100 compared with rs1740828 (r2 between 0.2 and 0.5) overlap with putative enhancers defined by Hnisz (23) and PreSTIGE (24) for SOX4, CASC15 and CDKAL1 (Fig. 3). CDKAL1 encodes for a methylthiotransferase and is a known type 2 diabetes susceptibility gene (25–27). ENCODE data also show these SNPs mapped to regions displaying evidence of enhancer-specific histone modification (mono-methylation of H3 lysine 4 (H3K4Me1) and H3 lysine 27 acetylation (H3K27Ac)), DNAseI hypersensitivity sites representative of open chromatin, and regions bound by transcription factors.
Expression quantitative trait loci analysis
In order to identify potential biological mechanisms underlying the association between the 6p22.3 locus and EC risk, we performed expression quantitative trait loci (eQTL) analysis using publicly available mRNA expression, somatic copy-number variation and methylation data of 408 EC tumor tissues and 30 adjacent normal endometrial tissues from The Cancer Genome Atlas (TCGA). Expression levels of SOX4, CASC15 and CDKAL1, identified as potential target genes by cross reference to Hnisz and PreSTIGE data, were assessed in the analysis. After adjusting for multiple comparisons, no significant associations were seen between SNPs in the risk loci region (Chr6:21549085–21749085) and expression levels of any of these three genes (Supplementary Material, Table S2a and b). Associations between SNPs and gene expression were also explored using uterine-specific Genotype-Tissue Expression project data (www.gtexportal.org). Similarly, no significant associations were observed between risk SNPs and expression levels of the target genes (data not shown).
Discussion
Our EC GWAS meta-analysis, the largest discovery data set for EC yet, identified one new susceptibility locus at 6p22.3 and confirmed previously discovered loci at 6q22.31 and 13q22.1. The new locus at 6p22.3, represented by rs1740828, lies between two genes, SOX4 and CASC15.
Assuming a log-additive association with risk, these four loci are estimated to account for ∼4.4% of the familial relative risk of EC in women of European ancestry. This fraction is less than what has been discovered in studies with comparable sample sizes for cancers such as colorectal (28) and pancreatic cancer (29). It is likely that additional common variants with more modest effect sizes, as well as copy-number variants, rare variants and indels not tagged by current genotyping arrays, have yet to be discovered, and will contribute to explaining familial EC risk. Our meta-analysis was ≥80% powered to detect an association of the magnitude of rs1740828 for SNPs with minor allele frequency (MAF) > 0.21, suggesting that even larger sample sizes would be needed to detect modest effects from lower frequency variants.
Functional annotation suggests that SNPs in LD with rs1740828 overlap putative enhancers for SOX4, CASC15 and CDKAL1. Our eQTL results do not support regulation of these particular genes by SNPs falling within 100 kb of the lead SNP of the 6p22.3 locus that we identified. However, this may be due to the lack of substantial eQTL data available for adjacent normal endometrial tissue or because eQTLs are context-dependent and may only be expressed in certain stages of cancer development or only when under particular stimuli. Comprehensive studies involving fine-mapping as well as functional analysis are needed to identify biological processes underlying our observed GWAS-identified risk signal at 6p22.3.
Of note, existing data suggest that the 6p22.3 region is relevant to cancer susceptibility in general, summarized in a review of genetic and biological studies reporting on the associations of CASC15, CDKAL1 and SOX4 SNPs and gene expression with cancer risk and prognosis (Supplementary Material, Table S3). In larger studies (21,30), SNPs in/near CASC15 have been associated with neuroblastoma (P < 10−9), and increased CASC15 expression has been implicated in melanoma progression (31). A GWAS of bladder cancer provided suggestive evidence of increased risk in the CDKAL1 region (lead SNP rs4510656, P = 6.98 × 10 −7) (32). Given the established associations between EC risk and body mass index (BMI) (33) and diabetes (34), it is no that the CDKAL1 region is also associated with diabetes risk and BMI (35). Furthermore, although the SOX4 region has yet to be associated with cancer risk by GWAS to date, SOX4 overexpression has been implicated in malignancy and poor prognosis in a variety of cancers, including chondrosarcoma (36) and cancers of the lung (37–39), prostate (40,41), breast (42,43) and endometrium (44). A meta-analysis of 10 studies with >1000 cancer patients reported that SOX4 tumor overexpression is modestly correlated with poor overall survival (45).
In summary, our study has identified a new EC risk locus at 6p22.3. Given previously published associations of SNPs in this region at either genome-wide or no levels of significance (P < 10−6) with other cancer types, our results also highlight this region as a potential general cancer susceptibility locus. Extensive fine-mapping and functional studies are required to identify the biological basis of cancer risk at this region.
Materials and Methods
Datasets
Four large genotyping studies, the ANECS, E2C2, NSECG and SEARCH, contributed a total of 16 852 women (4907 cases, 11 945 controls) of European ancestry with confirmed EC diagnosis to the meta-analysis. We did not restrict by EC subtype in this analysis. Details of the participating studies and genotyping platforms used are provided in Supplementary Material, Table S4.
Briefly, 606 cases from ANECS (16) were compared to 3083 Australian controls from the Brisbane Adolescent Twin Study (QIMR Controls) (46,47) (n = 1846) and the Hunter Community Study (48) (n = 1237). E2C2 (49) is an NCI-supported international consortium of more than 45 studies created to investigate the etiology of EC. As previously described (15), four US-based cohort studies, two US-based case-control studies and one Poland-based case-control study from the consortium contributed 2695 cases and 2777 controls to this analysis. Cases from NSECG (17) (n = 925) were compared with 895 controls from the UK1/CORGI colorectal cancer study (50). Cases from SEARCH (16) (n = 681) were compared to 5190 controls from the Wellcome Trust Case-Control Consortium (51).
Genotyping and imputation
Within each study, genotyping was performed on specific Illumina platforms, as detailed in Supplementary Material, Table S4. Quality control methods agreed upon by all studies were implemented. Briefly, this involved exclusion of SNPs with call rates <95%, MAFs <1%, Hardy–Weinberg violation of at least P <10−12 for cases and P <10−7 for controls, or individuals who are genetically male, first-degree cryptic relations or duplicates, or with call rates <95%. All genotypes were imputed to the positive strand of the 1000 Genomes Project v3, phase 1 dataset with either Minimac (52) or IMPUTE2 (53).
Statistical analysis
Primary association analyses of single variants with EC risk were performed separately in each study using logistic regression implemented with SNPTEST v2 (54) or ProbABEL (55), adjusting for relevant principal components and variables specific to the study. Summary statistics reported from each study were combined using fixed-effect meta-analysis with inverse variance weights in METAL (56). The P-value threshold to reach genome-wide significance in the meta-analysis was set to 5 × 10−8. Heterogeneity across studies was assessed using Cochran’s Q statistic. Conditional and joint analysis of summary-level associations, performed with GCTA (57), was used to determine the presence of secondary associations within chromosomal regions of size <500 kb. The power to detect an association of equal magnitude to rs1740828, the most significant result in the meta-analysis, was calculated using QUANTO 1.2 (58).
Functional annotation
SNPs in LD, defined as r2 > 0.2 in the European 1000 Genomes data, with the most significant SNP (rs1740828) were annotated using HaploregV2 (59) and data from ENCODE (60) including promoter and enhancer histone marks, DNaseI hypersensitivity sites, bound proteins and altered motifs. Additionally, enhancer-gene pairs reported by Hnisz (23) and PreSTIGE (24) were cross-referenced against risk loci to identify likely enhancers overlapping SNPs in LD (r2 > 0.2) with rs1740828.
eQTL analysis
To examine tissue-specific eQTLs, data from EC patients were accessed from TCGA (61). Normalized RNA-Seq, copy-number and methylation data were downloaded through the Cancer Browser (https://genome-cancer.ucsc.edu, last accessed April 1, 2016). Germline SNP genotypes (Affymetrix 6.0 arrays) were downloaded through the TCGA controlled access portal (https://tcga-data.nci.nih.gov/tcga/, last accessed April 1, 2016) and QC performed. SNPs were excluded for call rate <95%, MAF <1% or deviations from Hardy-Weinberg equilbrium significant at 10 −4. Samples were excluded for low overall call rate (<95%), heterozygosity >3 standard deviations from the mean and non-female sex status (X-chromosome homozygosity rate >0.2). For duplicate samples or samples identified as close relatives by Identity-By-State probabilities >0.85, the sample with the lower call rate was excluded. To assess untyped SNPs, we imputed genotypes present in the 1000 Genomes dataset Phase 3v5 in the risk locus region (±100 kb of the lead SNP, rs1740828) for SNPs that were not genotyped by the Affymetrix 6.0 platform. Haplotypes were phased using the MaCH program (62) before running minimac for genotype imputation (53,52), using the recommended parameters (20 iterations of the Markov sampler and 200 states). SNPs imputed with an r2 > 0.3 and MAF > 0.01 were included in the eQTL analysis. Associations were assessed after Bonferroni correction for the total number of tests performed (number of SNP investigated = 2088, number of genes assessed= 3 and number of sample sets = 2), with a P-value < 4.0 × 10 −6 required for statistical significance.
Thirty cancer tissue samples had adjacent normal endometrial tissues available with complete genotype and RNA-Seq data. Since gene expression in tumors is affected by acquired somatic alterations, we accounted for somatic copy-number variation and methylation in eQTL analysis of EC tissue. In total, 366 TCGA patients had complete genotype, RNA-Seq, copy-number and methylation data available for the analysis. Expression of SOX4, CASC15 and CDKAL1 (which were identified as target genes by cross-reference to Hnisz and PreSTIGE data) were adjusted for sequencing platform (Illumina GA or Illumina HiSeq) in adjacent normal EC, and adjusted for sequencing platform, copy-number variation and methylation in EC tissue. The associations between genotype and residual gene expression were evaluated using linear regression models by the mach2qtl program (62,63).
Contribution to familial risk
Contribution of known SNPs to familial relative risk under a multiplicative model was computed using the formula detailed in Eeles et al. (64). We assumed the observed familial risk to first-degree relatives of EC cases was 2-fold, the loci had a log-additive association with risk and the loci were not in LD.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
ANECS, SEARCH, QIMR, HCS, NSECG and UK1/CORGI thank the many individuals who participated in the research studies. We also thank Kaltin Ferguson, Felicity Lose, Shahana Ahmed, Catherine Healey, Kyriaki Michailidou, Ella Barclay, Lynn Martin and the ANECS research team, the Eastern Cancer Registration and Information Centre and the SEARCH research team, the National Cancer Research Network, the numerous institutions and their staff who supported recruitment. We thank Nick Martin, Grant Montgomery, Dale Nyholt and Anjali Henders for access to GWAS data from QIMR Controls. We thank and acknowledge the contribution of our clinical and scientific collaborators and their staff. See (http://www.anecs.org.au/, last accessed April 4, 2016) for full listing of the ANECS Group and other contributors to ANECS. SEARCH collaborators include Mitul Shah, Caroline Baynes, Don Conroy, Bridget Curzon, Patricia Harrington, Sue Irvine, Clare Jordan, Craig Luccarini, Rebecca Mayes, Hannah Munday, Barbara Perkins, Radka Platte, Anabel Simpson, Anne Stafford and Judy West. The NSECG Group comprises: M. Adams, A. Al-Samarraie, S. Anwar, R. Athavale, S. Awad, A. Bali, A. Barnes, G. Cawdell, S. Chan, K. Chin, P. Comes, M. Crawford, J. Cullimore, S. Ghaem-Maghami, R. Gomall, J. Green, M. Hall, M. Harvey, J. Hawe, A. Head, J. Herod, M. Hingorani, M. Hocking, C. Holland, T. Hollingsworth, J. Hollingworth, T. Ind, R. Irvine, C. Irwin, M. Katesmark, S. Kehoe, G. Kheng-Chew, K. Lankester, A. Linder, D. Luesley, C. B-Lynch, V. McFarlane, R. Naik, N. Nicholas, D. Nugent, S. Oates, A. Oladipo, A. Papadopoulos, S. Pearson, D. Radstone, S. Raju, A. Rathmell, C. Redman, M. Rymer, P. Sarhanis, G. Sparrow, N. Stuart, S. Sundar, A. Thompson, S. Tinkler, S. Trent, A. Tristram, N. Walji and R. Woolas. QIMR thanks Margie Wright, Lisa Bowdler, Sara Smith, Megan Campbell and Scott Gordon for control sample collection and data collection. The authors would like to thank the participants and staff of the Nurses’ Health Study for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. The authors assume full responsibility for analyses and interpretation of these data. The authors would also like to thank Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School. Finally, the authors would also like to acknowledge Pati Soule and Hardeep Ranu for their laboratory assistance.
Conflict of interest statement. None declared.
Funding
ANECS recruitment was supported by project grants from the National Health and Medical Research Council of Australia (ID 339435), The Cancer Council Queensland (ID 4196615) and Cancer Council Tasmania (ID 403031 and ID 457636). SEARCH recruitment was funded by a programme grant from Cancer Research UK [C490/A10124]. Case genotyping was supported by the National Health and Medical Research Council (ID 552402). Control data were generated by the Wellcome Trust Case Control Consortium (WTCCC), and a full list of the investigators who contributed to the generation of the data is available from the WTCCC website. We acknowledge use of DNA from the British 1958 Birth Cohort collection, funded by the Medical Research Council grant G0000934 and the Wellcome Trust grant 068545/Z/02—funding for this project was provided by the Wellcome Trust under award 085475. NSECG was supported by the EU FP7 CHIBCHA grant and CORGI by Cancer Research UK. Recruitment of the QIMR controls was supported by the National Health and Medical Research Council of Australia (NHMRC). The University of Newcastle, the Gladys M Brawn Senior Research Fellowship scheme, The Vincent Fairfax Family Foundation, the Hunter Medical Research Institute and the Hunter Area Pathology Service all contributed towards the costs of establishing the Hunter Community Study. T.O’M and J.N.P. are supported by NHMRC project grant (ID APP1031333). A.B.S. is supported by the National Health and Medical Research Council (NHMRC) Fellowship Scheme. D.F.E. is a Principal Research Fellow of Cancer Research UK. A.M.D. was supported by the Joseph Mitchell Trust. I.T. is supported by Cancer Research UK and the Oxford Comprehensive Biomedical Research Centre. M.M.C. is supported by training grant 5T32CA009001-38 from the NCI. The Nurses’ Health Study (NHS) is supported by the NCI, NIH Grants Number UM1 CA186107, P01 CA087969, R01 CA49449, 1R01 CA134958 and 2R01 CA082838. The Connecticut Endometrial Cancer Study was supported by NCI, NIH Grant Number 2R01 CA082838. The Fred Hutchinson Cancer Research Center (FHCRC) is supported by NCI, NIH Grant Number 2R01 CA082838, NIH RO1 CA105212, RO1 CA 87538, RO1 CA75977, RO3 CA80636, NO1 HD23166, R35 CA39779, KO5 CA92002 and funds from the Fred Hutchinson Cancer Research Center. The Multiethnic Cohort Study (MEC) is supported by the NCI, NHI Grants Number CA54281, CA128008 and 2R01 CA082838. The California Teachers Study (CTS) is supported by NCI, NIH Grant Number 2R01 CA082838, R01 CA91019 and R01 CA77398, and contract 97-10500 from the California Breast Cancer Research Fund. The Polish Endometrial Cancer Study (PECS) is supported by the Intramural Research Program of the NCI. The Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial (PLCO) is supported by the Extramural and the Intramural Research Programs of the NCI.
References
- 1. Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D.M., Forman D., Bray F. (2015) Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer, 136, E359–E386. [DOI] [PubMed] [Google Scholar]
- 2. Howlader N., Noone A.M., Krapcho M., Garshell J., Miller D., Altekruse S.F., Kosary C.L., Yu M., Ruhl J., Tatalovich Z. et al. (eds). (2014) SEER Cancer Statistics Review, 1975-2012. National Cancer Institute. Bethesda, MD.
- 3. Duncan M.E., Seagroatt V., Goldacre M.J. (2012) Cancer of the body of the uterus: trends in mortality and incidence in England, 1985-2008. BJOG, 119, 333–339. [DOI] [PubMed] [Google Scholar]
- 4. Wartko P., Sherman M.E., Yang H.P., Felix A.S., Brinton L.A., Trabert B. (2013) Recent changes in endometrial cancer trends among menopausal-age U.S. women. Cancer Epidemiol., 37, 374–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hemminki K., Vaittinen P., Dong C. (1999) Endometrial cancer in the family-cancer database. Cancer Epidemiol. Biomarkers Prev., 8, 1005–1010. [PubMed] [Google Scholar]
- 6. Lucenteforte E., Talamini R., Montella M., Dal Maso L., Pelucchi C., Franceschi S., La Vecchia C., Negri E. (2009) Family history of cancer and the risk of endometrial cancer. Eur. J. Cancer Prev., 18, 95–99. [DOI] [PubMed] [Google Scholar]
- 7. Win A.K., Reece J.C., Ryan S. (2015) Family history and risk of endometrial cancer: a systematic review and meta-analysis. Obstet. Gynecol., 125, 89–98. [DOI] [PubMed] [Google Scholar]
- 8. Gruber S.B., Thompson W.D. (1996) A population-based study of endometrial cancer and familial risk in younger women. Cancer and Steroid Hormone Study Group. Cancer Epidemiol. Biomarkers Prev., 5, 411–417. [PubMed] [Google Scholar]
- 9. Meyer L.A., Broaddus R.R., Lu K.H. (2009) Endometrial cancer and lynch syndrome: clinical and pathologic considerations. Cancer Control, 16, 14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schildkraut J.M., Risch N., Thompson W.D. (1989) Evaluating genetic association among ovarian, breast, and endometrial cancer: evidence for a breast/ovarian cancer relationship. Am. J. Hum. Genet., 45, 521–529. [PMC free article] [PubMed] [Google Scholar]
- 11. Lichtenstein P., Holm N.V., Verkasalo P.K., Iliadou A., Kaprio J., Koskenvuo M., Pukkala E., Skytthe A., Hemminki K. (2000) Environmental and heritable factors in the causation of cancer — analyses of cohorts of twins from Sweden, Denmark, and Finland. N. Engl. J. Med., 343, 78–85. [DOI] [PubMed] [Google Scholar]
- 12. Lu Y., Ek W.E., Whiteman D., Vaughan T.L., Spurdle A.B., Easton D.F., Pharoah P.D., Thompson D.J., Dunning A.M., Hayward N.K. et al. (2014) Most common ‘sporadic’ cancers have a significant germline genetic component. Hum. Mol. Genet., 23, 6112–6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L. et al. (2014) The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res., 42, D1001–D1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Long J., Zheng W., Xiang Y.B., Lose F., Thompson D., Tomlinson I., Yu H., Wentzensen N., Lambrechts D., Dörk T. et al. (2012) Genome-wide association study identifies a possible susceptibility locus for endometrial cancer. Cancer Epidemiol. Biomarkers Prev., 21, 980–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. De Vivo I., Prescott J., Setiawan V.W., Olson S.H., Wentzensen N., Australian National Endometrial Cancer Study Group. Attia J., Black A., Brinton L., Chen C. et al. (2014) Genome-wide association study of endometrial cancer in E2C2. Hum. Genet., 133, 211–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Spurdle A.B., Thompson D.J., Ahmed S., Ferguson K., Healey C.S., O’Mara T., Walker L.C., Montgomery S.B., Dermitzakis E.T., Australian National Endometrial Cancer Study Group. et al. (2011) Genome-wide association study identifies a common variant associated with risk of endometrial cancer. Nat. Genet., 43, 451–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Painter J.N., O’Mara T.A., Batra J., Cheng T., Lose F.A., Dennis J., Michailidou K., Tyrer J.P., Ahmed S., Ferguson K. et al. (2015) Fine-mapping of the HNF1B multicancer locus identifies candidate variants that mediate endometrial cancer risk. Hum. Mol. Genet., 24, 1478–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Thompson D.J., O’Mara T.A., Glubb D.M., Painter J.N., Cheng T., Folkerd E., Doody D., Dennis J., Webb P.M., Gorman M. et al. (2016) CYP19A1 fine-mapping and Mendelian randomisation: estradiol is causal for endometrial cancer. Endocr. Relat. Cancer, 23, 77–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Evangelou E., Ioannidis J.P.A. (2013) Meta-analysis methods for genome-wide association studies and beyond. Nat. Rev. Genet., 14, 379–389. [DOI] [PubMed] [Google Scholar]
- 20. Prior H.M., Walter M.A. (1996) SOX genes: architects of development. Mol. Med., 2, 405–412. [PMC free article] [PubMed] [Google Scholar]
- 21. Maris J.M., Mosse Y.P., Bradfield J.P., Hou C., Monni S., Scott R.H., Asgharzadeh S., Attiyeh E.F., Diskin S.J., Laudenslager M. et al. (2008) Chromosome 6p22 locus associated with clinically aggressive neuroblastoma. N. Engl. J. Med., 358, 2585–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Russell M.R., Penikis A., Oldridge D.A., Alvarez-Dominguez J.R., McDaniel L., Diamond M., Padovan O., Raman P., Li Y., Wei J.S. et al. (2015) CASC15-S is a tumor suppressor lncRNA at the 6p22 neuroblastoma susceptibility locus. Cancer Res., 75, 3155–3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-André V., Sigova A.A., Hoke H.A., Young R.A. (2013) Super-enhancers in the control of cell identity and disease. Cell, 155, 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Corradin O., Saiakhova A., Akhtar-Zaidi B., Myeroff L., Willis J., Cowper-Sallari R., Lupien M., Markowitz S., Scacheri P.C. (2014) Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res., 24, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Steinthorsdottir V., Thorleifsson G., Reynisdottir I., Benediktsson R., Jonsdottir T., Walters G.B., Styrkarsdottir U., Gretarsdottir S., Emilsson V., Ghosh S. et al. (2007) A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat. Genet., 39, 770–775. [DOI] [PubMed] [Google Scholar]
- 26. Wu Y., Li H., Loos R.J.F., Yu Z., Ye X., Chen L., Pan A., Hu F.B., Lin X. (2008) Common variants in CDKAL1, CDKN2A/B, IGF2BP2, SLC30A8, and HHEX/IDE genes are associated with type 2 diabetes and impaired fasting glucose in a Chinese Han population. Diabetes, 57, 2834–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Takeuchi F., Serizawa M., Yamamoto K., Fujisawa T., Nakashima E., Ohnaka K., Ikegami H., Sugiyama T., Katsuya T., Miyagishi M. et al. (2009) Confirmation of multiple risk Loci and genetic impacts by a genome-wide association study of type 2 diabetes in the Japanese population. Diabetes, 58, 1690–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Al-Tassan N.A., Whiffin N., Hosking F.J., Palles C., Farrington S.M., Dobbins S.E., Harris R., Gorman M., Tenesa A., Meyer B.F. et al. (2015) A new GWAS and meta-analysis with 1000 Genomes imputation identifies novel risk variants for colorectal cancer. Sci. Rep., 5, 10442.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wolpin B.M., Rizzato C., Kraft P., Kooperberg C., Petersen G.M., Wang Z., Arslan A.A., Beane-Freeman L., Bracci P.M., Buring J. et al. (2014) Genome-wide association study identifies multiple susceptibility loci for pancreatic cancer. Nat. Genet., 46, 994–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Diskin S.J., Capasso M., Schnepp R.W., Cole K.A., Attiyeh E.F., Hou C., Diamond M., Carpenter E.L., Winter C., Lee H. et al. (2012) Common variation at 6q16 within HACE1 and LIN28B influences susceptibility to neuroblastoma. Nat. Genet., 44, 1126–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lessard L., Liu M., Marzese D.M., Wang H., Chong K., Kawas N., Donovan N.C., Kiyohara E., Hsu S., Nelson N. et al. (2015) The CASC15 long intergenic noncoding RNA locus is involved in melanoma progression and phenotype switching. J. Invest. Dermatol., 135, 2464–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Figueroa J.D., Ye Y., Siddiq A., Garcia-Closas M., Chatterjee N., Prokunina-Olsson L., Cortessis V.K., Kooperberg C., Cussenot O., Benhamou S. et al. (2014) Genome-wide association study identifies multiple loci associated with bladder cancer risk. Hum. Mol. Genet., 23, 1387–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang Y., Liu H., Yang S., Zhang J., Qian L., Chen X. (2014) Overweight, obesity and endometrial cancer risk: results from a systematic review and meta-analysis. Int. J. Biol. Markers, 29, e21–e29. [DOI] [PubMed] [Google Scholar]
- 34. Liao C., Zhang D., Mungo C., Tompkins D.A., Zeidan A.M. (2014) Is diabetes mellitus associated with increased incidence and disease-specific mortality in endometrial cancer? A systematic review and meta-analysis of cohort studies. Gynecol. Oncol., 135, 163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wen W., Cho Y.S., Zheng W., Dorajoo R., Kato N., Qi L., Chen C.H., Delahanty R.J., Okada Y., Tabara Y. et al. (2012) Meta-analysis identifies common variants associated with body mass index in East Asians. Nat. Genet., 44, 307–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lu N., Lin T., Wang L., Qi M., Liu Z., Dong H., Zhang X., Zhai C., Wang Y., Liu L. et al. (2015) Association of SOX4 regulated by tumor suppressor miR-30a with poor prognosis in low-grade chondrosarcoma. Tumour Biol., 36, 3843–3852. [DOI] [PubMed] [Google Scholar]
- 37. Walter R.F.H., Mairinger F.D., Werner R., Ting S., Vollbrecht C., Theegarten D., Christoph D.C., Zarogoulidis K., Schmid K.W., Zarogoulidis P. et al. (2015) SOX4, SOX11 and PAX6 mRNA expression was identified as a (prognostic) marker for the aggressiveness of neuroendocrine tumors of the lung by using next-generation expression analysis (NanoString). Future Oncol., 11, 1027–1036. [DOI] [PubMed] [Google Scholar]
- 38. Wang D., Hao T., Pan Y., Qian X., Zhou D. (2015) Increased expression of SOX4 is a biomarker for malignant status and poor prognosis in patients with non-small cell lung cancer. Mol. Cell. Biochem., 402, 75–82. [DOI] [PubMed] [Google Scholar]
- 39. Zhou Y., Wang X., Huang Y., Chen Y., Zhao G., Yao Q., Jin C., Huang Y., Liu X., Li G. (2015) Down-regulated SOX4 expression suppresses cell proliferation, metastasis and induces apoptosis in Xuanwei female lung cancer patients. J. Cell. Biochem., 116, 1007–1018. [DOI] [PubMed] [Google Scholar]
- 40. Liu P., Ramachandran S., Ali Seyed M., Scharer C.D., Laycock N., Dalton W.B., Williams H., Karanam S., Datta M.W., Jaye D.L. et al. (2006) Sex-determining region Y box 4 is a transforming oncogene in human prostate cancer cells. Cancer Res., 66, 4011–4019. [DOI] [PubMed] [Google Scholar]
- 41. Wang L., Zhang J., Yang X., Chang Y.W.Y., Qi M., Zhou Z., Zhang J., Han B. (2013) SOX4 is associated with poor prognosis in prostate cancer and promotes epithelial-mesenchymal transition in vitro. Prostate Cancer Prostatic Dis., 16, 301–307. [DOI] [PubMed] [Google Scholar]
- 42. Zhang J., Liang Q., Lei Y., Yao M., Li L., Gao X., Feng J., Zhang Y., Gao H., Liu D.X. et al. (2012) SOX4 induces epithelial-mesenchymal transition and contributes to breast cancer progression. Cancer Res., 72, 4597–4608. [DOI] [PubMed] [Google Scholar]
- 43. Song G.D., Sun Y., Shen H., Li W. (2015) SOX4 overexpression is a novel biomarker of malignant status and poor prognosis in breast cancer patients. Tumour Biol., 36, 4167–4173. [DOI] [PubMed] [Google Scholar]
- 44. Huang Y.W., Liu J.C., Deatherage D.E., Luo J., Mutch D.G., Goodfellow P.J., Miller D.S., Huang T.H.M. (2009) Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 oncogene in endometrial cancer. Cancer Res., 69, 9038–9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen J., Ju H.L., Yuan X.Y., Wang T.J., Lai B.Q. (2016) SOX4 is a potential prognostic factor in human cancers: a systematic review and meta-analysis. Clin. Transl. Oncol., 18, 65–72. [DOI] [PubMed] [Google Scholar]
- 46. McGregor B., Pfitzner J., Zhu G., Grace M., Eldridge A., Pearson J., Mayne C., Aitken J.F., Green A.C., Martin N.G. (1999) Genetic and environmental contributions to size, color, shape, and other characteristics of melanocytic naevi in a sample of adolescent twins. Genet. Epidemiol., 16, 40–53. [DOI] [PubMed] [Google Scholar]
- 47. Painter J.N., Anderson C.A., Nyholt D.R., Macgregor S., Lin J., Lee S.H., Lambert A., Zhao Z.Z., Roseman F., Guo Q. et al. (2011) Genome-wide association study identifies a locus at 7p15.2 associated with endometriosis. Nat. Genet., 43, 51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. McEvoy M., Smith W., D’Este C., Duke J., Peel R., Schofield P., Scott R., Byles J., Henry D., Ewald B. et al. (2010) Cohort profile: The Hunter Community Study. Int. J. Epidemiol., 39, 1452–1463. [DOI] [PubMed] [Google Scholar]
- 49. Olson S.H., Chen C., De Vivo I., Doherty J.A., Hartmuller V., Horn-Ross P.L., Lacey J.V., Lynch S.M., Sansbury L., Setiawan V.W. et al. (2009) Maximizing resources to study an uncommon cancer: E2C2–Epidemiology of Endometrial Cancer Consortium. Cancer Causes Control, 20, 491–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Houlston R.S., Cheadle J., Dobbins S.E., Tenesa A., Jones A.M., Howarth K., Spain S.L., Broderick P., Domingo E., Farrington S. et al. (2010) Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13.33. Nat. Genet., 42, 973–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wellcome Trust Case Control Consortium (2007) Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature, 447, 661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fuchsberger C., Abecasis G.R., Hinds D.A. (2014) minimac2: faster genotype imputation. Bioinformatics, 31, 782–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Howie B.N., Donnelly P., Marchini J. (2009) A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet., 5, e1000529.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ferreira T., Marchini J. (2011) Modeling interactions with known risk loci-a Bayesian model averaging approach. Ann. Hum. Genet., 75, 1–9. [DOI] [PubMed] [Google Scholar]
- 55. Aulchenko Y.S., Struchalin M.V., van Duijn C.M. (2010) ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics, 11, 134.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Willer C.J., Li Y., Abecasis G.R. (2010) METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics, 26, 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yang J., Ferreira T., Morris A.P., Medland S.E., Genetic Investigation of ANthropometric Traits, (GIANT) Consortium, DIAbetes Genetics Replication, Meta-analysis, (DIAGRAM) et al. (2012) Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet., 44, 369–375, S1–S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gauderman W.J. (2002) Sample size requirements for association studies of gene-gene interaction. Am. J. Epidemiol., 155, 478–484. [DOI] [PubMed] [Google Scholar]
- 59. Ward L.D., Kellis M. (2011) HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res., 40, D930–D934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. ENCODE Project Consortium (2012) An integrated encyclopedia of DNA elements in the human genome. Nature, 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. The Cancer Genome Network (2013) Integrated genomic characterization of endometrial carcinoma. Nature, 497, 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Li Y., Willer C.J., Ding J., Scheet P., Abecasis G.R. (2010) MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol., 34, 816–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Li Y., Willer C., Sanna S., Abecasis G. (2009) Genotype imputation. Annu. Rev. Genomics Hum. Genet., 10, 387–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Eeles R.A., Al Olama A.A., Benlloch S., Saunders E.J., Leongamornlert D.A., Tymrakiewicz M., Ghoussaini M., Luccarini C., Dennis J., Jugurnauth-Little S. et al. (2013) Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat. Genet., 45, 385–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.