Abstract
Objective
Faecal microbiota transplantation (FMT) is effective for the treatment of recurrent Clostridium difficile infection (CDI). Studies have shown bacterial colonisation after FMT, but data on viral alterations in CDI are scarce. We investigated enteric virome alterations in CDI and the association between viral transfer and clinical outcome in patients with CDI.
Design
Ultra-deep metagenomic sequencing of virus-like particle preparations and bacterial 16S rRNA sequencing were performed on stool samples from 24 subjects with CDI and 20 healthy controls. We longitudinally assessed the virome and bacterial microbiome changes in nine CDI subjects treated with FMT and five treated with vancomycin. Enteric virome alterations were assessed in association with treatment response.
Results
Subjects with CDI demonstrated a significantly higher abundance of bacteriophage Caudovirales and a lower Caudovirales diversity, richness and evenness compared with healthy household controls. Significant correlations were observed between bacterial families Proteobacteria, Actinobacteria and Caudovirales taxa in CDI. FMT treatment resulted in a significant decrease in the abundance of Caudovirales in CDI. Cure after FMT was observed when donor-derived Caudovirales contigs occupied a larger fraction of the enteric virome in the recipients (p=0.024). In treatment responders, FMT was associated with alterations in the virome and the bacterial microbiome, while vancomycin treatment led to alterations in the bacterial community alone.
Conclusions
In a preliminary study, CDI is characterised by enteric virome dysbiosis. Treatment response in FMT was associated with a high colonisation level of donor-derived Caudovirales taxa in the recipient. Caudovirales bacteriophages may play a role in the efficacy of FMT in CDI.
Trial registration number
Keywords: BACTERIAL INFECTION, MUCOSAL INFECTION
Significance of this study.
What is already known on this subject?
Faecal microbiota transplantation (FMT) is highly effective for the treatment of recurrent Clostridium difficile infection (CDI).
Studies have shown bacterial colonisation after FMT, but data on viral alterations in CDI and the association between viral colonisation and treatment outcome are largely unknown.
What are the new findings?
CDI was characterised by a high abundance of Caudovirales bacteriophages and a low Caudovirales diversity, richness and evenness compared with healthy household controls.
Donor-derived Caudovirales taxa occupied a significantly larger fraction of the enteric virome in CDI subjects who responded to FMT compared with those who did not.
FMT was associated with alterations in the enteric virome and bacterial microbiome, while vancomycin treatment was associated with alterations of the bacterial microbiome only.
Recipients infused with donor faeces consisting of a higher richness of Caudovirales than that of recipient were all cured with FMT. CDI subjects who had restoration of bacteria community only were found to have disease recurrence.
How might it impact on clinical practice in the foreseeable future?
The restoration of virome community is as important as that of bacterial microbiome in FMT.
Donor selection based on virome characteristics should be considered in FMT practice.
Introduction
Clostridium difficile infection (CDI) is a leading nosocomial infection affecting half a million people in the USA.1 2 Antibiotic therapy is the first-line treatment, but up to one-third of patients do not achieve a durable response. Recently, faecal microbiota transplantation (FMT) with either fresh or frozen stool from healthy donor has been shown to be highly effective in patients with recurrent CDI with a cure rate of 85%–90%.3–5 The efficacy of FMT is mostly based on restoration of the phylogenetic diversity and bacterial microbiota to resemble that of a ‘healthy’ individual.6–8 However, the mechanism and long-term effects of FMT remain poorly understood.
Humans are also colonised by a large population of viruses, especially bacteriophages that may play a pivotal role in microbiome ecology.9–11 Limited studies have reported that bacteriophages can be transferred to recipients from the donor microbiota.12–15 In a study of three patients with UC, transfer of multiple viral lineages through FMT was reported.13 In a single case report of CDI, virome alterations over time after FMT largely consisted of Caudovirales.12 16 Recently, a preliminary investigation of five patients with CDI showed that transfer of sterile faecal filtrates from donor stool was effective to eliminate symptoms.15 Overall, these data suggest that other than bacterial components, bacteriophages or bacteriocins are likely contributing to the normal intestinal microenvironment in FMT. Besides, whether patients with CDI have alterations in the enteric virome and the extent of such changes have also not been studied. In this study, we postulate that bacteriophages may interact with host microbes and influence the clinical outcome after FMT. We herein performed ultra-deep virus-like particles’ (VLPs’) metagenomic sequencing and 16S rDNA sequencing to determine enteric virome and bacteriophages–bacteria interactions in subjects with CDI compared with healthy controls. We also serially assessed changes in the virome post FMT and examined whether bacteriophage transfer is associated with the clinical outcome of FMT. To our knowledge, this pilot study is the first and largest to date to characterise the enteric virome alterations in CDI and to elucidate changes in viral communities after FMT and its relatedness with treatment outcome.
Methods
Study subjects and treatment outcome
The current study was a substudy from a randomised controlled trial (RCT) of FMT versus vancomycin for patients with CDI. Consecutive CDI subjects enrolled into this RCT were invited to participate in a substudy of assessment of faecal microbiota. Patients were included if they had three or more loose or watery stools per day for at least two consecutive days, or eight or more soft or loose stools in 48 hours, and a positive stool test for C. difficile based on a two-step testing algorithm in our hospital, a positive GDH (glutamate dehydrogenase) screening test followed by a positive PCR test of C. difficile. A total of 24 subjects with CDI and 20 healthy household controls were recruited, and stool samples at baseline were obtained for analyses of virome and bacterial microbiome. Among them, 14 CDI subjects consented to have stool samples collected serially after treatment for microbiome analysis. Nine CDI subjects were treated with FMT and five were treated with vancomycin, and they were followed up at baseline and weeks 2, 4, 10 and 16 after treatment (see online supplementary figure 1). One patient FMT6 had recurrent CDI. This patient had five previous episodes of CDI prior to FMT. Subjects in the FMT group received 5 days of vancomycin followed by donor-infused stool via nasojejunal route, and those who had standard therapy (STD) received oral vancomycin 500 mg four times per day for 10 days. A computer-generated randomisation schedule was used to assign patients to the treatment sequences. All patients kept a stool diary and were questioned about stool frequency and consistency and medication use.
gutjnl-2017-313952supp001.pdf (13MB, pdf)
Treatment response was defined as an absence of diarrhoea or persistent diarrhoea that could be explained by other causes with a negative stool test for C. difficile toxin, while relapse was defined as diarrhoea with a positive stool test for C. difficile toxin. Treatment cure is defined as symptom resolution and a negative Clostridium difficile toxin in stool until the last follow-up (last follow-up was referred to as the last stool collection time point, as shown in supplementary figure 1). Six of the nine subjects who had FMT (FMT1–FMT6) and three of the five patients (STD3–STD5) who had vancomycin were cured of CDI (termed responders, table 1) at a median follow-up of 16 weeks. CDI recipients FMT8 and FMT9 shared the same donor, and this donor was termed ‘D8’. Clinical data of the subjects and collected stool samples are shown in online supplementary table S1. None of the patients had received antibiotics or proton pump inhibitors after FMT. The detailed study design and methods of donor recruitment were included in the online supplementary appendix.
Table 1.
Clinical characteristics and outcomes of subjects who received FMT or vancomycin for Clostridium difficile infections
| Subject | Sex | Age | Smoking | Severe/moderate | Duration of follow-up (weeks) | Outcome (till last follow-up) |
|---|---|---|---|---|---|---|
| FMT1 | M | 80 | Ex-smoker | Moderate | 16 | Cured |
| FMT2 | M | 52 | No | Severe | 27 | Cured |
| FMT3 | M | 38 | No | Moderate | 17 | Cured |
| FMT4 | F | 76 | No | Moderate | 18 | Cured |
| FMT5 | M | 63 | No | Severe | 18 | Cured |
| FMT6 | M | 88 | No | Severe | 23 | Cured |
| FMT7 | M | 45 | Ex-smoker | Severe | 20 | Recurrence at week 19 |
| FMT8 | F | 89 | No | Moderate | 11 | Recurrence at week 5 |
| FMT9 | F | 38 | No | Severe | 28 | Recurrence at week 28 |
| STD1 | M | 88 | Ex-smoker | Severe | 20 | Recurrence at week 12 |
| STD2 | M | 93 | No | Moderate | 7 | Recurrence at week 7 |
| STD3 | F | 78 | Smoker | Severe | 14 | Cured |
| STD4 | F | 83 | No | Severe | 17 | Cured |
| STD5 | F | 99 | No | Moderate | 26 | Cured |
FMT, faecal microbiota transplantation; STD, standard therapy (vancomycin).
VLPs enrichment and sequencing
VLPs were enriched from pulverised human stool, using a protocol according to previously described methods.11 17 The detailed procedure is included in the online supplementary appendix.
VLP DNAs were quantified (NanoDrop), and 1 μg of DNA was randomly fragmented by ultrasonication (Covaris) followed by library construction. The qualified libraries were amplified on cBot to generate the cluster on the flow cell (TruSeq PE Cluster Kit V3–cBot–HS, Illumina). The amplified libraries were sequenced paired end on the HiSeq 2000 System (TruSeq SBS KIT-HS V3, Illumina) (BGI, Shenzhen, China; standard 2×150 bp run), generating 20–60 million raw sequences (5–8G raw data) per sample (sequence statistics in supplementary table S2). Sequence processing and quality control, de novo contig assembly and taxonomy annotation are included in the online supplementary appendix.
Virome data analysis
To estimate contig abundance and calculate sequence diversity, all reads were aligned to the resulting contigs using Bowtie2 (V. 2.2.9).18 The mapped sequence counts, contig lengths and total sequence counts were used to normalise the sequence counts and represent the RPKM (reads per kilobase per million) of each sample to the contigs. These values were used to generate an operational taxonomic unit (OTU) relative abundance table, which was annotated with the taxonomy described above. The virome abundance data were imported into R 3.2.3. Richness, diversity and rarefaction calculation were performed using the estimated richness function of the phyloseq package. Diversity and richness plots were generated in GraphPad Prism (V. 6.0). Spearman correlations and their significance were calculated using the cor and cor.test functions in R, respectively. For the Caudovirales–bacterial taxa comparisons, Spearman correlations were calculated for the relative abundance of the 50 most abundant Caudovirales and the bacterial families determined to be significantly associated with disease by LEfSe analysis. Correlation plots were generated using the corrplot R package. Heat maps were generated using the pheatmap R package. LEfSe linear discriminant analysis and multivariate analysis are included in the online supplementary appendix.
Species and contig presence
To establish the presence of a species or a contig within a sample, the RPKM of a contig or a species is set to be >2 (a stringent criteria), so as to rule out the false-positive finding) for the contig or the species to be assured as present within a sample. In samples after FMT, if a contig or a species was not present in the recipient baseline sample but present in the corresponding donor baseline sample and in the recipient post-FMT sample, the contig or species was defined as ‘donor-derived’; if a contig or a species was not present in the corresponding donor baseline sample but detected both in the recipient baseline sample and in the recipient post-FMT sample, the contig or the species was defined as ‘recipient-exclusive’; if a contig or a species was present across the recipient baseline sample, the recipient post-FMT sample and the corresponding donor baseline sample, the contig or the species was defined as ‘donor-recipient coexisted’. To determine the abundance alteration of Caudovirales species after FMT, the species’ relative abundance (RA) fold change was defined as fc=log2(post-FMT last follow-up RA/pre-FMT baseline RA).
Results
Virome alterations in CDI compared with controls
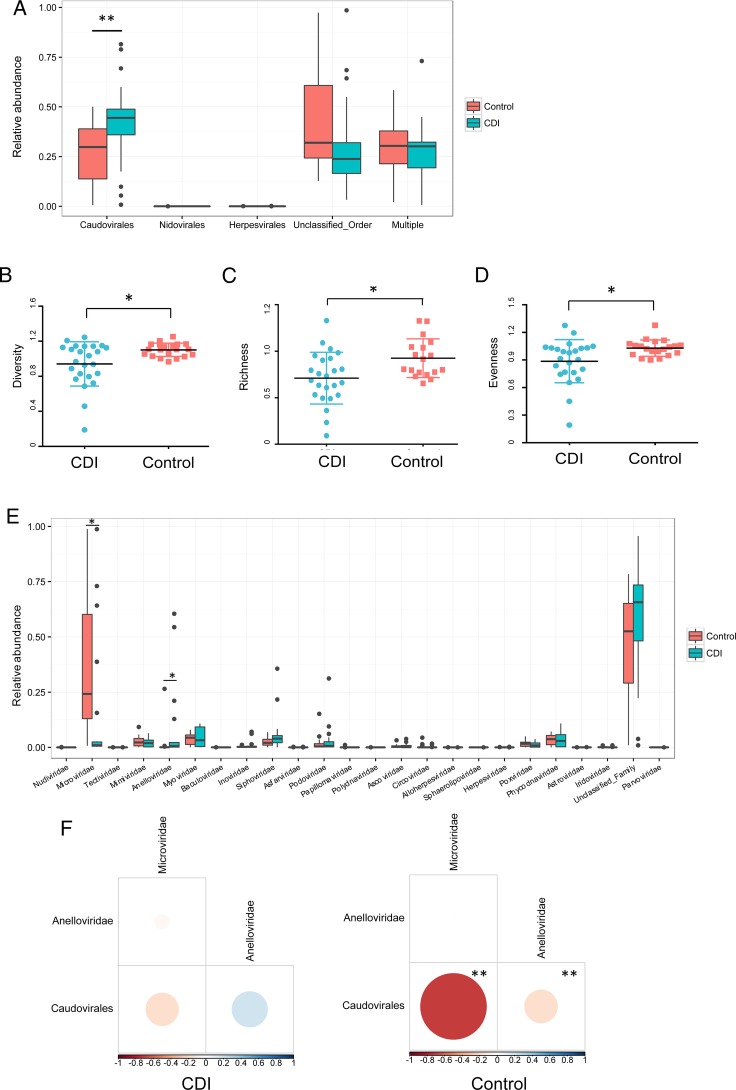
We first compared the faecal virome composition in subjects with CDI with that of healthy household controls. On average, 21 202 400 ±5 385 756 clean paired-end reads were obtained from the enriched faecal VLP preparations. Among the common viral orders detected, the most abundant in both CDI and healthy household controls was Caudovirales, one taxon of a consortium of temperate double-stranded DNA (dsDNA) bacteriophages. Compared with healthy individuals, CDI subjects had a significantly higher abundance of Caudovirales (figure 1A), but a decreased diversity of Caudovirales at the species level (figure 1B) and at the contig level (see online supplementary figure 2A). There was also a significant decrease in the richness and evenness of Caudovirales in CDI compared with healthy controls (figure 1C, D). In line with that, the rate of acquisition of new Caudovirales taxa in control samples rapidly outpaced new taxa acquisition in CDI samples, further demonstrating a lower Caudovirales richness in CDI than in controls (see online supplementary figure 2B). At the family level, CDI showed a decreased abundance of Microviridae compared with that of controls (figure 1E), as well as a decrease in Microviridae diversity (see online supplementary figure 2C). In contrast, the abundance of Anelloviridae was increased in CDI compared with controls (figure 1E). Using linear regression test, we found no significant correlation between the diversity or richness of Caudovirales and age, gender, household relationship or time of sample collection (see online supplementary table S1). After adjusting for multiple comparisons, no significant correlation was shown by MaAsLin between viral abundances and age, gender or time of sample collection. There was however a correlation between viral abundances and household relationship (False Discovery Rate (FDR) multiple comparison adjusted q<0.05) which suggests a household effect on the gut virome structure. In healthy controls, there was a significant inverse correlation between Caudovirales and Microviridae, Caudovirales and Anelloviridae, which was not seen in CDI (figure 1F). Overall, these findings indicate dysbiosis of the enteric virome in patients with CDI.
Figure 1.
Virome alterations in Clostridium difficile infection (CDI). (A) Comparison of the relative abundance of enteric virus in CDI subjects and healthy controls at the order level. The bars indicate the median and IQR. Statistical significance was determined by Mann-Whitney test. *p<0.05. Comparison of Caudovirales diversity (B), richness (C) and evenness (D) between CDI subjects and healthy controls at the species level. Statistical significance was determined by Mann-Whitney test. *p<0.05. (E) Relative abundance of viruses at the family level. Statistical significance was determined by LEfSe analysis with FDR correction comparing all samples with all samples. * q<0.05. (F) Spearman correlation plots of Caudovirales, Microvirida and Anelloviridae in CDI subjects and controls. Statistical significance was determined for all pairwise comparisons; those with values of p< 0.05 are shown. Positive values (blue circles) indicate positive correlations, and negative values (red circles) indicate inverse correlations. The size and shading of the circle indicate the magnitude of the correlation, whereby darker shades are more correlated than lighter shades.
We next assessed whether virome dysbiosis was specific to CDI diarrhoea by including an additional cohort of subjects with norovirus-associated infectious diarrhoea. We found that there was a significant decrease in both the richness and diversity of Caudovirales in subjects with norovirus infection compared with healthy controls (see online supplementary figure 3A), suggesting that altered virome richness and diversity may reflect a generic pathogen-driven event in subjects with acute infectious diarrhoea illness. However, norovirus subjects had a significantly lower abundance of Caudovirales and Anelloviridae (both p<0.05) compared with healthy controls, while no marked difference was observed in the relative abundance of Microviridae between norovirus subjects and controls (see online supplementary figure 3B). These data suggest that enteric virome dysbiosis in CDI is likely to be disease-specific.
Enteric virome alterations in patients with CDI after FMT
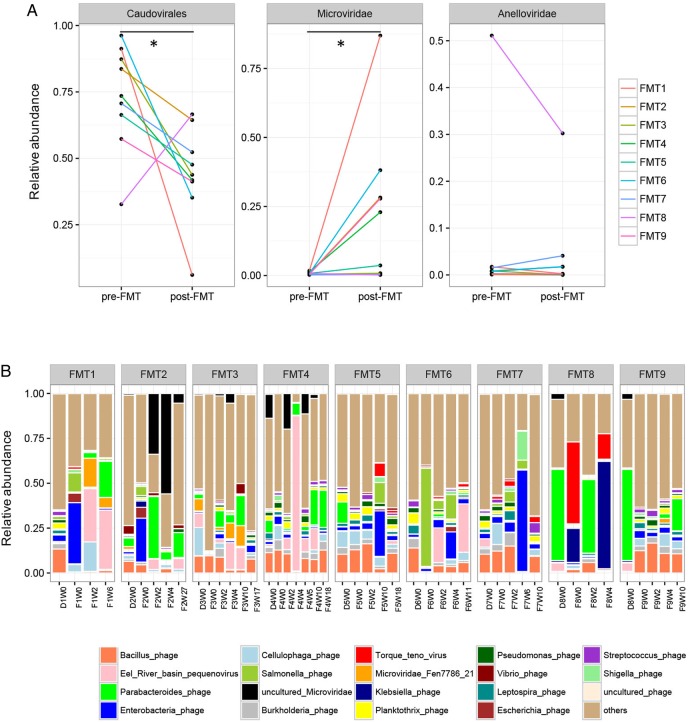
We next investigated whether the enteric virome changes after FMT. Nine CDI subjects who received FMT were followed up longitudinally at different time points (see online supplementary figure 1). CDI subjects showed a decrease in Caudovirales abundance and an increase in Microviridae abundance after FMT (paired Wilcoxon sign permutation test, p<0.05) (figure 2A). There were profound differences in the virome composition between the nine FMT subjects (figure 2B). Similar virome configurations were observed between donor baseline stool sample, recipient baseline stool sample and the follow-up stool samples of the recipient. As the donor and recipient shared the same family origin, except for FMT9, this observation, confirmed also by MaAsLin, further illustrated a significant household effect on the virome structure.
Figure 2.
Alterations in the enteric virome after faecal microbiota transplantation (FMT). (A) Relative abundance of Caudovirales, Microviridae, and Anelloviridae in pre-FMT samples and post-FMT samples collected at the last follow-up. Statistical significance was determined by paired Wilcoxon sign permutation test, *p<0.05. (B) Alterations in the relative abundance of viral species in the stool of Clostridium difficile infection subjects after FMT at different time points until the last follow-up. ‘F’ indicates FMT-treated subject. ‘D’ indicates FMT donor. ‘W’ indicates weeks post treatment.
In the nine CDI subjects who received FMT treatment, six subjects remained symptom-free with a negative stool C. difficile toxin at the last follow-up (responders, FMT1–FMT6), while three developed recurrence of CDI (non-responders, FMT7–FMT9) (table 1). We identified 15 species differentially enriched between FMT responders and non-responders via LEfSe analysis (see online supplementary figure 4). Among them, Eel River basin pequenovirus, a recently identified sister clade to the enterobacteria microviruses which may prey on Proteobacteria,19 was the most abundant and significant species harboured in the post-FMT stools of the responders.
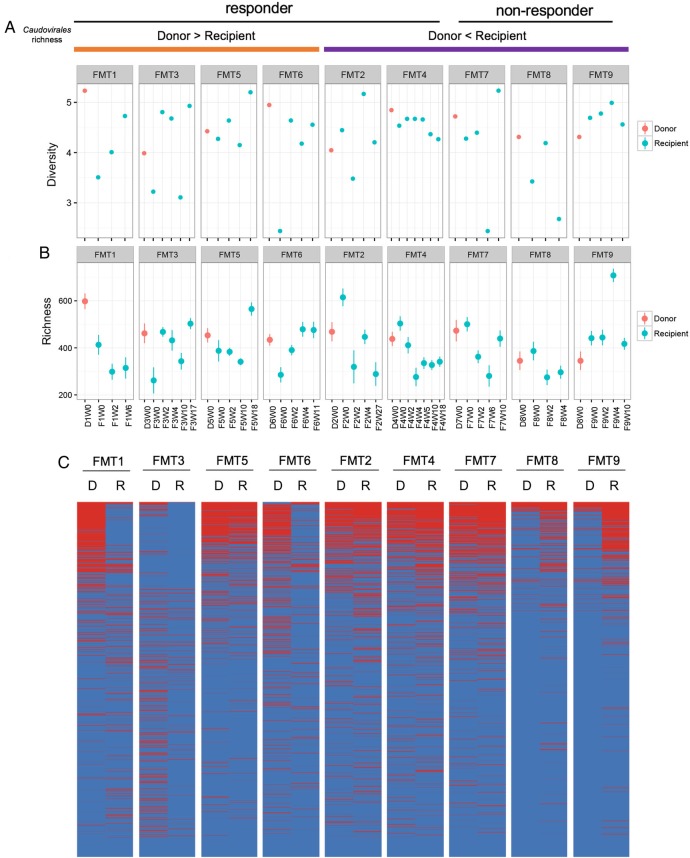
Donor Caudovirales richness and treatment response
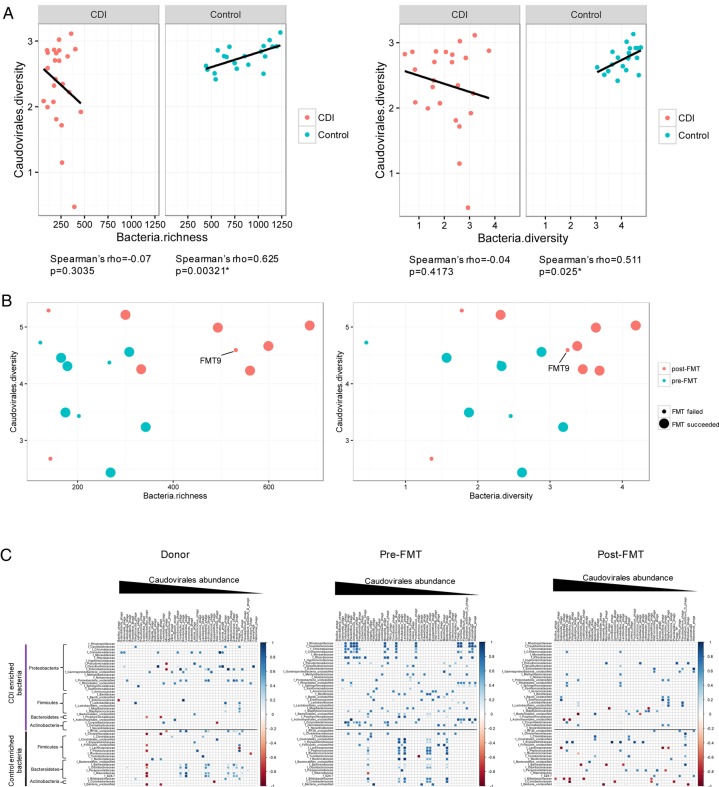
Given that Caudovirales was the most abundant and significantly changed viral taxon in patients with CDI, we focused on Caudovirales hereafter and studied the effect of donor Caudovirales richness on treatment response. In subjects FMT1, FMT3, FMT5 and FMT6, whereby the Caudovirales richness of the donor was higher than that of the recipient (figure 3A), all recipients were cured. These recipients also showed an increase in the diversity of Caudovirales after FMT, though it is not consistent in the richness change post FMT (figure 3A, B). In these four CDI subjects who responded to FMT, the Caudovirales contigs found in the donor baseline sample outnumbered that of the contigs in the recipient baseline stool sample (figure 3C), further substantiating that donor Caudovirales richness was higher than that of the recipient. In contrast, when the Caudovirales richness of the donor was lower than that of the recipient (FMT2, FMT4, FMT7, FMT8 and FMT9), the treatment outcome was not consistent across the recipients. FMT2 and FMT4 responded to treatment, but FMT7, FMT8 and FMT9 did not. The Caudovirales diversity and richness alterations post FMT were inconsistent among these subjects. The simultaneous treatment failure in subjects FMT8 and FMT9, where they shared the same donor, provided a valuable piece of evidence for a donor effect with regard to virome on FMT efficacy. Overall, we found that when the Caudovirales richness of the donor was higher than that of the recipient, all the CDI subjects achieved a response to FMT.
Figure 3.
Alterations of Caudovirales diversity and richness after faecal microbiota transplantation (FMT). Changes in the Caudovirales diversity (A) and Caudovirales richness (B) of the stool samples of donor and Clostridium difficile infection (CDI) subjects after FMT at different time points until the last follow-up. (C) Presence–absence heat map of Caudovirales contigs in the stool samples of CDI subjects and their corresponding donors. Only contigs with reads per kilobase per million >1 were shown. ‘D’ indicates FMT donor; ‘R’ indicates FMT recipient. ‘Donor > recipient’ indicates the Caudovirales richness in stool samples of donor were higher than that of the recipient. ‘Responder’ indicates CDI subjects who responded to FMT; ‘non-responders’ indicates CDI subjects who had disease recurrence after FMT.
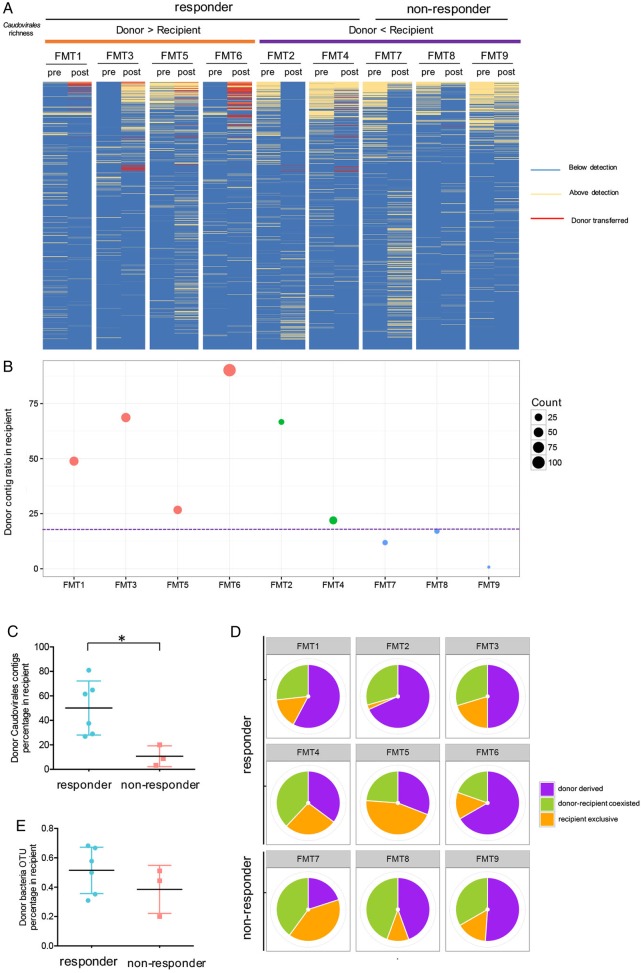
Transfer of Caudovirales bacteriophages from donor to recipient and treatment response
We next assessed the presence of donor-derived Caudovirales in relation to treatment response. A larger proportion of Caudovirales contigs was transferred from the donor to the recipient detected at the last follow-up stool in FMT responders than in FMT non-responders (figure 4A, B). Donor-derived Caudovirales occupied a significantly larger fraction (>20%) of the virome in the FMT responders than in the non-responders (<20%) (figure 4B,C, Mann-Whitney test, p<0.05). A similar and consistent pattern was also observed at the species level (see online supplementary figure 5B). Thus, the quantity and final proportion of donor-derived Caudovirales in the recipient appeared to be associated with treatment outcome of FMT.
Figure 4.
Transfer of Caudovirales bacteriophages and faecal microbiota transplantation (FMT) treatment outcome. (A) Presence–absence heat map of Caudovirales contigs in pre-FMT and post-FMT collected at the last follow-up for nine FMT recipients. Only contigs with reads per kilobase per million > 3 were shown to assure the colonisation of donor-derived contigs. Red lines indicate contigs transferred from the donor. (B) Percentage of donor-transferred Caudovirales contigs in FMT recipients at the last follow-up. The size of the circle indicates the count of Caudovirales contigs transferred from donor. The colour of the circle indicates the richness of Caudovirales in the recipient relating to the treatment response. (C) Comparison of the frequency of donor-derived Caudovirales contigs in FMT responders and in non-responders. Statistical significance was determined by Mann-Whitney test. *p<0.05. (D) Presence of bacterial operational taxonomic unit (OTUs) in FMT recipients at the last follow-up. The colour of the bar indicates the origin of the bacterial OTUs. Purple indicates donor-derived OTUs colonised in the recipient, orange indicates OTUs exclusively present in recipient but not in the donor, while green indicates OTUs present both in donor and in recipient. (E) Comparison of the frequency of donor-derived bacterial OTUs in FMT responders and in non-responders. Statistical significance was determined by Mann-Whitney test.
We also investigated bacterial transfer after FMT and correlated that with treatment outcome. We found increased frequency of bacterial families in the stool of CDI subjects after FMT, which included Lachnospiraceae and Ruminococcaceae (see online supplementary figure 6A,B). However, there was no significant difference in the frequency of donor-transferred bacterial OTUs between FMT responders and FMT non-responders (figure 4D,E). In two subjects who had early CDI recurrence (FMT7 and FMT8), a lower abundance of donor-transferred bacteria and a lack of increase in the bacterial diversity and richness were observed after FMT (see online supplementary figure 6B,C). These two subjects also had a low Caudovirales colonisation. Hence, early disease recurrence in FMT7 and FMT8 after FMT might be related to a lack of colonisation of both bacteria and viruses. Subject FMT9 experienced disease recurrence at week 28. This subject had low levels of Caudovirales taxa colonisation throughout the post-FMT follow-up stool samples even though there was an increase in bacterial diversity and richness and substantial bacteria colonisation after FMT (see online supplementary figure 6B,C). Overall, FMT responders had a lower colonisation of Caudovirales bacteriophages than non-responders. These data suggest a role of bacteriophages in the efficacy of long-term FMT outcomes.
We further assessed the presence of donor-derived contigs within each Caudovirales species along with the change in the abundance of these species after FMT. FMT responders acquired more donor-derived contigs within the Caudovirales species when compared with non-responders (see online supplementary figure 7A). Not all the relative abundance of Caudovirales species was significantly enhanced with the colonisation of corresponding Caudovirales contigs from the donor (see online supplementary figure 7A), which indicates that the newly formed Caudovirales ecosystem post FMT was based on a more intricate interaction network. Furthermore, not all transfer of Caudovirales contigs from donor was dependent on a high donor-to-recipient ratio of abundance of species (see online supplementary figure 7B).
Virome alterations in patients with CDI after vancomycin treatment
We assessed the impact of vancomycin treatment (STD) on the virome and bacterial microbiome in five patients with CDI (figure 1A, table 1). In subjects who responded to vancomycin (STD3, STD4, STD5), there was no significant change in the Caudovirales diversity or richness (see online supplementary figure 8A), suggesting that antibiotics may have minimal effect on Caudovirales community. In subjects who did not respond to vancomycin (STD1 and STD2), there was substantial fluctuation in Caudovirale diversity and richness as well as bacterial diversity and richness over the course of vancomycin treatment. A significant correlation between bacterial diversity and richness was observed in vancomycin responders. In FMT responders, a correlation was found between Caudovirales diversity and richness as well as a correlation between bacterial diversity and richness (supplementary figure 8B). Moreover, marginally significant inverse correlations were observed between Caudovirales richness and bacterial diversity, and between Caudovirales richness and bacterial richness in patients after FMT but not after vancomycin treatment. These data suggest that FMT has a synergistic effect on both the bacterial and viral component of the gut microbiome whereas vancomycin may predominantly affect the bacterial microbiome. We performed taxonomical analysis to further elaborate the effects of vancomycin on the virome community. In CDI subjects who responded to vancomycin, the virome structure was not significantly altered over time, whereas marked oscillation of the virome was seen in subjects who did not respond to vancomycin (supplementary figure 8C). Moreover, Caudovirales abundance was increased after vancomycin treatment, but decreased after FMT (supplementary figure 8D). To define the differentially enriched viral species between FMT and vancomycin responders, we implemented a LEfSe analysis across all the follow-up samples of treatment responders. A multitude of overpresented viral species was observed in the vancomycin samples, and nearly half of them belonged to Caudovirales (supplementary figure 9).
Virome and bacterial microbiome interactions in patients with CDI before and after FMT
To characterise the relationship between the configuration of virome and that of the bacterial microbiome, we evaluated the correlation of the diversity and richness of Caudovirales with that of bacterial communities in household controls and in patients with CDI. In controls, there was a significant correlation between Caudovirales diversity and bacterial diversity (Spearman's ρ=0.511, p<0.05), and between Caudovirales diversity and bacterial richness (Spearman's ρ=0.625, p<0.05) (figure 5A). However, these correlations were distorted in CDI. After FMT, we demonstrated a shift of the microbiome community from a low to high bacterial richness and diversity, and from a low to a high Caudovirales diversity (figure 5B). The microbiome of FMT non-responders changed less when compared with FMT responders. The temporal restoration of bacterial structure over the period of follow-up in subject FMT9 indicated bacterial reset, but disease recurrence at week 28 may be associated with unaltered virome structure after FMT. We further assessed the correlation of Caudovirales species with bacterial families in CDI subjects and controls. More positive correlations, particularly of bacterial families Proteobacteria and Actinobacteria with Caudovirales species, were found in CDI than in controls (see online supplementary figure 10). This corresponded with an increase in the relative abundance of these bacterial taxa in patients with CDI together with an overpresentation of Caudovirales taxa compared with controls. Among the Caudovirales taxa positively related to Proteobacteria, predominant bacteriophage species comprise Burkholderia phage, Planktothrix phage, Pseudomonas phage, Moraxella phage and Halomonas phage.
Figure 5.
Bacteria–Caudovirales relationship pattern (A) The correlation of bacteria richness with Caudovirales diversity and bacteria diversity with Caudovirales diversity in Clostridium difficile infection (CDI) and controls, respectively. Linear regression±95% CI, Spearman correlation coefficient and p value are shown. (B) The correlation of bacteria richness with Caudovirales diversity and bacteria diversity with Caudovirales diversity before and after faecal microbiota transplantation (FMT). (C) Bacteria–Caudovirales correlation pattern during FMT treatment. Spearman correlation plots of the relative abundances of Caudovirales species and bacterial families identified to be significantly associated with CDI and controls, in donor, pre-FMT and post-FMT samples. Statistical significance was determined for all pairwise comparisons; significant correlations (p<0.05) are displayed with asterisk. Blue circles and positive values indicate positive correlations, and red circles and negative values indicate inverse correlations. The size and shading indicate the magnitude of the correlation, where darker shades are more correlated than lighter ones.
We lastly investigated alterations in the bacteria–Caudovirales relationship in FMT responders. Pre-FMT samples showed a mutualistic relationship, particularly of Proteobacteria–Caudovirales and Actinobacteria–Caudovirales (figure 5C). However, a contraction of the number of significant correlations after FMT was observed between CDI-enriched bacteria and Caudovirales taxa. Interestingly, a few inverse correlations emerged after FMT, further implicating the importance of a ‘favourable’ virome–bacterial microbiome relationship. These results suggest a distinct virome–bacterial microbiome relationship before and after FMT treatment.
Discussion
To date, this study represents the most in-depth human gut virome study of FMT. In this pilot observational study, we showed that patients with CDI exhibited a state of enteric virome dysbiosis, characterised by an increased Caudovirales abundance and a decreased Caudovirales diversity, richness and evenness. The mechanism underlying the efficacy of FMT has been attributed to improved microbial diversity with re-establishment of a ‘normal’ bacterial microbiota as a host defence against C. difficile.2 3 Recently, Ott and colleagues showed that sterile faecal filtrate transfer was effective in treating five subjects with CDI, which highlights the therapeutic potential of other components including bacteriophages within the faecal matters other than bacteria.15 In this preliminary study, the observation that disease cure was associated with Caudovirales bacteriophage colonisation and donor Caudovirales richness provides new insights into the potential importance of virome derived from donor that may influence treatment outcome. Our data further highlight a new concept to FMT in that bacteriophages may be critical components of FMT. Bacteriophages have the potential to alter the composition and function of host microbiota and influence treatment outcomes. In particular, correlations between specific bacteriophages and bacteria shown in this study appeared to be associated with the outcome of FMT in CDI.
Among the enteric virome, Caudovirales is the most abundant taxon.17 20 Studies have shown that enteric Caudovirales were associated with intestinal inflammation although the underlying mechanisms remain largely unknown.17 21 Patients with IBD have been shown to have a significantly higher Caudovirales richness than healthy household controls,17 which is in contrast with the decreased Caudovirales richness observed in CDI subjects. Virome dysbiosis seen in CDI appeared to be disease-specific and was not observed in stool of subjects with norovirus-associated diarrhoea. Unlike the high cure rate of FMT in CDI, only up to a quarter of patients with UC, a subtype of IBD, achieved a sustained response with FMT.22–24 One plausible hypothesis is that a higher Caudovirales richness in donor than diseased recipient is important for FMT efficacy. In line with this hypothesis, we found that when donor Caudovirales richness was higher than that of the recipient, all the recipients responded to FMT, while more than half of the recipients whose Caudovirales richness was higher than that of the donor (three out of the five) experienced disease recurrence after FMT treatment. This finding suggests a donor effect of FMT efficacy and highlights that future FMT therapy should take into consideration detailed characterisation of donor and recipient faecal virome, although these findings need to be confirmed in a larger cohort. As this study has a modest sample size and we were unable to prove causation between donor Caudovirales richness, transfer of bacteriophages and FMT treatment outcome, it remains unclear whether altered bacteriophages represent a primary or secondary phenomenon. Understanding how Caudovirales bacteriophage transfer and donor virome richness affect FMT responses in CDI could help us understand their effects for treatments of other human diseases.
Activated phages can drive disease by reducing bacterial diversity and spreading virulence factors and antibiotic resistance.25 The expansion of Caudovirales abundance in CDI could arise from the induction of prophage from commensal microbes, according to the ‘predator–prey’ model by Lotka-Volterra in which bacterial phage prey on bacteria resulting in the release of bacteriophages after bacteria enter into the lytic cycle.26 27 Disparately expanded Caudovirales taxa may lead to decreased diversity and evenness in CDI compared with that in controls. Correlation of CDI-enriched bacteria with Caudovirales species in CDI and a reset of correlation after FMT indicate a ‘kill-the-winner’ dynamics, where upregulated Caudovirales bacteriophages were observed with the overpresentation of certain bacterial taxa enriched in CDI.28 Our data showed that restoration of a balanced relationship between bacterial microbiome and virome is important after FMT. It is notable that virome alterations in concert with bacterial changes after FMT may influence the efficacy of FMT in CDI.
Many patients with CDI often undergo multiple rounds of antibiotic therapy before undergoing FMT. This may lead to phage activation and act as a confounder. However, this scenario is unlikely as we included subjects on vancomycin as a control group and reported that the virome showed little variation over the course of treatment in subjects who responded to vancomycin.
This observational study had a modest sample size. However, to our knowledge, it represents the largest study to date that comprehensively followed CDI subjects after FMT and assessed in-depth virome alterations in association with clinical outcome. Importantly, our study provides detailed insight into the dynamics of interactions between viruses and bacteria in the intestine during FMT therapy. Sustained intestinal viral dysbiosis after FMT, due to limited amount of Caudovirales transfer from the donor, could be a potential factor that predisposes patients towards disease recurrence. These findings contribute to our knowledge on ‘optimal’ donor selection and highlight that future FMT therapy may consider detailed characterisation of donor and recipient faecal virome.
In conclusion, this pilot study showed that treatment response in FMT was associated with a high colonisation level of donor-derived Caudovirales taxa in the recipient, especially when Caudovirales richness in the donor was higher than that of the recipient. Our data suggest that establishing a bacteria and virome structure that more closely resembles that of healthy controls through FMT may be important to eradicate CDI.
Accession codes: Sequence data have been deposited to the NCBI Sequence Read Archive under BioProject accession number PRJNA353587 and PRJNA3535980.
Footnotes
Contributors: TZ performed the experiments, metagenomic analysis and drafted the manuscript. SHW performed FMT in clinic, helped virome analysis and revised the manuscript. KC collected the clinical samples and data. JYLC managed the clinical practice and clinical database. MC provided the virome sequencing data of patients with norovirus infection. PKSC, JCYW, KL and RL assisted in FMT and revised the manuscript. FKLC, JY and JJYS provided critical comments on the manuscript. SN designed the study, supervised the study and wrote the manuscript.
Funding: Hong Kong Society of Gastroenterology.
Competing interests: None declared.
Patient consent: Obtained.
Ethics approval: The Joint Chinese University of Hong Kong, New Territories East Cluster Clinical Research Ethics Committee.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Lessa FC, Winston LG, McDonald LC, et al. . Burden of Clostridium difficile Infection in the United States REPLY. N Engl J Med 2015;372:2369–70. 10.1056/NEJMc1505190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khoruts A, Dicksved J, Jansson JK, et al. . Changes in the composition of the human fecal microbiome after bacteriotherapy for recurrent clostridium difficile-associated diarrhea. J Clin Gastroenterol 2010;44:354–60. 10.1097/MCG.0b013e3181c87e02 [DOI] [PubMed] [Google Scholar]
- 3.van Nood E, Vrieze A, Nieuwdorp M, et al. . Duodenal Infusion of Donor Feces for Recurrent Clostridium difficile. N Engl J Med 2013;368:407–15. 10.1056/NEJMoa1205037 [DOI] [PubMed] [Google Scholar]
- 4.Drekonja D, Reich J, Gezahegn S, et al. . Fecal microbiota transplantation for clostridium difficile infection a systematic eeview. Ann Intern Med 2015;162:630–U230. 10.7326/M14-2693 [DOI] [PubMed] [Google Scholar]
- 5.Lee CH, Steiner T, Petrof EO, et al. . Frozen vs fresh fecal microbiota transplantation and clinical resolution of diarrhea in patients with recurrent clostridium difficile infection a randomized clinical trial. Jama-J Am Med Assoc 2016;315:142–9. [DOI] [PubMed] [Google Scholar]
- 6.Khoruts A, Sadowsky MJ. Therapeutic transplantation of the distal gut microbiota. Mucosal Immunol 2011;4:4–7. 10.1038/mi.2010.79 [DOI] [PubMed] [Google Scholar]
- 7.Manichanh C, Reeder J, Gibert P, et al. . Reshaping the gut microbiome with bacterial transplantation and antibiotic intake. Genome Res 2010;20:1411–19. 10.1101/gr.107987.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rea MC, Dobson A, O'Sullivan O, et al. . Effect of broad- and narrow-spectrum antimicrobials on Clostridium difficile and microbial diversity in a model of the distal colon. Proc Natl Acad Sci USA 2011;108(Suppl 1):4639–44. 10.1073/pnas.1001224107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barr JJ, Auro R, Furlan M, et al. . Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc Natl Acad Sci USA 2013;110:10771–6. 10.1073/pnas.1305923110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duerkop BA, Clements CV, Rollins D, et al. . A composite bacteriophage alters colonization by an intestinal commensal bacterium. Proc Natl Acad Sci USA 2012;109:17621–6. 10.1073/pnas.1206136109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reyes A, Wu M, McNulty NP, et al. . Gnotobiotic mouse model of phage-bacterial host dynamics in the human gut. Proc Natl Acad Sci USA 2013;110:20236–41. 10.1073/pnas.1319470110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Broecker F, Klumpp J, Schuppler M, et al. . Long-term changes of bacterial and viral compositions in the intestine of a recovered Clostridium difficile patient after fecal microbiota transplantation. Mol Case Stud 2016;2:a000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chehoud C, Dryga A, Hwang Y, et al. . Transfer of viral communities between human individuals during Fecal Microbiota rransplantation. MBio 2016;7:e00322 10.1128/mBio.00322-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Broecker F, Klumpp J, Moelling K. Long-term microbiota and virome in a Zürich patient after fecal transplantation against Clostridium difficile infection. Ann N Y Acad Sci 2016;1372:29–41. 10.1111/nyas.13100 [DOI] [PubMed] [Google Scholar]
- 15.Ott SJ, Waetzig GH, Rehman A, et al. . Efficacy of sterile fecal filtrate transfer for treating patients with clostridium difficile infection. Gastroenterology 2017;152:799–811.e7. 10.1053/j.gastro.2016.11.010 [DOI] [PubMed] [Google Scholar]
- 16.Broecker F, Russo G, Klumpp J, et al. . Stable core virome despite variable microbiome after fecal transfer. Gut Microbes 2016:1–7. 10.1080/19490976.2016.1265196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Norman JM, Handley SA, Baldridge MT, et al. . Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 2015;160:447–60. 10.1016/j.cell.2015.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9:357–U54. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bryson SJ, Thurber AR, Correa AMS, et al. . A novel sister clade to the enterobacteria microviruses (family Microviridae) identified in methane seep sediments. Environ Microbiol 2015;17:3708–21. 10.1111/1462-2920.12758 [DOI] [PubMed] [Google Scholar]
- 20.Reyes A, Haynes M, Hanson N, et al. . Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 2010;466:334–U81. 10.1038/nature09199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang JY, Kim MS, Kim E, et al. . Enteric viruses ameliorate gut inflammation via toll-like receptor 3 and toll-like receptor 7-mediated interferon-beta production. Immunity 2016;44:889–900. 10.1016/j.immuni.2016.03.009 [DOI] [PubMed] [Google Scholar]
- 22.Colman RJ, Rubin DT. Fecal microbiota transplantation as therapy for inflammatory bowel disease: a systematic review and meta-analysis. J Crohns Colitis 2014;8:1569–81. 10.1016/j.crohns.2014.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Leon LM, Watson JB, Kelly CR. Transient flare of ulcerative colitis after fecal microbiota transplantation for recurrent clostridium difficile infection. Clin Gastroenterol H 2013;11:1036–8. [DOI] [PubMed] [Google Scholar]
- 24.Kelly CR, Kahn SA, Kashyap P. Update on fecal microbiota transplantation 2015: indications, methodologies, mechanisms, and outlook. Gastroenterology 2015;149:223–37. 10.1053/j.gastro.2015.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Virgin HW. The Virome in Mammalian Physiology and Disease. Cell 2014;157:142–50. 10.1016/j.cell.2014.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cortez MH, Weitz JS. Coevolution can reverse predator-prey cycles. Proc Natl Acad Sci USA 2014;111:7486–91. 10.1073/pnas.1317693111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thingstad TF. Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol Oceanogr 2000;45:1320–8. [Google Scholar]
- 28.Rodriguez-Valera F, Martin-Cuadrado AB, Rodriguez-Brito B, et al. . OPINION Explaining microbial population genomics through phage predation. Nat Rev Microbiol 2009;7:828–36. 10.1038/nrmicro2235 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2017-313952supp001.pdf (13MB, pdf)