Abstract
Various phytohormones control plant growth and development and mediate biotic and abiotic stress responses. Gretchen Hagen 3 (GH3) acyl acid amido synthetases are plant enzymes that typically conjugate amino acids to indole-3-acetic acid (IAA) or jasmonic acid (JA) to inactivate or activate these phytohormones, respectively; however, the physiological and biological roles of many of these enzymes remain unclear. Using a biochemical approach, we found that the Arabidopsis thaliana GH3.15 (AtGH3.15) preferentially uses indole-3-butyric acid (IBA) and glutamine as substrates. The X-ray crystal structure of the AtGH3.15·AMP complex, modeling of IBA in the active site, and biochemical analysis of site-directed mutants provide insight on active site features that lead to AtGH3.15's preference for IBA. Assay-based in planta analysis of AtGH3.15-overexpressing lines indicated that their root elongation and lateral root density were resistant to IBA treatment but not to treatment with either IAA or JA. These findings suggest that AtGH3.15 may play a role in auxin homeostasis by modulating the levels of IBA for peroxisomal conversion to IAA. Analysis of AtGH3.15 promoter-driven yellow fluorescent protein reporter lines revealed that AtGH3.15 is expressed at significant levels in seedlings, roots, and parts of the siliques. We conclude that AtGH3.15 is unique in the GH3 protein family for its role in modifying IBA in auxin homeostasis and that it is the first GH3 protein shown to primarily modify a plant growth regulator other than IAA and JA.
Keywords: Arabidopsis thaliana, enzyme kinetics, auxin, plant hormone, X-ray crystallography, indole-3-butyric acid, jasmonic acid, phytohormone, auxin homeostasis
Introduction
Plants control growth and development and respond to biotic and abiotic stresses through the regulation of various plant hormones (1). Maintaining appropriate hormone levels and bioactive forms of these molecules is crucial for coordinating phytohormone responses. In these processes, enzymes involved in the synthesis, degradation, and modification of plant hormones help regulate their perception and downstream responses (2). For example, conjugation of plant hormones with amino acids can produce either active or inactive forms of these molecules, depending on the hormone and amino acid used. Formation of the bioactive jasmonate hormone (+)-7-iso-jasmonyl-l-isoleucine requires conjugation of jasmonic acid (JA)4 with isoleucine, whereas conjugation of indole-3-acetic acid (IAA), the primary auxin, with a variety of amino acids produces inactive and/or storage forms (3–5).
Gretchen Hagen 3 (GH3) acyl acid amido synthetases that conjugate amino acids to carboxylic acid-containing phytohormones, such as JA and IAA, contribute to regulating different aspects of plant growth, seed development, light signaling, and pathogen responses (3, 6–10). Biochemical and structural studies of GH3 proteins from Arabidopsis thaliana and other plants reveal that these enzymes catalyze a two-step reaction sequence requiring adenylation of the carboxylate substrate to form an acyl-AMP intermediate, which undergoes nucleophilic attack by an amino acid to yield a conjugated acyl acid–amino acid product (11–14). GH3 proteins are widespread in plants and typically found as multigene families (13, 15).
In A. thaliana, 19 genes encode GH3 proteins, but the physiological roles for many of these enzymes remain unclear. Early bioinformatic studies suggested that GH3 proteins group into JA-specific, IAA-specific, and benzoate-specific families (15), but structural studies of these enzymes and reanalysis using residues in the phytohormone (or acyl acid)-binding site led to delineation of seven groups for the Arabidopsis GH3 proteins (13). Group 1 consists of the (+)-7-iso-jasmonyl-l-isoleucine biosynthesis enzyme AtGH3.11/JAR1. The eight IAA-conjugating enzymes AtGH3.1, AtGH3.2, AtGH3.3, AtGH3.4, AtGH3.5/WES1, AtGH3.6/DFL1, AtGH3.9, and AtGH3.17/VAS2 form group 2. To date, physiological substrates for the remaining 10 GH3 proteins (group 3, AtGH3.10/DFL2; group 4, AtGH3.7 and AtGH3.12/PBS3; group 5, AtGH3.8; group 6, AtGH3.13, AtGH3.14, AtGH3.15, and AtGH3.16; group 7, AtGH3.19) are unknown. A non-physiological substrate for AtGH3.12/PBS3 has been used for activity assays (16). In an effort to elucidate potential biological roles for the uncharacterized Arabidopsis GH3 proteins, we used a biochemical screening approach with AtGH3.15, a representative member of group 6.
Here, we identify AtGH3.15 as an acyl acid amido synthetase specific for indole-3-butyric acid (IBA). Although several IAA-using GH3 proteins accept IBA as a substrate (8, 12), AtGH3.15 is unique among the characterized GH3 proteins as the first to modify IBA with amino acids at a high catalytic efficiency compared with IAA. Kinetic analysis of wildtype and mutant AtGH3.15 and the three-dimensional structure of the AtGH3.15·AMP complex provide molecular details that distinguish this protein from other structurally characterized GH3 proteins. In planta examination of Arabidopsis AtGH3.15 knockout and overexpression lines and expression analysis indicate a role for the enzyme throughout growth and development. Overall, this study reveals a broader role for GH3 proteins beyond the conjugation of JA and IAA in plant hormone homeostasis.
Results
Biochemical analysis of AtGH3.15 substrate specificity
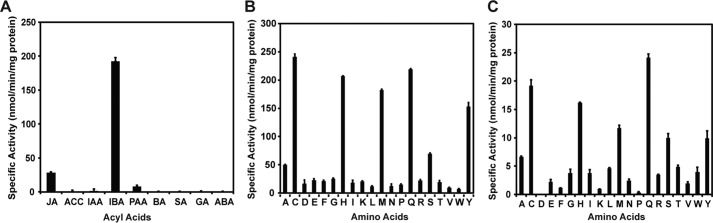
To identify a biochemical function for AtGH3.15, the protein was expressed in bacteria and purified with an N-terminal His tag. Screening for activity used JA, 1-aminocyclopropane-1-carboxylic acid, IAA, IBA, phenylacetic acid, benzoic acid, salicylic acid, gibberillic acid, and abscisic acid as potential acyl acid substrates, each with all 20 amino acids in a coupled enzyme assay (12). AtGH3.15 had the highest specific activity with IBA and glutamine as substrates (193 nmol min−1 mg−1 protein) but also displayed moderate activity with JA and glutamine (28 nmol min−1 mg−1 protein) (Fig. 1A). To confirm enzymatic formation of the IBA-Gln conjugate in the in vitro assay, QTRAP mass spectrometry analysis was performed. Incubation of AtGH3.15 with IBA, ATP, and glutamine led to formation of IBA-Gln (deprotonated molecular ion (M-H)− m/z = 330.3; Fig. S1). Assays in the absence of protein or any one substrate did not yield a product peak corresponding to the IBA-Gln conjugate. As reported for other GH3 proteins (8, 12–13), AtGH3.15 accepted a range of amino acids with both IBA (Fig. 1B) and JA (Fig. 1C), with the highest specific activities observed for cysteine, histidine, methionine, glutamine, and tyrosine. The amino acid profile of AtGH3.15 differs from previously studied GH3 proteins (3, 8, 17), which typically use either acidic amino acids with IAA or isoleucine and some other apolar amino acids with JA.
Figure 1.
Acyl acid and amino acid screening for AtGH3.15 activity. A, acyl acid activity screen of AtGH3.15 with each phytohormone (2.5 mm) at invariant ATP (1 mm) and glutamine (25 mm) concentrations. B, amino acid screen of AtGH3.15 with each amino acid (10 mm) at invariant IBA (1 mm) and ATP (1 mm) concentrations. C, amino acid screen of AtGH3.15 with each amino acid (10 mm) and invariant JA (1 mm) and ATP (1 mm) concentrations. All assays were performed using a coupled assay system as described under “Experimental procedures.” Average values ± S.D. (error bars) (n = 3) are shown.
Determination of steady-state kinetic parameters for AtGH3.15 confirms the ∼5-fold higher catalytic efficiency (kcat/Km) for IBA versus JA as the acyl acid substrate (Table 1). Comparison of the kinetic parameters for the amino acid substrates indicates that glutamine and cysteine displayed comparable catalytic efficiencies that were each 4–7-fold higher than the kcat/Km values for histidine, methionine, or tyrosine (Table 1). The up to 10-fold higher Km values for histidine, methionine, and tyrosine indicate that these are probably not preferred amino acid substrates. The kinetic parameters of AtGH3.15 for ATP (Table 1) were similar to those reported for other GH3 proteins, and Mg2+ was required for enzymatic activity (12, 18).
Table 1.
Steady-state kinetic analysis of AtGH3.15
Assays were performed with varied concentrations of one substrate and constant concentrations of the other two substrates. Fixed concentrations were as follows: IBA and JA (10 mm), ATP (1 mm), and Gln (25 mm). Initial velocity data were fit to the Michaelis–Menten equation using SigmaPlot. Average values ± S.D. (n = 3) are shown for steady-state kinetic parameters.
| Varied substrate | Fixed substrates | kcat | Km | kcat/Km |
|---|---|---|---|---|
| min−1 | μm | m−1 s−1 | ||
| IBA | Gln, ATP | 9.9 ± 0.2 | 527 ± 43 | 313 |
| JA | Gln, ATP | 4.4 ± 0.1 | 1,120 ± 72 | 65 |
| Gln | IBA, ATP | 17.0 ± 0.7 | 1,780 ± 190 | 159 |
| Gln | JA, ATP | 4.0 ± 0.1 | 2,530 ± 160 | 26 |
| Cys | IBA, ATP | 16.0 ± 0.3 | 2,080 ± 190 | 128 |
| His | IBA, ATP | 21.0 ± 0.4 | 12,300 ± 640 | 28 |
| Met | IBA, ATP | 13.0 ± 0.5 | 5,670 ± 650 | 38 |
| Tyr | IBA, ATP | 25.0 ± 0.6 | 18,600 ± 3,050 | 22 |
| ATP | IBA, Gln | 13.0 ± 0.5 | 84 ± 12 | 2,580 |
In the initial substrate screen, AtGH3.15 preferred an indole with a four-carbon side chain (i.e. IBA) versus an indole with a two-carbon side chain (i.e. IAA). To further examine whether side chain length influenced AtGH3.15 activity, assays using indoles with acidic side chains 2–6 carbons in length were performed (Table 2). AtGH3.15 used IAA, but less efficiently than IBA. Although IAA and IBA have similar Km values, the turnover rate was 14-fold slower with IAA. The three-carbon indole-3-propionic acid substrate displayed a 6-fold lower kcat/Km compared with IBA. Indole-3-valeric acid with a five-carbon side chain was used; however, saturation was not observed with indole-3-valeric acid (IVA; 250 μm maximum). An estimate of catalytic efficiency based on specific activity suggests comparable activity with IBA. No detectable activity was observed with the 6-carbon side-chain indole-3-caproic acid. AtGH3.15 accepts indole compounds with 2–5 carbon side chains with IBA as the preferred substrate.
Table 2.
Kinetic analysis of AtGH3.15 using indoles with varied side-chain length
Steady-state kinetic parameters were determined for IAA, IPA, and IBA as described under “Experimental procedures.” With IVA, saturation was not observed (maximum [S] = 250 μm), and an estimate of catalytic efficiency based on linear slope of initial velocity (if Km ≫ [S], then v = kcat/Km) is noted. Assays were performed with varied concentrations of the indole compound (0–10 mm), 1 mm ATP, and 10 mm glutamine. Average values ± S.D. (n = 3) are shown. Data were fit to the Michaelis–Menten equation using SigmaPlot. IPA, indole-3-propionic acid.
| Indole compound (side-chain length) | kcat | Km | kcat/Km |
|---|---|---|---|
| min−1 | μm | m−1 s−1 | |
| IAA (C2) | 0.76 ± 0.09 | 556 ± 160 | 23 |
| IPA (C3) | 10.2 ± 0.5 | 3,430 ± 360 | 50 |
| IBA (C4) | 9.9 ± 0.2 | 527 ± 43 | 313 |
| IVA (C5) | ∼540 |
Comparison of IBA versus IAA substrate preference in AtGH3 proteins
Other GH3 proteins from Arabidopsis (AtGH3.1, AtGH3.2, AtGH3.5/WES1, and AtGH3.17/VAS2) use IAA as the primary acyl acid substrate but can also conjugate IBA with varied specific activities (8, 17). To compare the activities of AtGH3.1, AtGH3.2, AtGH3.5/WES1, AtGH3.17/VAS2, and AtGH3.15 with IBA and IAA, each protein was expressed, purified, and assayed (Table 3). IAA data for AtGH3.1, AtGH3.2, AtGH3.5/WES1, and AtGH3.17/VAS2 were reported previously (17). Comparison of the catalytic efficiencies of AtGH3.1, AtGH3.2, AtGH3.5/WES1, and AtGH3.17/VAS2 shows 5–20-fold preferences for IAA over IBA for each enzyme (Fig. 2). In contrast, AtGH3.15 prefers IBA 15-fold over IAA. Moreover, the kcat/Km of AtGH3.15 for IBA is comparable with the catalytic efficiencies of AtGH3.1, AtGH3.2, AtGH3.5/WES1, and AtGH3.17/VAS2 for IAA. Biochemical analysis of AtGH3.15 identifies this as the first GH3 protein with robust IBA-conjugating activity.
Table 3.
Comparison kinetic parameters of Arabidopsis GH3 proteins with IAA and IBA
AtGH3.15 values for IAA and IBA are from Tables 1 and 2, respectively. Assays were performed as described under “Experimental procedures” with varied concentrations of either IAA or IBA and fixed concentrations of ATP (1 mm) and Asp (10 mm for AtGH3.5 and AtGH3.2), Asn (10 mm for AtGH3.1), Glu (10 mm for AtGH3.17), or Gln (10 mm for AtGH3.15). Initial velocity data was fit to the Michaelis–Menten equation using SigmaPlot. Average values ± S.D. (n = 3–9) are shown for steady-state kinetic parameters. IAA data for AtGH3.1, AtGH3.2, AtGH3.5/WES1, AtGH3.17/VAS2 were previously reported (17).
| kcatIAA | KmIAA | kcat/KmIAA | kcatIBA | KmIBA | kcat/KmIBA | |
|---|---|---|---|---|---|---|
| min−1 | μm | m−1 s−1 | min−1 | μm | m−1 s−1 | |
| AtGH3.15 | 0.76 ± 0.09 | 556 ± 160 | 23 | 9.9 ± 0.2 | 527 ± 43 | 313 |
| AtGH3.1 | 5.7 ± 0.6 | 530 ± 150 | 179 | 17 ± 20 | 17,000 ± 12,500 | 17 |
| AtGH3.2 | 17 ± 1.6 | 510 ± 105 | 556 | 17 ± 1.8 | 2,190 ± 390 | 129 |
| AtGH3.5 | 14.5 ± 1.4 | 770 ± 110 | 314 | 72 ± 40 | 16,500 ± 9,630 | 73 |
| AtGH3.17 | 2.7 ± 0.1 | 68.0 ± 6.0 | 662 | 1.2 ± 0.1 | 1,600 ± 160 | 13 |
Figure 2.
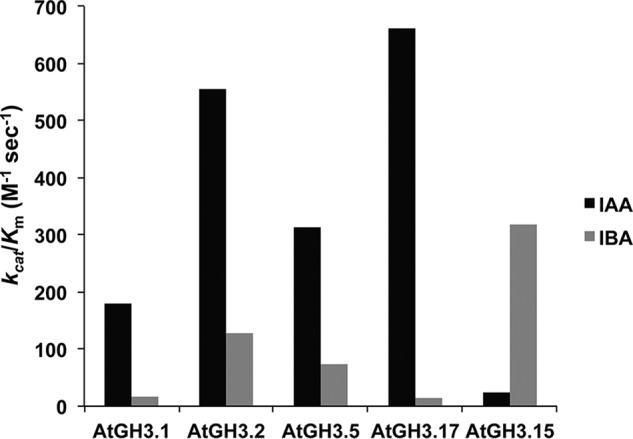
Comparison of catalytic efficiencies of AtGH3 proteins for IAA and IBA. The kcat/Km values of the indicated AtGH3 proteins with either IAA (black) or IBA (gray) as acyl acid substrates are shown. Values are summaries from the kinetic data in Table 3. IAA data for AtGH3.1, AtGH3.2, AtGH3.5/WES1, and AtGH3.17/VAS2 were published previously (17).
X-ray crystal structure of AtGH3.15 and modeling of IBA binding
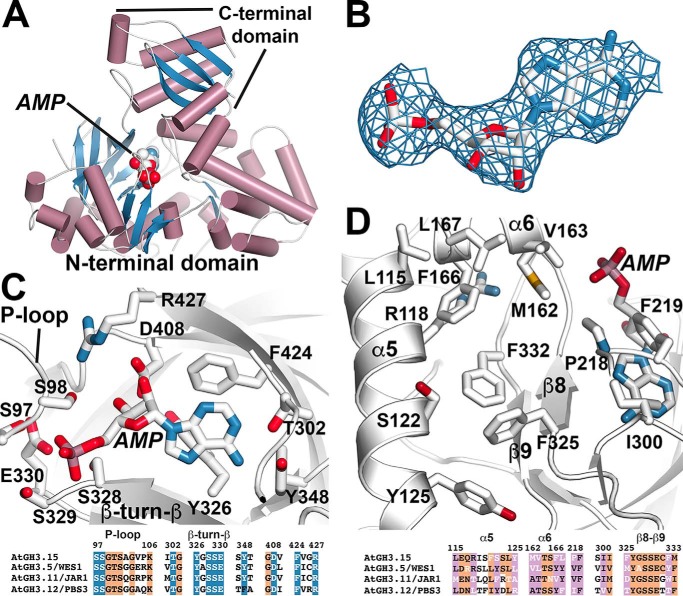
The GH3 acyl acid amido synthetases are a chemically versatile protein family because of their adaptable scaffold, which allows for recognition of IAA, JA, and benzoates, like salicylic acid and benzoic acid (13–14, 17, 19). As a step toward understanding the molecular basis for IBA specificity in AtGH3.15, the 3.0 Å resolution X-ray crystal structure of the AtGH3.15·AMP complex was solved by molecular replacement (Fig. 3 and Table 4). The overall fold of AtGH3.15 (Fig. 3A) is similar to other structurally characterized GH3 proteins, including AtGH3.5/WES1 (17), AtGH3.11/JAR1 (13), AtGH3.12/PBS3 (13, 19), and grape VvGH3.1 (14), with root mean square deviations of ∼1.3–1.5 Å2 for 590 Cα atoms. GH3 proteins contain a large N-terminal domain (∼450 amino acids) and a smaller conformationally flexible C-terminal domain (∼160 amino acids), which can rotate 180° to form open (adenylation reaction) and closed (conjugation reaction) active-site conformations (13). AtGH3.15 crystallized in the open conformation with clear electron density for AMP observed in the active site (Fig. 3B). The X-ray structure reveals a conserved ATP/AMP nucleotide binding site defined by a phosphate-binding loop (P-loop) and a β-turn-β motif (13) (Fig. 3C). Residues in this site are nearly invariant across GH3 proteins (13–14, 17, 19) (Fig. 3C, bottom). Although AtGH3.15 did not crystallize in the presence of an acyl acid substrate, previously reported GH3 protein structures (13–14, 17, 19) define the acyl acid–binding site as bordered by α5 (Leu-115, Arg-118, Ser-122, and Tyr-125), α6 (Met-162, Val-163, Phe-166, and Leu-167), β8 (Phe-325), and β9 (Phe-332), along with other residues (Pro-218, Phe-219, and Ile-300) (Fig. 3D). Comparison of the amino acids forming the putative IBA binding site of AtGH3.15 to the IAA, JA, and benzoate binding sites of AtGH3.5/WES1, AtGH3.11/JAR1, and AtGH3.12/PBS3, respectively, shows sequence variation that may account for the substrate preference of AtGH3.15 (Fig. 3D, bottom).
Figure 3.
X-ray crystal structure of AtGH3.15. A, overall three-dimensional structure. The ribbon diagram shows the N- and C- terminal domains with α-helices (rose) and β-strands (blue). Bound AMP is shown as a space-filling model. B, electron density for AMP is shown as a 2Fo − Fc omit map (1.5 σ). C, AtGH3.15 nucleotide binding site. Side chains are shown as stick models. Regions corresponding to the P-loop and the β-turn-β motif are indicated. A targeted sequence comparison of selected GH3 proteins from A. thaliana is shown below with numbering corresponding to AtGH3.15. Residues with side chains shown above are colored in blue with white text. Other conserved positions are highlighted in orange. D, AtGH3.15 acyl acid–binding site. Side chains of residues in the acyl acid–binding site are shown with α5, α6, and the β8-turn-β9 labeled. A targeted sequence comparison of selected GH3 proteins from A. thaliana is shown below with numbering corresponding to AtGH3.15. Residues with side chains shown above are colored in pink with white text indicating varied positions. Other conserved positions are highlighted in orange.
Table 4.
Summary of X-ray crystallographic statistics for the AtGH3.15·AMP complex
| Data collection | |
| Space group | F222 |
| Cell dimensions | a = 180.9 Å; b = 191.6 Å; c = 319.4 Å |
| Wavelength | 0.979 Å |
| Resolution (highest shell) | 47.9–3.01 Å (3.09–3.01 Å) |
| Reflections (total / unique) | 402,828/54,417 |
| Completeness (highest shell) | 100% (100%) |
| 〈I/σ〉 (highest shell) | 14.9 (2.6) |
| Rsym (highest shell) | 11.7% (75.9%) |
| Refinement | |
| Rcryst/Rfree | 18.6%/25.3% |
| No. of protein atoms/ligand atoms | 17,233/92 |
| r.m.s.d.,a bond lengths (Å) | 0.011 |
| r.m.s.d.,a bond angles (degrees) | 1.370 |
| Average B-factor: protein/ligand (Å2) | 52.4/36.8 |
| Stereochemistry: most favored, allowed, disallowed (%) | 96.6, 3.1, 0.3 |
a r.m.s.d., root mean square deviation.
To develop a model of how IBA binds to AtGH3.15, the substrate was computationally docked into the acyl acid–binding site of the AtGH3.15·AMP complex structure (Fig. 4). The solution most consistent with the known reaction chemistry orients the IBA toward the phosphate group of AMP, which would facilitate the first half-reaction that leads to adenylation of IBA and formation of the IBA-AMP reaction intermediate (12). The side chain of Arg-118 is positioned to potentially form a hydrogen bond with the indole nitrogen of IBA. This orientation also results in π-π stacking between Phe-332 and the IBA indole ring. In addition, Phe-325 may provide an edge-to-face interaction with the indole group of IBA. The resulting model of IBA binding in the AtGH3.15 active site is similar to the crystallographically determined binding of IAA in the AtGH3.5/WES1 and VvGH3.1 crystal structures (14, 17).
Figure 4.
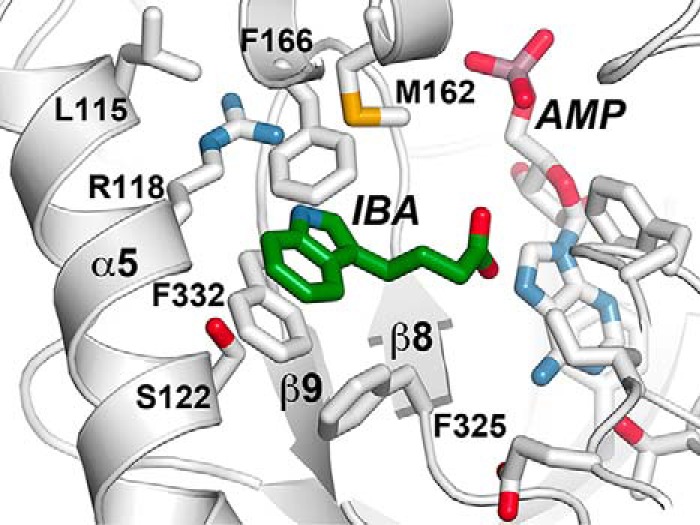
Structural model of IBA in the acyl acid–binding site of AtGH3.15. The three-dimensional structure of the AtGH3.15·AMP complex was used to computationally dock IBA into the acyl acid–binding site. Side chains of residues in the acyl acid–binding site of AtGH3.15 are shown with α5 and β8-turn-β9 noted. IBA is shown in green and AMP in white.
Mutations in the acyl acid–binding site of AtGH3.15 alter activity with IBA
To examine how acyl acid–binding site residues contribute to activity with IBA, site-directed mutations (S122F, S122I, S122L, S122W, S122Y, M162V, F166V, G216W, G216Y, P218V, F325M, F332V, and F332Y) were generated using PCR. One set of changes (S122Y, M162V, P218V, F325M, and F332Y) was chosen based on sequence alignments of acyl acid–binding site residues of IAA-specific AtGH3.1, AtGH3.2, AtGH3.5/WES1, and AtGH3.17/VAS2 versus AtGH3.15 (Fig. S2). Other mutations were selected based on the X-ray crystal structure of AtGH3.15 and the docking of IBA into the structure (Figs. 3 and 4). The G216W, G216Y, P218V, F325M, and F332Y mutants did not express to sufficient levels or were insoluble; however, the remaining mutant proteins were expressed and purified for biochemical assays (Table 5). Substitutions in the AtGH3.15 active site that change individual residues to those observed in the IAA-preferring enzymes generally resulted in modest activity changes and did not alter acyl acid preference (Table 5). For example, the S122Y and M162V mutants displayed catalytic efficiencies with IBA that were less than 3-fold different from wild type with IBA. Mutations at other positions in the active site resulted in slight decreases in catalytic efficiency with IBA (i.e. F166V and F332V); however, substitutions of Ser-122 proved interesting. Changes of Ser-122 to larger apolar residues improved the catalytic efficiency (kcat/Km) of AtGH3.15 with IBA (Table 5). The most substantial changes were observed with the S122I and S122L mutants that enhanced kcat/Km by 8- and 9.3-fold, respectively. Increasing side-chain size at this position may limit the space available for the indole ring of IBA to conformationally sample, which enhances IBA preference. However, if too large a side chain was introduced, such as in the S122W mutant, the kcat/Km value decreased by 12-fold. None of the point mutants exhibited improvements in kinetic parameters with IAA, which suggests that the IBA versus IAA preference probably requires a combination of multiple changes in the acyl acid site.
Table 5.
Steady-state kinetics of AtGH3.15 acyl acid–binding site mutants
Values for WT AtGH3.15 from Tables 1 and 2 and are shown for comparison with the mutant enzymes. Assays were performed as described under “Experimental procedures.” Average values ± S.D. (n = 3) for kinetic parameters are shown.
| kcatIBA | KmIBA | kcat/KmIBA | kcatIAA | KmIAA | kcat/KmIAA | |
|---|---|---|---|---|---|---|
| min−1 | μm | m−1 s−1 | min−1 | μm | m−1 s−1 | |
| WT | 9.9 ± 0.2 | 527 ± 43 | 313 | 0.76 ± 0.09 | 556 ± 160 | 23 |
| S122F | 24.0 ± 0.3 | 411 ± 28 | 983 | 8.4 ± 2.4 | 5,320 ± 1,930 | 26 |
| S122I | 15.0 ± 0.2 | 101 ± 8.0 | 2,550 | 3.2 ± 0.8 | 3,720 ± 1,240 | 14 |
| S122L | 17.0 ± 0.3 | 96 ± 8 | 2,950 | 3.6 ± 1.4 | 3,640 ± 1,970 | 16 |
| S122W | 0.8 ± 0.1 | 532 ± 101 | 26 | |||
| S122Y | 27.0 ± 0.4 | 620 ± 38 | 714 | 2.6 ± 0.4 | 2,710 ± 620 | 16 |
| M162V | 33.0 ± 0.4 | 487 ± 24 | 1,110 | 0.68 ± 0.03 | 294 ± 50 | 39 |
| F166V | 21.0 ± 0.7 | 1,360 ± 150 | 262 | 0.94 ± 0.06 | 441 ± 80 | 36 |
| F332V | 9.6 ± 0.2 | 1,290 ± 100 | 124 | 3.9 ± 5.3 | 12,800 ± 1,890 | 5.1 |
Phenotypic analysis of AtGH3.15 knockout and overexpression lines in Arabidopsis
To investigate the in planta role of AtGH3.15, T-DNA knockout lines were ordered from the Arabidopsis Biological Resource Center. Homozygous knockout lines of AtGH3.15 (SALK_108265C and SALK_079153) were obtained and confirmed via genotyping and RT-PCR (Fig. S3, A and B). Three independent A. thaliana Col-0 lines overexpressing N-terminal FLAG-tagged AtGH3.15 under control of the cauliflower mosaic virus 35S promoter were generated (35S:FLAG-AtGH3.15 1-5, 35S:FLAG-AtGH3.15 2-7, and 35S:FLAG-AtGH3.15 8-2). Overexpression was confirmed by RT-PCR (Fig. S3B) and α-FLAG immunoblot (Fig. S3C). Seedlings and adult plants of the knockout and overexpression lines showed no obvious phenotypes compared with wildtype.
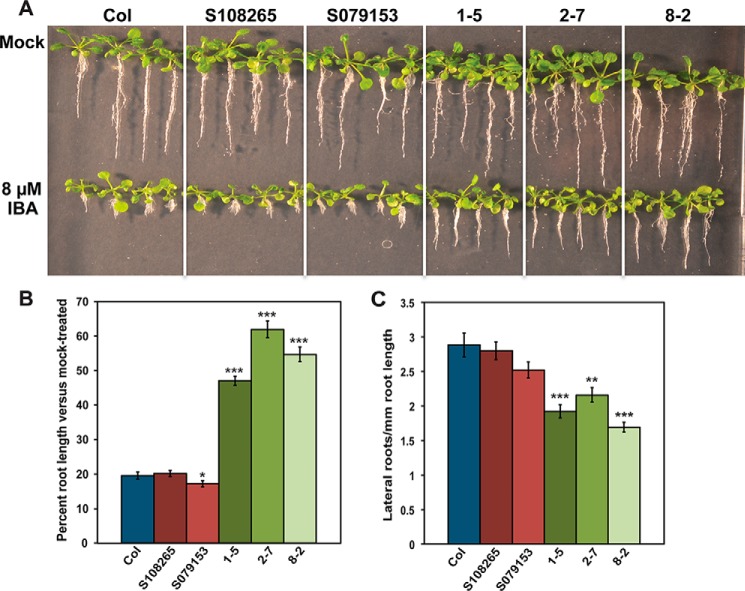
Treatment of wildtype Arabidopsis seedlings with IBA results in decreased primary root elongation and increased lateral root density (20). Perturbation of in planta IBA levels can lead to either hypersensitivity or resistance to treatment by IBA in root elongation assays (20). To assess whether conjugation of IBA by AtGH3.15 influences the sensitivity of AtGH3.15 knockout and overexpression lines to IBA treatment, root elongation assays on IBA were performed with wildtype Col-0, SALK_108265C, SALK_079153, 35S:FLAG-AtGH3.15 1-5, 35S:FLAG-AtGH3.15 2-7, and 35S:FLAG-AtGH3.15 8-2 lines for 18 days on 8 μm IBA (Fig. 5, A and B). The SALK 079153 knockout line showed a slight reduction in root length when grown on media containing IBA. In contrast, three overexpression lines displayed 50–65% longer roots than wildtype following IBA treatment, indicating resistance to IBA. Treatment with IBA results in both knockout lines displaying sensitivity comparable with wildtype and each overexpression line showing resistance. To determine whether lateral root density was also affected by either knockout or overexpression of AtGH3.15, wildtype Col-0, SALK_108285C, SALK_079153, 35S:FLAG-AtGH3.15 1-5, 35S:FLAG-AtGH3.15 2-7, and 35S:FLAG-AtGH3.15 8-2 were grown on 10 μm IBA for 10 days. Primary root length was measured, and the total lateral roots were counted to determine lateral root density (Fig. 5C). Both T-DNA knockout lines were comparable with wildtype in terms of lateral root density when grown under IBA treatment. In contrast, the overexpression lines showed 1.3–1.7-fold reductions in lateral root density under the same conditions.
Figure 5.
AtGH3.15 knockout and overexpression results in hypersensitivity and resistance to IBA in root elongation and lateral root density assays. A, seedlings of wildtype Col-0 (Col), SALK knockout lines (S108265 and S079153), and the 35S:FLAG-AtGH3.15 overexpression lines (1-5, 2-7, and 8-2) were grown under continuous yellow-filtered light for 18 days at 22 °C on medium supplemented with ethanol (mock) or 8 μm IBA. B, comparison of percentage root length change in 8 μm IBA-treated seedlings versus mock-treated seedlings grown for 18 days at 22 °C (see A). Percentage length of treated versus mock-treated was calculated as follows: (average root length IBA-treated seedling)/(average root length mock-treated seedling) × 100. C, effect of IBA treatment on lateral root density. Seedlings were grown under continuous yellow-filtered light for 10 days at 22 °C on medium supplemented with ethanol (mock) or 10 μm IBA. Lateral root density was calculated as number of lateral roots per mm root length. For B and C, error bars represent mean ± S.E. (error bars) (n = 20). *, p < 0.05; **, p < 0.001; and ***, p < 0.0001 versus wildtype.
When seedlings are grown on IAA, there is also a reduction in primary root elongation (20). As AtGH3.15 displayed a preference for IBA in biochemical assays (Figs. 1 and 2 and Tables 1 and 3) and is the first GH3 protein to do so, primary root elongation assays were performed on IAA-treated seedlings to confirm the selectivity of AtGH3.15 for IBA in planta (Fig. S4A). There were no differences between wildtype Col-0, SALK_108285C, SALK_079153, 35S:FLAG-AtGH3.15 1-5, 35S:FLAG-AtGH3.15 2-7, and 35S:FLAG-AtGH3.15 8-2 seedlings grown on 25, 50, or 100 nm IAA for 10 days. As with IBA and IAA, JA can also inhibit primary root elongation in Arabidopsis seedlings (3, 21). To determine whether the low activities shown with JA in vitro were physiologically relevant, wildtype Col-0, SALK_108285C, SALK_079153, 35S:FLAG-AtGH3.15 1-5, 35S:FLAG-AtGH3.15 2-7, and 35S:FLAG-AtGH3.15 8-2 seedlings were grown on 10 μm JA for 10 days, and root lengths were measured (Fig. S4B). There were no differences between the lines grown on JA compared with wildtype Col-0.
Expression pattern of AtGH3.15 in Arabidopsis
To determine when and where AtGH3.15 is expressed in planta, pAtGH3.15:YFP Arabidopsis lines were generated. These plants express yellow fluorescent protein (YFP) under control of the upstream promoter region of AtGH3.15. Seedlings at various stages through adult silique-producing plants were imaged using a Leica upright microscope. Two-day-old seedlings showed expression of YFP throughout the seedling, including root, shoot, and cotyledons (Fig. 6, A and B). Four-day-old (Fig. 6, C–E) and 5-day-old seedlings (Fig. 6F) had expression of YFP in the primary root, shoot, and cotyledons, but not in the root epidermal cells or root hairs. Nine-day-old seedlings displayed expression of YFP in the cotyledons and the shoot, but not in the true leaves (Fig. 6, G–I). There was continued YFP expression in the primary root and continued absence of expression in the root epidermal cells and root hairs; however, there was expression in lateral roots with a high level of expression in the tips. This pattern of root expression continued for 16- and 20-day-old plants (Fig. 6, J–L). No further changes were noted even after 30 days of growth. YFP expression in adult plants was greatest in the roots near the root/shoot junction but faded farther away from the root/shoot junction. At day 16, ∼75% of the root from shoot to root tip shows expression of YFP. At day 20, ∼50–75% of the root system showed expression of YFP. At day 28, mature siliques were observed to show YFP expression at the tip of the silique in the style just below the stigma (Fig. 7, left) and at the base of the silique in the replum and abscission zone of siliques (Fig. 7, right). Expression in above-ground tissue in adult plants was limited to these areas in the silique.
Figure 6.
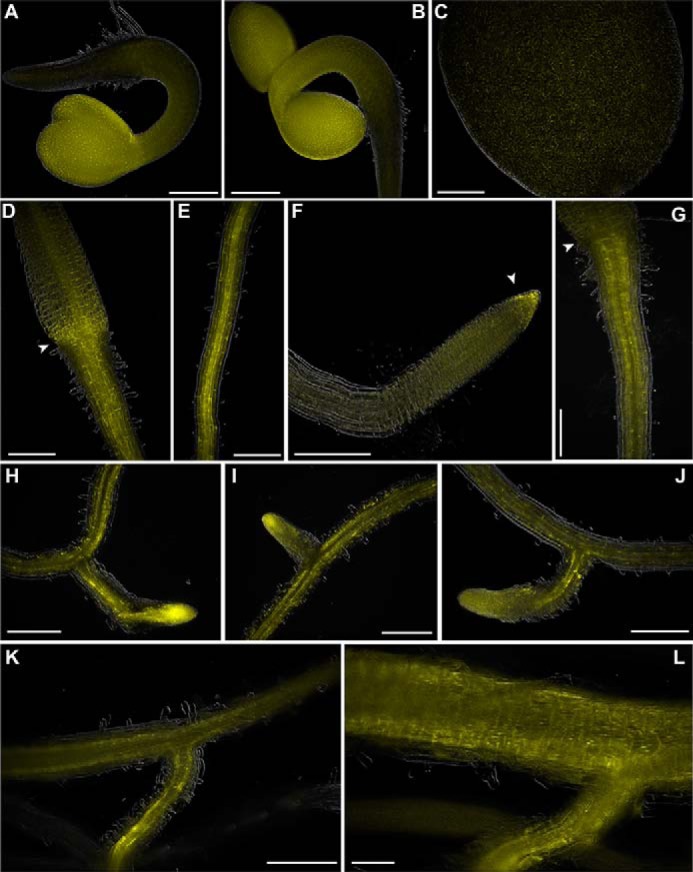
pAtGH3.15:YFP expression in Arabidopsis roots. Seedlings expressing YFP driven by the promoter region of AtGH3.15 (pAtGH3.15:YFP) were grown under continuous light for the indicated number of days at 22 °C and imaged using a Leica upright microscope. A and B, 2-day-old seedlings; C–E, 4-day-old seedlings; F, 5-day-old seedlings; G–I, 9-day-old seedlings; J, 11-day-old seedlings; K, 16-day-old plants; L, 20-day-old plants. Scale bar, 250 μm in A–K and 100 μm in L. Arrows in D and G indicate the root/shoot junction, and the arrow in F indicates the root tip.
Figure 7.

pAtGH3.15:YFP expression in mature siliques of Arabidopsis. Plants expressing YFP driven by the promoter region of AtGH3.15 (pAtGH3.15:YFP) were grown under continuous light for 28 days at 22 °C and imaged using a Leica upright microscope. Mature wildtype Col-0 silique is shown for reference. Left inset, pAtGH3.15-driven expression of YFP at the top of the silique in the style just below the stigma. Right inset, pAtGH3.15-driven expression of YFP at the base of the silique in the replum and abscission zone. Scale bars, 100 μm.
Discussion
Although IAA is the major auxin in plants, the biological roles of other endogenous auxins, such as IBA, are only beginning to be explored (22–24). Since the 1930s, IBA has been used for the rooting of plant cuttings, to promote root regeneration when transplanting plants, and to improve grafting success because it induces the formation of adventitious roots and roots from other organs, such as a stem and leaf (25–26). IBA is a metabolic precursor to IAA (27–28) (Fig. 8). Following import of IBA into the peroxisome, a process mimicking fatty acid β-oxidation converts IBA to IAA (29). Analysis of mutations in the peroxisomal β-oxidation pathway indicates a role for IBA in Arabidopsis seedling development (28, 30–34). Conversion of IBA to IAA also drives cell expansion in Arabidopsis seedlings, with mutants in the pathway exhibiting small cotyledons, decreased root hair expansion, fewer lateral roots, and delayed development (28, 30–34).
Figure 8.
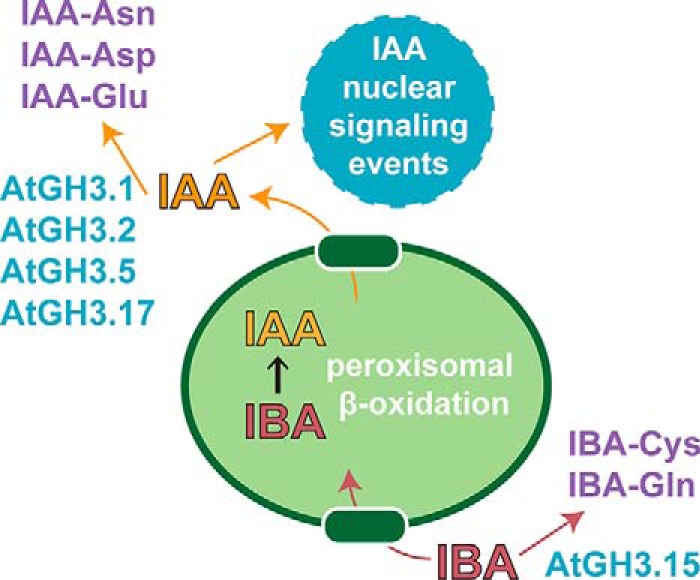
Overview of IBA to IAA conversion and conjugation with amino acids by GH3 proteins in Arabidopsis. IBA is imported into the peroxisome, where it undergoes peroxisomal β-oxidation to IAA. Export of IAA out of the peroxisome leads to nuclear signaling events. AtGH3.1, AtGH3.2, AtGH3.5/WES1, and AtGH3.17/VAS2 are IAA-conjugating enzymes that produce IAA-Asn (AtGH3.1), IAA-Asp (AtGH3.2 and AtGH3.5/WES1), and AtGH3.17/VAS2 (IAA-Glu). AtGH3.15 conjugates IBA. Conjugation of IAA and IBA lead to inactivation and/or storage forms of these auxins.
As with IAA, IBA conjugates have been described and may have a role in seedling development (20, 35–36); however, little information about the enzymes modifying IBA is known. The biochemical and physiological roles of GH3 acyl acid amido synthetases that modify JA and IAA by amino acid conjugation are well established (3, 8, 13, 23, 24, 36, 37). For example, multiple GH3 proteins in Arabidopsis, including AtGH3.1, AtGH3.2, AtGH3.5/WES1, and AtGH3.17/VAS2, generate various IAA-amino acid conjugates as part of the system that modulates auxin responses in plants (Fig. 8). Initial phylogenetic studies suggested that the 19 GH3 proteins in Arabidopsis formed three subfamilies with JA-conjugating, IAA-conjugating, and unknown conjugating activities, respectively (15). Later, crystallographic studies of AtGH3.11/JAR1, AtGH3.12/PBS3, and AtGH3.5/WES led to a re-examination based on residues forming the acyl acid–binding site, which suggested seven subgroups of GH3 proteins in Arabidopsis (13). Two of these subgroups corresponded to the previously identified JA- and IAA-specific GH3 proteins, but the physiological substrates of the other five groups were undefined.
In this study, we used an enzymatic screening approach to identify potential substrates for a member of one of these GH3 protein subfamilies of unknown biochemical function. In vitro analyses using a panel of 180 acyl acid/amino acid combinations (Fig. 1), mass spectrometry of the IBA-Gln conjugate (Fig. S1), and subsequent steady-state kinetic studies (Tables 1 and 2) identified AtGH3.15 as an IBA-specific acyl acid amido synthetase. Although other GH3 proteins were previously reported to have activity with IBA (8, 12), comparison of the catalytic efficiencies of the AtGH3.1, AtGH3.2, AtGH3.5/WES1, AtGH3.17/VAS2, and AtGH3.15 demonstrates clear differences in IAA versus IBA preferences (Fig. 2 and Table 3). AtGH3.15 is the first example of a GH3 protein that conjugates a phytohormone other than JA or IAA as the primary substrate.
To understand the molecular basis for the IBA preference of AtGH3.15, its X-ray crystal structure was determined (Fig. 3 and Table 4). AtGH3.15 shares a similar three-dimensional fold with other structurally characterized GH3 proteins, including AtGH3.5/WES1, AtGH3.11/JAR1, AtGH3.12/PBS3, and VvGH3.1 (13–14, 17, 19). This conservation of fold retains a highly conserved nucleotide binding site in the N-terminal domain and a conformationally flexible C-terminal domain that can rotate between open and closed states for the adenylation and transferase steps, respectively, of the chemical reaction sequence. Importantly, the structure of the AtGH3.15·AMP complex provides a detailed view of the acyl acid–binding site that leads to a preference for IBA over IAA. Comparison of the acyl acid–binding sites of AtGH3.15 (IBA-specific), AtGH3.11/JAR1 (JA-specific), AtGH3.5/WES1 (IAA-specific), and AtGH3.12/PBS3 (benzoate-using) highlights the distinct sets of residues in each active site that contribute to phytohormone specificity. Moreover, comparison of AtGH3.15 with the IAA-specific GH3 proteins in Arabidopsis emphasizes the differences in the IBA-binding site versus the conserved IAA specific pockets of AtGH3.1, AtGH3.2, AtGH3.5/WES1, and AtGH3.17/VAS2 (Fig. S2).
Modeling of IBA into the AtGH3.15 acyl acid–binding site highlights some aspects of the pocket that may influence IBA specificity (Fig. 4). The site is large enough to position the indole ring of IBA into the pocket bounded by residues that form potential van der Waals (Phe-325 and Phe-332) and hydrogen bond (Arg-118) interactions and to orient the four-carbon carboxylate side chain toward the AMP phosphate. Point mutations of residues in the AtGH3.15 acyl acid–binding site to those found in the IAA-specific GH3 proteins (Fig. S2) resulted in proteins with expression problems (P218V, F325M, and F332Y) or mutants (i.e. S122Y and M162V) that did not significantly alter substrate preference (Table 5). This suggests that multiple mutations are probably required for changes in activity and that additional substitutions may be needed to ensure proper protein folding. Follow-up mutagenesis identified Ser-122 as an interesting position in the acyl acid site, as mutations to apolar aliphatic side-chain residues (i.e. S122I and S122L) resulted in improved activity with IBA, whereas introduction of tryptophan at this position severely impacted catalytic function (Table 5). Although the specific determinants for IBA versus IAA preference in the GH3 proteins remain to be identified, the X-ray structure and initial mutagenesis of AtGH3.15 indicate that conversion of acyl acid preference probably involves changes across the acyl acid site.
Physiologically, the role of IAA-conjugating GH3 proteins has, thus far, largely been limited to inactivation of IAA in plants (5–10). As IBA is a precursor to the active auxin IAA, we explored the effects of in planta AtGH3.15 activity. Knockout and overexpression lines of AtGH3.15 did not show any obvious developmental phenotype (Fig. S3A, top). When grown on IBA, T-DNA insertion lines displayed root elongation inhibition and lateral root density similar to wildtype. In contrast, AtGH3.15 overexpression lines showed resistance to root elongation inhibition by IBA and decreased lateral root density (Fig. 5). These results suggest that AtGH3.15 may play a role in auxin homeostasis by modulating the levels of IBA for peroxisomal conversion to IAA (Fig. 8).
Treatment of plants with exogenous IBA, which is converted to IAA by the peroxisomal β-oxidation pathway, mimics the effect of IAA application (i.e. short root length and enhanced lateral root formation). Several peroxisomal enzymes dedicated to IBA conversion were identified through genetic mutations, including ech2, ibr1, ibr3, and ibr10, that confer IBA resistance without altering other IAA responses (28, 31). In each case, the mutation blocks conversion of IBA to IAA. Under normal growth conditions, only the ech2 mutant results in modestly shorter root growth, with the other mutants comparable with wildtype (18, 31). For the mutants, application of exogenous IBA results in longer roots and decreased lateral root density because of decreased conversion to IAA. Knockout of AtGH3.15, which conjugates IBA to amino acids, would be expected to not substantially alter seedling growth, as any exogenous IBA could still be converted by the peroxisomal pathway to IAA and result in seedlings with phenotypes that are similar to wildtype, which is what was observed here (Fig. 5). In contrast, overexpression of AtGH3.15 would enhance formation of IBA conjugates to reduce flux of IBA into the peroxisome and limit its conversion to IAA. This would provide resistance to IBA, which is exhibited by longer root length and reduced lateral root density (Fig. 5). Moreover, the IAA, JA, and phenylacetic acid responses of the AtGH3.15 T-DNA insertion and overexpression lines were similar to wildtype and indicate that AtGH3.15 does not significantly contribute to altering growth on those phytohormones. Overall, the observed phenotypes for the knockout and overexpression lines are consistent with AtGH3.15-catalyzed modification of IBA changing availability to the peroxisomal β-oxidation pathway (Fig. 8). At this point, it is not clear whether conjugation of IBA with amino acids leads to degradation of the hormone, as is the case with IAA-Asp, or to storage forms, such as IAA-Ala (23–26).
Consistent with the importance of IBA in seedling and root development (28, 30–34), we found AtGH3.15 promoter-driven expression of YFP in seedlings and the root (Fig. 6). These findings are also consistent with the previously reported possibility of IBA conjugates having a role in controlling levels of IBA in seedling development (20, 35–36). Based on the localization of AtGH3.15 reporter transcription, further investigation into the root architecture of knockout and overexpression lines of AtGH3.15 may yield phenotypes not seen in adult plants grown in soil. Expression of the YFP-reporter in the roots of plants past the seedling stage suggests that regulation of IBA levels is important not only at the seedling stage, but also perhaps throughout the growth and development life cycle of the plant. Besides seedlings, there was one above-ground organ where pAtGH3.15:YFP transcription was detected: the silique (Fig. 7). This is consistent with AtGH3.2 and AtGH3.6 expression in the abscission zone of siliques (7). Recent work has shown that the levels of IAA along the valve margins of siliques as well as the abscission zone are critical in organ shedding (38–39). Given the biochemical and physiological function of AtGH3.15, its localization suggests a potential role in shedding of mature seeds and siliques.
Determination of AtGH3.15 as an IBA-specific acyl acid amido synthetase involved in generating inactive and/or storage forms of IBA in the seedling, root, and silique contributes to the overall understanding of IBA function. Unraveling the fine-tuned network of IBA/IAA interplay will help in understanding the complex web of plant hormone homeostasis in which the GH3 proteins appear to play broader roles than previously thought. For example, multiple GH3 proteins are established as modulating free IAA, and the results presented here indicate that AtGH3.15 is associated with IBA, which would alter access to the peroxisomal pathway that produces IAA. Along with the structural data that expanded the classification of the GH3 proteins, these findings help to elucidate the complexity of the GH3 family. Further determination of the substrates of other unknown groups in the family will probably further expand the influence of the GH3 acyl acid amido synthetases on plant hormone homeostasis.
Experimental procedures
Cloning, protein expression, protein purification, and site-directed mutagenesis
The coding region for AtGH3.15 (At5g13370) was PCR-amplified from an ORF clone inserted in pENTR223 from the Arabidopsis Information Resource (accession number G60233) and ligated into pET-28a (Novagen) using the NdeI and BamHI restriction sites. The pET28a-AtGH3.15 construct was transformed into E. coli Rosetta (DE3) cells and grown at 37 °C in Terrific broth containing 50 μg ml−1 kanamycin and 34 μg ml−1 chloramphenicol to A600 nm ∼0.6–0.8. Protein expression was induced with 0.75 mm isopropyl β-d-1-thiogalactopyranoside overnight at 18 °C. Cells were pelleted (3,300 × g; 15 min), resuspended in lysis buffer (50 mm Tris, pH 8.0, 20 mm imidazole, 500 mm NaCl, 10% (v/v) glycerol, and 1% (v/v) Tween 20), and lysed by sonication. Cell debris was removed via centrifugation (35,000 × g; 1 h). Recombinant protein was purified using Ni2+-affinity chromatography. The supernatant was passed over a 1–2-ml Ni2+-nitrilotriacetic acid column, washed with buffer (50 mm Tris, pH 8.0, 10% (v/v) glycerol, 20 mm imidazole, and 500 mm NaCl). Protein was eluted from the column using 50 mm Tris, pH 8.0, 10% (v/v) glycerol, 250 mm imidazole, and 500 mm NaCl. For enzyme assays, protein was frozen in 100-μl aliquots and stored at −80 °C. For protein crystallization, the His-tagged protein was incubated in Spectra/Por 1 dialysis membrane with thrombin (1:2,000 ratio) in wash buffer overnight at 4 °C and then passed over a mixed Ni2+-nitrilotriacetic acid/benzamidine-Sepharose column to remove uncut protein and the protease. The flow-through was further purified by size-exclusion chromatography on a Superdex-75 26/60 HiLoad FPLC column equilibrated with 25 mm Tris, pH 8.0, and 100 mm NaCl. For all samples, protein concentration was determined by a Bradford assay with BSA as the standard. Expression and purification of other AtGH3 proteins for comparative assays were performed as described previously (17). Point mutations in AtGH3.15 were generated using the QuikChange PCR method with the resulting proteins expressed and purified as above.
Biochemical and steady-state kinetic analysis
Enzyme assays of AtGH3.15 used a coupled-enzyme assay to monitor the conversion of NADH to NAD+ with a loss of absorbance at 340 nm (12). Standard assay conditions were 50 mm Tris (pH 8.0), 3 mm MgCl2, 1 mm ATP, 1 mm phosphoenolpyruvate, 0.2 mm NADH, 2 units myokinase, 4 units pyruvate kinase, 4 units lactate dehydrogenase, and 10 μg of His-tagged protein in a 200-μl reaction. For the hormone screen, hormone substrates were assayed at 2.5 mm and amino acids at 20 mm. For amino acid screens, hormone substrates were assayed at 1 mm and amino acids at 10 mm. Kinetic analysis of hormones used 10 mm amino acid with varied concentrations of hormone up to 10 mm. Kinetic analysis of amino acids used 2.5 mm hormone and varied concentrations of amino acid up to 10 mm. Assays were performed on a Tecan Infinite 200 96-well plate reader with initial velocity data fit to the Michaelis–Menten equation using SigmaPlot.
Mass spectrometry of the IBA-Gln conjugate
Reactions were performed in the presence and absence of ∼20 μg of AtGH3.15 with 50 mm Tris (pH 8), 3 mm MgCl2, 1 mm ATP, 1 mm IBA, and 1 mm glutamine in a 200-μl volume. Reactions were allowed to react for 10 min at room temperature and then placed at −20 °C. The reactions were directly infused into the mass spectrometer. The MS1 (Q1) scan was acquired with the 6500-QTRAP (Sciex) in low mass using electrospray ionization in negative ion mode at a capillary voltage of −4,500 and a mass range of 50–500 m/z.
X-ray crystallography of AtGH3.15
Crystals of AtGH3.15 in complex with AMP were grown by vapor diffusion in hanging drops of a 1:1 mixture of protein (13 mg ml−1) and crystallization buffer (25% (v/v) PEG 1500, 0.1 m MIB (sodium malonate, imidazole, boric acid, pH 5.0), and 2 mm AMP. Crystals were frozen in liquid nitrogen with mother liquor supplemented with 25% (v/v) glycerol as a cryoprotectant. Diffraction data were collected at the SBC-19ID beamline of the Argonne National Laboratory Advanced Photon Source with indexing and scaling performed using HKL3000 (40). Molecular replacement was performed using PHENIX (41) with the N-terminal domain of AtGH3.12/JAR1 (13) (Protein Data Bank code 4ELQ; 37% amino acid sequence identity) as a search model. After molecular replacement, the C-terminal domain of AtGH3.15 was manually built, and side chains of the search model were changed to match the AtGH3.15 sequence. Model building and refinement were performed with COOT (42) and PHENIX, respectively. Crystallographic statistics are summarized in Table 4. Coordinates and structure factors for the AtGH3.15AtGH3.15·AMP complex were deposited in the Protein Data Bank (entry 6AVH).
Computational docking of IBA into the AtGH3.15 active site
Molecular docking of IBA into the three-dimensional structure of the AtGH3.15·AMP complex was performed using AutoDock Vina (version 1.1.2) (43). The ligand was generated using ChemDraw 3D and was energy-minimized. For docking of the ligand into the active site, a grid box of 20 × 20 × 20 Å was set with the level of exhaustiveness at 8.
Generation of Arabidopsis AtGH3.15 overexpression lines
A. thaliana ecotype Col-0 seeds were obtained from the Arabidopsis Biological Research Center (Ohio State University, Columbus, OH). The coding region was PCR-amplified from pET28a-AtGH3.15 and cloned into the pENTR/D-TOPO vector (Invitrogen). The coding region was then transferred into the pEarleyGate 202 vector (44) containing the cauliflower mosaic virus strong, constitutive 35S promoter and an N-terminal FLAG tag using the Gateway LR Clonase II enzyme mix kit (Invitrogen). The pEarleyGate 202-AtGH3.15 vector was electroporated into Agrobacterium tumefaciens LBA4404. A. thaliana plants were transformed via floral dip, and seeds were collected. Seeds were selected on Murashige and Skoog (MS) agar plates supplemented with 10 μg ml−1 glufosinate ammonium. Glufosinate-resistant seedlings were transferred to soil and grown to maturity. T2 lines were grown on MS agar plates supplemented with 10 μg ml−1 glufosinate ammonium, and lines displaying a 3:1 segregation ratio were used for isolation of independent homozygous lines. Three homozygous, single insertion lines were selected for phenotypic analysis. Leaves/seedlings were collected, frozen in liquid nitrogen, and ground. Extracts were centrifuged (28,000 × g; 5 min; 4 °C), and 30 μl of supernatant was removed and mixed with 1 μl of 1 m DTT. After the addition of loading dye, samples were boiled at 100 °C for 5 min and used to load two SDS-polyacrylamide gels, one for total protein staining with Coomassie Blue and one for immunoblotting. SDS-polyacrylamide gels for Western blotting were used to transfer proteins to either a polyvinylidene difluoride or nitrocellulose membrane using a semi-dry transfer method. Membranes were blocked with 5% (w/v) milk-PBST for 30–60 min and incubated rocking overnight at 4 °C with 5 ml of blocking solution and 1:5,000 primary antibody (α-FLAG). Membranes were rinsed three times for 5 min each with PBST and then incubated for 1 h with 5 ml of blocking solution and either 1:5,000 or 1:10,000 secondary antibody (α-rabbit IgG-alkaline phosphatase). Membranes were then rinsed as before. 5-Bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium was added and incubated until background started to change color, ∼10 min, and then rinsed with water.
Arabidopsis AtGH3.15 knockout lines
T-DNA insertion lines in At5g13370 were obtained from the Arabidopsis Information Resource. SALK_108265C and SALK_071953 are both insertions in the first exon of AtGH3.15. SALK_108265C was genotyped with primers LP (5′-dGCTTGTTTGTTAGGCACTTCG-3′) and RP (5′-dTACCCTAACATTTCGCCACAC-3′). SALK_071953 was genotyped with primers LP (5′-dGAGCTACATGAGCTGCATTCC-3′) and RP (5′-dTTCGTAGGTCACAACGGGTAC-3′). Both lines were genotyped using primer 5′-dATTTTGCCGATTTCGGAAC-3′, which is specific for the T-DNA insertion. Reduction in AtGH3.15 transcript was confirmed using RT-PCR with forward primer specific to exon 3 (5′-dCATTAAACAGACCAACTCATTTCTTTGGGATAGC-3′) and reverse primer specific to exon 5 (5′-CCAGCTTCTTGATGGTCTTTCTCGACCTCTAAG-3′). Actin transcript was used as a control and amplified with primers 5′-dACCGACCTTAATCTTCATGCTGC-3′ and 5′-dTACGCCAGTGGTCGTACAAC-3′. Plants identified as homozygous insertions with reduced AtGH3.15 transcript were used for subsequent experiments.
Root elongation assays
Seeds were surface-sterilized with 70% (v/v) ethanol for 5 min and 90% (v/v) ethanol for 1 min and resuspended in 0.1% sterile agar. Surface-sterilized seeds were stratified at 4 °C for 2–4 days, plated on MS plates with 0.6% (w/v) agar, and supplemented with 0.5% (v/v) sucrose. Plants were treated with either 8–10 μm IBA in 70% (v/v) ethanol or 70% ethanol as a control. Plates were sealed with 3M micropore tape and incubated at 22 °C for the indicated number of days. Seedlings grown on IBA or IAA were grown under continuous illumination through yellow long-pass filters to slow breakdown of the indolic compounds (45). Seedlings were excised from media and measured using a ruler. The percentage root length versus mock-treated was calculated using the equation (root length of treated seedlings)/(average root length of mock-treated seedlings) × 100. Treatment with IAA used 25, 50, or 100 nm IAA. JA treatments used 10 μm JA.
Lateral root density assay
Seeds were surface-sterilized, stratified, and plated as in the root elongation assays. Treatments used 10 μm IBA with mock-treated plates receiving equivalent amounts of 70% (v/v) ethanol (IBA was dissolved in 70% ethanol). Plates were sealed with 3M micropore tape and incubated at 22 °C for 10 days under continuous illumination through yellow long-pass filters to slow indolic compound breakdown (45). Seedlings were excised from the media, and root length was measured using a ruler. Lateral roots were then counted using a dissecting microscope. Lateral roots per mm of root length were calculated using the equation (number of lateral roots)/(root length (mm)).
Vector construction and transformation of pAtGH3.15:YFP into A. thaliana Col-0
The upstream region of AtGH3.15 was amplified using primers upstream315F (5′-dTTTTTGAATTCAACCGCCCTCACAATTTTTTTTTCTAG-3′) and upstream315R (5′-dTTTTCTCGAGGCTTGCTTACTGCTTTCTATTGTCTGC-3′). An 875-bp product, spanning the region between the beginning of AtGH3.15 and the upstream gene, was cloned into the pCR4 vector (Life Technologies, Inc.) to create pCR4-AtGH3.15upstream. The AtGH3.15 upstream region was excised from pCR4-AtGH3.15upstream using the restriction enzymes EcoRI and XhoI and subcloned into pMCS:YFP-GW to create pAtGH3.15:YFP-GW vector (46). The pAtGH3.15:YFP-GW vector was electroporated into A. tumefaciens GV3101. A. thaliana Col-0 plants were transformed via floral dip, and seeds were collected. Seeds were selected on MS agar plates supplemented with 10 μg ml−1 glufosinate ammonium. Glufosinate ammonium-resistant seedlings were transferred to soil and grown to maturity. T2 lines were grown on MS agar plates supplemented with 10 μg ml−1 glufosinate ammonium, and lines displaying a 3:1 segregation ratio were used for isolation of independent homozygous lines.
Imaging pAtGH3.15:YFP seedlings and adult plants
For seedling images, seeds were surface-sterilized with 70% ethanol for 5 min and 90% ethanol for 1 min and resuspended in 0.1% sterile agar. Surface-sterilized seeds were stratified at 4 °C for 2–4 days and plated on MS plates with 0.6% (w/v) agar and supplemented with 0.5% (v/v) sucrose. Plates were sealed with 3M micropore tape and incubated at 22 °C under continuous white light for the respective number of days. Seedlings were transferred to soil when too big for the plate. Plants were then grown at 22 °C under continuous white light and imaged at the respective number of days. To image roots of soil-transplanted plants, plants were gently removed from soil and rinsed in water to remove soil. Seedlings and adult plants were imaged using a Leica upright microscope or a Leica MZ10F fluorescence stereomicroscope with a YFP filter. Settings were determined when no fluorescence could be seen in wildtype Col-0 plants.
Author contributions
A.M.S., C.S.W., and J.M.J. conceptualization; A.M.S. and S.A. data curation; A.M.S., C.S.W., and S.A. formal analysis; A.M.S., C.S.W., and S.A. investigation; A.M.S., C.S.W., and S.A. methodology; A.M.S. and J.M.J. writing-original draft; A.M.S., C.S.W., S.A., and J.M.J. writing-review and editing; J.M.J. supervision; J.M.J. funding acquisition; J.M.J. validation; J.M.J. project administration.
Supplementary Material
Acknowledgments
The mass spectrometry measurement was performed at the Proteomics and Mass Spectrometry Facility at the Danforth Plant Science Center based upon work supported by National Science Foundation Grant DBI-1427621 for acquisition of the 6500 QTRAP LC-MS/MS. Portions of this research were carried out at the Argonne National Laboratory Structural Biology Center of the Advanced Photon Source, a national user facility operated by the University of Chicago for the Department of Energy Office of Biological and Environmental Research under Grant DE-AC02-06CH11357.
This work was supported by National Science Foundation (NSF) Grant NSF-MCB-1614539 (to J. M. J.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S4.
The atomic coordinates and structure factors (code 6AVH) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- JA
- jasmonic acid
- IAA
- indole-3-acetic acid
- GH3
- Gretchen Hagen 3
- IBA
- indole-3-butyric acid
- IVA
- indole-3-valeric acid
- YFP
- yellow fluorescent protein
- MS
- Murashige and Skoog.
References
- 1. Santner A., Calderon-Villalobos L. I., and Estelle M. (2009) Plant hormones are versatile chemical regulators of plant growth. Nat. Chem. Biol. 5, 301–307 10.1038/nchembio.165 [DOI] [PubMed] [Google Scholar]
- 2. Westfall C. S., Muehler A. M., and Jez J. M. (2013) Enzyme action in the regulation of plant hormone responses. J. Biol. Chem. 288, 19304–19311 10.1074/jbc.R113.475160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Staswick P. E., Tiryaki I., and Rowe M. L. (2002) Jasmonate response locuse JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14, 1405–1415 10.1105/tpc.000885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fonseca S., Chini A., Hamberg M., Adie B., Porzel A., Kramell R., Miersch O., Wasternack C., and Solano R. (2009) (+)-7-iso-Jasmonoyl-l-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5, 344–350 10.1038/nchembio.161 [DOI] [PubMed] [Google Scholar]
- 5. LeClere S., Tellez R., Rampey R. A., Matsuda S. P., and Bartel B. (2002) Characterization of a family of IAA-amino acid conjugate hydrolases from Arabidopsis. J. Biol. Chem. 277, 20446–20452 10.1074/jbc.M111955200 [DOI] [PubMed] [Google Scholar]
- 6. Nakazawa M., Yabe N., Ichikawa T., Yamamoto Y. Y., Yoshizumi T., Hasunuma K., and Matsui M. (2001) DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J. 25, 213–221 10.1046/j.1365-313x.2001.00957.x [DOI] [PubMed] [Google Scholar]
- 7. Takase T., Nakazawa M., Ishikawa A., Kawashima M., Ichikawa T., Takahashi N., Shimada H., Manabe K., and Matsui M. (2004) ydk1-D, an auxin-responsive GH3 mutant that is involved in hypocotyl and root elongation. Plant J. 37, 471–483 10.1046/j.1365-313X.2003.01973.x [DOI] [PubMed] [Google Scholar]
- 8. Staswick P. E., Serban B., Rowe M., Tiryaki I., Maldonado M. T., Maldonado M. C., and Suza W. (2005) Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17, 616–627 10.1105/tpc.104.026690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Park J. E., Park J. Y., Kim Y. S., Staswick P. E., Jeon J., Yun J., Kim S. Y., Kim J., Lee Y. H., and Park C. M. (2007) GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J. Biol. Chem. 282, 10036–10046 10.1074/jbc.M610524200 [DOI] [PubMed] [Google Scholar]
- 10. Nobuta K., Okrent R. A., Stoutemyer M., Rodibaugh N., Kempema L., Wildermuth M. C., and Innes R. W. (2007) The GH3 acyl adenylase family member PBS3 regulates salicylic acid-dependent defense responses in Arabidopsis. Plant Physiol. 144, 1144–1156 10.1104/pp.107.097691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen Q., Zhang B., Hicks L. M., Wang S., and Jez J. M. (2009) A liquid chromatography-tandem mass spectrometry-based assay for indole-3-acetic acid-amido synthetases. Anal. Biochem. 390, 149–154 10.1016/j.ab.2009.04.027 [DOI] [PubMed] [Google Scholar]
- 12. Chen Q., Westfall C. S., Hicks L. M., Wang S., and Jez J. M. (2010) Kinetic basis for the conjugation of auxin by a GH3 family indole acetic acid-amido synthetase. J. Biol. Chem. 285, 29780–29786 10.1074/jbc.M110.146431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Westfall C. S., Zubieta C., Herrmann J., Kapp U., Nanao M. H., and Jez J. M. (2012) Structural basis for pre-receptor modulation of plant hormones by GH3 family proteins. Science 336, 1708–1711 10.1126/science.1221863 [DOI] [PubMed] [Google Scholar]
- 14. Peat T. S., Böttcher C., Newman J., Lucent D., Cowieson N., and Davies C. (2012) Crystal structure of an indole-3-acetic acid amido synthetase from grapevine involved in auxin homeostasis. Plant Cell 24, 4525–4538 10.1105/tpc.112.102921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Terol J., Domingo C., and Talón M. (2006) The GH3 family in plants: genome wide analysis in rice and evolutionary history based on EST analysis. Gene 371, 279–290 10.1016/j.gene.2005.12.014 [DOI] [PubMed] [Google Scholar]
- 16. Okrent R. A., Brooks M. D., and Wildermuth M. C. (2009) Arabidopsis GH3.12 (PBS3) conjugates amino acids to 4-substituted benzoates and is inhibited by salicylate. J. Biol. Chem. 284, 9742–9754 10.1074/jbc.M806662200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Westfall C. S., Sherp A. M., Zubieta C., Alvarez S., Schraft E., Marcellin R., Ramirez L., and Jez J. M. (2016) Arabidopsis thaliana GH3.5 acyl acid amido synthetase mediates metabolic crosstalk in auxin and salicylic acid homeostasis. Proc. Natl. Acad. Sci. U.S.A. 113, 13917–13922 10.1073/pnas.1612635113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Westfall C. S., Herrmann J., Chen Q., Wang S., and Jez J. M. (2010) Modulating plant hormone levels by enzyme action: the GH3 family of acyl acid amido synthetases. Plant Signal. Behav. 5, 1607–1612 10.4161/psb.5.12.13941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Round A., Brown E., Marcellin R., Kapp U., Westfall C. S., Jez J. M., and Zubieta C. (2013) Determination of GH3.12 protein conformation through on-line HPLC-integrated SAXS measurements combined with X-ray crystallography. Acta Crystallogr. D Biol. Crystallogr. 69, 2072–2080 10.1107/S0907444913019276 [DOI] [PubMed] [Google Scholar]
- 20. Woodward A. W., and Bartel B. (2005) Auxin: regulation, action, and interaction. Ann. Bot. 95, 707–735 10.1093/aob/mci083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sugawara S., Mashiguchi K., Tanaka K., Hishiyama S., Sakai T., Hanada K., Kinoshita-Tsujimura K., Yu H., Dai X., Takebayashi Y., Takeda-Kamiya N., Kakimoto T., Kawaide H., Natsume M., Estelle M., et al. (2015) Distinct characteristics of indole-3-acetic acid and phenyl- acetic acid, two common auxins in plants. Plant Cell Physiol. 56, 1641–1654 10.1093/pcp/pcv088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Simon S., and Petrášek J. (2011) Why plants need more than one type of auxin. Plant Sci. 180, 454–460 10.1016/j.plantsci.2010.12.007 [DOI] [PubMed] [Google Scholar]
- 23. Korasick D. A., Enders T. A., and Strader L. C. (2013) Auxin biosynthesis and storage forms. J. Exp. Bot. 64, 2541–2555 10.1093/jxb/ert080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Enders T. A., and Strader L. C. (2015) Auxin activity: past, present, and future. Am. J. Bot. 102, 180–196 10.3732/ajb.1400285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kroin J. (1992) Advances using indole-3-butyric acid (IBA) dissolved in water for rooting cuttings, transplanting, and grafting. Intl. Plant Prop. Soc. 42, 489–492 [Google Scholar]
- 26. Ludwig-Müller J. (2011) Auxin conjugates: their role for plant development and the evolution of land plants. J. Exp. Bot. 62, 1757–1773 10.1093/jxb/erq412 [DOI] [PubMed] [Google Scholar]
- 27. Zolman B. K., Yoder A., and Bartel B. (2000) Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics 156, 1323–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Strader L. C., Culler A. H., Cohen J. D., and Bartel B. (2010) Conversion of endogenous indole-3-butyric acid to indole-3-acetic acid drives cell expansion in Arabidopsis seedlings. Plant Physiol. 153, 1577–1586 10.1104/pp.110.157461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Spiess G. M., and Zolman B. K. (2013) Peroxisomes as a source of auxin signaling molecules. Subcell Biochem. 69, 257–281 10.1007/978-94-007-6889-5_14 [DOI] [PubMed] [Google Scholar]
- 30. Bartel B., LeClere S., Magidin M., and Zolman B. K. (2001) Inputs to the active indole-3-acetic acid pool: de novo synthesis, conjugate hydrolysis, and indole-3-butyric acid β-oxidation. J. Plant Growth Regul. 20, 198–216 10.1007/s003440010025 [DOI] [Google Scholar]
- 31. Zolman B. K., Martinez N., Millius A., Adham A. R., and Bartel B. (2008) Identification and characterization of Arabidopsis indole-3-butyric acid response mutants defective in novel peroxisomal enzymes. Genetics 180, 237–251 10.1534/genetics.108.090399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Strader L. C., Wheeler D. L., Christensen S. E., Berens J. C., Cohen J. D., Rampey R. A., and Bartel B. (2011) Multiple facets of Arabidopsis seedling development require indole-3-butyric acid-derived auxin. Plant Cell 23, 984–999 10.1105/tpc.111.083071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Strader L. C., and Bartel B. (2011) Transport and metabolism of the endogenous auxin precursor indole-3-butyric acid. Mol. Plant 4, 477–486 10.1093/mp/ssr006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schlicht M., Ludwig-Müller J., Burbach C., Volkmann D., and Baluska F. (2013) Indole-3-butyric acid induces lateral root formation via peroxisome-derived indole-3-acetic acid and nitric oxide. New Phytol. 200, 473–482 10.1111/nph.12377 [DOI] [PubMed] [Google Scholar]
- 35. Bajguz A., and Piotrowska A. (2009) Conjugates of auxin and cytokinin. Phytochemistry 70, 957–969 10.1016/j.phytochem.2009.05.006 [DOI] [PubMed] [Google Scholar]
- 36. Ludwig-Müller (2000) Indole-3-butyric acid in plant growth and development. J. Plant Growth Regul. 32, 219–230 10.1023/A:1010746806891 [DOI] [Google Scholar]
- 37. Zheng Z., Guo Y., Novák O., Chen W., Ljung K., Noel J. P., and Chory J. (2016) Local auxin metabolism regulates environment-induced hypocotyl elongation. Nat. Plants 2, 16025 10.1038/nplants.2016.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Basu M. M., González-Carranza Z. H., Azam-Ali S., Tang S., Shahid A. A., and Roberts J. A. (2013) The manipulation of auxin in the abscission zone cells of Arabidopsis flowers reveals that indoleacetic acid signaling is a prerequisite for organ shedding. Plant Physiol. 162, 96–106 10.1104/pp.113.216234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. van Gelderen K., van Rongen M., Liu A., Otten A., and Offringa R. (2016) An INDEHISCENT-controlled auxin response specifies the separation layer in early Arabidopsis fruit. Mol. Plant 9, 857–869 10.1016/j.molp.2016.03.005 [DOI] [PubMed] [Google Scholar]
- 40. Minor W., Cymborowski M., Otwinowski Z., and Chruszcz M. (2006) HKL-3000: the integration of data reduction and structure solution: from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 62, 859–866 10.1107/S0907444906019949 [DOI] [PubMed] [Google Scholar]
- 41. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 43. Trott O., and Olson A. J. (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Earley K. W., Haag J. R., Pontes O., Opper K., Juehne T., Song K., and Pikaard C. S. (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45, 616–629 10.1111/j.1365-313X.2005.02617.x [DOI] [PubMed] [Google Scholar]
- 45. Stasinopoulos T. C., and Hangarter R. P. (1990) Preventing photochemistry in culture media by long-pass light filters alters growth of cultured tissues. Plant Physiol. 93, 1365–1369 10.1104/pp.93.4.1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Michniewicz M., Frick E. M., and Strader L. C. (2015) Gateway-compatible tissue-specific vectors for plant transformation. BMC Res. Notes 8, 63 10.1186/s13104-015-1010-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.