Abstract
Dihydroxyacid dehydratase (DHAD) is the third enzyme required for branched-chain amino acid biosynthesis in bacteria, fungi, and plants. DHAD enzymes contain two distinct types of active-site Fe–S clusters. The best characterized examples are Escherichia coli DHAD, which contains an oxygen-labile [Fe4S4] cluster, and spinach DHAD, which contains an oxygen-resistant [Fe2S2] cluster. Although the Fe–S cluster is crucial for DHAD function, little is known about the cluster-coordination environment or the mechanism of catalysis and cluster biogenesis. Here, using the combination of UV-visible absorption and circular dichroism and resonance Raman and electron paramagnetic resonance, we spectroscopically characterized the Fe–S center in DHAD from Arabidopsis thaliana (At). Our results indicated that AtDHAD can accommodate [Fe2S2] and [Fe4S4] clusters. However, only the [Fe2S2] cluster–bound form is catalytically active. We found that the [Fe2S2] cluster is coordinated by at least one non-cysteinyl ligand, which can be replaced by the thiol group(s) of dithiothreitol. In vitro cluster transfer and reconstitution reactions revealed that [Fe2S2] cluster–containing NFU2 protein is likely the physiological cluster donor for in vivo maturation of AtDHAD. In summary, AtDHAD binds either one [Fe4S4] or one [Fe2S2] cluster, with only the latter being catalytically competent and capable of substrate and product binding, and NFU2 appears to be the physiological [Fe2S2] cluster donor for DHAD maturation. This work represents the first in vitro characterization of recombinant AtDHAD, providing new insights into the properties, biogenesis, and catalytic role of the active-site Fe–S center in a plant DHAD.
Keywords: Arabidopsis thaliana, iron-sulfur protein, enzyme catalysis, electron paramagnetic resonance (EPR), circular dichroism (CD), dihydroxyacid dehydratase, iron-sulfur cluster trafficking, NFU protein
Introduction
Biosynthesis of branched-chain amino acids (BCAAs),4 namely leucine, isoleucine, and valine, is of particular research interest, not only because they are among the essential amino acids required by mammals, but also because the enzymes along their biosynthetic pathways are crucial in fermentative production of important industrial products (1). Dihydroxyacid dehydratase (DHAD, EC 4.2.1.9) catalyzes the third step of the BCAA biosynthesis, i.e. dehydration and tautomerization of 2,3-dihydroxyisovalerate and 2,3-dihydroxy-3-methylvalerate to the corresponding 2-keto acids, and an active-site Fe–S cluster has been shown to play a central role in catalysis (2–4).
DHADs have been identified in bacteria, archaea, fungi, and plants (2, 3, 5, 6) and are best characterized in Escherichia coli and spinach (Spinacia oleracea). As-isolated E. coli DHAD contains a [Fe4S4]2+ cluster that is probably coordinated by a non-cysteinate ligand at a specific iron site, as indicated by resonance Raman (RR) spectroscopy (3). As is the case for other Fe–S cluster–containing (de)hydratases, such as aconitase (7–9), the [Fe4S4]2+ cluster in E. coli DHAD is readily degraded, particularly in the presence of O2 or reactive oxygen species (ROS). As a result, even when purified under anaerobic conditions, the enzyme contained traces of cubane and linear [Fe3S4]1+ cluster species, which correspond to the initial O2-induced degradation products of the [Fe4S4] cluster (3). DHAD activity loss resulting from O2-induced [Fe4S4] cluster degradation has been studied both in vivo and in vitro (4, 10). Moreover, the NO-induced cluster degradation of bacterial DHADs has been implicated in the mammalian immune response against pathogens (11–13). In contrast, DHAD as-isolated from spinach has been shown to accommodate a [Fe2S2]2+ cluster, which is rare among the large family of Fe–S cluster–containing (de)hydratases (2). Dithionite-reduced spinach DHAD containing a [Fe2S2]1+ cluster was found to have 15–20% of the oxidized enzyme activity and exhibited EPR signals that undergo significant changes on binding substrate or product. In contrast to the [Fe4S4]2+ center in E. coli DHAD, the [Fe2S2]2+ cluster in spinach DHAD is stable in the presence of O2. The mechanism of the O2 resistance has yet to be determined, but it is suspected to be associated, at least in part, with the intrinsic oxygen stability of the all ferric [Fe2S2]2+ clusters compared with oxidized forms of the mixed-valence [Fe4S4]2+clusters (10).
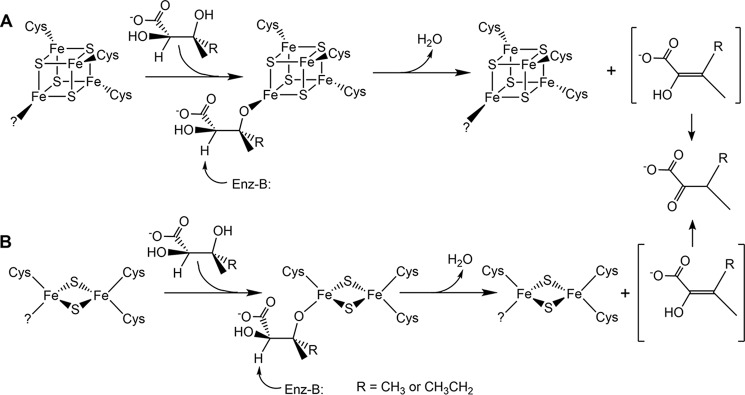
The catalytic mechanisms of DHADs are not yet fully understood, but have been shown to involve an enol intermediate with loss of a proton on the α-carbon during the tautomerization process (14). The substrate specificity and stereochemistry of DHADs have been studied in various organisms. It appears that both hydroxyl groups and the R-configuration at the α-carbon are required for the binding and recognition of the substrate, whereas configuration at the β-carbon is less important (15–17). The configuration of the β-carbon was retained after turnover, i.e. the hydrogen was added on the same side where the hydroxyl group was removed, implying that the enol intermediate was not released until the tautomerization was completed (14). Based on spectroscopic data and substrate specificity studies, it is postulated that the Fe–S cluster acts as a Lewis acid to activate the 3-hydroxy group of the substrate during the catalytic cycle (2, 16). The proposed mechanisms for both the [Fe2S2] and [Fe4S4] cluster–bound enzymes are depicted in Fig. 1 (18).
Figure 1.
Postulated catalytic mechanisms for E. coli DHAD (A) and spinach DHAD (B). In both mechanisms, DHAD is depicted as binding to a unique iron site of the cluster solely by the 3-hydroxyl group of the substrate. However, the possibility that the substrate carboxyl and/or 2-hydroxyl groups are also bound to the cluster cannot be excluded. Adapted from reference (18).
Thus far, studies on DHADs have been focused on the enzymology perspective, whereas little is known concerning active-site maturation and mechanisms of activation/inactivation. Over the last 2 decades, a number of highly conserved biosynthetic machineries dedicated to the biogenesis of Fe–S clusters have been discovered (19, 20). In vivo maturation of Fe–S proteins involves sulfur removal from cysteine by cysteine desulfurase, specific sulfur and iron delivery to the scaffold proteins, cluster assembly on the scaffold proteins, and intact cluster transfer to target enzymes either directly or via intermediate cluster carrier proteins (19, 20). However, the immediate cluster donor for [Fe4S4] or [Fe2S2] cluster–containing DHADs has yet to be determined. Insight into the mechanism of inactivation/activation of [Fe4S4] DHADs came from the observation that the loss in DHAD activity observed when E. coli cells are exposed to hyperbaric O2 levels is restored upon incubation in ambient air, without elevated protein expression levels (4). Subsequent in vitro reconstitution of inactive DHAD using E. coli crude cell extract identified four proteins that were involved in activation of DHAD, including IscS, a cysteine desulfurase, and three other enzymes involved in the cysteine metabolism pathway (21, 22). Other proteins that may be involved in the activation or maturation of E. coli DHAD are IscA and SufA, potential iron or Fe–S cluster carrier proteins (23–26) that have been shown to be required for bacterial [Fe4S4] cluster biogenesis under oxidative stress conditions (27). By incubating apo E. coli DHAD with iron-loaded E. coli IscA/SufA in the presence of IscS and cysteine, Ding and co-workers (27) successfully restored DHAD activity. Moreover, Lill and co-workers (28) have performed in vivo studies to show that yeast (Saccharomyces cerevisiae) Isa1/Isa2, homologs of bacterial IscAs, are essential for the maturation of [Fe4S4]2+ centers in mitochondrial proteins but are not required for the maturation of DHAD. Based on this observation and phylogenetic analyses, it was proposed that yeast DHAD contains a [Fe2S2] cluster, as is the case in plant enzymes (28).
In plants, BCAAs are synthesized in plastids (29, 30). Consequently, it is highly likely that plant DHADs are also located in plastids. Numerous proteins have been implicated in the biogenesis of Fe–S clusters in plastids, which utilize the SUF (sulfur utilization factors) machinery, including SUFBCD, SUFA1, SUFS, and SUFE, in addition to other proteins such as three NFU proteins (NFU1, NFU2, and NFU3), two monothiol glutaredoxins (GRXS14 and GRXS16), and the high chlorophyll fluorescence 101 (HCF101) protein (31, 32). Among these proteins, only NFU2 (33–36), GRXS14/S16 (37), and SUFA1 (38, 39) have been shown to bind [Fe2S2] clusters and therefore could potentially serve as Fe–S cluster donors for the maturation of plant DHAD. In this work, we present a comprehensive spectroscopic characterization of recombinant Arabidopsis thaliana (At) DHAD expressed in E. coli and an in vitro characterization of the immediate plastidial cluster donor for maturation of apo-DHAD.
Results
Spectroscopic characterization of as-purified AtDHAD
DHAD purified from spinach leaves was reported to contain stoichiometric [Fe2S2] clusters (2). Although AtDHAD heterologously expressed in E. coli and purified under aerobic conditions was found to exclusively contain [Fe2S2] clusters, the spectroscopic results discussed below and the iron and protein analyses (0.64 ± 0.06 iron per protein monomer) indicate sub-stoichiometric [Fe2S2] cluster content. Purification under strictly anaerobic conditions did not significantly increase the [Fe2S2] cluster content, indicating that the [Fe2S2] cluster is not lost during aerobic purification. Hence, it is likely that the cluster was not fully loaded by the E. coli host cells. In E. coli, maturation of the [Fe4S4] cluster active site of DHAD probably requires a distinct pathway for cluster assembly. Therefore, E. coli may lack the specific cluster donor that is required for effective cluster loading of AtDHAD, yielding only sub-stoichiometric cluster-bound forms of the plant enzyme.
Both UV-visible absorption and CD spectroscopy of as-purified recombinant AtDHAD indicate binding of a [Fe2S2]2+ cluster. The absorption spectrum is uniquely characteristic of a [Fe2S2]2+ center, with sulfur-to-Fe(III) charge transfer bands centered at 320, 420, and 460 nm and a broad shoulder ∼550 nm, with extinction coefficients of 6.6, 3.0, 2.9, and 1.0 mm−1 cm−1, respectively (Fig. 2). The extinction coefficients are indicative of ∼0.33 [Fe2S2]2+ clusters per DHAD monomer (36), in agreement with the iron and protein determinations that indicate 0.32 ± 0.03 [Fe2S2]2+ clusters per DHAD monomer. The CD spectrum is dominated by positive–negative–positive–negative–positive bands, centered at 320, 385, 467, 538, and 620 nm, respectively. Upon incubation with a 20-fold excess DTT, both the absorption and CD spectra underwent remarkable changes over the entire wavelength region. The absorption spectrum became more intense and less well-resolved throughout the UV-visible region, and the CD spectrum was completely different. These changes are not the result of cluster reduction, because dithionite reduction results in UV-visible absorption/CD and EPR spectra characteristic of a valence-localized S = 1/2 [Fe2S2]1+ center (Figs. 2 and 3), and the DTT-treated sample exhibits no EPR signal. Therefore, the spectroscopic changes induced by DTT appear to result from DTT binding to the [Fe2S2]2+ cluster as a unidentate or bidentate thiolate ligand. In accord with this interpretation, the absorption and CD spectra revert to those of the as-purified [Fe2S2]2+ cluster on extensive dialysis to remove excess DTT.
Figure 2.
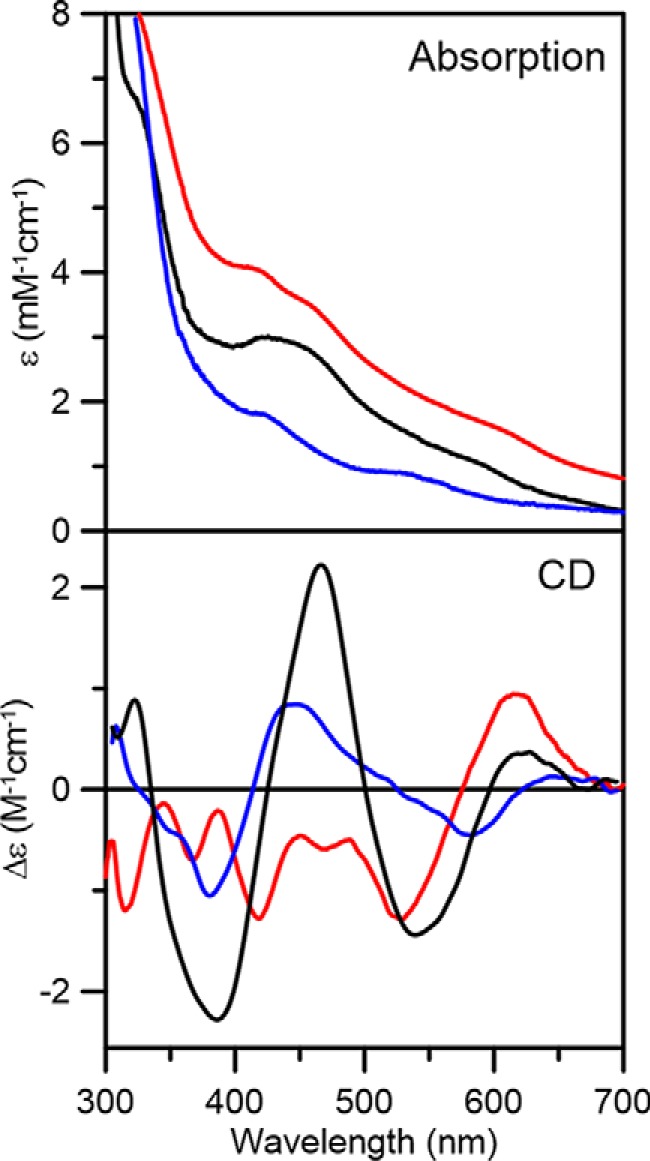
Room temperature UV-visible absorption and CD spectra of AtDHAD, as-purified (black lines), dithionite-reduced (blue lines), and as-purified after addition of DTT (red lines). All ϵ and Δϵ values were calculated based on the concentration of DHAD monomer.
Figure 3.
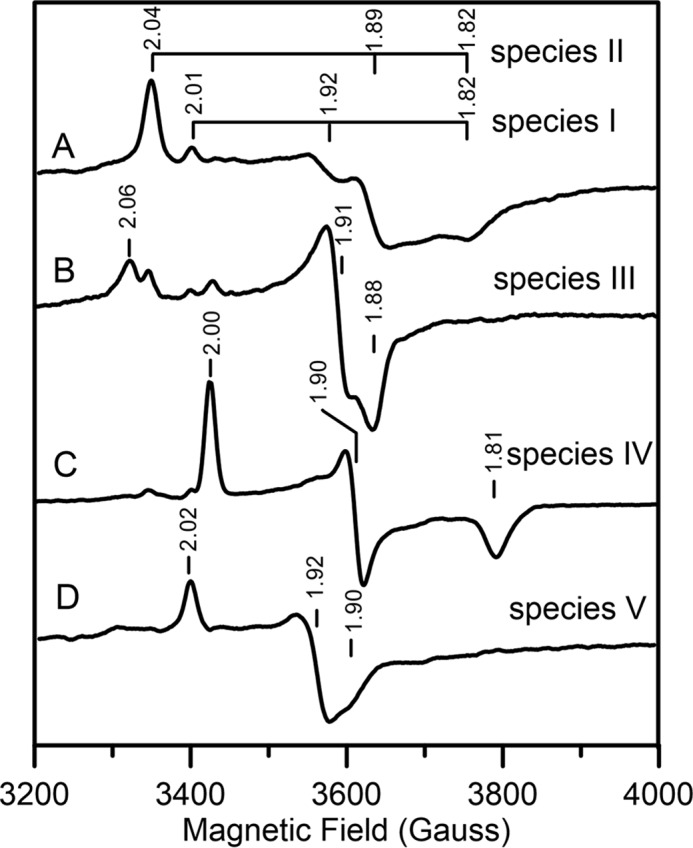
X-band EPR spectra of reduced [Fe2S2]1+ center in AtDHAD. All spectra presented were recorded at 35 K using 9.60 GHz microwave frequency, 6.4 G modulation amplitude, and a microwave power of 5 mW, except that B used 1 mW microwave power. A, dithionite-reduced DHAD; B, dithionite-reduced DHAD frozen immediately after addition of substrate; C, sample (B) after incubation with substrate at room temperature for 30 min; D, dithionite-reduced DHAD after incubating with DTT. The principal g-values corresponding to the major component(s) in each spectrum are indicated.
Further evidence for a [Fe2S2]2+ cluster that binds DTT in AtDHAD was provided by RR studies in the Fe–S stretching region (Fig. 4). RR spectroscopy provides information on the vibrational properties and coordination environment of Fe–S clusters and is sensitive to cluster ligation. The bands observed for as-purified DHAD are readily assigned based on normal mode calculations and 32S/34S isotope shift data for previously characterized synthetic and biological [Fe2S2] centers (40–42). Two predominantly Fe–S(Cys) terminal stretching modes, B3ut and Agt have been shown to be particularly useful in identifying non-cysteinyl ligands. As-purified AtDHAD exhibited these two bands at 302 and 352 cm−1, respectively, which are above the ranges established for all-cysteinate coordinated [Fe2S2]2+ clusters (281–295 and 326–340 cm−1, respectively), but are in the range established for the clusters with an oxygenic ligand (289–302 and 332–356 cm−1, respectively) (36, 40, 42–46). Moreover, the absence of bands below 280 cm−1 rules out the possibility of partial histidyl cluster ligation. Addition of the substrate did not significantly perturb the RR spectrum, suggesting that bound substrate or product also binds via an oxygenic ligand. In contrast, the presence of DTT significantly lowered the frequencies of the two bands to 292 and 332 cm−1, respectively, which is consistent with [Fe2S2]2+ centers with complete thiolate ligation. Therefore, the RR results indicate that DTT replaces an oxygenic ligand at the substrate-binding site of the [Fe2S2]2+ cluster.
Figure 4.
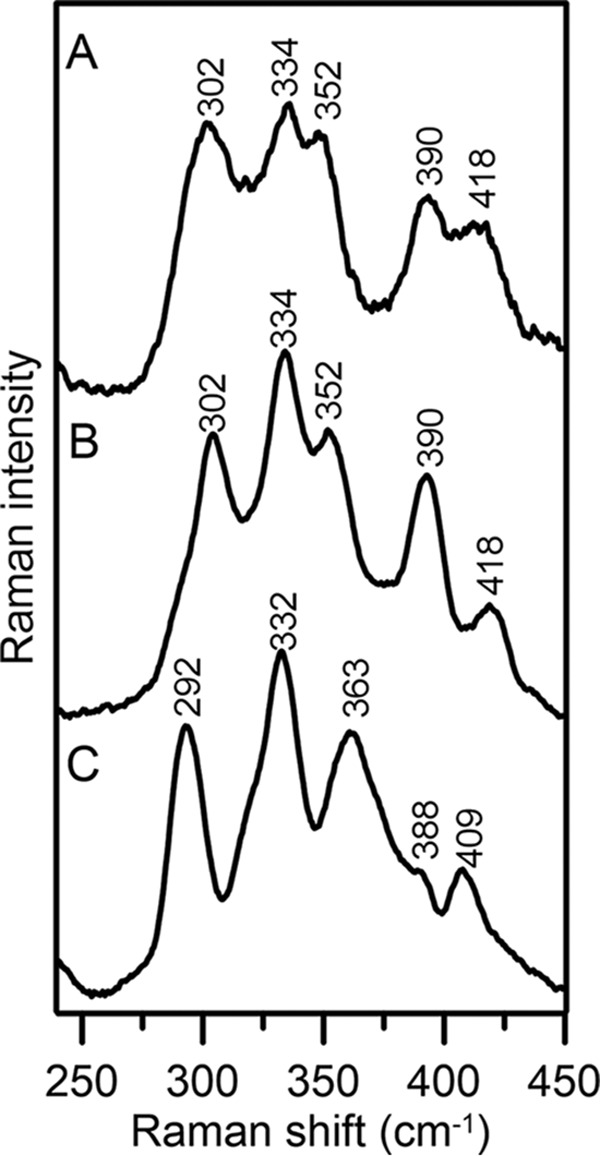
Low temperature (17 K) resonance Raman spectra of as-purified AtDHAD using 488-nm laser excitation line. A, as-purified DHAD; B, as-purified DHAD after addition of substrate; C, as-purified DHAD incubated with DTT. Bands resulting from the frozen buffer solution have been subtracted from all spectra.
As-purified AtDHAD did not exhibit an EPR signal attributable to a Fe–S cluster center, in accord with a S = 0 [Fe2S2]2+ center. However, on dithionite reduction to the valence-localized [Fe2S2]1+ reduced state, S = 1/2 resonances very similar to those previously characterized for reduced spinach DHAD were observed (Fig. 3) (2). Reduction of as-purified DHAD resulted in a complex resonance resulting from two overlapping S = 1/2 species (Fig. 3A): a minor rhombic species with g-values of 2.01, 1.92, and 1.82 (species I, gav = 1.92) and a dominant rhombic species with g-values of 2.04, 1.89, and 1.82 (species II, gav = 1.92). These resonances can be separated by microwave power and temperature dependence studies based on differences in relaxation behavior. Compared with species I, which undergoes no significant broadening at 70 K, species II is faster relaxing and is significantly broadened above 50 K. These resonances show increased g-value anisotropy and decreased gav values compared with the all-cysteine–ligated [Fe2S2]1+ centers. However, similar EPR spectra have been observed for Cys-to-Ser variants at the reducible site of all-cysteine–ligated [Fe2S2]1+ centers in ferredoxins (45–49) and of [Fe2S2]1+ centers with one or two His ligands at the reducible site (50–54).
Samples of dithionite-reduced DHAD frozen immediately after addition of substrate gave rise to a dominant faster relaxing species with g-values of 2.06, 1.91, and 1.88 (species III, gav = 1.95) and a minor slower relaxing species with g-values of 2.00, 1.90, and 1.81 (species IV, gav = 1.90) (Fig. 3B). Species III is attributed to the substrate-bound form, because thawing the sample and incubating at room temperature for 30 min resulted exclusively in the product-bound species IV (Fig. 3C). Evidence that species IV is a product-bound species also comes from the observation that an identical EPR signal was observed when reduced spinach DHAD was treated with the product 2-keto-isovalerate (2). Addition of DTT results in an almost axial spectrum, g-values of 2.02, 1.92, and 1.90 (species V, gav = 1.95), that is similar to those reported for fully cysteinyl-ligated clusters (Fig. 3D) (48, 51, 52). Taken together with the UV-visible absorption/CD and RR studies of the oxidized enzyme, the EPR results indicate that DTT binds to both the oxidized and reduced [Fe2S2]2+,1+ centers in DHAD.
IscS-mediated in vitro Fe–S cluster reconstitution of AtDHAD
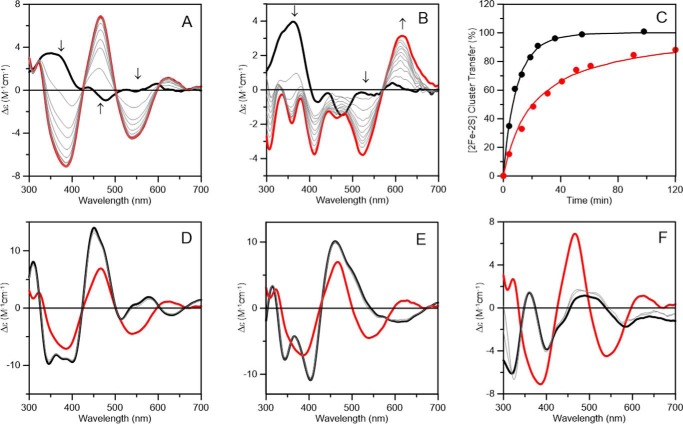
Because recombinant AtDHAD is expressed in E. coli with sub-stoichiometric [Fe2S2] clusters, IscS-mediated cluster reconstitution was attempted to obtain samples with maximal cluster content. Apo-DHAD was anaerobically incubated with excess l-cysteine, ferrous ammonium sulfate, and a catalytic amount of IscS for 4 h. Unexpectedly, repurification of the reconstitution mixture yielded a colored fraction that contained a different type of cluster, based on comparison of the UV-visible absorption and CD spectra with as-purified DHAD (Fig. 5). The reconstituted DHAD contained 3.9 ± 0.4 iron per monomer and exhibited a broad shoulder centered around 400 nm with an extinction coefficient of 16.0 mm−1 cm−1, indicative of a DHAD sample containing approximately one [Fe4S4]2+ cluster/monomer. Reconstituted DHAD contained a trace of residual [Fe2S2] cluster–bound form, as evidenced by the CD spectrum of the oxidized and dithionite-reduced forms (Fig. 5). The contribution of [Fe4S4]2+,1+ clusters to the CD spectrum is negligible. Dithionite reduction of reconstituted DHAD resulted in fast-relaxing S = 1/2 resonance with g-values of 2.05, 1.91, and 1.82 (gav = 1.93) accounting for 0.3 ± 0.1 spin cluster (Fig. 6). The sub-stoichiometric spin quantification is attributed to a low-potential [Fe4S4]2+,1+ cluster, which is only partially reduced by dithionite at pH 8 to yield a S = 1/2 [Fe4S4]1+ cluster. This conclusion is based on the absorption changes on reduction and the lack of significant low-field EPR features at low temperatures and high powers indicative of a S = 3/2 [Fe4S4]1+ cluster (55, 56). The EPR signal broadens with increasing temperature and is almost unobservable above 40 K, relaxation properties that are indicative of an S = 1/2 [Fe4S4]1+ cluster. The EPR signal was not perturbed by addition of the substrate, even after rapid freezing, suggesting that the assembled [Fe4S4] cluster is not able to bind substrate.
Figure 5.
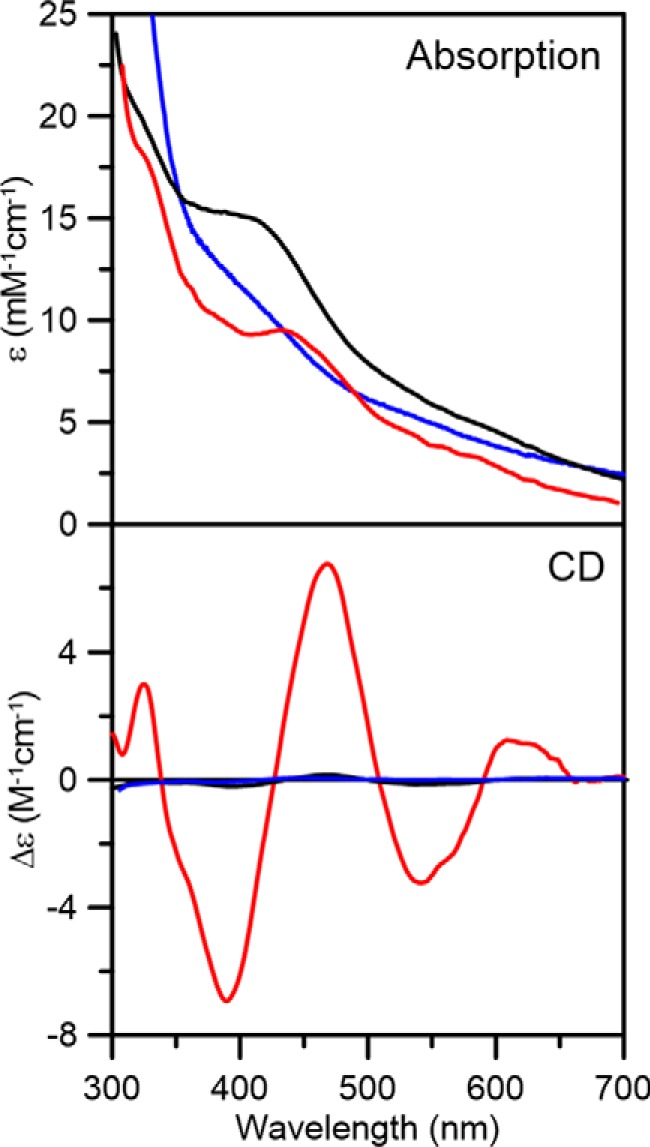
Room temperature UV-visible absorption and CD spectra of reconstituted AtDHAD (black lines), dithionite-reduced reconstituted DHAD (blue lines), and DHAD reconstituted in the presence of NFU2 (red lines). ϵ and Δϵ values were calculated based on the concentration of DHAD monomer.
Figure 6.
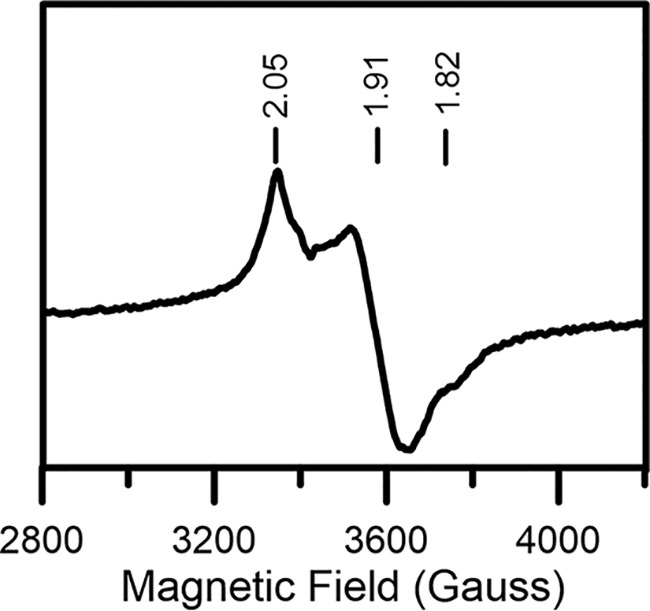
X-band EPR spectra of dithionite-reduced [Fe4S4] cluster–bound reconstituted AtDHAD. Spectrum recorded at 10 K using 9.60 GHz microwave frequency, 6.4 G modulation amplitude, and 5 mW microwave power.
We subsequently investigated the nature of the cluster reconstituted on DHAD in the presence of the plastidial carrier protein AtNFU2. NFU2 was chosen for three reasons. First, early studies provided indirect in vivo evidence for the involvement of NFU2 in the maturation of both [Fe2S2] and [Fe4S4]-containing proteins in plastids (33, 34). Second, recent in vivo studies have shown that BCAA synthesis is affected in an A. thaliana nfu2 mutant.5 Third, in vitro studies have shown that NFU2 is able to sequentially reconstitute both [Fe2S2]2+ and [Fe4S4]2+ clusters in the absence of DTT and to transfer these clusters rapidly and quantitatively to appropriate plastidial Fe–S proteins (36). After repurification, DHAD reconstituted in the presence of NFU2 contained near-stoichiometric [Fe2S2]2+ clusters based on UV-visible absorption and CD extinction coefficients (Fig. 5) and iron/protein determinations, 1.8 ± 0.3 Fe/monomer. [Fe4S4] cluster–bound NFU2 came off in a separate fraction. Moreover, there was no evidence, based on UV-visible absorption/CD and EPR studies, for the presence of [Fe4S4] cluster–bound DHAD in the sample reconstituted in the presence of NFU2. These results strongly implicate a role for AtNFU2 in the biogenesis of [Fe2S2] clusters in DHAD.
Maturation of AtDHAD via cluster transfer
The above results prompted an investigation into the rate of in vitro cluster transfer to apo DHAD using NFU2 pre-loaded with [Fe2S2]2+ clusters. The marked difference in the UV-visible CD spectra of the [Fe2S2] cluster-replete donor and acceptor proteins enables quantitative monitoring of the rate of cluster transfer (Fig. 7A). Upon addition of a 3-fold excess of apo DHAD, with respect to the [Fe2S2] cluster content of NFU2, the CD spectrum converts to that of holo DHAD with a single set of isosbestic points. This indicates intact [Fe2S2]2+ cluster transfer from NFU2 to DHAD, which is >90% complete within 24 min. Intact cluster transfer was also demonstrated by the observation that the rate of cluster transfer was not perturbed by the presence of 1 mm EDTA. The rate of cluster transfer was assessed by monitoring the change in CD intensity at 466 nm (Fig. 7C). Fits to second-order kinetics based on the initial concentration of [Fe2S2] clusters on the AtNFU2 donor (42 μm) and the concentration of the monomeric AtDHAD acceptor (126 μm) indicate a second-order rate constant of 1000 ± 100 m−1 min−1.
Figure 7.
Time course of cluster transfer from potential plastidial [Fe2S2]2+ cluster donor proteins to apo AtDHAD monitored by UV-visible CD spectroscopy. All Δϵ values are based on the concentrations of [Fe2S2]2+ clusters on the donor proteins in the reaction mixture, i.e. buffer A in the absence of 1 mm DTT unless otherwise as indicated. Arrows indicated the direction of change in CD intensity with time at selected wavelengths. The thick red lines and black lines in A, B, and D–F indicate the CD spectra of DHAD replete with [Fe2S2]2+ clusters and the CD spectra of the [Fe2S2]2+ cluster donors in the reaction mixture, prior to initiating the reaction by the addition of DHAD, respectively. A, CD spectra of a reaction mixture containing AtNFU2 (42 μm in [Fe2S2]2+ clusters) and apo AtDHAD (126 μm) at 4, 8, 13, 19, 24, 36, 55, and 100 min (thin gray lines). B, CD spectra of a reaction mixture containing AtNFU2 (42 μm in [Fe2S2]2+ clusters), apo AtDHAD (42 μm), and 1 mm DTT at 4, 13, 21, 31, 41, 51, 61, 91, and 120 min (thin gray lines). C, kinetic simulations of cluster transfer from [Fe2S2]-NFU2 to apo DHAD in the absence (black line) and presence (red line) of 1 mm DTT, based on second-order kinetics and the initial concentrations of [Fe2S2] clusters on NFU2 and of apo DHAD. Percent cluster transfer in the absence of DTT was assessed at 466 nm (black circles) and simulated with a second-order rate constant of 1000 m−1 min−1. Percent cluster transfer in the presence of DTT was assessed at 620 nm (red circles) and simulated with a second-order rate constant of 1050 m−1 min−1. D, CD spectra of a reaction mixture containing AtGRXS14 (45 μm in [Fe2S2]2+ clusters) and apo AtDHAD (45 μm in monomer) at 4 and 50 min (thin gray lines). E, CD spectra of a reaction mixture containing AtGRXS16 (45 μm in [Fe2S2]2+ clusters) and apo AtDHAD (45 μm in monomer) at 4 and 50 min (thin gray lines). F, CD spectra of a reaction mixture containing AtSUFA1 (42 μm in [Fe2S2]2+ clusters) and apo AtDHAD (42 μm in monomer) at 4 and 50 min (thin gray lines).
[Fe2S2]2+ cluster transfer from NFU2 to apo DHAD also occurred in the presence of 1 mm DTT to yield the DTT-bound form of DHAD, using a 1:1 cluster acceptor/donor ratio (Fig. 7B). However, the lack of a single set of isosbestic points and the rapid loss of the CD signal of the NFU2 [Fe2S2]2+ center during the first 4 min, without commensurate cluster assembly on DHAD, suggest a biphasic reaction most likely involving a DTT-bound [Fe2S2]2+ cluster intermediate. The second-order rate constant for the cluster insertion step (1050 ± 100 m−1 min−1) was assessed at 620 nm, where [Fe2S2] NFU2 has negligible CD intensity, and was found to be similar to that observed in the absence of DTT (Fig. 7C). Interestingly, the UV-visible absorption and CD spectra of [Fe2S2]2+ cluster–bound NFU2 are not significantly perturbed by the 1 mm DTT (compare Fig. 7, A and B). This suggests that the binding of [Fe2S2]2+ cluster–bound NFU2 to apo DHAD exposes the cluster, making it accessible to ligation by DTT. Such DTT-mediated cluster transfers are unlikely to be physiologically relevant.
There are several other potential carrier proteins that have been implicated in the biogenesis of [Fe2S2] cluster–containing proteins in plastids, namely GRXS14, GRXS16, and SUFA1. To assess the capability of these proteins to transfer clusters to DHAD, we also carried out parallel in vitro experiments using cluster-loaded forms of these alternative donors. Negligible (<10%) [Fe2S2]2+ cluster transfer to apo AtDHAD was observed after 50 min using [Fe2S2]2+ cluster–bound GRXS14 as the donor with a 1:1 cluster acceptor/donor ratio (Fig. 7D). Analogous results were observed using [Fe2S2]2+ cluster–bound GRXS16 in place of GRXS14 as the donor (Fig. 7E). In addition, no evidence for any [Fe2S2]2+ cluster transfer to apo AtDHAD cluster transfer was observed after 50 min using [Fe2S2]2+ cluster–bound SUFA1 as the donor with a 1:1 acceptor/donor ratio (Fig. 7D). Clearly, in vitro cluster transfer experiments point to [Fe2S2]2+ cluster–bound AtNFU2 as the probable physiological cluster donor for maturation of AtDHAD.
Activity studies of AtDHAD samples
As shown in Fig. 8, the specific activities of AtDHAD samples correlate well with the level of [Fe2S2]2+ cluster incorporation and demonstrate that the samples reconstituted with [Fe4S4]2+ clusters are inactive. These results are in accord with the EPR results presented above that show that the reduced [Fe2S2]1+ clusters in DHAD can bind both the substrate and product at a unique iron site, whereas the EPR signal of the reduced [Fe4S4]1+ is not perturbed by the substrate. The spectroscopic results also show that DTT can bind to both the oxidized and reduced [Fe2S2]2+,1+ centers in DHAD. This was confirmed by the activity results, which show a 63% decrease in activity on addition of a 20-fold excess of DTT that is largely reversed on removal of DTT by dialysis. This is consistent with DTT acting as a competitive inhibitor for DHAD.
Figure 8.
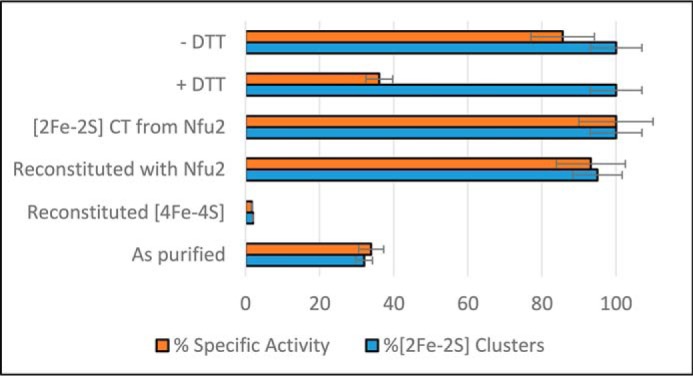
Specific activities and [Fe2S2]2+ cluster-content of different types of AtDHAD samples. Specific activity and [Fe2S2]2+ cluster-content are presented as a % of maximal values. The samples from bottom-to-top are as follows: as-purified DHAD containing substoichiometric [Fe2S2]2+ clusters; reconstituted DHAD containing near-stoichiometric [Fe4S4]2+ clusters and only trace amounts of [Fe2S2]2+ clusters; DHAD reconstituted in the presence of apo-NFU2 containing near-stoichiometric [Fe2S2]2+ clusters; DHAD reconstituted with stoichiometric [Fe2S2]2+ clusters by cluster transfer from [Fe2S2]2+ cluster–bound NFU2; DHAD with stoichiometric [Fe2S2]2+ clusters treated with 20-fold excess of DTT (+ DTT); and DHAD with stoichiometric [Fe2S2]2+ clusters treated with 20-fold excess of DTT followed by dialysis to remove DTT (−DTT).
Discussion
DHAD catalyzes a key step in the BCAA biosynthetic pathway in plant plastids, and in vivo studies in A. thaliana have shown that DHAD is essential and highly expressed in most vegetative and reproductive tissues (57). In addition, reduced expression of AtDHAD results in a short-root phenotype due to reduction of all three BCAAs in plant roots (57). This work describes the first in vitro characterization of recombinant AtDHAD expressed in E. coli and provides new insights into the nature, properties, biogenesis and catalytic role of the active-site Fe–S center. In particular, AtDHAD was shown to bind either one [Fe4S4] or one [Fe2S2] cluster, with only the latter being catalytically competent and capable of binding substrate and product. Moreover, in vitro cluster transfer studies with potential plastidial [Fe2S2] cluster donors and Fe–S cluster reconstitutions carried out in the presence of AtNFU2 implicate NFU2 as the physiological [Fe2S2] cluster donor for maturation of DHAD. These observations, coupled with the finding that the short-root phenotype associated with DHAD and NFU2 depletion (33, 57) can be rescued by the addition of BCAAs,5 provide compelling in vitro and in vivo evidence that NFU2 plays a role in DHAD maturation.
Coordination environment of Fe–S clusters in AtDHAD
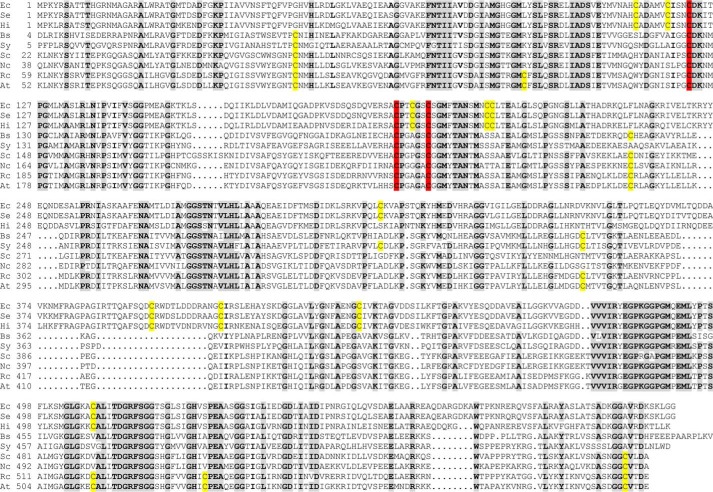
As a member of the Fe–S cluster–containing (de)hydratase family, the AtDHAD Fe–S cluster would be expected to be coordinated by three cysteine residues and have a water- or hydroxide-bound site for substrate binding and activation (7). However, as a rare example of a (de)hydratase that utilizes a [Fe2S2] cluster active site, plant DHADs may have unique cluster coordination compared with [Fe4S4] cluster–containing (de)hydratases. Indeed, EPR experiments on spinach DHAD in H217O buffer showed negligible broadening due to 17O (I = 5/2) binding, which argues against a water- or hydroxide-bound site (2). In addition, DHAD enzymes from different organisms assemble either [Fe4S4] or [Fe2S2] clusters, and the work presented herein shows for the first time that a DHAD can accommodate both types of cluster. Hence, it is of interest to compare sequence alignments of AtDHAD with other DHAD enzymes that contain [Fe4S4] or [Fe2S2] cluster active sites based on spectroscopic characterization and/or phylogenetic analysis. Sequence alignments of AtDHAD with eight other sequences from bacteria, fungi, and plants (top three with [Fe4S4] active sites and bottom five with [Fe2S2] active sites, based on phylogenetic analysis (28)) reveal three strictly conserved cysteine residues, Cys-173, Cys-239, and Cys-245 (numbered according to A. thaliana DHAD) (Fig. 9). Although there are nine additional conserved cysteines for the [Fe4S4] DHADs and one additional conserved cysteine for the [Fe2S2] DHADs, the three that are conserved for all DHADs are the best candidates for being cluster ligands. In addition, several serine or aspartate residues are fully conserved or conserved only in [Fe2S2] DHADs, making them candidates for oxygenic cluster ligands in the resting AtDHAD enzyme. The intriguing questions concerning the nature of the fourth ligand and how AtDHAD can accommodate either a [Fe2S2] or a [Fe4S4] cluster, with only the former being functional in catalysis, will need to be addressed by structural studies using X-ray crystallography. The ability of other [Fe2S2] cluster–containing DHADs to accommodate [Fe4S4] clusters, and vice versa, is another interesting question that needs to be addressed.
Figure 9.
Sequence alignment of DHADs from various organisms: Ec, E. coli; Se, S. enterica subsp. enterica sv. Javiana strain GA_MM04042433; Hi, H. influenzae Rd KW20; Bs, Bacillus subtilis sp. NRRL B-14911; Sy, Synechococcus sp. JA-2-3′a(2-13); Sc, S. cerevisiae; Nc, Neurospora crassa OR74A; Rc, Ricinus communis; and At, A. thaliana. Residues conserved for at least eight of nine sequences are highlighted in gray. Cysteine residues conserved in all sequences are highlighted in red, and those partially conserved are highlighted in yellow. Based on spectroscopic studies and/or phylogenetic analysis, the top three (E. coli, S. enterica, and H. influenzae) contain [Fe4S4] cluster active sites and the bottom six (B. subtilis, Synechococcus, S. cerevisiae, N. crassa, R. communis, and A. thaliana) contain [Fe2S2] cluster active sites.
The spectroscopic studies of the reduced and oxidized [Fe2S2]2+,1+ centers in AtDHAD indicate binding of substrate, product, and the competitive inhibitor DTT at a unique iron site of the [Fe2S2] cluster. Importantly, the RR studies rule out the possibility of partial histidyl cluster ligation for the oxidized [Fe2S2]2+ cluster and support ligation by three cysteinates and an oxygenic ligand in the as-purified, substrate-bound, and product-bound forms of the enzyme and three cysteinates and one DTT thiolate in the DTT-inhibited enzyme. The reduced enzyme has ∼15% of the oxidized enzyme activity (2) and exhibits distinct EPR spectra as purified and in the presence of substrate, product, and the competitive inhibitor DTT (Fig. 3). The g-value anisotropy and gav value of [Fe2S2]1+ centers depends on distortions or changes in the coordination at the Fe(II) site of a localized valence [Fe2S2]1+ cluster (48, 51, 52). Consequently, the observation of two species (I and II) with large g-value anisotropy and low gav values (1.92) in the reduced resting enzyme is attributed to heterogeneity at the Fe(II) site, possibly due monodentate/bidentate aspartate ligation or serine/serinate ligation. The product-bound form, species IV, is homogeneous with g-value anisotropy and a gav value (1.90) similar to the resting enzyme species I, implying monodentate or bidentate product ligation at the Fe(II) site of the reduced cluster. In contrast, the substrate-bound form, species III, has decreased g-value anisotropy and a gav value (1.95) similar to the DTT-bound form, species V (1.95). This implies that substrate binding as a monodentate or bidentate ligand induces a change in the site of reduction, so that the substrate is bound to the Fe(III) site and the Fe(II) site has two thiolate ligands. This is in accord with the observation of some activity for the reduced enzyme, because Fe(III) is a much stronger Lewis acid catalyst than Fe(II) for activating the 3-hydroxyl group on the substrate and facilitating the elimination reaction (Fig. 1). Clearly structural studies and/or EPR/ENDOR studies involving isotopically labeled substrate and product will be required for a complete understanding of the cluster environment, mode of substrate/product binding, and detailed catalytic mechanism of DHAD.
Maturation of apo DHAD
In vitro cluster transfer studies involving apo AtDHAD and four potential plastidial [Fe2S2] cluster donors, i.e. NFU2, GRXS14, GRXS16, and SUFA1, clearly implicate NFU2 as a viable physiological [Fe2S2] cluster donor. Although the rate constant for in vitro cluster transfer is relatively slow compared with the extremely fast [Fe4S4]2+ cluster transfer from AtNFU2 to AtAPR1 (150,000 m−1 min−1) (36), it is similar to that observed for [Fe2S2]2+ cluster transfer from AtNFU2 to AtGRXS16 (7000 m−1 min−1) (36), human NFU1 to human FDX1/2 (3600–5500 m−1 min−1) (58), and other intact in vitro cluster transfers that have been documented in the literature under comparable conditions (37, 59). Other plastidial NFUs may serve for maturation of DHAD in vivo. For instance, the study of nfu2 and nfu3 mutants indicated partial functional redundancy in terms of the roles of AtNFU2 and AtNFU3 in the maturation of [Fe4S4] cluster–containing subunits in photosystem I (34, 60) and complementation of NFU2 function by NFU3 in roots.5 Hence, it seems likely that both AtNFU2 and NFU3 are capable of binding and transferring both [Fe2S2] and [Fe4S4] clusters, and we cannot rule out the possibility that AtNFU2 and NFU3 have redundant functions for maturation of DHAD in plant shoots. However, recombinant AtNFU3 expressed in E. coli is purified as an apo protein under aerobic and anaerobic conditions. Also, in contrast to a recent report (60), anaerobic cluster reconstitution of AtNFU3 has failed to result in a stable [Fe4S4] or [Fe2S2] cluster–bound form in our laboratories. Although there is still much to be learned about the roles and specificity of the diverse types of NFU proteins in Fe–S cluster biogenesis, this work provides the first compelling evidence that NFU proteins are involved in the maturation of [Fe2S2] centers as well as [Fe4S4] centers (33, 34, 36). A role for NFU proteins in [Fe2S2] cluster trafficking may be confined to plastids and photosynthetic bacteria with high O2 levels, because bacterial and mitochondrial NFU proteins generally appear to be limited to [Fe4S4] cluster trafficking (61–66). The only exception is human mitochondrial NFU1, which has been shown to be capable of [Fe2S2] cluster trafficking in vitro (58), but there is as yet no in vivo evidence in support of this role.
Recent studies by Barondeau and co-workers (67, 68) have demonstrated the possibility of DTT-mediated, non-physiological Fe–S cluster transfer reactions. The [Fe2S2] cluster transfer from AtNFU2 to apo DHAD in the presence of DTT provides another example of a DTT-mediated cluster transfer and demonstrates that CD spectroscopy can identify such reactions. The CD time course of the cluster transfer is clearly a two-step process in the presence of DTT, with the first step involving DTT-induced removal of the [Fe2S2]2+ cluster from NFU2 in NFU2–DHAD transient complex, followed by insertion of the DTT-bound cluster into apo DHAD. Although pretreatment of the apo acceptor protein with DTT is commonly used to cleave protein disulfides, it is clearly necessary to remove DTT prior to conducting cluster transfer reactions under strictly anaerobic conditions.
Comments on the nature and the origin of the plant DHAD [Fe2S2] cluster active site
Among the many (de)hydratase enzymes reported, plant DHADs are a rare example of the [Fe2S2] cluster–containing member of this class of enzymes. The most obvious explanation for this is that the [Fe2S2] cluster–containing plant DHADs appear to be O2-tolerant and -resistant to ROS (2), whereas most of the regular [Fe4S4] cluster–containing (de)hydratases, including E. coli DHAD, are highly susceptible to oxygen and readily lose activity upon O2 exposure (7, 10). Because chloroplasts are O2-evolving organelles, stability in an aerobic environment is obviously an attractive proposal for the use of the less O2-sensitive [Fe2S2] cluster as the active site in plant DHADs. The only other dehydratase reported to bind a [Fe2S2] cluster is E. coli 2-methylcitrate dehydratase (PrpD), but this cluster was sensitive to oxygen and not as well-characterized (69). However, the reason why DHADs from different organisms utilize different types of clusters is still a mystery and is likely to be evolutionarily or metabolically relevant.
Some clues are provided by two recent phylogenetic studies on DHAD. By studying archaeal DHADs, Kim and Lee (5) suggested that bacterial DHADs and eukaryotic DHADs are genetically more closely related to Euyarchaeota and Crenarchaeota, respectively. So the differentiation point for different types of DHAD may have occurred early on the evolutionary time scale (5). Based on a more comprehensive phylogenetic tree involving 73 organisms, Lill and co-workers (28) classified DHADs from most of the eukaryotes and aerobic or chemoorganotrophic bacteria as [Fe2S2] cluster–containing, whereas the [Fe4S4] cluster–containing branch consisted primarily of facultative anaerobic bacteria, indicating that the types of clusters may be a result of aerobic growth. A sequence alignment of nine DHADs (five from bacteria, two from fungi, and two from higher plants) is shown in Fig. 9. Only 21% of residues are identical throughout all sequences. If only prokaryotes or eukaryotes are compared, the total number of identical residues increases to 29 and 46%, respectively. Among the 10 conserved cysteine residues for plants, three (Cys-173, Cys-239, and Cys-245 in Arabidopsis) are conserved for all nine sequences and potentially serve as cluster ligands. In addition, three more cysteine residues (Cys-100, Cys-282, and Cys-604 in Arabidopsis) are conserved for all the eukaryotic DHADs, and these may be involved in an oxygen-resistance mechanism. Another feature worth noting is that sequences of the three facultative anaerobic bacteria (E. coli, Salmonella enterica and Haemophilus influenzae) have an insertion of about 33 amino acid residues starting at around position 368 of the E. coli sequence. In fact, all the sequences of the [Fe4S4] cluster branch of the phylogenetic tree built by Lill and co-workers (28) have the same insertion, and the sequence identity for this group is close to 80%, which separate them from the other branch. Taken together, the appearance of [Fe2S2] cluster–containing DHADs may have resulted from an early evolutionary splitting as a strategy for coping with the gradual change from an anaerobic to an aerobic atmosphere on earth due to the production of O2 by cyanobacteria, the evolutionary precursors of chloroplasts.
Experimental procedures
All the chemicals and supplies used in this work, unless otherwise specified, were purchased from Sigma or Thermo Fisher Scientific. Anaerobic operations were performed inside a Vacuum Atmospheres glovebox under argon atmosphere with oxygen levels <2 ppm. Chromatography instruments and columns were purchased from GE Healthcare.
Plasmid construction
The AtDHAD (At3g23940) coding sequence was amplified by PCR from rosette cDNAs using the following primers:AtDHAD forward (5′CCCCCCATGGCTACTGACACCAATAAGCTC3′) and AtDHAD reverse (5′CCCCGGATCCTTACTCGTCAGTCACACA′). The PCR product was digested with NcoI and BamHI, cloned into the pET3d vector, and verified by DNA sequencing. The amplified sequence encodes a protein lacking the first 53 amino acids, corresponding to the putative plastid targeting sequence. Because of the use of the NcoI restriction, a codon for an alanine was added in the primer to keep the sequence in-frame.
Heterologous expression and purification of enzymes
Azotobacter vinelandii (Av)IscS (70), AtNFU2 (36), AtSUFA1 (39), and AtGRXS14/S16 (37) were heterologously expressed and purified as described previously. The plasmid containing the AtDHAD coding sequence was used for expression into the E. coli BL21(DE3) strain. An overnight LB-agar plate-grown colony was used to inoculate 1 liter of LB containing 100 μg/ml ampicillin and grown at 37 °C until reaching exponential phase. The culture was then treated with 0.5% (v/v) ethanol and kept at 4 °C for 2 h before induction by 200 μg/ml isopropyl 1-thio-β-d-galactopyranoside. After an overnight incubation at 20 °C, the cells were harvested by centrifuging at 6690 × g at 4 °C for 5 min and stored at −80 °C for later use.
In a routine purification of DHAD, 15 g of cell paste were resuspended in 40 ml of 100 mm Tris-HCl buffer, pH 8.0, containing 2 milliunits/ml DNase (Roche Applied Science), 0.5 μg/ml RNase (Roche Applied Science), 150 μg/ml phenylmethanesulfonyl fluoride, and 0.05% (v/v) polyethyleneimine. The cells were lysed by intermittent sonication using an ultrasound dismembrator model 500 (Thermo Fisher Scientific) and centrifuged at 39,800 × g at 4 °C for 1 h to remove insoluble components. The supernatant was subjected to 40% saturation of ammonium sulfate treatment, and the precipitate was removed by centrifugation at 39,800 × g for 20 min. The clear supernatant was then loaded onto a 70-ml phenyl-Sepharose column pre-equilibrated with 100 mm Tris-HCl buffer, pH 8.0, containing 1.0 m ammonium sulfate. Fractions containing DHAD eluted between 300 and 100 mm ammonium sulfate as determined by SDS-PAGE. The fractions were pooled and concentrated by Amicon ultrafiltration using a YM30 membrane (EMD Millipore) and loaded onto a 25-ml Q-Sepharose column. DHAD (>90% pure as judged by SDS-PAGE) eluted between 300 and 400 mm NaCl.
Preparation of apo DHAD and IscS-mediated in vitro reconstitution of DHAD
As-isolated DHAD in 100 mm Tris-HCl buffer, pH 8.0 (buffer A), was incubated with at least 50-fold excess of DTT, sodium dithionite, and EDTA for 3 h and desalted using a gel-filtration column. The resulting samples of apo AtDHAD contained <0.03 [Fe2S2] clusters per monomer, as judged by UV-visible absorption and CD spectroscopies. In vitro reconstitution of DHAD was initiated by addition of a catalytic amount of AvIscS to the reaction mixture containing 0.5–1.0 mm apo DHAD, 10-fold excess of ferrous ammonium sulfate, and 10-fold excess of l-cysteine under strictly anaerobic conditions in a glove box under argon. The reaction was monitored by absorption and CD spectroscopies for 3–5 h, and the cluster–bound protein was repurified using a 15-ml HiTrap Q HP column inside the glove box. A stoichiometric amount of apo AtNFU2 was added to the reaction mixture prior to the addition of IscS for anaerobic reconstitution of DHAD in the presence of NFU2.
Spectroscopic and analytical methods
All samples for spectroscopic studies were prepared under strictly anaerobic conditions. UV-visible absorption spectra were recorded using a Shimadzu UV-3101 PC scanning spectrophotometer. CD spectra were recorded using a JASCO J-715 spectropolarimeter. Septa-sealed quartz cuvette cells with either a 1-mm or 1-cm pathlength were used for both absorption and CD spectroscopies. Resonance Raman spectra were acquired using a Ramanor U1000 scanning spectrometer (Instruments SA, Edison, NJ) fitted with a cooled photomultiplier tube and photon-counting electronics (Instruments SA, Edison, NJ), using excitation lines from a Sabre argon laser (Coherent, Santa Clara, CA). A droplet of concentrated sample (∼2 mm in Fe–S clusters) was frozen at 17 K on a gold-plated sample holder mounted to a cold finger of a Displex Model CSA-202E closed cycle helium refrigerator (Air Products, Allentown, PA). Each RR spectrum is a sum of 80–100 scans with each scan involving photon counting for 1 s at 0.5-cm−1 increments with 7-cm−1 spectral resolution. X-band (∼9.6 GHz) EPR spectra were recorded using a Bruker ESP-300D spectrometer equipped with an ER-4116 dual mode cavity and an Oxford ESR 900 flow cryostat.
Protein concentrations were determined by DC protein assay (Bio-Rad) using bovine serum albumin as standard (71). Iron concentrations were determined colorimetrically with bathophenanthroline under reducing conditions using 1000 ppm atomic absorption iron as standards, after digesting proteins with KMnO4/HCl (72).
Cluster transfer from [Fe2S2] cluster–bound proteins to apo DHAD
The [Fe2S2] cluster–bound forms of AtNFU2, GRXS14, GRXS16, and SUFA1 were prepared by IscS-mediated reconstitution as described previously (36, 37, 39). Prior to the initiation of the cluster transfer reaction, apo DHAD was incubated with 1 mm DTT or 5 mm TCEP for at least 30 min. DTT or TCEP was then removed by dialysis prior to cluster transfer experiments, unless otherwise indicated. The cluster transfer experiments were carried out in buffer A and initiated by addition of stoichiometric or excess apo DHAD to cluster–bound forms of NFU2, GRXS14, GRXS16, or SUFA1, each of which had [Fe2S2]2+ cluster concentrations between 40 and 50 μm. The time course of cluster transfer was monitored by CD spectroscopy at room temperature for up to 120 min. Kinetic data for cluster transfer experiments were analyzed using the Chemical Kinetics Simulator software package (IBM).
Enzymatic activity of DHAD
DHAD was assayed spectrophotometrically by a modification of methods described by Kiritani and Wagner (6). The substrate (2,3-dihydroxyisovalerate) was synthesized using the procedure described by Nielsen et al. (73). The assay was conducted in buffer A containing 5 mm MgCl2. The formation of the 2,4-dinitrophenylhydrazone (from the keto product of the enzymatic reaction) was measured by absorption at 550 nm. The activity was recorded as specific activity (unit/mg of protein), where 1 unit is defined as amount of enzyme that converts 1 μmol of substrate to product per min at 37 °C.
Author contributions
H. G. and M. K. J. data curation; H. G., T. A., S. R., and M. K. J. formal analysis; H. G., N. R., and M. K. J. supervision; H. G., T. A., S. R., and J. C. investigation; H. G., J. C., and M. K. J. methodology; H. G. and M. K. J. writing-original draft; H. G., T. A., S. R., J. C., N. R., and M. K. J. writing-review and editing; N. R. and M. K. J. conceptualization; N. R. and M. K. J. resources; N. R. and M. K. J. funding acquisition; N. R. and M. K. J. project administration.
This work was supported by National Institutes of Health Grant R37GM62524 (to M. K. J.) and by a grant from the French National Research Agency as part of the “Investissements d'Avenir” Program (ANR-11-LABX-0002-01, Laboratory of Excellence (ARBRE)). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
B. Touraine, F. Vignols, J. Przybyla-Toscano, T. Dhalleine, H.-C. Wu, C. Magno, N. Berger, J. Couturier, S. Caffarri, M. Havaux, N. Rouhier, and F. Gaymard, unpublished results.
- BCAA
- branched-chain amino acids
- At
- Arabidopsis thaliana
- DHAD
- dihydroxyacid dehydratase
- ROS
- reactive oxygen species
- RR
- resonance Raman
- TCEP
- tris-(2-carboxyethyl) phosphine
- mW
- milliwatt.
References
- 1. Binder S. (2010) Branched-chain amino acid metabolism in Arabidopsis thaliana. Arabidopsis Book 8, e0137 10.1199/tab.0137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Flint D. H., and Emptage M. H. (1988) Dihydoxy acid dehydratase from spinach contains a [2Fe-2S] cluster. J. Biol. Chem. 263, 3558–3564 [PubMed] [Google Scholar]
- 3. Flint D. H., Emptage M. H., Finnegan M. G., Fu W., and Johnson M. K. (1993) The role and properties of the iron-sulfur cluster in Escherichia coli dihydroxy-acid dehydratase. J. Biol. Chem. 268, 14732–14742 [PubMed] [Google Scholar]
- 4. Flint D. H., Smyk-Randall E., Tuminello J. F., Draczynska-Lusiak B., and Brown O. R. (1993) The inactivation of dihydroxy-acid dehydratase in Escherichia coli treated with hyperbaric oxygen occurs because of the destruction of its Fe–S cluster, but the enzyme remains in the cell in a form that can be reactivated. J. Biol. Chem. 268, 25547–25552 [PubMed] [Google Scholar]
- 5. Kim S., and Lee S. B. (2006) Catalytic promiscuity in dihydroxy-acid dehydratase from the thermoacidophilic archaeon Sulfolobus solfataricus. J. Biochem. 139, 591–596 10.1093/jb/mvj057 [DOI] [PubMed] [Google Scholar]
- 6. Kiritani K., and Wagner R. P. (1970) α,β-Dihydroxy acid dehydratase (Neurospora crassa and spinach). Methods Enzymol. 17, 755–764 10.1016/0076-6879(71)17276-X [DOI] [Google Scholar]
- 7. Beinert H., Kennedy M. C., and Stout C. D. (1996) Aconitase as iron-sulfur protein, enzyme, and iron-regulatory protein. Chem. Rev. 96, 2335–2374 10.1021/cr950040z [DOI] [PubMed] [Google Scholar]
- 8. Kennedy M. C., Kent T. A., Emptage M., Merkle H., Beinert H., and Münck E. (1984) Evidence for the formation of a linear [3Fe-4S] cluster in partially unfolded aconitase. J. Biol. Chem. 259, 14463–14471 [PubMed] [Google Scholar]
- 9. Richards A. J., Thomson A. J., Holm R. H., and Hagen K. S. (1990) The magnetic circular dichroism spectra of the linear trinuclear clusters [Fe3S4(SR)4]3− in purple aconitase and in a synthetic model. Spectrochim. Acta 46A, 987–993 [Google Scholar]
- 10. Flint D. H., Tuminello J. F., and Emptage M. H. (1993) The inactivation of Fe–S cluster containing hydro-lyases by superoxide. J. Biol. Chem. 268, 22369–22376 [PubMed] [Google Scholar]
- 11. Hyduke D. R., Jarboe L. R., Tran L. M., Chou K. J., and Liao J. C. (2007) Integrated network analysis identifies nitric oxide response networks and dihydroxyacid dehydratase as a crucial target in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 104, 8484–8489 10.1073/pnas.0610888104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ren B., Zhang N., Yang J., and Ding H. (2008) Nitric oxide-induced bacteriostasis and modification of iron-sulphur proteins in Escherichia coli. Mol. Microbiol. 70, 953–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Duan X., Yang J., Ren B., Tan G., and Ding H. (2009) Reactivity of nitric oxide with the [4Fe-4S] cluster of dihydroxyacid dehydratase from Escherichia coli. Biochem. J. 417, 783–789 10.1042/BJ20081423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Arfin S. M. (1969) Evidence for an enol intermediate in the enzymatic conversion of α,β-dihydroxyisovalerate to α-ketoisovalerate. J. Biol. Chem. 244, 2250–2251 [PubMed] [Google Scholar]
- 15. Hill R. K., Yan S., and Arfin S. M. (1973) Enzymatic discrimination between diastereotopic enol faces in the dehydrase step of valine biosynthesis. J. Am. Chem. Soc. 95, 7857–7859 10.1021/ja00804a048 [DOI] [PubMed] [Google Scholar]
- 16. Pirrung M. C., Holmes C. P., Horowitz D. M., and Nunn D. S. (1991) Mechanism and stereochemistry of α,β-dihydroxyacid dehydratase. J. Am. Chem. Soc. 113, 1020–1025 10.1021/ja00003a042 [DOI] [Google Scholar]
- 17. Flint D. H., and Nudelman A. (1993) Studies on the active site of dihydroxy-acid dehydratase. Bioorg. Chem. 21, 367–385 10.1006/bioo.1993.1031 [DOI] [Google Scholar]
- 18. Flint D. H., and Allen R. M. (1996) Iron-sulfur proteins with non-redox functions. Chem. Rev. 96, 2315–2334 10.1021/cr950041r [DOI] [PubMed] [Google Scholar]
- 19. Johnson D. C., Dean D. R., Smith A. D., and Johnson M. K. (2005) Structure, function and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 74, 247–281 10.1146/annurev.biochem.74.082803.133518 [DOI] [PubMed] [Google Scholar]
- 20. Lill R. (2009) Function and biogenesis of iron-sulphur proteins. Nature 460, 831–838 10.1038/nature08301 [DOI] [PubMed] [Google Scholar]
- 21. Flint D. H., Tuminello J. F., and Miller T. J. (1996) Studies on the synthesis of the Fe–S cluster of dihydroxy-acid dehydratase in E. coli crude extract. J. Biol. Chem. 271, 16053–16067 10.1074/jbc.271.27.16053 [DOI] [PubMed] [Google Scholar]
- 22. Flint D. H. (1996) E. coli contains a protein homologous in function and N-terminal sequence to the protein encoded by the nifS gene in A. vinelandii and that can participate in the synthesis of the Fe–S cluster of dihydroxy-acid dehydratase. J. Biol. Chem. 271, 16068–16074 [PubMed] [Google Scholar]
- 23. Ding H., and Clark R. J. (2004) Characterization of iron-binding in IscA, an ancient iron-sulfur cluster assembly protein. Biochem. J. 379, 433–440 10.1042/bj20031702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Krebs C., Agar J. N., Smith A. D., Frazzon J., Dean D. R., Huynh B. H., and Johnson M. K. (2001) IscA, an alternative scaffold for Fe–S cluster biosynthesis. Biochemistry 40, 14069–14080 10.1021/bi015656z [DOI] [PubMed] [Google Scholar]
- 25. Mapolelo D. T., Zhang B., Naik S. G., Huynh B. H., and Johnson M. K. (2012) Spectroscopic and functional characterization of iron-bound forms of Azotobacter vinelandii NifIscA. Biochemistry 51, 8056–8070 10.1021/bi300664j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mapolelo D. T., Zhang B., Naik S. G., Huynh B. H., and Johnson M. K. (2012) Spectroscopic and functional characterization of iron-sulfur cluster–bound forms of Azotobacter vinelandii NifIscA. Biochemistry 51, 8071–8084 10.1021/bi3006658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tan G., Lu J., Bitoun J. P., Huang H., and Ding H. (2009) IscA/SufA paralogues are required for the [4Fe-4S] cluster assembly in enzymes of multiple physiological pathways in Escherichia coli under aerobic growth conditions. Biochem. J. 420, 463–472 10.1042/BJ20090206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mühlenhoff U., Richter N., Pines O., Pierik A. J., and Lill R. (2011) Specialized function of yeast Isa1 and Isa2 in the maturation of mitochondrial [4Fe-4S] proteins. J. Biol. Chem. 286, 41205–41216 10.1074/jbc.M111.296152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Singh B. K., and Shaner D. L. (1995) Biosynthesis of branched chain amino acids: from test tube to field. Plant Cell 7, 935–944 10.1105/tpc.7.7.935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Singh B. K. (1999) in Plant Amino Acids: Biochemistry and Biotechnology (Singh B. K., ed) pp. 227–247, Marcel Dekker, New York [Google Scholar]
- 31. Couturier J., Touraine B., Briat J.-F., Gaymard F., and Rouhier N. (2013) The iron-sulfur cluster assembly machineries in plants: Current knowledge and open questions. Front. Plant Sci. 4, 259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Balk J., and Schaedler T. A. (2014) Iron cofactor assembly in plants. Annu. Rev. Plant Biol. 65, 125–153 10.1146/annurev-arplant-050213-035759 [DOI] [PubMed] [Google Scholar]
- 33. Yabe T., Morimoto K., Kikuchi S., Nishio K., Terashima I., and Nakai M. (2004) The Arabidopsis chloroplast NifU-like protein CnfU, which can act as an iron-sulfur cluster scaffold protein, is required for the biogenesis of ferredoxin and photosystem I. Plant Cell 16, 993–1007 10.1105/tpc.020511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Touraine B., Boutin J. P., Marion-Poll A., Briat J. F., Peltier G., and Lobréaux S. (2004) Nfu2: a scaffold protein required for [4Fe-4S] and ferredoxin iron-sulfur cluster assembly in Arabidopsis chloroplasts. Plant J. 40, 101–111 10.1111/j.1365-313X.2004.02189.x [DOI] [PubMed] [Google Scholar]
- 35. Nishio K., and Nakai M. (2000) Transfer of iron-sulfur cluster from NifU to apoferredoxin. J. Biol. Chem. 275, 22615–22618 10.1074/jbc.C000279200 [DOI] [PubMed] [Google Scholar]
- 36. Gao H., Subramanian S., Couturier J., Naik S. G., Kim S.-H., Leustek T., Knaff D. B., Wu H.-C., Vignols F., Huynh B. H., Rouhier N., and Johnson M. K. (2013) Arabidopsis thaliana Nfu2 accommodates [2Fe-2S] or [4Fe-4S] clusters and is competent for in vitro maturation of chloroplast [2Fe-2S] and [4Fe-4S] cluster-containing proteins. Biochemistry 52, 6633–6645 10.1021/bi4007622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bandyopadhyay S., Gama F., Molina-Navarro M. M., Gualberto J. M., Claxton R., Naik S. G., Huynh B. H., Herrero E., Jacquot J.-P., Johnson M. K., and Rouhier N. (2008) Chloroplast monothiol glutaredoxins as scaffold proteins for the assembly and delivery of [2Fe-2S] clusters. EMBO J. 27, 1122–1133 10.1038/emboj.2008.50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Abdel-Ghany S. E., Ye H., Garifullina G. F., Zhang L., Pilon-Smits E. A., and Pilon M. (2005) Iron-sulfur cluster biogenesis in chloroplasts. Involvement of the scaffold protein CpIscA. Plant Physiol. 138, 161–172 10.1104/pp.104.058602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mapolelo D. T., Zhang B., Randeniya S., Albetel A.-N., Li H., Couturier J., Outten C. E., Rouhier N., and Johnson M. K. (2013) Monothiol glutaredoxins and A-type proteins: partners in Fe–S cluster trafficking. Dalton Trans. 42, 3107–3115 10.1039/c2dt32263c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Han S., Czernuszewicz R. S., and Spiro T. G. (1989) Vibrational spectra and normal mode analysis for [2Fe-2S] protein analogues using 34S, 54Fe, and 2H substitution: Coupling of Fe–S stretching and S-C-C bending modes. J. Am. Chem. Soc. 111, 3496–3504 10.1021/ja00192a002 [DOI] [Google Scholar]
- 41. Han S., Czernuszewicz R. S., Kimura T., Adams M. W., and Spiro T. G. (1989) Fe2S2 protein resonance Raman revisited: Structural variations among adrenodoxin, ferredoxin, and red paramagnetic protein. J. Am. Chem. Soc. 111, 3505–3511 10.1021/ja00192a003 [DOI] [Google Scholar]
- 42. Fu W., Drozdzewski P. M., Davies M. D., Sligar S. G., and Johnson M. K. (1992) Resonance Raman and magnetic circular dichroism studies of reduced [2Fe-2S] proteins. J. Biol. Chem. 267, 15502–15510 [PubMed] [Google Scholar]
- 43. Crouse B. R., Sellers V. M., Finnegan M. G., Dailey H. A., and Johnson M. K. (1996) Site-directed mutagenesis and spectroscopic characterization of human ferrochelatase: identification of residues coordinating the [2Fe-2S] cluster. Biochemistry 35, 16222–16229 10.1021/bi9620114 [DOI] [PubMed] [Google Scholar]
- 44. Sellers V. M., Wang K.-F., Johnson M. K., and Dailey H. A. (1998) Evidence that the fourth ligand to the [2Fe-2S] cluster in animal ferrochelatase is a cysteine. Characterization of the enzyme from Drosophila melanogaster. J. Biol. Chem. 273, 22311–22316 10.1074/jbc.273.35.22311 [DOI] [PubMed] [Google Scholar]
- 45. Fujinaga J., Gaillard J., and Meyer J. (1993) Mutated forms of a [2Fe-2S] ferredoxin with serine ligands to the iron-sulfur cluster. Biochem. Biophys. Res. Commun. 194, 104–111 10.1006/bbrc.1993.1791 [DOI] [PubMed] [Google Scholar]
- 46. Meyer J., Fujinaga J., Gaillard J., and Lutz M. (1994) Mutated forms of the [2Fe-2S] ferredoxin from Clostridium pasteurianum with noncysteinyl ligands to the iron-sulfur cluster. Biochemistry 33, 13642–13650 10.1021/bi00250a014 [DOI] [PubMed] [Google Scholar]
- 47. Cheng H., Xia B., Reed G. H., and Markley J. L. (1994) Optical, EPR, and 1H NMR spectroscopy of serine-ligated [2Fe-2S] ferredoxins produced by site-directed mutagenesis of cysteine residues in recombinant Anabaena 7120 vegetative ferredoxin. Biochemistry 33, 3155–3164 10.1021/bi00177a003 [DOI] [PubMed] [Google Scholar]
- 48. Orio M., and Mouesca J.-M. (2008) Variation of average g values and effective exchange coupling constants among [2Fe-2S] clusters: a density functional theory study of the impact of localization (trapping forces) versus delocalization (double-exchange) as competing factors. Inorg. Chem. 47, 5394–5416 10.1021/ic701730h [DOI] [PubMed] [Google Scholar]
- 49. Subramanian S., Duin E. C., Fawcett S. E., Armstrong F. A., Meyer J., and Johnson M. K. (2015) Spectroscopic and redox studies of valence-delocalized [2Fe-2S]+ centers in thioredoxin-like ferredoxins. J. Am. Chem. Soc. 137, 4567–4580 10.1021/jacs.5b01869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rieske J. S., MacLennan D. H., and Coleman R. (1964) Isolation and properties of an iron-protein from the (reduced coenzyme Q)-cytochrome c reductase complex of the respiratory chain. Biochem. Biophys. Res. Commun. 15, 338–344 10.1016/0006-291X(64)90171-8 [DOI] [Google Scholar]
- 51. Bertrand P., and Gayda J.-P. (1979) A theoretical interpretation of the variations of some physical parameters within the [2Fe-2S] ferredoxin group. Biochim. Biophys. Acta 579, 107–121 10.1016/0005-2795(79)90091-6 [DOI] [PubMed] [Google Scholar]
- 52. Bertrand P., Guigliarelli B., Gayda J.-P., Beardwood P., and Gibson J. F. (1985) A ligand-field model to describe a new class of 2Fe-2S clusters in proteins and their synthetic analogs. Biochim. Biophys. Acta 831, 261–266 10.1016/0167-4838(85)90044-5 [DOI] [Google Scholar]
- 53. Paddock M. L., Wiley S. E., Axelrod H. L., Cohen A. E., Roy M., Abresch E. C., Capraro D., Murphy A. N., Nechushtai R., Dixon J. E., and Jennings P. A. (2007) MitoNEET is a uniquely folded 2Fe2S outer mitochondrial membrane protein stabilized by pioglitazone. Proc. Natl. Acad. Sci. U.S.A. 104, 14342–14347 10.1073/pnas.0707189104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Li H., Mapolelo D. T., Dingra N. N., Naik S. G., Lees N. S., Hoffman B. M., Riggs-Gelasco P. J., Huynh B. H., Johnson M. K., and Outten C. E. (2009) The yeast iron regulatory proteins Grx3/4 and Fra2 form heterodimeric complexes containing a [2Fe-2S] cluster with cysteinyl and histidyl ligation. Biochemistry 48, 9569–9581 10.1021/bi901182w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lindahl P. A., Day E. P., Kent T. A., Orme-Johnson W. H., and Münck E. (1985) Mössbauer, EPR, and magnetization studies of Azotobacter vinelandii Fe protein: evidence for a [4Fe-4S]1+ cluster with spin S = 3/2. J. Biol. Chem. 260, 11160–11173 [PubMed] [Google Scholar]
- 56. Conover R. C., Kowal A. T., Fu W. G., Park J.-B., Aono S., Adams M. W., and Johnson M. K. (1990) Spectroscopic characterization of the novel iron-sulfur cluster in Pyrococcus furiosus ferredoxin. J. Biol. Chem. 265, 8533–8541 [PubMed] [Google Scholar]
- 57. Zhang C., Pang Q., Jiang L., Wang S., Yan X., Chen S., and He Y. (2015) Dihydroxyacid dehydratase is important for gametophyte development and disruption causes increased susceptibility to salinity stress in Arabidopsis. J. Exp. Bot. 66, 879–888 10.1093/jxb/eru449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wachnowsky C., Fidai I., and Cowan J. A. (2016) Iron–sulfur cluster exchange reactions mediated by the human Nfu protein. J. Biol. Inorg. Chem. 21, 825–836 10.1007/s00775-016-1381-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chandramouli K., and Johnson M. K. (2006) HscA and HscB stimulate [2Fe-2S] cluster transfer from IscU to apoferredoxin in an ATP-dependent reaction. Biochemistry 45, 11087–11095 10.1021/bi061237w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nath K., Wessendorf R. L., and Lu Y. (2016) A nitrogen-fixing subunit essential for accumulating 4Fe-4S-containing photosystem I core proteins. Plant Physiol. 172, 2459–2470 10.1104/pp.16.01564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tong W.-H., Jameson G. N., Huynh B. H., and Rouault T. A. (2003) Subcellular compartmentalization of human Nfu, an iron-sulfur cluster scaffold protein and its ability to assemble a [4Fe-4S] cluster. Proc. Natl. Acad. Sci. U.S.A. 100, 9762–9767 10.1073/pnas.1732541100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Smith A. D., Jameson G. N., Dos Santos P. C., Agar J. N., Naik S., Krebs C., Frazzon J., Dean D. R., Huynh B. H., and Johnson M. K. (2005) NifS-mediated assembly of [4Fe-4S] clusters in the N- and C-terminal domains of the NifU scaffold protein. Biochemistry 44, 12955–12969 10.1021/bi051257i [DOI] [PubMed] [Google Scholar]
- 63. Bandyopadhyay S., Naik S. G., O'Carroll I. P., Huynh B. H., Dean D. R., Johnson M. K., and Dos Santos P. C. (2008) A proposed role for the Azotobacter vinelandii NfuA protein as an intermediate iron-sulfur cluster carrier. J. Biol. Chem. 283, 14092–14099 10.1074/jbc.M709161200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Angelini S., Gerez C., Ollagnier-de Choudens S., Sanakis Y., Fontecave M., Barras F., and Py B. (2008) NfuA, a new factor required for maturing Fe/S proteins in Escherichia coli under oxidative stress and iron starvation conditions. J. Biol. Chem. 283, 14084–14091 10.1074/jbc.M709405200 [DOI] [PubMed] [Google Scholar]
- 65. Py B., Gerez C., Angelini S., Planel R., Vinella D., Loiseau L., Talla E., Brochier-Armanet C., Garcia Serres R., Latour J.-M., Ollagnier-de Choudens S., Fontecave M., and Barras F. (2012) Molecular organization, biochemical function, cellular role and evolution of NfuA, an atypical Fe–S carrier. Mol. Microbiol. 86, 155–171 10.1111/j.1365-2958.2012.08181.x [DOI] [PubMed] [Google Scholar]
- 66. McCarthy E. L., and Booker S. J. (2017) Destruction and reformation of an iron-sulfur cluster during catalysis by lipoyl synthase. Science 358, 373–377 10.1126/science.aan4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Vranish J. N., Russell W. K., Yu L. E., Cox R. M., Russell D. H., and Barondeau D. P. (2015) Fluorescent probes for tracking the transfer of iron–sulfur cluster and other metal cofactors in biosynthetic reaction pathways. J. Am. Chem. Soc. 137, 390–398 10.1021/ja510998s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Fox N. G., Chakrabarti M., McCormick S. P., Lindahl P. A., and Barondeau D. P. (2015) The human iron–sulfur assembly complex catalyzes the synthesis of [2Fe-2S] clusters on ISCU2 that can be transferred to acceptor molecules. Biochemistry 54, 3871–3879 10.1021/bi5014485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Blank L., Green J., and Guest J. R. (2002) AcnC of Escherichia coli is a 2-methyl citrate dehydratase (PrpD) that can use citrate and isocitrate as substrates. Microbiology 148, 133–146 10.1099/00221287-148-1-133 [DOI] [PubMed] [Google Scholar]
- 70. Zheng L., Cash V. L., Flint D. H., and Dean D. R. (1998) Assembly of iron-sulfur clusters. Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J. Biol. Chem. 273, 13264–13272 10.1074/jbc.273.21.13264 [DOI] [PubMed] [Google Scholar]
- 71. Brown R. E., Jarvis K. L., and Hyland K. J. (1989) Protein measurement using bicinchoninic acid: elimination of interfering substances. Anal. Biochem. 180, 136–139 10.1016/0003-2697(89)90101-2 [DOI] [PubMed] [Google Scholar]
- 72. Fish W. W. (1988) Rapid colorimetric micromethod for the quantitation of complexed iron in biological samples. Methods Enzymol. 158, 357–364 10.1016/0076-6879(88)58067-9 [DOI] [PubMed] [Google Scholar]
- 73. Nielsen B. E., Larsen P. K., and Lemmich J. (1969) The stereochemistry of 1,2,3-trihydroxy-3-methylbutane and 2,3-dihydroxy-3-methylbutyric acid. Acta Chem. Scand. 23, 967–970 10.3891/acta.chem.scand.23-0967 [DOI] [PubMed] [Google Scholar]