Abstract
Epithelial cell polarity is controlled by components of the Scribble polarity module, and its regulation is critical for tissue architecture and cell proliferation and migration. In Drosophila melanogaster, the adaptor protein Guk-holder (Gukh) binds to the Scribbled (Scrib) and Discs Large (Dlg) components of the Scribble polarity module and plays an important role in the formation of neuromuscular junctions. However, Gukh's role in epithelial tissue formation and the molecular basis for the Scrib–Gukh interaction remain to be defined. We now show using isothermal titration calorimetry that the Scrib PDZ1 domain is the major site for an interaction with Gukh. Furthermore, we defined the structural basis of this interaction by determining the crystal structure of the Scrib PDZ1–Gukh complex. The C-terminal PDZ-binding motif of Gukh is located in the canonical ligand-binding groove of Scrib PDZ1 and utilizes an unusually extensive network of hydrogen bonds and ionic interactions to enable binding to PDZ1 with high affinity. We next examined the role of Gukh along with those of Scrib and Dlg in Drosophila epithelial tissues and found that Gukh is expressed in larval-wing and eye-epithelial tissues and co-localizes with Scrib and Dlg at the apical cell cortex. Importantly, we show that Gukh functions with Scrib and Dlg in the development of Drosophila epithelial tissues, with depletion of Gukh enhancing the eye- and wing-tissue defects caused by Scrib or Dlg depletion. Overall, our findings reveal that Scrib's PDZ1 domain functions in the interaction with Gukh and that the Scrib–Gukh interaction has a key role in epithelial tissue development in Drosophila.
Keywords: cell polarity, Drosophila, drosophila genetics, isothermal titration calorimetry (ITC), X-ray crystallography, gukholder, scribble
Introduction
Cell polarity is a key property of tissue development and manifests itself as the asymmetric organization of cellular components, such as proteins and lipids into distinct cellular domains. Correct establishment of cell polarity is important for tissue architecture, cell proliferation, cell migration, and cellular fate, and its dysregulation has been recognized as a cancer hallmark, with ∼70% of epithelial cancers displaying defects in polarity regulation (1–4). Cell polarity is regulated by the interplay between three key polarity modules, Scribble, PARtitioning-defective (PAR),5 and Crumbs (5). Epithelial cell polarity (apico-basal polarity) is established and maintained by the antagonistic interactions between the Scribble module and the PAR and Crumbs complexes, resulting in the restriction of PAR and Crumbs complex components to the apical cortex and the Scribble module components to the basolateral cortex. In apico-basal cell polarity, the three polarity modules function to specify the apical and basal membrane domains and to position the adherens and tight junctions, which are required for cell–cell contact, cell communication, epithelial tissue coherence, and tissue growth regulation (6).
The Scribble module comprises three tumor suppressor proteins, Scribbled (Scrib), Disc large (Dlg), and Lethal-2-giant larvae (Lgl), which are highly conserved in structure and function from the vinegar fly, Drosophila melanogaster, to humans (7). Genetic analyses in Drosophila have provided valuable insight into their in vivo function (8, 9) and revealed that Scrib, Dlg, and Lgl, in addition to their role in cell polarity, are also involved in cell proliferation, differentiation, and migration (8–12). In addition to the core Scribble module components, studies in Drosophila neuromuscular junctions revealed that the interaction between Scrib and Dlg is mediated by an adaptor protein, termed GUK-holder (Gukh) (13). However, at a molecular level the precise interactions between these three tumor suppressors, and indeed the role of Gukh, are not well-defined.
Scrib is a scaffold protein belonging to the LAP (LRR and PDZ) protein family and contains 16 Leucine-rich repeats (LRRs), two LAP-specific domains (LAPSADa and LAPSADb), and four PDZ domains. Whereas the N-terminal LRR domain is critical for Scrib's cortical localization (14), the four PDZ domains are required for cell–cell junction localization (13) and are the major mediators of Scrib interactions with other proteins (15). hScrib (human Scrib) PDZ domains have been shown to bind a diverse set of cellular interactors, including β-PIX, MCC, GIT1, and β-catenin (16–20), enabling Scrib to integrate a range of cellular cues for the establishment of apico-basal polarity, cell migration, and cell signaling.
Dlg is a member of the membrane-associated guanylate kinase homolog (MAGUK) scaffolding protein family and comprises three PDZ domains, an Src homology 3 (SH3) domain, a Hook domain, and a guanylate kinase-like (GUK) domain at its C terminus. Four Dlg homologs have been found in mammalian systems, named: Dlg1 (hDlg/SAP97), Dlg2 (PSD-93/Chapsyn-110), Dlg3 (NE-Dlg/SAP102), and Dlg4 (PSD-95/SAP90) (3). In D. melanogaster, the Dlg PDZ2 domain is required for its localization at basolateral (septate) junctions, whereas the SH3 and Hook domains are necessary for precise localization of Dlg to the cell membrane (21, 22). The GUK domain (765–960 amino acids) is catalytically inactive and in Drosophila regulates Dlg–Scrib interactions via interaction with Gukh (1, 13).
Gukh was identified in a yeast two-hybrid screen as a protein that bound to the Drosophila Dlg GUK domain (13). Two orthologs of Drosophila Gukh have been identified in humans, Nance-Horan syndrome (NHS) and NHSL1 (23). NHS is an uncommon X-linked disorder characterized by congenital nuclear cataracts, dental irregularities, and craniofacial dysmorphisms, with mental deficiencies also occurring in ∼30% of the cases (24). The molecular details of how NHS mutations cause the NHS are unclear, although recent studies have implicated it in regulation of epithelial junctions (25) as well as actin remodeling (23, 26).
Drosophila Gukh contains a GUK-holding domain at its C terminus, which directly binds the Dlg GUK domain (13, 27). Additionally, another study showed that the binding of Gukh to the GUK domain of Dlg occurs in a mutually exclusive manner via the PDZ domain, only permitting Gukh interaction when the PDZ remains unbound (28). However, others have reported that interactions between Gukh and Dlg require the SH3–GUK domain of Dlg (29, 30). This interaction is regulated via interdomain interactions of PDZ3–SH3–GUK via a PDZ3-binding motif in a linker region enabling dynamic regulation of ligand binding to Dlg PDZ3 (29, 30).
Furthermore, in yeast two-hybrid assays the Gukh C-terminal region is able to engage Scrib PDZ2 but not PDZ3-4 (13). Moreover, co-immunoprecipitation analysis from Drosophila larval muscles showed that Scrib can form a complex with Dlg and Gukh, and the interaction with Dlg is reduced in a Gukh mutant (13). These genetic data suggest that Gukh is important for the formation of a ternary complex between Scrib, Dlg, and Gukh. Consistent with this notion, all three proteins co-localize at Drosophila neuromuscular junctions, and Dlg and Gukh are required for the correct localization of Scrib (13). This interaction is likely to be evolutionarily conserved, as a Zebrafish Gukh ortholog, Nhs1b, also physically interacts with Scrib and Dlg in cultured cells, and nhs1b and scrib genetically interact as shown by the observation that heterozygotes show a strong neural cell migration defect relative to single heterozygotes (31).
Although Drosophila Gukh interacts with Dlg and Scrib in neuromuscular junctions (13), its role in epithelial tissue formation and the molecular basis for the Scrib–Gukh interaction remain to be defined. Here, we identify the Scrib PDZ1 domain as the major interacting PDZ domain with the Gukh C-terminal peptide. Using X-ray crystallography, we then define the structural basis of Scrib PDZ1 interactions with the Gukh C-terminal peptide. Furthermore, our studies reveal a novel role for Gukh in epithelial development. We show that Gukh is expressed in larval-wing and eye-epithelial tissue and co-localizes with Scrib and Dlg at the apical cell cortex. Importantly, we show that Gukh functions with Scrib and Dlg in Drosophila epithelial tissues, with depletion of Gukh enhancing the eye- and wing-tissue defects caused by Scrib or Dlg depletion. These findings provide the first evidence for a role for Gukh in the Scribble module in the control of epithelial cell polarity and provide structural and mechanistic insights into the Scrib–Gukh interaction in Drosophila.
Results
Molecular and structural basis of Scrib–Gukh interaction
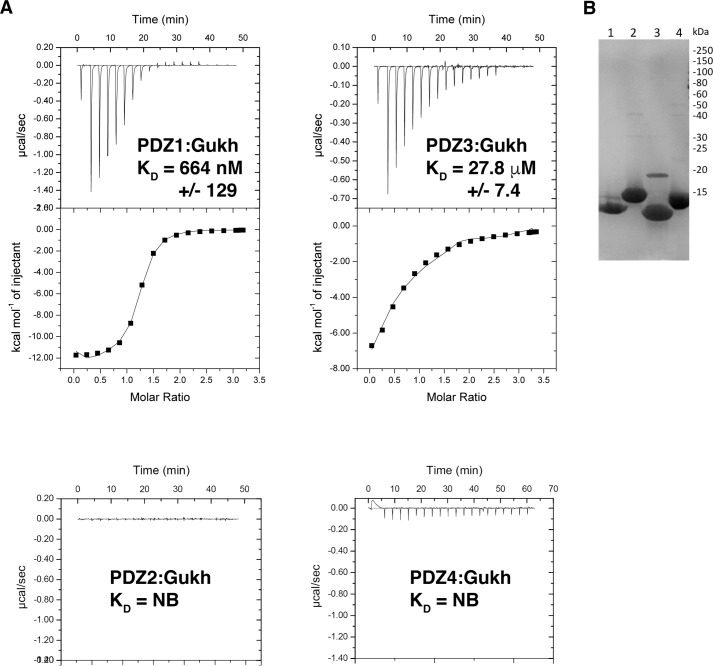
To understand the molecular basis of the reported interaction between Scrib and Gukh, we performed protein–ligand interaction studies using individual recombinant PDZ domains from Scrib and C-terminal peptides derived from Gukh, and we determined binding affinities using isothermal titration calorimetry (ITC) (Fig. 1 and Table 1). ITC experiments were conducted using purified Drosophila Scrib PDZ1, PDZ2, PDZ3, and PDZ4 domains with wildtype Gukh C-terminal peptides. Raw heats of titrations obtained for PDZ1 with GukhWT peptide revealed a tight interaction with a calculated KD of 664 nm, whereas PDZ3 engaged GukhWT peptide with only modest affinity with a KD of 27.8 μm. In contrast, PDZ2 and PDZ4 did not show any detectable binding to GukhWT peptide, thus rendering the Scrib PDZ1 domain as the primary functional interaction site for the Gukh C terminus.
Figure 1.
Interactions between Scrib PDZ domains and the C terminus of Gukh. A, raw heats of titration were measured using isothermal titration calorimetry for interactions between Scrib PDZ1, PDZ2, PDZ3, and PDZ4 domains with an 8-mer peptide encoding the C-terminal PDZ-binding motif of Gukh (LPSFETAL, Gukh residues 1781–1788). All experiments were performed in triplicate, error is S.D. NB denotes no binding. B, SDS-PAGE analysis of wildtype Scrib PDZ domains 1–4 (lanes 1–4). All experiments were performed in triplicate, error is S.D. NB, denotes no binding.
Table 1.
Summary of thermodynamic binding parameters for Scrib PDZ domain interactions with Gukh peptide measured at pH 7.5 and 25 °C
| KD | −ΔH | TΔS | N | |
|---|---|---|---|---|
| nm | kcal/mol | cal/mol/K | ||
| PDZ1 | 664 ± 129 | 12.5 ± 0.8 | −13.8 ± 2.5 | 0.91 ± 0.18 |
| PDZ2 | NBa | NB | NB | NB |
| PDZ3 | 27800 ± 7400 | 2.8.3 ± 1.0 | −12.7 ± 3.4 | 0.88 ± 0.05 |
| PDZ4 | NB | NB | NB | NB |
a NB denotes no binding.
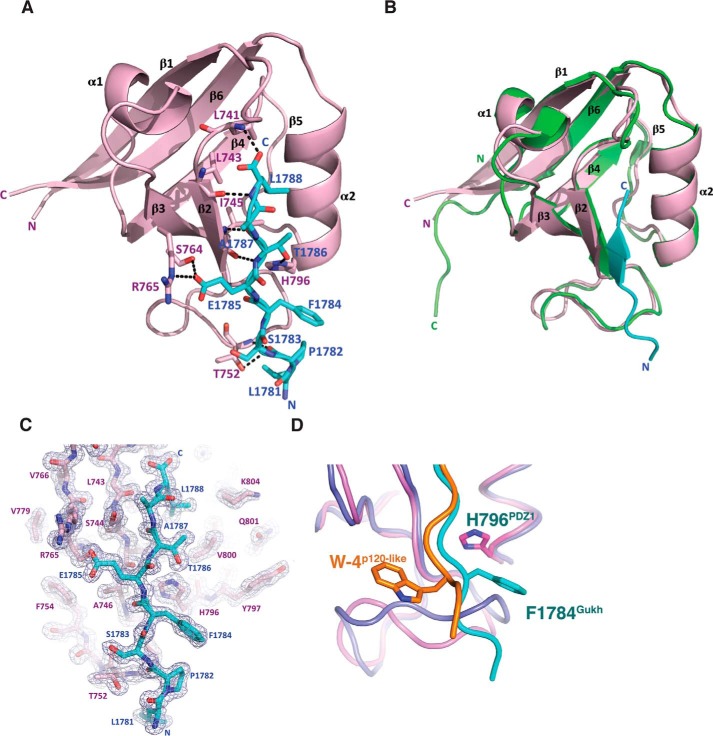
We then examined the structural basis of the PDZ1–Gukh interaction by determining the crystal structure of a Scrib PDZ1–Gukh C-terminal peptide complex (Table 2). Drosophila Scrib PDZ1 adopts the typical PDZ fold consisting of six β-strands and two α-helices that form a β-sandwich structure (Fig. 2A) and is highly similar to the previously determined human PDZ1 (hsPDZ1) structure (PDB code 2W4F). The PDZ1 domain from the PDZ1–Gukh complex superimposes with the apo hsPDZ1 domain with an r.m.s.d. of 0.8995 Å over 92 Cα atoms, indicating that the binding of the Gukh peptide to the PDZ1 domain does not substantially change the overall fold (Fig. 2B).
Table 2.
Crystallographic data collection and refinement statistics
| PDZ1/Gukh peptide | |
|---|---|
| Data collection | |
| Space group | C2221 |
| No. of molecules in asymmetric unit | 1 + 1 |
| Cell dimensions | |
| a, b, c (Å) | 36.43, 53.88, 94.22 |
| α, β, γ (°) | 90.00, 90.00, 90.00 |
| Wavelength (Å) | 0.9537 |
| Resolution (Å)a | 31.42–1.55 (1.58–1.55) |
| Rsym or Rmergea | 0.094 (0.580) |
| I/σIa | 18.4 (4.3) |
| CC(1/2) | 0.99 (0.92) |
| Completeness (%)a | 98.2 (94.6) |
| Redundancya | 13.8 (13.4) |
| Wilson B-factor | 11.8 |
| No. of unique reflections | 13,578 (617) |
| No. of observed reflections | 187,879 (8282) |
| Refinement | |
| Resolution (Å) | 31.42–1.55 |
| Rwork/Rfree | 0.197/0.224 |
| No. non-hydrogen atoms | |
| Protein | 766 |
| Ligand/ion | 38 |
| Water | 135 |
| B-factors | |
| Protein | 13.42 |
| Ligand/ion | 18.45 |
| Water | 23.39 |
| r.m.s.d. | |
| Bond lengths (Å) | 0.004 |
| Bond angle (°) | 0.68 |
| Ramachandran plot (%) | |
| Favored | 97.98 |
| Allowed | 2.02 |
| Disallowed | 0 |
| MolProbity clash score | 1.2 (99th percentile) |
| Poor rotamers | 1 |
| MolProbity score | 0.9 (100th percentile) |
a Values in parentheses are for the highest resolution shell.
Figure 2.
Crystal structures of D. melanogaster Scrib PDZ1 bound to a Gukh peptide. The Gukh peptide engages the PDZ1 domain via a shallow groove located between the β2 and α2. A, PDZ1 (light pink) is shown as a cartoon with β-PIX peptide (cyan) represented as sticks. Side chains of the residues involved in interactions (shown as dashed black lines) are displayed as sticks and are labeled. B, overlay of cartoons of Drosophila PDZ1 (light pink, residues 726–819, 1.65 Å resolution) bound to Gukh peptide (cyan) and human PDZ1 (green, residues 724–819, 1.91 Å resolution). C, simulated anneal composite omit electron density map encompassing the binding groove of Scrib PDZ1 in complex with Gukh. PDZ1 is shown as light pink sticks, and Gukh is shown as cyan sticks. The electron density map is shown as a blue mesh contoured at 1.0 σ and was calculated by omitting the entire Gukh peptide. D, cartoon representation of Scrib PDZ1–Gukh complex (light pink and cyan) with the Erbin PDZ domain bound to a synthetic p120-like peptide (dark blue and orange, PDB code 1N7T). Residue Trp-4 from p120-like peptide engages a pocket on the β2–3 loop and is critical for the high-affinity interaction with Erbin. Gukh does not utilize a Trp residue to achieve a high-affinity interaction, and instead it utilizes Phe-1784 for hydrophobic interactions with Scrib PDZ1 His-796 on the opposite side of the ligand-binding groove.
In the PDZ1–Gukh structure, PDZ1 features an atypical β5 displaying increased flexibility in its geometry. PDZ1 interacts with Gukh peptide via its canonical ligand-binding groove formed by the β2 strand and helix α2 (Fig. 2, A and C). Gukh binding to PDZ1 buries a combined total of 937 Å2 of solvent-accessible surface area, with the interface having a shape complementarity score of 0.78, indicating a very good fit. In the complex, the Gukh peptide faces the β2 strand in an anti-parallel manner, with its N-terminal solvent exposed and its C terminus stabilized by the PDZ1 β1–2 loop. The complex is achieved via an extensive network of hydrogen bonds formed by Leu-741PDZ1–Leu-1788Gukh, Leu-743PDZ1–Leu-1788Gukh, Ile-745PDZ1–Thr-1786Gukh, Thr-752PDZ1–Pro-1782Gukh, Ser-764PDZ1–Glu-1785Gukh, His-796PDZ1–Thr-1786Gukh, as well as an ionic interaction between Arg-765PDZ1–Glu-1785Gukh. Furthermore, Gukh Leu-1788 is accommodated in a hydrophobic pocket comprising Leu-741, Leu-743, Val-800, and Leu-803. Hydrophobic interactions are also present between Gukh Phe-1784 and dmPDZ1 Gly-747 and Ser-751, with Phe-1784Gukh being further stabilized by π-stacking with His-796PDZ1.
To validate the crystal structure, we performed site-directed mutagenesis to target two key interactions, Arg-765PDZ1–Glu-1785Gukh and His-796PDZ1–Phe-1784Gukh, as well as the floor of the canonical PBM-binding site via G747W (Table 3). Mutation of Arg-765 to an Ala resulted in a 5-fold reduction in affinity (KD = 5.1 μm), whereas the H796A displayed no affinity for WT Gukh, suggesting that both residues make important contributions for Scrib PDZ1–Gukh interactions. Furthermore, introduction of the large hydrophobic Trp at Gly-747PDZ1 also completely ablates the ability of PDZ1 to bind to the PBM of Gukh (Table 3).
Table 3.
Summary of binding parameters for wildtype and mutants of Scrib PDZ1 domain interactions with Gukh peptide measured at pH 7.5 and 25 °C
| Scribble | KD | N |
|---|---|---|
| nm | ||
| WT PDZ1 | 664 ± 129 | 0.91 ± 0.18 |
| PDZ1 R765A | 5100 ± 2220 | 0.89 ± 0.04 |
| PDZ1 H796A | NBa | NB |
| PDZ1 G747W | NB | NB |
a NB denotes no binding.
Gukh is expressed in wing and eye-epithelial tissue and co-localizes with Dlg and Scrib in the apical cortex
Considering the previously established role for the Scrib–Gukh interaction in neuromuscular junction development, we next examined whether Scrib and Gukh are involved in epithelial tissue development. To determine whether Gukh was expressed in Drosophila larval epithelial tissues, we stained developing eye and wing epithelia from third instar larvae with an anti-Gukh antibody (13). Staining with the anti-Gukh antibody, and using expression of a gukh RNAi line in the wing epithelium or clones of a gukh loss-of-function P transposable-element mutant in the eye epithelium as controls, revealed that Gukh was expressed in the developing wing (Fig. 3, A, quantified in C) and eye (Fig. 3, B, quantified in D) tissues. We then examined the cellular distribution of Gukh relative to Scrib and Dlg (Fig. 4). In the larval-wing epithelium, Gukh was localized cortically and enriched apically, co-localizing with Dlg and Scrib in the apical cortex; however, cross-sections revealed that Gukh extended further apically and basolaterally than did Dlg and Scrib (Fig. 4A). A similar co-localization between Gukh and Scrib or Dlg was observed in the larval eye epithelia, although Gukh exhibited a stronger staining of the apical cortex of photoreceptor cells, which are enriched in F-actin (Fig. 4B). Thus, Gukh shows overlapping localization with Scrib and Dlg in two larval epithelial tissues, although Gukh is also distributed more generally around the cell cortex, which is consistent with Gukh possessing a WH1 F-actin–binding domain at its N terminus (13, 26).
Figure 3.
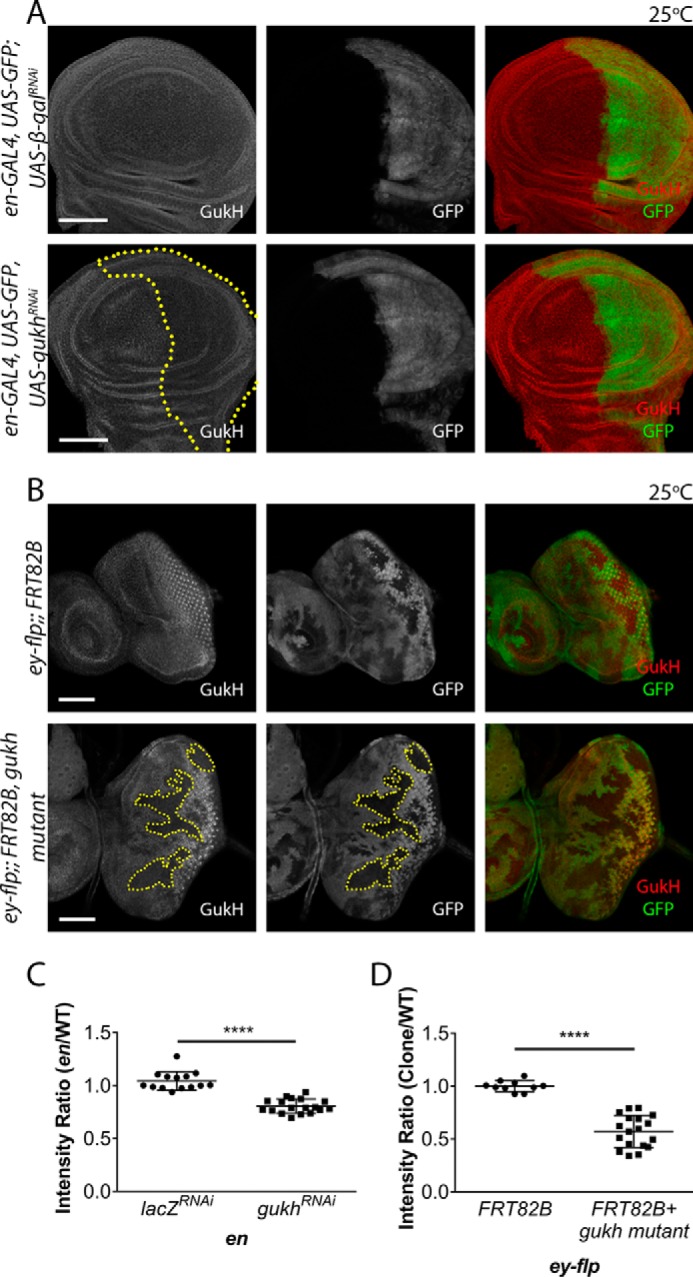
Gukh is expressed in epithelial tissues. A, confocal images of immunofluorescently stained third instar larval imaginal wing discs with an anti-Gukh antibody (red). The engrailed (en)-GAL4 driver was used to drive expression of either a UAS-β-galRNAi control or UAS-gukhRNAi in the En domain indicated by the GFP marked posterior compartment of the wing disc (green). In en-driven gukhRNAi discs down-regulation of Gukh staining was observed compared with the uniform staining seen in the control disc. B, generation of ey-FLP clones in otherwise wildtype imaginal eye discs by crossing gukhBG02660 FRT82B flies to ey-FLP FRT82B Ubi-GFP at 25 °C. The absence of GFP represents patches of mutant tissue and reveals down-regulation of Gukh staining. C, quantification of the intensity ratio in the wing samples (from A), which is measured by the intensity of Gukh staining in GFP domains compared with the non-GFP domain. There is a significant decrease (by ∼25%) in the intensity ratio when gukhRNAi is expressed (n = 14–18 wing discs). D, quantification of the intensity ratio in the eye samples (from B), showing that the intensity ratio of Gukh staining in non-GFP mutant tissue compared with GFP-marked tissue was significantly decreased by ∼50% (n = 10–18 eye discs). Images were taken at ×40 (A) and ×20 (B) magnification. Scale bars, 50 μm. Error bars represent mean ± S.D. Student's t test used to test for significance. ****, p ≤ 0.0001.
Figure 4.
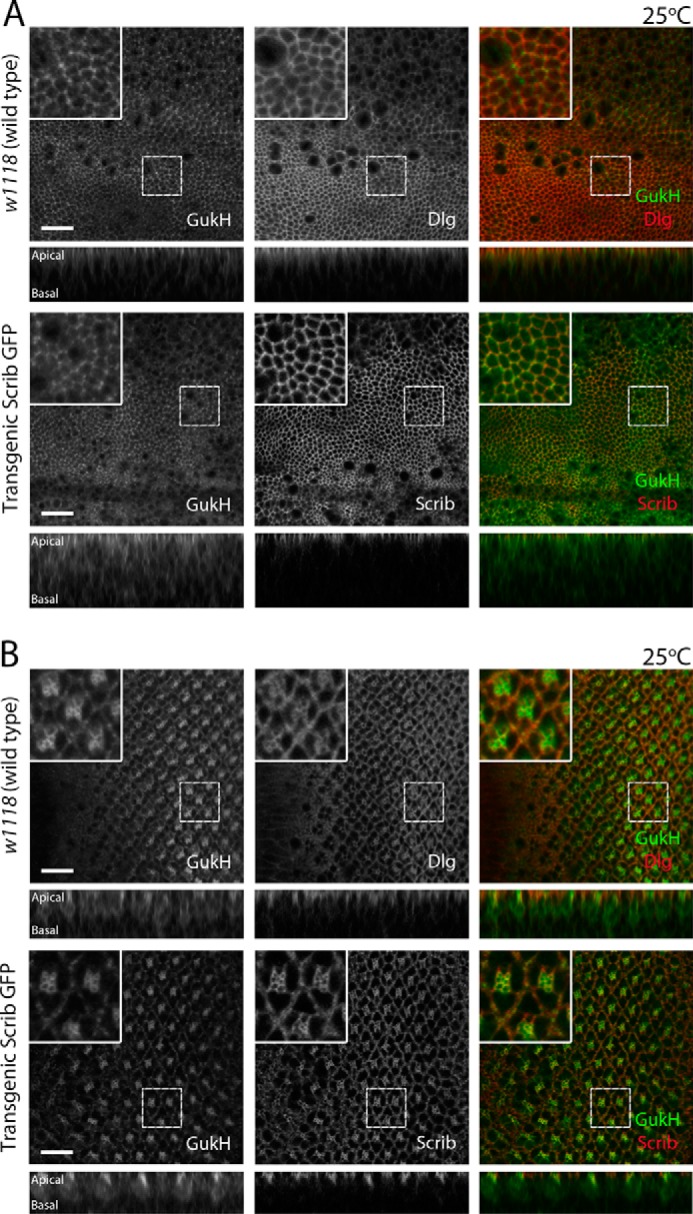
Gukh co-localizes with Scrib and Dlg in Drosophila third instar larval epithelia. Representative confocal images of third instar larval eye and wing epithelial tissue immunofluorescently stained with anti-Gukh and anti-Dlg antibodies are shown. A Scrib-GFP protein trap (homozygous viable), where GFP is inserted in the endogenous gene, allowed visualization of Scrib protein localization. Each row shows a planar (top) and a cross-sectional (bottom) image of the disc. Stainings are shown in grayscale and in merge (right panel) with Gukh shown in green and Dlg or Scrib in red. A, in wing epithelia, Scrib and Dlg localize to the cell cortex, mainly at the apical region of the cells. Gukh shows a strong cytoplasmic localization in comparison but is still highly concentrated at the cortex of cells at the apical region. B, in eye epithelia, Gukh shows similar localization to that observed in wing epithelia, additionally showing enrichment in the apical region of photoreceptor cells. Images were taken at ×63 magnification. Scale bars, 10 μm.
Gukh genetically interacts with Scrib and Dlg in epithelial tissues
Previous studies revealed that the Gukh GUK-holder domain binds to the Dlg GUK domain and that the C-terminal PDZ-binding motif of Gukh interacted with Scrib PDZ domains (13). However, whether Gukh has a functional role with Scrib or Dlg in epithelial tissue is currently unknown. Consequently, we examined the genetic interaction between scrib and gukh in Drosophila eye and wing epithelial tissues. To manipulate the expression of several transgenes in a tissue-specific manner, the UAS/GAL4 system was used to selectively knock down scrib, dlg, or both genes using UAS-RNAi lines in the Drosophila eye or wing epithelial tissues, using the ey-GAL4 or dpp-GAL4 drivers, respectively. Genetic interactions with gukh expression/function knockdown were then examined relative to a control transgene (UAS-lacZ or UAS-GFP, to control for UAS element number).
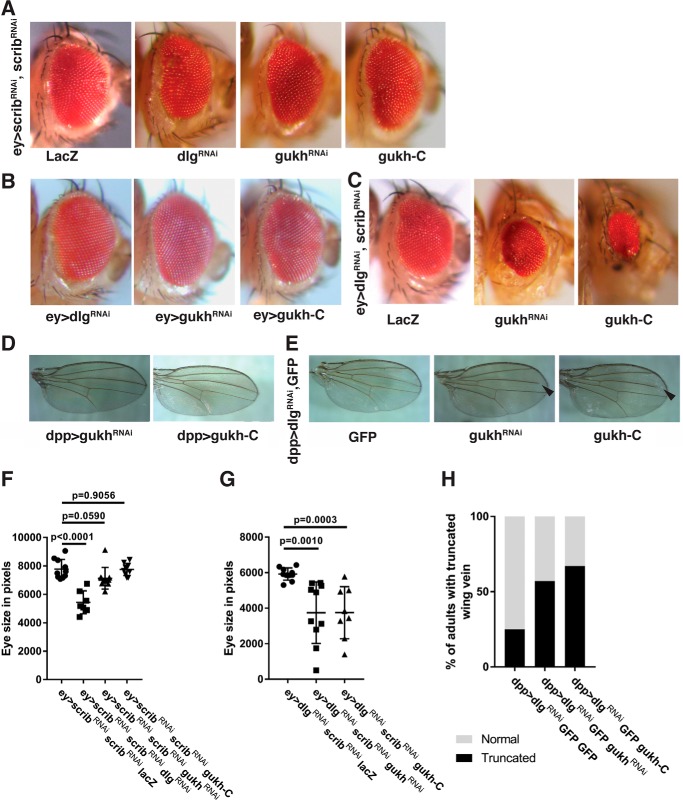
The scrib knockdown using two RNAi transgenes (one on the 2nd chromosome and the other on the 3rd chromosome) via the ey-GAL4 driver in the Drosophila developing eye resulted in a slightly reduced and disorganized eye phenotype (termed a rough eye phenotype, Fig. 5A) relative to control adult eyes (Fig. 5B). To verify that this phenotype was modifiable, we knocked down Dlg using RNAi, which showed, as expected, a robust enhancement of the eye roughness (Fig. 5A) and a reduction in eye size (Fig. 5F). Importantly, knockdown of Gukh using RNAi enhanced the Scrib knockdown rough eye phenotype (Fig. 5A) and led to a slight reduction in eye size (although this was not statistically significant below p < 0.05). We then examined the effect on the Scrib knockdown eye phenotype upon expression of the Gukh–C-terminal region (Gukh-C), which lacks the important F-actin–binding WH1/EVH1 domain at the N terminus, and it is expected to act in a dominant-negative manner (13). When gukh-C was overexpressed with concurrent scrib knockdown, an enhancement of the eye roughness was also observed (Fig. 5A). In contrast, individual overexpression of gukh-C or gukhRNAi expression showed normal eye size and arrangement of ommatidia relative to the wildtype; however, dlgRNAi showed a slight decrease in eye size relative to the other genotypes (Fig. 5B). Altogether, these results reveal a genetic interaction between scrib and gukh in the eye, although not as strong as that observed between scrib and dlg.
Figure 5.
Gukh genetically interacts with Dlg and Scrib. A–C, representative images of male adult eyes are shown for the genotypes indicated. A, interactions with the Scrib knockdown phenotype using ey>scribRNAi (2nd) scribRNAi (3rd). Knockdown of Scrib using the ey-GAL4 driver results in a small rough eye phenotype, which is strongly enhanced upon knockdown of Dlg and mildly enhanced when Gukh function is impaired, using either RNAi or the dominant-negative transgene (gukh-C). B, control phenotypes of crosses to ey-GAL4, showing normal eye phenotypes. C, knockdown of Scrib together with Dlg using the ey-GAL4 driver results in a small rough eye phenotype, which is strongly enhanced upon impairment of Gukh function, using either RNAi or the dominant-negative transgene, gukh-C. D and E, representative images of adult female wings are shown for the genotypes indicated. D, control phenotypes of gukhRNAi or gukh-C crossed to dpp-GAL4, showing normal wing phenotypes. E, interactions with the Dlg knockdown wing phenotype using dpp>dlgRNAi. The Dpp driver results in expression along between the 3rd and 4th wing veins. Knockdown of Dlg resulted in wing vein truncation in 25% of progeny; however, impairment of Gukh function using gukhRNAi or the gukh-C dominant-negative transgene resulted in truncation of the 3rd wing vein (arrowhead) in the majority of flies (∼60–70%). F, graph of the comparison of eye size between genotypes (as indicated) shown in A. G, graph of the comparison of eye size between genotypes (as indicated) shown in C. Values are the mean eye size, and error bars represent mean ± S.D. Student's t test used to test for significance. H, quantitation of wing phenotypes observed in indicated fly genotypes shown in D and E. Values are % of flies displaying vein truncation. Number of flies scored: UAS-GFP, 68; UAS-gukh RNAi, 60; UAS-gukh-C, 54.
We next examined whether gukh genetically interacts with dlg. Because Dlg knockdown via the ey driver did not produce a strong phenotype (Fig. 5B), we examined whether Gukh knockdown could modify the small rough eye phenotype observed upon Scrib and Dlg co-knockdown (Fig. 5, C and G). Strikingly, both gukhRNAi and gukh-C expression resulted in a strong enhancement of the small rough eye phenotypes of ey>scribRNAi dlgRNAi (Fig. 5, C and G) that were statistically significant (p = 0.001 and 0.0003, respectively). Thus, Gukh genetically interacts with Dlg and Scrib in the eye, suggesting that Scrib, Dlg, and Gukh function in the same genetic process in eye-epithelial development.
To extend these results, we used the dpp-GAL4 driver to knock down scrib or dlg in another epithelial tissue, the wing epithelium, and we examined the interactions with gukhRNAi and gukh-C. The Dpp driver is expressed along the anterior–posterior boundary in the developing wing, which constitutes the region between the 3rd and 4th wing vein of the adult wing. Expression of gukh-RNAi or gukh-C via the dpp driver did not affect the wing phenotype (Fig. 5D), but dpp>scribRNAi resulted in a severe wing phenotype and less than 6% survival (data not shown); therefore, the interaction with gukh could not be examined. However, dpp>dlgRNAi resulted in a normal wing phenotype, except for the truncation of the 3rd wing vein in 25% of cases (Fig. 5E). Importantly, knockdown of gukh or overexpression of gukh-C together with dlgRNAi resulted in a truncation of the 3rd wing vein in 57 and 67% (respectively) of flies examined. This was greater than a 2-fold increase in comparison with the 25% seen in the GFP control (Fig. 5, E and H). Thus, gukh genetically interacts with dlg in the wing epithelium, suggesting that Gukh and Dlg function in a common genetic pathway in wing development.
Discussion
Epithelial tissues are highly polarized, and the Scribble polarity module is a critical regulator of epithelial tissue organization and polarization in Drosophila and in mammals. In addition to the core components of the Scribble module, Scrib, Dlg, and Lgl, an important regulatory role has been emerging for the adaptor protein Gukh. Notably, in Drosophila neuromuscular junctions (13) Gukh is a crucial adaptor protein that enables assembly of a functional ternary complex of Scrib, Dlg, and Gukh, thereby allowing correct synaptic localization of Scrib. However, neither the molecular basis for Scrib–Gukh interactions nor a role for Gukh in epithelial tissue structure or function has previously been described. We now show that the Scrib PDZ1 domain is the major high-affinity interaction site for Gukh, and we define the molecular basis for this interaction. Furthermore, we now provide the first description of Gukh expression and function in Drosophila larval epithelial tissues. We show that Gukh is expressed in the larval-wing and eye-epithelial tissues and that Gukh is generally cortically localized and overlaps with Scrib and Dlg at the apical cell cortex. Importantly, our studies have revealed that Gukh together with Scrib and Dlg are key mediators of epithelial tissue development in Drosophila, with loss of Gukh in combination with loss of Scrib and Dlg leading to morphological and differentiation defects in eye and wing tissues.
Expression and localization of Gukh in epithelial tissue
Our expression analysis of Gukh protein in the eye and wing epithelium revealed co-localization with Scrib and Dlg at the apical cortex; however, Gukh was also distributed more apically as well as basolaterally around the cell cortex, and in the differentiated region of the eye epithelium strong staining was observed in the apical region of the photoreceptor cells, where F-actin accumulates. Because Gukh/NHS1, via its WH1 domain, regulates the WAVE/SCAR-ARP2/3-branched F-actin pathway (26, 31), Gukh's general cortical localization might be commensurate with this role in F-actin biogenesis, which conceivably might also function independently of its function with Dlg and Scrib. However, our genetic data in Drosophila shows that expression of Gukh-C (which can bind to Dlg and Scrib but lacks the F-actin regulatory WH1 domain) phenocopies knockdown of gukh in its interaction with scrib and dlg in the eye and wing. This suggests that the Gukh WH1 domain and regulation of F-actin is essential for Scribble module function in epithelial development.
Function of Gukh in larval epithelial tissues
In the eye, knockdown of Scrib or Scrib with Dlg via the ey driver gave rise to tissue growth and patterning defects, most likely due to the disruption of epithelial cell polarity and the deregulation of the Hippo- and JNK-signaling pathways (14, 32). The enhancement of the Scrib and Scrib Dlg phenotypes by impairment of Gukh function (via RNAi or the dominant-negative transgene) suggests that Gukh functions in the same genetic pathway as Scrib and Dlg in epithelial cell polarity and cell signaling. The more robust enhancement observed by Gukh impairment in the background of Scrib and Dlg knockdown is consistent with the proteins functioning in a tripartite complex in epithelial tissues, as has been shown in the neuromuscular junctions (13). Indeed, Gukh and Dlg are both required for Scrib localization in the neuromuscular junctions, with greater Scrib mislocalization being observed in the double mutant (13). It remains to be determined whether this co-regulation of Scrib localization by Dlg and Gukh also occurs in epithelial tissues; however, this function is consistent with the stronger enhancement of the Scrib knockdown phenotype by dual knockdown of Dlg and Gukh.
In the wing, knockdown of Gukh together with Dlg resulted in a pronounced truncation of the 3rd wing vein. Because EGFR-Ras signaling is a key pathway involved in wing vein formation (33), and Scrib has been previously shown to repress this pathway (18, 34), we first considered whether Gukh might also function together with Dlg and Scrib in regulating EGFR-Ras signaling. However, because knockdown of Scrib function would be expected to increase Ras signaling, and enhanced Ras signaling is associated with ectopic wing veins (35), which was not observed in dlg gukh impaired wing tissue, it is unlikely that the Ras pathway is up-regulated by Dlg knockdown in this context. Conversely, knockdown of Dpp (transforming growth factor-β/BMP) signaling results in truncated wing veins (36), and therefore the generation of a similar phenotype by dlg and gukh impairment suggests Dlg and Gukh might positively regulate the Dpp pathway in this context. Consistent with this notion, Scrib via its LRR domain has been shown to bind to the BMP type I receptor, Tkv, type II receptor, Pnt, and the phosphorylated (active) Mad transcription factor in Drosophila wing posterior cross vein development, which is thought to facilitate BMP receptor signaling (37, 38). Additionally, the Scribble module protein, Lgl, has been implicated in the regulation of Dpp secretion in embryonic ectodermal cells (37, 38). Further studies are required to determine whether Dlg, Scrib, and Gukh interact to affect Dpp signaling during wing development.
Molecular interaction of Gukh with Scrib
To establish a molecular basis for the observed genetic interaction between Gukh and Scrib, we performed a biochemical analysis of individual PDZ domains of Drosophila Scrib with peptides encoding for the C-terminal PDZ-binding motif in Gukh. Our binding data demonstrated that the major site of interaction between Gukh and Scrib is the Scrib PDZ1 domain, thus for the first time establishing a definitive molecular basis for this interaction. These findings are in contrast to that of Mathew et al. (13), who showed that Scrib PDZ2 but not PDZ3 and PDZ4 had strong interaction with the Gukh C-terminal peptide in yeast two-hybrid assays; however, they were unable to draw conclusions regarding PDZ1 in their experiments due to high background activity with these constructs (13). Thus, our findings have revealed a previously unexplored role for Scrib PDZ1 in binding to Gukh. However, in contrast to the Mathew et al. report (13), our study did not reveal a role for Scrib PDZ2 in binding the Gukh C-terminal peptide, which might be due to differences in construct design, post-translational modification in the yeast system, or to the inherent propensity for false positives using the yeast two-hybrid system, due to heterologous protein expression and inappropriate cell localization. Importantly, our findings provide a rationale for the observation that the Scrib truncation mutant scrib5 (39), which results in the loss of Scrib PDZ3 and PDZ4 domains, is functionally active and displays normal adherens junction and basolateral/septate junction (SJ) formation, whereas the scrib4 mutant, which lacks active PDZ domains, results in disrupted SJ formation. These data, together with our results, indicate that the presence of an intact and active PDZ1 domain in Scrib should be necessary and sufficient for normal adherens junction and septate junction formation. Moreover, because we have shown that Gukh interacts with Scrib PDZ1, and Gukh genetically interacts with Scrib and Dlg in epithelial tissues, this suggests that Gukh could contribute to the role of Scrib–PDZ1 in epithelial structure as well as potentially in directed epithelial cell migration (39).
Interestingly, our measurements of Scrib PDZ–Gukh interactions identified the PDZ1–Gukh interaction as unusually tight for PDZ domain interactions with an endogenous ligand, with tight nanomolar interactions typically found in PDZ-synthetic ligand complexes such as those of Erbin or ZO-1 interactions with peptides derived from phage display. To better understand the structural basis for this, we determined the crystal structure of Scrib PDZ1–Gukh. Superimposition of the PDZ1 domain from Drosophila bound to Gukh over the human Scrib PDZ1 domain (PDZ 2W4F) reveals no significant structural changes in the ligand-binding groove upon Gukh binding (Fig. 2B), which has previously been observed for the GRIP1 PDZ6–peptide complex (40). Examination of the interface of the PDZ1–Gukh complex revealed an extensive net of hydrogen bonds and ionic interactions, which supplement the insertion of the C-terminal Gukh Leu-1788 into a hydrophobic pocket. In particular, the ionic interaction between Gukh Glu-1785 and Scrib Arg-765 is reminiscent of Erbin Atg-49–p120-catenin-like peptide E4 (41). Loss of this ionic interaction in a Scrib R765A mutant leads to an ∼8-fold loss of Gukh binding to Scrib, suggesting that the Arg-765PDZ1–Glu-1785Gukh salt bridge is important. Interestingly, despite displaying nanomolar affinity, Gukh does not contain any Trp residues in its PDZ-binding motif. In the case of Erbin, a Trp in the −1 position has been shown to be important for binding, with a second Trp in position −4 position being critical for the high-affinity interaction by engaging the β2–3 loop (Fig. 2D). A similar key role is played by a Trp in the −6 position of a synthetic peptide in complex with ZO1 (42, 43), which has been shown to contribute substantially to the binding of ZO1 by inserting into the β2–3 loop in a similar location as the −4 Trp in the Erbin complex (43). In contrast, Gukh harbors a Phe in the −4 position rather than a Trp, which nonetheless makes significant contact with the Scrib PDZ1 domain via π-stacking with His-796. Thus, the −4 position in Gukh is still able to contribute substantially to binding to PDZ1 by exploiting the opposite side of the ligand-binding groove. Indeed, mutation of His-796 to an Ala abrogates binding to Gukh, supporting the notion that the His-796PDZ–Phe-1784Gukh π-stacking is important for the Scrib–Gukh interaction. Furthermore, Gukh only forms a single hydrogen bond with the β2–3 loop via the main chain carbonyl of Pro-1782, indicating that the engagement of the β2–3 loop is not necessary for a high affinity nanomolar interaction.
In conclusion, our study has revealed novel roles and regulatory mechanisms for Gukh in epithelial development. Our discovery of a novel role for Drosophila Scrib PDZ1 in the interaction with Gukh, and the important function of Gukh together with Scrib and Dlg in epithelial tissue morphogenesis and differentiation, increases our understanding of a previously poorly-studied protein. It will now be important to investigate whether the vertebrate Gukh orthologs also interact with Scrib and Dlg in a similar manner in epithelial tissue development.
Experimental procedures
Scrib PDZ domain expression and purification
Protein expression constructs encoding the PDZ domains of Drosophila Scrib (Uniprot accession numbers Q7KRY7: PDZ1(726–820); PDZ2(929–1019); PDZ3(1237–1328); and PDZ4(1335–1427)) were obtained as the synthetic cDNA codons optimized for Escherichia coli expression and cloned into the pGex-6P3 vector (Bioneer). Mutants PDZ1 R765A and H796A were obtained as the synthetic cDNA codon optimized for E. coli expression and cloned into the pGex-6P1 vector (Genscript). Individual PDZ expression plasmids were transformed into E. coli BL21 (DE3) pLysS. Bacterial cells were grown in Super Broth media (3.2% w/v tryptone, 2% w/v yeast extract, 0.5% w/v NaCl, and 5 mm NaOH) supplemented with 0.2 mg/ml ampicillin. Protein overexpression was induced by the autoinduction method (44) by supplementing the growth media with 100 mm NaCl, 50 × 5052 (25% glycerol, 10% lactose, and 2.5% glucose), 10 mm Tris, pH 7.6, and 1 mm MgSO4. Bacterial cultures were incubated at 16 °C with shaking at 160 rpm for 72 h. Bacterial cells were harvested by centrifugation at 3724 × g for 15 min at 4 °C using the Avanti J-E centrifuge (Beckman Coulter), JLA-9.1000 rotor. Bacterial pellets were resuspended in GST purification buffer (50 mm Tris, pH 8.0, 150 mm NaCl, 1 mm EDTA) and lysed using TS series benchtop cell disruptor (Constant Systems, Ltd.) at 25,000 p.s.i. at 4 °C. Bacterial cell lysates were clarified by centrifugation at 48,384 × g for 20 min at 4 °C using the Avanti J-E centrifuge (Beckman Coulter), JA-25.50 rotor. Supernatants containing the target proteins were subjected to affinity chromatography using glutathione-Sepharose 4B (GE Healthcare), and target proteins were liberated from the column overnight at 4 °C by addition of HRV 3C protease (1:10 weight ratio). Flow-through containing target proteins was concentrated using centrifugal concentrators (Millipore) with a 3-kDa molecular mass cutoff. Concentrated protein was subjected to size-exclusion chromatography using a HiLoad 16/600 Superdex 75 column (GE Healthcare) mounted on an AKTA Pure (GE Healthcare) equilibrated with 25 mm HEPES, pH 7.0, for DM-PDZ2, DM-PDZ1, and DM-PDZ4, HEPES, pH 8.0, for DM-PDZ3, supplemented with 150 mm NaCl.
ITC
Purified Drosophila Scrib PDZ domains were used in titration experiments against 8-mer peptides spanning the C terminus of Drosophila Gukh isoform-A (LPSFETAL, GenScript). Raw heats were measured using a Microcal NanoITC200 system (GE Healthcare) at 25 °C. Because of a lack of useful aromatic amino acids in PDZ domain proteins, protein concentrations were calculated using the Scope method (45) by measuring absorbance at 205 and 280 nm using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). As controls, a non-binding Gukh mutant peptide (LPSFEAAA, GenScript) and a superpeptide (RSWFETWV, GenScript) engineered to harbor pan-PDZ binding activity (46) as positive control were used. Binding isotherms were analyzed using Origin 7.0E (MicroCal).
dmPDZ1–Gukh complex crystallization and data collection
The complex of dmPDZ1 with Gukh peptide was reconstituted by mixing protein and peptide at a 1:2 molar ratio. The dilute protein complex was then concentrated to 20 mg/ml using a 3-kDa molecular mass cutoff centrifugal concentrator (Millipore), flash-cooled, and stored under liquid nitrogen. Crystallization trials were carried out using 96-well sitting-drop trays (Swissci) and vapor diffusion at 20 °C either in-house or at the CSIRO C3 Collaborative Crystallization Centre, Melbourne, Australia. A 0.15-μl dmPDZ1–Gukh peptide complex was mixed with 0.15 μl of various crystallization conditions using a Phoenix nanodispenser robot (Art Robbins). Commercially available screening kits (PACT Suite and JCSG-plus Screen) were used for the initial crystallization screening, with hit optimization performed using a 96-well plate at the CSIRO C3 Centre. Crystals of dmPDZ1 in complex with Gukh peptide were obtained at 20 mg/ml in 0.2 m zinc acetate dehydrate, 0.1 m sodium cacodylate, pH 6.5, and 10% (v/v) propanol. The crystals were cryo-protected using 30% (v/v) ethylene glycol and flash-cooled at 100 K using liquid nitrogen. Hexagonal rod crystals were obtained belonging to space group P3212. Unfortunately, structural determination failed with those crystals, which were subsequently used in a cross-seeding experiment into the Shotgun screen at CSIRO C3. After 2 months, the crystals of dmPDZ1 in complex with Gukh peptide were obtained at 20 mg/ml in 30% (v/v) PEG 4000, 0.2 m sodium acetate, and 0.1 m Tris chloride, pH 8.5. The crystals were cryo-protected using 30% (v/v) ethylene glycol and flash-cooled at 100 K using liquid nitrogen. All diffraction data were collected on the MX1 beamline at the Australian Synchrotron using ADSC Quantum 315r CCD detector (Area Detector Systems Corp., Poway, CA) with an oscillation range of 1.0° per frame using a wavelength of 0.9537 Å. Diffraction data were integrated using XDSme (47) and scaled using AIMLESS (48). The structure of dmPDZ1–Gukh peptide was solved by molecular replacement using Phaser (49) with the structure of hsPDZ1 (PDB code 5VWC) as a search model. The final TFZ and LLG values were 19.1 and 471, respectively. The solution produced by Phaser was manually rebuilt over multiple cycles using Coot (50) and refined using PHENIX (51). Data collection and refinement statistics details are summarized in Table 1. MolProbity scores were obtained from the MolProbity web server (52). Shape complementarity was calculated using the program SC (53). Coordinate files have been deposited in the Protein Data Bank under the accession code 5WOU. All images were generated using the PyMOL Molecular Graphics System, Version 1.8, Schrödinger, LLC. All software was accessed using the SBGrid suite (54). All raw diffraction images were deposited on the SBGrid Data Bank (55) using accession number 5WOU.
D. melanogaster stocks and genetic analysis
w1118 (wildtype with white eyes), gukhRNAi (P[TRiP.GL01345]),ey-GAL4, dppBLK-GAL4 (dpp-GAL4), en-GAL4, UAS-lacZ, UAS-GFP, UAS-GAL80ts, ey-FLP, hsp70-FLP, FRT82B Ubi-GFP, FRT82B Ubi-GFP, and gukhBG02660 (the gukh P element mutant) stocks were obtained from the Bloomington Drosophila Stock Centre. UAS-β-galRNAi was obtained from K. Harvey. The Scrib-GFP protein trap (CA07683, homozygous viable) was obtained from the Flytrap collection (L. Cooley). The following Vienna Drosophila Resource Centre stocks were used: UAS-scribRNAi (101128, VDRC stock 105412) and UAS-scribRNAi (11663 C2V, gift from B. Dickson, 2nd chromosome);UAS-scribRNAi (11663 C3S, gift from B. Dickson, 3rd chromosome) (VDRC stock 45555); and UAS-dlgRNAi (4689 C2V, gift from B. Dickson, 2nd chromosome) (VDRC stock 31134). The UAS-gukh-C (C-terminal transgene, missing the N-terminal actin binding domain) was obtained from V. Budnik (13). The efficacy of dlg 4699 C2V and scrib 11663 RNAi lines in targeting these proteins has been previously confirmed (11), and we have confirmed that the scrib 101128 RNAi efficiently knocks down the Scrib protein (data not shown). We also confirmed the efficacy of the gukh-RNAi line by testing its ability to decrease Gukh levels when expressed via engrailed-GAL4 in the larval-wing epithelium, and we verified the specificity of the Gukh antibody by showing that Gukh immunoreactively was decreased in gukhBG02660 mutant clones in the eye epithelium (Fig. 3).
Stocks of ey-GAL4 and UAS-scribRNAi 11663 C2V and UAS-scribRNAi 11663 C3S or ey-GAL4 and UAS-dlgRNAi 4689 C2V and UAS-scribRNAi 11663 C3S were generated and balanced over CyO and TM6B. Stocks of dpp-GAL4 UAS-GFP with UAS-scribRNAi 11663 C2V or UAS-dlgRNAi 4689 C2V were generated and maintained at 18 °C (where expression of the transgenes is low).
For induction of gukh clones in the eye epithelium, gukhBG02660 FRT82B flies were crossed to ey-FLP FRT82B Ubi-GFP at 25 °C, and the eye-antennal discs were dissected from third instar larvae.
Fly crosses were performed at 25 or 29 °C (as indicated), and they were grown on standard fly media (molasses 93.3 g/liter, agar 5.6 g/liter, glucose 10.6 g/liter, fresh yeast 60 g/liter, semolina (coarse) 66 mg/liter, acid mix 0.92%, v/v).
Analysis of genetic interactions in the Drosophila eye and wing tissues
For analysis of genetic interactions in Drosophila eyes, crosses of ey>scribRNAi (2nd) scribRNAi (3rd); ey>dlgRNAi (2nd) scribRNAi (3rd) to UAS-gukh-C, UAS-gukhRNAi, UAS-dlgRNAi or UAS-lacZ (control) were conducted at 29 °C. Crosses of ey-GAL4 to each UAS-RNAi or UAS-transgene were performed as controls. Drosophila adults were collected 7–8 days after crossing at 29 °C, and at least 50 progeny were examined from each cross, and photographs were obtained for at least six samples for each genotype. Eye size was measured using Adobe Photoshop Extended tools.
For analysis of genetic interactions in Drosophila wings, dpp>scribRNAi 11663 C2V (2nd) or dpp>dlgRNAi 4689 C2V (2nd) were crossed to UAS-gukh-C, UAS-gukhRNAi, UAS-dlgRNAi, or UAS-GFP (control) at 25 °C. Crosses of dpp-GAL4 to each UAS-RNAi or UAS-transgene were performed as controls. Scoring was performed based on the presence or absence of the truncated wing vein phenotype. At least 50 individual Drosophila adults were scored from each cross. For imaging, wings from 10 adult flies/sample were carefully detached from the torso and mounted on glass slides using a mixture of methyl salicylate and Canada Balsam (Sigma) in ratio of 1:1 and were left to dry overnight. Microscopy images of Drosophila eyes and wings were obtained at ×2.5 and ×3.2 magnification, respectively, on an Olympus SZX7 microscope equipped with an INFINITY-1 camera and images were processed using INFINITY capture software.
Immunofluorescent staining, confocal microscopy, and quantification
Primary antibodies were rabbit anti-Gukh antibody (1:500 (V. Budnik) raised to the N terminus of Gukh) and mouse anti-Dlg (Developmental Studies Hybridoma Bank, 4F3). Antibody staining was performed using similar methodology as described previously (11). The secondary antibodies were goat anti-rabbit AlexaFluor-568 (Invitrogen) and goat anti-mouse AlexaFluor-633 (Molecular Probes). Wing and/or eye imaginal discs were mounted onto glass slides in one drop of ProLong® GOLD antifade mountant (Molecular Probes, catalog no. P36934) or 80% glycerol in PBS and covered with a glass coverslip. All confocal images were taken on either a confocal Leica TCS (true confocal scanner) SP5 (Leica Microsystems, Germany) or a confocal Zeiss ELYRA (Carl Zeiss, Germany) microscope. Staining intensity and cell migration were quantified using the Fiji (ImageJ) image analysis software. Quantification of Pixel intensity was determined for Gukh staining after RNAi-mediated knockdown (marked by the expression of GFP) or in gukh mutant clones (marked by the absence of GFP expression) versus the wildtype tissue using Fuji software. Adult eye size was determined by drawing a region of interest followed by area measurement (size = 0-infinity pixel units, circularity = 0.00–0.10). Statistical analysis was conducted with Student's t test using Graphpad Prism, where p < 0.05 (version 6.00 for Windows, GraphPad Software, La Jolla, CA).
Author contributions
S. C., C. M. M., and T. S. designed and performed experiments, analyzed the data, and wrote the manuscript. M. P., K. Y. B. L., J. Y. H., and B. Z. S. designed and performed experiments and analyzed the data. P. O. H., H. E. R., and M. K. conceived the project, designed the experiments, analyzed the data, and wrote the manuscript. All authors reviewed and commented on the final manuscript.
Acknowledgments
We thank the staff at the MX beamlines at the Australian Synchrotron for help with X-ray data collection, the CSIRO C3 Collaborative Crystallization Centre for assistance with crystallization, and the Comprehensive Proteomics Platform and LIMS BioImaging Facility at La Trobe University for core instrument support. We thank everyone in our laboratories for supplying Drosophila stocks or reagents and for helpful discussions on this project.
This work was supported in whole or part by National Health and Medical Research Council Australia Project Grant APP1103871 (to M. K., P. O. H., and H. E. R.), Fellowship APP1079133 (to P. O. H.), and APP1020056 (to H. E. R.); Australian Research Council Fellowship FT130101349 (to M. K.); Cancer Council Victoria grant funding (to M. P.) and Project Grant APP1041817 (to H. E. R.); La Trobe University Scholarship (to C. M. M. and K. Y. B. L.), and La Trobe University and La Trobe School of Molecular Science funding (to H. E. R.). The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (code 5WOU) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- PAR
- PARtitioning-defective (PAR LRR, leucine-rich repeat
- PDB
- Protein Data Bank
- ITC
- isothermal titration calorimetry
- SH
- Src homology
- NHS
- Nance-Horan syndrome
- BMP
- bone morphogenetic protein
- EGFR
- epidermal growth factor receptor
- GUK
- guanylate kinase-like
- r.m.s.d.
- root mean square deviation
- SJ
- septate junction
- PBM
- PDZ-binding domain.
References
- 1. Elsum I., Yates L., Humbert P. O., and Richardson H. E. (2012) The Scribble-Dlg-Lgl polarity module in development and cancer: from flies to man. Essays Biochem. 53, 141–168 10.1042/bse0530141 [DOI] [PubMed] [Google Scholar]
- 2. Xu X., Farach-Carson M. C., and Jia X. (2014) Three-dimensional in vitro tumor models for cancer research and drug evaluation. Biotechnol. Adv. 32, 1256–1268 10.1016/j.biotechadv.2014.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Humbert P. O., Grzeschik N. A., Brumby A. M., Galea R., Elsum I., and Richardson H. E. (2008) Control of tumourigenesis by the Scribble/Dlg/litergl polarity module. Oncogene 27, 6888–6907 10.1038/onc.2008.341 [DOI] [PubMed] [Google Scholar]
- 4. Hanahan D., and Weinberg R. A. (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 5. Tepass U. (2012) The apical polarity protein network in Drosophila epithelial cells: regulation of polarity, junctions, morphogenesis, cell growth, and survival. Annu. Rev. Cell Dev. Biol. 28, 655–685 10.1146/annurev-cellbio-092910-154033 [DOI] [PubMed] [Google Scholar]
- 6. Yap A. S., Gomez G. A., and Parton R. G. (2015) Adherens junctions revisualized: organizing cadherins as nanoassemblies. Dev. Cell 35, 12–20 10.1016/j.devcel.2015.09.012 [DOI] [PubMed] [Google Scholar]
- 7. Stephens R., Lim K. Y., Portela M., Kvansakul M., Humbert P. O., and Richardson H. E. (2018) The scribble cell polarity module in the regulation of cell signalling in tissue development and tumourigenesis. J. Mol. Biol. 10.1016/j.jmb.2018.01.011 [DOI] [PubMed] [Google Scholar]
- 8. Bilder D., Li M., and Perrimon N. (2000) Cooperative regulation of cell polarity and growth by Drosophila tumor suppressors. Science 289, 113–116 10.1126/science.289.5476.113 [DOI] [PubMed] [Google Scholar]
- 9. Brumby A. M., and Richardson H. E. (2003) Scribble mutants cooperate with oncogenic Ras or Notch to cause neoplastic overgrowth in Drosophila. EMBO J. 22, 5769–5779 10.1093/emboj/cdg548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Parsons L. M., Portela M., Grzeschik N. A., and Richardson H. E. (2014) Lgl regulates Notch signaling via endocytosis, independently of the apical aPKC-Par6-Baz polarity complex. Curr. Biol. 24, 2073–2084 10.1016/j.cub.2014.07.075 [DOI] [PubMed] [Google Scholar]
- 11. Grzeschik N. A., Parsons L. M., Allott M. L., Harvey K. F., and Richardson H. E. (2010) Lgl, aPKC, and Crumbs regulate the Salvador/Warts/Hippo pathway through two distinct mechanisms. Curr. Biol. 20, 573–581 10.1016/j.cub.2010.01.055 [DOI] [PubMed] [Google Scholar]
- 12. Grzeschik N. A., Amin N., Secombe J., Brumby A. M., and Richardson H. E. (2007) Abnormalities in cell proliferation and apico-basal cell polarity are separable in Drosophila lgl mutant clones in the developing eye. Dev. Biol. 311, 106–123 10.1016/j.ydbio.2007.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mathew D., Gramates L. S., Packard M., Thomas U., Bilder D., Perrimon N., Gorczyca M., and Budnik V. (2002) Recruitment of scribble to the synaptic scaffolding complex requires GUK-holder, a novel DLG binding protein. Curr. Biol. 12, 531–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Leong G. R., Goulding K. R., Amin N., Richardson H. E., and Brumby A. M. (2009) Scribble mutants promote aPKC and JNK-dependent epithelial neoplasia independently of Crumbs. BMC Biol. 7, 62 10.1186/1741-7007-7-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Santoni M. J., Pontarotti P., Birnbaum D., and Borg J. P. (2002) The LAP family: a phylogenetic point of view. Trends Genet. 18, 494–497 10.1016/S0168-9525(02)02738-5 [DOI] [PubMed] [Google Scholar]
- 16. Arnaud C., Sebbagh M., Nola S., Audebert S., Bidaut G., Hermant A., Gayet O., Dusetti N. J., Ollendorff V., Santoni M. J., Borg J. P., and Lécine P. (2009) MCC, a new interacting protein for Scrib, is required for cell migration in epithelial cells. FEBS Lett. 583, 2326–2332 10.1016/j.febslet.2009.06.034 [DOI] [PubMed] [Google Scholar]
- 17. Audebert S., Navarro C., Nourry C., Chasserot-Golaz S., Lécine P., Bellaiche Y., Dupont J. L., Premont R. T., Sempéré C., Strub J. M., Van Dorsselaer A., Vitale N., and Borg J. P. (2004) Mammalian Scribble forms a tight complex with the βPIX exchange factor. Curr. Biol. 14, 987–995 10.1016/j.cub.2004.05.051 [DOI] [PubMed] [Google Scholar]
- 18. Dow L. E., Kauffman J. S., Caddy J., Zarbalis K., Peterson A. S., Jane S. M., Russell S. M., and Humbert P. O. (2007) The tumour-suppressor Scribble dictates cell polarity during directed epithelial migration: regulation of Rho GTPase recruitment to the leading edge. Oncogene 26, 2272–2282 10.1038/sj.onc.1210016 [DOI] [PubMed] [Google Scholar]
- 19. Nola S., Sebbagh M., Marchetto S., Osmani N., Nourry C., Audebert S., Navarro C., Rachel R., Montcouquiol M., Sans N., Etienne-Manneville S., Borg J. P., and Santoni M. J. (2008) Scrib regulates PAK activity during the cell migration process. Hum. Mol. Genet. 17, 3552–3565 10.1093/hmg/ddn248 [DOI] [PubMed] [Google Scholar]
- 20. Sun Y., Aiga M., Yoshida E., Humbert P. O., and Bamji S. X. (2009) Scribble interacts with β-catenin to localize synaptic vesicles to synapses. Mol. Biol. Cell 20, 3390–3400 10.1091/mbc.E08-12-1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Humbert P., Russell S., and Richardson H. (2003) Dlg, Scribble and Lgl in cell polarity, cell proliferation and cancer. Bioessays 25, 542–553 10.1002/bies.10286 [DOI] [PubMed] [Google Scholar]
- 22. Hough C. D., Woods D. F., Park S., and Bryant P. J. (1997) Organizing a functional junctional complex requires specific domains of the Drosophila MAGUK Discs large. Genes Dev. 11, 3242–3253 10.1101/gad.11.23.3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brooks S. P., Ebenezer N. D., Poopalasundaram S., Lehmann O. J., Moore A. T., and Hardcastle A. J. (2004) Identification of the gene for Nance-Horan syndrome (NHS). J. Med. Genet. 41, 768–771 10.1136/jmg.2004.022517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Burdon K. P., McKay J. D., Sale M. M., Russell-Eggitt I. M., Mackey D. A., Wirth M. G., Elder J. E., Nicoll A., Clarke M. P., FitzGerald L. M., Stankovich J. M., Shaw M. A., Sharma S., Gajovic S., Gruss P., et al. (2003) Mutations in a novel gene, NHS, cause the pleiotropic effects of Nance-Horan syndrome, including severe congenital cataract, dental anomalies, and mental retardation. Am. J. Hum. Genet. 73, 1120–1130 10.1086/379381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sharma S., Ang S. L., Shaw M., Mackey D. A., Gécz J., McAvoy J. W., and Craig J. E. (2006) Nance-Horan syndrome protein, NHS, associates with epithelial cell junctions. Hum. Mol. Genet. 15, 1972–1983 10.1093/hmg/ddl120 [DOI] [PubMed] [Google Scholar]
- 26. Brooks S. P., Coccia M., Tang H. R., Kanuga N., Machesky L. M., Bailly M., Cheetham M. E., and Hardcastle A. J. (2010) The Nance-Horan syndrome protein encodes a functional WAVE homology domain (WHD) and is important for co-ordinating actin remodeling and maintaining cell morphology. Hum. Mol. Genet. 19, 2421–2432 10.1093/hmg/ddq125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Garcia J. D., Dewey E. B., and Johnston C. A. (2014) Dishevelled binds the Discs large 'Hook' domain to activate GukHolder-dependent spindle positioning in Drosophila. PLoS ONE 9, e114235 10.1371/journal.pone.0114235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Qian Y., and Prehoda K. E. (2006) Interdomain interactions in the tumor suppressor discs large regulate binding to the synaptic protein GukHolder. J. Biol. Chem. 281, 35757–35763 10.1074/jbc.M607057200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang J., Lewis S. M., Kuhlman B., and Lee A. L. (2013) Supertertiary structure of the MAGUK core from PSD-95. Structure 21, 402–413 10.1016/j.str.2012.12.014 [DOI] [PubMed] [Google Scholar]
- 30. McCann J. J., Zheng L., Rohrbeck D., Felekyan S., Kühnemuth R., Sutton R. B., Seidel C. A., and Bowen M. E. (2012) Supertertiary structure of the synaptic MAGuK scaffold proteins is conserved. Proc. Natl. Acad. Sci. U.S.A. 109, 15775–15780 10.1073/pnas.1200254109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Walsh G. S., Grant P. K., Morgan J. A., and Moens C. B. (2011) Planar polarity pathway and Nance-Horan syndrome-like 1b have essential cell-autonomous functions in neuronal migration. Development 138, 3033–3042 10.1242/dev.063842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Doggett K., Grusche F. A., Richardson H. E., and Brumby A. M. (2011) Loss of the Drosophila cell polarity regulator Scribbled promotes epithelial tissue overgrowth and cooperation with oncogenic Ras-Raf through impaired Hippo pathway signaling. BMC Dev. Biol. 11, 57 10.1186/1471-213X-11-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Blair S. S. (2007) Wing vein patterning in Drosophila and the analysis of intercellular signaling. Annu. Rev. Cell Dev. Biol. 23, 293–319 10.1146/annurev.cellbio.23.090506.123606 [DOI] [PubMed] [Google Scholar]
- 34. Nagasaka K., Pim D., Massimi P., Thomas M., Tomaić V., Subbaiah V. K., Kranjec C., Nakagawa S., Yano T., Taketani Y., Myers M., and Banks L. (2010) The cell polarity regulator hScrib controls ERK activation through a KIM site-dependent interaction. Oncogene 29, 5311–5321 10.1038/onc.2010.265 [DOI] [PubMed] [Google Scholar]
- 35. Lesokhin A. M., Yu S. Y., Katz J., and Baker N. E. (1999) Several levels of EGF receptor signaling during photoreceptor specification in wildtype, Ellipse, and null mutant Drosophila. Dev. Biol. 205, 129–144 10.1006/dbio.1998.9121 [DOI] [PubMed] [Google Scholar]
- 36. Inaki M., Kojima T., Ueda R., and Saigo K. (2002) Requirements of high levels of Hedgehog signaling activity for medial-region cell fate determination in Drosophila legs: identification of pxb, a putative Hedgehog signaling attenuator gene repressed along the anterior-posterior compartment boundary. Mech. Dev. 116, 3–18 10.1016/S0925-4773(02)00119-3 [DOI] [PubMed] [Google Scholar]
- 37. Arquier N., Perrin L., Manfruelli P., and Sémériva M. (2001) The Drosophila tumor suppressor gene lethal(2)giant larvae is required for the emission of the Decapentaplegic signal. Development 128, 2209–2220 [DOI] [PubMed] [Google Scholar]
- 38. Gui J., Huang Y., and Shimmi O. (2016) Scribbled optimizes BMP signaling through its receptor internalization to the Rab5 endosome and promote robust epithelial morphogenesis. PLoS Genet. 12, e1006424 10.1371/journal.pgen.1006424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zeitler J., Hsu C. P., Dionne H., and Bilder D. (2004) Domains controlling cell polarity and proliferation in the Drosophila tumor suppressor Scribble. J. Cell Biol. 167, 1137–1146 10.1083/jcb.200407158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Im Y. J., Park S. H., Rho S. H., Lee J. H., Kang G. B., Sheng M., Kim E., and Eom S. H. (2003) Crystal structure of GRIP1 PDZ6-peptide complex reveals the structural basis for class II PDZ target recognition and PDZ domain-mediated multimerization. J. Biol. Chem. 278, 8501–8507 10.1074/jbc.M212263200 [DOI] [PubMed] [Google Scholar]
- 41. Skelton N. J., Koehler M. F., Zobel K., Wong W. L., Yeh S., Pisabarro M. T., Yin J. P., Lasky L. A., and Sidhu S. S. (2003) Origins of PDZ domain ligand specificity. Structure determination and mutagenesis of the Erbin PDZ domain. J. Biol. Chem. 278, 7645–7654 10.1074/jbc.M209751200 [DOI] [PubMed] [Google Scholar]
- 42. Zhang Y., Yeh S., Appleton B. A., Held H. A., Kausalya P. J., Phua D. C., Wong W. L., Lasky L. A., Wiesmann C., Hunziker W., and Sidhu S. S. (2006) Convergent and divergent ligand specificity among PDZ domains of the LAP and zonula occludens (ZO) families. J. Biol. Chem. 281, 22299–22311 10.1074/jbc.M602902200 [DOI] [PubMed] [Google Scholar]
- 43. Appleton B. A., Zhang Y., Wu P., Yin J. P., Hunziker W., Skelton N. J., Sidhu S. S., and Wiesmann C. (2006) Comparative structural analysis of the Erbin PDZ domain and the first PDZ domain of ZO-1. Insights into determinants of PDZ domain specificity. J. Biol. Chem. 281, 22312–22320 10.1074/jbc.M602901200 [DOI] [PubMed] [Google Scholar]
- 44. Studier F. W. (2005) Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 10.1016/j.pep.2005.01.016 [DOI] [PubMed] [Google Scholar]
- 45. Scopes R. K. (1974) Measurement of protein by spectrophotometry at 205 nm. Anal. Biochem. 59, 277–282 10.1016/0003-2697(74)90034-7 [DOI] [PubMed] [Google Scholar]
- 46. Tonikian R., Zhang Y., Sazinsky S. L., Currell B., Yeh J. H., Reva B., Held H. A., Appleton B. A., Evangelista M., Wu Y., Xin X., Chan A. C., Seshagiri S., Lasky L. A., Sander C., et al. (2008) A specificity map for the PDZ domain family. PLoS Biol. 6, e239 10.1371/journal.pbio.0060239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Storoni L. C., McCoy A. J., and Read R. J. (2004) Likelihood-enhanced fast rotation functions. Acta Crystallogr. D Biol. Crystallogr. 60, 432–438 10.1107/S0907444903028956 [DOI] [PubMed] [Google Scholar]
- 50. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 51. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Williams C. J., Headd J. J., Moriarty N. W., Prisant M. G., Videau L. L., Deis L. N., Verma V., Keedy D. A., Hintze B. J., Chen V. B., Jain S., Lewis S. M., Arendall W. B. 3rd, Snoeyink J., Adams P. D., Lovell S. C., Richardson J. S., and Richardson D. C. (2017) MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lawrence M. C., and Colman P. M. (1993) Shape complementarity at protein/protein interfaces. J. Mol. Biol. 234, 946–950 10.1006/jmbi.1993.1648 [DOI] [PubMed] [Google Scholar]
- 54. Morin A., Eisenbraun B., Key J., Sanschagrin P. C., Timony M. A., Ottaviano M., and Sliz P. (2013) Collaboration gets the most out of software. Elife 2, e01456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Meyer P. A., Socias S., Key J., Ransey E., Tjon E. C., Buschiazzo A., Lei M., Botka C., Withrow J., Neau D., Rajashankar K., Anderson K. S., Baxter R. H., Blacklow S. C., Boggon T. J., et al. (2016) Data publication with the structural biology data grid supports live analysis. Nat. Commun. 7, 10882 10.1038/ncomms10882 [DOI] [PMC free article] [PubMed] [Google Scholar]