Abstract
Regulation of major histocompatibility complex class II (MHC-II) expression is important not only to maintain a diverse pool of MHC-II–peptide complexes but also to prevent development of autoimmunity. The membrane-associated RING-CH (March) E3 ubiquitin ligase March-I regulates ubiquitination and turnover of MHC-II–peptide complexes in resting dendritic cells (DCs) and B cells. However, activation of either cell type terminates March-I expression, thereby stabilizing MHC-II–peptide complexes. Despite March-I's important role in the biology of antigen-presenting cells (APCs), how expression of March-I mRNA is regulated remains unknown. We now show that both DCs and B cells possess a distinct isoform of March-I whose expression is regulated by a promoter located within the March-I gene. Using March-I promoter fragments to drive expression of GFP, we also identified a core promoter for expression of March-I in DCs and B cells, but not in fibroblasts, kidney cells, or epithelial cells, that contains regulatory regions that down-regulate March-I expression upon activation of DCs. Curiously, we found downstream sequence elements, present in the first coding exon of March-I in APCs, that confer regulation of March-I expression in activated APCs. In summary, our study identifies regulatory regions of the March-I gene that confer APC-specific expression and activation-induced modulation of March-I expression in DCs and B cells.
Keywords: major histocompatibility complex (MHC), antigen presentation, ubiquitin ligase, dendritic cell, gene regulation, intragenic promoter
Introduction
Activation of naïve CD4 T cells requires effective antigen processing and presentation by MHC class II-positive (MHC-II+) antigen-presenting cells (APCs).4 APCs, including dendritic cells (DCs) and B cells, internalize potential antigens and degrade them into peptide fragments that bind to MHC-II molecules in endo/lysosomal compartments (1). These peptide–MHC-II (pMHC-II) complexes then move to the APC surface where they can be surveyed by T-cell receptors (TCRs) expressed on CD4 T cells (2, 3). Resting DCs are continually generating new and different pMHC-II complexes as part of their “sentinel” function in the immune system, and under steady-state conditions pMHC-II generation and degradation rates are identical (4). Not only must the pMHC-II complexes on the APC surface present the appropriate pMHC-II to the TCR on an antigen-specific T cell, but sustained engagement by pMHC-II with the TCR is necessary for complete T-cell activation (5). DC activation suppresses the process of pMHC-II degradation, effectively “fixing” the pMHC-II repertoire on the DC surface and increasing the likelihood of effective pMHC-II–TCR interactions necessary for T cell activation (6, 7).
MHC-II turnover in APCs, including DCs and B cells, is regulated by ubiquitination of MHC-II by the E3 ubiquitin ligase March-I (8). March-I is a transmembrane ubiquitin ligase that preferentially targets internalized pMHC-II for lysosomal degradation (9). Not only is degradation of “old” pMHC-II important to allow T cells to sample APCs for “new” pMHC-II, but the kinetics of pMHC-II turnover can affect affinity maturation of germinal center B cells (10), negative selection of CD4 T cells in the thymus (11), and generation of Foxp3-expressing regulatory T cells (11).
March-I is unusual among ubiquitin ligases because of its unique tissue distribution and expression pattern. March-I is expressed in resting DCs and B cells, and activation of either cell type by a variety of TLR ligands terminates expression of March-I, reduces pMHC-II ubiquitination, and prolongs the half-life of pMHC-II (12, 13). March-I is not expressed in non-hematopoietic APCs, including thymic epithelial cells, highlighting lineage-specific regulation of March-I expression in hematopoietic APCs (14, 15). Even among APCs, March-I expression can be up-regulated as monocytes and macrophages express almost no March-I unless the cells are treated with interferon-γ and IL-10 (16–19). In addition to controlling pMHC-II expression, March-I also regulates the APC expression of CD86 (12, 20, 21), a costimulatory protein that is important for the generation of immunogenic, and not tolerogenic, T cells. However, it is likely that most of the biologically significant effects of March-I reside in its ability to regulate pMHC-II turnover as mutant mice expressing non-ubiquitinatable forms of MHC-II (but still containing endogenous March-I and CD86) show phenotypes that are nearly indistinguishable from those of March-I–deficient mice (22).
Despite the well-documented role of March-I in controlling pMHC-II and CD86 expression in APCs (and thereby influencing immunological consequences of pMHC-II/CD86 dysregulation), the mechanisms regulating tissue-specific expression of March-I and TLR-mediated down-regulation of March-I mRNA expression have not been addressed. March-I protein has a very short half-life (23), and for this reason it is likely that March-I expression is regulated primarily at the transcriptional level. In this study, we have examined the March-I gene, identified the March-I isoform present in APCs, and identified the regulatory sequences within the March-I gene that confer tissue-specific expression and activation-induced repression of March-I transcription in DCs.
Results and discussion
March-I variant 2 is the primary form of March-I found in DCs
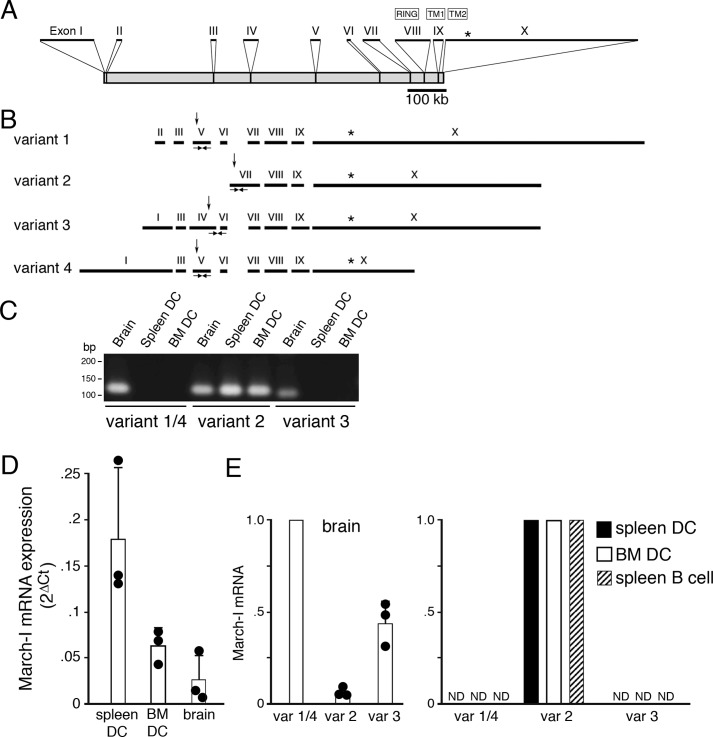
March-I was originally identified using a BLAST search of GenBankTM for human RING-CH domain–containing E3 ligases (20). Both the Vega (24) and Ensembl (25) gene annotation systems indicate that two variants of human March-I and four variants of mouse March-I exist; however, the relative abundance of these March-I variants in professional APCs has not been determined. The organization of the March-I gene as annotated in the Ensembl database is shown in Fig. 1A. The location of the E3 ligase RING domain and transmembrane domains 1 and 2 (present in exons 8, 9, and 10, respectively) are indicated and are common to each variant of March-I.
Figure 1.
March-I v2 is the predominant March-I variant expressed in DCs. A, schematic representation of the March-I gene as described in UCSC Genome Browser. The positions of each exon (I–X), their relative size, and their location within the March-I gene is indicated. The location of the translation stop codon in exon X is indicated by an asterisk. B, the compositions of March-I variant 1, variant 2, variant 3, and variant 4 (as described in Ensembl) are indicated. The position of the E3 ligase RING domain, transmembrane domain 1 (TM1), and transmembrane domain 2 (TM2) are indicated. The relative position of the translation initiation codon in each variant is indicated by an arrow, and the common translation stop codon in exon X is indicated by an asterisk. The locations of specific PCR primers that specifically amplify March-I fragments specific to variant 1/4, variant 2, and variant 3 are indicated. C–E, PCR primers specific for March-I variant 1/4, variant 2, or variant 3 were used to amplify mRNA from spleen DCs, BM DCs, and brain. C, representative gel showing the presence of March-I variant 2 in all tissues and only small amounts of variant 1/4 and variant 3 in the brain. Forty cycles of PCR amplification for each primer pair were performed, and aliquots of the PCR were analyzed on an agarose gel. D, quantitation of the amount of total March-I mRNA (using a primer set common to all March-I variants) was performed by RT-PCR, and data were normalized to expression of GAPDH in each sample. Data are shown as the 2ΔCt (ΔCt = CtGAPDH − CtMarch-I) value for each condition. The results shown are the average (error bars represent S.D.) of three independent experiments. E, the relative amount of each March-I isoform present in spleen DCs, BM DCs, spleen B cells, and brain was determined by RT-PCR, and the 2ΔCt (ΔCt = CtGAPDH − CtMarch-I) value for each sample was determined. The amount of mRNA for the most abundant March-I variant present in spleen DCs, BM DCs, B cells, and brain was arbitrarily designated a value of 1. The results shown are the average (error bars represent S.D.) of three independent experiments. var, variant; ND, not detectable.
In an attempt to quantitate March-I mRNA expression in DCs and in mouse brain (a tissue in which ESTs for each March-I variant have been identified), we designed PCR primers that selectively amplify March-I variants 1/4, 2, and 3 (Fig. 1B). It should be noted that March-I variants 1 and 3 contain a truncated form of exon 7, and variants 1 and 2 contain a truncated form of exon 9, most likely due to the recognition of internal splice-acceptor sequences in these exons (26). Mouse brain contained transcripts encoding at least three March-I isoforms (our exon 5 primer sets cannot distinguish between variants 1 and 4); however, spleen DCs and bone marrow–derived DCs (BM DCs) only contained March-I variant 2 (March-I v2) mRNA (Fig. 1C). Quantitative RT-PCR using RING-domain primers common to each variant demonstrated that the absolute amount of March-I in brain was quite low as compared with that present in spleen DCs (Fig. 1D) and that the small amount of March-I mRNA in brain consisted primarily of variants 1/4 and 3 (Fig. 1E). These variants are capable of down-regulating expression of MHC-II in transfected cells;5 however, whether their function is regulation of MHC-II expression in the brain remains to be determined. Unlike the results in brain, RT-PCR revealed that March-I v2 was the only form of March-I detected in DCs and B cells. These data demonstrate that March-I v2 is the primary, if not the only, isoform of March-I present in DCs and B cells. It is important to note that March-I v2 was the isoform originally identified by Bartee et al. (20) and is the variant that has been used in all March-I overexpression studies published to date.
LPS signaling does not affect March-I v2 mRNA stability
March-I mRNA expression in a variety of APCs is rapidly reduced upon exposure of the cells to the TLR4 ligand LPS (12, 13). We explored the possibility that reduced expression of March-I v2 mRNA in DCs exposed to LPS was a consequence of enhanced degradation of pre-existing March-I v2 mRNA. Actinomycin D is an inhibitor of RNA synthesis when used at low concentrations (27), and for this reason actinomycin D treatment has been used as a method to measure mRNA half-life. DCs were left untreated or pretreated with LPS for 1 h before addition of actinomycin D, and the amount of March-I v2 mRNA present over time was determined by quantitative RT-PCR. In agreement with previous findings (12, 19), 1-h pretreatment of DCs resulted in a modest but detectable reduction in March-I v2 mRNA present at the time of actinomycin D addition (Fig. 2A). However, when normalizing the amount of March-I v2 mRNA present in each sample to that present at time 0, the rate of March-I v2 mRNA degradation was unaffected by LPS pretreatment of DCs (Fig. 2B). These data strongly suggest that down-regulation of March-I v2 mRNA expression is regulated transcriptionally and not by post-transcriptional degradation of March-I v2 mRNA.
Figure 2.
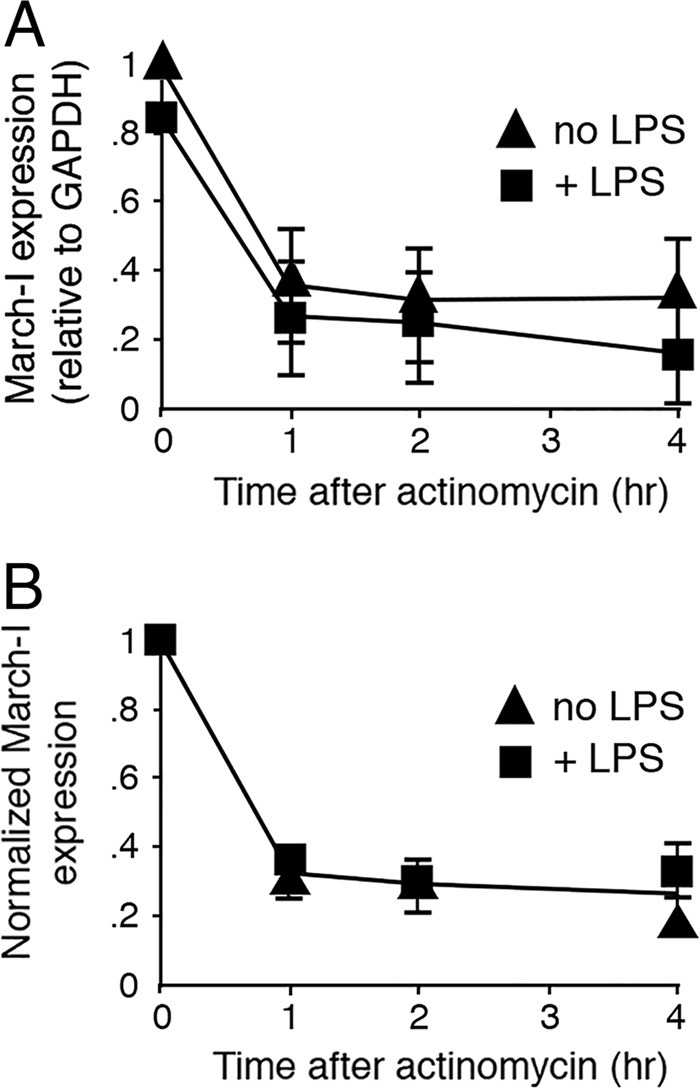
DC activation does not promote March-I v2 mRNA degradation. DCs were pretreated (or not) with LPS for 1 h, washed, and then incubated in complete medium containing actinomycin D (5 μg/ml) for the indicated time. Cells were harvested, mRNA was isolated, and March-I v2 expression was determined by RT-PCR. The data from each condition were normalized to the expression of GAPDH at each time point. A, March-I v2 mRNA expression in each condition was plotted as a function of time incubated in actinomycin. B, March-I v2 mRNA expression in either untreated or LPS-treated DCs at each time point was normalized to March-I v2 expression at t = 0 (time of addition of actinomycin D). The results shown are the average (error bars represent S.D.) of three independent experiments.
Analysis of the March-I v2 promoter
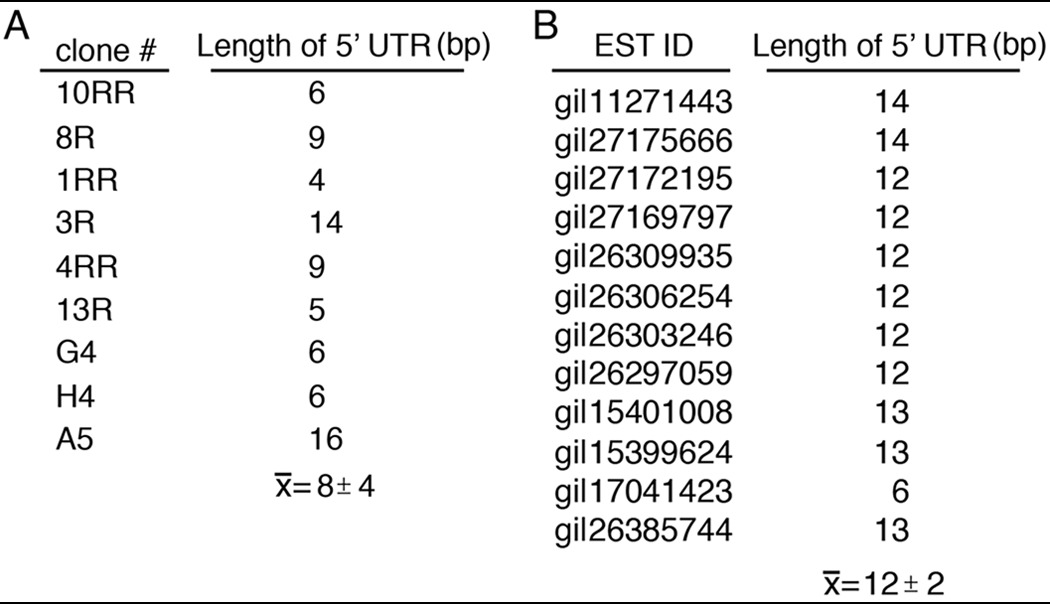
We performed 5′ RACE and EST database searches to identify the transcription start site(s) (TSS) of March-I v2. 5′ RACE analysis revealed that the 5′-UTR of mouse March-I v2 was extremely short and was located only 8.9 ± 4.2 bp upstream of the translation start site in exon 7 (Table 1). Examination of the mouse EST database confirmed our 5′ RACE results and confirmed that the 5′-UTR of mouse March-I v2 was less than 20 bp upstream of the translation start site.
Table 1.
March-I v2 has a very short 5′-UTR
A, 5′ RACE was performed using mRNA isolated from mouse spleen DCs. The length of the 5′-UTR (relative to the initiation codon) for nine independent clones was identified by DNA sequence analysis. B, 12 different March-I ESTs were aligned to the March-I v2 coding sequence, and the length of the 5′-UTR (relative to the initiation codon) was determined. The GenBank ID for each EST is indicated. The average ± S.D. for each set of analyses is shown.
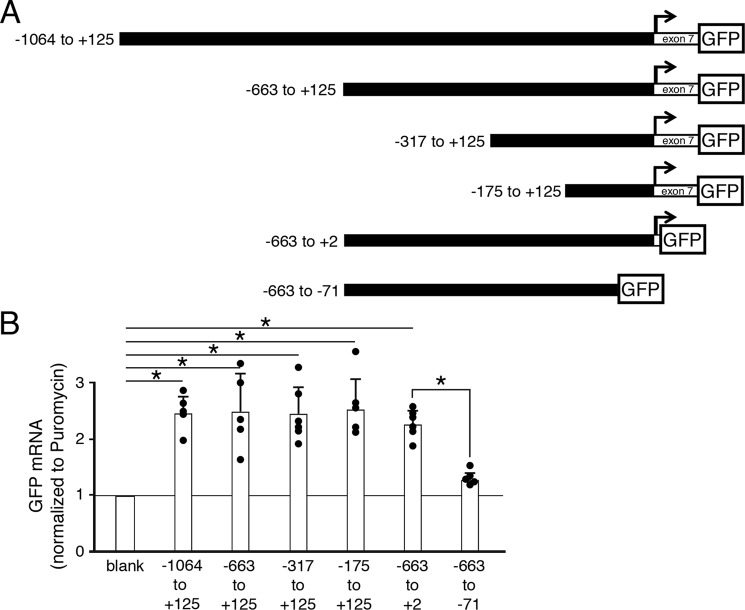
Based on our identification of the March-I v2 TSS, we generated March-I v2 promoter constructs containing up to 1 kb upstream of the TSS driving expression of GFP in lentiviral vectors (Fig. 3A). Because regulatory elements are often present in the first coding exon of genes (28), we included the first 125 bp of exon 7 in some of our promoter constructs. As a negative control, we used the lentiviral vector containing no promoter elements (but still retaining the GFP open reading frame). Because March-I v2 is only expressed in professional APCs (including DCs) and March-I v2 expression is down-regulated in activated APC lines, bone marrow cells were transduced with lentivirus prior to differentiation of the cells into DCs in vitro using GM-CSF. Because we were primarily interested in monitoring March-I v2 promoter activity in our studies, we simultaneously monitored expression of mRNA for endogenous March-I v2 and March-I v2 promoter–dependent expression of GFP in each sample. Each lentivirus encoded puromycin N-acetyltransferase (puromycin; whose expression was dependent on the phosphoglycerate kinase promoter), and for that reason transduction efficiency in each condition was normalized to the amount of puromycin mRNA present in the sample. Each of the promoter constructs expressed GFP mRNA in DCs well above promoterless control lentivirus levels (Fig. 3B). Sequences in the first 125 bp of exon 7 were not required for basal expression of GFP as expression of the March-I v2 (−663 to +125)-GFP construct was nearly indistinguishable from that of the shorter March-I v2 (−663 to +2)-GFP construct. Importantly, deletion of 71 bp upstream of the TSS almost completely prevented March-I v2 promoter activity, highlighting an important role for sequences present within this region for March-I v2 core promoter activity. From these data, we conclude that March-I v2 promoter constructs containing as little as 175 bp upstream of the TSS allow March-I v2 expression in DCs and that sequences present in exon 7 are not required for basal March-I v2 promoter activity.
Figure 3.
Identification of an internal March-I v2 promoter in DCs. A, the transcription start site of March-I v2 is indicated by an arrow, and promoter constructs consisting of DNA sequences 5′ of the TSS with (or without) 125 bp of exon 7 fused to GFP were generated. B, DCs were transduced with lentivirus encoding the indicated promoter construct, and expression of GFP mRNA and lentivirus-encoded puromycin was determined by RT-PCR. The “blank” viral construct encoded GFP but did not contain any March-I promoter sequences. GFP mRNA expression was normalized to the amount of puromycin mRNA present in the sample (as described under “Experimental procedures”) and expressed relative to the amount of mRNA detected in the promoterless blank construct. The results shown are the average (error bars represent S.D.) of multiple independent experiments. *, p < 0.05.
TLR signaling regulates March-I v2 promoter activity
Activation of APCs leads to down-regulation of March-I expression (12, 13, 29), and we have shown that, after as little as 2 h of stimulation with LPS, March-I mRNA is reduced by ∼50% (19). To directly examine whether activation of DCs affects the function of our March-I v2 promoter-GFP reporter constructs, we stimulated immature DCs transduced with March-I v2 promoter-GFP lentivirus with the TLR4 ligand LPS or the TLR9 ligand CpG for 2 h prior to isolating mRNA and measuring expression of GFP in our March-I v2 promoter-GFP constructs. Both LPS and CpG down-regulated expression of the March-I v2 (−663 to +125), March-I v2 (−385 to +125), and March-I v2 (−175 to +125) promoter constructs (Fig. 4A), demonstrating that these promoter constructs contained sequences that respond to DC activation signals. Curiously, GFP expression was not altered by LPS or CpG treatment of cells expressing the March-I v2 (−663 to +2) promoter construct. Monitoring endogenous March-I v2 expression in these transduced cells confirmed that the cells responded to these activation stimuli like non-transduced cells (Fig. 4B). These data reveal a role for sequences present in March-I v2 exon 7 in the response of the March-I v2 promoter to DC activation by LPS or CpG.
Figure 4.
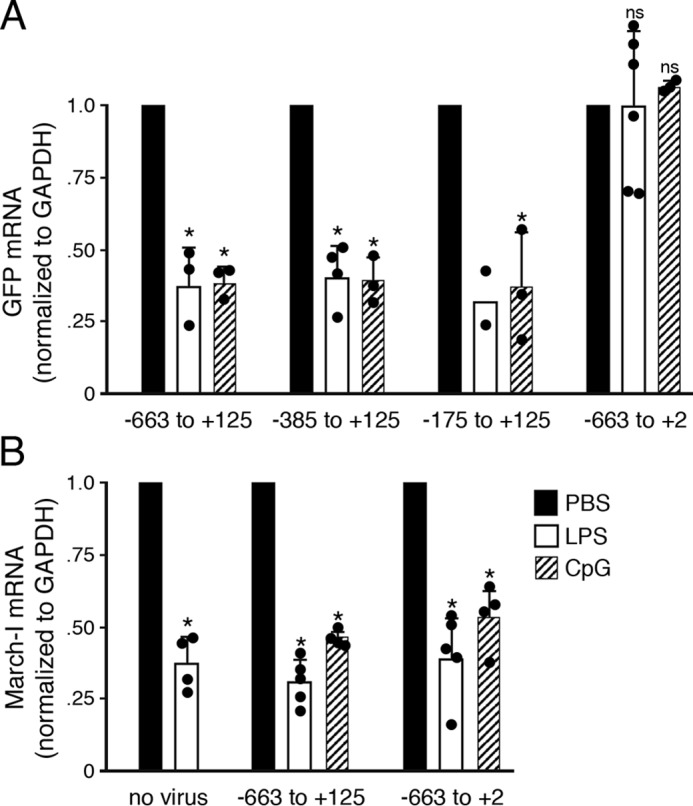
The LPS- and CpG-responsive element in the March-I v2 promoter resides in exon 7. DCs were left untreated or were transduced with lentivirus encoding the indicated promoter construct and incubated in the presence of PBS (as a control), LPS, or CpG for 2 h. Cells were harvested, and expression of endogenous March-I v2 or GFP mRNA and GAPDH was determined by RT-PCR. GFP mRNA expression (A) or endogenous March-I v2 expression (B) in each sample was normalized to the amount of GAPDH mRNA present in the sample and expressed relative to the amount of mRNA detected in the PBS-treated control cells for each lentiviral construct as described under “Experimental procedures.” March-I RING domain primers were used to detect endogenous March-I. The results shown are the average (error bars represent S.D.) of multiple independent experiments. *, p < 0.05; ns, not significant.
The March-I v2 promoter is only active in APCs
March-I v2 is expressed primarily in APCs (14, 15), so we set out to determine whether the March-I v2 (−663 to +125) promoter construct we identified resulted in cell type–specific expression of our GFP reporter. Whereas we could reliably detect CMV promoter–driven expression of GFP in epithelial cells, kidney cells, and fibroblasts, we were unable to detect any activity of the March-I v2 promoter construct in these cells types above background levels (Fig. 5). By contrast, both the CMV and March-I v2 promoter constructs resulted in expression of March-I v2 mRNA in DCs, although the March-I v2 promoter was significantly weaker than the CMV promoter. The March-I v2 promoter construct was also expressed well in spleen B cells. (Human CMV promoter activity in lentivirus is weak in mouse B cells (30), and for this reason March-I v2 promoter activity seems high as compared with CMV promoter activity.) Although it is not possible to examine March-I v2 promoter activity in every cell type in the mouse, these data strongly suggest that the March-I v2 promoter identified in this study contains the DNA elements necessary for cell-specific expression of March-I v2 in APCs.
Figure 5.
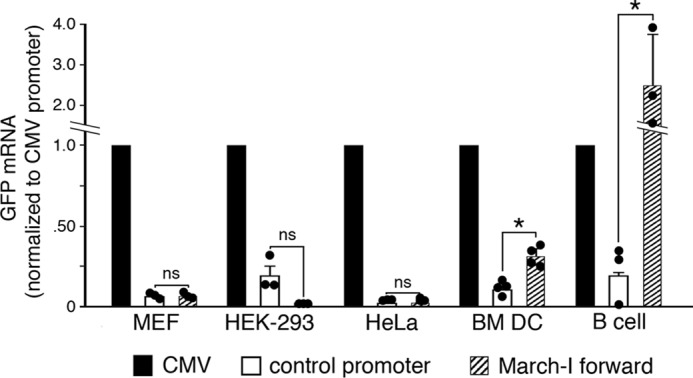
The March-I v2 promoter is inactive in non-APCs. Epithelial (HeLa) cells, kidney (HEK-293) cells, mouse embryonic fibroblasts, and BM DCs were transduced with lentivirus encoding GFP using the CMV promoter, the March-I v2 (−663 to +125) “forward” promoter, or a negative “control promoter.” Expression of GFP, puromycin, and GAPDH mRNA was determined by RT-PCR. Data using HEK-293 cells, HeLa cells, BM DCs, and spleen B cells were normalized to the amount of puromycin mRNA present in the sample, and the control was the promoterless lentivirus. Data using MEFs were normalized to the amount of GAPDH mRNA present in the sample, and the control was lentivirus with the March-I v2 (−663 to +125) promoter inserted into the expression vector in the backward orientation. In each sample, the amount of GPF mRNA was expressed relative to the amount of mRNA detected using the CMV promoter construct (arbitrarily designated a value of 1). Control experiments using RNA from MEFs, HEK-293 cells, HeLa cells, or BM DCs that were not reverse transcribed revealed negligible contamination of cDNA with host cell genomic DNA or lentivirus DNA. Spleen B-cell cDNA did contain contaminating lentivirus DNA, and therefore RT-PCR data were calculated as 2ΔCt (ΔCt = Ctplus RT − Ctminus RT). The results shown are the average (error bars represent S.D.) of three independent experiments. The expression of GFP driven by the March-I v2 promoter in each cell type was compared with that using the control promoter. *, p < 0.05; ns, not significant.
In this report, we show that, of the isoforms of March-I annotated in Vega and Ensembl databases, only March-I v2 is expressed in DCs and B cells. Whereas other March-I variants are expressed in brain, RT-PCR demonstrated that the overall expression of March-I in brain is quite low as compared with spleen DCs. Having said that, the amount of March-I v2 mRNA expressed in spleen DCs is also quite small as we have quantitated the amount of March-I v2 mRNA present in resting spleen DCs (using plasmid DNA as a standard) and found that in three independent spleen DC samples there is ∼1 molecule of March-I v2 mRNA per cell.6 Such a finding is consistent with the nearly undetectable expression of endogenous March-I protein in APCs reported by others (8, 12, 23).
Because March-I v2 cannot, by definition, be the product of alternative splicing, we reasoned that nucleotide sequences regulating expression of March-I in APCs would be present upstream of the variant 2 TSS. Within only a few hundred bp of the TSS were intragenic promoter sequences that could drive expression of a GFP reporter in DCs but not in kidney cells, epithelial cells, or fibroblasts, suggesting that this region of the March-I v2 promoter contained the sequences necessary for cell type–specific expression of March-I.
DC activation by a variety of TLR ligands leads to down-regulation of March-I expression (12, 13). Analysis of March-I v2 mRNA half-life in DCs demonstrated that DC activation did not destabilize existing March-I v2 mRNA, suggesting there are regulatory sequences present in the March-I v2 promoter that directly regulate March-I v2 mRNA expression. Like endogenous March-I v2, expression of March-I v2 promoter-GFP reporter constructs was also down-regulated in LPS- and CpG-activated DCs. The sequences responsible for down-regulation of March-I v2 promoter activity were present in March-I exon 7. Regulatory regions are often present in exons of genes (28), and exon 7 contains numerous DNA elements, including downstream control elements and downstream promoter elements (28) that could be responsible for the regulation of March-I v2 expression in APCs. Future studies using mutant mice lacking these regulatory regions could reveal the importance of terminating March-I v2 expression in APCs and how dysregulation of March-I v2 expression affects immunity.
Experimental procedures
Cell isolation and culture
C57BL/6 mice were bred and maintained in house at the NCI-Frederick animal facility. All mice were cared for in accordance with National Institutes of Health guidelines and approved by the National Cancer Institute Animal Care and Use Committee. HEK-293 cells, HeLa cells, and mouse embryonic fibroblasts (MEFs) were cultured in DMEM containing 10% fetal bovine serum and 10 mm Hepes, pH 7.4. Cells were subcultured every 2nd or 3rd day and maintained at subconfluent levels. DCs and B cells from mouse spleens were purified by positive selection or negative selection, respectively, using Miltenyi Biotec kits. BM DCs were generated by differentiating mouse bone marrow cells in BM DC medium (RPMI 1640 medium containing 10% fetal bovine serum, 50 μm β-mercaptoethanol, 10 mm Hepes, pH 7.4, 25 μg/ml gentamycin, and 20 ng/ml GM-CSF) as described previously (29).
Lentiviral promoter constructs
The various DNA fragments spanning the putative March-I v2 promoter were cloned into the promoterless lentiviral plasmid pLV-unsGFP-PGK-Puro (Cellomics). This plasmid encodes destabilized GFP (whose expression is regulated by an experimentally determined promoter) as well as puromycin (whose expression is regulated by the phosphoglycerate kinase promoter). The bp numbering for each promoter construct is shown relative to the March-I v2 transcription start site identified by 5′ RACE (as described in this report). LV-102 (−1064 to +125), LV-115 (−663 to +125), LV-309 (−317 to +125), and LV-314 (−175 to +125) were constructed first by generating the PCR fragments using C57BL/6 mouse genomic DNA as template and different forward primers, BIG-S-F, SM-S-F, SMSF-400, and SMSF-300, respectively, and the common reverse primer P4S-R. The antisense construct LV-123 (+125 to −663) was constructed in a similar fashion by amplifying the DNA fragment using the forward primer SM-AS-F and the reverse primer P4AS-R. The resultant PCR fragment was digested with BamHI and XhoI and cloned into pLV-unsGFP-PGK-Puro. LV-26 (−385 to +125) and LV-27 (+125 to −385) were constructed by first PCR-amplifying a 758-bp fragment using PR4-F3 and PR4-R1 as primers and mouse genomic DNA as template and then cloning the PCR product in TA cloning vector pCR4-TOPO (Invitrogen). This plasmid was digested with EcoRI, and a 513-bp EcoRI fragment from this digestion was cloned into pLV-unsGFP-PGK-Puro. LV-306 (−663 to −71) and LV-665 (−663 to +7) were constructed using the common forward primer SM-S-F and reverse primers P4SR-120 and P4SR-ATG, respectively. The resultant PCR fragments were digested with XhoI and BamHI and cloned in these sites in pLV-unsGFP-PGK-Puro. The nucleotide sequences of all March-I promoter lentiviral constructs were confirmed by DNA sequence analysis.
Promoter construct primer sequences
The promoter construct primer sequences used were: BIG-S-F, gagagactcgagccacattttggacaccagcgttttgag; SM-S-F, gagagactcgaggccacaggataagtccaatagtg; SMSF-400, gagagactcgaggggccttgttgctaactcaaggtg; SMSF-300, gagagactcgagcccaggcaaattgagctattccac; P4S-R, gagagaggatcccaagttagataatttggcatcttg; SM-AS-F, gagagaggatccgccacaggataagtccaatagtg; P4AS-R, gagagactcgagcaagttagataatttggcatcttg; PR1-F3, ggtataacatggcttctac; PR4-R1, caagttagataatttggcatc; P4SR-120, gagagaggatcctgcactgccctgctgttgcttcac; P4SR-ATG, gagagaggatcctgaatctgggaagtcagggggcttg.
RT-PCR primer sequences
The oligonucleotides used for measuring relative transcript levels in this study were: GFP, 5′-caacagccacaacgtctatatcat-3′ and 5′atgttgtggcggatcttgaag-3′; puromycin, 5′-gttcgccgactaccccg-3′ and 5′-agagttcttgcagctcggtg; and March-I RING domain, 5′-aagagagcccactcatcacacc-3′ and 5′-atctggagcttttcccacttcc-3′. The oligonucleotides used selectively in the RT-PCR assays for March-I variants were: variant 1/4, 5′-gatgtggccgatgcctctcaa-3′ and 5′-tcgttgggctgcttgcttttg-3′; variant 2, 5′-acttgtttctccaggcaagc-3′ and 5′-agtgggctctcttcatctcct-3′; and variant 3, 5′-gcaatgggagaagttcaatggg-3′ and 5′-gttgggctgcttgcttttgata-3′.
Generation of lentiviral stocks
Lentiviral stocks were generated by transfecting subconfluent HEK-293T cells with lentiviral promoter plasmid, EZ-LentiPACK Packaging plasmid, and EZ-Transfx reagent as described in the EZ-LentiPACK Packaging System kit (Cellomics Technology). Lentiviral supernatants were harvested 48 and 72 h after transfection. Viral supernatants were centrifuged briefly (500 × g, 10 min) to remove cellular debris. The approximate titer of viral supernatants was quantitated using Lentivirus Titration XpressCards (Cellomics Technology).
Lentiviral transduction
Lentivirus (10–100 μl of HEK-293 viral supernatant) was added to bone marrow cultures on day 2 of culture in BM DC medium containing 10 μg/ml protamine sulfate (Sigma). BM DC medium was changed every other day. Day 7 BM DC cultures were cultured in the presence or absence of 1 μg/ml LPS (Sigma) and 2 μg/ml CpG-1668 (Invivogen) for 2 h. BM DCs were then washed and harvested at 4 °C, and the cell pellet was immediately resuspended in TRIzol (Ambion) and stored at −80 °C.
Primary spleen B cells were isolated by negative selection using Miltenyi Biotec kits and transduced with lentivirus by spinoculation. Lentivirus (250 μl of HEK-293 viral supernatant) was added to 8 × 106 B cells in 2 ml of RPMI 1640 medium containing 10% fetal bovine serum, non-essential amino acids, the B cell–activating factor BAFF (10 ng/ml), 10 mm Hepes, pH 7.4, and 10 μg/ml Polybrene (Sigma) in a tissue culture plate. After 15-min incubation at room temperature, the cultures were subjected to centrifugation at 1100 × g for 90 min and then placed in a 37 °C, 5% CO2 incubator. After 2 days, cells were harvested at 4 °C and washed, and the cell pellet was immediately resuspended in TRIzol (Ambion) and stored at −80 °C.
Adherent MEFs, HEK-293 cells, and HeLa cells were cultured in RPMI 1640 medium containing 10% fetal bovine serum and 10 mm Hepes, pH 7.4. Cells were transduced using 10–100 μl of HEK-293 viral supernatant in the above medium containing 10 μg/ml protamine sulfate, and the medium was changed after 1 day. The next day cells were harvested at 4 °C and washed, and the cell pellet was immediately resuspended in TRIzol (Ambion) and stored at −80 °C.
RNA Isolation, cDNA synthesis, and RT-PCR
Total RNA was isolated from the TRIzol suspensions according to the manufacturer's instructions. cDNA was synthesized using 1–2 μg of total RNA and the Superscript III First-Strand Synthesis kit (Invitrogen). RT-PCR was performed using a QuantiStudio6 Flex sequence detection system (Applied Biosystems) and the Quantitect SYBR Green PCR kit (Qiagen).
Quantitation of March-I v2 promoter activity
In experiments monitoring expression of March-I v2 promoter activity in lentivirus-transduced BM DCs, RT-PCR was used to quantitate the amount of GFP and puromycin mRNA present in the sample. Ct values for GFP and puromycin were determined, and data are shown as 2ΔCt (ΔCt = Ctpuromycin − CtGFP). 2ΔCt for the promoterless (blank) vector control was arbitrarily set at 1.0, and all values are expressed relative to the blank vector control.
In experiments monitoring regulation of March-I v2 promoter activity in transduced BM DCs treated with either PBS, LPS, or CpG, RT-PCR was used to quantitate the amount of GFP and GAPDH mRNA present in the sample. Ct values for GFP and GAPDH were determined, and data are shown as 2ΔCt (ΔCt = CtGAPDH − CtGFP). In experiments using the March-I v2 (−663 to +125) and March-I v2 (−663 to +2) sense constructs, the control virus was a March-I v2 (+125 to −663) antisense construct, and in experiments using the March-I v2 (−385 to +125) and March-I v2 (−175 to +125) sense constructs, the control virus was a March-I v2 (+125 to −385) antisense construct. The data shown are 2ΔCt for the sense construct/2ΔCt for the antisense construct. The amount of GFP mRNA present after treatment of each sample with LPS or CpG was expressed as a fraction of that present in the PBS-treated control sample.
Bioinformatics
Human and mouse March-I variant annotations are described in Ensembl Genome Browser 90 (http://ensembl.org)7 and Vega Genome Browser Release 68 (http://vega.archive.ensembl.org).7 The entire 10-exon mouse March-I gene is located on chromosome 8 between bp 65,618,086 and 66,471,637 (Vega Genome Browser numbering). The Vega Genome Browser transcript identifiers for mouse March-I are OTTMUST00000040256 (variant 1), OTTMUST00000040267 (variant 2), OTTMUST00000040269 (variant 3), and OTTMUST00000040271 (variant 4). The Vega Genome Browser transcript identifiers for human March-I are ENST00000503008 (variant 1) and ENST00000339875 (variant 2). The mouse EST database in GenBank (NCBI) was screened using a Basic Local Alignment Search Tool (BLAST) search of nucleotide sequences present in mouse March-I exon 7 that are not present in the 5′-truncated forms of exon 7 present in March-I variants 1, 3, and 4.
Statistical analyses
Results were analyzed using two-tailed Student's t tests. p values <0.05 were considered statistically significant. p values are indicated by * for p < 0.05, and non-significant differences are indicated by ns.
Author contributions
S. K. and S. K. M. data curation; S. K. and S. K. M. formal analysis; S. K. investigation; S. K. and P. A. R. writing-original draft; S. K., S. K. M., and P. A. R. writing-review and editing; S. K. M. and P. A. R. conceptualization; P. A. R. supervision; P. A. R. validation; P. A. R. project administration.
This work was supported by the Intramural Research Program of the National Institutes of Health. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
S. K. Mittal, unpublished observation.
S. Kaul, unpublished observation.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party-hosted site.
- APC
- antigen-presenting cell
- DC
- dendritic cell
- March
- membrane-associated RING-CH
- March-I v2
- March-I variant 2
- MEF
- mouse embryonic fibroblast
- puromycin
- puromycin N-acetyltransferase
- TCR
- T-cell receptor
- TSS
- transcription start site(s)
- pMHC-II
- peptide–MHC-II
- TLR
- toll-like receptor
- EST
- expressed sequence tag
- BM DC
- bone marrow–derived DC
- RACE
- rapid amplification of cDNA ends.
References
- 1. Roche P. A., and Furuta K. (2015) The ins and outs of MHC class II-mediated antigen processing and presentation. Nat. Rev. Immunol. 15, 203–216 10.1038/nri3818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Turley S. J., Inaba K., Garrett W. S., Ebersold M., Unternaehrer J., Steinman R. M., and Mellman I. (2000) Transport of peptide-MHC class II complexes in developing dendritic cells. Science 288, 522–527 10.1126/science.288.5465.522 [DOI] [PubMed] [Google Scholar]
- 3. Inaba K., Turley S., Iyoda T., Yamaide F., Shimoyama S., Reis e Sousa C., Germain R. N., Mellman I., and Steinman R. M. (2000) The formation of immunogenic major histocompatibility complex class II-peptide ligands in lysosomal compartments of dendritic cells is regulated by inflammatory stimuli. J. Exp. Med. 191, 927–936 10.1084/jem.191.6.927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wilson N. S., El-Sukkari D., and Villadangos J. A. (2004) Dendritic cells constitutively present self antigens in their immature state in vivo and regulate antigen presentation by controlling the rates of MHC class II synthesis and endocytosis. Blood 103, 2187–2195 10.1182/blood-2003-08-2729 [DOI] [PubMed] [Google Scholar]
- 5. Iezzi G., Karjalainen K., and Lanzavecchia A. (1998) The duration of antigenic stimulation determines the fate of naive and effector T cells. Immunity 8, 89–95 10.1016/S1074-7613(00)80461-6 [DOI] [PubMed] [Google Scholar]
- 6. Cella M., Engering A., Pinet V., Pieters J., and Lanzavecchia A. (1997) Inflammatory stimuli induce accumulation of MHC class II complexes on dendritic cells. Nature 388, 782–787 10.1038/42030 [DOI] [PubMed] [Google Scholar]
- 7. Pierre P., Turley S. J., Gatti E., Hull M., Meltzer J., Mirza A., Inaba K., Steinman R. M., and Mellman I. (1997) Developmental regulation of MHC class II transport in mouse dendritic cells. Nature 388, 787–792 10.1038/42039 [DOI] [PubMed] [Google Scholar]
- 8. Matsuki Y., Ohmura-Hoshino M., Goto E., Aoki M., Mito-Yoshida M., Uematsu M., Hasegawa T., Koseki H., Ohara O., Nakayama M., Toyooka K., Matsuoka K., Hotta H., Yamamoto A., and Ishido S. (2007) Novel regulation of MHC class II function in B cells. EMBO J. 26, 846–854 10.1038/sj.emboj.7601556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Furuta K., Walseng E., and Roche P. A. (2013) Internalizing MHC class II-peptide complexes are ubiquitinated in early endosomes and targeted for lysosomal degradation. Proc. Natl. Acad. Sci. U.S.A. 110, 20188–20193 10.1073/pnas.1312994110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bannard O., McGowan S. J., Ersching J., Ishido S., Victora G. D., Shin J. S., and Cyster J. G. (2016) Ubiquitin-mediated fluctuations in MHC class II facilitate efficient germinal center B cell responses. J. Exp. Med. 213, 993–1009 10.1084/jem.20151682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Oh J., Wu N., Baravalle G., Cohn B., Ma J., Lo B., Mellman I., Ishido S., Anderson M., and Shin J. S. (2013) MARCH1-mediated MHCII ubiquitination promotes dendritic cell selection of natural regulatory T cells. J. Exp. Med. 210, 1069–1077 10.1084/jem.20122695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. De Gassart A., Camosseto V., Thibodeau J., Ceppi M., Catalan N., Pierre P., and Gatti E. (2008) MHC class II stabilization at the surface of human dendritic cells is the result of maturation-dependent MARCH I down-regulation. Proc. Natl. Acad. Sci. U.S.A. 105, 3491–3496 10.1073/pnas.0708874105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Walseng E., Furuta K., Goldszmid R. S., Weih K. A., Sher A., and Roche P. A. (2010) Dendritic cell activation prevents MHC class II ubiquitination and promotes MHC class II survival regardless of the activation stimulus. J. Biol. Chem. 285, 41749–41754 10.1074/jbc.M110.157586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu H., Jain R., Guan J., Vuong V., Ishido S., La Gruta N. L., Gray D. H., Villadangos J. A., and Mintern J. D. (2016) Ubiquitin ligase MARCH 8 cooperates with CD83 to control surface MHC II expression in thymic epithelium and CD4 T cell selection. J. Exp. Med. 213, 1695–1703 10.1084/jem.20160312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. von Rohrscheidt J., Petrozziello E., Nedjic J., Federle C., Krzyzak L., Ploegh H. L., Ishido S., Steinkasserer A., and Klein L. (2016) Thymic CD4 T cell selection requires attenuation of March8-mediated MHCII turnover in cortical epithelial cells through CD83. J. Exp. Med. 213, 1685–1694 10.1084/jem.20160316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thibodeau J., Bourgeois-Daigneault M. C., Huppé G., Tremblay J., Aumont A., Houde M., Bartee E., Brunet A., Gauvreau M. E., de Gassart A., Gatti E., Baril M., Cloutier M., Bontron S., Früh K., et al. (2008) Interleukin-10-induced MARCH1 mediates intracellular sequestration of MHC class II in monocytes. Eur. J. Immunol. 38, 1225–1230 10.1002/eji.200737902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tze L. E., Horikawa K., Domaschenz H., Howard D. R., Roots C. M., Rigby R. J., Way D. A., Ohmura-Hoshino M., Ishido S., Andoniou C. E., Degli-Esposti M. A., and Goodnow C. C. (2011) CD83 increases MHC II and CD86 on dendritic cells by opposing IL-10-driven MARCH1-mediated ubiquitination and degradation. J. Exp. Med. 208, 149–165 10.1084/jem.20092203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hunt D., Wilson J. E., Weih K. A., Ishido S., Harton J. A., Roche P. A., and Drake J. R. (2012) Francisella tularensis elicits IL-10 via a PGE2-inducible factor, to drive macrophage MARCH1 expression and class II down-regulation. PLoS One 7, e37330 10.1371/journal.pone.0037330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mittal S. K., Cho K. J., Ishido S., and Roche P. A. (2015) Interleukin 10 (IL-10)-mediated immunosuppression: March-I induction regulates antigen presentation by macrophages but not dendritic cells. J. Biol. Chem. 290, 27158–27167 10.1074/jbc.M115.682708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bartee E., Mansouri M., Hovey Nerenberg B. T., Gouveia K., and Früh K. (2004) Downregulation of major histocompatibility complex class I by human ubiquitin ligases related to viral immune evasion proteins. J. Virol. 78, 1109–1120 10.1128/JVI.78.3.1109-1120.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Baravalle G., Park H., McSweeney M., Ohmura-Hoshino M., Matsuki Y., Ishido S., and Shin J. S. (2011) Ubiquitination of CD86 is a key mechanism in regulating antigen presentation by dendritic cells. J. Immunol. 187, 2966–2973 10.4049/jimmunol.1101643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ohmura-Hoshino M., Matsuki Y., Mito-Yoshida M., Goto E., Aoki-Kawasumi M., Nakayama M., Ohara O., and Ishido S. (2009) Cutting edge: requirement of MARCH-I-mediated MHC II ubiquitination for the maintenance of conventional dendritic cells. J. Immunol. 183, 6893–6897 10.4049/jimmunol.0902178 [DOI] [PubMed] [Google Scholar]
- 23. Jabbour M., Campbell E. M., Fares H., and Lybarger L. (2009) Discrete domains of MARCH1 mediate its localization, functional interactions, and posttranscriptional control of expression. J. Immunol. 183, 6500–6512 10.4049/jimmunol.0901521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ashurst J. L., Chen C. K., Gilbert J. G., Jekosch K., Keenan S., Meidl P., Searle S. M., Stalker J., Storey R., Trevanion S., Wilming L., and Hubbard T. (2005) The vertebrate genome annotation (Vega) database. Nucleic Acids Res. 33, D459–D465 10.1093/nar/gki135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aken B. L., Ayling S., Barrell D., Clarke L., Curwen V., Fairley S., Fernandez Banet J., Billis K., García Girón C., Hourlier T., Howe K., Kähäri A., Kokocinski F., Martin F. J., Murphy D. N., et al. (2016) The Ensembl gene annotation system. Database 2016, baw093 10.1093/database/baw093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. De Conti L., Baralle M., and Buratti E. (2013) Exon and intron definition in pre-mRNA splicing. Wiley Interdiscip. Rev. RNA 4, 49–60 10.1002/wrna.1140 [DOI] [PubMed] [Google Scholar]
- 27. Aivasashvilli V. A., and Beabealashvilli R. S. (1983) Sequence-specific inhibition of RNA elongation by actinomycin D. FEBS Lett. 160, 124–128 10.1016/0014-5793(83)80950-8 [DOI] [PubMed] [Google Scholar]
- 28. Roy A. L., and Singer D. S. (2015) Core promoters in transcription: old problem, new insights. Trends Biochem. Sci. 40, 165–171 10.1016/j.tibs.2015.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cho K. J., Walseng E., Ishido S., and Roche P. A. (2015) Ubiquitination by March-I prevents MHC class II recycling and promotes MHC class II turnover in antigen-presenting cells. Proc. Natl. Acad. Sci. U.S.A. 112, 10449–10454 10.1073/pnas.1507981112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lutzko C., Senadheera D., Skelton D., Petersen D., and Kohn D. B. (2003) Lentivirus vectors incorporating the immunoglobulin heavy chain enhancer and matrix attachment regions provide position-independent expression in B lymphocytes. J. Virol. 77, 7341–7351 10.1128/JVI.77.13.7341-7351.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]