Abstract
Kinesin-2s are major transporters of cellular cargoes. This subfamily contains both homodimeric kinesins whose catalytic domains result from the same gene product and heterodimeric kinesins with motor domains derived from two different gene products. In this Minireview, we focus on the progress to define the biochemical and biophysical properties of the kinesin-2 family members. Our understanding of their mechanochemical capabilities has been advanced by the ability to identify the kinesin-2 genes in multiple species, expression and purification of these motors for single-molecule and ensemble assays, and development of new technologies enabling quantitative measurements of kinesin activity with greater sensitivity.
Keywords: microtubule, processivity, intracellular trafficking, ATPase, single-molecule biophysics, pre-steady-state kinetics, cilia, cytoskeleton, molecular motor, motor protein, neuron, tubulin
Introduction
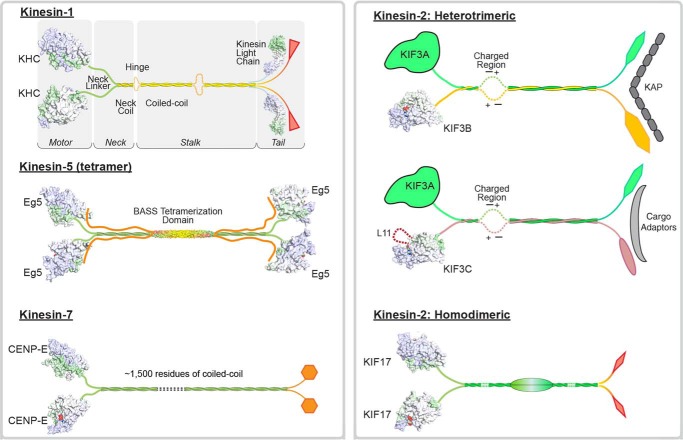
Kinesins constitute a superfamily of microtubule-based molecular motor enzymes that couple the chemical energy from ATP turnover to force production for diverse cellular functions (1–12). Kinesins are classified into 15 different subfamilies, yet they share a structurally conserved kinesin motor domain (1, 3, 13–16). However, key amino acid residue changes can confer unique mechanochemical properties to each kinesin, which in turn specify cellular function. The N-terminal kinesins are composed of an N-terminal motor domain connected to a long α-helical region that dimerizes into a coiled-coil stalk that ends with a C-terminal domain that may interact with specific adaptor proteins for cargo linkage (Fig. 1). N-kinesin subfamilies include conventional kinesin-1, kinesin-2, kinesin-3, kinesin-5 Eg5/KSP, and kinesin-7 CENP-E, and all are best known for their roles in intracellular transport.
Figure 1.
Molecular organization of kinesin-1, -2, -5, and -7 processive motors. These processive kinesins all contain two molecular motor domains, although the molecular organization of the remaining and associated polypeptide chains differs within and between kinesin subfamilies. The depictions shown here include representative space-filling models for domains whose three-dimensional structures are known and cartoons for those segments whose structures are yet to be determined. The lengths for the coiled-coiled and globular domains, whose structures have not been defined, are not drawn to scale. The X-ray coordinates used to generate this figure include the motor domains for kinesin-1, 3KIN; Eg5, 4PXU; CENP-E, 1T5C; KIF3B, 3B6U; KIF3C, 3B6V; and KIF17, 2VVG. The coordinates for the Eg5 BASS tetramerization domain and kinesin-1 light chain are 4PXU and 3CEQ, respectively.
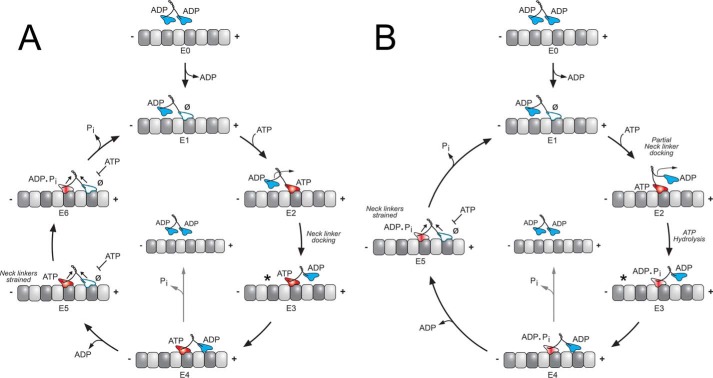
N-kinesins carry cargo directionally toward the plus-end of microtubules, which are polymerized from αβ-tubulin subunits to form a cylindrical polymer of 13 protofilaments. The kinesins are able to “read” the polarity of the microtubule because of the structural asymmetry of αβ-tubulin subunits. The movement of N-kinesins is designated as “processive,” which implies that upon microtubule collision, a single dimeric kinesin steps continuously toward the microtubule plus-end in an asymmetric hand-over-hand manner hydrolyzing one ATP per 8-nm step for hundreds of steps (17–22). The 8-nm step size results from the distance between adjacent αβ-tubulin dimers along the microtubule lattice. As Fig. 2 illustrates, a processive kinesin binds the microtubule and then goes through a series of structural transitions, each modulated by nucleotide state. To maintain a processive run with continuous stepping, the ATPase cycle of each head remains out-of-phase with the other to avoid premature release if both heads exist in a microtubule weak binding state simultaneously. The degree of processivity, quantified by “run length,” varies between kinesin subfamilies and is regulated by a series of “gating” mechanisms in which a chemical and/or mechanical requirement must be satisfied to proceed forward. There has been significant effort to define the determinants of processivity structurally and mechanistically. The framework has been shaped by work on kinesin-1 (recent advances include Refs. 23–34) and applied to kinesin-2.
Figure 2.
KIF3 stepping models. Two variations of a kinesin stepping cycle are presented: a front-head–gated model (A), and a revised rear-head–gated model (B). Each cycle begins as one motor head collides with the microtubule; ADP is released, and the asymmetry of the ATPase cycle on each motor domain is established (E0–E1). The E1 intermediate is tightly bound to the microtubule with its leading head nucleotide-free and the trailing head detached as the weak-binding ADP state. A, ATP binds to the leading head and generates a structural transition transmitted through the neck-linker motif (E2–E3). The ATP-induced structural transition is designated neck-linker docking (*) and shifts the lagging unbound head forward by 16 nm to the next microtubule-binding site toward the microtubule plus-end (E2–E4). ADP is subsequently released (E4–E5). Both heads are bound to the microtubule with the leading head now nucleotide-free and tightly bound to the microtubule. ATP hydrolysis on the rear head (E5) results in another series of structural transitions in which phosphate is released; the trailing head transitions into a weakly bound ADP state and detaches from the microtubule to form the E1 intermediate. The first 8-nm step of the cycle is coupled to one ATP turnover and positions the new leading head to begin the second step of the processive run waiting for ATP (E1). The front-head–gated model (A) proposes that ATP binding promotes neck-linker docking that is coupled with a structural step (E2–E5) and that in the two-head bound state (E5), and ATP binding on the leading head is inhibited. In contrast, the revised rear-head–gated model (B) proposes that ATP binding partially docks the neck linker onto the catalytic core but posits that ATP hydrolysis (E2–E4) occurs while the tethered head is in its diffusional search for its microtubule-binding site with ATP hydrolysis required to completely dock the neck linker. The rear-head–gated model proposes that the E3–E4 intermediate is in a kinetic race for the front head to bind tightly to the microtubule before phosphate is released on the rear head (E4–E5).
Kinesin-2 subfamily
Kinesin-2 was initially discovered in sea urchin eggs (35, 36). The purified protein promoted microtubule plus-end–directed microtubule gliding in motility assays. Yet, unlike kinesin-1, this kinesin was heterotrimeric with two different motor polypeptides and a non-motor accessory protein designated kinesin-associated polypeptide (KAP).3 Soon thereafter, heterodimeric and heterotrimeric kinesins were identified in multiple eukaryotic species, and like kinesin-1 they were associated with long distance cargo transport (3–7, 11).
In mammals there are four kinesin-2 genes: KIF3A, KIF3B, KIF3C, and KIF17. KIF17 as well as its Caenorhabditis elegans homolog OSM-3 form homodimers that function as fast, highly processive motors and operate in multiple cell types, including neurons (5, 6, 37–40). In contrast, KIF3A associates with either KIF3B or KIF3C to form heterodimeric KIF3AB and KIF3AC motors (41–45). KIF3B does not form heterodimers with KIF3C (45–47). Moreover, multiple studies showed that heterodimerization was preferential over homodimer formation (43, 46, 48, 49), although there is evidence for an injury-specific homodimer of KIF3CC in neurons (45, 47, 50, 51).
KIF3AB forms a heterotrimeric complex by association with KAP (Fig. 1) (36, 52–54). KAP is a distinctive adaptor largely composed of armadillo repeats (55, 56), and it is these motifs that provide specificity of the interaction between KIF3AB and KAP and between KIF3AB–KAP and its cargo. Note also that KIF3A, KIF3B, and KAP are all essential genes (57–62). Knockout mice for KIF3A or KIF3B show the absence of nodal cilia that are crucial for proper mesodermal patterning during embryogenesis and thus have a randomized left–right body axis (57–59). Other studies have linked KIF3AB–KAP to cilia-dependent signal transduction pathways, including the Hedgehog-signaling pathway (63, 64). KIF3AB–KAP transports multimeric protein complexes (designated IFT particles) into the cilium, and its transport role for ciliogenesis is considered the reason that KIF3AB–KAP is essential for development. It also appears to be the basis of similarity between KIF3AB–KAP and other kinesin-2 heterotrimeric orthologs.
In contrast, there is not strong experimental evidence that mammalian KIF3AC binds KAP (5, 52, 55, 65). Another observation to rule out an association of KAP with KIF3AC is the lack of sequence similarity of KIF3C at the putative KAP-binding region of KIF3AB. Note also that a universal KIF3AC adaptor for cargo linkage has not yet been identified.
KIF3C exhibits a signature motif that is conserved in mammals, a 25-residue insert in loop L11 of the catalytic motor domain enriched in glycines and serines (Fig. 3) (38, 44, 66, 67). When this insert was deleted from loop L11 of KIF3C (designated KIF3CΔL11), the run length of KIF3ACΔL11 increased from 1.23 μm for KIF3AC to 1.55 μm and therefore was similar to the run length of KIF3AB at 1.62 μm. These results suggested that this motif regulated processivity (67). The KIF3C L11 insert was also implicated in the ability of homodimeric KIF3CC to be targeted to microtubule plus-ends and act as a potent catastrophe factor to promote microtubule dynamics. KIF3CCΔL11 lost the ability to promote catastrophe, suggesting that the loop L11 extension, unique to KIF3C, is a structural motif required for regulation of microtubule dynamics by KIF3CC (51).
Figure 3.
Species-specific neck-linker length analysis and KIF3C loop L11 sequence motif. A, neck-linker sequence comparison for processive kinesins and neck-linker length predictions based on structural analysis (80). B, species-specific alignment of loop L11 sequences between KIF3A, KIF3B, and KIF3C in comparison with other processive kinesins. Red sequence represents the extended KIF3C-specific residues.
Yet, despite similarities in sequence and structure, KIF3AC appears to function specifically in neurons, whereas KIF3AB–KAP and KIF17 appear more ubiquitously expressed (4, 5, 12, 38). This Minireview will focus specifically on the properties of KIF3AC and KIF3AB.
Neck linker hypothesis for kinesin-2 processivity
The neck-linker domain was identified for kinesin-1 as the critical determinant of processivity (23, 68–71). Based on these studies, it was proposed that a longer or shorter neck linker affected the communication and therefore the coordination between the motor domains. The impact observed by elongating the neck linker was a decrease in run length because of the higher probability of both motor heads reaching the ADP weak binding state at the same time. Initial studies with Xenopus laevis kinesin-2 Xklp3A/3B revealed that it was not as processive as kinesin-1 and detached from the microtubule at low hindering loads rather than stall (72–75). The next question ahead was to understand mechanistically and structurally why kinesin-2 was sensitive to force resulting in shorter run lengths and why it was so different from kinesin-1.
A series of publications were released beginning in 2009 that explored the single-molecule behavior of mammalian KIF3AB to define the mechanistic basis of the shorter run lengths observed for Xklp3A/3B (76–78). The authors proposed that the shortened run length observed for KIF3AB was due in part to its longer neck linker (Fig. 3A). In comparison with kinesin-1, the neck linker of KIF3AB appeared to be extended by three residues (Asp-Ala-Leu, DAL) at its C terminus and prior to helix α7 (Fig. 3A) (76–78). Initial studies showed that the run length of homodimeric Drosophila kinesin-1 was 1.76 μm, yet the run length of kinesin-2 KIF3AB was 0.45 μm consistent with this hypothesis (76). Furthermore, when the neck linker of kinesin-1 was engineered and extended by the DAL motif of kinesin-2 (Fig. 3A), the kinesin-1 + DAL run length decreased to 0.35 μm, thus providing additional evidence for the hypothesis that an extended neck linker shortened the run length observed for kinesin-2.
As an extension of these studies, Shastry and Hancock (78) examined other N-terminal processive kinesins, including kinesin-3 (C. elegans Unc104), kinesin-5 (X. laevis Eg5/KSP), and kinesin-7 (X. laevis CENP-E) in comparison with Drosophila kinesin-1 and murine KIF3AB (78). Based on the sequence alignment of helix α6, the neck-linker peptide, and helix α7, kinesin-1 exhibited a 14-residue neck linker, kinesin-2, and kinesin-3 each with 17-residue neck linkers, and kinesin-5 and kinesin-7 each with 18-residue neck-linker domains (Fig. 3A). For these studies, a similar motor design approach was used in which the kinesin-specific motor domain with neck linker was fused to the neck-coil beginning at helix α7 of the proximal coiled-coil region of Drosophila kinesin-1. The results for these hybrid motors showed that processivity based on run length scaled with neck-linker length except for CENP-E, which was highly processive with its 18-residue neck linker and became less processive as the neck linker was shortened.
The hypothesis that neck-linker length controlled the efficiency of gating and therefore run length came under scrutiny immediately. Düselder et al. (79) pursued a study with X. laevis kinesin-5 Eg5 in which they varied the length of the neck linker from 9 to 21 residues. Their results showed that native Eg5 motors with neck linkers down to 12 residues were highly processive, but notably, the run lengths were maximal when the neck linker was close to that of the native X. laevis Eg5. The authors argued that there was no optimal neck-linker length, but they proposed instead that the optimal neck linker was kinesin-specific (79). Guzik-Lendrum et al. (67) designed KIF3 constructs that when expressed contained the N-terminal native sequence of each motor domain, neck linker, and native helix α7, followed by a dimerization motif to stabilize the native coiled-coil. Using total internal reflection fluorescence microscopy, KIF3AC–Qdot complexes were found to be highly processive with run lengths of 1.23 μm, matching the run length of kinesin-1 (67). The authors concluded that the 17-residue neck linker of KIF3AC clearly did not impede processive stepping. Moreover, the newly designed KIF3AB exhibited a run length of 1.62 μm, exceeding the run length of KIF3AC and kinesin-1, and this run length was significantly greater than the run length published previously (76, 77).
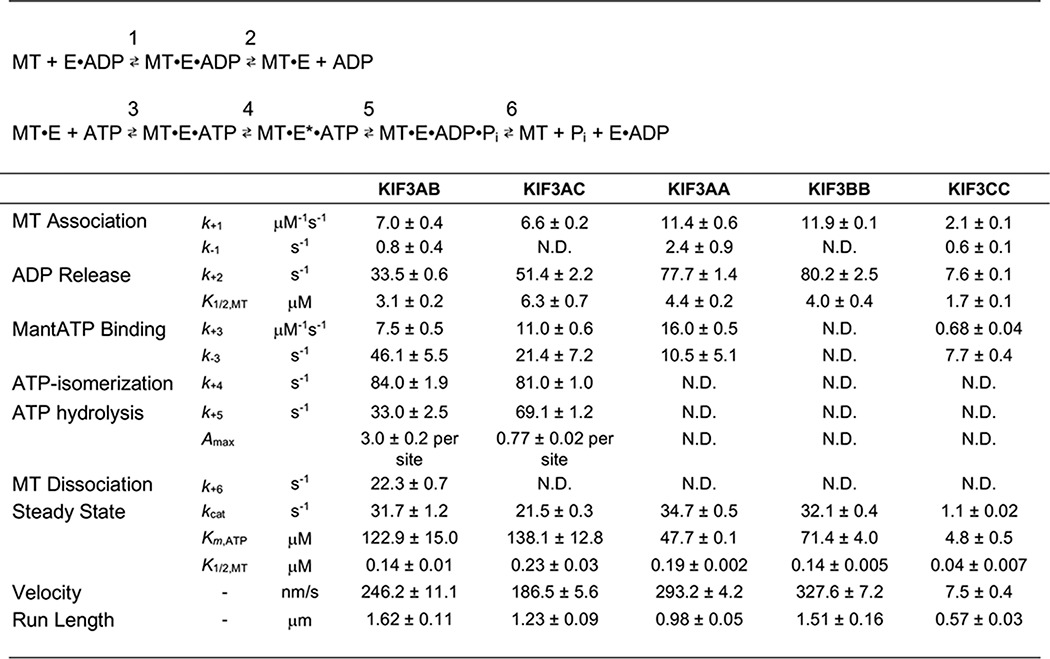
Guzik-Lendrum et al. (67) proposed that for each of the kinesin-2 motors studied, the significant difference in the run lengths observed was due to inclusion of native helix α7 to initiate the correct start of the coiled-coil. These studies clearly showed that in single-molecule assays without a hindering load, KIF3AC and KIF3AB were highly processive (Table 1).
Table 1.
ATPase scheme and experimentally determined constants
Constants were reported previously in Ref. 67, 88–90. MT is microtubule; mant-ATP is 2′-(or 3′)-O-(N-methylanthraniloyl)-ATP.
Structural studies show that the coiled-coil predictions were not accurate
The concept of neck-linker length is inevitably tied to the question of the location of the start of the coiled-coil. Phillips et al. (80) initiated a comprehensive structural study to address this question using X-ray crystallography. The assumption has been that coiled-coil algorithms such as COILS (a position-specific scoring matrix model) or MARCOIL (a Hidden Markov Model) were good predictors of coiled-coil domains (81, 82). The original estimates of the kinesin neck-linker length assumed that the coiled-coil would begin on a hydrophobic residue in either the a or d position of the coiled-coil heptad repeat (83). Yet, predictions of the first residue to adopt a helical conformation in any coiled-coil are ambiguous, although these algorithms do recognize the heptad repeat within a coiled-coil domain. As Phillips et al. (80) showed, the beginning of the coiled-coil in kinesin-2 was much more difficult to predict than that of kinesin-1.
There are two structures for a dimeric N-terminal kinesin, rat kinesin-1 (PDB code 3KIN) and Drosophila kinesin 1 (PDB code 2Y5W), and these provided the true α7 start and neck-linker length in the context of a dimeric kinesin (84, 85). Phillips et al. (80) determined seven X-ray crystal structures of kinesin homodimers without their motor domains but included the neck-linker motif followed by helix α7 that is the start of the coiled-coil stalk. Although the prediction of kinesin-1 was accurate with helix α7 beginning at Ala-345, those for KIF3A, KIF3C, Eg5, and CENP-E were predicted inaccurately (Fig. 3A). The predictions suggested that the kinesin-2 helix α7 would begin at Leu-360 in KIF3A and Leu-382 in KIF3C, yet in the crystal structures helix α7 begins five residues earlier at Pro-355 and Pro-377 in KIF3A and KIF3C, respectively. Therefore, the neck linker is shortened from 17 residues to 12 residues, thus shorter than the neck linker of kinesin-1 at 14 residues. The crystal structures of both Eg5 and CENP-E reveal much shorter neck linkers than predicted (80). Helix α7 of Eg5 begins at Lys-371 instead of Ile-375, thus shortening the neck linker from the predicted 18 residues to 14 residues. Moreover, the coiled-coil of CENP-E begins at Asp-341 rather than Leu-345 resulting in a neck linker that is 14 residues rather than 18 residues as predicted. Structures of KIF17 have not yet been determined, but as Fig. 3A shows, the sequence of the KIF17 neck linker and helix α7 are almost identical to those of KIF3A, KIF3B, and KIF3C, suggesting that its coiled-coil will also begin at the proline of PKDAL as determined for KIF3A and KIF3C.
These results provided evidence that neck-linker length in the context of the native sequence did not determine processivity. Moreover, this study reinforced the importance of conjoining the native sequences of both the native neck linker and helix α7 for engineered constructs to study the motile properties of kinesin family members (80). In addition, the structural study by Phillips et al. (80) revealed similar disparities in the coiled-coil predictions for a wide variety of non-motor proteins in the Protein Data Bank.
Andreasson et al. (86) clarified the run length debate using full-length kinesin-2 motors expressed in Sf9 cells. Using full-length KIF3AB motors, the authors showed that the run lengths in the absence of hindering load were quite long and approached run lengths of kinesin-1. However, against any appreciable external hindering load (∼1 pN), stepping was disrupted and the processive run terminated. This study was critically important because it framed the run length discussion in the context of response to hindering load separated from unloaded processivity. Moreover, the authors (86) stated that there was no evidence that force sensitivity was encoded in the neck linker. In addition, Milic et al. (87) reported that kinesin-2 KIF17 continues to step under a 6-pN hindering load, whereas KIF3AB detaches from the microtubule at these conditions. Note that the neck-linker motif of KIF17 is almost identical in sequence to that of KIF3A and KIF3B (Fig. 3A), thereby weakening the argument that kinesin-2 motors are inherently less processive than other processive kinesins because of the neck-linker length.
Presteady-state kinetics reveal unexpected properties of heterodimeric KIF3AC and KIF3AB
The unusual response to force by heterotrimeric kinesin-2 in combination with the series of publications about the role of neck-linker length in processivity motivated a comprehensive analysis of KIF3AC and KIF3AB using presteady-state kinetics methodologies (stopped-flow and chemical quench flow) to probe the ATPase cycle (88–90). The KIF3 constructs used to generate the kinesin-2 motors included the native motor domain, neck linker, and helix α7 followed by a dimerization motif to stabilize the helix α7 coiled-coil (67, 88). The steady-state ATPase parameters were consistent with the predictions from the single-molecule results (Table 1). What was surprising from these initial studies was that KIF3AB and KIF3AC were similar in their single-molecule velocity as well as the velocities of homodimeric KIF3AA and KIF3BB. However, the single-molecule velocity of homodimeric KIF3CC was exceptionally slow at 7.5 nm/s with the steady-state kcat at 1.1 s−1. These initial results led the authors (67) to propose that KIF3AA and KIF3BB were intrinsically fast in comparison with KIF3CC, and thus, during stepping KIF3A accelerates KIF3C and KIF3C slows KIF3A.
Entry into the processive run
To pursue mechanistic studies, the experiments were designed based on the ATPase cycle in Fig. 2A and Table 1 (88–90). The series of experiments for entry into the processive run (Fig. 2, E0–E1) revealed that microtubule association for KIF3AC and KIF3AB was similar at ∼7 μm−1 s−1 followed by ADP release for KIF3AC at 51 s−1 and for KIF3AB at 33.5 s−1 (Table 1). In contrast, these constants for KIF3AA and KIF3BB were significantly faster at ∼11–13 μm−1 s−1 with ADP release also fast at ∼80 s−1, yet the parameters for KIF3CC were very slow with microtubule association at 2.1 μm−1 s−1 and ADP release at 7.6 s−1. These results reinforced the conclusion that KIF3AA and KIF3BB are both catalytically fast and similar to each other, yet KIF3CC is intrinsically extremely slow (67, 86).
Additional experiments were pursued to test the hypothesis that the rate constant for microtubule association was a function of heterodimerization of KIF3AB and KIF3AC rather than the intrinsic properties of each motor domain. The microtubule association experiments were repeated with mixtures of KIF3AA + KIF3BB or KIF3AA + KIF3CC at the same motor concentration as KIF3AB or KIF3AC. The results clearly showed that regardless of the mixture composition, the KIF3AB transient could not be recapitulated by any sum of KIF3AA + KIF3BB, and similarly the KIF3AC transient could not be captured by mixtures of KIF3AA and KIF3CC (89, 90). The authors concluded by proposing that although the processive run may begin by either KIF3A or KIF3B/KIF3C, the kinetics observed were an emergent property due to intermolecular communication within the heterodimer rather than the intrinsic catalytic capability of each motor head.
One important conclusion resulting from these studies is that the catalytic properties of KIF3AB and KIF3AC are well suited for cargo transport where they may readily detach from the microtubule track. Because the microtubule association constants are so high, there would be a high probability of motor rebinding the microtubule rapidly. The rate of rebinding of KIF3–ADP is determined by the local microtubule concentration, which was estimated previously at ∼1 mm near the microtubule lattice (91). Therefore, as long as KIF3AB or KIF3AC remain in close proximity to the microtubule lattice, the rebinding rate would be ∼7,000 s−1 resulting in a very short detachment time, ∼143 μs. The authors concluded that heterodimeric KIF3AB and KIF3AC are optimized for rapid rebinding to the microtubule to continue transport of their cargoes (89, 90).
ATP binding and ATP hydrolysis
The second-order rate constant for ATP binding was measured by preforming the microtubule–kinesin complex and rapidly mixing with the fluorescent analog mant-ATP in the stopped-flow instrument (Fig. 2A, E1–E2). The results showed that this constant is quite fast for KIF3AB, KIF3AC, KIF3AA, and KIF3BB yet is significantly slower for KIF3CC (Table 1) (88–90, 92). Pulse-chase experiments using the chemical quench-flow instrument revealed that the rate constant for the ATP-promoted isomerization that occurs after ATP binding was similar for both KIF3AC and KIF3AB at ∼82 s−1 (88, 89). The ATP-promoted isomerization has traditionally been viewed as representing the series of structural transitions that include neck-linker docking and orientation of the active-site residues around MgATP to form the intermediate poised for ATP hydrolysis (71, 93–95).
When ATP hydrolysis was measured directly, the rate constant for KIF3AC was determined at 69 s−1. The authors concluded that the constants for ATP binding and ATP hydrolysis were not limiting the single-molecule rate of stepping for KIF3AC at 186 nm/s or 23 s−1 per 8-nm step (89). In contrast, the rate of ATP hydrolysis determined for KIF3AB was 33 s−1, but the amplitude of the burst indicated that there were three ATP turnovers per active site collapsed into one (88). Therefore, the ATP hydrolysis constant must be significantly faster as pointed out by Chen et al. (92). Andreasson et al. (86) modeled the ATP-promoted transition for KIF3AB, including neck-linker docking at a very fast rate, >500 s−1, with much slower ATP hydrolysis at ∼80 s−1. Because ATP binding followed by the ATP-promoted structural transitions are coupled with ATP hydrolysis, these are linked where one is fast and the other is slow. Experimentally, the slow step is quantified, but its identity cannot be determined from the experiments. Therefore, both the presteady-state kinetics and the load-dependent single-molecule experiments identify a rate-limiting transition for stepping at ∼80 s−1 and a very fast rate that was not limiting the ATPase cycle (86, 89, 90, 92).
Andreasson et al. (86) also reported that the run length was more sensitive to load than the velocity of stepping, leading to the hypothesis that the KIF3AB load-dependent processivity could result from a faster dissociation from the one-head–bound state or slower binding of the tethered head under load or both (Fig. 2B, E2–E5). This would imply that relative to kinesin-1, the strict coordination of the ATPase cycle for KIF3A and KIF3B is not well maintained resulting in motor detachment from the microtubule at hindering loads as low as ∼1 pN (86).
Is kinesin-2 processivity controlled by front-head or rear-head gating?
Processive stepping continues because the chemical and structural transitions on one head are inhibited until the partner head proceeds through its mechanochemical cycle (Fig. 2). The front-head gating model proposes that when both heads are bound to the microtubule (Fig. 2A, E5), ATP binding on the front head is inhibited until ATP hydrolysis occurs on the lagging head, followed by phosphate release, and motor head detachment (Fig. 2A, E5–E1) (23, 96–98). This model previously posited that strain within the two-head bound state inhibited the ability of ATP to bind at the active site of the front head. However, more recently, Dogan et al. (29) have shown for kinesin-1 that it is not strain per se but the backward orientation of the front head neck linker. As Fig. 2, A and B, shows when both heads are bound to the microtubule, the front head's neck linker is undocked and pointed backward, whereas the neck linker of the rear head is docked onto the catalytic core pointed to the plus-end of the microtubule.
In contrast, the rear-head gating model (Fig. 2B) proposes that binding of the front head accelerates detachment of the trailing head from the microtubule (99, 100). More recently, the rear-head gating model has been refined to propose that ATP hydrolysis at E2–E3 (Fig. 2B) occurs while the ADP head is in its diffusional search to find its next microtubule-binding site (Fig. 2B, E3–E5). Therefore, it becomes a kinetic race for the ADP head to step forward, bind to the microtubule, and release ADP before phosphate release from the bound head occurs to form an ADP weak-binding state (28, 31, 101). These two models are not necessarily mutually exclusive of each other, thus making it difficult to design definitive experiments that distinguish one model from the other especially for KIF3AB and KIF3AC (33, 86, 87, 92).
The rear-head gating model has been difficult to test because it requires capturing the transient intermediate states during the ATPase cycle. To tackle this question, Mickolajczyk and Hancock (33) have used a new imaging method designated “iSCAT,” in which interference reflection dark-field microscopy in combination with laser illumination is able to achieve extremely high spatial (nanometer) and temporal (<1 ms) resolution of unloaded kinesin motility. A 30-nm gold particle was attached to one head of the KIF3AA–kinesin-1 hybrid motor resulting in nanometer precision for tracking steps (33). The authors argue that they can distinguish one-head versus two-head bound states and can manipulate the kinetics of each state using KIF3AA–kinesin-1 hybrid motors with different length neck linkers. The authors propose that greater processivity is correlated with faster attachment of the tethered head prior to detachment of post-ATP hydrolysis one-head vulnerable ADP–Pi state (Fig. 2B, E2–E4). Therefore, processivity is maintained through a race for tethered head attachment at its next microtubule-binding site before the one-head bound E3 ADP–Pi intermediate detaches from the microtubule (Fig. 2B). These experiments are technically challenging and depend on ATP analogs and neck-linker insertions to specifically affect the duration of the one-head bound state but not the two-head bound state. Moreover, it is difficult to reconcile this new model with high resolution X-ray crystallography and cryo-electron microscopy studies (27, 102). These structures indicate that ATP binding induces a large structural change within the catalytic motor domain that drives docking of the neck linker and therefore immediately results in a forward step.
The recent publications in support of the rear-head gating model will no doubt motivate the motility field to design new types of experiments with technological advances to provide additional support for the front head and/or rear head models.
Concluding remarks and outstanding questions
Kinesin-2 KIF3AB and KIF3AC are fascinating molecular motors especially when one considers how similar their catalytic motor domain sequences are to each other and to kinesin-1. Outstanding questions include how is force sensitivity encoded structurally for kinesin-2s and whether the response to force by KIF3AC is similar to KIF3AB and therefore a key principle for kinesin-2s. Although there is a much greater understanding of the behavior of heterotrimeric kinesin-2 in intraflagellar transport, there are significant gaps in our understanding of KIF3AC transport in neurons. For example, its adaptors for linkage to cargo have yet to be identified as well as the identity of the KIF3AC-specific dendritic organelles. Moreover, the catalytic properties of KIF3C remain puzzling. Why is KIF3C so slow and how is this property encoded? Finally, we do not yet know whether KIF3AB and KIF3AC read the tubulin code differently for selective transport to axons versus dendrites (12, 38, 103). There are many discoveries ahead waiting for novel experiments and innovative technologies.
Acknowledgments
We are indebted to the past and present members of our laboratories for their many contributions, which are in part reviewed here.
This work was supported by National Institutes of Health Grant R37-GM054141 (to S. P. G.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- KAP
- kinesin-associated polypeptide
- PDB
- Protein Data Bank
- pN
- piconewton
- mant-ATP
- 2′-(or 3′)-O-(N-methylanthraniloyl)-ATP.
References
- 1. Vale R. D. (2003) The molecular motor toolbox for intracellular transport. Cell 112, 467–480 10.1016/S0092-8674(03)00111-9 [DOI] [PubMed] [Google Scholar]
- 2. Hirokawa N., and Noda Y. (2008) Intracellular transport and kinesin superfamily proteins, KIFs: structure, function, and dynamics. Physiol. Rev. 88, 1089–1118 10.1152/physrev.00023.2007 [DOI] [PubMed] [Google Scholar]
- 3. Hirokawa N., Noda Y., Tanaka Y., and Niwa S. (2009) Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol. 10, 682–696 10.1038/nrm2774 [DOI] [PubMed] [Google Scholar]
- 4. Hirokawa N., Niwa S., and Tanaka Y. (2010) Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron 68, 610–638 10.1016/j.neuron.2010.09.039 [DOI] [PubMed] [Google Scholar]
- 5. Verhey K. J., Kaul N., and Soppina V. (2011) Kinesin assembly and movement in cells. Annu. Rev. Biophys. 40, 267–288 10.1146/annurev-biophys-042910-155310 [DOI] [PubMed] [Google Scholar]
- 6. Scholey J. M. (2013) Kinesin-2: a family of heterotrimeric and homodimeric motors with diverse intracellular transport functions. Annu. Rev. Cell Dev. Biol. 29, 443–469 10.1146/annurev-cellbio-101512-122335 [DOI] [PubMed] [Google Scholar]
- 7. Maday S., Twelvetrees A. E., Moughamian A. J., and Holzbaur E. L. (2014) Axonal transport: cargo-specific mechanisms of motility and regulation. Neuron 84, 292–309 10.1016/j.neuron.2014.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hirokawa N., and Tanaka Y. (2015) Kinesin superfamily proteins (KIFs): various functions and their relevance for important phenomena in life and diseases. Exp. Cell Res. 334, 16–25 10.1016/j.yexcr.2015.02.016 [DOI] [PubMed] [Google Scholar]
- 9. Lu W., and Gelfand V. I. (2017) Moonlighting motors: kinesin, dynein, and cell polarity. Trends Cell Biol. 27, 505–514 10.1016/j.tcb.2017.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. He M., Agbu S., and Anderson K. V. (2017) Microtubule motors drive Hedgehog signaling in primary cilia. Trends Cell Biol. 27, 110–125 10.1016/j.tcb.2016.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Prevo B., Scholey J. M., and Peterman E. J. (2017) Intraflagellar transport: mechanisms of motor action, cooperation, and cargo delivery. FEBS J. 284, 2905–2931 10.1111/febs.14068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bentley M., and Banker G. (2016) The cellular mechanisms that maintain neuronal polarity. Nat. Rev. Neurosci. 17, 611–622 10.1038/nrn.2016.100 [DOI] [PubMed] [Google Scholar]
- 13. Lawrence C. J., Dawe R. K., Christie K. R., Cleveland D. W., Dawson S. C., Endow S. A., Goldstein L. S., Goodson H. V., Hirokawa N., Howard J., Malmberg R. L., McIntosh J. R., Miki H., Mitchison T. J., Okada Y., et al. (2004) A standardized kinesin nomenclature. J. Cell Biol. 167, 19–22 10.1083/jcb.200408113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Miki H., Setou M., Kaneshiro K., and Hirokawa N. (2001) All kinesin superfamily protein, KIF, genes in mouse and human. Proc. Natl. Acad. Sci. U.S.A. 98, 7004–7011 10.1073/pnas.111145398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kull F. J., Sablin E. P., Lau R., Fletterick R. J., and Vale R. D. (1996) Crystal structure of the kinesin motor domain reveals a structural similarity to myosin. Nature 380, 550–555 10.1038/380550a0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marx A., Müller J., and Mandelkow E. (2005) The structure of microtubule motor proteins. Adv. Protein Chem. 71, 299–344 10.1016/S0065-3233(04)71008-6 [DOI] [PubMed] [Google Scholar]
- 17. Hua W., Young E. C., Fleming M. L., and Gelles J. (1997) Coupling of kinesin steps to ATP hydrolysis. Nature 388, 390–393 10.1038/41118 [DOI] [PubMed] [Google Scholar]
- 18. Schnitzer M. J., and Block S. M. (1997) Kinesin hydrolyses one ATP per 8-nm step. Nature 388, 386–390 10.1038/41111 [DOI] [PubMed] [Google Scholar]
- 19. Svoboda K., Schmidt C. F., Schnapp B. J., and Block S. M. (1993) Direct observation of kinesin stepping by optical trapping interferometry. Nature 365, 721–727 10.1038/365721a0 [DOI] [PubMed] [Google Scholar]
- 20. Asbury C. L., Fehr A. N., and Block S. M. (2003) Kinesin moves by an asymmetric hand-over-hand mechanism. Science 302, 2130–2134 10.1126/science.1092985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kaseda K., Higuchi H., and Hirose K. (2003) Alternate fast and slow stepping of a heterodimeric kinesin molecule. Nat. Cell Biol. 5, 1079–1082 10.1038/ncb1067 [DOI] [PubMed] [Google Scholar]
- 22. Yildiz A., Tomishige M., Vale R. D., and Selvin P. R. (2004) Kinesin walks hand-over-hand. Science 303, 676–678 10.1126/science.1093753 [DOI] [PubMed] [Google Scholar]
- 23. Yildiz A., Tomishige M., Gennerich A., and Vale R. D. (2008) Intramolecular strain coordinates kinesin stepping behavior along microtubules. Cell 134, 1030–1041 10.1016/j.cell.2008.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Asenjo A. B., and Sosa H. (2009) A mobile kinesin-head intermediate during the ATP-waiting state. Proc. Natl. Acad. Sci. U.S.A. 106, 5657–5662 10.1073/pnas.0808355106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gigant B., Wang W., Dreier B., Jiang Q., Pecqueur L., Plückthun A., Wang C., and Knossow M. (2013) Structure of a kinesin-tubulin complex and implications for kinesin motility. Nat. Struct. Mol. Biol. 20, 1001–1007 10.1038/nsmb.2624 [DOI] [PubMed] [Google Scholar]
- 26. Cao L., Wang W., Jiang Q., Wang C., Knossow M., and Gigant B. (2014) The structure of apo-kinesin bound to tubulin links the nucleotide cycle to movement. Nat. Commun. 5, 5364 10.1038/ncomms6364 [DOI] [PubMed] [Google Scholar]
- 27. Shang Z., Zhou K., Xu C., Csencsits R., Cochran J. C., and Sindelar C. V. (2014) High-resolution structures of kinesin on microtubules provide a basis for nucleotide-gated force-generation. Elife 3, e04686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mickolajczyk K. J., Deffenbaugh N. C., Arroyo J. O., Andrecka J., Kukura P., and Hancock W. O. (2015) Kinetics of nucleotide-dependent structural transitions in the kinesin-1 hydrolysis cycle. Proc. Natl. Acad. Sci. U.S.A. 112, E7186–E7193 10.1073/pnas.1517638112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dogan M. Y., Can S., Cleary F. B., Purde V., and Yildiz A. (2015) Kinesin's front head is gated by the backward orientation of its neck linker. Cell Rep. 10, 1967–1973 10.1016/j.celrep.2015.02.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Andreasson J. O., Milic B., Chen G. Y., Guydosh N. R., Hancock W. O., and Block S. M. (2015) Examining kinesin processivity within a general gating framework. Elife 4, e07403 10.7554/eLife.07403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Isojima H., Iino R., Niitani Y., Noji H., and Tomishige M. (2016) Direct observation of intermediate states during the stepping motion of kinesin-1. Nat. Chem. Biol. 12, 290–297 10.1038/nchembio.2028 [DOI] [PubMed] [Google Scholar]
- 32. Liu D., Liu X., Shang Z., and Sindelar C. V. (2017) Structural basis of cooperativity in kinesin revealed by 3D reconstruction of a two-head-bound state on microtubules. Elife 6, e24490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mickolajczyk K. J., and Hancock W. O. (2017) Kinesin processivity is determined by a kinetic race from a vulnerable one-head-bound state. Biophys. J. 112, 2615–2623 10.1016/j.bpj.2017.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang Q., Diehl M. R., Jana B., Cheung M. S., Kolomeisky A. B., and Onuchic J. N. (2017) Molecular origin of the weak susceptibility of kinesin velocity to loads and its relation to the collective behavior of kinesins. Proc. Natl. Acad. Sci. U.S.A. 114, E8611–E8617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cole D. G., Cande W. Z., Baskin R. J., Skoufias D. A., Hogan C. J., and Scholey J. M. (1992) Isolation of a sea urchin egg kinesin-related protein using peptide antibodies. J. Cell Sci. 101, 291–301 [DOI] [PubMed] [Google Scholar]
- 36. Cole D. G., Chinn S. W., Wedaman K. P., Hall K., Vuong T., and Scholey J. M. (1993) Novel heterotrimeric kinesin-related protein purified from sea urchin eggs. Nature 366, 268–270 10.1038/366268a0 [DOI] [PubMed] [Google Scholar]
- 37. Setou M., Nakagawa T., Seog D. H., and Hirokawa N. (2000) Kinesin superfamily motor protein KIF17 and mLin-10 in NMDA receptor-containing vesicle transport. Science 288, 1796–1802 10.1126/science.288.5472.1796 [DOI] [PubMed] [Google Scholar]
- 38. Huang C. F., and Banker G. (2012) The translocation selectivity of the kinesins that mediate neuronal organelle transport. Traffic 13, 549–564 10.1111/j.1600-0854.2011.01325.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hammond J. W., Blasius T. L., Soppina V., Cai D., and Verhey K. J. (2010) Autoinhibition of the kinesin-2 motor KIF17 via dual intramolecular mechanisms. J. Cell Biol. 189, 1013–1025 10.1083/jcb.201001057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sirajuddin M., Rice L. M., and Vale R. D. (2014) Regulation of microtubule motors by tubulin isotypes and post-translational modifications. Nat. Cell Biol. 16, 335–344 10.1038/ncb2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Aizawa H., Sekine Y., Takemura R., Zhang Z., Nangaku M., and Hirokawa N. (1992) Kinesin family in murine central nervous system. J. Cell Biol. 119, 1287–1296 10.1083/jcb.119.5.1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kondo S., Sato-Yoshitake R., Noda Y., Aizawa H., Nakata T., and Matsuura Y., and Hirokawa N. (1994) KIF3A is a new microtubule-based anterograde motor in the nerve axon. J. Cell Biol. 125, 1095–1107 10.1083/jcb.125.5.1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yamazaki H., Nakata T., Okada Y., and Hirokawa N. (1995) KIF3A/B: A heterodimeric kinesin superfamily protein that works as a microtubule plus end-directed motors for membrane organelle transport. J. Cell Biol. 130, 1387–1399 10.1083/jcb.130.6.1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sardella M., Navone F., Rocchi M., Rubartelli A., Viggiano L., Vignali G., Consalez G. G., Sitia R., and Cabibbo A. (1998) KIF3C, a novel member of the kinesin superfamily: sequence, expression, and mapping to human chromosome 2 at 2p23. Genomics 47, 405–408 10.1006/geno.1997.5123 [DOI] [PubMed] [Google Scholar]
- 45. Muresan V., Abramson T., Lyass A., Winter D., Porro E., Hong F., Chamberlin N. L., and Schnapp B. J. (1998) KIF3C and KIF3A form a novel neuronal heteromeric kinesin that associates with membrane vesicles. Mol. Biol. Cell 9, 637–652 10.1091/mbc.9.3.637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yang Z., and Goldstein L. S. (1998) Characterization of the KIF3C neural kinesin-like motor from mouse. Mol. Biol. Cell 9, 249–261 10.1091/mbc.9.2.249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Davidovic L., Jaglin X. H., Lepagnol-Bestel A. M., Tremblay S., Simonneau M., Bardoni B., and Khandjian E. W. (2007) The fragile X mental retardation protein is a molecular adaptor between the neurospecific KIF3C kinesin and dendritic RNA granules. Hum. Mol. Genet. 16, 3047–3058 10.1093/hmg/ddm263 [DOI] [PubMed] [Google Scholar]
- 48. Yang Z., Roberts E. A., and Goldstein L. S. (2001) Functional analysis of mouse kinesin motor Kif3C. Mol. Cell. Biol. 21, 5306–5311 10.1128/MCB.21.16.5306-5311.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chana M. S., Tripet B. P., Mant C. T., and Hodges R. (2005) Stability and specificity of heterodimer formation for the coiled-coil neck regions of the motor proteins Kif3A and Kif3B: the role of unstructured oppositely charged regions. J. Pept. Res. 65, 209–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gumy L. F., Chew D. J., Tortosa E., Katrukha E. A., Kapitein L. C., Tolkovsky A. M., Hoogenraad C. C., and Fawcett J. W. (2013) The kinesin-2 family member KIF3C regulates microtubule dynamics and is required for axon growth and regeneration. J. Neurosci. 33, 11329–11345 10.1523/JNEUROSCI.5221-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Guzik-Lendrum S., Rayment I., and Gilbert S. P. (2017) Homodimeric kinesin-2 KIF3CC promotes microtubule dynamics. Biophys. J. 113, 1845–1857 10.1016/j.bpj.2017.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yamazaki H., Nakata T., Okada Y., and Hirokawa N. (1996) Cloning and characterization of KAP3: a novel kinesin superfamily-associated protein of KIF3A/3B. Proc. Natl. Acad. Sci. U.S.A. 93, 8443–8448 10.1073/pnas.93.16.8443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wedaman K. P., Meyer D. W., Rashid D. J., Cole D. G., and Scholey J. M. (1996) Sequence and submolecular localization of the 115-kD accessory subunit of the heterotrimeric kinesin-II (KRP85/95) complex. J. Cell Biol. 132, 371–380 10.1083/jcb.132.3.371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shimizu K., Shirataki H., Honda T., Minami S., and Takai Y. (1998) Complex formation of SMAP/KAP3, a KIF3A/B ATPase motor-associated protein, with a human chromosome-associated polypeptide. J. Biol. Chem. 273, 6591–6594 10.1074/jbc.273.12.6591 [DOI] [PubMed] [Google Scholar]
- 55. Doodhi H., Ghosal D., Krishnamurthy M., Jana S. C., Shamala D., Bhaduri A., Sowdhamini R., and Ray K. (2009) KAP, the accessory subunit of kinesin-2, binds the predicted coiled-coil stalk of the motor subunits. Biochemistry 48, 2248–2260 10.1021/bi8018338 [DOI] [PubMed] [Google Scholar]
- 56. Gindhart J. G. Jr., and Goldstein L. S. (1996) Armadillo repeats in the SpKAP115 subunit of kinesin-II. Trends Cell Biol. 6, 415–416 10.1016/S0962-8924(96)20037-6 [DOI] [PubMed] [Google Scholar]
- 57. Nonaka S., Tanaka Y., Okada Y., Takeda S., Harada A., Kanai Y., Kido M., and Hirokawa N. (1998) Randomization of left–right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell 95, 829–837 10.1016/S0092-8674(00)81705-5 [DOI] [PubMed] [Google Scholar]
- 58. Takeda S., Yonekawa Y., Tanaka Y., Okada Y., Nonaka S., and Hirokawa N. (1999) Left-right asymmetry and kinesin superfamily protein KIF3A: new insights in determination of laterality and mesoderm induction by kif3A−/− mice analysis. J. Cell Biol. 145, 825–836 10.1083/jcb.145.4.825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Marszalek J. R., Ruiz-Lozano P., Roberts E., Chien K. R., and Goldstein L. S. (1999) Situs inversus and embryonic ciliary morphogenesis defects in mouse mutants lacking the KIF3A subunit of kinesin-II. Proc. Natl. Acad. Sci. U.S.A. 96, 5043–5048 10.1073/pnas.96.9.5043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Marszalek J. R., Liu X., Roberts E. A., Chui D., Marth J. D., Williams D. S., and Goldstein L. S. (2000) Genetic evidence for selective transport of opsin and arrestin by kinesin-II in mammalian photoreceptors. Cell 102, 175–187 10.1016/S0092-8674(00)00023-4 [DOI] [PubMed] [Google Scholar]
- 61. Lin F., Hiesberger T., Cordes K., Sinclair A. M., Goldstein L. S., Somlo S., and Igarashi P. (2003) Kidney-specific inactivation of the KIF3A subunit of kinesin-II inhibits renal ciliogenesis and produces polycystic kidney disease. Proc. Natl. Acad. Sci. U.S.A. 100, 5286–5291 10.1073/pnas.0836980100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Teng J., Rai T., Tanaka Y., Takei Y., Nakata T., Hirasawa M., Kulkarni A. B., and Hirokawa N. (2005) The KIF3 motor transports N-cadherin and organizes the developing neuroepithelium. Nat. Cell Biol. 7, 474–482 10.1038/ncb1249 [DOI] [PubMed] [Google Scholar]
- 63. Huangfu D., Liu A., Rakeman A. S., Murcia N. S., Niswander L., and Anderson K. V. (2003) Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature 426, 83–87 10.1038/nature02061 [DOI] [PubMed] [Google Scholar]
- 64. Drummond I. A. (2012) Cilia functions in development. Curr. Opin. Cell Biol. 24, 24–30 10.1016/j.ceb.2011.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Brunnbauer M., Mueller-Planitz F., Kösem S., Ho T. H., Dombi R., Gebhardt J. C., Rief M., and Okten Z. (2010) Regulation of a heterodimeric kinesin-2 through an unprocessive motor domain that is turned processive by its partner. Proc. Natl. Acad. Sci. U.S.A. 107, 10460–10465 10.1073/pnas.1005177107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rank K. C., and Rayment I. (2013) Functional asymmetry in kinesin and dynein dimers. Biol. Cell 105, 1–13 10.1111/boc.201200044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Guzik-Lendrum S., Rank K. C., Bensel B. M., Taylor K. C., Rayment I., and Gilbert S. P. (2015) Kinesin-2 KIF3AC and KIF3AB can drive long-range transport along microtubules. Biophys. J. 109, 1472–1482 10.1016/j.bpj.2015.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Romberg L., Pierce D. W., and Vale R. D. (1998) Role of the kinesin neck region in processive microtubule-based motility. J. Cell Biol. 140, 1407–1416 10.1083/jcb.140.6.1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Thorn K. S., Ubersax J. A., and Vale R. D. (2000) Engineering the processive run length of the kinesin motor. J. Cell Biol. 151, 1093–1100 10.1083/jcb.151.5.1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tomishige M., and Vale R. D. (2000) Controlling kinesin by reversible disulfide cross-linking: identifying the motility-producing conformational change. J. Cell Biol. 151, 1081–1092 10.1083/jcb.151.5.1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Rice S., Lin A. W., Safer D., Hart C. L., Naber N., Carragher B. O., Cain S. M., Pechatnikova E., Wilson-Kubalek E. M., Whittaker M., Pate E., Cooke R., Taylor E. W., Milligan R. A., and Vale R. D. (1999) A structural change in the kinesin motor protein that drives motility. Nature 402, 778–784 10.1038/45483 [DOI] [PubMed] [Google Scholar]
- 72. Levi V., Serpinskaya A. S., Gratton E., and Gelfand V. (2006) Organelle transport along microtubules in Xenopus melanophores: evidence for cooperation between multiple motors. Biophys. J. 90, 318–327 10.1529/biophysj.105.067843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hendricks A. G., Perlson E., Ross J. L., Schroeder H. W. 3rd., Tokito M., and Holzbaur E. L. (2010) Motor coordination via a tug-of-war mechanism drives bidirectional vesicle transport. Curr. Biol. 20, 697–702 10.1016/j.cub.2010.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Schroeder H. W. 3rd, Hendricks A. G., Ikeda K., Shuman H., Rodionov V., Ikebe M., Goldman Y. E., and Holzbaur E. L. (2012) Force-dependent detachment of Kinesin-2 biases track switching at cytoskeletal filament intersections. Biophys. J. 103, 48–58 10.1016/j.bpj.2012.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hendricks A. G., Holzbaur E. L., and Goldman Y. E. (2012) Force measurements on cargoes in living cells reveal collective dynamics of microtubule motors. Proc. Natl. Acad. Sci. U.S.A. 109, 18447–18452 10.1073/pnas.1215462109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Muthukrishnan G., Zhang Y., Shastry S., and Hancock W. O. (2009) The processivity of kinesin-2 motors suggests diminished front-head gating. Curr. Biol. 19, 442–447 10.1016/j.cub.2009.01.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Shastry S., and Hancock W. O. (2010) Neck linker length determines the degree of processivity in Kinesin-1 and Kinesin-2 motors. Curr. Biol. 20, 939–943 10.1016/j.cub.2010.03.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Shastry S., and Hancock W. O. (2011) Interhead tension determines processivity across diverse N-terminal kinesins. Proc. Natl. Acad. Sci. U.S.A. 108, 16253–16258 10.1073/pnas.1102628108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Düselder A., Thiede C., Schmidt C. F., and Lakämper S. (2012) Neck-linker length dependence of processive Kinesin-5 motility. J. Mol. Biol. 423, 159–168 10.1016/j.jmb.2012.06.043 [DOI] [PubMed] [Google Scholar]
- 80. Phillips R. K., Peter L. G., Gilbert S. P., and Rayment I. (2016) Family-specific kinesin structures reveal neck-linker length based on initiation of the coiled-coil. J. Biol. Chem. 291, 20372–20386 10.1074/jbc.M116.737577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Delorenzi M., and Speed T. (2002) An HMM model for coiled-coil domains and a comparison with PSSM-based predictions. Bioinformatics 18, 617–625 10.1093/bioinformatics/18.4.617 [DOI] [PubMed] [Google Scholar]
- 82. Lupas A., Van Dyke M., and Stock J. (1991) Predicting coiled coils from protein sequences. Science 252, 1162–1164 10.1126/science.252.5009.1162 [DOI] [PubMed] [Google Scholar]
- 83. Hariharan V., and Hancock W. O. (2009) Insights into the mechanical properties of the kinesin neck linker domain from sequence analysis and molecular dynamics simulations. Cell. Mol. Bioeng. 2, 177–189 10.1007/s12195-009-0059-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kozielski F., Sack S., Marx A., Thormählen M., Schönbrunn E., Biou V., Thompson A., Mandelkow E. M., and Mandelkow E. (1997) The crystal structure of dimeric kinesin and implications for microtubule-dependent motility. Cell 91, 985–994 10.1016/S0092-8674(00)80489-4 [DOI] [PubMed] [Google Scholar]
- 85. Kaan H. Y., Hackney D. D., and Kozielski F. (2011) The structure of the kinesin-1 motor-tail complex reveals the mechanism of autoinhibition. Science 333, 883–885 10.1126/science.1204824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Andreasson J. O., Shastry S., Hancock W. O., and Block S. M. (2015) The mechanochemical cycle of mammalian kinesin-2 KIF3A/B under load. Curr. Biol. 25, 1166–1175 10.1016/j.cub.2015.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Milic B., Andreasson J. O. L., Hogan D. W., and Block S. M. (2017) Intraflagellar transport velocity is governed by the number of active KIF17 and KIF3AB motors and their motility properties under load. Proc. Natl. Acad. Sci. U.S.A. 114, E6830–E6838 10.1073/pnas.1708157114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Albracht C. D., Rank K. C., Obrzut S., Rayment I., and Gilbert S. P. (2014) Kinesin-2 KIF3AB exhibits novel ATPase characteristics. J. Biol. Chem. 289, 27836–27848 10.1074/jbc.M114.583914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Zhang P., Rayment I., and Gilbert S. P. (2016) Fast or slow, either head can start the processive run of kinesin-2 KIF3AC. J. Biol. Chem. 291, 4407–4416 10.1074/jbc.M115.705970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Albracht C. D., Guzik-Lendrum S., Rayment I., and Gilbert S. P. (2016) Heterodimerization of kinesin-2 KIF3AB modulates entry into the processive run. J. Biol. Chem. 291, 23248–23256 10.1074/jbc.M116.752196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Gilbert S. P., Webb M. R., Brune M., and Johnson K. A. (1995) Pathway of processive ATP hydrolysis by kinesin. Nature 373, 671–676 10.1038/373671a0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Chen G. Y., Arginteanu D. F., and Hancock W. O. (2015) Processivity of the kinesin-2 KIF3A results from rear head gating and not front head gating. J. Biol. Chem. 290, 10274–10294 10.1074/jbc.M114.628032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Gilbert S. P., and Johnson K. A. (1994) Pre-steady-state kinetics of the microtubule-kinesin ATPase. Biochemistry 33, 1951–1960 10.1021/bi00173a044 [DOI] [PubMed] [Google Scholar]
- 94. Auerbach S. D., and Johnson K. A. (2005) Alternating site ATPase pathway of rat conventional kinesin. J. Biol. Chem. 280, 37048–37060 10.1074/jbc.M502984200 [DOI] [PubMed] [Google Scholar]
- 95. Rosenfeld S. S., Jefferson G. M., and King P. H. (2001) ATP reorients the neck linker of kinesin in two sequential steps. J. Biol. Chem. 276, 40167–40174 10.1074/jbc.M103899200 [DOI] [PubMed] [Google Scholar]
- 96. Rosenfeld S. S., Fordyce P. M., Jefferson G. M., King P. H., and Block S. M. (2003) Stepping and stretching: how kinesin uses internal strain to walk processively. J. Biol. Chem. 278, 18550–18556 10.1074/jbc.M300849200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Klumpp L. M., Hoenger A., and Gilbert S. P. (2004) Kinesin's second step. Proc. Natl. Acad. Sci. U.S.A. 101, 3444–3449 10.1073/pnas.0307691101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Guydosh N. R., and Block S. M. (2006) Backsteps induced by nucleotide analogs suggest the front head of kinesin is gated by strain. Proc. Natl. Acad. Sci. U.S.A. 103, 8054–8059 10.1073/pnas.0600931103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Crevel I. M., Nyitrai M., Alonso M. C., Weiss S., Geeves M. A., and Cross R. A. (2004) What kinesin does at roadblocks: the coordination mechanism for molecular walking. EMBO J. 23, 23–32 10.1038/sj.emboj.7600042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Schief W. R., Clark R. H., Crevenna A. H., and Howard J. (2004) Inhibition of kinesin motility by ADP and phosphate supports a hand-over-hand mechanism. Proc. Natl. Acad. Sci. U.S.A. 101, 1183–1188 10.1073/pnas.0304369101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Milic B., Andreasson J. O., Hancock W. O., and Block S. M. (2014) Kinesin processivity is gated by phosphate release. Proc. Natl. Acad. Sci. U.S.A. 111, 14136–14140 10.1073/pnas.1410943111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Wang W., Cao L., Wang C., Gigant B., and Knossow M. (2015) Kinesin, 30 years later: recent insights from structural studies. Protein Sci. 24, 1047–1056 10.1002/pro.2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Tas R. P., Chazeau A., Cloin B. M. C., Lambers M. L. A., Hoogenraad C. C., and Kapitein L. C. (2017) Differentiation between oppositely oriented microtubules controls polarized neuronal transport. Neuron 96, 1264–1271 10.1016/j.neuron.2017.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]