Abstract
During neurogenesis, neural patterning is a critical step during which neural progenitor cells differentiate into neurons with distinct functions. However, the molecular determinants that regulate neural patterning remain poorly understood. Here we optimized the “dual SMAD inhibition” method to specifically promote differentiation of human pluripotent stem cells (hPSCs) into forebrain and hindbrain neural progenitor cells along the rostral–caudal axis. We report that neural patterning determination occurs at the very early stage in this differentiation. Undifferentiated hPSCs expressed basal levels of the transcription factor orthodenticle homeobox 2 (OTX2) that dominantly drove hPSCs into the “default” rostral fate at the beginning of differentiation. Inhibition of glycogen synthase kinase 3β (GSK3β) through CHIR99021 application sustained transient expression of the transcription factor NANOG at early differentiation stages through Wnt signaling. Wnt signaling and NANOG antagonized OTX2 and, in the later stages of differentiation, switched the default rostral cell fate to the caudal one. Our findings have uncovered a mutual antagonism between NANOG and OTX2 underlying cell fate decisions during neural patterning, critical for the regulation of early neural development in humans.
Keywords: cell biology, cell differentiation, human, neural stem cell (NSC), neurodifferentiation, Wnt signaling, NANOG, OTX2
Introduction
Embryonic neurodevelopment is a spatiotemporally regulated process during which distinct cell fates are progressively restricted based on spatial regions (1, 2). At the early stage of neurogenesis, the specified neural ectoderm divides into functionally distinct cell fates along the anterior–posterior (A-P)2 and dorsal–ventral (D-V) axes (3). This neural patterning process is a critical step to specify different neural precursors such as the forebrain, midbrain, hindbrain, and spinal cord. It has been known that neural patterning is induced by the temporal and special morphogen gradients along the A-P and D-V axes (4, 5). These morphogens, including BMPs, WNTs, FGFs, RA, and sonic hedgehog, coordinate and form gradients to specify regionally transcriptional programs and distinct neural progenitors (6–10). However, the precise timing and mechanisms underlying morphogen-induced neuraxial patterning has not been fully elucidated in mammals, especially in humans.
Human pluripotent stem cells (hPSCs), including embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs), can differentiate into neuroepithelial cells and regionally specified neural precursor cells (11–16), thus providing a valuable model to investigate the molecular determinants of neural patterning in a human background. The most used method to induce neural differentiation in hPSCs is through suppression of both transforming growth factor β and BMP signaling (17, 18). Dual inhibition of SMAD-dependent TGF-β and BMP signaling by their inhibitors (SB431542 and Noggin) or SB431542 and Dorsomorphin can efficiently trigger hPSC differentiation into NPCs (19–21). Dual SMAD inhibition-triggered NPCs are believed to be more close to the anterior forebrain fate (22, 23). Interestingly, the anterior fate has also been considered as a “default” fate for hPSCs to initiate neural differentiation (24, 25). To initiate the caudal fate, other morphogens, such as WNTs, need to be applied on the basis of dual SMAD inhibition. Indeed, regional NPCs along the A-P axis could be specified from hPSCs via dose-dependent activation of WNT signaling by the GSK3β inhibitor CHIR combined with dual SMAD inhibition (26). However, how GSK3β inhibition coordinates with other signaling to regulate early neural patterning in hPSCs remains unclear.
In this study, we investigated the molecular determinants that regulate neural patterning in hPSC differentiation. We demonstrated that neural patterning is committed at a very early stage of differentiation. The basal amount of OTX2 actively drives hPSCs into the default anterior fate right after the exit from pluripotency. Depending on WNT activation, CHIR treatment at the same stage temporally sustains NANOG and further represses OTX2 and switches the default rostral fate to the caudal one in later differentiation.
Results
Generation of regionally specified forebrain and hindbrain NPCs from hPSCs
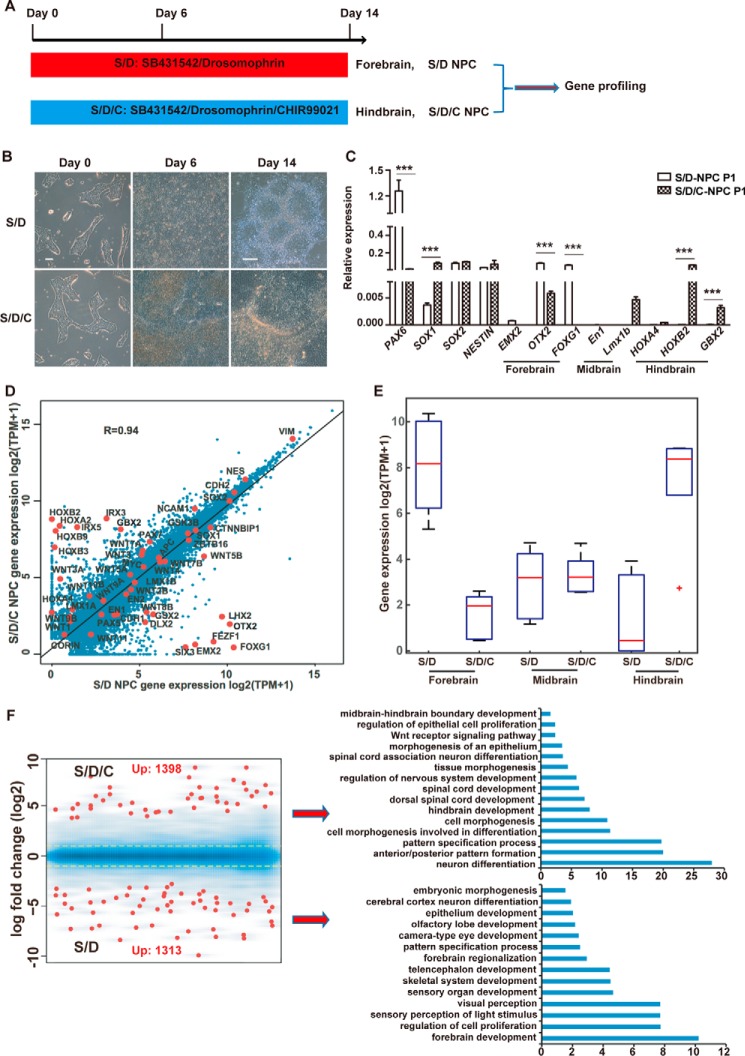
HPSCs could be differentiated into regionally neural cells by applying different morphogenetic cues, such as WNT, FGF, or RA activation (27–29). To investigate the molecular determinants of neural patterning, we optimized a monolayer and defined a condition to induce neutralization of hPSCs by applying dual SMAD inhibition combined with or without GSK3β inhibition (Fig. 1A). Human ESCs (30) or uterine cell–derived iPSCs (31) treated with dual SMAD inhibition alone (SB431542/Dorsomorphin (SD)) or combined with GSK3β inhibition (SB431542/Dorsomorphin/CHIR99021 (SDC)) showed obvious but different morphology changes (Fig. 1B). Consistently, we observed a significant difference in the expression of regionally specified neural markers between SD- and SDC-treated cells (Fig. 1C). Forebrain marker genes such as EMX2, OTX2, and FOXG1 were highly expressed in SD cells, whereas hindbrain marker genes like HOXB2, GBX2, and Lmx1b were highly expressed in SDC cells. These data indicate that the SD-triggered NPCs were of rostral fate, whereas the SDC NPCs were of caudal fate. Consistently, the pan-NPC marker genes SOX2 and NESTIN were highly expressed in both SD and SDC NPCs. Notably, other neural factors, PAX6 and SOX1, also showed a remarkable difference in expression between SD NPCs and SDC NPCs (Fig. 1C). We further performed a whole-genome transcriptome analysis on SD and SDC NPCs. Pearson correlation analysis showed distinct expression profiling between SD and SDC NPCs (Fig. 1D). The selected forebrain marker genes were highly expressed in SD NPCs, whereas the hindbrain genes were highly expressed in SDC NPCs (Fig. 1E). Furthermore, gene ontology analysis showed that the up-regulated genes in SDC NPCs were more related to anterior/posterior region formation and hindbrain/spinal cord development, whereas genes in SD NPCs were more related to forebrain and telencephalon development (Fig. 1F). Several signaling pathways that were reported to be critical to regulate neural patterning, such as WNTs, SMADs, and RA, also showed different expression between SD and SDC NPCs (Table S3). For example, the expression level of WNT5A was relatively higher whereas that of WNT5B was lower in SDC NPCs. Genes that are related to SMAD, WNT, and RA signaling pathways are summarized in Table S3.
Figure 1.
Differentiation of forebrain or hindbrain NPCs from hPSCs. A, schematic of neural differentiation protocols to induce forebrain and hindbrain NPCs from hPSCs. B, cell morphologic of hPSCs treated with SD or SDC at the indicated times. Scale bars = 100 μm. C, QPCR analysis of the indicated marker genes in SD- or SDC-treated cells. D, Pearson correlation of the whole-genome transcriptome between SDC NPCs and SD NPCs. The red dots represent the expression levels of the indicated marker genes. The R value represents Pearson's correlation coefficient. E, box plot of the RNA levels of selected marker genes based on RNA sequencing data. F, gene ontology analysis of the differentially expressed genes in SD- and SDC-treated cells. Yellow lines correspond to a 2-fold change. ***, p < 0.001.
An immunostaining assay confirmed that both NPCs maintained in vitro are SOX2-, NESTIN-, and KI67-positive (Fig. S1D) and could differentiate into astrocytes and subtype neurons, including GABAergic neurons, glutamatergic neurons, dopaminergic neurons, and motor neurons (Fig. S1E). Neurons differentiated from SDC or SD NPCs exhibit functional electrophysiological properties, including robust but similar Na+ and K+ currents and repetitive action potentials (Fig. S1F). In sum, we demonstrated that SD or SDC conditions induced different regionally specific NPCs; i.e. SD induced forebrain-specific NPCs, whereas SDC induced NPCs close to the hindbrain region. Both SDC-triggered caudal and SD-triggered rostral NPCs hold the potency to differentiate various subtype neural cells.
Rostral–caudal patterning occurs at the early stage of neural differentiation
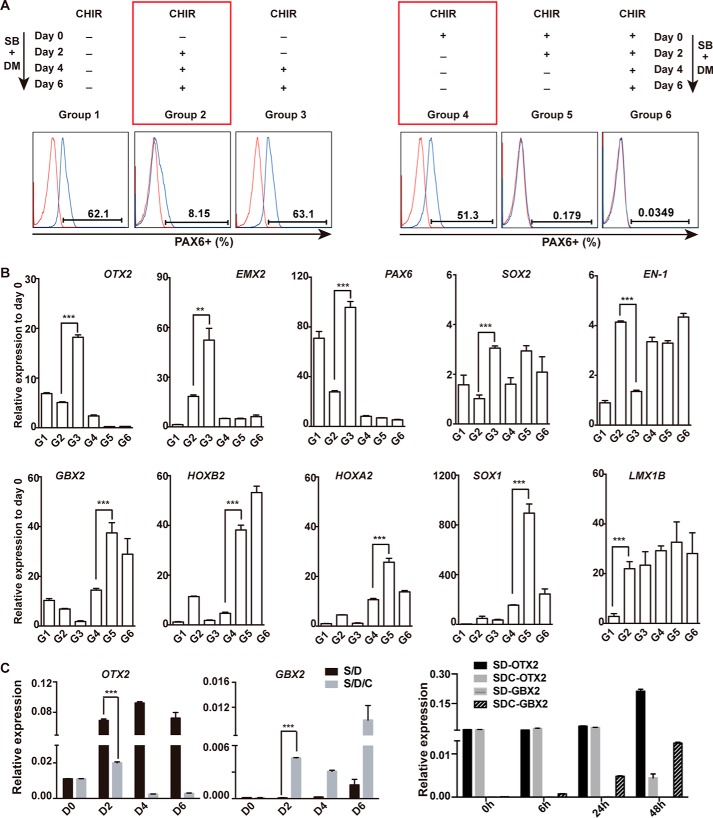
The rostral neural fate is usually considered a default fate in neural differentiation of hPSCs (32). We were interested in investigating how and when GSK3β inhibition coordinates with dual SMAD inhibition to switch the default rostral fate to the caudal one. We first designed experiments to examine the timing of CHIR treatment to switch the SD-triggered rostral fate in hPSCs. In this experiment, CHIR was added or withdrawn on day 2 or day 4 during SD- or SDC-treated differentiation (Fig. 2A). Cell regional characters were checked at day 6. We chose PAX6 as a cell fate indicator to assess different treatments because it has been reported as a critical factor in the fore- and midbrain (33, 34) and is also suppressed by CHIR in our experiments (Fig. S1, A–C). Other regional marker genes, such as the forebrain markers FOXG1, EMX2, and OTX2, the midbrain markers Lmx1b and EN-1, and the hindbrain markers GBX2, HOXA2, and HOXB2, were also examined by QPCR to confirm the different regional fate in different treatments (Fig. 2B). As shown in Fig. 2A (left panel), CHIR added as early at day 2 of SD treatment could significantly suppress PAX6 expression and switch the regional neural fate (Fig. 2A, Group 2). However, applying CHIR at later time points, on day 4, failed to affect the SD-trigged rostral fate (Fig. 2, A and B, Group 3). On the other hand, CHIR treatment for the first 4 days could be sufficient to switch the SD-trigged rostral fate to the caudal hindbrain one (Fig. 2, A and B, Group 5). In contrast, CHIR treatment only for the first 2 days showed no effect on cell fate transition (Fig. 2A, right panel, Group 4). Furthermore, based on the analysis of several known critical factors for A-P neural patterning, we found that OTX2 and GBX2, two known essential factors in fore-, and mid-, and hindbrain development (35–37), exhibited significantly differential activation between SD- and SDC-treated cells at 48 hr (Fig. 2C and Fig. S2A), highlighting that day 2 is the critical time point for irreversible rostral or caudal regional specification in neural differentiation of hPSCs.
Figure 2.
Commitment assay on rostral or caudal NPCs in hPSC differentiation. A, CHIR was added or withdrawn at the indicted time points during SD-induced neural differentiation of hPSCs. PAX6 was examined in cells from each group by FACS. SB, SB431542; DM, Dorsomorphin. B, the expression of other indicated marker genes in cells from each group were analyzed by QPCR. C, OTX2 and GBX2 were the first responders on day 2 of differentiation. **, p < 0.01; ***, p < 0.001.
OTX2 dominantly triggers rostral fate differentiation when hPSCs exit pluripotency
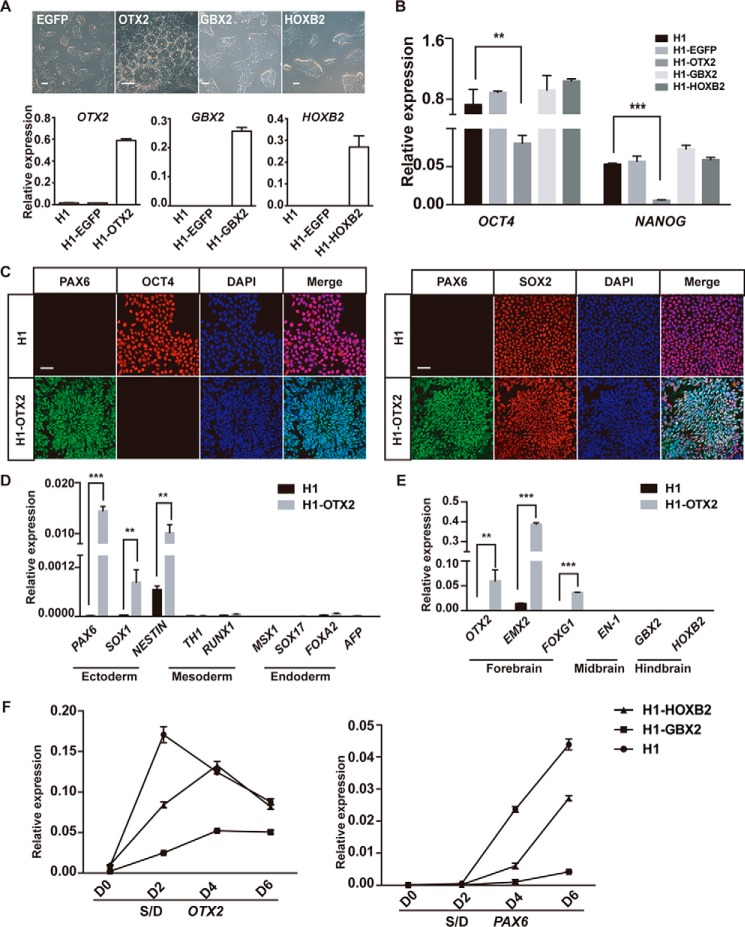
Our data showed that OTX2 and GBX2, but not other tested factors, exhibited significant differential activation between SD- and SDC-treated cells at day 2, the critical time window for neural patterning in differentiation (Fig. 2B), indicating that they might play essential roles in neural patterning in hPSC differentiation, as reported for other model systems. To investigate the precise roles of these factors, we overexpressed OTX2, GBX2, or HOXB2 in hESCs through a lentiviral approach (Fig. 3A). Interestingly, hESCs with OTX2 overexpression displayed a typical neural rosette–like phenotype, even when maintained in normal hPSC medium, which supports self-renewal and suppresses differentiation (Fig. 3A). In contrast, hESCs with GBX2 or HOXB2 overexpression kept the undifferentiated morphology (Fig. 3A). Consistently, pluripotent genes such as OCT4 and NANOG were suppressed in OTX2-overexpressing hESCs but well maintained in GBX2- or HOXB2-overexpressing hESCs (Fig. 3B). Through immunostaining and Western blotting, we confirmed that cells with OTX2 overexpression are positive for NPC markers such as PAX6 and SOX2 but negative for OCT4 (Fig. 3C and Fig. S2B). Similarly, neural ectoderm genes, but not other germ layer genes, are highly activated in OTX2-overexpressing cells (Fig. 3D). For regionally restricted neural marker genes, the forebrain genes EMX2 and FOXG1, but not midbrain and hindbrain genes, are significantly up-regulated in OTX2-overexpressing hESCs (Fig. 3E). These data demonstrate that OTX2 dominantly triggers neural differentiation of hPSCs toward the preferential forebrain fate.
Figure 3.
OTX2 play a dominated role in neural patterning during hPSC differentiation. A, cell morphology of hESCs overexpressing OTX2, GBX2, or HOXB2. Scale bars = 100 μm. Expression levels of OTX2, GBX2, and HOXB2 were analyzed by QPCR. B, expression of OCT4 and NANOG in each indicated cell line. C, immunofluorescence results of PAX6, OCT4, and SOX2 in OTX2-overexpressing cell lines. Scale bars = 50 μm. D and E, QPCR analysis of the expression of the indicated lineage genes in wildtype hESCs and hESCs with OTX2 expression. F, expression of OTX2 and PAX6 under SD induction in GBX2 or HOXB2-overexpressing cells. **, p < 0.01; ***, p < 0.001.
To examine whether GBX2 could affect the regional cell fate at later neural differentiation, we triggered differentiation of GBX2- or HOXB2-expressing hESCs by SD. As shown, GBX2 overexpression significantly suppressed forebrain genes such as PAX6 and OTX2 induced by SD treatment, whereas HOXB2 showed no similar suppression effect (Fig. 3F). In all, our data indicate that both OTX2 and GBX2 are involved in A-P neural patterning in hPSC differentiation but function differently. OTX2 is a dominant trigger and could drive hPSCs into the forebrain neural fate alone, whereas GBX2 could repress the forebrain fate and switch it to the hindbrain fate at a later differentiation stage.
GSK3β inhibition sustains NANOG and represses OTX2 to switch the rostral–caudal fate decision
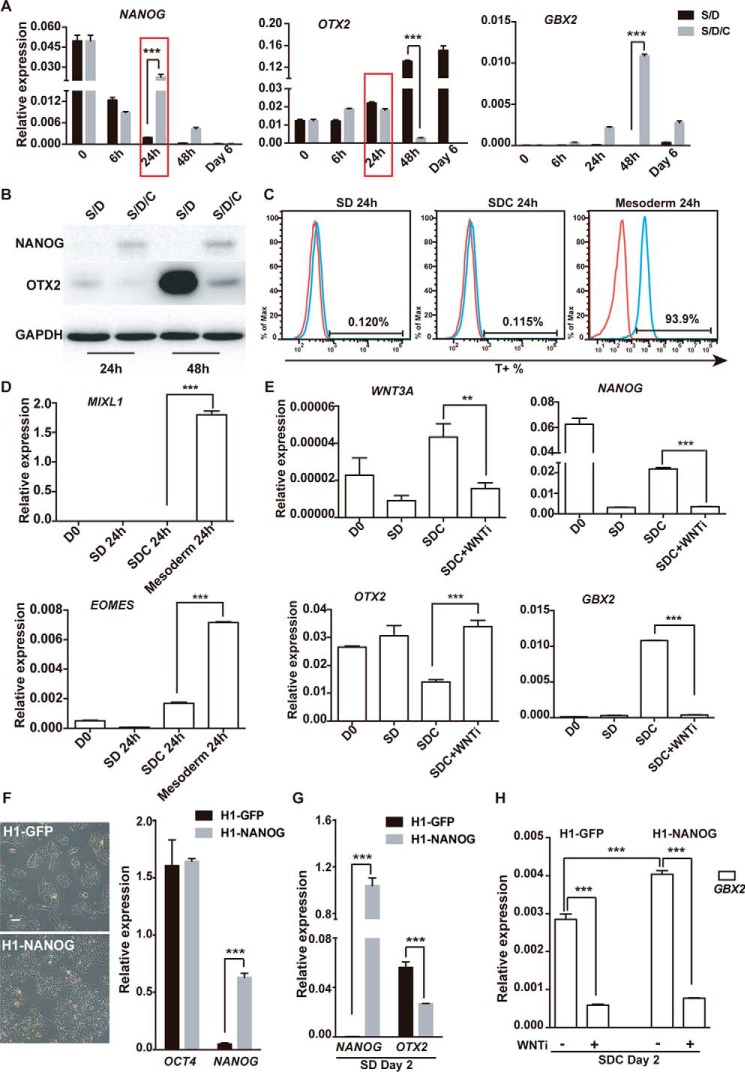
To investigate the role of GSK3β inhibition in neural fate decision, we examined the detailed expression of several critical factors during the first 2 days of differentiation triggered by SD or SDC (Fig. 4A). We identified that NANOG exhibits significantly higher expression in SDC-treated hPSCs than in SD-treated cells at 24 h of differentiation (Fig. 4A). OTX2 showed a similar level between the two treatments at 24 h but was dramatically suppressed at later time points in SDC-treated cells (Fig. 4A). Further, by Western blotting, we confirmed that, at the 24 h or 48 h time points, a certain level of NANOG protein was maintained in SDC- but not SD-treated hESCs (Fig. 4B). These data indicate that the temporal expression of NANOG in SDC cells might play important roles in neural fate decision at an early stage.
Figure 4.
GSK3β inhibition sustains NANOG and suppresses OTX2 in hPSC differentiation. A, QPCR analysis of the indicated genes in time course experiments of SD- or SDC-treated hPSC differentiation. B, Western blot analysis of NANOG and OTX2 in time course experiments of SD- or SDC-treated hPSC differentiation at 24 h and 48 h. GAPDH served as the loading control. C and D, the primitive streak markers T, MIXL1, and EOMES were analyzed through FACS and QPCR. E, expression of WNT3A and NANOG in hESCs treated with SD, SDC, and SDC with WNT inhibitor at 24 h of differentiation. F, cell morphology and QPCR analysis of H1 ESCs with NANOG or GFP overexpression. Scale bar = 100 μm. G, expression of OTX2 in H1 hESCs with NANOG or GFP overexpression treated with SD or SDC. H, expression of GBX2 in hESCs with NANOG or GFP overexpression treated with SDC with or without WNT inhibitor. **, p < 0.01; ***, p < 0.001.
To determine the effect of NANOG on caudal induction, we prepared hESCs with overexpression of NANOG through a lentiviral approach. NANOG-overexpressing hESCs maintained a typical undifferentiated phenotype and showed no significant difference compared with control hESCs with overexpression of GFP (Fig. 4C). We then treated them with SD or SDC to trigger rostral or caudal neural differentiation and examined the expression of OTX2 and GBX2 at day 2. We showed that NANOG overexpression significantly suppressed OTX2 expression in SD-triggered rostral fate differentiation (Fig. 4D). In contrast, GBX2 was not suppressed but up-regulated in NANOG-overexpressing cells during SDC-induced caudal fate differentiation (Fig. 4E). Because GBX2 was reported to be a direct target of WNT signaling (38), the activation of GBX2 in CHIR-treated cells might be due to the activation of WNT signaling. Then, by using a WNT inhibitor (XAV939, 0.5 μm) (39), we confirmed that GBX2 expression in CHIR-treated cells is indeed WNT signaling–dependent (Fig. 4E). Similarly, the temporal expression of NANOG in SDC cells was also due to the activation of WNT signaling by CHIR (Fig. 4F). In all, these data suggest that CHIR treatment temporarily maintains NANOG at an early stage via WNT signaling to repress OTX2 and activate GBX2 to initiate the caudal fate.
Previous reports showed that NANOG also plays roles in primitive streak and mesendoderm differentiation in early mouse development (40–44). However, we failed to detect significant up-regulation of known primitive streak or mesendoderm genes such as T, MIXL1, and EMOES in SD- and SDC-treated cells, indicating a non-mesendoderm fate in SDC-treated hPSCs (Fig. 4G.)
NANOG antagonize OTX2 to balance the cell fate between pluripotency and the default forebrain in hPSCs
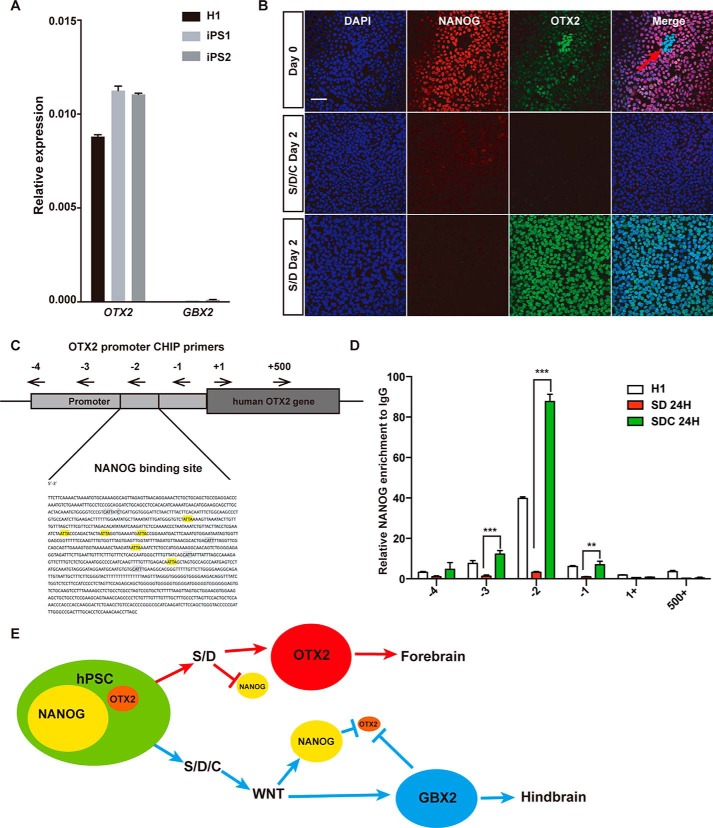
When we examined the expression of OTX2, we found that a certain amount of OTX2 expression, but not GBX2 expression, could be detected in undifferentiated hPSCs (Fig. 5A). The expression of OTX2 in hPSCs might explain the default forebrain fate differentiation for hPSCs under non-optimal conditions to support self-renewal; for example, inhibition of SMAD signaling. We then performed double immunostaining on NANOG and OTX2 in both undifferentiated and SD- or SDC-treated hESCs (Fig. 5B). Indeed, a significant level of OTX2 could be detected in undifferentiated hESCs and further increased in SD- but not SDC-treated cells (Fig. 5B). Consistent with the data shown in Fig. 4, a low amount of NANOG could be detected in SDC- but not SD-treated cells (Fig. 5B). Interestingly, we observed a mutually exclusive pattern between NANOG and OTX2 in some hESCs cells maintained under undifferentiated conditions (Fig. 5B, arrow), indicating a reciprocal antagonism between NANOG and OTX2 in balancing the cell fate between pluripotency and the default forebrain. To further confirm direct suppression of OTX2 by NANOG, we examined the proximal region of the OTX2 promoter and identified a couple of known NANOG binding motifs (45): CAAT and ATTA (Fig. 5C). Further, by ChIP assay, we showed that binding of NANOG on the OTX2 promoter region is significantly enriched not only in undifferentiated hESCs but also in 24 h of SDC differentiation, demonstrating that NANOG directly binds the OTX2 promoter region in hESCs, especially in caudal initiation (Fig. 5D). Taken together, we demonstrate that NANOG is an important regulator underlying rostral–caudal patterning at an early stage of neural differentiation in hPSCs, as shown in the schematic in Fig. 5E.
Figure 5.
NANOG antagonizes OTX2 to regulate neural patterning in hESCs. A, QPCR analysis of the expression levels of OTX2 or GBX2 in undifferentiated H1 hESCs and two iPSCs. B, dual immunostaining of NANOG and OTX2 in untreated or SD- or SDC-treated hESCs. Scale bar = 50 μm. C, NANOG binding motifs (highlighted) in the proximal region of the human OTX2 promoter. Four pairs of primers to detect NANOG binding in the ChIP assay are shown. D, ChIP-QPCR assay to detect NANOG binding in the OTX2 gene region by the designed primer pairs. Goat IgG served as a negative control. E, schematic of the role of NANOG in fate decision during neural patterning in hESCs. ***, p < 0.001.
Discussion
Generation of expandable region-specific NPCs from hPSCs is important for investigation of the molecular determinants of neural patterning and obtaining clinically relevant region-specific NPCs for regenerative medicine. Neural patterning is a critical step in neural development as it starts to specify neural precursors with different functions. Studies of model systems such as mice, Drosophila, etc. propose that neural patterning usually initiates with the specification of rostral–forebrain precursors (32, 46). The caudalizing morphogens subsequently respecify regional identity to establish other subdivisions of caudal neural precursors (47). Consistently, the rostral forebrain is also considered as a default cell fate during neural differentiation of hPSCs (19, 22, 23, 48). However, the detailed regulation and mechanisms underlying neural patterning at an early stage have not yet been fully elucidated. Here we show that OTX2, a known rostral marker gene, exhibits a relatively high level of expression in undifferentiated hPSCs and is a dominant trigger for hPSC neural differentiation. GSK3β inhibition via CHIR at an early stage of differentiation can switch the default rostral to the caudal fate. Mechanistically, we showed that NANOG was transiently sustained by CHIR at the early differentiation stage through WNT signaling. Furthermore, NANOG served as a direct repressor for OTX2 during differentiation to regulate the specification of the regional neural fate. Our findings provide new insight into understanding the molecular mechanisms underlying cell fate choice during neural morphogenesis, particularly in the human model.
It has been reported that Wnt activation regulates pluripotency in both human and mouse PSCs (49, 50) and that NANOG is a key factor of pluripotency. We confirmed that NANOG was sustained by GSK3β inhibition at an early stage of differentiation via WNT signaling (Fig. 4E). Wnt signaling is also important to regulate neural patterning through forming morphogen gradients with other signaling factors (7, 51–53). Our data then extend the role of NANOG in patterning the regional specified neural precursors at an early stage of differentiation. Interestingly, OTX2 was also reported to interact with NANOG in a different stage of development (45, 54, 55). Our findings in the hPSC model then extend studies showing that NANOG and OTX2 form a mutual regulatory loop to regulate lineage decisions when hPSCs determine to exit pluripotency and initiate differentiation.
Experimental procedures
Cell culture and neural induction
Human embryonic stem cells (H1 and H9, passages 40–50, Wicell, Madison, WI) and an iPSC line (UC5C1, passages 15–25) were maintained on 6-well plates coated with Matrigel (BD) in mTeSR1 (STEMCELL Technologies). The cells were passaged at a 1:4 split ratio every 3 or 4 days by 0.5 mm EDTA–Na2 dissociation.
Neural differentiation was performed as described previously (12, 19). Pluripotent stem cells were passaged normally on Matrigel and changed into neural induction medium (NIM) the following day. NIM contained N2B27 medium (Dulbecco's modified Eagle's medium/F12:Neuralbasal (1:1), 0.5× N2, 0.5× B27, 1% Glutamax (Gibco), 1% non-essential amino acids (Gibco), and inhibitors. SD-NIM contained 5 μm SB431542 (Sigma) and 1 μm Dorsomorphin (Sigma). SDC-NIM included 3 μm CHIR99021 (Sigma) with SB431542 and Dorsomorphin. After 6 days of induction, cells were passaged on a new Matrigel plate using 0.2 mg/ml dispase at a 1:2 ratio. Induction continued to day 14, changing NIM every 2 days. Cells were dissociated to small clumps and suspended in neural stem cell medium containing N2B27 medium, 20 ng/ml epidermal growth factor, and 20 ng/ml basic fibroblast growth factor (bFGF).
Neural differentiation in vitro
SD NPCs and SDC NPCs were cultured in neural stem cell medium, expanding in vitro in the form of single cells dissociated by Accutase (Sigma). For pan-neural differentiation, NSC spheres were plated on Matrigel-coated glass coverslips and cultured in N2B27 medium with brain-derived neurotrophic factor and glial cell line-derived neurotrophic factor (GDNF) (both at 10 ng/ml, Peprotech) and 1 μm cAMP (Sigma). The medium was changed every 2–3 days. Neuron markers were examined by immunofluorescence after 4 weeks of differentiation.
RNA extraction and QPCR
TRIzol (Life Technologies) was used to extract RNA according to the manufacturer's protocol. Real-time PCR was carried out using a CFX-96 real-time PCR detection system (Bio-Rad). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used to normalize the measured transcript. Primer sequences are listed in the supporting information.
Fluorescence-activated cell sorting
Human pluripotent stem cells or differentiated derivatives were dissociated into single cells using Accutase, centrifuged, resuspended with BD Fixation Buffer (catalog number 554655) for 20 min at room temperature, washed in PBS, and then permeabilized with dilute permeate buffer (BD Biosciences, 11×) for 10 min at 4 °C. The straight marked antibodies mouse anti-PAX6 (1:200, BD Biosciences) and goat anti-T (1:200, R&D Systems) were diluted with BD Permeate Buffer, incubated for 1 h at room temperature, washed and suspended with PBS, and then sorted using a BD C6 analyzer. All primary antibody information is listed in Table S2.
Immunostaining
Cells were fixed in 4% paraformaldehyde for 20 min at room temperature and washed briefly three times with PBS. Primary antibodies were diluted in buffer containing 10% serum and 0.3% Triton X-100 in PBS. Then cells were incubated for 16–18 h at 4 °C. Primary antibodies were washed three times with PBS. Secondary antibodies diluted in PBS were incubated for 1 h at room temperature. After washing three times with PBS, 4′,6-diamidino-2-phenylindole (0.2 μg/ml) was incubated for 10 min at room temperature. Then the coverslips were washed twice with PBS and mounted in Fluoromount-G (Life Technologies).
Western blot and phospho-Western blot analyses
After removing the medium, cells were washed using PBS. Then whole-cell protein was extracted by lysing cells in complete Lysis-M, an EDTA-free kit containing Complete Protease Inhibitor Mixture or adding Phos-STOP (Roche, Indianapolis, IN). Cell lysate was collected and centrifuged at 14,000 × g for 5–10 min. The supernatants containing soluble proteins were used for further analysis. The extracted protein was fractionated by SDS-polyacrylamide gel electrophoresis on a 12% acrylamide gel and electroblotted onto a polyvinylidene difluoride membrane (Millipore, Billerica, MA). The membranes were reacted with the following primary and secondary antibodies: GAPDH, PAX6 (Developmental Studies Hybridoma Bank, Iowa City, IA), GSK3β mouse mAb (Cell Signaling Technology), phospho-GSK3β (Ser-9) rabbit mAb (Cell Signaling Technology), anti-rabbit IgG horseradish peroxidase (HRP)-conjugated secondary antibody (Cell Signaling Technology), and anti-mouse IgG HRP-conjugated secondary antibody (Cell Signaling Technology). HRP was detected by Hyperfilm electrochemiluminescence (Invitrogen) and visualized with a gel imaging system (SmartChemiTM II, Sagecreation).
Electrophysiological analysis (patch-clamp recording)
Whole-cell patch clamp recording techniques were used to analyze the physiological properties of induced NSC-derived neurons in culture using a MultiClamp700B amplifier (Molecular Devices). Individual coverslips transferred to the recording chamber were perfused with external solution containing 145 mm NaCl, 3 mm KCl, 2 mm MgCl2, 3 mm CaCl2, 10 mm HEPES, and 10 mm glucose (pH with NaOH to 7.3). Patch pipettes with a resistance between 8–10 megaohm were pulled from borosilicate glass and filled with intracellular solution with the following composition: 136.5 mm K-gluconate, 0.2 mm EGTA, 10 mm HEPES, 5 mm NaCl, 1 mm MgCl2, 4 mm Mg-ATP, and 0.3 Na-GTP, adjusted with KOH to pH 7.2, 285 osmol/liter. The spontaneous postsynaptic current and current response to exogenous focal application of glutamate and GABA were recorded at a holding potential of −70 mV. Pressure ejection was used to deliver 1 mm glutamate (10 pounds/square inch, 100 ms) and 1 mm GABA (10 pounds/square inch, 100 ms) through a puffing electrode (3–4 megaohm) placed near the recorded neuron. Pipettes were filled with solutions containing (in mm): 145 potassium gluconate, 0.2 EGTA, 10 HEPES, 5 NaCl, 1 MgCl2, 4 Mg-ATP, and 0.3 Na-GTP (pH 7.2 with KOH, 285 osmol/liter). The action potentials were elicited with injected current under the current clamp mode. Signals were digitized with a Digidata 1440 and acquired with pClamp 10 software. Off-line data analysis was performed using Clampfit 10 (Molecular Devices).
RNA sequencing
Total RNA was prepared with the Direct-zol RNA MiniPrep kit (Zymo Research) following the manufacturer's protocol. RNA was then purified, fragmented, reverse-transcribed, labeled, and amplified to generate a sequencing-ready cDNA library with the TruSeq RNA Sample Prep Kit (Illumina). A size selection step was included to purify cDNA libraries to enrich for 250- to 300-bp fragments instead of AMPure XP bead purification. The DNA was recovered from each gel slice using the QIAquick gel extraction kit (Qiagen). The cDNA library concentration was determined with the QubitdsDNA HS Assay kit (Invitrogen). An additional sample concentrating step was included when the library concentration fell below the required loading amount. The samples were run on a MiSeq and Nexseq500 system with a MiSeq Reagent Kit v2 (50 cycles) and NextSeq 500/550 High Output v2 (150 cycles) (Illumina). In data analysis, during the correlation analysis, to avoid TPM divide by zero errors and log (0) errors, 1 was added to the TPM value and then the expression value was log-transformed. In short, the correlation analysis was calculated based on log(TPM+1). In differential expression profile analysis, the up-regulated genes in SDC NPC and SD NPC samples were those with a -fold change >2, and the down-regulated genes in samples were those with a -fold change <½.
ChIP-QPCR
ChIP assays were performed as described elsewhere (56) with goat anti-NANOG and normal goat IgG. The sequences of all ChIP primers used in this study are given in Table S2. The results were normalized to the IgG control.
Statistical analysis
In general, experiments were done from three biological repeats. Data are presented as mean ± S.D. calculated using Prism. Data were compared by using standard or repeated measures analysis of variance. Pairwise comparisons were performed with two-tailed Student's t test (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
Author contributions
G. P., Z. S., and B. L. conceived the hypotheses and designed the experiments. G. P., Z. S., and X. Z. wrote the article. Z. S. and Y. Z. performed the experiments and generated data for all figures. L. W. and Y. G. provided assistance for the research strategy. X. C. participated in experiments and analysis for Fig. 1. H. W. completed and analyzed the results of the electrophysiology. Y. S. performed experiments for the ChIP assay.
Supplementary Material
Acknowledgments
We thank the laboratory members at the Guangzhou Institutes of Biomedicine and Health for help.
This work was supported by the National Key Research and Development Program of China, Stem Cell and Translational Research (2017YFA0102601); Frontier and Key Technology Innovation Special Grants from the Department of Science and Technology of Guangdong Province (2014B020225006, 2014B020225002, 2014B050504008, 2015B020228003, 2016B030230002, and 2016B030229008); the Natural Science Foundation of Guangdong Province, China (2016A030313167); the National Basic Research Program of China, 973 Program of China (2015CB964901); the National Natural Science Foundation of China (31371514 and 31421004); Cooperation Grants of the Natural Science Foundation of Guangdong Province (2014A030313801, 2014A030312012, and 2015A030310229); Science and Information Technology of Guangzhou Key Project (201508020258, 201506010092, and 2015A030310254); the Youth Innovation Promotion Association of the Chinese Academy of Sciences (2015293); the “Hundred Talents Program” of the Chinese Academy of Science (to X. Z.); the Science and Technology Planning Project of Guangdong Province, China (2014B030301058); and the Guangdong Province Special Program for Elite Scientists in Science and Technology Innovation (2014TQ01R746 to B. L. and 2015TX01R203 to G. P.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1 and S2 and Tables S1–S3.
- A-P
- anterior–posterior
- D–V
- dorsal–ventral
- FGF
- fibroblast growth factor
- RA
- retinoic acid
- hPSC
- human pluripotent stem cell
- ESC
- embryonic stem cell
- iPSC
- induced pluripotent stem cell
- CHIR
- CHIR99021
- SD
- SB431542/Dorsomorphin
- SDC
- SB431542/Dorsomorphin/CHIR99021
- QPCR
- quantitative PCR
- NIM
- neural induction medium
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- HRP
- horseradish peroxidase
- BMP
- bone morphogenic protein.
References
- 1. Wurst W., and Bally-Cuif L. (2001) Neural plate patterning: upstream and downstream of the isthmic organizer. Nat. Rev. Neurosci. 2, 99–108 10.1038/35053516 [DOI] [PubMed] [Google Scholar]
- 2. Götz M., and Huttner W. B. (2005) The cell biology of neurogenesis. Nat. Rev. Mol. Cell Biol. 6, 777–788 10.1038/nrm1739 [DOI] [PubMed] [Google Scholar]
- 3. Kicheva A., and Briscoe J. (2015) Developmental pattern formation in phases. Trends Cell Biol. 25, 579–591 10.1016/j.tcb.2015.07.006 [DOI] [PubMed] [Google Scholar]
- 4. Bier E., and De Robertis E. M. (2015) Embryo development: BMP gradients: a paradigm for morphogen-mediated developmental patterning. Science 348, aaa5838–aaa5838 10.1126/science.aaa5838 [DOI] [PubMed] [Google Scholar]
- 5. Pera E. M., Acosta H., Gouignard N., Climent M., and Arregi I. (2014) Active signals, gradient formation and regional specificity in neural induction. Exp. Cell Res. 321, 25–31 10.1016/j.yexcr.2013.11.018 [DOI] [PubMed] [Google Scholar]
- 6. Liu A., and Niswander L. A. (2005) Bone morphogenetic protein signalling and vertebrate nervous system development. Nat. Rev. Neurosci. 6, 945–954 10.1038/nrn1805 [DOI] [PubMed] [Google Scholar]
- 7. Ciani L., and Salinas P. C. (2005) Signalling in neural development: WNTS in the vertebrate nervous system: from patterning to neuronal connectivity. Nat. Rev. Neurosci. 6, 351–362 10.1038/nrn1665 [DOI] [PubMed] [Google Scholar]
- 8. Fuccillo M., Joyner A. L., and Fishell G. (2006) Morphogen to mitogen: the multiple roles of hedgehog signalling in vertebrate neural development. Nat. Rev. Neurosci. 7, 772–783 10.1038/nrn1990 [DOI] [PubMed] [Google Scholar]
- 9. Sánchez-Arrones L., Ferrán J. L., Rodríguez-Gallardo L., and Puelles L. (2009) Incipient forebrain boundaries traced by differential gene expression and fate mapping in the chick neural plate. Dev. Biol. 335, 43–65 10.1016/j.ydbio.2009.08.012 [DOI] [PubMed] [Google Scholar]
- 10. Zhou S., Ochalek A., Szczesna K., Avci H. X., Kobolák J., Varga E., Rasmussen M., Holst B., Cirera S., Hyttel P., Freude K. K., and Dinnyés A. (2016) The positional identity of iPSC-derived neural progenitor cells along the anterior-posterior axis is controlled in a dosage-dependent manner by bFGF and EGF. Differentiation 92, 183–194 10.1016/j.diff.2016.06.002 [DOI] [PubMed] [Google Scholar]
- 11. Zhang S. C., Wernig M., Duncan I. D., Brüstle O., and Thomson J. A. (2001) In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat. Biotechnol. 19, 1129–1133 10.1038/nbt1201-1129 [DOI] [PubMed] [Google Scholar]
- 12. Gerrard L., Rodgers L., and Cui W. (2005) Differentiation of human embryonic stem cells to neural lineages in adherent culture by blocking bone morphogenetic protein signaling. Stem Cells 23, 1234–1241 10.1634/stemcells.2005-0110 [DOI] [PubMed] [Google Scholar]
- 13. Koch P., Opitz T., Steinbeck J. A., Ladewig J., and Brüstle O. (2009) A rosette-type, self-renewing human ES cell-derived neural stem cell with potential for in vitro instruction and synaptic integration. Proc. Natl. Acad. Sci. U.S.A. 106, 3225–3230 10.1073/pnas.0808387106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Du Z.-W., Chen H., Liu H., Lu J., Qian K., Huang C.-L., Zhong X., Fan F., and Zhang S.-C. (2015) Generation and expansion of highly pure motor neuron progenitors from human pluripotent stem cells. Nat. Commun. 6, 6626 10.1038/ncomms7626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ma L., Hu B., Liu Y., Vermilyea S. C., Liu H., Gao L., Sun Y., Zhang X., and Zhang S.-C. (2012) Human embryonic stem cell-derived GABA neurons correct locomotion deficits in quinolinic acid-lesioned mice. Cell Stem Cell 10, 455–464 10.1016/j.stem.2012.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Krencik R., Weick J. P., Liu Y., Zhang Z.-J., and Zhang S.-C. (2011) Specification of transplantable astroglial subtypes from human pluripotent stem cells. Nat. Biotechnol. 29, 528–534 10.1038/nbt.1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tao Y., and Zhang S.-C. (2016) Neural subtype specification from human pluripotent stem cells. Cell Stem Cell 19, 573–586 10.1016/j.stem.2016.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang S.-C. (2006) Neural subtype specification from embryonic stem cells. Brain Pathol. 16, 132–142 10.1111/j.1750-3639.2006.00008.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chambers S. M., Fasano C. A., Papapetrou E. P., Tomishima M., Sadelain M., and Studer L. (2009) Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 27, 275–280 10.1038/nbt.1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shi Y., Kirwan P., and Livesey F. J. (2012) Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat. Protoc. 7, 1836–1846 10.1038/nprot.2012.116 [DOI] [PubMed] [Google Scholar]
- 21. Shi Y., Kirwan P., Smith J., Robinson H. P. C., and Livesey F. J. (2012) Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nat. Neurosci. 15, 477–486, S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Surmacz B., Fox H., Gutteridge A., Fish P., Lubitz S., and Whiting P. (2012) Directing differentiation of human embryonic stem cells toward anterior neural ectoderm using small molecules. Stem Cells 30, 1875–1884 10.1002/stem.1166 [DOI] [PubMed] [Google Scholar]
- 23. LaVaute T. M., Yoo Y. D., Pankratz M. T., Weick J. P., Gerstner J. R., and Zhang S.-C. (2009) Regulation of neural specification from human embryonic stem cells by BMP and FGF. Stem Cells 27, 1741–1749 10.1002/stem.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lupo G., Bertacchi M., Carucci N., Augusti-Tocco G., Biagioni S., and Cremisi F. (2014) From pluripotency to forebrain patterning: an in vitro journey astride embryonic stem cells. Cell. Mol. Life Sci. 71, 2917–2930 10.1007/s00018-014-1596-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zirra A., Wiethoff S., and Patani R. (2016) Neural conversion and patterning of human pluripotent stem cells: a developmental perspective. Stem Cells Int. 10.1155/2016/8291260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kirkeby A., Grealish S., Wolf D. A., Nelander J., Wood J., Lundblad M., Lindvall O., and Parmar M. (2012) Generation of regionally specified neural progenitors and functional neurons from human embryonic stem cells under defined conditions. Cell Rep. 1, 703–714 10.1016/j.celrep.2012.04.009 [DOI] [PubMed] [Google Scholar]
- 27. Henrique D., Abranches E., Verrier L., and Storey K. G. (2015) Neuromesodermal progenitors and the making of the spinal cord. Development 142, 2864–2875 10.1242/dev.119768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maury Y., Côme J., Piskorowski R. A., Salah-Mohellibi N., Chevaleyre V., Peschanski M., Martinat C., and Nedelec S. (2015) Combinatorial analysis of developmental cues efficiently converts human pluripotent stem cells into multiple neuronal subtypes. Nat. Biotechnol. 33, 89–96 10.1038/nbt.3049 [DOI] [PubMed] [Google Scholar]
- 29. Kelava I., and Lancaster M. A. (2016) Stem cell models of human brain development. Cell Stem Cell 18, 736–748 10.1016/j.stem.2016.05.022 [DOI] [PubMed] [Google Scholar]
- 30. Thomson J. A., Itskovitz-Eldor J., Shapiro S. S., Waknitz M. A., Swiergiel J. J., Marshall V. S., and Jones J. M. (1998) Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 10.1126/science.282.5391.1145 [DOI] [PubMed] [Google Scholar]
- 31. Xue Y., Cai X., Wang L., Liao B., Zhang H., Shan Y., Chen Q., Zhou T., Li X., Hou J., Chen S., Luo R., Qin D., Pei D., and Pan G. (2013) Generating a non-integrating human induced pluripotent stem cell bank from urine-derived cells. PLoS ONE 8, e70573–7 10.1371/journal.pone.0070573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stern C. D., Charité J., Deschamps J., Duboule D., Durston A. J., Kmita M., Nicolas J.-F., Palmeirim I., Smith J. C., and Wolpert L. (2006) Head-tail patterning of the vertebrate embryo: one, two or many unresolved problems? Int. J. Dev. Biol. 50, 3–15 10.1387/ijdb.052095cs [DOI] [PubMed] [Google Scholar]
- 33. Simpson T. I., and Price D. J. (2002) Pax6; A pleiotropic player in development. BioEssays 24, 1041–1051 10.1002/bies.10174 [DOI] [PubMed] [Google Scholar]
- 34. Zhang X., Huang C. T., Chen J., Pankratz M. T., Xi J., Li J., Yang Y., Lavaute T. M., Li X.-J., Ayala M., Bondarenko G. I., Du Z.-W., Jin Y., Golos T. G., and Zhang S.-C. (2010) Pax6 is a human neuroectoderm cell fate determinant. Cell Stem Cell 7, 90–100 10.1016/j.stem.2010.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sunmonu N. A., Li K., Guo Q., and Li J. Y. (2011) Gbx2 and Fgf8 are sequentially required for formation of the midbrain-hindbrain compartment boundary. Development 138, 725–734 10.1242/dev.055665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Carletti B., and Rossi F. (2008) Neurogenesis in the cerebellum. Neuroscientist 14, 91–100 [DOI] [PubMed] [Google Scholar]
- 37. Li J. Y., and Joyner A. L. (2001) Otx2 and Gbx2 are required for refinement and not induction of mid-hindbrain gene expression. Development 128, 4979–4991 [DOI] [PubMed] [Google Scholar]
- 38. Funa N. S., Schachter K. A., Lerdrup M., Ekberg J., Hess K., Dietrich N., Honoré C., Hansen K., and Semb H. (2015) β-Catenin regulates primitive streak induction through collaborative interactions with SMAD2/SMAD3 and OCT4. Cell Stem Cell 16, 639–652 10.1016/j.stem.2015.03.008 [DOI] [PubMed] [Google Scholar]
- 39. Huang S.-M., Mishina Y. M., Liu S., Cheung A., Stegmeier F., Michaud G. A., Charlat O., Wiellette E., Zhang Y., Wiessner S., Hild M., Shi X., Wilson C. J., Mickanin C., Myer V., et al. (2009) Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461, 614–620 10.1038/nature08356 [DOI] [PubMed] [Google Scholar]
- 40. Hart A. H., Hartley L., Ibrahim M., and Robb L. (2004) Identification, cloning and expression analysis of the pluripotency promoting Nanog genes in mouse and human. Dev. Dyn. 230, 187–198 10.1002/dvdy.20034 [DOI] [PubMed] [Google Scholar]
- 41. Hatano S.-Y., Tada M., Kimura H., Yamaguchi S., Kono T., Nakano T., Suemori H., Nakatsuji N., and Tada T. (2005) Pluripotential competence of cells associated with Nanog activity. Mech. Dev. 122, 67–79 10.1016/j.mod.2004.08.008 [DOI] [PubMed] [Google Scholar]
- 42. Osorno R., Tsakiridis A., Wong F., Cambray N., Economou C., Wilkie R., Blin G., Scotting P. J., Chambers I., and Wilson V. (2012) The developmental dismantling of pluripotency is reversed by ectopic Oct4 expression. Development. 139, 2288–2298 10.1242/dev.078071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vallier L., Mendjan S., Brown S., Chng Z., Teo A., Smithers L. E., Trotter M. W., Cho C. H., Martinez A., Rugg-Gunn P., Brons G., and Pedersen R. A. (2009) Activin/Nodal signalling maintains pluripotency by controlling Nanog expression. Development. 136, 1339–1349 10.1242/dev.033951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang Z., Oron E., Nelson B., Razis S., and Ivanova N. (2012) Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell 10, 440–454 10.1016/j.stem.2012.02.016 [DOI] [PubMed] [Google Scholar]
- 45. Gifford C. A., Ziller M. J., Gu H., Trapnell C., Donaghey J., Tsankov A., Shalek A. K., Kelley D. R., Shishkin A. A., Issner R., Zhang X., Coyne M., Fostel J. L., Holmes L., Meldrim J., et al. (2013) Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell 153, 1149–1163 10.1016/j.cell.2013.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Andoniadou C. L., and Martinez-Barbera J. P. (2013) Developmental mechanisms directing early anterior forebrain specification in vertebrates. Cell. Mol. Life Sci. 70, 3739–3752 10.1007/s00018-013-1269-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Becker C. G., and Diez Del Corral R. (2015) Neural development and regeneration: it's all in your spinal cord. Development 142, 811–816 10.1242/dev.121053 [DOI] [PubMed] [Google Scholar]
- 48. Ozair M. Z., Noggle S., Warmflash A., Krzyspiak J. E., and Brivanlou A. H. (2013) SMAD7 directly converts human embryonic stem cells to telencephalic fate by a default mechanism. Stem Cells 31, 35–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sato N., Meijer L., Skaltsounis L., Greengard P., and Brivanlou A. H. (2004) Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat. Med. 10, 55–63 10.1038/nm979 [DOI] [PubMed] [Google Scholar]
- 50. Ying Q.-L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., and Smith A. (2008) The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 10.1038/nature06968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Leung A. W., Murdoch B., Salem A. F., Prasad M. S., Gomez G. A., and García-Castro M. I. (2016) WNT/ -catenin signaling mediates human neural crest induction via a pre-neural border intermediate. Development 143, 398–410 10.1242/dev.130849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yaguchi J., Takeda N., Inaba K., and Yaguchi S. (2016) Cooperative Wnt-Nodal signals regulate the patterning of anterior neuroectoderm. PLoS Genet. 12, e1006001–27 10.1371/journal.pgen.1006001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Caronia-Brown G., Anderegg A., and Awatramani R. (2016) Expression and functional analysis of the Wnt/β-catenin induced mir-135a-2 locus in embryonic forebrain development. Neural Dev. 11, 9 10.1186/s13064-016-0065-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tsankov A. M., Gu H., Akopian V., Ziller M. J., Donaghey J., Amit I., Gnirke A., and Meissner A. (2015) Transcription factor binding dynamics during human ES cell differentiation. Nature 518, 344–349 10.1038/nature14233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Acampora D., Omodei D., Petrosino G., Garofalo A., Savarese M., Nigro V., Di Giovannantonio L. G., Mercadante V., and Simeone A. (2016) Loss of the Otx2-binding site in the Nanog promoter affects the integrity of embryonic stem cell subtypes and specification of inner cell mass-derived epiblast. Cell Rep. 15, 2651–2664 10.1016/j.celrep.2016.05.041 [DOI] [PubMed] [Google Scholar]
- 56. Chen J., Liu H., Liu J., Qi J., Wei B., Yang J., Liang H., Chen Y., Chen J., Wu Y., Guo L., Zhu J., Zhao X., Peng T., Zhang Y., et al. (2013) H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat. Genet. 45, 34–42 10.1038/ng.2491 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.