Abstract
GlcUAβ1–3GalNAc(4S,6S) (E unit)–rich domains have been shown to play key roles in various biological functions of chondroitin sulfate (CS). However, an enzyme that can specifically isolate such domains through the selective digestion of other domains in polysaccharides has not yet been reported. Here, we identified a glycosaminoglycan lyase from a marine bacterium Vibrio sp. FC509. This enzyme efficiently degraded hyaluronic acid (HA) and CS variants, but not E unit–rich CS-E, into unsaturated disaccharides; therefore, we designated this enzyme a CS-E–resisted HA/CS lyase (HCLase Er). We isolated a series of resistant oligosaccharides from the final product of a low-sulfated CS-E exhaustively digested by HCLase Er and found that the E units were dramatically accumulate in these resistant oligosaccharides. By determining the structures of several resistant tetrasaccharides, we observed that all of them possessed a Δ4,5HexUAα1–3GalNAc(4S,6S) at their non-reducing ends, indicating that the disulfation of GalNAc abrogates HCLase Er activity on the β1–4 linkage between the E unit and the following disaccharide. Δ4,5HexUAα1–3GalNAc(4S,6S)β1–4GlcUAβ1–3GalNAc(4S,6S) was most strongly resistant to HCLase Er. To our knowledge, this study is the first reporting a glycosaminoglycan lyase specifically inhibited by both 4-O- and 6-O-sulfation of GalNAc. Site-directed and truncation mutagenesis experiments indicated that HCLase Er may use a general acid–base catalysis mechanism and that an extra domain (Gly739–Gln796) is critical for its activity. This enzyme will be a useful tool for structural analyses and for preparing bioactive oligosaccharides of HA and CS variants, particularly from E unit–rich CS chains.
Keywords: enzyme degradation, glycosaminoglycan, chondroitin sulfate, hyaluronan, oligosaccharide, proteoglycan, lyase
Introduction
CS2 is a negatively charged polysaccharide synthesized as the glycosaminoglycan (GAG) side chain of proteoglycans and is widely distributed on cell surfaces and in the extracellular matrix (1–3). CS participates in various important biological processes, such as neuronal growth (4–6), morphogenesis (7), inflammation (8, 9), tumor progression (10–12), virus infection (13–15), and cytokinesis (16, 17). These various functions of CS are attributed to its structural diversity.
The backbone of the CS chain is composed of repeating disaccharide units of glucuronic acid (GlcUA) and N-acetyl-d-galactosamine (GalNAc) and is further modified by specific sulfotransferases at C-2 of GlcUA/l-iduronic acid and/or C-4 and/or C-6 of GalNAc to yield substantial structural diversity via differential sulfation patterns (18, 19). Moreover, some GlcUA residues are epimerized into l-iduronic acid residues (IdoUA) by glucuronyl C-5 epimerase, and correspondingly, the chain containing repeating disaccharide units of -IdoUA-GalNAc- is designated dermatan sulfate (DS) (20, 21). Thus, CS and DS often exist in a single chain in a hybrid form (CS-DS) and are usually periodically distributed in a cell/tissue-specific manner. Several different sulfation modifications by a number of sulfotransferases result in the creation of a variety of CS disaccharide units, such as the O unit (GlcUAβ1–3GalNAc), the A unit (GlcUAβ1–3GalNAc(4S)), the C unit (GlcUAβ1–3GalNAc(6S)), the D unit (GlcUA(2S)β1–3GalNAc(6S)), the E unit (GlcUAβ1–3GalNAc(4S,6S)), and the T unit (GlcUA(2S)β1-3GalNAc(4S,6S)), where 2S, 4S, and 6S represent 2-O-, 4-O-, and 6-O-sulfate groups, respectively.
CS chains containing the E unit have been found in mammalian tissues, including brain, liver, spleen, kidney, and malignant tumors, and they have been shown to be involved in various physiological and pathological processes through interactions with various growth factors or chemokines (22, 23). Interestingly, these interactions are specifically inhibited by CS or DS chains containing the E unit and iE unit (IdoUA-GalNAc(4S,6S) where “i” represents IdoUA), respectively (1). Furthermore, E unit–rich CS polysaccharides, such as CS-E from squid cartilage, show remarkable biological activities, such as promotion of neurite outgrowth (24–26), inhibition of viral infection (13, 27, 28), and osteoanabolism induced by estrogen (29). Studies of the structure–function relationship of E unit-containing polysaccharides have shown that E unit–rich domains in these CS/DS chains play key roles in various biological functions, and the minimal essential structures for various bioactivities of CS-E have been identified (11, 30–32). Compared with the complexity of polysaccharides, CS-E oligosaccharides are much more homogeneous in size and structure, which will greatly exclude the ambiguity of polysaccharides in a specific bioactivity, and they are more easily absorbed and utilized due to their low molecular mass. Thus, the ability to selectively prepare E unit–rich oligosaccharides from CS-E polysaccharides will be critical for the development of CS therapeutic agents.
Various CS/DS-degrading enzymes from bacteria have been used for the structure–function studies and oligosaccharide preparations of CS/DS polysaccharides (33–35). However, to date, no enzyme has been found that can effectively digest CS/DS domains/chains without degrading E unit–rich sequences and thereby selectively preserve E unit–rich oligosaccharides in the digests of polysaccharides. In this study, an endolytic GAG lyase was identified from a marine bacterium Vibrio sp. FC509. This enzyme efficiently digests HA, CS-A, CS-C, and CS-D into disaccharides but only partially degrades CS-E. Further study showed that the presence of an E unit, and particularly consecutive E units, is a strong resistant structure of the activity of this enzyme. Notably, this enzyme is the first to be identified with activity that is specifically inhibited by both 4-O- and 6-O-sulfation of GalNAc, and it will serve as a very useful tool for structure–function studies and bioactive oligosaccharide preparations of E unit-containing CS/DS.
Results
Information related to the HCLase Er gene and protein sequence
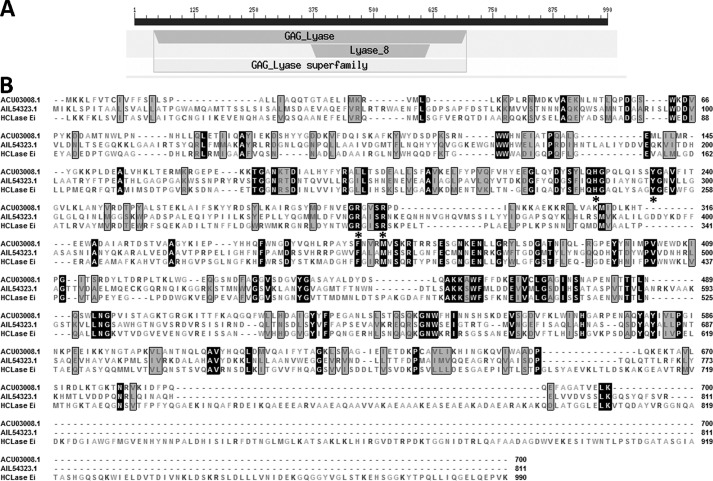
The putative GAG-degrading gene named hclase er (GenBankTM accession number MF458894) was 2973 bp in length and had a GC content of 50.6%. The predicted full-length protein was composed of 990 amino acid residues with a theoretical molecular mass of 108.4 kDa and an isoelectric point of 5.34. The Simple Modular Architecture Research Tool (SMART), SignalP 4.0, and LipoP 1.0 analyses indicated that HCLase Er contained a type I signal peptide composed of 21 amino acid residues at its N terminus. Carbohydrate-active enZYme database (http://www.cazy.org/)3 (55), Simple Modular Architecture Research Tool, and BLASTp search showed that the HCLase Er protein contained a GAG_lyase superfamily module (Glu41–Pro695) and a lyase_8 module (Lys369–Pro617), suggesting that the enzyme is a member of polysaccharide lyase 8 family (Fig. 1A). A BLASTp search showed that HCLase Er shared a sequence identity of higher than 30% with a number of putative chondroitinases from bacteria such as the Vibrio, Bacteroides, Indibacter, Flavobacterium, and Pedobacter strains. However, of the proteins identified, HCLase Er shared the highest percentages of sequence identity (34%) with a chondroitinase AC lyase from Flavobacterium heparinum (36) and followed by xanthan lyase (27%) from Bacillus sp. GL1 (37) and HCLase lyase (25%) from Vibrio sp. FC509 (34). Notably, compared with the two identified GAG lyases, chondroitinase AC (ACU03008.1) and HCLase (AIL54323.1), there are two long extra fragments (Gly739–Gln796 and Val816–Lys990) at the C terminus of HCLase Er protein (Fig. 1B).
Figure 1.
Sequence properties of the HCLase Er from Vibrio sp. FC509. A, module organization of HCLase Er. The GAG_ lyase module is a putative catalytic domain (Glu41–Pro695), and the module Lyase_8 (Lys369–Pro617) is conservative among polysaccharide lyase 8 family. B, amino acid sequence alignment of HCLase Er to two other chondroitinases. The asterisk below the residues indicates the critical catalytic residues, and amino acid residues with homologies of ≥75% are shaded in black frames and gray background. The figures on the right indicate the residue number of each sequence. GenBankTM accession number ACU03008.1, chondroitinase AC lyase from F. heparinum; GenBankTM accession number AIL54323.1, HCLase from Vibrio sp. FC509.
Heterologous expression of HCLase Er in Escherichia coli
The full-length sequence of the HCLase Er open reading frame was amplified directly from the genomic DNA of Vibrio sp. FC509. The PCR product was recovered and cloned into the pET-30a(+) vector after a T7 promoter. In this HCLase Er expression vector (pE30a-rHCLase Er), a His6 tag was added at the C terminus. SDS-PAGE analysis indicated that after induction with isopropyl 1-thio-β-d-galactopyranoside, E. coli BL21(DE3) cells harboring the pE30-rHCLase Er plasmid could form soluble products (∼200 mg/liter) with a correct molecular mass of 108 kDa.
The crude enzymes were extracted from the cultures of host cells by sonication and centrifugation. The recombinant enzymes were further purified by nickel-nitrilotriacetic acid–affinity chromatography. As shown in Fig. 2, SDS-PAGE showed that the rHCLase Er protein could be eluted from the nickel-nitrilotriacetic acid column using a gradient of imidazole concentrations ranging from 50 to 250 mm. The molecular mass of the purified protein was consistent with the theoretical molecular mass, and the purified protein had a purity of >99%.
Figure 2.
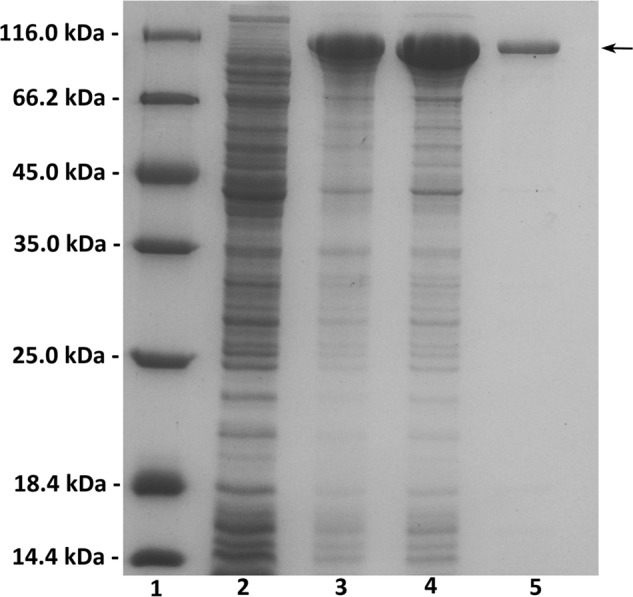
Purification of recombinant HCLase Er from E. coli by Ni2+-chelation chromatography. Enzyme purity following each fractionation step was assessed by SDS-PAGE using 13.2% polyacrylamide gels followed by staining with Coomassie Brilliant Blue. Lane 1, unstained protein molecular weight marker SM 0431 (Thermo Fisher Scientific); lane 2, uninduced cell lysate; lane 3, induced cell lysate; lane 4, supernatant fluid of the induced cell lysate; lane 5, purified recombinant HCLase Er. Ten microliters of corresponding sample containing about 10 μg of protein was individually loaded on lanes 1–4, and 1 μg of purified enzyme was loaded on lane 5. Molecular mass markers and their corresponding masses are indicated.
Enzymatic characteristics of rHCLase Er
The purified rHCLase Er was able to degrade HA, CS-A, CS-C, CS-D, and CS-E to produce oligosaccharides with an absorbance at 232 nm, which is characteristic of typical unsaturated oligosaccharides. However, rHCLase Er had no effect on DS and heparin (data not shown). These results showed that the HCLase Er protein was a GAG lyase with specificity to HA and CS.
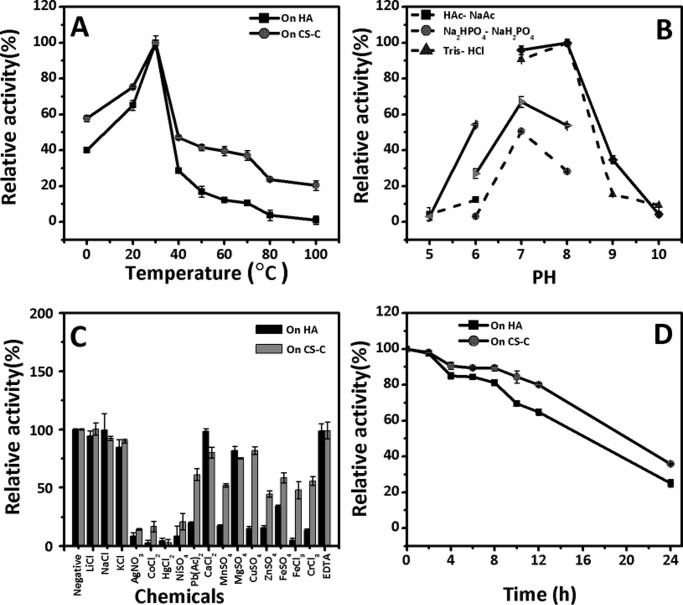
The rHCLase Er enzyme showed optimal activity at a temperature of 30 °C when either HA or CS-C was used as the substrate, and its activity rate rapidly decreased at temperatures higher than 30 °C, suggesting that this enzyme is temperature-sensitive, especially in the high-temperature region from 40 to 100 °C (Fig. 3A). The effects of pH on the reaction rate of rHCLase Er were investigated at the optimal temperature of 30 °C, and the results showed that the optimal pH was 8.0 when either HA or CS-C was used as the substrate, whereas the rate of enzymatic activity was much higher in 50 mm Tris-HCl buffer than in 50 mm NaH2PO4·Na2HPO4 buffer (Fig. 3B). Moreover, the enzyme also exhibited pH sensitivity, and the activity was completely abolished at the outlying pH values of 5.0 and 10.0. To determine the effects of metal ions on the enzymatic activity, various metal ions were added to the basic reaction buffer (50 mm Tris-HCl (pH 8.0)), and the reaction rate of rHCLase Er was measured at 30 °C. As shown in Fig. 3C, no metal ion exhibited a significant enhancing effect; however, most of the tested metal ions, such as Ag+, Co2+, Hg2+, Ni2+, Mn2+, Zn2+, and Pb3+, strongly inhibited the enzymatic activity. Additionally, the chelating regent EDTA exhibited no significant effect. The thermostability of this enzyme was assayed under the optimal conditions; the rate of HCLase Er activity gradually declined from 100 to 60% during 12 h, indicative of mild stability (Fig. 3D). Interestingly, this enzyme did not show a halophilic characteristic as previously found for HCLase (34), and even its activity was significantly inhibited when the concentration of NaCl was higher than 500 mm (data not shown). Under the optimal conditions of 50 mm Tris-Cl (pH 8.0) at 30 °C, the specific activity of rHCLase Er was measured using HA and various types of commercial CS variants as described under “Experimental procedures.” The specific activity of rHCLase Er using HA, CS-A, CS-C, CS-D, and CS-E was 13.8, 7.2, 6.7, 5.9, and 0.76 units/mg protein, respectively (Table 1). Notably, the activity of rHCLase Er against CS-E was relatively low compared with its activity against other CS variants. This result indicates that CS-E contains structures that confer resistance against digestion by rHCLase Er.
Figure 3.
Biochemical reaction conditions for recombinant HCLase Er. A, effects of temperature. The enzyme activities of rHCLase Er were measured using HA and CS-C as substrates in the 50 mm Tris-HCl buffer (pH 8.0) at different temperatures for 2 h. Data are shown as the percentage of the activity of that obtained at 30 °C (100%) for rHCLase Er. B, effects of pH. The activities of rHCLase Er against HA (dashed lines) and CS-C (solid lines) were measured in buffers with varying pH values from 5 to 10 at 30 °C for 2 h. Data are shown as the percentage of the activity of that obtained in the 50 mm Tris-HCl buffer at pH 8.0. C, effects of metal ions. The activities of rHCLase Er against HA and CS-C were measured in Tris-HCl buffer (pH 8.0) containing a 5 mm concentration of various metal ions at 30 °C for 2 h. Data are shown as the percentage of the activity of that obtained in the buffer without tested metal ions (Negative). D, thermostability of rHCLase Er. The enzyme in 50 mm Tris-HCl buffer (pH 8.0) was preincubated for 0–24 h at the optimal temperature, and the residual activity was determined in the optimal conditions. Data are shown as the percentage of that obtained for untreated HCLase Er. Error bars represent means of triplicates ±S.D.
Table 1.
Activity analysis for rHCLase Er and its mutants
| rHCLase Er | rHCLase Er-T815 | rHCLase Er-H243A, -Y252A, -R306A, -R310A, -T738, -T756 | |
|---|---|---|---|
| units/mg | units/mg | units/mg | |
| HA | 13.8 | 12.9 | NDa |
| CS-A | 7.2 | 7.3 | ND |
| CS-C | 6.7 | 6.6 | ND |
| CS-D | 5.9 | 5.7 | ND |
| CS-E | 0.76 | 0.63 | ND |
a ND means not detected.
Digestion pattern of polysaccharides by rHCLase Er
To determine the action pattern of rHCLase Er against polysaccharides, the degradation of HA or CS-A (1 mg/ml) by rHCLase Er (0.03 unit/ml) was monitored at 30 °C. The digestion of HA (Fig. 4A) and CS-A (Fig. 4B) was tested at 0, 5, 10, and 30 min and 1 and 2 h. The digests were loaded onto a SuperdexTM peptide 10/300GL column and monitored by measuring the absorbance at 232 nm. Whenever HA or CS-A was used as the substrate, rHCLase Er initially produced higher molecular mass oligosaccharides followed by smaller oligomers with strong absorbance at 232 nm, suggesting that the HCLase Er protein is a GAG endolyase.
Figure 4.
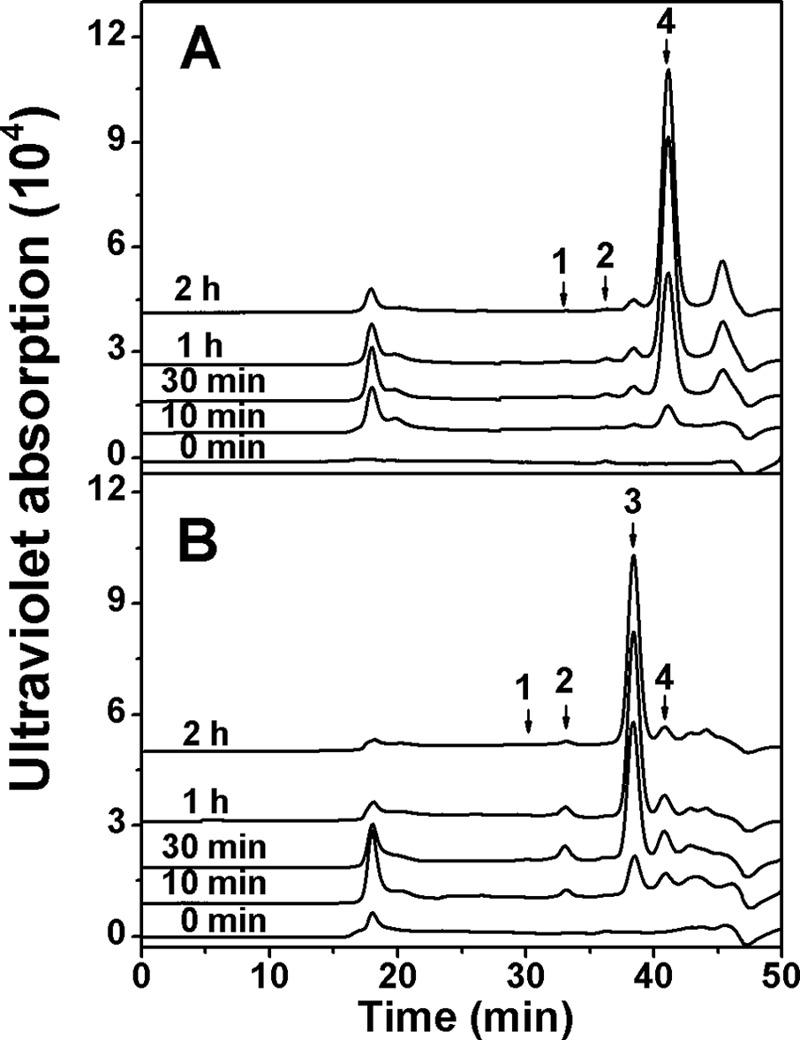
Time-course experiments of degradation of HA or CS-A by rHCLase Er. HA (A) or CS-A (B) (1 mg/ml) was treated with rHCLase Er (0.03 unit/ml), and a 20-μg aliquot was taken at different time points for gel-filtration analysis as described under “Experimental procedures.” The elution positions of the following standard oligosaccharides are indicated by arrows: 1, CS or HA hexasaccharide fraction; 2, CS or HA tetrasaccharide; 3, monosulfated CS disaccharide; 4, non-sulfated CS or HA disaccharide.
Furthermore, commercial HA, CS-A, CS-C, CS-D, and CS-E were exhaustively digested with rHCLase Er to determine their final products. The resulting oligosaccharides were analyzed and assigned by gel-filtration analysis using HA- and CS-derived authentic unsaturated oligosaccharides. As shown in Fig. 5, non-sulfated and monosulfated disaccharides were the main final products for HA (Fig. 5A) and CS-A (Fig. 5B), respectively. The digests of CS-C (Fig. 5C) and CS-D (Fig. 5D) resulted in two main disaccharide peaks corresponding to monosulfated and disulfated disaccharides. However, only a small amount of monosulfated disaccharide as well as larger oligosaccharides was detected from the digest of commercial CS-E (Fig. 5E), which suggested that specific structures in CS-E might inhibit the enzymatic activity. Furthermore, the main final products of HA and CS-A were collected and analyzed by MS spectrometry as described under “Experimental procedures.” The detected major signals at m/z at 378.1029 and 458.0557 could be assigned by mass calculation to non-sulfated unsaturated Δ4,5HexUA-GlcNAc and monosulfated Δ4,5HexUA-GalNAc(4S), respectively, which confirmed that HCLase Er is a lyase cleaving GAG chains via β-elimination mechanism.
Figure 5.
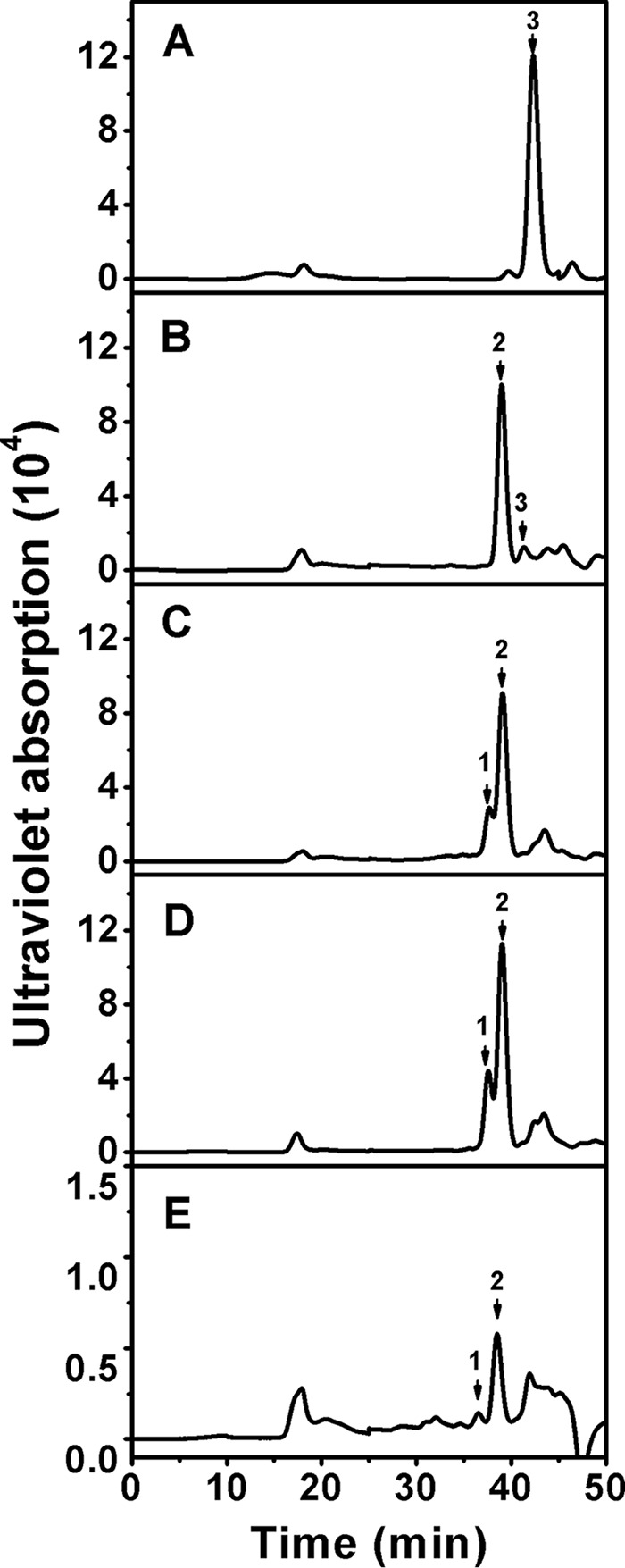
Analysis of the final products of HA and CS variants digested by rHCLase Er. Twenty micrograms each of HA (A), CS-A (B), CS-C (C), CS-D (D), or CS-E (E) were exhaustively digested with rHCLase Er and then separated into its constituent oligosaccharides by gel-filtration chromatography on a SuperdexTM peptide column as described under “Experimental procedures.” The elution positions of the following standard oligosaccharides are indicated by arrows: 1, disulfated CS disaccharides; 2, monosulfated CS disaccharides; 3, non-sulfated CS or HA disaccharides.
To confirm the finding that the specific structure in CS-E might be resistant to the enzyme activity, commercial CS-E was degraded by commercial CSase ABC and a new HA/CS lyase HCLase identified from the same bacterium Vibrio sp. FC509 (34), respectively. The results clearly showed that CS-E could be completely digested into disaccharides with one or two sulfates by CSase ABC and HCLase but not by HCLase Er (Fig. 6). The E units (GlcUAβ1–3GalNAc(4S,6S)) are the major component (>60%) in the disaccharide composition of CS-E from squid cartilage (Seikagaku Corp.). Considering this unique structural feature, we speculated that E units might play a key role in the resistance of CS-E to HCLase Er action. To test this speculation, a preparation of low-sulfated CS-E polysaccharide from Dosidicus gigas cartilage, which contained ∼39% of E units (Table 2), was used as the substrate for digestion by HCLase Er. In a time-course experiment, we could see that the low-sulfated CS-E was gradually cut into a series of low-molecular-mass oligosaccharides over time (Fig. 7). However, we noted that the profile of the chromatogram showed no significant change after digestion for 48 h, and it still showed many peaks, indicating the presence of oligosaccharides larger in size than disaccharides even after digestion for 72 h. This result indicated that the final digests of the low-sulfated CS-E contained several HCLase Er-resistant sequences of different sizes, which provided ample material for the structural analysis of HCLase Er-resistant oligosaccharides.
Figure 6.
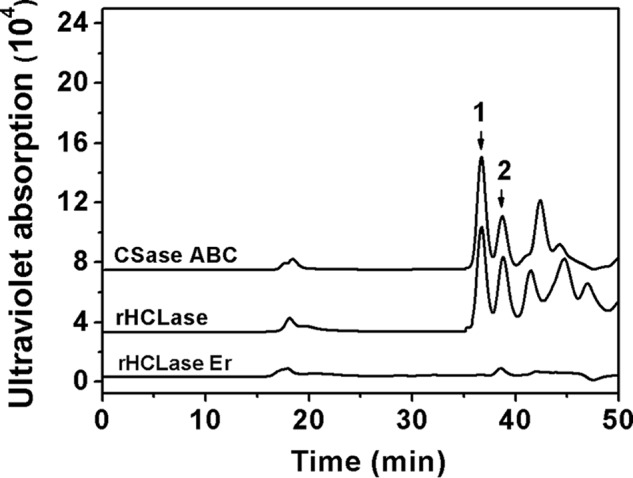
Analysis of the products of commercial CS-E digested by different CS lyases. Twenty micrograms of CS-E were digested with CSase ABC, rHCLase, or rHCLase Er in the optimal condition for 24 h and then analyzed by gel-filtration chromatography on a SuperdexTM peptide column as described under “Experimental procedures.” The elution positions of the following standard oligosaccharides are indicated by arrows: 1, disulfated CS disaccharides; 2, monosulfated CS disaccharides.
Table 2.
Disaccharide compositions of rHCLase Er-resistant oligosaccharide fractions and low sulfated CS-E polysaccharide from D. gigas cartilage
| Oligosaccharidea/polysaccharide | ΔOb | ΔC | ΔA | ΔE |
|---|---|---|---|---|
| mol %c | ||||
| Fraction 1 | NDd | 6.94 | 9.18 | 83.88 |
| Fraction 2 | ND | 8.97 | 13.44 | 77.59 |
| Fraction 3 | ND | 10.40 | 13.29 | 76.31 |
| Fraction 4 | ND | 5.72 | 19.42 | 74.86 |
| Fraction 5 | 5.37 | 6.52 | 12.04 | 76.07 |
| Fraction 6 | ND | 4.94 | 4.17 | 90.86 |
| Fraction 7 | 12.10 | 8.94 | 30.0 | 48.96 |
| Fraction 8 | ND | ND | ND | ND |
| Fraction 9 | 2.2 | 27.07 | 58.47 | 12.34 |
| Low sulfated CS-E polysaccharide | 11.5 | 13.43 | 35.74 | 39.33 |
a The names refer to the peaks and fractions designated in Fig. 7.
b The following abbreviations are used for unsaturated disaccharide units: ΔO (Δ4,5HexUAα1–3GalNAc; ΔC (Δ4,5HexUAα1–3GalNAc(6S)); ΔA (Δ4,5HexUAα1–3GalNAc(4S)); ΔE (Δ4,5HexUAα1–3GalNAc(4S,6S)).
c The molar ratio of the disaccharides was calculated in percentage terms from the peaks area obtained by digestion with chondroitinase ABC on YMC-Pack PA-G column.
d ND means not determined.
Figure 7.
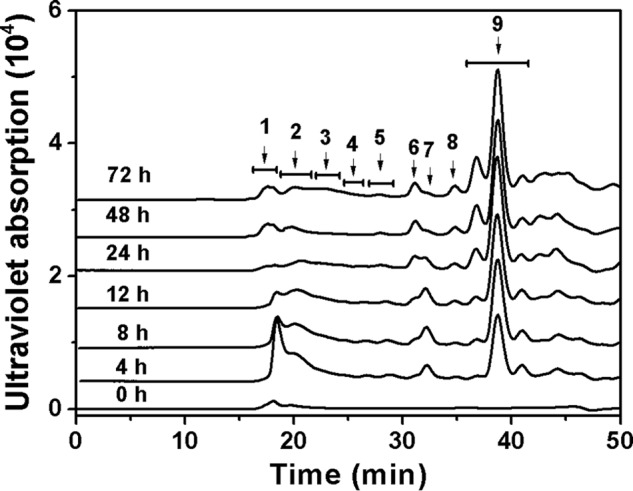
Time-course experiments of degradation of D. gigas cartilage CS-E by rHCLase Er. CS-E (1 mg/ml) was exhaustively treated with rHCLase Er, and a 20-μg aliquot was taken at different time points for gel-filtration analysis as described under “Experimental procedures.” In an enlarged experiment, the eluted fractions of the final product from digestion for 72 h were collected as indicated by arrows for further analysis as described under “Experimental procedures.”
Disaccharide composition analysis of the resistant oligosaccharides
To investigate whether E units were accumulated in the CS-E structures resistant to HCLase Er, the products of low-sulfated CS-E (10 mg) after exhaustive digestion by HCLase Er for 72 h were fractionated and collected according to the molecular size of the oligosaccharides based on the profile of chromatography (72 h), as indicated in Fig. 7. A disaccharide composition analysis showed that compared with the amount (∼39%) in the parental polysaccharide, the proportion of E units in each resistant fraction was dramatically increased and even reached levels of 84 and 90% in fractions 1 and 6, respectively (Table 2). These results strongly support our speculation that the E unit is the key inhibitory factor for the digestion of CS-E by HCLase Er.
Notably, the fraction 8 located between the CS/DS disaccharides (fraction 9) and low-sulfated tetrasaccharides (fraction 7) could not be digested by CSase ABC, indicating that it was not a CS tetra/trisaccharide fraction (data not shown). Furthermore, to exclude the possibility that it may be the CS/DS-derived unsaturated trisulfated disaccharide (Δ4,5HexUA(2S)α1-3GalNAc(4S,6S), ΔT unit), this fraction was treated with a 4-O-endosulfatase, which can effectively remove the 4-O-sulfate group from the ΔT unit (38), followed by an analysis by anion-exchange HPLC. The results showed that fraction 8 was also resistant to digestion by the 4-O-endosulfatase (data not shown), indicating it was not the ΔT unit. Thus, we speculated that fraction 8 might be the linkage regions of CS chains covalently attached to the core proteins of proteoglycans (39–41).
Sequence of the resistant tetrasaccharides from low-sulfated CS-E
To investigate the minimal essential structure of the HCLase Er-resistant oligosaccharides, the tetrasaccharide fractions (fractions 6 and 7) resistant to digestion by HCLase Er were collected as indicated in Fig. 7 and then individually subfractionated by anion-exchange chromatography on a YMC-Pack PA-G column.
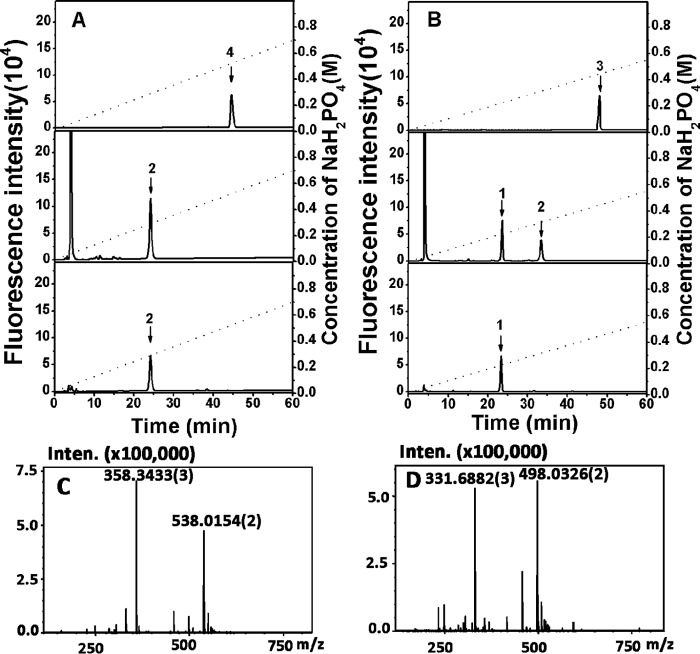
A major subfraction, accounting for 91% of the total oligosaccharides in fraction 6 (Table 3), was detected and collected for the subsequent characterization. To confirm the homogeneity of this major fraction, an aliquot (5 pmol) of the purified sample was labeled with 2-AB and analyzed by anion-exchange HPLC again, and a single peak was again detected, confirming its high purity (Fig. 8A, top panel). To further analyze the structure of this oligosaccharide, 5 pmol of this fraction was digested with CSase ABC followed by 2-AB labeling for a disaccharide composition assay by anion-exchange HPLC. The result showed that only ΔE unit (Δ4,5HexUAα1–3GalNAc(4S,6S)) was detected in the 2-AB-labeled digest (Fig. 8A, middle panel), indicating that the HCLase Er-resistant tetrasaccharide should be ΔE-E (Δ4,5HexUAα1–3GalNAc(4S,6S)β1–4GlcUAβ1-3GalNAc(4S,6S)). To confirm the identity of the tetrasaccharide, the same amount (5 pmol) of oligosaccharides as used for the disaccharide assay was first labeled with 2-AB. This process was followed by digestion with an exo-CSase from the same bacterium Vibrio sp. FC509, which could effectively cleave 2-AB-labeled CS tetrasaccharides. An anion-exchange HPLC assay showed that a single peak at the position corresponding to the 2-AB-labeled ΔE unit was detected (Fig. 8A, bottom panel). The peak area was approximately half the size of that in Fig. 8A, middle panel, indicating that the E unit at the reducing end accounted for half of all of the disaccharides contained in this oligosaccharide and thus further confirming that this major HCLase Er-resistant oligosaccharide in fraction 6 is ΔE-E. Furthermore, this oligosaccharide was analyzed by MS spectrometry in a negative ion mode. Two major signals at m/z 358.3433 [tetra-ΔEE-3H]3− and 538.0154 [tetra-ΔEE-2H]2− are observed in the MS spectrum (Fig. 8C) and can be assigned to an unsaturated CS tetrasaccharide with four sulfate groups, which has the same theory mass (1078.05 Da) as that of ΔE-E.
Table 3.
Sequences of tetrasaccharides isolated from the final product of D. gigas cartilage CS-E exhaustively digested by rHCLase Er
| Fraction | Sequence | Percentage |
|---|---|---|
| mol % | ||
| Fraction 6 | ΔE-E | 91 |
| Fraction 7 | ΔE-C | 16 |
| ΔE-A | 56 | |
| ΔE-O | 19 | |
| ΔE-E | 4 |
Figure 8.
Sequencing analysis and time-of-flight mass spectra analysis of two HCLase Er-resistant tetrasaccharides purified from fractions 6 and 7. A, sequencing of the major HCLase Er-resistant tetrasaccharide in fraction 6. Top panel, the purity of this resistant tetrasaccharide was confirmed through 2-AB labeling followed by anion-exchange HPLC; middle panel, the disaccharide composition of this resistant tetrasaccharide was determined by comparing the elution positions of the 2-AB-labeled unsaturated disaccharides produced by digestion using CSase ABC with those of authentic 2-AB-derivatized unsaturated CS disaccharides indicated by arrows; bottom panel, the disaccharide moiety at the reducing end of the resistant tetrasaccharide was identified by digesting the 2-AB-labeled tetrasaccharide with a novel CSase from Vibrio sp. FC509. B, sequencing of a main HCLase Er-resistant tetrasaccharide in fraction 7. The purity (top panel), disaccharide composition (middle panel), and reducing-end disaccharide (bottom panel) of this resistant tetrasaccharide were analyzed as described above. All samples were analyzed by HPLC on a YMC-Pack PA-G column using a NaH2PO4 gradient as indicated by the dashed line. 1, Δ4,5HexUAα1–3GalNAc(4S); 2, Δ4,5HexUAα1–3GalNAc(4S,6S); 3, resistant tetrasaccharide from fraction 7; 4, resistant tetrasaccharide from fraction 6. C, mass spectrum of the major HCLase Er-resistant tetrasaccharide in fraction 6 at different charge states. D, mass spectrum of the major HCLase Er-resistant tetrasaccharide in fraction 7 at different charge states. Inten., intensity.
Compared with the major HCLase Er-resistant tetrasaccharide fraction 6, fraction 7 significantly accumulated in the early stages of polysaccharide digestion and then decreased gradually to the point of nearly disappearing as treatment with HCLase Er continued (Fig. 7). This result suggested that this tetrasaccharide fraction showed some degree of resistance to this enzyme, although not to the same extent as the significantly resistant tetrasaccharide ΔE-E. To investigate the structural features of the oligosaccharides in this fraction, the collected fraction 7 was subfractionated by anion-exchange chromatography as mentioned above, and four significant subfractions were detected and collected for further structural analysis (Table 3). The homogeneity, disaccharide composition, and reducing end disaccharide assays were carried out as described for the tetrasaccharide ΔE-E above, and the sequencing data for the predominant component ΔE-A in fraction 7 are presented as a representative sample in Fig. 8B. To further confirm the structure, ΔE-A was analyzed by MS spectrometry, too. Two major peaks at m/z 331.6884 [tetra-ΔEA-3H]3− and 498.0326 [tetra-ΔEA-2H]2− can be assigned to a unsaturated trisulfated CS tetrasaccharide with a theory mass 998.09 (Fig. 8D), which is consistent with the calculated mass of ΔE-A. From data shown in Table 3, we can see that with the exception of the minor subfraction ΔE-E, which must be present due to an overlap of fraction 6 with fraction 7, the other three subfractions (ΔE-A, ΔE-O, and ΔE-C) all contain a ΔE unit at the non-reducing end of these tetrasaccharides.
Taken together, all of the results from the structural analysis of the HCLase Er-resistant tetrasaccharides suggested that the presence of an E unit in the CS chain would become resistant to the cutting of the β1–4 linkage between the non-reducing end E unit and the reducing end disaccharide, such as an O, A, C, or E unit. Continuous E units such as E-E had much stronger resistant character against the digestion by HCLase Er. These results indicate that these resistant CS-E tetrasaccharides may act as inhibitors to HCLase Er. In an inhibition assay, the CS-E tetrasaccharide dose-dependently inhibited the digestion of HA or CS-A by HCLase Er, but the inhibitory activity was not strong enough (Fig. 9), suggesting that CS-E and its oligosaccharides were poor substrates rather than inhibitors for HCLase Er.
Figure 9.
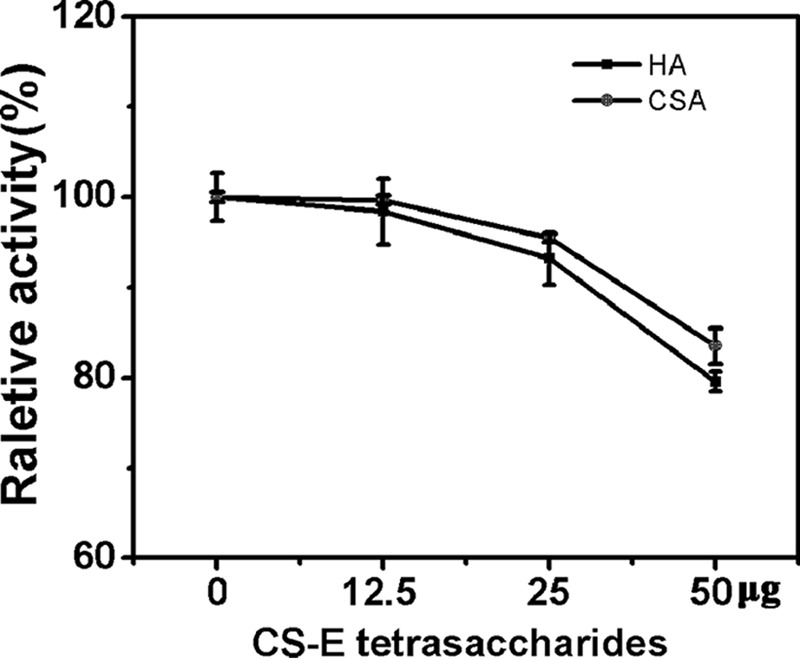
Effect of resistant tetrasaccharide on the digestion of HA and CS-A by rHCLase Er. The inhibitory capacity of CS-E tetrasaccharide to the digestion of HA/CS-A by HCLase Er was evaluated as described under “Experimental procedures.” Data are shown as the percentage of the activity obtained from the reaction without adding resistant tetrasaccharide. Error bars represent means of triplicates ± S.D.
Effect of 4-O-endosulfatase on the digestion of tetrasaccharide ΔE-E by rHCLase Er
From the sulfation pattern of the E unit, we can see that the 4-O- and 6-O-positions of the GalNAc residue were both sulfated. Thus, we speculated that the disulfation of GalNAc in the E unit might play a key role in conferring the resistance against CS digestion by rHCLase Er. To test this speculation, the major rHCLase Er-resistant tetrasaccharide ΔE-E was treated first with a 4-O-endosulfatase, which could specifically remove 4-O-sulfate from the E unit to form a C unit, and then was digested with rHCLase Er. As we can see in Fig. 10, ΔE-E was hardly digested by HCLase Er only (Fig. 10B), but after treatment with the 4-O-endosulfatase (Fig. 10C), it could be easily degraded by this enzyme to produce two ΔC units (Δ4,5HexUAα1-3GalNAc(6S)) (Fig. 10D). These results are consistent with our hypothesis.
Figure 10.
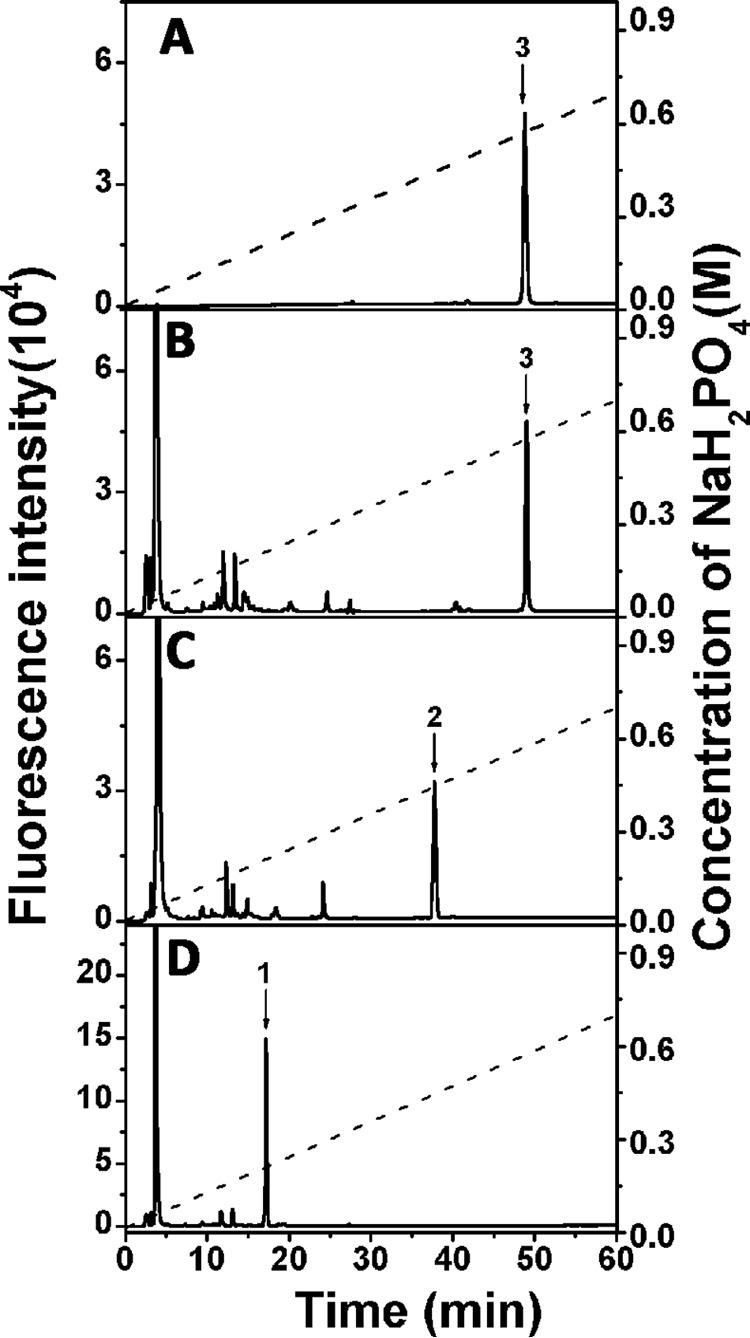
Effect of 4-O-endosulfatase treatment on the digestion of ΔE-E by rHCLase Er. The major HCLase Er-resistant tetrasaccharide ΔE-E was treated with no enzyme (A), HCLase Er only (B), 4-O-endosulfatase only (C), or 4-O-endosulfatase followed by HCLase Er (D). All the products were 2-AB-labeled and analyzed by anion-exchange HPLC on a YMC-Pack PA-G column using a NaH2PO4 gradient (indicated by the dashed line). The elution positions of the following standard oligosaccharides are indicated by numbered arrows: 1, Δ4,5HexUAα1–3GalNAc(6S); 2, Δ4,5HexUAα1–3GalNAc(6S)β1–4GlcUAβ1–3GalNAc(6S); and 3, Δ4,5HexUAα1–3GalNAc(4S,6S)β1–4GlcUAβ1–3GalNAc(4S,6S).
Site-directed and truncated mutagenesis of HCLase Er
To preliminarily investigate the action mechanism of HCLase Er, conservative amino acid residues His243, Tyr252, Arg306, and Arg310 were individually replaced by alanine (Ala) using site-directed mutagenesis. The capacity of these mutants degrading HA/CS was estimated by gel-filtration chromatography. As shown in Fig. 11, the mutants rHCLase Er-H243A, rHCLase Er-Y252A, rHCLase Er-R306A, and rHCLase Er-R310A were no longer able to degrade HA and CS-E, indicating that these four conservative residues play crucial roles in the catalytic mechanism of HCLase Er digesting HA/CS.
Figure 11.
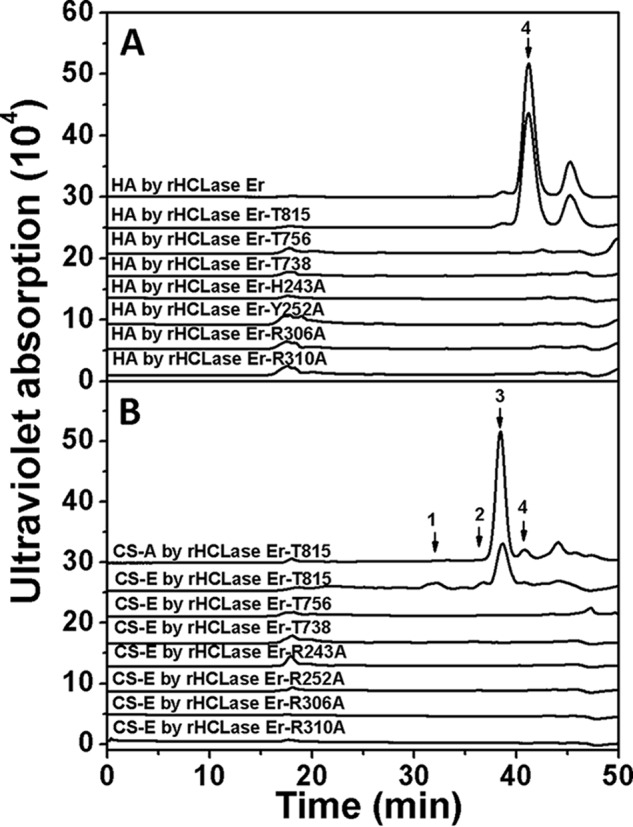
HPLC of HA treated with rHCLase Er mutant proteins. Twenty micrograms of HA (A) or CS-A (B) were exhaustively digested by each rHCLase Er mutant protein (2 μg) in 50 mm Tris-HCl buffer (pH 8.0) at 30 °C for 24 h, and then the final product was analyzed by gel-filtration chromatography on the SuperdexTM peptide column as described under “Experimental procedures.” The elution positions of the following standard oligosaccharides are indicated by arrows: 1, low-sulfated CS-E tetrasaccharide; 2, disulfated CS disaccharides; 3, monosulfated CS disaccharides; 4, non-sulfated CS or HA disaccharides.
To investigate the effects of the two extra domains (Gly739–Gln796 and Val816–Lys990) on enzyme activity, three truncated mutants of HCLase Er, rHCLase Er-T815 with Val816–Lys990 domain deletion, rHCLase Er-T738 with Gly739-Lys990 domain deletion, and rHCLase Er-T756 with Gly739-Gln796 and Val816–Lys990 domain deletion, were prepared based on the sequence alignment as discussed above. The activity of these mutants degrading HA was investigated by gel-filtration chromatography. The results showed that rHCLase Er-T815 could effectively degrade HA (Fig. 11A), CS-A and CS-E (Fig. 11B), and its activities against HA/CS were slightly affected by the deletion of the extra C-terminal domain (Val816–Lys990) (Table 1), and in contrast rHCLase Er-T738 and rHCLase Er-T756, both of which had a deletion mutant of Gly739–Gln796 domain, completely lost the HA/CS-degrading activity (Fig. 11 and Table 1). These results indicate that the extra domain of Gly739–Gln796 but not Gly816–Lys990 is essential for the activity of HCLase Er.
Discussion
In this study, an HA/CS lyase, HCLase Er, was identified from a marine bacterium Vibrio sp. FC509. This enzyme shows a relatively low sequence similarity to previously identified GAG lyases, of which the chondroitinase AC lyase (CSase AC I) from F. heparinum shares the highest sequence identity (34%) (36). Notably, HCLase Er shares only 25% identity with the HCLase identified from Vibrio sp. FC509 (34), indicating likely differences in enzymatic properties, although they were from the same bacterium. In fact, compared with other identified HA/CS lyases, HCLase Er shows a unique substrate degradation characteristic: its activity is specifically resisted by CS-E but not by HA, CS-A, CS-C, or CS-D. To the best of our knowledge, this enzyme is the first HA/CS-degrading that does not act on the E unit and thus will be very useful for structure–function studies and functional domain preparations of E unit–rich CS/DS.
The effects of temperature, pH, and metal ions on the activity of HCLase Er were investigated in detail, and the optimal conditions were determined. This enzyme exhibits the maximal activity toward both HA and CS in Tris-HCl buffer (pH 8.0) at 30 °C and is very sensitive to changes in temperature and pH. Moreover, no observations were made regarding the activation of the HCLase Er by alkali metal ions such as Na+ in seawater, which is different from the halophilic HCLase (34). Furthermore, this enzyme exhibits much higher activity in Tris-HCl buffer than in phosphate buffer at the optimal pH of 8.0, which is also different from HCLase (34) but is the same as what has been observed for CSase ABC from Proteus vulgari and CSase AC from F. heparinum (42, 43).
With the exception of CSase ABC, which can cleave almost all types of CS/DS variants with various sulfation patterns, the ability of most enzymes to digest CS has limitations in the susceptible structures. Hyaluronidases from either bacteria or animals are typically able to digest non- or low-sulfated CS but not highly sulfated CS (44). CSase AC I and HCLase cannot cleave the galactosaminidic bond bound to the D unit (GlcUA(2S)β1–3GalNAc(6S)) (34, 45). By contrast, HCLase Er can effectively digest various CS forms, including CS-D, to primarily generate disaccharides as the final products while negligibly degrading CS-E from squid cartilage (Fig. 5). To investigate the resistant structures in CS-E chains, a series of oligosaccharide fractions with different polymerization degrees were isolated from the product of a low-sulfated CS-E after exhaustive digestion by HCLase Er. The disaccharide composition analysis showed that E units dramatically accumulated in these resistant oligosaccharides, indicating the key role of the E unit in the resistance of CS-E to HCLase Er. Furthermore, several E unit-containing tetrasaccharides (ΔE-O, ΔE-C, ΔE-A, and ΔE-E) were isolated and identified as the minimal essential structures necessary for resistance against HCLase Er. Our structural analyses indicated that one ΔE unit was always located at the non-reducing end of these resistant tetrasaccharides, suggesting that the E unit could interfere with the cleavage of the galactosaminidic bond to the disaccharide at the reducing end. Among these tetrasaccharides, ΔE-E showed much more resistance to the action of HCLase Er during an exhaustive digestion (Fig. 7), which suggested that CS chains composed of repeating E units were more difficult to be digested by HCLase Er. Based on the structure of the E unit, we speculate that the disulfation of GalNAc hinders the enzyme's ability to act on the β1–4 linkage between the E unit and the sequential disaccharide. This result is similar to the observation that the sulfation of GlcUA in the D unit interferes with the abilities of CSase AC I and HCLase to cleave the bond before the D unit (45). This speculation was proven by the desulfation of ΔE-E using a 4-O-endosulfatase identified by our group (38), which showed that the selectively desulfated substrate could be easily digested by HCLase Er.
There is growing evidence that the E unit is a rare but important component for the various biological functions of CS/DS chains expressed in animal tissues/organs (1, 46, 47). Previous studies have shown that some E unit–rich domains in CS/DS chains play important roles in these biological events (10, 13, 46, 48). Various CS/DS-degrading enzymes such as CSase ABC and hyaluronidase were used to investigate the structure–function relationship of these E unit-containing CS/DS chains (10, 31), but as we mentioned above, these enzymes lack selectivity for substrates. By contrast, the enzyme HCLase Er does not act on the E unit but on the other common disaccharides, even disulfated D unit, and thus can be used to digest CS/DS chains to prepare the E unit–rich functional domains.
Based on the comparison of homologous amino acid sequences and site-directed mutagenesis analysis, four conserved amino acids, His243, Tyr252, Arg306, and Arg310, were identified as key residues for the activity of HCLase Er. In the case of chondroitinase AC from F. heparinum, these four conserved residues were shown to form a catalytic tetrad in a general acid–base catalysis (49). Additionally, compared with chondroitinase AC and HCLase, HCLase Er has two long extra domains Gly739–Gln796 and Gly816–Lys990 at the C terminus. Deletion mutation assay showed that the long tail (Gly816–Lys990) had almost no effect on the activity and substrate-degrading pattern of HCLase Er, whereas the inserted extra domain (Gly739–Gln796) was critical for the enzyme activity. However, the reason why this enzyme does not act on glucuronic acid–N-acetylgalactosamine (4,6-O-disulfate) is still unclear and needs to be revealed through structural biology technology in future studies.
In conclusion, HCLase Er is a unique HA/CS lyase isolated from a marine bacterium. Compared with the previously reported GAG lyases, it is the first identified HA/CS-degrading enzyme that does not act on E unit in CS chains. HCLase Er will be a very useful tool for structural analyses and bioactive oligosaccharide preparations of HA and CS variants, especially E unit-containing CS chains.
Experimental procedures
Materials and bacterial strains
Standard unsaturated disaccharides, CS-C and CS-D from shark cartilage, CS-E from squid cartilage, and DS from porcine skin, were purchased from Seikagaku Corp. (Tokyo, Japan). 2-Aminobenzamide (2-AB), cyanoborohydride (NaBH3CN), HA from Streptococcus equi (15–30 kDa), CS-A from bovine trachea, heparin from porcine intestinal mucosa, and chondroitinase ABC (CSase ABC) (EC 4.2.2.4) were obtained from Sigma. Low-sulfated CS-E from D. gigas cartilage was prepared as described previously (50). The bacterial strains and plasmid vector used in this study are listed in Table 4. Prime STARTM HS DNA polymerase, restriction endonuclease, T4 DNA ligase, and other genetic engineering enzymes were purchased from Takara Inc. (Dalian, China). All of the other chemicals and reagents were of the highest quality available.
Table 4.
Bacterial strains, plasmids, and primers used for sequencing in this study
Restriction enzyme sites are underlined; Kanr, kanamycin-resistant.
| Description | Source | |
|---|---|---|
| Strain | ||
| Vibrio sp. FC509 | A GAG-degrading marine bacterium (patented as CGMCC8913) | This study |
| E. coli BL21(DE3) | F−, ompT, hsdSB (rB-, mB-), dcm, gal,λ (DE3), pLysS, Cmr | Novagen |
| Plasmid | ||
| pET30a | Expression vector; Kanr | Novagen |
| pET30a-HCLase Er | pET30a carrying an amplified NdeI–XhoI fragment encoding the recombinant protein of HCLase Er fused with a His6 tag at the C terminus | This study |
| Sequencing primers | ||
| HCLase Er-F | 5′-GGGAATTCCATATGATAATCAAAGAAGTCGAAAATC-3′ | |
| HCLase Er-R | 5′-CCGCTCGAGTTTCACCGGCTCTTGCAAC-3′ | |
| HCLase Er-H243A-F | 5′-GCCGGCGCTCAACTGTACAGCGCAG-3′ | |
| HCLase Er-H243A-R | 5′-CTGGTGGAAAGAGTAATCCGCCTG-3′ | |
| HCLase Er-Y252A-F | 5′-GCTGGCGAAGTCTGGTTTGGCGC-3′ | |
| HCLase Er-Y252A-R | 5′-GCCTGCGCTGTACAGTTGAGCG-3′ | |
| HCLase Er-R306A-F | 5′-GCCGGTATCAGCCGCAGCAAACCAG-3′ | |
| HCLase Er-R306A-R | 5′-TCCCCAGGTGTTGTAGTCCAGACGG-3′ | |
| HCLase Er-R310A-F | 5′-GCCAGCAAACCAGAACTGACTCCTC-3′ | |
| HCLase Er-R310A-R | 5′-GCTGATACCGCGTCCCCAGGTGTTG-3′ | |
| HCLase Er-T815-R | 5′-CCGCTCGAGACCGCGCACATAAGCGTCTTG-3′ | |
| HCLase Er-T738-R | 5′-CCGCTCGAGTTGGTAGAACGGGAAGGTTACGC-3′ | |
| HCLase Er-T756-R | 5′-CCGCTCGAGACCGCGCACATAAGCGTCTTGAGTCACTTTCAGCTCTAGTCCGCCAGTTGCCAGATCTTGGTAGAACGGGAAGGTTAC-3′ | |
Sequence analyses of the chondroitin lyase gene and protein
The GC content (G + C%) calculation of the opening reading frame and multiple sequence alignment were carried out using Bio-Edit version 7.0.5.3. The protein features were analyzed as described previously (34). Briefly, the protein sequence similarity search was performed using the BLASTp algorithm on NCBI. The protein molecular mass was estimated using the peptide mass tool on the ExPASy server of the Swiss Institute of Bioinformatics. The protein modules and domains were identified using the SMART database. The identities and types of secretion signal peptides were determined by using the SMART and SignalP 4.0 server, LipoP 1.0 server.
Heterologous expression of the HCLase Er gene
Using the genomic DNA from Vibrio sp. FC509 as template, the full-length gene of HCLase Er (GenBankTM accession number MF458894) without the signal peptide sequence was amplified. Primer pairs with restriction enzyme sites NdeI–XhoI (underlined in Table 4) were designed based on the gene sequence and the insertion-site sequences of the expression plasmid pET-30a(+) (Novagen), with a His6 tag at the C terminus of the recombinant protein. The gel-recovered PCR product was ligated into the expression vector pET-30a(+). The recombined expression plasmid (pE30a-rHCLase Er) was amplified in E. coli DH5α cells and then transformed into E. coli BL21(DE3) cells for protein expression. The constructed expression plasmid was sequenced to confirm the integrity of the inserted gene.
Recombinant HCLase Er was expressed and purified essentially as described previously (34). Briefly, E. coli cells harboring pE30a-rHCLase Er were cultured in LB broth containing 50 μg/ml kanamycin, and the target protein expression was induced by adding 0.1 mm isopropyl 1-thio-β-d-galactopyranoside. After culturing for another 24 h at 16 °C, cells were collected by centrifugation and disrupted by sonication in ice-cold buffer A (50 mm Tris-HCl, 150 mm NaCl (pH 8.0)). After centrifugation at 15,000 × g for 30 min, the supernatant was collected for further purification of rHCLase Er.
Purification of recombinant protein rHCLase Er
The supernatant containing rHCLase Er was loaded onto a column packed with nickel-SepharoseTM 6 Fast Flow resin (GE Healthcare), and after removing impurities by washing with buffer A containing 50 mm imidazole, rHCLase Er was eluted with a gradient concentration of imidazole, ranging from 50 to 250 mm. The presence and purity of the proteins in each fraction were assessed by SDS-PAGE according to the method reported by Sambrook and Russell (51). The purified rHCLase Er was collected and quantified using the BCA (bicinchoninic acid) method.
Assay of rHCLase Er activity toward various polysaccharide substrates
To determine the substrates of rHCLase Er, 10 μl of stock solution (10 mg/ml) of each polysaccharide (CS-A, CS-C, CS-D, CS-E, DS, heparin, or hyaluronic acid) was mixed with 20 μl of 250 mm NaH2PO4·Na2HPO4 buffer (pH 7.0), 60 μl of water, and 10 μl of the appropriately diluted enzyme and then incubated at 37 °C for 12 h. The reaction was stopped by heating at 100 °C for 10 min and then cooled in ice-cold water for 10 min. After centrifugation at 15,000 × g for 15 min, the supernatant was collected and analyzed by measuring the absorbance at 232 nm and by gel-filtration chromatography as described previously (52).
Biochemical characterization of the recombinant protein rHCLase Er
To determine the optimal pH for the rate of rHCLase Er, HA and CS-C (1 mg/ml) were individually digested at 30 °C for 2 h with 2 μg of rHCLase Er in three different buffer systems: 50 mm NaAc-HAc buffer (pH 5.0–6.0); NaH2PO4·Na2HPO4 buffer (pH 6.0–8.0); and Tris-HCl buffer (pH 7.0–10.0) in a total volume of 100 μl. After the optimal pH was determined, the activity of rHCLase Er at various temperatures (0–100 °C) was investigated in the optimal buffer (50 mm Tris-HCl buffer (pH 8.0)) for 2 h. The effects of metal ions/chelating reagents (5 mm) on the rate of rHCLase Er degrading HA/CS-C were further investigated at the optimal buffer (50 mm Tris-HCl buffer (pH 8.0)) and temperature (30 °C). To determine the thermostability, the enzyme was preincubated in 50 mm Tris-HCl buffer (pH 8.0) for 0–24 h at the optimal temperature (30 °C), and then the residual activity was determined under the optimal conditions (50 mm Tris-HCl (pH 8.0), at 30 °C). The activity of the enzyme was estimated by measuring the absorbance at 232 nm, and all of the reactions were performed in triplicate.
The activity assay of rHCLase Er
The activities of rHCLase Er against HA and CS variants were measured according to the method provided by Yamagata et al. (43). Briefly, rHCLase Er (20 μg) was added to 1 mg/ml substrate in 50 mm Tris-HCl buffer (pH 8.0) in a total volume of 1 ml. The reaction mixture was incubated at 30 °C. At various time intervals (up to 5 min), aliquots of 100 μl were withdrawn in duplicate, boiled for 10 min, and then cooled in ice-cold water for 10 min. After being centrifuged at 15,000 × g for 15 min, the supernatant was collected, diluted three times, and analyzed by reading the absorbance at 232 nm. One unit of enzyme was defined as the amount of enzyme that produced 1 μmol of unsaturated carbon bonds per min.
Digestion pattern of polysaccharides by rHCLase Er
To investigate the substrate-degrading pattern of rHCLase Er, HA or CS-A (1 mg/ml) was digested by rHCLase Er (0.03 unit/ml) at 30 °C for various time intervals. Aliquots of the digests were removed for time-course experiments and analyzed using gel filtration by monitoring the absorbance at 232 nm (52).
To further determine the final products, 20 μg of commercial HA, CS-A, CS-C, CS-D, or CS-E was digested with excessive rHCLase Er (2 μg) in 50 mm Tris-HCl (pH 8.0) buffer at 30 °C for 24 h, and then the digest was analyzed on a SuperdexTM peptide 10/300 GL column by monitoring the absorbance at 232 nm. To confirm that CS-E is resistant to the HCLase Er activity, commercial CS-E from squid cartilage was individually digested by commercial CSase ABC and HCLase from Vibrio sp. FC509 (34) for 24 h in their optimal environment, and the final products were analyzed as described above.
To investigate the digestion profile of the low-sulfated CS-E from D. gigas cartilage, the CS-E sample (1 mg/ml) was exhaustively digested with rHCLase Er to determine the action pattern and final products under optimal conditions with the addition of fresh enzyme every 24 h. Aliquots (20 μg) of the degradation products were taken for the time-course experiment assay as described for HA and CS-A above.
To determine the molecular weight and structural characteristics of the final product of HA/CS, 1 ml (1 mg/ml) of HA or CS-A was exhaustively digested by HCLase Er, and the final product was fractionated by SuperdexTM peptide 10/300 GL column. The major disaccharide fraction was freeze-dried and identified by electrospray ionization MS on an ion-trap TOF hybrid mass spectrometer (LCMS-IT-TOF, Shimadzu, Japan) with negative-ionization modes as described previously (34).
Oligosaccharide preparation of D. gigas cartilage-derived CS-E by digestion with rHCLase Er
To further determine the structural characteristics of the degradation product of CS-E from D.>igas cartilage, 1 ml of CS-E polysaccharides (10 mg/ml) was digested using rHCLase Er (1 unit/ml) for 72 h at 30 °C with the addition of fresh enzyme every 24 h. The final reaction mixture was heated in boiling water for 10 min and subsequently cooled to 4 °C. After being centrifuged at 15,000 × g for 30 min, the supernatant was loaded onto a pre-equilibrated SuperdexTM peptide 10/300 GL column. Fractionated oligosaccharide samples were collected by on-line monitoring at 232 nm and freeze-dried repeatedly to remove NH4HCO3 for further identification.
Disaccharide composition assay of low-sulfated CS-E polysaccharide and oligosaccharide fractions
To determine the disaccharide composition of the CS-E oligosaccharide fractions prepared by digestion with rHCLase Er as well as their parental polysaccharide, each oligosaccharide fraction (10 pmol) or low-sulfated CS-E (2 μg) from D. gigas cartilage was digested with CSase ABC (Sigma) and labeled with 2-AB and sodium cyanoborohydride reagents as described by Bigge et al. (53). After the free 2-AB was removed by extraction with chloroform, the final products of the oligosaccharide fractions were individually analyzed by anion-exchange HPLC on a YMC-Pack PA-G column (YMC, Kyoto, Japan), eluted with a linear gradient from 16 to 460 mm NaH2PO4 over a 60-min period, and monitored using a fluorescence detector. The on-line monitoring and data analysis (e.g. molar ratio determination) were performed using the software LC solution version 1.25.
Sequencing of the rHCLase Er-resistant tetrasaccharide from CS-E
The rHCLase Er-resistant tetrasaccharide fraction was further subfractionated by anion-exchange HPLC on a YMC-Pack PA-G column. The tetrasaccharide fraction was loaded onto the column, equilibrated with 16 mm NaH2PO4, and then eluted at room temperature with a linear gradient from 16 to 700 mm NaH2PO4 over 60 min at a flow rate of 1.0 ml/min. The eluates were monitored by measuring the absorbance at 232 nm, and the major peaks were collected and desalted with a SuperdexTM peptide column as described above.
The purified subfractions from the tetrasaccharide fractions were sequenced at a low picomole level by chondroitinase digestion and HPLC. Briefly, first, to analyze the disaccharide composition of the rHCLase Er-resistant tetrasaccharide, an aliquot (5 pmol) of the tetrasaccharide fraction was digested with CSase ABC (Sigma) and labeled with the 2-AB and sodium cyanoborohydride reagents. Second, to identify the reducing end, the subfractions (50 pmol) used for sequencing were labeled with 2-AB and purified by paper chromatography, and an aliquot (5 pmol) of the 2-AB-labeled tetrasaccharide subfraction was then treated with a novel exo-CSase from Vibrio sp. FC509 (54). All of these preparations were individually analyzed by anion-exchange HPLC on a YMC-Pack PA-G column, eluted with a linear gradient of NaH2PO4 as indicated over a 60-min period, and monitored using a fluorescence detector.
To further determine the molecular weight and structural characteristics of the HCLase Er-resistant tetrasaccharides, the two major subfractions purified from fractions 6 and 7 were analyzed by MS spectrometry as described above. The mass acquisition range was set at 200–1500.
To confirm the resistant tetrasaccharide inhibited the enzyme activity, 0, 12.5, 25, or 50 μg of HCLase Er-resistant tetrasaccharide (fraction 6) was added into 50 μg of HA/CS-A dissolved in 50 mm Tris-HCl buffer (pH 8.0), and then 2 μg of HCLase Er was added to the above substrate in a total volume of 100 μl. The reaction mixture was incubated at 30 °C for 5 min followed by measuring the absorbance of each resulting product at 232 nm.
Effect of 4-O-endosulfatase treatment on the digestion of tetrasaccharide ΔE-E by rHCLase Er
To investigate whether the desulfation by 4-O-endosulfatase promotes the digestion of the resistant tetrasaccharide by rHCLase Er, the activities of rHCLase Er against ΔE-E in the absence or presence of 4-O-endosulfatase were compared. Briefly, 10 pmol of ΔE-E was pretreated with or without 4-O-endosulfatase in 50 mm Tris-HCl (pH 8.0) at 30 °C for 12 h, and then an aliquot (5 pmol) of each pretreated sample was further digested with or without rHCLase Er (2 μg) for 2 h at the optimal conditions. All of the final products were labeled by 2-AB and analyzed by anion-exchange HPLC on a YMC-Pack PA-G column using a NaH2PO4 gradient from 16 to 700 mm over a 60-min period and monitored using a fluorescence detector, as described above.
Expression and characterization of mutant rHCLase Er
Based on the similarity analysis of protein sequences above, four amino acids His243, Tyr252, Arg306, and Arg310 were identified as the putative catalytic residues of HCLase Er. To determine the role of these residues, they were individually mutated into alanine by site-directed mutagenesis. Briefly, mutants were produced by pET30-rHCLase Er plasmid denaturation and annealing of complementary oligonucleotide primers containing the desired mutation, and then a series of gene mutation products were formed by using rapid PCR amplification with high-fidelity DNA polymerase of Prime STAR Max Premix (Takara Inc.). Primer sequences for mutagenesis studies are presented in Table 4. Amplified gene products were individually phosphorylated at the 5′-end, circulated, and transformed into E. coli DH5α and E. coli BL21(DE3) cells, and protein expression was then induced. GAG-degrading activities of these mutants were measured as described above.
To investigate the possible function of the two redundant domains at the C terminus of HCLase Er, three reversed primers, HCLase Er-T815-R, HCLase Er-T738-R, and HCLase Er-T756-R (as listed in Table 4) with the XhoI restriction site, were designed to generate the truncated versions of the gene. PCR was run using Vibrio sp. FC509 genomic DNA as the template. The amplified gene products were individually ligated into the expression vector pET-30a(+). The truncated protein rHCLase Er-T815 (Ile22–Gly815), rHCLase Er-T738 (Ile22–Glu738), or rHCLase Er-T756 (Ile22–Glu738 and Asp797–Gly815) was expressed in E. coli BL21(DE3) as described above. Then, the HA/CS-degrading activities were measured as for the wildtype enzyme.
Author contributions
C. P. and F. L. conceptualization; C. P. and F. L. data curation; C. P. and F. L. software; C. P. and F. L. formal analysis; C. P., Q. W., W. H., and F. L. supervision; C. P., Q. W., S. W., W. W., R. J., and F. L. validation; C. P., R. J., and F. L. investigation; C. P. visualization; C. P., Q. W., S. W., W. W., R. J., and F. L. methodology; C. P. writing-original draft; C. P. and F. L. project administration; C. P., W. H., and F. L. writing-review and editing; Q. W., S. W., W. W., R. J., and F. L. resources; F. L. funding acquisition.
This work was supported by National Natural Science Foundation of China Grants 31570071 and 31300664, Science and Technology Special Project of Shandong Province Grant 2015zdjs04002, Science and Technology Development Project of Shandong Province Grant 2016GGH4502, Independent Innovation Plan of Colleges and Universities in Jinan Grant 201401242, and the Fundamental Research Funds of Shandong University Grant 2015JC002. The authors declare that they have no conflicts of interest with the contents of this article.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) MF458894.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- CS
- chondroitin sulfate
- DS
- dermatan sulfate
- E unit
- GlcUAβ1–3GalNAc(4S,6S)
- HA
- hyaluronic acid
- GAG
- glycosaminoglycan
- HexUA
- hexuronic acid
- Δ4,5HexUA
- Δ4,5-unsaturated hexuronic acid
- 2S
- 2-O-sulfate
- 4S
- 4-O-sulfate
- 6S
- 6-O-sulfate
- CSase
- chondroitinase
- 2-AB
- 2-aminobenzamide
- IdoUA
- l-iduronic acid
- HCLase
- HA and CS lyase
- HCLase Er
- CS-E–resisted HA/CS lyase
- rHCLase Er
- recombinant HCLase Er.
References
- 1. Sugahara K., Mikami T., Uyama T., Mizuguchi S., Nomura K., and Kitagawa H. (2003) Recent advances in the structural biology of chondroitin sulfate and dermatan sulfate. Curr. Opin. Struct. Biol. 13, 612–620 10.1016/j.sbi.2003.09.011 [DOI] [PubMed] [Google Scholar]
- 2. Iozzo R. V. (1998) Matrix proteoglycans: from molecular design to cellular function. Annu. Rev. Biochem. 67, 609–652 10.1146/annurev.biochem.67.1.609 [DOI] [PubMed] [Google Scholar]
- 3. Miyata S., and Kitagawa H. (2017) Formation and remodeling of the brain extracellular matrix in neural plasticity: roles of chondroitin sulfate and hyaluronan. Biochim. Biophys. Acta 1861, 2420–2434 10.1016/j.bbagen.2017.06.010 [DOI] [PubMed] [Google Scholar]
- 4. Faissner A., Clement A., Lochter A., Streit A., Mandl C., and Schachner M. (1994) Isolation of a neural chondroitin sulfate proteoglycan with neurite outgrowth promoting properties. J. Cell Biol. 126, 783–799 10.1083/jcb.126.3.783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schnädelbach O., Mandl C., and Faissner A. (1998) Expression of DSD-1-PG in primary neural and glial-derived cell line cultures, upregulation by TGF-β, and implications for cell-substrate interactions of the glial cell line Oli-neu. Glia 23, 99–119 10.1002/(SICI)1098-1136(199806)23:2%3C99::AID-GLIA2%3E3.0.CO%3B2-Z [DOI] [PubMed] [Google Scholar]
- 6. Smith P. D., Coulson-Thomas V. J., Foscarin S., Kwok J. C., and Fawcett J. W. (2015) “GAG-ing with the neuron”: The role of glycosaminoglycan patterning in the central nervous system. Exp. Neurol. 274, 100–114 10.1016/j.expneurol.2015.08.004 [DOI] [PubMed] [Google Scholar]
- 7. Klüppel M., Wight T. N., Chan C., Hinek A., and Wrana J. L. (2005) Maintenance of chondroitin sulfation balance by chondroitin-4-sulfotransferase 1 is required for chondrocyte development and growth factor signaling during cartilage morphogenesis. Development 132, 3989–4003 10.1242/dev.01948 [DOI] [PubMed] [Google Scholar]
- 8. Taylor K. R., and Gallo R. L. (2006) Glycosaminoglycans and their proteoglycans: host-associated molecular patterns for initiation and modulation of inflammation. FASEB J. 20, 9–22 10.1096/fj.05-4682rev [DOI] [PubMed] [Google Scholar]
- 9. Pomin V. H. (2015) Sulfated glycans in inflammation. Eur. J. Med. Chem. 92, 353–369 10.1016/j.ejmech.2015.01.002 [DOI] [PubMed] [Google Scholar]
- 10. Li F., Ten Dam G. B., Murugan S., Yamada S., Hashiguchi T., Mizumoto S., Oguri K., Okayama M., van Kuppevelt T. H., and Sugahara K. (2008) Involvement of highly sulfated chondroitin sulfate in the metastasis of the Lewis lung carcinoma cells. J. Biol. Chem. 283, 34294–34304 10.1074/jbc.M806015200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sugahara K. N., Hirata T., Tanaka T., Ogino S., Takeda M., Terasawa H., Shimada I., Tamura J., Ten Dam G. B., van Kuppevelt T. H., and Miyasaka M. (2008) Chondroitin sulfate E fragments enhance CD44 cleavage and CD44-dependent motility in tumor cells. Cancer Res. 68, 7191–7199 10.1158/0008-5472.CAN-07-6198 [DOI] [PubMed] [Google Scholar]
- 12. Iida J., Meijne A. M., Knutson J. R., Furcht L. T., and McCarthy J. B. (1996) Cell surface chondroitin sulfate proteoglycans in tumor cell adhesion, motility and invasion. Semin. Cancer Biol. 7, 155–162 10.1006/scbi.1996.0021 [DOI] [PubMed] [Google Scholar]
- 13. Bergefall K., Trybala E., Johansson M., Uyama T., Naito S., Yamada S., Kitagawa H., Sugahara K., and Bergström T. (2005) Chondroitin sulfate characterized by the E-disaccharide unit is a potent inhibitor of herpes simplex virus infectivity and provides the virus binding sites on gro2C cells. J. Biol. Chem. 280, 32193–32199 10.1074/jbc.M503645200 [DOI] [PubMed] [Google Scholar]
- 14. Uyama T., Ishida M., Izumikawa T., Trybala E., Tufaro F., Bergström T., Sugahara K., and Kitagawa H. (2006) Chondroitin 4-O-sulfotransferase-1 regulates E disaccharide expression of chondroitin sulfate required for herpes simplex virus infectivity. J. Biol. Chem. 281, 38668–38674 10.1074/jbc.M609320200 [DOI] [PubMed] [Google Scholar]
- 15. Williams R. K., and Straus S. E. (1997) Specificity and affinity of binding of herpes simplex virus type 2 glycoprotein B to glycosaminoglycans. J. Virol. 71, 1375–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mizuguchi S., Uyama T., Kitagawa H., Nomura K. H., Dejima K., Gengyo-Ando K., Mitani S., Sugahara K., and Nomura K. (2003) Chondroitin proteoglycans are involved in cell division of Caenorhabditis elegans. Nature 423, 443–448 10.1038/nature01635 [DOI] [PubMed] [Google Scholar]
- 17. Izumikawa T., Kitagawa H., Mizuguchi S., Nomura K. H., Nomura K., Tamura J., Gengyo-Ando K., Mitani S., and Sugahara K. (2004) Nematode chondroitin polymerizing factor showing cell-/organ-specific expression is indispensable for chondroitin synthesis and embryonic cell division. J. Biol. Chem. 279, 53755–53761 10.1074/jbc.M409615200 [DOI] [PubMed] [Google Scholar]
- 18. Mikami T., and Kitagawa H. (2017) Sulfated glycosaminoglycans: their distinct roles in stem cell biology. Glycoconj. J. 34, 725–735 10.1007/s10719-016-9732-9 [DOI] [PubMed] [Google Scholar]
- 19. Kusche-Gullberg M., and Kjellén L. (2003) Sulfotransferases in glycosaminoglycan biosynthesis. Curr. Opin. Struct. Biol. 13, 605–611 10.1016/j.sbi.2003.08.002 [DOI] [PubMed] [Google Scholar]
- 20. Silbert J. E., and Sugumaran G. (2002) Biosynthesis of chondroitin/dermatan sulfate. IUBMB Life 54, 177–186 10.1080/15216540214923 [DOI] [PubMed] [Google Scholar]
- 21. Maccarana M., Olander B., Malmström J., Tiedemann K., Aebersold R., Lindahl U., Li J. P., and Malmström A. (2006) Biosynthesis of dermatan sulfate: chondroitin-glucuronate C5-epimerase is identical to SART2. J. Biol. Chem. 281, 11560–11568 10.1074/jbc.M513373200 [DOI] [PubMed] [Google Scholar]
- 22. Sugahara K., and Mikami T. (2007) Chondroitin/dermatan sulfate in the central nervous system. Curr. Opin. Struct. Biol. 17, 536–545 10.1016/j.sbi.2007.08.015 [DOI] [PubMed] [Google Scholar]
- 23. Deepa S. S., Umehara Y., Higashiyama S., Itoh N., and Sugahara K. (2002) Specific molecular interactions of oversulfated chondroitin sulfate E with various heparin-binding growth factors. J. Biol. Chem. 277, 43707–43716 10.1074/jbc.M207105200 [DOI] [PubMed] [Google Scholar]
- 24. Valcarcel J., Novoa-Carballal R., Pérez-Martín R. I., Reis R. L., and Vázquez J. A. (2017) Glycosaminoglycans from marine sources as therapeutic agents. Biotechnol. Adv. 35, 711–725 10.1016/j.biotechadv.2017.07.008 [DOI] [PubMed] [Google Scholar]
- 25. Purushothaman A., Fukuda J., Mizumoto S., ten Dam G. B., van Kuppevelt T. H., Kitagawa H., Mikami T., and Sugahara K. (2007) Functions of chondroitin sulfate/dermatan sulfate chains in brain development. critical roles of E and iE disaccharide units recognized by a single chain antibody GD3G7. J. Biol. Chem. 282, 19442–19452 10.1074/jbc.M700630200 [DOI] [PubMed] [Google Scholar]
- 26. Clement A. M., Sugahara K., and Faissner A. (1999) Chondroitin sulfate E promotes neurite outgrowth of rat embryonic day 18 hippocampal neurons. Neurosci. Lett. 269, 125–128 10.1016/S0304-3940(99)00432-2 [DOI] [PubMed] [Google Scholar]
- 27. Kato D., Era S., Watanabe I., Arihara M., Sugiura N., Kimata K., Suzuki Y., Morita K., Hidari K. I., and Suzuki T. (2010) Antiviral activity of chondroitin sulphate E targeting dengue virus envelope protein. Antiviral Res. 88, 236–243 10.1016/j.antiviral.2010.09.002 [DOI] [PubMed] [Google Scholar]
- 28. Jinno-Oue A., Tanaka A., Shimizu N., Mori T., Sugiura N., Kimata K., Isomura H., and Hoshino H. (2013) Inhibitory effect of chondroitin sulfate type E on the binding step of human T-cell leukemia virus type 1. AIDS Res. Hum. Retroviruses 29, 621–629 10.1089/aid.2012.0156 [DOI] [PubMed] [Google Scholar]
- 29. Koike T., Mikami T., Shida M., Habuchi O., and Kitagawa H. (2015) Chondroitin sulfate-E mediates estrogen-induced osteoanabolism. Sci. Rep. 5, 8994 10.1038/srep08994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sotogaku N., Tully S. E., Gama C. I., Higashi H., Tanaka M., Hsieh-Wilson L. C., and Nishi A. (2007) Activation of phospholipase C pathways by a synthetic chondroitin sulfate-E tetrasaccharide promotes neurite outgrowth of dopaminergic neurons. J. Neurochem. 103, 749–760 10.1111/j.1471-4159.2007.04849.x [DOI] [PubMed] [Google Scholar]
- 31. Deepa S. S., Kalayanamitra K., Ito Y., Kongtawelert P., Fukui S., Yamada S., Mikami T., and Sugahara K. (2007) Novel sulfated octa- and decasaccharides from squid cartilage chondroitin sulfate E: sequencing and application for determination of the epitope structure of the monoclonal antibody MO-225. Biochemistry 46, 2453–2465 10.1021/bi602374m [DOI] [PubMed] [Google Scholar]
- 32. Tully S. E., Mabon R., Gama C. I., Tsai S. M., Liu X., and Hsieh-Wilson L. C. (2004) A chondroitin sulfate small molecule that stimulates neuronal growth. J. Am. Chem. Soc. 126, 7736–7737 10.1021/ja0484045 [DOI] [PubMed] [Google Scholar]
- 33. Sugahara K., Nadanaka S., Takeda K., and Kojima T. (1996) Structural analysis of unsaturated hexasaccharides isolated from shark cartilage chondroitin sulfate D that are substrates for the exolytic action of chondroitin ABC lyase. Eur. J. Biochem. 239, 871–880 10.1111/j.1432-1033.1996.0871u.x [DOI] [PubMed] [Google Scholar]
- 34. Han W., Wang W., Zhao M., Sugahara K., and Li F. (2014) A novel eliminase from a marine bacterium that degrades hyaluronan and chondroitin sulfate. J. Biol. Chem. 289, 27886–27898 10.1074/jbc.M114.590752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nandini C. D., Mikami T., Ohta M., Itoh N., Akiyama-Nambu F., and Sugahara K. (2004) Structural and functional characterization of oversulfated chondroitin sulfate/dermatan sulfate hybrid chains from the notochord of hagfish. J. Biol. Chem. 279, 50799–50809 10.1074/jbc.M404746200 [DOI] [PubMed] [Google Scholar]
- 36. Féthière J., Eggimann B., and Cygler M. (1999) Crystal structure of chondroitin AC lyase, a representative of a family of glycosaminoglycan degrading enzymes. J. Mol. Biol. 288, 635–647 10.1006/jmbi.1999.2698 [DOI] [PubMed] [Google Scholar]
- 37. Hashimoto W., Nankai H., Mikami B., and Murata K. (2003) Crystal structure of Bacillus sp. GL1 xanthan lyase, which acts on the side chains of xanthan. J. Biol. Chem. 278, 7663–7673 10.1074/jbc.M208100200 [DOI] [PubMed] [Google Scholar]
- 38. Wang W., Han W., Cai X., Zheng X., Sugahara K., and Li F. (2015) Cloning and characterization of a novel chondroitin sulfate/dermatan sulfate 4-O-endosulfatase from a marine bacterium. J. Biol. Chem. 290, 7823–7832 10.1074/jbc.M114.629154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kim B. T., Tsuchida K., Lincecum J., Kitagawa H., Bernfield M., and Sugahara K. (2003) Identification and characterization of three Drosophila melanogaster glucuronyltransferases responsible for the synthesis of the conserved glycosaminoglycan-protein linkage region of proteoglycans. J. Biol. Chem. 278, 9116–9124 10.1074/jbc.M209344200 [DOI] [PubMed] [Google Scholar]
- 40. Yamada S., Okada Y., Ueno M., Iwata S., Deepa S. S., Nishimura S., Fujita M., Van Die I., Hirabayashi Y., and Sugahara K. (2002) Determination of the glycosaminoglycan-protein linkage region oligosaccharide structures of proteoglycans from Drosophila melanogaster and Caenorhabditis elegans. J. Biol. Chem. 277, 31877–31886 10.1074/jbc.M205078200 [DOI] [PubMed] [Google Scholar]
- 41. Yamada S., Oyama M., Kinugasa H., Nakagawa T., Kawasaki T., Nagasawa S., Khoo K. H., Morris H. R., Dell A., and Sugahara K. (1995) The sulphated carbohydrate-protein linkage region isolated from chondroitin 4-sulphate chains of inter-α-trypsin inhibitor in human plasma. Glycobiology 5, 335–341 10.1093/glycob/5.3.335 [DOI] [PubMed] [Google Scholar]
- 42. Hamai A., Hashimoto N., Mochizuki H., Kato F., Makiguchi Y., Horie K., and Suzuki S. (1997) Two distinct chondroitin sulfate ABC lyases. An endoeliminase yielding tetrasaccharides and an exoeliminase preferentially acting on oligosaccharides. J. Biol. Chem. 272, 9123–9130 10.1074/jbc.272.14.9123 [DOI] [PubMed] [Google Scholar]
- 43. Yamagata T., Saito H., Habuchi O., and Suzuki S. (1968) Purification and properties of bacterial chondroitinases and chondrosulfatases. J. Biol. Chem. 243, 1523–1535 [PubMed] [Google Scholar]
- 44. Stern R., and Jedrzejas M. J. (2006) Hyaluronidases: their genomics, structures, and mechanisms of action. Chem. Rev. 106, 818–839 10.1021/cr050247k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mizumoto S., Murakoshi S., Kalayanamitra K., Deepa S. S., Fukui S., Kongtawelert P., Yamada S., and Sugahara K. (2013) Highly sulfated hexasaccharide sequences isolated from chondroitin sulfate of shark fin cartilage: insights into the sugar sequences with bioactivities. Glycobiology 23, 155–168 10.1093/glycob/cws137 [DOI] [PubMed] [Google Scholar]
- 46. Sugahara K., and Mizumoto S. (2012) ISCSM2011 chondroitin sulfate E-type structure at tumor cell surface is involved in experimental metastasis. Adv. Exp. Med. Biol. 749, 33–45 10.1007/978-1-4614-3381-1_3 [DOI] [PubMed] [Google Scholar]
- 47. Basappa Murugan S., Sugahara K. N., Lee C. M., ten Dam G. B., van Kuppevelt T. H., Miyasaka M., Yamada S., and Sugahara K. (2009) Involvement of chondroitin sulfate E in the liver tumor focal formation of murine osteosarcoma cells. Glycobiology 19, 735–742 10.1093/glycob/cwp041 [DOI] [PubMed] [Google Scholar]
- 48. Avirutnan P., Zhang L., Punyadee N., Manuyakorn A., Puttikhunt C., Kasinrerk W., Malasit P., Atkinson J. P., and Diamond M. S. (2007) Secreted NS1 of dengue virus attaches to the surface of cells via interactions with heparan sulfate and chondroitin sulfate E. PLoS Pathog. 3, e183 10.1371/journal.ppat.0030183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Huang W., Boju L., Tkalec L., Su H., Yang H. O., Gunay N. S., Linhardt R. J., Kim Y. S., Matte A., and Cygler M. (2001) Active site of chondroitin AC lyase revealed by the structure of enzyme-oligosaccharide complexes and mutagenesis. Biochemistry 40, 2359–2372 10.1021/bi0024254 [DOI] [PubMed] [Google Scholar]
- 50. Li F., Shetty A. K., and Sugahara K. (2007) Neuritogenic activity of chondroitin/dermatan sulfate hybrid chains of embryonic pig brain and their mimicry from shark liver. J. Biol. Chem. 282, 2956–2966 10.1074/jbc.M609296200 [DOI] [PubMed] [Google Scholar]
- 51. Sambrook J., and Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., pp. A8.40-A8.47, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 52. Li F., Nandini C. D., Hattori T., Bao X., Murayama D., Nakamura T., Fukushima N., and Sugahara K. (2010) Structure of pleiotrophin- and hepatocyte growth factor-binding sulfated hexasaccharide determined by biochemical and computational approaches. J. Biol. Chem. 285, 27673–27685 10.1074/jbc.M110.118703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bigge J. C., Patel T. P., Bruce J. A., Goulding P. N., Charles S. M., and Parekh R. B. (1995) Nonselective and efficient fluorescent labeling of glycans using 2-amino benzamide and anthranilic acid. Anal. Biochem. 230, 229–238 10.1006/abio.1995.1468 [DOI] [PubMed] [Google Scholar]
- 54. Wang W., Cai X., Han N., Han W., Sugahara K., and Li F. (2017) Sequencing of chondroitin sulfate oligosaccharides using a novel exolyase from a marine bacterium that degrades hyaluronan and chondroitin sulfate/dermatan sulfate. Biochem. J. 474, 3831–3848 10.1042/BCJ20170591 [DOI] [PubMed] [Google Scholar]
- 55. Lombard V., Golaconda Ramulu H., Drula E., Coutinho P. M., and Henrissat B. (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495 10.1093/nar/gkt1178 [DOI] [PMC free article] [PubMed] [Google Scholar]