Abstract
The human enzyme histone acetyltransferase binding to ORC1 (HBO1) regulates DNA replication, cell proliferation, and development. HBO1 is part of a multiprotein histone acetyltransferase (HAT) complex that also contains inhibitor of growth family member (ING) 4/5, MYST/Esa1-associated factor (MEAF) 6, and the scaffolding proteins Jade family PHD finger (JADE) 1/2/3 or bromodomain and PHD finger-containing protein (BRPF) 2/3 to acetylate histone H4 H4K5/8/12 or H3K14, respectively. Within this four-protein complex, JADE1 determines histone H4 substrate specificity of the HBO1–HAT complex. However, the mechanism by which JADE1 controls the H4-specific acetyltransferase activity of HBO1 is unknown. Here we used recombinant proteins in vitro to dissect the specific regions and activities of HBO1 and JADE1 that mediate histone H3–H4 acetylation via the HBO1-HAT domain. We found that JADE1 increases the catalytic efficiency of HBO1 acetylation of an H3–H4 substrate by about 5-fold through an N-terminal, 21-residue HBO1- and histone-binding domain and a nearby second histone core–binding domain. We also demonstrate that HBO1 contains an N-terminal histone-binding domain (HBD) that makes additional contacts with H3–H4 independent of JADE1 interactions with histones and that the HBO1 HBD does not significantly contribute to HBO1's overall HAT activity. Experiments with JADE1 deletions in vivo recapitulated these in vitro interactions and their roles in HBO1 histone acetylation activity. Together, these results indicate that the N-terminal region of JADE1 functions as a platform that brings together the catalytic HBO1 subunit with its cognate H3–H4 substrate for histone acetylation.
Keywords: acetyltransferase, chromatin regulation, histone acetylation, epigenetics, protein complex
Introduction
Histone acetylation, a covalent modification on lysine residues of histones, predominantly in the N-terminal tail regions, regulates local and global chromatin dynamics, thus playing an important role in modulating DNA accessibility and gene expression in all eukaryotic organisms. Generally, hyperacetylation and hypoacetylation are observed in transcriptionally active and inactive regions of the chromatin, respectively (1–5). Histone acetylation has been shown to promote transcription in at least two ways. First, acetylation neutralizes the lysine positive charge, thus altering its physiochemical properties and making the local chromatin environment more accessible for gene transcription activation (1). Second, lysine acetylation generates docking recognition sites for various gene-activating proteins containing acetyl-lysine recognition domains such as bromodomains (5–9). Therefore, the tight regulation of acetylation on lysine residues within histones, as well as the enzymes that mediate these modifications, are critical to maintain normal gene expression and regulation. The enzymes that mediate histone acetylation are histone acetyltransferases (HATs),3 whereas the enzymes that convert acetyl-lysine residues within histone back to lysine are histone deactylases (HDACs) (10). HDACs are grouped into four classes according to their functional properties and sequence conservation, which fall into two conserved structural folds (classes I, II, and IV and the class III sirtuins). HATs show considerably more sequence and structural diversity than HDACs, forming at least five different families: Gcn5/PCAF, MYST, p300/CBP, Rtt109, and HAT1. HATs typically function within multiprotein complexes in vivo, where their substrate specificities and activities are modulated by the other subunits of the respective HAT complexes (11, 12).
Human HBO1 (also referenced as KAT7 or MYST2), a member of the MYST family, regulates DNA replication (13, 14), cell proliferation, and development (15). Consistent with their role in gene expression, HBO1–HAT complexes are localized at transcription start sites and coding regions of active genes in the genome (16) and are responsible for the majority of H4 (14, 17, 18) and H3K14 (19, 20) acetylation. HBO1 has also been shown to play more specific roles in activating the expression of key genes in development and embryonic patterning (19). The aberrant activity of HBO1 has been correlated with several cancers. HBO1 has been shown to be overexpressed in MCF7 and Saos-2 established cancer cell lines and in a subset of human primary cancers such as carcinomas of the testis, breast, and ovary (21, 22). Finally, a HBO1 catalytically defective mutant inhibits MCM complex loading onto the origin, indicating that HBO1 functions as a DNA replication co-activator in addition to its transcriptional roles (5, 13, 18).
HBO1 functions in the context of two multiprotein histone acetyltransferase complexes containing ING4/5, MEAF6, and the paralogs JADE1/2/3 for targeting the H4 tail or BRPF1/2/3 for H3K14 acetylation (11, 20). The activity and substrate selectivity of HBO1 is dependent on the proteins within the HBO1 HAT complex, which also have other functions in various cellular proliferation and tumor suppression pathways. For example, JADE1 was initially identified as a protein partner of the von Hippel–Lindau tumor suppressor to regulate cellular oxygen sensing (23). A recent study demonstrated that an HBO1 complex containing the BRPF1 protein in place of JADE1 has specificity toward H3 tail acetylation rather than the H4 tail acetylation preferred by the JADE1-containing complex. This suggests that JADE1 contributes specifically toward H4 tail acetylation by the HBO1 complex (5, 11, 15). Another member of the HBO1 complex, ING, is an inhibitor of growth protein responsible for growth regulation in eukaryotic organisms. Mutation or down-regulation of ING genes leads to tumor developmental pathways (24, 25). The HBO1–HAT complex contains three PHD fingers responsible for recognizing various methylated states of H3K4. JADE1 contains two of these motifs (termed PZP), which cooperate to enable acetylation of nucleosome particles and recognition of unmethylated H3K4 (11, 15, 16). ING4 contains the third PHD finger, which recognizes trimethylated H3K4 (H3K4me3). The H3K4me3 modification state has been shown to be correlated with the ability of the HBO1–HAT complex to acetylate H4 (5, 11, 14, 16, 17). Through its recognition of H3K4me3, ING4 has been proposed to play a lynchpin role in the acetylation process. Thus, ING can link its aforementioned tumor suppressor function with chromatin binding, transcriptional activity, and cellular proliferation by nucleating the complex recruitment process (15, 24, 25). This is further substantiated by the activity of JADE1 spliced variants. Specifically, although the longer JADE1L form harbors tumor suppressor activity, the shorter JADE1 variant that lacks the ING4 binding domain but still binds to HBO1, called JADE1S, does not function as a tumor suppressor (15). JADE1 in the HBO1–HAT complex has been proposed to function as an intermediary to link HBO1–HAT catalytic activity to ING tumor suppressor activity through cross-talk between two different epigenetic modifications of methylation and acetylation (16). The absence of any of the HBO1–HAT complex subunits leads to diminished acetylation activity on cognate substrates, implying that these HBO1–HAT complex subunits play important regulatory roles (11, 15, 16).
JADE1 has been demonstrated to act as a platform to assemble the tetrameric HBO1–HAT complex for histone H4 acetylation, with a region of JADE1 N-terminal to the JADE1-PHD fingers (called region I) shown to recruit HBO1 and a region of JADE1 C-terminal to the JADE1-PHD fingers (called region II) shown to recruit the ING–MEAF6 subcomplex (15). It has also been shown that a modified tetrameric complex in which BRPF replaces JADE has specificity for H3K14 acetylation (11). These studies highlight the importance of BRPF and JADE subunits in dictating histone substrate specificity. Although a recent study has revealed the molecular basis for how BRPF mediates H3K14-specific acetylation by HBO1 (26), the molecular basis for how JADE potentiates the H4-specific activity of HBO1 is not known. Given this gap in knowledge, we set out to dissect the specific regions and activities of JADE1 that mediate histone H4 acetylation by the HBO1–HAT complex. We found that JADE1 contains specific HBO1 and histone binding regions that serve to increase the catalytic efficiency of HBO1 acetylation of histone H4. The implications of these studies for the differential regulation of HBO1 by JADE1 and BRPF1 paralogs are discussed.
Results
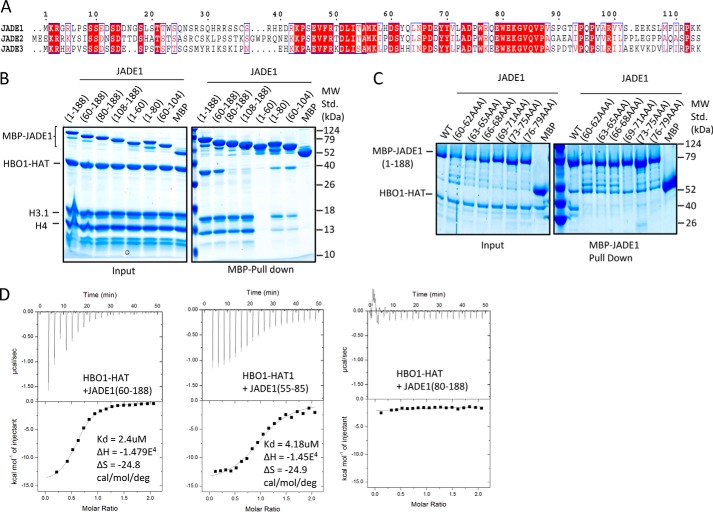
A 21-residue segment of JADE1 directly contacts the HBO1-HAT domain and histone substrate to facilitate acetylation of free histones
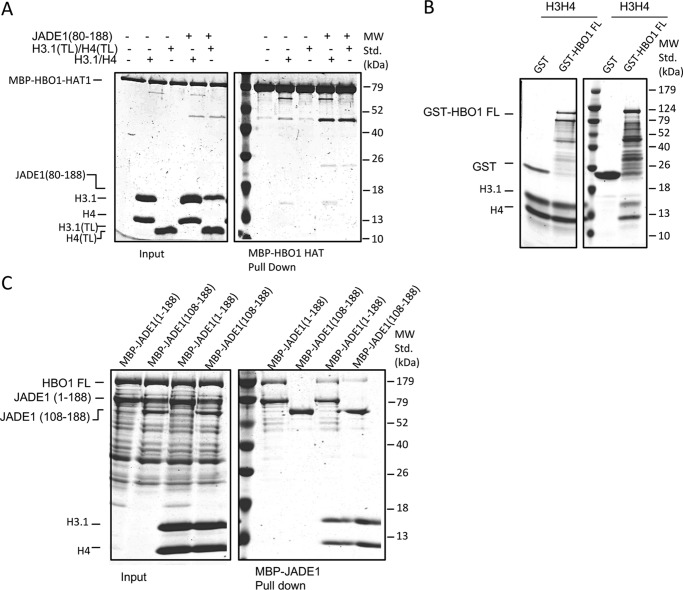
It was previously demonstrated that an N-terminal region of JADE1 between residues 90–199 participated in HBO1 catalytic activity (15). This region of JADE1 contains sequence conservation within residues 115–179 of yeast to human proteins associated with ING–MYST–containing HAT complexes (11, 15). We also noted an additional region of sequence conservation among the JADE proteins just N-terminal to this region (Fig. 1A). To map the region of the JADE1 N terminus that directly interacts with the HBO1-HAT domain, we used pulldown experiments with the HBO1-HAT and N-terminal truncation constructs of JADE1 (Fig. 1B). These experiments revealed that MBP-JADE1 (1–188) and MBP-JADE1 (60–188) were able to pull down the HBO1-HAT complex whereas MBP-JADE1 (80–188) was not. These data revealed the importance of residues 60–80 of JADE1 for the direct interaction with the HBO1-HAT complex. To further narrow down the key residue(s) within this 21-residue segment critical for the interaction with the HBO1-HAT domain, we introduced triple alanine mutations spanning residues 60 to 80 within the context of MBP-JADE1 (1–188). These studies revealed that each of the mutants resulted in no detectable HBO1-HAT pulldown (Fig. 1C). The observation that each of the JADE1 mutants disrupted HBO1-HAT interaction suggested that either the entire JADE1 (60–80) domain makes important contacts to the HBO1-HAT or that the domain contains a binding surface and tertiary structure that are both important for HBO1-HAT interaction. To more quantitatively assess the binding of the JADE N-terminal region to the HBO1-HAT, we performed isothermal titration calorimetry (ITC) using various constructs of JADE1. The ITC results were consistent with the pulldown data, demonstrating that JADE1 (60–188) showed significant binding to the HBO1-HAT with a Kd of ∼2.4 μm and a stoichiometry of ∼1:1, whereas JADE1 (80–188) showed no detectable binding (Fig. 1D). Strikingly, a peptide of residues 55–85 of JADE1 showed binding with a comparable dissociation constant (Kd = 4.18 μm) to JADE1 (60–188), demonstrating that residues 60–80 of JADE1 are essential for HBO1-HAT binding.
Figure 1.
An N-terminal region of JADE1 binds HBO1. A, sequence alignment of the N-terminal region of JADE1/2/3 paralogs, with the highest degree of conservation shaded in red and conservative substitutions colored in red. The HBO1 and HHBD are highlighted. B, MBP-JADE1 pulldowns with various N-terminal and C-terminal deletion constructs of the HBO1-HAT domain and H3/H4 complex with results resolved on SDS-PAGE. MW Std., molecular weight standard. C, MBP-JADE1 pulldowns of the HBO-HAT with JADE1 triple alanine mutations with results resolved on SDS-PAGE. D, ITC studies of JADE1 constructs titrated into the HBO1-HAT, showing the heat profile (top) and the calculated binding isotherm (bottom). Thermodynamic data calculated from this data are shown at the right.
We next asked whether residues of 60–80 of JADE1 are also able to interact with histones to potentially bridge the HBO1-HAT acetylation of histones. To do this, we carried out pulldown studies of MBP-JADE1 N- and C-terminal deletion constructs with HBO1-HAT plus H3–H4 (Fig. 1B). These studies revealed that only JADE1 constructs harboring residues 60–80 of JADE1 were able to interact with both the HBO1-HAT and H3–H4, confirming that JADE1 residues 60–80 bridge HBO1-HAT–histone interactions. Notably, JADE1 constructs starting at residue 60 and extending to residue 188 showed more robust pulldown with HBO1-HAT and histones than JADE1 constructs harboring only residues 60–80, suggesting that additional regions of JADE1 between residues 108 and 188 also contribute to HBO1–histone interactions (Fig. 1B). Based on the HBO1-HAT and histone binding properties of residues 60–80 of JADE1, we refer to it as the HBO1 and histone binding domain (JADE1-HHBD).
We then asked whether the minimal region of JADE1 that facilitates HBO1 and histone binding is sufficient to potentiate HBO1-HAT activity toward histone tail acetylation. To do this, we titrated JADE1(55–85) and JADE1(1–188) into the HBO1-HAT and H3–H4 complex and assayed for histone acetyltransferase activity (Fig. 2A). This analysis revealed that titration of JADE1(55–85) in vast excess resulted in only marginal effects on HBO1 histone acetyltransferase activity, whereas titration of JADE1(1–188) showed significantly more potentiation of HBO1 histone acetyltransferase activity, to a level of about 6-fold relative to no added titrant. Taken together with the previous data showing that residues 60–80 of JADE1 bridge HBO-HAT–histone interaction, these data demonstrate that residues 60–80 of JADE1 contribute to but are not sufficient to mediate HBO1 acetylation of histones.
Figure 2.
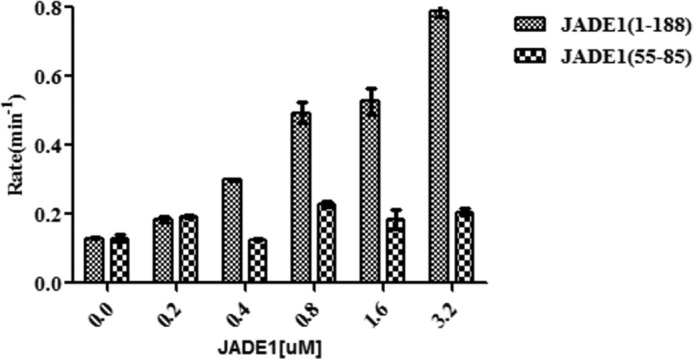
JADE1 requires an additional N-terminal region for H3/H4 binding and HBO1 HAT potentiation. Shown is the titration of JADE1 constructs (55–85 and 1–188) from 0- to 16-fold molar excess of HBO1 HAT domain tested for acetyltransferase activity on the H3/H4 complex. Radioactive counts are converted to enzyme rate as described under “Experimental procedures.”
JADE1 contains a second histone binding domain that contributes to HBO1 acetylation of histone H4 substrate by anchoring the HBO1–JADE1 complex to the histone core
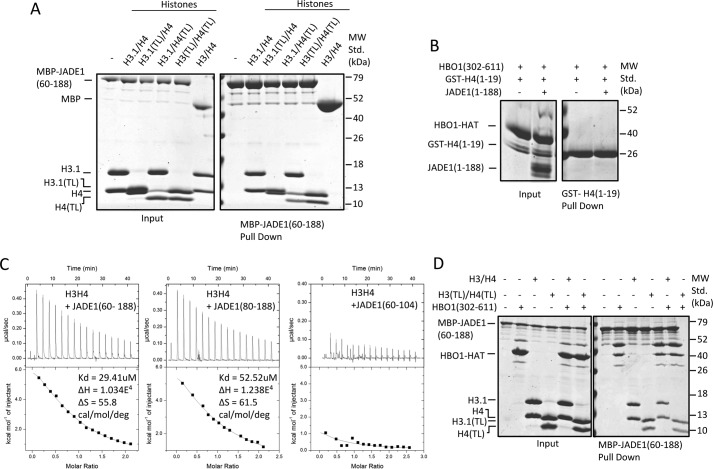
Inspection of the pulldown experiments of Fig. 1B reveals that an additional region of JADE1, within residues 108–188, interacts with histones but not the HBO1-HAT, and the histone acetyltransferase activity assay of Fig. 2A further suggests that regions within JADE1 residues 80–188 are also required to fully potentiate HBO1-HAT histone acetyltransferase activity. Based on these studies, we carried out further studies to define the regions of JADE1 and histones that mediate this interaction. To specifically probe the requirement for the histone tails for JADE1 interaction, we carried out MBP-JADE1(60–188) pulldown with H3–H4 with either full-length or tailless H3 and/or H4 subunits. These studies revealed that JADE1(60–188) pulled down all of the histone complexes, including the completely tailless complexes (H3(TL)–H4(TL)) (Fig. 3A). These pulldown experiments demonstrated that the primary JADE1(60–188) binding site exists on the core of the H3/H4 complex.
Figure 3.
The additional JADE1 N-terminal HCBD specifically binds the histone core region. A, MBP-JADE1 pulldowns of four different H3/H4 complexes (H3/H4, H3(TL)/H4, H3/H4(TL), H3(TL)/H4(TL)) with results resolved on SDS-PAGE. MW Std., molecular weight standard. B, GST-H4(1–19) pulldowns of HBO1-HAT in the presence and absence of JADE1(1–188) with results resolved on SDS-PAGE. C, ITC studies of JADE1 constructs titrated into the H3/H4 complex showing the heat profile (top) and the calculated binding isotherm (bottom). Thermodynamic data calculated from these data are shown at the right. D, MBP-JADE1 pulldowns of HBO1-HAT and H3/H4 complex in the presence and absence of each other, with results resolved on SDS-PAGE.
Previous studies indicate that JADE1 facilitates HBO1 H4–specific tail acetylation (11). For this reason, we asked whether JADE1 makes an additional direct or HBO1-dependent contact with the H4 tail. To test this, we prepared GST-tagged H4 tail 1 to 19 and performed a GST pulldown with JADE1(1–188) in the presence of the HBO1-HAT. This experiment revealed that HBO1-HAT is unable to interact directly with the isolated H4 histone tail in the absence or presence of JADE1(1–188) (Fig. 3B), consistent with the conclusion that JADE1 histone core binding is required for HBO1-HAT acetylation of the histone H4 tail. We refer to the domain of JADE1 that mediates H3–H4 core binding as the histone core binding domain (JADE1-HCBD).
MBP pulldowns with JADE1 deletion constructs and H3–H4 (Fig. 1B) suggest that JADE1 contains two histone binding sites, JADE1(60–80) and JADE1(104–188). To more quantitatively evaluate the histone binding properties of these two regions of JADE1, we performed ITC with H3–H4 using various constructs of JADE1. The ITC data showed that JADE1(80–188) binds H3/H4 with a dissociation constant of 52.52 μm (Fig. 3C), whereas JADE1(60–188), containing both histone binding sites, showed H3–H4 binding with a dissociation constant of about 2-fold higher, 29.41 μm (Fig. 3C). These studies indicate that the HHBD and HCBD regions of JADE1 bind additively to H3–H4.
We also carried out MBP-JADE(1–188) pulldown studies with H3–H4 (with or without tails) in the presence or absence of the HBO-HAT, revealing that the histone binding properties of JADE1(1–188) are independent of HBO1-HAT binding (Fig. 3D).
JADE1-HCBD shows selectivity for free histone H3–H4 over H2A–H2B and nucleosomes
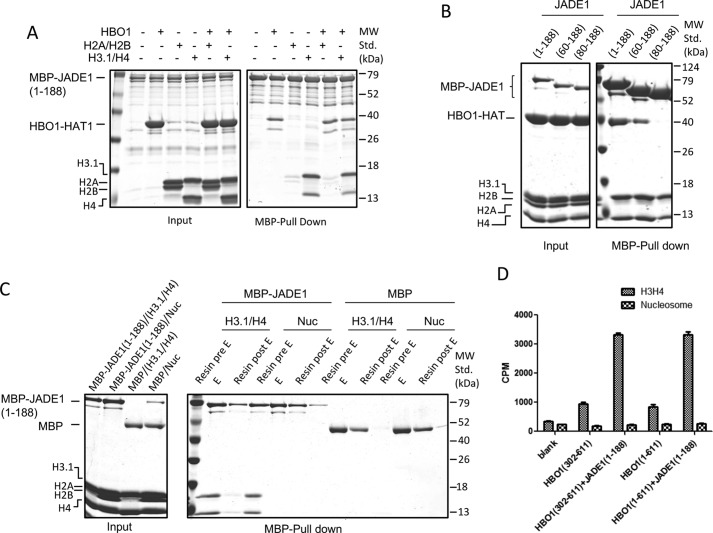
To further probe the histone binding specificity of JADE1-HCBD, we performed pulldown experiments with MBP-JADE1(1–188) and histones H3–H4 and H2A–H2B. These studies demonstrated that, although JADE1(1–188) interacts with histone H3–H4 in the presence or absence of the HBO1-HAT, it does not interact with histone H2A–H2B (Fig. 4A). Moreover, when we carried out similar pulldown studies with a mixture of H3–H4 and H2A–H2B complexes, JADE1(1–188) specifically selects for H3–H4 over H2A–H2B, suggesting that JADE1-HCBD does not interact with the H3–H4 core through a nonspecific charge interaction (Fig. 4B).
Figure 4.
JADE1 selectively binds to H3/H4 over H2A/H2B and the nucleosome core particle. A, MBP-JADE1 pulldowns of HBO1 HAT and (H3/H4 or H2A/H2B) in the presence or absence of each other, with results resolved on SDS-PAGE. MW Std., molecular weight standard. B, MBP-JADE1 pulldowns of HBO1 HAT and H3/H4, H2A/H2B and mixtures of the two with results resolved on SDS-PAGE. C, MBP-JADE1 pulldowns of H3/H4 or nucleosome core particles with results resolved on SDS-PAGE. D, activity comparison of HBO1 HAT and HBO1 full-length in the presence and absence of a saturating concentration of JADE1(1–188) as either H3H4 complex or nucleosome core particles as the substrate. The activity of these proteins on various substrates was compared using radioactive counts.
It has been demonstrated previously that the H3 tail is essential for the HBO1-HAT complex to interact with chromatin (16). JADE1 contains two PHD finger domains (PHD–Zn knuckle–PHD or PZP domain) C-terminal to the JADE-HCBD that recognize specific methylation states of the H3 tail and allow the HBO1–HAT complex to bind and acetylate nucleosomes (15–17). Given this information, we wanted to further investigate whether the JADE-HCBD can bind to H3–H4 when it is assembled into a nucleosome. Pulldown studies with the assembled nucleosome revealed the absence of an interaction (Fig. 4C), suggesting that the octamer or the DNA is excluding the JADE1-HCBD binding surface to H3–H4 (Fig. 4C). To further substantiate these findings, we compared the acetylation activity of HBO1-HAT and HBO1-FL with or without JADE1 (1–188) against H3–H4 and nucleosomes. These studies revealed that JADE1(1–188) potentiated the activity of HBO1-HAT and HBO1-FL against H3–H4 but not nucleosomes (Fig. 4D). These data suggest that other regions of JADE1 are required for HBO1 targeting to nucleosomes. This is consistent with previously reported data that the PZP, PHD1, and PHD2 domains of JADE1, located C-terminal to the JADE1 HHBD and HCBD domains, contribute to HBO1-mediated acetylation of nucleosomes (11, 15, 16). In contrast, it appears that the HHBD and HCBD domains of JADE1 play an important role in capturing free histones (H3–H4) during transcription (or replication) for further acetylation of the tails.
JADE1-HHBD and JADE1-HCBD bind independently to their respective partners but cooperate to potentiate HBO1-HAT acetylation
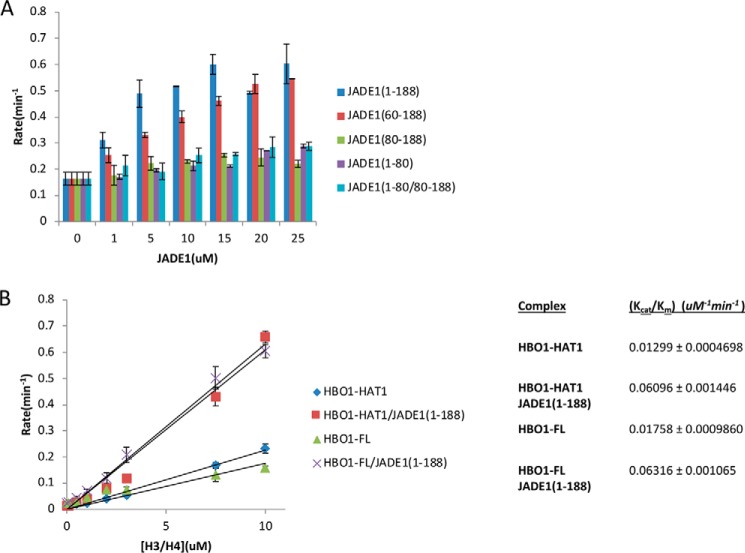
After establishing that the HHBD and HCBD regions of JADE1 bind independently to their respective histone and/or HBO1-HAT binding partners, we set out to determine the contribution of these JADE1 regions to HBO1 acetyltransferase activity. We prepared four JADE1 deletion constructs spanning the HHBD and/or HCBD regions: 1–188(HHBD-HCBD), 60–188(HHBD-HCBD), 80–188(HCBD), and 1–80(HHBD). We assayed HBO1-HAT activity in the presence of a 25-fold molar excess of each of these JADE1 constructs. This experiment revealed that only JADE1 construct spanning both the HHBD and HCBD showed potentiating effects of HBO1-HAT activity (Fig. 5A). Moreover, titration with separate fragments of JADE1 1–80(HHBD) and JADE1 80–188(HCBD) did not potentiate HBO1-HAT acetylation activity (Fig. 5A). These results demonstrate that the JADE1-HHBD and HCBD regions work cooperatively to potentiate HBO1 HAT activity and that these two regions of JADE1 must be connected to mediate this activity.
Figure 5.
JADE1-HHBD and -HCBD work cooperatively to activate HBO1 HAT activity. A, titration of JADE1 constructs (1–188, 60–188, 80–188, 1–80, and 1–80/80–188) from 0 to 25 molar excess over the HBO1 HAT domain tested for acetyltransferase activity on the H3/H4 complex. Radioactive counts are converted to enzyme rate as described under “Experimental procedures.” B, plot of the linear range of the Michaelis–Menten plot of HBO1-HAT and HBO1-FL ± JADE1 with H3/H4 as the substrate to calculate the catalytic efficiency (slope, Kcat/Km). Radioactive counts are converted to enzyme rate as described under “Experimental procedures.”
To more quantitatively assess the contribution of JADE1 to HBO1-HAT catalytic activity, we carried out bisubstrate kinetics. Because of a significant chemical acetylation that occurs on the core of histones at high concentration, we were not able to carry out a full Michaelis–Menten plot. Instead, we measured the catalytic efficiency (Kcat/Km) using the slope of the linear range in the Michaelis–Menten plot. We found that JADE1(1–188) increased the catalytic efficiency of HBO1 by ∼5-fold (Fig. 5B). Together, these data demonstrate the importance of JADE1 in contributing to the overall catalytic activity of the HBO1–HAT complex by anchoring the complex to the substrate and by properly orienting the HBO1-HAT domain.
HBO1 contains a region N-terminal to the HAT domain that binds H3–H4 independent of JADE1 binding to H3–H4
Having established that the HHBD and HCBD regions of JADE1 contact H3/H4, we asked whether other regions of HBO1 also contribute to histone binding. To address this, we first carried out pulldown studies with MBP-HBO1-HAT, JADE1(80–188), and H3–H4 to demonstrate that the HBO1-HAT is unable to directly pull down H3–H4 (Fig. 6A). Although HBO1-HAT does not make stable direct contacts with the histones in the ground state, we asked whether the rest of HBO1 has any contribution toward H3–H4 binding independent of JADE1. To address this possibility, we carried out H3–H4 pulldown studies with either GST-HBO1-FL or GST alone. These data demonstrate that, unlike HBO1-HAT, HBO1-FL is able to pull down H3–H4, indicating that HBO1 also contains an H3–H4 binding site outside of the HAT domain independent of JADE1 (Fig. 6B). Revealing this independent histone binding property of HBO1-FL, we asked whether JADE1 and HBO1 can interact with H3–H4 in the presence of each other. To do this, we carried out additional MBP-JADE1 pulldown studies with HBO1-FL and H3–H4 in the presence and absence of JADE1-HHBD. As expected, JADE1(1–188) was able to pull down HBO1-FL independent of H3–H4 binding at the HCBD. However, JADE1(108–188) was only able to pull down HBO1-FL in the presence of H3–H4 (Fig. 6C). These data suggest that, unlike HBO1-HAT, HBO1-FL is able to pull down H3–H4, indicating that HBO1 also contains an H3–H4 binding site outside of the HAT domain independent of JADE1 (Fig. 6B). Although we favor the interpretation that H3–H4 is being bound by the full-length HBO1, given the presence of additional bands in the preparation, we cannot rule out that some of the interactions are through the smaller fragments.
Figure 6.
HBO1-FL contains an N-terminal H3/H4 binding domain that binds histones independently of JADE1. A, MBP-HBO1-HAT pulldown of H3/H4 of H3(TL)/H4(TL) in the presence and absence of JADE1-HCBD, with results resolved on SDS-PAGE. MW Std., molecular weight standard. B, GST-HBO-FL and GST control pulldown of the H3/H4 complex, with results resolved on SDS-PAGE. Note that JADE1(80–188) and H3.1 co-migrate on SDS-PAGE. C, MBP-JADE1(1–188) and (108–188) pulldown of HBO1-FL ± H3/H4 complex, with results resolved on SDS-PAGE.
To investigate the role of the additional histone binding domain N-terminal to the HBO1-HAT region (HBO1-HBD), we asked whether the full-length HBO1 shows higher enzymatic efficiency toward the substrate. When compared, the catalytic efficiencies of HBO1-HAT and HBO1-FL are comparable with each other. This analysis demonstrates that, despite the additional histone binding property of HBO1-FL, HBO1-HBD does not have a significant contribution to the overall HBO1 catalytic efficiency toward H3–H4 and requires JADE1(1–188) to potentiate its acetyltransferase activity (Fig. 5B). The fact that HBO1-HAT and HBO1-FL have comparable catalytic efficiencies suggests that the additional H3–H4 interactions mediated by HBO1-FL relative to HBO1-HAT are not productive for catalysis.
Cellular experiments with JADE1 deletions recapitulate the role of JADE1 in histone and HBO1 interactions and HBO1 acetyltransferase activity
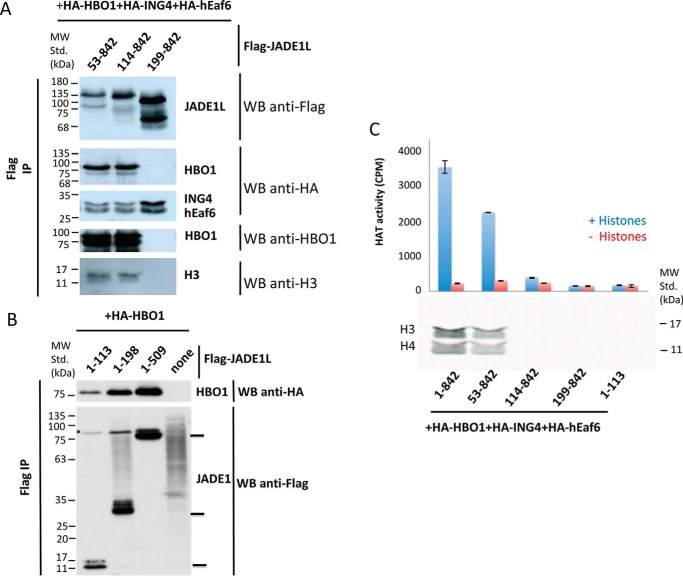
To support the conclusions drawn by the in vitro interaction studies, we performed co-transfections in HEK 293T cells with vectors expressing different truncation constructs of JADE1, followed by anti-FLAG immunoprecipitation and peptide elution of the complexes. Western blot analysis of the purified fractions with progressive N-terminal truncations of JADE1 indicates that deletion of the first 113 amino acids, which includes the HHBD, does not disrupt the interaction of JADE1 with the HBO1–H3–H4 complex in cells (Fig. 7A), whereas the additional deletion of the JADE1-HCBD abolishes JADE1 interaction with the HBO1–H3–H4 complex, as reported previously (15). The observation that JADE1 constructs that contain residues C-terminal to the HHBD (53–842, 114–842, and 199–842) immunoprecipitate with ING4–EAF6 is also consistent with our earlier studies (15). We performed a similar experiment using progressive C-terminal deletions of JADE1, demonstrating that the region containing the HHBD is also sufficient to interact with HBO1 in cells (Fig. 7B).
Figure 7.
JADE1 HHBD and HCBD are both required for HBO1 acetyltransferase activity on histones in cells. A, Western blot (WB) analysis of JADE1 complexes with HBO1, H3, ING4, and EAF6 with different N-terminal JADE1 truncations. An anti-HA immunoblot was done first to visualize the HA-tagged subunits of the FLAG-JADE1 complex overexpressed in HEK 293T cells. The membrane was then reprobed with the HBO1 antibody (showing endogenous HBO1 association beside HA-HBO1) and finally with the anti-FLAG antibody. IP, immunoprecipitation; MW Std., molecular weight standard. B, HBO1 association with the C-terminal truncated forms of JADE1. Anti-HA was followed by anti-FLAG immunoblot on FLAG-JADE1 complex overexpressed in 293T cells (the asterisk denotes the remaining HBO1 HA signal in the anti-FLAG blot). C, HAT assay of immunopurified complexes from A. HAT assays were performed on free histones (0.5 μg) with JADE1 complex using the same ratio as for the immunoblotting. Shown is a graphical representation of counts per minute, measured by scintillation, and a fluorogram of the radioactive HAT assay on free histones.
Consistent with the in vitro studies, acetylation assays of cell-immunoprecipitated JADE1 deletion constructs demonstrated that acetylation activity requires both the HHBD and HCBD regions of JADE1 (Fig. 7C). These results demonstrate that, as observed in vitro, although the JADE1-HCBD is required to stabilize the HBO1 complex, the JADE1-HHBD region is required to enable HBO1 acetylation activity toward histones.
Discussion
Histone tail acetylation events have been linked to many important cellular processes through controlling chromatin dynamics to mediate various DNA transactions, including DNA replication and gene transcription (2, 4). Although the structure and catalytic activity of HATs have been well delineated, their mode of substrate-specific acetylation has been less rigorously analyzed. GCN5 has been reported to mediate preferential H3K14 acetylation through making extensive direct contact with the residues of the H3 tail flanking the H3K14 target lysine residue using sequence-based recognition (27). On the other hand, numerous HATs function in multisubunit complexes where the catalytic efficiency and substrate specificities are determined by subunits outside of the catalytic domain (11). A recent structural study of the NuA4 HAT complex revealed a sequence/position double recognition mechanism for substrate specificity whereby the HAT complex binds to the nucleosome through a subunit outside of the catalytic domain to position the H4 tail proximal to the catalytic domain, which then employs sequence-based recognition for sequence-specific acetylation (28). Additionally, BRFP2 was recently reported to potentiate and mediate H3-specific acetylation by HBO1 through a short N-terminal fragment that bridges interaction between the HAT domain and H3–H4 (26).
In this study, we dissected the role of JADE1 in HBO1 HAT activity toward free histones (H3–H4) based on previous studies suggesting that JADE1 plays a key role in HBO1 complex H4 substrate–specific acetylation. The in vitro and cellular data presented here demonstrate that robust histone H3–H4 acetylation by the HBO1 requires the HBO1-HAT domain (residues 311–611) in addition to a 21-residue N-terminal JADE1 region (HHBD, residues 60–80) that bridges HBO1-HAT and H3–H4 contacts and a second N-terminal JADE1 region (HCBD, residues 108–188) that makes additional H3–H4 contacts within the histone core region (Fig. 8A). The N-terminal region of JADE1 therefore functions as a platform to bring the catalytic HBO1 subunit and the free histone H3–H4 substrate tail together for catalysis.
Figure 8.
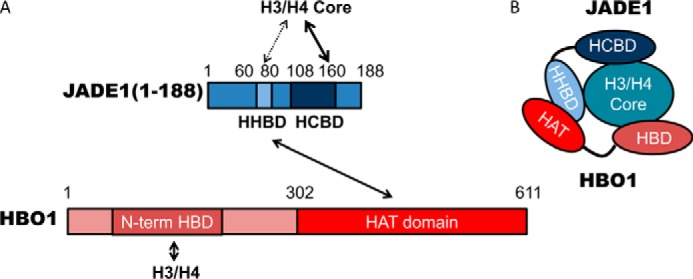
Schematic of the HBO1, JADE1, H3/H4 complex based on the data obtained in this study. A, schematic of the JADE1 and HBO1 domains. B, schematic of JADE1-HBO1 domain interactions.
The mechanism of JADE1 activation of the HBO1–HAT complex has similarities and differences to that of BRFP2. Both JADE1 and BRFP2 contain short N-terminal fragments (HHBD in the case of JADE1) that bind to both HBO1 and H3–H4, thus bridging the appropriate HBO1-histone interaction for substrate-specific acetylation. Unlike BRFP2, JADE1 contains a second N-terminal HCBD that also binds to H3–H4 and cooperates with the HHBD to significantly increase the catalytic efficiency of HBO1 for H4 acetylation.
In addition to the role of the N-terminal region of JADE1 in HBO1 acetylation described here, the role of two C-terminal PHD fingers of JADE1 have been characterized previously. These tandem PHD fingers cooperate to recognize the H3 tail and facilitate the targeting of the HBO1–HAT complex to chromatin for nucleosome binding (17). Previous studies have suggested that the HBO1–HAT complex is heavily localized to transcriptionally active regions ahead of polymerase to facilitate nucleosome disassembly, consistent with HBO1 complex copurification with the FACT (facilitates chromatin transcription) complex (16). In the context of transcription within chromatin, JADE1-PHD finger–mediated recognition of methylated histones may facilitate the subsequent JADE1-mediated HBO1 acetylation of histone H4 to contribute to nucleosome disassembly through increasing chromatin dynamics after nucleosome disassembly and/or provide recognition marks for other proteins involved in transcription elongation. JADE1 has also been linked to HBO1-mediated histone H4 acetylation during DNA replication (18), although the relative roles of the HHBD, HCBD, and other C-terminal JADE1 domains in this process remain to be determined.
Experimental procedures
JADE1/2/3 N-terminal sequence alignment
Sequence alignment for JADE1/2/3 was carried out using ClustalW2 (29) and further formatted using ESPript (3.0) for more optimal sequence homology of JADE homologs (30, 31).
Recombinant HBO1 and JADE1 protein production
HBO1 and JADE1 protein constructs were generated by PCR amplification of the corresponding DNAs, which were cloned into pRSF or pDB.His.MBP vectors using Sacl/NotI sites with cleavable N-terminal affinity tags (GST, His6, and MBP). These constructs were grown and expressed using BL21(DE3) cells in Luria Broth (LB). Cells were induced for protein overexpression at an optical density of ∼0.6 with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside for 16–18 h at 18 °C. Cells were harvested by centrifugation at 4000 rpm and lysed by sonicating in lysis buffer (20 mm Tris (pH 8). 500 mm NaCl, 5 mm β-mercaptoethanol (BME), and 0.1 mg/ml phenylmethylsulfonyl fluoride). After lysis, the sample was spun down at 20,000 rpm for 30 min. and the soluble fraction of the post-sonicated sample was applied to either Pierce glutathione–agarose resin (Thermo), amylose resin (New England Biolabs), or nickel-nitrilotriacetic acid resin (Biosciences) for batch binding at 4 °C for 1 h. Samples were washed with 20 column volumes of wash buffer (20 mm Tris (pH 8), 500 mm NaCl, and 5 mm BME) and then eluted with eluant with either 30 mm reduced l-glutathione (GST), 20 mm maltose monohydrate (MBP), or 300 mm imidazole (His6). Affinity tags were cleaved with tobacco etch virus protease or were left on for affinity tag pulldowns. Proteins were further purified to homogeneity by either a HiTrapQ (GE Healthcare) anion exchange column for JADE1 constructs or HiTrapHP (GE Healthcare) cation exchange for HBO1 constructs. Ion exchange chromatography was performed using an NaCl gradient from 100 mm to 1 m over 30 column volumes. To ensure the proper fold of recombinant proteins, they were subjected to size exclusion chromatography using a Superdex 200 column in 20 mm Tris (pH 8), 500 mm NaCl, and 5 mm BME. MBP-JADE1(1–188) mutant constructs were generated through site-directed mutagenesis using Q5 Hot Start Polymerase (New England Biolabs). All of the mutant constructs were verified by DNA sequencing and were expressed and purified using the same protocol mentioned previously for wildtype constructs.
Isothermal titration calorimetry
Interaction of HBO1–JADE1 and histone–JADE1 with various JADE1 constructs was quantitatively analyzed using a MicroCal ITC200 (Malvern). All proteins were prepared and dialyzed to 20 mm HEPES (pH 7.5), 500 mm NaCl, and 5 mm BME before the analysis. All ITC experiments were conducted by injecting 800 μm JADE1 deletion constructs into a calorimetry cell either containing 40 μm HBO1-HAT or H3–H4. JADE1 deletion constructs were injected in 2-μl increments every 2 min at 20 °C. ITC data were analyzed using MicroCal ITC-ORIGIN (Malvern).
Histone and nucleosome core particle production
Each of the histones (H3, H4, H2A, and H2B) were expressed and purified as inclusion bodies. Refolding of H2A–H2B and H3–H4 and their purification were performed as described previously (31, 32). The tailless constructs H3 (36–135) and H4(20–104) were prepared in the same way as the wildtype. These refolded histone components, in addition to the 601-nucleosome positioning sequence, were used to form nucleosome core particles as described previously (32).
Pulldown assay (GST/MBP)
2 μm tagged protein and 10 μm untagged protein were incubated together with 50 μl of resin for 1 h at 4 °C. The reactions were carried out in sizing buffer (20 mm Tris (pH 8), 500 mm NaCl, and 5 mm BME). Resin was then washed extensively with the same buffer and collected for analysis on SDS-PAGE gel. All of the pulldowns were done side by side with the tag alone under the same conditions as controls for background interactions.
HAT activity assay
HBO1-HAT activity was measured using a previously described HAT assay with 14C-labeled Ac-CoA (50–60 mCi/mmol, Moravek) (33). A time course for the reaction was initially performed to determine the linear range of enzyme activity at an enzyme concentration of 200 nm (except that 2 μm was used in Fig. 5A) and saturating concentrations of Ac-CoA (300 μm) and H3–H4 (or nucleosome core particles) (100 μm). The HAT reaction was done with 20 mm Tris (pH 7.5), 500 mm NaCl, and 0.25 mg/ml BSA for 1 h at 20 °C in a volume of 50 μl. The reaction was spotted on P81 filter paper. The positively charged and C14-labeled H4 peptide portion of the histone complex (H3–H4) was captured on the filter paper, whereas free 14C-labeled Ac-CoA was washed away. The paper was analyzed using liquid scintillation (34). The amount of 14C-labeled H4 peptide bound to the paper is directly proportional to the amount of acetyl group transferred from the cofactor to peptide, thus the data were used to evaluate HBO1 acetyltransferase activity (34). All reactions were done in duplicate. Using the known concentration series of Ac-CoA, a standard curve was generated. The standard curve was used to convert radioactive counts to molar units.
JADE1–HBO1 complex cotransfection in 293T cells
JADE1L, HBO1, hEAF6, and ING4 constructs were generated by PCR amplification of the corresponding DNAs, which were cloned into the pcDNA3 vector encoding an N-terminal FLAG tag using BglII/Xho1 sites or a HA tag. These plasmids were then used to cotransfected 293T cells with the calcium phosphate method for protein complex overexpression with HA-HBO1–HA-ING–/HA-hEAF6 and full-length or N-terminal truncated 3× FLAG-JADE1L: amino acids 1–842, 53–842, 114–842, 199–842, and 1–113. A second set of cotransfection was done using C-terminal truncated 1× FLAG JADE1; the first 113, 198, or 509 amino acids were overexpressed in combination with HA-HBO1.
Cells were harvested 48 h post-cotransfection and lysed in high-salt buffer (450 mm NaCl, 50 mm Tris-HCl (pH 8.0), 1% Triton X-100, 2 mm MgCl2, 0.1 mm ZnCl2, 2 mm EDTA, and 10% glycerol) supplemented with protease inhibitor mixture. The NaCl concentration was reduced to 225 mm, and the whole-cell extract was centrifuged at 14,000 rpm for 30 min. The FLAG JADE1 was purified from the soluble fraction using anti-FLAG M2 affinity beads (Sigma) for 2 h at 4°C on rotating wheel. FLAG beads were next washed with 225 mm NaCl buffer and eluted with 400 μg/ml of 3× or 1× FLAG peptide (Sigma) for the N-terminal or C-terminal truncations, respectively.
Immunoblotting
The amounts of the different JADE1 constructs were normalized empirically following SDS-PAGE and transferred onto a nitrocellulose membrane. Anti-FLAG M2 conjugated to horseradish peroxidase (Sigma) was used at a 1:10,000 dilution, and the immunoblots were visualized using a Western Lightning Plus ECL reagent (PerkinElmer Life Sciences). Anti-HA 3F10 (Roche, rat polyclonal) and anti-HBO1 (Novus, rabbit polyclonal) antibodies were used at a 1:1000 dilution. Horseradish peroxidase–conjugated secondary anti-rat and anti-rabbit antibodies (Sigma) were used at a 1:10,000 dilution.
HAT activity assays on purified JADE1 complexes
HBO1 activity in the purified JADE1 complexes was measured with 0.125 μCi of 3H-labeled Ac-CoA (2.1 Ci/mmol, PerkinElmer Life Sciences). The amounts of the different purified JADE1 complexes used in the HAT assays were normalized based on the immunoblot data. The HAT reactions were performed in a volume of 15 or 20 μl using 0.5 μg of human free histones or native short oligonucleosomes as substrate, respectively, in HAT buffer (50 mm Tris-HCl (pH 8), 50 mm KCl, 10 mm sodium butyrate, 5% glycerol, 0.1 mm EDTA, and 1 mm dithiothreitol) for 1 h at 30°C. The reactions were then captured on P81 filter paper. The free 3H-labeled Ac-CoA was washed away, and the paper was analyzed using liquid scintillation. The 3H counts were used to evaluate HBO1 acetyltransferase activity in the complexes containing different JADE1 constructs.
Author contributions
J. H., C. L., M. D. R., C. E. M., M. G., B. E. B., J. C., and R. M. conceptualization; J. H., C. L., M. D. R., C. E. M., M. G., and J. C. data curation; J. H., C. L., M. D. R., C. E. M., and J. C. formal analysis; J. H. writing-original draft; C. L. investigation; C. L. methodology; M. D. R., C. E. M., M. G., B. E. B., J. C., and R. M. writing-review and editing; B. E. B., J. C., and R. M. funding acquisition; J. C. and R. M. supervision; J. C. validation; J. C. and R. M. project administration; R. M. resources.
Acknowledgments
We thank Rob Magin and Adam Olia for helpful discussions.
This work was supported by National Institutes of Health Grants R01 GM060293 and R35 GM118090 and the Ovarian Cancer Research Fund Alliance (to R. M.), Canadian Institutes of Health Research Grant FDN-143314 (to J. C.), National Institutes of Health Grant R01 GM082989 (to B. E. B.), and the DNA Sequencing Facility at the Perelman School of Medicine, University of Pennsylvania (NIH P30 CA016520). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- HAT
- histone acetyltransferase
- HDAC
- histone deacetylase
- H3K4me3
- trimethylated H3K4
- PCAF
- p300/CBP–associated factor
- CBP
- CREB-binding protein
- CREB
- cAMP-response element-binding protein
- PHD
- plant homeodomain
- MBP
- maltose-binding protein
- BME
- β-mercaptoethanol
- ITC
- isothermal titration calorimetry
- HHBD
- HBO1 and histone binding domain
- TL
- tailless
- GST
- glutathione S-transferase
- HCBD
- histone core binding domain
- PZP
- PHD–Zn knuckle–PHD
- FL
- full-length
- Ac-CoA
- acetyl-CoA
- HA
- hemagglutinin.
References
- 1. Sapountzi V., and Côté J. (2011) MYST-family histone acetyltransferases: beyond chromatin. Cell. Mol. Life Sci. 68, 1147–1156 10.1007/s00018-010-0599-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Strahl B. D., and Allis C. D. (2000) The language of covalent histone modifications. Nature 403, 41–45 10.1038/47412 [DOI] [PubMed] [Google Scholar]
- 3. Kornberg R. D., and Lorch Y. (1999) Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98, 285–294 10.1016/S0092-8674(00)81958-3 [DOI] [PubMed] [Google Scholar]
- 4. Thomas T., and Voss A. K. (2007) The diverse biological roles of MYST histone acetyltransferase family proteins. Cell Cycle 6, 696–704 10.4161/cc.6.6.4013 [DOI] [PubMed] [Google Scholar]
- 5. Havasi A., Haegele J. A., Gall J. M., Blackmon S., Ichimura T., Bonegio R. G., and Panchenko M. V. (2013) Histone acetyl transferase (HAT) HBO1 and JADE1 in epithelial cell regeneration. Am. J. Pathol. 182, 152–162 10.1016/j.ajpath.2012.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Taverna S. D., Li H., Ruthenburg A. J., Allis C. D., and Patel D. J. (2007) How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 14, 1025–1040 10.1038/nsmb1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Turner B. M. (2000) Histone acetylation and an epigenetic code. BioEssays 22, 836–845 10.1002/1521-1878(200009)22:9%3C836::AID-BIES9%3E3.0.CO%3B2-X [DOI] [PubMed] [Google Scholar]
- 8. Peterson C. L., and Laniel M. A. (2004) Histones and histone modifications. Curr. Biol. 14, R546–551 10.1016/j.cub.2004.07.007 [DOI] [PubMed] [Google Scholar]
- 9. Jenuwein T., and Allis C. D. (2001) Translating the histone code. Science 293, 1074–1080 10.1126/science.1063127 [DOI] [PubMed] [Google Scholar]
- 10. Allis C. D., Berger S. L., Cote J., Dent S., Jenuwien T., Kouzarides T., Pillus L., Reinberg D., Shi Y., Shiekhattar R., Shilatifard A., Workman J., and Zhang Y. (2007) New nomenclature for chromatin-modifying enzymes. Cell 131, 633–636 10.1016/j.cell.2007.10.039 [DOI] [PubMed] [Google Scholar]
- 11. Lalonde M. E., Avvakumov N., Glass K. C., Joncas F. H., Saksouk N., Holliday M., Paquet E., Yan K., Tong Q., Klein B. J., Tan S., Yang X. J., Kutateladze T. G., and Côté J. (2013) Exchange of associated factors directs a switch in HBO1 acetyltransferase histone tail specificity. Genes Dev. 27, 2009–2024 10.1101/gad.223396.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lalonde M. E., Cheng X., and Côté J. (2014) Histone target selection within chromatin: an exemplary case of teamwork. Genes Dev. 28, 1029–1041 10.1101/gad.236331.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Miotto B., and Struhl K. (2008) HBO1 histone acetylase is a coactivator of the replication licensing factor Cdt1. Genes Dev. 22, 2633–2638 10.1101/gad.1674108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Doyon Y., Cayrou C., Ullah M., Landry A. J., Côté V., Selleck W., Lane W. S., Tan S., Yang X. J., and Côté J. (2006) ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol. Cell 21, 51–64 10.1016/j.molcel.2005.12.007 [DOI] [PubMed] [Google Scholar]
- 15. Avvakumov N., Lalonde M. E., Saksouk N., Paquet E., Glass K. C., Landry A. J., Doyon Y., Cayrou C., Robitaille G. A., Richard D. E., Yang X. J., Kutateladze T. G., and Côté J. (2012) Conserved molecular interactions within the HBO1 acetyltransferase complexes regulate cell proliferation. Mol. Cell. Biol. 32, 689–703 10.1128/MCB.06455-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Saksouk N., Avvakumov N., Champagne K. S., Hung T., Doyon Y., Cayrou C., Paquet E., Ullah M., Landry A. J., Côté V., Yang X. J., Gozani O., Kutateladze T. G., and Côté J. (2009) HBO1 HAT complexes target chromatin throughout gene coding regions via multiple PHD finger interactions with histone H3 tail. Mol. Cell 33, 257–265 10.1016/j.molcel.2009.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Foy R. L., Song I. Y., Chitalia V. C., Cohen H. T., Saksouk N., Cayrou C., Vaziri C., Côté J., and Panchenko M. V. (2008) Role of Jade-1 in the histone acetyltransferase (HAT) HBO1 complex. J. Biol. Chem. 283, 28817–28826 10.1074/jbc.M801407200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Miotto B., and Struhl K. (2010) HBO1 histone acetylase activity is essential for DNA replication licensing and inhibited by Geminin. Mol. Cell 37, 57–66 10.1016/j.molcel.2009.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kueh A. J., Dixon M. P., Voss A. K., and Thomas T. (2011) HBO1 is required for H3K14 acetylation and normal transcriptional activity during embryonic development. Mol. Cell. Biol. 31, 845–860 10.1128/MCB.00159-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Feng Y., Vlassis A., Roques C., Lalonde M. E., González-Aguilera C., Lambert J. P., Lee S. B., Zhao X., Alabert C., Johansen J. V., Paquet E., Yang X. J., Gingras A. C., Côté J., and Groth A. (2016) BRPF3-HBO1 regulates replication origin activation and histone H3K14 acetylation. EMBO J. 35, 176–192 10.15252/embj.201591293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Duong M. T., Akli S., Macalou S., Biernacka A., Debeb B. G., Yi M., Hunt K. K., and Keyomarsi K. (2013) Hbo1 is a cyclin E/CDK2 substrate that enriches breast cancer stem-like cells. Cancer Res. 73, 5556–5568 10.1158/0008-5472.CAN-13-0013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Iizuka M., Takahashi Y., Mizzen C. A., Cook R. G., Fujita M., Allis C. D., Frierson H. F. Jr, Fukusato T., and Smith M. M. (2009) Histone acetyltransferase Hbo1: catalytic activity, cellular abundance, and links to primary cancers. Gene 436, 108–114 10.1016/j.gene.2009.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhou M. I., Wang H., Ross J. J., Kuzmin I., Xu C., and Cohen H. T. (2002) The von Hippel-Lindau tumor suppressor stabilizes novel plant homeodomain protein Jade-1. J. Biol. Chem. 277, 39887–39898 10.1074/jbc.M205040200 [DOI] [PubMed] [Google Scholar]
- 24. Piche B., and Li G. (2010) Inhibitor of growth tumor suppressors in cancer progression. Cell. Mol. Life Sci. 67, 1987–1999 10.1007/s00018-010-0312-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ythier D., Larrieu D., Brambilla C., Brambilla E., and Pedeux R. (2008) The new tumor suppressor genes ING: genomic structure and status in cancer. Int. J. Cancer 123, 1483–1490 10.1002/ijc.23790 [DOI] [PubMed] [Google Scholar]
- 26. Tao Y., Zhong C., Zhu J., Xu S., and Ding J. (2017) Structural and mechanistic insights into regulation of HBO1 histone acetyltransferase activity by BRPF2. Nucleic Acids Res. 45, 5707–5719 10.1093/nar/gkx142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rojas J. R., Trievel R. C., Zhou J., Mo Y., Li X., Berger S. L., Allis C. D., and Marmorstein R. (1999) Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide. Nature 401, 93–98 10.1038/43487 [DOI] [PubMed] [Google Scholar]
- 28. Xu P., Li C., Chen Z., Jiang S., Fan S., Wang J., Dai J., Zhu P., and Chen Z. (2016) The NuA4 core complex acetylates nucleosomal histone H4 through a double recognition mechanism. Mol. Cell 63, 965–975 10.1016/j.molcel.2016.07.024 [DOI] [PubMed] [Google Scholar]
- 29. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gouet P., Robert X., and Courcelle E. (2003) ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 31, 3320–3323 10.1093/nar/gkg556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ricketts M. D., Frederick B., Hoff H., Tang Y., Schultz D. C., Singh Rai T., Grazia Vizioli M., Adams P. D., and Marmorstein R. (2015) Ubinuclein-1 confers histone H3.3-specific-binding by the HIRA histone chaperone complex. Nat. Commun. 6, 7711 10.1038/ncomms8711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Luger K., Rechsteiner T. J., and Richmond T. J. (1999) Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 304, 3–19 10.1016/S0076-6879(99)04003-3 [DOI] [PubMed] [Google Scholar]
- 33. Yan Y., Harper S., Speicher D. W., and Marmorstein R. (2002) The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate. Nat. Struct. Biol. 9, 862–869 [DOI] [PubMed] [Google Scholar]
- 34. Berndsen C. E., and Denu J. M. (2005) Assays for mechanistic investigations of protein/histone acetyltransferases. Methods 36, 321–331 10.1016/j.ymeth.2005.03.002 [DOI] [PubMed] [Google Scholar]