Abstract
Recent advances in genetics present unique opportunities for enhancing knowledge about human physiology and disease susceptibility. Understanding this information at the individual gene level is challenging and requires extracting, collating, and interpreting data from a variety of public gene repositories. Here, I illustrate this challenge by analyzing the gene for human insulin-like growth factor 2 (IGF2) through the lens of several databases. IGF2, a 67-amino acid secreted peptide, is essential for normal prenatal growth and is involved in other physiological and pathophysiological processes in humans. Surprisingly, none of the genetic databases accurately described or completely delineated human IGF2 gene structure or transcript expression, even though all relevant information could be found in the published literature. Although IGF2 shares multiple features with the mouse Igf2 gene, it has several unique properties, including transcription from five promoters. Both genes undergo parental imprinting, with IGF2/Igf2 being expressed primarily from the paternal chromosome and the adjacent H19 gene from the maternal chromosome. Unlike mouse Igf2, whose expression declines after birth, human IGF2 remains active throughout life. This characteristic has been attributed to a unique human gene promoter that escapes imprinting, but as shown here, it involves several different promoters with distinct tissue-specific expression patterns. Because new testable hypotheses could lead to critical insights into IGF2 actions in human physiology and disease, it is incumbent that our fundamental understanding is accurate. Similar challenges affecting knowledge of other human genes should promote attempts to critically evaluate, interpret, and correct human genetic data in publicly available databases.
Keywords: insulin-like growth factor (IGF), genetics, genomics, gene mapping, gene expression, gene regulation, human genetics, gene annotation, gene variation, genetic database, insulin-like growth factor 2
Introduction
Insulin-like growth factor 2 (IGF2),2 a conserved 67-amino acid, single-chain secreted protein, is intimately involved in several critical physiological processes in humans and other mammals (1–6). IGF2, along with IGF1 and insulin, also comprises a gene family that is found in most mammalian species (7–9). In humans, IGF2 plays a central role in normal pre-natal growth, and changes in its expression have been implicated in a variety of developmental growth syndromes (10, 11). For example, individuals with Silver-Russell syndrome produce diminished amounts of IGF2, have reduced fetal and post-natal somatic growth, and exhibit a variety of dysmorphic features and bodily asymmetry (10, 11). In contrast, in Beckwith-Wiedemann syndrome, which is characterized by asymmetric overgrowth (10, 11), overexpression of IGF2 is seen.
IGF2 also plays an important role in growth biology in other mammals. Targeted Igf2 gene knockout in mice caused impaired fetal but normal post-natal growth (12). IGF2 also was identified as a key quantitative trait locus for muscle mass in pigs, with more heavily muscled animals bearing a single nucleotide polymorphism in an IGF2 gene promoter (13), that was responsible for enhancing gene activity by interfering with binding of a transcriptional repressor (14, 15).
Igf2/IGF2 gene regulation is complicated. In mice, humans, and presumably in other mammalian species, Igf2/IGF2 is part of a conserved autosomal linkage group that undergoes parental imprinting (16). Igf2/IGF2 is expressed from the paternally derived chromosome, with the adjacent H19 gene being activated on the maternal chromosome via the epigenetic actions of an imprinting control region (ICR) found in intergenic chromatin (Fig. 1A) (16). DNA within the ICR contains sites for the nuclear protein, CTCF (16–18). CTCF binds to the ICR in maternally-derived chromatin in mice, and it facilitates access of distal enhancers to the H19 promoter while preventing interactions with Igf2 promoters (17, 18). In paternal chromatin, DNA in the ICR is methylated, and CTCF cannot bind, thus allowing the same enhancers to associate with Igf2 (17, 18). It is likely that similar mechanisms control both IGF2 and H19 expressions in humans, although experimental evidence is more limited (19).
Figure 1.
Organization of the mouse Igf2–H19 and human IGF2–H19 loci. A, map of the mouse Igf2–H19 locus on chromosome 7 and the human IGF2–H19 locus on chromosome 11p15.5 with chromosomal coordinates. For Igf2 and IGF2, individual exons are depicted as boxes and introns as horizontal lines. A scale bar is shown. Other genes in each locus are shown as single boxes and include tyrosine hydroxylase (TH), insulin (Ins2, INS), H19, mitochondrial ribosomal protein L23 (Mrpl23, MRPL23), and troponin T3, fast skeletal type (Tnnt3, TNNT3). Horizontal arrows indicate the direction of transcription. The purple and green circles depict the location of the ICR, which is found just 5′ to H19 (40–42), and the orange circles indicate distal enhancers 1–10, and their human homologues (43). B, close-up view of the human TH, INS, and IGF2 genes. All exons are shown as boxes, and introns and flanking DNA as horizontal lines. Horizontal arrows indicate the direction of transcription; a scale bar is shown. All TH exons are gray, and INS exons are in blue. For IGF2, non-coding exons are black, and coding regions are red.
In mice, the Igf2 gene is composed of eight exons, and gene expression is regulated by three adjacent promoters, termed P1–P3, and an upstream promoter, P0, each with its own unique non-coding 5′ leader exon, whereas exons 6–8 encode the IGF2 precursor protein (Fig. 1A) (20–22). The human IGF2 gene is more complicated, and based on the published literature, it contains an additional upstream promoter with its associated non-coding exon and another coding exon (Fig. 1) (23–26). Because Igf2 expression declines precipitously during the early post-natal period in rodents (27–30), but is maintained through adult life in humans (24, 31), it has been assumed that the extra IGF2 gene promoter in humans is responsible for these species-specific differences in gene regulation (32). Moreover, as it has been shown that this distinctive human promoter is expressed in both alleles in some human tissues and thus in certain circumstances does not undergo parental imprinting (33), it has been hypothesized that lack of imprinting control also may be a key feature of maintenance of IGF2 production in adult humans (33). In both cases, the experimental evidence supporting these contentions is limited.
Major advances in genetics and genomics in recent years now present unique opportunities for enhancing our understanding of human physiology, and a wealth of information is now available in publicly accessible databases (34) with the potential for contributing new insights into human variation, disease pre-disposition, and human origins and evolution (35–37). To date, understanding these data at the individual gene level has been challenging, in part because both the extraction of the information and the evaluation of its quality can be difficult. Here, I have used publicly available genomic and gene expression databases to examine the human IGF2 gene. This study was prompted in part by recent publications potentially linking IGF2 DNA polymorphisms with a variety of traits, including differential susceptibility to type 2 diabetes (38) and association with athletic performance (39).
Results and discussion
Topography of the human IGF2-H19 locus
The human IGF2-H19 locus on chromosome 11p15.5 shares many features with the corresponding mouse locus on chromosome 7 (26), including gene order, and the presence of an imprinting control region (ICR), located just 5′ to H19 and containing binding sites for the boundary transcription factor, CTCF (40, 41), although in the human genome the segment differs from the mouse by consisting of two repeat units rather than a single element (Fig. 1A) (42). Also, as revealed here by homology searches, in the human IGF2-H19 locus there are DNA sequences similar to distal enhancers that are responsible in the mouse for both developmental and cell-lineage–specific regulation of Igf2 and H19 (Fig. 1A and Table 1) (43). However, in the human genome only 9 of the 10 elements defined in the mouse could be detected, with conserved segment 3 being absent (Table 1). In fact, with the exception of human conserved segment 8, in which DNA sequence similarity extended throughout the 277-bp mouse segment, DNA homology was limited to 50% or less of the corresponding mouse element (Table 1). In addition, although in the mouse genome these enhancers are located in intergenic DNA, on the corresponding human chromosomal region, 7/9 elements are found within the MRPL23 gene (Fig. 1A). Moreover, no experiments have yet shown that any of these putative enhancers are functional in human tissues, in contrast to studies using transgenic mice and other experimental models that have defined several chromosomal segments that could direct gene activity to endodermal- or mesenchymal-derived tissues (43–49). Other differences between the mouse Igf2-H19 and human IGF2-H19 loci include the separation of Th in the former species to 226 kb from Ins2 versus 2.6 kb from INS in humans (Ins2 in mice is the homologue of human INS), a shorter intergenic distance between Igf2 and H19 (72 kb in mice versus 128 kb in humans) and a longer distance between Ins2 and Igf2 (12 kb in mice versus 1.4 kb in humans) (Fig. 1A). The anatomical relationship in the human genome among TH, INS, and IGF2 is depicted in Fig. 1B.
Table 1.
Nucleotide identity of human DNA with mouse Igf2-H19 locus enhancers
See Fig. 1 for locations. Nomenclature is as defined by Ishihara et al. (43); CS indicates conserved segment; ND means no DNA sequence identity detected.
| Enhancer | % identity (no. of base pairs aligned) |
|---|---|
| CS1 (218 bp) | 95 (76) |
| CS2 (472 bp) | 89 (251) |
| CS3 (214 bp)a | ND |
| CS4 (385 bp)a | 84 (81) |
| CS5 (385 bp) | 86 (92) |
| CS6 (360 bp) | 87 (95) |
| CS7 (231 bp) | 95 (74) |
| CS8 (277 bp) | 85 (277) |
| CS9 (486 bp) | 84 (112) |
| CS10 (286 bp) | 93 (106) |
a Overlap with endodermal enhancers defined by Yoo-Warren et al. (77).
Human IGF2 gene
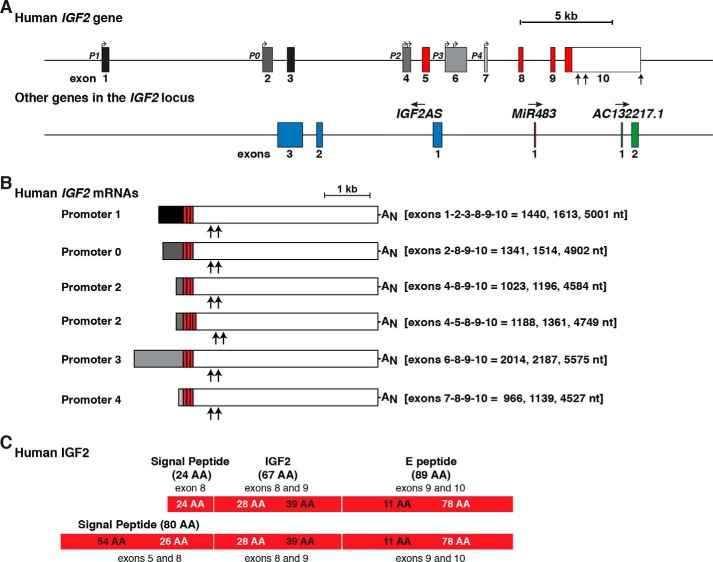
Based on compilation of primary data (23–26, 50–53), the human IGF2 gene spans ∼29.3 kb of chromosomal DNA and contains five promoters and 10 exons (Fig. 2A). This information is not fully delineated in Ensembl, UCSC, or Gencode Genome Browsers or in other databases. In fact, the annotation of human genome assembly GRCh38/hg38 in each browser only lists three of five human IGF2 promoters (P2–4) and seven of 10 exons (exons 4–10). Accurate information is included in these databases about variable transcription start sites in individual promoters and alternative polyadenylation in exon 10. Also, all three genome browsers do correctly identify read-through transcripts composed of the INS promoter, some INS exons, and some IGF2 exons (INS-IGF2 mRNAs). By contrast, the Genotype-Tissue Expression project (GTEx) denotes four human IGF2 promoters (P1–4) and eight exons (exons 1 and 4–10).
Figure 2.
Human IGF2 gene structure and expression. A, close-up view of the human IGF2 gene, illustrating promoters P0 and P1–P4, and exons 1–10. All 5′-noncoding exons are in black or shades of gray; coding regions are in red, and the 3′ UTR of exon 10 is in white. Bent arrows signify transcription start sites, and vertical arrows delineate different polyadenylation sites in exon 10. Below IGF2 are three other non-coding genes that are embedded within the boundaries of human IGF2. Their direction of transcription is indicated. For all genes, exons are represented as boxes, and introns as horizontal lines. A scale bar is shown. All IGF2AS exons are blue; the MiR483 exon is dark red, and AC132217.1 exons are green. B, diagram of the six major classes of human IGF2 mRNAs. The responsible promoters are listed, with their 5′ non-coding exons in black or gray; coding exons are in red, and the non-coding part of exon 10 is in white to match A. The lengths of each class of mRNAs are indicated in nucleotides (nt), and exons found in each transcript are listed. The locations of polyadenylation sites of shorter transcripts are depicted by vertical arrows (23, 24), with AN representing the polyadenylic acid tail at the 3′ end of the longest mRNAs. C, diagram of the two human IGF2 protein precursors, showing the derivation of each segment from different IGF2 exons. The contributions of different exons to amino acids in different parts of each protein are shown in alternating black and white text. Mature, 67-amino acid IGF2 (center), presumptive signal peptides (left), and the 89-residue COOH-terminal E-peptide (right) are each labeled.
The five human IGF2 gene promoters described in the published literature and partially identified in genome browsers govern the production of six different major classes of IGF2 transcripts with five distinct 5′ UTRs and two types of coding segments (Fig. 2, A and B) (25, 26, 50–53). P2 is responsible for a pair of IGF2 mRNAs that differ by the presence or absence of alternatively spliced exon 5. Transcripts directed by P1 include three untranslated exons, exons 1–3 (Fig. 2B). Exon 2 in this context is truncated at its 5′ end compared with when it is the sole 5′-untranslated exon in mRNAs regulated by P0 (Fig. 2B). P3 and P4 each direct expression of IGF2 mRNAs with single 5′-untranslated exons (Fig. 2B). There also is variation in the lengths of 3′ UTRs affecting most IGF2 mRNAs, secondary to alternative polyadenylation, as noted above (see Fig. 2B). The four transcripts controlled by P1, P0, P3, and P4 encode the same 180-amino acid IGF2 precursor proteins (Fig. 2, B and C). This precursor is composed of a 24-amino acid signal peptide, a 67-residue mature IGF2 molecule, and an 89-amino acid COOH-terminal extension peptide, or E-domain (Fig. 2C). The signal peptide is encoded by exon 8, mature IGF2 by exons 8 and 9, and the E-region by exons 9 and 10 (Fig. 2C). The presence of exon 5 in some mRNAs directed by P2 leads to a second predicted IGF2 protein precursor of 236 residues. This molecule is composed of the same 67-residue IGF2 and 89-amino acid E-peptide, but it contains an 80-amino acid presumptive signal peptide (54 codons from exon 5 and 26 from exon 8, with the additional two 5′ codons in exon 8 resulting from maintenance of the open reading frame after splicing of exons 5 and 8) (Fig. 2C).
Three non-coding genes are located within the boundaries of the human IGF2 gene. All are annotated in Ensembl, UCSC, and Gencode Genome Browsers. Human IGF2 antisense (IGF2AS) is a 3-exon gene that is transcribed in the opposite direction from IGF2. One of its exons overlaps IGF2 exon 3 (Fig. 2A). MiR483 is composed of a single exon and is found between IGF2 exons 8 and 9 (Fig. 2A). It has been shown to be up-regulated in human Wilms tumors (54) and has been found in experimental studies to increase transcription from IGF2 P2, P3, and P4, and IGF2AS, although by unknown mechanisms (54). Both of these genes are located in corresponding positions within the mouse Igf2 locus. AC132217.1 is a predicted non-coding gene, with limited functional annotation, and does not appear to be present in the mouse genome.
Few studies have been performed examining the molecular mechanisms controlling the activity of each of the five human IGF2 promoters. Limited cell-based experiments have shown that human promoters P1, P3, P4, and P0 can function in transfected cells, whereas P2 cannot (25, 53, 58, 59). Transcription factors C/EBPα, C/EBPβ, and SP1 have been found to activate P1 through potential proximal promoter control elements (32, 60), whereas AP2 was shown to stimulate P3 (61), and the Wilms tumor repressor, WT1, was found to inhibit P3, potentially through many binding sites in proximal promoter DNA (62). In vivo, P0 has been shown to be active in several human fetal tissues, but only in adult skeletal muscle (25), whereas other promoters are expressed in a variety of different organs and tissues (23–26, 50–53).
There have been several analyses of parental imprinting of IGF2 in humans, which potentially controls access of distal transcriptional regulatory elements and their associated transcription factors to all IGF2 gene promoters (19, 33). Investigations have identified individuals with deletion mutations in the human ICR, which leads to bi-allelic expression of IGF2 and silencing of H19 (42), and also have found bi-allelic expression of IGF2 in several human cancers in which excess production of IGF2 may play a pathogenic role (55–57).
New insights regarding human IGF2 gene expression
GTEx has collected gene expression data by RNA-sequencing from many non-diseased human tissues (63–65), with 80–388 donors per tissue (GTEX release 7). Presented in Fig. 3 are the compiled data for IGF2. These results are based on information about isoform-specific transcripts in GTEx and have been quantified here according to the IGF2 promoter being used, which was determined by the presence of a distinct promoter-specific 5′-untranslated exon or exons. In these different organs and tissues, the overall abundance of IGF2 transcripts varied over a 34-fold range, with the highest levels of mRNAs seen in visceral adipose tissue, endo-cervix, and fallopian tube (103, 93, and 92 transcripts for kilobase million reads (TPM), respectively, Fig. 3A), and the lowest values in brain regions, pancreas, and stomach (2.9, 3.2, and 3.8 TPM, respectively, Fig. 3B). The vast majority of IGF2 mRNAs in this collection of human tissues and organs were regulated by P3 and P4, which together comprised 76 to >99% of total transcripts (Fig. 3). IGF2 mRNAs controlled by P1 were measurable only in liver (∼15% of total), and the contributions of P0 and P2 were negligible (1% or less; some of the IGF2 mRNAs in the GTEx dataset did not contain a promoter-specific exon and may have arisen from endonucleolytic processing of larger IGF2 transcripts (66, 67)). Taken together, the GTEx results on IGF2 expression in human liver as presented here clearly undercut prior hypotheses about P1 being the major driver of IGF2 production in that organ (32, 33), because it appears to be a minor species in this dataset. In fact, because mRNAs governed by P3 and P4 are the predominant active human promoters in adult liver and in other organs and tissues (Fig. 3), the results also argue against the idea that P1, which is not found in the mouse genome, is responsible for maintaining IGF2 gene transcription in human adults (33). Thus, the GTEx data collectively indicate that the cause of pervasive IGF2 mRNA biosynthesis and protein production throughout life in humans remains unknown and that the reasons for differences with mice, in which Igf2 expression is attenuated during the early post-natal period, also are undefined. These results therefore present a new opportunity to elucidate the molecular and biochemical mechanisms responsible for temporal control of IGF2/Igf2 gene activity in different mammalian species. Of note, analysis of mouse Igf2 promoter usage via the FANTOM5 promoter expression atlas (68, 69) demonstrates that P2 (homologue of human P3) and P3 (homologue of human P4) predominate in fetal and neonatal organs and tissues, although mouse P1 (homologue of human P2) is more active than P2 in neonatal murine liver (68, 69). Thus, some patterns of tissue- and organ-specific Igf2/IGF2 promoter activity appear to be similar between mice and humans.
Figure 3.
Expression of IGF2 mRNAs in different human tissues. Data on organ- and tissue-specific human IGF2 gene expression was obtained from the GTEx portal and graphed as TPM. The information shown has been derived from the exon expression module of GTEx, and the displayed results have been quantified by the abundance of promoter-specific 5′-untranslated exons. A, 18 organs and tissues with the highest levels of IGF2 mRNAs are depicted. B, 20 organs and tissues with the lowest IGF2 mRNAs are shown. The data from human brain consist of the average levels of IGF2 mRNAs found in 12 different sub-regions (cortex, frontal cortex, anterior cingulate cortex, amygdala, hypothalamus, hippocampus, substantia nigra, putamen, caudate, nucleus accumbens, cerebellum, and cerebellar hemisphere).
Predicted variation in IGF2 in human populations
The Exome Aggregation Consortium (ExAC) contains DNA sequence information from the exons of 60,706 people representing different population groups from around the world (70). The data have revealed substantial variation within the coding regions of genes in this large cohort, but the data have also shown that most alterations were uncommon, as more than half were detected in a single allele and over 99% were seen in less than 1% of the study group (70). In addition, the vast majority of this variation has consisted of synonymous nucleotide changes and amino acid substitutions (70). Alterations in the IGF2 gene were seen in 2.5% of the 121,412 alleles, with 80% of this being reflected in a single nucleotide change mapping to the splice acceptor site at the intron 4 to exon 5 junction (71) (found in ∼2% of the total population; also see single nucleotide polymorphism rs149483638). Nearly all of the remaining ∼0.5% consisted of 78 different predicted amino acid substitutions (71). However, the single change of arginine 157 to histidine in the E-peptide region predominated and accounted for ∼75% of all such substitutions (71). The impact of these variants on IGF2 biology is unknown, and they thus collectively present new research opportunities for obtaining insights into aspects of human physiology (see below).
Potential alterations in IGF2 in cancer
The Cancer Genome Atlas represents a compendium of DNA sequencing results from genomic DNA derived from a variety of human neoplasms (see https://portal.gdc.cancer.gov/). Potential mutations at 149 different locations in the IGF2 gene have been found in 24 different cancers. The prevalence of these changes has ranged from over 7% of uterine endometrial carcinomas to ≤0.5% of invasive breast cancers, adenocarcinomas of the prostate, renal cancers, gliomas, or glioblastomas, among others (see https://portal.gdc.cancer.gov/genes/ENSG00000167244). Of the 149 different alterations identified in the IGF2 gene in these human cancers, 87 were found in IGF2 exons and 62 in introns (Tables 2 and 3). Considering just changes within exons that mapped to mature IGF2 transcripts, 36 predicted missense mutations, frameshifts, or the addition of a stop codon to IGF2 (Table 3). Twelve of these locations were identified as human population variants in ExAC, although their prevalence was extremely low and ranged from 1 to 11 alleles (<0.01%, Table 3). At present, the roles of these modified transcripts and IGF2 proteins in cancer biology are undefined, and the potential is thus high for gaining insights into aspects of carcinogenesis or metastasis by studying these mutations.
Table 2.
Human IGF2 gene mutations in cancer
| Nucleotide modification | No. of examples |
|---|---|
| Missense | 31 |
| Frame-shift; stop codon | 5 |
| Synonymous | 11 |
| 5′ non-translated region | 8 |
| 3′ non-translated region | 32 |
| Intron | 62 |
Table 3.
Predicted cancer-associated amino acid mutations in IGF2
#fs indicates frameshift.
| Mutation | Population variant | ExAC prevalence |
|---|---|---|
| M5I | M5R | 1 allele |
| S20I | S20L, S20W | 11 alleles, 1 allele |
| A24T | None | |
| A25V | None | |
| R27H | None | |
| G35R | None | |
| F43L | None | |
| R48H | R48L | 1 allele |
| Y52stop | None | |
| S53R | None | |
| R61H | R61H | 1 allele |
| R64C, R64H | R64C | 1 allele |
| V67I | None | |
| R73C, R73H | None | |
| E91K | None | |
| S95L | None | |
| P98L | P98L, P98Q | 1 allele, 1 allele |
| P102L | P102L | 6 alleles |
| F114L | None | |
| R127C, R127H, R127S | None | |
| G129S | None | |
| L133V | None | |
| #R137Afs*22 | None | |
| K153I | None | |
| H155D | None | |
| L161V | None | |
| T163L | None | |
| D165Y | D165E | 1 allele |
| #G170Afs*30 | G170D, G170S | 2 alleles, 1 allele |
| A171T | A171V | 1 allele |
| #P173Qfs*27 | P173A, P173Q, P173R, P173S | 4 alleles, 1 allele 4 alleles, 3 alleles |
| #E174Rfs*50 | E174D | 1 allele |
Challenges inherent in genome-based predictions about putative human IGF2 proteins
IGF2 mRNAs derived from P1, P0, P3, and P4 and the smaller transcript originating from P2 all encode a predicted secreted IGF2 protein precursor of 180 residues (Fig. 2C), which closely resembles IGF2 proteins found in other mammalian species (7, 8). Once these mRNAs are translated, the 24-amino acid NH2-terminal signal peptide is co-translationally cleaved from the rest of the protein progenitor, and the 89-residue COOH-terminal E-peptide is post-translationally processed to release the fully bioactive 67-residue IGF2 (6, 31). The fate or function of the putative 236-amino acid IGF2 precursor molecule originating from the larger transcript governed by P2 is unknown (Fig. 2C). Moreover, the potential 80-amino acid signal peptide in this predicted protein is far longer than other described mammalian signal sequences (72, 73), and in the absence of any experimental evidence, it is not known whether an authentic IGF2 pro-protein or 67-amino acid mature IGF2 can be generated from this molecule or even whether the mRNA is stable in human cells.
The listing of the larger transcript regulated by IGF2 P2, and the encoded putative 236-amino acid IGF2 precursor protein as a major product of the human IGF2 gene in genome databases such as ExAC and others, has the potential to cause confusion for users searching for actionable information about IGF2. Moreover, this inference is wrong, based on GTEx gene expression data, which, as noted above, indicates that this is at best a very minor IGF2 mRNA species in multiple human tissues. One example of the problems created by this type of incorrect information may be found in the recently published analysis of a family with apparent Silver-Russell syndrome, including both pre- and post-natal growth deficiencies (74). DNA sequencing revealed that the genomes of all affected members harbored a heterozygous nonsense mutation in IGF2 that prevented production of the IGF2 protein, and indeed, accumulation of IGF2 in the blood of the subjects was substantially reduced from normal concentrations (74). The mutation, which consisted of a single Cys → Ala change within IGF2 exon 8 in all subjects, was described as altering amino acid serine 64 to a termination codon (TCG>TAG) in the 80-amino acid presumptive signal peptide (thus named p.Ser64Ter). More accurately, this modification should be represented as p.Ser8Ter, reflecting its location in the signal sequence encoded by major IGF2 transcripts.
How can we use genomic databases to accurately understand IGF2 actions in humans?
A recent publication identified the single nucleotide change mapping to and disrupting the IGF2 intron 4 to exon 5 splice acceptor site as being prevalent in a cohort of 8668 individuals of Mexican origin (average minor allele frequency (MAF) of 0.17 (38)). As noted above, this identical alteration was found with a MAF of 0.02 in the ExAC study group, which consisted of different populations, with 60% of individuals with European origins, ∼8% with Hispanic origins, and ∼8% from Africa (70). This new analysis also showed that the allele that perturbs the splice acceptor site was rare in other groups (MAF of 0.001 in Africans and 0.0002 in Europeans), and thus that the contribution of the Hispanic cohorts was likely primarily responsible for the overall observed ExAC population prevalence of ∼2% (71). By studying individuals with and without type 2 diabetes, the authors additionally concluded that the minor allele was protective against this disease, with heterozygous individuals having a 22% and homozygotes a 40% diminished risk (38). Although intriguing, it remains unclear how modifications in this genomic location could alter disease risk, because transcripts directed by IGF2 P2 are normally minimally expressed in human tissues (Fig. 3). One possibility is that the minor allele can alter levels of INS-IGF2 read-through transcripts (25), although the abundance of these latter mRNAs and their functions in different human tissues has not been determined. Moreover, they are not found in GTEx except in liver, where expression levels are very low (≤1 TPM for each of 4 mRNA species). In addition, because the genetics of Mexico are very complicated, with region-specific sub-groups being identified (75), it is likely that the MAF of 0.17 identified in the study cohort (38) will vary significantly among different populations of Mexican origin once a larger and more thorough genetic analysis is completed.
The statistical interpretation of the population genetics data and the associated claim of a new disease risk allele (38) noted above illustrate one of the major quandaries and challenges of modern genetics: how to connect clinical and basic research to accurately and mechanistically understand disease causation (34). As analyzed here for the human IGF2 gene, the difficulties in attaining this goal are possibly even more complicated. The complexity of the gene, with its multiple promoters, alternative splicing, overarching regulation by parental imprinting, and the possibility of two distinct classes of protein precursors, yet a single 67-amino acid biologically active IGF2 peptide, presents its own challenges, and these are amplified by the paucity of complete or correct information about IGF2 gene structure and expression currently available in gene repositories. Because new testable hypotheses derived from analysis of human IGF2 in data from genomic and gene expression sequencing projects could lead to novel insights about the biology of IGF2 and its actions in both human physiology and disease, it is incumbent that these genetic databases thoroughly describe the fundamentals of IGF2 gene structure and gene activity. As similar opportunities for translation of genetic information to medical practice will exist for other human genes and gene families, there is a need for ongoing critical review and correction of data in public gene repositories to ensure future users that the primary information is both accurate and complete.
Experimental procedures
Database searches and analyses
Mouse and human genomic information was obtained from Ensembl Genome Browser (https://www.ensembl.org/index.html),3 the UCSC Genome Browser (https://genome.ucsc.edu),3 (78) and the Gencode Browser (http://www.gencodegenes.org/).3 Searches were performed using BlastN under normal sensitivity, with mouse Igf2 or H19 gene DNA segments, or regions near the two loci (Mus musculus, genome assembly GRCm38) or human IGF2 DNA segments (Homo sapiens, genome assembly GRCh38) as queries. Information on human IGF2 also was obtained from several primary publications (23–26, 50–53). Data on human IGF2 gene expression was derived from the Portal for the Genotype-Expression Project (GTEx version 7, https://www.gtexportal.org/home/),3 using the exon expression module and analyzing isoform-specific transcripts, which were categorized by the presence of a promoter-specific 5′-untranslated exon or exons. Protein sequences were from the Uniprot browser (http://www.uniprot.org/)3 and GENCODE/Ensembl databases and the National Center for Biotechnology Information Consensus CDS Protein Set (https://www.ncbi.nlm.nih.gov/CCDS/). Information on variation in human IGF2 was from the ExAc genome browser (http://exac.broadinstitute.org/)3 (79), which contains the results of sequencing of the exons from 60,706 individuals (76). Other databases examined included Online Mendelian Inheritance in Man (OMIM, https://www.omim.org/),3 the Type 2 Diabetes Knowledge Portal (http://www.type2diabetesgenetics.org/),3 and the Cancer Genome Atlas (https://cancergenome.nih.gov/) and its data portal (https://portal.gdc.cancer.gov/).
Author contributions
P. R. conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; writing-original draft; project administration; writing-review and editing.
This work was supported by National Institutes of Health Research Grant R01 DK042748-28 (to P. R.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- IGF2
- insulin-like growth factor 2
- CTCF
- CCTC-binding factor
- IGF2AS
- IGF2 anti-sense
- C/EBP
- CCAT enhancer-binding protein
- GTEx
- Genotype-Tissue Expression project
- ExAC
- Exome Aggregation Consortium
- MAF
- minor allele frequency
- ICR
- imprinting control region
- TPM
- transcripts for kilobase million reads.
References
- 1. Le Roith D., Bondy C., Yakar S., Liu J. L., and Butler A. (2001) The somatomedin hypothesis: 2001. Endocr. Rev. 22, 53–74 10.1210/edrv.22.1.0419 [DOI] [PubMed] [Google Scholar]
- 2. LeRoith D. (2008) Clinical relevance of systemic and local IGF-I: lessons from animal models. Pediatr. Endocrinol. Rev. 5, Suppl. 2, 739–743 [PubMed] [Google Scholar]
- 3. Gems D., and Partridge L. (2013) Genetics of longevity in model organisms: debates and paradigm shifts. Annu. Rev. Physiol 75, 621–644 10.1146/annurev-physiol-030212-183712 [DOI] [PubMed] [Google Scholar]
- 4. Pollak M. (2012) The insulin and insulin-like growth factor receptor family in neoplasia: an update. Nat. Rev. Cancer 12, 159–169 10.1038/nrc3215 [DOI] [PubMed] [Google Scholar]
- 5. Livingstone C. (2013) IGF2 and cancer. Endocr. Relat. Cancer 20, R321–R339 10.1530/ERC-13-0231 [DOI] [PubMed] [Google Scholar]
- 6. Livingstone C., and Borai A. (2014) Insulin-like growth factor-II: its role in metabolic and endocrine disease. Clin. Endocrinol. 80, 773–781 10.1111/cen.12446 [DOI] [PubMed] [Google Scholar]
- 7. Wallis M. (2009) New insulin-like growth factor (IGF)-precursor sequences from mammalian genomes: the molecular evolution of IGFs and associated peptides in primates. Growth Horm. IGF Res. 19, 12–23 10.1016/j.ghir.2008.05.001 [DOI] [PubMed] [Google Scholar]
- 8. Rotwein P. (1999) in The IGF System (Rosenfeld R. G., and Roberts C. T. Jr., eds) pp. 19–35, Humana Press, Totowa, NJ [Google Scholar]
- 9. Schwartz T. S., and Bronikowski A. M. (2016) Evolution and function of the insulin and insulin-like signaling network in ectothermic reptiles: some answers and more questions. Integr. Comp. Biol. 56, 171–184 10.1093/icb/icw046 [DOI] [PubMed] [Google Scholar]
- 10. Eggermann T., Begemann M., Spengler S., Schröder C., Kordass U., and Binder G. (2010) Genetic and epigenetic findings in Silver-Russell syndrome. Pediatr. Endocrinol. Rev. 8, 86–93 [PubMed] [Google Scholar]
- 11. Azzi S., Abi Habib W., and Netchine I. (2014) Beckwith-Wiedemann and Russell-Silver Syndromes: from new molecular insights to the comprehension of imprinting regulation. Curr. Opin. Endocrinol. Diabetes Obes. 21, 30–38 10.1097/MED.0000000000000037 [DOI] [PubMed] [Google Scholar]
- 12. DeChiara T. M., Robertson E. J., and Efstratiadis A. (1991) Parental imprinting of the mouse insulin-like growth factor II gene. Cell 64, 849–859 10.1016/0092-8674(91)90513-X [DOI] [PubMed] [Google Scholar]
- 13. Van Laere A. S., Nguyen M., Braunschweig M., Nezer C., Collette C., Moreau L., Archibald A. L., Haley C. S., Buys N., Tally M., Andersson G., Georges M., and Andersson L. (2003) A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig. Nature 425, 832–836 10.1038/nature02064 [DOI] [PubMed] [Google Scholar]
- 14. Markljung E., Jiang L., Jaffe J. D., Mikkelsen T. S., Wallerman O., Larhammar M., Zhang X., Wang L., Saenz-Vash V., Gnirke A., Lindroth A. M., Barrés R., Yan J., Strömberg S., De S., et al. (2009) ZBED6, a novel transcription factor derived from a domesticated DNA transposon regulates IGF2 expression and muscle growth. PLoS Biol. 7, e1000256 10.1371/journal.pbio.1000256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Butter F., Kappei D., Buchholz F., Vermeulen M., and Mann M. (2010) A domesticated transposon mediates the effects of a single-nucleotide polymorphism responsible for enhanced muscle growth. EMBO Rep. 11, 305–311 10.1038/embor.2010.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Edwards C. A., and Ferguson-Smith A. C. (2007) Mechanisms regulating imprinted genes in clusters. Curr. Opin. Cell Biol. 19, 281–289 10.1016/j.ceb.2007.04.013 [DOI] [PubMed] [Google Scholar]
- 17. Wallace J. A., and Felsenfeld G. (2007) We gather together: insulators and genome organization. Curr. Opin. Genet. Dev. 17, 400–407 10.1016/j.gde.2007.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Phillips J. E., and Corces V. G. (2009) CTCF: master weaver of the genome. Cell 137, 1194–1211 10.1016/j.cell.2009.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Giannoukakis N., Deal C., Paquette J., Goodyer C. G., and Polychronakos C. (1993) Parental genomic imprinting of the human IGF2 gene. Nat. Genet. 4, 98–101 10.1038/ng0593-98 [DOI] [PubMed] [Google Scholar]
- 20. Rotwein P., and Hall L. J. (1990) Evolution of insulin-like growth factor II: characterization of the mouse IGF-II gene and identification of two pseudo-exons. DNA Cell Biol. 9, 725–735 10.1089/dna.1990.9.725 [DOI] [PubMed] [Google Scholar]
- 21. Moore T., Constancia M., Zubair M., Bailleul B., Feil R., Sasaki H., and Reik W. (1997) Multiple imprinted sense and antisense transcripts, differential methylation and tandem repeats in a putative imprinting control region upstream of mouse Igf2. Proc. Natl. Acad. Sci. U.S.A. 94, 12509–12514 10.1073/pnas.94.23.12509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Constância M., Hemberger M., Hughes J., Dean W., Ferguson-Smith A., Fundele R., Stewart F., Kelsey G., Fowden A., Sibley C., and Reik W. (2002) Placental-specific IGF-II is a major modulator of placental and fetal growth. Nature 417, 945–948 10.1038/nature00819 [DOI] [PubMed] [Google Scholar]
- 23. Sussenbach J. S., Steenbergh P. H., and Holthuizen P. (1992) Structure and expression of the human insulin-like growth factor genes. Growth Regul. 2, 1–9 [PubMed] [Google Scholar]
- 24. Sussenbach J. S., Rodenburg R. J., Scheper W., and Holthuizen P. (1993) Transcriptional and post-transcriptional regulation of the human IGF-II gene expression. Adv. Exp. Med. Biol. 343, 63–71 [DOI] [PubMed] [Google Scholar]
- 25. Monk D., Sanches R., Arnaud P., Apostolidou S., Hills F. A., Abu-Amero S., Murrell A., Friess H., Reik W., Stanier P., Constância M., and Moore G. E. (2006) Imprinting of IGF2 P0 transcript and novel alternatively spliced INS-IGF2 isoforms show differences between mouse and human. Hum. Mol. Genet. 15, 1259–1269 10.1093/hmg/ddl041 [DOI] [PubMed] [Google Scholar]
- 26. Nordin M., Bergman D., Halje M., Engström W., and Ward A. (2014) Epigenetic regulation of the Igf2/H19 gene cluster. Cell Prolif. 47, 189–199 10.1111/cpr.12106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee J. E., Pintar J., and Efstratiadis A. (1990) Pattern of the insulin-like growth factor II gene expression during early mouse embryogenesis. Development 110, 151–159 [DOI] [PubMed] [Google Scholar]
- 28. Soares M. B., Ishii D. N., and Efstratiadis A. (1985) Developmental and tissue-specific expression of a family of transcripts related to rat insulin-like growth factor II mRNA. Nucleic Acids Res. 13, 1119–1134 10.1093/nar/13.4.1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Brown A. L., Graham D. E., Nissley S. P., Hill D. J., Strain A. J., and Rechler M. M. (1986) Developmental regulation of insulin-like growth factor II mRNA in different rat tissues. J. Biol. Chem. 261, 13144–13150 [PubMed] [Google Scholar]
- 30. Graham D. E., Rechler M. M., Brown A. L., Frunzio R., Romanus J. A., Bruni C. B., Whitfield H. J., Nissley S. P., Seelig S., and Berry S. (1986) Coordinate developmental regulation of high and low molecular weight mRNAs for rat insulin-like growth factor II. Proc. Natl. Acad. Sci. U.S.A. 83, 4519–4523 10.1073/pnas.83.12.4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Daughaday W. H., and Rotwein P. (1989) Insulin-like growth factors I and II. Peptide, messenger ribonucleic acid and gene structures, serum, and tissue concentrations. Endocr. Rev. 10, 68–91 10.1210/edrv-10-1-68 [DOI] [PubMed] [Google Scholar]
- 32. Rodenburg R. J., Holthuizen P. E., and Sussenbach J. S. (1997) A functional Sp1 binding site is essential for the activity of the adult liver-specific human insulin-like growth factor II promoter. Mol. Endocrinol. 11, 237–250 10.1210/mend.11.2.9888 [DOI] [PubMed] [Google Scholar]
- 33. Vu T. H., and Hoffman A. R. (1994) Promoter-specific imprinting of the human insulin-like growth factor-II gene. Nature 371, 714–717 10.1038/371714a0 [DOI] [PubMed] [Google Scholar]
- 34. Manolio T. A., Fowler D. M., Starita L. M., Haendel M. A., MacArthur D. G., Biesecker L. G., Worthey E., Chisholm R. L., Green E. D., Jacob H. J., McLeod H. L., Roden D., Rodriguez L. L., Williams M. S., Cooper G. M., et al. (2017) Bedside back to bench: building bridges between basic and clinical genomic research. Cell 169, 6–12 10.1016/j.cell.2017.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Acuna-Hidalgo R., Veltman J. A., and Hoischen A. (2016) New insights into the generation and role of de novo mutations in health and disease. Genome Biol. 17, 241 10.1186/s13059-016-1110-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Katsanis N. (2016) The continuum of causality in human genetic disorders. Genome Biol. 17, 233 10.1186/s13059-016-1107-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Quintana-Murci L. (2016) Understanding rare and common diseases in the context of human evolution. Genome Biol. 17, 225 10.1186/s13059-016-1093-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mercader J. M., Liao R. G., Bell A. D., Dymek Z., Estrada K., Tukiainen T., Huerta-Chagoya A., Moreno-Macías H., Jablonski K. A., Hanson R. L., Walford G. A., Moran I., Chen L., Agarwala V., Ordoñez-Sánchez M. L., et al. (2017) A loss-of-function splice acceptor variant in IGF2 is protective for type 2 diabetes. Diabetes 66, 2903–2914 10.2337/db17-0187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ben-Zaken S., Meckel Y., Nemet D., and Eliakim A. (2017) High prevalence of the IGF2 rs680 GG polymorphism among top-level sprinters and jumpers. Growth Horm. IGF Res. 37, 26–30 10.1016/j.ghir.2017.10.001 [DOI] [PubMed] [Google Scholar]
- 40. Bell A. C., and Felsenfeld G. (2000) Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405, 482–485 10.1038/35013100 [DOI] [PubMed] [Google Scholar]
- 41. Hark A. T., Schoenherr C. J., Katz D. J., Ingram R. S., Levorse J. M., and Tilghman S. M. (2000) CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405, 486–489 10.1038/35013106 [DOI] [PubMed] [Google Scholar]
- 42. Sparago A., Cerrato F., Vernucci M., Ferrero G. B., Silengo M. C., and Riccio A. (2004) Microdeletions in the human H19 DMR result in loss of IGF2 imprinting and Beckwith-Wiedemann syndrome. Nat. Genet. 36, 958–960 10.1038/ng1410 [DOI] [PubMed] [Google Scholar]
- 43. Ishihara K., Hatano N., Furuumi H., Kato R., Iwaki T., Miura K., Jinno Y., and Sasaki H. (2000) Comparative genomic sequencing identifies novel tissue-specific enhancers and sequence elements for methylation-sensitive factors implicated in Igf2/H19 imprinting. Genome Res. 10, 664–671 10.1101/gr.10.5.664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Leighton P. A., Ingram R. S., Eggenschwiler J., Efstratiadis A., and Tilghman S. M. (1995) Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature 375, 34–39 10.1038/375034a0 [DOI] [PubMed] [Google Scholar]
- 45. Ainscough J. F., John R. M., Barton S. C., and Surani M. A. (2000) A skeletal muscle-specific mouse Igf2 repressor lies 40 kb downstream of the gene. Development 127, 3923–3930 [DOI] [PubMed] [Google Scholar]
- 46. Kaffer C. R., Srivastava M., Park K. Y., Ives E., Hsieh S., Batlle J., Grinberg A., Huang S. P., and Pfeifer K. (2000) A transcriptional insulator at the imprinted H19/Igf2 locus. Genes Dev. 14, 1908–1919 [PMC free article] [PubMed] [Google Scholar]
- 47. Kaffer C. R., Grinberg A., and Pfeifer K. (2001) Regulatory mechanisms at the mouse Igf2/H19 locus. Mol. Cell. Biol. 21, 8189–8196 10.1128/MCB.21.23.8189-8196.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Eun B., Sampley M. L., Good A. L., Gebert C. M., and Pfeifer K. (2013) Promoter cross-talk via a shared enhancer explains paternally biased expression of Nctc1 at the Igf2/H19/Nctc1 imprinted locus. Nucleic Acids Res. 41, 817–826 10.1093/nar/gks1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Eun B., Sampley M. L., Van Winkle M. T., Good A. L., Kachman M. M., and Pfeifer K. (2013) The Igf2/H19 muscle enhancer is an active transcriptional complex. Nucleic Acids Res. 41, 8126–8134 10.1093/nar/gkt597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gray A., Tam A. W., Dull T. J., Hayflick J., Pintar J., Cavenee W. K., Koufos A., and Ullrich A. (1987) Tissue-specific and developmentally regulated transcription of the insulin-like growth factor 2 gene. DNA 6, 283–295 10.1089/dna.1987.6.283 [DOI] [PubMed] [Google Scholar]
- 51. de Pagter-Holthuizen P., Jansen M., van Schaik F. M., van der Kammen R., Oosterwijk C., Van den Brande J. L., and Sussenbach J. S. (1987) The human insulin-like growth factor II gene contains two development-specific promoters. FEBS Lett. 214, 259–264 10.1016/0014-5793(87)80066-2 [DOI] [PubMed] [Google Scholar]
- 52. Holthuizen P., van der Lee F. M., Ikejiri K., Yamamoto M., and Sussenbach J. S. (1990) Identification and initial characterization of a fourth leader exon and promoter of the human IGF-II gene. Biochim. Biophys. Acta 1087, 341–343 10.1016/0167-4781(90)90010-Y [DOI] [PubMed] [Google Scholar]
- 53. Ikejiri K., Wasada T., Haruki K., Hizuka N., Hirata Y., and Yamamoto M. (1991) Identification of a novel transcription unit in the human insulin-like growth factor-II gene. Biochem. J. 280, 439–444 10.1042/bj2800439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liu M., Roth A., Yu M., Morris R., Bersani F., Rivera M. N., Lu J., Shioda T., Vasudevan S., Ramaswamy S., Maheswaran S., Diederichs S., and Haber D. A. (2013) The IGF2 intronic miR-483 selectively enhances transcription from IGF2 fetal promoters and enhances tumorigenesis. Genes Dev. 27, 2543–2548 10.1101/gad.224170.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rainier S., Johnson L. A., Dobry C. J., Ping A. J., Grundy P. E., and Feinberg A. P. (1993) Relaxation of imprinted genes in human cancer. Nature 362, 747–749 10.1038/362747a0 [DOI] [PubMed] [Google Scholar]
- 56. Ogawa O., Eccles M. R., Szeto J., McNoe L. A., Yun K., Maw M. A., Smith P. J., and Reeve A. E. (1993) Relaxation of insulin-like growth factor II gene imprinting implicated in Wilms' tumour. Nature 362, 749–751 10.1038/362749a0 [DOI] [PubMed] [Google Scholar]
- 57. Zhan S., Shapiro D. N., and Helman L. J. (1994) Activation of an imprinted allele of the insulin-like growth factor II gene implicated in rhabdomyosarcoma. J. Clin. Invest. 94, 445–448 10.1172/JCI117344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. van Dijk M. A., van Schaik F. M., Bootsma H. J., Holthuizen P., and Sussenbach J. S. (1991) Initial characterization of the four promoters of the human insulin-like growth factor II gene. Mol. Cell. Endocrinol. 81, 81–94 10.1016/0303-7207(91)90207-9 [DOI] [PubMed] [Google Scholar]
- 59. Raizis A. M., Eccles M. R., and Reeve A. E. (1993) Structural analysis of the human insulin-like growth factor-II P3 promoter. Biochem. J. 289, 133–139 10.1042/bj2890133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rodenburg R. J., Teertstra W., Holthuizen P. E., and Sussenbach J. S. (1995) Postnatal liver-specific expression of human insulin-like growth factor-II is highly stimulated by the transcriptional activators liver-enriched activating protein and CCAAT/enhancer binding protein-α. Mol. Endocrinol. 9, 424–434 10.1210/mend.9.4.7659086 [DOI] [PubMed] [Google Scholar]
- 61. Rietveld L. E., Koonen-Reemst A. M., Sussenbach J. S., and Holthuizen P. E. (1999) Dual role for transcription factor AP-2 in the regulation of the major fetal promoter P3 of the gene for human insulin-like growth factor II. Biochem. J. 338, 799–806 [PMC free article] [PubMed] [Google Scholar]
- 62. Drummond I. A., Madden S. L., Rohwer-Nutter P., Bell G. I., Sukhatme V. P., and Rauscher F. J. 3rd. (1992) Repression of the insulin-like growth factor II gene by the Wilms tumor suppressor WT1. Science 257, 674–678 10.1126/science.1323141 [DOI] [PubMed] [Google Scholar]
- 63. Battle A., Brown C. D., Engelhardt B. E., and Montgomery S. B. (2017) Genetic effects on gene expression across human tissues. Nature 550, 204–213 10.1038/nature24277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ward M. C., and Gilad Y. (2017) Human genomics: Cracking the regulatory code. Nature 550, 190–191 10.1038/550190a [DOI] [PubMed] [Google Scholar]
- 65. eGTEx Project. (2017) Enhancing GTEx by bridging the gaps between genotype, gene expression, and disease. Nat. Genet. 49, 1664–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Scheper W., Meinsma D., Holthuizen P. E., and Sussenbach J. S. (1995) Long-range RNA interaction of two sequence elements required for endonucleolytic cleavage of human insulin-like growth factor II mRNAs. Mol. Cell. Biol. 15, 235–245 10.1128/MCB.15.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. van Dijk E. L., Sussenbach J. S., and Holthuizen P. E. (2000) Distinct RNA structural domains cooperate to maintain a specific cleavage site in the 3′-UTR of IGF-II mRNAs. J. Mol. Biol. 300, 449–467 10.1006/jmbi.2000.3856 [DOI] [PubMed] [Google Scholar]
- 68. FANTOM Consortium and the RIKEN PMI and CLST (DGT), Forrest A. R., Kawaji H., Rehli M., Baillie J. K., de Hoon M. J., Haberle V., Lassmann T., Kulakovskiy I. V., Lizio M., Itoh M., Andersson R., Mungall C. J., Meehan T. F., Schmeier S., et al. (2014) A promoter-level mammalian expression atlas. Nature 507, 462–470 10.1038/nature13182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lizio M., Harshbarger J., Abugessaisa I., Noguchi S., Kondo A., Severin J., Mungall C., Arenillas D., Mathelier A., Medvedeva Y. A., Lennartsson A., Drabløs F., Ramilowski J. A., Rackham O., Gough J., et al. (2017) Update of the FANTOM web resource: high resolution transcriptome of diverse cell types in mammals. Nucleic Acids Res. 45, D737–D743 10.1093/nar/gkw995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lek M., Karczewski K. J., Minikel E. V., Samocha K. E., Banks E., Fennell T., O'Donnell-Luria A. H., Ware J. S., Hill A. J., Cummings B. B., Tukiainen T., Birnbaum D. P., Kosmicki J. A., Duncan L. E., Estrada K., et al. (2016) Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Rotwein P. (2017) Large scale analysis of variation in the insulin-like growth factor family in humans reveals rare disease links and common polymorphisms. J. Biol. Chem. 292, 9252–9261 10.1074/jbc.M117.783639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. von Heijne G. (1985) Signal sequences. The limits of variation. J. Mol. Biol. 184, 99–105 10.1016/0022-2836(85)90046-4 [DOI] [PubMed] [Google Scholar]
- 73. von Heijne G. (1990) The signal peptide. J. Membr. Biol. 115, 195–201 10.1007/BF01868635 [DOI] [PubMed] [Google Scholar]
- 74. Begemann M., Zirn B., Santen G., Wirthgen E., Soellner L., Büttel H. M., Schweizer R., van Workum W., Binder G., and Eggermann T. (2015) Paternally inherited IGF2 mutation and growth restriction. N. Engl. J. Med. 373, 349–356 10.1056/NEJMoa1415227 [DOI] [PubMed] [Google Scholar]
- 75. Moreno-Estrada A., Gignoux C. R., Fernández-López J. C., Zakharia F., Sikora M., Contreras A. V., Acuña-Alonzo V., Sandoval K., Eng C., Romero-Hidalgo S., Ortiz-Tello P., Robles V., Kenny E. E., Nuño-Arana I., Barquera-Lozano R., et al. (2014) Human genetics. The genetics of Mexico recapitulates Native American substructure and affects biomedical traits. Science 344, 1280–1285 10.1126/science.1251688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Karczewski K. J., Weisburd B., Thomas B., Solomonson M., Ruderfer D. M., Kavanagh D., Hamamsy T., Lek M., Samocha K. E., Cummings B. B., Birnbaum D., The Exome Aggregation Consortium, Daly M. J., and MacArthur D. G. (2017) The ExAC browser: displaying reference data information from over 60 000 exomes. Nucleic Acids Res. 45, D840–D845 10.1093/nar/gkw971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Yoo-Warren H., Pachnis V., Ingram R. S., and Tilghman S. M. (1988) Two regulatory domains flank the mouse H19 gene. Mol. Cell. Biol. 8, 4707–4715 10.1128/MCB.8.11.4707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kent W. J., Sugnet C. W., Furey T. S., Roskin K. M., Pringle T. H., Zahler A. M., and Haussler D. (2002) The human genome browser at UCSC. 2002;12: 996–1008 10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lek M., Karczewski K. J., Minikel E. V., Samocha K. E., Banks E., Fennell T., O'Donnell-Luria A. H., Ware J. S., Hill A. J., Cummings B. B., Tukiainen T., Birnbaum D. P., Kosmicki J. A., Duncan L. E., Estrada K., et al. (2016) Analysis of protein-coding genetic variation in 60,706 humans. bioRxiv 10.1101/030338 [DOI] [PMC free article] [PubMed] [Google Scholar]