Abstract
Reduction of Methyl Red (MR) and Orange II (Or II) by 26 human skin bacterial species was monitored by a rapid spectrophotometric assay. The analysis indicated that skin bacteria, representing the genera Staphylococcus, Corynebacterium, Micrococcus, Dermacoccus and Kocuria, were able to reduce MR by 74–100 % in 24 h, with only three species unable to reduce completely the dye in that time. Among the species tested, only Corynebacterium xerosis was unable to reduce Or II to any degree by 24 h, and only Staphylococcus delphini, Staphylococcus sciuri subsp. sciuri and Pseudomonas aeruginosa were able to reduce completely this dye within 24 h. MR reduction started with early-exponential growth in Staphylococcus aureus and Staphylococcus epidermidis, and around late-exponential/early-stationary growth in P. aeruginosa. Reduction of Or II, Ponceau S and Ponceau BS started during late-exponential/early-stationary growth for all three species. Using liquid chromatography/electrospray ionization mass spectrometry analyses, MR metabolites produced by Staph. aureus, Staph. epidermidis and P. aeruginosa were identified as N,N-dimethyl-p-phenylenediamine and 2-aminobenzoic acid. Searches of available genomic and proteomic data revealed that at least four of the staphylococci in this study, Staphylococcus haemolyticus, Staph. epidermidis, Staphylococcus cohnii and Staphylococcus saprophyticus, have hypothetical genes with 77, 76, 75 and 74 % sequence identity to azo1 encoding an azoreductase from Staph. aureus and hypothetical proteins with 82, 80, 72 and 74 % identity to Azo1, respectively. In addition, Staphylococcus capitis has a protein with 79 % identity to Azo1. Western analysis detected proteins similar to Azo1 in all the staphylococci tested, except Staph. delphini, Staph. sciuri subsp. sciuri and Staphylococcus auricularis. The data presented in this report will be useful in the risk assessment process for evaluation of public exposure to products containing these dyes.
INTRODUCTION
Azo dyes are characterized by one or more azo bonds (R–N=N–R) that allow visible light to be absorbed by the dyes. These dyes are used in a wide variety of consumer products including textile dyes, tattoo inks and cosmetics (Moller & Wallin, 2000). These products directly contact human skin for several hours on a daily basis (Golka et al., 2004). In 2007, approximately 95 million US women used cosmetics. Estimates indicate that over 45 million US citizens currently have some type of tattooing (tattoo or permanent make-up).
Azo dyes may be biotransformed into colourless aromatic amines that are more easily absorbed by the skin. Some of these metabolites may be less toxic than the original dye (Collier et al., 1993), while others, such as arylamines and free radicals, are potentially carcinogenic (Chung, 1983; Mason et al., 1977; Nakayama et al., 1983). Thousands of azo dyes are available and more than 500 contain potentially carcinogenic aromatic amines (Platzek et al., 1999). A cancer risk assessment for the National Institute of Public Health and the Environment of The Netherlands found carcinogenic aromatic amines in commonly used products coloured by azo dyes including textile toys, bed sheets and watch straps (Zeilmaker et al., 2000).
While azo dyes are generally considered to be persistent pollutants because they are typically recalcitrant to aerobic biotransformation (Chen, 2006; Stolz, 2001), they may be metabolized by azoreductases from intestinal microorganisms and from mammalian liver cells (Levine, 1991). Azo dye metabolism has been described in anaerobic and aerobic bacteria (Levine, 1991; Xu et al., 2007).
Two types of azoreductases have been identified in bacteria: (1) monomeric flavin-free enzymes containing a putative NAD(P)H binding motif and (2) polymeric flavin-dependent enzymes (Chen, 2006). Although anaerobic conditions have been considered more common for azo dye reduction (Stolz, 2001), aerobic azoreductases have been characterized from both aerobic and anaerobic members of the human microbiome (Chen et al., 2004, 2005, 2008; Liu et al., 2007). A tetrameric, flavin-dependent, NADPH-dependent azoreductase (Azo1) from Staphylococcus aureus (ATCC 25923) metabolizes Methyl Red (MR) to the colourless products 2-aminobenzoic acid and N,N-dimethyl-p-phenylenediamine, and also metabolizes the azo dyes Orange II (Or II), Amaranth, Ponceau BS and Ponceau S (Chen et al., 2005).
At least several hundred species of bacteria are residents of the human skin (Gao et al., 2007). Staphylococcus, Micrococcus and Corynebacterium are among the most prevalent aerobic species that colonize the skin (Kawamura et al., 1998; Kloos & Musselwhite, 1975). A number of species that were originally classified as micrococci have been reclassified into the Kocuria, Kytococcus and Dermacoccus genera (Stackebrandt et al., 1995). Any number of human skin microbiota species potentially express azoreductase(s) and may contribute to the metabolism of azo dyes with which they come into contact (Chen et al., 2005; Platzek et al., 1999).
The current report examines the reduction of azo dyes by specific skin bacteria from nine different genera, including Staphylococcus, Dermacoccus, Kocuria, Micrococcus, Kytococcus and Corynebacterium. This study also detects proteins with some similarity to Azo1 that are expressed by many of the species examined.
METHODS
Bacterial strains
Bacterial strains NCH 281–302 (Table 1) were obtained from Taxonometrics. Staphylococcus epidermidis (NTH 118) and Staph. aureus (NTH 125) were kindly provided by Dr Mark Hart (US Food and Drug Administration). The remaining strains were from the NCTR collection. All strains were maintained aerobically in brain heart infusion (BHI) medium at 37 °C for 16–18 h, except Kytococcus sedentarius (NCH 301), which was maintained at 26 °C for at least 42 h.
Table 1.
Azo dye reduction by skin bacteria
| Isolate ID | Name | MR reduction | Or II reduction | Western* | Isolation source (ATCC no.) |
|---|---|---|---|---|---|
| NCH 281 | Dermacoccus nishinomiyaensis | 74 % by 24 h | 15 % by 24 h | Multi | Water (29093) |
| NCH 282 | Kocuria kristinae | 100 % by 7 h | 25 % by 24 h | + | Human skin (27570) |
| NCH 283 | Micrococcus luteus | 100 % by 14.5 h | 35 % by 24 h | − | ATCC 4698 |
| NCH 284 | Micrococcus lylae | 100 % by 12.5 h | 35 % by 24 h | Multi | Human skin (27566) |
| NCH 285 | Staphylococcus auricularis | 100 % by 9 h | 33 % by 24 h | − | Human external auditory canal (33753) |
| NCH 286 | Staphylococcus capitis subsp. capitis | 96.4 % by 7 h† | 21 % by 24 h | + | Human skin (27840) |
| NCH 287 | Staphylococcus caprae | 100 % by 1.25 h | 58 % by 24 h | + | Human (35538) |
| NCH 289 | Staphylococcus chromogenes | 100 % by 1.25 h | 97 % by 24 h | + | Pig skin (43764) |
| NCH 290 | Staphylococcus cohnii subsp. cohnii | 100 % by 9 h | 19 % by 24 h | + | Human skin (29974) |
| NCH 291 | Staphylococcus delphini | 100 % by 0.5 h | 100 % by 21 h | − | Dolphin purulent matter (49171) |
| NCH 292 | Staphylococcus hominis subsp. hominis | 100 % by 8 h | 41 % by 24 h | + | Human skin (27844) |
| NCH 294 | Staphylococcus lugdunensis | 100 % by 4.25 h | 58 % by 24 h | + | Axillary lymph node (43809) |
| NCH 295 | Staphylococcus saprophyticus | 100 % by 5 h | 34 % by 24 h | + | Human (15305) |
| NCH 297 | Staphylococcus sciuri subsp. sciuri | 100 % by 1 h | 100 % by 23 h | − | Eastern grey squirrel skin (29062) |
| NCH 298 | Staphylococcus simulans | 100 % by 1.25 h | 86 % by 24 h | + | Human skin (27848) |
| NCH 299 | Staphylococcus warneri | 100 % by 4.5 h | 43 % by 24 h | + | Human skin (27836) |
| NCH 300 | Staphylococcus xylosus | 100 % by 3.5 h | 68 % by 24 h | + | Human skin (29971) |
| NCH 301 | Kytococcus sedentarius‡ | 58 % by 48 h | 12 % by 48 h | − | Bull (14392) |
| NCH 302 | Staphylococcus haemolyticus | 100 % by 5.5 h | 40 % by 24 h | + | Human skin (29970) |
| NCH 303 | Corynebacterium xerosis | 100 % by 13.5 h | 0 % by 24 h | Multi | Ear discharge of a child (373) |
| NCH 304 | Pseudomonas aeruginosa | 100 % by 12 h | 100 % by 14.5 h | Multi | Blood culture (27853) |
| NCH 305 | Pseudomonas putida | 100 % by 12 h | 62 % by 24 h | Multi | ATCC 23974 |
| NCH 307 | Serratia liquefaciens | 100 % by 4.25 h | 22 % by 24 h | − | Milk, Cork, Ireland (27592) |
| NCH 309 | Streptococcus pyogenes | 100 % by 11 h | 16 % by 24 h | − | Pharynx of a child (19615) |
| NTH 118 | Staphylococcus epidermidis | 100 % by 4.5 h | 67 % by 24 h | + | ATCC 12228 |
| NTH 125 | Staphylococcus aureus | 100 % by 1.5 h | 88 % by 24 h | + | ATCC 25923 |
Multi, multiple proteins were detected by the polyclonal anti-Azo1 serum.
By 24 h, only 97.2 % had been reduced.
Kytococcus sedentarius required 48 h at 26 °C for proper growth, so reduction experiments were conducted under these conditions.
Azo dye reduction assay
Spectrophotometric monitoring of the reduction of 200 μM MR in BHI by Staph. epidermidis (NTH 118) at 430 nm for 3 h indicated no difference in the reduction pattern between samples with and without cells present.
To establish optimal dye concentrations, BHI supplemented with 25–200 μM MR was added (180 or 190 μl per well) to a 96-well plate (Becton Dickinson), with BHI (blank) or overnight Staph. epidermidis culture bringing volumes to 200 μl (each in triplicate). MR reduction was monitored in a SpectraMax Plus 384 plate reader (Molecular Devices) at 37 °C for 3 h, with 10 s of shaking before readings. The mean optical density (OD) of cells+BHI was taken from the mean OD of cells+MR. This method was used for Or II (482 nm), Ponceau S (520 nm) and Ponceau BS (502 nm).
The optimum OD of the initial inoculum was determined using initial OD550 of 0.005–1.0 with Staph. aureus, Staph. epidermidis and Pseudomonas aeruginosa. An OD of 0.1 allowed growth and ensured no significant lag in dye reduction. An initial inoculum OD of 0.05 was used in some experiments to observe dye reduction closely during early growth.
Reduction assays were performed twice (each in triplicate) for each bacterial species with MR and with Or II. Results in Table 1 reflect the mean of at least four wells. Bacterial growth was monitored at 550 nm, or at 600 nm for Ponceau S (absorption maximum=520 nm).
Bacterial species with inconsistent reduction patterns from the plate reader, Staphylococcus capitis subsp. capitis (NCH 286) and Serratia liquefaciens (NCH 307), were assayed manually in 100 ml. Aliquots were taken periodically from MR cultures and at 24 h from Or II cultures. Aliquots were centrifuged (10 000 g, 1 min) and supernatants were assayed.
Analysis of MR metabolites
Cultures of Staph. aureus, Staph. epidermidis and P. aeruginosa were cultivated overnight in BHI broth supplemented with 200 μM MR. The same volume of absolute ethanol was added to 10 ml of the cultures, the mixtures were vortexed, and then centrifuged at 12 000 g for 10 min. Supernatants were filtered using a 0.2 μm filter and analysed by liquid chromatography/electrospray ionization mass spectrometry (LC/ESI-MS). The samples were diluted 1/10 with aqueous 0.1 % formic acid, mixed by hand, and filtered using integral 0.2 μm PTFE filters in Whatman filtration vials. The samples and a mixture containing 10 μg ml−1 each of authentic N,N-dimethyl-p-phenylenediamine and 2-aminobenzoic acid were similarly analysed by LC/ESI-MS. A TSQ Quantum Ultra mass spectrometer (ThermoFinnigan) coupled with an 1100 Agilent HPLC system was operated in the positive-ion electrospray mode with the capillary temperature 270 °C, capillary voltage 4.0 kV, CID offset 14 V, sheath gas 20, sweep gas 4.0 and aux gas 0. With argon collision gas at 1.5 mTorr and 20 eV collision energy, product ion spectra of m/z 137 and 138 were acquired while scanning Q3 from m/z 30 to 150 every 0.25 s for a total cycle time of 0.5 s. A Curosil-PFP column 5 μm × 2 mm × 250 mm (Phenomenex) was maintained at 30 °C with a column heater. The mobile phase, delivered at 200 μl min−1, was aqueous 0.1 % formic acid for 10 min, followed by a linear 20 min gradient to 94.9 % acetonitrile/5 % H2O with 0.1 % formic acid, and held another 20 min.
Antibodies, cell lysates and Western analysis
Polyclonal antibodies against Staph. aureus Azo1 were obtained by subcutaneous inoculation of female rabbits with 0.5 mg purified recombinant Azo1 in 0.5 ml PBS emulsified with 1.5 ml Freund’s complete adjuvant (Chen et al., 2005). Animals were inoculated with 0.25 mg twice at 5-week intervals after initial immunization. Two weeks after the last administration, the rabbits were bled through the ear vein and the sera were tested for anti-Azo1 activity by dot-blot, ELISA and Western blots.
Overnight cultures in BHI supplemented with 200 μM MR were centrifuged at 5000 g for 20 min at 4 °C, then washed and suspended in 1 ml chilled PBS. Glass beads (0.1 ml, 212–300 μm; Sigma) were added and the cells were disrupted by: (1) vigorous vortexing (VORTEX-GENIE; Scientific Industries) for 20 min at 4 °C, (2) storage at −80 °C for 2 h and (3) thawing. Steps 1–3 were performed three times. Cell debris was pelleted by centrifugation at 12 000 g at 4 °C for 10 min. The supernatant was transferred to pre-chilled tubes and protein concentrations were determined using a bicinchoninic acid assay kit (Pierce) with BSA standards. Cell lysate (50 μg) from each of the bacteria was separated by 12.5 % (w/v) SDS-PAGE (Bio-Rad) and transferred to nitrocellulose membranes (Pierce). The membranes were treated for 2 h at room temperature with Superblock (Pierce), and then overnight at 4 °C with polyclonal rabbit anti-Staph. aureus Azo1 (1 : 2500). Next, the membranes were washed and incubated at room temperature with peroxidase-labelled goat anti-rabbit IgG (1 : 20 000) (Thermo) for 1 h. Signals on the membrane were detected by SuperSignal West Pico Chemiluminescent Substrate (Thermo).
BLAST sequences
In a BLASTN search (Altschul et al., 1997), the DNA sequence of Staph. aureus (ATCC 25923) azo1, accession number AY545994, returned results including 14 additional Staph. aureus strains, along with Staphylococcus haemolyticus (AP006716), four Staph. epidermidis strains (including ATCC 12228; AE015929), Staphylococcus cohnii (EU849488) and Staphylococcus saprophyticus (ATCC 15305; AP008934).
In a BLASTP search (Altschul et al., 1997, 2005), the protein sequence of Staph. aureus (ATCC 25923) Azo1, accession number AY545994, returned results including Staph. haemolyticus (YP_254347), Staph. epidermidis (ATCC 12228; NP_763885), Staph. capitis (ZP_ 03612986), Staph. saprophyticus (ATCC 15305; YP_302244) and Staph. cohnii (ACF54629).
RESULTS
Azo dye reduction by bacterial cultures
The disappearance of MR and Or II, indicative of azoreductase activity, was measured spectrophotometrically in the presence of various bacterial species (Table 1).
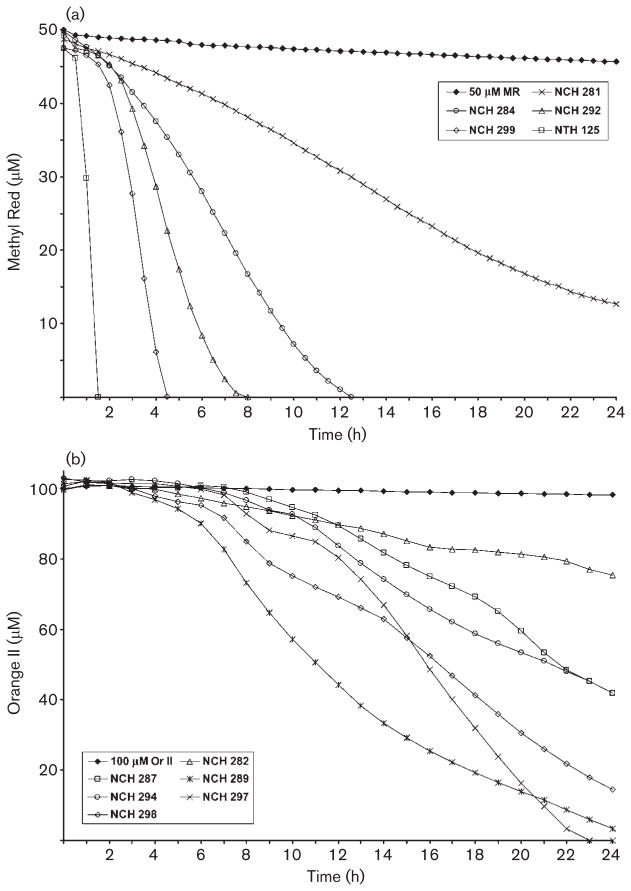
MR reduction times varied among the bacteria (Fig. 1a). Only three of the 26 species did not completely reduce MR by 14.5 h. Staph. capitis subsp. capitis (NCH 286) reduced MR by 96.4 % at 7 h, and by 97.2 % at 24 h. Dermacoccus nishinomiyaensis (NCH 281) reduced MR by 74 % at 24 h. Kyt. sedentarius (NCH 301) required 48 h at 26 °C for growth, and reduced MR by 58 % under those conditions.
Fig. 1.
Representative dye reduction curves. (a) MR reduction. Among the species that completely reduced 50 μM MR within 24 h, six required 1.5 h or less (represented by Staph. aureus, NTH 125); seven required between 3.5 and 5.5 h (represented by Staphylococcus warneri, NCH 299); four required between 7 and 9 h (represented by Staphylococcus hominis subsp. hominis, NCH 292); and six required between 11 and 14.5 h (represented by M. lylae, NCH 284). Of the 26 bacterial species tested, only three did not completely reduce MR by 24 h (represented by D. nishinomiyaensis, NCH 281). (b) Or II reduction. Only three species were able to completely reduce Or II within 24 h (represented by Staph. sciuri subsp. sciuri, NCH 297). Among the remaining species, no distinct patterns of reduction were observed.
Or II reduction also varied among the tested species, with no distinct groupings based on reduction times (Fig. 1b). Half (13) of the bacterial species were only able to reduce between 15 and 43 % of Or II by 24 h (Table 1). Kyt. sedentarius had reduced only 12 % of Or II by 48 h. By 24 h, five species had reduced between 58 and 68 % and six species had reduced more than 85 % of Or II. Only three species, Staphylococcus delphini (NCH 291), Staphylococcus sciuri subsp. sciuri (NCH 297) and P. aeruginosa (NCH 304), completely reduced Or II within 24 h. Corynebacterium xerosis (NCH 303) was unable to reduce Or II at all.
Bacterial growth during azo dye reduction
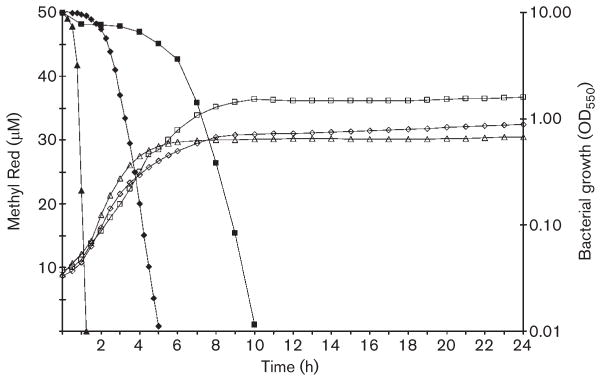
To examine the growth phase during which azo dye reduction occurred, Staph. aureus (NTH 125), Staph. epidermidis (NTH 118) and P. aeruginosa were grown in the presence and absence of MR (Fig. 2), Or II, Ponceau S and Ponceau BS. Azo dye disappearance and bacterial growth were measured simultaneously.
Fig. 2.
Bacterial growth phase during reduction of MR. Reduction of 50 μM MR (filled shapes) was measured at 430 nm and bacterial growth (open shapes) was measured at 550 nm simultaneously. Representative reduction and growth curves are shown for Staph. aureus (NTH 125, triangles), Staph. epidermi-dis (NTH 118, diamonds) and P. aeruginosa (NCH 304, squares). The representative curves shown were from an initial inoculum of OD550 0.05.
Reduction of MR by Staph. aureus and Staph. epidermidis started in early-exponential phase and was complete by 5 h (Fig. 2). Staph. aureus and Staph. epidermidis Or II reduction began around late-exponential/early-stationary phase and was not complete by 24 h. The two species did not begin Ponceau S or Ponceau BS reduction until late-exponential/early-stationary phase. At 24 h, Staph. aureus had reduced Ponceau S and Ponceau BS by 37 and 70 %, and Staph. epidermidis had reduced the dyes by 57 and 87 %, respectively.
P. aeruginosa reduction of MR, Or II, Ponceau BS and Ponceau S started at approximately the same time during growth, late-exponential/early-stationary phase (Fig. 2). P. aeruginosa completely reduced these dyes by 15 h.
Identification of MR metabolites by bacterial cultures
MR metabolites, produced in cultures of Staph. aureus, Staph. epidermidis and P. aeruginosa, were directly analysed by LC/ESI-MS. The retention times and product ion spectra of the MR metabolites were compared to those of authentic standards for identification. The metabolite eluting at 3.62 min had a protonated molecule at m/z 137 that fragmented at 20 eV to give product ions at m/z 122, 121, 107, 93 and 80. It was identified as N,N-dimethyl-p-phenylenediamine, since it eluted at the same retention time as did the standard and had an identical product ion spectrum. The metabolite eluting at 25.11 min had a protonated molecule at m/z 138 that fragmented at 20 eV to give product ions at m/z 120, 92 and 65. It was identified as 2-aminobenzoic acid since it eluted at the same retention time as did the standard and had an identical product ion spectrum.
Western analysis using polyclonal anti-Azo1
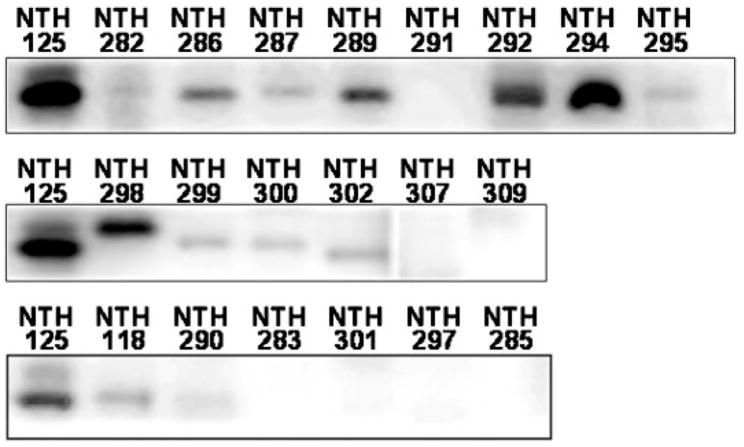
A polyclonal antibody to Staph. aureus Azo1 was used in Western analyses to detect similar proteins in skin bacteria cultured in the presence of MR. Among the 15 staphylococci examined, the antibody bound to a protein in each, except Staphylococcus auricularis (NCH 285), Staph. sciuri subsp. sciuri and Staph. delphini (Fig. 3).
Fig. 3.
Western blotting of bacterial protein using a polyclonal anti-Azo1 antiserum. Lanes are labelled using the isolate identification numbers in Table 1. Staph. aureus (NTH 125) protein was used as a positive control for each set of blots. Blots for NCH 281, 284, 303, 304 and 305 are not shown due to non-specific binding of the Azo1 antibody to multiple proteins in these samples.
Among the non-staphylococcal species, only Kocuria kristinae (NCH 282) produced a protein that was specifically detected by the Staph. aureus Azo1 antibody (Fig. 3). The antibody bound non-specifically to multiple proteins from P. aeruginosa, Pseudomonas putida (NCH 305), Micrococcus lylae, D. nishinomiyaensis and C. xerosis.
DISCUSSION
Azo dye reduction on the surface of skin could potentially lead to the formation of carcinogenic aromatic amines that are more readily absorbed by the skin than the original dyes (Platzek et al., 1999). Investigation of the ability of human skin microbiota to reduce azo dyes used in cosmetics, tattoo inks and other products that routinely contact skin is essential for evaluation of potential health risks involved in using these products (Chen et al., 2005). The results of this study suggest that at least a portion of the human skin microbiota is capable of azo dye reduction.
All of the bacteria studied were able to reduce MR to some extent within 24 h. Fifteen staphylococcal species completely reduced MR by 9 h. Staph. capitis subsp. capitis was the only staphylococcus that did not completely reduce MR by 24 h. However, this species had achieved 96.4 % reduction by 7 h without significant further reduction. Among the other genera, only Kyt. sedentarius, which required a longer incubation time, and D. nishinomiyaensis had not completely reduced MR by 24 h.
Although Or II reduction was generally not as rapid or as complete as that of MR, only C. xerosis was unable to reduce Or II to any degree. Only Staph. delphini, Staph. sciuri subsp. sciuri and P. aeruginosa completely reduced Or II within 24 h.
Bacterial growth varied among the species, but was not affected by the dyes. The slowest growers, including D. nishinomiyaensis, M. lylae, Kyt. sedentarius and C. xerosis, were among the slowest to reduce MR. Staph. delphini, the first to finish MR reduction, was among the fastest growing species. Although adequate growth was likely required for dye reduction, growth did not appear to directly correlate with MR reduction among many of the species.
MR metabolites from Staph. aureus, Staph. epidermidis and P. aeruginosa cultures were identified as N,N-dimethyl-p-phenylenediamine and 2-aminobenzoic acid, indicating cleavage of the MR azo bond to form aromatic amines (Chen et al., 2005; Sugiura et al., 1999).
BLASTN and BLASTP searches using azo1 and Azo1 sequences, respectively, revealed that the staphylococcal species for which genomic and proteomic sequences were available, Staph. haemolyticus, Staph. epidermidis, Staph. cohnii and Staph. saprophyticus, have similar hypothetical genes and proteins. These species have hypothetical genes with 77, 76, 75 and 74 % identity to azo1, and hypothetical proteins with 82, 80, 72 and 74 % identity to Azo1, respectively. In addition, Staph. capitis has a protein with 79 % identity to Azo1.
Proteins similar to Azo1 were detected by Western blotting in most of the staphylococci tested, confirming BLAST data. Although the amount of protein used in the Western analyses was the same for each species, some protein bands were more intensely detected than others, likely due to the polyclonal nature of the Azo1 antiserum.
Azoreduction is not performed by a single bacterial enzyme (Brige et al., 2008). Azo1-like proteins were not detected in Staph. delphini or Staph. sciuri subsp. sciuri, the first two species to completely reduce MR in assays. No obvious pattern of dye reduction among the species was associated with the presence of an Azo1-like protein. In addition, Staph. aureus and Staph. epidermidis began MR reduction much earlier during growth than for reduction of the other dyes, while P. aeruginosa reduced all dyes at approximately the same point in growth. Together, these observations suggest that additional bacterial enzymes are likely involved in azo dye reduction.
This study has provided data evaluating the role of skin microbiota in the metabolism of azo dyes. This information will be essential in the risk assessment process to evaluate public exposure to products containing azo dyes.
Acknowledgments
We thank Drs Mohamed S. Nawaz and Jinhui Feng for critical review of the manuscript, and Dr Mark Hart for strains NTH 125 and NTH 118 and helpful discussions. This study was funded by the Office of Women’s Health and the National Center for Toxicological Research, United States Food and Drug Administration, and was supported in part by appointments (R. L. S. and W. Z.) to the Postgraduate Research Program at the National Center for Toxicological Research, administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the US Department of Energy and the US Food and Drug Administration. The views presented in this article do not necessarily reflect those of the US Food and Drug Administration.
Abbreviations
- LC/ESI-MS
liquid chromatography/electrospray ionization mass spectrometry
- MR
Methyl Red
- Or II
Orange II
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Wootton JC, Gertz EM, Agarwala R, Morgulis A, Schäffer AA, Yu YK. Protein database searches using compositionally adjusted substitution matrices. FEBS J. 2005;272:5101–5109. doi: 10.1111/j.1742-4658.2005.04945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brige A, Motte B, Borloo J, Buysschaert G, Devreese B, Van Beeumen JJ. Bacterial decolorization of textile dyes is an extracellular process requiring a multicomponent electron transfer pathway. Microbial Biotechnol. 2008;1:40–52. doi: 10.1111/j.1751-7915.2007.00005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. Recent advances in azo dye degrading enzyme research. Curr Protein Pept Sci. 2006;7:101–111. doi: 10.2174/138920306776359786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Wang RF, Cerniglia CE. Molecular cloning, overexpression, purification, and characterization of an aerobic FMN-dependent azoreductase from Enterococcus faecalis. Protein Expr Purif. 2004;34:302–310. doi: 10.1016/j.pep.2003.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Hopper SL, Cerniglia CE. Biochemical and molecular characterization of an azoreductase from Staphylococcus aureus, a tetrameric NADPH-dependent flavoprotein. Microbiology. 2005;151:1433–1441. doi: 10.1099/mic.0.27805-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Xu H, Kweon O, Chen S, Cerniglia CE. Functional role of Trp-105 of Enterococcus faecalis azoreductase (AzoA) as resolved by structural and mutational analysis. Microbiology. 2008;154:2659–2667. doi: 10.1099/mic.0.2008/019877-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung KT. The significance of azo-reduction in the mutagenesis and carcinogenesis of azo dyes. Mutat Res. 1983;114:269–281. doi: 10.1016/0165-1110(83)90035-0. [DOI] [PubMed] [Google Scholar]
- Collier SW, Storm JE, Bronaugh RL. Reduction of azo dyes during in vitro percutaneous absorption. Toxicol Appl Pharmacol. 1993;118:73–79. doi: 10.1006/taap.1993.1011. [DOI] [PubMed] [Google Scholar]
- Gao Z, Tseng CH, Pei Z, Blaser MJ. Molecular analysis of human forearm superficial skin bacterial biota. Proc Natl Acad Sci U S A. 2007;104:2927–2932. doi: 10.1073/pnas.0607077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golka K, Kopps S, Myslak ZW. Carcinogenicity of azo colorants: influence of solubility and bioavailability. Toxicol Lett. 2004;151:203–210. doi: 10.1016/j.toxlet.2003.11.016. [DOI] [PubMed] [Google Scholar]
- Kawamura Y, Hou XG, Sultana F, Hirose K, Miyake M, Shu SE, Ezaki T. Distribution of Staphylococcus species among human clinical specimens and emended description of Staphylococcus caprae. J Clin Microbiol. 1998;36:2038–2042. doi: 10.1128/jcm.36.7.2038-2042.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloos WE, Musselwhite MS. Distribution and persistence of Staphylococcus and Micrococcus species and other aerobic bacteria on human skin. Appl Microbiol. 1975;30:381–395. doi: 10.1128/am.30.3.381-395.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine WG. Metabolism of azo dyes: implication for detoxication and activation. Drug Metab Rev. 1991;23:253–309. doi: 10.3109/03602539109029761. [DOI] [PubMed] [Google Scholar]
- Liu ZJ, Chen H, Shaw N, Hopper SL, Chen L, Chen S, Cerniglia CE, Wang BC. Crystal structure of an aerobic FMN-dependent azoreductase (AzoA) from Enterococcus faecalis. Arch Biochem Biophys. 2007;463:68–77. doi: 10.1016/j.abb.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Mason RP, Peterson FJ, Holtzman JL. The formation of an azo anion free radical metabolite during the microsomal azo reduction of sulfonazo III. Biochem Biophys Res Commun. 1977;75:532–540. doi: 10.1016/0006-291x(77)91505-4. [DOI] [PubMed] [Google Scholar]
- Moller P, Wallin H. Genotoxic hazards of azo pigments and other colorants related to 1-phenylazo-2-hydroxynaphthalene. Mutat Res. 2000;462:13–30. doi: 10.1016/s1383-5742(99)00090-3. [DOI] [PubMed] [Google Scholar]
- Nakayama T, Kimura T, Kodama M, Nagata C. Generation of hydrogen peroxide and superoxide anion from active metabolites of naphthylamines and aminoazo dyes: its possible role in carcinogenesis. Carcinogenesis. 1983;4:765–769. doi: 10.1093/carcin/4.6.765. [DOI] [PubMed] [Google Scholar]
- Platzek T, Lang C, Grohmann G, Gi US, Baltes W. Formation of a carcinogenic aromatic amine from an azo dye by human skin bacteria in vitro. Hum Exp Toxicol. 1999;18:552–559. doi: 10.1191/096032799678845061. [DOI] [PubMed] [Google Scholar]
- Stackebrandt E, Koch C, Gvozdiak O, Shumann P. Taxonomic dissection of the genus Micrococcus: Kocuria gen. nov., Nesterenkonia gen. nov., Kytococcus gen. nov., Dermacoccus gen. nov., and Micrococcus Cohn 1872 gen. emend. Int J Syst Bacteriol. 1995;45:682–692. doi: 10.1099/00207713-45-4-682. [DOI] [PubMed] [Google Scholar]
- Stolz A. Basic and applied aspects in the microbial degradation of azo dyes. Appl Microbiol Biotechnol. 2001;56:69–80. doi: 10.1007/s002530100686. [DOI] [PubMed] [Google Scholar]
- Sugiura W, Miyashita T, Yokoyama T, Arai M. Isolation of azo-dye-degrading microorganisms and their application to white discharge printing of fabric. J Biosci Bioeng. 1999;88:577–581. doi: 10.1016/s1389-1723(00)87680-x. [DOI] [PubMed] [Google Scholar]
- Xu H, Heinze TM, Chen S, Cerniglia CE, Chen H. Anaerobic metabolism of 1-amino-2-naphthol-based azo dyes (Sudan dyes) by human intestinal microflora. Appl Environ Microbiol. 2007;73:7759–7762. doi: 10.1128/AEM.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeilmaker MJ, van Kranen HJ, van Veen MP, Janus JA. Project 601503, General Assistance for the National Policy towards Substances. Bilthoven, The Netherlands: Research for Man and Environment, National Institute of Public Health and the Environment; 2000. Cancer risk assessment of azo dyes and aromatic amines from tattoo bands, folders of paper, toys, bed clothes, watch straps and ink; pp. 1–45. [Google Scholar]