Abstract
Long non-coding RNAs (lncRNAs) have been implicated in the occurrence and progression of multiple cancers. In the present study, we investigated the role of lncRNA X inactive-specific transcript (XIST) in the development and progression of pancreatic cancer (PC). Firstly, we found that lncRNA XIST was markedly upregulated in PC tissues and PC cell lines, respectively. Overexpression of XIST significantly promoted the proliferation, migration and invasion, and suppressed cell apoptosis of BxPC-3 cells; knockdown of XIST significantly inhibited the proliferation, migration and invasion, and accelerated cell apoptosis of PANC-1 cells. Furthermore, BxPC-3 and PANC-1 cells transfected with different vectors were injected subcutaneously into nude mice to explore tumor formation. We found that XIST promoted tumor formation in vivo. Subsequently, we found that microRNA-34a-5p (miR-34a-5p) was downregulated in PC tissues, and predicted a poor prognosis in PC patients. In addition, the results indicated that miR-34a-5p is a target gene of XIST and was significantly negatively correlated with XIST. More importantly, we found that miR-34a-5p rescued the facilitation of malignant behavior mediated by XIST. These results indicated that XIST and miR-34a-5p may be potential effective therapeutic targets for PC.
Keywords: long non-coding RNA XIST, pancreatic cancer, miR-34a-5p, migration, invasion, proliferation
Introduction
Pancreatic cancer (PC) is a malignant tumor with a high mortality rate and has become the leading cause of cancer-related death (1). At present, the prognosis of patients with PC remains poor and the 5-year survival rate of PC patients is still low (2,3). Therefore, it is urgent to understand the molecular mechanism of PC carcinogenesis in order to unearth reliable diagnostic and therapeutic targets for PC.
Long non-coding RNAs (lncRNAs) are non-protein-coding transcripts longer than 200 nucleotides and serve important roles in tumorigenesis (4,5). In addition, mounting evidence has shown that lncRNAs are functional in every stage of tumor progression (6), and are involved in tumor growth, invasion, metastasis of multiple cancers including PC (7–9). Previous studies have shown that the lncRNAs ATB, MALAT1, HOTAIR1, H19 and HOTTIP are associated with the development and progression of PC (10–14). lncRNA X inactive-specific transcript (XIST) (18 kb) is important for inactivation of X chromosome in the development of female mammals and is a prototype of gene-silencing lncRNAs (15). Studies have indicated that lncRNA XIST is involved in the progression of glioblastoma, non-small cell lung and gastric cancer, and hepatocellular carcinoma (16–20). However, the role of lncRNA XIST in PC is not fully understood.
MicroRNAs (miRNAs) are small (~20 nucleotides in length) non-coding RNA molecules that regulate gene expression by inhibiting translation or degrading mRNA transcripts (21). Research has shown that miRNAs participate in the occurrence and development of various diseases (22). There is a growing body of research showing that miRNAs are involved in the regulation of various biological processes including proliferation, differentiation, apoptosis, migration and invasion (23–26). Therefore, miRNAs are new and effective biomarkers for human cancer diagnostics. MicroRNA-34a-5p (miR-34a-5p) plays critical roles in the progression of various diseases. However, the biological functions and molecular mechanisms of miR-34a-5p in PC are not entirely clear.
Recently, in molecular biology, competing endogenous RNAs (abbreviated ceRNAs) regulate other RNA transcripts by competing for shared microRNAs. This has been proposed and defined as the crosstalk of RNA transcripts with miRNA response elements (MREs) (27,28). An increasing number of studies have found that the interaction between lncRNAs and miRNAs occurs in many different types of cancers (29–32), adding more puzzles for miRNA and lncRNA regulatory networks. For example, lncRNA-UCA1 was found to serve as a ceRNA, promoting the cell proliferation of esophageal cancer (33); lncRNA H19 functions as a ceRNA to promote epithelial-to-mesenchymal transition of colorectal cancer (30); lncRNA HOTAIR serves as a ceRNA to regulate HER2 expression by targeting miR-331-3p in gastric cancer (34); lncRNA FER1L4 inhibits cancer cell proliferation by functioning as a ceRNA (35).
In the present study, we investigated the expression and function of XIST in PC. We found that XIST expression was markedly upregulated in PC tissues and cells. Overexpression of XIST promoted PC cell growth, migration, invasion and metastasis; knockdown of XIST suppressed PC cell growth, migration, invasion and metastasis, implying a possible role of lncRNA XIST as an oncogene in PC. Moreover, we demonstrated that XIST was involved in the proliferation, migration, invasion and progression of PC by targeting miR-34a-5p.
Materials and methods
Ethics statement and specimens
Paired PC (n=139) and corresponding non-tumor control tissues were collected from the Japan Union Hospital of Jilin University from 2013 to 2015. The present study was approved by the Ethics Committee of Japan Union Hospital of Jilin University, and written informed consent was obtained from all patients. All tissue samples were immediately conserved at −80°C after washing with sterile phosphate-buffered saline (PBS).
Cell culture
Six PC cell lines (PANC-1, ASPC-1, MIA PaCa-2, HPAC, CFAPC-1 and BxPC-3), and human pancreatic ductal epithelial (HPDE), and 293T cells were purchased from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). Cells were grown in Dulbecco's modified Eagles medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (both from Invitrogen Life Technologies, Carlsbad, CA, USA) and 100 U/ml penicillin and 100 µg/ml streptomycin in an atmosphere of 5% CO2 at 37°C.
Lentiviral vector construction
Human XIST full-length DNAs were amplified by PCR from mRNA of BxPC-3 cells. Then, the cDNAs were inserted into a pcDNA3.1 vector. Enhanced green fluorescent protein (EGFP) served as a control. Specific shRNA against XIST and luciferase shRNA (Luc-shRNA; control) were synthesized and validated effective by RiboBio Co. (Guangzhou, China). The VSV-G pseudotyped lentiviruses were produced by co-transfecting 293T cells with lentivirus expression plasmid and packaging plasmids (pMD2.G, pMDL-G/P-RRE and pRSV-REV). Cells (5×104) were then transduced with the lentiviruses in the presence of 8 µg/ml Polybrene (Sigma-Aldrich, St. Louis, MO, USA).
Cell transfection
For plasmid transfection, BxPC-3 cells (2×105 cells/well) were seeded in 6-well plates, and transfected with XIST or the control using Lipofectamine™ 3000 (Invitrogen, Carlsbad, CA, USA) respectively according to the manufacturer's protocols. Likewise, PANC-1 cells were transfected with shXIST or control, respectively.
For miRNA transfection, miR-34a-5p mimics, inhibitor and negative control (miR-34a-5p NC) were purchased from GenePharma Co., Ltd. (Shanghai, China). Cells were transfected with miR-34a-5p mimics, miR-34a-5p inhibitors and miR-34a-5p NC respectively using Lipofectamine™ 3000, respectively, according to the manufacturer's protocols.
RNA isolation and quantitative real-time PCR (qRT-PCR)
Total RNA from PC tissues or cells was isolated using TRIzol reagent (Invitrogen) after treatments. Then, reverse transcription reaction was performed with Revert Aid First Strand cDNA Synthesis kit (Thermo Fisher Scientific Inc., Rockford, IL, USA) and random primers to provide cDNA products. The thermocycling conditions were 25°C for 5 min; 42°C for 60 min; 70°C for 10 min for the reverse transcription. qRT-PCR assays were performed using SYBR Premix Ex Taq (Takara, Tokyo, Japan), primers and cDNA templates on the Applied Biosystems 7500 Real-Time PCR system [Applied Biosystems Inc. (ABI) Carlsbad, CA, USA). Each individual sample was performed in triplicate and the expression levels were quantified using the comparative cycle threshold (CT) method. Results were normalized to GAPDH expression and RNA enrichments were calculated using the equation 2−ΔΔCt (36). Specific primers for XIST and miR-34a-5p were designed and synthesized by RiboBio Co. The primer sequences used in the present study are shown in Table I.
Table I.
Primer sequences for qRT-PCR analysis.
| Gene name | Primer sequences |
|---|---|
| miR-34a-5p | Forward 5′-GGGGTGGCAGTGTCTTAGC-3′ |
| Reverse 5′-CAGTGCGTGTCGTGGAGT-3′ | |
| U6 | Forward 5′-CTCGCTTCGGCAGCACA-3′ |
| Reverse 5′-AACGCTTCACGAATTTGCGT-3′ | |
| XIST | Forward 5′-AGCTCCTCGGACAGCTGTAA-3′ |
| Reverse 5′-CTCCAGATAGCTGGCAACC-3′ | |
| GAPDH | Forward 5′-TGTTCGTCATGGGTGTGAAC-3′ |
| Reverse 5′-ATGGCATGGACTGTGGTCAT-3′ |
miR-34a-5p, microRNA-34a-5p; XIST, X inactive-specific transcript.
Luciferase reporter assay
293T cells (1×105 cells/well) were placed in a 24-well plate, and co-transfected 200 ng of either pGL3-XIST-wt or pGL3-XIST-Mut vector and 80 ng of either miR-34a-5p or miR-NC. After 48 h for transfection, cells were harvested and luciferase activities were measured with the Dual-Luciferase Reporter Assay System (Promega, Wisconsin, WI, USA). All transfection experiments were conducted in triplicate.
Cell viability
Cell viability was assessed by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) assay. The treated cells (4,000 cells/well) were seeded in a 96-well plate and cultured in complete medium for 1, 2, 3, 4 and 5 days. MTT (0.5 mg/ml) was added to each well and incubation was carried out at 37°C for 4 h. Afterward, the supernatant was carefully aspirated and 100 µl of dimethyl sulfoxide (DMSO) was added. The absorbance was measured at 490 nm using a microplate reader. All experiments were repeated ≥3 times.
Colony formation assay
For the colony formation assay, the treated BxPC-3 and PANC-1 cells were seeded in 6-well plates at a density of 500 cells/well after transfection with different vectors. After 14 days, the cells were fixed with a 4% paraformaldehyde solution and stained with crystal violet. The total number of colonies in each plate from three independent transfections was counted under an inverted microscope to evaluate the colony formation ability.
Wound-healing assay
BxPC-3 or PANC-1 cells were transfected with different vectors and seeded in 6-well plates. Small linear wounds were created by removing a line of cells with a disinfected Eppendorf tip. After removing cell debris by washing with FBS-free medium, the wound areas were photographed under a microscope. Three different positions of distance between the two edges of the wound were calculated and analyzed by image analysis software (National Institute of Health, Bethesda, MD, USA).
Migration and invasion assays
Cell migration and invasion assays were performed using the Transwell assay according to the manufacturer's instructions. BxPC-3 or PANC-1 cells (5×104 cells/well) were transfected with different vectors and seeded in the upper compartment of the Transwell and incubated in serum-free media, and the lower compartment was filled with complete medium supplemented with 10% FBS. After 48 h of incubation at 37°C, non-invading cells were removed. The migratory and invasive cells on the bottom surface of the filters were fixed using 4% paraformaldehyde, and stained with 0.1% crystal violet solution. Four randomly selected fields of the fixed cells were counted for each group. The experiments were performed in triplicate.
Flow cytometric analysis
Treated cells were washed with 1X PBS, trypsinized and fixed with 70% ethanol for 30 min on ice. RNA was degraded with 20 mg/ml RNase (Sigma-Aldrich) for 1 h at 37°C. DNA was then labeled with 20 mg/ml propidium iodide (PI; Sigma-Aldrich). The cell cycle images were obtained and analyzed using FACSCalibur (BD Biosciences, Franklin Lakes, NJ, USA) and FlowJo software (Tree Star, Inc., Ashland, OR, USA).
Tumor formation assay in nude mice
The BxPC-3 cells were transfected with control plasmid or XIST plasmid. The PANC-1 cells were infected with control or shXIST plasmid, respectively. Four-week-old male nude mice were purchased from the National Laboratory Animal Center (Shanghai, China) and divided into four groups for subcutaneous injection using BxPC-3 or PANC-1 cells. Animals were sacrificed 40 days after injection and tumors were collected for measurement of the volume every 10 days. The tumor volume (V) was calculated by the formula: V (mm3) = length × width2/2. All experiments were performed strictly in accordance with a protocol approved by the Administrative Panel on Laboratory Animal Care of Jilin University.
Statistical analysis
The data were analyzed by the Student's t-test and one-way analysis of variance (ANOVA) using SPSS 15.0 software (SPSS, Inc., Chicago, IL, USA). Pearson's correlation coefficient was used to calculate the correlation between miR-34a-5p and XIST. The Kaplan-Meier method test was utilized for survival analysis. Each experiment was repeated at least three times. All results were summarized and are presented as means ± SD. P<0.05 was considered statistically significant.
Results
Upregulation of lncRNA-XIST predicts a poor prognosis in PC patients
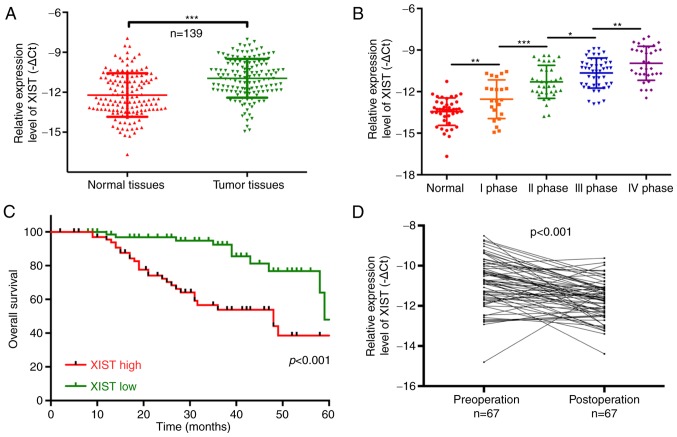
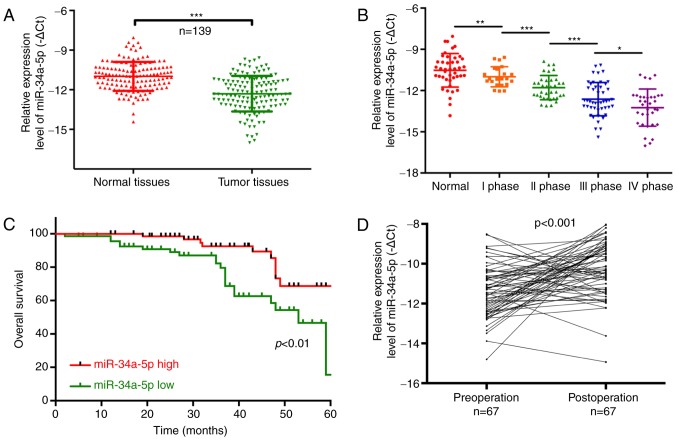
In the present study, we determined lncRNA-XIST expression in 139 PC patients and analyzed the relationship between XIST expression and clinicopathological characteristics [age, sex, lymphatic metastasis, distal metastasis and TNM stage] of the PC patients. The results showed that lncRNA-XIST was significantly upregulated in tumor tissues compared with that noted in the matched adjacent normal tissues (P<0.001; Fig. 1A). We also found that lncRNA-XIST expression was significantly related to the tumor-node-metastasis (TNM) stage, and was higher in the TNM I, II, III and IV stages than that noted in the normal group (P<0.05, P<0.01, P<0.001; Fig. 1B). Therefore, our results indicated that lncRNA-XIST expression is correlated with the malignant degree of PC. The Chi-square analysis indicated that the expression level of lncRNA-XIST was positively correlated with TNM stage (P<0.05) and distal metastasis (P<0.01), suggesting that lncRNA-XIST may be a potential biomarker for PC (Table II). In addition, PC patients with a high expression of XIST had a shorter overall survival than patients with low XIST expression (P<0.001; Fig. 1C). The results also indicated that lncRNA-XIST expression was significantly downregulated in 67 post-operation patients compared with pre-operation patients (P<0.001; Fig. 1D). All the above results demonstrated that a high expression level of XIST is associated with poor prognosis.
Figure 1.
Upregulation of lncRNA-XIST predicts the poor prognosis of PC patients. (A) The expression level of lncRNA-XIST was detected using qRT-PCR assay in PC tissues and their corresponding adjacent samples (n=139, ***P<0.001). (B) lncRNA-XIST expression was determined by qRT-PCR assay in tissues with different tumor-node-metastasis (TNM) stages (*P<0.05, **P<0.01, ***P<0.001). (C) Patients with high lncRNA-XIST expression had a worse prognosis when compared with patients with low lncRNA-XIST expression (P<0.001). (D) Expression of lncRNA-XIST was analyzed using qRT-PCR assay in 67 patients with PC pre-operation and post-operation (P<0.001). PC, pancreatic cancer; lncRNA, long non-coding RNA; XIST, X inactive-specific transcript.
Table II.
Relationship between lncRNA-XIST expression level (∆Ct) and clinicopathological characteristics of the PC patients.
| lncRNA-XIST | |||
|---|---|---|---|
| Characteristics | n (%) | Mean ± SD | P-value |
| Total no. of patients | 139 | ||
| Age (years) | 0.101 | ||
| >60 | 59 (42.4) | 11.85±1.73 | |
| ≤60 | 80 (57.6) | 11.19±2.69 | |
| Sex | 0.232 | ||
| Male | 89 (64.0) | 11.59±1.75 | |
| Female | 50 (36.0) | 12.02±2.45 | |
| Lymphatic metastasis | 0.235 | ||
| N0 | 94 (67.6) | 12.05±1.49 | |
| N1-N2 | 45 (32.4) | 11.71±1.73 | |
| Distal metastasis | 0.003b | ||
| M0 | 105 (75.5) | 11.98±1.06 | |
| M1 | 34 (24.5) | 11.14±2.16 | |
| TNM stage | 0.012a | ||
| 0, I, II | 91 (65.5) | 11.72±1.58 | |
| III, IV | 48 (34.5) | 10.96±1.83 |
P<0.05
P<0.01. PC, pancreatic cancer; lncRNA, long non-coding RNA; XIST, X inactive-specific transcript; TNM, tumor-node-metastasis.
XIST promotes PC tumor cell growth in vitro
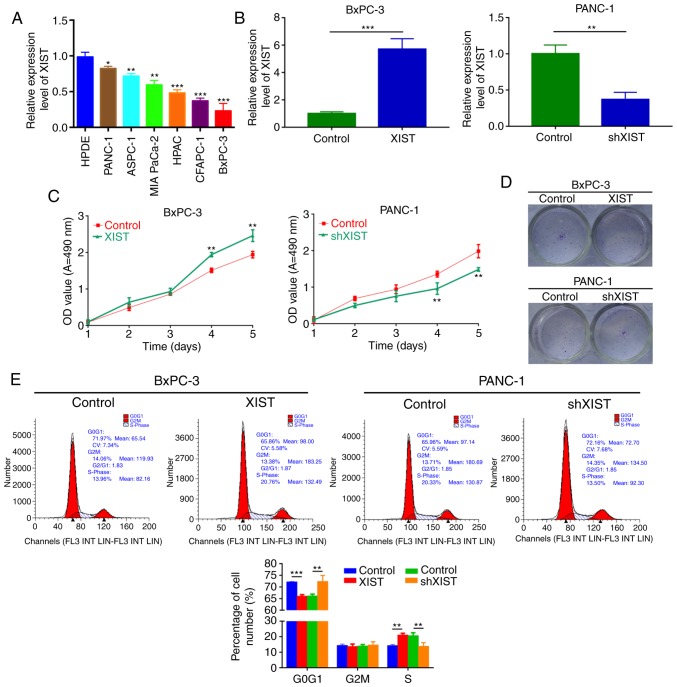
To further explore the oncogenic roles of XIST on PC in vitro, the expression level of lncRNA-XIST was detected by qRT-PCR in human pancreatic ductal epithelial (HPDE) cells and PC cell lines (PANC-1, ASPC-1, MIA PaCa-2, HPAC, CFAPC-1, and BxPC-3). The results revealed that lncRNA-XIST expression was increased in the PC cell lines compared with that noted in the HPDE cells (P<0.05, P<0.01, P<0.001; Fig. 2A). According to the lncRNA-XIST expression level in PC cells, we established PC cell lines (BxPC-3 and PANC-1) with XIST stable overexpression or knockdown (shXIST), respectively. The results indicated that lncRNA-XIST may play a critical role in the progression of PC. As shown in Fig. 2B, the expression level of XIST was increased in the BxPC-3 cells transfected with XIST-overexpressing plasmid compared with the control. XIST expression was decreased in PANC-1 cells transfected with shXIST compared with the control. Overexpression of XIST significantly promoted BxPC-3 cell viability, and knockdown of XIST significantly inhibited PANC-1 cell viability (P<0.01, P<0.001; Fig. 2C and D). Then, we evaluated the effect of XIST on cell cycle distribution of the PC cells. In agreement with the above results, we found that overexpression of XIST observably decreased the percentage of cells in the G0/G1 stage and increased the percentage of cells in the S and G2/M stages; knockdown of XIST significantly induced cell cycle arrest in the G0/G1 stage and decreased the percentage of cells in the S and G2/M stages (P<0.05, P<0.01; Fig. 2E).
Figure 2.
XIST promotes PC cell growth in vitro. (A) Expression level of lncRNA-XIST was detected by qRT-PCR assay in human pancreatic ductal epithelial (HPDE) cells and PC cell lines (PANC-1, ASPC-1, MIA PaCa-2, HPAC, CFAPC-1 and BxPC-3). Each assay was performed for at least three biological replicates (*P<0.05, **P<0.01, ***P<0.001). (B) XIST expression was examined by qRT-PCR assay in BxPC-3 cells transfected with XIST plasmid and control, and PANC-1 cells transfected with shXIST and control. Relative expression was normalized to GAPDH expression (**P<0.01). (C) MTT assay was performed to measure the viability of BxPC-3 and PANC-1 cells treated as in B (**P<0.01). (D) Colony formation assay was used to detect the proliferation ability of the transfected cells. (E) Cell Cycle distribution was analyzed by flow cytometry in BxPC-3 and PANC-1 cells after transfection (**P<0.01, ***P<0.001). PC, pancreatic cancer; lncRNA, long non-coding RNA; XIST, X inactive-specific transcript.
XIST accelerates the migration and invasion abilities of PC
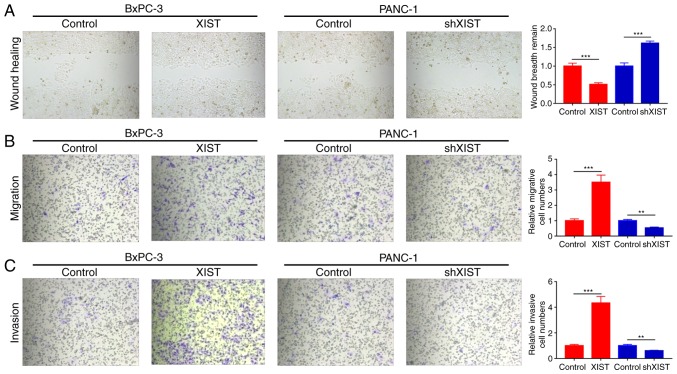
We further studied whether XIST functions as an oncogene. Wound healing assays found that overexpression of XIST significantly promoted the wound healing ability and knockdown of XIST significantly decreased this ability (P<0.001; Fig. 3A). Moreover, we used Transwell assay to explore the effects of XIST on the migration and invasion of BxPC-3 and PANC-1 cells. The results indicated that XIST obviously promoted the migration and invasion abilities of the BxPC-3 cells; knockdown of XIST markedly inhibited the migration and invasion abilities of the PANC-1 cells (P<0.01, P<0.001; Fig. 3B and C).
Figure 3.
XIST accelerates the migration and invasion abilities of PC. BxPC-3 cells were transfected with XIST or control, and PANC-1 cells were transfected with shXIST or control, respectively. (A) Wound healing assay was used to detect the migration capability (***P<0.001). (B and C) Transwell assays were performed to detect the migration and invasion abilities (**P<0.01, ***P<0.001 compared with the control group). PC, pancreatic cancer; XIST, X inactive-specific transcript;
XIST promotes tumor formation in vivo
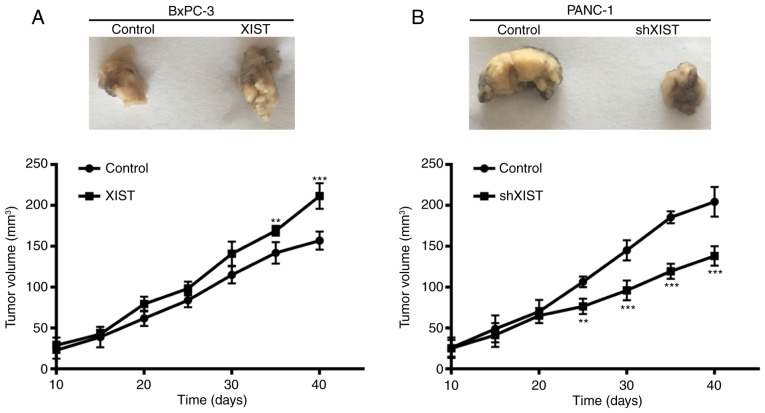
To further assess the growth effect of XIST on PC, we examined the tumorigenicity in nude mice. The result showed that overexpression of XIST in BxPC-3 cells accelerated tumor growth in the nude mouse model, and knockdown of XIST in PANC-1 cells inhibited tumor growth in the nude mouse model (P<0.01, P<0.001; Fig. 4). At 10, 15, 20, 25, 30, 35 and 40 days after injection, the tumors were removed. The tumor volume was accordance with above observation. From the above results, we suggest that XIST is an oncogene and promotes tumor formation.
Figure 4.
XIST promotes tumor formation in vivo. BxPC-3 cells were transfected with XIST or control, and PANC-1 cells were transfected with shXIST or control, respectively. The transfected cells were inoculated into the nude mouse for 40 days. Finally, the nude mice were sacrificed and the tumors excised. Dissected tumors were photographed every 10 days. The tumor volume was also measured (**P<0.01, ***P<0.001 compared with the control group). XIST, X inactive-specific transcript.
Downregulation of miR-34a-5p predicts a poor prognosis in PC patients
Similarly, we found that miR-34a-5p was significantly decreased in tumor tissues compared with that noted in the matched adjacent normal tissues (P<0.001; Fig. 5A). The results also showed that miR-34a-5p expression was significantly related to TNM stage, and was lower in the TNM I, II, III and IV stage than that in the normal group (P<0.05, P<0.01, P<0.001, Fig. 5B). The Chi-square analysis also indicated that miR-34a-5p expression was related to lymphatic metastasis (P=0.006) and TNM stage (P=0.009), suggesting that miR-34a-5p may be a potential biomarker for PC (Table III). Furthermore, high-expression of miR-34a-5p in patients with PC had a longer overall survival than patients with low miR-34a-5p expression (P<0.01; Fig. 5C). miR-34a-5p expression was significantly increased in 67 post-operation patients compared with that noted in pre-operation patients (P<0.001; Fig. 5D).
Figure 5.
Downregulation of miR-34a-5p predicts a poor prognosis in PC patients. (A) The expression level of miR-34a-5p was analyzed using qRT-PCR assay in PC tissues and their corresponding adjacent samples (n=139, ***P<0.001). (B) miR-34a-5p expression was measured by qRT-PCR assay in different tumor-node-metastasis (TNM) stages (*P<0.05, **P<0.01, ***P<0.001). (C) Patients with low miR-34a-5p expression had a worse prognosis than the patients with high miR-34a-5p expression (P<0.01). (D) miR-34a-5p expression was measured by qRT-PCR assay in 67 patients with PC in pre-operation and post-operation (P<0.001). PC, pancreatic cancer.
Table III.
Correlation between miR-34a-5p expression level (∆Ct) and clinicopathological characteristics of the PC patients.
| miR-34a-5p | |||
|---|---|---|---|
| Characteristics | n (%) | Mean ± SD | P-value |
| Total no. of patients | 139 | ||
| Age (years) | 0.785 | ||
| >60 | 59 (42.4) | 11.48±1.75 | |
| ≤60 | 80 (57.6) | 11.39±2.03 | |
| Sex | 0.202 | ||
| Male | 89 (64.0) | 11.45±1.79 | |
| Female | 50 (36.0) | 11.04±1.84 | |
| Lymphatic metastasis | 0.006a | ||
| N0 | 94 (67.6) | 11.06±1.64 | |
| N1-N2 | 45 (32.4) | 11.91±2.52 | |
| Distal metastasis | 0.089 | ||
| M0 | 105 (75.5) | 11.24±1.93 | |
| M1 | 34 (24.5) | 11.94±2.46 | |
| TNM stage | 0.009a | ||
| 0 & I & II | 91 (65.5) | 11.58±1.74 | |
| III & IV | 48 (34.5) | 12.35±1.39 |
P<0.01. TNM, tumor-node-metastasis. PC, pancreatic cancer.
miR-34a-5p abrogates the facilitation of malignant behavior mediated by XIST
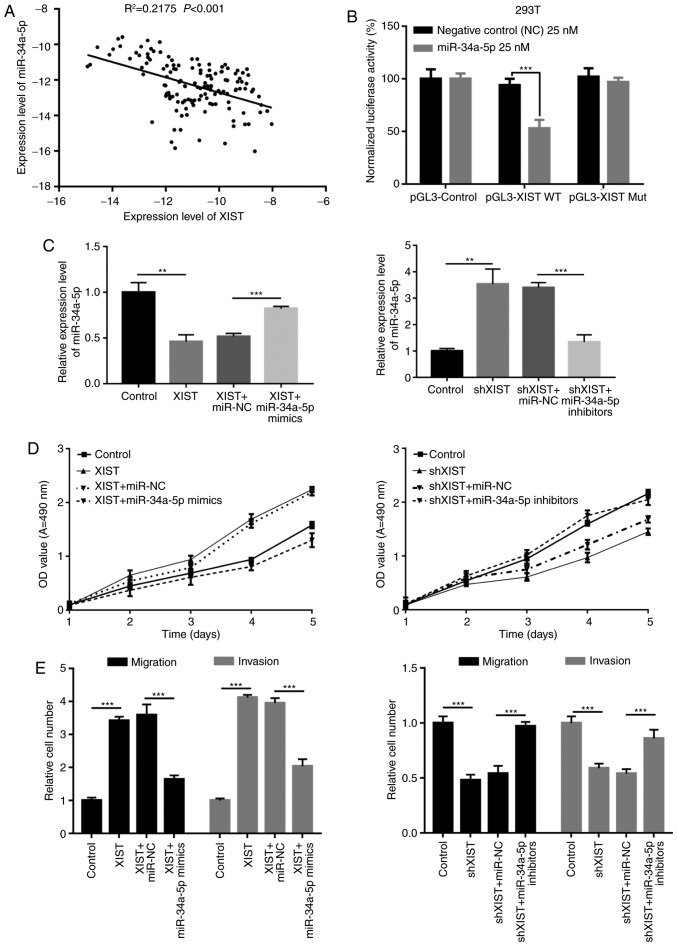
Furthermore, Pearson's correlation coefficient indicated that miR-34a-5p was significantly negatively correlated with XIST (R2=0.2175, P<0.001; Fig. 6A). We hypothesized that miR-34a-5p may play roles in organisms by targeting XIST. 293T cells were co-transfected with luciferase construct (pGL3-Control, pGL3-XIST WT or pGL3-XIST-Mut) and negative control (miR-control) or miR-34a-5p mimics, respectively. Luciferase reporter assay was performed to detect the regulatory relationship between miR-34a-5p and XIST. The results showed that miR-34a-5p markedly decreased the relative fluorescence value in Luc-wt reporter constructs, suggesting that miR-34a-5p was a target gene of XIST (P<0.001; Fig. 6B). In addition, we found that upregulation of XIST expression significantly decreased miR-34a-5p expression in BxPC-3 cells, and miR-34a-5p mimics significantly increased miR-34a-5p expression inhibited by XIST. Meanwhile, knockdown of XIST significantly increased miR-34a-5p expression in PANC-1 cells, and miR-34a-5p inhibitors significantly decreased miR-34a-5p expression induced by XIST (P<0.01, P<0.001; Fig. 6C). Furthermore, we used Transwell chamber inserts to explore the effects of miR-34a-5p and XIST on the migration and invasion of BxPC-3 and PANC-1 cells. The results indicated that overexpression of XIST promoted cell proliferation, while miR-34a-5p mimics inhibited cell proliferation induced by XIST; knockdown of XIST decreased cell proliferation, and miR-34a-5p inhibitors blocked this decrease mediated by XIST (Fig. 6D). Meanwhile, we also found that overexpression of XIST accelerated cell migration and invasion, while miR-34a-5p mimics inhibited this acceleration induced by XIST; knockdown of XIST decreased cell migration and invasion, and miR-34a-5p inhibitors blocked this decrease mediated by XIST (P<0.001; Fig. 6E).
Figure 6.
miR-34a-5p abrogates the facilitation of malignant behavior mediated by XIST. (A) The correlation between miR-34a-5p and XIST was determined by Pearson's correlation coefficient in clinical samples (R2=0.2175, P<0.001). (B) Luciferase reporter assay was performed in 293T cells co-transfected with luciferase construct (pGL3-Control, pGL3-XIST WT or pGL3-XIST-Mut) and negative control (miR-control) or miR-34a-5p mimics, respectively (***P<0.001). (C) BxPC-3 cells were transfected with control, XIST, XIST plus miR-NC or XIST plus miR-34a-5p mimics; PANC-1 cells were transfected with control, shXIST, shXIST plus miR-NC or shXIST plus miR-34a-5p inhibitors, respectively. qRT-PCR assay was used to detect miR-34a-5p expression. GAPDH was used as control (**P<0.01, ***P<0.001). (D) MTT assay was performed to determine the viability of BxPC-3 and PANC-1 cells treated as in C. (E) The migration and invasion abilities were detected by Transwell assays in BxPC-3 and PANC-1 cells treated as in C (***P<0.001). XIST, X inactive-specific transcript;
Discussion
Pancreatic cancer (PC) has been the main cause of cancer-related death worldwide for several decades (37,38). Mortality of PC is projected to surpass breast and colorectal cancer by 2030 in the US (39,40). The prognosis for patients with PC is poor with a reduced 5-year overall survival, and the median survival of patients with untreated PC is only 6 months with an extremely low percentage of long-term-surviving patients (41–43). However, at present, effective therapeutic strategies for patients with PC have been difficult to identify.
Emerging studies have identified numerous genes which are involved in the pathogenesis of human PC, and lncRNAs are important (12,14,44–46). Numerous studies have indicated that lncRNAs participate in the biological processes of various cancer cells, including cell proliferation, development, apoptosis and metastases (47–49). For example, IRAIN was found to promote proliferation and suppress the apoptosis of PC cells (48); MALAT-1 accelerates cell growth, migration and invasion in PC (47). Nevertheless, the mechanisms of long non-coding RNA XIST in PC are not clear.
MicroRNAs (miRNAs) have been demonstrated to be involved in the pathogenesis of many diseases, including cancers (50), infections (51,52) and diabetes (53). In addition, the differences in miRNA expression have regulatory functions in post-transcriptional modification or degradation of their target genes by binding to complementary regions in the 3′-untranslated region (UTR) of their target mRNA transcripts (54,55). Previous studies have demonstrated that miRNAs play a crucial role in pancreatic development and function. For example, miR-34a-5p inhibits colorectal cancer metastasis and is related to patient recurrence (56); miR-34a-5p enhances the multi-drug resistance of osteosarcoma (57); miR-34a-5p promotes chemoresistance of osteosarcoma (58).
In the present study, we found that XIST was significantly upregulated in human PC tissues and PC cell lines, compared with that noted in the adjacent normal tissues and HBE normal lung epithelial cell line. Our result also indicated that XIST expression was markedly higher at later stages of tumor development and in pre-operation patients with PC. In addition, XIST significantly increased PC cell viability, G1-G0 phase arrest, cell proliferation, migration and invasion and inhibited cell apoptosis in vitro, while XIST knockdown had opposite effects. The in vivo studies demonstrated that XIST downregulation suppressed tumor growth. All of these data indicate that XIST plays an important role in the development of PC. However, the underlying mechanism by which XIST mediates gene expression and participates in tumorigenesis remains to be clarified. Recently, the ceRNA hypothesis proposed that lncRNAs communicate with other protein-coding RNA transcripts via shared common miRNA binding sites (32). According to the ceRNA hypothesis, miRNA complementary base pairing with XIST was predicted by starBase and TargetScan, and miR-34a-5p was identified. Quantitative real-time PCR showed that the expression of miR-34a-5p in BxPC-3 cells was decreased upon overexpresion of XIST, however increased by transfected with miR-34a-5p mimic. In PANC-1 cells, the expression of miR-34a-5p was increased upon knockdown of XIST; however the expression of miR-34a-5p was suppressed by miR-34a-5p inhibitor. Therefore, we suggested that XIST is a direct target of miR-34a-5p and there was interactive suppression between them. Furthermore, we used Transwell chamber inserts to explore the effects of miR-34a-5p and XIST on the migration and invasion of BxPC-3 and PANC-1 cells. The results showed that the transfection of the miR-34a-5p mimic abrogated the XIST-promoted BxPC-3 cell migration and invasion. However, transfection of the miR-34a-5p inhibitor hampered shXIST-decreased PANC-1 cell migration and invasion.
lncRNA XIST functions a tumor-promoting gene in PC. It promotes cell proliferation and invasion in PC by directly targeting and suppressing tumor-suppressor miR-34a-5p. Consequently, XIST could be a potential target for the prevention of the metastasis of PC.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med. 2014;371:1039–1049. doi: 10.1056/NEJMra1404198. [DOI] [PubMed] [Google Scholar]
- 2.Egawa S, Toma H, Ohigashi H, Okusaka T, Nakao A, Hatori T, Maguchi H, Yanagisawa A, Tanaka M. Japan pancreatic cancer registry; 30th year anniversary: Japan pancreas society. Pancreas. 2012;41:985–992. doi: 10.1097/MPA.0b013e318258055c. [DOI] [PubMed] [Google Scholar]
- 3.Luo J, Xiao L, Wu C, Zheng Y, Zhao N. The incidence and survival rate of population-based pancreatic cancer patients: Shanghai Cancer Registry 2004–2009. PLoS One. 2013;8:e76052. doi: 10.1371/journal.pone.0076052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boon RA, Jae N, Holdt L, Dimmeler S. Long non-coding RNAs: From clinical genetics to therapeutic targets? J Am Coll Cardiol. 2016;67:1214–1226. doi: 10.1016/j.jacc.2015.12.051. [DOI] [PubMed] [Google Scholar]
- 5.Qiu MT, Hu JW, Yin R, Xu L. Long non-coding RNA: An emerging paradigm of cancer research. Tumour Biol. 2013;34:613–620. doi: 10.1007/s13277-013-0658-6. [DOI] [PubMed] [Google Scholar]
- 6.Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: A new frontier of translational research? Oncogene. 2012;31:4577–4587. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang X, Zhi X, Gao Y, Ta N, Jiang H, Zheng J. LncRNAs in pancreatic cancer. Oncotarget. 2016;7:57379–57390. doi: 10.18632/oncotarget.10545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peng W, Gao W, Feng J. Long non-coding RNA HULC is a novel biomarker of poor prognosis in patients with pancreatic cancer. Med Oncol. 2014;31:346. doi: 10.1007/s12032-014-0346-4. [DOI] [PubMed] [Google Scholar]
- 9.Zhan HX, Wang Y, Li C, Xu JW, Zhou B, Zhu JK, Han HF, Wang L, Wang YS, Hu SY. LincRNA-ROR promotes invasion, metastasis and tumor growth in pancreatic cancer through activating ZEB1 pathway. Cancer Lett. 2016;374:261–271. doi: 10.1016/j.canlet.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 10.Cheng Y, Jutooru I, Chadalapaka G, Corton JC, Safe S. The long non-coding RNA HOTTIP enhances pancreatic cancer cell proliferation, survival and migration. Oncotarget. 2015;6:10840–10852. doi: 10.18632/oncotarget.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, Kim S, Safe S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616–1625. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu JH, Chen G, Dang YW, Li CJ, Luo DZ. Expression and prognostic significance of lncRNA MALAT1 in pancreatic cancer tissues. Asian Pac J Cancer Prev. 2014;15:2971–2977. doi: 10.7314/APJCP.2014.15.7.2971. [DOI] [PubMed] [Google Scholar]
- 13.Ma C, Nong K, Zhu H, Wang W, Huang X, Yuan Z, Ai K. H19 promotes pancreatic cancer metastasis by derepressing let-7's suppression on its target HMGA2-mediated EMT. Tumour Biol. 2014;35:9163–9169. doi: 10.1007/s13277-014-2185-5. [DOI] [PubMed] [Google Scholar]
- 14.Qu S, Yang X, Song W, Sun W, Li X, Wang J, Zhong Y, Shang R, Ruan B, Zhang Z, et al. Downregulation of lncRNA-ATB correlates with clinical progression and unfavorable prognosis in pancreatic cancer. Tumour Biol. 2016;37:3933–3938. doi: 10.1007/s13277-015-4252-y. [DOI] [PubMed] [Google Scholar]
- 15.Smola MJ, Christy TW, Inoue K, Nicholson CO, Friedersdorf M, Keene JD, Lee DM, Calabrese JM, Weeks KM. SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the Xist lncRNA in living cells; Proc Natl Acad Sci USA; 2016; pp. 10322–10327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tantai J, Hu D, Yang Y, Geng J. Combined identification of long non-coding RNA XIST and HIF1A-AS1 in serum as an effective screening for non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8:7887–7895. [PMC free article] [PubMed] [Google Scholar]
- 17.Yao Y, Ma J, Xue Y, Wang P, Li Z, Liu J, Chen L, Xi Z, Teng H, Wang Z, et al. Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Lett. 2015;359:75–86. doi: 10.1016/j.canlet.2014.12.051. [DOI] [PubMed] [Google Scholar]
- 18.Chen DL, Ju HQ, Lu YX, Chen LZ, Zeng ZL, Zhang DS, Luo HY, Wang F, Qiu MZ, Wang DS, et al. Long non-coding RNA XIST regulates gastric cancer progression by acting as a molecular sponge of miR-101 to modulate EZH2 expression. J Exp Clin Cancer Res. 2016;35:142. doi: 10.1186/s13046-016-0420-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang J, Sun CC, Gong C. Long non-coding RNA XIST acts as an oncogene in non-small cell lung cancer by epigenetically repressing KLF2 expression. Biochem Biophys Res Commun. 2016;478:811–817. doi: 10.1016/j.bbrc.2016.08.030. [DOI] [PubMed] [Google Scholar]
- 20.Zhuang LK, Yang YT, Ma X, Han B, Wang ZS, Zhao QY, Wu LQ, Qu ZQ. MicroRNA-92b promotes hepatocellular carcinoma progression by targeting Smad7 and is mediated by long non-coding RNA XIST. Cell Death Dis. 2016;7:e2203. doi: 10.1038/cddis.2016.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayes EL, Lewis-Wambi JS. Mechanisms of endocrine resistance in breast cancer: An overview of the proposed roles of non-coding RNA. Breast Cancer Res. 2015;17:40. doi: 10.1186/s13058-015-0542-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Ann Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 23.Muluhngwi P, Klinge CM. Roles for miRNAs in endocrine resistance in breast cancer. Endocr Relat Cancer. 2015;22:R279–R300. doi: 10.1530/ERC-15-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Filipska M, Skrzypski M, Bigda JJ, Jassem J. Biological role of prognostic microRNAs (miRNAs) in squamous lung cancer cell lines. J Thorac Oncol. 2015;10:S391–S391. [Google Scholar]
- 25.Kara M, Yumrutas O, Ozcan O, Celik OI, Bozgeyik E, Bozgeyik I, Tasdemir S. Differential expressions of cancer-associated genes and their regulatory miRNAs in colorectal carcinoma. Gene. 2015;567:81–86. doi: 10.1016/j.gene.2015.04.065. [DOI] [PubMed] [Google Scholar]
- 26.Zhang P, Zuo Z, Wu A, Shang W, Bi R, Jin Q, Wu J, Jiang L. miR-600 inhibits cell proliferation, migration and invasion by targeting p53 in mutant p53-expressing human colorectal cancer cell lines. Oncol Lett. 2017;13:1789–1796. doi: 10.3892/ol.2017.5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ergun S, Oztuzcu S. Oncocers: ceRNA-mediated cross-talk by sponging miRNAs in oncogenic pathways. Tumour Biol. 2015;36:3129–3136. doi: 10.1007/s13277-015-3346-x. [DOI] [PubMed] [Google Scholar]
- 28.Su X, Xing J, Wang Z, Chen L, Cui M, Jiang B. microRNAs and ceRNAs: RNA networks in pathogenesis of cancer. Chin J Cancer Res. 2013;25:235–239. doi: 10.3978/j.issn.1000-9604.2013.03.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo LL, Song CH, Wang P, Dai LP, Zhang JY, Wang KJ. Competing endogenous RNA networks and gastric cancer. World J Gastroenterol. 2015;21:11680–11687. doi: 10.3748/wjg.v21.i41.11680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang WC, Fu WM, Wong CW, Wang Y, Wang WM, Hu GX, Zhang L, Xiao LJ, Wan DC, Zhang JF, Waye MM. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6:22513–22525. doi: 10.18632/oncotarget.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: Possible functions and clinical implications. J Med Genet. 2015;52:710–718. doi: 10.1136/jmedgenet-2015-103334. [DOI] [PubMed] [Google Scholar]
- 32.Xia T, Liao Q, Jiang X, Shao Y, Xiao B, Xi Y, Guo J. Long non-coding RNA associated-competing endogenous RNAs in gastric cancer. Sci Rep. 2014;4:6088. doi: 10.1038/srep06088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiao C, Song Z, Chen J, Zhong J, Cai W, Tian S, Chen S, Yi Y, Xiao Y. LncRNA-UCA1 enhances cell proliferation through functioning as a ceRNA of Sox4 in esophageal cancer. Oncol Rep. 2016;36:2960–2966. doi: 10.3892/or.2016.5121. [DOI] [PubMed] [Google Scholar]
- 34.Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD, Kong R, Xia R, Lu KH, Li JH, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92. doi: 10.1186/1476-4598-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xia T, Chen S, Jiang Z, Shao Y, Jiang X, Li P, Xiao B, Guo J. Long non-coding RNA FER1L4 suppresses cancer cell growth by acting as a competing endogenous RNA and regulating PTEN expression. Sci Rep. 2015;5:13445. doi: 10.1038/srep13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 37.Alzheimer's Association, corp-author. 2013 Alzheimer's disease facts and figures. Alzheimers Dement. 2013;9:208–245. doi: 10.1016/j.jalz.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 38.Malvezzi M, Bertuccio P, Levi F, La Vecchia C, Negri E. European cancer mortality predictions for the year 2013. Ann Oncol. 2013;24:792–800. doi: 10.1093/annonc/mdt010. [DOI] [PubMed] [Google Scholar]
- 39.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 40.No authors listed: Cancer statistics JAMA. 2013;310:982. doi: 10.1001/jama.2013.5289. [DOI] [PubMed] [Google Scholar]
- 41.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 42.Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362:1605–1617. doi: 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- 43.Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. Pancreatic cancer. Lancet. 2011;378:607–620. doi: 10.1016/S0140-6736(10)62307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li X, Deng SJ, Zhu S, Jin Y, Cui SP, Chen JY, Xiang C, Li QY, He C, Zhao SF, et al. Hypoxia-induced lncRNA-NUTF2P3-001 contributes to tumorigenesis of pancreatic cancer by derepressing the miR-3923/KRAS pathway. Oncotarget. 2016;7:6000–6014. doi: 10.18632/oncotarget.6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Müller S, Raulefs S, Bruns P, Afonso-Grunz F, Plötner A, Thermann R, Jäger C, Schlitter AM, Kong B, Regel I, et al. Next-generation sequencing reveals novel differentially regulated mRNAs, lncRNAs, miRNAs, sdRNAs and a piRNA in pancreatic cancer. Mol Cancer. 2015;14:94. doi: 10.1186/s12943-015-0401-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ye S, Yang L, Zhao X, Song W, Wang W, Zheng S. Bioinformatics method to predict two regulation mechanism: TF-miRNA-mRNA and lncRNA-miRNA-mRNA in pancreatic cancer. Cell Biochem Biophys. 2014;70:1849–1858. doi: 10.1007/s12013-014-0142-y. [DOI] [PubMed] [Google Scholar]
- 47.Jiao F, Hu H, Yuan C, Wang L, Jiang W, Jin Z, Guo Z, Wang L. Elevated expression level of long non-coding RNA MALAT-1 facilitates cell growth, migration and invasion in pancreatic cancer. Oncol Rep. 2014;32:2485–2492. doi: 10.3892/or.2014.3518. [DOI] [PubMed] [Google Scholar]
- 48.Lian Y, Wang J, Feng J, Ding J, Ma Z, Li J, Peng P, De W, Wang K. Long non-coding RNA IRAIN suppresses apoptosis and promotes proliferation by binding to LSD1 and EZH2 in pancreatic cancer. Tumour Biol. 2016;37:14929–14937. doi: 10.1007/s13277-016-5380-8. [DOI] [PubMed] [Google Scholar]
- 49.Zheng S, Chen H, Wang Y, Gao W, Fu Z, Zhou Q, Jiang Y, Lin Q, Tan L, Ye H, et al. Long non-coding RNA LOC389641 promotes progression of pancreatic ductal adenocarcinoma and increases cell invasion by regulating E-cadherin in a TNFRSF10A-related manner. Cancer Lett. 2016;371:354–365. doi: 10.1016/j.canlet.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 50.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2006;94:776–780. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sullivan CS, Ganem D. MicroRNAs and viral infection. Mol Cell. 2005;20:3–7. doi: 10.1016/j.molcel.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 52.Staedel C, Darfeuille F. MicroRNAs and bacterial infection. Cell Microbiol. 2013;15:1496–1507. doi: 10.1111/cmi.12159. [DOI] [PubMed] [Google Scholar]
- 53.Kato M, Castro NE, Natarajan R. MicroRNAs: Potential mediators and biomarkers of diabetic complications. Free Radic Biol Med. 2013;64:85–94. doi: 10.1016/j.freeradbiomed.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 55.Filipowicz W. RNAi: The nuts and bolts of the RISC machine. Cell. 2005;122:17–20. doi: 10.1016/j.cell.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 56.Gao J, Li N, Dong Y, Li S, Xu L, Li X, Li Y, Li Z, Ng SS, Sung JJ, et al. miR-34a-5p suppresses colorectal cancer metastasis and predicts recurrence in patients with stage II/III colorectal cancer. Oncogene. 2015;34:4142–4152. doi: 10.1038/onc.2014.348. [DOI] [PubMed] [Google Scholar]
- 57.Pu Y, Zhao F, Wang H, Cai W, Gao J, Li Y, Cai S. MiR-34a-5p promotes the multi-drug resistance of osteosarcoma by targeting the CD117 gene. Oncotarget. 2016;7:28420–28434. doi: 10.18632/oncotarget.8546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pu Y, Zhao F, Li Y, Cui M, Wang H, Meng X, Cai S. The miR-34a-5p promotes the multi-chemoresistance of osteosarcoma via repression of the AGTR1 gene. BMC Cancer. 2017;17:45. doi: 10.1186/s12885-016-3002-x. [DOI] [PMC free article] [PubMed] [Google Scholar]