Abstract
Previous studies have revealed that HURP (also known as DLGAP5 or KIAA0008) is overexpressed in many types of human cancers, such as hepatocellular carcinoma, squamous cell bladder cancer, and transitional cell carcinoma, indicating that HURP is a putative oncoprotein that promotes carcinogenesis through various molecular mechanisms. However, the role of HURP in the pathogenesis of non-small cell lung cancer (NSCLC) has not been reported. In the present study, we investigated the prognostic value of HURP among NSCLC patients through the GEO database. The online tool of KM-plotter was used to identify the correlation of HURP expression and the survival of NSCLC patients. We found the HURP expression at the mRNA level was correlated with the clinicopathologic characteristics and prognosis of NSCLC patients. HURP was highly expressed in aggressive NSCLC cells, and its higher expression was associated with shorter survival. Further cytological experiments revealed that the silencing of HURP caused cell cycle arrest and inhibited the proliferation of NSCLC cells. Transwell assay showed that HURP shRNA inhibited cell migration and invasion in vitro. The bioinformatic analysis suggests that HURP promotes carcinogenesis in multiple manners. Taken together, we revealed the prognostic value of HURP in NSCLC patients and HURP may be a potential therapeutic target for NSCLC.
Keywords: non-small cell lung cancer, hepatoma upregulated protein, HURP, biomarker, bioinformatics, GEO database
Introduction
Non-small cell lung cancer (NSCLC) is one of the leading causes of cancer-related death, with 221,200 estimated new cases annually in the United States (1). During the past few years, the incidence and mortality rates of NSCLC have increased rapidly. The mortality rate of NSCLC Chinese patients has increased from 0.07% in the 1970s to 0.4% in 2000 (2). Most lung cancer patients are in an advanced stage at initial diagnosis. Platinum-based doublet chemotherapy is the standard first-line treatment for advanced NSCLC patients and has a response rate of 30%. However, the efficacy of chemotherapy has reached a plateau. An understanding of the molecular pathology of NSCLC could open doors to new therapeutic techniques.
Hepatoma upregulated protein (HURP) (also known as DLGAP5 or KIAA0008) is a microtubule-associated protein and is a mitotic phosphorylated substrate of Aurora-A. Through bioinformatic analysis, Tsou et al (3) demonstrated that HURP is differentially expressed in human hepatocellular carcinoma and is also under cell cycle regulation. Furthermore, elevated HURP in a stable cell line resulted in anchorage-independent growth and low serum-dependent cell growth. In addition, the relationship of HURP with proliferation has been confirmed by the elevation of HURP expression in regenerating liver (3), generative cells (4), and stem cells (5). Overexpression of HURP has been detected in many types of human cancers, such as hepatocellular carcinoma (6–8), squamous cell bladder cancer (9), and transitional cell carcinoma (10), suggesting that HURP may take part in carcinogenesis. HURP is highly expressed in the G2/M phase and decreased in the G1 phase. It has been confirmed that HURP functions in stabilizing spindle (11), promoting spindle assembly (12), and forming a connection between the kinetochore and centrosome (13). Except for cell cycle modulation, HURP is able to enter the nucleus and engage in the regulation of cyclin. Yu et al (6) found that HURP could shuttle from the cytoplasm to the nucleus to avoid degradation. Chen et al (14) further confirmed that HURP enters into the nucleus through the nuclear localization signal and is engaged in the regulation of cyclin E1 expression as a co-transcription factor. From the above findings, HURP was confirmed as a putative oncoprotein that promotes carcinogenesis through various molecular mechanisms. Shi et al (15) identified HURP as a promising biomarker for the early detection of lung cancer and the prognosis of lung cancer patients through genome-wide mRNA expression data. However, the mechanism of HURP in NSCLC remains unclear.
In the present study, we aimed to validate the role of HURP in NSCLC carcinogenesis and to identify a new potential therapeutic target for NSCLC.
Materials and methods
Cell lines, cell culture and antibodies
The NSCLC cell lines, A549, H1975, 95D and H1299 were maintained in Dulbecco's modified Eagle's medium (DMEM; Corning, Corning, NY, USA) supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine, and 100 units penicillin/streptomycin (all from Invitrogen, Carlsbad, CA, USA) in 5% CO2 at 37°C in a humidified chamber. Rabbit anti-HURP monoclonal antibody (cat. no. ab107646) was purchased from Abcam (Cambridge, UK). Mouse anti-GAPDH monoclonal antibody (cat. no. sc-32233) and secondary polyclonal antibody, rabbit IgG (cat. no. sc-2004) and mouse IgG (cat. no. sc-2005), were obtained from Santa Cruz Biotechnolgy (Santa Cruz, CA, USA).
RNA isolation and real-time quantitative PCR
Total RNA was isolated from cell lines using Invitrogen™ Trizol reagents (Thermo Fisher Scientific, Inc., Waltham, MA, USA) following the manufacturer's instructions. cDNA was synthesized from 1 µg of total RNA by M-MLV Reverse Transcriptase (Promega, Madison, WI, USA). The primer sequences for HURP and GAPDH are as follows: HURP forward, 5′-AAGTGGGTCGTTATAGACCTGA-3′ and reverse, 5′-TGCTCGAACATCACTCTCGTTAT-3′; GAPDH forward, 5′-TGACTTCAACAGCGACACCCA-3′ and reverse, 5′-CACCCTGTTGCTGTAGCCAAA-3′.
The real-time quantitative polymerase chain reaction (RT-qPCR) analyses were performed with SYBR-Green Master Mixture (Takara, Shiga, Japan) on LightCycler 480 instrument (Roche Diagnostics GmbH, Mannheim, Germany). Each of the 12 µl quantitative PCR mixture contained 6 µl SYBR-Green Master Mixture, 0.6 µl cDNA product, 0.3 µl each of the 5 µM forward and reverse primers, and 5.1 µl RNase-free H2O. All these quantitative PCR experiments were performed in triplicate. The housekeeping gene GAPDH was used as an internal control. The 2−ΔΔCt (Ct is the threshold cycle) method was used to calculate relative target gene expression.
RNA interference
For the knockdown of human HURP, HURP-specific shRNAs were designed and purchased commercially (GeneChem, Shanghai, China). The target sequences of shRNAs were as follows: HURP-shRNA#1, GCAATGAGAGAGAGAATTA; HURP-shRNA#2, GGATATAAGTACTGAAATG and HURP-shRNA#3, TTGAAAGAGCAGAGAGAGA.
The shRNA constructs were inserted into the vector pGCSIL-GFP
Lentiviral particles were packaged in 293T cells by co-transfecting shRNA vectors, or control shRNA, together with the packaging plasmids.
Western blot analysis
Briefly, the NSCLC cell lines were lysed with RIPA buffer containing protease inhibitor. Following determination of the protein concentrations using the BCA Protein Assay kit (Beyotime Institute of Biotechnology, Haimen, China), equal amounts of protein per sample (20 µg) were separated on 10% sodium dodecyl sulfate (SDS) polyacrylamide gels for 2 h and transferred to nitrocellulose membranes. The membranes were subsequent blocked with 5% low-fat milk in TBST for 1 h and incubated with primary antibodies overnight at 4°C. The primary antibodies included: anti-HURP (1:2,000; Abcam) and GAPDH (1:2,000; Santa Cruz Biotechnology). This was followed by incubation with secondary antibodies (1:5,000; Santa Cruz Biotechnology) for 1.5 h at room temperature and rinsed four times with TBST for 8 min each. Reactions were visualized using the ECL system (Thermo Fisher Scientific, Waltham, MA, USA).
MTT assay
The cells were seeded in 96-well plates with DMEM containing 10% FBS for 1 to 5 days, followed by the MTT assay. Briefly, 20 µl of 5 mg/ml MTT solution was added and incubated with the cells for 4 h at room temperature. Then the medium was removed and 100 µl dimethyl sulfoxide (DMSO) was added for 5 min to extract the colored product catalyzed by the living cells. The end product was quantified with a spectrophotometer (Tecan Infinite, Mölnda, Sweden) at 490 nm wavelength. The final data were normalized with the OD490 value on day 1.
Apoptosis analysis
The NSCLC cells were trypsinized and washed twice with cold phosphate-buffered saline (PBS), and then stained with Annexin V-FITC/PI in the dark for 15 min at 37°C following the manufacturer's instructions. Binding buffer (400 µl) was added to a test tube and the proportion of apoptotic cells was quantified with a flow cytometer (Merck Millipore, Darmstadt, Germany).
Cell cycle analysis by flow cytometry
Cells under a log phase of growth were trypsinized and washed twice with cold PBS. After centrifugation, the cells were fixed with 100% ice-cold methanol at 4°C overnight. Then the cells were stained with 50 mg/ml propidium iodide (PI) and Rnase staining buffer in PBS for 30 min in the dark at room temperature. BD FACS (Becton Dickinson, San Jose, CA, USA) was used for analyzing cell cycle stage. A total of 10,000 cells were subjected to cell cycle analysis using a flow cytometer (Merck Millipore).
Cell migration and invasion assays
Transwell chambers (8-µm pore size) (Corning, Corning, NY, USA) were used in the cell migration and invasion assays. The cells were suspended in serum-free DMEM. At first, a total of 100 µl DMEM containing 104 cells was plated into the upper chamber of Transwell chambers, and 600 µl DMEM containing 10% FBS was added to the lower chamber. Then the cells were cultured at 37°C with 5% CO2 for 16 h. The cells in the upper surface were removed with cotton swabs. Cells that had adhered to the lower surface were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. Images were captured under a microscope (Olympus Corp., Tokyo, Japan) at ×100 magnification. The protocol for the invasion assay was the same, except that the Transwell chambers were precoated with Matrigel (BD Biosciences, San Jose, CA, USA), and the cells were also cultured for 16 h. Each experiment was performed in triplicate, and representative images are shown.
Gene Set Enrichment Analysis
Gene Set Enrichment Analysis (GSEA) was used to compare the gene expression level of each indicated geneset between the high HURP expression group and low HURP expression group. We divided GSE68465 and GSE30219 into two groups according to HURP expression in order to explore the biological process potentially modulated by HURP. The gene sets with normal P-value <0.05 and a false discovery rate (FDR) value <0.25 after 1,000 permutations were considered significantly enriched, and the Molecular Signatures Database was used to download the genesets.
Pathway enrichment analysis
The original array data underwent background correction and were normalized to the base-2 logarithm by Robust Multi-Array Average and Linear Models for Microarray. Then the differentially expressed genes (DEGs) between the low HURP expression group and high HURP expression group were identified. The absolute value of log2-fold change ≥1.5 and P-value <0.001 were considered as the threshold value. To assess the prospective functions of DEGs, we utilized the Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.kegg.jp/kegg/) using the Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov/), which provides a comprehensive set of functional annotation tools for investigators to understand biological meaning behind a large list of genes. The Search Tool for the Retrieval of Interacting Genes (STRING, https://string-db.org/) was used to investigate the interaction between these DEGs. The Gene Ontology (GO) enrichment analysis of DEGs was performed by BiNGO plugin for Cytoscape (http://apps.cytoscape.org/apps/bingo. The supplier is Steven Maere (VIB Department of Plant Systems Biology, UGent).
Validation of gene expression by immunochemistry
The Human Protein Atlas (HPA, http://www.proteinatlas.org/) is a public database that builds on gene expression data that includes quantitative transcriptomics data and spatial proteomics data (16). It curates histological images of 44 normal human tissues and the 20 most common types of cancer. In addition, HPA also provides functional analyses of proteomes and serves as an advantageous online tool in protein expression analysis and medical diagnosis. The expression of HURP in lung cancer tissues was obtained from the data deposited in the HPA. The expression level of HURP was defined as high, medium or low relative to that of normal lung tissues.
Statistical analysis
We investigated the prognostic value of HURP among NSCLC patients. The NSCLC mRNA expression data and corresponding clinical data were obtained from Gene Expression Omnibus (GEO) database. The GSE33532 and GSE19188 were gene expression microarrays providing primary tumors and matched distant normal lung tissue or adjacent normal tissue from NSCLC patients. The GSE30219 and GSE68465 were included in the analysis because of complete clinical data and large sample size. We defined the low expression cases as those whose HURP mRNA expression was less than or equal to the median value in order to explore the association between HURP and clinical characteristics. The χ2 test was applied to estimate the relationship between HURP expression and clinical parameters, where appropriate. Overall survival and recurrence-free survival were estimated with the Kaplan-Meier method. Survival differences were further validated by KM-plotter (http://kmplot.com/analysis/index.php?p=background) (17). The expression level of HURP in NSCLC cell lines was detected in Metabolic gEne RApid Visualizer (MERAV, http://merav.wi.mit.edu/SearchByGenes.html) and further validated by RT-qPCR and western blot analysis. Differences between the gene expression levels within different groups of cells were analyzed using ANOVA analysis followed by Tukey's multiple conparison. Each assay was performed three times for averaging replicates, and the unpaired Student's t-test was applied to evaluate the differences between the control and treatment group. All P-values are two-tailed with a significant level at 0.05. The above statistical analyses were performed using SPSS V19.0 (SPSS, Inc., Chicago, IL, USA) and GraphPad Prism (V.6.0) (GraphPad Software, Inc., La Jolla, CA, USA).
Results
Expression level of HURP is correlated with the clinicopathologic characteristics of the NSCLC patients
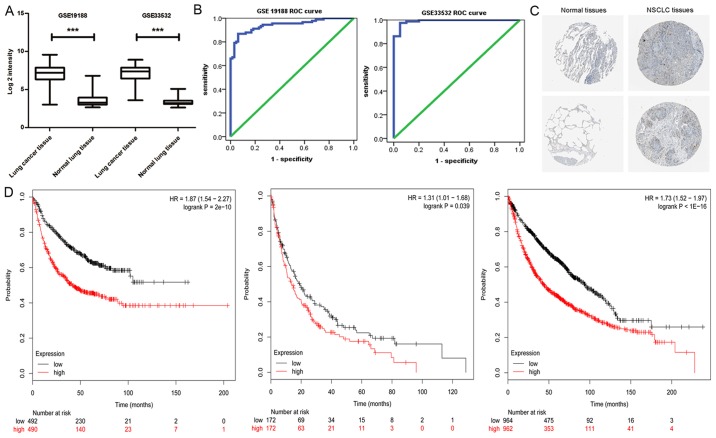
At first, we observed HURP expression at the mRNA level of NSCLC patients from GSE33532 and GSE19188. As shown in Fig. 1A, HURP expression was significantly upregulated in tumor tissues when compared with that noted in the distant normal lung tissues. Furthermore, HURP was able to provide high accuracy in regards to NSCLC tissue classification as estimated by ROC curve analysis (Fig. 1B). Therefore, we assessed the Human Protein Atlas database in order to analyze the expression of HURP in lung cancer and their normal counterparts. As shown in Fig. 1C, in accordance with the microarray analysis data, positive immunostaining of HURP in lung cancer tissues was observed (8 positive cases from 12 analyzed tumor samples). Furthermore, we assessed the association between HURP mRNA levels and the clinicopathologic characteristics in two independent cohorts of NSCLC patients from the GEO datasets. We categorized NSCLC patients into two groups based on HURP mRNA levels and defined the low expression cases as whose with HURP mRNA expression less than or equal to the median value. As shown in Table I, higher HURP expression was correlated with higher American Joint Committee on Cancer (AJCC) T stage (P<0.01) and N stage (P<0.01) in GSE30219. There was no significant difference between the high and low HURP expression group in regards to age (P=0.13) distribution. The multivariate analyses revealed that the progression-free survival (PFS) and overall survival (OS) of patients with high expression of HURP were significantly shorter than patients with high expression of HURP after adjusting for age, sex, clinical stage and pathology (PFS: HR=2.71, 95% CI 1.53–4.80, P<0.01; OS: HR=1.75, 95% CI 1.18–2.61, P<0.01) (data not shown).
Figure 1.
HURP is highly expressed in NSCLC tissues. (A) mRNA level of HURP was highly expressed in NSCLC tissues compared with that observed in the matched normal lung tissues (data from GSE33532 and GSE19188). (B) An ROC curve built on a univariate classification based on HURP expression across GSE19188 and GSE33532 for predicting NSCLC. (C) Immunohistochemical analysis of HURP was collected from the Human Atlas for normal and NSCLC tissues. (D) Survival analysis of NSCLC patients with low and high HURP expression using Kaplan-Meier Plotter website. ***P<0.05.
Table I.
Correlations of HURP with clinical characteristics of lung cancer patients (GSE30219).
| HURP | ||||
|---|---|---|---|---|
| Clinical characteristics | Low expression | High expression | χ2 | P-value |
| Sex | 10.16 | <0.01 | ||
| Male | 82 | 168 | ||
| Female | 25 | 18 | ||
| Age (years) | 2.34 | 0.13 | ||
| ≤60 | 49 | 69 | ||
| >60 | 57 | 117 | ||
| T stage | 23.31 | <0.01 | ||
| 1–2 | 102 | 113 | ||
| 3–4 | 4 | 48 | ||
| N stage | 11.46 | <0.01 | ||
| 0–1 | 101 | 150 | ||
| 2–3 | 5 | 35 | ||
P-values were calculated by the Pearson χ2 test.
We also downloaded and analyzed the clinical information from GSE68465
More high HURP expression patients presented with a higher clinical stage (P<0.01) and poorer differentiated carcinoma (P<0.01; Table II), and the results were similar to those obtained from GSE30219. Although no association between HURP and PFS was observed in the multivariate analyses, the relevance of HURP with OS was significant with adjustment for age, sex and clinical stage (HR=1.46, 95% CI 1.10–1.93, P<0.01). These results suggest that high expression of HURP may cause rapid tumor cell proliferation and a highly aggressive phenotype (data not shown).
Table II.
Correlations of HURP with clinical characteristics of lung cancer patients (GSE68465).
| HURP | ||||
|---|---|---|---|---|
| Clinical characteristics | Low expression | High expression | χ2 | P-value |
| Sex | 1.75 | 0.19 | ||
| Male | 122 | 101 | ||
| Female | 134 | 86 | ||
| Age (years) | 0.65 | 0.42 | ||
| ≤60 | 81 | 66 | ||
| >60 | 175 | 121 | ||
| T stage | 16.59 | <0.01 | ||
| 1–2 | 244 | 157 | ||
| 3–4 | 11 | 29 | ||
| N stage | 2.76 | 0.09 | ||
| 0–1 | 229 | 158 | ||
| 2 | 25 | 28 | ||
| Differentiation | 54.16 | <0.01 | ||
| Low | 61 | 108 | ||
| High and middle | 193 | 76 | ||
P-values were calculated by the Pearson χ2 test.
The KM-plotter was further utilized to validate the survival differences for all NSCLC patients in 14 microarray datasets with 1928 patients. The results indicated that the first progression time, post progression survival time and overall survival time were significantly different between the high and low HURP expression groups in the NSCLC patients. Higher HURP expression was associated with shorter survival time (Fig. 1D).
Silencing of HURP results in cell cycle arrest and inhibits the proliferation of NSCLC cells
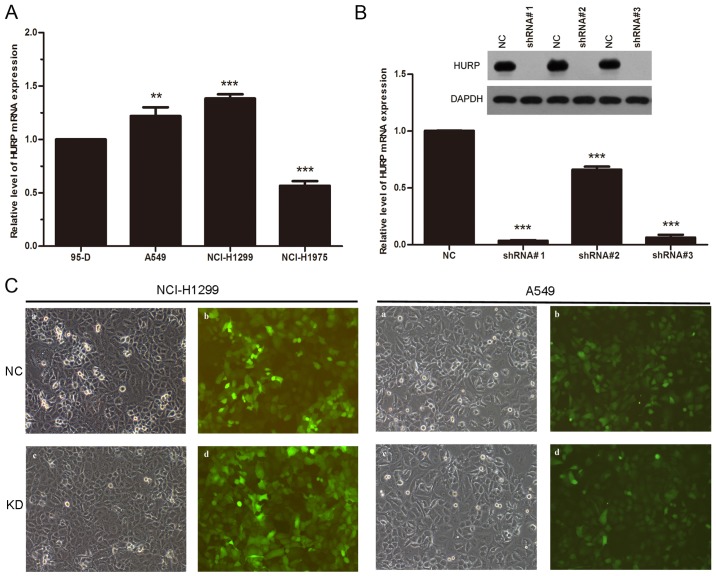
At first, we analyzed the expression level of HURP in 95-D, A549, H1299 and NCI-H1975 cell lines. The MERAV database indicated that the expression of HURP was upregulated in all four NSCLC cell lines. Futhermore, RT-qPCR revealed that A549 and H1299 cells expressed higher HURP compared to the 95-D cells (Fig. 2A). To investigate whether HURP is involved in the proliferation of NSCLC cells, we infected H1299 and A549 cells with a lentivirus to silence HURP. Three shRNAs were designed to suppress the expression of HURP and we detected the mRNA and protein levels of HURP in the different groups. The mRNA levels of HURP were significantly reduced in cells infected with shRNA#1 and shRNA#3 (P<0.01). Accordingly, the protein levels of HURP in the shRNA groups were also greatly decreased compared with levels noted in the negative control group (Fig. 2B). shRNA#1 was chosen for the following experiments due to the better efficacy. Expression of green fluorescence protein was observed in each group (Fig. 2C).
Figure 2.
(A) The mRNA level of HURP in 4 NSCLC cell lines. The levels were normalized to GAPDH. (B) The mRNA and protein level of HURP were significantly reduced in the H1299 cells after HURP silencing. (C) H1299 and A549 cells were transfected with empty vector or shRNA#1. Expression of green fluorescence protein was observed in the each group (ANOVA analysis followed by the post hoc Tukey's comparison; **P<0.05, ***P<0.01). NC, negative control cells; KD, knockdown cells.
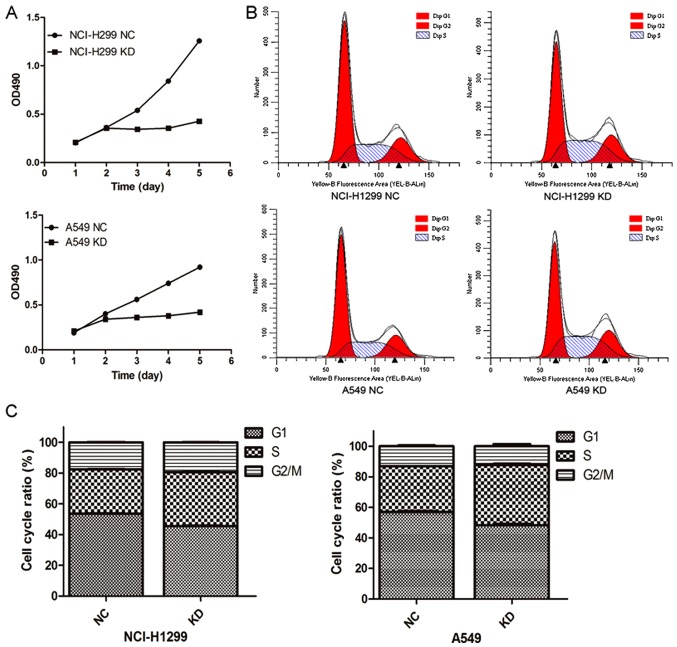
To explore the potential cause of HURP in regulating the proliferation of NSCLC cells, MTT assays were conducted to assess cell viability. As shown in Fig. 3A, after a 48-h post infection in control or shRNA groups, the cell viability was markedly decreased in the cells transfected with the HURP-targeting shRNA. The optical density (OD) values at 490 nm of the HURP shRNA H1299 group (0.34±0.01, 0.36±0.01, 0.43±0.01) were significantly lower than those in the negative control group (0.54±0.07, 0.84±0.01 and 1.26±0.01) from day 3 to day 5 (P<0.05). Meanwhile, similar results were found in the A549 cells. The OD values at 490 nm of the HURP shRNA A549 group were 0.36±0.05, 0.38±0.04 and 0.42±0.02 compared to 0.56±0.02, 0.74±0.06, and 0.92±0.03 in the negative control group from day 3 to day 5 (P<0.05).
Figure 3.
Knockdown of HURP in NSCLC cells inhibits cell proliferation. (A) Silencing of HURP significantly inhibited cell viability of the H1299 KD and A549 KD cells. (B and C) Flow cytometry illustrated that the cell cycle was arrested in the G1/S stage in H1299 KD and A549 cells KD. Data presented were representative of three independent experiments. NC, negative control cells; KD, knockdown cells.
Cell cycle assays detected by flow cytometry revealed that the cells were mainly distributed in the S stage in the HURP-null group (HURP shRNA H1299 group, 35.09±0.32%; negative control group, 28.83±0.10%, P<0.05). Accordingly, the percentage of HURP shRNA A549 cells in the S phase (39.12±0.43%) was significantly higher than that of the negative control group (32.03±0.25%, P<0.05; Fig. 3B and C). The results suggested that HURP not only regulates cell division through spindle assembly, but also promotes cell proliferation through other means.
We then detected the effects of HURP on apoptosis. However, there was no significant difference in apoptosis rates between the control group and HURP-knockdown group (data not shown). Collectively, HURP may modulate the cell cycle to regulate cell proliferation.
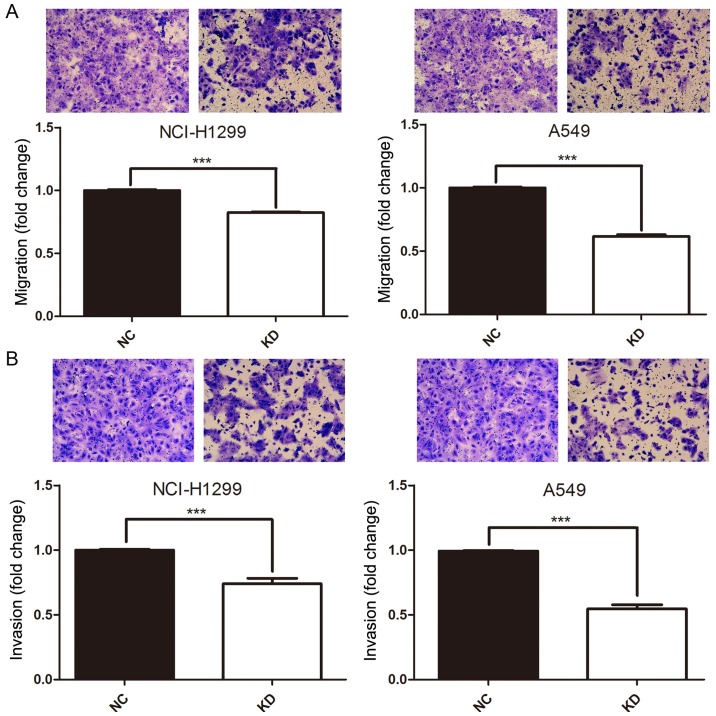
Depletion of HURP inhibits NSCLC cell migration and invasion in vitro
To evaluate whether HURP plays a role in cell migration and invasion, the Transwell assay was performed. A549 and H1299 cells were pretreated with either shNC or shRNA#1. After 16 h of incubation, the cells that transmigrated to the lower chamber were significantly decreased in the shRNA group compared to that noted in the negative control group (P<0.05, Fig. 4A). Furthermore, the Transwell matrix penetration assay was conducted to assess the effects of HURP on cell invasion. As shown in Fig. 4B, after 16-h post infection of shNC or shRNA#1, the cells with downregulated HURP expression demonstrated statistically significant decreased invasion ability in the A549 and H1299 cells (P<0.05). Altogether, HURP modulates NSCLC migration and invasion in vitro.
Figure 4.
(A) Cell migration was assessed by Transwell assay in H1299 and A549 cells after depletion of HURP for 16 h. The cells migrated into the lower chamber were stained. (B) Cell invasion was evaluated by Transwell assay with Matrigel in H1299 and A549 cells after depletion of HURP for 16 h (by t-test analysis, ***P<0.05). KD, knockdown cells; NC, negative control cells.
HURP regulates multiple biological processes in NSCLC cells
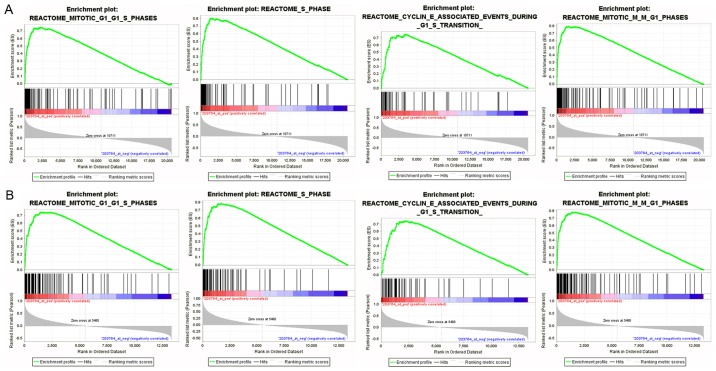
After elucidating the role of HURP in the modulation of proliferation, cell cycle regulation and migration, we aimed to ascertain other potential functions of HURP in NSCLC cells. In order to corroborate the biological process potentially modulated by HURP, we performed GSEA analysis using the microarray datasets of GSE30219 and GSE68465. The enriched expression of genesets included cell cycle, cell mitosis, cell cycle checkpoints, and mitotic spindle regulation. In particular, HURP was involved in G1/S phase transition, cyclin E-associated events during G1/S transition and M/G1 transition. Furthermore, HURP may participate in activation of NF-κB, P53 independent DNA damage checkpoint and transcription process (Fig. 5).
Figure 5.
Gene set enrichment analyses of GSE30219 (A) and GSE68465 (B) indicated that the genes associated with cell cycle, cyclin E-associated events during G1/S transition, activation of NFκB, P53-independent DNA damage checkpoint and transcription process were enriched in the groups with high HURP expression.
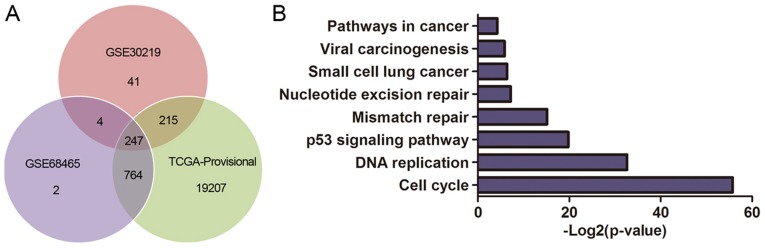
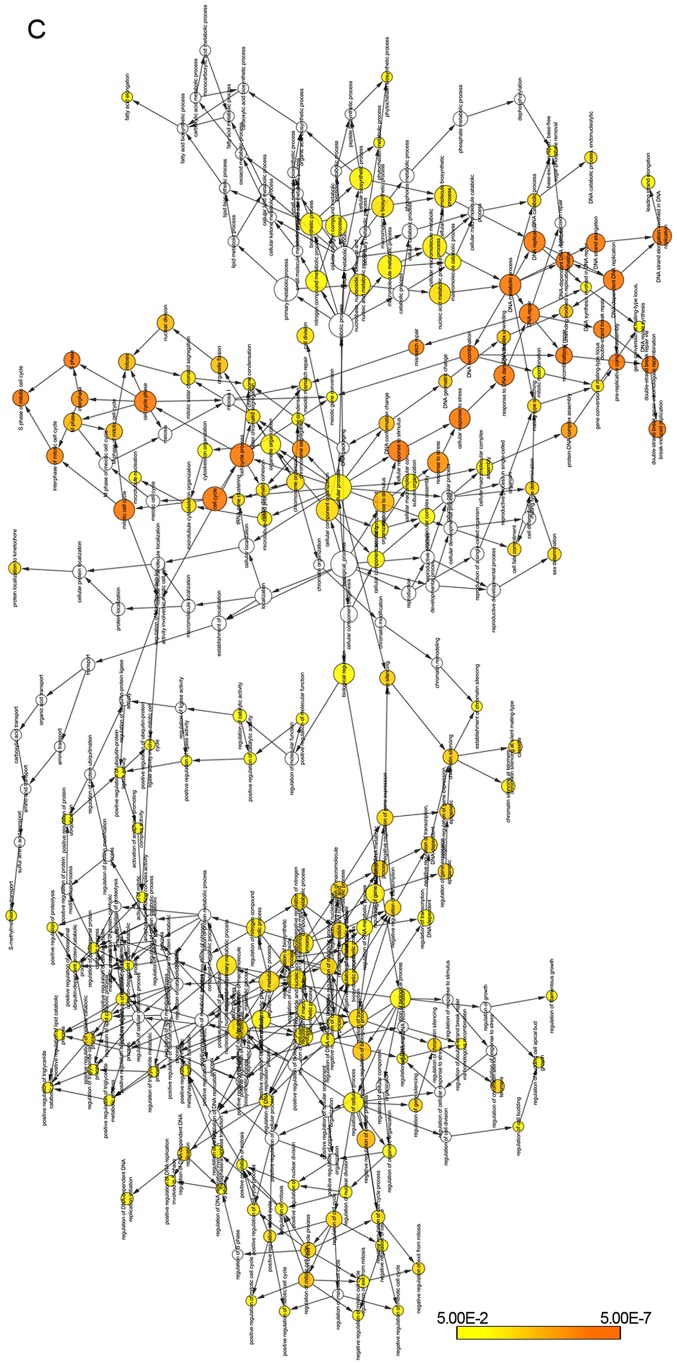
The DEGs were obtained from GSE30219, GSE68465 and TCGA database, and 247 genes are summarized in total (Fig. 6A). We performed KEGG pathway analysis to clarify the potential biological functions of HURP. The top KEGG pathways enriched for DEGs included cell cycle, p53 signaling pathway, small cell lung cancer, pathways in cancer and viral carcinogenesis (Fig. 6B). In addition, the DEGs were mainly enriched in the GO biological process related to cell cycle regulation (Fig. 6C) and the top 50 significant GO biological process terms are shown in Table III.
Figure 6.
(A) Venn diagrams of DEGs between the high HURP expression group and low HURP expression group extracted from GSE30219, GSE68465 and TCGA-Provisional. (B) KEGG pathways enriched for DEGs were mainly cancer-related pathways. (C) The GO biological terms enriched by DEGs were performed by BiNGO plugin for Cytoscape. DEGs, differentially expressed genes.
Table III.
The top 50 significant Gene Ontology biological process terms enriched by DEGs.
| GO ID | Adjusted P-value | Description |
|---|---|---|
| 43933 | 2.07E-02 | Macromolecular complex subunit organization |
| 65003 | 2.14E-02 | Macromolecular complex assembly |
| 32268 | 6.68E-01 | Regulation of cellular protein metabolic process |
| 32434 | 3.25E-02 | Regulation of proteasomal ubiquitin-dependent protein catabolic process |
| 65007 | 2.61E-02 | Biological regulation |
| 50789 | 2.20E-02 | Regulation of biological process |
| 44237 | 1.07E-01 | Cellular metabolic process |
| 34641 | 2.07E-02 | Cellular nitrogen compound metabolic process |
| 19222 | 1.96E-03 | Regulation of metabolic process |
| 6259 | 1.94E-09 | DNA metabolic process |
| 6310 | 8.88E-08 | DNA recombination |
| 22403 | 3.94E-11 | Cell cycle phase |
| 7126 | 8.74E-02 | Meiosis |
| 279 | 1.09E-04 | M phase |
| 51726 | 1.04E-03 | Regulation of cell cycle |
| 45787 | 2.15E-03 | Positive regulation of cell cycle |
| 101 | 5.76E-02 | Sulfur amino acid transport |
| 15806 | 1.37E-02 | S-methylmethionine transport |
| 10564 | 6.41E-04 | Regulation of cell cycle process |
| 7096 | 1.87E-02 | Regulation of exit from mitosis |
| 51641 | 2.92E-01 | Cellular localization |
| 70058 | 2.15E-03 | tRNA gene clustering |
| 51321 | 9.33E-02 | Meiotic cell cycle |
| 51327 | 8.74E-02 | M phase of meiotic cell cycle |
| 51340 | 7.13E-02 | Regulation of ligase activity |
| 51351 | 4.11E-02 | Positive regulation of ligase activity |
| 34622 | 9.16E-03 | Cellular macromolecular complex assembly |
| 65004 | 8.00E-06 | Protein-DNA complex assembly |
| 22414 | 1.60E-01 | Reproductive process |
| 48610 | 1.14E-01 | Reproductive cellular process |
| 7533 | 9.00E-04 | Mating type switching |
| 60968 | 1.19E-03 | Regulation of gene silencing |
| 31935 | 1.19E-03 | Regulation of chromatin silencing |
| 9058 | 1.37E-02 | Biosynthetic process |
| 8610 | 5.56E-01 | Lipid biosynthetic process |
| 10556 | 1.09E-02 | Regulation of macromolecule biosynthetic process |
| 45449 | 1.93E-03 | Regulation of transcription |
| 10604 | 1.43E-01 | Positive regulation of macromolecule metabolic process |
| 51247 | 2.24E-01 | Positive regulation of protein metabolic process |
| 44283 | 8.54E-01 | Small molecule biosynthetic process |
| 7088 | 2.51E-02 | Regulation of mitosis |
| 30071 | 1.99E-02 | Regulation of mitotic metaphase/anaphase transition |
| 51488 | 2.37E-02 | Activation of anaphase-promoting complex activity |
| 7092 | 2.37E-02 | Activation of mitotic anaphase-promoting complex activity |
| 22402 | 8.44E-15 | Cell cycle process |
| 7127 | 9.96E-02 | Meiosis I |
| 45786 | 2.20E-02 | Negative regulation of cell cycle |
| 31329 | 9.12E-02 | Regulation of cellular catabolic process |
| 61136 | 3.25E-02 | Regulation of proteasomal protein catabolic process |
| 16053 | 5.86E-01 | Organic acid biosynthetic process |
DEGs, differentially expressed genes.
Discussion
HURP, also known as DLGAP5 or KIAA0008, has been reported to be overexpressed in many types of human cancers, including hepatocellular carcinoma (6–8), bladder cell carcinoma (9), and transitional cell carcinoma (10). However, no clear evidence has been established to explore the role of HURP in NSCLC.
Initially identified as a potentially regulatory gene involved in the carcinogenesis of hepatocellular carcinoma (3), HURP is a microtubule-associated protein that functions in inducing novel tubulin sheet formation (18), facilitating spindle formation (19), and promoting the capture of spindle by kinetochore (20). As a cell cycle-regulated gene, the expression level of HURP changes periodically during the cell cycle, and reaches a peak at the G2/M phase and subsequently decreases in G1 (21). Degradation is modulated by Cdc2/cyclinB and SCF complex at the end of mitosis phase (21). On the other hand, HURP is phosphorylated by AURKA and remains stable from degradation when the cells progress from M to G1 (6).
Tsou et al (3) firstly reported that overexpression of HURP resulted in low serum-dependent and anchorage-independent growth, indicating that HURP plays a role in the carcinogenesis of cancer cells. Liao et al (22) showed that HURP is upregulated in hepatocellular cancer specimens when compared with adjacent liver tissues. Moreover, silencing of HURP expression resulted in suppressed cell growth, colony formation and migration in vitro, suggesting that HURP strongly promotes the malignant phenotype of hepatocellular cells. We investigated the prognostic value of HURP in NSCLC patients using bioinformatics. GSE33532 and GSE19188 revealed that HURP was overexpressed in lung tumor tissues when compared with the level in normal lung tissues. Analyses of GSE68465 and GSE30219 showed that the HURP expression level was correlated with pathological characteristics, reflecting the relation of HURP and the development of NSCLC. Importantly, there was a correlation between HURP and the prognosis of NSCLC patients. The higher the HURP expression, the shorter was the survival time of the patients. The results were consistent with Shi et al (15), who found that the expression level of HURP was negatively correlated with overall survival and relapse-free survival of NSCLC patients and could robustly distinguish lung cancer patients form normal subjects. Schneider et al (23) analyzed the expression of HURP in an independent large cohort of NSCLC patients and demonstrated that HURP was associated with poor overall survival in NSCLC patients, and arrived at the same conclusion as us.
To explore the potential role of HURP in modulating the proliferation of NSCLC cells, we silenced HURP by shRNA. MTT assays revealed that the cell viability was significantly reduced in the HURP-null group. Furthermore, cell cycle analysis and apoptosis analysis demonstrated that HURP regulated cell proliferation by cell cycle modulation instead of apoptosis regulation. There are numerous studies that have shown that disruption of cell cycle regulation is the most critical and frequent occurrence among the many altered pathways during lung carcinogenesis (24). In normal cells, cell division is a tightly regulated sophisticated process comprised of five stages: G0, G1, S, G2 and M stage. In other words, cell cycle regulation that requires the balance of growth factors and growth inhibitor factors, determines if cells enter the correct next stage. Deregulation of cell cycle leads to uncontrolled malignant proliferation and the molecules involved in cell cycle regulation have been reported as prognostic biomarkers and potential antitumor targets (25,26). Sterlacci et al (27) summarized various cell cycle-regulated molecules and their prognostic value in NSCLC. Except cyclins, which are the most investigated molecules participating in cell cycle regulation, other promising cell cycle-related factors are currently under research, including Aurora and Polo-like kinases (28). As the phosphorylated substrate of AURKA, HURP is involved in cell cycle regulation by inducing the novel tubulin sheet formation and facilitating spindle formation. Apart from regulating the spindle in the M stage, we found that HURP is involved in G1/S phase transition and M/G1 transition through GSEA analysis.
The cell migration and invasion assays demonstrated that depletion of HURP inhibited NSCLC migration and invasion in vitro, indicating that HURP promoted epithelial to mesenchymal transition and enhanced the invasive capacity of the NSCLC cells. Collectively, our clinical data and in vitro studies suggested that HURP may promote carcinogenesis in multiple ways. These findings corroborated the initial conclusions of Chen et al (14). The authors found that HURP was phosphorylated by AURKA and shuttled from the cytoplasm to the nucleus and engaged in the regulation of cyclin E1 expression by combining with NFκB. These observations revealed that the phosphorylated HURP combined with NFκB and activated the NFκB signaling pathway as a transcription co-regulator. Kuo et al (29) found that HURP induced malignant transformation through degradation of p53 and accumulation of gankyrin. HURP knockdown did not affect the proliferation of H1299 cells with depletion of p53. Furthermore, the expression of p53 in H1299 was decreased following overexpression of HURP, and these cells were resistant to cisplatin. Above all, HURP might promote carcinogenesis through different signaling pathways, and further studies should focus on the mechanism of HURP in NSCLC tumorigenicity.
In conclusion, we revealed the potential mechanism of HURP in facilitating the malignant phenotype of NSCLC cells and the prognostic value of HURP in NSCLC patients, indicating that HURP might be a potential therapeutic target of NSCLC.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Li L, Lu F, Zhang S. Analyses of variation trend and short-term detection of Chinese malignant tumor mortality during twenty years. Zhonghua Zhong Liu Za Zhi. 1997;19:3–9. (In Chinese) [PubMed] [Google Scholar]
- 3.Tsou AP, Yang CW, Huang CY, Yu RC, Lee YC, Chang CW, Chen BR, Chung YF, Fann MJ, Chi CW, et al. Identification of a novel cell cycle regulated gene, HURP, overexpressed in human hepatocellular carcinoma. Oncogene. 2003;22:298–307. doi: 10.1038/sj.onc.1206129. [DOI] [PubMed] [Google Scholar]
- 4.Segal NH, Blachere NE, Shiu HY, Leejee S, Antonescu CR, Lewis JJ, Wolchok JD, Houghton AN. Antigens recognized by autologous antibodies of patients with soft tissue sarcoma. Cancer Immun. 2005;5:4. [PubMed] [Google Scholar]
- 5.Gudmundsson KO, Thorsteinsson L, Sigurjonsson OE, Keller JR, Olafsson K, Egeland T, Gudmundsson S, Rafnar T. Gene expression analysis of hematopoietic progenitor cells identifies Dlg7 as a potential stem cell gene. Stem Cells. 2007;25:1498–1506. doi: 10.1634/stemcells.2005-0479. [DOI] [PubMed] [Google Scholar]
- 6.Yu CT, Hsu JM, Lee YC, Tsou AP, Chou CK, Huang CY. Phosphorylation and stabilization of HURP by Aurora-A: Implication of HURP as a transforming target of Aurora-A. Mol Cell Biol. 2005;25:5789–5800. doi: 10.1128/MCB.25.14.5789-5800.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao L, Qin LX, Ye QH, Zhu XQ, Zhang H, Wu X, Chen J, Liu YK, Tang ZY. KIAA0008 gene is associated with invasive phenotype of human hepatocellular carcinoma-a functional analysis. J Cancer Res Clin Oncol. 2004;130:719–727. doi: 10.1007/s00432-004-0595-2. [DOI] [PubMed] [Google Scholar]
- 8.Chang ML, Lin SM, Yeh CT. HURP expression-assisted risk scores identify prognosis distinguishable subgroups in early stage liver cancer. PLoS One. 2011;6:e26323. doi: 10.1371/journal.pone.0026323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang YL, Chiu AW, Huan SK, Wang YC, Ju JP, Lu CL. Prognostic significance of hepatoma-up-regulated protein expression in patients with urinary bladder transitional cell carcinoma. Anticancer Res. 2003;23:2729–2733. [PubMed] [Google Scholar]
- 10.Chiu AW, Huang YL, Huan SK, Wang YC, Ju JP, Chen MF, Chou CK. Potential molecular marker for detecting transitional cell carcinoma. Urology. 2002;60:181–185. doi: 10.1016/S0090-4295(02)01672-2. [DOI] [PubMed] [Google Scholar]
- 11.Tedeschi A, Ciciarello M, Mangiacasale R, Roscioli E, Rensen WM, Lavia P. RANBP1 localizes a subset of mitotic regulatory factors on spindle microtubules and regulates chromosome segregation in human cells. J Cell Sci. 2007;120:3748–3761. doi: 10.1242/jcs.009308. [DOI] [PubMed] [Google Scholar]
- 12.Uehara R, Goshima G. Functional central spindle assembly requires de novo microtubule generation in the interchromosomal region during anaphase. J Cell Biol. 2010;191:259–267. doi: 10.1083/jcb.201004150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu JM, Chen CT, Coumar MS, Lin WH, Chen ZJ, Hsu JT, Peng YH, Shiao HY, Lin WH, Chu CY, et al. Aurora kinase inhibitors reveal mechanisms of HURP in nucleation of centrosomal and kinetochore microtubules; Proc Natl Acad Sci USA; 2013; pp. E1779–E1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen JM, Chiu SC, Wei TY, Lin SY, Chong CM, Wu CC, Huang JY, Yang ST, Ku CF, Hsia JY, Yu CT. The involvement of nuclear factor-kappaB in the nuclear targeting and cyclin E1 upregulating activities of hepatoma upregulated protein. Cell Signal. 2015;27:26–36. doi: 10.1016/j.cellsig.2014.09.020. [DOI] [PubMed] [Google Scholar]
- 15.Shi YX, Yin JY, Shen Y, Zhang W, Zhou HH, Liu ZQ. Genome-scale analysis identifies NEK2, DLGAP5 and ECT2 as promising diagnostic and prognostic biomarkers in human lung cancer. Sci Rep. 2017;7:8072. doi: 10.1038/s41598-017-08615-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 17.Győrffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koffa MD, Casanova CM, Santarella R, Köcher T, Wilm M, Mattaj IW. HURP is part of a Ran-dependent complex involved in spindle formation. Curr Biol. 2006;16:743–754. doi: 10.1016/j.cub.2006.03.056. [DOI] [PubMed] [Google Scholar]
- 19.Santarella RA, Koffa MD, Tittmann P, Gross H, Hoenger A. HURP wraps microtubule ends with an additional tubulin sheet that has a novel conformation of tubulin. J Mol Biol. 2007;365:1587–1595. doi: 10.1016/j.jmb.2006.10.064. [DOI] [PubMed] [Google Scholar]
- 20.Casanova CM, Rybina S, Yokoyama H, Karsenti E, Mattaj IW. Hepatoma up-regulated protein is required for chromatin-induced microtubule assembly independently of TPX2. Mol Biol Cell. 2008;19:4900–4908. doi: 10.1091/mbc.E08-06-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsu JM, Lee YC, Yu CT, Huang CY. Fbx7 functions in the SCF complex regulating Cdk1-cyclin B-phosphorylated hepatoma up-regulated protein (HURP) proteolysis by a proline-rich region. J Biol Chem. 2004;279:32592–32602. doi: 10.1074/jbc.M404950200. [DOI] [PubMed] [Google Scholar]
- 22.Liao W, Liu W, Yuan Q, Liu X, Ou Y, He S, Yuan S, Qin L, Chen Q, Nong K, et al. Silencing of DLGAP5 by siRNA significantly inhibits the proliferation and invasion of hepatocellular carcinoma cells. PLoS One. 2013;8:e80789. doi: 10.1371/journal.pone.0080789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schneider MA, Christopoulos P, Muley T, Warth A, Klingmueller U, Thomas M, Herth FJ, Dienemann H, Mueller NS, Theis F, Meister M. AURKA, DLGAP5, TPX2, KIF11 and CKAP5: Five specific mitosis-associated genes correlate with poor prognosis for non-small cell lung cancer patients. Int J Oncol. 2017;50:365–372. doi: 10.3892/ijo.2017.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singhal S, Amin KM, Kruklitis R, DeLong P, Friscia ME, Litzky LA, Putt ME, Kaiser LR, Albelda SM. Alterations in cell cycle genes in early stage lung adenocarcinoma identified by expression profiling. Cancer Biol Ther. 2003;2:291–298. doi: 10.4161/cbt.2.5.514. [DOI] [PubMed] [Google Scholar]
- 25.Sterlacci W, Tzankov A, Veits L, Zelger B, Bihl MP, Foerster A, Augustin F, Fiegl M, Savic S. A comprehensive analysis of p16 expression, gene status, and promoter hypermethylation in surgically resected non-small cell lung carcinomas. J Thorac Oncol. 2011;6:1649–1657. doi: 10.1097/JTO.0b013e3182295745. [DOI] [PubMed] [Google Scholar]
- 26.Sterlacci W, Fiegl M, Hilbe W, Jamnig H, Oberaigner W, Schmid T, Augustin F, Auberger J, Obermann EC, Tzankov A. Deregulation of p27 and cyclin D1/D3 control over mitosis is associated with unfavorable prognosis in non-small cell lung cancer, as determined in 405 operated patients. J Thorac Oncol. 2010;5:1325–1336. doi: 10.1097/JTO.0b013e3181e77efc. [DOI] [PubMed] [Google Scholar]
- 27.Sterlacci W, Fiegl M, Tzankov A. Prognostic and predictive value of cell cycle deregulation in non-small-cell lung cancer. Pathobiology. 2012;79:175–194. doi: 10.1159/000336462. [DOI] [PubMed] [Google Scholar]
- 28.Tomonaga T, Nomura F. Chromosome instability and kinetochore dysfunction. Histol Histopathol. 2007;22:191–197. doi: 10.14670/HH-22.191. [DOI] [PubMed] [Google Scholar]
- 29.Kuo TC, Chang PY, Huang SF, Chou CK, Chao CC. Knockdown of HURP inhibits the proliferation of hepa-cellular carcinoma cells via downregulation of gankyrin and accumulation of p53. Biochem Pharmacol. 2012;83:758–768. doi: 10.1016/j.bcp.2011.12.034. [DOI] [PubMed] [Google Scholar]