Abstract
Advanced gastric cancer (GC) has a poor prognosis and its treatment strategies are not very efficient. Heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1) has emerged as a plausible GC marker, however the role and molecular mechanism of hnRNPA1 in cell invasion and migration remains unknown. In the present study, the gene expression across normal and tumor tissue (GENT) database was used to evaluate the mRNA expression of hnRNPA1 in various types of cancer. Western blot analysis (WB) and immunohistochemistry (IHC) were performed to detect the protein expression of hnRNPA1 in GC tissues and adjacent non-tumor tissues. The expression of multiple oncogenes was detected by western blot analysis and quantitative RT-PCR in hnRNPA1 overexpressing GC cells. Soft agar colony formation, EdU incorporation, wound healing and invasion assays were applied to verify the role of hnRNPA1 in anchorage-independent cell growth, migration and invasion in GC cells. Epithelial-to-mesenchymal transition (EMT) markers were detected by immunofluorescence, western blot analysis and IHC in vitro. A nude mice model of metastasis carcinoma was established to confirm the role of hnRNPA1 during EMT in vivo. Our results revealed that hnRNPA1 was significantly upregulated in GC tissue. HnRNPA1 overexpression significantly induced cell growth, migration and invasion ability in GC cells. In addition, hnRNPA1 promoted EMT of GC cells in vitro and in vivo. These findings indicated that hnRNPA1 is highly expressed in GC and promoted invasion by inducing EMT transition in GC cells. Thus, hnRNPA1 may be a potential therapeutic target for GC.
Keywords: hnRNPA1, oncogene, epithelial-to-mesenchymal transition, therapeutic target, gastric cancer
Introduction
Gastric cancer (GC) is the fourth most common cancer and advanced GC has a poor prognosis and high mortality rate worldwide (1,2). GC progresses through a multistep process, whereby cells acquire a series of genetic and epigenetic alterations in key growth regulatory genes that endow them with proliferative and survival advantages (3,4). Early stage GC patients usually do not present with specific symptoms. Thus, most GC patients are diagnosed at an advanced stage with lymph node or organ metastasis, resulting in a low 5-year survival rate, ranging from 20 to 30% (5,6). Conventional treatments for GC include surgery, radiotherapy and chemotherapy. However, there is a need for more efficacious therapies and treatment strategies.
Heterogeneous nuclear ribonucleoproteins (hnRNPs) are a large family of RNA-binding nuclear proteins in mammalian cells. There are more than 20 members in hnRNPs (7–9), all of which are associated with precursor mRNAs and many of which influence pre-mRNA processing and other functions of mRNA. The best-characterized protein in this family is hnRNPA1 (10,11), which is aberrantly expressed in various cancers (12,13). Liu et al (14) found that hnRNPA1 protein was overexpressed in lung cancer (LC), whereas knockdown of hnRNPA1 inhibited cell viability and colony formation of LC cells and arrested the cell cycle in the G0/G1 phase. Furthermore, another study revealed the upregulated expression and aberrant cytoplasmic localization of hnRNPA1 in cervical squamous cell cancer (15). Recently, hnRNPA1 has emerged as a plausible GC biomarker (16). However, the role and molecular mechanism of hnRNPA1 in GC invasion and migration are not well defined.
Based on these previous studies, we hypothesized that hnRNPA1 is important in the development of GC. Therefore, bioinformatics combined with in vivo and in vitro experiments were performed to characterize the effect of hnRNPA1 on the invasive biological behavior of GC.
Materials and methods
Cell lines and tissue specimens
Mouse anti-hnRNPA1 (4B10; cat. no. sc-32301) and rabbit anti-E-cadherin (H-108; cat. no. sc-7870) were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). Rabbit anti-vimentin (Ag0489; cat. no. 65039-1-Ig) and mouse anti-human GAPDH (cat. no. HRP-60004) were purchased from ProteinTech Group, Inc. (Wuhan, China). Rabbit anti-Snail (cat. no. ab110490) was purchased from Abcam (Cambridge, UK). Human gastric carcinoma cell lines AGS (American Type Culture Collection, Rockville, MD, USA) and BGC-823 cell lines (Beijing Institute of Cancer Research, Beijing, China) were maintained in Dulbecco's modified Eagle's medium (DMEM) basic media containing 10% fetal calf serum (FCS; Invitrogen Life Technologies, Carlsbad, CA, USA) at 37°C and 5% CO2. Seven pairs of GC tissues and normal gastric tissues were collected. In radical resection of GC, the surgeon usually resects at least 5 cm away from the tumor as the cut edge to ensure that the boundary is pathologically negative. Subsequently, the tissue is stained with hematoxylin and eosin (H&E) and the pathologist judges whether the cut edge has been invaded by tumor cells. The tissue we used to perform western blotting was chosen by this criterion and it was clinically pathologically confirmed. All tissue slides used to perform H&E staining and IHC analysis were examined independently by two experienced pathologists. All patients signed informed consents for the use of their tissues and the present study was approved by the Institutional Review Board of the Gannan Medical University.
RNA isolation and quantitative real-time PCR
The cells were harvested and total RNA was extracted using TRIzol reagent (Gibco-BRL; Thermo Fischer Scientific, Gaithersburg, MD, USA) as previously described (17,18). The primer sequences in RT-PCR were as follows: hnRNPA1 forward, 5′-TGCCCAGAAAATGAAAAAGG-3′ and reverse, 5′-GTGTATGTGGCAATGCGTTC-3′ (201 bp); GAPDH forward, 5′-GTCAACGGATTTGGTCGTATTG-3′ and reverse, 5′-CTCCTGGAAGATGGTGATGGG-3′ (216 bp); c-jun forward, 5′-CCCCAAGATCCTGAAACAGA-3′ and reverse, 5′-CCGTTGCTGGACTGGATTAT-3′ (168 bp); Survivin forward, 5′-TGTCTTGAAAGTGGCACCAG-3′ and reverse, 5′-GCCTTCTTCCCCCTCACTT-3′ (154 bp); SNAI1 forward, 5′-TTTACCTTCCAGCAGCCCTA-3′ and reverse, 5′-CCCACTGTCCTCATCTGACA-3′ (207 bp); Cyclin D1 forward, 5′-CGTGGCCTCTAAGATGAAGG-3′ and reverse, 5′-CTGGCATTTTGGAGAGGAAG-3′ (185 bp); ZEB1 forward, 5′-CGCTTTACCTCTCTGAAAGAACA-3′ and reverse, 5′-TTACACCCAGACTGCGTCAC-3′ (170 bp).
Transient siRNA transfection
Knockdown of hnRNPA1 expression was performed by transfecting cells with small interfering RNA (siRNA) duplexes (sense strand: 632-GCCACAACTGTGAAGTTAGAA-653, synthesized by GenePharma Co., Shanghai, China) using Lipofectamine 2000 (Invitrogen; Thermo Fischer Scientific). Scrambled RNA (scr-siRNA) was used as a negative control. Subsequently, 48 h post-transfection, western blot analysis was performed.
Constructs and generation of stable transfectants
cDNA corresponding to full-length hnRNPA1 was obtained by RT-PCR amplification of normal human testis cDNA with primers specific to hnRNPA1. PCR aliquots were subcloned into the pcDNA3.1 mammalian expression vector (Invitrogen; Thermo Fischer Scientific). pcDNA 3.1 or pcDNA3.1-hnRNPA1 were transfected into BCG823 and AGS cells. Transfects were cultured by RPMI-1640 medium supplemented with Geneticin (G418; Calbiochem; Merck KGaA, Darmstadt, Germany) to generate stable cell lines.
Western blot analysis and immunofluorescence
Whole cell lysates were prepared as previously described (19,20). For western blot analysis, 30 µg whole protein lysates were used to detect the indicated protein. For the immunofluorescence assay, the cells on a cover glass were fixed and incubated with primary antibodies followed by Texas Red (cat. no. SAB3700022; Sigma-Aldrich, Darmstadt, Germany) or fluorescein isothiocyanate-conjugated secondary antibodies (cat. no. PA1-85440; Thermo Fisher Scientific, Inc., Waltham, MA, USA). Nuclei were stained with Hoechst 33258 nuclear staining dye. Coverslips were washed, mounted and then analyzed using a fluorescence microscope.
Anchorage-independent cell growth assay
Scr-siRNA and hnRNPA1-siRNA cells (5×103) were seeded into medium with 0.35% agar, plated in triplicate on plates containing 0.7% agar base. Colonies were stained with Coomassie Blue (cat. no. sc-24972; Santa Cruz Biotechnology, Inc.). Colonies containing at least 50 cells were counted using Photoshop software.
EdU incorporation assay
Each group of isolated tumor cells was seeded onto 96-well plates in triplicate and incubated for 48 h in normal medium, and then for an additional 2 h in medium containing 50 µM 5-ethynyl-2′-deoxyuridine (EdU; Guangzhou RiboBio, Co., Ltd., Guangzhou, China). After being washed, fixed and permeabilized, the cells were incubated with 1X Apollo reaction cocktail (Guangzhou RiboBio Co., Ltd.) for 30 min. DNA was incubated with Hoechst 33342 nuclear staining dye for 30 min and visualized with an inverted fluorescence microscope (Leica DM5500; Leica, Stuttgart, Germany). Five random fields were imaged at a ×100 magnification for each EdU experiment. Captured images were processed and analyzed with ImageJ software (National Institutes of Health, Bethesda, MD, USA). The number of EdU-positive cells was identified by Hoechst 33342 nuclei staining and expressed as a percentage of the total number of cells in each field.
Cell migration and invasion assays
Cell migration was assessed by wound healing assay as previously described (21). Briefly, indicated cells with 24-h culture were wounded with a pipette tip. The media was changed to remove cell debris and images were captured after 60 h. The cell invasion was assessed using the Matrigel Invasion Chamber (BD Biosciences, San Jose, CA, USA) according to the manufacturer's instructions. Twenty-four hour post-siRNA transfection, the cells were re-suspended in serum-free media and placed on each Transwell membrane filter insert, with the lower chamber filled with complete medium. After 24-h incubation, invaded cells were stained with crystal violet and counted under a microscope.
Construction of lentiviral vectors with hnRNPA1 short hairpin RNA
An hnRNPA1 RNAi lentiviral vector (pGCSIL-hnRNPA1-shRNA) was constructed (Shanghai GeneChem Co., Ltd., Shanghai, China) to investigate the effect of knockdown of hnRNPA1 expression on the metastasis of GC in vivo. Double-stranded oligonucleotides encoding human hnRNPA1-vshRNA (NM_002136, 5′-GCCACAACTGTGAAGTTAGAACTCGAGTTCTAACTTCACAGTTGTGGCTTTTT-3′) were annealed and inserted into the pGCSIL-GFP short hairpin RNA (shRNA) expression vector. A GFP-lentiviral vector (pGCSIL-GFP) was used as the negative control.
In vivo metastasis assays
Four- to 6-week-old BALB/c-nu/nu nude mice were obtained from the Laboratory Animal Unit, Southern Medical University (Baiyun, China). Four hundred thousand cells from each group (pcDNA3.1 shRNA and pcDNA3.1-hnRNPA1 shRNA lentiviral) were injected into mice through the tail vein (n=3 for each group). Following 35 days, the mice were sacrificed, the lungs were harvested and images were captured. Tissue sections were attained by traditional methods and H&E, IHC and qPCR assays were performed. All experiments performed followed the internationally recognized guidelines, as well as the local and national regulations for animal experiments. Institutional Review Board of the Gannan Medical University approved the experiments.
Statistical analysis
All data are presented as the mean ± standard deviation of at least three independent experiments, using SPSS 16.0 version (SPSS, Inc., Chicago, IL, USA). Groups with different treatments were compared using the two-tailed Student's t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
HnRNPA1 expression is higher in human GC tissues
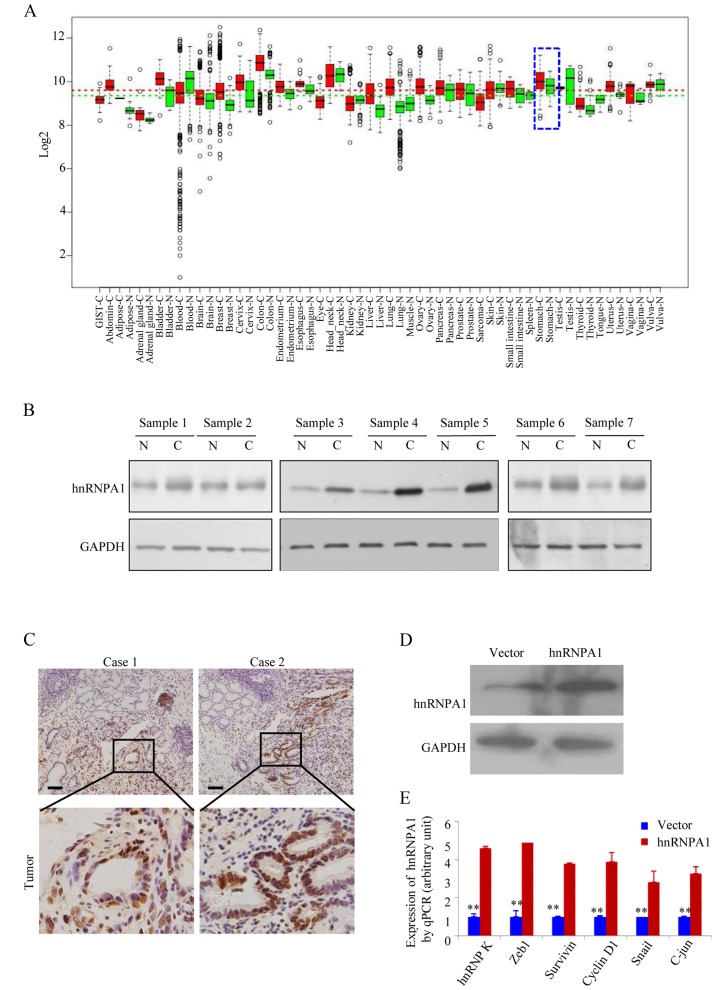
In the GENT database (available at http://medicalgenome.kribb.re.kr/GENT/ or http://genome.kobic.re.kr/GENT/), hnRNPA1 is upregulated in adrenal gland, bladder, breast, cervix uteri, colon, endometrium, esophagus, liver, lung, ovary, pancreas, prostate, small intestine, thyroid, uterus and stomach cancer compared with corresponding normal tissues (Fig. 1A). This finding indicated that hnRNPA1 may be associated with various types of cancer including gastric cancer. We examined the expression of hnRNPA1 in seven pairs of human GC tissues and matched non-cancerous colonic mucosa by western blotting. As displayed in Fig. 1B, all of the cancer tissues exhibited a higher expression level of hnRNPA1 in relation to their corresponding non-cancerous controls (Fig. 1B). Higher expression levels of hnRNPA1 protein in GC tissues from two patients were also confirmed by IHC (Fig. 1C). To assess the effect of constitutive hnRNPA1 expression on the malignant behavior of GC cells in vitro, we established stable transfectants with hnRNPA1-sense and vector plasmids (Fig. 1D) and confirmed the induced expression of five major oncogenes involved in the proliferation and transformation (Fig. 1E). The mRNA expression of endogenous ZEB1, survivin, cyclin D1, Snail and c-jun genes was upregulated in the stable hnRNPA1 transfectants of AGS cells. These results indicated that hnRNPA1 is overexpressed and is involved in GC tumorigenesis.
Figure 1.
HnRNPA1 expression in GC is higher than in normal cells. (A) GENT database was used to find the hnRNPA1 mRNA expression in various types of cancer. Boxes represent the median and 25th and 75th percentiles, dots represent outliers, red boxes represent tumor tissues, green boxes represent normal tissues and red and green dashed lines represent the average value of all tumor and normal tissues, respectively. HnRNPA1 mRNA expression in gastric tissue, represented by blue dotted lines. (B) Resected tumors and adjacent non-tumor tissue specimens were lysed and hnRNPA1 protein expression was detected by western blot analysis. C, gastric carcinoma; N, normal gastric tissues (n=7). (C) In two selected patient cases, higher expression of hnRNPA1 in tumor tissues was confirmed by IHC. Scale bars, 100 µm. (D) Western blotting was used to access the expression of hnRNPA1 in stable transfectants. (E) The expression of multiple oncogenes in vector and hnRNPA1 stable cells was detected by qPCR. **P<0.05.
HnRNPA1 enhances the malignant biological behavior of GC cells
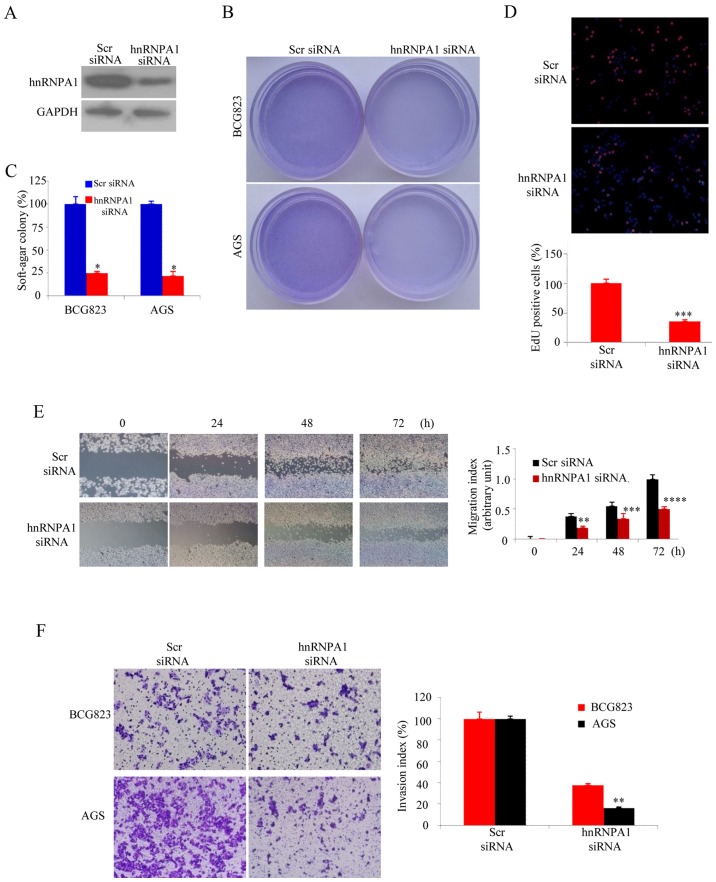
The soft agar assay was performed as anchorage-independent growth factor to evaluate hnRNPA1 function in malignant transformation (22). The siRNA-knockdown efficiency was confirmed by western blotting (Fig. 2A). As expected, hnRNPA1 downregualtion significantly inhibited the colony forming capacity of both BCG823 and AGS GC cells (Fig. 2B and C). The EdU incorporation assays revealed that hnRNPA1 supression was significantly lower than that of AGS cells containing scr-siRNA (Fig. 2D). In order to elevate hnRNPA1 downregulation on the metastatic potential of GC cells, wound healing assay was performed. Compared with the cells transfected with scr-siRNA, those transfected with hnRNPA1-siRNA exhibited significantly decreased migration (Fig. 2E). To examine cell invasion in vitro, we used matrix-coated cell culture inserts. After the downregulation of hnRNPA1, the invasiveness of BCG823 and AGS cells decreased by ~2.5- and ~6-fold, respectively, compared with the control cells (Fig. 2F). These data indicated that hnRNPA1 downregulation inhibited the malignant biological behavior of GC cells.
Figure 2.
HnRNPA1 facilitates the malignant biological behavior of GC cells. (A) HnRNPA1 expression was detected by western blot analysis in BCG823 and AGS cells. (B and C) HnRNPA1-silenced GC cells and scr siRNA cells were plated individually in soft agar culture dishes with complete culture medium. Cell colonies were visualized after 12 days. *P<0.05. (D) DNA synthesis of the BCG823 cells was assessed using an EdU incorporation assay after transfection at 48 h. ***P<0.01, between hnRNPA1 and Scr siRNA. (E) Monolayer BCG823 cells transfected with the indicated siRNA were wounded and images at different time-points were captured. **P<0.05, ***P<0.01, ****P<0.001. (F) The invasive potential of the BCG823 and AGS cells transfected with Scr siRNA or hnRNPA1 siRNA was evaluated by invasion chamber and assessed by invasion index. **P<0.05. These experiments were repeated at least three times.
HnRNPA1 upregulation promotes metastasis through EMT in GC
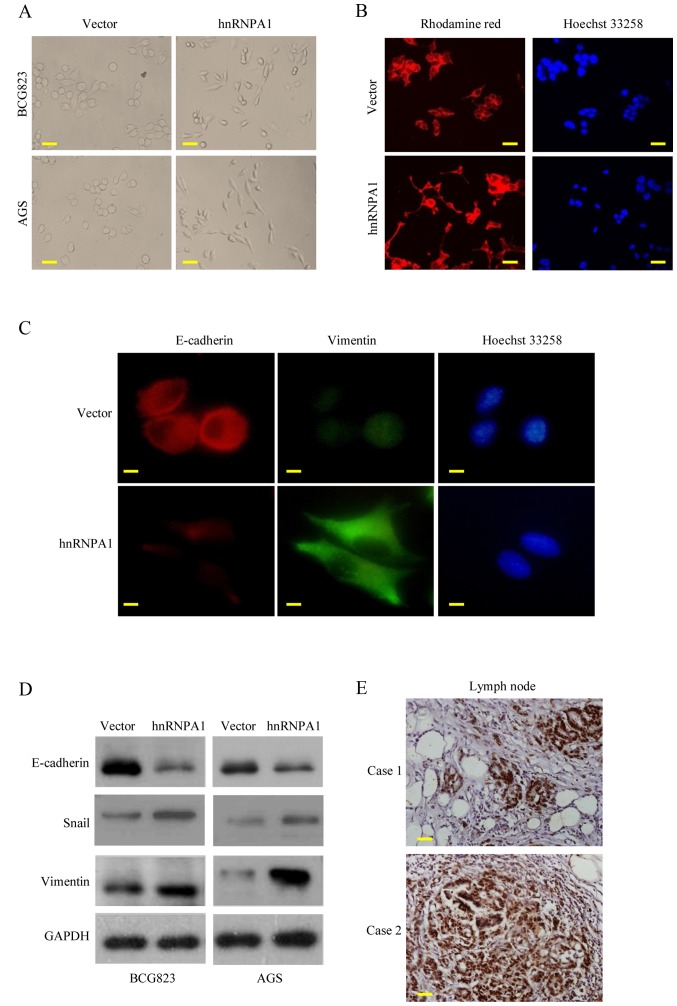
EMT is the key process driving cancer metastasis (23,24). To evaluate the role of hnRNPA1 in EMT, we examined the morphologic features of GC cells. HnRNPA1 overexpression induced the loss of cell-cell contact and a cobblestone-like phenotype. The cells became elongated, spindle-shaped and scattered. In contrast, empty vector transfectants had a round or flat morphology with a short cytoplasmic process (Fig. 3A). Furthermore, compared with vector-expressing cells, F-actin staining of the hnRNPA1-overexpressing cells was observed throughout the cytoplasm and at the rim zone of the protrusion. Concurrently, filopodia and lamellipodia were present on the hnRNPA1-overexpressing cell membrane surfaces and were required for actin polymerization and involved in the invasion and metastasis of cancer cells (Fig. 3B). Concurrently, the expression of EMT markers was assessed. E-cadherin expression was decreased, while vimentin expression was increased in the hnRNPA1-overexpressing cells, as determined by immunofluorescent analysis (Fig. 3C). HnRNPA1 overexpression was also confirmed by western blot analysis. EMT induction was demonstrated by a shift from the expression of epithelial markers (E-cadherin) to mesenchymal markers (vimentin and Snail) (Fig. 3D). To further confirm these findings in vivo, we detected the hnRNPA1 expression of regional lymph nodes in metastatic cancer tissues from two patients. High levels of hnRNPA1 in the nucleus of the cancer cells were investigated (Fig. 3E). These findings demonstrated that hnRNPA1 induced EMT and facilitated GC cell invasion.
Figure 3.
HnRNPA1 regulates EMT in vitro. (A) Morphology of stable vector and hnRNPA1 transfected cells (BCG823 and AGS) as visualized under a phase-contrast microscope. (B) BCG823 cells carried with hnRNPA1 or vector stable cells stained with rhodamine-phallotoxin were visualized by fluorescent microscopy to access the expression of F-actin. (C) BCG823 transfected with hnRNPA1 or vector cells were stained by E-cadherin (red) and vimentin (green) primary antibody and then second antibodies individually, fluorescence microscope visualized the expression changes of E-cadherin and vimentin. (D) E-cadherin, Snail and vimentin expression in BCG823 stable cells were detected by western blotting. (E) Two GC tissues with lymph node metastatic sites were stained by hnRNPA1 as the representative IHC results. These experiments were repeated three times with identical findings. Scale bars represent 50 µm in A and B; 20 µm in C and 100 µm in E.
HnRNPA1 induces EMT and metastasis in GC in vivo
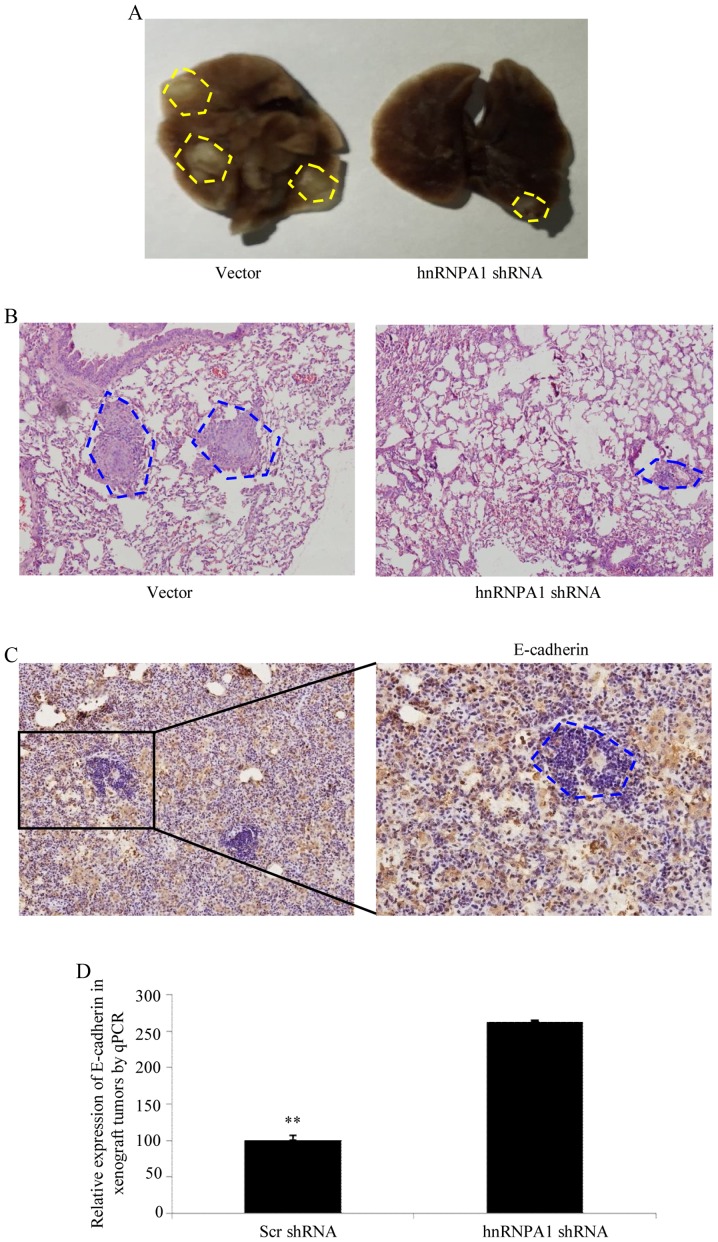
To verify the aforementioned results in vivo, shRNA-mediated silencing of hnRNPA1 was performed in BCG823 cells and an orthotopic lung cancer model was established in mice by tail vein injections (Fig. 4A). Compared with scr-shRNA transfection in BCG823 cells, cells transfected with hnRNPA1-shRNA exhibited a significant decrease in visible lung tumors, which correlated with a higher number of metastatic loci (Fig. 4B). The expression of E-cadherin in tumors was determined by IHC (Fig. 4C). Furthermore, orthotopic xenograft tumors with depletion of the hnRNPA1 expression exhibited increased E-cadherin levers compared with scr-shRNA at the mRNA level (Fig. 4D). Collectively, these results demonstrated that hnRNPA1 is important in EMT and metastasis progression in GC.
Figure 4.
HnRNPA1 promotes tumor metastasis in vivo. (A) Mice were orthotopically transplanted with the indicated cells (n=3 in each group). Representative images of metastatic loci in the lungs are displayed in yellow dotted lines. (B) H&E staining was used to verify the metastatic cancer tissues from sacrificed mice. (C) The expression of E-cadherin in tumors derived from BCG823 cells was determined by IHC. (D) qPCR was performed to evaluated the expression of E-cadherin in different group of tumor tissues. **P<0.01, Scr-shRNA vs. hnRNPA1-shRNA. Scale bars represent 200 µm in B and 100 µm in C.
Discussion
The present study characterized the role of hnRNPA1 in GC cell growth and invasion both in vivo and in vitro. Elevated levels of hnRNPA1 protein were detected in most GC tissues compared to normal tissues. Silencing of hnRNPA1 resulted in reduced cell growth, invasion, migration and reversal of EMT in GC cells. Collectively, these findings indicated that hnRNPA1 plays a pivotal role in GC invasion and metastasis.
HnRNPA1 is known to be involved in cancer progression and metastasis (25,26). However, few studies have focused on its role in GC. In the present study, we found that GC tissues had higher levels of hnRNPA1 compared with normal tissues and hnRNPA1 knockdown significantly inhibited anchorage-dependent growth in GC cells. These data indicated that hnRNPA1 was important to cell growth and progression of GC.
EMT is a crucial phenotypic conversion during cancer progression (27,28) and is a process by which cancer cells lose their polarized epithelial structures and acquire plastic and high motile mesenchymal properties (29,30). Clinicopathological studies revealed that EMT strongly correlated with poor histological differentiation, destruction of tissue integrity and metastasis (31,32). Therefore, EMT is considered to be an exclusive phenotypic switch for the invasion and metastasis of cancers including GC (33). EMT-gene signature included a decrease of epithelial molecules such as E-cadherin, an increase of mesenchymal molecules such as vimentin and an alteration of the localization and/or function of some transcription factors such as ZEB1 (33) and Snail (34). A previous study (27) demonstrated that hnRNPA1 promoted breast cancer progression through the induction of EMT and provided potential targets to develop anticancer therapies. In the present study, we revealed that GC cells with high hnRNPA1 expression exhibited amoeboid morphology and shifted changes of the EMT markers, indicating that hnRNPA1 may be a potent inducer of EMT and promote the invasion and metastasis in GC. Furthermore, recent studies have hypothesized that mesenchymal-epithelial transition (MET) was essential for the successful seeding and outgrowth of distant metastasis. Therefore, we investigated the expression of EMT markers in metastatic lung nodules from xenograft tumor models by IHC and found that MET was essential for hnRNPA1-induced lung metastasis as expected.
In conclusion, our results revealed a novel and unexpected regulatory function of hnRNPA1. HnRNPA1 may be a mediator or trigger of EMT and may cause dissemination and metastatic phenotypes in GC. Thus, the data of the present study provided evidence that hnRNPA1 played a vital role in GC growth and progression, and as such, may serve as a therapeutic target for GC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the NSFC of Jiangxi Province.
Availability of data and materials
The analyzed datasets generated during the study are available from the corresponding author on reasonable request.
Authors' contributions
YC, JL, WW and LX performed the experiments, JW, SL and ZG designed the study, HZ and ZG prepared and wrote the study. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Carcas LP. Gastric cancer review. J Carcinog. 2014;13:14. doi: 10.4103/1477-3163.146506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Houghton J, Fox JG, Wang TC. Gastric cancer: Laboratory bench to clinic. J Gastroenterol Hepatol. 2002;17:495–502. doi: 10.1046/j.1440-1746.2002.02770.x. [DOI] [PubMed] [Google Scholar]
- 3.Strickler JG, Zheng J, Shu Q, Burgart LJ, Alberts SR, Shibata D. p53 mutations and microsatellite instability in sporadic gastric cancer: When guardians fail. Cancer Res. 1994;54:4750–4755. [PubMed] [Google Scholar]
- 4.Zheng L, Wang L, Ajani J, Xie K. Molecular basis of gastric cancer development and progression. Gastric Cancer. 2004;7:61–77. doi: 10.1007/s10120-004-0277-4. [DOI] [PubMed] [Google Scholar]
- 5.Zeng H, Zheng R, Guo Y, Zhang S, Zou X, Wang N, Zhang L, Tang J, Chen J, Wei K, et al. Cancer survival in China, 2003–2005: A population-based study. Int J Cancer. 2015;136:1921–1930. doi: 10.1002/ijc.29227. [DOI] [PubMed] [Google Scholar]
- 6.Sasako M, Sano T, Yamamoto S, Kurokawa Y, Nashimoto A, Kurita A, Hiratsuka M, Tsujinaka T, Kinoshita T, Arai K, et al. D2 lymphadenectomy alone or with para-aortic nodal dissection for gastric cancer. N Engl J Med. 2008;359:453–462. doi: 10.1056/NEJMoa0707035. [DOI] [PubMed] [Google Scholar]
- 7.Chen HH, Chang JG, Lu RM, Peng TY, Tarn WY. The RNA binding protein hnRNP Q modulates the utilization of exon 7 in the survival motor neuron 2 (SMN2) gene. Mol Cell Biol. 2008;28:6929–6938. doi: 10.1128/MCB.01332-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ostareck-Lederer A, Ostareck DH, Cans C, Neubauer G, Bomsztyk K, Superti-Furga G, Hentze MW. c-Src-mediated phosphorylation of hnRNP K drives translational activation of specifically silenced mRNAs. Mol Cell Biol. 2002;22:4535–4543. doi: 10.1128/MCB.22.13.4535-4543.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou ZJ, Dai Z, Zhou SL, Fu XT, Zhao YM, Shi YH, Zhou J, Fan J. Overexpression of HnRNP A1 promotes tumor invasion through regulating CD44v6 and indicates poor prognosis for hepatocellular carcinoma. Int J Cancer. 2013;132:1080–1089. doi: 10.1002/ijc.27742. [DOI] [PubMed] [Google Scholar]
- 10.Sueoka E, Goto Y, Sueoka N, Kai Y, Kozu T, Fujiki H. Heterogeneous nuclear ribonucleoprotein B1 as a new marker of early detection for human lung cancers. Cancer Res. 1999;59:1404–1407. [PubMed] [Google Scholar]
- 11.Loh TJ, Moon H, Cho S, Jang H, Liu YC, Tai H, Jung DW, Williams DR, Kim HR, Shin MG, et al. CD44 alternative splicing and hnRNP A1 expression are associated with the metastasis of breast cancer. Oncol Rep. 2015;34:1231–1238. doi: 10.3892/or.2015.4110. [DOI] [PubMed] [Google Scholar]
- 12.Yu C, Guo J, Liu Y, Jia J, Jia R, Fan M. Oral squamous cancer cell exploits hnRNP A1 to regulate cell cycle and proliferation. J Cell Physiol. 2015;230:2252–2261. doi: 10.1002/jcp.24956. [DOI] [PubMed] [Google Scholar]
- 13.Torosyan Y, Dobi A, Glasman M, Mezhevaya K, Naga S, Huang W, Paweletz C, Leighton X, Pollard HB, Srivastava M. Role of multi-hnRNP nuclear complex in regulation of tumor suppressor ANXA7 in prostate cancer cells. Oncogene. 2010;29:2457–2466. doi: 10.1038/onc.2010.2. [DOI] [PubMed] [Google Scholar]
- 14.Liu X, Zhou Y, Lou Y, Zhong H. Knockdown of HNRNPA1 inhibits lung adenocarcinoma cell proliferation through cell cycle arrest at G0/G1 phase. Gene. 2016;576:791–797. doi: 10.1016/j.gene.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 15.Qing S, Tulake W, Ru M, Li X, Yuemaier R, Lidifu D, Rouzibilali A, Hasimu A, Yang Y, Rouziahong R, et al. Proteomic identification of potential biomarkers for cervical squamous cell carcinoma and human papillomavirus infection. Tumour Biol. 2017;39:1010428317697547. doi: 10.1177/1010428317697547. [DOI] [PubMed] [Google Scholar]
- 16.Hermann R, Hensel F, Müller EC, Keppler M, Souto-Carneiro M, Brändlein S, Müller-Hermelink HK, Vollmers HP. Deactivation of regulatory proteins hnRNP A1 and A2 during SC-1 induced apoptosis. Hum Antibodies. 2001;10:83–90. [PubMed] [Google Scholar]
- 17.Guo Z, Zhang W, Xia G, Niu L, Zhang Y, Wang X, Zhang Y, Jiang B, Wang J. Sp1 upregulates the four and half lim 2 (FHL2) expression in gastrointestinal cancers through transcription regulation. Mol Carcinog. 2010;49:826–836. doi: 10.1002/mc.20659. [DOI] [PubMed] [Google Scholar]
- 18.Wu Y, Guo Z, Zhang D, Zhang W, Yan Q, Shi X, Zhang M, Zhao Y, Zhang Y, Jiang B, et al. A novel colon cancer gene therapy using rAAV-mediated expression of human shRNA-FHL2. Int J Oncol. 2013;43:1618–1626. doi: 10.3892/ijo.2013.2090. [DOI] [PubMed] [Google Scholar]
- 19.Zhang W, Guo Z, Jiang B, Niu L, Xia G, Wang X, Cheng T, Zhang Y, Wang J. Identification of a functional p53 responsive element within the promoter of XAF1 gene in gastrointestinal cancer cells. Int J Oncol. 2010;36:1031–1037. doi: 10.3892/ijo_00000584. [DOI] [PubMed] [Google Scholar]
- 20.Guo Z, Zhou Y, Evers BM, Wang Q. Rictor regulates FBXW7-dependent c-Myc and cyclin E degradation in colorectal cancer cells. Biochem Biophys Res Commun. 2012;418:426–432. doi: 10.1016/j.bbrc.2012.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang W, Jiang B, Guo Z, Sardet C, Zou B, Lam CS, Li J, He M, Lan HY, Pang R, et al. Four-and-a-half LIM protein 2 promotes invasive potential and epithelial-mesenchymal transition in colon cancer. Carcinogenesis. 2010;31:1220–1229. doi: 10.1093/carcin/bgq094. [DOI] [PubMed] [Google Scholar]
- 22.Shin KH, Kang MK, Kim RH, Christensen R, Park NH. Heterogeneous nuclear ribonucleoprotein G shows tumor suppressive effect against oral squamous cell carcinoma cells. Clin Cancer Res. 2006;12:3222–3228. doi: 10.1158/1078-0432.CCR-05-2656. [DOI] [PubMed] [Google Scholar]
- 23.Gumireddy K, Li A, Gimotty PA, Klein-Szanto AJ, Showe LC, Katsaros D, Coukos G, Zhang L, Huang Q. KLF17 is a negative regulator of epithelial-mesenchymal transition and metastasis in breast cancer. Nat Cell Biol. 2009;11:1297–1304. doi: 10.1038/ncb1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang MH, Wu MZ, Chiou SH, Chen PM, Chang SY, Liu CJ, Teng SC, Wu KJ. Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat Cell Biol. 2008;10:295–305. doi: 10.1038/ncb1691. [DOI] [PubMed] [Google Scholar]
- 25.Park WC, Kim HR, Kang DB, Ryu JS, Choi KH, Lee GO, Yun KJ, Kim KY, Park R, Yoon KH, et al. Comparative expression patterns and diagnostic efficacies of SR splicing factors and HNRNPA1 in gastric and colorectal cancer. BMC Cancer. 2016;16:358. doi: 10.1186/s12885-016-2387-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou B, Wang Y, Jiang J, Jiang H, Song J, Han T, Shi J, Qiao H. The long noncoding RNA colon cancer-associated transcript-1/miR-490 axis regulates gastric cancer cell migration by targeting hnRNPA1. IUBMB Life. 2016;68:201–210. doi: 10.1002/iub.1474. [DOI] [PubMed] [Google Scholar]
- 27.Bonomi S, di Matteo A, Buratti E, Cabianca DS, Baralle FE, Ghigna C, Biamonti G. HnRNP A1 controls a splicing regulatory circuit promoting mesenchymal-to-epithelial transition. Nucleic Acids Res. 2013;41:8665–8679. doi: 10.1093/nar/gkt579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tauler J, Zudaire E, Liu H, Shih J, Mulshine JL. hnRNP A2/B1 modulates epithelial-mesenchymal transition in lung cancer cell lines. Cancer Res. 2010;70:7137–7147. doi: 10.1158/0008-5472.CAN-10-0860. [DOI] [PubMed] [Google Scholar]
- 29.Chaudhury A, Hussey GS, Ray PS, Jin G, Fox PL, Howe PH. TGF-beta-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nature Cell Biol. 2010;12:286–293. doi: 10.1038/ncb2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou ZJ, Dai Z, Zhou SL, Hu ZQ, Chen Q, Zhao YM, Shi YH, Gao Q, Wu WZ, Qiu SJ, et al. HNRNPAB induces epithelial-mesenchymal transition and promotes metastasis of hepatocellular carcinoma by transcriptionally activating SNAIL. Cancer Res. 2014;74:2750–2762. doi: 10.1158/0008-5472.CAN-13-2509. [DOI] [PubMed] [Google Scholar]
- 31.Ratz L, Laible M, Kacprzyk LA, Wittig-Blaich SM, Tolstov Y, Duensing S, Altevogt P, Klauck SM, Sültmann H. TMPRSS2:ERG gene fusion variants induce TGF-β signaling and epithelial to mesenchymal transition in human prostate cancer cells. Oncotarget. 2017;8:25115–25130. doi: 10.18632/oncotarget.15931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chandra Mangalhara K, Manvati S, Saini SK, Ponnusamy K, Agarwal G, Abraham SK, Bamezai RNK. ERK2-ZEB1-miR-101-1 axis contributes to epithelial-mesenchymal transition and cell migration in cancer. Cancer Lett. 2017;391:59–73. doi: 10.1016/j.canlet.2017.01.016. [DOI] [PubMed] [Google Scholar]
- 33.Bucay N, Bhagirath D, Sekhon K, Yang T, Fukuhara S, Majid S, Shahryari V, Tabatabai Z, Greene KL, Hashimoto Y, et al. A novel microRNA regulator of prostate cancer epithelial-mesenchymal transition. Cell Death Differ. 2017;24:1263–1274. doi: 10.1038/cdd.2017.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao J, Ou B, Han D, Wang P, Zong Y, Zhu C, Liu D, Zheng M, Sun J, Feng H, et al. Tumor-derived CXCL5 promotes human colorectal cancer metastasis through activation of the ERK/Elk-1/Snail and AKT/GSK3β/β-catenin pathways. Mol Cancer. 2017;16:70. doi: 10.1186/s12943-017-0629-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed datasets generated during the study are available from the corresponding author on reasonable request.