Abstract
The natural product neocarzinostatin (NCS), a protein–small molecule complex, exhibits potent antiproliferative activity in mammalian cells but has little apparent effect on the growth of the unicellular eukaryotic organism, Saccharomyces cerevisiae. Here, we show by whole-genome transcription profiling experiments that incubation of S. cerevisiae with NCS leads to dramatic and wide-ranging modifications in the expression profile of yeast genes. Approximately 18% of yeast transcripts are altered by 2-fold or more within 4 h of treatment with NCS. Analysis of the observed transcription profile provides evidence that yeast rapidly and continuously overexpress multiple DNA-damage repair genes during NCS exposure. Perhaps to meet the energetic requirements of continuous DNA-damage repair, yeast cells enter respiration upon prolonged exposure to NCS, although grown in nutrient-rich medium. The NCS protein component is readily transported into S. cerevisiae, as demonstrated by fluorescence microscopy of yeast treated with fluorescently labeled NCS. Transcription profiling experiments with neocarzinostatin protein alone implicate a specific resistance mechanism in yeast that targets the NCS protein component, one involving the nonclassical export pathway. These experiments provide a detailed picture of the effects of exposure to NCS upon yeast and the mechanisms they engage as a response to this protein–small molecule DNA-damaging agent.
The chromoprotein natural product neocarzinostatin (NCS) is composed of an 11-kDa protein (apo-NCS) complexed with a highly reactive small molecule (NCS chromophore) and displays antibiotic and antiproliferative activities (1, 2). Scientific interest in NCS stems primarily from its highly unusual chemical composition and its reactivity, notably the ability to cleave double-stranded DNA by a novel mechanistic pathway (3). NCS has shown some efficacy in the treatment of human cancers of the bladder (4), stomach (5), and liver (6).
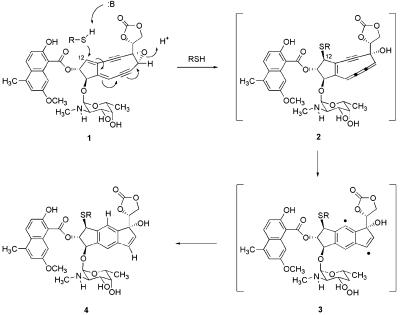
The small-molecule component of NCS, NCS chromophore, is bound noncovalently in a cleft of the binding protein (7) and is dissociable (8). When not bound to apo-NCS, the chromophore is highly reactive, particularly toward nucleophilic reagents. Nucleophilic activation of the chromophore, principally with thiols (9), initiates a sequence of reactions terminating in the formation of a biradical species (10) that can function as a DNA-damaging agent in vitro (Fig. 1). Both single- and double-stranded DNA-damage products have been identified in the reaction of NCS with double-stranded DNA upon thiol activation, and lesions arising from 5-prime (11), 4-prime (12), and 1-prime hydrogen-atom abstraction (13) reactions have been characterized. For in vitro activation with methyl thioglycolate, the pathway of Fig. 1 has been linked unequivocally with DNA cleavage; the cumulene intermediate 2 has been prepared and characterized at low temperature and shown to cleave a 35-mer duplex DNA with the same efficiency and specificity as that arising from incubation of the same duplex oligonucleotide with NCS chromophore and methyl thioglycolate (14). The pathway(s) of NCS activation in vivo are less well established. There is circumstantial evidence that a thiol-activation pathway may operate in vivo, with glutathione functioning as the nucleophilic activating agent, in that HeLa cells treated with the glutathione biosynthesis inhibitor buthionine sulfoximine gain resistance to growth inhibition by NCS treatment (15). Pathways of activation not involving thiols also have been identified in vitro (16–18), and an alternative nucleophilic activation pathway is known to occur with small thiols, involving addition to the chromophore before its dissociation from the protein complex (19, 20).
Figure 1.
Proposed mechanism for the nucleophilic activation of NCS chromophore (1) with thiols such as methyl thioglycolate (R = CH2CO2CH3) or glutathione (3). Thiol addition at C12 produces the cumulene intermediate 2. This intermediate then undergoes rapid unimolecular cycloaromatization to form the highly reactive biradical 3 (14), proposed to initiate DNA damage by hydrogen-atom abstraction from the ribose backbone of DNA (10).
The effects of NCS on the growth of mammalian cells in culture are profound (21). HeLa cells treated with NCS (≈20 nM) show delayed entry into and prolonged progression through S phase and do not undergo the G2-M transition (22). By contrast, the growth of yeast (23), fungi (24), and Gram-negative bacteria (21) seems to be essentially unaffected, even in the presence of millimolar concentrations of NCS. The basis of this resistance is not known. Drug impermeability, drug inactivation, and the operation of efficient DNA-repair mechanisms have been proposed as potential resistance factors in yeast and other organisms. For some time, our laboratory has been involved in studies designed to elucidate the details of the chemistry that occurs when living cells are treated with NCS. The determination of the mechanism of action of any small molecule in vivo is challenging under normal circumstances, but is especially complicated when the molecule is highly reactive, such as NCS, with multiple pathways of reaction that vary in response to small changes in the chemical environment. It is expected that the distribution of NCS-derived reaction products may vary as a function of cell type and environment and, thus, that the specific biological response also may vary. As an important component of NCS mechanistic analysis, we wanted to determine as precisely as possible the molecular changes in a specific living cell after drug treatment. Toward this end, we have conducted a series of experiments by using the unicellular eukaryotic organism, Saccharomyces cerevisiae, to analyze in detail the effects of NCS exposure, as determined at the level of transcription. With the advent of methods for whole-genome monitoring, the degree of detail now possible in such an analysis is extraordinary (25–29). Here, we describe transcription profiling experiments of S. cerevisiae grown in the presence of NCS, providing a detailed picture of the consequences to the organism of NCS exposure and the mechanisms by which it responds to this xenobiotic agent.
Materials and Methods
DNA Microarray Construction.
A set of 6,218 verified ORFs was obtained from Research Genetics (Huntsville, AL). Each double-stranded ORF contains common 19-bp sequences at the 5′ and 3′ ends and was amplified to levels required for preparation of DNA microarrays by the PCR (27). Successful production of PCR products was confirmed by agarose gel electrophoresis for 98% of the ORFs. Longer ORFs that were not adequately amplified in the first PCR experiment were amplified with the GIBCO/BRL Elongase Amplification Kit (Life Technologies, Rockville, MD; 40 cycles of the sequence: denaturation for 1 min at 95°C, annealing for 1 min at 55°C, and elongation for 10 min at 68°C). For array production, the amplified DNA was precipitated with isopropanol, washed with 70% EtOH, and resuspended in 25 μl of Micro Spotting Solution (Telechem, Sunnyvale, CA). DNA solutions were spotted on CMT-GAPS slides (Corning) by using a microarraying robot with a 16-pin head constructed from a design by Patrick O. Brown (http://cmgm.stanford.edu/pbrown/). Postprocessing of the slides was accomplished according to published procedures (30).
Neocarzinostatin.
Neocarzinostatin was generously provided by Kayaku (Tokyo) and was purified according to the published procedure (14), described in detail in Supporting Materials and Methods, which is published on the PNAS web site, www.pnas.org.
Growth of S. cerevisiae Cultures.
A single colony of S. cerevisiae strain BY4741 (MATa his3Δ1 leu2Δ1 met15Δ0 ura3Δ0) was used to inoculate 50 ml of YPD medium (2% glucose/2% peptone/1% yeast extract) for expression-profiling experiments with NCS or apo-NCS. The cells were grown at 30°C on a shaker (225 rpm) overnight to a density of 1 × 108 cells/ml, the growth medium was diluted 200-fold in YPD medium prewarmed to 30°C, and incubation was continued to a density of 3 × 106 cells/ml (A600 = 0.3). The cells were treated with solutions of holo-NCS or apo-NCS in sterile water (10 mg/ml, final concentration 50 μg/ml), and 40-ml aliquots of the culture medium were harvested at the indicated times. Cells from each aliquot were pelleted by centrifugation (3,000 × g) for 5 min at 30°C and then were flash-frozen in liquid nitrogen and stored at −80°C for later extraction of RNA. Each experiment with NCS and apo-NCS was conducted in duplicate.
Extraction of mRNA.
Total RNA was extracted from flash-frozen pellets of cultured yeast cells by using the acidic phenol method (31). Poly(A) RNA was isolated from total RNA by using an oligo(dT) resin (Oligotex, Qiagen, Chatsworth, CA).
Preparation of cDNA and Hybridizations.
Fluorescent DNA probes were prepared from poly(A) RNA isolated from control or NCS-treated yeast populations by using an oligo(dT)-primed reverse transcriptase (GIBCO/BRL, Life Technologies). Transcription reactions used Cy3-dUTP or Cy5-dUTP (Amersham Pharmacia) and poly(A) RNA (1.8 μg) isolated from control or treated yeast populations, respectively, essentially as described (31). Competitive hybridizations were performed in duplicate for each experiment. Because each experiment was also run in duplicate, each time point is therefore represented by no fewer than four successful competitive hybridizations.
Data Acquisition and Analysis.
Fluorescent DNA bound to the microarray was detected with a GenePix 4000A array scanner (Axon Instruments, Foster City, CA), by using GENEPIX 3.0 software to locate individual spots, quantitate the Cy3 and Cy5 fluorescence intensity at each spot, and determine background signal intensities. Data from spots that were determined to be the result of hybridization anomalies or microarray errors were excluded from further analysis. Fluorescence intensity values were determined by subtraction of the local background from the foreground. Only those signal intensities greater than two standard deviations above the average background intensity (calculated over the entire array) were considered for further analysis to avoid errors as a result of low signal intensity. The ratio (total signal from all Cy3 channels)/(total signal from all Cy5 channels) was calculated to provide a scaling factor for normalization between the channels; this factor was then applied uniformly to each spot. Only genes with reproducible (≥4 hybridizations) expression differences of 2-fold or more were considered in our analysis. GENESPRING software (Silicon Genetics, Redwood City, CA) was used for data analysis and gene clustering. Data sets from profiling experiments and a detailed hybridization protocol can be obtained at http://www.chem.harvard.edu/groups/myers/myers_research_group.htm.
Synthesis of Fluorescently Labeled NCS (NCS-Fl) and Fluorescence Microscopy of S. cerevisiae Treated with NCS-Fl.
The synthesis of NCS-Fl and fluorescence microscopy experiments are described in detail in Supporting Materials and Methods.
Results and Discussion
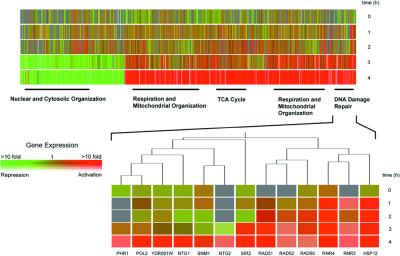
Yeast grown in YPD at 30°C and exposed to NCS at a concentration of 50 μg/ml showed little evidence of growth inhibition relative to nontreated controls over the 4-h course of our experiments, consistent with the prior observations of Mousstacchi and Favaudon (23). NCS-treated yeast cells were enlarged relative to untreated controls after 12 h, a phenotype equated with aging (32). Transcription profiling experiments showed that extensive modification of the expression pattern of yeast genes occurred during NCS exposure, with progressively greater variation as a function of time, over the time course examined (1, 2, 3, and 4 h). After 4 h, expression levels of 780 genes had increased more than 2-fold; of these, expression of 355 genes was enhanced by more than 3-fold (Fig. 2). By contrast, expression levels of 488 genes were repressed more than 2-fold relative to untreated controls, and expression of 133 genes was repressed more than 3-fold. Together, these changes represent a modification in the expression levels of ≈18% of yeast transcripts.
Figure 2.
Time course of expression modification of S. cerevesiae treated with NCS (50 μg/ml) at 1-, 2-, 3-, and 4-h time points. Only genes whose expression was enhanced or repressed by ≥2-fold are shown, clustered hierarchically by using the genespring software package. Rows are used for each time point, and columns for individual genes. Color coding is used to represent the fold-change in expression, as indicated within the Inset color bar, and saturation of the degree of fluorescence signal intensity. Red indicates genes with enhanced expression levels in the treated cells relative to untreated controls, and green indicates genes with reduced expression levels. In total, 1,288 genes exhibited modified mRNA transcript levels (2-fold or more). Functional categories (nuclear and cytosolic organization, respiration and mitochondrial organization, TCA cycle genes, and DNA-damage repair genes) are those of the Munich Information Center for Protein Sequences (http://www.mips.biochem.mpg.de/). Genes identified that fall within these categories are indicated by a solid black bar and provide the following P values: nuclear and cytosolic organization, P = 1.2 × 10−5; respiration and mitochondrial organization, P = 1.4 × 10−22; TCA cycle, P = 6.1 × 10−14; DNA-damage repair, P = 2.1 × 10−3 (P values were calculated by using the genespring software package). The highlighted section shows in detail a representative hierarchical clustering of enhanced genes, these for DNA-damage repair.
Analysis of the expression profile by a hierarchical clustering algorithm (GENESPRING software) allowed many of the modified transcripts to be grouped by their functions, as well as temporally. General functional categories (Munich Information Center for Protein Sequences, http://www.mips.biochem.mpg.de/) identified include DNA-damage repair and detoxification, mitochondrial organization and respiration (subdivided by temporal distribution, as shown in Fig. 2), genes of the tricarboxylic acid cycle, and nuclear and cytosolic organization. Of genes overexpressed >2-fold at 4 h, 209 were of unknown function; 164 genes whose expression was diminished >2-fold were also of unknown function. Discussion of selected specific, transcriptionally modified genes follows, organized according to the general categories listed.
DNA-Damage Repair Genes.
Prominent among DNA-damage repair genes up-regulated by the first time point (1 h) and continuously throughout the 4-h incubation period are RNR2, RNR3, and RNR4 (2.2-, 2.2-, and 2.4-fold, respectively, after 1 h). The protein products of these genes are subunits of ribonucleotide reductase, an enzyme critical for the maintenance of nucleotide pools in DNA replication and repair (33). At 3 h and, to a greater degree, at 4 h, transcript levels of members of the RAD52 epistasis group of DNA repair genes, RAD50, RAD51, and RAD52, were observed to increase by more than 2-fold. These genes code for proteins involved in recombination and the recombinational repair of double-strand breaks in DNA. Yeast RAD52-deletion mutants display extraordinary sensitivity to ionizing radiation and double-strand breaks in particular. It has been calculated that a single double-strand break in a rad52-deletant can be lethal to a cell (34). The RAD50 gene product is required for DNA repair by nonhomologous end-joining (35). The RAD51 and RAD52 protein products are known to interact and are involved in the repair of double-strand DNA lesions by homologous recombination (36). Double-strand DNA breaks have been documented in in vitro experiments of oligonucleotides with NCS (thiol activated); however, they represent a minority of the total DNA-damage products identified, and their significance in vivo has not been established with certainty (37).
Genes known to code for proteins involved with the repair of types of DNA damage other than double-strand breaks were also overexpressed, but only after 4 h of NCS treatment (>2-fold). We observed increased transcript levels of NTG1 and NTG2; nucleotide excision repair genes encoding proteins associated with the repair of oxidatively damaged bases (e.g., 8-oxoguanine, 5-hydroxycytosine) (38); PSO2, a gene encoding a protein for the repair of interstrand crosslinks produced upon exposure of DNA to UV light; nitrogen mustards, or cis-diaminedichloroplatinum (39); and PHR1, coding a DNA photolyase that repairs pyrimidine dimers (40). Neither oxidatively damaged bases nor interstrand crosslinks have been identified among NCS-induced DNA-damage products. The up-regulation of genes encoding proteins known to repair such lesions here may signal the formation of previously unrecognized types of DNA-damage products upon NCS exposure. Alternatively, it may be that the protein products of the overexpressed genes are not restricted in their activities to the repair of, e.g., oxidatively damaged bases or that these genes are overexpressed as a result of coregulation with another gene. In this regard, it is interesting to note that both NTG1 and NTG2 have previously been shown to be up-regulated upon exposure of yeast to methyl methanesulfonate, a DNA-alkylating agent with no apparent ability to produce oxidatively damaged DNA (41).
Although not a DNA-damage repair gene, per se, it is perhaps significant in the context of this discussion that the gene SIR2 is observed to be overexpressed upon NCS exposure (1.1-, 0.8-, 2.4-, and 3.1-fold at 1, 2, 3, and 4 h, respectively). SIR2 is a pleiotropic gene believed to be of central importance in aging in yeast, as well as higher organisms (42). Sir2p is an NAD-dependent histone deacetylase involved in gene silencing (43). It also has been proposed to play a role in DNA-damage repair (44). In one model, the Sir2,3,4p complex is proposed to undergo relocation from silenced chromatin to sites of DNA damage, where it interacts with the yKu heterodimeric protein complex. The relocation of the Sir2,3,4p complex depends on the checkpoint proteins Rad9p and Mec1p, whose gene transcripts also were observed to be up-regulated in our experiments, as discussed below (45).
DNA-Damage Checkpoint Genes.
After 4 h of exposure to NCS, increased transcript levels of the cell cycle control genes MEC1 and RAD9 were observed (2.4- and 2.6-fold, respectively). Both are checkpoint genes that cause cell cycle arrest in response to DNA damage. It has been shown that both genes are required for the repair of double-strand breaks by a process that involves the relocation of Sir proteins from a telomere-associated complex to the break site. The mitosis-entry checkpoint gene MEC1, a protein kinase, is homologous with the human gene ATM, whose mutation is associated with the disease ataxia telangiectasia (46). Characteristics of this disease include an increased sensitivity of cells to ionizing radiation and a predisposition toward malignancy. DNA-damage checkpoint genes maintain the integrity of the genome by recognizing sites of DNA damage, recruiting the necessary proteins for DNA repair, and arresting the cell cycle so that repair can occur (47). However, in a unicellular organism, prolonged arrest has been equated to cell death. It has been suggested that adaptation may occur in such situations (e.g., upon continuous production of nonrepairable double-strand DNA lesions) to allow for continued growth (48). This may account for our apparently conflicting observations of continued cell growth and simultaneous up-regulation of the DNA-damage checkpoint genes RAD9 and MEC1.
Genes Involved in the Stress Response.
Of the 780 genes overexpressed during the 4-h time course of our experiments, 180 contain the stress-responsive element (STRE) 5′-CCCCT, in their respective promoter sequences, within 700 bp of the start codon (49). The STRE promoter sequence has been shown to be transcriptionally activated by Msn2p and Msn4p by direct interaction of these proteins with the 5′-CCCCT sequence (50). Among stress-responsive genes up-regulated were HSP12, DDR2, and DDR48, functional reporters of stress associated with cellular damage (51). HSP12 was overexpressed >2-fold after 1 h, whereas DDR2 and DDR48 were not overexpressed until 3 h after treatment, attaining a maximum overexpression after 4 h (3-fold overexpression). HSP12 is a multistress response gene, overexpressed in response to many different forms of environmental stress. It codes for a protein that is predicted by its sequence to function as a chaperonin (52). Interestingly, we observed that HSP12 and the adjacent 3′-ORF YFL013W-A were overexpressed contemporaneously, and to the same degree. DDR2 and DDR48 are stress response genes that are overexpressed as a consequence of DNA damage (53). Not all genes containing the STRE in their promoter sequences were overexpressed in our experiments. For example, the cytosolic catalase gene CTT1, whose transcription is induced by oxidative stress (e.g., exposure to hydrogen peroxide; ref. 54), was not observed among genes transcriptionally enhanced upon NCS treatment.
Genes Involved in Respiration and Energy Production.
A clear and consistent pattern of overexpression of genes that encode proteins necessary for respiration and energy production was observed during the course of incubation with NCS. Evidence in this regard is the up-regulation of many of the genes in the tricarboxylic acid cycle (>2-fold within 4 h) and the concomitant down-regulation of genes involved with glycolysis and gluconeogenesis. These modifications are part of the transition from fermentation to respiration in yeast (25). This transition is known to be under the transcriptional control of the HAP genes (HAP2, HAP3, HAP4, and HAP5), which also showed increased transcript levels in our profile (2.2-, 2.3-, 6.2-, and 2.4-fold, respectively, at 4 h) (55). The HAP genes, in turn, are transcriptionally regulated by the protein Sip4p, subsequent to its phosphorylation by the kinase Snf1p (56). Significantly, both SIP4 and SNF1 were also overexpressed in our profile, the latter by 2-fold at the 2-h time point (sustained at this level at 3 and 4 h) and the former 3.4-fold after 4 h. Thus, many of the genes that regulate the transition from fermentation to respiration show increased transcript levels, and, chronologically, increased transcription of each gene is found to coincide with its position in the established sequence of genetic control. Thus, we observe up-regulation of SNF1 before up-regulation of SIP4 (whose protein product is a substrate of Snf1p) and the HAP genes and genes for the tricarboxylic cycle.
We also observed induction of several other genes transcriptionally controlled by the phosphorylated Sip4p protein (57) that are indicative of a shift toward oxidative metabolism. These genes include ACS1, FBP1, ICL1, IDP2, JEN1, MLS1, PCK1, and SFC1 (each up-regulated >10-fold at 4 h), encoding proteins required for the glyoxylate cycle and the conversion of ethanol to acetic acid. We also see elevated transcript levels of other members of the SNF family of genes: SNF2, coding for a chromatin remodeling transcription factor (58); SNF3, a receptor involved in glucose sensing (59); and RIS1, a DNA-dependent ATPase involved in remodeling protein-DNA structure (2.7-, 2.8-, and 2.7-fold increase, respectively, at 4 h) (60).
Snf1p is well established to function in the derepression of glucose-repressed genes (61) and is a necessary component for carbon utilization under conditions of stress, such as elevated temperature (62). It functions as a protein kinase after phosphorylation, which is proposed to occur in response to an elevation in the ratio of AMP to ATP in cells (63), a primary indicator of stress, such as DNA damage. It may also be of significance that SNF gene products are active in chromatin remodeling, a process that provides access to nucleosomal DNA in transcription and, it is proposed, DNA-damage repair. It is interesting in the context of the present work that SNF1 was also observed to be up-regulated upon exposure of yeast to the DNA alkylating agent methyl methanesulfonate (41). The factors that regulate the expression of SNF1 are presently not known.
Detoxification Genes: Identification of an NCS Protein-Specific Response in Yeast.
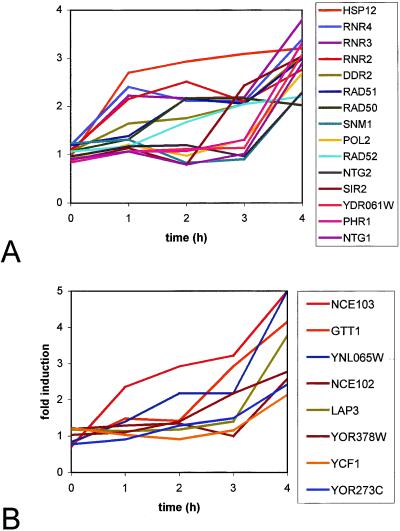
A number of genes associated with cellular detoxification were overexpressed as a result of NCS exposure. Genes encoding the multidrug-resistance proteins YOR273C, YOR378W, and YNL065W showed a time-dependent increase in expression levels over the course of the treatment, reaching levels of 2.4-, 2.8-, and 5.2-fold, respectively, at 4 h (64). LAP3, coding a cysteine protease with nucleic acid binding affinity, is overexpressed 3.8-fold at 4 h. Lap3p has been shown to be important for the detoxification of bleomycin (65). YJL068C, overexpressed upon exposure of yeast to formaldehyde and methyl methanesulfonate, was overexpressed 2.7-fold at 4 h (66). YCF1, coding a protein homologous to the human multidrug resistance protein hMRP1, was overexpressed 2.1-fold at 4 h (67). In addition, the gene encoding glutathione S-transferase, GTT1, was overexpressed 4.1-fold at 4 h (68). Expression of this gene is induced upon exposure of yeast to 1-chloro-2,4-dinitrobenzene, methyl methanesulfonate, and other electrophilic agents. GTT1 is considered to be a common environmental response gene and contains three copies of the STRE in its promoter (Fig. 3).
Figure 3.
Graphical representation of the time course of expression change (in units of fold-induction) for several key genes involved in (A) DNA-damage repair, and (B) detoxification and nonclassical protein export. Functional categories are those of the Munich Information Center for Protein Sequences (http://www.mips.biochem.mpg.de/).
We also observed immediate and sustained induction of the gene NCE103 (2.4-, 2.9-, 2.9-, and 6.6-fold at 1, 2, 3, and 4 h, respectively) and, later, increased transcript levels of NCE102 (2.6-fold, 4 h). Like GTT1, these genes are considered to be common environmental response genes, overexpressed upon exposure of yeast to many stressors. NCE103 is a poorly characterized protein in terms of its biochemical function. Deletion of NCE103 in yeast produces a viable strain with marked sensitivity to growth under aerobic conditions (69). The basis for this sensitivity is not understood. Its name derives from a putative role in a pathway for the nonclassical export of proteins in yeast. NCE103 was first identified in a screen of yeast mutants that were modified to express the foreign protein galectin, toxic in these strains at high expression levels. Cleves et al. (70) proposed that a potential physiological role for the nonclassical protein export pathway is to remove toxic proteins from the cytoplasm by a mechanism not involving the endoplasmic reticulum or Golgi apparatus, an intriguing suggestion in light of the present study. NCE103, a 25-kDa hydrophilic protein, was proposed to be an endogenous substrate in this nonclassical export pathway (70).
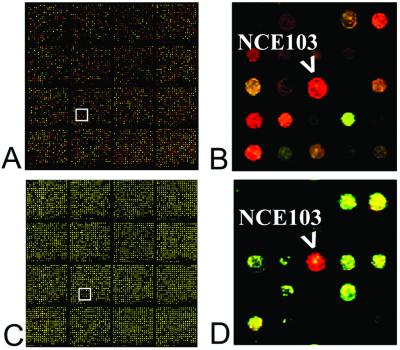
Our data suggest that the induction of NCE103 may be a more specific response to the exposure of yeast to NCS. This evidence comes in the form of a control experiment that is somewhat unique to the chromoprotein antiproliferative agents. X-ray crystal structure determination has shown that NCS (protein–chromophore complex) and apo-NCS (binding protein alone) are nearly identical in their three-dimensional shapes, differing only in the positioning of a single phenylalanine residue. To learn whether the protein component plays any role in the response of yeast to NCS, we conducted expression-profiling studies with apo-NCS alone (Fig. 4). The expression profiles at 1 h and thereafter were remarkable in that a single gene, NCE103, showed modified expression (4-fold).
Figure 4.
Pseudo-color images of competitive hybridization experiments using DNA microarrays. Competitive hybridizations were performed by using Cy5-Labeled cDNA, prepared from mRNA isolated from treated cells, and Cy3-labeled cDNA, prepared from mRNA isolated from controls. (A) DNA microarray after competitive hybridization, yeast treated with NCS (50 μg/ml) for 4 h at 30 °C. (B) Magnification of the boxed region of A, containing the ORF corresponding to the gene NCE103. The red color is indicative of enhanced expression of the gene in the treated cells relative to controls. (C) DNA microarray after competitive hybridization, yeast treated with apo-NCS (50 μg/ml) for 4 h at 30 °C. (D) Magnification of the boxed region of C, containing the ORF corresponding to the gene NCE103.
In light of the finding that the nonclassical export protein NCE103p is overexpressed uniquely upon apo-NCS exposure, we sought to determine whether the NCS-associated protein actually penetrated yeast cells. A clear role for the protein component in NCS activity is to bind and stabilize the unstable chromophore component; the role of the protein in the cellular penetration of the chromophore is less clear. In prior work, exposure of yeast to 125I-labeled NCS produced a radioactive cellular sedimentation fraction. It was not determined whether the radioactivity was associated with the cell wall or if internalization of the protein had occurred. To address this question, we synthesized NCS with a fluorescence label (NCS-Fl) and conducted fluorescence-imaging experiments with yeast exposed to the labeled complex. The fluorescence label was introduced by conjugation with a commercial reagent (N-hydroxy succinimide ester) containing the Alexa Fluor 350 chromophore (Molecular Probes). The labeled complex was purified by ion-exchange chromatography and the purified material was shown by UV-visible spectroscopy to have incorporated two fluorophores per protein molecule, as expected if labeling of the amino terminus (an alanine residue) and the single lysine residue of NCS had occurred. Fluorescence microscopy of exponentially growing S. cerevisiae cells exposed to NCS-Fl for 1 h, after fixing with formaldehyde and washing, showed that intracellular incorporation did occur, with apparently uniform dissemination throughout the cell (Fig. 5). Our experiments parallel results from a prior study of the incorporation of fluorescein-labeled NCS into human bladder cancer cells, where uniform intracellular distribution of the fluorescence label was observed (71). Our results show that NCS is also capable of penetrating the less permeable yeast cell wall. With the knowledge that the protein component achieves cellular penetration, and in light of data from transcription profiling of the NCS protein component alone, a dynamic process is envisioned to occur, whereby yeast treated with NCS internalize the protein–chromophore complex (by an unknown mechanism) and counter the absorption of this toxic agent by expulsion through the nonclassical export pathway.
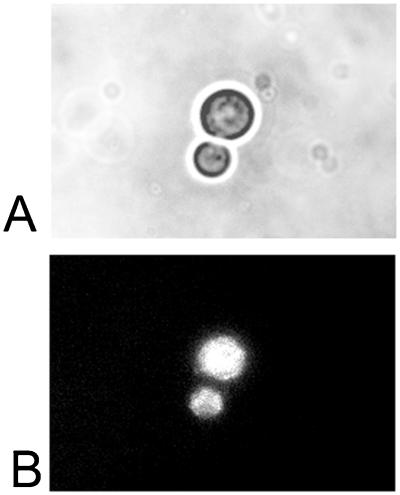
Figure 5.
Phase-contrast (A) and fluorescence (B) microscope images (magnification ×1,000) showing budding yeast after incubation with NCS-Fl (NCS bearing a 350-nm fluorescence label) for 1 h at 30 °C. The fluorescence image shows uniform intracellular distribution of the labeled drug, providing evidence that the NCS protein component is internalized in yeast. Fluorescence imaging was conducted with excitation between 340 and 380 nm by using a 400-nm emission filter.
It is evident from our experiments that expression of the yeast genome is subject to substantive and far-ranging modification as a result of NCS treatment. Analysis of the expression profile provides a self-consistent picture of the effects of NCS exposure upon the organism and the mechanisms it mounts in response to this treatment. Certainly, evidence for the occurrence of DNA damage, and double-strand damage in particular, is compelling. A number of DNA-damage repair proteins and DNA-damage checkpoint control genes are overexpressed, in addition to genes associated with the generalized stress response. A large proportion of the expression modification that we see can be attributed to a shift from fermentative growth to respiration, perhaps to meet the increased energy requirements for DNA-damage repair and the stress response. A picture also emerges of a dynamic process of cellular penetration by the protein–chromophore complex with, it is proposed, competitive export involving the nonclassical protein export pathway. The identification of cell-specific responses and resistance mechanisms to a small-molecule antiproliferative agent, such as NCS, provides an important component of mechanistic understanding and may also assist in predicting therapeutic profiles with different (cancer) cell types.
Supplementary Material
Acknowledgments
We thank Professor Stuart Schreiber for many insightful discussions. This work would not have been possible without the technical expertise of Dr. Jeff Tong, Dr. James Hardwick, Paul Grosu, and Jeffrey Townsend. We are indebted to the staff of the Harvard University Center for Genomics Research for their assistance. We express our gratitude to the members of the Schreiber research group, Harvard University, for the use of their equipment and for their technical expertise and advice. This research was supported by a grant from the National Institutes of Health. D.C. is the recipient of fellowship support from the Italian Government. S.E.S. is the recipient of a fellowship from the National Institutes of Health.
Abbreviations
- NCS
neocarzinostatin
- NCS-Fl
fluorescently labeled neocarzinostatin
- STRE
stress-responsive element
References
- 1.Goldberg I H, Kappan L S. In: Endiyne Antibiotics as Antitumor Agents. Borders D B, Doyle T W, editors. New York: Dekker; 1995. pp. 327–362. [Google Scholar]
- 2.Maeda H, Edo K, Ishida N, editors. Neocarzinostatin: The Past, Present, and Future of an Anticancer Drug. New York: Springer; 1997. [Google Scholar]
- 3.Myers A G. Tetrahedron Lett. 1987;28:4493–4496. [Google Scholar]
- 4.Sakamoto S, Ogta J, Ikegami K, Maeda H. Cancer Treat Rep. 1979;62:453–455. [PubMed] [Google Scholar]
- 5.Takahashi M, Toriyama K, Maeda M, Kikuchi M, Kumagai K, Ishida N. Tohoku J Exp Med. 1969;98:273–280. doi: 10.1620/tjem.98.273. [DOI] [PubMed] [Google Scholar]
- 6.Ikeda K, Saitoh S, Kobayashi M, Suzuki Y, Suzuki F, Tsubota A, Arase Y, Chayama K, Murashima N, Kumada H. J Gastroenterol. 2000;35:353–360. doi: 10.1007/s005350050360. [DOI] [PubMed] [Google Scholar]
- 7.Kim K H, Kwon B M, Myers A G, Rees D C. Science. 1993;262:1042–1046. doi: 10.1126/science.8235619. [DOI] [PubMed] [Google Scholar]
- 8.Napier M A, Holmquist B, Strydom D J, Goldberg I H. Biochem Biophys Res Commun. 1979;89:635–642. doi: 10.1016/0006-291x(79)90677-6. [DOI] [PubMed] [Google Scholar]
- 9.Beerman T A, Poon R, Goldberg I H. Biochim Biophys Acta. 1977;475:294–306. doi: 10.1016/0005-2787(77)90020-x. [DOI] [PubMed] [Google Scholar]
- 10.Kappen L S, Goldberg I H. Nucleic Acids Res. 1985;13:1637–1648. doi: 10.1093/nar/13.5.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kappen L S, Goldberg I H, Liesch J M. Proc Natl Acad Sci USA. 1982;79:744–748. doi: 10.1073/pnas.79.3.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saito I, Kawabata H, Fujiwara T, Sugiyama H, Matsuura T. J Am Chem Soc. 1989;111:8302–8303. [Google Scholar]
- 13.Kappen L S, Goldberg I H. Biochemistry. 1989;28:1027–1032. doi: 10.1021/bi00429a016. [DOI] [PubMed] [Google Scholar]
- 14.Myers A G, Cohen S B, Kwon B M. J Am Chem Soc. 1994;116:1670–1682. [Google Scholar]
- 15.Degraff W G, Mitchell J B. Cancer Res. 1985;45:4760–4762. [PubMed] [Google Scholar]
- 16.Hensens O D, Dewey R S, Liesch J M, Napier M A, Reamer R A, Smith J L, Albers-Schonberg G, Goldberg I H. Biochem Biophys Res Commun. 1983;113:538–547. doi: 10.1016/0006-291x(83)91759-x. [DOI] [PubMed] [Google Scholar]
- 17.Gomibuchi T, Masahiro H. J Am Chem Soc. 1995;48:738–740. [Google Scholar]
- 18.Kappen L S, Goldberg I H. Science. 1993;261:1319–1321. doi: 10.1126/science.8362243. [DOI] [PubMed] [Google Scholar]
- 19.Sugiyama H, Yamashita K, Nishi M, Saito I. Tetrahedron Lett. 1992;33:515–518. [Google Scholar]
- 20.Myers A G, Arvedson S P, Lee R W. J Am Chem Soc. 1996;118:4725–4726. [Google Scholar]
- 21.Ishida N, Miyazaki K, Kumagai K, Rikimaru M. J Antibiot. 1965;18:68–76. [PubMed] [Google Scholar]
- 22.Berry D E, Collins J M. Cancer Res. 1980;40:2405–2410. [PubMed] [Google Scholar]
- 23.Mousstacchi E, Favaudon V. Mutat Res. 1982;104:87–94. doi: 10.1016/0165-7992(82)90125-7. [DOI] [PubMed] [Google Scholar]
- 24.DeGraff W G. Mutat Res. 1984;128:127–135. doi: 10.1016/0027-5107(84)90099-x. [DOI] [PubMed] [Google Scholar]
- 25.DeRisi J L, Iyer V R, Brown P O. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 26.Marton M J, DeRisi J L, Bennett H A, Iyer V R, Meyer M R, Roberts C J, Stoughton R, Burchard J, Slade D, Dai H, et al. Nat Med. 1998;4:1293–1301. doi: 10.1038/3282. [DOI] [PubMed] [Google Scholar]
- 27.Hardwick J S, Kuruvilla F G, Tong J K, Shamji A F, Schreiber S L. Proc Natl Acad Sci USA. 1999;96:14866–14870. doi: 10.1073/pnas.96.26.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bernstein B E, Tong J T, Schreiber S L. Proc Natl Acad Sci USA. 2000;97:13708–13713. doi: 10.1073/pnas.250477697. . (First Published November 28, 2000; 10.1073/pnas.250477697) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shamji A F, Kuruvilla F G, Schreiber S L. Curr Biol. 2000;10:1574–1581. doi: 10.1016/s0960-9822(00)00866-6. [DOI] [PubMed] [Google Scholar]
- 30.Eisen M B, Brown P O. Methods Enzymol. 1999;303:179–205. doi: 10.1016/s0076-6879(99)03014-1. [DOI] [PubMed] [Google Scholar]
- 31.Schmitt M E, Brown T A, Trumpower B L. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laun P, Pichova A, Madeo F, Fuchs J, Ellinger A, Kohlwein S, Dawes I, Frolich K-U, Breitenbach M. Mol Microbiol. 2001;39:1166–1173. [PubMed] [Google Scholar]
- 33.Nguyen H-H T, Ge J, Perlstein D L, Stubbe J A. Proc Natl Acad Sci USA. 1999;96:12339–12344. doi: 10.1073/pnas.96.22.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weiffenbach B, Haber J E. Mol Cell Biol. 1981;1:522–534. doi: 10.1128/mcb.1.6.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Usui T, Ohta T, Oshiumi H, Tomizawa J, Ogawa H, Ogawa T. Cell. 1998;95:705–716. doi: 10.1016/s0092-8674(00)81640-2. [DOI] [PubMed] [Google Scholar]
- 36.Shinohara A, Ogawa T. Nature (London) 1998;391:404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- 37.Dedon P C, Goldberg I H. J Biol Chem. 1990;265:14713–14716. [PubMed] [Google Scholar]
- 38.Sentürker S, Kemp P A, You H J, Doetsch P W, Dizdaroglu M, Boiteux S. Nucleic Acids Res. 1998;26:5270–5276. doi: 10.1093/nar/26.23.5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolter R, Siede W, Brendel M. Mol Gen Genet. 1996;250:162–168. doi: 10.1007/BF02174175. [DOI] [PubMed] [Google Scholar]
- 40.Suter B, Livingstone-Zatchej M, Thoma F. EMBO J. 1997;16:2150–2160. doi: 10.1093/emboj/16.8.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jelinski S A, Samson L D. Proc Natl Acad Sci USA. 1999;96:1486–1491. doi: 10.1073/pnas.96.4.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guarente L. Genes Dev. 2000;14:1021–1026. [PubMed] [Google Scholar]
- 43.Imai S, Armstrong C M, Kaeberlein M, Guarente L. Nature (London) 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 44.Haber J E. Mutat Res. 2000;451:53–69. doi: 10.1016/s0027-5107(00)00040-3. [DOI] [PubMed] [Google Scholar]
- 45.Mills K D, Sinclair D A, Guarente L. Cell. 1999;97:609–620. doi: 10.1016/s0092-8674(00)80772-2. [DOI] [PubMed] [Google Scholar]
- 46.Jaspers N G J, Gatti R A, Baan C, Linssen P C M L, Bootsma D. Cytogenet Cell Genet. 1988;49:259–263. doi: 10.1159/000132673. [DOI] [PubMed] [Google Scholar]
- 47.Zhou B-B S, Elledge S J. Nature (London) 2000;408:433–438. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 48.Toczyski D P, Galgoczy D J, Hartwell L H. Cell. 1997;90:1097–1106. doi: 10.1016/s0092-8674(00)80375-x. [DOI] [PubMed] [Google Scholar]
- 49.Moskvina E, Schüller C, Maurer C T C, Mager W H, Ruis H. Yeast. 1998;14:1041–1050. doi: 10.1002/(SICI)1097-0061(199808)14:11<1041::AID-YEA296>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 50.Gorner W, Durchschlag E, Martinez-Pastor M T, Estruch F, Ammerer G, Hamilton B, Ruis H, Schuller C. Genes Dev. 1998;12:586–597. doi: 10.1101/gad.12.4.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gasch A P, Spellman P T, Kao C M, Carmel-Harel O, Eisen M B, Storz G, Botstein D, Brown P O. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Costanzo M C, Crawford M E, Hirschman J E, Kranz J E, Olsen P, Robertson L S, Skrzypek M S, Braun B R, Hopkins K L, Kondu P, et al. Nucleic Acids Res. 2001;29:75–79. doi: 10.1093/nar/29.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McClanahan T, McEntee K. Mol Cell Biol. 1984;4:2356–2363. doi: 10.1128/mcb.4.11.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Causton H C, Ren B, Koh S S, Harbison C T, Kanin E, Jennings E G, Lee T I, True H L, Lander E S, Young R A. Mol Biol Cell. 2001;12:323–337. doi: 10.1091/mbc.12.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blom J, DeMattos M J T, Grivell L A. Appl Environ Microbiol. 2000;66:1970–1973. doi: 10.1128/aem.66.5.1970-1973.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sudarsanam P, Iyer V R, Brown P O, Winston F. Proc Natl Acad Sci USA. 2000;97:3364–3369. doi: 10.1073/pnas.050407197. . (First Published March 21, 2000; 10.1073/pnas.050407197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lesage P, Yang X, Carlson M. Mol Cell Biol. 1996;16:1921–1928. doi: 10.1128/mcb.16.5.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Logie C, Peterson C L. EMBO J. 1997;16:6772–6782. doi: 10.1093/emboj/16.22.6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ozcan S, Johnston M. Microbiol Mol Biol Rev. 1997;63:554–569. doi: 10.1128/mmbr.63.3.554-569.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Z, Buchman A R. Mol Cell Biol. 1997;17:5461–5472. doi: 10.1128/mcb.17.9.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Celenza J L, Carlson M. Mol Cell Biol. 1984;4:49–53. doi: 10.1128/mcb.4.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thompson-Jaeger S, Francois J, Gaughran J P, Tatchell K. Genetics. 1991;129:697–706. doi: 10.1093/genetics/129.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wilson W A, Hawley S A, Hardie D G. Curr Biol. 1996;6:1426–1434. doi: 10.1016/s0960-9822(96)00747-6. [DOI] [PubMed] [Google Scholar]
- 64.Goffeau A, Park J, Paulsen I T, Jonniaux J L, Dinh T, Mordant P, Saier M H. Yeast. 1997;13:43–54. doi: 10.1002/(SICI)1097-0061(199701)13:1<43::AID-YEA56>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 65.Zheng W, Johnston S A. Mol Cell Biol. 1998;18:3580–3585. doi: 10.1128/mcb.18.6.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Degrassi G, Uotila L, Klima R, Venturi V. Appl Environ Microbiol. 1999;65:3470–3472. doi: 10.1128/aem.65.8.3470-3472.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li Z S, Szczypka M, Lu Y P, Thiele D J, Rea P A. J Biol Chem. 1996;271:6509–6517. doi: 10.1074/jbc.271.11.6509. [DOI] [PubMed] [Google Scholar]
- 68.Choi J H, Lou W, Vancura A. J Biol Chem. 1998;273:29915–29922. doi: 10.1074/jbc.273.45.29915. [DOI] [PubMed] [Google Scholar]
- 69.Gotz R, Gnann A, Zimmermann F K. Yeast. 1999;15:855–864. doi: 10.1002/(SICI)1097-0061(199907)15:10A<855::AID-YEA425>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 70.Cleves A E, Cooper D N, Barondes S H, Kelly R B. J Cell Biol. 1996;133:1017–1026. doi: 10.1083/jcb.133.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sakamoto S, Maeda H, Ogata J. Experentia. 1979;35:1233–1235. doi: 10.1007/BF01963309. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.