Abstract
Recent genome-wide association studies identified over 100 genetic loci that significantly associate with schizophrenia (SZ). A top candidate gene, ZNF804A, was robustly replicated in different populations. However, its neural functions are largely unknown. Here we show in mouse that ZFP804A, the homolog of ZNF804A, is required for normal progenitor proliferation and neuronal migration. Using a yeast two-hybrid genome-wide screen, we identified novel interacting proteins of ZNF804A. Rather than transcriptional factors, genes involved in mRNA translation are highly represented in our interactome result. ZNF804A co-fractionates with translational machinery and modulates the translational efficiency as well as the mTOR pathway. The ribosomal protein RPSA interacts with ZNF804A and rescues the migration and translational defects caused by ZNF804A knockdown. RNA immunoprecipitation–RNAseq (RIP-Seq) identified transcripts bound to ZFP804A. Consistently, ZFP804A associates with many short transcripts involved in translational and mitochondrial regulation. Moreover, among the transcripts associated with ZFP804A, a SZ risk gene, neurogranin (NRGN), is one of ZFP804A targets. Interestingly, downregulation of ZFP804A decreases NRGN expression and overexpression of NRGN can ameliorate ZFP804A-mediated migration defect. To verify the downstream targets of ZNF804A, a Duolink in situ interaction assay confirmed genes from our RIP-Seq data as the ZNF804A targets. Thus, our work uncovered a novel mechanistic link of a SZ risk gene to neurodevelopment and translational control. The interactome-driven approach here is an effective way for translating genome-wide association findings into novel biological insights of human diseases.
Introduction
Schizophrenia (SZ) is a chronic, debilitating disease affects ~1% of the population worldwide,1 and only partially effective, symptomatic treatments are available. The lifetime risk of suicide reaches 10–15% in SZ, and 5% of patients die from suicide.2 It ranks one of the top 10 leading causes of disability (WHO). The complexity of the symptoms affecting multiple emotional and cognitive functions results in disability for a large proportion of SZ patients. The annual cost for SZ in 2013 was $155.7 billion in the US.3 Thus, SZ is responsible for an enormous burden of disability, personal suffering and economic costs.
No single genetic or environmental factor accounts for most cases. Recent meta-analysis of genome-wide association studies identified more than 100 genetic loci as associated with SZ.4 One of the top associations was ZNF804A. rs1344706 is an intronic single-nucleotide polymorphism in the ZNF804A gene on chromosome 2q32.1 that was first associated with combined SZ and bipolar disorder.5 rs1344706 is associated with SZ and other psychiatric diseases in different populations.6, 7, 8, 9 Other single-nucleotide polymorphisms near the ZNF804A gene are associated with SZ,10, 11 suggesting that this gene contributes to disease risk. Structural variants at the ZNF804A locus occur in multiple psychiatric diseases, including copy number variations,8, 12 inversions or translocations.13, 14 Besides structural variations, the risk variant rs1344706 has also been reported to be associated with ZNF804A expression during fetal brain development15 and in different psychiatric diseases.16 Thus, ZNF804A gene dosage may be critical for normal brain function, and ZNF804A may participate in multiple psychiatric disorders.
ZNF804A affects brain structure and function.17, 18, 19 In humans, ZNF804A protein levels increase in early fetal stages (10–13 post conceptual weeks) and peak in the early-mid fetal stage (13–16 post conceptual weeks) when neuronal migration is raising in the brain.16, 20, 21 Overexpressing ZNF804A in human cells altered expression of genes involved in TGFβ signaling, which is critical for brain development.22 ZNF804A knockdown in human neuronal progenitor cells (NPCs) altered expression of genes that regulate cell adhesion,23 suggesting that it is involved in neuronal migration. Consistently, ZNF804A regulates neurite outgrowth, dendritic spine maintenance and activity-dependent structural plasticity.24 The ZNF804A protein sequence is predicted to have a C2H2-type zinc-finger (ZnF) domain characteristic of the classic ZnF family. ZNF804A chromatin-immunoprecipitation identified SZ risk genes (for example, COMT and PRSS16) as likely targets.25 Although these data support its role in brain development, its cellular function is unknown.
We sought to determine the function of ZNF804A. We found that its expression peaks at early embryonic day (E)12–E16 and modulates NPC proliferation and neuronal migration. An unbiased interactomic yeast two-hybrid (Y2H) screen identified a series of protein interactors of ZNF804A. Remarkably, many of these newly identified interactors are involved in translation and cell adhesion, suggesting a novel role of ZNF804A in protein synthesis. We further confirmed multiple interactions in an immunoprecipitation (IP) assay. Furthermore, ZNF804A co-fractionates with ribosomal proteins and modulates translational efficiency and mTOR signaling pathway. The ribosomal protein RPSA functionally interacts with ZNF804A and ameliorates ZNF804A-mediated defects in neural migration. RPSA reverses translational suppression caused by ZNF804A knockdown. Mechanistically, ZNF804A stimulates protein translation and interacts with mRNAs of a large number of genes, particularly those involved in mitochrondria functions and translation control. Interestingly, NGRN, another SZ risk gene, is a target of ZNF804A and counteracts the migration defect mediated by ZNF804A. In summary, our results reveal a novel role of the SZ risk gene, ZNF804A, in regulating neurodevelopment and RNA translational control.
Materials and methods
Animals
C57BL/6 N male and female mice from Taconic were housed (3–5 mice per cage) in a room with a 12-h light/dark cycle (lights on at 0700) and provided ad libitum access to food and water. All procedures were performed according to standards and requirements of the IACUC at the Pennsylvania State University.
Cell culture
Mouse neuroblastoma cell line, CAD, and human HEK293 (from ATCC, Manassas, VA, USA, mycoplasma free) were cultured in DMEM medium containing 10% FBS, L-glutamine and penicillin/streptomycin. Primary cortical neurons were cultured as described previously.26
DNA constructs
Human full-length ZNF804A cDNA (reference sequence: NM_194250.1) was PCR amplified from mRNA of SH-SY5Y human neuroblastoma cells and subcloned into the lentiviral vector pLV-3FLAG3HA-T2A-GFP at Nhe-I and BamH-I sites. Mouse Zfp804a cDNA (reference sequence: NM_175513.3) was PCR amplified from mRNA of mouse brain and subcloned into pcDNA3 vector with 3FLAG tag using primers: 5′-ATGGAGTGTTACTACATTGTCATCAGC-3′ and 5′-CTAGAAGAGGGGCTGGAGGGGAATGA-3′. The sequences for shRNAs targeting mouse Zfp804a are as follows: control shRNA: 5′-GGCTCCCGTGAATTGGAATCC-3′ against firefly luciferase; shRNA-1: 5′-CAGAGAGAATTTGCTCGAAATG-3′ shRNA-2: 5′-TCCTTTGCATTTCCAAAGAAAG-3′ in pLB2-CPGm vector. The sequences for shRNAs targeting mouse Rpsa are as follows: shRNA-3: 5′-GTGACGGTATCTACATCATAAA-3′ shRNA-4: 5′-GCCTGATCTTTACTTCTACAGA-3′, in pLKO vector (Sigma, St. Louis, MO, USA). Human siRNAs used in paper are as follows: siControl: 5′-rCrUrUrCrCrUrCrUrCrUrUrUrCrUrCrUrCrCrCrUrUrGrUGA, siRNA-1: 5′-rGrCrArGrArUrCrArArArUrArCrCrArCrUrA, siRNA-2: 5′-rGrGrU rUrUrA rUrArCrArArCrUrArCrGrArA. Other constructs expressing interacting proteins were obtained from the DNASU Plasmid Consortium.
ZNF804A antibody generation and immunoblot
To generate rabbit anti-ZNF804A polyclonal antibody, the first 450 nucleotides of ZNF804A coding sequence were cloned into pGEX-4T2. To make the recombinant proteins, GST was added in its N-terminus, and a 6HIS tag was added at its C-terminus. Expression of the recombinant proteins was induced with 1 mM IPTG in BL21 (DE3) Escherichia coli after the cell density at OD600 reached 0.4–0.6. Levels of the fusion protein were tested each hour after induction. Induced bacteria were collected and lysed with TNT lysis buffer, containing 5 mm imidazole, 20 μg ml−1 lysozyme, 10 μg ml−1 RNase A and 5 μg ml−1 DNase I. The lysate was alternately sonicated for 30 s and allowed to rest for 30 s for 5 min total. Clear lysates were centrifuged at 14000 r.p.m., 4 °C for 20 min. The supernatant was then collected and incubated with glutathione-agarose resins at 4 °C for 4 h with rolling. Purified fusion protein was collected with 10 mm glutathione.
Purified GST-ZNF804A-N150 protein was sent for generating polyclonal antibodies by Covance (Covance, Princeton, NJ, USA). Antibodies were purified from immune rabbit serum with ZNF804A-N150-conjugated protein A Sepharose beads (Invitrogen, Carlsbad, CA, USA, catalog number: 101041) at 4 °C for 4 h with rolling. The proteins were eluted with 0.1 m glycine, pH 2.8, and neutralized with 1 m Tris-Cl, pH 8.0, and the pH of the elution fractions was adjusted to pH 7.0, and NaN3 was added to 0.1%. Purified antibodies were preserved in −20 freezer with 50% glycerol. The specificity of homemade antibodies was tested by blotting with ZNF804A antibody, ZNF804A antibody with GST or ZNF804A antibody with GST-ZNF804A-N150 antigen (Supplementary Figure S1).
In utero electroporation
E15 mice were used in in utero electroporation for overexpression, and E14.5 mice were used for knockdown experiments as previously described.27 Pregnant mice were deeply anesthetized with 20 mg ml−1 Avertin in 20 μl g−1 body weight ratio and put on a heating pad. The uterine horns were carefully exposed through a midline abdominal incision. A DNA solution was prepared with the same concentration of control and experimental in PBS containing 0.01% fast green as a tracer. DNA (2 μl) was injected through the uterine wall into the lateral ventricle of the embryos with a glass micropipette made from a microcapillary tube. After injection, electroporation (five 50 ms square pulses of 35 V with 950 ms intervals) was carried out with the positive end on the cortex side of the embryo. Then, uterine horns were placed back into the abdominal cavity, and the cavity was filled with warm PBS. The abdominal wall of the pregnant mouse was sutured.
Embryos were harvested 3 days after DNA electroporation. The brains were dissected out, fixed in 4% paraformaldehyde overnight. For expression validation, the brains were transferred to PBS for another 24 h, and sectioned by Vibratome in 35 μm thickness. Rabbit anti HA antibody (Cell signaling, Danvers, MA, USA) was used to confirm ZNF804A overexpression, and ZNF804A N150 antibody was used to confirm shRNA knockdown. Meanwhile, the brain electroporated with ZNF804A overexpression construct (3:1 mixed with GFP-expressing construct) was dissected out under Zeiss Axio Zoom. V16 florescence dissection microscope. GFP-expressing tissue and three random pieces of non-GFP tissue was lysed and tested in western blot. For functional studies, the PFA fixed brains were dehydrated in 30% sucrose in PBS overnight in 4 °C. After that, the brains were embedded in OCT compound (Tissue-Tek* O.C.T. Compound, VWR 25608-930, Radnor, PA, USA), frozen on dry ice and stored at -80 °C until use. Cryosections at 14 μm thickness were made on a cryostat and examined by immunohistochemistry.
Y2H library screening and pairwise assays
A pre-made cDNA library derived from HeLa cell and brain containing 9.6 × 106 primary clones were screened using ZNF804A protein bait. The screens were performed in yeast strain L40, in which transcription of LexA-driven reporters results in the production of beta-galactosidase and also allows growth of yeast cells on medium lacking histidine. Approximately 5 × 106 clones were screened with the bait. Library-derived cDNAs were PCR-amplified from His+, beta-galactosidase+ yeast colonies and analyzed by DNA sequencing.
Plasmid pHybLex-ZNF804A-Zeo was constructed by subcloning full-length human ZNF804A cDNA into the Y2H bait plasmid pHybLexZeo (Invitrogen) to generate hybrid proteins with the LexA DNA binding domain appended to the N-terminus of ZNF804A protein.
Y2H screening and assays were performed according to the manufacturer's instructions for the Hybrid Hunter Y2H system (Invitrogen) in yeast strain L40 in which LexA-driven transcription results in expression of both histidine and beta-galactosidase reporters. Briefly, yeast cells were transformed with the bait plasmid pHybLex-ZNF804A-Zeo, and selection was maintained with Zeocin. Yeast cells harboring the bait plasmids were transformed with a pre-made HeLa cell-derived Y2H cDNA library constructed in plasmid pJG4-5 in which preys are fused with B42 transcription activation domain (Invitrogen). For screening, transformants were plated on synthetic medium lacking tryptophan, uracil, lysine and histidine and containing 300 μg ml−1 Zeocin. After incubation for 3 days at 30 °C, growing colonies were selected as primary positives and further tested using a beta-galactosidase colony filter assay, as instructed by the manufacturer. The positive yeast clones were confirmed by DNA sequencing analysis.
In pairwise assays to confirm binding in yeast, prey plasmids were isolated from His+, beta-galactosidase+ yeast clones. Prey plasmids were re-transformed into L40 cells harboring pHybLex-PIV5 M-Zeo, pHyb-PIV5 M-LexZeo, or the empty bait plasmid pHybLexZeo. Beta-galactosidase activity was measured in replicates of four using a colony filter assay.
Polyribosome sucrose gradient
Cells were grown in 100-mm dish to 80–90% confluence within 24 h of seeding. At 1 h before collection, 100 μg ml−1 cycloheximide (CHX) were added to culture medium. The cell monolayers were washed three times with ice-cold PBS containing 100 μg ml−1 CHX, and 3–5 × 106 cells were lysed in 0.5 ml buffer containing 5 mM Tris-HCl, pH 7.5, 2.5 mM MgCl2, 1.5 mM KCl, 100 μg ml−1 CHX, 0.5% Triton X-100, 0.5% sodium deoxycholate, 1 mM phenylmethyl sulfonyl fluoride (PMSF), 1 mM Na3VO4, 1 mM NaF and 10 μl ml−1 protease inhibitor mixture P8340 (Sigma). The EDTA treatment group was treated with 30 mm EDTA after cell lysis. After centrifugation at 13,000 r.p.m. for 30 min, the supernatant was collected as cytoplasmic extracts, which were then analyzed by sedimentation velocity in 7.5–30% (w/w) linear sucrose gradients made up in 20 mm HEPES, 100 mm KCl, 100 mm MgCl2 and 100 μg ml−1 CHX. After centrifugation in a Beckman SW55-Ti rotor for 2 h at 48 000 r.p.m. at 4 °C, 14 fractions were collected from top to bottom. All steps were carried out at 4 °C. 6 × protein loading buffer was added to each fraction. After boiling in 100 °C for 5 min, 14 samples were ready to be tested in the western blot assay.
SUnSET assay
To test protein translation efficiency, the SUnSET method was applied using a puromycin labeling technique.28 HEK293 cell was transfected with either ZNF804A expressing constructs or siRNAs targeting endogenous ZNF804A. At 48 h post transfection, the negative control cells were first treated with 50 μg ml−1 CHX for 20 min and then with 10 μg ml−1 puromycin and 50 μg ml−1 CHX. The experimental groups were treated with 10 μg ml−1 puromycin alone for 20 min. After the puromycin pulse, the old medium was replaced with fresh medium, and the chasing step took 1 h. Cell lysates were collected and analyzed by western blot. Nascent proteins were detected by a specific mouse anti-puromycin antibody.
Bioorthogonal non-canonical amino acid tagging (BONCAT)
CAD cells were transfected with DNA constructs for 48 h before changing to 4 mm l-azidohomoalanine (AHA)-containing HEPES-buffered saline for 2 h and then collected in PBS (pH 7.8) supplemented with protease inhibitors without EDTA. Total protein concentrations were determined, and samples were diluted so that each sample contained an equal amount of total protein. Nascent proteins were tagged by incubating protein extracts with Cy7-alkyne, as described.29 After an overnight incubation, the same volume of each sample was separated by SDS page gel and transferred to a western blot membrane. Signal was detected using an Odyssey CLx infrared imager (LI-COR Bioscience, Lincoln, NE, USA) at a wavelength of 800 nm. AHA and Cy7-alkyne were purchased from Click Chemistry Tools (AHA, cat. #1066; Cy7-alkyne, cat. #1053).
Duolink in situ interaction assay
The interaction between RPSA and ZFP804A in CAD cells was detected using Duolink In Situ Orange Starter Kit Mouse/Rabbit (DUO92102, Sigma), according to the instructions of the manufacturer. CAD cells were plated on the coverslips coated with poly-d-lysine (PDL, Sigma, 100 μg ml−1) or mouse laminin (BD, Franklin Lakes, NJ, USA, 25 μg ml−1). After incubation of primary antibodies against RPSA (1:100, mouse) and ZFP804A (1:200, rabbit), two proximity ligation assay (PLA) probes (mouse and rabbit), which were oligonucleotide labeled secondary antibodies, were added. The PLAs generated a signal only when they were bound in close proximity (<40 nm).30 The resulting positive signals were visualized as bright fluorescent dots, each representing one reaction spot. The cells were visualized with a microscope system (Apotome.2, Zeiss, Oberkochen, Germany).
Newly synthesized proteins can be labeled with puromycin and visualized in situ with PLA fluorescent tags (fluorescence non-canonical amino acid tagging; FUNCAT).30 To determine if ZNF804A affects translation efficiency of associated transcript, CAD cells expressing vector or ZNF804A construct (GFP positive) were transiently labeled with puromycin for 30 min as described in the SUnSET assay. The translation efficiency was monitored by the PLA signals from both puromycin and the specific protein, such as USMG5 and RPL7A. The specificity of this assay was assessed by omitting the incubation with puromycin; thus, no positive signals were obtained even in the presence of the two primary antibodies.
RNA immunoprecipitation–RNAseq (RIP-Seq)
The RIP was conducted as previously described.31, 32 Two buffers were used: polysome lysis buffer (100 mm KCl, 5 mm MgCl2, 10 mm HEPES, pH 7.0, 0.5% NP40, 1 mm DTT, 100 units per ml RNase Out, 400 μM ribonucleoside-vanadyl complex (New England Biolab, Ipswich, MA, USA), and protease inhibitor cocktail) and NT2 Buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm MgCl2 and 0.05% NP40). WT C57/B6 mouse brains at E18 were homogenized in 1 ml of polysome lysis buffer. Cell lysates were centrifuged at 15 000 r.p.m. for 15 min to remove any debris. The supernatant of the lysate contained isolated ribonucleoproteins. Protein-G Sepharose beads (BioVision, Milpitas, CA, USA) were incubated in 2.5 ml of NT2 buffer with 5% BSA for 1 h at 4 °C. Protein-G beads were pulse centrifuged, and rabbit anti-ZNF804A or pre-immune rabbit normal IgG was incubated directly to the bead pellet overnight at 4 °C with constant tumbling. The following day, the beads were washed with 1 ml of cold NT2 buffer for a total of five washes, and the pellet was re-suspended in 850 μl of cold NT2 buffer supplemented with protease inhibitors. Next, 100 μl of homogenized brain lysate was added directly to each sample of antibody-conjugated beads (ZNF804A and control serum). Samples were thoroughly mixed and tumbled for 2 h at room temperature. After the incubation, the beads were washed with 1 ml of NT2 buffer for a total of five washes before re-suspending the pellet with 100 μl NT2 buffer. RNA associated with ZFP804A was isolated from the beads with TRIzol (Invitrogen) and sent for RNAseq analysis with the Illumina TruSeq Stranded mRNA Kit (NIH NHLBI Sequencing Core). One lane of 50 × 50 paired-end reads was generated by the Illumina Hi-Seq 2500 platform at the NHLBI Genomics Core and mapped to the human genome (UCSC build hg19). The resulting paired-end sequencing reads were aligned to mouse genome (mm10) using STAR aligner v2.4.33 Sequencing quality control metrics were calculated using Picard. Reads uniquely mapped to annotated gene features (GENCODE vM6) were counted at gene-level with the FeatureCount software package.34 Genes enriched by anti-ZNF804A immunoprecipitation over control serum were called using the edgeR and voom r packages with experimental batches adjusted as covariates.35, 36 Over-represented functional categories among the ZNF804A targets were identified using PANTHER GO Enrichment Analysis.37 Brain cell-type-specific gene signatures (that is, genes with greater than eightfold expression on average in one cell type over other cell types) were derived from published mouse brain cell transcriptome data set.38 RIP-Seq raw data were uploaded to NIH Genebank with project ‘PRJNA362465’.
Statistical analysis
For the quantitation experiments, data were from at least three independent experiments. Two-tailed unpaired Student’s t-test was used to compare the data of immunohistochemical studies between the control and experimental groups. D'Agostino and Pearson normality tests were used to verify the normal distribution of variables. Two-group comparisons of data, for which the normality assumption could not be ascertained (Duolink assays), were analyzed by Mann–Whitney U-test. Multiple comparisons were analyzed by one-way ANOVA (Tukey's multiple comparisons test when compares every mean with every other mean, and Dunnett’s test when compares every mean to a control mean). In all analyses, P<0.05 was considered statistically significant. All statistical analysis was performed in PRISM software (Graphpad, La Jolla, CA, USA). No randomization method was used to allocate animals in each group. No blinding analysis was performed for the animal allocation. The exact number of samples indicates biological replicates and are indicated in each figure legend.
Results
ZNF804A expression in developing and adult brain
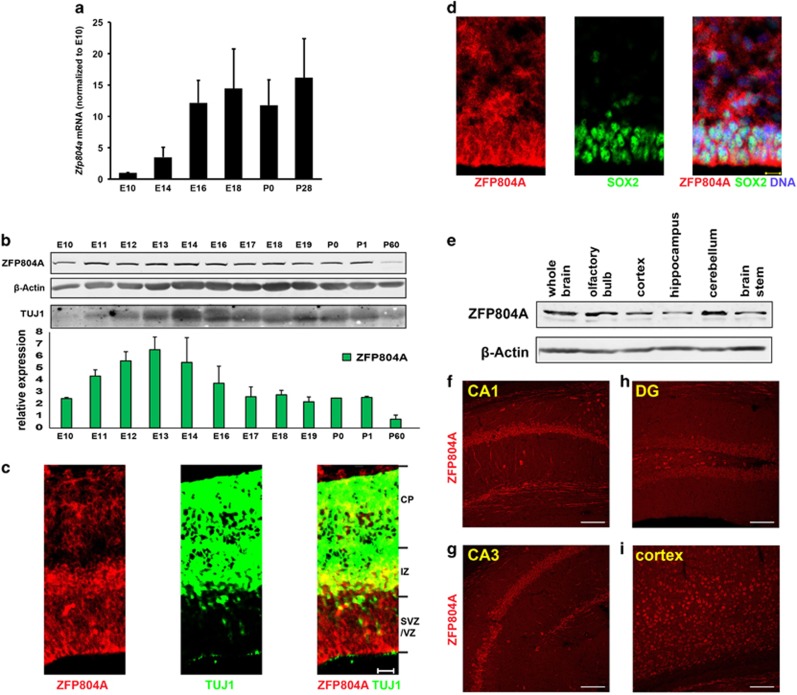
The expression of ZNF804A in human brain increases from embryonic to early fetal stage and reaches a peak around early-mid fetal stage. It decreases afterwards and stays at a constant level till late adulthood.20 Endogenous mouse homolog of ZNF804A (ZFP804A) is expressed in E11 NPCs.25 We examined ZFP804A in mouse brain. Quantitative PCR with reverse transcription showed that Zfp804a transcript peaks in the embryonic day 16 (E16) and postnatal day 0 (P0) were 10-fold greater than at E10 (Figure 1a). Human ZNF804A expression peaks in early-mid fetal development.20 To measure protein levels, we generated a rabbit polyclonal antibody to the N-terminal 150 amino acids of ZNF804A, which is highly conserved as ZFP804A protein. The affinity-purified antibody recognized exogenous and endogenous ZNF804A protein from human or mouse cells (Supplementary Figure S1A) and purified recombinant GST-ZNF804A protein fragment (Supplementary Figure S1B). The ZNF804A band could be competed off by the recombinant fragment (GST-ZNF804A-N150), but not by GST, confirming antibody specificity (Supplementary Figure S1A). Moreover, our ZNF804A antibody recognizes both exogenous ZNF804A with HA tag (high molecular weight with red) and endogenous ZFP804A (low molecular weight with green) (Supplementary Figure S1C). The ZNF804A band completely overlapped with a mouse ZNF804A monoclonal antibody from Millipore, further supporting its specificity (Supplementary Figure S1D). Our antibody was specific as it recognized a strong band on a western blot from mouse brain and HEK293 cells (Supplementary Figure S1E).
Figure 1.
ZFP804A expression during brain development. (a) qRT-PCR shows Zfp804a mRNA is increased during brain development. n=2, mean ±s.e.m. (b) Endogenous ZFP804A expression increases from E10 to E16 and peaks at E13, at the whole brain level. n=2, mean ±s.e.m. (c) Cortical staining of ZFP804A in E14 brain with TUJ1, a new-born neuron marker. Green, TUJ1; red, ZFP804A. Scale bar, 20 μm. (d) Cortical staining of ZFP804A at E14 brain with SOX2, an NPC marker. Green, SOX2; red, ZFP804A. Scale bar, 20 μm. (e) ZFP804A is highly expressed in the cortical and hippocampal regions of the mouse brain. (f–i) ZFP804A expression in the CA1 (f), the CA3 (g), the DG (h) and the cortex (i). Scale bar, 50 μm. DG, dental gyrus; NPC, neuronal progenitor cell; qRT-PCR, quantitative PCR with reverse transcription.
Using the antibody, we found increased ZFP804A protein in embryonic brain from E11 to E14, when neurogenesis and neuronal migration peak, and decreased levels in adult brain (Figure 1b). Next, we tested ZFP804A distribution by immunohistochemistry. ZFP804A is expressed throughout the neocortex at E14, with particularly high levels in the subventricular zone (SVZ), intermediate zone (IZ) and upper layer of the cortical plate (CP, Figure 1c). ZFP804A expression was detected in SOX2-positive NPCs in vitro (Supplementary Figure S2) and in utero in E14 brains (Figure 1d). ZFP804A was mainly localized in the cytosol and extensions of cultured primary SOX2-positive NPCs with low levels in the nucleus (Supplementary Figure S2), despite ZNF804A contains a ZnF domain. In adult brain, olfactory bulb and cerebellum expressed higher levels of ZFP804A than other regions (Figure 1e). ZFP804A was detected in cortical and hippocampal neurons (Figures 1f–i). It labeled dendritic tracts ventral to the CA1 (Figure 1f), suggesting a role in developing and mature brains.
ZNF804A regulates neuronal migration and NPC proliferation
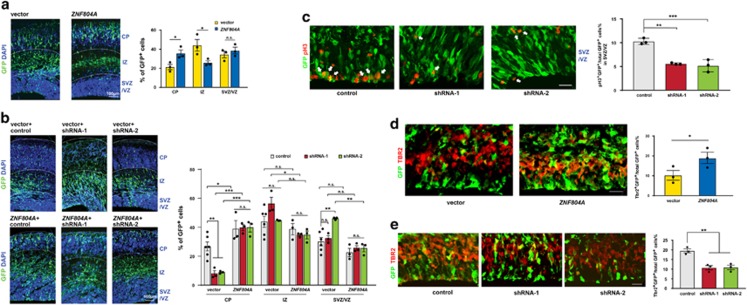
To determine the function of ZNF804A, an overexpression construct was used to express the full-length ZNF804A coding region fused with 3HA and 3FLAG tags (Supplementary Figure S3A). To confirm the overexpression of ZNF804A in utero, vector and ZNF804A construct were electroporated into E15 mouse brains. Three days later, brains were analyzed. Overexpressed ZNF804A in vivo was clearly detected by immunohistochemistry staining (Supplementary Figure S3B). Moreover, the electroporated brain was dissected to collect brain lysate for GFP-positive tissue and neighboring GFP negative portion. The HA–tagged ZNF804A can be detected by western blot only in GFP+ brain lysate (Supplementary Figure S3C). Overexpressing ZNF804A led to about twofold more cells migrating to the CP (Figure 2a), and cells migrating to the IZ dropped from 44.1% in the vector group to 25.8% in the overexpression group. Yet the cells in the SVZ were not affected by ZNF804A, suggesting that ZNF804A overexpression selectively stimulates cells in the IZ migrating to the CP.
Figure 2.
ZNF804A regulates neuronal migration and NPC proliferation. (a) ZNF804A overexpression enhances neural migration in E18 neocortex. Vector or ZNF804A plasmids were electroporated into E15 embryonic mouse brains, and mice were killed at E18. Percentage of GFP cells in each region. n=3 for each group, *P<0.05, n.s., mean ±s.e.m., t-tests. Scale bar, 100 μm. (b) Zfp804a knockdown induces migration defects of neural stem cells in E17.5 neocortex, which are rescued by ZNF804A overexpression. Control or Zfp804a shRNA plasmids were electroporated into E14.5 embryonic mouse brains alone or with ZNF804A, and mice were killed at E17.5. Percentage of GFP cells in each region. n=6 for vector+control and n=3 for other groups, *P<0.05, **P<0.01, ***P<0.001, n.s., not significant, mean ±s.e.m., one-way ANOVA with Tukey's multiple comparison test. Scale bar, 100 μm. (c) Brain slices of embryonic brains electroporated with control and shRNA constructs are stained with pH3. The SVZ/VZ area is shown. Percentages of pH3 and GFP double-positive cells in total GFP-positive cells are measured. n=3 for each group, ***P<0.001, mean ±s.e.m., t-tests. Scale bar, 20 μm. White arrows indicate GFP and pH3 double-positive cells. (d) Embryonic brains were electroporated with vector and ZNF804A OE constructs at E15 and harvested at E18. Coronal sections stained with intermediate neural progenitor marker, TBR2 (red). Percentages of TBR2 and GFP double-positive cells in total GFP-positive cells are shown. n=3, *P<0.05, mean ±s.e.m., t-tests. Scale bar, 20 μm. (e) Embryonic brains were electroporated with control and Zfp804a shRNAs constructs at E14.5 and harvested at E17.5. Brain sections stained with TBR2 (red). Percentages of TBR2 and GFP double-positive cells in total GFP-positive cells are shown. n=3 for each group, **P<0.01, mean±s.e.m., t-tests. scale bar, 20 μm. ANOVA, analysis of variance; CP, cortical plate; GFP, green fluorescent protein; n.s., not significant; IZ, the intermediate zone; SVZ/VZ, the subventricular zone/ventricular zone.
To assess the knockdown effect, two specific shRNAs against mouse Zfp804a were used. Their efficiency to knockdown the exogenous and endogenous Zfp804a expression (Supplementary Figures S4A and B, respectively) was confirmed in western blot. To manipulate the ZFP804A expression level in vivo, the shRNA or control shRNA was electroporated into E14.5 mouse brains, which were collected and processed 3 days after. The knockdown effect in utero was confirmed by immunostaining with ZNF804A-N150 antibody (Supplementary Figure S4C). Interestingly, Zfp804a silencing resulted in a fourfold fewer cells in the CP (Figure 2b). Thus, Zfp804a knockdown causes a specific migration defect because wild-type ZNF804A, which is resistant to mouse shRNAs (as shown in Supplementary Figures S4D and E), rescues the defect (Figure 2b).
As ZFP804A is expressed in SOX2-positive NPCs, we examined the effect of ZFP804A on cell proliferation in vitro. Zfp804a shRNAs were transfected into CAD cells. BrdU incorporation and a mitotic marker, phospho-histone H3 at serine 10 (pH3), were measured (Supplementary Figures S5A and B). Both Zfp804a shRNAs reduced BrdU incorporation and mitotic index, suggesting Zfp804a is required for normal cell proliferation. We further determined if Zfp804a regulates NPC proliferation in utero with pH3. Zfp804a knockdown in utero decreased cell proliferation (Figure 2c). These results indicated that Zfp804a is critical in cell proliferation.
Since ZFP804A modulates NPC proliferation, cell fate may be affected. We tested if ZFP804A modulates the population of intermediate neural progenitors. E14.5 mouse brains were electroporated with ZNF804A cDNA or Zfp804a shRNA constructs, and at E17.5, brains were analyzed by immunohistochemistry with GFP and TBR2 antibodies (Figures 2d and e). Overexpression of ZNF804A increased TBR2-positive intermediate neural progenitors twofold in the cortex (Figure 2d), whereas Zfp804a knockdown decreased TBR2-positive intermediate neural progenitors by twofold (Figure 2e), suggesting that a correct level of ZNF804A is critical to maintain cell identity. In addition, BRN2-positive upper-layer cortical neurons were increased by 15% in ZNF804A-overexpressing cells (Supplementary Figure S6A). However, Zfp804a silencing did not changed the BRN2-positive neuronal population (Supplementary Figure S6B).
Y2H screen reveals novel ZNF804A-interacting proteins
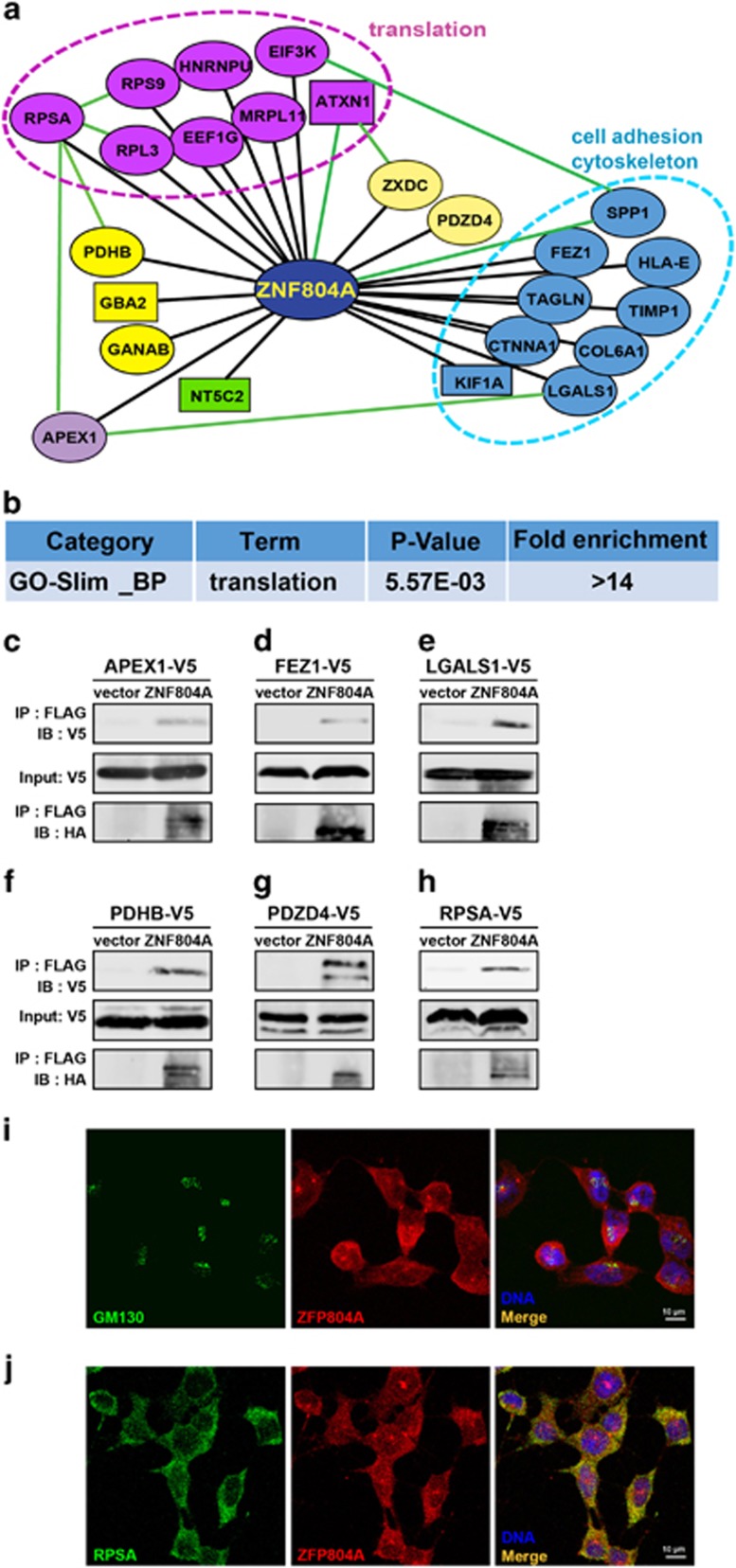
To determine the underlying molecular mechanisms that mediate ZNF804A neural functions, we next identified its interacting proteins in an unbiased Y2H screen. Using full-length human ZNF804A, we found 22 new interacting proteins besides the known binding proteins, ATAXIN139 and SPP140 (Supplementary Table S1). Our simplified interactomic network, based on our Y2H results and published databases (Figure 3a), shows that several new ZNF804A-interacting proteins bind each other (green connections) from other studies.41, 42 For example, RPSA interacts with RPL3, PDHB, APEX1 and RPS9,41, 42 and now our study reveals that they interact with ZNF804A (black connections), supporting the notion that ZNF804A brings multiple proteins together. We have run PANTHER Overrepresentation Test using PANTHER GO-Slim Biological Process Data Set (http://www.geneontology.org/page/go-enrichment-analysis) on our interactomic genes and identified functional process in translation (P=0.00557, fold enrichment=14.11, from 248 Biological Processes and 20972 human genes, Figure 3b and Supplementary Table S2).
Figure 3.
ZNF804A interacts with genes involved in translation and cell adhesion. (a) Biological processes of ZNF804A-interacting proteins. Black lines indicate new interactions identified here. Green lines indicate interactions reported by NCBI. Purple cluster represents genes in translational control. Blue cluster includes genes involved in cell adhesion and cytoskeleton. (b) Gene Ontology analysis shows enrichment of genes involved in translational regulation. (c–h) Immunoprecipitation studies confirm that ZNF804A associates with APEX1, FEZ1, LGALS1, PDHB, PDZD4 and RPSA. Each experiment has been repeated twice to confirm the results. (i) ZFP804A (red) is co-localized with GM130 (green) in CAD cells. Scale bar, 10 μm. (j) ZFP804A (red) is co-localized with endogenous RPSA (green) in CAD cells. Scale bar,10 μm.
Some new ZNF804A-interacting proteins are implicated in disorders (Supplementary Table S1). Mutations in GBA2, NT5C2 and KIF1A cause hereditary spastic paraplegia.43, 44, 45 FEZ1 interacts with the SZ risk gene, DISC1 and regulates axonal outgrowth.46, 47 RPSA is a component of ribosomal proteins and a non-integrin laminin receptor, which regulates cell adhesion and migration.48 To confirm the Y2H results, we obtained multiple constructs expressing V5-tagged APEX1, FEZ1, LGALS1, PDHB1, PDZD4 and RPSA, and our immunoprecipitation (IP) study showed that these proteins physically interact with ZNF804A (Figures 3c–h). ZNF804A interacts with ribosomal proteins and was enriched in an organelle close to the nucleus, but a weaker signal was also found in the cytosol and nucleus (Supplementary Figure S2A). Moreover, ZFP804A was co-localized with GM130, a marker for cis-Golgi body (Figure 3i and Supplementary Movie 1), suggesting a role for ZFP804A in protein translation. Among ZNF804A-interacting proteins, RPSA was co-localized with ZFP804A in the cytosol (Figure 3j), confirming that they are in the same complex.
ZNF804A controls translational efficiency
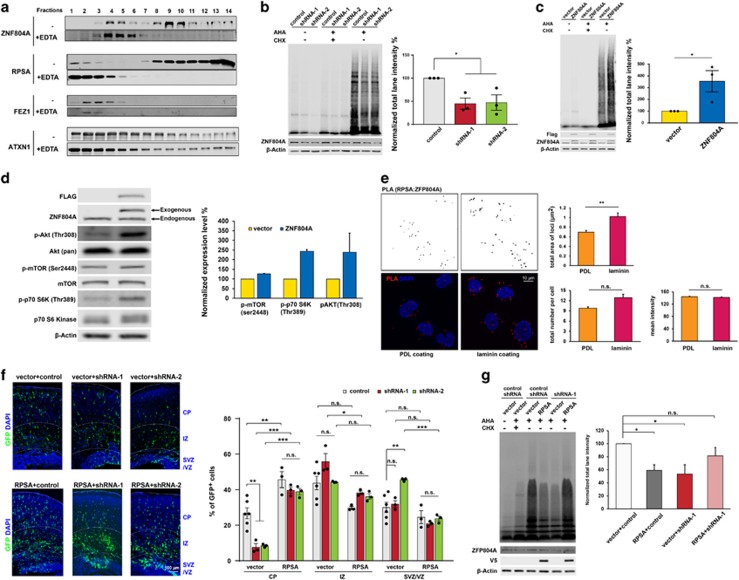
As ZnF domains function as RNA-binding motifs,49 we hypothesized that ZNF804A is involved in the mRNA translational machinery. Many RNA-binding proteins, such as fragile X mental retardation protein (FMRP), are present in ribonucleoprotein (RNP) complexes associated with heavier sedimenting polyribosomes (HSP). Functional clustering of our Y2H results showed that ZNF804A is involved in translational regulation (Figures 3a and b). To test whether the distribution of ZNF804A was similar to that of FMRP, a lysate of HEK293 cells was analyzed by sucrose gradient centrifugation. ZNF804A was found in two fractions (Figure 4a). A heavier fraction corresponding to HSP overlapped with the RPSA fraction and a lighter one overlapped with the expression of FEZ1, suggesting that ZNF804A forms multiple functional complex in cells. When EDTA was added to disrupt polyribosomes and RNP complexes, ZNF804A dissociated from polyribosomes and switched to the fractions at the top of the gradient similar to RPSA. These results suggest that ZNF804A participates in ribosomal functions.
Figure 4.
ZNF804A controls protein translation efficiency. (a) ZNF804A sedimented in two fractions of polyribosomes prepared from HEK293 cells. EDTA treatment disassociated ZNF804A from the high-to-low molecular mass fractions. (b) AHA-labeled nascent proteins decrease with Zfp804a shRNAs in mouse CAD cells. n=3 for each group, *P<0.05, mean±s.e.m., t-tests. (c) AHA-labeled nascent proteins increase with ZNF804A overexpression in mouse CAD cells. n=3 for each group, *P<0.05, mean±s.e.m., t-tests. (d) ZNF804A overexpression increases phosphorylation of AKT, mTOR and p70 S6K. n=2, mean±s.e.m. (e) Laminin stimulates ZNF804A and RPSA interaction. Duolink assay detects close interaction of ZNF804A and RPSA in response to PDL and laminin. Quantifications show total area of the loci (unit= μm2), total number of loci each cell, and mean intensity (AU). n=99 for PDL coating and n=107 for laminin coating, **P<0.01, mean±s.e.m., Mann–Whitney U-test. Scale bar, 10 μm. (f) Co-electroporation of RPSA with Zfp804a shRNAs in E14.5 embryonic brain rescues the migration deficit caused by ZNF804A knockdown. Percentage of GFP cells in each region. n=6 for vector+control and n=3 for other groups, *P<0.05, **P<0.01, ***P<0.001, mean ±s.e.m., one-way ANOVA with Tukey's multiple comparison test. Scale bar, 100 μm. (g) Deficiency of AHA-labeled nascent protein caused by Zfp804a shRNAs can be rescued by RPSA in mouse CAD cells. n=5, *P<0.05, **P<0.01, mean±s.e.m., one-way ANOVA with Dunnett's multiple comparison test. AHA, azidohomoalanine; ANOVA, analysis of variance; AU, arbitrary unit; GFP, green fluorescent protein; n.s., not siginificant PDL, poly-d-lysine.
We further hypothesized that ZNF804A modulates translational efficiency. To test the overall translational efficiency in CAD cells, bioorthogonal non-canonical amino acid, AHA, was used to tag newly synthesized peptides.29 Interestingly, BONCAT assay showed that ZFP804A knockdown decreased the efficiency of nascent protein translation by 60% (Figure 4b). Conversely, when ZNF804A was overexpressed, the newly synthesized proteins increased threefold over control (Figure 4c). To further confirm that this observation is specific to ZNF804A function, but is not cell type- or chemical-dependent, we changed to HEK293 cells and used puromycin to label nascent protein synthesis.28 Consistently, overexpression of ZNF804A enhanced translation rate (Supplementary Figure S7A), whereas ZNF804A knockdown reduced translation (Supplementary Figure S7B). As ZNF804A increases translation efficiency and the mTOR signaling pathway has an important role in the control of protein synthesis, we next tested activities of several key regulators in the mTOR pathway (Figure 4d). Remarkably, ZNF804A overexpression stimulated phosphorylation of AKT on Thr308, p70 S6 kinase on Thr389 and mTOR on Ser2448, suggesting an increase in the kinase activity. We double confirmed that ZNF804A overexpression in mouse primary cortical neuron increased p-AKT and p-p70 S6K (Supplementary Figure S8). These results reveal a novel role of ZNF804A in translational modulation.
Among the proteins interacting with ZNF804A, RPSA, also known as laminin receptor 1 (LAMR1), is conserved throughout evolution50 and is required for the assembly and/or stability of the 40S ribosomal subunit. It affects cell adhesion, differentiation and migration.50 As RPSA co-localizes with ZFP804A inside of cells (Figure 3j), we further determined how the extracellular matrix affects their interaction and how close RPSA localizes with ZFP804A. We plated CAD cells on the PDL and laminin-coated coverslips. The interacting loci between RPSA and ZFP804A that are in close proximity (<40 nm), can be detected using the PLA probes30 (Supplementary Figure S13A). We found that larger total area of reaction spots was present in laminin-coated than PDL-coated coverslips (Figure 4e). These data further confirm that ZFP804A interacts with RPSA at very close distance (<40 nm) and suggest that laminin stimulates the ZFP804A-RPSA interaction.
To determine if RPSA rescues the cell migration deficit caused by Zfp804a knockdown, RPSA expression plasmids were co-electroporated into E14.5 brains with shRNA-1, shRNA-2 or control constructs and a GFP-expressing vector. Compared to vector groups at E17.5, RPSA overexpression stimulated neuronal migration and ameliorated the migration defect caused by Zfp804a knockdown in the CP (Figure 4f). However, this rescue effect is specific to neuronal migration, as RPSA cannot restore NPC proliferation that is decreased by ZFP804A knockdown as detected by pH3 (Supplementary Figure S9). Rescue of neural migration deficit by RPSA suggested that RPSA mediates ZNF804A’s role in translation. Since RPSA rescues migration defects of Zfp804a knockdown and RPSA also functions as a component of ribosome subunits, we further tested if the translational defect could be rescued by RPSA overexpression. Interestingly, both RPSA silencing (Supplementary Figure S10A) and overexpression (Supplementary Figure S10B) decreased translational efficiency. However, when co-expressing with Zfp804a shRNA, RPSA reversed the translational defects (Figure 4g). These data suggest that RPSA rescues ZFP804A-mediated migration defect by restoring the translational efficiency.
ZNF804A binds mRNAs of multiple genes and regulates gene expression
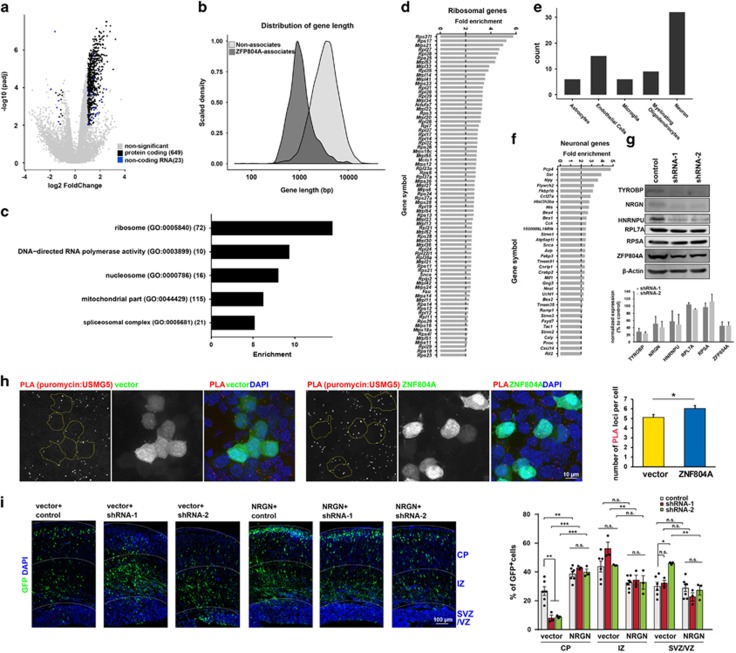
We examined the ability of ZNF804A to interact with RNA and identified its target genes by RIP-Seq. Target RNAs bound by ZFP804A in wild-type (WT) C57/B6 mouse brains at E18 were co-precipitated by anti-ZFP804A antibody followed by deep sequencing (Supplementary Figure S11). Compared to controls, 655 genes (637 protein-coding genes and 18 noncoding RNAs, Supplementary Table S3) were enriched (false discovery rate<0.01, fold change>2) in the ZFP804A IP samples (Figure 5a). The ZFP804A-associated transcripts tend to be short (median length=980 base pairs (bp), vs 3284 bp for all expressed genes in the study). This trend was noted in ubiquitously expressed and cell-type-specific ZFP804A-bound genes (Figure 5b). Few genes showed reductions, indicating that these enriched transcripts were specifically pulled down by ZFP804A.
Figure 5.
RNA-IP sequencing identified a large number of RNAs bound by ZFP804A. (a) Volcano plot of RIP-seq results showing target genes enriched by ZFP804A IP. (b) Gene length distribution of ZFP804A target genes versus background distribution. (c) Major functional categories enriched in ZFP804A target genes. (d) Enrichment of ribosomal genes by ZFP804A IP. (e) Brain cell-type-specific genes among the ZFP804A target genes. (f) Enrichment of neuron-specific genes, including several interneuron marker genes. (g) Western blot confirms that TYROBP and NRGN are downregulated when ZFP804A is knocked down. n=2, mean±s.e.m. (h) CAD cells expressing vector or ZNF804A construct (GFP+) were transiently labeled with puromycin. The translation efficiency was monitored by the PLA signals amplified from antibodies against puromycin and USMG5. Duolink assay showed that USMG5 translation is significantly increased by ZNF804A. n=105 with vector and n=114 with ZNF804A overexpression, *P<0.05, mean±s.e.m., Mann–Whitney U-test. Scale bar, 10 μm. (i) Co-electroporation of NGRN with Zfp804a shRNAs in E14.5 embryonic brain rescues the migration deficit caused by Zfp804a knockdown. Percentage of GFP cells in each region. n=6 for vector+control, n=7 for NRGN+control, n=3 for other groups, *P<0.05, **P<0.01, ***P<0.001, one-way ANOVA with Tukey's multiple comparison test. Scale bar, 100 μm. ANOVA, analysis of variance; IP, immunoprecipitates; GFP, green fluorescent protein; n.s., not significant.
By GO analysis, ZFP804A-bound genes were highly enriched for essential biological processes and cellular components, including two large groups of genes: ribosome (72 genes, P=1.96E−58) and mitochondria (112 genes, P=3.1E−52), and smaller groups of housekeeping processes (Figures 5c and d). ZFP804A was co-localized with mitochondrial proteins when labeling with MitoTracker (Supplementary Figure S12 and Supplementary Movie 2). When cross-examined with published gene expression data sets of mouse brain cell types, a few genes showed strong cell-type-specific gene expression. Among them, 35 ZFP804A target genes were highly specifically expressed in neurons and involved in a broad range of neuronal functions (for example, members of the Bex gene family involved in neuronal development). Additional target genes specific to non-neuronal cell types were also observed (for example, Tyrobp for microglia, Mgp for endothelial cells) (Figure 5e). A few interneuron-specific genes (that is, Pcp4, Sst, Npy, Cck, Tac1) were also recognized by ZFP804A (Figure 5f). Reductions in some interneuron genes were seen in SZ postmortem brain samples.51, 52 The interaction of ZNF804A with interneuron genes could provide further support for the causal link between the interneuron deficiency and SZ etiology.
Next, we knocked down ZFP804A in CAD cells and measured the protein level of ZFP804A bound genes (Figure 5g). TYROBP, which is implicated in dementia,53 bipolar disorder and SZ,54 and NRGN, which is encoded by SZ risk gene NRGN,4 showed 60% less protein than control shRNA, supporting the idea that ZNF804A controls expression of other SZ risk genes. Furthermore, we used a Duolink assay, combined with FUNCAT30 (Supplementary Figure S13A), to detect to what extend that ZFP804A affects the newly synthesized proteins identified from RIP-RNAseq data. In this assay, puromycin was used to trace labeled nascent proteins for half hour. Combination of the gene-specific antibody and puromycin antibody together with PLA probes can be used to specifically tag and quantify newly synthesized proteins in a cell. Interestingly, translational efficiency of a ZFP804A-associated transcript, Usmg5, was increased by ZFP804A (Figure 5h). However, this increase is not ubiquitous for every gene. Consistent with result in Figure 5g, ZNF804A did not affect the translation efficiency for another ribosomal gene, Rpl7a (Supplementary Figure S13B). Moreover, to determine the functional significance of Nrgn gene in ZNF804A-mediated roles, human NRGN expression plasmids were co-electroporated into E14.5 brains with control, shRNA-1 or shRNA-2 constructs, and a GFP-expressing vector and brains were analyzed at E17.5. Strikingly, similar to RPSA, NRGN promotes migration and reverses ZFP804A-mediated migration defects (Figure 5i). However, NRGN has no effect on NPC proliferation (Supplementary Figure S14). These results revealed a novel role of NRGN involved in neuronal migration.
Discussion
Since 2008, ZNF804A has been identified as a SZ risk gene in multiple genome-wide association studies.5, 6, 8, 9, 55 However, the functions of ZNF804A have not been fully elucidated. Here we identified a novel role of ZNF804A (as well as its mouse homolog ZFP804A) in neurodevelopment. ZNF804A is highly expressed in embryonic brain and present in neurogenic niches. Its knockdown leads to proliferation and migration defects. We identified multiple interactors of ZNF804A that are implicated in RNA translation and cell adhesion. The interactions were confirmed by IP. We showed ZNF804A is required for efficient translation. RPSA rescued a migration deficit due to reduced ZFP804A. RIP-Seq provided evidence that ZFP804A associates with mRNAs involved in translation and mitochondrial functions. To the best of our knowledge, this is the first interactome study to reveal a novel function of ZNF804A, and to provide molecular evidence on how this risk gene contributes to psychiatric disorders.
We found that ZFP804A is highly expressed in embryonic brain where it regulates NPC proliferation and neuronal migration. Our results are consistent with recent two studies,24, 56 supporting a critical role of ZFP804A in early brain development. Suppression of ZFP804A expression decreased NPC proliferation and migration of cortical neurons, suggesting that the proper expression of ZNF804A is critical for neural development, and that the disrupted expression may lead to disorders. Although SZ symptoms appear in adulthood, genetic mutations affecting early embryonic development could embed risk for future behavioral changes. NPCs proliferate and differentiate into mature neurons, and are essential for brain structural and functional integrity. The disturbances of early brain development may cause NPCs to deviate from their typical developmental path, resulting in alterations in neuronal circuitry and eventually resulting in the emergence behavioral deficits seen in adult psychiatric patients. The neurodevelopmental hypothesis of SZ proposes that a brain defect is inherited or sustained early in life, but is not fully expressed until adolescence.57, 58 Consistently, gestational disruption of brain development in rats leads to a reduction of the prefrontal cortex and hippocampus size, resulting in schizophrenic behavior changes.59 Decreases in neural stem cell proliferation have been reported in SZ patients.60 Evidence from longitudinal in vivo imaging studies of patients with childhood-onset-SZ61, 62, 63, 64, 65 revealed that progressive structural changes in brain precede the onset of SZ, and moreover, that these changes continue to progress after the onset of psychosis. Progressive changes in certain brain structures and neuronal circuits precede disease manifestation in SZ patients. Additionally, NPCs and neurons obtained through reprogramming of fibroblasts from SZ patients have shown aberrant differentiation,66 defective Wnt activation67 and abnormal neuronal connectivity.67 Thus, our data provide further evidence for the neurodevelopmental hypothesis.
Our results showed that ZFP804A knockdown leads to a migration deficit of cortical neurons. Many other proteins related to SZ affect neuronal migration of cortical neurons. For example, knockdown of DISC1 by RNAi leads to drastic neuronal migration deficits.68, 69 Consistently, microarray analysis of ZNF804A silencing in neurons showed that ZNF804A modulates genes that are involved in cell adhesion.23 Neuronal migration defect could link to the brain structural deficits observed in psychiatric patients carrying ZNF804A risk allele.17, 18, 70, 71 Although not reaching statistical significance, ZNF804A interacted with multiple proteins that modulate cell adhesion, also suggesting a role in cell adhesion. Consistent with this notion, it has been reported that ZNF804A localizes to the synapses and regulates neurite formation and dendritic spines maintenance in an activity-dependent manner in human NPC-derived neurons.24
We showed that RPSA rescues the migration deficit from Zfp804a knockdown. RPSA, also known as laminin receptor 1 (LAMR1), is conserved throughout evolution.50 LAMR1 was first identified as a 67-kDa-binding protein for laminin and later as a 37-kDa RPS2 family ribosomal component.72 Our results show, for the first time, that RPSA rescues a neuronal migration deficit caused by Zfp804a knockdown. That rescue could be achieved by balancing ribosomal function and recovery of the normal translation rate in cells. To test this, our data show that RPSA itself is required for normal translational efficiency. When co-expressing RPSA with Zfp804a shRNA, RPSA can restore the efficacy of translation to normal level, suggesting that RPSA rescues the migration defect through regulation of translation. However, as RPSA also functions as a laminin receptor, it may also modulate migration through cell-adhesion molecules. Using the Duolink technique that only detects protein–protein interaction in very close proximity (<40 nm), we observed that laminin-facilitated ZFP804A-RPSA interaction, suggesting that the extracellular matrix molecule may be involved in this process. Thus, both translation and extracellular matrix-dependent mechanisms play a role in ZNF804A-dependent migration.
ZNF804A is a key factor modulating protein synthesis. Many proteins that interact with ZNF804A are involved in translation, and our RIP-Seq results suggest that ZFP804A binds to RNA. Similar proteins include FMRP, which forms a messenger ribonucleoprotein complex via interacting with RNA transcripts (hairpin structure) and proteins. FMRP-messenger ribonucleoprotein is transported out of the nucleus by the FMRP nuclear export signal. In the cytoplasm, FMRP-messenger ribonucleoprotein complex associates with ribosomes and regulates protein synthesis in soma or dendrites. Like FMRP, ZNF804A associates with polyribosomes in an RNA-dependent manner. FMRP suppresses translation,73 whereas our data show that ZNF804A enhances translation. We also found that ZFP804A interacts with Tyrobp and Nrgn mRNA, the human homologs of which are risk genes associated with SZ,74 and modulates their expression, suggesting that they are direct targets of ZNF804A. RPSA was not changed by downregulating ZFP804A, suggesting that it is not a general translational modulator for all proteins. Importantly, NRGN can rescue the migration defect, but not proliferation defect caused by ZFP804A knockdown (Figure 5i and Supplementary Figure S14), suggesting that ZNF804A controls translation of different genes to mediate different cellular functions.
ZNF804A regulates both NPC proliferation and neuronal migration. Although NPC proliferation is tightly linked to neuronal migration, our data suggest that ZNF804A regulates these processes through different mechanisms. The first evidence is that RPSA, as a direct interacting protein of ZNF804A, can rescue ZFP804A-mediated migration defect (Figure 4f), but cannot restore the NPC proliferation. The second evidence is that Nrgn, a ZNF804A downstream gene, can also rescue ZFP804A-mediated migration defect (Figure 5i), but not the NPC proliferation. These results provide compelling evidences that NPC proliferation and neuronal migration, although both are affected by ZNF804A, are controlled by different pools of downstream genes.
In summary, we uncovered a novel role of the SZ risk gene, ZNF804A, in modulating neurodevelopment and translation. Our results provide new insight into the underlying molecular mechanisms of psychiatric disorders.
Acknowledgments
We thank Wenyuan Wang (MIT) for pLV-3FLAG3HA-T2A-GFP construct. We thank Young Investigator Grant of NARSAD and Scientist Development Grant of American Heart Association to Yingwei Mao. This work is supported by NIH 1R21MH108983. We thank Jun Zhu at NHLBI and Craig Praul at the Genomic Core facility of Pennsylvania State University for technical support of RIP-Seq. We thank Richard Ordway, Fumiko Kawasaki, Gong Chen, Bernhard Luscher, Timothy Jegla, Aimin Liu, Douglas Cavener, Siying Zhu, Xueqiao Jiang and Colleen McSweeney for technical support. L Liu is partially supported by the Research Foundation of National University of Defense Technology (JC 14-02-01). We thank Gary Wood for critical reading on our manuscript.
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
The authors declare no conflict of interest.
Supplementary Material
References
- International-Schizophrenia-Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature 2008; 455: 237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer BA, Pankratz VS, Bostwick JM. The lifetime risk of suicide in schizophrenia: a reexamination. Arch Gen Psychiatry 2005; 62: 247–253. [DOI] [PubMed] [Google Scholar]
- Cloutier M, Aigbogun MS, Guerin A, Nitulescu R, Ramanakumar AV, Kamat SA et al. The economic burden of schizophrenia in the United States in 2013. J Clin Psychiatry 2016; 77: 764–771. [DOI] [PubMed] [Google Scholar]
- Consortium SWGotPG. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014; 511: 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donovan MC, Craddock N, Norton N, Williams H, Peirce T, Moskvina V et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet 2008; 40: 1053–1055. [DOI] [PubMed] [Google Scholar]
- Zhang R, Lu SM, Qiu C, Liu XG, Gao CG, Guo TW et al. Population-based and family-based association studies of ZNF804A locus and schizophrenia. Mol Psychiatry 2011; 16: 360–361. [DOI] [PubMed] [Google Scholar]
- Williams HJ, Norton N, Dwyer S, Moskvina V, Nikolov I, Carroll L et al. Fine mapping of ZNF804A and genome-wide significant evidence for its involvement in schizophrenia and bipolar disorder. Mol Psychiatry 2011; 16: 429–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg S, Mors O, Borglum AD, Gustafsson O, Werge T, Mortensen PB et al. Expanding the range of ZNF804A variants conferring risk of psychosis. Mol Psychiatry 2011; 16: 59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley B, Thiselton D, Maher BS, Bigdeli T, Wormley B, McMichael GO et al. Replication of association between schizophrenia and ZNF804A in the Irish case-control study of schizophrenia sample. Mol Psychiatry 2010; 15: 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Valenzuela RK, Lu S, Meng L, Guo T, Du X et al. Is the conserved mammalian region of ZNF804A locus associated with schizophrenia? A population-based genetics analysis. Schizophr Res 2011; 133: 159–164. [DOI] [PubMed] [Google Scholar]
- Zhang F, Chen Q, Ye T, Lipska BK, Straub RE, Vakkalanka R et al. Evidence of sex-modulated association of ZNF804A with schizophrenia. Biol Psychiatry 2011; 69: 914–917. [DOI] [PubMed] [Google Scholar]
- Griswold AJ, Ma D, Cukier HN, Nations LD, Schmidt MA, Chung RH et al. Evaluation of copy number variations reveals novel candidate genes in autism spectrum disorder-associated pathways. Hum Mol Genet 2012; 21: 3513–3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkowski ME, Rosenfeld JA, Blumenthal I, Pillalamarri V, Chiang C, Heilbut A et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell 2012; 149: 525–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake J, Riddell A, Theiss S, Gonzalez AP, Haase B, Jauch A et al. Sequencing of a patient with balanced chromosome abnormalities and neurodevelopmental disease identifies disruption of multiple high risk loci by structural variation. PLoS One 2014; 9: e90894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill MJ, Bray NJ. Evidence that schizophrenia risk variation in the ZNF804A gene exerts its effects during fetal brain development. Am J Psychiatry 2012; 169: 1301–1308. [DOI] [PubMed] [Google Scholar]
- Tao R, Cousijn H, Jaffe AE, Burnet PW, Edwards F, Eastwood SL et al. Expression of ZNF804A in human brain and alterations in schizophrenia, bipolar disorder, and major depressive disorder: a novel transcript fetally regulated by the psychosis risk variant rs1344706. JAMA Psychiatry 2014; 71: 1112–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassink TH, Epping EA, Rudd D, Axelsen M, Ziebell S, Fleming FW et al. Influence of ZNF804a on brain structure volumes and symptom severity in individuals with schizophrenia. Arch Gen Psychiatry 2012; 69: 885–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Li M, Kang Z, Li L, Diao F, Zhang R et al. ZNF804A rs1344706 is associated with cortical thickness, surface area, and cortical volume of the unmedicated first episode schizophrenia and healthy controls. Am J Med Genet B Neuropsychiatr Genet 2015; 168B: 265–273. [DOI] [PubMed] [Google Scholar]
- Esslinger C, Walter H, Kirsch P, Erk S, Schnell K, Arnold C et al. Neural mechanisms of a genome-wide supported psychosis variant. Science 2009; 324: 605. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M et al. Spatio-temporal transcriptome of the human brain. Nature 2011; 478: 483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayatti N, Moss JA, Sun L, Ambrose P, Ward JFH, Lindsay S et al. A molecular neuroanatomical study of the developing human neocortex from 8 to 17 postconceptional weeks revealing the early differentiation of the subplate and subventricular zone. Cerebral Cortex 2008; 18: 1536–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi K, Hashimoto R, Yamamori H, Yasuda Y, Fujimoto M, Umeda-Yano S et al. The impact of the genome-wide supported variant in the cyclin M2 gene on gray matter morphology in schizophrenia. Behav Brain Funct 2013; 9: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill MJ, Jeffries AR, Dobson RJ, Price J, Bray NJ. Knockdown of the psychosis susceptibility gene ZNF804A alters expression of genes involved in cell adhesion. Hum Mol Genet 2012; 21: 1018–1024. [DOI] [PubMed] [Google Scholar]
- Deans PJ, Raval P, Sellers KJ, Gatford NJ, Halai S, Duarte RR et al. Psychosis risk candidate ZNF804A localizes to synapses and regulates neurite formation and dendritic spine structure. Biol Psychiatry 2016; 82: 49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girgenti MJ, LoTurco JJ, Maher BJ. ZNF804a regulates expression of the schizophrenia-associated genes PRSS16, COMT, PDE4B, and DRD2. PloS One 2012; 7: e32404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou D, Chen L, Deng D, Jiang D, Dong F, McSweeney C et al. DREADD in parvalbumin interneurons of the dentate gyrus modulates anxiety, social interaction and memory extinction. Curr Mol Med 2016; 16: 91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Ge X, Frank CL, Madison JM, Koehler AN, Doud MK et al. Disrupted in schizophrenia 1 regulates neuronal progenitor proliferation via modulation of GSK3beta/beta-catenin signaling. Cell 2009; 136: 1017–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods 2009; 6: 275–277. [DOI] [PubMed] [Google Scholar]
- Dieterich DC, Lee JJ, Link AJ, Graumann J, Tirrell DA, Schuman EM. Labeling, detection and identification of newly synthesized proteomes with bioorthogonal non-canonical amino-acid tagging. Nat Protoc 2007; 2: 532–540. [DOI] [PubMed] [Google Scholar]
- Dieterich DC, Hodas JJ, Gouzer G, Shadrin IY, Ngo JT, Triller A et al. In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nat Neurosci 2010; 13: 897–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alachkar A, Jiang D, Harrison M, Zhou Y, Chen G, Mao Y. An EJC factor RBM8a regulates anxiety behaviors. Curr Mol Med 2013; 13: 887–899. [DOI] [PubMed] [Google Scholar]
- Consortium ISGCatWTCC. Genome-wide association study implicates HLA-C*01:02 as a risk factor at the major histocompatibility complex locus in schizophrenia. Biol Psychiatry 2012; 72: 620–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013; 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014; 30: 923–930. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010; 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 2014; 15: R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res 2016; 44: D336–D342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O'Keeffe S et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 2014; 34: 11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Hao T, Shaw C, Patel AJ, Szabo G, Rual JF et al. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell 2006; 125: 801–814. [DOI] [PubMed] [Google Scholar]
- Long P, Samnakay P, Jenner P, Rose S. A yeast two-hybrid screen reveals that osteopontin associates with MAP1A and MAP1B in addition to other proteins linked to microtubule stability, apoptosis and protein degradation in the human brain. Eur J Neurosci 2012; 36: 2733–2742. [DOI] [PubMed] [Google Scholar]
- Wan C, Borgeson B, Phanse S, Tu F, Drew K, Clark G et al. Panorama of ancient metazoan macromolecular complexes. Nature 2015; 525: 339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havugimana PC, Hart GT, Nepusz T, Yang H, Turinsky AL, Li Z et al. A census of human soluble protein complexes. Cell 2012; 150: 1068–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin E, Schule R, Smets K, Rastetter A, Boukhris A, Loureiro JL et al. Loss of function of glucocerebrosidase GBA2 is responsible for motor neuron defects in hereditary spastic paraplegia. Am J Hum Genet 2013; 92: 238–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebe S, Lossos A, Azzedine H, Mundwiller E, Sheffer R, Gaussen M et al. KIF1A missense mutations in SPG30, an autosomal recessive spastic paraplegia: distinct phenotypes according to the nature of the mutations. Eur J Hum Genet 2012; 20: 645–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novarino G, Fenstermaker AG, Zaki MS, Hofree M, Silhavy JL, Heiberg AD et al. Exome sequencing links corticospinal motor neuron disease to common neurodegenerative disorders. Science 2014; 343: 506–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K, Honda A, Baba K, Taniguchi M, Oono K, Fujita T et al. Disrupted-In-Schizophrenia 1, a candidate gene for schizophrenia, participates in neurite outgrowth. Mol Psychiatry 2003; 8: 685–694. [DOI] [PubMed] [Google Scholar]
- Kang E, Burdick KE, Kim JY, Duan X, Guo JU, Sailor KA et al. Interaction between FEZ1 and DISC1 in regulation of neuronal development and risk for schizophrenia. Neuron 2011; 72: 559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea VE, Rossi FW, De Paulis A, Ragno P, Selleri C, Montuori N. 67 kDa laminin receptor: structure, function and role in cancer and infection. Infez Med 2012; 20: 8–12. [PubMed] [Google Scholar]
- Draper DE. Protein-RNA recognition. Annu Rev Biochem 1995; 64: 593–620. [DOI] [PubMed] [Google Scholar]
- DiGiacomo V, Meruelo D. Looking into laminin receptor: critical discussion regarding the non-integrin 37/67-kDa laminin receptor/RPSA protein. Biol Rev Camb Philos Soc 2016; 91: 288–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellios N, Huang HS, Baker SP, Galdzicka M, Ginns E, Akbarian S. Molecular determinants of dysregulated GABAergic gene expression in the prefrontal cortex of subjects with schizophrenia. Biol Psychiatry 2009; 65: 1006–1014. [DOI] [PubMed] [Google Scholar]
- Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci 2016; 19: 1442–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paloneva J, Kestila M, Wu J, Salminen A, Bohling T, Ruotsalainen V et al. Loss-of-function mutations in TYROBP (DAP12) result in a presenile dementia with bone cysts. Nat Genet 2000; 25: 357–361. [DOI] [PubMed] [Google Scholar]
- de Baumont A, Maschietto M, Lima L, Carraro DM, Olivieri EH, Fiorini A et al. Innate immune response is differentially dysregulated between bipolar disease and schizophrenia. Schizophr Res 2015; 161: 215–221. [DOI] [PubMed] [Google Scholar]
- Ripke S, O'Dushlaine C, Chambert K, Moran JL, Kahler AK, Akterin S et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet 2013; 45: 1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang EH, Kirtley A, Chandon TS, Borger P, Husain-Krautter S, Vingtdeux V et al. Postnatal neurodevelopmental expression and glutamate-dependent regulation of the ZNF804A rodent homologue. Schizophr Res 2015; 168: 402–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger DR. Implications of normal brain development for the pathogenesis of schizophrenia. Arch Gen Psychiatry 1987; 44: 660–669. [DOI] [PubMed] [Google Scholar]
- Murray RM, Lewis SW. Is schizophrenia a neurodevelopmental disorder? Brit Med J (Clin Res Ed) 1987; 295: 681–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagstad P, Mork A, Glenthoj BY, van Beek J, Michael-Titus AT, Didriksen M. Disruption of neurogenesis on gestational day 17 in the rat causes behavioral changes relevant to positive and negative schizophrenia symptoms and alters amphetamine-induced dopamine release in nucleus accumbens. Neuropsychopharmacology 2004; 29: 2052–2064. [DOI] [PubMed] [Google Scholar]
- Reif A, Fritzen S, Finger M, Strobel A, Lauer M, Schmitt A et al. Neural stem cell proliferation is decreased in schizophrenia, but not in depression. Mol Psychiatry 2006; 11: 514–522. [DOI] [PubMed] [Google Scholar]
- Keller A, Jeffries NO, Blumenthal J, Clasen LS, Liu H, Giedd JN et al. Corpus callosum development in childhood-onset schizophrenia. Schizophr Res 2003; 62: 105–114. [DOI] [PubMed] [Google Scholar]
- Giedd JN, Jeffries NO, Blumenthal J, Castellanos FX, Vaituzis AC, Fernandez T et al. Childhood-onset schizophrenia: progressive brain changes during adolescence. Biol Psychiatry 1999; 46: 892–898. [DOI] [PubMed] [Google Scholar]
- Gogtay N, Sporn A, Clasen LS, Nugent TF 3rd, Greenstein D, Nicolson R et al. Comparison of progressive cortical gray matter loss in childhood-onset schizophrenia with that in childhood-onset atypical psychoses. Arch Gen Psychiatry 2004; 61: 17–22. [DOI] [PubMed] [Google Scholar]
- Frazier JA, Alaghband-Rad J, Jacobsen L, Lenane MC, Hamburger S, Albus K et al. Pubertal development and onset of psychosis in childhood onset schizophrenia. Psychiatry Res 1997; 70: 1–7. [DOI] [PubMed] [Google Scholar]
- Mattai AA, Weisinger B, Greenstein D, Stidd R, Clasen L, Miller R et al. Normalization of cortical gray matter deficits in nonpsychotic siblings of patients with childhood-onset schizophrenia. J Am Acad Child Adolesc Psychiatry 2011; 50: 697–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robicsek O, Karry R, Petit I, Salman-Kesner N, Muller FJ, Klein E et al. Abnormal neuronal differentiation and mitochondrial dysfunction in hair follicle-derived induced pluripotent stem cells of schizophrenia patients. Mol Psychiatry 2013; 18: 1067–1076. [DOI] [PubMed] [Google Scholar]
- Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature 2011; 473: 221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka K, Kamiya A, Oh EC, Kanki H, Seshadri S, Robinson JF et al. DISC1-dependent switch from progenitor proliferation to migration in the developingcortex. Nature 2011; 473: 92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya A, Kubo K, Tomoda T, Takaki M, Youn R, Ozeki Y et al. A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat Cell Biol 2005; 7: 1167–1178. [DOI] [PubMed] [Google Scholar]
- Schultz CC, Nenadic I, Riley B, Vladimirov VI, Wagner G, Koch K et al. ZNF804A and cortical structure in schizophrenia: in vivo and postmortem studies. Schizophr Bull 2014; 40: 532–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nenadic I, Maitra R, Basmanav FB, Schultz CC, Lorenz C, Schachtzabel C et al. ZNF804A genetic variation (rs1344706) affects brain grey but not white matter in schizophrenia and healthy subjects. Psychol Med 2015; 45: 143–152. [DOI] [PubMed] [Google Scholar]
- Ardini E, Pesole G, Tagliabue E, Magnifico A, Castronovo V, Sobel ME et al. The 67-kDa laminin receptor originated from a ribosomal protein that acquired a dual function during evolution. Mol Biol Evol 1998; 15: 1017–1025. [DOI] [PubMed] [Google Scholar]
- Zhang YQ, Bailey AM, Matthies HJ, Renden RB, Smith MA, Speese SD et al. Drosophila fragile X-related gene regulates the MAP1B homolog Futsch to control synaptic structure and function. Cell 2001; 107: 591–603. [DOI] [PubMed] [Google Scholar]
- Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D et al. Common variants conferring risk of schizophrenia. Nature 2009; 460: 744–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.