Chlamydomonas CDKA and CYCA1 are dispensable for mitosis, but the plant-specific CDKB, probably in complex with CYCB1, promotes most events of mitosis and downregulates CDKA following cell division.
Abstract
The cyclin-dependent kinase CDK1 is essential for mitosis in fungi and animals. Plant genomes contain the CDK1 ortholog CDKA and a plant kingdom-specific relative, CDKB. The green alga Chlamydomonas reinhardtii has a long G1 growth period followed by rapid cycles of DNA replication and cell division. We show that null alleles of CDKA extend the growth period prior to the first division cycle and modestly extend the subsequent division cycles, but do not prevent cell division, indicating at most a minor role for the CDK1 ortholog in mitosis in Chlamydomonas. A null allele of cyclin A has a similar though less extreme phenotype. In contrast, both CDKB and cyclin B are essential for mitosis. CDK kinase activity measurements imply that the predominant in vivo complexes are probably cyclin A-CDKA and cyclin B-CDKB. We propose a negative feedback loop: CDKA activates cyclin B-CDKB. Cyclin B-CDKB in turn promotes mitotic entry and inactivates cyclin A-CDKA. Cyclin A-CDKA and cyclin B-CDKB may redundantly promote DNA replication. We show that the anaphase-promoting complex is required for inactivation of both CDKA and CDKB and is essential for anaphase. These results are consistent with findings in Arabidopsis thaliana and may delineate the core of plant kingdom cell cycle control that, compared with the well-studied yeast and animal systems, exhibits deep conservation in some respects and striking divergence in others.
INTRODUCTION
Cell cycle research in fungi and animals has resulted in a consensus model for control of the cell cycle (Morgan, 2007). Central components are the cyclin-dependent kinase CDK1, its cyclin activators, and the anaphase-promoting complex (APC) E3 ubiquitin ligase. The core circuitry is a negative feedback loop in which cyclin-CDK1 promotes mitotic entry, including spindle assembly and APC activation. APC, in turn, mediates degradation of the anaphase inhibitor securin, and additionally the degradation of cyclins, turning off CDK1. In fungi, CDK1 is activated by early-expressed B-type cyclins and promotes DNA replication as well as subsequent mitosis. In animals, CDK2, a close relative of CDK1, is activated by cyclins A or E and is the primary activator of DNA replication. In fungi and animals, CDK1 is the primary activator of mitosis, and it is the sole CDK regulating the cell cycle in fungi. In mice, CDK1 can carry out all cell cycle-specific CDK function in the absence of CDK2, 4, 5, and 6 (Santamaría et al., 2007). Although almost all fungi and animals contain multiple cyclin genes, with diversified function (Bloom and Cross, 2007), in fission yeast, a single B-type cyclin is sufficient for viability (Fisher and Nurse, 1996), and two cyclins (one G1 cyclin and one B-type cyclin) will support viability in budding yeast (Rahi et al., 2016).
Fungi and animals comprise the Opisthokont clade. Other eukaryotic kingdoms diverged from Opisthokonts early in eukaryotic evolution. The plant kingdom, consisting of algae and land plants, diverged close to the root of the eukaryotic tree (Rogozin et al., 2009). Therefore, features highly conserved among Opisthokonts could be specific to that lineage and altogether absent in the plant kingdom, and vice versa. The central importance of the plant kingdom for terrestrial life means that it is of great significance to understand such divergences. On the other hand, features conserved between Opisthokonts and plants may reflect features of their last common ancestor.
Cell cycle control in land plants exhibits much conservation but also very significant divergence compared with Opisthokonts (Harashima et al., 2013), due in part to apparent rewiring of regulatory circuitry (Dissmeyer et al., 2009; Nowack et al., 2012). Remarkably, CDKA, the plant ortholog of CDK1, is dispensable in Arabidopsis, although proliferation is markedly reduced in its absence (Nowack et al., 2012). CDKB kinases may provide essential functions in the absence of CDKA (Nowack et al., 2012).
CDKB is a plant-specific CDK. Best reciprocal BLAST analysis (Remm et al., 2001) shows consistent integrity of the CDKA and CDKB families across plant genomes (Supplemental Figure 1). CDKA is the best-reciprocal BLAST partner of Opisthokont CDK1, but CDKB lacks a similar partner in Opisthokonts. CDKB may have arisen in the plant lineage early after separation from Opisthokonts. Alternatively, it may have been present in their last common ancestor and was lost early in the Opisthokont lineage.
Land plant lineages underwent repeated whole-genome duplications, resulting in variable but frequently very high copy number for some genes (Vanneste et al., 2014). The rampant gene duplication in land plant genomes afforded regulatory opportunities: For example, Arabidopsis thaliana contains ∼30 A-, B-, and D-type cyclins, with different members responding to environmental, developmental, or hormonal signals in order to attain precise control of proliferation in distinct cell lineages (Lorenz et al., 2003; Dewitte et al., 2007; Sozzani et al., 2010; Sanz et al., 2011; Vanneste et al., 2011). However, gene duplicates also introduce a high level of genetic redundancy that poses a significant challenge to genetic analysis.
The green alga Chlamydomonas reinhardtii is a microbial member of Viridiplantae with numerous advantages for analyzing cell cycle control. The whole-genome duplications in land plants occurred after their divergence from green algae. As a consequence, most Chlamydomonas genes are single copy (Merchant et al., 2007), simplifying genetic analysis. Additionally, because Chlamydomonas grows as a haploid, the phenotypic consequences of single gene mutations are exposed immediately. Its cell division cycle is rapid and continuous, and cultures can be synchronized. These features allowed isolation of temperature-sensitive lethal mutations inactivating diverse components in cell cycle control and execution (Tulin and Cross, 2014; Breker et al., 2016). Among the mutated genes were CDKA1 and CDKB1. The cdka1-1 temperature-sensitive allele resulted in a long delay in cell cycle initiation at the restrictive temperature, but ultimately mutant cells were able to execute complete rounds of DNA replication and mitosis, forming slow-growing colonies. In contrast, cdkb1-1 mutant cells completed a single round of DNA replication with a moderate delay but then arrested without forming a mitotic spindle.
Chlamydomonas has a long cell growth period without DNA replication during which cells can increase in size more than 10-fold. This is followed by rapid sequential cycles of DNA replication, nuclear division, and cytokinesis (Cross and Umen, 2015). After three to four divisions, newborn cells hatch from the mother cell wall and reenter the growth phase. Commitment to division occurs early in the growth phase and may involve cell-size-dependent inactivation of the Chlamydomonas homolog of the retinoblastoma (Rb) protein (Umen and Goodenough, 2001; Fang et al., 2006; Olson et al., 2010). CDK activation occurs several hours after commitment (Bisova et al., 2005) but the mechanistic connection is unknown.
Here, we characterize CDKA1 and CDKB1 abundance, localization, and associated kinase activity through the cell cycle. We analyze reciprocal regulation of CDKA1 and CDKB1, their regulation by A- and B-type cyclins and the APC, and how these activities are connected to execution of DNA replication, spindle formation, and cytokinesis.
RESULTS
Chlamydomonas CDK1 Is Not Required for Mitosis: A Selective Genetic Screen Yields cdka1 Null Alleles That Delay but Do Not Block the Cell Division Cycle
The cdka1-1 mutation in the Chlamydomonas CDK1 ortholog results in a lengthy delay in cell cycle initiation at the restrictive temperature. However, mutant cells enter the cell cycle and are ultimately viable forming slow-growing colonies (Tulin and Cross, 2014). This suggested that CDKA1 might not be required for mitosis; however, leakage into divisions could also be due to partial function of cdka1-1 at high temperature.
Many cell cycle mutants in Chlamydomonas undergo cell lysis shortly after reaching their specific arrest points (Tulin and Cross, 2014). Consistent with the role of CDKA1 in promoting cell cycle initiation, cell lysis due to these mutations was strongly delayed in double mutants also containing cdka1-1 (Tulin and Cross, 2014), presumably because lysis depends on CDKA1-dependent initiation of the defective cell cycle. We hypothesized that this suppression of cell lysis would provide an effective selection for new mutations in genes required for cell cycle initiation. The div19-2 (topoisomerase II) mutation resulted in a sharp drop in viability after ∼12 h at the restrictive temperature (Figure 1A), approximately the time when the wild type is executing cell divisions under these conditions (Tulin and Cross, 2014) . This lethality may be due to attempted mitosis with catenated DNA; budding yeast topoisomerase II mutants exhibit a similar loss of viability as they attempt mitosis (Holm et al., 1989). Inclusion of cdka1-1 in the div19-2 background greatly extended viability at restrictive temperature (Figure 1A), presumably because cdka1-1 delays initiation of the mitotic program (Tulin and Cross, 2014).
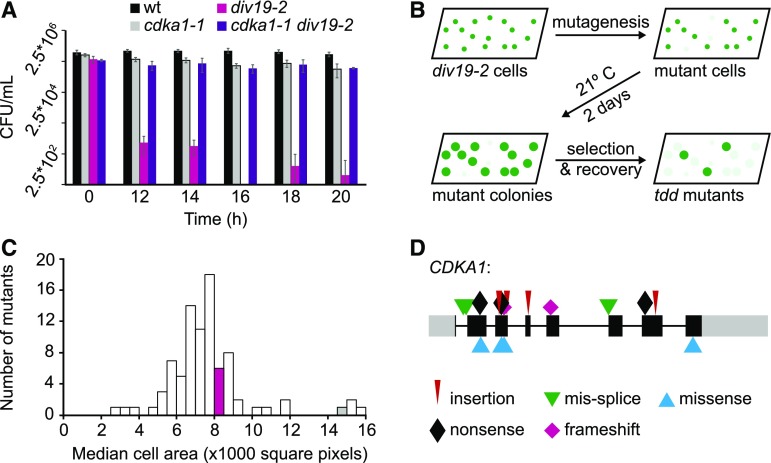
Figure 1.
Death Delay Screen and Mutant Characterization.
(A) Viability of the wild type, cdka1-1, div19-2, and cdka1-1 div19-2 after exposure to 33°C. Error bars represent the sd of four technical replicates.
(B) Schematic of the death delay screen.
(C) Distribution of median cell area for a representative set of tdd mutants after 20 h at 33°C. The magenta and gray bars indicate the cell area of the div19-2 and cdka1-1 div19-2 mutants, respectively.
(D) Mutations in CDKA1 in large-cell arresting tdd mutants (>12,000 square pixels). Colored symbols show the type of mutation. Gray boxes, 5′ and 3′ untranslated regions; black boxes, exons; gray lines, introns.
We used transient div19-2 inactivation to select for new mutations that delay cell cycle progression (Figure 1B). Many such mutants, which we have termed tdd (top2 death delay) mutants, were slow growing and are likely to indirectly delay division due to a cell growth/cell size requirement for cell cycle entry (Cross and Umen, 2015). However, cells of some of the mutants grew to a very large size before initiating mitosis, as does cdka1-1 (Figure 1C). All mutants recovered with the large cell size phenotype have CDKA1 mutations (Figure 1D).
Many such UV-induced cdka1 mutations were severe alterations: early nonsense, frameshift, insertion/deletion, etc. (Supplemental Table 1). For any one of these mutations, it is possible that nonsense suppression, frameshift suppression, or alternative splicing might rescue partial function. However, this seems unlikely for the entire group, since such mechanisms are typically inefficient. All lesions, even if suppressed in some way, will alter or remove coding sequence for highly conserved amino acids (e.g., identical in human CDK1). Using this selection, we also obtained insertional disruptions that place antibiotic resistance marker cassettes within highly conserved coding exons. Overall, we conclude that most of these cdka1 alleles are null for function and that, therefore, CDKA1 is inessential for viability (Supplemental Table 1). CDKA1 is also inessential in Arabidopsis, based on recovery of viable homozygous plants bearing insertional disruption alleles (Nowack et al., 2012). Thus, the CDK1 ortholog is not an essential gene in Viridiplantae, unlike in the Opisthokonts.
In Arabidopsis, the CDKA requirement for efficient proliferation is substantially abrogated by mutation of the RBR1 Rb homolog, suggesting that Rb inactivation may be an important function of CDKA (Nowack et al., 2012). The Chlamydomonas Rb homolog MAT3 regulates cell division timing (Umen and Goodenough, 2001). We constructed cdka1Δ mat3Δ strains and determined their first division time to be intermediate between times of wild-type and cdka1Δ strains, suggesting the possibility of a conserved regulatory circuit in Viridiplantae. In Chlamydomonas, MAT3 may be regulated by a different CDK, CDKG (Y. Li et al., 2016). It will be interesting to characterize the relationship of CDKG and CDKA1 activity in regulation of MAT3 and cell cycle initiation.
Chlamydomonas Cyclin A Is Nonessential but Regulates the Timing of Cell Divisions
Chlamydomonas has four D-type cyclins, one A-type cyclin, one B-type cyclin, and one “hybrid” A/B-type cyclin (Bisova et al., 2005). We searched the insertional disruptions in the library of Li et al. (2016) and found a disruption of cyclin A (Cre03.g207900). We confirmed the structure of this disruption, which inserts a paromomycin resistance cassette in the middle of conserved cyclin homology sequence. cyca1Δ strains were viable at low and high temperature. However, cyca1Δ caused a cell cycle delay, similar to (though less extreme than) that observed with cdka1Δ disruption.
We quantified this delay by time-lapse microscopy in single and double mutants (Figure 2; Supplemental Table 2). On average, wild-type cells required 11 h of incubation before the first division cycle. The subsequent two to three division cycles were rapid, averaging 0.6 h each. Disruption of either cyca1 or cdka1 caused long delays in the time to the first division cycle, with average increases of 2.3 and 5.3 h, respectively. More modest delays were also observed in subsequent cycles, with average increases of 0.2 and 0.4 h, respectively. cyca1Δ cdka1Δ double mutants were somewhat more severely affected than cdka1Δ single mutants. In addition, a substantial though variable proportion of cdka1Δ and cyca1Δ cdka1Δ double mutant cells grew very large but failed to initiate any division cycles, even after 35 h (Figure 2B; Supplemental Table 2). The phenotypes of both cyca1Δ and cdka1Δ mutants suggest that cell growth occurs independently of the cell division cycle, as was shown previously in yeast (Nurse et al., 1976, Johnston et al., 1977).
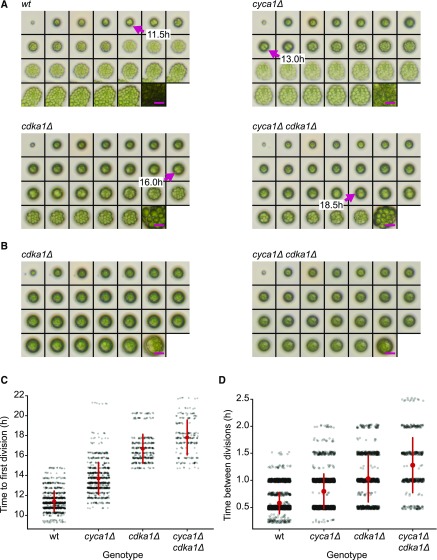
Figure 2.
Delay of Cell Division in cdka1Δ and cyca1Δ Mutants.
(A) and (B) Montages of time-lapse microscopy. Time points: 0-h, 10- to 22-h at 0.5-h intervals, and 35 h. Magenta arrows in (A) show the first divisions. Magenta bars = 25 μm. Hatching from the mother cell wall (see wild type 14.5-h and 15.0-h images) allows cells to spread, taking up more area in the image. Some cdka1Δ and cyca1Δ cdka1Δ cells grow very large (B), but fail to divide even after 35 h (Supplemental Table 2). Bars = 25 μm.
(C) Time to first division in wild-type, cyca1Δ, cdka1Δ, and cyca1Δ cdka1Δ cells. At time zero, saturated TAP cultures were spotted on a plate and transferred to 33°C. Each data point represents a single quantified division time. Red circle and bars indicate mean and sd. Differences between all groups are significant (Mann-Whitney U test, P < 0.001). Pooled results from two segregants of each genotype.
(D) Time between divisions following the first division in the wild type, cyca1Δ, cdka1Δ, and cyca1Δ cdka1Δ. Differences between all groups are significant (Mann-Whitney U test, P < 0.001). Red circle and bars indicate mean and sd.
These deleterious phenotypes of cdka1 disruption were most readily observed at 33°C (Supplemental Figure 2). Qualitatively similar phenotypes are detectable for cdka1Δ at 30°C (data not shown). We do not understand the basis for this temperature sensitivity of null alleles. Previous work (Tulin and Cross, 2014) indicates that the delay in the first cell cycle in cdka1-1 mutant cells occurs before DNA replication. However, we do not know what cell cycle phase(s) are extended in subsequent divisions. The delays in cyca1Δ mutant cells have also not been mapped to specific phase(s).
Cyclin B Is Required for Spindle Formation but Not DNA Replication or the Initiation of Cytokinesis
Chlamydomonas contains a single B-type cyclin CYCB1 (Cross and Umen, 2015). The cycb1-5 mutation (p.Glu325>Lys; F. Tulin and F.R. Cross, unpublished data) caused a strong temperature-sensitive lethal phenotype. At the restrictive temperature, synchronized cultures of cycb1-5 replicated DNA, though with a delay compared with wild-type cultures. Mutant cells arrested after one round of replication (Figure 3A) and exhibited aberrant initiation of cytokinesis at approximately the same time that wild-type cells performed their first complete cell division (Supplemental Figure 3A). cycb1-5 mutant cells rarely contained mitotic spindles (Figure 3B; Supplemental Table 3). This arrest was very similar to that of cdkb1-1 (Tulin and Cross, 2014; Supplemental Figure 3B). Thus, CYCB1 and CDKB1 were specifically required for spindle formation but not for DNA replication or initiation of cytokinesis.
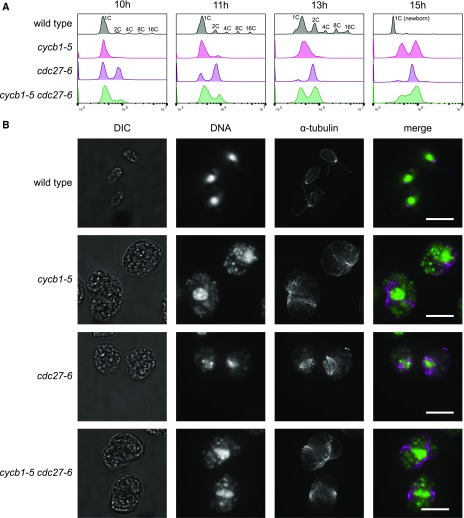
Figure 3.
Dependence of DNA Replication and Spindle Formation on CYCB1 and the APC.
(A) and (B) Cells were blocked by nitrogen deprivation and released at 33°C.
(A) DNA content analyzed by flow cytometry. Wild-type cells undergo multiple rounds of DNA synthesis and mitosis within the mother cell wall, producing cell clusters with 2, 4, 8, or 16C DNA content, before hatching to produce small 1C newborn cells. The apparent DNA content of these cells compared with predivision 1C cells is shifted to the left (Tulin and Cross, 2014).
(B) From a separate experiment, cells at 13.5 h after release were processed for anti-α-tubulin immunofluorescence and DNA localization by SYTOX staining. DIC, differential interference contrast image. Bars = 10 μm. Quantification is shown in Supplemental Table 3.
The delay in DNA replication in cycb1 mutants indicates that CYCB1 is a significant, though nonessential, activator of DNA replication. Perhaps it is nonessential for replication due to functional overlap with other cyclins. Indeed, cycb1-5 cyca1Δ double mutant strains did not exhibit DNA replication over an extended time course (Supplemental Figure 4), and formation of cleavage furrows was substantially attenuated. Thus, in the absence of CYCB1, CYCA1 was required to promote DNA replication and for efficient initiation of cytokinesis.
Previously, we showed that DNA replication and initiation of cytokinesis in cdkb1-1 mutant cells was dependent on CDKA1 (Tulin and Cross, 2014), suggesting that these events, which signal the initiation of the cell division program, were largely under specific control of CDKA1 and CDKB1, activated (in some combination) by CYCA1 and CYCB1.
The APC Is Required for Anaphase, Cytokinesis, and DNA Rereplication
In fungi and animals, the APC is required for anaphase, exit from mitosis, and relicensing of origins of DNA replication. We reported previously on the phenotypes of mutations in APC components APC6 and CDC27 (Tulin and Cross, 2014). These mutants arrested with one or two nuclei and with 2C or 4C DNA content. Many, but not all, cells had metaphase spindles. We speculated that limited mitotic progression was due to partial function of the temperature-sensitive alleles. Indeed, here, we found that the cdc27-6 allele (p.Met884>Ile, Gly885>Ser) caused a uniform arrest with once-replicated DNA and metaphase spindles (Figures 3A and 3B; Supplemental Table 3). Arrested cells completely lacked cleavage furrows (Supplemental Figures 3A and 3B). This result demonstrates the absolute requirement of the APC for anaphase and mitotic exit in Chlamydomonas. APC subunits are required for gametogenesis in Arabidopsis (Capron et al., 2003; Kwee and Sundaresan, 2003; Wang et al., 2012), consistent with an essential role throughout the plant kingdom.
cycb1-5 mutant cells arrested without spindles and formed cleavage furrows that failed to progress to full cell division (see above), and cdc27-6 cells arrested with the opposite phenotype. cycb1-5 was strongly epistatic to cdc27-6: Double mutants lacked mitotic spindles, and cells exhibited aberrant cleavage furrows that accumulated with similar timing to those in cycb1-5 single mutants (Figure 3B; Supplemental Figure 3A). Essentially similar results were obtained with cdkb1-1 cdc27-6 double mutants (Supplemental Figure 3B). These results indicate that the late mitotic arrest phenotype of cdc27-6 requires activity of CYCB1 and CDKB1.
In Opisthokonts, the APC has two critical functions: degradation of mitotic cyclins and degradation of securin, the anaphase inhibitor that maintains sister chromatid cohesion. If CYCB1 is similarly an important target of the APC in Chlamydomonas, this could account for epistasis of cycb1-5 and cdkb1-1 to cdc27-6, since mutational inactivation of CYCB1-CDKB1 could bypass the APC requirement for cyclin degradation.
Functional Tagging of CDKA1 and CDKB1
To gain insight into biochemical regulation of CDK activity, we constructed strains expressing mCherry-tagged CDKA1 and CDKB1 transgenes. The native sequence with introns and promoter was included (Figure 4A) to increase the likelihood of similar levels and regulation of expression of the transgene compared with the endogenous gene.
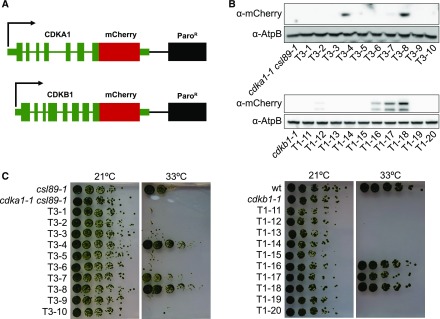
Figure 4.
Generation of Functional mCherry-Tagged CDKA1 and CDKB1.
(A) DNA used for transformation. The native sequences, including promoter, terminator, and introns were used for both CDKA1 and CDKB1. The sequence encoding the mCherry tag was added at the 5′ end of the coding sequence following a 3x GGGGS linker sequence. Paromomycin was used to select for transformants.
(B) Immunoblot showing expression of the tagged CDK transgenes in random transformants. AtpB was used as a loading control.
(C) Complementation of temperature sensitivity of cdka1-1 csl89-1 and cdkb1-1 in the transformants from (B).
mCherry-tagged constructs were transformed into temperature-sensitive mutants that depend on functional CDKA1 or CDKB1 at the restrictive temperature. For CDKA1, we used a mutant background, csl89-1, in which CDKA1 is essential at high temperature. csl89-1 was isolated in a screen for enhancers of cdka1-1 (our unpublished data). The mutation inactivates a predicted tRNA methyltransferase (Cre05.g244236). For unknown reasons, csl89-1 mutants are viable at high temperature in a CDKA1 background but inviable in a cdka1 background. This provided a background for selection of functional tagged CDKA1 transgene insertions. For CDKB1, we used the cdkb1-1 temperature-sensitive-lethal allele (Tulin and Cross, 2014).
The viability of transformants was tested at 33°C. For both constructs, expression of the mCherry-tagged CDKs correlated with complementation of the temperature-sensitive mutations (Figures 4B and 4C). Overall, we noted no significant defects in cell cycle progression upon complementation by the transgenes. Functional CDKA1-mCherry insertions isolated in the cdka1-1 csl89-1 background were crossed away from csl89-1. cdka1-1 CDKA1:mCherry strains from this cross did not show the cell cycle delay of cdka1-1 strains at 33°C, indicating complementation of cdka1-1.
CDKA1 May Localize to Nuclei and Basal Bodies
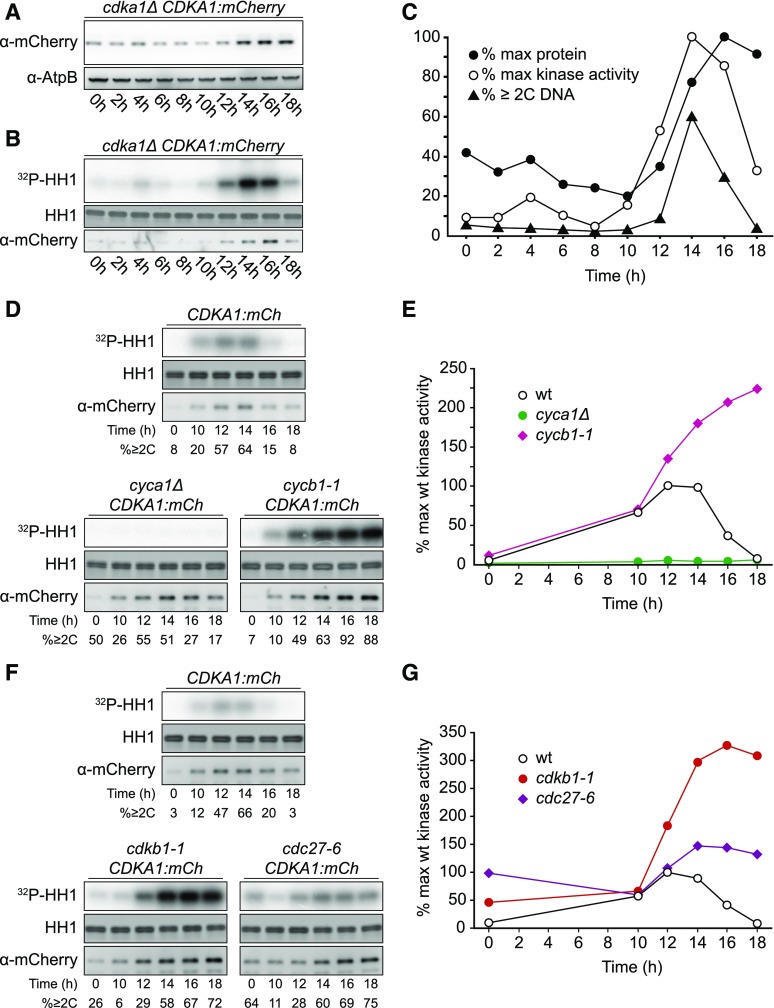
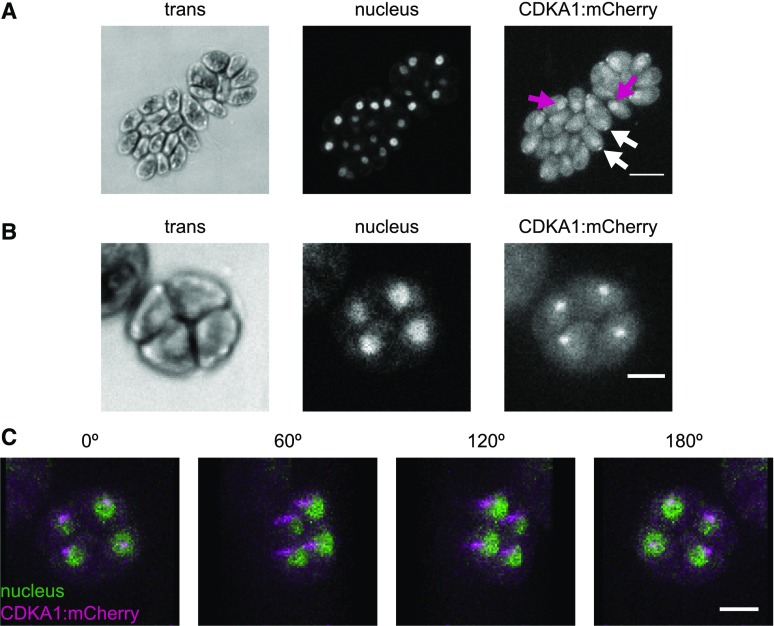
CDKA1 was present throughout the cell cycle; however, its levels increased during the multiple division phase (Figures 5A and 5C), reflecting CDKA1 RNA abundance (Bisova et al., 2005; Tulin and Cross, 2015; Zones et al., 2015). Subcellular localization of CDKA1:mCherry was determined by confocal microscopy of live, cycling cells. Nuclei were visualized using a nuclear-targeted ble:GFP construct (Y. Li et al., 2016). In newborn cells, CDKA1 is enriched both in nuclei and in focused puncta adjacent to the nuclei at the apical end of the cell body (Figure 6A), approximately at the base of the flagella. CDKA1 and CYCA1 were previously identified by mass spectrometry as components of the ciliary transition zone between the basal bodies and the flagellum (Diener et al., 2015). In dividing cells lacking flagella, CDKA1 was also localized to focused puncta, which appear to be located just adjacent to nuclei (Figures 6B and 6C). The ciliary cycle is integrated in a complex manner with the mitotic cycle because basal bodies detach from flagella and serve as spindle pole bodies in mitosis (Dutcher and O’Toole, 2016); we speculate that CDKA1-CYCA1 may directly regulate early steps in this process. Colocalization of CDKA with mitotic microtubule structures was reported in Arabidopsis (Weingartner et al., 2001). All localizations of CDKA1:mCherry described above were specific to tagged cells. (By contrast, general cytoplasmic background in these images was essentially identical in tagged and untagged cells [data not shown].)
Figure 5.
Regulation of CDKA1 Abundance and Kinase Activity.
Strains of the indicated genotypes were synchronized by nitrogen deprivation and released at 33°C. The contrast was adjusted such that images can be compared within but not between subfigures.
(A) Immunoblot detection of CDKA1-mCherry.
(B) Kinase activity of immunoprecipitated CDKA1-mCherry on a histone H1 substrate. Note: Background kinase activity in immunoprecipitates from untagged cells was low and varied little through the cell cycle (data not shown).
(C) Quantification of (A) and (B) and the fraction of cells/cell clusters with ≥2C DNA content as determined by flow cytometry.
(D) and (E) Cyclin dependence of CDKA1-associated kinase activity. See also Supplemental Figure 7A.
(F) and (G) CDKB1 and APC negatively regulate CDKA1-associated kinase activity.
Figure 6.
CDKA1 Is Localized to the Nucleus and the Base of Flagella.
Confocal micrographs from a population of live, cycling cdka1ΔCDKA1:mCherry cells. Contrast was adjusted separately for each image, so intensities should not be compared across images. Cytoplasmic background fluorescence in the mCherry channel was also observed in cells lacking the transgene (data not shown).
(A) CDKA1 localization in newborn cells. CDKA1 was found in nuclei (magenta arrows) and in small puncta at the apical end of the cell body (white arrows), possibly at the base of the flagella. Images are average intensity z-projections. Bar =10 μm.
(B) In dividing cells, CDKA1 was primarily found in small puncta. Bar = 5 μm. Images are average intensity z-projections.
(C) CDKA1 puncta in dividing cells were adjacent to nuclei. Images are rotations from a brightest point 3D projection of the cell cluster in (B). Bar = 5 μm.
CDKB Is a Nuclear Protein Present Specifically during Cell Division Cycles
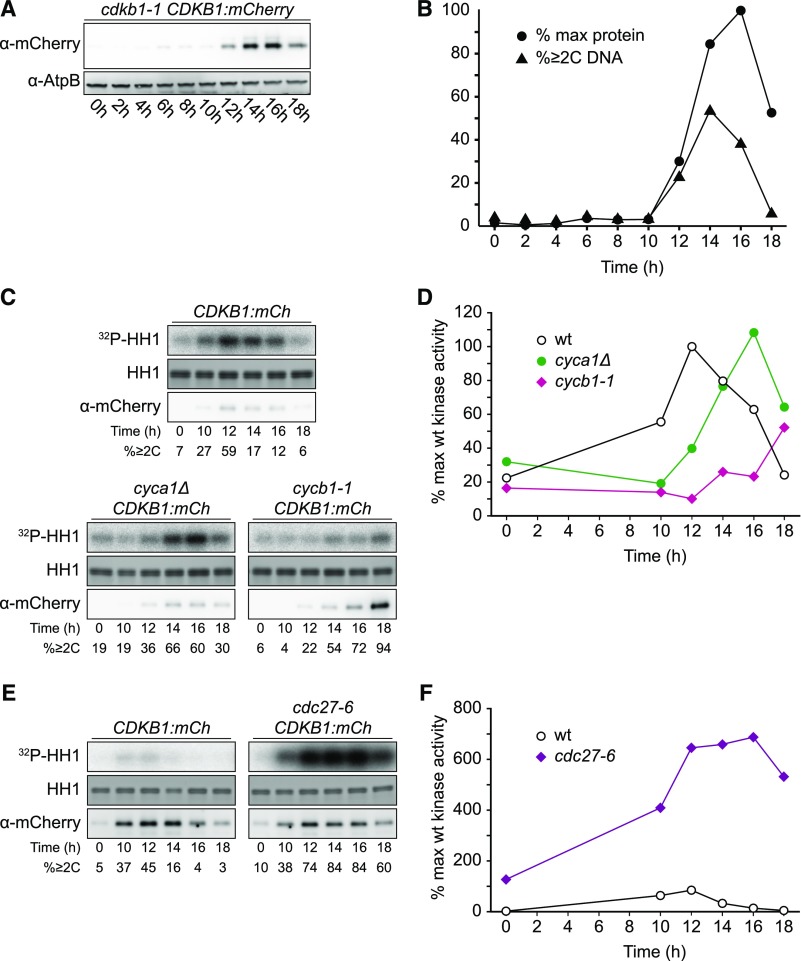
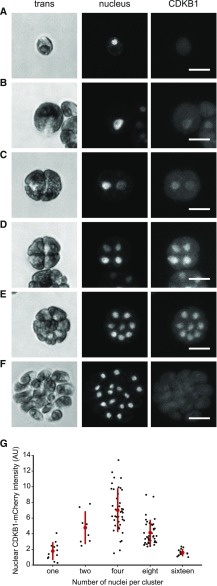
CDKB1 was essentially absent during the extended growth period, was strongly induced coincident with the onset of cell division, and was degraded as cells exited the cell cycle (Figures 7A and 7B), reflecting CDKB1 RNA abundance (Bisova et al., 2005; Tulin and Cross, 2015; Zones et al., 2015). In live-cell confocal microscopy, newborn cells showed no detectable CDKB1 (Figure 8A). After a period of cell growth, CDKB1 was detectable, with predominantly nuclear localization through multiple division cycles (Figures 8B to 8E). When cells had completed their final divisions, CDKB1 was no longer detectable (Figure 8F), suggesting rapid degradation before cells enter the next growth phase. This nuclear signal was not observed in untagged cells analyzed in parallel (data not shown).
Figure 7.
Regulation of CDKB1 Abundance and Associated Kinase Activity
Strains of the indicated genotypes were synchronized by nitrogen deprivation and released at 33°C. Contrast was adjusted such that images can be compared within but not between subfigures.
(A) Immunoblot detection of CDKB1-mCherry.
(B) Quantification of (A) and fraction of cells with ≥2C DNA content as determined by flow cytometry.
(C) and (D) Cyclin dependence of CDKB1-associated kinase activity. See also Supplemental Figure 7B.
(E) and (F) APC negatively regulates CDKB1-associated kinase activity. Note: Background kinase activity in immunoprecipitates from untagged cells was low and varied little through the cell cycle (data not shown).
Figure 8.
CDKB1 Is a Nuclear Protein during Division Cycles.
Confocal micrographs from a population of live cells expressing CDKB1:mCherry and ble-GFP as a nuclear marker (Y. Li et al., 2016). All images are average intensity z-projections. Contrast was adjusted separately for each trans and nuclear (GFP) image for clarity. CDKB1 images are contrast adjusted to the maximum of the group, so intensities can be compared. The non-nuclear signal in the mCherry channel was observed at similar levels in control strains lacking the transgene (data not shown).
(A) Newborn cell with no detectable CDKB1.
(B) to (E) The 1-, 2-, 4-, and 8-cell clusters, respectively, with CDKB1 in all nuclei. Bars = 10 μm.
(F) The 16-cell cluster of postmitotic newborn cells (just before hatching) lack CDKB1.
(G) Nuclear concentration of CDKB1 as a function of number of cells per cluster (see Supplemental Methods). Bar = 10 μm.
We have so far been unable to reliably detect CDKB1 by live-cell time-lapse imaging, so we do not know whether CDKB1 is degraded in each cell cycle or, instead, is degraded only upon completion of all approximately three to four divisions in the multiple fission cycle (Cross and Umen, 2015). Analysis of variability of CDKB1 levels in cell clusters with varying numbers of nuclei (Figure 8G) showed variable signals but no clearly bimodal distribution, as would be expected in the case of a significant period of CDKB1 instability, suggesting that CDKB1 may be present during most or all of the multiple fission cycle.
cdka1Δ cells exhibited highly delayed division (see above). Nuclear CDKB1 was not detectable in these cells until they became much larger than wild-type cells and had begun division cycles (Supplemental Figure 5), consistent with the finding that CDKA1 is required for efficient accumulation of CDKB1 RNA (Tulin and Cross, 2014). A similar conclusion was reached in Arabidopsis (Nowack et al., 2012), suggesting a conserved Viridiplantae-specific regulatory circuit.
CDKA1 and CDKB1-Associated Kinase Activity Peaks in Dividing Cells
We examined CDKA1- and CDKB1-associated protein kinase activity in immunoprecipitates using histone H1 as a substrate. For both kinases, activity was specifically induced during cell divisions (Figures 5 and 7). As noted above, cells at all points in the cycle contained CDKA1 protein, but only dividing cells had significant associated kinase activity. CDKB1 was present and active only during cell divisions (Figure 7).
CDKB1-associated activity was much lower than CDKA1-associated activity (Supplemental Figure 6). Many factors could explain this behavior, including differential substrate preference or sensitivity of the kinases to the conditions of the assay. Importantly, even though CDKB1-associated kinase activity measured in vitro is low, kinase activity is biologically relevant, since a kinase-dead (K33R) version of CDKB1 fails to rescue the cdkb1-1 mutant (Supplemental Table 4).
CDKA1-Associated Kinase Activity Is Largely Dependent on Cyclin A and Is Independent of Cyclin B
CDKs are expected to be tightly dependent on cyclin binding for enzymatic activity (Jeffrey et al. 1995). We assessed CDKA1 activity in mutant backgrounds with either cyclin A or cyclin B inactivated (cyca1Δ or cycb1-1 [p.Leu288>Pro]). Activation of CDKA1 kinase was largely eliminated in the cyca1Δ background (Figures 5D and 5E), even though these cells replicated DNA and executed mitotic divisions (Figures 2 and 5D). The phenotype of the cyca1Δ mutant was considerably less severe than that of the cdka1Δ mutant (Figures 2C and 2D), indicating that CDKA1 must have CYCA1-independent biological activity. This activity may have been due to activation of CDKA1 by other cyclins, but if this is the case, these complexes must have low biochemical activity.
By contrast, CDKA1 kinase activity rose with similar timing in the cycb1-1 mutant compared with the wild type, with no diminution in amount despite the strong inhibition of progression to mitosis in this mutant (Figures 5D and 5E). We therefore conclude that the normal activation of CDKA1 is largely dependent on CYCA1 and independent of CYCB1. A simple model to explain this is that CYCA1 is the primary biochemical activator of CDKA1. We confirmed efficient CYCA1-dependent activation of CDKA1 kinase in cycb1-deficient backgrounds with the cycb1-3 (p.Glu325>Lys) allele (Supplemental Figure 7A), which gave a tighter cell cycle arrest than cycb1-1 (M. Breker, K. Lieberman, and F.R. Cross, unpublished data).
Elimination of CDKA1-Associated Kinase Activity Depends on CDKB1, CYCB1, and the APC
In fungi and animals, the APC targets mitotic cyclins for destruction, eliminating CDK kinase activity at the end of the cell cycle. This is essential to allow mitotic exit and relicensing of DNA replication origins. We asked whether the APC is required for inactivation of CDKA1 in Chlamydomonas by assessing CDKA1-associated kinase activity in the cdc27-6 APC mutant. The initial rise in CDKA1 kinase activity was similar to the wild type in the cdc27-6 mutant. Activity fell in the wild type as cell division cycles were completed. In contrast, a high level of activity was maintained in the cdc27-6 mutant (Figures 5F and 5G). We conclude that the APC is required for inactivation of CDKA1.
The APC targets the destruction box motif (Glotzer et al., 1991; King et al., 1996; He et al., 2013). Of the Chlamydomonas cyclins, only cyclin A and cyclin B contain canonical destruction boxes (Supplemental Figure 8). Because CDKA1 activity was independent of cyclin B (see above), the APC may inactivate CDKA1 at the end of the cell cycle by targeting cyclin A for destruction. Consistently, in Arabidopsis, mutation of APC subunits slows degradation of a destruction box reporter (Kwee and Sundaresan, 2003; Wang et al., 2012).
In addition to the APC, CDKB1 is required for inactivation of CDKA1-associated kinase (Figures 5F and 5G). This finding is consistent with the previous report that bulk CDK-associated kinase activity remains at a high level in arrested cdkb1-1 mutants (Tulin and Cross, 2014). One interpretation of this result is that competition between CDKA1 and CDKB1 for cyclins was reduced in the absence of functional CDKB1. To test this, we examined the effect of inactivating CYCB1 on CDKA1-associated kinase. The results were almost identical to the results of inactivating CDKB1 (Figures 5D and 5E), arguing against relief of cyclin competition as the mechanism for increasing CDKA1 activity (because in this experiment CDKB1 was the wild type, and available cyclins were reduced by inactivation of CYCB1). Overall, these results suggest that CYCB1 and CDKB1 may inactivate the CYCA1-CDKA1 complex.
CDKB-Associated Kinase Activity Is Dependent on Cyclin B, Is Independent of Cyclin A, and Is Strongly Repressed by the APC
Similarly, we examined cyclin dependence of CDKB1-associated kinase activity. Consistent with the delayed onset of division in the cyca1Δ mutant (Figures 2C and 2D), the onset of peak CDKB1 protein and kinase activity was also delayed, but eventually rose to normal levels (Figures 7C and 7D). By contrast, in the cycb1-1 mutant, CDKB1 kinase activity was greatly diminished, even though CDKB1 protein accumulated to very high levels (Figures 7C and 7D). We conclude that CDKB1 kinase activity is largely dependent on cyclin B and, aside from timing, is independent of cyclin A. Notably, this is the opposite pattern of cyclin dependence to that of CDKA1, which was dependent on CYCA1 but independent of CYCB1.
CDKB1 kinase activity was strikingly elevated in the cdc27-6 mutant (Figures 7E and 7F), indicating that the APC is critical for restricting CDKB1 activity. Because CDKB1 was largely dependent on CYCB1 but not CYCA1 for activity, and CYCB1 (but not CDKB1) contains a canonical destruction box (Supplemental Figure 8), the APC most likely inhibits CDKB1 by targeting CYCB1 for destruction. CYCB1 may be the major biochemical activator of CDKB1 kinase.
Although APC prevents the loss of CDKA1 kinase activity (that is, levels go to slightly above normal peak and do not decline), APC inactivation has a much greater effect on CDKB1 activity, whose levels increase ∼10-fold above normal). We lack a mechanistic explanation for this difference at present.
DISCUSSION
The Opisthokont Mitotic Inducer CDK1 Is Replaced by CDKB in the Plant Kingdom
In Opisthokonts, CDK1 is an essential inducer of mitosis. In Arabidopsis, the CDK1 homolog CDKA is nonessential, although cdka plants are severely compromised in cell proliferation and development (Nowack et al., 2012). In Chlamydomonas, a temperature-sensitive cdka1-1 mutant is delayed for cell cycle entry at restrictive temperature (Tulin and Cross, 2014). Here, by the isolation of null alleles, we demonstrated that as in Arabidopsis, the CDK1 homolog in Chlamydomonas was not essential for viability. CDKA1 was nevertheless required for timely cell cycle initiation, both for the first division after the extended growth period, and also for the multiple subsequent divisions that follow, before the terminal hatched newborn cells reenter the growth phase. Despite timing defects, mutants lacking CDKA1 eventually performed all the events of the cell cycle successfully.
Thus, in Chlamydomonas, CDKA1 is not required for mitosis, but instead has a primary role early in the cell cycle of promoting entry into the division program. Similar results in Arabidopsis (Nowack et al., 2012) suggest divergence of CDK function after the split of the plant kingdom from the Opisthokonts. One possibility is that evolution of the plant-specific CDKB may have allowed for subfunctionalization (Lynch and Force, 2000; Lynch et al., 2001): Once CDKB gained the ability to regulate mitosis, then CDKA, no longer an essential gene, lost the ability to promote mitosis and became specialized in the promotion of cell cycle initiation. A possible alternative is that the last ancestor of Opisthokonts and plants employed CDKA for cell cycle initiation and CDKB for mitosis, and CDKB was lost in the Opisthokont lineage, with CDKA broadening its function to include regulating mitosis. Because CDKB is plant kingdom specific, this alternative requires either early branching of plants from all other eukaryotes (Rogozin et al., 2009) or independent loss of CDKB in multiple lineages.
Mutation of CDKA1 results in a moderate delay in later cycles, in addition to strong delay in initiating the first cell cycle (Figure 2). Cell cycle synchrony is not good enough for accurate analysis of events within these later cell cycles. Therefore, at present, we do not know whether this reflects an expanded period before DNA replication in these later cycles due to the absence of CDKA1 or to elongation of other cell cycle phases.
In plants, many cells outside of the proliferating meristems undergo endoreduplication (many rounds of DNA replication without mitosis), and CDKB downregulation is a likely mechanism to accomplish this (Boudolf et al., 2004, 2009). Endoreduplication is probably critical for making very large cells (Mendell et al., 2008; Melaragno et al., 1993; Chevalier et al., 2014), including the endosperm of grass seeds, which provide the bulk of human food. Specialization of CDKB for mitotic control may facilitate endoreduplication because blocking CDKB transcription would leave the replication-promoting CDKA still present and active (even hyperactive; Figures 5F and 5G). Thus, replacement of CDK1 with CDKB could be an important modification that allowed subsequent evolution of developmentally regulated endoreduplication in land plant lineages. However, cdkb1-1 mutants undergo only a single round of DNA replication (Tulin and Cross, 2014), indicating the requirement for additional regulatory modification to produce endocycles.
CYCA1 Is Probably the Main Activator of CDKA1
In animals, A-type cyclins, in association with CDK1 or its close relative CDK2, play an important role both in the promotion of S phase (Pagano et al., 1992) and in early events in mitotic entry (Pagano et al., 1992; Gong and Ferrell, 2010). Arabidopsis has 10 A-type cyclins that fall into three separate families (Vandepoele et al., 2002). The various cyclins are expressed in distinct but overlapping patterns in different tissues and developmental stages. Some A-type cyclins respond specifically to certain hormones or developmental transitions (Vanneste et al., 2011; Polko et al., 2015). A-type cyclins also regulate transitions from mitotic cycles to endocycles of DNA replication (Imai et al., 2006).
Chlamydomonas has only one A-type cyclin, CYCA1, providing a simplified model for cyclin A function in the plant kingdom. CYCA1 is nonessential, though cyca1Δ cells exhibit significant delays in cell division. CYCA1 is specifically required for full biochemical activation of CDKA1, which may explain division delays in cyca1Δ cells. However, cdka1Δ cells exhibit much greater division delays than cyca1Δ cells (Figure 2; Supplemental Table 2), so either CDKA1 has additional cyclin activators, or it has cyclin-independent activities. Arabidopsis CDKA1 can bind to multiple cyclins, and it is tightly dependent on cyclin binding for biochemical activity (Harashima and Schnittger, 2012), so the former explanation may be more likely. At present, there are no available mutations in the four Chlamydomonas D-type cyclins, or in the hybrid A/B-type cyclin, to test whether these provide CDKA1 activation “backing up” CYCA1.
CYCB1-CDKB1 Is Probably the Main Inducer of Mitotic Entry
In fungi, B-type cyclins regulate both DNA replication and mitosis (Bloom and Cross, 2007). In the animal kingdom, B-type cyclins, in association with CDK1, promote entry to mitosis, and their degradation by the APC is essential for completion of the cell cycle (Morgan, 2007; Murray and Kirschner, 1989; Murray et al., 1989). Degradation of cyclin B is also critical in plants, since cyclin B with a mutated destruction box inhibits late mitotic events (Weingartner et al., 2004). In Arabidopsis, the CYCB1 family (comprising four of the twelve Arabidopsis B-type cyclins) is thought to be involved in DNA repair, possibly in conjunction with members of the CDKB1 family (Weimer et al., 2016), in addition to having a role in regulating mitosis (Schnittger et al., 2002).
Because the Chlamydomonas genome encodes only one B-type cyclin, CYCB1, it is a simple system for analyzing the role of B-type cyclins in the plant kingdom. We found that CYCB1 was an essential gene required for the formation of mitotic spindles. It was not essential for DNA replication, although replication was delayed in its absence. These phenotypes are very similar to those of cdkb1-1 (Tulin and Cross, 2014). Consistently, cycb1-5 and cdkb1-1 were similarly epistatic to phenotypes of APC inactivation (Figure 3B; Supplemental Figure 3). Both CYCB1 and CDKB1 were required for inactivation of CYCA1-CDKA1 kinase activity (Figures 5D to 5G). Finally, CYCB1 but not CYCA1 was required for robust activation of in vivo CDKB1 kinase activity (Figures 7C and 7D). Taken together, these results strongly imply that CYCB1 is the main activator of CDKB1 and that the CYCB1-CDKB1 complex is the predominant mitotic inducer in Chlamydomonas.
A direct test of the hypothesis that CYCA1 and CYCB1 are specific biochemical activators of CDKA1 and CDKB1, respectively, must await production of functional tagged CYCA1 and CYCB1. CYCA1 interacted with CDKA1, and CYCB1 with CDKB1, in yeast two-hybrid assays (Y. Li et al., 2016), consistent with this idea, but the other pairings were not tested. In addition, preliminary mass spectrometry experiments (S. Obado, K.C. Atkins, F.R. Cross, and M. Rout, unpublished data) showed that CYCA1 but not CYCB1 coimmunoprecipitated with CDKA1, and CYCB1 (but not CYCA1) coimmunoprecipitated with CDKB1, supporting this simple biochemical model.
The APC Has Cell Cycle Regulatory Roles That Are Conserved among Eukaryotes
In Opisthokonts, the APC is essential both for releasing sister chromatid cohesion via degradation of the separase inhibitor securin and for degradation of mitotic cyclins, which is required for mitotic exit. Furthermore, these are the sole essential roles of the APC (Thornton and Toczyski, 2003). APC subunits are essential for plant gametogenesis (Capron et al., 2003; Kwee and Sundaresan, 2003; Wang et al., 2012), but APC cellular roles in plants are relatively uncharacterized. Our results suggest conservation of APC functions between Opisthokonts and the plant kingdom. The APC mutant cdc27-6 arrests in the first division cycle with metaphase spindles, condensed and unseparated chromatin, and high CDK activity, and this cdc27-6 mitotic arrest phenotype is dependent on CYCB1 and CDKB1, consistent with the idea that CYCB1 is a critical APC target. There is at present no candidate for securin in the plant kingdom (securin is poorly conserved even among Opisthokonts). Plants, including Chlamydomonas, have a clear separase homolog, which is essential and implicated in chromosome segregation (Tulin and Cross, 2014; Liu and Makaroff, 2006). Consistent with a role for APC in separase activation, some temperature-sensitive alleles of separase (DIV30) are synthetically lethal with cdc27-6 at permissive temperature (F.R. Cross, unpublished data).
Although almost all eukaryotic genomes contain clear sequence homologs for APC components, Giardia intestinalis has none, and cyclin B degradation may occur by entirely different mechanisms (Gourguechon et al., 2013). This led to the suggestion that the ancestral eukaryote might have lacked the APC, a conclusion that depends on the deepest rooting of the eukaryotic tree. However, if the plant kingdom diverged very early from other eukaryotic lineages (Rogozin et al., 2009), then presence of APC in plants supports ancestral presence of this complex. Our functional results in Chlamydomonas support the deep conservation of APC function as a central mitotic regulator. In Opisthokonts, the APC activity cycle is interdigitated with the cyclin-CDK cycle, with complex conserved regulatory mechanisms ensuring out-of-phase oscillations of CDK and APC activity (Morgan, 2007). It is an open question how the plant cyclin-CDK cycle is integrated with the APC cycle, given the divergent displacement of CDK1 by CDKB as the primary mitotic inducer.
Sequential CDK Function in Control of the Chlamydomonas Cell Cycle
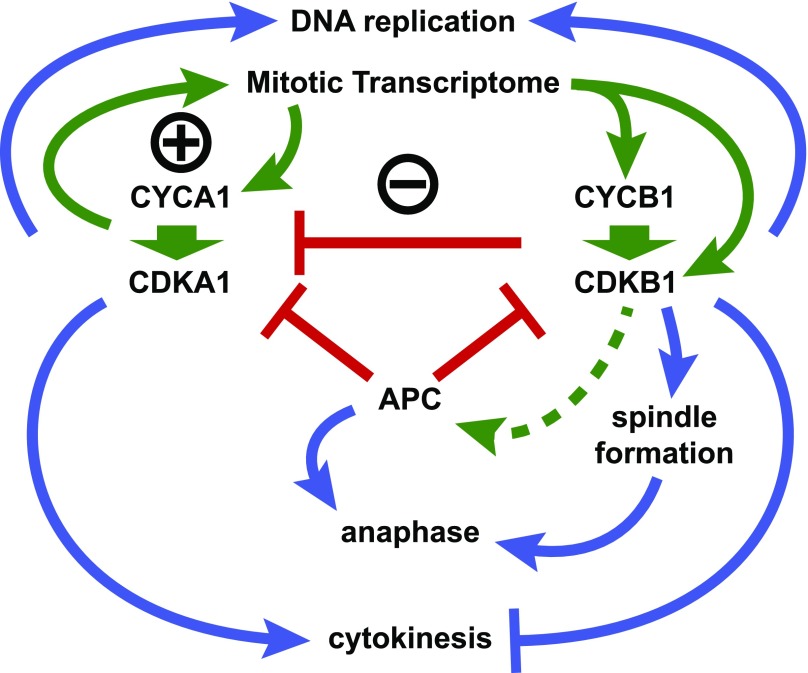
We propose a working model for Chlamydomonas cell cycle control (Figure 9). This model incorporates the main features of the models of Tulin and Cross (2014, 2015) along with findings from this work. CDKA1 is proposed to initiate cell division, at least in part, by induction of the mitotic transcriptome. Approximately 1000 genes are tightly and coordinately induced as cells enter the replicative cycle (Zones et al., 2015, Tulin and Cross, 2015). Maximal expression of a subset of these is strongly dependent on CDKA1 (Tulin and Cross, 2015). Included in the CDKA1-dependent subset are CYCA1, CYCB1, and CDKB1. CDKA1 expression, in contrast, is independent of CDKA1. This circuitry suggests a potential positive feedback loop for CDKA1 activation.
Figure 9.
Proposed Model for Chlamydomonas Cell Cycle Regulation by CYCA1-CDKA1 and CYCB1-CDKB1.
We hypothesize that the main in vivo cyclin-CDK complexes are CYCA1-CDKA1 and CYCB1-CDKB1. The APC negatively regulates CYCA1-CDKA1 and CYCB1-CDKB1; CYCB1-CDKB1 also negatively regulates CYCA1-CDKA1 (red bars). We propose CYCA1 and CYCB1 as predominant, specific in vivo activators of CDKA1 and CDKB1, respectively (heavy green arrows). CDKA1 activation of CYCA1 transcription represents a possible positive feedback loop (circled +); CDKA1 activation of CYCB1-CDKB1 with ensuing repression of CYCA1-CDKA1 by CYCB1-CDKB1 represents a possible negative feedback loop (circled −). Blue arrows, promotion of cell division cycle events; blue bars, inhibition of cell division cycle events; green arrows, activation of control circuitry; red bars, inhibition of control circuitry; dashed green line, possible activation of APC by CYCB1-CDKB1.
CDKA1 activation of transcription of CYCB1 and CDKB1 with ensuing repression of CYCA1-CDKA1 by CYCB1-CDKB1 represents a possible negative feedback loop. We have tested a transgene with the CDKA1 promoter driving CDKB1 transcription. The transgene can rescue cdkb1-1 lethality, but shows little rescue of the delayed cell cycle initiation phenotype of cdka1 disruption using the assay shown in Figure 2. This could be either because CDKA1 has functions beyond transcriptional induction or because CYCB1 (and perhaps other genes) is not efficiently expressed in the absence of CDKA1 (Tulin and Cross, 2015), preventing robust activation of CYCB1-CDKB1.
The circuit architecture of a negative feedback loop reinforced by positive feedback stabilizing alternative states has been proposed for mitotic control in Xenopus laevis (Pomerening et al., 2003) and budding yeast (Cross, 2003), suggesting conservation of, or convergence to, an effective topology despite the use of different molecules and mechanisms (Cross et al., 2011). In Opisthokonts, APC-CDC20 is activated by cyclin B-CDK1; we speculate that CYCB1-CDKB1 may similarly activate the APC, resulting in both CYCA1 and CYCB1 degradation. This would explain why CDKA1- and CDKB1-associated kinase activities are elevated in the absence of APC function (Figures 5F, 5G, 7E, and 7F). However, we do not understand why APC inactivation has a hugely greater quantitative effect on CDKB1-associated kinase than on CDKA1-associated kinase.
We propose that DNA replication initiation is redundantly regulated by CDKA1 and by CYCB1-CDKB1. Single mutations in cdka1, cdkb1, and cycb1 delay but do not block DNA replication (Tulin and Cross, 2014; see above). By contrast, double mutations in cdka1 and cdkb1 (Tulin and Cross, 2014) or cyca1 and cycb1 (Supplemental Figure 4) effectively block replication. In Opisthokonts, CDK phosphorylation of factors including Sld2 and Sld3 links CDK activation to DNA replication (Morgan, 2007). Conversely, replication origin reloading is inhibited in fungi by CDK phosphorylation of origin binding proteins such as ORC and CDC6 (Nguyen et al., 2001), and inhibited in animals by geminin, which is degraded by the APC (McGarry and Kirschner, 1998). In Chlamydomonas, APC inactivation leads to arrest after a single round of DNA replication (Figure 3). We do not know whether additional rounds of replication in arrested cells are blocked due to the accumulation of a geminin-like molecule, to CDK-mediated inhibition of origin components, or to some other mechanism.
CYCB1 and CDKB1 are required for spindle formation (Figure 3; Tulin and Cross, 2014), suggesting that CYCB1-CDKB1 phosphorylate spindle-regulatory components. In contrast, the ability of cyca1 and cdka1 mutants to complete divisions implies that neither is required for spindle formation. CDK1 is required in fungi and animals for spindle assembly, but the mechanism is complex and still not completely understood.
Cytokinetic initiation (formation of cleavage furrows) is delayed in cyca1 and cdka1 mutants, suggesting an important though nonessential role in promoting initiation of cytokinesis. Cytokinesis probably initiates after anaphase and is dependent on microtubules, probably those in the phycoplast structure that forms along (and may direct) the cleavage furrow (Ehler and Dutcher, 1998; Cross and Umen, 2015). Both wild-type metaphase cells and arrested apc mutant cells have microtubules almost exclusively in the mitotic spindle, and they lack evident phycoplasts (Doonan and Grief, 1987; Tulin and Cross, 2014; Figure 3). Intriguingly, cycb1 and cdkb1 mutants, which never form mitotic spindles, nevertheless form phycoplast-like structures and initiate cleavage furrows (Tulin and Cross, 2014; Figure 3). Thus, CYCB1-CDKB1 may repress cleavage furrow formation. Restoration of furrows in cdc27-6 mutants by additional inactivation of CYCB1 or CDKB1 (see above) is consistent with this idea. We speculate that APC-mediated inactivation of CYCB1 as cells complete anaphase may relieve inhibition of furrow formation, resulting in the appropriate sequence of nuclear and cellular division. Mitotic CDK probably inhibits cytokinesis in yeast and animals (Drapkin et al., 2009; Potapova et al., 2006), though mechanisms likely differ.
The great evolutionary distance between Opisthokonts and plants implies the likelihood of significant divergence even in core cellular functions. In addition to mitotic replacement of CDK1 by CDKB, other examples of divergence are known (Harashima et al., 2013), and more will doubtless be found. The central importance of the plant kingdom to life on earth makes it very important to understand both conservation and divergence of biological mechanisms compared with the well-studied fungal and animal lineages.
METHODS
Strains and Alleles
Parental strains were isoloM and isoloP from S. Dutcher (congenic cc124 background). The disruption allele in CYCA1 (from the library of X. Li et al. [2016]) was confirmed by PCR and backcrossed to isoloM/P at least five times before use in experiments.
We discovered ts-lethal mutations mapping to sequences annotated as the 3′ untranslated region of the gene Cre08.g370450 (M. Breker, K. Lieberman, F.R. Cross, unpublished data). This gene is the neighbor of Cre08.g370401, annotated as an N-terminal fragment of cyclin B (https://phytozome.jgi.doe.gov). Examination of the sequence showed that the annotated 3′ untranslated region of Cre08.g370450 contained the C-terminal coding sequences for cyclin B, and RNA evidence indicated that the actual transcript covered the entire coding sequence without interruption (https://phytozome.jgi.doe.gov). The Cre08.g370450 gene model should be corrected as follows: The last exon (chromosome_8 2310083:2310188) should be removed and substituted with the correct final exon in chromosome_8 (2310321:2311564). cycb1-5 (p.Glu325>Lys) was provided by F. Tulin (unpublished data); cycb1-1 (p.Leu288>Pro) and cycb1-3 (p.Glu325>Lys; same as cycb1-5) were provided by M. Breker (unpublished data). The cdc27-6 mutation (div23-6; Tulin and Cross, 2014) is p.Met884>Ile, Gly885>Ser in Cre17.g740510. cdka1-1 and cdkb1-1 were described previously (Tulin and Cross, 2014). The disruption allele of CDKA1 used here (cdka1Δ) inserts Aph7” downstream of codon 93. We confirmed all results using multiple meiotic segregants for both single and double mutants, following multiple rounds of backcrossing to a congenic background.
Tagging CDKA1 and CDKB1
Plasmids pKA11 and pKA1 were constructed using one-step isothermal assembly with the NEBuilder HiFi DNA Assembly Master Mix (New England Biolabs), using fragments amplified from genomic DNA (primers in Supplemental Table 5) and pSI103 as vector (Chlamydomonas Resource Center), cut with PsiI. Chlamydomonas reinhardtii codon-optimized mCherry was amplified from pBR29-mCherry (Rasala et al., 2013). A (3X GGGGS)3 linker was included between CDK and mCherry. CDK-mCherry constructs were transformed by electroporation into cdka1-1 csl89 or cdkb1-1, for CDKA1 and CDKB1, respectively. Expression of integrated mCherry-tagged transgenes was assessed by immunoblot in synchronized cultures. Complementation of temperature sensitivity was assessed by serial dilution of transformants.
Death Delay Screen and Mutant Identification
For insertional mutagenesis, a div19-2 strain was transformed with the AphVIII cassette (conferring resistance to paromomycin) amplified from pSI103. For UV mutagenesis, ∼2*10^6 div19-2 cells were spread on 100-mm agar plates and mutagenized by UV as described (Tulin and Cross, 2014). To select for death delay mutants, plates with transformant/UV-irradiated colonies were transferred through the following sequence (two cycles of selection and recovery): 33°C 20 h, 21°C 48 h, 33°C 20 h, and 21°C 8 days, all with continuous illumination (for UV mutagenesis, 16 h 33°C selection periods were used). Surviving colonies were collected manually. Mutant cell size was examined after growth on TAP agar for 20 h at 33°C by quantification of pixel area of bright-field images.
The genomic location of insertions was determined using either the RATE technique (Karlyshev et al., 2000) or the Hairpin PCR technique (Plecenikova et al., 2014). UV-induced mutations in CDKA1 were determined by Sanger sequence of amplified PCR fragments.
Synchronization
We synchronized cultures by nitrogen deprivation. Cell suspension (0.25 mL; OD750 of ∼8) was spread on a 100-mm 1.5% TAP agar plate with 1/10 normal ammonium chloride and incubated at 21°C with illumination for 24 h. Most cells performed two divisions, then arrested due to nitrogen deprivation. Cells were resuspended, spread on TAP agar with full ammonium chloride, and incubated at 33°C with illumination. In this protocol, there was little or no DNA replication or cell division for ∼10 h; wild-type cells then undergo three to four rapid (∼0.6 h) sequential divisions. Almost all wild-type cells complete divisions by ∼15 h. Synchrony was evaluated microscopically and by flow cytometry, as described (Tulin and Cross, 2014). In some experiments, the time-zero sample (before release) contained numerous fully divided cell clusters that had not completely hatched from the mother cell wall, giving a spuriously high ≥2C peak in DNA flow cytometry. Such cell clusters hatched within half an hour of release.
Time-Lapse Microscopy
Suspensions of cells blocked by nitrogen deprivation were plated on agar and placed at 33°C with illumination. At intervals, the plates were transferred to a heated, illuminated chamber on a Zeiss Axioskop 40 tetrad dissection microscope stage with calibrated positions, and bright-field images were captured. Custom MATLAB software was used to contrast-enhance images, segment individual cells, and present an interface for manual scoring of time points by which divisions or division attempts occurred. Matlab code, and a complete description of the scoring scheme, is available upon request to F.R.C.
Immunostaining and Fluorescence Microscopy
Indirect antitubulin immunofluorescence and DNA detection (SYTOX green) were performed on methanol-fixed cells as described (Tulin and Cross, 2014). Images were collected using DeltaVision deconvolution microscopy. Differential interference contrast images are from one in-focus z-slice. DNA images (SYTOX green) were blurred by Gaussian smoothing (σ = 6) and presented as maximum intensity z-projections. Gaussian smoothing reduced patchy appearance from maximum intensity projections due to occasional very bright pixels in some sections. α-Tubulin images are maximum intensity z-projections without smoothing. All images are individually contrast-adjusted for clarity, so intensities should not be compared across images.
Live-Cell Confocal Fluorescence Microscopy
Synchronized cycling populations of cells were placed on thin TAP + 1.5% low melt agarose slabs for imaging. Confocal laser microscopy was performed using an inverted TCS SP8 confocal microscope (Leica) equipped with three HyD detectors and one PMT detector (scan speed 600 Hz, resolution 1024 × 1024 pixels). Objective lens used was HC PL APO CS2 63×/1.40 oil with 0.75× optical zoom. A time gating of 0.3 to 12.0 ns was used to eliminate chloroplast autofluorescence (Kodama, 2016). mCherry images were acquired with excitation from the 587-nm laser light and xyz scanning with the hybrid detector at 597 to 660 nm with 4× line accumulation. Nuclear ble-GFP was imaged with excitation from the 489-nm laser light and xyz scanning with the hybrid detector at 497 to 561 nm with line averaging of 2×. Bright-field images were obtained with the PMT trans detector. ImageJ was used for average Z-projections and contrast adjustment as described in figures.
Nuclear concentration of CDKB1 (Figure 8G) was measured using confocal micrographs of live cdkb1-1 CDKB1-mCherry ble-GFP. Individual nuclei were partitioned from ble-GFP images using ImageJ. Within the nuclear boundaries, mCherry pixel intensities were summed through the z-stack, and nuclear concentration determined as the sum of the pixel intensities divided by nuclear pixel number. Background fluorescence was subtracted from the signal using a correction factor determined from images of the ble-GFP (no CDKB1:mCherry) strain.
Immunoblotting
Twenty micrograms of protein in total cell lysates was used for immunoblot, according to standard methods. Anti-mCherry antibody (Thermo Fisher; M11217) was used at a dilution of 1:1000. Anti-AtpB antibody (Agrisera; AS05 085) and HRP-conjugated secondary antibodies (Thermo-Fisher, A18865; GE Healthcare, NA934) were used at a 1:2000 dilution. Blots were imaged with an ImageQuant LAS 4000 imager (GE Healthcare). Quantification and contrast adjustment were performed using ImageJ.
Immunoprecipitation and Protein Kinase Assay
LaM-2 nanobody against mCherry (Fridy et al., 2014) was kindly provided by Peter Fridy and was conjugated to Dynabeads M-270 Epoxy (Invitrogen; 14302D) as described (Fridy et al., 2014). Cells were scraped from plates, collected by centrifugation, and resuspended in 1 mL of ice-cold LSHN buffer (10 mM HEPES, pH 7.5, 50 mM NaCl, and 10% [v/v] glycerol), pelleted again by centrifugation, and stored at −80°C. Approximately 10^8 cells at time zero, or OD-equivalent cell amounts at later time points, were harvested. Two hundred microliters of acid-washed glass beads (Sigma-Aldrich; G8772) were added to the cell pellet followed by addition of 250 μL of RIPA buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% [v/v] Nonidet P-40, 0.5% [v/v] sodium deoxycholate, and 0.1% [w/v] SDS) containing phosphatase inhibitors (10 mM sodium pyrophosphate) and protease inhibitors (10% aprotinin [Sigma-Aldrich; A6279], 0.5 mM phenylmethylsulfonyl fluoride, 10 μg/mL of leupeptin, and 10 μg/mL pepstatin). Cells were broken at 4°C using a FastPrep FP120 Cell Disruptor (Thermo Electron) with two 20-s cycles (setting 5). Lysate was clarified by 5 min microcentrifugation. Two hundred micrograms of protein (Pierce BCA Protein Assay Kit) was diluted in RIPA buffer plus inhibitors to 200 μL and added to 10 μL nanobody-conjugated Dynabeads, followed by 1 h rotation at 4°C. Beads were washed four times with 1 mL of RIPA buffer, two times with 1 mL HNN buffer (10 mM HEPES, pH 7.5, 250 mM NaCl, 10% [v/v] glycerol, and 0.1% [v/v] Nonidet P-40), and once with 1 mL kinase buffer (10 mM HEPES, pH 7.5, 50 mM KCl, 10 mM MgCl2, and 1 mM DTT). Beads were suspended to final 60 μL in kinase buffer, and 15 μL added to a mix of 1 μL of 2 mg/mL histone H1 (EMD Millipore; 382150), 2 μL of 50 μM ATP, 1 μL of [γ-32P]ATP (Perkin-Elmer; NEG502A001MC), and 1 μL of water. Reactions were incubated at 30°C for 15 min, mixing every 3 min. Controls established that the reaction was approximately linear through this time. Reactions were stopped with SDS sample buffer. A polyacrylamide gel was run with 10 μL of the kinase reaction. The free [γ-32P]ATP was cut from the gel front and gels stained with GelCode Blue Stain Reagent (Thermo Scientific). Quantification of radioactivity was by storage phosphor screen (Molecular Dynamics), imaged with a Typhoon 9400 Variable Imager (Amersham Biosciences). Control experiments established that background kinase from immunoprecipitates from untagged cells was low and not significantly altered through the cell cycle.
Accession Numbers
Sequence data from this article can be found in the Phytozome database under the following accession numbers: Cr-CDKA1 (Cre10.g465900), Cr-CDKB1 (Cre08.g3725000), Cr-CYCA1 (Cre03.g207900), Cr-CYCB1 (Cre08.g370401/Cre08.g370450; see Methods for discussion of amended CYCB1 gene model fusing these models), CSL89 (Cre05.g244236), At-CDKA;1 (AT3G48750.1), At-CDKB1;1 (AT3G54180.1), Bd-CDKA (Bradi3g02270.1), Bd-CDKB1 (Bradi4g25980.1), At-CYCA2;4 (AT1G80370.1), and At-CYCB2;4 (AT1G76310.1). Accession numbers from NCBI are as follows: Mm-CDK1 (NP_031685.2) and Mm-CDK2 (NP_058036.1).
Supplemental Data
Supplemental Figure 1. Relationship of Chlamydomonas CDKA1 and CDKB1 to land plant and animal CDKs.
Supplemental Figure 2. Temperature sensitivity of the cdka1Δ mutant.
Supplemental Figure 3. Formation of cleavage furrows is dependent on APC-dependent inhibition of CYCB1-CDKB1.
Supplemental Figure 4. Formation of cleavage furrows and DNA replication in the absence of CYCB1 depends on CYCA1.
Supplemental Figure 5. Expression of CDKB1:mCherry is delayed and is correlated to cell division in the cdka1Δ mutant.
Supplemental Figure 6. Activity of CDKA1 against histone H1 is higher than that of CDKB1.
Supplemental Figure 7. Cyclin dependence of CDKA1 and CDKB1 protein kinase activity.
Supplemental Figure 8. Overall sequence and destruction box alignments for CYCA1 and CYCB1: CYCA1 is an ortholog of land plant cyclin A; CYCB1 is an ortholog of land plant cyclin B; both contain strong destruction box consensus sequences.
Supplemental Table 1. Mutations in CDKA1 in UV-mutagenized tdd mutants.
Supplemental Table 2. Summary statistics for cell cycle delay in the cyca1Δ, cdka1Δ, and cyca1Δ cdka1Δ mutants.
Supplemental Table 3. Formation of mitotic spindles and cleavage furrows in the cycb1, cdc27, and cycb1 cdc27 mutants.
Supplemental Table 4. Kinase dead (K33R) CDKB1:mCherry fails to rescue temperature-sensitive lethality of the cdkb1-1 mutant.
Supplemental Table 5. Oligonucleotides used for plasmid construction and sequencing.
Acknowledgments
We thank F. Tulin for providing the cdc27-6 and cycb1-5 mutants, and M. Breker and K. Lieberman for correction of the reference CYCB1 annotations and providing additional CYCB1 mutants. We thank K. Pecani for assistance with DNA flow cytometry and antimicrotubule immunofluorescence. We thank M. Breker, K. Lieberman, K. Pecani, and F. Tulin for useful discussions. We thank Alison North (Rockefeller Bioimaging Resource Center) for critical advice and assistance with time-resolved confocal microscopy. The work was supported by NIH grant RO1 GM07853 to F.R.C. and by support from the Rockefeller University to F.R.C. and K.C.A.
AUTHOR CONTRIBUTIONS
F.R.C. and K.C.A. collaborated in designing and performing research, analyzing data, writing the text of the manuscript, and designing figures.
Footnotes
Articles can be viewed without a subscription.
References
- Bisova K., Krylov D.M., Umen J.G. (2005). Genome-wide annotation and expression profiling of cell cycle regulatory genes in Chlamydomonas reinhardtii. Plant Physiol. 137: 475–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom J., Cross F.R. (2007). Multiple levels of cyclin specificity in cell-cycle control. Nat. Rev. Mol. Cell Biol. 8: 149–160. [DOI] [PubMed] [Google Scholar]
- Boudolf V., et al. (2009). CDKB1;1 forms a functional complex with CYCA2;3 to suppress endocycle onset. Plant Physiol. 150: 1482–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudolf V., Vlieghe K., Beemster G.T.S., Magyar Z., Torres Acosta J.A., Maes S., Van Der Schueren E., Inzé D., De Veylder L. (2004). The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 16: 2683–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breker M., Lieberman K., Tulin F., Cross F.R. (2016). High-throughput robotically assisted isolation of temperature-sensitive lethal mutants in Chlamydomonas reinhardtii. J. Vis. Exp. 118: e54831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capron A., Serralbo O., Fülöp K., Frugier F., Parmentier Y., Dong A., Lecureuil A., Guerche P., Kondorosi E., Scheres B., Genschik P. (2003). The Arabidopsis anaphase-promoting complex or cyclosome: molecular and genetic characterization of the APC2 subunit. Plant Cell 15: 2370–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier C., Bourdon M., Pirrello J., Cheniclet C., Gévaudant F., Frangne N. (2014). Endoreduplication and fruit growth in tomato: evidence in favour of the karyoplasmic ratio theory. J. Exp. Bot. 65: 2731–2746. [DOI] [PubMed] [Google Scholar]
- Cross F.R. (2003). Two redundant oscillatory mechanisms in the yeast cell cycle. Dev. Cell 4: 741–752. [DOI] [PubMed] [Google Scholar]
- Cross F.R., Buchler N.E., Skotheim J.M. (2011). Evolution of networks and sequences in eukaryotic cell cycle control. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366: 3532–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross F.R., Umen J.G. (2015). The Chlamydomonas cell cycle. Plant J. 82: 370–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte W., Scofield S., Alcasabas A.A., Maughan S.C., Menges M., Braun N., Collins C., Nieuwland J., Prinsen E., Sundaresan V., Murray J.A. (2007). Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc. Natl. Acad. Sci. USA 104: 14537–14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener D.R., Lupetti P., Rosenbaum J.L. (2015). Proteomic analysis of isolated ciliary transition zones reveals the presence of ESCRT proteins. Curr. Biol. 25: 379–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissmeyer N., Weimer A.K., Pusch S., De Schutter K., Alvim Kamei C.L., Nowack M.K., Novak B., Duan G.-L., Zhu Y.-G., De Veylder L., Schnittger A. (2009). Control of cell proliferation, organ growth, and DNA damage response operate independently of dephosphorylation of the Arabidopsis Cdk1 homolog CDKA;1. Plant Cell 21: 3641–3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doonan J., Grief C. (1987). Microtubule cycle in Chlamydomonas reinhardtii: an immunofluorescence study. Cell Motil. Cytoskeleton 7: 381–392. [Google Scholar]
- Drapkin B.J., Lu Y., Procko A.L., Timney B.L., Cross F.R. (2009). Analysis of the mitotic exit control system using locked levels of stable mitotic cyclin. Mol. Syst. Biol. 5: 328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher S.K., O’Toole E.T. (2016). The basal bodies of Chlamydomonas reinhardtii. Cilia 5: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehler L.L., Dutcher S.K. (1998). Pharmacological and genetic evidence for a role of rootlet and phycoplast microtubules in the positioning and assembly of cleavage furrows in Chlamydomonas reinhardtii. Cell Motil. Cytoskeleton 40: 193–207. [DOI] [PubMed] [Google Scholar]
- Fang S.-C., de los Reyes C., Umen J.G. (2006). Cell size checkpoint control by the retinoblastoma tumor suppressor pathway. PLoS Genet. 2: e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher D.L., Nurse P. (1996). A single fission yeast mitotic cyclin B p34cdc2 kinase promotes both S-phase and mitosis in the absence of G1 cyclins. EMBO J. 15: 850–860. [PMC free article] [PubMed] [Google Scholar]
- Fridy P.C., Li Y., Keegan S., Thompson M.K., Nudelman I., Scheid J.F., Oeffinger M., Nussenzweig M.C., Fenyö D., Chait B.T., Rout M.P. (2014). A robust pipeline for rapid production of versatile nanobody repertoires. Nat. Methods 11: 1253–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M., Murray A.W., Kirschner M.W. (1991). Cyclin is degraded by the ubiquitin pathway. Nature 349: 132–138. [DOI] [PubMed] [Google Scholar]
- Gong D., Ferrell J.E. Jr. (2010). The roles of cyclin A2, B1, and B2 in early and late mitotic events. Mol. Biol. Cell 21: 3149–3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourguechon S., Holt L.J., Cande W.Z. (2013). The Giardia cell cycle progresses independently of the anaphase-promoting complex. J. Cell Sci. 126: 2246–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harashima H., Dissmeyer N., Schnittger A. (2013). Cell cycle control across the eukaryotic kingdom. Trends Cell Biol. 23: 345–356. [DOI] [PubMed] [Google Scholar]
- Harashima H., Schnittger A. (2012). Robust reconstitution of active cell-cycle control complexes from co-expressed proteins in bacteria. Plant Methods 8: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Chao W.C.H., Zhang Z., Yang J., Cronin N., Barford D. (2013). Insights into degron recognition by APC/C coactivators from the structure of an Acm1-Cdh1 complex. Mol. Cell 50: 649–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm C., Stearns T., Botstein D. (1989). DNA topoisomerase II must act at mitosis to prevent nondisjunction and chromosome breakage. Mol. Cell. Biol. 9: 159–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai K.K., Ohashi Y., Tsuge T., Yoshizumi T., Matsui M., Oka A., Aoyama T. (2006). The A-type cyclin CYCA2;3 is a key regulator of ploidy levels in Arabidopsis endoreduplication. Plant Cell 18: 382–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey P.D., Russo A.A., Polyak K., Gibbs E., Hurwitz J., Massagué J., Pavletich N.P. (1995). Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature 376: 313–320. [DOI] [PubMed] [Google Scholar]
- Johnston G.C., Pringle J.R., Hartwell L.H. (1977). Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp. Cell Res. 105: 79–98. [DOI] [PubMed] [Google Scholar]
- Karlyshev A.V., Pallen M.J., Wren B.W. (2000). Single-primer PCR procedure for rapid identification of transposon insertion sites. Biotechniques 28: 1078, 1080, 1082. [DOI] [PubMed] [Google Scholar]
- King R.W., Glotzer M., Kirschner M.W. (1996). Mutagenic analysis of the destruction signal of mitotic cyclins and structural characterization of ubiquitinated intermediates. Mol. Biol. Cell 7: 1343–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama Y. (2016). Time gating of chloroplast autofluorescence allows clearer fluorescence imaging in planta. PLoS One 11: e0152484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwee H.-S., Sundaresan V. (2003). The NOMEGA gene required for female gametophyte development encodes the putative APC6/CDC16 component of the Anaphase Promoting Complex in Arabidopsis. Plant J. 36: 853–866. [DOI] [PubMed] [Google Scholar]
- Li X., Zhang R., Patena W., Gang S.S., Blum S.R., Ivanova N., Yue R., Robertson J.M., Lefebvre P.A., Fitz-Gibbon S.T., Grossman A.R., Jonikas M.C. (2016). An indexed, mapped mutant library enables reverse genetics studies of biological processes in Chlamydomonas reinhardtii. Plant Cell 28: 367–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Liu D., López-Paz C., Olson B.J., Umen J.G. (2016). A new class of cyclin dependent kinase in Chlamydomonas is required for coupling cell size to cell division. eLife 5: e10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Makaroff C.A. (2006). Arabidopsis separase AESP is essential for embryo development and the release of cohesin during meiosis. Plant Cell 18: 1213–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz S., Tintelnot S., Reski R., Decker E.L. (2003). Cyclin D-knockout uncouples developmental progression from sugar availability. Plant Mol. Biol. 53: 227–236. [DOI] [PubMed] [Google Scholar]
- Lynch M., Force A. (2000). The probability of duplicate gene preservation by subfunctionalization. Genetics 154: 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., O’Hely M., Walsh B., Force A. (2001). The probability of preservation of a newly arisen gene duplicate. Genetics 159: 1789–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry T.J., Kirschner M.W. (1998). Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell 93: 1043–1053. [DOI] [PubMed] [Google Scholar]
- Melaragno J.E., Mehrotra B., Coleman A.W. (1993). Relationship between dndopolyploidy and cell size in epidermal tissue of Arabidopsis. Plant Cell 5: 1661–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell J.E., Clements K.D., Choat J.H., Angert E.R. (2008). Extreme polyploidy in a large bacterium. Proc. Natl. Acad. Sci. USA 105: 6730–6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant S.S., et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan D. (2007). The Cell Cycle: Principles of Control. (London: New Science Press).
- Murray A.W., Kirschner M.W. (1989). Cyclin synthesis drives the early embryonic cell cycle. Nature 339: 275–280. [DOI] [PubMed] [Google Scholar]
- Murray A.W., Solomon M.J., Kirschner M.W. (1989). The role of cyclin synthesis and degradation in the control of maturation promoting factor activity. Nature 339: 280–286. [DOI] [PubMed] [Google Scholar]
- Nguyen V.Q., Co C., Li J.J. (2001). Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature 411: 1068–1073. [DOI] [PubMed] [Google Scholar]
- Nowack M.K., Harashima H., Dissmeyer N., Zhao X., Bouyer D., Weimer A.K., De Winter F., Yang F., Schnittger A. (2012). Genetic framework of cyclin-dependent kinase function in Arabidopsis. Dev. Cell 22: 1030–1040. [DOI] [PubMed] [Google Scholar]
- Nurse P., Thuriaux P., Nasmyth K. (1976). Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 146: 167–178. [DOI] [PubMed] [Google Scholar]
- Olson B.J.S.C., Oberholzer M., Li Y., Zones J.M., Kohli H.S., Bisova K., Fang S.-C., Meisenhelder J., Hunter T., Umen J.G. (2010). Regulation of the Chlamydomonas cell cycle by a stable, chromatin-associated retinoblastoma tumor suppressor complex. Plant Cell 22: 3331–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano M., Pepperkok R., Verde F., Ansorge W., Draetta G. (1992). Cyclin A is required at two points in the human cell cycle. EMBO J. 11: 961–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plecenikova A., Slaninova M., Riha K. (2014). Characterization of DNA repair deficient strains of Chlamydomonas reinhardtii generated by insertional mutagenesis. PLoS One 9: e105482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polko J.K., et al. (2015). Ethylene-mediated regulation of A2-type CYCLINs modulates hyponastic growth in Arabidopsis. Plant Physiol. 169: 194–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerening J.R., Sontag E.D., Ferrell J.E. Jr (2003). Building a cell cycle oscillator: hysteresis and bistability in the activation of Cdc2. Nat. Cell Biol. 5: 346–351. [DOI] [PubMed] [Google Scholar]
- Potapova T.A., Daum J.R., Pittman B.D., Hudson J.R., Jones T.N., Satinover D.L., Stukenberg P.T., Gorbsky G.J. (2006). The reversibility of mitotic exit in vertebrate cells. Nature 440: 954–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahi S.J., Pecani K., Ondracka A., Oikonomou C., Cross F.R. (2016). The CDK-APC/C oscillator predominantly entrains periodic cell-cycle transcription. Cell 165: 475–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala B.A., Barrera D.J., Ng J., Plucinak T.M., Rosenberg J.N., Weeks D.P., Oyler G.A., Peterson T.C., Haerizadeh F., Mayfield S.P. (2013). Expanding the spectral palette of fluorescent proteins for the green microalga Chlamydomonas reinhardtii. Plant J. 74: 545–556. [DOI] [PubMed] [Google Scholar]
- Remm M., Storm C.E., Sonnhammer E.L. (2001). Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 314: 1041–1052. [DOI] [PubMed] [Google Scholar]
- Rogozin I.B., Basu M.K., Csürös M., Koonin E.V. (2009). Analysis of rare genomic changes does not support the unikont-bikont phylogeny and suggests cyanobacterial symbiosis as the point of primary radiation of eukaryotes. Genome Biol. Evol. 1: 99–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaría D., Barrière C., Cerqueira A., Hunt S., Tardy C., Newton K., Cáceres J.F., Dubus P., Malumbres M., Barbacid M. (2007). Cdk1 is sufficient to drive the mammalian cell cycle. Nature 448: 811–815. [DOI] [PubMed] [Google Scholar]
- Sanz L., et al. (2011). The Arabidopsis D-type cyclin CYCD2;1 and the inhibitor ICK2/KRP2 modulate auxin-induced lateral root formation. Plant Cell 23: 641–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger A., Schöbinger U., Stierhof Y.D., Hülskamp M. (2002). Ectopic B-type cyclin expression induces mitotic cycles in endoreduplicating Arabidopsis trichomes. Curr. Biol. 12: 415–420. [DOI] [PubMed] [Google Scholar]
- Sozzani R., Cui H., Moreno-Risueno M.A., Busch W., Van Norman J.M., Vernoux T., Brady S.M., Dewitte W., Murray J.A.H., Benfey P.N. (2010). Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature 466: 128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton B.R., Toczyski D.P. (2003). Securin and B-cyclin/CDK are the only essential targets of the APC. Nat. Cell Biol. 5: 1090–1094. [DOI] [PubMed] [Google Scholar]
- Tulin F., Cross F.R. (2014). A microbial avenue to cell cycle control in the plant superkingdom. Plant Cell 26: 4019–4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulin F., Cross F.R. (2015). Cyclin-dependent kinase regulation of diurnal transcription in Chlamydomonas. Plant Cell 27: 2727–2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen J.G., Goodenough U.W. (2001). Control of cell division by a retinoblastoma protein homolog in Chlamydomonas. Genes Dev. 15: 1652–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K., Raes J., De Veylder L., Rouzé P., Rombauts S., Inzé D. (2002). Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14: 903–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S., et al. (2011). Developmental regulation of CYCA2s contributes to tissue-specific proliferation in Arabidopsis. EMBO J. 30: 3430–3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste K., Baele G., Maere S., Van de Peer Y. (2014). Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous-Paleogene boundary. Genome Res. 24: 1334–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Hou Y., Gu H., Kang D., Chen Z., Liu J., Qu L.-J. (2012). The Arabidopsis APC4 subunit of the anaphase-promoting complex/cyclosome (APC/C) is critical for both female gametogenesis and embryogenesis. Plant J. 69: 227–240. [DOI] [PubMed] [Google Scholar]
- Weimer A.K., et al. (2016). The plant-specific CDKB1-CYCB1 complex mediates homologous recombination repair in Arabidopsis. EMBO J. 35: 2068–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingartner M., Binarova P., Drykova D., Schweighofer A., David J.P., Heberle-Bors E., Doonan J., Bögre L. (2001). Dynamic recruitment of Cdc2 to specific microtubule structures during mitosis. Plant Cell 13: 1929–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingartner M., Criqui M.-C., Mészáros T., Binarova P., Schmit A.-C., Helfer A., Derevier A., Erhardt M., Bögre L., Genschik P. (2004). Expression of a nondegradable cyclin B1 affects plant development and leads to endomitosis by inhibiting the formation of a phragmoplast. Plant Cell 16: 643–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zones J.M., Blaby I.K., Merchant S.S., Umen J.G. (2015). High-resolution profiling of a synchronized diurnal transcriptome from Chlamydomonas reinhardtii reveals continuous cell and metabolic differentiation. Plant Cell 27: 2743–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]