A study of U. maydis gene expression provides unexpected new leads concerning fungal nutrition, defense suppression, and tumor induction during plant colonization.
Abstract
The maize smut fungus Ustilago maydis is a model organism for elucidating host colonization strategies of biotrophic fungi. Here, we performed an in depth transcriptional profiling of the entire plant-associated development of U. maydis wild-type strains. In our analysis, we focused on fungal metabolism, nutritional strategies, secreted effectors, and regulatory networks. Secreted proteins were enriched in three distinct expression modules corresponding to stages on the plant surface, establishment of biotrophy, and induction of tumors. These modules are likely the key determinants for U. maydis virulence. With respect to nutrient utilization, we observed that expression of several nutrient transporters was tied to these virulence modules rather than being controlled by nutrient availability. We show that oligopeptide transporters likely involved in nitrogen assimilation are important virulence factors. By measuring the intramodular connectivity of transcription factors, we identified the potential drivers for the virulence modules. While known components of the b-mating type cascade emerged as inducers for the plant surface and biotrophy module, we identified a set of yet uncharacterized transcription factors as likely responsible for expression of the tumor module. We demonstrate a crucial role for leaf tumor formation and effector gene expression for one of these transcription factors.
INTRODUCTION
Plant pathogenic fungi have adopted discrete lifestyles in interaction with their hosts ranging from biotrophy, where the plant needs to be kept alive to sustain fungal growth, to necrotrophy, where infected plant tissue is actively killed and the fungus feeds on the dead material. In between these extremes are hemibiotrophs, which initially establish a biotrophic phase and then at some point switch to necrotrophic development. To promote the respective colonization strategy pathogens secrete a large arsenal of effector proteins. In recent years the study of lifestyle transitions in plant pathogenic fungi by time resolved transcriptome analyses through RNA-seq provided deep insights into the processes associated with stages of fungal development on and inside the host (Kawahara et al., 2012; O’Connell et al., 2012; Hacquard et al., 2013; Jupe et al., 2013; Dong et al., 2015; Fondevilla et al., 2015; Kong et al., 2015; Rudd et al., 2015; Dobon et al., 2016; Thatcher et al., 2016; Copley et al., 2017; Wang et al., 2017; Zeng et al., 2017; Massonnet et al., 2018). These and other studies (Toruño et al., 2016) have shown that different sets of effectors are synthesized and presumably needed during discrete developmental stages in fungal pathogens.
The biotrophic fungus Ustilago maydis causes smut disease in maize (Zea mays). This system has advanced to a model for biotrophic pathogens mainly because of the ease by which fungal genes can be manipulated through reverse genetic techniques (Kämper, 2004; Dean et al., 2012; Schuster et al., 2016). In U. maydis and related smut fungi, the process of plant colonization is intimately coupled with sexual development. Plant colonization is initiated when haploid cells of a compatible mating type recognize each other on the leaf surface via a pheromone-receptor system (Bölker et al., 1992), fuse, and develop a dikaryotic filament. Filament formation and subsequent pathogenic development are controlled by the heterodimeric bE/bW (bEast/bWest) homeodomain transcription factor, which is formed after cells with different alleles of the b locus have fused (Gillissen et al., 1992). The dikaryon is cell cycle arrested (Castanheira et al., 2014) and is able to invade the plant via a specialized infection structure, the appressorium. Appressoria allow direct invasion of epidermal plant cells in a process that is likely aided by plant cell wall-degrading enzymes and plant cell wall loosening. During this stage, the fungus remains intracellular and becomes completely encased by the plasma membrane of the host, forming a tight and extended interaction zone. After penetration, the cell cycle block is released and mitotic growth of the dikaryotic form is observed, using characteristic clamp-like structures for sorting the nuclei (Scherer et al., 2006). After reaching the mesophyll layer and the veins, U. maydis cells grow along and inside of the veins, presumably to forage nutrients. During this stage, discrete plant cell types enlarge and resume mitotic divisions (Matei et al., 2018), leading to the formation of tumors, the most conspicuous signs of maize smut disease. In these tumors, extracellular hyphae form large aggregates in cavities between plant tumor cells, their dikaryotic nuclei fuse, and massive proliferation ensues followed by hyphal fragmentation and spore formation (Vollmeister et al., 2012; Matei and Doehlemann, 2016; Tollot et al., 2016; Lanver et al., 2017; Redkar et al., 2017; Snetselaar and McCann, 2017). This entire developmental cycle strictly depends on the plant and remains biotrophic throughout.
U. maydis is predicted to encode 476 secreted proteins of which 215 lack any known structural or functional domains, preventing conclusions to be drawn about their molecular functions (Schuster et al., 2017). Many of these potential effector genes reside in clusters in the genome (Kämper et al., 2006; Schirawski et al., 2010), are expressed specifically in tumor tissue compared with axenic culture conditions (Kämper et al., 2006), and contribute to virulence (Kämper et al., 2006; Müller et al., 2008; Schirawski et al., 2010; Schilling et al., 2014; Stirnberg and Djamei, 2016). So far, the molecular function of only five U. maydis effectors has been elucidated (Djamei et al., 2011; Hemetsberger et al., 2012; Mueller et al., 2013; Tanaka et al., 2014; Redkar et al., 2015).
The bE/bW transcription factor triggers a regulatory cascade including several transcription factor genes as well as 38 potential effector genes (Heimel et al., 2010a, 2010b). The expression of the majority of these genes further requires the bE/bW-regulated transcription factor rbf1 (regulator of b-filament; Heimel et al., 2010b). Early infection-related development of U. maydis up to the stage of appressorium formation can be mimicked by stimulation with hydroxy fatty acids and exposing cells to a hydrophobic surface (Mendoza-Mendoza et al., 2009). An array study of these stages revealed that two transmembrane proteins, the U. maydis homologs of Sho1p (synthetic high osmolarity sensitive) and Msb2p (multicopy suppressor of a budding defect) from Saccharomyces cerevisiae, are specifically responsible for plant surface cue-induced expression of 41 potential effectors (Lanver et al., 2014). In addition, two transcription factors, hdp2 (homeodomain transcription factor 2) and biz1 (b-dependent zinc finger protein), which belong to the rbf1-induced genes (Heimel et al., 2010b) and have critical roles in virulence (Flor-Parra et al., 2006; Lanver et al., 2014), are transcriptionally induced by Sho1 and Msb2 providing indirect evidence that these transcription factors may induce the expression of early effectors. After induction of tumors, the WOPR transcription factor Ros1 (regulator of sporogenesis 1) induces nuclear fusion, subsequent proliferation, and initiates spore production (Tollot et al., 2016). Ros1 also emerged as an important regulator of effector gene expression. Seventy genes encoding secreted proteins require Ros1 for full expression and 128 genes encoding secreted proteins are downregulated by Ros1. It is speculated that U. maydis can afford the downregulation of so many effectors, including some of the essential effectors expressed early during pathogenic development because Ros1 is also required for the generation of a mucilaginous matrix which may shield hyphae from plant defense molecules (Tollot et al., 2016).
The transcriptional response of maize plants during infection with U. maydis has been studied in detail using genome-wide array analysis. These studies have revealed that U. maydis triggers early plant defense responses when on the leaf surface, presumably through the perception of microbe-associated molecular patterns. These responses are subsequently suppressed during the early colonization stages, likely through the action of early effectors (Doehlemann et al., 2008b). U. maydis also induces transcription of plant cell death suppressors like cystatins and Bax-inhibitor 1, induces jasmonate signaling, and prevents the transition from sink to source leaves (Doehlemann et al., 2008b). All these studies were performed with the engineered solopathogenic haploid strain SG200 (Kämper et al., 2006) that can infect plants without a mating partner. The transcriptome of the U. maydis SG200 strain as well as of SG200 mutants during pathogenic development has been investigated in several studies using array technology (Kämper et al., 2006; Zheng et al., 2008; Skibbe et al., 2010; Gao et al., 2013; Schuler et al., 2015; Rabe et al., 2016). A few studies also analyzed the transcriptome of compatible wild-type strains at discrete stages after infection (Wahl et al., 2010; Zahiri et al., 2010; Tollot et al., 2016). However, most global analyses of the U. maydis transcriptome during colonization were so far restricted to stages of biotrophic development where significant amounts of fungal biomass have accumulated. A comprehensive, time-resolved transcriptional profiling of the plant-associated stages of the U. maydis life cycle is lacking so far. We have recently noted that SG200 behaves differently from compatible haploid strains during the late stages of pathogenic development. While fungal biomass significantly increases after karyogamy in infections with compatible wild-type strains (Tollot et al., 2016), fungal biomass of SG200 decreased at the corresponding time points (S. Tanaka, P. Erchinger, S. Krombach, and R. Kahmann, personal communication). In this study, we have therefore performed an RNA-seq analysis of compatible haploid U. maydis strains starting at 12 h after infection of maize seedlings, i.e., when cells have mated, switched to filamentous growth, and begun to form appressoria, to 12 d postinfection (dpi) when tumors contain mature spores. This offered an unprecedented view of the changes in the fungal transcriptome associated with the passage through the biotrophic life cycle. Deep sequencing enabled us to resolve also the very early stages of infection where fungal biomass is low and fungal transcripts are heavily underrepresented. We focus our attention on fungal metabolism, nutritional strategies, secreted effectors, and regulatory networks. Based on uncovering discrete gene expression waves successively following the developmental stages of U. maydis, we expect this data set to become a highly valuable resource for future studies in this biotrophic fungal model as well as related host-pathogen systems.
RESULTS AND DISCUSSION
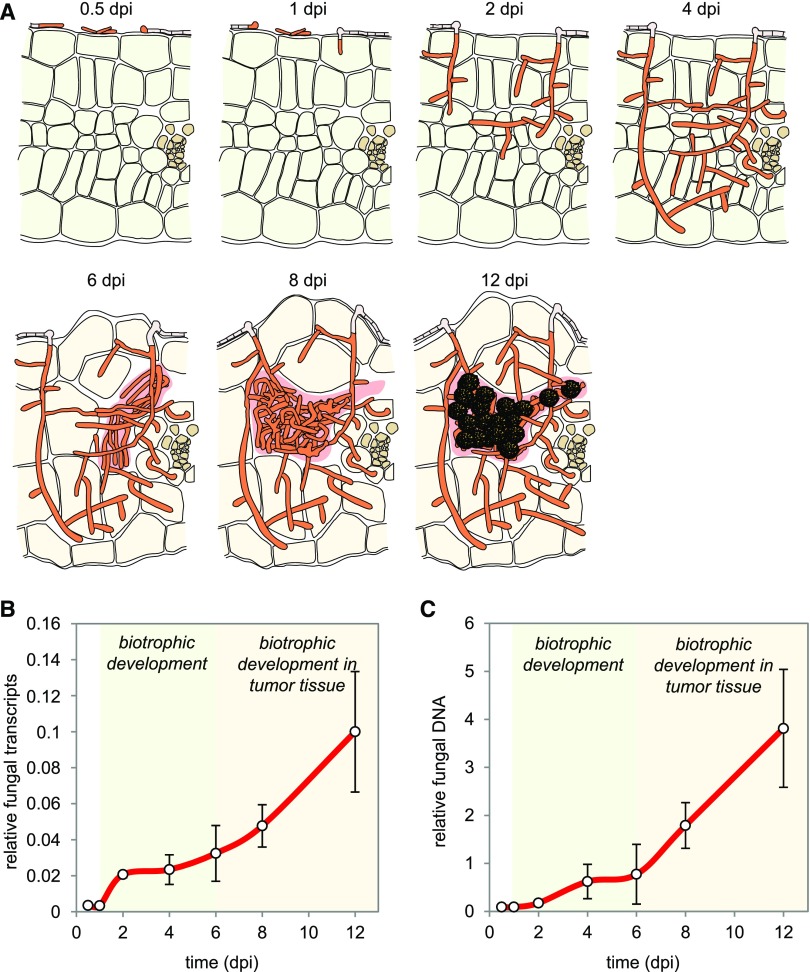
To analyze the transcriptional changes during plant-associated stages of U. maydis, maize seedlings of the variety Early Golden Bantam were infected with the compatible U. maydis wild-type strains FB1 and FB2 (Banuett and Herskowitz, 1989), and infected tissue was collected over a period of 12 d (Figure 1A). The samples represented the following developmental stages of U. maydis: filaments and appressoria in the prepenetration phase (0.5 dpi), penetrating appressoria (1 dpi), dikaryotic biotrophic hyphae with clamp connections (2 dpi), proliferating hyphae and aggregated hyphae (4 and 6 dpi), fragmented hyphae (8 dpi), and mature spores (12 dpi; Figure 1A). For each time point, three biological replicates were generated. As an additional reference, we included FB1 and FB2 grown exponentially in YEPSL medium.
Figure 1.
Changes in the Amount of Fungal Transcripts and Fungal Biomass during Infection.
(A) Schematic view of cross sections of U. maydis infected maize leaves illustrating the stages of fungal development as well as plant tumor formation at the time points analyzed by RNA-seq-based transcriptional profiling. U. maydis infection is not synchronized, and each sample thus contains fungal transcripts from different developmental stages. Green, plant leaf tissue; brown, vascular tissue; orange, fungal cytoplasm; gray, empty fungal hyphae separated by septa; beige, plant tumor cells; rose, matrix; ornamented in black, fungal spores.
(B) Amount of fungal transcripts based on the RNA-seq analysis. For each time point (0.5, 1, 2, 4, 6, 8, and 12 dpi) the ratio of reads uniquely mapped to the U. maydis genome relative to the total number of uniquely mapped reads (U. maydis and maize) was determined. Error bars denote sd of three biological replicates.
(C) Fungal biomass determination based on the amount of genomic DNA. A qPCR with plant-specific (GAPDH) and fungus-specific (ppi) primers was performed using the same infected plant material that was used for the RNA-seq analysis. Data points give mean ratios of fungal DNA to plant DNA (2−ΔCt). Error bars denote sd of three biological replicates.
By Illumina sequencing of mRNA libraries, we created in total more than two billion paired-end reads from all samples (Supplemental Data Set 1). Prior to mapping them to the U. maydis genome, reads mapping to the maize genome were filtered. These data are not discussed here but have been deposited in the NCBI Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE103876. For the early time points of 0.5 dpi and 1 dpi in particular, reads mapping to the fungal genome amounted to <0.5%, i.e., were heavily underrepresented (Figure 1B; Supplemental Data Set 1). In the total paired-end reads from all samples, the sequencing depths of the 0.5 and 1 dpi samples has been adjusted to reach ∼500 thousand-read counts that mapped to the fungal genome and in the remaining samples to reach about one million or more fungal read counts (Supplemental Data Set 1). In each sample, >75% of the 6766 U. maydis genes were represented with more than 10 fragments per kilobase of exon per million fungal reads, indicating that we have efficiently detected fungal gene expression across all time points. Correlated with the increasing fungal biomass, the fungal reads steadily increased during later time points and reached ∼10% at 12 dpi (Figures 1B and 1C). We observed a linear correlation between the number of fungal transcripts and increase in fungal biomass (Supplemental Figure 1). However, the steep increase in fungal transcripts from 1 to 2 dpi (Figure 1B), that is statistically significant (Supplemental Data Set 2), was not accompanied by a comparable change in fungal biomass (Figure 1C). This suggests that the transcriptional activity of U. maydis cells after recovery from cell cycle arrest (2 dpi) is higher than at the stage before penetration or during later biotrophic development.
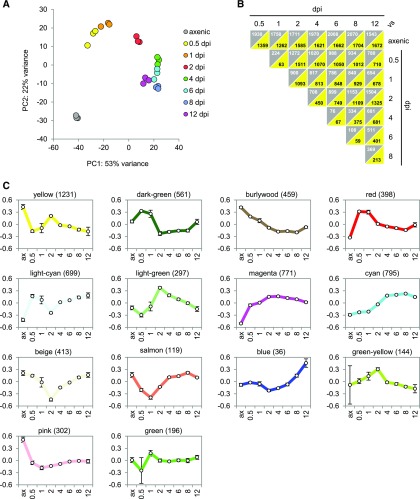
To enable comparisons between samples, all read counts were normalized by DESeq2 (differential expression analysis for sequence count data 2) (Love et al., 2014) (Supplemental Data Set 3). To assess variability among the samples, we performed a principal component analysis (Figure 2A). The three biological replicates formed distinct clusters, indicating time point-specific expression patterns and an acceptable variation between replicates at any of the chosen time points. To analyze differential gene expression, we compared expression in all 28 possible pairs of the eight different time points. This analysis revealed that in total 5759 genes (85% of all U. maydis genes) were differentially expressed (log2 fold change > 0.5 and adjusted P value < 0.01) in at least one of the 28 comparisons (Figure 2B; Supplemental Data Set 4). To validate the expression data, we randomly picked nine genes expressed at different levels and performed RT-qPCR using all 24 generated RNA samples. We observed a strong linear correlation (r = 0.90) between the RT-qPCR and RNA-seq data (Supplemental Figure 2). To further strengthen the bioinformatics analysis, EdgeR was used as an alternative tool to normalize the data and to identify differentially expressed genes. This analysis yielded results comparable to the DESeq2 analysis (Supplemental Data Set 5). In the following analyses, we refer to the DESeq2 results.
Figure 2.
Assessment of the RNA-Seq Data Set of U. maydis during Infection.
(A) Principal component analysis of RNA-seq data. The replicates of each developmental stage of U. maydis (axenic, 0.5, 1, 2, 4, 6, 8, and 12 dpi) form distinct clusters.
(B) The expression data of the eight analyzed developmental stages of U. maydis (axenic, 0.5, 1, 2, 4, 6, 8, and 12 dpi) was the basis to extract all 28 possible contrasts. Genes with a log2 fold change > 0.5 and adjusted P value < 0.01 were considered differentially expressed. Gray triangles depict the number of genes expressed at higher levels at the stages denoted by the horizontal labels, and yellow triangles depict the number of genes expressed at higher levels at the vertically labeled stages. In total, 5759 of the 6766 U. maydis genes were differentially expressed.
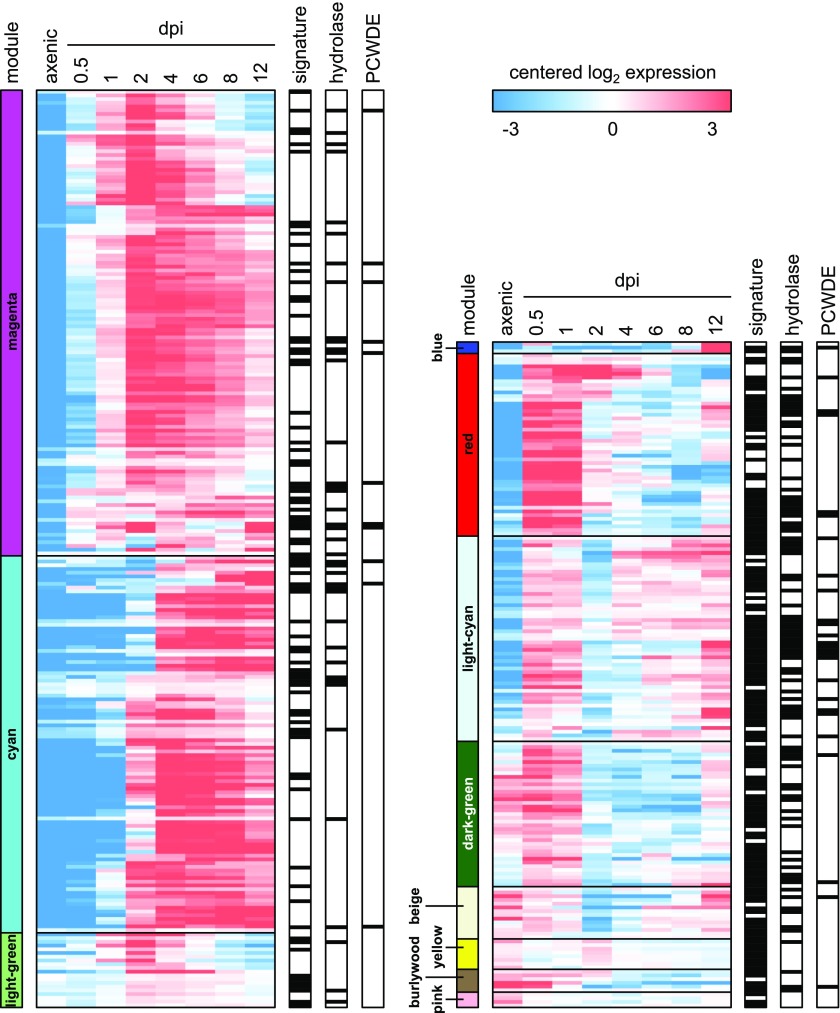
(C) Modules of coexpressed genes during pathogenic development of U. maydis. The RNA-seq expression data set was subjected to WGCNA to detect modules of coexpressed genes. Each graph shows the expression of the module eigengene, which can be considered as the representative gene of the respective coexpression module. The vertical axes indicate log2 expression values relative to the mean expression across all stages. The horizontal axes indicate the stages, i.e., axenic (ax), 0.5, 1, 2, 4, 6, 8, and 12 dpi. Error bars indicate sd of three biological replicates. The modules are named according to their color, and the number of genes residing in each module is given in parentheses.
The most dramatic changes in gene expression were observed in pairwise comparisons including axenic culture conditions (Figure 2B). However, even if comparisons with the axenic culture sample, which may be unrelated to the conditions on or inside the plant with respect to nutrient availability, were excluded from the analysis, 4586 genes (68% of the U. maydis genes) remained differentially expressed (Figure 2B). Such a high proportion of genes showing differential expression during the plant-associated developmental stages distinguishes U. maydis from other pathogenic fungi where similar RNA-seq studies have been performed. In Zymoseptoria tritici, Colletotrichum higginsianum, and Puccinia striiformis f. sp tritici, 28%, 44%, and 50% of the genes were differentially expressed, respectively (O’Connell et al., 2012; Rudd et al., 2015; Dobon et al., 2016). We consider it likely that the coupling between pathogenic and sexual development in U. maydis and the associated morphological changes contribute to this high percentage of differentially expressed U. maydis genes. For the other three examples given, asexual reproduction cycles were studied. However, rather than assuming that the discretely different lifestyles of these pathogens contribute to the percentage in differentially expressed genes, variation in sequencing depth and numbers of analyzed samples in the different studies cannot be excluded as cause. Overall, the large number of differentially expressed U. maydis genes underscores the comprehensiveness of our analysis and indicates a complex transcriptional regulation during all stages of biotrophic development.
In the next step of the analysis, we used the expression data of all stages to perform a weighted gene coexpression network analysis (WGCNA; Supplemental Figure 3). This analysis identifies modules of coexpressed genes and represents the modules by their centrally located genes, referred to as module eigengenes (Zhang and Horvath, 2005; Langfelder and Horvath, 2008). We identified 14 modules that were color-coded and ranged in size from 36 genes (blue module) to 1231 genes (yellow module; Supplemental Data Set 6). The expression profiles of the respective module eigengenes are depicted in Figure 2C. Some of the modules reflect distinct stages during fungal development. The red module was expressed solely on the plant surface (0.5–1 dpi). The light-green module was expressed during penetration and early biotrophic development (1–2 dpi) and ceased afterwards. The magenta module was strongly induced from 0.5 to 2 dpi and expression was largely maintained up to 12 dpi. This module therefore correlated with the establishment and maintenance of biotrophy. The cyan module was induced after establishment of biotrophy just at the onset of tumor induction (2–4 dpi), and expression stayed high also at later time points. Thus, the cyan module represented a tumor module. The blue module was specific for spore development (8–12 dpi).
To confirm that the observed expression patterns are not a product of the specific bioinformatics tool, we performed k-mean clustering of the gene expression data, sorting the genes into six clusters. Five of these clusters were highly correlated (>0.9) with a distinct module of the coexpression obtained by WGCNA (Supplemental Data Set 7). These included the three virulence modules red, magenta, and cyan, as well as the yellow and light-cyan module. One k-mean cluster correlated with two, the dark-green and the burlywood module, which are highly correlated modules (Supplemental Figure 3B). There was thus a substantial overlap between the respective gene sets obtained by WGCNA and k-mean clustering with overlap coefficients ranging from 0.7 to 1.0 (Supplemental Data Set 7). All subsequent analyses refer to the modules obtained by WGCNA.
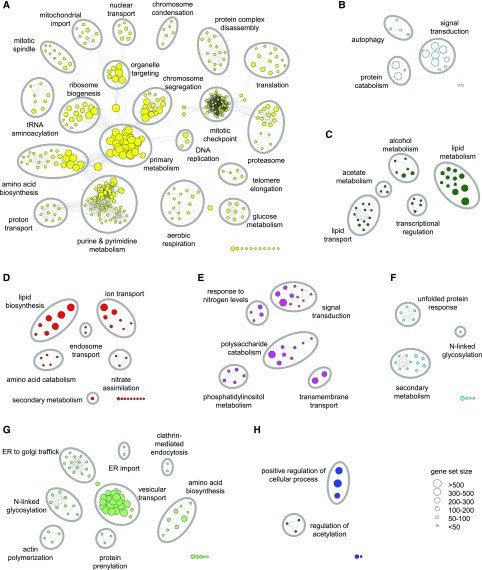
To generate a concise picture of the biological processes associated with pathogenic development, each module was subjected to an enrichment analysis for Gene Ontology (GO) terms (Supplemental Data Set 8) (Ashburner et al., 2000; The Gene Ontology Consortium, 2017). We visualized the respective enriched gene sets in a weighted similarity network, which facilitated the identification of the predominant processes in any given module (Figure 3; Supplemental Figure 4). In the sections that follow, we make use of these functionally annotated modules to gain a better understanding of the virulence strategies adopted by U. maydis.
Figure 3.
Biological Processes Enriched in Selected Coexpression Modules.
GO enrichment analysis for the yellow (A), light-cyan (B), dark-green (C), red (D), magenta (E), cyan (F), light-green (G), and blue (H) modules. Only biological process terms were considered in the analysis. Each significantly enriched gene set (hypergeometric P value < 0.005) is represented by a node. Node sizes are proportional to the number of genes within the respective gene set, and the edges indicate overlapping member genes. Highly similar gene sets tend to form clusters, which were manually circled and labeled with appropriate summarizing terms. Gene sets that have no overlap with other enriched GO terms are shown in the rightmost corner and are not labeled despite two exceptions, one in (D) (secondary metabolism) and one in (F) (N-linked glycosylation). See Supplemental Data Set 8 to retrieve all GO terms of the enriched gene sets.
General Changes in Fungal Metabolism throughout the Infection Cycle
Looking for modules that reflect high metabolic and cellular activity identified the yellow module (Figure 3A; Supplemental Data Set 9). This module is enriched for genes involved in translation, ribosome biogenesis, amino acid and nucleic acid biosynthesis, cell division, primary metabolism, and respiration. The highest expression level of the yellow module was observed during U. maydis growth in axenic culture (Figure 2C), a condition in which the doubling time of a cell was around 2 h, which likely exceeds the growth rate in all plant-associated stages. The yellow module shows lowest expression at 0.5 and 1 dpi, i.e., the stages where mating occurs and the cell cycle-arrested dikaryon is formed. At 2 dpi, the module displays a second expression peak, likely reflecting the release of the cell cycle block after penetration and early biotrophic growth. This expression pattern resembles the response of starving cells to nutrient repletion (Conway et al., 2012). From 2 dpi until 12 dpi, genes of the yellow module were progressively downregulated (Figure 2C), and instead genes involved in protein catabolism and autophagy were induced. Such genes are located in the light-cyan module (Figure 3B; Supplemental Data Set 10), which is almost perfectly negatively correlated with the yellow module (r = −0.99, Pearson correlation; Figure 2C). Autophagy is an important mechanism by which eukaryotic cells degrade cytosolic macromolecules and recycle them for the synthesis of new macromolecules or use them as energy source. In addition to induction of autophagy, expression of genes involved in lipid transport and lipid metabolism increased after 2 dpi. The respective genes are located in the dark-green module (Figure 3C; Supplemental Data Sets 8 and 9). These findings suggest that autophagy-mediated cellular recycling as well as degradation of fatty acids become important during the later biotrophic interaction. Increased fatty acid metabolism has also been associated with the biotrophic growth of Z. tritici (Rudd et al., 2015) and arbuscular mycorrhizal fungi. The latter were recently shown to take up lipids from their host to sustain colonization (Rudd et al., 2015; Jiang et al., 2017; Keymer et al., 2017). The synchronous upregulation of autophagy and lipid metabolism genes and downregulation of ribosome biogenesis genes are typical expression patterns during slowed growth and during starvation (Gasch et al., 2000). We also observed that nit2 (nitrogen catabolic enzyme regulatory protein) and snf1 (sucrose nonfermenting1), the nitrogen and carbon catabolite derepressors (Nadal et al., 2010; Horst et al., 2012), have increased transcript levels during tumor formation compared with early biotrophic growth (Supplemental Data Sets 3 and 4), indicating that carbon and nitrogen sources may be limiting. Previous studies indicated that tumor tissue is a strong sink tissue with an efficient supply of organic nutrients from systemic source leaves (Billet and Burnett, 1978; Doehlemann et al., 2008b; Horst et al., 2008, 2010). Metabolome profiling of U. maydis infected tissue revealed that free hexoses and amino acids are highly abundant in tumor tissue and even reach the levels of juvenile sink tissue (Horst et al., 2010). This latter finding seems to contradict our RNA-seq-based assessment that U. maydis may be starved for carbon and nitrogen in tumor tissue. However, the metabolome analysis (Horst et al., 2010) was conducted with SG200, a haploid solopathogenic U. maydis strain. In contrast to infections with compatible wild-type strains in which fungal biomass continuously increases within tumor tissue (Figure 1C) (Tollot et al., 2016), SG200 does not increase its biomass late during infection (S. Tanaka, P. Erchinger, S. Krombach, and R. Kahmann, personal communication). We speculate that the free hexoses and amino acids detected in SG200-induced tumor tissue (Horst et al., 2010) may not accumulate to the same extent in tumors induced by wild-type strains because they may be taken up to support the continuous fungal proliferation (Billet and Burnett, 1978; Doehlemann et al., 2008b; Horst et al., 2008, 2010). The proliferation within tumors of U. maydis wild-type strains that are most likely diploid at that stage (Tollot et al., 2016) may therefore resemble chemostat growth, in which cells grow slowly due to nutrient limitation but, due to a constant nutrient flow toward tumor tissue, are not starving for essential nutrients. A detailed metabolic profiling of tumors induced by a mixture of compatible U. maydis wild-type strains will have to be done to monitor the dynamics of the available nutrients. It will be interesting to see how this can then be linked to the transcriptome data presented here, which indicate nutrient-limited fungal growth within tumors. We consider it likely that nutrient limitation could also contribute to the induction of the developmental program for aggregate formation and teliospore formation inside the tumors.
Nitrogen Transporters
The utilization of complex nitrogen sources in fungi is regulated by specific transcription factors, which derepress the expression of genes needed for the degradation and uptake of various nitrogen compounds in situations where the most favorable nitrogen sources ammonia and glutamine are scarce (Marzluf, 1997). In many plant pathogenic fungi, including U. maydis, mutants of the nitrogen catabolite derepressors (nit2 in U. maydis) show reduced virulence (Pellier et al., 2003; Divon et al., 2006; Thomma et al., 2006; Divon and Fluhr, 2007; Kim and Woloshuk, 2008; Horst et al., 2012). The importance of nitrogen availability in biotrophic associations is further corroborated by the observation that nitrogen fertilizers generally increase the susceptibility of plants to biotrophs, whereas they decrease the susceptibility of plants to necrotrophs (Snoeijers et al., 2000; Dordas, 2008; Ballini et al., 2013). Indeed, U. maydis is known to grow on various nitrogen sources and has the ability to generate all proteinogenic amino acids (Holliday, 1961; McCann and Snetselaar, 2008).
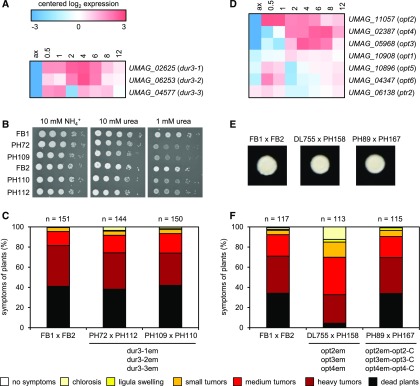
To obtain more insights into nutrient assimilation during biotrophic growth, we searched for transporters that are highly induced in the plant environment compared with growth in YEPSL medium (2 dpi versus axenic). The top five induced transporters were two putative urea permeases dur3-1 (UMAG_02625) and dur3-2 (UMAG_06253), two putative oligopeptide transporters (OPTs) opt2 (UMAG_11057) and opt4 (UMAG_02387), and the candidate methylammonium permease (MEP) ump2 (UMAG_05889) (Supplemental Data Set 11; Figures 4A and 4D). In fungi, related transporter families are required for nitrogen utilization from peptides, urea and ammonium, respectively (ElBerry et al., 1993; Lorenz and Heitman, 1998; Hauser et al., 2001; Abreu et al., 2010; Hartmann et al., 2011; Navarathna et al., 2011; Dunkel et al., 2013), while OPTs can also mediate sulfur utilization by taking up glutathione (Bourbouloux et al., 2000). All five transporters are located in the magenta expression module (Figure 2C) and are thus linked to biotrophy (Supplemental Data Set 11).
Figure 4.
Expression Pattern and Virulence Function of Nitrogen Compound Transporters.
(A) The heat map shows the expression profiles of the U. maydis urea transporters; log2 expression values are visualized relative to the mean expression across all stages.
(B) U. maydis dur3 transporters are important for nitrogen utilization from urea. Serial dilutions of FB1 and FB2 wild-type strains and the respective dur3-1,2,3 triple mutants in FB1 (PH72 and PH109) and FB2 (PH110 and PH112) were spotted on minimal medium with ammonium or urea as sole nitrogen source in the indicated final concentrations.
(C) The indicated mixtures of compatible strains were injected into maize seedlings and symptoms were scored 12 d after infection according to severity; the color code for each category is given below. Three independent experiments were performed and the average values are expressed as percentage of the total number of infected plants (n), which is given above each column.
(D) The heat map shows the expression profile of the U. maydis peptide transporters; log2 expression values are shown relative to the mean expression across all stages.
(E) The indicated mixtures of compatible haploid strains were spotted on charcoal-containing agar plates. FB1 and FB2 are wild-type strains, DL755 and PH158 are compatible opt2,3,4 mutants, and PH89 and PH167 are opt2,3,4 mutants simultaneously complemented with wild-type opt2,3,4 genes. The occurrence of white mycelium indicates the formation of dikaryotic hyphae.
(F) The indicated mixtures of compatible strains were injected into maize seedlings and symptoms were scored 12 d after infection according to severity as described in (C). For (C) and (F), the gene name followed by “em” indicates that the respective gene was inactivated by CRISPR-Cas9. The gene name followed by “-C” indicates that a single copy of the respective gene was introduced into the indicated strains to test for complementation. Please note that dead plants, which represent the most severe symptom category, are a result of the artificial virulence assay that is based on young maize seedlings and a high inoculum. U. maydis is a strict biotroph and does not kill plants under natural conditions.
Besides the two urea permeases dur3-1 and dur3-2, which are highly induced during biotrophic development, U. maydis possesses dur3-3 (UMAG_04577), a likely third urea permease. While dur3-1 and dur3-2 are not induced under nitrogen depletion (Horst et al., 2012; Sánchez-Arreguin et al., 2017), dur3-3 expression depends on nit2 under nitrogen starvation conditions (Horst et al., 2012). In line with this observation, our expression analysis placed both dur3-3 and nit2 into the light-cyan expression module likely involved in the response to limiting nutrients (Figure 2C).
To study the contribution of all three urea transporters to virulence and to exclude redundancy, we made use of the recently established CRISPR-Cas9 system in U. maydis (Schuster et al., 2016, 2017) and generated frameshift mutations near the 5′ends of the respective three genes in haploid FB1 and FB2 strains. The resulting dur3-1,2,3 triple mutants were affected during growth on medium with urea as sole nitrogen source (Figure 4B), but they were not affected in virulence (Figure 4C), suggesting that urea uptake is not important for biotrophic development. However, we did not investigate the effects on virulence when plants are grown on nitrogen-poor soil and we therefore cannot exclude that the urea transporters become virulence factors when the overall nitrogen supply of the plant is lower.
The high affinity ammonium transporter Ump2 has been characterized previously (Smith et al., 2003). Besides mediating ammonium acquisition, this transporter has a signaling function and initiates filamentous growth under nitrogen starvation (Smith et al., 2003). The ump2 gene is partially subject to nitrogen catabolite repression, i.e., is regulated by Nit2 (Horst et al., 2012). The high expression of ump2 during early biotrophic growth, which we observed here (Supplemental Data Set 11), suggests additional regulation by plant signals. While ump2 mutants were unaffected in virulence (Smith et al., 2003), an ump1 (with a defect in the gene encoding a low affinity ammonium transporter) ump2 double mutant was severely reduced in virulence (M. Perlin, personal communication). Our expression analysis thus reinforces the importance of ammonium uptake and its regulation for biotrophic development of U. maydis.
From the seven U. maydis peptide transporters, none was demonstrated to be regulated by Nit2 in response to nitrogen starvation (Horst et al., 2012). According to our transcriptional profiling, three OPTs, opt2, opt4, and opt3 (UMAG_05968), were highly induced during biotrophic development (Figure 4D). opt5 (UMAG_10896) and opt6 (UMAG_04347) are placed in the dark-green module (Figure 2C) based on their expression pattern (Figure 4D) and may therefore be coupled to nutritional regulation (see previous section), while opt1 (UMAG_10908) was constitutively expressed (Figure 4D). U. maydis possesses only one member of the dipeptide/tripeptide transporter (PTR) family, ptr2 (UMAG_06138), and this gene also showed little variation in expression during fungal development (Figure 4D). To study whether the biotrophy-coupled induction of OPT transporters is important for virulence, we introduced frameshift mutations by CRISPR-Cas9 in the 5′ regions of opt2, opt3, and opt4 genes in the FB1 and FB2 strain backgrounds. Plant infections with compatible mixtures of the respective triple mutants revealed that these transporters are important for virulence. While mating and filament formation were not affected (Figure 4E), severe disease symptoms such as heavy tumors and plant death were drastically reduced in infections with these mutants (Figure 4F). The virulence defect of the triple mutant could be completely restored by introducing single copies of all three opt genes into the ip locus of the triple mutant strains (Figure 4F). Full complementation makes it unlikely that truncated gene products were produced due to the CRISPR-Cas9-induced frameshifts, which could cause the attenuated virulence in the triple mutant. The full restoration of virulence by complementation also makes off-target mutations of Cas9 with unwanted side effects highly unlikely. The virulence function of the oligopeptide transporters suggests that peptides produced from extracellular proteins are important nutrient sources for U. maydis during biotrophic growth. It has been previously demonstrated in the yeast Candida albicans that synchronous production of aspartic proteases and OPTs enables growth on proteins as sole nitrogen source (Martínez and Ljungdahl, 2005). Interestingly, two secreted aspartic proteases from U. maydis (UMAG_05097 and UMAG_12330) are located in the magenta module (Figure 2C) and are thus coexpressed with the highly induced OPTs. Given the specific expression pattern of the described subset of OPTs and aspartic proteases, we propose that extracellular proteolysis and subsequent uptake of peptides may be intrinsically tied to the plant-associated developmental program of U. maydis. With respect to utilization of carbohydrates, the uptake of sucrose follows a similar scheme (Wahl et al., 2010): The high-affinity sucrose transporter srt1 (UMAG_02374), an important virulence factor of U. maydis, is transcriptionally induced specifically during pathogenic development, but not by the presence of sucrose under axenic culture conditions or under carbon starvation (Wahl et al., 2010). The trigger for induction during the biotrophic phase is unknown, but srt1 as well as the nitrogen-related transporters opt2, opt4, dur3-1, dur3-2, and ump2 are all induced by the plant surface cues hydrophobicity and 16-hydroxy hexadecanoic acid (Lanver et al., 2014). This corroborates that the early developmental stages like filamentation and appressorium formation on the plant surface prepare U. maydis not only with respect to effectors that can suppress plant defenses, but also with respect to nutrient utilization systems important for growth inside the plant environment. In the wheat stripe rust fungus, OPT transporters have been shown to be highly expressed in haustoria, the biotrophic fungal feeding structures (Garnica et al., 2013). Interestingly, an OPT of the hemibiotroph Colletotrichum gloeosporioides was identified as an auxin-induced gene (Chagué et al., 2009). In the U. maydis-maize system, auxin levels are highly induced during infection (Turian and Hamilton, 1960; Basse et al., 1996; Reineke et al., 2008), and it will be interesting to test whether auxin controls opt expression also in U. maydis.
Overall, our data suggest that nitrogen utilization in U. maydis has a high level of complexity involving nutrient level dependent as well as independent regulation. Both utilization modes contribute to virulence and it may be this two-pronged strategy that allows U. maydis to proliferate efficiently inside the infected plant tissue.
The Machinery for Protein Secretion
From the GO enrichment analysis (Figure 3), we found the light-green module (Figure 2C) enriched in various processes related to protein secretion, e.g., ER-to-Golgi trafficking, vesicular transport, and N-glycosylation (Figure 3; Supplemental Data Sets 8 and 10). Genes of the unfolded protein response (UPR) also mainly reside in the light-green module (Supplemental Data Set 10). The light-green secretion machinery module is induced during penetration of the plant surface (1 dpi) and peaks at 2 dpi (Figure 2C). A similar expression pattern was observed for many genes encoding secreted proteins (discussed below), and in all likelihood this reflects the high demand for the secretion machinery during plant colonization. The observed strong increase in expression of the light-green module from 1 to 2 dpi is challenging to interpret. Since the 1 dpi time point represents a mixture of mostly filamentous hyphae on the leaf surface and only a small percentage of hyphae that have developed appressoria and have penetrated (Figure 1A), it is conceivable that the actual expression levels of the secretion machinery and the secretome components in the few penetrated hyphae are as high as the levels observed in branching mycelium at 2 dpi. The lower overall expression values at 1 dpi might thus simply reflect that only a small percentage of the inoculum has mated and penetrated. This interpretation is in line with previous studies showing that penetrated hyphae activate the UPR through the Cib1 (Clp1 interacting bZip1) transcription factor (Heimel et al., 2013). The UPR affects secretion as well as expression of effectors (Heimel et al., 2013; Hampel et al., 2016; Lo Presti et al., 2016).
In contrast to N-glycosylation, components of the O-glycosylation pathway are most strongly expressed in axenic culture as well as on the plant surface (yellow and burlywood module; Figure 2C; Supplemental Data Set 10). Both processes, O-mannosylation and N-glycosylation, are necessary for virulence of U. maydis (Fernández-Álvarez et al., 2009, 2013). Previous studies could trace back the virulence defect of mutants in the O-mannosylation pathway to a failure in appressoria formation, mainly explained by the reduced glycosylation of the plant surface cue sensing receptor Msb2 (Fernández-Álvarez et al., 2012). By contrast, mutants defective in components of the N-glycosylation pathway are able to penetrate the plant, but induce strong defense responses, e.g., an oxidative burst, indicative of a malfunction of effectors (Schirawski et al., 2005; Fernández-Álvarez et al., 2013). Our expression data strongly support the model that O-mannosylation is particularly important for early fungal development on the plant surface, while N-glycosylation is mainly necessary inside the plant tissue to produce functional effectors. For Magnaporthe oryzae it has been demonstrated that N-glycosylation of an effector is necessary for its virulence promoting function (Chen et al., 2014). For U. maydis effectors, this has yet to be demonstrated.
Development-Associated Changes of the Secretome
The U. maydis genome contains 467 genes encoding putatively secreted proteins (Schuster et al., 2017), and 215 of the predicted secreted proteins are lacking any predicted structural or functional domain (Schuster et al., 2017). Some of those proteins have been previously found to act as important virulence effectors (Lanver et al., 2017). Secreted proteins are distributed over 12 of the 14 expression modules (Figure 5; Supplemental Data Set 12), i.e., their expression occurs in waves following the characteristic expression profile of the respective module during the course of an infection. This suggests that certain groups of secreted proteins are only needed during specific periods of an infection cycle. A search for modules in which secreted proteins are significantly overrepresented identified only three modules: the red module (P value 5.71E-5, Fisher exact test), the magenta module (P value 8.46E-23, Fisher exact test), and the cyan module (P value 3.42E-10, Fisher exact test; Figure 2C; Supplemental Data Set 13), i.e., the modules specific for the plant surface, biotrophic development and tumor formation, respectively. These three modules have in common that they are almost completely off during the axenic culture condition (Figure 5). Thus, they likely represent specific virulence modules.
Figure 5.
Expression of the U. maydis Secretome.
The heat map shows expression of differentially expressed genes encoding putative secreted proteins. For each module indicated on the left, genes were hierarchically clustered, and log2 expression values are visualized relative to the mean expression across all stages. Modules are colored according to Figure 2C. Black bars on the right indicate for each gene the presence of known signatures (based on InterPro scan), the predicted hydrolytic capabilities, and more specifically the predicted ability to degrade the plant cell wall (PCWDE).
The secreted proteins of the red module (Figure 2C) predominantly have predicted hydrolytic functions (Figure 5; Supplemental Data Set 12) and contain among others cutinases, lipases, and peptidases (Supplemental Data Set 12). Thus, the red module harbors key enzymes potentially involved in the degradation of the cuticle. It is conceivable that some of the secreted hydrolases also have antimicrobial activity like a putative lysozyme (UMAG_06218). In this context, it may thus not be a coincidence that this module also contains genes from secondary metabolite clusters like mannosylerythritol lipid and ustilagic acid biosynthesis (Supplemental Data Set 14) (Teichmann et al., 2007). Mannosylerythritol lipids are highly potent surface-active substances (Fluharty and O’Brien, 1969), while ustilagic acid has broad antimicrobial activity (Haskins and Thorn, 1951) and was shown to aid against microbial competitors like the necrotrophic fungus Botrytis cinerea in coinfection experiments (Teichmann et al., 2007).
After penetrating the cuticle, the fungal hyphae need to breach the polysaccharide plant cell wall, and most likely this is accomplished by the secretion of plant cell wall degrading enzymes (PCWDEs) (Schirawski et al., 2005; Lanver et al., 2014) of which U. maydis is predicted to encode 40 (Figure 5; Supplemental Data Set 12) (Lo Presti et al., 2015). Notably, PCWDEs were not enriched in the red module but in the light-cyan module (Figure 2C), a module that is otherwise rather scarce in secreted proteins (Figure 5; Supplemental Data Set 12). We implicated the light-cyan module in the response to nutrient limitation during growth on the leaf surface as well as during late biotrophic interaction (see previous sections), suggesting that most PCWDEs mediate utilization of carbon from plant cell walls at these stages. The expression pattern of the PCWDEs also correlated with the colonization strategy of U. maydis, in which plant cell walls must be penetrated at the beginning of the infection to enable initial intracellular growth, and plant cell walls must be loosened again later during infection to allow enlargement of cells during tumor formation. The expression data do not allow discrimination between a role for these enzymes in nutrition and/or breaching plant cell walls. Previous studies in other plant pathogenic fungi suggest a direct link between carbon utilization and plant cell wall penetration (Tonukari et al., 2000; Ospina-Giraldo et al., 2003). In U. maydis, this needs to be followed up by functional studies of candidate genes.
Interestingly, our expression data show that few PCWDEs are uncoupled from nutritional regulation and are instead tied to biotrophic development (i.e., they are found in the magenta module). In this group of PCWDEs are all three U. maydis GH45 cellulases, five potential hemicellulases, including two arabinofuranosidases, and the sole predicted pectin lyase (Supplemental Data Set 12). While virulence functions of the GH45 family and the pectin lyase were so far not demonstrated (Schauwecker et al., 1995; Doehlemann et al., 2008a; Lanver et al., 2014), the arabinofuranosidase UMAG_01829 contributes to virulence (Lanver et al., 2014; Schilling et al., 2014). Our expression data therefore suggest that carbon supply from complex sources, e.g., plant cell walls, might be ensured via a two-pronged strategy consisting of nutrient level-dependent and independent regulation.
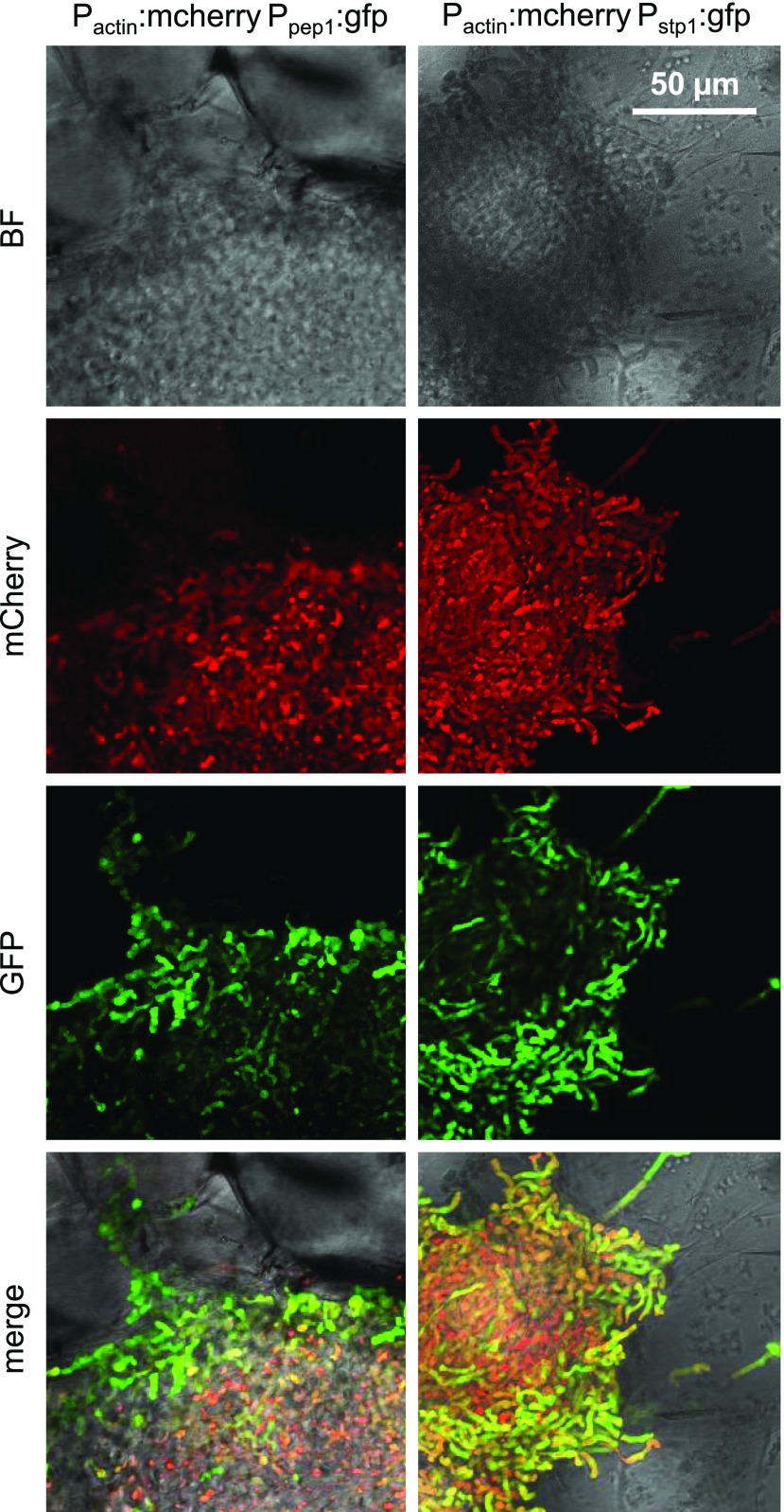
The magenta module (Figure 2C) predominantly contains genes encoding secreted proteins lacking known functional domains (P value 2.79E-8, Fisher exact test; Supplemental Data Sets 12 and 13) (Schuster et al., 2017).This module is the only module overrepresented for core effectors lacking known domains which are conserved in five sequenced smut fungi (Schuster et al., 2017) (Supplemental Data Sets 12 and 13): Of the 24 core effector families (plus 30 putatively paralogous genes in U. maydis), the magenta module harbors 28 genes including pep1 (protein essential during penetration1), pit2 (protein important for tumor2), and stp1 (stop after penetration1), the three effectors known to be essential for establishing biotrophy (Supplemental Data Sets 12 and 13) (Doehlemann et al., 2009, 2011; Schipper, 2009; Mueller et al., 2013). We therefore speculate that these core effectors, which represent 14 different effector families (Schuster et al., 2017), may contain the key determinants for establishing biotrophy in smut fungi. Another effector found in the magenta module but not belonging to the core effectors is see1 (seedling efficient effector1). This effector has been shown to contribute to the reactivation of plant DNA synthesis in leaves, which is crucial for tumor formation (Redkar et al., 2015). The early expression peak of see1 observed at 2 dpi (Supplemental Data Set 12) suggests that this activation of plant DNA synthesis either has to happen only in a narrow time window or that maintaining replication within tumors is controlled also by other factors. The fact that expression of genes encoding secreted proteins in the magenta module is decreasing during later stages of infection (Figure 5) is puzzling, given the immunity suppressing function of several of these effectors (Doehlemann et al., 2009; Schipper, 2009; Mueller et al., 2013). To shed light on the underlying mechanism, we generated two haploid U. maydis strains in which promoters from two of the effectors from the magenta module, pep1 and stp1, were fused to gfp. In addition, these strains expressed cytoplasmic mCherry constitutively under the control of the actin promoter. These strains were crossed with untagged compatible haploid strains and the developmental stage was analyzed when downregulation of the module takes place. At this time point, fungal aggregates are the predominant structures in the infected tissue (Tollot et al., 2016). While mCherry was rather evenly distributed in cells forming the aggregate, pep1 and stp1 promoter activity was mainly restricted to cells at the surface of the aggregates (Figure 6). To exclude that this spatial expression pattern is specific for the gfp reporter, we also placed gfp under the control of the actin promoter and found the signal rather evenly distributed within the aggregates (Supplemental Figure 5). This illustrates that while the majority of cells in the aggregates have ceased to express pep1 and stp1, fungal cells that are in direct contact with the infected plant tissue maintain high expression levels of these genes, which in all likelihood is sufficient to downregulate plant defenses. How this heterogeneity in effector gene expression is achieved and to which extent the production of matrix provides additional protection is currently unknown.
Figure 6.
Heterogenous Effector Gene Expression in Fungal Aggregates.
Maize seedlings were infected with mixtures of FB1 carrying the indicated reporter constructs (top) and FB2 wild-type strains. Seven days postinoculation, fungal aggregates within the tumor tissue were visualized by confocal microscopy. The fluorescence of GFP and mCherry was monitored and merged with the respective bright-field (BF) image. All images are projections of multiple z-stacks. GFP fluorescence indicative of effector gene expression was mainly detected at the surface of the aggregates, while mCherry control fluorescence was rather evenly distributed throughout the aggregates.
The cyan module is induced after establishment of biotrophy (2 dpi), which coincides with the development of tumors and fungal aggregates. Like the magenta module, the cyan module is enriched for secreted proteins lacking functional annotation (P value 1.92E-7, Fisher exact test; Supplemental Data Set 13).The cyan and magenta modules together contain 153 of the 215 secreted proteins lacking functional signatures (Figure 5; Supplemental Data Set 12). None of the candidate effector proteins belonging to the cyan module have been characterized so far. Due to their tumor-specific expression profile, we hypothesize that effectors in the cyan module might directly be involved in inducing the plant cell developmental switch to tumor cells. It is interesting that the cyan module also contains a nonribosomal peptide synthase (UMAG_10543), as well as three of the six U. maydis polyketide synthases (UMAG_10532, UMAG_06414, and UMAG_06418; Supplemental Data Set 14). While UMAG_06414 is involved in spore melanin biosynthesis (Islamovic et al., 2015), the functions of the other polyketide synthases remain to be elucidated. The third wave of secreted proteins is thus accompanied by active secondary metabolism. It is not clear whether the yet unknown secondary metabolites produced are used for communication with the plant or represent molecules needed for development of U. maydis.
Taken together, the transcriptome reveals the modular expression of putatively secreted proteins while U. maydis is on the plant surface, during biotrophic development and during tumor formation. Consecutive waves of effector gene expression linked to the transition from the biotrophic lifestyle to necrotrophy have also been observed in the hemibiotrophic fungi Colletotrichum higgensianum, Colletotrichum graminicola, Z. tritici, and Leptosphaeria maculans (Kleemann et al., 2012; O’Connell et al., 2012; Mirzadi Gohari et al., 2015; Gervais et al., 2017). In C. higgensianum, the induced transcriptome of early infection stages is dominated by genes for secondary metabolism, leading to the speculation also here that they may actually function in host manipulation (O’Connell et al., 2012). In the biotrophic fungi Blumeria graminis f. sp hordei, Blumeria graminis f. sp tritici, and P. striiformis f. sp tritici, effector expression waves have also been described (Hacquard et al., 2013; Dobon et al., 2016; Zeng et al., 2017), reinforcing the idea that the need for certain effectors changes during host colonization and that this is a conserved feature of different pathosystems.
Identification of Potential Transcriptional Regulators of Effector Waves
To shed more light into the regulation of genes encoding secreted proteins, we visualized a weighted coexpression network of all differentially regulated genes encoding secreted proteins and all differentially regulated predicted transcription factors which are connected to at least one of these (Figures 7A and 7B). We then looked for transcription factors that have the strongest connectivity to the respective modules in which secreted proteins reside. Such intramodular hub genes are likely the key drivers of a given module (Mason et al., 2009). Through this analysis, we detected three transcription factor genes with strong connectivity (>0.9) to the red module: the homeodomain protein UMAG_10544, the TEA/ATTS transcription factor UMAG_02835, and the bHLH transcription factor UMAG_11235 (Figures 7B and 7C; Supplemental Data Set 15). We also detected rbf1 (UMAG_03172), the central transcriptional regulator downstream of the bE/bW complex (Heimel et al., 2010b), with a high intramodular connectivity of 0.9 (Figures 7B and 7C). Rbf1 is responsible for the bE/bW-induced filamentation and cell cycle arrest (Scherer et al., 2006; Heimel et al., 2010b). The previously observed downregulation of rbf1 after penetration has been suggested to be a prerequisite to resume the cell cycle after entering the plant (Heimel et al., 2010a). In line with our prediction of rbf1 as potential driver of the red module, the previously identified rbf1-induced genes (Heimel et al., 2010b) are highly overrepresented in the red module (Supplemental Data Set 13). To what extent the other three transcription factors (UMAG_10544, UMAG_02835, and UMAG_11235; Figure 7C) detected here drive expression of the red module needs to be investigated.
Figure 7.
Association of Transcription Factors with Modules Containing Secreted Proteins.
(A) and (B) Network of gene expression profiles of differentially regulated genes encoding secreted proteins (A) and differentially regulated transcription factors (B) belonging to the respective modules in which the secreted proteins reside. The weighted network is based on the topological overlap matrix of the expression data, edges were included when the pairwise overlap was greater than 0.2, and genes are colored according to their modular membership. Selected transcription factors are labeled with their respective names.
(C) Connectivity of transcription factors to the red (left panel), magenta (middle panel), and cyan module (right panel). Depicted are all transcription factors having an intramodular connectivity of greater than 0.9 to any of the three modules. Color intensity indicates connectivity strength.
By the same type of analysis we found hdp2 (UMAG_04928) (Heimel et al., 2010b), biz1 (UMAG_02549) (Flor-Parra et al., 2006), and the two mating type genes bE (UMAG_12052) and bW (UMAG_00578) (Gillissen et al., 1992) to be the transcription factors with strongest connectivity to the magenta module, i.e., the second wave of effectors (Figure 7B; Supplemental Data Set 15). Those transcription factors have previously been suggested to be the main inducers of early effectors (Heimel et al., 2010b; Lanver et al., 2014). Furthermore, our analysis suggests UMAG_11658, UMAG_00501, mzr1 (mig2-5 zinc finger regulator1, UMAG_05804) (Zheng et al., 2008), and UMAG_02104 (Figure 7C) as being involved in early effector gene expression. They all have a reasonably strong connectivity to the magenta module of >0.9 (Figure 7C). Consistently, mzr1 has been demonstrated to be involved in the expression of several effector genes (Zheng et al., 2008), which we now place in the magenta module (Supplemental Data Set 12). However, in contrast to hdp2 and biz1 (Flor-Parra et al., 2006; Heimel et al., 2010b; Lanver et al., 2014), mzr1 is not a major virulence factor (Zheng et al., 2008), most likely because biz1 and hdp2 can compensate for the lack of mzr1 during infection.
Genes for which induced expression has been observed after artificial overexpression of rbf1 (Heimel et al., 2010b) are not only found in the red module as discussed above, but also in the magenta module. The latter group includes hdp2, biz1, and 11 potential effectors (Supplemental Data Sets 6 and 12). In our analysis of intramodular connectivity, which relies solely on the coexpression of transcription factors and their targets under natural expression conditions, we were thus unable to identify the contribution of rbf1 to expression of the magenta module. Overall, our analysis delivered those transcription factors which were expected to control early effectors, and this made us confident that the analysis would also yield promising candidate transcriptional regulators for the third expression wave of effectors, i.e., the cyan module.
For the cyan module, three potential drivers were identified, UMAG_05601, UMAG_02765, and UMAG_04778, which have intramodular connectivities of 0.98, 0.97, and 0.96, respectively (Figures 7B and 7C; Supplemental Data Set 15). In addition, several other transcription factors (UMAG_05721, UMAG_11138, fox1 [forkhead box1, UMAG_01523; Zahiri et al., 2010], UMAG_06257, UMAG_06308, and UMAG_01456) showed intramodular connectivity at slightly lower values of between 0.9 and 0.93 (Figures 7B and 7C; Supplemental Data Set 15) and were thus also candidates for transcriptional regulators of the cyan module. Except for fox1 (Zahiri et al., 2010), none of these transcription factors had so far been functionally analyzed. fox1 mutants displayed reduced virulence, and transcriptional profiling of this mutant revealed a set of 29 potential effector genes that required fox1 for full expression (Zahiri et al., 2010). These putative fox1 target genes are significantly overrepresented in the cyan module (Supplemental Data Set 13). This shows that fox1 contributes to the regulation of the cyan module, but according to our data and the intramodular connectivity analysis (Figures 7B and 7C), fox1 may not be the main driver of the third wave of effector gene expression.
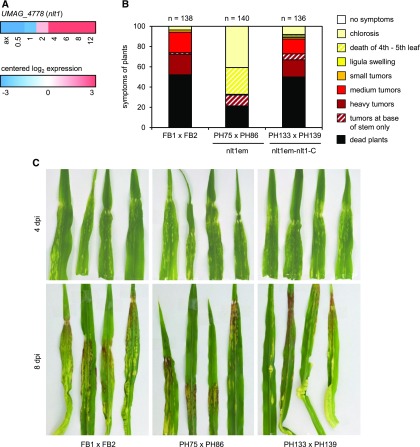
To analyze a possible contribution of the yet uncharacterized potential regulators of the cyan module to virulence and tumor formation, we picked the APSES-type transcription factor gene UMAG_04778 (Figures 7C and 8A) and generated a targeted knockout mutant in the compatible FB1 and FB2 strains. Interestingly, the introduced frameshift mutation in UMAG_04778 caused a strong reduction of virulence, with tumor formation in leaves being completely abolished (Figure 8B). We therefore named the gene nlt1 (no leaf tumors1). The virulence defect of the mutants was almost fully complemented by introduction of a single copy of nlt1 into the ip locus of the respective mutant strains (Figure 8B). This links the mutant phenotype to the inactivation of the nlt1 gene and makes additional off-target effects of Cas9 unlikely. While chlorotic spots observed at 4 dpi were comparable after infections with nlt1 mutants, the wild type, and complemented strains, the latter two had induced leaf tumors at 8 dpi, while the nlt1 mutant failed to do so (Figure 8C). Even at 12 dpi when tumors induced by wild-type and complemented strains had reached their maximum size and started to turn black due to spore formation, no leaf tumors were detected in infections with the nlt1 mutants (Figure 8B; Supplemental Figure 6A). Strong anthocyanin formation in leaves infected with nlt1 mutants (Figure 8C) showed that the mutants were able to successfully establish a biotrophic interaction. Anthocyanin induction requires expression of the tin2 (tumor inducing 2) effector gene (Tanaka et al., 2014), a gene placed in the magenta module (Supplemental Data Set 12). nlt1 mutants displayed two additional phenotypes rarely seen in wild-type infections: death of the 4th or 5th leaf only in otherwise viable plants in ∼25% of the cases (Figure 8B; Supplemental Figure 6A) and late spore-filled tumors (detected later than 8 dpi) restricted to the base of the stem in ∼8% of the infected plants (Figure 8B; Supplemental Figure 6B), sometimes associated with death of the 4th or 5th leaf. The presence of basal stem tumors in nlt1 mutant infections indicates that the ability to induce tumors is not completely abolished and can occur in meristematic stem tissue. We speculate that the dead leaf phenotype and the appearance of late stem tumors may be connected, i.e., formation of the basal stem tumors may affect the nutrient supply to the 4th or 5th leaf.
Figure 8.
Role of U. maydis nlt1 in Virulence.
(A) The heat map shows the expression profile of the U. maydis nlt1 gene; log2 expression values are visualized relative to the mean expression across all stages.
(B) The indicated mixtures of strains were injected into maize seedlings and symptoms were scored 12 d after infection according to severity; the color code for each category is given on the right. The nlt1 mutants were unable to induce tumors in leaves. nlt1em indicates that the nlt1 gene was inactivated by CRISPR-Cas9. nlt1-C indicates that a single copy of nlt1 was introduced into the indicated strains to test for complementation. Three independent experiments were performed and the average values are expressed as a percentage of the total number of infected plants (n), which is given above each column. Please note that dead plants, which represent the most severe symptom category, are the result of the virulence assay that is based on young maize seedlings and high cell density of the inoculum. U. maydis is a strict biotroph and does not kill plant tissue under natural conditions.
(C) Representative leaves of infections with wild-type strains, nlt1 mutants, and the complemented strains at 4 and 8 dpi are shown. Examples of stem tumors and dead leaves observed in infections with nlt1 mutants are depicted in Supplemental Figure 6.
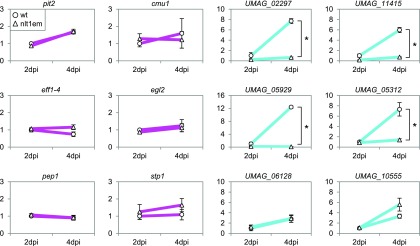
The ability of the nlt1 mutants to establish a biotrophic interaction is in line with our expectations, i.e., the third effector wave, which we consider being controlled by nlt1, temporally follows the establishment of biotrophy mediated by the second effector wave. To verify that nlt1 contributes to the induction of the third wave of effectors, we measured expression of six potential effectors of the cyan module during plant colonization (2 and 4 dpi) in the nlt1 mutants and compared this with the expression in wild-type infections. We found that four of the six cyan genes chosen were highly dependent on nlt1 for induction (Figure 9). As a negative control, we included in this analysis also six effectors of the magenta module, and none of these genes required nlt1 for expression (Figure 9). These data demonstrate that nlt1 is indeed a driver of the cyan module. The observation that two of the tested effectors of the cyan module did not show any dependence on nlt1 suggests a sharp division of labor between nlt1 and the other transcription factors in the cyan module. Thus, nlt1 likely drives the expression of a specific subset of effectors in this module. The two Zn2Cys6 proteins UMAG_05601 and UMAG_02765, which are highly connected to the cyan module (Figure 7C), are interesting candidates for the expression of other subsets of genes of the cyan module. Such a proposed division of labor may also hold true for transcription factors of the magenta module and this requires experimental clarification.
Figure 9.
Regulation of Effector Gene Expression by nlt1.
The expression of selected effector genes during pathogenic development in crosses of FB1 and FB2 wild-type strains (circles) as well as in crosses of compatible nlt1 mutants (triangles) was measured via RT-qPCR. Six effector genes of the magenta module (leftmost two panels) and six potential effectors of the cyan module (rightmost two panels) were tested. In each graph, the expression of the respective gene in the wild-type 2 dpi samples was set to 1 and relative expression is depicted. Significant expression differences (P value < 0.01, Student’s t test) between nlt1 mutants and wild-type strains are indicated with an asterisk if applicable. Error bars denote sd of three biological replicates. Four of six genes of the cyan module required nlt1 for expression while none of the six effector genes of the magenta module was regulated by nlt1.
Recently, the central regulator of spore formation in U. maydis, ros1 (UMAG_05853), has been identified (Tollot et al., 2016). Ros1 not only induces spore formation but also regulates many effector genes (Tollot et al., 2016). We identified Ros1 as being part of the cyan module with a medium high intramodular connectivity of 0.82 (Supplemental Table 15). Indeed, Ros1-induced genes are significantly overrepresented in the cyan module (P value = 3.83E-21, Fisher exact test; Supplemental Data Set 13). However, a substantial number of genes of the cyan module are also repressed by Ros1 (Supplemental Data Sets 12 and 13). We therefore conclude that Ros1 may to some extent contribute to the expression of the third wave of effectors, but is clearly not a driver of this module. We also searched for modules in which Ros1-repressed genes were significantly overrepresented and Ros1-induced genes were significantly underrepresented. Two modules, red and magenta, fulfilled these criteria (P value < 0.001, Fisher exact test; Supplemental Data Set 13). Both modules have in common that they are expressed early upon contact with the plant, corroborating the previous finding that Ros1 is a repressor of effector genes required early during pathogenic development (Tollot et al., 2016).
Our approach of using intramodular connectivity to identify regulators of effector gene expression has identified three sets of transcription factors likely responsible for the plant-associated expression of the secretome. The first and second sets of transcription factors largely consist of components of the b-cascade. This corroborates previous studies that emphasized the impact of the b-cascade for early pathogenic development and the establishment of biotrophy (Brachmann et al., 2001; Heimel et al., 2010b; Lanver et al., 2014). The third set of transcription factors that we identified here consists mainly of yet uncharacterized genes. It will be an interesting future task to study the contribution of these transcription factors to the expression of the third wave of effectors, which is expected to fulfill virulence functions after the early biotrophic phases have been established. Studying in detail which effector groups are regulated by these transcription factors and linking this information with the physiology of plants infected by the respective transcription factor mutants could be a key to identifying novel, host-manipulating fungal strategies that go beyond suppression of the plant immune system.
This large-scale transcriptome analysis has provided a detailed temporal view of gene expression in U. maydis throughout its biotrophic life cycle. The analysis has allowed us to formulate novel hypotheses concerning fungal nutrition in the plant environment and to visualize the deployment of certain groups of secreted effectors as well as connected transcription factors during discrete stages of colonization. This is expected to fuel, stimulate, and direct future functional studies of the identified U. maydis genes, as well as provide a new resource for comparative studies in related fungal pathogens.
METHODS
Bacterial and Fungal Strains and Growth Conditions
The Escherichia coli strain Top10 (Life Technologies) was used for cloning purposes. Ustilago maydis strains used in this study are listed in Supplemental Data Set 16. They are derived from haploid strains FB1 and FB2 (Banuett and Herskowitz, 1989). DNA from the solopathogenic strain SG200, which was derived from FB1 (Kämper et al., 2006), was used for gene amplifications. U. maydis strains were grown in liquid YEPSL (0.4% yeast extract, 0.4% peptone, and 2% sucrose) at 28°C on a rotary shaker at 200 rpm. To assess the ability of compatible strains to mate and form dikaryotic hyphae, FB1 and FB2 wild-type strains and the respective mutants were grown in YEPSL to an OD600 of ∼1, harvested by centrifugation (1700g, 10 min at room temperature), and resuspended in water to an OD600 of 1. Compatible strains were mixed in a 1:1 ratio and 4 μL of this mixture was spotted on potato dextrose plates containing 1% (w/v) activated charcoal. The plates were incubated at room temperature for 48 h. Strains which have mated produce dikaryotic hyphae, visible as white mycelium against the black background. To test growth of haploid U. maydis strains on different nitrogen sources, FB1 and FB2 wild-type strains and the respective dur3-1,2,3 triple mutants were grown in YEPSL to an OD600 of ∼1, harvested by centrifugation (1700g, 10 min at room temperature), washed two times with water, and resuspended in water to an OD600 = 1. Serial dilutions (OD600 = 1 to 10−4) were prepared in water and 2.5 μL of each dilution was spotted on minimal medium plates (Holliday, 1961), supplemented with 2% glucose and either 10 mM ammonium or 10 or 1 mM urea. The plates were incubated for 3 d at 28°C.
Plasmid and Strain Construction
All strains generated in this study, plasmids, primers, and double-stranded DNA fragments, are listed in Supplemental Data Sets 16 to 19. To generate U. maydis mutants, the recently established CRISPR-Cas9 multiplex system was used (Schuster et al., 2017). The nonintegrative, self-replicating backbone plasmids of this system, pDL272 and pMS73, contain cas9 either under the control of the strong constitutive otef promoter (pDL272) or under control of the even stronger hsp70 promoter (pMS73). All target sequences for the guide RNA constructs were designed using the E-CRISP tool (www.e-crisp.org) (Heigwer et al., 2014) with medium stringency settings. pDL272 and pMS73 contain one copy of the U6 promoter for fusion with the first guide RNA construct. Additional guide RNA constructs are then fused to distinct tRNA promoters, and all components are finally cloned into one single plasmid (Schuster et al., 2017).This yielded the plasmids described below.
To generate the plasmid pDL286 for inactivation of the three opt genes, UMAG_11057, UMAG_05968, and UMAG_02387, the respective double-stranded DNA fragments fDL12, fDL13, and fDL14 listed in Supplemental Data Set 18 were synthesized (gBLOCKs from IDT) and cloned into Acc65I-linearized pDL272 using isothermal assembly (Gibson et al., 2009). To generate the plasmid pDL287 for inactivation of the three urea transporters UMAG_02625, UMAG_04577, and UMAG_06253, the respective double-stranded DNA fragments fDL15, fDL16, and fDL17 were synthesized and cloned into Acc65I-linearized pMS73. As the resulting plasmid proved inefficient for inactivation of all three genes except in strain PH72, a second plasmid, pPH22 was generated that is identical to pDL287 but contains new target sequences for the inactivation of UMAG_02625 and UMAG_06253. To this end, fPH1 and fPH2 were generated by PCR using oPH163 and oPH160 as primers on gBLOCK fDL15, and oPH161 and oPH162 as primers on gBLOCK fDL16 (Supplemental Data Sets 17 and 18). pPH22 was then assembled by Gibson cloning using fPH1, fPH2, fDL17, and Acc65I-linearized pMS73. To generate the plasmid pPH20 for inactivation of the transcription factor gene UMAG_04778, the double-stranded fragment fPH3 listed in Supplemental Data Set 18 was generated by PCR with primers oPH137 and oPH138 using gBLOCK fDL17 as template. This fragment was cloned via Gibson assembly into Acc65I-linearized pMS73 to yield the final plasmid.
For complementation analysis, we made use of the genome integrative p123 plasmid (Aichinger et al., 2003). To generate the plasmid for complementation of the opt triple mutant, plasmid pPH19 was constructed. UMAG_11057 was amplified from genomic DNA of SG200 using primers oPH131 and oPH132, UMAG_02387 was amplified using primers oPH133 and oPH134, and UMAG_05968 was amplified using primers oPH135 and oPH136. The three fragments were inserted into the backbone of p123 cleaved with Acc65I and EcoRV. In pPH19 all three genes carry their native promoters and termination regions. The plasmid was linearized with SspI prior to transformation in U. maydis.
To generate the plasmid for complementation of the nlt1 mutant, pPH23 was generated. To this end the nlt1gene was amplified from SG200 DNA with primers oPH164 and oPH165. The amplified fragment was Gibson assembled into the backbone of p123 cleaved with Acc65I and EcoRV. In pPH23, nlt1 is driven by its native promoter and termination region. The plasmid was linearized with PsrI prior to transformation.
For the generation of U. maydis strains carrying fluorescence reporter constructs, we constructed the following integrative plasmids based on the backbone of p123. To place mcherry under the control of the actin promoter, we amplified 2.0 kb of the promoter of the U. maydis actin gene (UMAG_11232) using primers oDL575 and oDL576. The product was digested with HindIII and NcoI and subjected to a three fragment ligation with a 0.9-kb NcoI/NsiI mcherry fragment from pMF5-15g (plasmid collection of M. Feldbrügge; www.mikrobiologie.hhu.de/ustilago-community) and the HindIII/NsiI-digested p123 plasmid backbone yielding pPact-mcherry (pDL252). The mcherry fragment of pPact-mcherry was replaced with a 0.9-kb NcoI/NsiI gfp fragment from p123 to generate pPact-gfp (pDL289). To place gfp under the control of effector promoters, we amplified 1.5 kb of the promoter of pep1 (UMAG_01987) and 0.6 kb of the promoter of stp1 (UMAG_02475) using primer pairs oDL746/oDL747 and oDL744/oDL745, respectively. In addition, we amplified a gfp-Tnos fragment with the primers oDL742 and oDL743 using p123 as template. The gfp-Tnos fragment, the respective effector promoter fragment, and the HindIII linearized pPact-mcherry plasmid were Gibson assembled to yield pPact-mcherry-Ppep1-gfp (pDL281) and pPact-mcherry-Pstp1-gfp (pDL290). The plasmids containing fluorescence reporter constructs were linearized with AgeI prior to transformation.
Transformation of U. maydis was performed as described previously (Schulz et al., 1990), and the transformants were selected on carboxin containing media (2 µg/mL). To identify strains carrying Cas9-induced mutations, we followed the established screening protocol (Schuster et al., 2017). The respective loci were amplified and sequenced with gene-specific primers (Supplemental Table 14). The stable integration of p123-based plasmids was verified by DNA gel blot as described previously (Loubradou et al., 2001). All complementation plasmids were integrated in single copy into the U. maydis ip locus. Fluorescence reporter constructs were integrated in multiple copies (>3) into the ip locus.
Plant Infections and Collection of Samples
For the transcriptional profiling, the compatible haploid strains FB1 and FB2 were grown separately in YEPSL to an OD600 of 1.0. Cells were collected by centrifugation (5 min at 1250g at room temperature) and resuspended in the original volume of sterile water. Compatible strains were mixed in equal amounts prior to injection into 6-d-old maize (Zea mays) seedlings of the variety Early Golden Bantam (UrbanFarmers) as described previously (Tollot et al., 2016). The plants were kept in growth chambers (Percival AR95-HIL) with a 16-h-light (30 kLux)/8-h-dark cycle, with 28°C/20°C and 40%/60% humidity. To generate biological replicates, three plant infections were performed with independently cultivated fungal strains on different days. For each replicate, more than 100 plants were infected with a mixture of compatible wild-type strains and the same number of plants was mock infected with water. Samples of 10 infected maize seedlings were collected at 0.5, 1, 2, 4, 6, 8, 10, and 12 dpi. For early time points, where symptoms are not yet visible (0.5, 1, and 2 dpi) 2-cm sections of leaf blade of the 2nd and 3rd leaves were excised 0.5 cm below the infection holes. These areas were shown by microscopy to be colonized at these time points. At later time points (4, 6, 8, and 12 dpi) all visibly infected areas, including the leaf blades to ∼1 cm below the injection holes on the 2nd, 3rd, and 4th leaf and the attached leaf sheets were harvested. At each time point, comparable parts of seedlings were harvested for the mock infected control plants. For each sample, the collected material from 10 different plants was pooled, immediately frozen in liquid nitrogen, and stored at −80°C. For the axenic culture samples, FB1 and FB2 cells separately grown in YEPSL to an OD600 of 1.0 in three independent biological replicates were harvested by centrifugation (5 min at 3000g), directly frozen in liquid nitrogen, and stored at −80°C until RNA extraction.
All virulence assays were performed in the greenhouse under controlled conditions with a light-dark cycle of 28°C for 14 h and 20°C for 10 h. During the day phase, the illumination intensity was at least 25 to 30 kLux (with additional sunlight up to 90 kLux). U. maydis was grown and injected into 6-d-old maize plants as described above. Except for nlt1 mutants, the OD600 of the injected suspension for virulence assays was adjusted to 1.0. To assess virulence of nlt1 mutants and respective wild-type strains the OD600 was adjusted to 0.2. Virulence symptoms were scored 12 dpi using the disease rating scheme developed previously (Kämper et al., 2006). When scoring disease symptoms of infections with nlt1 mutants, two new disease categories were introduced. Hatched was a category where plants developed late tumors at the base of the stem only, sometimes associated with death of the 4th or 5th leaf, as well as a category where only the 4th or 5th leaf died in otherwise healthy maize seedlings (see Supplemental Figure 6 for examples).
RNA Extraction and Sequencing
Pooled plant material corresponding to individual samples was ground to a fine powder in liquid nitrogen using a Retsch CryoMill with a 50-mL grinding beaker and a 20-mm grinding ball. For each sample, the machine was precooled for 30 s followed by 60 s of grinding at 20 Hz. Approximately 500 mg of powder was resuspended in 1 mL TRIzol reagent (Life Technologies) and total RNA was extracted according to the manufacturer’s recommendation. For RNA extraction from U. maydis derived from axenic culture, glass beads (0.4–0.6 mm) together with 1 mL of TRIzol reagent (Life Technologies) were added to the frozen cell pellet followed by vigorous shaking (IKA Vibrax VXR, 2000 rpm, 10 min) and further processing. To eliminate genomic DNA contamination, the Ambion Turbo DNA-free kit (Life Technologies) was used. The total RNA was further purified using the RNeasy Mini Kit (Qiagen) and subjected to quality control with an Agilent 2100 Bioanalyzer according to the instructions of the Agilent RNA 6000 Nano Assay Protocol. Libraries were prepared using the Illumina TruSeq stranded RNA sample preparation Kit and 1 µg of total RNA as input. After poly(A) selection (using poly-T oligo-attached magnetic beads), mRNA was purified and fragmented using divalent cations under elevated temperature. The RNA fragments underwent reverse transcription using random primers. This was followed by second strand cDNA synthesis with DNA Polymerase I and RNase H. After end repair and A-tailing, indexing adapters were ligated. The products were then purified and amplified (14 PCR cycles, 10 μL template) to create the final cDNA libraries. After library validation and quantification (Agilent 2100 Bioanalyzer), up to six libraries were pooled. Pools were quantified using the Peqlab KAPA library quantification kit and the Applied Biosystems 7900HT sequence detection system. One pool per lane was sequenced using an Illumina TruSeq PE Cluster Kit v3 and an Illumina TruSeq SBS Kit v3-HS on an Illumina HiSeq 2000 sequencer with a paired-end (101 × 7 × 101 cycles) protocol. The raw data have been deposited in the NCBI Gene Expression Omnibus (GEO; Edgar et al., 2002) and are accessible through GEO Series accession number GSE103876 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE103876).
Read Mapping, Normalization, and Statistical Analysis of Differential Gene Expression
All reads were imported into CLC genomics workbench (v8.5) and quality-filtered to keep only sequences with PhredScores higher than 20 (quality limit 0.01). The reads were trimmed for the presence of TruSeq adapters and, according to a FastQC quality control, 12 nucleotides at the 5′ end were statically removed. Reads smaller than 50 nucleotides were discarded. The paired end reads were merged to single reads, if the overlap was more than 10 nucleotides. Depending on the sample, reads from different sequencing runs (but from the same Truseq libraries) were pooled. For the axenic culture samples, the separately generated reads from FB1 and FB2 strains were pooled and analyzed in one batch. All reads were filtered against the annotated maize genes (90% identity over 80% of the sequence) and all unmapped reads were then mapped against the annotated U. maydis genes (80% identity over 60% of the sequence). For expression analysis, only uniquely mapping exon read counts were considered. Since highly expressed genes make up a substantial fraction of the total library, causing the remaining genes to be undersampled and possibly making them falsely appear to be downregulated in that sample, it is important to normalize for the library composition. We chose to normalize our data set using the relative log expression method implemented in the DESeq2 package (v1.10) (Love et al., 2014) in R (www.r-project.org). Alternatively, we used the trimmed mean of M-values method implemented in the EdgeR package (v3.12) (Robinson et al., 2010; Robinson and Oshlack, 2010). Both methods are very efficient in normalizing for library composition effects (Dillies et al., 2013). Pairwise comparisons were made with EdgeR and DESeq2 using the conditional maximum likelihood (qCML) method and the Cox-Reid profile-adjusted likelihood method, respectively. In both analyses, genes with a log2 fold change > 0.5 and Benjamini and Hochberg-adjusted P value < 0.01 were considered differentially expressed (Eisen et al., 1998). All count data have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE103876 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE103876). This also includes all count data mapped to the maize genome, which are not discussed here.
Coexpression Analysis
For the unsupervised WGCNA, only genes that have at least for one time point over all three replicates a DESeq2-normalized read count of at least 10 were considered. A total of 6421 genes passed these filtering criteria. Log2-transformed DESeq2-normalized counts then served as input for the network analysis. We used the function blockwiseModules, implemented in the R (www.r-project.org) WGCNA package (v1.51) (Langfelder and Horvath, 2008), to create a signed network of a Pearson correlated matrix. All genes were treated in a single block. The signed network ensured that only positive correlations were considered. We have chosen a soft power threshold of 7 because this was the lowest power needed to reach scale-free topology (R2 = 0.92). Module detection was performed with default settings (mergeCutHeight of 0.15 and enabled PAMstage). The minimal module size was set to 25 genes. After module detection, highly correlated modules were merged using a cut height of 0.3. This step reduced the number of modules from 22 to 14 (Supplemental Figure 4). For each module, the expression profile of the module eigengene was calculated, which is defined as the first principal component of the module’s expression data. For each gene, the intramodular connectivity (kME) was calculated, which represents Pearson correlation of the individual gene with the respective module eigengenes. To visualize weighted coexpression networks, the topological overlap matrix was exported to Cytoscape (v3.3) (Shannon et al., 2003) with an adjacency cutoff of <0.2. The k-mean clustering was performed in MeV (v4.9) (http://mev.tm4.org) using the same DESeq2 normalized and log2 transformed expression data set that was used for the WGCNA (see above). To determine a suitable number of clusters, we used the figure of merit analysis and chose to cluster the genes into six clusters. The k-mean clusters were completed after 20 iterations using Pearson correlation as distance measure. Average linkage hierarchical clustering of individual modules and gene sets was performed with Cluster (v3.0) (Eisen et al., 1998) and heat maps were visualized with Java Treeview (v1.1.6).
GO Term Enrichment Analysis
GO term enrichment analysis (Ashburner et al., 2000; The Gene Ontology Consortium, 2017) was performed with the GOseq package (v1.22) (Young et al., 2010) in R (www.r-project.org) using hypergeometric testing and an overrepresented P value cutoff of < 0.005. For the enrichment analysis of the individual modules, only genes were considered that had an intramodular connectivity (kME) of greater than 0.5. Visualization of GO term networks was performed with the Cytoscape Enrichment Map plug-in (v2.1; www.baderlab.org/Software/EnrichmentMap) (Merico et al., 2010) using a Jaccard coefficient cutoff of >0.2. Clusters of gene sets were manually marked and named according to their GO annotation using the Cytoscape Word Cloud plug-in (v3.1).
RT-qPCR and Quantitative Real-Time PCR
For gene expression analysis via RT-qPCR, plants were infected with the indicated strains adjusted to a cell density at OD600 of 1.0. Then, 1 to 2 µg of isolated total RNA was reverse-transcribed with oligo(dT) primers using the First-Strand Super Script III cDNA Synthesis Kit (Life Technologies). Per RT-qPCR reaction, cDNA derived from 10 to 50 ng RNA was deployed. The RT-qPCR was performed in a Bio-Rad iCycler using SYBR Green RT-qPCR SuperMix-UDG (Life Technologies). Cycling conditions were 1 min 30 s at 95°C, followed by 45 cycles of 15 s at 95°C, 30 s at 60°C, and 30 s at 72°C. Primers are listed in Supplemental Data Set 17. Two control genes were used: the previously established peptidylprolyl isomerase (ppi) UMAG_03726 plus the rab7 GTPase UMAG_05511, which was according to the data set created here one of the most constantly expressed genes (coefficient of variation of 14.2). The arithmetic mean of the two reference Ct values served as normalization. For the expression of fold changes of RT-qPCR data, the 2−ΔΔCt method was used (Livak and Schmittgen, 2001).
To determine fungal biomass, the same plant material that was used for the RNA-seq analysis was utilized. DNA was extracted from the ground powder using a phenol-based protocol (Hoffman and Winston, 1987). Then, 150 ng total DNA was subjected to qPCR (quantitative real-time PCR) analysis with plant-specific GAPDH (oDL833/oDL834) and fungal-specific ppi primer pairs (oPH231/oPH232; Supplemental Data Set 14), and the ratios of fungal DNA to plant DNA (2−ΔCt) were calculated for each sample. This analysis allowed us to relate for each sample the number of fungal transcripts to the amount of fungal DNA.
Microscopy
To visualize fungal aggregates in tumor tissue, 7-dpi tumors were collected, and thin slices of the tumors were prepared using a razor blade. The slices were analyzed by a TCS-SP5 confocal microscope (Leica Microsystems). GFP was excited at 488 nm and emitted fluorescence was detected at 495 to 530 nm. mCherry was excited at 561 nm and emission was detected at 580 to 630 nm. Images were processed using LAS-AF software (Leica Microsystems).
Accession Numbers, Genome References, and Annotations
The maize reference genome was downloaded from Ensembl (http://plants.ensembl.org/info/website/ftp/index.html, Zea_mays.AGPv3.28.dna.toplevel). The U. maydis reference genome was downloaded from NCBI (www.ncbi.nlm.nih.gov/www.ncbi.nlm.nih.gov, accession numbers NC_026478.1 - NC_026500.1, NW_011929455.1 - NW_011929458.1, and NC_008368.1). GO categories and InterPro motifs of U. maydis genes were extracted from the pedant database (http://pedant.helmholtz-muenchen.de/). Membrane transport proteins were defined by filtering the InterPro annotation of U. maydis for any of the following keywords: porter, porting, P-type ATPase, facilitator, permease, channel, and aquaporin. Secreted hydrolases were classified by searching the InterPro descriptions of the 467 secreted proteins for the keywords hydrolase, protease, peptidase, nuclease, lipase, esterase, lyase, hydrolysis, glucanase, and lytic. Carbohydrate active enzymes (CAZymes) were categorized according to the CAZy database (http://www.cazy.org). CAZymes potentially affecting the plant cell wall were defined previously (Lo Presti et al., 2015). To identify and classify U. maydis transcription factors, we used a previously defined list of 82 InterPro signatures known to be involved in fungal transcriptional regulation (Park et al., 2008), and supplemented this list with the InterPro signatures IPR001092, IPR011598 representing bHLH, and IPR018608 representing WOPR transcription factors, respectively. The InterPro annotation of U. maydis was then filtered for the presence of any of those 85 signatures. The RNA-seq data are available in the GEO database under accession number GSE103876.
Supplemental Data
Supplemental Figure 1. Correlation between amount of fungal transcripts and fungal biomass.
Supplemental Figure 2. Correlation between RNA-seq and RT-qPCR derived expression data.
Supplemental Figure 3. Detection of gene coexpression modules of U. maydis during pathogenic development.
Supplemental Figure 4. Enriched biological processes in coexpression modules.
Supplemental Figure 5. Expression of GFP under control of the actin promoter in fungal aggregates.
Supplemental Figure 6. Disease categories specific to infections with nlt1 mutants.
Supplemental Data Set 1. Overview of read counts mapping to U. maydis and Z. mays.
Supplemental Data Set 2. Statistics of changes of fungal biomass based on DNA and transcripts.
Supplemental Data Set 3. DESeq2-normalized U. maydis read counts.
Supplemental Data Set 4. Differentially expressed U. maydis genes throughout the infection.
Supplemental Data Set 5. Overlap between differentially expressed gene sets identified by DESeq2 and EdgeR.
Supplemental Data Set 6. Gene coexpression modules of U. maydis and intramodular connectivities of the genes.
Supplemental Data Set 7. Overlap between modules detected by WGCNA and clusters detected by k-mean clustering.
Supplemental Data Set 8. GO-term enrichment analysis of coexpression modules.
Supplemental Data Set 9. Expression of primary metabolism pathways.
Supplemental Data Set 10. Expression of autophagy, glycosylation, and ER quality control pathways.
Supplemental Data Set 11. Expression of membrane transporters.
Supplemental Data Set 12. Expression of the secretome.
Supplemental Data Set 13. Enrichment of diverse gene sets in coexpression modules.
Supplemental Data Set 14. Expression of secondary metabolism pathways.
Supplemental Data Set 15. Intramodular connectivities of transcription factors with coexpression modules.
Supplemental Data Set 16. U. maydis strains used in this study.
Supplemental Data Set 17. Oligonucleotides used in this study.
Supplemental Data Set 18. Double-stranded DNA fragments used in this study.
Supplemental Data Set 19. Plasmids used in this study.
Acknowledgments
We thank Armin Djamei and Florian Raths for providing preliminary results on U. maydis oligopeptide transporters. We thank Michael Bölker for sharing his interpretation of fungal growth within tumors. We thank Nicole Ludwig for helping with sampling of infected plant material. We thank Kai Heimel for listing U. maydis UPR components and Gertrud Mannhaupt and Ulrich Güldener for providing gene annotations. We enjoyed helpful discussions with Frank Hochholdinger and Kristian K. Ullrich and all members of the Kahmann group. This work was supported by generous funds from the Max Planck Society.
AUTHOR CONTRIBUTIONS
R.K. conceived the project. A.N.M. planned and conducted plant infections and generated samples for RNA-seq analysis. M.F. and J.A. performed RNA sequencing. D.L. and G.S. performed data analysis. F.B.H. and S.A.R. provided critical input in data analysis and quality control. P.H. and D.L. generated mutants. P.H. performed RT-qPCR analyses and virulence assays. A.N.M. and C.P. contributed to the finalization of the data set. D.L. and R.K. led the study and wrote the manuscript with help from S.R.
Footnotes
Articles can be viewed without a subscription.
References
- Abreu C., Sanguinetti M., Amillis S., Ramon A. (2010). UreA, the major urea/H+ symporter in Aspergillus nidulans. Fungal Genet. Biol. 47: 1023–1033. [DOI] [PubMed] [Google Scholar]
- Aichinger C., Hansson K., Eichhorn H., Lessing F., Mannhaupt G., Mewes W., Kahmann R. (2003). Identification of plant-regulated genes in Ustilago maydis by enhancer-trapping mutagenesis. Mol. Genet. Genomics 270: 303–314. [DOI] [PubMed] [Google Scholar]
- Ashburner M., et al. ; The Gene Ontology Consortium (2000). Gene ontology: tool for the unification of biology. Nat. Genet. 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballini E., Nguyen T.T., Morel J.B. (2013). Diversity and genetics of nitrogen-induced susceptibility to the blast fungus in rice and wheat. Rice (N.Y.) 6: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banuett F., Herskowitz I. (1989). Different a alleles of Ustilago maydis are necessary for maintenance of filamentous growth but not for meiosis. Proc. Natl. Acad. Sci. USA 86: 5878–5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basse C.W., Lottspeich F., Steglich W., Kahmann R. (1996). Two potential indole-3-acetaldehyde dehydrogenases in the phytopathogenic fungus Ustilago maydis. Eur. J. Biochem. 242: 648–656. [DOI] [PubMed] [Google Scholar]
- Billet E.E., Burnett J.H. (1978). The host-parasite physiology of the maize smut fungus Ustilago maydis. 11. Translocation of 14C-labelled assimilates in smutted maize plants. Plant Pathol. 12: 102–112. [Google Scholar]
- Bölker M., Urban M., Kahmann R. (1992). The a mating type locus of U. maydis specifies cell signaling components. Cell 68: 441–450. [DOI] [PubMed] [Google Scholar]
- Bourbouloux A., Shahi P., Chakladar A., Delrot S., Bachhawat A.K. (2000). Hgt1p, a high affinity glutathione transporter from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 275: 13259–13265. [DOI] [PubMed] [Google Scholar]
- Brachmann A., Weinzierl G., Kämper J., Kahmann R. (2001). Identification of genes in the bW/bE regulatory cascade in Ustilago maydis. Mol. Microbiol. 42: 1047–1063. [DOI] [PubMed] [Google Scholar]
- Castanheira S., Mielnichuk N., Pérez-Martín J. (2014). Programmed cell cycle arrest is required for infection of corn plants by the fungus Ustilago maydis. Development 141: 4817–4826. [DOI] [PubMed] [Google Scholar]
- Chagué V., Maor R., Sharon A. (2009). CgOpt1, a putative oligopeptide transporter from Colletotrichum gloeosporioides that is involved in responses to auxin and pathogenicity. BMC Microbiol. 9: 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.L., Shi T., Yang J., Shi W., Gao X., Chen D., Xu X., Xu J.R., Talbot N.J., Peng Y.L. (2014). N-glycosylation of effector proteins by an α-1,3-mannosyltransferase is required for the rice blast fungus to evade host innate immunity. Plant Cell 26: 1360–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway M.K., Grunwald D., Heideman W. (2012). Glucose, nitrogen, and phosphate repletion in Saccharomyces cerevisiae: common transcriptional responses to different nutrient signals. G3 (Bethesda) 2: 1003–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copley T.R., Duggavathi R., Jabaji S. (2017). The transcriptional landscape of Rhizoctonia solani AG1-IA during infection of soybean as defined by RNA-seq. PLoS One 12: e0184095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean R., Van Kan J.A., Pretorius Z.A., Hammond-Kosack K.E., Di Pietro A., Spanu P.D., Rudd J.J., Dickman M., Kahmann R., Ellis J., Foster G.D. (2012). The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13: 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillies M.A., et al. ; French StatOmique Consortium (2013). A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief. Bioinform. 14: 671–683. [DOI] [PubMed] [Google Scholar]
- Divon H.H., Fluhr R. (2007). Nutrition acquisition strategies during fungal infection of plants. FEMS Microbiol. Lett. 266: 65–74. [DOI] [PubMed] [Google Scholar]
- Divon H.H., Ziv C., Davydov O., Yarden O., Fluhr R. (2006). The global nitrogen regulator, FNR1, regulates fungal nutrition-genes and fitness during Fusarium oxysporum pathogenesis. Mol. Plant Pathol. 7: 485–497. [DOI] [PubMed] [Google Scholar]
- Djamei A., et al. (2011). Metabolic priming by a secreted fungal effector. Nature 478: 395–398. [DOI] [PubMed] [Google Scholar]
- Dobon A., Bunting D.C., Cabrera-Quio L.E., Uauy C., Saunders D.G. (2016). The host-pathogen interaction between wheat and yellow rust induces temporally coordinated waves of gene expression. BMC Genomics 17: 380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehlemann G., Reissmann S., Assmann D., Fleckenstein M., Kahmann R. (2011). Two linked genes encoding a secreted effector and a membrane protein are essential for Ustilago maydis-induced tumour formation. Mol. Microbiol. 81: 751–766. [DOI] [PubMed] [Google Scholar]
- Doehlemann G., Wahl R., Vranes M., de Vries R.P., Kämper J., Kahmann R. (2008a). Establishment of compatibility in the Ustilago maydis/maize pathosystem. J. Plant Physiol. 165: 29–40. [DOI] [PubMed] [Google Scholar]
- Doehlemann G., van der Linde K., Assmann D., Schwammbach D., Hof A., Mohanty A., Jackson D., Kahmann R. (2009). Pep1, a secreted effector protein of Ustilago maydis, is required for successful invasion of plant cells. PLoS Pathog. 5: e1000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehlemann G., Wahl R., Horst R.J., Voll L.M., Usadel B., Poree F., Stitt M., Pons-Kuhnemann J., Sonnewald U., Kahmann R., Kamper J. (2008b). Reprogramming a maize plant: transcriptional and metabolic changes induced by the fungal biotroph Ustilago maydis Plant J. 56: 181–195. [DOI] [PubMed] [Google Scholar]
- Dong Y., et al. (2015). Global genome and transcriptome analyses of Magnaporthe oryzae epidemic isolate 98-06 uncover novel effectors and pathogenicity-related genes, revealing gene gain and lose dynamics in genome evolution. PLoS Pathog. 11: e1004801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dordas C. (2008). Role of nutrients in controlling plant diseases in sustainable agriculture. Agron. Sustain. Dev. 28: 33–46. [Google Scholar]
- Dunkel N., Hertlein T., Franz R., Reuß O., Sasse C., Schäfer T., Ohlsen K., Morschhäuser J. (2013). Roles of different peptide transporters in nutrient acquisition in Candida albicans. Eukaryot. Cell 12: 520–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A.E. (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen M.B., Spellman P.T., Brown P.O., Botstein D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95: 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ElBerry H.M., Majumdar M.L., Cunningham T.S., Sumrada R.A., Cooper T.G. (1993). Regulation of the urea active transporter gene (DUR3) in Saccharomyces cerevisiae. J. Bacteriol. 175: 4688–4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Alvarez A., Elías-Villalobos A., Ibeas J.I. (2009). The O-mannosyltransferase PMT4 is essential for normal appressorium formation and penetration in Ustilago maydis. Plant Cell 21: 3397–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Álvarez A., Elías-Villalobos A., Jiménez-Martín A., Marín-Menguiano M., Ibeas J.I. (2013). Endoplasmic reticulum glucosidases and protein quality control factors cooperate to establish biotrophy in Ustilago maydis. Plant Cell 25: 4676–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Álvarez A., Marín-Menguiano M., Lanver D., Jiménez-Martín A., Elías-Villalobos A., Pérez-Pulido A.J., Kahmann R., Ibeas J.I. (2012). Identification of O-mannosylated virulence factors in Ustilago maydis. PLoS Pathog. 8: e1002563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor-Parra I., Vranes M., Kämper J., Pérez-Martín J. (2006). Biz1, a zinc finger protein required for plant invasion by Ustilago maydis, regulates the levels of a mitotic cyclin. Plant Cell 18: 2369–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluharty A.L., O’Brien J.S. (1969). A mannose- and erythritol-containing glycolipid from Ustilago maydis. Biochemistry 8: 2627–2632. [DOI] [PubMed] [Google Scholar]
- Fondevilla S., Krezdorn N., Rotter B., Kahl G., Winter P. (2015). In planta identification of putative pathogenicity factors from the chickpea pathogen Ascochyta rabiei by se novo rranscriptome aequencing using RNA-seq and massive analysis of cDNA ends. Front. Microbiol. 6: 1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., Kelliher T., Nguyen L., Walbot V. (2013). Ustilago maydis reprograms cell proliferation in maize anthers. Plant J. 75: 903–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnica D.P., Upadhyaya N.M., Dodds P.N., Rathjen J.P. (2013). Strategies for wheat stripe rust pathogenicity identified by transcriptome sequencing. PLoS One 8: e67150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch A.P., Spellman P.T., Kao C.M., Carmel-Harel O., Eisen M.B., Storz G., Botstein D., Brown P.O. (2000). Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11: 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gervais J., Plissonneau C., Linglin J., Meyer M., Labadie K., Cruaud C., Fudal I., Rouxel T., Balesdent M.H. (2017). Different waves of effector genes with contrasted genomic location are expressed by Leptosphaeria maculans during cotyledon and stem colonization of oilseed rape. Mol. Plant Pathol. 18: 1113–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A. III, Smith H.O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6: 343–345. [DOI] [PubMed] [Google Scholar]
- Gillissen B., Bergemann J., Sandmann C., Schroeer B., Bölker M., Kahmann R. (1992). A two-component regulatory system for self/non-self recognition in Ustilago maydis. Cell 68: 647–657. [DOI] [PubMed] [Google Scholar]
- Hacquard S., Kracher B., Maekawa T., Vernaldi S., Schulze-Lefert P., Ver Loren van Themaat E. (2013). Mosaic genome structure of the barley powdery mildew pathogen and conservation of transcriptional programs in divergent hosts. Proc. Natl. Acad. Sci. USA 110: E2219–E2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampel M., Jakobi M., Schmitz L., Meyer U., Finkernagel F., Doehlemann G., Heimel K. (2016). Unfolded protein response (UPR) regulator Cib1 controls expression of genes encoding secreted virulence factors in Ustilago maydis. PLoS One 11: e0153861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann T., Cairns T.C., Olbermann P., Morschhäuser J., Bignell E.M., Krappmann S. (2011). Oligopeptide transport and regulation of extracellular proteolysis are required for growth of Aspergillus fumigatus on complex substrates but not for virulence. Mol. Microbiol. 82: 917–935. [DOI] [PubMed] [Google Scholar]
- Haskins R.H., Thorn J.A. (1951). Biochemistry of the Ustilaginales: VII. Antibiotic activity of ustilagic acid. Can. J. Bot. 29: 585–592. [Google Scholar]
- Hauser N.C., Fellenberg K., Gil R., Bastuck S., Hoheisel J.D., Pérez-Ortín J.E. (2001). Whole genome analysis of a wine yeast strain. Comp. Funct. Genomics 2: 69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heigwer F., Kerr G., Boutros M. (2014). E-CRISP: fast CRISPR target site identification. Nat. Methods 11: 122–123. [DOI] [PubMed] [Google Scholar]
- Heimel K., Scherer M., Schuler D., Kämper J. (2010a). The Ustilago maydis Clp1 protein orchestrates pheromone and b-dependent signaling pathways to coordinate the cell cycle and pathogenic development. Plant Cell 22: 2908–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimel K., Freitag J., Hampel M., Ast J., Bölker M., Kämper J. (2013). Crosstalk between the unfolded protein response and pathways that regulate pathogenic development in Ustilago maydis. Plant Cell 25: 4262–4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimel K., Scherer M., Vranes M., Wahl R., Pothiratana C., Schuler D., Vincon V., Finkernagel F., Flor-Parra I., Kämper J. (2010b). The transcription factor Rbf1 is the master regulator for b-mating type controlled pathogenic development in Ustilago maydis. PLoS Pathog. 6: e1001035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemetsberger C., Herrberger C., Zechmann B., Hillmer M., Doehlemann G. (2012). The Ustilago maydis effector Pep1 suppresses plant immunity by inhibition of host peroxidase activity. PLoS Pathog. 8: e1002684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman C.S., Winston F. (1987). A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57: 267–272. [DOI] [PubMed] [Google Scholar]
- Holliday R. (1961). Induced mitotic crossing-over in Ustilago maydis. Genet. Res. 2: 231–248. [DOI] [PubMed] [Google Scholar]
- Horst R.J., Engelsdorf T., Sonnewald U., Voll L.M. (2008). Infection of maize leaves with Ustilago maydis prevents establishment of C4 photosynthesis. J. Plant Physiol. 165: 19–28. [DOI] [PubMed] [Google Scholar]
- Horst R.J., Zeh C., Saur A., Sonnewald S., Sonnewald U., Voll L.M. (2012). The Ustilago maydis Nit2 homolog regulates nitrogen utilization and is required for efficient induction of filamentous growth. Eukaryot. Cell 11: 368–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst R.J., Doehlemann G., Wahl R., Hofmann J., Schmiedl A., Kahmann R., Kämper J., Sonnewald U., Voll L.M. (2010). Ustilago maydis infection strongly alters organic nitrogen allocation in maize and stimulates productivity of systemic source leaves. Plant Physiol. 152: 293–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islamovic E., García-Pedrajas M.D., Chacko N., Andrews D.L., Covert S.F., Gold S.E. (2015). Transcriptome analysis of a Ustilago maydis ust1 deletion mutant uncovers involvement of laccase and polyketide synthase genes in spore development. Mol. Plant Microbe Interact. 28: 42–54. [DOI] [PubMed] [Google Scholar]
- Jiang Y., Wang W., Xie Q., Liu N., Liu L., Wang D., Zhang X., Yang C., Chen X., Tang D., Wang E. (2017). Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science 356: 1172–1175. [DOI] [PubMed] [Google Scholar]
- Jupe J., Stam R., Howden A.J., Morris J.A., Zhang R., Hedley P.E., Huitema E. (2013). Phytophthora capsici-tomato interaction features dramatic shifts in gene expression associated with a hemi-biotrophic lifestyle. Genome Biol. 14: R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kämper J. (2004). A PCR-based system for highly efficient generation of gene replacement mutants in Ustilago maydis. Mol. Genet. Genomics 271: 103–110. [DOI] [PubMed] [Google Scholar]
- Kämper J., et al. (2006). Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 444: 97–101. [DOI] [PubMed] [Google Scholar]
- Kawahara Y., Oono Y., Kanamori H., Matsumoto T., Itoh T., Minami E. (2012). Simultaneous RNA-seq analysis of a mixed transcriptome of rice and blast fungus interaction. PLoS One 7: e49423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keymer A., et al. (2017). Lipid transfer from plants to arbuscular mycorrhiza fungi. eLife 6: e29107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Woloshuk C.P. (2008). Role of AREA, a regulator of nitrogen metabolism, during colonization of maize kernels and fumonisin biosynthesis in Fusarium verticillioides. Fungal Genet. Biol. 45: 947–953. [DOI] [PubMed] [Google Scholar]
- Kleemann J., Rincon-Rivera L.J., Takahara H., Neumann U., Ver Loren van Themaat E., van der Does H.C., Hacquard S., Stüber K., Will I., Schmalenbach W., Schmelzer E., O’Connell R.J. (2012). Sequential delivery of host-induced virulence effectors by appressoria and intracellular hyphae of the phytopathogen Colletotrichum higginsianum. PLoS Pathog. 8: e1002643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong W., Chen N., Liu T., Zhu J., Wang J., He X., Jin Y. (2015). Large-scale transcriptome analysis of cucumber and Botrytis cinerea during infection. PLoS One 10: e0142221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P., Horvath S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanver D., Berndt P., Tollot M., Naik V., Vranes M., Warmann T., Münch K., Rössel N., Kahmann R. (2014). Plant surface cues prime Ustilago maydis for biotrophic development. PLoS Pathog. 10: e1004272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanver D., Tollot M., Schweizer G., Lo Presti L., Reissmann S., Ma L.S., Schuster M., Tanaka S., Liang L., Ludwig N., Kahmann R. (2017). Ustilago maydis effectors and their impact on virulence. Nat. Rev. Microbiol. 15: 409–421. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Lo Presti L., López Díaz C., Turrà D., Di Pietro A., Hampel M., Heimel K., Kahmann R. (2016). A conserved co-chaperone is required for virulence in fungal plant pathogens. New Phytol. 209: 1135–1148. [DOI] [PubMed] [Google Scholar]
- Lo Presti L., Lanver D., Schweizer G., Tanaka S., Liang L., Tollot M., Zuccaro A., Reissmann S., Kahmann R. (2015). Fungal effectors and plant susceptibility. Annu. Rev. Plant Biol. 66: 513–545. [DOI] [PubMed] [Google Scholar]
- Lorenz M.C., Heitman J. (1998). The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J. 17: 1236–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loubradou G., Brachmann A., Feldbrügge M., Kahmann R. (2001). A homologue of the transcriptional repressor Ssn6p antagonizes cAMP signalling in Ustilago maydis. Mol. Microbiol. 40: 719–730. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez P., Ljungdahl P.O. (2005). Divergence of Stp1 and Stp2 transcription factors in Candida albicans places virulence factors required for proper nutrient acquisition under amino acid control. Mol. Cell. Biol. 25: 9435–9446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluf G.A. (1997). Genetic regulation of nitrogen metabolism in the fungi. Microbiol. Mol. Biol. Rev. 61: 17–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason M.J., Fan G., Plath K., Zhou Q., Horvath S. (2009). Signed weighted gene co-expression network analysis of transcriptional regulation in murine embryonic stem cells. BMC Genomics 10: 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massonnet M., Morales-Cruz A., Figueroa-Balderas R., Lawrence D.P., Baumgartner K., Cantu D. (2018). Condition-dependent co-regulation of genomic clusters of virulence factors in the grapevine trunk pathogen Neofusicoccum parvum. Mol. Plant Pathol. 19: 21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matei A., Ernst C., Gunl M., Thiele B., Altmuller J., Walbot V., Usadel B., Doehlemann G. (2018). How to make a tumour: cell type specific dissection of Ustilago maydis-induced tumour development in maize leaves. New Phytol. 217: 1681–1695. [DOI] [PubMed] [Google Scholar]
- Matei A., Doehlemann G. (2016). Cell biology of corn smut disease-Ustilago maydis as a model for biotrophic interactions. Curr. Opin. Microbiol. 34: 60–66. [DOI] [PubMed] [Google Scholar]
- McCann M.P., Snetselaar K.M. (2008). A genome-based analysis of amino acid metabolism in the biotrophic plant pathogen Ustilago maydis. Fungal Genet. Biol. 45 (suppl. 1): S77–S87. [DOI] [PubMed] [Google Scholar]
- Mendoza-Mendoza A., Berndt P., Djamei A., Weise C., Linne U., Marahiel M., Vranes M., Kämper J., Kahmann R. (2009). Physical-chemical plant-derived signals induce differentiation in Ustilago maydis. Mol. Microbiol. 71: 895–911. [DOI] [PubMed] [Google Scholar]
- Merico D., Isserlin R., Stueker O., Emili A., Bader G.D. (2010). Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One 5: e13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzadi Gohari A., Ware S.B., Wittenberg A.H., Mehrabi R., Ben M’Barek S., Verstappen E.C., van der Lee T.A., Robert O., Schouten H.J., de Wit P.P., Kema G.H. (2015). Effector discovery in the fungal wheat pathogen Zymoseptoria tritici. Mol. Plant Pathol. 16: 931–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller A.N., Ziemann S., Treitschke S., Aßmann D., Doehlemann G. (2013). Compatibility in the Ustilago maydis-maize interaction requires inhibition of host cysteine proteases by the fungal effector Pit2. PLoS Pathog. 9: e1003177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller O., Schreier P.H., Uhrig J.F. (2008). Identification and characterization of secreted and pathogenesis-related proteins in Ustilago maydis. Mol. Genet. Genomics 279: 27–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadal M., Garcia-Pedrajas M.D., Gold S.E. (2010). The snf1 gene of Ustilago maydis acts as a dual regulator of cell wall degrading enzymes. Phytopathology 100: 1364–1372. [DOI] [PubMed] [Google Scholar]
- Navarathna D.H., Das A., Morschhäuser J., Nickerson K.W., Roberts D.D. (2011). Dur3 is the major urea transporter in Candida albicans and is co-regulated with the urea amidolyase Dur1,2. Microbiology 157: 270–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell R.J., et al. (2012). Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nat. Genet. 44: 1060–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ospina-Giraldo M.D., Mullins E., Kang S. (2003). Loss of function of the Fusarium oxysporum SNF1 gene reduces virulence on cabbage and Arabidopsis. Curr. Genet. 44: 49–57. [DOI] [PubMed] [Google Scholar]
- Park J., et al. (2008). FTFD: an informatics pipeline supporting phylogenomic analysis of fungal transcription factors. Bioinformatics 24: 1024–1025. [DOI] [PubMed] [Google Scholar]
- Pellier A.L., Laugé R., Veneault-Fourrey C., Langin T. (2003). CLNR1, the AREA/NIT2-like global nitrogen regulator of the plant fungal pathogen Colletotrichum lindemuthianum is required for the infection cycle. Mol. Microbiol. 48: 639–655. [DOI] [PubMed] [Google Scholar]
- Rabe F., Seitner D., Bauer L., Navarrete F., Czedik-Eysenberg A., Rabanal F.A., Djamei A. (2016). Phytohormone sensing in the biotrophic fungus Ustilago maydis - the dual role of the transcription factor Rss1. Mol. Microbiol. 102: 290–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redkar A., Matei A., Doehlemann G. (2017). Insights into host cell modulation and induction of new cells by the corn smut Ustilago maydis. Front. Plant Sci. 8: 899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redkar A., Hoser R., Schilling L., Zechmann B., Krzymowska M., Walbot V., Doehlemann G. (2015). A secreted effector protein of Ustilago maydis guides maize leaf cells to form tumors. Plant Cell 27: 1332–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke G., Heinze B., Schirawski J., Buettner H., Kahmann R., Basse C.W. (2008). Indole-3-acetic acid (IAA) biosynthesis in the smut fungus Ustilago maydis and its relevance for increased IAA levels in infected tissue and host tumour formation. Mol. Plant Pathol. 9: 339–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., Oshlack A. (2010). A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd J.J., et al. (2015). Transcriptome and metabolite profiling of the infection cycle of Zymoseptoria tritici on wheat reveals a biphasic interaction with plant immunity involving differential pathogen chromosomal contributions and a variation on the hemibiotrophic lifestyle definition. Plant Physiol. 167: 1158–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Arreguin J.A., Hernandez-Oñate M.A., León-Ramirez C.G., Ruiz-Herrera J. (2017). Transcriptional analysis of the adaptation of Ustilago maydis during growth under nitrogen fixation conditions. J. Basic Microbiol. 57: 597–604. [DOI] [PubMed] [Google Scholar]
- Schauwecker F., Wanner G., Kahmann R. (1995). Filament-specific expression of a cellulase gene in the dimorphic fungus Ustilago maydis. Biol. Chem. Hoppe Seyler 376: 617–625. [DOI] [PubMed] [Google Scholar]
- Scherer M., Heimel K., Starke V., Kämper J. (2006). The Clp1 protein is required for clamp formation and pathogenic development of Ustilago maydis. Plant Cell 18: 2388–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling L., Matei A., Redkar A., Walbot V., Doehlemann G. (2014). Virulence of the maize smut Ustilago maydis is shaped by organ-specific effectors. Mol. Plant Pathol. 15: 780–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schipper K. (2009). Charakterisierung eines Ustilago maydis Genclusters, das für drei neuartige sekretierte Effektoren kodiert. PhD dissertation (Marburg, Germany: Philipps Universität).
- Schirawski J., Böhnert H.U., Steinberg G., Snetselaar K., Adamikowa L., Kahmann R. (2005). Endoplasmic reticulum glucosidase II is required for pathogenicity of Ustilago maydis. Plant Cell 17: 3532–3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirawski J., et al. (2010). Pathogenicity determinants in smut fungi revealed by genome comparison. Science 330: 1546–1548. [DOI] [PubMed] [Google Scholar]
- Schuler D., Wahl R., Wippel K., Vranes M., Münsterkötter M., Sauer N., Kämper J. (2015). Hxt1, a monosaccharide transporter and sensor required for virulence of the maize pathogen Ustilago maydis. New Phytol. 206: 1086–1100. [DOI] [PubMed] [Google Scholar]
- Schulz B., Banuett F., Dahl M., Schlesinger R., Schäfer W., Martin T., Herskowitz I., Kahmann R. (1990). The b alleles of U. maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain-related motif. Cell 60: 295–306. [DOI] [PubMed] [Google Scholar]
- Schuster M., Schweizer G., Kahmann R. (2017). Comparative analyses of secreted proteins in plant pathogenic smut fungi and related basidiomycetes. Fungal Genet. Biol., 10.1016/j.fgb.2016.12.003. [DOI] [PubMed]
- Schuster M., Schweizer G., Reissmann S., Kahmann R. (2016). Genome editing in Ustilago maydis using the CRISPR-Cas system. Fungal Genet. Biol. 89: 3–9. [DOI] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbe D.S., Doehlemann G., Fernandes J., Walbot V. (2010). Maize tumors caused by Ustilago maydis require organ-specific genes in host and pathogen. Science 328: 89–92. [DOI] [PubMed] [Google Scholar]
- Smith D.G., Garcia-Pedrajas M.D., Gold S.E., Perlin M.H. (2003). Isolation and characterization from pathogenic fungi of genes encoding ammonium permeases and their roles in dimorphism. Mol. Microbiol. 50: 259–275. [DOI] [PubMed] [Google Scholar]
- Snetselaar K., McCann M. (2017). Ustilago maydis, the corn smut fungus, has an unusual diploid mitotic stage. Mycologia 109: 140–152. [DOI] [PubMed] [Google Scholar]
- Snoeijers S.S., Pérez-García A., Joosten M.H.A.J., De Wit P.J.G.M. (2000). The effect of nitrogen on disease development and gene expression in bacterial and fungal plant pathogens. Eur. J. Plant Pathol. 106: 493–506. [Google Scholar]
- Stirnberg A., Djamei A. (2016). Characterization of ApB73, a virulence factor important for colonization of Zea mays by the smut Ustilago maydis. Mol. Plant Pathol. 17: 1467–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Brefort T., Neidig N., Djamei A., Kahnt J., Vermerris W., Koenig S., Feussner K., Feussner I., Kahmann R. (2014). A secreted Ustilago maydis effector promotes virulence by targeting anthocyanin biosynthesis in maize. eLife 3: e01355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichmann B., Linne U., Hewald S., Marahiel M.A., Bölker M. (2007). A biosynthetic gene cluster for a secreted cellobiose lipid with antifungal activity from Ustilago maydis. Mol. Microbiol. 66: 525–533. [DOI] [PubMed] [Google Scholar]
- Thatcher L.F., Williams A.H., Garg G., Buck S.G., Singh K.B. (2016). Transcriptome analysis of the fungal pathogen Fusarium oxysporum f. sp. medicaginis during colonisation of resistant and susceptible Medicago truncatula hosts identifies differential pathogenicity profiles and novel candidate effectors. BMC Genomics 17: 860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium (2017). Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 45: D331–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma B.P., Bolton M.D., Clergeot P.H., DE Wit P.J. (2006). Nitrogen controls in planta expression of Cladosporium fulvum Avr9 but no other effector genes. Mol. Plant Pathol. 7: 125–130. [DOI] [PubMed] [Google Scholar]
- Tollot M., Assmann D., Becker C., Altmüller J., Dutheil J.Y., Wegner C.E., Kahmann R. (2016). The WOPR protein Ros1 is a master regulator of sporogenesis and late effector gene expression in the maize pathogen Ustilago maydis. PLoS Pathog. 12: e1005697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonukari N.J., Scott-Craig J.S., Walton J.D. (2000). The Cochliobolus carbonum SNF1 gene is required for cell wall-degrading enzyme expression and virulence on maize. Plant Cell 12: 237–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toruño T.Y., Stergiopoulos I., Coaker G. (2016). Plant-pathogen effectors: cellular probes interfering with plant defenses in spatial and temporal manners. Annu. Rev. Phytopathol. 54: 419–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turian G., Hamilton R.H. (1960). Chemical detection of 3-indolylacetic acid in Ustilago zeae tumors. Biochim. Biophys. Acta 41: 148–150. [DOI] [PubMed] [Google Scholar]
- Vollmeister E., Schipper K., Baumann S., Haag C., Pohlmann T., Stock J., Feldbrügge M. (2012). Fungal development of the plant pathogen Ustilago maydis. FEMS Microbiol. Rev. 36: 59–77. [DOI] [PubMed] [Google Scholar]
- Wahl R., Wippel K., Goos S., Kämper J., Sauer N. (2010). A novel high-affinity sucrose transporter is required for virulence of the plant pathogen Ustilago maydis. PLoS Biol. 8: e1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.D., Yan N., Wang Z.H., Zhang X.H., Zhang J.Z., Xue H.M., Wang L.X., Zhan Q., Xu Y.P., Guo D.P. (2017). RNA-seq analysis provides insight into reprogramming of culm development in Zizania latifolia induced by Ustilago esculenta. Plant Mol. Biol. 95: 533–547. [DOI] [PubMed] [Google Scholar]
- Young M.D., Wakefield M.J., Smyth G.K., Oshlack A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahiri A., Heimel K., Wahl R., Rath M., Kämper J. (2010). The Ustilago maydis forkhead transcription factor Fox1 is involved in the regulation of genes required for the attenuation of plant defenses during pathogenic development. Mol. Plant Microbe Interact. 23: 1118–1129. [DOI] [PubMed] [Google Scholar]
- Zeng F.S., Menardo F., Xue M.F., Zhang X.J., Gong S.J., Yang L.J., Shi W.Q., Yu D.Z. (2017). Transcriptome analyses shed new insights into primary metabolism and regulation of Blumeria graminis f. sp. tritici during conidiation. Front. Plant Sci. 8: 1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Horvath S. (2005). A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 4: article17. [DOI] [PubMed] [Google Scholar]
- Zheng Y., Kief J., Auffarth K., Farfsing J.W., Mahlert M., Nieto F., Basse C.W. (2008). The Ustilago maydis Cys2His2-type zinc finger transcription factor Mzr1 regulates fungal gene expression during the biotrophic growth stage. Mol. Microbiol. 68: 1450–1470. [DOI] [PubMed] [Google Scholar]