The SnRK1 kinase-controlled C/S1 bZIP transcription factor network initiates substantial transcriptional reprogramming to ensure plant survival under stress.
Abstract
Sustaining energy homeostasis is of pivotal importance for all living organisms. In Arabidopsis thaliana, evolutionarily conserved SnRK1 kinases (Snf1-RELATED KINASE1) control metabolic adaptation during low energy stress. To unravel starvation-induced transcriptional mechanisms, we performed transcriptome studies of inducible knockdown lines and found that S1-basic leucine zipper transcription factors (S1-bZIPs) control a defined subset of genes downstream of SnRK1. For example, S1-bZIPs coordinate the expression of genes involved in branched-chain amino acid catabolism, which constitutes an alternative mitochondrial respiratory pathway that is crucial for plant survival during starvation. Molecular analyses defined S1-bZIPs as SnRK1-dependent regulators that directly control transcription via binding to G-box promoter elements. Moreover, SnRK1 triggers phosphorylation of group C-bZIPs and the formation of C/S1-heterodimers and, thus, the recruitment of SnRK1 directly to target promoters. Subsequently, the C/S1-bZIP-SnRK1 complex interacts with the histone acetylation machinery to remodel chromatin and facilitate transcription. Taken together, this work reveals molecular mechanisms underlying how energy deprivation is transduced to reprogram gene expression, leading to metabolic adaptation upon stress.
INTRODUCTION
Mitochondrial respiration provides most of the cellular energy used by eukaryotic organisms. Sugars, the predominant substrates in this process, are stored to ensure a constant energy supply. In photosynthetic organisms such as plants, this storage function is performed by starch, providing resources during the night (Stitt and Zeeman, 2012). Nevertheless, if plants are subjected to a short extension of the dark period or are cultivated under low-light conditions, this energy buffer is rapidly depleted, resulting in energy starvation (Usadel et al., 2008) and the induction of alternative pathways to generate ATP from noncarbohydrate resources such as proteins, fatty acids, or chlorophyll (Araújo et al., 2011a, 2011b). These alternative routes are therefore crucial for plant survival under stress. Along these lines, several studies have proposed a relationship between energy availability and stress tolerance, survival, cell growth, and longevity (Baena-González and Sheen, 2008). Hence, regulatory circuits are required that precisely match energy supply and metabolic demand.
The plant kinases SnRK1 (Snf1-RELATED KINASE1), as well as their homologs AMPK (AMP-ACTIVATED PROTEIN KINASE) in mammals and Snf1 (SUCROSE-NON-FERMENTING1) in yeast, have emerged as central metabolic regulators linking nutrient and energy availability with growth and development (Hardie, 2015; Li and Sheen, 2016). Although the sensing of metabolic signals by SnRK1s is not fully understood (Crozet et al., 2014; Broeckx et al., 2016), these kinases are proposed to support cellular functions ranging from adaptation to stress (e.g., oxygen-limiting conditions upon flooding) to various aspects of plant development (e.g., vegetative-to-reproductive phase transitions). Transcriptome studies of Arabidopsis thaliana protoplasts overexpressing a SnRK1 kinase have provided insights into their regulatory potential (Baena-González et al., 2007). The principal processes activated by SnRK1s are major energy-preserving catabolic pathways related to carbohydrate, lipid, and amino acid metabolism, as well as autophagy or general stress responses. In contrast, a large set of energy-consuming processes is repressed, such as ribosome biogenesis, protein translation, and cell wall biosynthesis. Supporting its major function in energy homeostasis, SnRK1-regulated genes were found to be positively correlated with stress and starvation-associated genes or negatively correlated with genes activated by sucrose or glucose (Baena-González and Sheen, 2008).
SnRK1/AMPK/Snf1 kinases operate as heterotrimeric complexes consisting of α-catalytic and β- and γ-regulatory subunits. In Arabidopsis, the catalytic subunits are encoded by a small gene family with three members, namely, SnRK1α1 (AKIN10), SnRK1α2 (AKIN11), and SnRK1α3 (AKIN12). Whereas the latter is hardly expressed, the others possess partially redundant functions, although they differ considerably in their expression profiles (Williams et al., 2014; Baena-González et al., 2007). A stable double mutant in SnRK1α1 and SnRK1α2 appears to be lethal.
SnRK1s control energy metabolism through direct regulation of enzymes, transcription factors (TFs) and microRNAs (Confraria et al., 2013; Nukarinen et al., 2016; Sheen, 2014). Although several TF targets of SnRK1 have been proposed, the detailed regulatory mechanisms of downstream gene regulation remain largely elusive (Kleinow et al., 2009; Lin et al., 2014; O’Brien et al., 2015; Tsai and Gazzarrini, 2012). Basic leucine zipper (bZIP) TFs of group S1 (bZIP1, bZIP2, bZIP11, bZIP44, and bZIP53) (Jakoby et al., 2002) were proposed to be targets of SnRK1s, although direct phosphorylation has not been demonstrated (Baena-González et al., 2007). Indeed, S1-bZIPs play an important role in transcriptional reprogramming of carbon and nitrogen metabolism in response to stress, growth, and hormonal control as well as during various developmental processes such as seed maturation (Alonso et al., 2009; Dietrich et al., 2011; Hartmann et al., 2015; Ma et al., 2011; Weiste et al., 2017; Weiste and Dröge-Laser, 2014; Baena-González et al., 2007). Importantly, dark-induced (DIN) genes regulated by SnRK1s, such as ASPARAGINE SYNTHETASE1 (ASN1), are directly controlled by S1-bZIPs via their G-box cis-elements. Notably, due to the high level of redundancy among these bZIPs, most of the data were derived from overexpression rather than loss-of-function studies (Baena-González et al., 2007; Dietrich et al., 2011).
S1-bZIPs are controlled transcriptionally and/or posttranscriptionally by sucrose, indicating that their function is highly correlated with the energy status of the cell (Weltmeier et al., 2009; Wiese et al., 2004). Moreover, S1-bZIPs preferentially form heterodimers with group C bZIPs (bZIP9, bZIP10, bZIP25, and bZIP63) (Ehlert et al., 2006; Weltmeier et al., 2006). Of these, bZIP63 controls gene expression in response to starvation and was recently identified as the first in vivo TF target of SnRK1 (Mair et al., 2015). SnRK1 phosphorylates bZIP63 on three serine (Ser) residues, promoting the formation of C/S1 heterodimers and, hence, constituting an important regulatory mechanism in gene expression control.
Taken together, although several players in the plant starvation response have been proposed, many open questions remain. Do S1-bZIPs transduce SnRK1 responses? If so, what are the target genes and how is their expression related to the plant’s adaptation to low energy stress? Do S1-bZIPs act redundantly or do they control specific functions? If S1-bZIPs are not directly targeted by the SnRK1 kinase, how do they transduce information about cellular energy supply? Finally, how do C/S1 heterodimers control transcription?
In this study, we devised a combinatorial strategy using genetics, genomics, and cell-based analyses to dissect the signaling functions of SnRK1 and S1-bZIP in response to extended darkness, which mimics low energy stress in plants. We uncovered a distinct overlap between the genome-wide transcriptional responses exerted by SnRK1s and S1-bZIPs, which supports the view that S1-bZIPs act downstream of SnRK1s. Strikingly, both signaling components control the expression of genes involved in branched-chain amino acid (BCAA) catabolism, an alternative respiratory pathway that supports respiration under carbohydrate-limiting conditions. ELECTRON-TRANSFER FLAVOPROTEIN: UBIQUINONE OXIDO-REDUCTASE (ETFQO) is an important component of this alternative pathway in both plants and mammals (Watmough and Frerman, 2010; Ishizaki et al., 2005, 2006). Based on comprehensive promoter analysis, we propose a mechanism by which starvation-induced C/S1-bZIP heterodimers recruit SnRK1 directly to the ETFQO promoter, which in turn leads to altered histone acetylation and subsequently activates transcription. Taken together, our data provide a detailed mechanistic model linking SnRK1-mediated low energy signaling to gene regulation and cellular adaptation.
RESULTS
SnRK1s and S1-bZIPs Control Shared and Distinct Sets of Genes in Response to Short-Term Extended Darkness
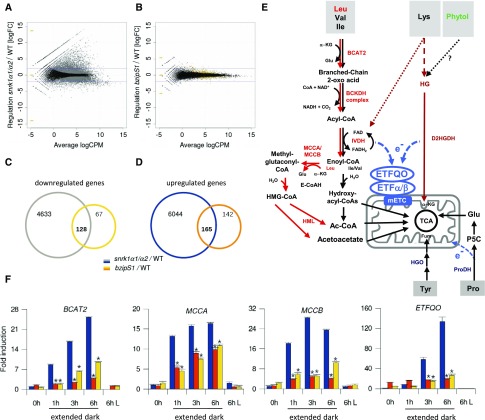
S1-bZIPs are thought to mediate transcriptional responses driven by SnRK1s upon energy deprivation. To decipher the SnRK1-S1-bZIP signaling network, we performed genome-wide expression profiling of wild-type, snrk1α1/α2, and bzipS1 knockdown Arabidopsis lines under starvation conditions provoked by 6 h of extended darkness (Figures 1A to 1D).
Figure 1.
SnRK1 and S1-bZIPs Control Shared and Distinct Sets of Genes in Response to Extended Darkness.
(A) and (B) RNA-seq transcriptome analysis of rosette leaves derived from wild-type and snrk1α1/α2 (A) or wild-type and bzipS1 (B) plants. Experiments were performed in triplicate using Est induction conditions as outlined in Methods. Smear plots of DEGs identified after 6 h of extended darkness. Gray and yellow dots represent genes with significantly differential expression with p-adjust (“BH correction”) < 0.01. Blue lines are at logFC = ±2. CPM, counts per million.
(C) and (D) Venn diagram displaying the number of down- and upregulated genes in snrk1α1/α2 (gray and blue) and bzipS1 (yellow and orange). The respective overlap provides the number of DEGs shared by both mutants. No logFC filter was applied (gene lists are given in Supplemental Data Set 1).
(E) Schematic representation of alternative pathways feeding into the mETC. Degradation of BCAAs (Leu, Val, and Ile). The sub-branch mediating Leu catabolism is marked in red. α-KG, α-ketoglutarate; BCKDH, BRANCHED-CHAIN KETOACID DEHYDROGENASE; IVDH, ISOVALERYL-COA DEHYDROGENASE; E-CoAH, ENOYL-COA HYDRATASE; HMG-CoA, 3-hydroxy-3-methyl-glutaryl-CoA; HML, 3-HYDROXYL-3- METHYLGLUTARYL-COA LYASE; Fum, fumarate; Glu, glutamate. Pro catabolism: P5C, Pyrroline-5-Carboxylate. Dotted lines: Phytol derived from thylakoid degradation is most likely not a source for the ETF/ETFQO pathway; Lys degradation pathway is not fully established (Hildebrandt et al., 2015; Peng et al., 2015).
(F) RT-qPCR validation of the expression of BCAT2, ETFQO, MCCA, and MCCB in 4-week-old wild-type (blue), snrk1α1/α2 (orange), and bzipS1 (yellow) rosette leaves. 0 h is defined as the end of the dark period; 1, 3, and 6 h time points correspond to extended dark, and “6 h L” refers to plants cultivated for 6 h in the light. Given are mean expression levels (±sd); (n = 3) relative to the wild type at 0 h. Student’s t test of the wild type at the same time point, *P < 0.01.
As constitutive snrk1α1/α2 double mutants are lethal (Baena-González et al., 2007), we generated an inducible knockdown line by transforming a snrk1α1 T-DNA insertion line with a β-estradiol (Est)-inducible artificial microRNA (amiRNA), which specifically targets SnRK1α2. Given the partial redundancy of S1-bZIPs, a multiple loss-of-function approach was required to target all five members of this subgroup. To this end, we generated bzipS1 by transforming a bzip1/bzip53 T-DNA insertion line with an Est-inducible amibZIP2/11/44 construct (Weiste and Dröge-Laser, 2014). The molecular characterization of these plant lines and the conditions of Est treatment are provided in Supplemental Figure 1.
RNA sequencing (RNA-seq) experiments comparing snrk1α1/α2 plants and the wild type (both after 6 h of extended darkness) showed strong alterations in their transcriptional responses. Applying a filter (p-adjust < 0.01; log fold change [logFC] >2), a total of 2717 and 747 genes were differentially up- and downregulated, respectively (Supplemental Data Sets 1D and 1E). Processes controlled by SnRK1s were similar to those previously identified by overexpressing SnRK1α1 in Arabidopsis protoplasts (Baena-González et al., 2007). A comparison of both data sets and GO (Gene Ontology) analysis are provided in Supplemental Data Set 2. For example, the snrk1α1/α2 knockdown line was unable to induce catabolic pathways that provide alternative sources of energy and to repress highly energy-demanding anabolic processes, such as ribonucleoprotein complex biogenesis.
The impact of S1-bZIPs on gene expression in response to extended darkness was less pronounced. In particular, 92% of the differentially expressed genes (DEGs) showed only minor changes in expression (logFC +2 to −2) (Figure 1B). This finding may be explained by the residual presence of bZIP2, bZIP11, and bZIP44 in the bzipS1 plants, as the use of Est-inducible amiRNA leads only to partially reduced bZIP expression (Supplemental Figure 1B). Hence, for subsequent analyses, we extracted all DEGs (p-adjust < 0.01) without further selection by FC magnitude. Compared with the wild type, overall, 307 and 195 genes were up- or downregulated, respectively, in the bzipS1 lines (Supplemental Data Sets 1F and 1G). Among the DEGs, 293 were regulated by both SnRK1s and S1-bZIPs (Figures 1C and 1D; Supplemental Data Sets 1H and 1I). Nevertheless, 209 genes were specifically deregulated only in bzipS1 plants, indicating that other pathways besides SnRK1 signaling converge on S1-bZIPs. Of the genes commonly downregulated in snrk1α1/α2 and bzipS1 knockdown plants, 86% were induced by extended dark treatment in wild-type plants, as determined in a comparison with publicly available microarray data (Supplemental File 1). GO analyses of shared DEGs revealed an overrepresentation of catabolic processes for downregulated genes and enrichment in anabolic processes with respect to upregulated genes (Supplemental Figures 2B to 2E and Supplemental Data Sets 1J and 1K).
Genes Involved in Starvation-Induced BCAA Degradation Are Tightly Regulated by SnRK1s and S1-bZIPs
Energy-limiting conditions induce alternative metabolic pathways to generate ATP from cellular resources (Araújo et al., 2011b; Hildebrandt et al., 2015; Hörtensteiner and Kräutler, 2011; Hüdig et al., 2015; Kunz et al., 2009; Miyashita and Good, 2008; Barros et al., 2017). In this respect, RNA-seq data obtained with the bzipS1 lines showed changes specifically in genes involved in amino acid catabolism, but not in other alternative pathways such as lipid or chlorophyll degradation. Importantly, several genes involved in degradation of the BCAAs Leu, Val, and Ile (Supplemental Data Set 1; Figure 1E) were significantly downregulated after 6 h of starvation. We extracted selected DEGs from the RNA-seq data set and validated their expression pattern via detailed RT-qPCR time-course experiments. Within 6 h of extended darkness, BRANCHED CHAIN TRANSAMINASE2 (BCAT2), METHYLCROTONYL-COA CARBOXYLASE (MCCA and MCCB), and ETFQO were strongly induced in wild-type plants, whereas their expression was significantly reduced in snrk1α1/α2 and bzipS1 (Figure 1F). To further support our hypothesis, we grew the plants under low-light conditions, which provided an additional experimental system to mimic low-energy related environmental conditions that naturally occur during the plant life cycle (Supplemental Figure 3). Transcription of the set of BCAA catabolic genes (BCAT2, IVDH, MCCA, MCCB, and ETFQO) was found to be induced after a shift to low light conditions and importantly, was partially impaired in the bzipS1 knockdown line. In line with these findings, tight coexpression of BCAA catabolic genes was observed in public transcriptome data sets (Supplemental Figure 4A). Gene induction correlated specifically with stress situations corresponding to energy limitation (e.g., extended darkness, low light, pathogen infection, and hypoxia). In contrast, this pathway was not responsive to stresses in general (e.g., drought and cold) and was not induced when carbohydrates were present.
Besides BCAAs, other amino acids can also be used to fuel the mitochondrial electron transport chain (mETC) via ETF/ETFQO during starvation (Figure 1E). In line with public expression data (Araújo et al., 2010, 2011a; 2011b; Engqvist et al., 2011; Hüdig et al., 2015), the central gene in Lys catabolism D2HGDH (D2-HYDROXYGLUTARATE DEHYDROGENASE), was not found among the DEG in the RNA-seq data set, as it was neither coregulated with BCAA catabolic genes nor controlled by S1-bZIPs when analyzed by RT-qPCR (Supplemental Figure 3). These data support the assumption that S1-bZIPs specifically control BCAA, but not Lys catabolic genes. However, based on the RNA-seq data, a broader impact on amino acid catabolism can be observed due to the regulation of Tyr (HOMOGENTISATE 1,2-DIOXYGENASE [HGO]) and Pro (PROLINE DEHYDROGENASE1 [ProDH1]) catabolism (Supplemental Data Set 1).
Gene regulation by TFs can be accomplished indirectly or directly by binding to target promoters. Mining the genome-wide in vitro DNA binding data sets by O’Malley et al. (2016), we found that bZIP11 or bZIP2 directly interact with most promoters of BCAA catabolic genes, which are activated by energy starvation (Supplemental Figures 4A and 4B). However, there were a few exceptions, such as IVDH, where no correlation between starvation-induced gene activation and bZIP binding was observed, which might be due to distinct differences in regulation. Using chromatin immunoprecipitation coupled to PCR (ChIP-PCR), we confirmed the in vivo binding of bZIP2 to selected BCAA catabolic genes (Supplemental Figure 5). In contrast, promoters of genes that do not respond to energy-limiting conditions, such as ETFα, were not bound by bZIP2 in vitro or in vivo (Supplemental Figures 3 to 5). Moreover, we confirmed the direct in vivo binding of bZIP2 to ProDH1 and HGO, indicating that the BCAA, Pro, and Tyr metabolic pathways are directly targeted by group S1-bZIPs. Indeed, the latter were recently proposed to be regulated by C/S1-bZIPs (Hartmann et al., 2015; Dietrich et al., 2011).
Metabolic Studies of snrk1α1/α2 and bzipS1 Lines Reveal Shared Alterations in Primary Metabolism
Our transcriptome analyses suggested that SnRK1 and S1-bZIPs have an impact on primary metabolism, particularly amino acid metabolism. We therefore quantified primary metabolites in wild-type, snrk1α1/α2, and bzipS1 plants that were grown side-by-side with the plants used for RNA-seq studies (Supplemental Figures 6A and 6B and Supplemental Data Set 3).
Consistent with its crucial function as a central metabolic regulator, the repression of SnRK1 dramatically affected steady state levels of many primary metabolites, whereas changes in bzipS1 plants were less pronounced. In both plant lines, the Glc-to-Suc ratio was elevated, supporting the notion that SnRK1 and S1-bZIPs have a major impact on carbohydrate metabolism. This assumption was further underlined by an increase in myo-inositol and trehalose levels in these lines. Glycolytic metabolite levels remained unchanged, indicating that glycolysis was functional. We detected a decrease in maltose levels only in the snrk1α1/α2 line. Specifically, the content of tricarboxylic acid (TCA) cycle intermediates was strongly affected in snrk1α1/α2 plants, as malate, fumarate, and succinate levels increased, whereas citrate levels decreased. These findings reflect the crucial function of SnRK1 in metabolic control. This might be explained by progressive upregulation of the TCA cycle during extended darkness to compensate for the reduced availability of respiratory substrates. However, it should be noted that no transcriptional changes in TCA-related genes were observed in the RNA-seq data set.
Whereas etfqo plants also displayed increased levels of the TCA cycle intermediates fumarate, succinate, and malate (Ishizaki et al., 2005), a limited change was observed in bzipS1. Reduced expression of BCAA catabolic genes should be reflected in the measured amino acid levels. Indeed, snrk1α1/α2 and particularly bzipS1 plants showed increased Leu levels but no changes in Ile or Val levels. This might be due to a major impact of S1-bZIPs on gene regulation related to Leu catabolism (MCCA and MCCB) (Figure 1E). Previous studies on amino acid levels in etfqo and mutants of BCAA catabolic genes revealed highly increased levels of BCAA as well as Asn, Arg, Trp, and Phe (Ishizaki et al., 2006; Peng et al., 2015). It should be noted that the moderate changes observed in bzipS1 plants may be due to the partial knockdown of the transcriptional regulators and that measurements were performed soon after 6 h dark treatment instead of several days in the etfqo mutant. Consistent with the transcription data, Lys levels were unaltered in both snrk1α1/α2 and bzipS1 plants. Whereas an increase in Pro levels was observed only in snrk1α1/α2 plants, reduced expression of the HGO gene was reflected in the Tyr levels in both snrk1α1/α2 and bzipS1 plants. Furthermore, only in snrk1α1/α2, the levels of amino acid in the glutamate and aspartate families increased, supporting the pronounced impact of the upstream kinase on the TCA cycle.
In line with the transcriptome data, analyses of primary metabolites suggest that SnRK1s and S1-bZIPs support adaptation processes upon energy deprivation. Whereas SnRK1 dramatically influences primary metabolism, S1-bZIPs execute a limit but well-defined subset of SnRK1 responses.
SnRK1s and S1-bZIPs Regulate Mitochondrial Metabolism, Particularly via the Electron Transfer Protein ETFQO
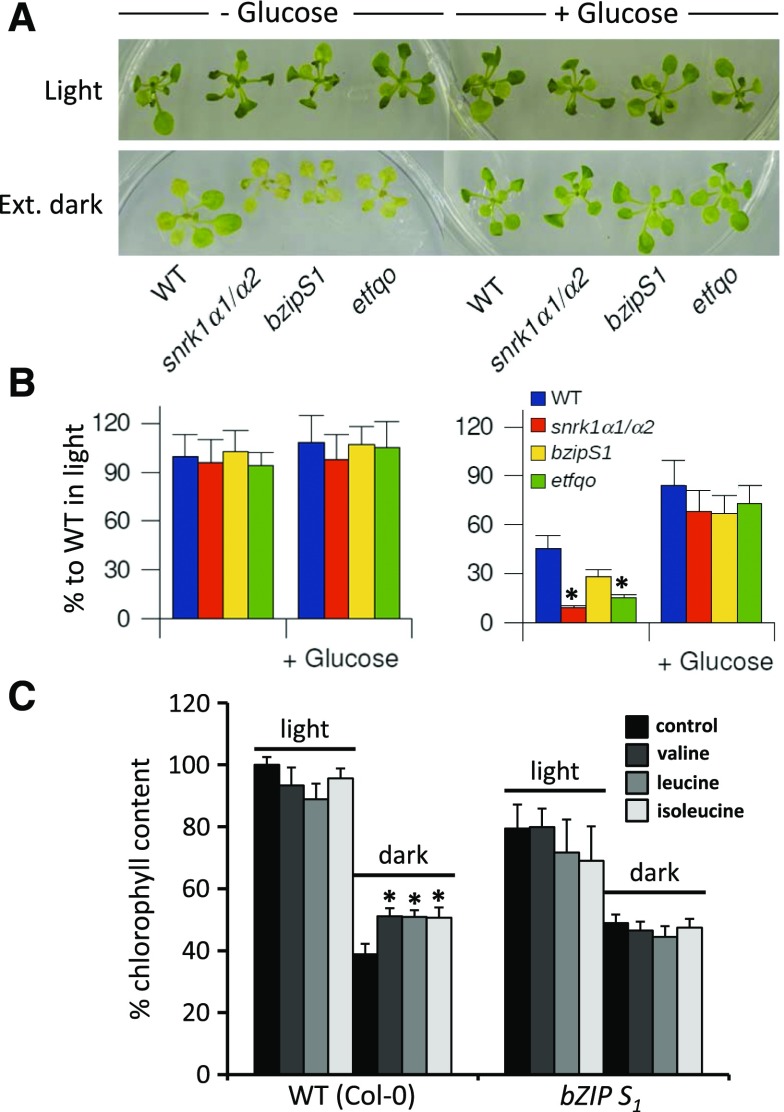
To explore the functional connection between SnRK1s, S1-bZIPs, and energy metabolism on the level of gene regulation, we focused on one potential target as a model gene. We selected the ETFQO gene for several reasons: (1) ETFQO has been found to be starvation induced (Baena-González et al., 2007; Cookson et al., 2016); (2) its expression is strongly impaired in snrk1α1/α2 and bzipS1 plants; (3) ETFQO is a single-copy gene, facilitating functional mutant analysis; and (4) etfqo mutants have been intensively studied, displaying an impaired response to extended darkness and an early senescence phenotype (Ishizaki et al., 2006, 2005). Under the normal day/night regime, we observed no visible differences between snrk1α1/α2, bzipS1, and etfqo compared with wild-type plants using chlorophyll content as a readout for plant viability and survival (Figures 2A and 2B). However, 6 d of extended darkness resulted in a strong reduction in chlorophyll content in wild-type plants. Similarly, all three mutant and knockdown lines displayed even less chlorophyll and reduced growth compared with the wild type, as previously described for etfqo and several mutants with disrupted alternative metabolic pathways in the starvation response (Ishizaki et al., 2005, 2006).
Figure 2.
Phenotypical Responses to Extended Darkness.
(A) Phenotypes of wild-type, snrk1α1/α2, bzipS1, and etfqo plants. Plants were grown for 2 weeks on MS medium with Est under a 12/12-h light/dark cycle without (left) or with supplementation of 2% Glc (right). This culture was continued (upper panel, light) or the plants were transferred to darkness for 6 d (lower panel, ext. dark).
(B) Chlorophyll measurements of the plants shown in (A) after 6 d of treatment: wild type (blue), snrk1α1/α2 (red), bzipS1 (yellow), and etfqo (green). Given are mean values from chlorophyll measurements (±se; n > 17) expressed as percentage to wild-type plants growing under 12/12-h light/dark photoperiod. Student’s t test of the wild type, *P < 0.05.
(C) Plant survival in darkness after feeding with BCAA. Wild-type and bzipS1 plants were grown on MS medium for 2 weeks. After transfer to medium supplemented with 25 μM of the indicated amino acid, plants were grown for 10 d under a normal day-light regime (light) or constant darkness (dark). Given is the chlorophyll content relative to wild-type plants in the light (100%). Student’s t test of untreated plants, *P < 0.05.
To further strengthen the link between group S1-bZIPs, the BCAA catabolic pathway, and electron transfer via ETFQO, we performed metabolite-feeding experiments. The application of 2% Glc as carbon source rescued plant growth and chlorophyll content in all genotypes, indicating that SnRK1, S1-bZIPs, and ETFQO are particularly important under starvation conditions. Whereas exogenously supplied Leu, Val, or Ile increased plant survival, this was not the case for bzipS1 plants (Figure 2C). Altogether, these phenotypic studies strongly support the impact of the BCAA catabolic pathway on energy homeostasis and plant survival during starvation and highlight its dependency on SnRK1-S1-bZIP signaling.
The ETFQO Promoter Is Directly Regulated by S1-bZIPs
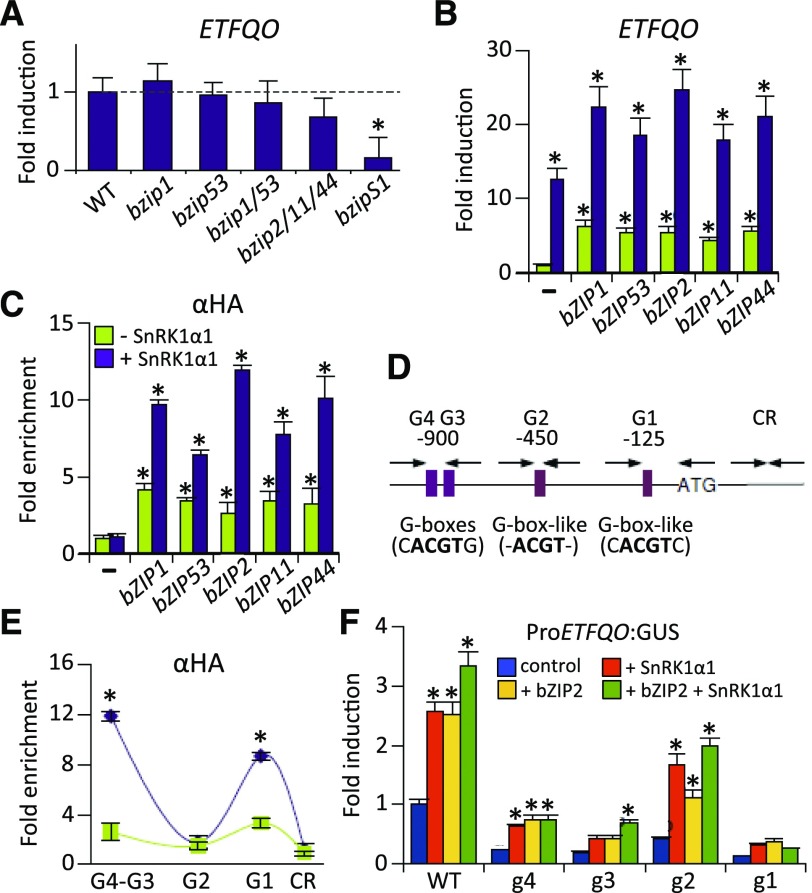
Based on the results of the transcriptome analysis, the ETFQO gene is perfectly suited to study the mechanistic connection between SnRK1s, S1-bZIPs, and mitochondrial-dependent energy metabolism. The expression of ETFQO was only slightly altered in single and multiple S1-bZIP mutants, although it was strongly impaired in the quintuple bzipS1 background (Figure 3A). This is likely due to functional redundancy between S1-bZIPs, as previously described (Dietrich et al., 2011). Consistent with this notion, protoplast transactivation assays using RT-qPCR to quantify ETFQO transcript levels demonstrated that overexpression of any of the five S1-bZIPs was sufficient to activate the target gene (Figure 3B). Interestingly, coexpression with SnRK1α1 strongly enhanced ETFQO transcript abundance and the activation potential of all group S1-bZIPs. These results are in line with the observation that overexpression of SnRK1α1 in protoplasts mimics energy deprivation conditions (Baena-González et al., 2007).
Figure 3.
Group S1-bZIPs Directly Regulate the Expression of ETFQO.
(A) RT-qPCR experiments to assess the expression of ETFQO in rosette leaves of different S1-bZIP loss-of-function mutant plants. Expression was normalized to that of wild-type plants exposed to 6 h of extended darkness. Student’s t test of the wild type (n = 3), *P < 0.05.
(B) RT-qPCR experiment to assess the transcript abundance of ETFQO in response to overexpression of S1-bZIPs in leaf-derived protoplasts (green) or after coexpression with SnRK1α1 (purple). Values are normalized to controls transfected with the reporter only (–).
(C) ChIP-PCR in protoplasts: binding of HA-tagged S1-bZIPs to the ETFQO promoter. An HA-tag antibody and primers specific for the G3-G4 sites (see [D]) were used. In (B) and (C), fold enrichment is given relative to the nontransformed control (−) (mean ± sd, n = 3). Student’s t test of promoter background activity, *P < 0.05.
(D) Schematic representation of the ETFQO promoter: Positions of G-box and G-box-like motives and primer binding sites are highlighted relative to the start of translation (ATG). CR: control corresponding to the coding region.
(E) ChIP-PCR of HA-bZIP2 in Arabidopsis protoplasts using the indicated ETFQO-specific primers and an α-HA-specific antibody to detect binding at the G-boxes G1-G4. Fold enrichment is given relative to the nontransformed control (mean values ± sd, n = 3), Student’s t test, *P < 0.01.
(F) Activation of mutated versions (g1–g4) of the ProETFQO:GUS reporter by SnRK1α1 (red), HA-bZIP2 (yellow), or a combination of both (green). Given values are mean values (±sd; n = 3) relative to transfections with reporter vector only (control, blue). Student’s t test of respective control samples, *P < 0.05.
S1-bZIPs are thought to preferentially bind to G-boxes (CACGTG) and ACGT-core sequences (O’Malley et al., 2016). Analysis of the ETFQO promoter revealed the presence of a G-box like (CACGTC) motif at –125 bp (G1), an ACGT-core motif at –450 bp (G2), and tandemly repeated perfect G-boxes around –900 bp (G3 and G4) relative to the translational start site. ChIP-PCR performed in transiently transformed protoplasts demonstrated that all S1-bZIPs directly bind to the ETFQO promoter (Figures 3C and 3D) and that their recruitment could be enhanced by coexpression with SnRK1α1. The finding that overexpression of TFs can lead to less specific binding could explain why all five S1-bZIPs bound to the ETFQO promoter in our experiments. Despite the ability of all S1-bZIPs to activate the expression of ETFQO, bZIP2 alone and in cooperation with SnRK1α1 showed the strongest binding and activation potential in protoplasts. Hence, we performed promoter-scanning ChIP experiments to fine-map bZIP2 binding to the ETFQO promoter. This analysis confirmed the binding of HA-tagged bZIP2 to the promoter in the region of the G1 and G3-4 sites (Figure 3E). Due to their close vicinity, G3 and G4 elements could not be discriminated by ChIP analysis. Importantly, this binding was strongly enhanced by SnRK1α1. These findings were supported by mutational analysis targeting the G-boxes in the ETFQO promoter. Indeed, the integrity of the G1, G3, and G4 binding motifs was necessary for the activation of the ProETFQO:GUS reporter by both bZIP2 and SnRK1 (Figure 3F). The G2 element had no or only a very limited impact on bZIP binding and SnRK1-dependent ETFQO activation. Taken together, both SnRK1 and S1-bZIP-mediated activation of the ETFQO promoter are facilitated via G-boxes.
Multiple bZIP Heterodimers of Group S1 and Group C Control the ETFQO Promoter
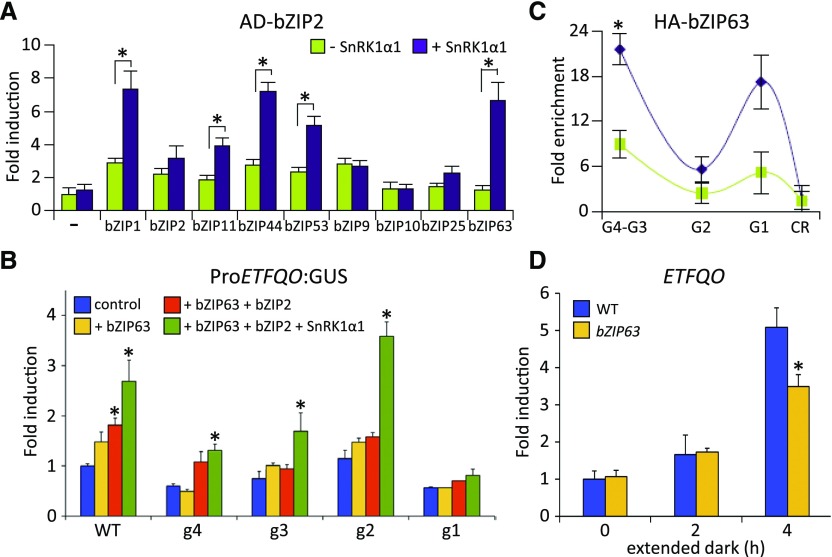
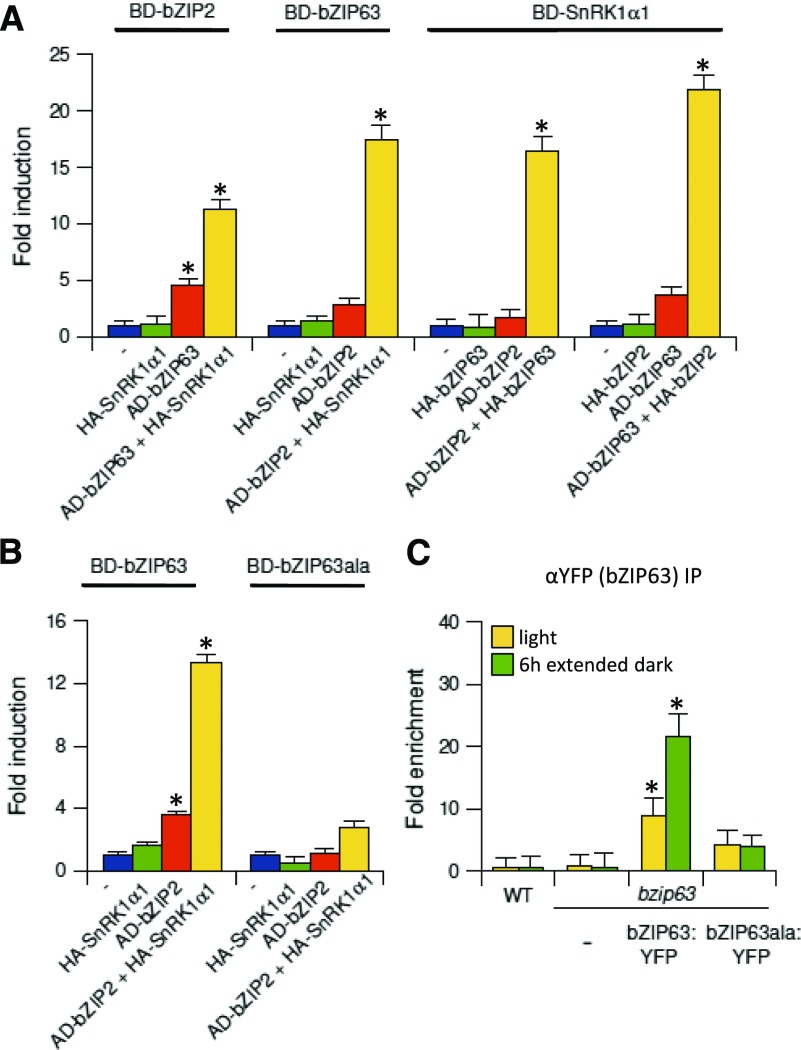
Using a semiquantitative protoplast two-hybrid (P2H) screening, we detected S1-specific dimer formation, which further increased in response to coexpression with SnRK1 (Supplemental Figure 7). In line with previous studies, S1-bZIPs preferentially formed heterodimers with C-bZIPs (Ehlert et al., 2006). As bZIP2 led to the strongest activation of the ETFQO promoter, we selected this S1 member for quantitative P2H assays (Figure 4A). Importantly, coexpression with SnRK1α1 strongly increased interactions with bZIP1, bZIP11, bZIP44, and bZIP53 (group S1), but particularly bZIP63 (group C). As the latter is directly phosphorylated and controlled by SnRK1 (Mair et al., 2015), we further focused on this member. Indeed, bZIP63 was able to induce the activation of the ProETFQO:GUS reporter in an SnRK1α1-dependent manner (Figure 4B). Mapping of promoter binding sites and ChIP-PCR studies in protoplasts revealed the same ETFQO promoter-specific binding pattern for bZIP63 and bZIP2 (Figures 4B and 4C). RT-qPCR analysis of a bzip63 mutant revealed a 30% decrease in starvation-induced ETFQO transcription, supporting a partial impact in the starvation response (Figure 4D). Taken together, in addition to S1-specific dimers, C/S1 heterodimers bind to the ETFQO promoter. As heterodimerization is increased by SnRK1α1 phosphorylation, these bZIP dimers provide additional input into ETFQO transcription.
Figure 4.
Regulation of the ETFQO Gene by bZIP Dimers of Group C and S1.
(A) P2H assays. Changes in the dimerization properties of BD-bZIP2 with AD-fusions of the bZIP TFs indicated (green), depending on coexpression with SnRK1α1 (purple). Given are mean values (±sd; leaf-derived protoplasts, n = 3) relative to transfections with reporter only (–), Student’s t test, *P < 0.05. BD, Gal4-DNA binding domain; AD, Gal4-DNA activation domain.
(B) Activation of mutated versions (g1–g4) of the ProETFQO:GUS reporter by HA-bZIP63 (yellow), HA-bZIP2 + HA-bZIP63 (orange), and HA-bZIP2 + HA-bZIP63 + SnRK1α1 (green). Given values are mean values (±sd; n = 3) relative to transfections with reporter vector only (control, blue). Student’s t test, *P < 0.05.
(C) ChIP-PCR of HA-bZIP63 in Arabidopsis protoplasts using the indicated ETFQO-specific primers and an α-HA-specific antibody to detect binding at the G-boxes G1-G4. CR: control corresponding to the coding region. Given values are mean values (±sd; n = 3) relative to enrichment of the control region, Student’s t test, *P < 0.01.
(D) RT-qPCR analysis of ETFQO expression in the wild type and bzip63 after extended darkness for the time points indicated (n = 3). Student’s t test of the wild type at the respective time points, *P < 0.05.
SnRK1α1, bZIP2, and bZIP63 Proteins Physically Interact
Although S1-bZIPs are crucial for the induction of ETFQO, they are not directly phosphorylated by and do not interact with SnRK1α1. However, bilateral physical interaction between SnRK1 and bZIP63 or bZIP63 and S1-bZIPs (Ehlert et al., 2006; Kang et al., 2010) has been well documented using various in vitro and in vivo methods. Hence, it is tempting to speculate that a ternary complex between all three partners may be formed. Using a protoplast three-hybrid (P3H) approach, we demonstrated that the interaction of bZIP2 and bZIP63 is promoted when coexpressed with SnRK1α1 (Figure 5A). Moreover, the previously described interaction between bZIP63 and SnRK1α1 was also stronger in the presence of bZIP2. Surprisingly, we observed an interaction between bZIP2 and SnRK1α1, but only when they were coexpressed with bZIP63. These results suggest that bZIP63 forms a bridge between bZIP2 and SnRK1 proteins. Importantly, dimerization between bZIP63 and bZIP2 was largely abolished when a mutated version of bZIP63 (bZIP63ala) was assayed using P3H analysis (Figure 5B). This bZIP63 variant harbored Ser-to-Ala mutations at three amino acid positions (29, 294, and 300) that are specifically phosphorylated by SnRK1α1 in vivo. In line with these results, ChIP-PCR experiments demonstrated that the integrity of the SnRK1α1 phosphorylation sites within the bZIP63 protein is indispensable for its recruitment to the ETFQO promoter in planta (Figure 5C). For this experiment, we complemented a bzip63 mutant line with a genomic ProbZIP63:bZIP63:YFP construct (bZIP63:YFP) or the corresponding mutant harboring the Ser-to-Ala exchanges (bZIP63ala:YFP) (Mair et al., 2015). Taken together, these data suggest that an SnRK1-bZIP63-bZIP2 complex forms on the ETFQO promoter, which depends on the SnRK1-specific phosphorylation of bZIP63.
Figure 5.
Complex Formation between SnRK1α1, bZIP2, and bZIP63.
(A) P3H assay. Interaction between the BD- and AD-fused proteins indicated in the presence or absence of the third partner (HA-fusion protein).
(B) P3H assay. Interaction between AD-bZIP2 with BD-bZIP63 or BD-bzip63ala (S/A exchange mutant of the SnRK1-specific phosphorylation sites: S29/294/300A). Values are calculated as fold induction relative to transfections with the reporter plasmid only (–) (mean values ± sd, n = 3; Student’s t test of promoter background activity, *P < 0.05).
(C) ChIP-PCR in 4-week-old bzip63 knockout (Ws) plants and lines complemented with bZIP63:YFP or bZIP63ala:YFP under the control of the endogenous promoter. Specific primers detecting the G3-G4 site on the ETFQO promoter and an αGFP-antibody were used. Plant material was harvested after 6 h in the light (yellow) or 6 h in extended darkness (green). Given is fold enrichment (mean values ± sd, n = 3) relative to the wild type in the light. Student’s t test of the wild type in light, *P < 0.05.
SnRK1 Is Recruited to Chromatin via bZIP Adapters
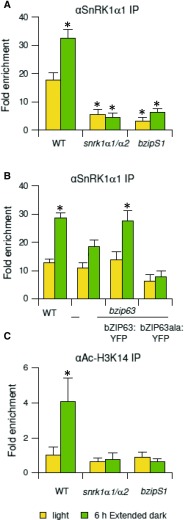
The above findings prompted us to test whether bZIP63 and bZIP2 assist in recruiting SnRK1 to chromatin in planta. SnRK1s were shown to be localized in the nucleus (Bitrián et al., 2011). Remarkably, ChIP-PCR analyses demonstrated that SnRK1α1 is associated with the ETFQO promoter (Figure 6A). The recruitment of SnRK1α1 increased within 6 h of extended dark treatment, supporting the notion that SnRK1α1 is recruited to chromatin in response to low energy stress. S1-bZIPs and bZIP63 are both crucial for SnRK1α1 recruitment (Figures 6A and 6B). However, whereas the quintuple group S1 knockdown completely abolished binding to the ETFQO promoter, the bzip63 knockout reduced binding by ∼40%. This is in line with the reduction in ETFQO transcript abundance in bZIP63 (Figure 4D) and may be explained by the participation of redundant group C members (Dietrich et al., 2011; Hartmann et al., 2015). Moreover, we demonstrated that a fully functional bZIP63 is indispensable for SnRK1α1 recruitment, as SnRK1α1 did not bind to the ETFQO promoter in a bzip63 mutant line complemented with a genomic bZIP63:YFP construct harboring Ala exchange mutations (bZIP63ala:YFP) (Figure 6B). It is important to note that the differences in ChIP results observed between the knockout and complemented line (bZIP63ala:YFP) may be due to the absence or presence of the protein, which can still bind DNA and compete as a protein interaction partner.
Figure 6.
Recruitment of SnRK1 to the ETFQO Promoter.
(A) ChIP-PCR of SnRK1α1 in wild-type, snrk1α1/α2 and bzipS1 plants using primers amplifying the G3-G4 site of the ETFQO promoter and an SnRK1α1-specific antibody.
(B) ChIP-PCR of SnRK1α1 in wild type, bzip63 mutant, and bzip63 complemented with a genomic bZIP63 fragment fused to YFP (bZIP63:YFP) (all ecotype Ws) or the corresponding construct carrying S/A exchange mutations in the SnRK1-specific phosphorylation sites (bZIP63ala:YFP) using an SnRK1α1-specific antibody.
(C) Acetylation of the ETFQO promoter. ChIP-PCR using an Ac-H3K14 antibody. All analyses are performed using three biological replicates of 3-week-old rosette leaves in the light (yellow) or 6 h extended darkness (green). Given are mean values (±sd) calculated relative to the input in the light (set to 1). Student’s t test of wild-type samples in light, *P < 0.05.
SnRK1 Is Required for Histone Acetylation of the ETFQO Promoter
In yeast, the SnRK1 homolog Snf1 also binds to promoters and regulates chromatin structure via histone acetyltransferases (HATs), which modify histones due to acetylation (Abate et al., 2012; Lo et al., 2001). Acetylation of histone 3 lysine 14 (H3K14) is a general mark associated with a euchromatic state and a high level of transcription (Lee and Workman, 2007). In wild-type plants, we found a 4-fold increased level of H3K14 acetylation of the ETFQO promoter within 6 h of extended dark treatment, which was abolished in both snrk1α1/α2 and bzipS1 knockdown plants (Figure 6C). In contrast, this increase in acetylation was not detected for ETFα, which is not induced by extended night treatment (Supplemental Figure 8). Taken together, the protein interaction and ChIP data support a mechanism in which recruitment of SnRK1 to the ETFQO promoter via bZIP adapters is crucial for chromatin remodeling to initiate transcription.
DISCUSSION
Plants possess an impressive degree of metabolic flexibility to survive starvation conditions. This work unravels a molecular mechanism by which energy starvation is transduced to reprogram gene expression to support plant survival.
Using an inducible loss-of-function approach, we demonstrated the importance of SnRK1 as a central regulator of transcriptional networks involved in stress and energy signaling on a genome-wide level. This transcriptome approach has several advantages compared with previous studies (Baena-González et al., 2007), as whole plants instead of protoplast cultures and inducible loss-of-function instead of gain-of-function lines were studied under energy-deprived conditions. In general, SnRK1s activate catabolic processes and inactivate energy-consuming anabolic processes. Importantly, these functions proposed for plant SnRK1 largely match those described for orthologous kinases in other species, indicating that these crucial metabolic regulators are evolutionarily conserved (Baena-González and Sheen, 2008; Hardie, 2015). In contrast, downstream TF targets of Snf1 in yeast and AMPK in mammals appear not to be evolutionarily conserved in plants. Although several downstream TFs have been suggested, unambiguous proof is largely missing (Kleinow et al., 2009; Lin et al., 2014; O’Brien et al., 2015; Tsai and Gazzarrini, 2012). Whereas C- and S1-bZIPs have already been proposed to regulate typical SnRK1 target genes (Baena-González et al., 2007), our transcriptome analyses strongly supports the function of S1-bZIPs downstream of SnRK1. As only a subset of SnRK1-regulated genes is controlled by S1-bZIPs, other TFs are likely needed to execute the complete SnRK1 response. Despite the huge transcriptional reprogramming initiated by SnRK1 upon extended darkness, we also identified a substantial number of genes regulated by S1-bZIPs that do not overlap with those of SnRK1. The C/S1 heterodimerization network could hence serve as an important hub, where different input signals for cellular energy status and environmental conditions converge to reprogram the plant transcriptome. In line with this assumption, the C/S1 network has been shown to be involved in a plethora of biological functions, such as responses to biotic and abiotic stresses, nutritional responses, seed development, and auxin signaling.
Several alternative pathways likely acting in parallel have been described that support ATP production when energy is sparse, e.g., by the consumption of fatty acids (Kunz et al., 2009) and chlorophyll (Hörtensteiner and Kräutler, 2011) or by NAD(H)-dependent glutamate dehydrogenase activity feeding intermediates into the TCA cycle (Miyashita and Good, 2008). The transcriptome and mutant data presented here support the view that both SnRK1s and S1-bZIPs control alternative metabolic pathways, which via the ETF/ETFQO proteins transport electrons to the ubiquinone pool of the mETC. Whereas ETF/ETFQO in the mammalian system uses electrons derived from at least 11 enzymes (Watmough and Frerman, 2010), plants are fed by only two sources, the catabolism of Lys and BCAA (Araújo et al., 2010, 2011a, 2011b; Hüdig et al., 2015; Ishizaki et al., 2005, 2006; Peng et al., 2015). However, only BCAA but no Lys catabolic genes are affect by SnRK1-S1-bZIP signaling.
In line with our data, recently published transcriptome and metabolome studies analyzing multiple stress conditions revealed the tightly coordinated transcriptional regulation of BCAA catabolic genes (Caldana et al., 2011). Whereas these genes are not generally induced by stresses (e.g., high light, heat, cold, and drought), energy-limiting conditions (e.g., low light, night extension, and hypoxia) lead to their activation. This study extends this view, as most of the genes involved in BCAA catabolism including ETFQO are upregulated by night extension or growth under low light and depend on both SnRK1 and S1-bZIPs. Moreover, in support of recent studies on progressive C depletion and C resupply (Cookson et al., 2016), Glc feeding suppresses BCAA degradation genes and restores the growth of snrk1α1/α2 and bzipS1 mutants, even when cultivated in extended darkness. Nevertheless, it should be noted that all well-accepted experimental conditions that provoke energy starvation might to a certain degree also affect other inputs such as light perception.
Feeding with BCAAs Leu, Val, and Ile partially rescued plant survival under extended darkness, but not in bzipS1 plants. These findings further emphasize the impact of these S1-bZIPs on alternative ATP generation. Besides BCAA, Pro and Tyr catabolic genes were found to be regulated by S1-bZIPs, proposing a larger impact of these TFs on distinct amino acid catabolic pathways. Importantly, most of the promoters of starvation-induced amino acid catabolic genes bind S1-bZIPs in vitro (O’Malley et al., 2016). In vivo binding was confirmed for selected promoters such as ETFQO, MCCB, ProDH1, and HGO but not for promoters of transcriptionally nonregulated genes, such as ETFα. Taken together, these findings suggest that a coordinated gene regulatory network is controlled by S1-bZIPs via direct binding to their promoters.
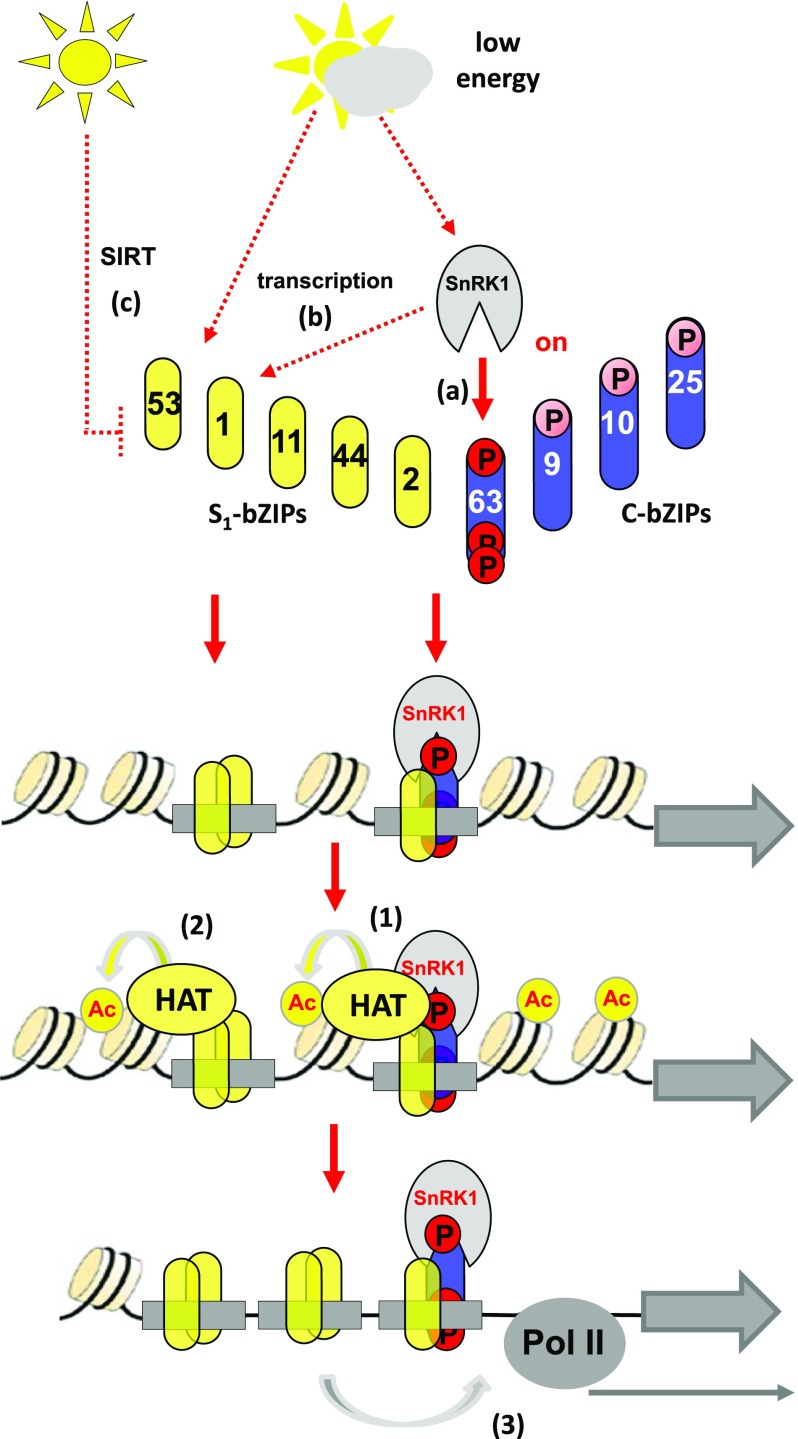
Although loss-of-function data support the view that group S1-bZIPs are crucial for controlling a subset of SnRK1-dependent genes, none of the S1-bZIPs were found to be directly phosphorylated by SnRK1 kinases. Based on previous findings, several nonexclusive mechanisms likely operate in parallel to explain the energy dependency of S1-bZIPs (Figure 7): (1) The translation of all S1-bZIPs is negatively controlled by sugar supply due to a system of upstream open reading frames named SIRT (sucrose-induced repression of translation), leading to increased translation of S1-bZIPs under energy-limiting conditions (Wiese et al., 2004). However, the energy-sensing mechanism is not yet elucidated. (2) The transcription of the S1-member bZIP1 is induced by extended darkness (Dietrich et al., 2011) in a SnRK1-dependent manner (Supplemental Data Set 1). As homo- or heterodimers are formed within the S1-group, and as they bind to and activate the transcription of various genes, these mechanisms are sufficient to explain starvation-induced gene regulation. (3) Moreover, bZIP63 is phosphorylated by SnRK1 to boost C/S1 heterodimerization. This additional mechanism may also link S1-bZIPs to SnRK1 signaling. While S1-bZIPs do not physically interact with SnRK1, three-hybrid studies in plant-derived protoplasts suggest that bZIP63 bridges SnRK1 to S1-bZIPs, pointing to the formation of a ternary complex. Importantly, complex formation depends on the integrity of SnRK1-specific phosphorylation sites that are crucial for bZIP63 function (Mair et al., 2015). As experiments aimed at isolating this complex via coimmunoprecipitation techniques failed and as it was only detected using in vivo methods requiring DNA, we propose that the postulated complex transiently forms when bound to DNA. Indeed, highly sensitive methods such as ChIP-PCR and DNA-protein cross-linking are needed to provide indirect evidence for the presence of the postulated complex. Recently, bZIP1 was shown to alter gene expression after transient binding (“hit”) and subsequent mobilization to a second binding site (“run”) (Para et al., 2014). This hit-and-run model may explain the transient nature of the complex; however, whether a related mechanism is active for the other S1-bZIPs is presently unresolved.
Figure 7.
Working Model Summarizing Starvation-Induced Transcriptional Control via the SnRK1-C/S1-bZIP Pathway.
Energy-limiting conditions are transmitted into activation of the C/S1-bZIP network via several mechanisms: posttranslational activation of group C bZIP63 via phosphorylation (Mair et al., 2015) (A), SnRK1-dependent transcriptional activation of bZIP1 (this work) (B), or translational regulation of all group S1 members via SIRT (Wiese et al., 2004) (C). Transcriptional activation by C/S1-bZIPs is mediated by several nonexclusive mechanisms: (1) formation of a ternary SnRK1-C/S1-bZIP complex induces histone acetylation (Ac) (this work), (2) bZIP11, bZIP2, and bZIP44 recruit HATs independently of SnRK1 (Weiste and Dröge-Laser, 2014), and (3) another yet undefined transactivation mechanism. Pol II, polymerase II.
Although the functional impact of bZIP63 in cooperation with S1-bZIPs in the starvation response was previously demonstrated, our results further support this model by directly linking SnRK1 and C/S1-bZIPs to the ETFQO promoter. Analysis of primary metabolites (Mair et al., 2015) and the reduced ETFQO transcription in bzip63 further extend this model, as shown in Figure 7. The limited but significant impact of bZIP63 on ETFQO transcription is in line with the notion that S1-dimers might regulate ETFQO independently of C-bZIPs. Moreover, functional redundancy within group C has been described, and due to the presence of conserved SnRK1-specific phosphorylation sites, a similar control mechanism is expected (Mair et al., 2015).
Indeed, bZIP63 itself did not display strong activation properties in protoplasts. Hence, we propose that it specifically functions as an adapter that is phosphorylated by SnRK1 and supports complex formation on target DNA. In contrast to bZIP63, several group S1-members show strong transcriptional activation properties. We recently showed that the N termini of bZIP2, bZIP11, and bZIP44 interact with the adaptor protein Ada2b, which is part of a multiprotein HAT complex related to yeast SAGA (Spt-Ada-Gcn5-Histone-Acetyltransferase) (Weiste and Dröge-Laser, 2014). However, this interaction does not apply for bZIP1, suggesting that several parallel transactivation mechanisms function within the C/S1 network (Figure 7). Chromatin remodeling is an important step required for transcriptional initiation. These findings are in line with starvation-induced histone acetylation of the ETFQO promoter, which depends on the presence of SnRK1 and S1-bZIPs. It should be noted that in contrast to ETFQO, other genes involved in BCAA catabolism are already highly acetylated in the noninduced state (Supplemental Figure 8B). Hence, genome-wide fine mapping approaches are needed to access whether H3K14 acetylation is of general importance in dark-induced transcription. In yeast, the Snf1 kinase is also recruited to chromatin to phosphorylate histones (H3S10) (Abate et al., 2012; Lo et al., 2001). This modification is recognized by HAT complexes that facilitate histone acetylation as a secondary chromatin mark. Thus, SnRK1s may contribute to modifications of chromatin status via several nonexclusive mechanisms. Kinase interactions are usually transient. Hence, it is conceivable that SnRK1, being present in the chromatin, possesses additional structural functions. We therefore propose that SnRK1 acts as a moonlighting protein, functioning as both a kinase and a scaffolding protein to link low-energy signaling to gene expression (Copley, 2012).
As demonstrated by ETFQO promoter mapping, there are three functional bZIP binding sites. The most important G1 sequence (CACGTC) is in line with recently published cistrome data for S1-bZIPs (O’Malley et al., 2016), whereas G3 and G4 (CACGTG) encode classical G-boxes. Interestingly, mutations in all boxes abolished both bZIP- and SnRK1-dependent activation of the ETFQO promoter in a nonadditive fashion. Hence, these data suggest the presence of regulatory interplay between these cis-elements, e.g., via protein interaction between the bound TFs. In contrast to ETFQO, the ASN1 promoter harbors only one functional G-box (Baena-González et al., 2007). It is tempting to speculate that the promoter context, as well as its occupancy with particular bZIP dimers, substantially alters the transcriptional properties of a particular promoter. Unfortunately, adequate methods still need to be established to follow TF interactions and dynamic promoter occupancy patterns in vivo on a single-promoter basis.
Taken together, this study provides mechanistic insights into how SnRK1-mediated low-energy signaling is transduced into gene regulation and cellular adaptation. This knowledge is of crucial importance for obtaining a basic understanding of plant metabolic control as well as for plant breeding for sustainable agriculture. Moreover, BCAAs represent essential amino acid in the human diet. Hence, understanding the regulatory network for their biosynthesis and catabolism might open avenues for future crop improvement (Peng et al., 2015). Finally, ETFQO-related proteins are well conserved from plants to mammals and humans (Watmough and Frerman, 2010). Further studies are needed to address the evolutionarily conserved regulation of mitochondrial metabolism by SnRK1 and its homologs.
METHODS
Plant Materials
Arabidopsis thaliana ecotype Col-0 was used as the wild type with the exception of bzip63 (ecotype Wassilewskija [Ws]). To generate the snrk1α1/α2 knockdown plants, an amiRNA targeting SnRK1α2 transcripts was transformed into the snrk1α1-3 mutant (GABI KAT GABI_579E09) (Mair et al., 2015) via Agrobacterium tumefaciens-mediated floral-dip transformation (Weigel and Glazebrook, 2002). Similarly, bzip1/bzip53 double mutants (Salk 059343; 069883) (Dietrich et al., 2011) were transformed with an amiRNA construct targeting bZIP2/11/44 (Weiste and Dröge-Laser, 2014). Furthermore, etfqo (Ishizaki et al., 2005), single and multiple bzip mutants (Alonso et al., 2009; Dietrich et al., 2011) were used. The bzip63 mutant and its complemented lines are in the Ws background and were previously described (Mair et al., 2015). XVE-bZIP2 plants are described by Weiste and Dröge-Laser (2014).
Plant Growth Conditions
Unless indicated otherwise, all plants were grown in a plant growth incubator (Binder, 2010; OSRAM L 30W/865, Lumilux, cool daylight) under a 12-h-light (120 µmol m−2 s−1)/12-h-dark photoperiod, at 22°C/20°C and a humidity of 60%. For sterile culture, plants were grown on Murashige and Skoog (MS)-agar medium (MS: M0222, Duchefa-Biochemie; agar: P1003, Duchefa-Biochemie) under the same light conditions. Extended dark treatment was performed by prolonging the dark period into the subsequent light phase for 6 h. The temperature was kept at 22°C. For low-light cultivation, plants were grown for 2 weeks on MS-agar medium under a long-day regime (16 h light/8 h dark) and subsequently shifted to short day/low light (8 h light/16 h dark; illumination 30 µmol m−2 s−1). For Leu/Ile/Val feeding, plants were grown on MS medium under a long-day regime for 2 weeks. As required, 25 µM of the respective amino acid (Sigma-Aldrich) was supplied to the medium and the plants were compared after 10 d in the light or darkness.
Induction of amiRNA expression was performed by supplementing the 1× MS-agar medium with Est after sterilization (T <40°C) to a final concentration of 10 μM. Growth conditions and Est treatment for RNA-seq experiments were optimized as outlined in Supplemental Figure 1. Due to the differences in protein stability, 14-d-old wild-type and snrk1α1/α2 plants were pretreated with Est for 6 d, whereas wild-type and bzipS1 plants were grown for an additional 5 d on MS and subsequently transferred to medium with Est for 1d. Subsequently, all plants were cultured for 6 h in extended night and harvested simultaneously. All RNA-seq experiments were performed with three biological replicates under extended night conditions. As wild-type plants with or without Est treatment did not display significant differential gene expression and as we were focusing on gene expression regulated by SnRK1 and S1-bZIP, nontreated samples were not included in the analysis. Plants for metabolome studies (four to six replicates) were grown side-by-side with those used for the RNA-seq experiment.
RT-qPCR
Rosettes were pooled for RNA preparation. One microgram of plant total RNA isolated as previously described (Dietrich et al., 2011) was used for cDNA synthesis. First-strand synthesis was performed using DNaseI (EN0521; Thermo Fisher Scientific) and RevertAid H Minus Reverse Transcriptase (Thermo Fisher Scientific) according to the manufacturer’s protocol. RT-qPCR was performed with BIOTAQ DNA polymerase (BIO-line) using the following cycling conditions: 10 min at 95°C, 40 cycles of 20 s at 95°C, 10 s at 55°C, and 30 s at 72°C. Amplification products were visualized by SYBR green. The UBIQUITIN5 gene (At3g62250) was used as an internal standard for relative quantification. Calculations are based on three to five independently grown sets of plants (biological replicates; depicted in the figure legends as n = 3–5). The primers used in this study are given in Supplemental Data Set 4.
RNA-Seq and Data Analysis
Total plant RNA was cleaned up using an RNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol. Five micrograms of total RNA was used for library preparation. mRNA was isolated using Sera-Mag Magnetic Oligo(dT) Particles (Thermo Fisher Scientific), and the cDNA library was prepared using the NEBNext mRNA Library Prep Master Mix Set for Illumina (New England Biolabs) in combination with NEBNext Multiplex Oligos for Illumina (New England Biolabs). The quality of the RNA and fragmentation size were checked using an Experion RNA HighSens Analysis Kit (Bio-Rad). The quality of the cDNA at the end of the library preparation was checked using Experion DNA Chips (Bio-Rad). During library preparation, products were isolated using a QIAquick PCR purification kit (Qiagen). For sequencing, six libraries were pooled for each lane of the Illumina Chip. High-throughput sequencing was performed on an Illumina GAIIx platform following the manufacturer’s instructions. Quality control of the sequencing data was done using fastQC (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/). Mapping of the reads was performed using Bowtie 0.12.8 (Lawrence et al., 2009) onto the Arabidopsis genome release TAIR9. The resulting BAM files were then sorted and indexed using samtools 0.1.18. For DEG analysis, R was used with GenomicRanges, rtracklayer (Lawrence et al., 2009), samtools, and edgeR (Robinson et al., 2010) libraries. Only genes with a p-adjust (“BH” correction according to Benjamini and Hochberg) <0.01 were used for further analysis. DAVID was used for GO enrichment analysis (Jiao et al., 2012) to obtain the list of genes commonly regulated by SnRK1s and S1-bZIPs.
Chlorophyll Content Measurement
One hundred milligrams of frozen material was pulverized using a Mixer Mill (MM400; Retsch) and metal beads in a 2-mL reaction tube. One milliliter of methanol was used for the extraction. The extract was incubated at 60°C for 30 min and for 10 min at room temperature (RT). The supernatant was clarified by centrifugation in a bench-top centrifuge. Absorption of a 1:10 dilution of the clarified supernatant was measured at 650 and 665 nm in a spectrophotometer. Total chlorophyll content was obtained using the formula: 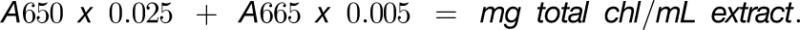
Protoplast Transformation
Protoplast transformation was performed according to Weiste and Dröge-Laser (2014). Protoplasts were obtained from rosette leaves of 3-week-old plants grown on soil. One hour after dawn, the leaves were cut into tiny stripes and digested for 30 to 60 min under a vacuum and for 3 h at atmospheric pressure with enzyme solution (1.25% [w/v] Cellulase R-10, 0.3% Macerozyme R-10, 0.4 M mannitol, 20 mM KCl, 10 mM CaCl2, and 20 mM MES, pH 5.7). The protoplast suspension was filtered through a metal mesh to remove leaf debris and washed twice with 10 mL of W5 solution (2 mM MES, 154 mM NaCl, 125 mM CaCl2, and 5 mM KCl, pH 5.7). Afterwards, the protoplasts were resuspended in 10 mL of W5, incubated on ice for at least 1 h, and subsequently resuspended to a final concentration of 105 cell/mL in MMg buffer (4 mM MES, 0.4 M mannitol, and 15 mM MgCl2, pH 5.7). Two hundred microliters of protoplast suspension was gently mixed with DNA in a 2-mL reaction tube. PEG (220 μL; 40% PEG4000, 0.2 M mannitol, and 100 mM CaCl2) was added to the reaction tube and gently mixed, followed by 10 min of incubation at RT. W5 buffer (800 μL) was used to wash the protoplasts, followed by centrifugation (300g) for 1 min. A syringe was used to remove the supernatant. The protoplasts were incubated for 16 h in 200 μL of WI solution (4 mM MES, 0.5 M mannitol, and 20 mM KCl, pH 5.7) in the growth incubator under a 12/12-h diurnal regime.
For the P2H/P3H assays (Ehlert et al., 2006), 10 μg of effector plasmid, 7 μg of reporter (ProGal4:GUS or ProGal4:LUC), and 3 μg of transfection control reporter (Pro35S:NAN or Pro35S:RENILLA) (Ehlert et al., 2006) were used. For the ChIP experiments, 10 μg of effector plasmid was used. If not stated otherwise, three independent transfections (n = 3) were used for each data point.
ChIP Using Protoplasts
ChIP assays with protoplasts were performed using a modified protocol according to Weiste and Dröge-Laser (2014). For ChIP assays, the incubation time was reduced to 8 h to prevent unspecific binding of the proteins. Twelve samples of transformed protoplasts (each sample containing 106–107 protoplasts) were pooled, centrifuged for 1 min at 300g, and resuspended in 200 μL of WI solution. Formaldehyde was added to a final concentration of 1% and the suspension was incubated for 10 min at RT. The addition of 250 μL of 2.5 M glycine was followed by 5 min incubation at RT. Protoplasts were washed twice with 800 μL of ice-cold W5 and resuspended in 500 μL of extraction buffer 1 (1 M hexylenglycol, 50 mM PIPES-KOH, pH 7.2, 10 mM MgCl2, 5 mM β-mercaptoethanol, and one tablet per 10 mL complete protease inhibitor cocktail tablets; Roche). After 20 min of incubation on ice, the extraction buffer was removed by centrifugation for 5 min at 150g. The pellet was resuspended in 500 μL of RIPAF buffer (50 mM HEPES, pH 7.9, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate, and 0.1% SDS) and incubated on ice for 10 min. The lysate was sonicated (28 times for 15 s, 100 Hz) on ice. Chromatin was cleared by centrifugation for 15 min at 11,000g at 4°C. The DNA fragment size was checked on a 2% agarose gel after de-crosslinking and chloroform/phenol extraction. The chromatin was incubated with 2 μg of ChIP grade HA-antibody (ab9110; AbCam) for 6 h at 4°C and subsequently overnight after the addition of 70 μL of protein-A-coated magnetic beads dissolved in BSA-PBS (5 mg/mL) (Invitrogen). Subsequently, the beads were washed with 1 mL of wash buffers 1 (20 mM Tris-HCl, pH 8.1, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, and 0.1% SDS), 2 (20 mM Tris-HCl, pH 8.1, 500 mM NaCl, 2 mM EDTA, 1% Triton X-100, and 0.1% SDS), 3 (20 mM Tris-HCl, pH 8.1, 250 mM LiCl, 1 mM EDTA, 1% Nonidet P-40, and 1% Na-deoxycholate), and 1 mL of TE buffer (5 mM Tris-HCl, pH 8.5). Each washing step was performed for 5 min at 4°C. The chromatin was eluted twice using 150 μL of elution buffer (1% SDS and 100 mM NaHCO3) each time for 15 min at RT. Finally, the DNA was purified by phenol/chloroform extraction and quantified by RT-qPCR using the input DNA for normalization. The primers used are given in Supplemental Data Set 4.
ChIP from Plant Material
Five grams of 3-week-old rosette leaves was incubated for 30 min under vacuum in 20 mL cross-linking buffer (50 mM KH2PO4/K2HPO4 buffer, pH 5.8, 1% [v/v] formaldehyde). Cross-linking was stopped by incubating the samples in 20 mL of 2.5 M glycine for 15 min under vacuum. The plant material was then washed twice with ice-cold water and subsequently frozen and pulverized. For nuclei isolation, the plant material was resuspended in 10 mL ice-cold extraction buffer (1 M hexylenglycol, 50 mM PIPES-KOH, pH 7.2, 10 mM MgCl2, 5 mM β-mercaptoethanol, and protease inhibitor cocktail; Roche) and filtered through two layers of Miracloth. Then, 0.5 mL of 25% Triton X-100 was added to the extract, followed by stirring for 15 min at 4°C. Nuclei were isolated by density gradient centrifugation using 35% Percoll solution. From this step on, the material was treated as described in the protocol for ChIP from protoplasts. The primers used are given in Supplemental Data Set 4. The following antibodies were used: α-HA-antibody (ab9110; Abcam), α-SnRK1.1 (AS10 919; Agrisera), α-SnRK1.2 (AS10 920; Agrisera), α-Ac-H3K14 (AB4729; Abcam), and α-GFP/YFP (Roche 11814460001). Data were calculated based on three to four biological replicates.
Immunoblot Analysis
Immunoblot analysis was performed as described by Weiste and Dröge-Laser (2014) making use of the primary antibodies mentioned above (from rabbit: 1:700 dilution; from mouse: 1:1000 dilution) and the corresponding secondary anti-rabbit (1:10,000 dilution) (catalog no. NA934) or anti-mouse (1:7500 dilution) (catalog no. RPN4201) immunoglobulin G conjugated with a horseradish peroxidase (GE Healthcare).
GC-MS Metabolite Analysis
Metabolite extraction was performed by adding 1 mL of cold (−20°C) methanol/chloroform/water (2.5/1/0.5) mixture to 50 to 80 mg of ground plant material. The samples were vortexed, incubated on ice for 8 to 10 min, and centrifuged for 4 min at 4°C. Five hundred microliters of water was added to the supernatant, followed by brief vortexing and 2 min of centrifugation. The polar phase was split into two equal aliquots and 10 μL of a 0.1 g L−1 solution of C13-labeled sorbitol was added as an internal standard. Samples were dried for derivatization. The dried pellets were resolved at 30°C for 90 min in 20 μL of a 40 mg/mL methoxyamine hydrochloride in pyridine solution. Eighty microliters of MSTFA spiked with 30 µL/mL of a mix of even-numbered alkanes was added, and the samples were incubated for 30 min at 37°C under constant shaking, followed by 2 min of centrifugation. The supernatant was transferred into a glass vial for measurement. GC-MS measurements were performed on an Agilent 6890 gas chromatograph coupled to a LECO Pegasus 4D GCxGC-TOF mass spectrometer (LECO Corporation). For GC analysis, the initial oven temperature was set to 70°C for 1 min, followed by a 9°C/min ramp up with a 350°C ending temperature, which was set constant for 8 min. For MS, the data acquisition rate was set to 20 spectra/second at a detector voltage of 1550 V. The acquisition delay was set to 5.5 min and the detected mass range was set from 40 to 600 m/z. Raw data were processed with LECO Chroma-TOF software (LECO Corporation). Peak areas were normalized by the peak area of the internal standard and by the sample fresh weight.
Statistics
Statistical tests were all performed using R (https://www.r-project.org/).
Accession Numbers
RNA-seq data from this article can be found in the Gene Expression Omnibus (GEO:GSE109388). Arabidopsis Genome Initiative identifiers for the genes mentioned in this article are as follows: SnRK1α1/KIN10 (At3g01090), SnRK1α2/KIN11 (At3g29160), bZIP53 (At3g62420), bZIP1 (At5g49450), bZIP2 (At2g18160), bZIP11 (At4g34590), bZIP44 (At1g75390), bZIP63 (At5g28770), bZIP10 (At4g02640), bZIP25 (At3g54620), bZIP9 (At5g24800), ASN1 (At3g47340), BCAT2 (At1g10070), MCCA (At1g03090), MCCB (At4G34030), IVDH (At3g45300), ETFQO (At2g43400), ETFα (At1g50940), ETFβ (At5g43430), D2HGDH (At4g36400), ProDH1 (At3g30775), HGO (At5g54080), UBI5 (At3g62250), ACTIN7 (At5g09810), and ACTIN8 (At1g49240).
Supplemental Data
Supplemental Figure 1. Characterization of the inducible snrk1α1/α2 and bzipS1 knockdown lines.
Supplemental Figure 2. Analyses of RNA-seq data.
Supplemental Figure 3. The BCAA degradation pathway is controlled by energy starvation due to low-light cultivation and group S1 bZIPs.
Supplemental Figure 4. Most BCAA catabolic genes are coordinately induced by low energy conditions and harbor S1-bZIP binding sites.
Supplemental Figure 5. In vivo binding of bZIP2 to selected target promoters.
Supplemental Figure 6. Impact of SnRK1 and S1-bZIPs on metabolic adjustment.
Supplemental Figure 7. SnRK1 expression enhances heterodimerization between particular bZIP members of groups C and S1.
Supplemental Figure 8. ChIP data on H3K14 histone acetylation.
Supplemental Data Set 1. Transcriptome analyses (RNA-seq) performed with snrk1α1/α2 and bzipS1 seedlings upon 6 h extended night treatment.
Supplemental Data Set 2. Comparison snrk1α1/α2 RNA-seq data with microarray data using protoplasts expressing SnRK1α1.
Supplemental Data Set 3. Summary of metabolome analysis performed with snrk1α1/α2 and bzipS1 seedlings upon 6 h extended night treatment.
Supplemental Data Set 4. Primers used in this study.
Supplemental File 1. Comparison of snrk1α1/α2 RNA-seq data with public data sets on energy starvation.
Acknowledgments
We thank J. Göttler for valuable technical assistance and S. Gillig for proofreading. We thank N.H. Chua (Rockefeller University), B. Weisshaar (University of Bielefeld, Germany), and K. Ishizaki (Kobe University, Japan) for sharing seeds and vector constructs. The research was supported by European Commission FP7 Marie Curie ITN MERIT (GA 264474) and the Austrian Science Fund (FWF) with projects P 23435 and P 26342.
AUTHOR CONTRIBUTIONS
L.P. performed most of the experiments. C.W. performed ProETFQO:GUS activation assays, BCAA feeding, and low-light experiments as well as ChIP-PCR of XVE-bZIP2 plants. K.D. designed the amiRNA construct for SnRK1α2 and generated the snrk1α1/α2 plant line. RNA-seq experiments were done under the supervision of E.W. F.L. prepared the RNA library for RNA-seq. T.N. and W.W. performed the metabolic measurements and their analysis. A.M. and M.T. provided unpublished plant lines and vector constructs. E.B.-G. actively supported the conceptual work. L.P., C.W., and W.D.-L. designed the research and wrote the manuscript.
References
- Abate G., Bastonini E., Braun K.A., Verdone L., Young E.T., Caserta M. (2012). Snf1/AMPK regulates Gcn5 occupancy, H3 acetylation and chromatin remodelling at S. cerevisiae ADY2 promoter. Biochim. Biophys. Acta 5: 419–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso R., Oñate-Sánchez L., Weltmeier F., Ehlert A., Diaz I., Dietrich K., Vicente-Carbajosa J., Dröge-Laser W. (2009). A pivotal role of the basic leucine zipper transcription factor bZIP53 in the regulation of Arabidopsis seed maturation gene expression based on heterodimerization and protein complex formation. Plant Cell 21: 1747–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo W.L., Ishizaki K., Nunes-Nesi A., Larson T.R., Tohge T., Krahnert I., Witt S., Obata T., Schauer N., Graham I.A., Leaver C.J., Fernie A.R. (2010). Identification of the 2-hydroxyglutarate and isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. Plant Cell 22: 1549–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo W.L., Ishizaki K., Nunes-Nesi A., Tohge T., Larson T.R., Krahnert I., Balbo I., Witt S., Dörmann P., Graham I.A., Leaver C.J., Fernie A.R. (2011a). Analysis of a range of catabolic mutants provides evidence that phytanoyl-coenzyme A does not act as a substrate of the electron-transfer flavoprotein/electron-transfer flavoprotein:ubiquinone oxidoreductase complex in Arabidopsis during dark-induced senescence. Plant Physiol. 157: 55–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo W.L., Tohge T., Ishizaki K., Leaver C.J., Fernie A.R. (2011b). Protein degradation - an alternative respiratory substrate for stressed plants. Trends Plant Sci. 16: 489–498. [DOI] [PubMed] [Google Scholar]
- Baena-González E., Rolland F., Thevelein J.M., Sheen J. (2007). A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942. [DOI] [PubMed] [Google Scholar]
- Baena-González E., Sheen J. (2008). Convergent energy and stress signaling. Trends Plant Sci. 13: 474–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros J.A.S., Cavalcanti J.H.F., Medeiros D.B., Nunes-Nesi A., Avin-Wittenberg T., Fernie A.R., Araújo W.L. (2017). Autophagy deficiency compromises alternative pathways of respiration following energy deprivation. Plant Physiol. 175: 62–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder S. (2010). Branched-chain amino acid metabolism in Arabidopsis thaliana. Arabidopsis Book 8: e0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitrián M., Roodbarkelari F., Horváth M., Koncz C. (2011). BAC-recombineering for studying plant gene regulation: developmental control and cellular localization of SnRK1 kinase subunits. Plant J. 65: 829–842. [DOI] [PubMed] [Google Scholar]
- Broeckx T., Hulsmans S., Rolland F. (2016). The plant energy sensor: evolutionary conservation and divergence of SnRK1 structure, regulation, and function. J. Exp. Bot. 67: 6215–6252. [DOI] [PubMed] [Google Scholar]
- Caldana C., Degenkolbe T., Cuadros-Inostroza A., Klie S., Sulpice R., Leisse A., Steinhauser D., Fernie A.R., Willmitzer L., Hannah M.A. (2011). High-density kinetic analysis of the metabolomic and transcriptomic response of Arabidopsis to eight environmental conditions. Plant J. 67: 869–884. [DOI] [PubMed] [Google Scholar]
- Confraria A., Martinho C., Elias A., Rubio-Somoza I., Baena-González E. (2013). miRNAs mediate SnRK1-dependent energy signaling in Arabidopsis. Front. Plant Sci. 4: 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson S.J., Yadav U.P., Klie S., Morcuende R., Usadel B., Lunn J.E., Stitt M. (2016). Temporal kinetics of the transcriptional response to carbon depletion and sucrose readdition in Arabidopsis seedlings. Plant Cell Environ. 39: 768–786. [DOI] [PubMed] [Google Scholar]
- Copley S.D. (2012). Moonlighting is mainstream: paradigm adjustment required. BioEssays 34: 578–588. [DOI] [PubMed] [Google Scholar]
- Crozet P., Margalha L., Confraria A., Rodrigues A., Martinho C., Adamo M., Elias C.A., Baena-González E. (2014). Mechanisms of regulation of SNF1/AMPK/SnRK1 protein kinases. Front. Plant Sci. 5: 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich K., Weltmeier F., Ehlert A., Weiste C., Stahl M., Harter K., Dröge-Laser W. (2011). Heterodimers of the Arabidopsis transcription factors bZIP1 and bZIP53 reprogram amino acid metabolism during low energy stress. Plant Cell 23: 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlert A., Weltmeier F., Wang X., Mayer C.S., Smeekens S., Vicente-Carbajosa J., Dröge-Laser W. (2006). Two-hybrid protein-protein interaction analysis in Arabidopsis protoplasts: establishment of a heterodimerization map of group C and group S bZIP transcription factors. Plant J. 46: 890–900. [DOI] [PubMed] [Google Scholar]
- Engqvist M.K.M., Kuhn A., Wienstroer J., Weber K., Jansen E.E.W., Jakobs C., Weber A.P.M., Maurino V.G. (2011). Plant D-2-hydroxyglutarate dehydrogenase participates in the catabolism of lysine especially during senescence. J. Biol. Chem. 286: 11382–11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie D.G. (2015). AMPK: positive and negative regulation, and its role in whole-body energy homeostasis. Curr. Opin. Cell Biol. 33: 1–7. [DOI] [PubMed] [Google Scholar]
- Hartmann L., et al. (2015). Crosstalk between Two bZIP signaling pathways orchestrates salt-induced metabolic reprogramming in Arabidopsis roots. Plant Cell 27: 2244–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt T.M., Nunes Nesi A., Araújo W.L., Braun H.-P. (2015). Amino acid catabolism in plants. Mol. Plant 8: 1563–1579. [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S., Kräutler B. (2011). Chlorophyll breakdown in higher plants. Biochim. Biophys. Acta 1807: 977–988. [DOI] [PubMed]
- Hüdig M., Maier A., Scherrers I., Seidel L., Jansen E.E.W., Mettler-Altmann T., Engqvist M.K.M., Maurino V.G. (2015). Plants possess a cyclic mitochondrial metabolic pathway similar to the mammalian metabolic repair mechanism involving malate dehydrogenase and l-2-hydroxyglutarate dehydrogenase. Plant Cell Physiol. 56: 1820–1830. [DOI] [PubMed] [Google Scholar]
- Ishizaki K., Larson T.R., Schauer N., Fernie A.R., Graham I.A., Leaver C.J. (2005). The critical role of Arabidopsis electron-transfer flavoprotein:ubiquinone oxidoreductase during dark-induced starvation. Plant Cell 17: 2587–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki K., Schauer N., Larson T.R., Graham I.A., Fernie A.R., Leaver C.J. (2006). The mitochondrial electron transfer flavoprotein complex is essential for survival of Arabidopsis in extended darkness. Plant J. 47: 751–760. [DOI] [PubMed] [Google Scholar]
- Jakoby M., Weisshaar B., Dröge-Laser W., Vicente-Carbajosa J., Tiedemann J., Kroj T., Parcy F.; bZIP Research Group (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7: 106–111. [DOI] [PubMed] [Google Scholar]
- Jiao X., Sherman B.T., Huang W., Stephens R., Baseler M.W., Lane H.C., Lempicki R.A. (2012). DAVID-WS: a stateful web service to facilitate gene/protein list analysis. Bioinformatics 28: 1805–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S.G., Price J., Lin P.-C.C., Hong J.C., Jang J.-C.C. (2010). The arabidopsis bZIP1 transcription factor is involved in sugar signaling, protein networking, and DNA binding. Mol. Plant 3: 361–373. [DOI] [PubMed] [Google Scholar]
- Kleinow T., Himbert S., Krenz B., Jeske H., Koncz C. (2009). NAC domain transcription factor ATAF1 interacts with SNF1-related kinases and silencing of its subfamily causes severe developmental defects in Arabidopsis. Plant Sci. 177: 360–370. [Google Scholar]
- Kunz H.H., Scharnewski M., Feussner K., Feussner I., Flügge U.I., Fulda M., Gierth M. (2009). The ABC transporter PXA1 and peroxisomal beta-oxidation are vital for metabolism in mature leaves of Arabidopsis during extended darkness. Plant Cell 21: 2733–2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M., Gentleman R., Carey V. (2009). rtracklayer: an R package for interfacing with genome browsers. Bioinformatics 25: 1841–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.K., Workman J.L. (2007). Histone acetyltransferase complexes: one size doesn’t fit all. Nat. Rev. Mol. Cell Biol. 8: 284–295. [DOI] [PubMed] [Google Scholar]
- Li L., Sheen J. (2016). Dynamic and diverse sugar signaling. Curr. Opin. Plant Biol. 33: 116–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.-R., Lee K.-W., Chen C.-Y., Hong Y.-F., Chen J.-L., Lu C.A., Chen K.-T., Ho T.-H.D., Yu S.-M. (2014). SnRK1A-interacting negative regulators modulate the nutrient starvation signaling sensor SnRK1 in source-sink communication in cereal seedlings under abiotic stress. Plant Cell 26: 808–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo W.S., Duggan L., Emre N.C., Belotserkovskya R., Lane W.S., Shiekhattar R., Berger S.L. (2001). Snf1--a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science 293: 1142–1146. [DOI] [PubMed] [Google Scholar]
- Ma J., Hanssen M., Lundgren K., Hernández L., Delatte T., Ehlert A., Liu C.-M., Schluepmann H., Dröge-Laser W., Moritz T., Smeekens S., Hanson J. (2011). The sucrose-regulated Arabidopsis transcription factor bZIP11 reprograms metabolism and regulates trehalose metabolism. New Phytol. 191: 733–745. [DOI] [PubMed] [Google Scholar]
- Mair A., et al. (2015). SnRK1-triggered switch of bZIP63 dimerization mediates the low-energy response in plants. eLife 4: e05828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita Y., Good A.G. (2008). NAD(H)-dependent glutamate dehydrogenase is essential for the survival of Arabidopsis thaliana during dark-induced carbon starvation. J. Exp. Bot. 59: 667–680. [DOI] [PubMed] [Google Scholar]
- Nukarinen E., Nägele T., Pedrotti L., Wurzinger B., Mair A., Landgraf R., Börnke F., Hanson J., Teige M., Baena-Gonzalez E., Dröge-Laser W., Weckwerth W. (2016). Quantitative phosphoproteomics reveals the role of the AMPK plant ortholog SnRK1 as a metabolic master regulator under energy deprivation. Sci. Rep. 6: 31697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien M., Kaplan-Levy R.N., Quon T., Sappl P.G., Smyth D.R. (2015). PETAL LOSS, a trihelix transcription factor that represses growth in Arabidopsis thaliana, binds the energy-sensing SnRK1 kinase AKIN10. J. Exp. Bot. 66: 2475–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley R.C., Huang S.C., Song L., Lewsey M.G., Bartlett A., Nery J.R., Galli M., Gallavotti A., Ecker J.R. (2016). Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165: 1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Para A., et al. (2014). Hit-and-run transcriptional control by bZIP1 mediates rapid nutrient signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: 10371–10376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C., Uygun S., Shiu S.-H., Last R.L. (2015). The impact of the branched-chain ketoacid dehydrogenase complex on amino acid homeostasis in Arabidopsis. Plant Physol. 169: 1807– 1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. (2014). Master regulators in plant glucose signaling networks. J. Plant Biol. 57: 67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M., Zeeman S.C. (2012). Starch turnover: pathways, regulation and role in growth. Curr. Opin. Plant Biol. 15: 282–292. [DOI] [PubMed] [Google Scholar]
- Tsai A.Y.-L., Gazzarrini S. (2012). AKIN10 and FUSCA3 interact to control lateral organ development and phase transitions in Arabidopsis. Plant J. 69: 809–821. [DOI] [PubMed] [Google Scholar]
- Usadel B., Bläsing O.E., Gibon Y., Retzlaff K., Höhne M., Günther M., Stitt M. (2008). Global transcript levels respond to small changes of the carbon status during progressive exhaustion of carbohydrates in Arabidopsis rosettes. Plant Physiol. 146: 1834–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watmough N.J., Frerman F.E. (2010). The electron transfer flavoprotein: ubiquinone oxidoreductases. Biochim. Biophys. Acta 1797: 1910–1916. [DOI] [PubMed] [Google Scholar]
- Weigel R., Glazebrook J. (2002). Arabidopsis: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbour Laboratory Press; ). [Google Scholar]
- Weiste C., Dröge-Laser W. (2014). The Arabidopsis transcription factor bZIP11 activates auxin-mediated transcription by recruiting the histone acetylation machinery. Nat. Commun. 5: 3883. [DOI] [PubMed] [Google Scholar]
- Weiste C., Pedrotti L., Selvanayagam J., Muralidhara P., Fröschel C., Novák O., Ljung K., Hanson J., Dröge-Laser W. (2017). The Arabidopsis bZIP11 transcription factor links low-energy signalling to auxin-mediated control of primary root growth. PLoS Genet. 13: e1006607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weltmeier F., et al. (2009). Expression patterns within the Arabidopsis C/S1 bZIP transcription factor network: availability of heterodimerization partners controls gene expression during stress response and development. Plant Mol. Biol. 69: 107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weltmeier F., Ehlert A., Mayer C.S., Dietrich K., Wang X., Schütze K., Alonso R., Harter K., Vicente-Carbajosa J., Dröge-Laser W. (2006). Combinatorial control of Arabidopsis proline dehydrogenase transcription by specific heterodimerisation of bZIP transcription factors. EMBO J. 25: 3133–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese A., Elzinga N., Wobbes B., Smeekens S. (2004). A conserved upstream open reading frame mediates sucrose-induced repression of translation. Plant Cell 16: 1717–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S.P., Rangarajan P., Donahue J.L., Hess J.E., Gillaspy G.E. (2014). Regulation of Sucrose non-Fermenting Related Kinase 1 genes in Arabidopsis thaliana. Front. Plant Sci. 5: 324. [DOI] [PMC free article] [PubMed] [Google Scholar]