Maize GIF1 promotes meristematic function in leaves and stems but restricts indeterminacy in the inflorescence by regulating the expression of several known inflorescence architecture-related genes.
Abstract
Plant architecture results from a balance of indeterminate and determinate cell fates. Cells with indeterminate fates are located in meristems, comprising groups of pluripotent cells that produce lateral organs. Meristematic cells are also found in intercalary stem tissue, which provides cells for internodes, and at leaf margins to contribute to leaf width. We identified a maize (Zea mays) mutant that has a defect in balancing determinacy and indeterminacy. The mutant has narrow leaves and short internodes, suggesting a reduction in indeterminate cells in the leaf and stem. In contrast, the mutants fail to control indeterminacy in shoot meristems. Inflorescence meristems are fasciated, and determinate axillary meristems become indeterminate. Positional cloning identified growth regulating factor-interacting factor1 (gif1) as the responsible gene. gif1 mRNA accumulates in distinct domains of shoot meristems, consistent with tissues affected by the mutation. We determined which GROWTH REGULATING FACTORs interact with GIF1 and performed RNA-seq analysis. Many genes known to play roles in inflorescence architecture were differentially expressed in gif1. Chromatin immunoprecipitation identified some differentially expressed genes as direct targets of GIF1. The interactions with these diverse direct and indirect targets help explain the paradoxical phenotypes of maize GIF1. These results provide insights into the biological functions of gif1.
INTRODUCTION
Plant architecture results from the activity of meristems, comprising populations of dividing cells that are totipotent and indeterminate. The shoot apical meristem is an indeterminate meristem, initiating primordia indefinitely. Meristematic cells are also found in growing organs such as stems or leaves, contributing to the height of the plant or width of a leaf (Alvarez et al., 2016; Tsuda et al., 2017). Meristematic cells lose their indeterminacy as their fate becomes determined. Floral meristems are considered determinate, as they terminate after the production of floral organs. Although leaf initiation is often considered to involve a switch from indeterminate to determinate cell fates, populations of indeterminate cells are included in that leaf initiation event. Every leaf has an axillary meristem, a group of totipotent cells located at the base of the leaf. The axillary meristem may become a branch, a flower, or remain dormant. Leaf initiation also includes cells for the internode, the stem tissue between leaves. The leaf, subtending internode, and axillary meristem unit is called a phytomer (Galinat, 1959; Howell, 1998; Chuck et al., 2010).
Most developmental mutants identified so far affect one or two parts of the phytomer and rarely play a role in multiple aspects of shoot architecture. We identified a pleiotropic maize (Zea mays) mutant that affects multiple meristems, but in different ways. Intercalary meristems of the stem often fail to proliferate and leaves are narrow. At the same time, inflorescence meristems are fasciated, with an overproliferation of meristematic cells, and determinacy is lost in normally determinate axillary meristems. The mutant was identified in two populations, one resulting from EMS mutagenesis and the other as a naturally occurring variant. We positionally cloned the gene in both populations and identified it as growth regulating factor-interacting factor1 (gif1).
Growth regulating factors (GRFs) are a class of plant-specific proteins involved in the regulation of stem and leaf development that mainly act as positive regulators of cell proliferation (Omidbakhshfard et al., 2015). The first member of the GRF family identified was OsGRF1 in rice (Oryza sativa). Its expression in the intercalary meristem of internodes is inducible by gibberellic acid (van der Knaap et al., 2000). In Arabidopsis thaliana, grf mutants develop smaller and narrower leaves compared with the wild type (Debernardi et al., 2014; Horiguchi et al., 2005; Kim et al., 2003). In rice, repression of GRF3, GRF4, and GRF5 results in dwarfism and delayed inflorescence development (Kuijt et al., 2014), while enhancing expression of GRFs results in significant increases in panicle length and grain length, width, and weight (Che et al., 2015; Duan et al., 2015; Li et al., 2016; Sun et al., 2016). Overexpressing a GRF1 resistant to the microRNA miR396a in maize increases the number of dividing cells, resulting in larger leaves, although the overall plant height is reduced (Nelissen et al., 2015).
GRFs function by interacting with members of the GRF-interacting factor (GIF) family in vivo to establish plant-specific transcriptional complexes (Hoe Kim and Tsukaya, 2015). In Arabidopsis, GIF1 interacts with GRF3 (Debernardi et al., 2014) and GRF5 (Horiguchi et al., 2005). gif1 mutants show a narrow-leaf phenotype due to a decrease in cell number. In contrast, overexpressing GIF1 enlarges leaf size by enhancing cell proliferation in leaf primordia (Horiguchi et al., 2005). The GIF family in maize is composed of three members, gif1, gif2, and gif3. In particular, the composition of the GIF1-interacting complex within the growing leaf is variable: GRF1/6/7/12/15/17 are significantly enriched in the division zone, while both GRF4 and GRF10 are enriched in the expansion zone of the growing leaf (Nelissen et al., 2015), indicating that growth and development of a given tissue and organ may be fine-tuned by a specific GIF-interacting complex.
In this study, we provide a description of a gif1 loss-of-function mutant in maize. gif1 promotes meristematic function in leaves and stems, but restricts indeterminacy in inflorescence meristems. We show that GIF1 binds to the inflorescence architecture gene unbranched3 (ub3) and regulates expression of several known inflorescence architecture-related genes. We hypothesize that the interaction with specific maize targets explains the different phenotypes observed in gif1.
RESULTS
gif1 Mutants Are Dwarf with Narrow Leaves and Sterile Florets
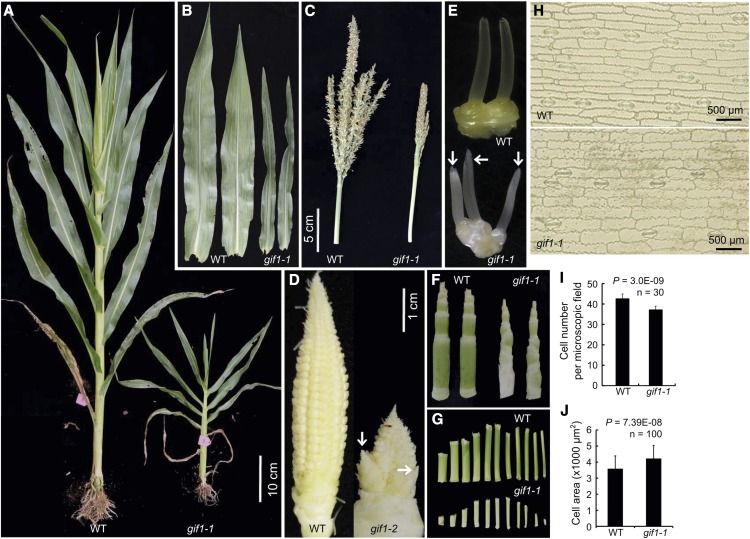
The gif1 mutation was found as both a naturally occurring mutation and an EMS-induced mutation. gif1-1 was identified in the BS238 family line derived from the elite Stiff Stalk Synthetic population BSSSC9, and gif1-2 was identified in an EMS mutagenized A619 F2 family and characterized after backcrosses to multiple inbreds. The phenotypes of both alleles are similar (Figures 1 and 2). Most agronomically important traits, including plant height, ear height, length and width of ear leaf, tassel length, and branch number differed significantly from that in the wild type (Supplemental Figure 1). The most dramatic phenotypes were semidwarf plants (Figures 1A and 2A), narrow and small leaves (Figures 1B and 2B), and a reduced number of branches in the tassel (Figures 1C and 2C). Conversely, branches developed at the base of the ear (Figure 1D), and mutants developed fasciated ears (Figures 2D to 2N). Ears are normally surrounded by sterile husk leaves. The gif1 mutants often contained axillary shoots in the axils of husk leaves (Figures 2I to 2L), which contained fasciated secondary ears (Figures 2M and 2N). Wild-type female florets have a single pistil, known as the silk. In both gif1 alleles, multiple silks were found per floret (Figures 1E, 2O, and 2P). Nucellar tissue, which surrounds the female gametophyte and is normally confined within the fused carpels of the silk, proliferated in the gif1 mutants (Figure 2P), indicating that the floral meristem was also indeterminate. Both alleles were male and female sterile.
Figure 1.
Vegetative and Inflorescence Traits in gif1-1.
Phenotypes of gif1-1. In each, the wild type is on the left and gif1-1 is on the right (n ≥ 30).
(A) Whole plants during vegetative development.
(B) Excised mature leaf blades.
(C) Mature tassels.
(D) Immature, unpollinated ears. gif1-1 has ectopic branching at the ear base.
(E) Ear spikelets. Arrows indicate three rather than the two spikelets in gif1-1.
(F) Internodes subtending the tassel, with leaves removed.
(G) Progressive internode lengths with the topmost internode on the right.
(H) Cells of the abaxial epidermis are larger in gif1-1.
(I) Quantification of cell number per microscopic field. Error bars indicate the sd.
(J) Quantification of cell size. Error bars indicate the sd.
Figure 2.
Vegetative and Inflorescence Traits in gif1-2.
(A) Whole plants during vegetative development.
(B) Excised mature leaf blades.
(C) Mature tassels. gif1-2 has fewer tassel branches.
(D) Immature, unpollinated ears. gif1-2 has fasciated ears.
(E) to (H) Male and female inflorescence observed by scanning electron microscopy. Male (E) and female (G) inflorescences of the wild type, and male (F) and female (H) inflorescences of gif1-2; gif1-2 has a fasciated inflorescence meristem in the tassel and ear.
(I) Entire ear of gif1-2 (5xB73).
(J) Outer nine husk leaves excised. Most husk leaves enclosed small axillary ears (white asterisks).
(K) Remaining husk leaves and any axillary ears (asterisks) excised. The primary ear is marked with a red star.
(L) Close-up of primary ear.
(M) Close-up of an axillary ear.
(N) A second axillary husk ear.
(O) Pistil. gif1-2 generates multiple silks per floret.
(P) Unfused carpels of the silk of gif1-2. Arrow points to expanded nucellus.
(Q) Internodes with leaves removed. gif1-2 has short and asymmetric internodes.
Bars = 100 μm in (E) to (H) and (L) to (N).
To determine the cause of the dwarf phenotype, we counted leaf number and measured internode length. The leaf number was the same as nonmutant siblings (wild type), but internodes were often dramatically shortened (Figures 1F and 1G). The decrease in internode size of gif1-2 in the B73 background was highly irregular and differed from plant to plant. Short internodes were often asymmetric, with the side of the internode that is attached to the midrib shorter than the side attached to the margins (Figures 1F and 2Q). The decrease in height and leaf width was independent of background (Table 1).
Table 1. The Phenotypic Effects of gif1 Alleles on the Decrease in Plant Height and Leaf Width in Different Genetic Backgrounds.
| Inbred Line | Genotype | No. of Samples | Plant Height (cm) | % Reduction | Leaf Width (cm) | % Reduction |
|---|---|---|---|---|---|---|
| A619 | gif1-2 | 20 | 98 ± 14 | 51 | 5.8 ± 1.2 | 44 |
| +/gif1-2 | 13 | 198 ± 36 | 10.4 ± 1.4 | |||
| B73 | gif1-2 | 5 | 67 ± 11 | 64 | 5.2 ± 1.0 | 46 |
| +/gif1-2 | 5 | 185 ± 12 | 9.6 ± 0.9 | |||
| Mo17 | gif1-2 | 7 | 74 ± 20 | 58 | 4.5 ± 0.4 | 54 |
| +/gif1-2 | 6 | 177 ± 19 | 9.8 ± 1.7 | |||
| B73/W22 | gif1-mum2 | 9 | 88 ± 20 | 62 | 4.6 ± 1.1 | 58 |
| gif1-mum2/+ | 6 | 230 ± 34 | 11 ± 0.9 | |||
| A619/W22 | gif1-mum2/gif1-2 | 7 | 129 ± 47 | 37 | 6.1 ± 0.8 | 47 |
| gif1-mum2/gif1-mum3 | 9 | 139 ± 31 | 32 | 8.3 ± 2.2 | 28 | |
| +/gif1-2 | 16 | 204 ± 20 | 11.5 ± 1.1 | |||
| A619/W22 | gif1-mum3/gif1-2 | 10 | 160 ± 13 | 25 | 10.0 ± 1.0 | 12 |
| +/gif1-2 | 9 | 214 ± 14 | 11.3 ± 0.8 |
The phenotypic values of plant height and leaf width are shown as the mean value ± sd.
To determine if the small leaf size was due to the presence of fewer or smaller cells, we observed the lower epidermal cells of leaves. Cell number per microscopic field in the wild type was significantly more than in gif1-1 (P = 3.0E-09, n = 30; Figures 1H and 1I), but cells were larger in gif1-1 (P = 7.39E-08, n = 100; Figures 1H and 1J). This result points to supra-cellular control of leaf shape in which individual cells try to compensate for fewer cells by expanding more than normal. This phenomenon is also observed in Arabidopsis gif1 an3 mutants (Horiguchi et al., 2005).
Maize gif1 Mutants Have a Loss of Determinacy in Axillary Meristems
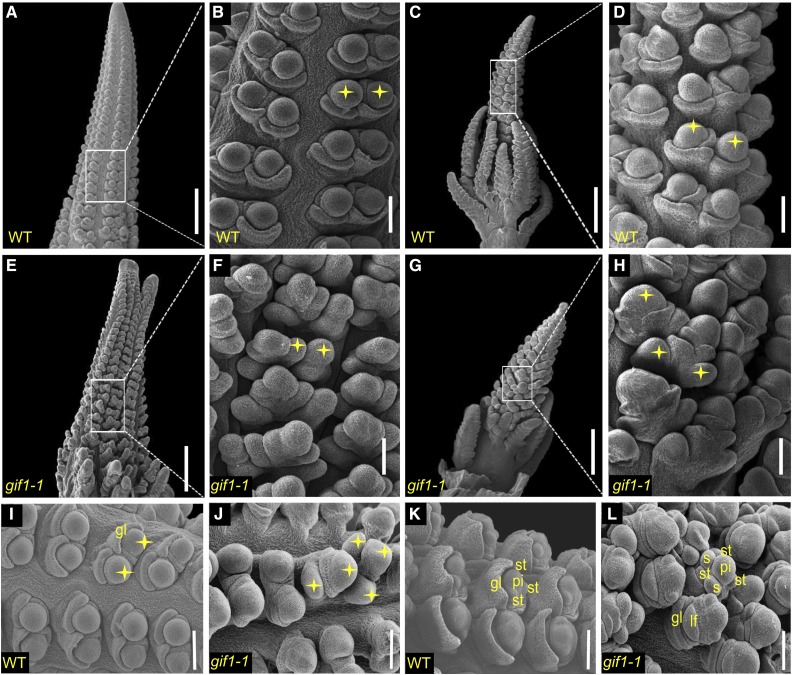
To better understand the ontogeny of the defective inflorescence architecture, we subjected the developing gif1-1 tassels and ears to scanning electron microscopy. The wild-type female inflorescence meristem (IM) forms rows of spikelet pair meristems (SPMs), which produce two spikelet meristems (SMs) (Figures 3A and 3B), and these initiate two floral meristems (FMs). All three meristem types (SPM, SM, and FM) are considered determinate, as they produce a defined number of organs and then terminate (Bortiri et al., 2006; Chuck et al., 2007; Laudencia-Chingcuanco and Hake, 2002). The lower floral meristem aborts, as do the stamens, to make a single female flower per spikelet. The male inflorescence meristem is similar but first produces branch meristems that themselves produce SPMs (Figures 3C and 3D). Pistils in the tassel abort, thereby producing male flowers.
Figure 3.
Male and Female Inflorescences Observed by Scanning Electron Microscopy.
(A), (B), and (I) SMs in developing ears of the wild type.
(C) and (D) SMs in developing tassels of the wild type.
(E) and (F) SMs in developing ears of gif1-1.
(G) and (H) SMs in developing tassels of gif1-1.
(J) Indeterminacy of tassel SPM shown by supernumerary SM.
(K) FM of wild-type ear.
(L) Indeterminacy of FM shown by supernumerary floral organs. Stars indicate SMs. st, stamen; pi, pistil; gl, outer glume; lf, lower floret; s, supernumerary organs.
Bars = 500 μm in (A), (C), (E), and (G) and 100 μm in (B), (D), (F), and (H) to (L).
In the gif1-1 mutant, the tips of both ear and tassel inflorescence meristems were fasciated (Figures 3E and 3G). The fasciation was also observed in the tassel and ear of gif1-2 and was quite severe in the ear (Figures 2D to 2H). The axillary meristems found inside the normally sterile husk leaves had small ears that were usually fasciated (Figures 2M and 2N). At the same time, SPMs were indeterminate, with more than two SMs generated from an SPM in both the tassel and ear (Figures 3F and 3H). We also observed reduced and developmentally delayed branch meristems in the tassel (Figure 3G), but increased short branches in the ear (Figure 3E) compared with the wild type (Figure 3A). Glume primordia produced from the SM of male florets were arrested (Figure 3L), and degeneration of the lower floret and stamen primordia of the female inflorescence was delayed (Figure 3J) compared with the wild type (Figures 3I and 3K).
These results suggest that the function of GIF1 is tissue specific. Determinate axillary meristems become indeterminate in the gif1 mutants and the indeterminate inflorescence meristems become fasciated. This loss of control contrasts with what happens in vegetative tissues, where leaves are narrow and internodes short, suggesting a loss of meristematic cells. Thus, GIF1 promotes determinacy in one tissue and promotes indeterminacy in others.
Loss of Function of gif1 Is Responsible for the Mutant Phenotypes
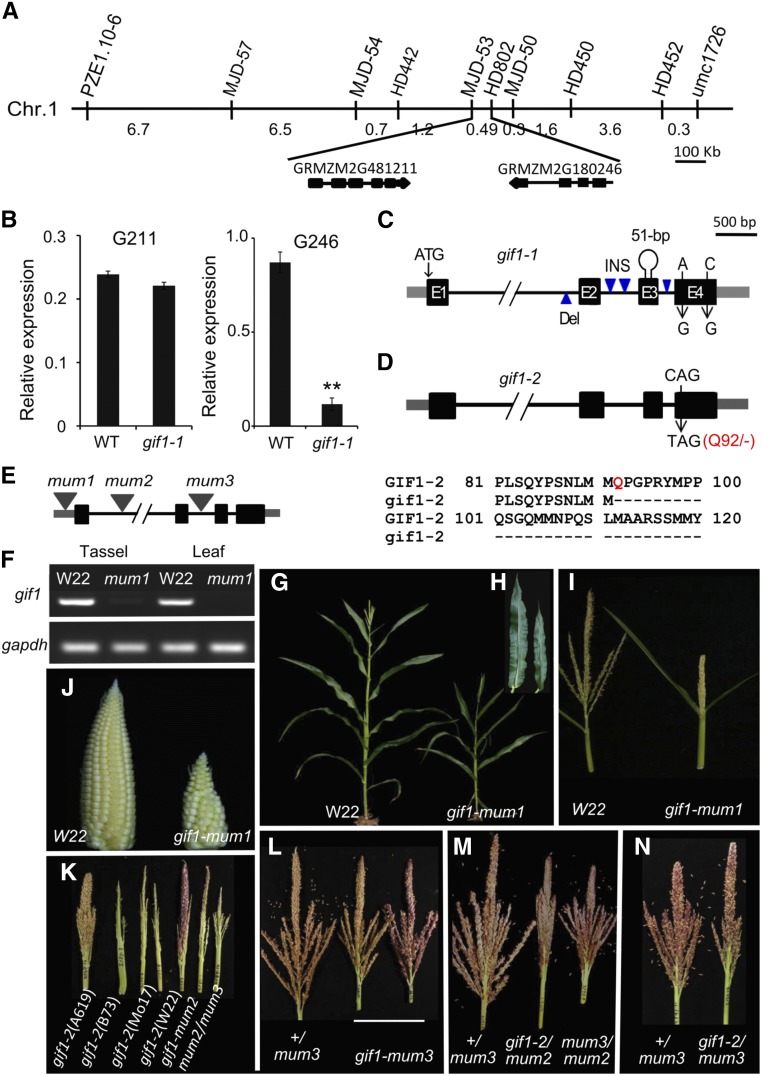
We mapped gif1-1 to the long arm of chromosome 1 by bulked segregant analysis and performed fine mapping by genotyping 4270 individuals from self-pollinated progenies of heterozygous plants (Figure 4A). Two newly developed markers (MJD-53 and HD802) delineated a 49-kb region containing two annotated genes, GRMZM2G481211 and GRMZM2G180246. The former encodes a CBS domain-containing protein that is expressed at a low level in immature tassels and did not show an expression difference between the wild type and gif1-1 (Figure 4B). The latter is homologous to AtGIF1/ANGUSTIFOLIA3 (AN3) and OsGIF1 (Supplemental Figure 2). Importantly, the expression of GRMZM2G180246 was significantly higher in the wild type compared with gif1-1 in immature tassels (Figure 4B). Furthermore, we sequenced the two genes and found that the DNA sequence of GRMZM2G481211 was conserved between the wild type and gif1-1, while GRMZM2G180246 exhibited sequence differences between the wild type and gif1-1, including four insertions/deletions in introns, two single nucleotide polymorphisms in exon 4, and a 51-bp insertion composed of a 43-bp Popin transposon and an 8-bp target site duplication in the third exon (Figure 4C). These results suggest that the 51-bp insertion leads to a loss of function of AN3/GIF1 in gif1-1.
Figure 4.
Molecular Cloning of gif1-1 and Complementation Test.
(A) Two markers delineated a 49-kb region containing two annotated genes: GRMZM2G481211 and GRMZM2G180246.
(B) Expression of GRMZM2G481211 (left) and GRMZM2G180246 (right) in immature tassels (∼5 mm) of wild type and gif1-1. The columns are the mean value of expression level detected in three separate experiments, each with three technical replicates. Error bar show the sd.
(C) Genic lesions of gif1-1. Exons are represented by black boxes; UTRs are indicated by gray boxes; introns are indicated by thin black lines.
(D) Genic lesion of gif1-2; the CAG (Q92) codon is mutated to TAG (stop).
(E) Diagram of gif1 Mutator insertion sites.
(F) gif1 expression in the wild type (W22) and gif1-mum1.
(G) A gif1-mum1 plant during vegetative development.
(H) Excised mature leaf blades of gif1-mum1.
(I) Tassel phenotype of gif1-mum1.
(J) Ear phenotype of gif1-mum1.
(K) Tassel phenotypes of mutants in different inbred backgrounds.
(L) Tassels of gif1-mum3 in the family 2189.
(M) Tassels of +/gif1-mum3, gif1-2/gif1-mum2, and gif1-mum2/gif1-mum3 segregating in the family 2193.
(N) Tassels of +/gif1-mum3 and gif1-2/gif1-mum3 in the family 2192.
gif1-2 was mapped to chromosome 1 using bulked segregant mapping. Analysis of DNA from 668 mutant plants identified 64 recombinants, narrowing the position to a 2.76-Mb region containing 70 genes. RNA-seq analysis was performed to identify the candidate gene. gif1 was the only gene in the interval with a T to C mutation, resulting in a CAG (Gln/Q) converted into a TAG (stop codon) (Figure 4D).
To further confirm the identity of the gif1 gene, three lines with UniformMu insertions in GRMZM2G180246 were obtained from the Maize Genetics Cooperation Stock Center (http://maizecoop.cropsci.uiuc.edu). A Mu7 element was located in the 5′-untranslated region (UTR) of gif1-mum1, while gif1-mum2 and gif1-mum3 contain Mutator elements located in the first and second intron of gif1, respectively (Figure 4E). The Mu insertion in gif1-mum1 led to a significant reduction in gif1 expression in immature tassels and developing leaves relative to that in W22 (the inbred background of UniformMu) (Figure 4F). Genotyping 202 individuals from self-pollinated progeny of +/gif1-mum1 identified 48 gif1-mum1 homozygous individuals. All gif1-mum1 plants developed a semidwarf phenotype (Figure 4G), with narrow and short leaves (Supplemental Table 1; Figure 4H), small female and male inflorescences (Figures 4I and 4J), and short pistils on the ear (Supplemental Figure 3A). Plants genotyped for the homozygous gif1-mum2 allele were similar, with reduced height, narrow leaves (Table 1), and tassels similar to those of gif1-1 and gif1-2 in multiple inbreds (Figure 4K).
We crossed gif1-2 to gif1-mum3 and gif1-mum2 for complementation tests. Although gif1-mum3 homozygotes are slightly shorter with smaller tassels (Figure 4L), gif1-mum3 failed to complement gif1-2 and gif1-mum2 (Figures 4K to 4N, Table 1; Supplemental Figures 3B and 3C). The double gif1-2/gif1-mum2 heterozygotes had inflorescence phenotypes including fewer upright tassel branches, increased spikelet density, branched ears, and axillary husk ears (Figure 4M; Supplemental Figures 3D to 3G). Collectively, these results confirm that mutations in GRMZM2G180246 are responsible for the gif1 phenotype and that the inflorescence phenotypes are a consistent trait in multiple gif1 alleles.
gif1 Is Expressed in Actively Dividing Cells of Shoot Apical and Inflorescence Meristems
We analyzed the expression of gif1 by qRT-PCR. gif1 was highly expressed in dividing tissues, including shoot apical meristems (SAMs), ∼5-mm tassels and 5- to 10-mm ears (Supplemental Figure 4A). This result was supported by RNA-seq data (Supplemental Figure 4B), showing that gif1 expression was higher in early stages (stage 1 and stage 2) than in later stage (stage 3) of tassel development and that its expression in IM and SPM was higher than in SM and FM of ears. RNA-seq data also showed that gif1 expression is reduced in stage 1 of all three ramosa (branched) mutants, ra1, ra2, and ra3 (Eveland et al., 2014).
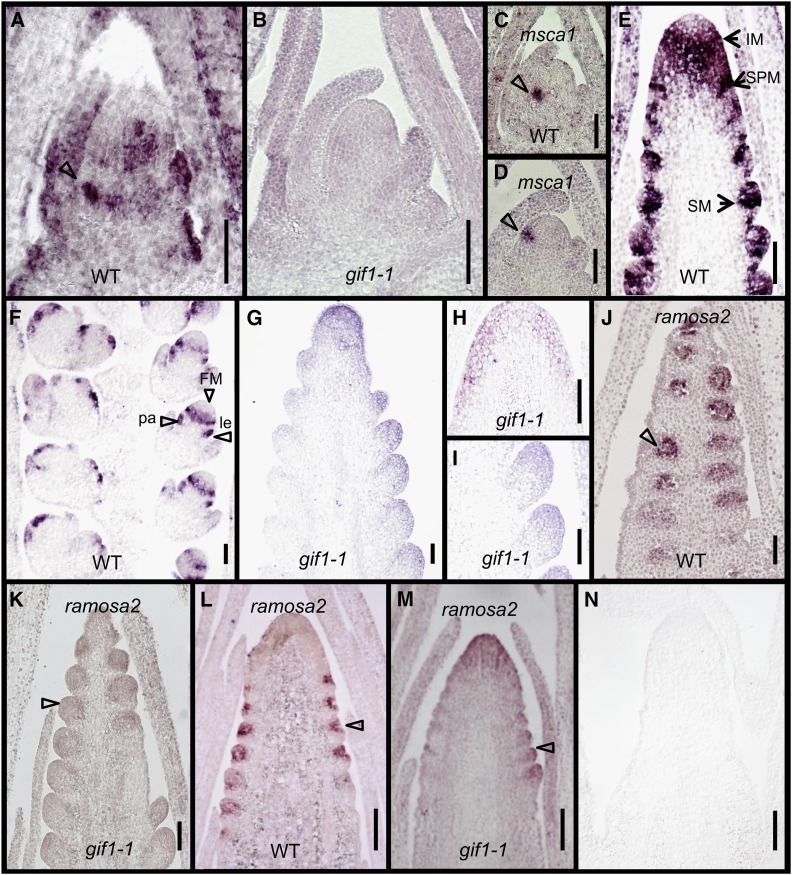
mRNA in situ hybridization showed that gif1 transcripts were enriched in the SAM but were excluded from the tip (Figures 5A and 5B). Strong expression was found in young leaf primordia and at the positions of leaf margins. Expression at the base of the SAM, adjacent to the recently initiated leaf, was detected at the site of the future internode. The SAM and leaf expression pattern was absent in gif1-1 (Figure 5B). We used the glutaredoxin gene msca1 (Yang et al., 2015) as a positive control for gif1-1 meristems. msca1 hybridized to the leaf initial cells of gif1-1 meristems in a similar pattern to that of wild-type meristems (Figures 5C and 5D). High levels of expression were seen in the IM except at the tip, and in or surrounding SPM and SM (Figure 5E). Expression patterns at slightly later stages include a ring around the FM and in the lemma and palea primordia (Figure 5F). Expression was not detected in the mutant (Figures 5G to 5I). The expression pattern overlapped with that of ra2 in the SPM and SM meristems (Figures 5J and 5L) (Bortiri et al., 2006). The expression of ra2 was greatly reduced in the mutant (Figures 5K and 5M). In control experiments, a 443-bp sense probe produced no signal (Figure 5N). The expression patterns of gif1 are consistent with the defects in internode elongation, leaf width, and inflorescence meristem determinacy in gif1-1.
Figure 5.
mRNA in Situ Hybridization Pattern of gif1.
(A), (B), and (E) to (I) Hybridizations with a gif1 antisense probe.
(A) Wild-type shoot meristem. Arrow at the margin of a leaf primordium.
(B) gif1-1 shoot meristem.
(C) and (D) Expression domains of msca1 in wild-type (C) and gif1-1 (D) shoot meristem. Arrow indicates the future leaf primordium expressing msca1.
(E) and (F) Immature tassels of the wild type.
(G) to (I) Immature tassels of gif1-1.
(J) to (M) Hybridizations with a ramosa2 (ra2) antisense probe.
(J) and (K) Immature tassels of wild type (J) and gif1-1 (K).
(L) and (M) Immature ears of wild type (L) and gif1-1(M).
(N) Hybridization of a wild-type inflorescence with a gif1 sense probe.
le, lemma; pa, palea. Bars = 100 μm.
GIF1 Protein Interacts with Multiple GRFs
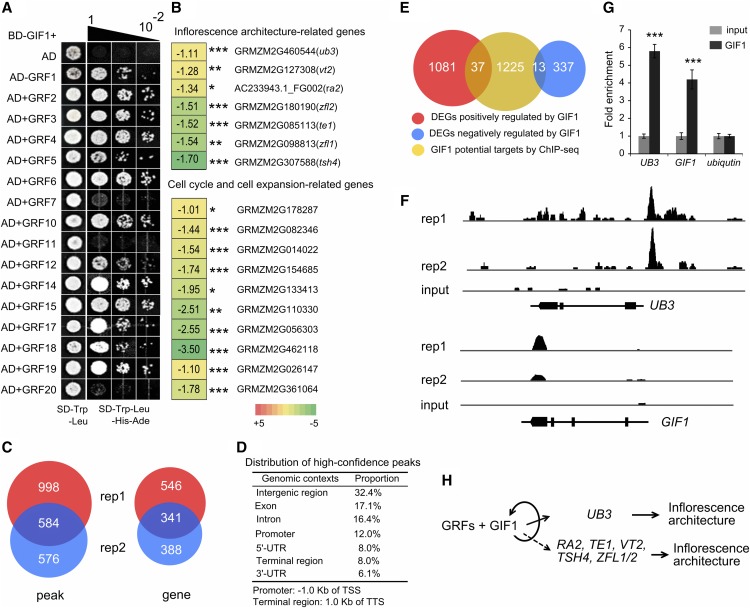
It is well established that GIF1 is a transcriptional coactivator that interacts with GRF1 in Arabidopsis (Kim and Kende, 2004). To identify the maize GRFs that interact with GIF1, we performed yeast two-hybrid assays. GIF1 interacted in vitro with 13 of 16 GRFs tested (Figure 6A). Only GRF7, GRF11, and GRF20 did not interact. The results were consistent with the findings of Nelissen et al. (2015), who performed tandem affinity purification followed by mass spectrometry using leaf and ear tissue, except for GRF11, which they found to be an interactor. Interaction of GIF1 with tissue-specific GRFs may explain the diversity of gif1 phenotypes, for example, why loss of indeterminacy is found in the leaf and stem, while gain of indeterminacy is found in the inflorescence. These tissue-specific GIF/GRF complexes may then regulate distinct downstream targets.
Figure 6.
GIF1-Interacting Proteins and Association Mapping.
(A) Identification of GIF1-interacting GRFs by yeast two-hybrid analysis. BD, binding domain; AD, activation domain.
(B) Expression changes for inflorescence architecture genes, and cell cycle and cell expansion genes in ∼5-mm tassels of gif1-1 compared with wild-type siblings. The number in the box represents expression alteration of the gene in log2 (RPKM in gif1-1/RPKM in the wild type). *P < 0.001, **P <0.0001, and ***P <0.00001.
(C) Venn diagram showing the number of peaks and genes identified in two biological replicates. Each replicate contained 10 tassels. rep, replicate.
(D) Distribution of high-confidence peaks. GIF1 binds in various genomic contexts, with high proportion of peaks located within genes. TSS, transcription start site; TTS, transcription termination site.
(E) Venn diagram representing a comparison between RNA-seq and ChIP-seq results.
(F) GIF1-bound regions in UB3 and GIF1. GIF1 can binds to its own fourth exon and 3′-UTR.
(G) PCR verification of GIF1-bound regions. The columns are the mean value of fold enrichment detected in three separate experiments, each with three technical replicates. Error bar show the sd. The statistical significance was estimated using a Student’s t test. *P < 0.05, **P < 0.01, and ***P < 0.001.
(H) A hypothetical model describing the GIF1 regulatory pathway in maize inflorescence development.
gif1 Regulates Expression of Cell Cycle, Inflorescence Development, and Hormone-Related Genes
To gain insight into transcriptional regulation by GIF1, we profiled genome-wide expression changes in the developing tassel of gif1-1 using RNA-seq. RNA-seq reads from two biological replicates of the wild type and gif1-1 were mapped separately to the maize reference genome (B73 RefGen_v3) and were used to quantify expression of the high-confidence filtered gene set. Each replica contained 10 tassels. We identified 1468 differentially expressed genes (DEGs) (q < 0.05). Among these, 350 (24%) DEGs were upregulated, while 1118 (76%) were downregulated in the gif1-1 tassel (Supplemental Data Set 1) relative to the wild type. We validated the expression patterns of 19 randomly selected DEGs using qRT-PCR (Supplemental Figure 5A). Gene Ontology analysis revealed that the downregulated genes were highly enriched for biological processes associated with transcriptional regulation, biosynthetic processes, metabolic processes, cellular processes, response to stimulus, and protein modification and were also enriched for molecular function categories related to regulation of transcription, transferase and catalytic activity (Supplemental Figure 5B).
Many downregulated genes in the gif1-1 mutant encode key regulators of axillary meristem identity and determinacy in maize, such as ub3, ra2, terminal ear1 (te1), Zea floricaula/leafy1 (zfl1), and zfl2 (Figure 6B; Supplemental Table 2). Auxin biosynthesis and signaling genes were also significantly downregulated in the tassel of gif1-1, as were auxin biosynthesis gene vanishing tassel2 (vt2) and auxin-responsive genes including a number of auxin-responsive GH3 family members and Aux/IAA gene family members. In addition to auxin, some brassinosteroid- and gibberellin-related genes were also downregulated in the gif1-1 tassel. Intriguingly, many of the genes that were significantly downregulated in gif1-1 are involved in the cell cycle and cell expansion, such as six cyclin-like protein-encoding genes and three expansin protein-encoding genes (Figure 6B; Supplemental Table 2). Of the GRF family members, grf3 was upregulated, while both grf7 and grf17 were significantly downregulated. The expression of gif2 and gif3 was not affected in gif1-1 tassels compared with the wild type.
To determine the targets occupied by GIF1, we performed chromatin immunoprecipitation sequencing (ChIP-seq) to immunoprecipitate GIF1-bound DNA regions in immature tassels (∼5 mm) of transgenic maize overexpressing GIF1-GFP using GFP antibody with two biological replicates. We identified 2158 high-confidence peaks by comparing significantly GIF1-enriched peaks with the input control (P < 1e-05) (Figure 6C). A total of 1582 peaks were detected in replicate 1, 1160 peaks were detected in replicate 2, and 584 peaks were shared between replicates. GIF1 bound in various genomic contexts, with a high proportion (47.6%) of binding within gene bodies and 12.0% of binding within 1.0-kb promoter regions upstream of the transcription start site (Figure 6D). Within 10 kb of high-confidence peaks from either replicate or both, we identified 1275 genes as putative targets of GIF1, with an overlap of 341 genes (Figure 6C; Supplemental Data Set 2). Putative GIF1 targets include a number of known transcriptional regulators, including UB3, ZMPLATZ5, ZMARR7, bHLH, and MYB family members (Supplemental Data Set 2). We found 50 differentially expressed genes in the gif1-1 mutant that were bound by GIF1 in either replicate or both, including 37 downregulated and 13 upregulated genes (Figure 6E), suggesting that GIF1 can act as an activator or repressor of gene expression. GIF1 could strongly bind to its own gene body, showing self-regulation at the transcriptional level (Figures 6F and 6G). ub3 is a strong candidate for a key target gene. ChIP-qPCR showed a 4-fold enrichment in binding over the Input control, and the RNA-seq showed a 50% decrease in expression. Like gif1 mutants, ub3 mutant plants have fasciated meristems with reduced tassel branches (Chuck et al., 2014). The expression patterns of gif1 and ub3 overlap in the shoot apical and inflorescence meristems. Four other genes were differentially expressed and bound by GIF1 in both replicates (Supplemental Figure 6).
DISCUSSION
The gif1 mutation affects both lateral organs and meristems: mutants have narrow leaves, short internodes, fewer tassel branches, and indeterminate axillary meristems compared with the wild type. Despite this pleiotropy, the phenotype is consistent between alleles and in different inbred backgrounds, unlike other pleiotropic mutants analyzed (Rosa et al., 2017). We positionally cloned the gene responsible for the mutant phenotype from two distinct populations and found that it encodes the maize ortholog of GIF1, also known as AN3. Analysis of the direct and indirect targets begins to explain these striking phenotypes.
The maize mutant shares some aspects of the Arabidopsis mutant phenotype, but affects many more tissues and processes. In Arabidopsis, loss of GIF1/AN3 function results in plants with small and narrow leaves caused by the repression of cell proliferation (Kim and Kende, 2004; Horiguchi et al., 2005). Unlike the Arabidopsis mutants, maize gif1 mutants are also dwarf due to short and asymmetric internodes. The narrow leaves and reduced stature correlate with the expression domain of gif1, which is highly expressed in presumptive leaf primordia and enriched at the base of the SAM in the future internode.
The Arabidopsis gif1 mutant is reported to be partially fertile, producing 30% fewer seeds than the wild type. This sterility is dependent on the dosage of grf mutations (Kim and Kende, 2004), indicating that the gif1 and grf mutations act synergistically to cause sterility in Arabidopsis. Four of the five maize alleles are completely male and female sterile in multiple genetic backgrounds. The female sterility may result from an indeterminate floral apex. Arabidopsis triple mutants (gif1, gif2, and gif3) are male and female sterile, but the female sterility appears to be due to failure of the carpel margin meristems and not to indeterminacy (Lee et al., 2014). These differences suggest that the GIFs function redundantly in Arabidopsis, whereas in maize, GIF1 alone plays a crucial role during reproduction. gif1 is expressed at much higher levels than either gif2 or gif3, which may explain the severe phenotype of the maize gif1 mutant. GIF1 also has novel roles in regulating determinacy because of the complexity of branching within maize inflorescences (Tanaka et al., 2013).
In addition to fertility defects, we found that inflorescence architecture is markedly affected in gif1 mutants, indicating the important role of gif1 in the regulation of inflorescence development. Tassel branch number is reduced, whereas short branches develop in ears. Both the tassel and ear apices are fasciated, revealing a failure to maintain a single, cohesive inflorescence meristem. The normally sterile husk leaves produce small axillary shoots that contain fasciated and branched ears. Spikelet pair meristems initiate more than two spikelets, indicating that the determinacy of axillary meristems is altered in gif1 mutants. Floral meristems are defective; the nucellus expands outside of the carpels and glumes arrest in all florets of gif1-1. Pistil degeneration in male florets and stamen degeneration in female florets is delayed. These findings uncover roles for GIF1 in promoting meristem determinacy within the inflorescence and regulating floral organ fate in maize.
Intriguingly, several well-characterized inflorescence architecture genes were significantly downregulated in the gif1-1 tassel. ub3 mutants have fewer tassel branches than wild-type and fasciated inflorescence meristems, which is consistent with the gif1 phenotype (Chuck et al., 2014). ramosa2 mutant tassels have indeterminate spikelet pair meristems, leading to branches in ears and increased indeterminacy in tassels (Bortiri et al., 2006). Downregulated expression of ramosa2 provides an explanation for the branching phenotype in the ear and the indeterminate spikelet pair meristems in the tassel. Although RA2 contains a LOB domain (Shuai et al., 2002), the phenotype is unique to maize. te1 is involved in the regulation of leaf initiation: Loss of its function causes an increase in the frequency of leaf primordia initiation and irregular internode length (Veit et al., 1998). The downregulation of te1 is consistent with the short and variable internodes of gif1. Double mutants of the maize LEAFY orthologs, zfl1 and zfl2, have the same ear phenotype as gif1, with axillary branches inside the husk leaves (Bomblies et al., 2003). The zfl1 zfl2 double mutant also has extra silks in the ear, like gif1. TE1 and LEAFY are found in other species, but their mutant phenotypes in maize are unique. Finally, VT2 is a grass-specific tryptophan aminotransferase that is involved in auxin biosynthesis and is required for vegetative and reproductive development. vt2 mutants are short, with barren tassels (Phillips et al., 2011). Similarly, gif1 mutants are short, with reduced tassel branch number. The downregulated expression of the key auxin biosynthesis gene vt2 and auxin-signaling genes in gif1-1 highlight the importance of auxin accumulation for inflorescence development and the regulation of GRFs-GIF1 complexes to auxin pathways. Thus, most of the inflorescence phenotypes of the maize gif1 mutant can be explained by the downregulation of putative target genes.
The inflorescence phenotypes of maize gif1 provide insights into GIF1-GRF complexes. In Arabidopsis, the miR396-GRF-GIF1 pathway controls leaf, seed, and SAM size (Debernardi et al., 2012; Gonzalez et al., 2010; Liang et al., 2014; Rodriguez et al., 2010). In rice, this regulatory pathway also modulates tissue and organ size (Che et al., 2015; Duan et al., 2015; Li et al., 2016). Intriguingly, overexpression of a recombinant GRF1 gene that is resistant to microRNA regulation led to larger leaves but shorter internodes (Nelissen et al., 2015). We show that maize GIF1 not only regulates organ size, but also the balance between indeterminacy and determinacy. The different aspects of the phenotype may be due to interaction of GIF1 with different GRFs or with the balance of either protein in a complex. GIF1 is highly expressed in meristematic tissue, whereas the expression levels of GRFs are more variable. Using tandem affinity purification followed by mass spectrometry, GIF1 copurified distinct GRFs in the ear compared with the leaf (Nelissen et al., 2015). Thus, the composition of GIF1-GRF complexes is likely to be highly dynamic based on the temporal and spatial patterns of GRF expression. We propose that diverse GIF1-GRF complexes are established in the SAM to promote leaf and internode development, in the IM to promote cohesive growth, and in axillary meristems to maintain determinacy (Figure 6H).
METHODS
Plant Material and Phenotypic Characterization
The maize (Zea mays) gif1-1 mutant was originally found in the BS238 family line, which was derived from the elite Stiff Stalk Synthetic population BSSSC9. Heterozygous siblings of BS238 (BS238H) were continuously self-pollinated to transmit the gif1-1 allele and to develop a segregating population for phenotyping and gene mapping. The selfed progeny showed a segregation ratio of 143 wild types to 49 gif1-1 phenotypes (mutants), which is consistent with the expected ratio of 3:1 (χ2 test, P < 0.87). The phenotypes of traits including plant height, ear height, total leaf number, length and width of ear leaf, leaf number above ear, tassel length, and tassel branch number were characterized in the S6 and S7 self-pollinated progeny of BS238H. The sample size for each phenotypic value was more than 30 individuals. In addition to the traits observed in mature plants, the height of more than 50 seedlings at 4 to 15 d after germination was measured to reveal the difference between the wild type and the gif1-1 mutant.
gif1-2 was identified in a screen of an EMS mutagenized A619 F2 population. Heterozygous, normal plants were crossed to B73 and self-pollinated. Homozygotes are sterile, so this process was repeated through three backcrosses before phenotypic analysis. Several plant traits were observed, including plant height, leaf number, length and width, and internode length. Observed post-flowering traits included: spikelet density of the tassel main spike, unfused carpels, and inflorescence fasciation in both the tassel and ear.
Three Mutator-mediated mutants, UFMu-05726 (gif1-mum1), UFMu-06440 (gif1-mum2), and UFMu-04889 (gif1-mum3) were requested from the Maize Genetics Cooperation Stock Center (http://maizecoop.cropsci.uiuc.edu). The Mutator insertion site was characterized using PCR with gif1-specific and Mutator-specific primers (Supplemental Table 3). Heterozygous individuals (+/gif1-mum1) were backcrossed to W22 for one generation. The heterozygous individuals (+/gif1-mum1) in the BC1 family were self-pollinated and then 202 individuals were developed for the progeny test. The phenotypes of plant, leaf and inflorescence architecture-related traits were measured at the mature plant stage.
To determine if the gif1-2 phenotype is caused by a mutation in GRMZM2G180246, we crossed gif1-2/+ by gif1-mum2 and gif1-mum3, as well as crossed gif1-mum2 by gif1-mum3, and then assayed the phenotype following genotyping. Traits measured included plant height, spikelet density, and gross morphology.
Cytological Observation
Immature male and female inflorescences (5–10 mm) of gif1-1, gif1-2, and the wild type were sampled at the same leaf age according to the description of Li et al. (2006). A sequential sampling procedure was performed from the beginning of the ninth leaf stage in order to observe the time course of inflorescence development. Inflorescence samples were fixed in a glutaraldehyde fixative solution (2.5% glutaraldehyde in 0.08 M phosphoric acid buffer) for 24 h at 4°C and then dehydrated through a graded series of ethanol from 30 to 90%. Samples were dried using a critical point dryer, sputter coated with gold palladium for 45 s, and observed on a Hitachi S-4700 scanning electron microscope at an accelerating voltage of 5 kV.
To measure cell size, mature leaves at the ear position of five individuals were sampled from gif1-1 and the wild type. The lower epidermal cells on the central region of the leaf were observed using a Nikon Eclipse 80i upright microscope on the bright-field setting. Two fields were observed for each leaf, and ∼20 to 30 cells per field were measured under ×10 magnification. The average length of measured cells from five leaves was used to represent cell size for each genotype.
Positional Cloning of gif1-1 and gif1-2
Preliminary mapping of gif1-1 was performed using bulked segregant analysis (Michelmore et al., 1991). Thirty gif1-1 and 30 wild-type individuals from BS238H selfed progeny were pooled. Genomic DNA was extracted using the CTAB method and quantified by NanoDrop 2000c (Thermo Scientific) to pool an equivalent amount from every sample. Genotyping analysis of two pools and all individuals in the F2 population were subjected to electrophoretic separation by 6% SDS-PAGE. Approximately 1000 pairs of SSR markers were used to identify the causal locus. Only one marker, umc1726 on the long arm of chromosome 1, was associated with the gif1-1 phenotype. To clone gif1-1, 14 newly developed markers (Supplemental Table 3) within a 10-Mb interval flanking umc1726 were used to further map gif1-1. Furthermore, a total of 4270 F2 individuals derived from the BS238H selfed progeny were genotyped to identify the recombinants that were then phenotyped at Wuhan (30°N, 114°E) and Sanya (18°N, 109°E), China in 2015.
Fertile, heterozygous sibs of gif1-2 were crossed to A188 and self-pollinated to create the F2 mapping population for bulked segregant analysis. Pools of DNA from 18 mutant and 18 normal individuals were sent to Iowa State for single nucleotide polymorphism genotype mapping. Linkage was found on chromosome 1L. Positional cloning using 668 mutants from a self-pollinated outcross to B73 was used to narrow this interval to 2.76 Mb containing 70 genes, at which point we found a lack of polymorphism. To narrow down the list of possible candidate genes responsible for the gif1-2 phenotype, an RNA-seq library was constructed from young gif1-2 ears, and likely EMS lesions were identified in the mapping interval. Total RNA was extracted from a pool of ten ears of gif1-2 and an RNA-seq library was prepared and indexed as described by Tsuda et al. (2014), except that 6 μg of total RNA was used rather than 2 μg. The library was sequenced on a NextSeq Illumina platform with PE75 (paired-end). Reads were aligned to the maize genome (AGPv3.31) using Tophat2-PE (2.0.9) (Trapnell et al., 2009) hosted in the Discovery Environment at Cyverse (Goff et al., 2011; Merchant et al., 2016). A GFF file containing coordinates of mRNA, exon, and coding sequence of nuclear-encoded genes for maize (AGPv3.31) was used to guide annotation. This and the genome file are available at Ensembl Genome Archive (ftp://ftp.ensemblgenomes.org/pub/plants/) (Kersey et al., 2016). Mapped reads were visualized using Integrative Genomics Viewer (IGV; Thorvaldsdóttir et al., 2013).
Gene Expression Analysis
To analyze the expression of gif1, developing tissues, including roots, shoots, and SAMs of seedlings at 14 d after germination, mature leaves, stems, immature ear (∼5 mm), and immature tassel (∼5 mm) were collected from gif1-1 and wild-type plants. Total RNA was extracted from plant tissues using Ambion Pure Link Plant RNA Reagent (Life Technologies) and reverse-transcribed with M-MLV reverse transcriptase (Life Technologies) according to the manufacturer’s instructions. qRT-PCR was performed using a SYBR Green qRT-PCR kit (Bio-Rad) according to the manufacturer’s instructions with three biological replicates, each replica contained 10 individuals. The maize gapdh gene (GRMZM2G046804) was used as the internal control. All reactions were performed on a CFX96 real-time system (Bio-Rad). Total RNA was extracted from immature tassels and mature leaves of gif1-mum1 and W22. All primers used for qRT-PCR and RT-PCR analysis are listed in Supplemental Table 3). Expression data for grfs and gifs in B73 were downloaded from the qTeller website (www.qteller.com) and the MaizeGDB website (www.maizegdb.org).
Phylogenetic Analysis
The GIF sequences of maize, rice (Oryza sativa), and Arabidopsis thaliana were retrieved from the MaizeGDB (http://maizegdb.org/), TIGR (http://www.tigr.org/), and TAIR (http://www.arabidopsis.org/) databases, respectively. The GIF sequences of poplar (Populus trichocarpa) and soybean (Glycine max) were retrieved from Gramene (http://www.gramene.org/). Multiple sequence alignment was performed by ClustalX 2.0 using default parameters. The phylogenetic analyses were constructed by MEGA software version 6.0 using the neighbor-joining method. Bootstrap analysis was performed with 1000 replicates, and bootstrap values are shown at each node. Protein sequences used to produce the phylogenetic tree are shown in Supplemental File 1.
Transcriptome Profiling
Ten immature tassels (∼5 mm) from homozygous gif1-1 mutant plants and from wild-type siblings were collected and pooled for each of two biological replicates. Fresh immature tassels were immediately frozen in liquid nitrogen. Total RNA was extracted from each pool using Trizol (Life Technologies, Invitrogen). After removing DNA with RQ1 DNase (Promega), 10 μg of the total RNA was used for RNA-seq library preparation. Polyadenylated mRNAs were purified and concentrated with oligo(dT)-conjugated magnetic beads (Life Technologies). Purified mRNAs were fragmented at 95°C for 1 min, followed by end repair and 5′ adaptor ligation. Reverse transcription was then performed with a RT primer harboring a 3′ adaptor sequence and randomized hexamer. The cDNA was purified and amplified, and PCR products corresponding to 200 to 500 bp were purified, quantified, and subjected to paired-end sequencing on the Illumina HiSeq 2000 system at BGI.
For RNA-seq analysis, clean reads were mapped to the maize reference genome (B73 RefGen_v2) using SOAPaligner/SOAP2 (Li et al., 2009) with no more than five mismatches allowed in the alignment. Gene expression levels were calculated using the RPKM method (reads per kilobase transcriptome per million mapped reads). The DEGs in gif1-1 and the wild type were identified as those showing at least 2-fold difference between gif1-1 and the wild type at an adjusted false discovery rate ≤0.001. The DEGs are listed in Supplemental Data Set 1. To confirm the DEGs, 19 randomly selected genes were analyzed by qRT-PCR with the gene-specific primers listed in Supplemental Table 3, using the maize GAPDH gene (GRMZM2G046804) as the internal control. Gene ontology analysis (by Cytoscape v3.2.1) and KEGG analysis (Kanehisa et al., 2008) was used to identify pathways enriched among the DEGs.
mRNA in Situ Hybridization
Shoot apical meristems of seedlings at 14 d after germination and developing inflorescences (∼5-mm tassel and 2- to 5-mm ear) from the gif1-1 mutant and the wild type were fixed in a solution containing 5% formalin, 50% ethanol, and 5% acetic acid for 16 h at 4°C, which was then replaced with 70% ethanol twice and dehydrated with an ethanol series, substituted with xylene, embedded in Paraplast Plus (Sigma-Aldrich), and sectioned to a thickness of 8 μm. To construct sense and antisense RNA probes, the primer set GIF1_F, 5′-TGAACCCGCAGTCGCTGATG-3′, and GIF1_R, 5′- TCAAAGCTACCTGCTAATCC-3′, was used to amplify a 443-bp fragment including the 75-bp 3′-UTR and 368-bp coding region of gif1. The amplification products were cloned into pSPT18 (Roche) and linearized with HindIII and EcoRI, respectively. Sense and antisense probes were then synthesized using SP6 and T7 RNA polymerase, respectively, with digoxigenin-UTP as a label. The msca1 and ramosa2 probes were prepared as described by Yang et al. (2015) and Bortiri et al. (2006), respectively. RNA hybridization and immunologic detection of the hybridized probes were performed as described previously (Greb et al., 2003), with the addition of 8% polyvinyl alcohol to the detection buffer to minimize diffusion of the reaction products. The slides were exposed for ∼12 to 15 h before mounting and imaging and visualized under a microscope (Nikon eclipse 80i). gif1 in situ hybridization was performed using sense and antisense probes, with the sense probe used for the control.
Yeast Two-Hybrid Assay
The yeast one-hybrid assay was performed using the Matchmaker Gold Yeast One-Hybrid Library Screening System (Clontech) and Yeastmaker Yeast Transformation System 2 (Clontech) according to the manufacturer’s instructions. Based on the amino acid sequence QLQ (Gln-Leu-Gln) and the WRC (Trp-Arg-Lys) domains of nine GRFs in Arabidopsis, 16 GRF family genes (ZmGRFs) in the maize genome were identified using the Hidden Markov Model and isolated by amplifying cDNA from inflorescence meristems. To investigate the interaction between the GIF1 protein and 16 GRFs, the full-length coding sequences of GIF1 and 16 GRFs were fused with the GAL4-BD and GAL4-AD domains, respectively, in vectors pGBKT7 and pGADT7 (Clontech). The yeast strains Y2H (MATa, trp1-901, leu2-3, 112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2:-GAL1UAS–Gal1TATA–His3, GAL2UAS–Gal2TATA–Ade2 URA3:MEL1UAS –Mel1TATA AUR1-C MEL1) and Y187 (MATa, ura3-52, his3-200, ade2-101, trp1-901, leu2-3, 112, gal4Δ, gal80Δ, met–, URA3:GAL1UAS–Gal1TATA –LacZ, MEL1) were transformed with bait and prey plasmids, mated overnight, and plated on selective dropout medium without Leu/Trp or Ade/His/Leu/Trp. The primers used to produce these constructs are listed in Supplemental Table 3.
ChIP-Seq and Data Analysis
Approximately 1 g immature tassel (∼5 mm) tissue was harvested from 35Spro:GIF1-GFP plants grown in the greenhouse with two biological replicates. Expression of the transformed target gene was verified by protein gel blot analysis using GFP antibody (AB290; Abcam), at a dilution of 1:1000 (v/v) in Tris-buffered saline buffer containing 5% nonfat milk powder. The inflorescences were immediately cross-linked in buffer containing 1% formaldehyde for 15 min under a vacuum, followed by the addition of glycine to a concentration of 0.1 M and infiltration for 5 min. After three washes with distilled water (4°C), the cross-linked tissues were dried with paper towels and flash frozen in liquid nitrogen. The frozen tissues were ground thoroughly to a fine powder, which was then transferred to a precooled 50-mL tube with 20 mL of cold complete extraction buffer 1 (0.4 M sucrose, 10 mM Tris-HCl pH 8.0, 10 mM MgCl2, 5 mM β-mercaptoethanol, and Plant Protease Inhibitor Cocktail). Homogenized tissues were centrifuged for 20 min at 1000g at 4°C. The pellets were washed five times with 5 mL complete extraction buffer 2 (0.25 M sucrose, 10 mM Tris-HCl, pH 8.0, 10 mM MgCl2, 1% Triton X-100, 5 mM β-mercaptoethanol, and Plant Protease Inhibitor Cocktail) and once with extraction buffer 3 (1.7 M sucrose, 10 mM Tris-HCl, pH 8.0, 2 mM MgCl2, 0.15% Triton X-100, 5 mM β-mercaptoethanol, and Plant Protease Inhibitor Cocktail). The washed pellets were resuspended in 300 μL of sonication buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, 1% SDS, and Plant Protease Inhibitor Cocktail), and the suspension was treated with a Bioruptor for 8 to 10 cycles with the settings 30 s ON/30 s OFF at 4°C. The sonicated sample was centrifuged for 10 min at 12,000g at 4°C, and the supernatant was collected and used for chromatin isolation. Chromatin extracted from 35Spro:GIF1-GFP plants was immunoprecipitated with anti-GFP antibody (A11122; Invitrogen) with a Plant ChIP-seq kit (Diagenode) according to the manufacturer’s instructions. Following de-cross-linking, isolation, and purification of the immunoprecipitated DNA, libraries were constructed using an Ovation Low Input DR kit (NuGEN Technologies). Two input and two IP libraries were subjected to sequencing on the Illumina HiSeq 2000 platform.
ChIP-seq reads were aligned to the maize reference genome (AGPv2) using Hisat2 (v.2.0.5; Kim et al., 2015). Only uniquely mapped reads were considered for further processing. PCR duplicates were removed using PICARD Picard MarkDuplicates (v.2.9.0; http://picard.sourceforge.net/). Peak calling was performed with MACS (v.1.4.2) (Zhang et al., 2008). Peaks were identified as significantly enriched (P < 1e-05) in each of the ChIP-seq libraries compared with input DNA. FGS gene model within 10 kb of the peak summit was considered as a putative target of GIF1. ChIP tracks showing GIF1-GFP fusion protein binding sites were visualized using IGV (Thorvaldsdóttir et al., 2013).
To detect specific DNA targets, ChIP-qPCR was performed to quantify DNA targets immunoprecipitated by anti-GFP antibodies relative to input DNA using SYBR Green qPCR Master Mix (Bio-Rad) with three biological replicates each with three technical replicates. The DNA target-specific primers used for the ChIP-PCR assay are listed in Supplemental Table 3. The abundance of a target was normalized to nonspecific genomic regions, and fold enrichment of the DNA target relative to the input sample was then calculated. Significance of differences was estimated by a Student’s t test.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: gif1, MG893020; ra2, AC233943.1_FG002; ub3, GRMZM2G460544; vt2, GRMZM2G127308; and te1, GRMZM2G085113. SRA accession numbers are as follows: SRP131943 containing SRR6660960, SRR66609601, and SRR66609602 for ChIP-seq, and SRR6660963, SRR6660964, SRR6660965, and SRR6660966 for RNA-seq.
Supplemental Data
Supplemental Figure 1. Phenotypes of agriculturally important traits in wild type and gif1-1.
Supplemental Figure 2. A phylogenetic tree of GRF-interacting factor1.
Supplemental Figure 3. Ear phenotypes of Mutator insertion alleles.
Supplemental Figure 4. Expression of maize growth-regulating factor family genes (grfs) and GRF-interacting factor family genes (gifs).
Supplemental Figure 5. Validation of RNA-seq differentially expressed genes and enriched GO terms.
Supplemental Figure 6. GIF1-bound regions identified by ChIP-seq and PCR verification of GIF1-bound regions in four genes that are differentially expressed.
Supplemental Table 1. Phenotypes of plant and inflorescence-related traits in W22 and gif1-mum1.
Supplemental Table 2. Representative categories of differentially expressed genes in immature tassel (∼5 mm) of wild type and gif1-1.
Supplemental Table 3. List of primer sequences used in this study.
Supplemental File 1. Alignment used to produce the phylogenetic tree shown in Supplemental Figure 2.
The following materials have been deposited in the DRYAD depository under accession number http://dx.doi.org/10.5061/dryad.5pf64.
Supplemental Data Set 1. Differentially expressed genes between wild type and the gif1-1 mutant.
Supplemental Data Set 2. Putative GIF1 target genes (GIF1 high-confidence peak within 10 kb).
Acknowledgments
This work was supported by the National Natural Science Foundation of China (91635303 and 31671700), the National Basic Research Program of China (2014CB138203), and Fundamental Research Funds for the Central Universities (52902-0900202929). R.S. was supported by a summer undergraduate research fellowship in 2016 and by a grant to Markus Pauly in 2017 (DE-SC001240). C.L. was supported by National Science Foundation IOS-1238202 and S.H. by ARS CRIS 5335-21000-013-00D.
AUTHOR CONTRIBUTIONS
S.H. and Z.Z. conceived and designed the experiments. D.Z., R.S., Y.Z., M.L., Z.C., W.S., and C.L. performed the experiments. Z.Z., S.H., and C.L. analyzed the data. W.S., Z.Z., S.H., and C.L. wrote the manuscript.
Footnotes
Articles can be viewed without a subscription.
References
- Alvarez J.P., Furumizu C., Efroni I., Eshed Y., Bowman J.L. (2016). Active suppression of a leaf meristem orchestrates determinate leaf growth. eLife 5: e15023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K., Wang R.L., Ambrose B.A., Schmidt R.J., Meeley R.B., Doebley J. (2003). Duplicate FLORICAULA/LEAFY homologs zfl1 and zfl2 control inflorescence architecture and flower patterning in maize. Development 130: 2385–2395. [DOI] [PubMed] [Google Scholar]
- Bortiri E., Chuck G., Vollbrecht E., Rocheford T., Martienssen R., Hake S. (2006). ramosa2 encodes a LATERAL ORGAN BOUNDARY domain protein that determines the fate of stem cells in branch meristems of maize. Plant Cell 18: 574–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che R., Tong H., Shi B., Liu Y., Fang S., Liu D., Xiao Y., Hu B., Liu L., Wang H., Zhao M., Chu C. (2015). Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat. Plants 2: 15195. [DOI] [PubMed] [Google Scholar]
- Chuck G., Meeley R., Irish E., Sakai H., Hake S. (2007). The maize tasselseed4 microRNA controls sex determination and meristem cell fate by targeting Tasselseed6/indeterminate spikelet1. Nat. Genet. 39: 1517–1521. [DOI] [PubMed] [Google Scholar]
- Chuck G., Whipple C., Jackson D., Hake S. (2010). The maize SBP-box transcription factor encoded by tasselsheath4 regulates bract development and the establishment of meristem boundaries. Development 137: 1243–1250. [DOI] [PubMed] [Google Scholar]
- Chuck G.S., Brown P.J., Meeley R., Hake S. (2014). Maize SBP-box transcription factors unbranched2 and unbranched3 affect yield traits by regulating the rate of lateral primordia initiation. Proc. Natl. Acad. Sci. USA 111: 18775–18780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daryanto S., Wang L., Jacinthe P.A. (2016). Global synthesis of drought effects on maize and wheat production. PLoS One 11: e0156362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debernardi J.M., Rodriguez R.E., Mecchia M.A., Palatnik J.F. (2012). Functional specialization of the plant miR396 regulatory network through distinct microRNA-target interactions. PLoS Genet. 8: e1002419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debernardi J.M., Mecchia M.A., Vercruyssen L., Smaczniak C., Kaufmann K., Inze D., Rodriguez R.E., Palatnik J.F. (2014). Post-transcriptional control of GRF transcription factors by microRNA miR396 and GIF co-activator affects leaf size and longevity. Plant J. 79: 413–426. [DOI] [PubMed] [Google Scholar]
- Duan P., Ni S., Wang J., Zhang B., Xu R., Wang Y., Chen H., Zhu X., Li Y. (2015). Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nat. Plants 2: 15203. [DOI] [PubMed] [Google Scholar]
- Eveland A.L., et al. (2014). Regulatory modules controlling maize inflorescence architecture. Genome Res. 24: 431–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinat W.C. (1959). The phytomer in relation to the floral homologies in the American Maydea. Bot. Mus. Lealf. Harv. Univ. 19: 1–32. [Google Scholar]
- Greb T., Clarenz O., Schafer E., Muller D., Herrero R., Schmitz G., Theres K. (2003). Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 17: 1175–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff S.A., et al. (2011). The iPlant Collaborative: cyberinfrastructure for plant biology. Front. Plant Sci. 2: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez N., et al. (2010). Increased leaf size: different means to an end. Plant Physiol. 153: 1261–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoe Kim J., Tsukaya H. (2015). Regulation of plant growth and development by the GROWTH-REGULATING FACTOR and GRF-INTERACTING FACTOR duo. J. Exp. Bot. 66: 6093–6107. [DOI] [PubMed] [Google Scholar]
- Howell S.H. (1998). Molecular Genetics of Plant Development. (Cambridge, UK: Cambridge University Press; ). [Google Scholar]
- Horiguchi G., Kim G.T., Tsukaya H. (2005). The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 43: 68–78. [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M., Katayama T., Kawashima S., Okuda S., Tokimatsu T., Yamanishi Y. (2008). KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36: D480–D484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey P.J., et al. (2016). Ensembl Genomes 2016: more genomes, more complexity. Nucleic Acids Res. 44: D574–D580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S.L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Choi D., Kende H. (2003). The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 36: 94–104. [DOI] [PubMed] [Google Scholar]
- Kim J.H., Kende H. (2004). A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 13374–13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijt S.J., et al. (2014). Interaction between the GROWTH-REGULATING FACTOR and KNOTTED1-LIKE HOMEOBOX families of transcription factors. Plant Physiol. 164: 1952–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudencia-Chingcuanco D., Hake S. (2002). The indeterminate floral apex1 gene regulates meristem determinacy and identity in the maize inflorescence. Development 129: 2629–2638. [DOI] [PubMed] [Google Scholar]
- Lee B.H., Wynn A.N., Franks R.G., Hwang Y.S., Lim J., Kim J.H. (2014). The Arabidopsis thaliana GRF-INTERACTING FACTOR gene family plays an essential role in control of male and female reproductive development. Dev. Biol. 386: 12–24. [DOI] [PubMed] [Google Scholar]
- Li R., Yu C., Li Y., Lam T.W., Yiu S.M., Kristiansen K., Wang J. (2009). SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25: 1966–1967. [DOI] [PubMed] [Google Scholar]
- Li S., et al. (2016). The OsmiR396c-OsGRF4-OsGIF1 regulatory module determines grain size and yield in rice. Plant Biotechnol. J. 14: 2134–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Ruf S., Bock R. (2006). Constancy of organellar genome copy numbers during leaf development and senescence in higher plants. Mol. Genet. Genomics 275: 185–192. [DOI] [PubMed] [Google Scholar]
- Liang G., He H., Li Y., Wang F., Yu D. (2014). Molecular mechanism of microRNA396 mediating pistil development in Arabidopsis. Plant Physiol. 164: 249–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machanick P., Bailey T.L. (2011). MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27: 1696–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore R.W., Paran I., Kesseli R.V. (1991). Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 88: 9828–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant N., Lyons E., Goff S., Vaughn M., Ware D., Micklos D., Antin P. (2016). The iPlant collaborative: cyberinfrastructure for enabling data to discovery for the life sciences. PLoS Biol. 14: e1002342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen H., et al. (2015). Dynamic changes in ANGUSTIFOLIA3 complex composition reveal a growth regulatory mechanism in the maize leaf. Plant Cell 27: 1605–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omidbakhshfard M.A., Proost S., Fujikura U., Mueller-Roeber B. (2015). Growth-Regulating Factors (GRFs): A small transcription factor family with important functions in plant biology. Mol. Plant 8: 998–1010. [DOI] [PubMed] [Google Scholar]
- Phillips K.A., Skirpan A.L., Liu X., Christensen A., Slewinski T.L., Hudson C., Barazesh S., Cohen J.D., Malcomber S., McSteen P. (2011). vanishing tassel2 encodes a grass-specific tryptophan aminotransferase required for vegetative and reproductive development in maize. Plant Cell 23: 550–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez R.E., Mecchia M.A., Debernardi J.M., Schommer C., Weigel D., Palatnik J.F. (2010). Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 137: 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa M., Abraham-Juárez M.J., Lewis M.W., Fonseca J.P., Tian W., Ramirez V., Luan S., Pauly M., Hake S. (2017). The maize MID-COMPLEMENTING ACTIVITY homolog CELL NUMBER REGULATOR13/NARROW ODD DWARF coordinates organ growth and tissue patterning. Plant Cell 29: 474–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai B., Reynaga-Peña C.G., Springer P.S. (2002). The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 129: 747–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun P., Zhang W., Wang Y., He Q., Shu F., Liu H., Wang J., Wang J., Yuan L., Deng H. (2016). OsGRF4 controls grain shape, panicle length and seed shattering in rice. J. Integr. Plant Biol. 58: 836–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka W., Pautler M., Jackson D., Hirano H.-Y. (2013). Grass meristems II: inflorescence architecture, flower development and meristem fate. Plant Cell Physiol. 54: 313–324. [DOI] [PubMed] [Google Scholar]
- Thorvaldsdóttir H., Robinson J.T., Mesirov J.P. (2013). Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14: 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K., Kurata N., Ohyanagi H., Hake S. (2014). Genome-wide study of KNOX regulatory network reveals brassinosteroid catabolic genes important for shoot meristem function in rice. Plant Cell 26: 3488–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K., Abraham-Juarez M.J., Maeno A., Dong Z., Aromdee D., Meeley R., Shiroishi T., Nonomura K.I., Hake S. (2017). KNOTTED1 cofactors, BLH12 and BLH14, regulate internode patterning and vein anastomosis in maize. Plant Cell 29: 1105–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Knaap E., Kim J.H., Kende H. (2000). A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. Plant Physiol. 122: 695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit B., Briggs S.P., Schmidt R.J., Yanofsky M.F., Hake S. (1998). Regulation of leaf initiation by the terminal ear 1 gene of maize. Nature 393: 166–168. [DOI] [PubMed] [Google Scholar]
- Yang F., Bui H.T., Pautler M., Llaca V., Johnston R., Lee B.H., Kolbe A., Sakai H., Jackson D. (2015). A maize glutaredoxin gene, abphyl2, regulates shoot meristem size and phyllotaxy. Plant Cell 27: 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]