Summary
Recent genomic analyses show that the earliest peoples reaching Remote Oceania – associated with Austronesian-speaking Lapita culture – were almost completely East Asian, without detectable Papuan ancestry. Yet Papuan-related genetic ancestry is found across present-day Pacific populations, indicating that peoples from Near Oceania have played a significant – but largely unknown – ancestral role. Here, new genome-wide data from 19 South Pacific individuals provide direct evidence of a so-far undescribed Papuan expansion into Remote Oceania starting ~2,500 years before present, far earlier than previously estimated and supporting a model from historical linguistics. New genome-wide data from 27 contemporary ni-Vanuatu demonstrate a subsequent and almost complete replacement of Lapita-Austronesian by Near Oceanian ancestry. Despite this massive demographic change, incoming Papuan languages did not replace Austronesian languages. Population replacement with language continuity is extremely rare – if not unprecedented – in human history. Our analyses show that rather than one large-scale event, the process was incremental and complex, with repeated migrations and sex-biased admixture with peoples from the Bismarck Archipelago.
Sahul – the continent comprising present-day Australia, Tasmania and New Guinea – was colonized by modern humans during the Pleistocene as early as 65,000 years before present1 (y BP). Yet it took more than 60,000 years for humans to move east of the Solomon Islands, from Near Oceania out into Remote Oceania2 (Fig. 1b). These seafaring Neolithic peoples, part of the Austronesian Expansion beginning ~5,500y BP, likely in present-day Taiwan and the nearby mainland3–5, carried farming technology and a major branch of the Austronesian languages6 into Island Southeast Asia, eventually reaching New Guinea and the Bismarck Archipelago and encountering indigenous Papuans. Here, at ~3,300y BP the Lapita Cultural Complex3,7 appeared – characterized by distinctive dentate-stamped pottery – and using the out-rigger sailing canoe, Lapita peoples expanded east, leap-frogging beyond the Solomon Islands8,9. They transported their landscapes3 and Oceanic languages out into Remote Oceania, first arriving in the Reef-Santa Cruz islands, Vanuatu10 and New Caledonia ~3,000y BP11, and rapidly navigated >800km of open ocean to Fiji, reaching western Polynesia by ~2,850y BP12.
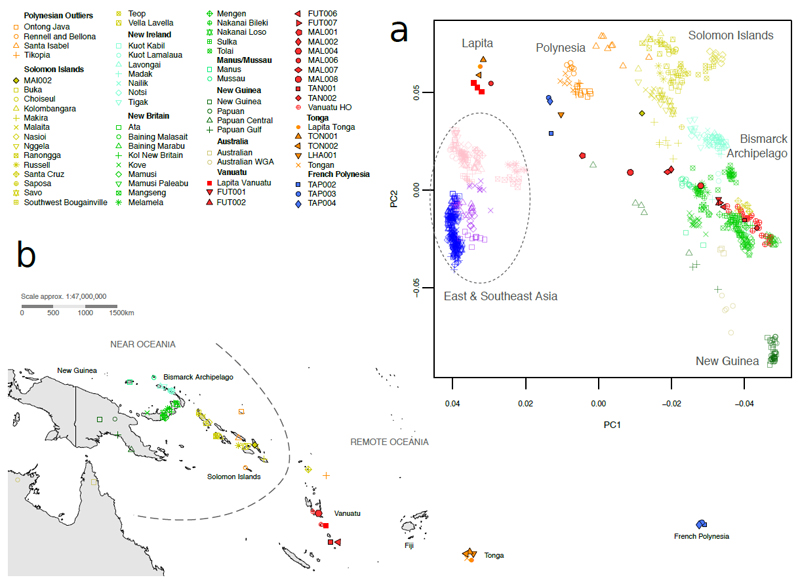
Fig. 1. Spatial and genetic distribution of ancient and present-day individuals.
(a) Principal component analysis of modern-day East Asian and Near and Remote Oceanian populations genotyped on the Affymetrix Human Origins Array, with 23 ancient individuals projected. Ancient samples are indicated by filled symbols – the new data from this study have a black border – and present-day samples are indicated by open symbols. (b) Regional map, showing locations of Near and Remote Oceanian sample populations and ancient individuals.
Uncovering the extent of interaction between incoming Austronesian-Lapita and indigenous Papuan peoples is critical to understanding all subsequent Pacific prehistory. ‘Papuan’ here refers to both the non-Austronesian languages found across New Guinea and a component of genetic ancestry, likely to have diverged from the ancestors of present-day East Asians at least 27,000y BP13. The linguistic, cultural and genetic diversity in New Guinea is immense, due to complex histories of differentiation since first arrival14. While the majority of Near Oceanians today speak Papuan languages, Remote Oceanians almost exclusively speak Oceanic languages of the Austronesian family15. Bayesian phylogenetic analyses of 400 of the >1,200 Austronesian languages5 broadly support the Express Train model of the Austronesian Expansion, whereby Austronesian-speaking groups had negligible cultural or genetic interaction with indigenous Papuans in Near Oceania before moving further into the Pacific. However, the genetic composition of the present-day South Pacific indicates a more complex history, comprising major East Asian-Austronesian and minor Papuan components of genome-wide ancestry (~79:21%16, ~87:13%13). Mitochondrial DNA (mtDNA)17 and Y-chromosome18,19 studies show that populations across Polynesia have maternal ancestry largely of Austronesian origin (>96%20) while the majority of their Y-chromosomes derive from Near Oceania (>60%20), confirmed in recent X-chromosome analyses13,21. This suggests that Oceanic-speaking populations – prior to or during the formation of the Lapita Cultural Complex – experienced significantly sex-biased admixture, involving women of Austronesian origin and Papuan men. This model requires that Lapita peoples, while maintaining Oceanic language(s), had admixed ancestry in Near Oceania prior to their eastward expansion into Remote Oceania. However, the first genome-wide ancient data from the region21 demonstrates – consistent with craniofacial analyses22 – that Papuan ancestry is largely absent in individuals from Lapita sites in both Vanuatu and Tonga. The present-day genetic ancestry of Remote Oceania can therefore only be explained by subsequent population expansion, carrying Papuan ancestry into the Pacific.
Vanuatu has been an important hub in the western Pacific23 from Lapita onwards. Uncovering the detailed demographic processes shaping the genetic and linguistic landscape of Vanuatu is thus crucial to understanding those of the wider Pacific. Here we provide the earliest direct evidence of Papuan genetic ancestry in Remote Oceania. Our results reveal that peoples from Near Oceania began arriving just a few centuries after the first Lapita settlements in Vanuatu. This was followed by an almost complete – yet incremental – replacement of Lapita-Austronesian by Bismarck Archipelago-like genetic ancestry.
Results
Ancient and modern genome-wide data
We recovered genome-wide and mitochondrial aDNA data from the bones or teeth of 19 individuals from archaeological sites 14C-dated to ~2,600-200y BP across Vanuatu (n=12), Tonga (n=3), French Polynesia (n=3) and the Solomon Islands (n=1) (Table 1, Supplementary table 1, Supplementary table 2, Methods). DNA was extracted24 and converted into double stranded genetic libraries25,26 in dedicated cleanroom facilities. Hybridization capture targeted the complete mitochondrial genome and ~1.24 million single nucleotide polymorphisms (SNPs) (1240K)27,28, followed by next generation sequencing. The isolated aDNA was authenticated based on the presence of typical deamination patterns, low levels of mtDNA contamination, X-chromosome contamination in males, and analyses were restricted, if necessary, to the likely endogenous deaminated sequences29 (Supplementary table 3, Supplementary table 4, Supplementary figure 1, Methods). The genome-wide aDNA was co-analyzed with four published Lapita samples21, 781 present-day Oceanian and East Asian samples genotyped for ~600K SNPs on the Affymetrix Human Origins (HO) Array21,30 and 308 high coverage genomes31. We also genotyped 27 ni-Vanuatu samples from the islands of Malakula and Efate (Methods, Supplementary figure 2) on the HO Array, with eight also shotgun sequenced (SG) at low coverage (0.6-3 fold) (Supplementary table 5). All newly generated data were analyzed alongside published genome-wide Illumina HumanCore-24 data from 754 individuals across Remote Oceania, including 610 from Vanuatu32 (Supplementary table 6).
Table 1.
Data description for the newly reported genome-wide data from 19 ancient individuals. Radiocarbon dating and ancient DNA summary statistics.
| Sample Name | Country, Island | Anatomical element | cal BP (AD/BC) 95.4% | Sex | mtDNA haplogroup | Y chromosome haplogroup | Damage restrict | Mean coverage e1240K | SNPs 1240K | Library type |
|---|---|---|---|---|---|---|---|---|---|---|
| FUT001 | Vanuatu, Futuna | L petrous | 1230-980 (720-970 AD) |
F | P1d2a | - | No | 1.289 | 647,595 | noUDG |
| FUT002 | Vanuatu, Futuna | R petrous | 1240-1000 (710-950 AD) |
F | M28b1 | - | No | 1.163 | 626,821 | UDGhalf |
| FUT006 | Vanuatu, Futuna | L petrous | 1270-1070 (680-880 AD) |
M | P1d2a | K2 | No | 0.748 | 453,192 | UDGhalf |
| FUT007 | Vanuatu, Futuna | R petrous | 1190-970 (760-980 AD) |
M | M28b1 | K2b1a3 | No | 0.596 | 392,622 | UDGhalf |
| LHA001 | Tonga, Tongatapu | Molar | 780-550 (1170-1400 AD) |
F | B4a1a1 | - | Yes | 0.048 | 37,058 | UDGhalf |
| MAI002 | Solomon Islands, Malaita | R Petrous | 540-480 (1410-1470 AD) |
F | B4a1a1a | - | No | 5.582 | 913,583 | noUDG |
| MAL001 | Vanuatu, Malakula | L petrous | 2330-2100 (380-150 BC) |
F | B4a1a1 | - | No | 0.089 | 78,100 | noUDG |
| MAL002 | Vanuatu, Malakula | L petrous | 2490-2200 (540-250 BC) |
F | B4a1a1a | - | No | 0.302 | 220,082 | UDGhalf |
| MAL004 | Vanuatu, Malakula | L petrous | 2690-2320 (740-370 BC) |
M | B4a1a1a | M1b | No | 1.751 | 697,939 | UDGhalf |
| MAL006 | Vanuatu, Malakula | L petrous | 2670-2320 (720-370 BC) |
F | B4a1a1a11 | - | Yes | 0.011 | 10,418 | noUDG |
| MAL007 | Vanuatu, Malakula | R petrous | 2140-1920 (190-30 BC) |
F | B4a1a1a | - | No | 0.609 | 394,207 | UDGhalf |
| MAL008 | Vanuatu, Malakula | L petrous | 2290-1940 (350 BC - 10AD) |
F | B4a1a1a | - | Yes | 0.025 | 22,381 | noUDG |
| TAN001 | Vanuatu, Tanna | L petrous | 260-0 (1690-1950 AD) |
M | P1d1 | O2a2b2a | No | 1.223 | 629,733 | UDGhalf |
| TAN002 | Vanuatu, Tanna | R petrous | 2630-2350 (680-400 BC) |
M | Q2a | K2b1 | No | 0.241 | 191,304 | UDGhalf |
| TAP002 | French Polynesia, Ra'iatea | Molar | 270- -10 (1680-1960 AD) |
M | B4a1a1m1 | n/a | Yes | 0.041 | 39,897 | noUDG |
| TAP003 | French Polynesia, Ra'iatea | Molar | 270- -10 (1680-1960 AD) |
M | B4a1a1c | CT | No | 0.158 | 137,660 | UDGhalf |
| TAP004 | French Polynesia, Ra'iatea | Molar | 240-10 (1710-1940 AD) |
M | B4a1a1+16126 | CT | No | 0.072 | 66,227 | noUDG |
| TON001 | Tonga, Tongatapu | R petrous | 2670-2320 (720-370 BC) |
F | B4a1a1a | - | Yes | 0.092 | 82,790 | noUDG |
| TON002 | Tonga, Tongatapu | L petrous | 2690-2350 (740-400 BC) |
M | B4a1a1 | O1a1a1a | Yes | 0.406 | 285,776 | noUDG |
Demographic history of Vanuatu
While early Lapita people in Vanuatu had largely East Asian-Austronesian ancestry21, principal component analysis (PCA) shows that – though diverse – the 27 present-day individuals fall instead within the Near Oceanic cline, in close proximity to Santa Cruz and New Britain populations (Fig. 1a,b), demonstrating an almost complete population turnover since initial settlement. Previous ALDER33 analysis estimated the time of Papuan admixture into Remote Oceania at 1,927-1,239y BP for Polynesian populations21, and our analyses on regional populations give similar estimates of ~2,000-1,500y BP (see below). Yet the 14C dates for the ancient samples demonstrate that Papuan ancestry was already in Vanuatu up to 1,000 years earlier, from ~2,500y BP. Both the earliest (TAN002) and latest (TAN001) ancient samples from Tanna (Supplementary figure 2) lay inside the distribution of the new present-day HO samples, but it is striking that ancient samples from Malakula and Futuna within this timeframe do not (Fig. 1a). The Malakula time-transect bridges much of the massive genetic distance between initial Lapita inhabitants and contemporary ni-Vanuatu. ADMIXTURE34 analyses on ancient and modern Vanuatu SG data support a complex population replacement. With K=5 ancestral components – allowing the distinction between Asian-Austronesian (blue) and Near Oceanian-Papuan (green) – Vanuatu demonstrates a general but heterogeneous trend of increasing Papuan ancestry through time (Fig. 2a), from largely Austronesian Lapita (ref. 21, and MAL006) to predominantly Papuan ni-Vanuatu ancestry.
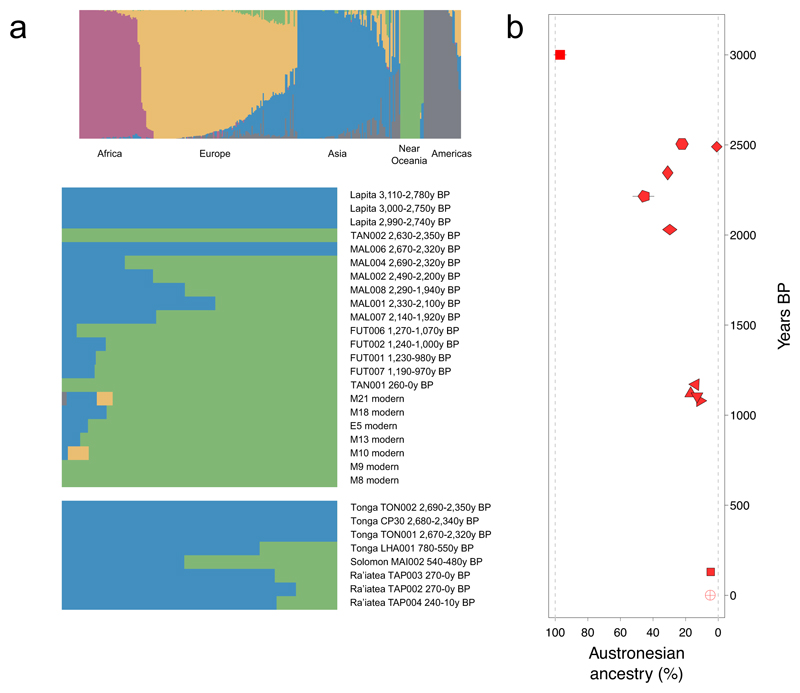
Fig. 2. Admixture proportions of Papuan- vs. Lapita-related ancestry in ancient and present-day populations using 1240K genome-wide data.
(a) Unsupervised ADMIXTURE analyses of present-day global populations and ancient Pacific individuals, with 5 ancestral components. (b) Austronesian ancestry proportion (modeled by indigenous Taiwanese population Ami) in ancient and present-day Vanuatu individuals estimated through qpAdm analyses. Symbol legend is given in Fig. 1, and standard errors are indicated by black lines if larger than the symbol (see also Supplementary table 8).
qpWave analysis35 determined that ancient Vanuatu could be modeled as a two-way admixture between Papuan and Austronesian populations (Supplementary table 7), using qpAdm36 to quantify the relative ancestry proportions (Fig. 2b, Supplementary table 8). The near-contemporaneous genetic heterogeneity in Malakula is striking. Over the ~500y period beginning ~2,500y BP Malakula was home to individuals with between 22 to 46% of their ancestry derived from ancestral Austronesians (Futuna samples ~1,100y BP have 11 to 17%). The earliest ancient individual, TAN002, is a male carrying both Papuan mtDNA and Y-chromosome haplogroups (Q2a and K21b, respectively), with autosomes consistent with having no Austronesian ancestry (Fig. 2b, Supplementary figure 3). We estimated the excess Austronesian X-chromosome ancestry relative to the autosomes across our time-transect, finding diverse levels of maternal ancestry within Malakula (Supplementary table 8). In particular, MAL004 – a male with typical Papuan Y-chromosome haplogroup M1b – carries as much as ~50% Austronesian maternal excess (and Polynesian mtDNA haplogroup B4a1a1a), providing the first direct snapshot of this sex-biased admixture in progress17–20. The latest ancient sample, TAN001, shows similar autosomal admixture proportions to contemporary ni-Vanuatu, and carries a Papuan mtDNA haplogroup and Polynesian Y-chromosome haplogroup (P1d1 and O2a2b2a, respectively).
To identify potential source populations of post-Lapita Near Oceanian ancestry we calculated D-statistics30 on the new ancient Vanuatu data, down-sampled to the more geographically extensive HO dataset (Supplementary table 9). Using the model D(Near Oceanian, New Guinea ; Vanuatu ancient, Mbuti), where Near Oceanian is drawn from all potential sources reported in ref. 21, we identified Baining Marabu and Baining Malasait in New Britain, Bismarck Archipelago (Fig. 1b) as the closest present-day proxy sources of Near Oceanian ancestry in the ancient Vanuatu individuals (Z>>0). One possible confounding factor is the significant difference in the levels of Austronesian ancestry in Baining populations compared to New Guinea Papuans shown by D(Baining Marabu or Baining Malasait, New Guinea ; Ami, Mbuti): Z=3.7 or 4.2. However, TAN002 does not show such an attraction to Ami, confirming that its affinity to Baining relative to Papuans is not explained by shared Austronesian ancestry (Supplementary table 9). Furthermore, although Denisovan admixture levels are observed to decline with increased Austronesian ancestry proportion37, the best-supported source populations have values consistent with New Guinea Papuans (D(Baining Marabu or Baining Malasait, New Guinea ; Denisovan, Mbuti): Z=-0.8 or -1.9). Thus, D-statistics confirm the close relationship observed in PCA between Baining populations and the earliest Vanuatu individual carrying Near Oceanian ancestry (TAN002), despite the immense geographical distance (Fig. 1a,b).
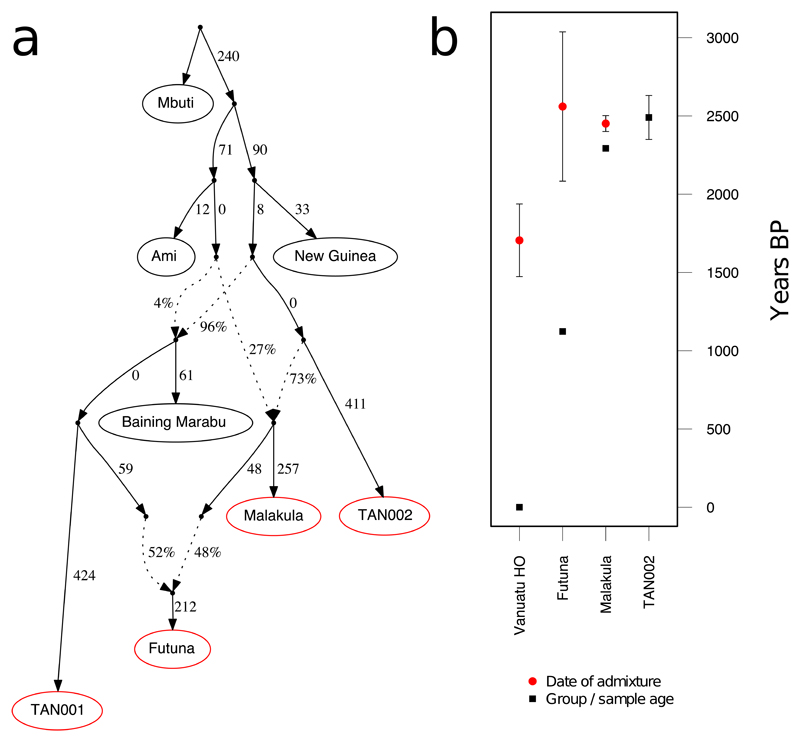
qpGraph30 analyses (Fig. 3a) showed that TAN002 could be modeled as an unadmixed individual descended from a population ancestral to modern Baining Marabu, before the latter receives a 4% Austronesian contribution. In Vanuatu, a population associated with TAN002 would admix with local Lapita people (proxied by Ami) giving rise to ancient Malakula individuals ~2,500-2,000y BP. Additional Papuan admixture is needed to account for the lower Austronesian proportion in the ~1,100y BP Futuna population (Fig. 2b, Supplementary table 8, Supplementary figure 3). The most recent ancient individual TAN001 can only be modeled as descended directly from a Baining-related population, suggesting complete local population replacement. We were unable to fit present-day Vanuatu HO alongside the new ancient samples in a single model (Supplementary figure 4), indicating that present-day ni-Vanuatu may carry an additional genetic component not found in ancient populations.
Fig. 3. Demographic history of ancient Vanuatu individuals.
(a) qpGraph model that fits observed allele frequency patterns with branch lengths representing drift in FST*1000 units and edge percentages indicating admixture proportions. Ancient samples or groups are indicated with a red border. (b) ALDER analyses estimating the date of Papuan and East Asian admixture, converted into years with a generation time of 28.1 years. Standard error bars are shown for date estimates, while sample ages for the two ancient groups (Futuna and Malakula) are averaged radiocarbon dating confidence interval (CI) midpoints. As the earliest ancient Vanuatu individual with unadmixed Near Oceanian ancestry, TAN002 is included for age comparison, with error bar indicating the 95.4% radiocarbon dating CI.
Different genetic trajectory in Polynesia
Analyses of two new Lapita individuals (TON001, TON002) from the Talasiu site in Tonga21, confirmed their genetic similarity to early peoples in Vanuatu (Fig. 1a). Notably, TON002 is a male carrying Y-chromosome haplogroup O1a1a1a, providing direct evidence that this clade – like the “Polynesian mtDNA motif” haplogroup B4a1a1a – was associated with the Austronesian expansion38. After Lapita settlement, the populations of Vanuatu and Tonga appear to follow a considerably different genetic trajectory; PCA analyses indicate that present-day Tongans fall between the East Asian and Near Oceanian clines (Fig. 1a, Supplementary figure 5), more specifically between Lapita individuals and Solomon Islanders. A newly sequenced ancient Tongan female sample (LHA001), from 780-550y BP, lay relatively close in PCA to modern Tongans, but its lower affinity to Solomon Islanders suggests that modern Tongan ancestry was not yet completely in place by this time (D(LHA001, Tongan; Savo, Mbuti): Z=-3).
We obtained genome-wide data from three individuals unearthed at the monumental site Taputapuātea (TAP002, TAP003, TAP004) on the island of Ra’iātea, French Polynesia dated to the time of European contact in the 18th century AD39. ADMIXTURE34 analyses (Fig. 2a) show these individuals have major Austronesian (blue) and minor Papuan (green) ancestry components, and both carry typical Polynesian mtDNA haplogroups (Table 1). In PCA space they fall in close proximity to the Tongan individual LHA001 – slightly more towards the East Asian cline – suggesting that the population expansion to East Polynesia ~900-800y BP40 may have originated in western Polynesia. ADMIXTURE analyses (K=4) on a subset of HO data – including 454 present-day and 13 ancient Near and Remote Oceanian individuals (Supplementary figure 5) – show that present-day ni-Vanuatu carry a heterogeneous proportion of three major components that are maximized in Near Oceanian populations (Papuan, Baining and Bougainville), with a minor Lapita-related component (Supplementary figure 5). Conversely, present-day Tongans have substantial Lapita ancestry, with a minor component of Near Oceanian admixture (with different proportions of Papuan, Baining and Bougainville) (Supplementary figure 5). qpAdm analyses further support modeling modern Tongans as a two-way admixture between ancestral Austronesians and a population ancestral to some present-day Solomon Island groups – such as Malaita and Makira – or represented by the ~500y BP Malaita individual (MAI002), even when Papuan and Bismarck are included as an additional outgroup (Supplementary table 10). Thus, Solomon Islanders alone can explain the Near Oceanian ancestry found in Tongans, without contribution from New Guinea Papuans. This higher affinity to Solomon Islanders provides evidence that, post-Lapita, Tonga likely received its Near Oceanian ancestry from a different source than did Vanuatu.
Genetic cline in present-day Vanuatu
We analyzed the new ancient and modern data alongside a dataset from Remote Oceania32, which includes 754 individuals from New Caledonia, Vanuatu, Fiji and Tonga (Supplementary table 6), genotyped on the HumanCore-24 BeadChip, with ~160K and ~50K SNP overlap with the 1240K and HO data, respectively. After removing individuals with genetic evidence of non-autochthonous ancestry, PCA and ADMIXTURE analyses (Supplementary figure 6 and Supplementary figure 7) demonstrated high genetic diversity in ni-Vanuatu from the islands of Santo and Maewo (north of Malakula, Supplementary figure 2), with these individuals laying on a cline running from close to New Britain, through Vanuatu, New Caledonia and Fiji, towards present-day Tonga. The new Vanuatu HO data from the islands of Malakula and Efate (Supplementary figure 2), and the most recent ancient Tanna individual (TAN001), lay overwhelmingly towards the New Britain end of this cline. Down-sampled to ~50K SNPs, the different trajectories for post-Lapita Vanuatu and Tonga populations identified in the HO analyses are less distinguishable. We used D-statistics to test whether this cline describes a separate demographic process to that which brought Bismarck-like ancestry to Vanuatu (Methods) but – at the resolution of currently available regional genotyping data – we are unable to distinguish between the two clines with confidence (Supplementary figure 8), suggesting that a Tongan-like ancestry may have played some role in the formation of present-day genetic diversity in Vanuatu. However, the HO analyses demonstrate that present-day Tongan ancestry, forming one end of this cline, was not fully in place prior to ~780-550y BP (LHA001), so this influence may be significantly later than the initial arrival of Bismarck ancestry in Malakula (~2,500y BP).
Austronesian-Papuan admixture date estimation
We performed ALDER33 analyses on both modern and ancient Vanuatu data to gain independent estimates of arrival times for the Papuan ancestry component. We obtain an estimate of 60.7±8.2 generations BP for the 27 HO Vanuatu individuals, which – assuming a 28.1 year generation-time21 – equates to 1,705±232y BP (Fig. 3b, Methods). Interestingly, admixture time estimates similarly obtained for ancient Vanuatu provided 51.2±17 generations for three Futuna individuals (FUT002, FUT006 and FUT007) and 5.6±1.8 generations for three ancient Malakula individuals (MAL002, MAL004 and MAL007). Accounting for ancient sample ages, the admixture date is estimated at 2,560±477y BP for Futuna and 2,451±51y BP for Malakula, coinciding with the latest presence of individuals in the new Vanuatu time-transect with unadmixed Papuan (TAN002) or Austronesian (MAL006) ancestry (Fig. 3b). ALDER analyses of the Parks et al.32 data gave dates ranging from 1,569±79y BP (Fiji) to 1,999±101y BP (Port Olry, Vanuatu), overlapping the interval proposed by Skoglund et al.21, yet still significantly later than the directly dated admixed ancient individuals in Malakula (Supplementary figure 9).
Discussion
The population history of Remote Oceania is relatively short but these early stages appear complex, particularly in Vanuatu. New genome-wide aDNA data directly demonstrates the presence of Papuan peoples in Remote Oceania far earlier than estimated with present-day regional genome-wide data (Supplementary figure 9, and ref. 21), with unadmixed Bismarck-like individuals apparent in Vanuatu as early as ~2,500y BP, possibly contemporaneous with the end of the Lapita horizon. The new HO data from contemporary Malakula and Efate shows that while Oceanic speaking Lapita peoples were genetically replaced by a population closely related to Papuan-speaking Baining people, present-day ni-Vanuatu continue to speak Oceanic languages. The almost complete replacement of a population’s genetic ancestry that leaves the original languages in situ is extremely rare – possibly without precedent – in human history and requires explanation. Alongside linguistic and archaeological evidence, our aDNA analyses provide a plausible and compelling model for this language continuity, namely an extended and incremental process of population replacement by peoples from the Bismarck Archipelago (Fig. 3a), rather than a single massive turnover event that would likely have brought a shift from Oceanic to Papuan languages.
The >120 languages spoken today in Vanuatu – per capita the most linguistically diverse place on Earth – are exclusively Oceanic14, yet many aberrant, seemingly Papuan, linguistic features are evident41. These include quinary numeral systems, rounded labial phonemes, dual exclusion of p and c phonemes, and serial verb construction42–45. These features are heterogeneously distributed across Vanuatu42–44, extremely rare or absent in other Austronesian languages and are shared almost exclusively with Papuan languages (e.g. Supplementary figure 10). A number of ethnographically attested cultural practices or artifacts also share this near exclusive distribution, including large nasal piercing ornaments, penis sheaths, head-binding and the rearing of full-circle tusker pigs46,42. These shared cultural and linguistic features provide further support for the Baining-Papuan genetic connection we identify. While some linguists argue for a single admixed expansion into Vanuatu from Near Oceania47, or Papuan involvement in initial Lapita settlement43, others propose a 2-wave model42, where an initial unadmixed proto-Oceanic-speaking population arrive, followed closely by a separate Papuan-speaking expansion. The latter42 is supported because the putative Papuan linguistic features found in Vanuatu cannot be reconstructed for proto-Oceanic, and their marked deviation from most other Oceanic languages suggests development within Vanuatu42–44. Some features can be reconstructed for the proto languages of Vanuatu – rounded labials and the p/c gap for Proto-North-Central Vanuatu48, and quinary numeral systems for Proto-Southern Vanuatu49 – pointing to their early development and strongly supporting early Papuan influence. An undifferentiated proto-Oceanic operating as a lingua franca for linguistically diverse Papuan migrant groups could explain42 the continuity of Oceanic languages in the face of secondary Papuan expansion.
Our aDNA analyses lend direct support to this historical linguistic model42. Indeed, some archaeologists have argued that the process by which Papuans made their way into Remote Oceania was strikingly different to the initial arrival of Lapita people23, suggesting a continuing process of long-distance interaction rather than a simple dispersal event. One element of this process – namely the sex-biased admixture inferred from present-day South Pacific populations e.g.13,21 – is already becoming clearer, with such genetically admixed ancient individuals (e.g. MAL004) observed shortly after the very earliest arrival of Near Oceanian peoples in Remote Oceania (Fig. 2b, Supplementary table 8). We show that initially genetically homogeneous Lapita peoples in Vanuatu and Tonga21 follow strikingly different post-Lapita population trajectories, reflected in the clear cultural separation seen in the archaeological record. As a defined stylistic horizon, Lapita lasted only a few hundred years after settlement – local differentiation in pottery design beginning ~2,700y BP suggests significant fragmentation of the previously well-connected Lapita peoples23. In central Vanuatu, the appearance of the incised Erueti ceramic complex ~2,550y BP50 seems to parallel a contemporaneous stylistic shift across island Melanesia post-Lapita, including both New Caledonia and the Bismarck Archipelago3. It is an intriguing possibility that the early arrival of Bismarck-like people we now directly observe in Vanuatu may have exacerbated – even triggered – the process of Lapita fragmentation23 and the ongoing long-distance interactions we uncover may also have influenced the convergent processes of stylistic diversification3,50 found in pottery sequences across the region.
Our analysis of present-day Remote Oceanian data32 suggests a possible Tongan-like influence on the genetic diversity of present-day eastern Melanesia, with populations in northern Vanuatu, New Caledonia and Fiji lying on a cline towards modern Tonga (Supplementary figure 6). Given the data resolution, we were unable to clearly distinguish this from the other cline formed by the post-Lapita population trajectory in Vanuatu (Fig. 1a), but the ancient Tongan individual LHA001 suggests that it formed later. One possibility is that this genetic structure was influenced by interactions with western Polynesia leading to the many Polynesian outlier communities – characterized by retention of various Polynesian linguistic features, cultural practices and genetic ancestry3 – distributed across Micronesia, New Guinea, the Solomon Islands, New Caledonia and Vanuatu. While the timing, scale and impact of this westward Polynesian migration is not yet precisely estimated, it likely coincided with the initial colonization of eastern Polynesia ~900-800y BP40.
In conclusion, our analyses of Vanuatu genome-wide data – both ancient and modern – combined with linguistic and archaeological evidence, strongly support a model of interaction and incremental admixture between Lapita-Austronesian peoples and incoming Bismarck Islanders that lead to an eventual population turnover, but left the pre-existing Oceanic languages in place. This multidisciplinary work has begun to uncover the complex, localized demographic processes that drove the initial colonization of the wider South Pacific and formed the enduring cultural and linguistic spheres that continue to shape the Pacific today.
Methods
Ancient and modern-day DNA processing
Ancient DNA sampling
All samples were processed in dedicated laboratories at the Max Planck Institute for the Science of Human History in Jena, Germany. Bone powder for DNA extraction was obtained from petrous bones by drilling the densest osseous matter around the cochlea and from teeth by cutting at the junction between root and crown and sampling the dental pulp. For detailed information on the analyzed samples, their archaeological context and radiocarbon age see Supplementary text, Supplementary table 1, Supplementary table 2, Fig. 1 and Supplementary figure 2.
Extraction
DNA from the 23 ancient individuals was extracted following established protocols24, negative and cave bear positive controls were included. To release DNA from 50-100mg of bone powder a solution of 900µl EDTA, 75µL H2O and 25µL Proteinase K was added. In a rotator, samples were digested for at least 16 hours at 37°C, followed by an additional hour at 56°C51. The suspension was then centrifuged and transferred into a binding buffer as previously described24. To bind DNA, silica columns for high volumes (High Pure Viral Nucleic Acid Large Volume Kit, Roche) were used. After two washing steps using the manufacturer’s wash buffer, DNA was eluted in TET (10mM Tris, 1mM EDTA and 0.05% Tween) in two steps for a final volume of 100µl.
Library Preparation
For aDNA authentication and contamination estimates screening DNA libraries were built from 20µl of DNA extract in the absence of uracil DNA glycosylase (non-UDG libraries), following a double stranded library preparation protocol25. After assessing human DNA contamination levels, one or two additional 25µl aliquots of DNA extract were transformed either into non-UDG libraries25 or into “UDG-half” double-stranded libraries with a protocol that makes use of the UDG enzyme to reduce but not eliminate the amount of deamination induced damage towards the end of aDNA fragments26. Negative and positive controls were carried out alongside each experiment. Libraries were quantified using the IS7 and IS8 primers25 in a quantification assay with DyNAmo SYBP Green qPCR kit (Thermo Scientific) on the Lightcycler 480 Roche. Each aDNA library was double indexed51 in one to four parallel 100µl reactions using PfuTurbo DNA Polymerase (Agilent Technologies). The indexed products for each library were pooled, purified over MinElute columns (Qiagen), eluted in 50µL TET and again quantified using the IS5 and IS6 primers25 with the quantification method described above. Four microliters of the purified product were amplified in multiple 100µl reactions using Herculase II Fusion Polymerase (Agilent) following the manufacturer’s specifications with 0.3µM of the IS5/IS6 primers. After another MinElute purification, the product was quantified using the Agilent 2100 Bioanalyzer DNA 1000 chip. An equimolar pool of all libraries was then prepared for shotgun sequencing on Illumina platforms.
Enrichment
Both UDG-half and non-UDG treated libraries were further amplified with IS5/IS6 primers to reach a concentration of 200-400ng/µl as measured on a NanoDrop™ spectrophotometer (Thermo Fisher Scientific). mtDNA capture27 was performed on screened libraries that after shotgun sequencing showed the presence of aDNA, highlighted by the typical CtoT and GtoA substitution pattern towards 5’ and 3’ molecule ends, respectively. Furthermore, samples with a percentage of human DNA in shotgun data around 0.1% or greater were enriched53 for a list of 1,237,207 targeted SNPs across the human genome (1240K capture)28.
Sequencing
The enriched DNA product was sequenced on an Illumina HiSeq 4000 instrument with 75 cycles single-end or 50 cycles pair-end runs (for TAN001 and FUT006) using the manufacturer's protocol. The output was de-multiplexed using bcl2fastq v2.17.1.14 and dnaclust v3.0.0.
Modern DNA sampling
Genetic sampling was carried out as part of a long-term linguistic and anthropological fieldwork project, directed by Prof. Russell Gray and Dr. Heidi Colleran at the Max Planck Institute for the Science of Human History (http://www.shh.mpg.de/456217/vanuatu-languages-lifeways). The saliva samples of 27 present-day ni-Vanuatu from the islands of Malakula and Efate were collected using the Oragene OG-500 saliva collection kit. Ethical approval for this work was granted by the Ethik-Kommission der Friedrich-Schiller-Universität in Jena, Germany, and we obtained research permission from the Vanuatu Kaljoral Senta, the institution that regulates all research in the country. Sampling was carried out in 5 communities that are already participating in the linguistic and anthropological project, and all participants gave documented informed consent and were provided the means to withdraw from the study if required.
Modern DNA extraction and library preparation
Extraction and library preparation were performed in the molecular biology laboratories of the Max Planck Institute for the Science of Human History in Jena, Germany. Modern-day DNA was extracted from the Oragene kit following the manufacturer's protocols with the only modifications that 600µl of sample volume was used accordingly adjusting the following reaction volumes. 10µl of eight modern-day DNA extracts (Supplementary table 5) were used to build double-stranded DNA libraries25. They were then indexed in one reaction following the same protocols mentioned above, pooled equimolarly and shotgun sequenced on an Illumina HiSeq 4000 instrument (75 cycles single-end run).
Genotyping of present-day humans
The company Atlas Biolabs in Berlin, Germany genotyped 27 modern DNA extracts on the Axiom Genome-Wide Human Origins array. After checking DNA quality and quantity on both a 1% Agarose gel and a NanoDrop, samples were adjusted to 20ng/µl using a Qubit high sensitivity kit (Thermo Fisher Scientific), loaded on the Axiom Genome-Wide Human Origins array (Affymetrix) and genotyped on a GeneTitan. Genotyping was performed using the Affymetrix Genotyping Console, and all individuals had >94% genotyping completeness.
Genomic data processing
Pre-processing of the sequenced reads was performed using EAGER v1.92.4454. Reads resulting from the sequencing of modern and ancient DNA libraries were clipped to remove residual adaptor sequences using Clip&Merge54 and AdapterRemoval v255, respectively. Clipped sequences were then mapped against the human reference genome hg19 using BWA56 turning seeding off and with the –n parameter set to 0.01. Duplicates were removed with DeDup54 that removes reads with identical start and end coordinates. Additionally a mapping quality filter of 30 was applied using samtools57. Alignment files were filtered for reads showing the presence of likely deaminated bases as the result of post-mortem damage (PMD) using pmdtools v0.6058. Both damage restricted and non-restricted sequences from either non-UDG or UDG-half libraries were trimmed for the first and last three positions in order to reduce the impact of deamination induced miss-incorporations during genotyping. Trimmed reads were genotyped using pileupCaller (https://github.com/stschiff/sequenceTools/tree/master/src-pileupCaller) a tool that randomly draws one allele at each of the 1240K targeted SNPs covered at least once. The generated pseudo-haloid calls for 19 ancient Pacific individuals (Table 1) were merged to a pull-down of the 1240K SNPs from the Simon Genome Diversity Project (SGDP)31, eight shotgun sequenced modern-day individuals from Vanuatu and four previously published 1240K captured individuals associated with the Lapita culture from Vanuatu and Tonga21. Moreover the newly generated capture data for the ancient individuals as well as 27 genotyped modern-day individuals (Supplementary table 5) were merged to the ~600K SNPs of the Human Origins (HO) dataset21,30.
Authentication of ancient DNA
In the field of aDNA several methods have been developed to assess authenticity of the retrieved DNA29. First, the typical features of aDNA were inspected with DamageProfiler (https://bintray.com/apeltzer/EAGER/DamageProfiler), e.g. short average fragment length (~40-70bp) and an increased proportion of miscoding lesions due to deamination at the molecule termini (Supplementary table 3). Sex determination was performed by comparing the coverage on the targeted X-chromosome SNPs (~50K positions within the 1240K capture) normalized by the coverage on the targeted autosomal SNPs to the coverage on the Y-chromosome SNPs (~30K), again normalized by the coverage on the autosomal SNPs59 (Table 1). Individuals falling in an intermediate position between male and female are assigned to undetermined sex and indicate the presence of present-day DNA contamination. For male individuals ANGSD was run to measure the rate of heterozygosity of polymorphic sites on the X-chromosome after accounting for sequencing errors in the flanking regions60. This provides an estimate of nuclear contamination in males that are expected to have only one allele at each site. For all male samples that exhibit X-chromosome contamination levels below 2% with at least 100 X-chromosome SNPs covered twice, all reads were retained for further analyses (Supplementary table 4). Otherwise only PMD fragments that are likely of endogenous origin were used61 (Table 1). For both male and female individuals mtDNA captured data was used to jointly reconstruct the mtDNA consensus sequence and estimate contamination levels with schmutzi62 (Supplementary table 11). For specimens where a relatively low proportion of mtDNA molecules compared to nuclear DNA (mt/nuclear DNA ratio) was observed (Supplementary table 11), mtDNA contamination estimate can be used as reliable predictor for nuclear contamination29. Population genetic analyses on samples presenting mtDNA levels of contamination above 4% were restricted to PMD fragments. Moreover, for each individual the positioning in PCA space was compared to the data after restriction to deaminated sequences21. Samples that were substantially displaced in PCA space (Supplementary figure 1) were restricted to PMD fragments for population genetic analyses.
Population genetic analyses
PCA were computed with present-day populations from the HO dataset composed of 781 Oceanians and East Asians21 and 27 modern-day Vanuatu individuals newly genotyped here, for a total of 808 individuals. Ancient individuals were projected onto the two first components using smartpca (v13050)63 with the options “lsqproject: YES” and “numoutlieriter: 0” (Fig. 1 and Supplementary figure 1). Another PCA was computed on the ~50K SNPs overlapping the HO dataset and a recently published Illumina HumanCore-24 dataset (typed on ~240K SNPs in total)32 (Supplementary figure 6). The same 808 modern-day Oceanians and East Asians were used to build the principal components on which 669 individuals across Remote Oceania (Supplementary table 6) and 15 ancient Pacific individuals with more than 6K SNPs were projected. The software ADMIXTURE v1.3.034 was run in unsupervised mode on high coverage genomes of 308 modern-day worldwide individuals31, eight shotgun sequenced present-day Vanuatu individuals and all 23 ancient Pacific individuals. Only transversions sites of the 1240K SNPs (~220K positions) were considered in order to reduce the impact on the clustering algorithm of residual damage still present in non-UDG treated libraries. An additional regional ADMIXTURE analysis was carried out also on the transversions subset of the HO data (~110K SNPs) including 13 ancient individuals from Vanuatu and Tonga (more than 15K SNPs) and 454 modern-day Oceanian individuals (Supplementary figure 5). Finally, ADMIXTURE was run on the overlapping SNPs between HO and Parks et al.32 datasets for the 27 newly genotyped present-day individuals from Malakula and Efate in Vanuatu (Supplementary table 5) in addition to 754 present-day individuals from New Caledonia, Vanuatu, Fiji and Tonga (Supplementary figure 7). From the latter dataset 85 individuals harboring more than 2% of non-local ancestry at K=5 were removed for a total of 669 individuals retained (Supplementary table 6). In the following analyses all SNPs were investigated for individuals with UDG-half libraries whereas only transversion SNPs were used for individuals with non-UDG libraries to avoid spurious results originating from leftover aDNA damage.
D-statistics were calculated with qpDstats v711 program from the ADMIXTOOL suite (https://github.com/DReichLab) in the form D(Pop1, Pop2 ; Pop3, Outgroup). A negative value implies that either Pop1 and Outgroup, or Pop2 and Pop3 share more alleles than expected under the null hypothesis of a symmetrical relationship between Pop1 and Pop2 (Supplementary table 9). To jointly observe the affinity of modern-day Fiji, Tonga, New Caledonia and Vanuatu individuals from Parks et al.32 and HO datasets as well as ancient Vanuatu individuals towards Ami and Tonga populations, we calculated two sets of D-statistics in the form A: D(Baining, X; Ami, Mbuti) and B: D(Baining, X; modern Tongan, Mbuti), where X is drawn from Fiji, Tonga, Maewo (Vanuatu), Port Olry (Vanuatu), Santo (Vanuatu) and New Caledonia from Parks et al.32, as well as the Vanuatu HO and ancient Malakula, Futuna and Tanna samples. Plotting A against B (Supplementary figure 8) shows that we cannot see a clear deviation between modern and ancient individuals, as all values do not appreciably differ from the straight line expected for no differential ancestry.
qpWave v40035 was implemented on the HO dataset in order to test if the ancient individuals are consistent with two sources of ancestry represented by modern-day Ami (as the best proxy for ancestral Austronesian) and Papuan individuals, with respect to a set of outgroups (Mbuti, Denisovan, Sardinian, English, Yakut, Chukchi, Mala, Japanese, Ju_hoan_North, Mixe, Onge, Yoruba). This is obtained when rank n-1 cannot be rejected (p>0.05) as shown for all our ancient Vanuatu individuals, as well as modern Vanuatu HO individuals despite a much lower p-value (Supplementary table 7). The same populations for both HO and 1240K datasets were then used in qpAdm v61036 to estimate admixture proportions for ancient and modern-day Vanuatu individuals (Supplementary figure 3, Fig. 2b and Supplementary table 8). qpAdm models each individual as a mixture of Ami and Papuan by fitting admixture proportions that match the observed matrix of f4-statistics and computing standard errors with a block jackknife. To evaluate potential sex bias admixture, qpAdm analysis, as described above, was run only on X-chromosome SNPs (option “chrom:23”) of the 1240K dataset. Differences in admixture proportions between autosomal and X-chromosome SNPs provide an indication of sex-biased admixture (Supplementary table 8).
Modern-day Tongans were modeled in qpAdm as resulting from a two-way admixture between Ami (as the best proxy for ancestral Austronesian) and ancient (MAI002) or modern-day Solomon Islanders from the island of Makira, Malaita and Bougainville (Naisoi and Choiseul populations). When selecting the 12 outgroups listed above, Tongans can successfully be modeled with p>0.05, using a block jackknife to calculate standard errors as indicated previously. qpAdm was re-run expanding the outgroup population list with Papuan and Baining Marabu. For present-day individuals from Makira, Malaita and the ancient individual from Malaita (MAI002) rank n-1 can still not be rejected, indicating that additional Papuan New Guinea or Bismarck ancestry is not necessary to model modern-day Tongans (Supplementary table 10).
Admixture dates were estimated based on linkage disequilibrium using ALDER33 on the ~160K overlapping SNPs between 1240K capture and Parks et al.32 datasets. As source populations, 20 Asian (Ami, Atayal, Igorot, Kinh, Dai, She, Lahu, Han) and 16 Papuan individuals were chosen. The estimated dates of admixture were converted into years assuming a generation time of 28.1 years21,64 for the 27 Vanuatu HO individuals (Fig. 3b) and for modern-day New Caledonia, Vanuatu, Fiji and Tonga populations32 (Supplementary figure 9). Admixture dates were also estimated for SNPs overlapping to the 1240K capture for three ancient Futuna individuals (FUT002, FUT006, FUT007) with average age set to 1,123y BP and three ancient Malakula individuals (MAL002, MAL004, MAL007) with average age set to 2,293y BP (Fig. 3b).
Admixture graphs on the HO dataset were fitted with qpGraph v521130,65 that matches a matrix of f-statistics testing the relationships between all analyzed populations at the same time. An initial backbone graph modern-day populations without signs of admixture were built into the tree (Mbuti, Ami, New Guinea). The differential proportion of Denisovan ancestry between Mbuti-Ami and New Guinea populations66 was not modeled here since this is accommodated in the graph by shifting the splitting point of the African Mbuti population. Baining Marabu was then incorporated as admixed between an Ami-related and a New Guinea-related lineage, as suggested from D-statistics analyses (Supplementary table 9). Ancient UDG-half individuals from Vanuatu (three Futuna individuals grouped, three Malakula individuals grouped and two Tanna individuals separately) were added chronologically one-by-one at each possible position of the graph reporting every time the highest D-statistic between the observed and fitted model and calculating the Z-score with a block jackknife. The graph reported in Fig. 3a is built with a total of 38,789 SNPs and fits the allele frequency relationships between modern-day and ancient individuals with all empirical f-statistics within the 3 standard error interval and only one significant D-statistic (Z=2.6). The modern-day Vanuatu HO population can be fitted as admixed between modern-day Baining Marabu and Ami-related populations but this relatively simple model with only four populations has already the worst Z-score, equal to 2.3 (Supplementary figure 4a). Moreover, we were unable to fit a modern-day HO Vanuatu population in the graph once ancient individuals are included, neither by replacing the ~200y BP TAN001 individual (Supplementary figure 4b) nor modeling Vanuatu HO as deriving part of its ancestry from the ~1,100y BP Futuna population (Supplementary figure 4c) with the worst Z-score of 6 and 5.2, respectively.
Haplogroup assignment for uniparental markers
After enrichment of the libraries for the mitochondrial genome (mtDNA capture) reads were pre-processed in EAGER v1.92.55 as described above and aligned to the mitochondrial reference genome (rCRS) using CircularMapper, a program that takes into account the circularity of the mtDNA54. Contamination was estimated while assembling the mitochondrial genome using schmutzi62 with the parameters “--notusepredC –uselength”. Present-day human contamination estimates were performed using a comparative database of 197 modern-day worldwide mtDNAs provided with the software package. For the resulting sequences we filtered positions with likelihood above 20 or 30 (Supplementary table 11) and used HaploGrep267 to assign the corresponding mtDNA haplogroup. For the FUT007 individual the mtDNA consensus sequence was reconstructed from the mtDNA off-target reads in the combined non-UDG and UDG-half 1240K capture data (Table 1 and Supplementary table 11). Sequenced reads overlapping the Y-chromosome SNPs present in the ISOGG database v11.349 (http://www.isogg.org/tree) were investigated to assign Y-chromosome haplogroups. ANGSD60 was used to count ancestral and derived allele occurrence and perform a majority call for positions covered at least once. For this analysis UDG-half and no-UDG data were combined for each sample (Supplementary table 3). To avoid miss-assignments due to DNA damage, CtoT and GtoA mutations required a minimum of two consistent nucleotides to be called. Haplogroup assignment was based on the most downstream SNP retrieved after evaluating the presence of upstream mutations along the related haplogroup phylogeny59.
Data Availability
All newly reported ancient DNA data including nuclear DNA alignment files and mtDNA sequences are archived at the European Nucleotide Archive database (accession number PRJEB24810). Newly reported SNP genotyping and shotgun sequence data will be made available on request to H.C. (colleran@shh.mpg.de) and A.P (powell@shh.mpg.de), subject to a signed agreement to restrict usage to anonymized non-medical studies of population history, as outlined in the ethics and consent documentation.
Supplementary Material
Acknowledgements
We would like to thank the communities in Malakula and Efate in Vanuatu who participated in this study, and particularly all sample donors. We are grateful to Mark Stoneking, Irina Pugach and Chuan-Chao Wang for comments, and to Guido Brandt, Raffaela Bianco and technicians at the Max Planck Institute for the Science of Human History for laboratory support. Archaeological investigations on Malakula, Vanuatu were funded by the Sasakawa Pacific Island Nations Fund, the Marsden Fund of the Royal Society of New Zealand (Fast-Start 9011/3602128; 04-U00–007), a National Geographic Scientific Research Grant (7738–04) and an Australian Research Council Discovery-Project Grant (DP0880789). Investigations on Tanna, Vanuatu, were supported by Australian Research Council Discovery-Project Grant (DP160103578). F.V. is funded by CNRS-UMR 7041, and A.P. is funded by European Research Council Starting Grant Waves (ERC758967).
Footnotes
Author Contributions
F.V., S.B., R.S., H.B., R.K., G.R.C., C.R., J.F., T.M., J.M., J.G. & L.K. contributed archaeological material and H.C., K.W.K. & A.P. contributed the 27 present-day Vanuatu samples. J.Z., F.P. & P.R. contributed isotopic data and radiocarbon date calibrations. M.W. & R.G. contributed linguistic interpretation, and F.V., S.B, J.M, F.P. & P.R. contributed text in the supplementary information. K.J.R., K.A., S.J.O., A.V.S.H. & A.J.M. contributed geographical labels for Parks et al. 2017 samples. C.P. & K.N. performed ancient DNA laboratory work, and C.P., K.N., C.J. & A.P. performed population genetic analyses. C.P., K.N., H.C. & A.P. wrote the paper with input from F.V., S.B., H.B., M.W., F.P., P.R., C.J., R.G. & J.K, and C.P. & A.P. created the figures. The study was conceived and coordinated by C.P., K.N., H.C., R.G., J.K. & A.P.
Competing Interests
The authors declare no competing financial interests.
References
- 1.Clarkson C, et al. Human occupation of northern Australia by 65,000 years ago. Nature. 2017;547:306–310. doi: 10.1038/nature22968. [DOI] [PubMed] [Google Scholar]
- 2.Pawley A, Green R. Dating the dispersal of the Oceanic languages. Ocean Linguist. 1973;12:1–67. [Google Scholar]
- 3.Kirch PV. On the road of the winds: An archaeological history of the Pacific islands before European contact. Revised and expanded edition. Berkeley: University of California Press; 2017. [Google Scholar]
- 4.Blust RA. The prehistory of the Austronesian-speaking peoples: A view from language. J World Prehist. 1995;9:453–510. [Google Scholar]
- 5.Gray RD, Drummond AJ, Greenhill SJ. Language phylogenies reveal expansion pulses and pauses in Pacific settlement. Science. 2009;323:479–483. doi: 10.1126/science.1166858. [DOI] [PubMed] [Google Scholar]
- 6.Blust RA. The Austronesian Languages. ANU, Canberra: Research School of Pacific and Asian Studies; 2009. [Google Scholar]
- 7.Summerhayes GR, et al. Tamuarawai (EQS): An early Lapita Site on Emirau, New Ireland, PNG. J Pacific Archaeology. 2010;1:62–75. [Google Scholar]
- 8.Sheppard PJ. Lapita colonization across the Near/Remote Oceania boundary. Curr Anthropol. 2011;52:799–840. [Google Scholar]
- 9.Pugach I, et al. The gateway from Near into Remote Oceania: New insights from genome-wide data. Mol Biol Evol. 2018 doi: 10.1093/molbev/msx333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petchey FJ, Spriggs M, Bedford S, Valentin F, Buckley H. Radiocarbon dating of burials from the Teouma Lapita cemetery, Efate, Vanuatu. J Archaeol Sci. 2014;50:227–242. [Google Scholar]
- 11.Sand C. Lapita Calédonien. Archéologie d’un premier peuplement insulaire océanien. Paris: Société des Océanistes; 2010. [Google Scholar]
- 12.Burley D, Weisler MI, Zhao J-X. High precision U/Th dating of first Polynesian settlement. PLoS ONE. 2012;7:e48769. doi: 10.1371/journal.pone.0048769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wollstein A, et al. Demographic history of Oceania inferred from genome-wide data. Curr Biol. 2010;20:1983–1992. doi: 10.1016/j.cub.2010.10.040. [DOI] [PubMed] [Google Scholar]
- 14.Pawley A, Attenborough R, Golson J, Hide R. Papuan pasts: cultural, linguistic and biological histories of Papuan-speaking peoples. ANU: Pacific Linguistics; 2005. [Google Scholar]
- 15.Lynch J, Ross M, Crowley T. The Oceanic languages. Richmond, UK: Curzon; 2002. [Google Scholar]
- 16.Kayser M, et al. Genome-wide analysis indicates more Asian than Melanesian ancestry of Polynesians. Am J Hum Genet. 2008;82:194–198. doi: 10.1016/j.ajhg.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melton T, et al. Polynesian genetic affinities with Southeast Asian populations as identified by mtDNA analysis. Am J Hum Genet. 1995;57:403–414. [PMC free article] [PubMed] [Google Scholar]
- 18.Kayser M, et al. Melanesian origin of Polynesian Y chromosomes. Curr Biol. 2000;10:1237–1246. doi: 10.1016/s0960-9822(00)00734-x. [DOI] [PubMed] [Google Scholar]
- 19.Hurles ME, et al. Y-chromosomal evidence for the origins of Oceanic-speaking peoples. Genetics. 2002;160:289–303. doi: 10.1093/genetics/160.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kayser M, et al. Melanesian and Asian origins of Polynesians: mtDNA and Y chromosome gradients across the Pacific. Mol Biol Evol. 2006;23:2234–2244. doi: 10.1093/molbev/msl093. [DOI] [PubMed] [Google Scholar]
- 21.Skoglund P, et al. Origins and genetic legacy of the first people in Remote Oceania. Nature. 2016;538:510–513. [Google Scholar]
- 22.Valentin F, Détroit F, Spriggs M, Bedford S. Early Lapita skeletons from Vanuatu show Polynesian craniofacial shape: Implications for Remote Oceanic settlement and Lapita origins. Proc Natl Acad Sci USA. 2015;113:292–297. doi: 10.1073/pnas.1516186113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bedford S, Spriggs M. The archaeology of Vanuatu: 3000 years of history across islands of ash and coral. In: Cochrane E, Hunt T, editors. The Oxford Handbook of Prehistoric Oceania. Oxford: Oxford University Press; 2014. [Google Scholar]
- 24.Dabney J, et al. Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc Natl Acad Sci USA. 2013;110:15758–15763. doi: 10.1073/pnas.1314445110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer M, Kircher M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc. 2010;6 doi: 10.1101/pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- 26.Rohland N, Harney E, Mallick S, Nordenfelt S, Reich D. Partial uracil–DNA–glycosylase treatment for screening of ancient DNA. Philos Trans Royal Soc B. 2015;370 doi: 10.1098/rstb.2013.0624. 20130624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu Q, et al. A revised timescale for human evolution based on ancient mitochondrial genomes. Curr Biol. 2013;23:553–559. doi: 10.1016/j.cub.2013.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu Q, et al. An early modern human from Romania with a recent Neanderthal ancestor. Nature. 2015;524:216–219. doi: 10.1038/nature14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Key FM, Posth C, Krause J, Herbig A, Bos KI. Mining metagenomic data sets for ancient DNA: Recommended protocols for authentication. Trends Genet. 2017;33:508–520. doi: 10.1016/j.tig.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 30.Patterson N, et al. Ancient admixture in human history. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mallick S, et al. The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature. 2016;538:201–206. doi: 10.1038/nature18964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parks T, et al. Association between a common immunoglobulin heavy chain allele and rheumatic heart disease risk in Oceania. Nat Commun. 2017 doi: 10.1038/ncomms14946. 14946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loh PR, et al. Inferring admixture histories of human populations using linkage disequilibrium. Genetics. 2013;193:1233–1254. doi: 10.1534/genetics.112.147330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reich D, et al. Reconstructing Native American population history. Nature. 2012;488:370–374. doi: 10.1038/nature11258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haak W, et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature. 2015;522:207–211. doi: 10.1038/nature14317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reich D, et al. Denisova admixture and the first modern human dispersals into Southeast Asia and Oceania. Am J Hum Genet. 2011;89:516–528. doi: 10.1016/j.ajhg.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mirabal S, et al. Increased Y-chromosome resolution of haplogroup O suggests genetic ties between the Ami aborigines of Taiwan and the Polynesian Islands of Samoa and Tonga. Gene. 2012;492:339–348. doi: 10.1016/j.gene.2011.10.042. [DOI] [PubMed] [Google Scholar]
- 39.Oliver DL. Ancient Tahitian Society: Ethnography. Honolulu, HI: The University Press of Hawaii; 1974. [Google Scholar]
- 40.Wilmshurst JM, Hunt TL, Lipo CP, Anderson AJ. High-precision radiocarbon dating shows recent and rapid initial human colonization of East Polynesia. Proc Natl Acad Sci USA. 2011;108:1815–1820. doi: 10.1073/pnas.1015876108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blust R. Review of Lynch, Ross, and Crowley, “The Oceanic Languages”. Ocean Linguist. 2005;44:544–548. [Google Scholar]
- 42.Blust R. Remote Melanesia: One history or two? An addendum to Donohue and Denham. Ocean Linguist. 2008;47:445–459. [Google Scholar]
- 43.Donohue M, Denham T. The language of Lapita: Vanuatu and an early Papuan presence in the Pacific. Ocean Linguist. 2008;47:365–376. [Google Scholar]
- 44.Lynch J. Melanesian diversity and Polynesian homogeneity: The other side of the coin. Ocean Linguist. 1981;20:95–129. [Google Scholar]
- 45.Tryon DT. Austronesian languages. In: May RJ, Nelson H, editors. Melanesia: Beyond diversity. ANU, Canberra: Research School of Pacific Studies; 1982. [Google Scholar]
- 46.Speiser F. Ethnology of Vanuatu. An early twentieth century study. Bathurst: Crawford House Press; 1996. [Google Scholar]
- 47.Pawley A. Explaining the aberrant Austronesian languages of Southeast Melanesia: 150 years of debate. J Polyn Soc. 2006;115:215–258. [Google Scholar]
- 48.Clark R. *Leo Tuai: A comparative lexical study of north and central Vanuatu languages. ANU, Canberra: Pacific Linguistics; 2009. [Google Scholar]
- 49.Lynch J. The linguistic history of southern Vanuatu. ANU, Canberra: Pacific Linguistics; 2001. [Google Scholar]
- 50.Bedford S. Pieces of the Vanuatu Puzzle: Archaeology of the North, South and Centre. Vol. 23. ANU Press; 2006. pp. 157–192. [Google Scholar]
- 51.Rohland N, Hofreiter M. Ancient DNA extraction from bones and teeth. Nat Protoc. 2007;2:1756–1762. doi: 10.1038/nprot.2007.247. [DOI] [PubMed] [Google Scholar]
- 52.Kircher M, Sawyer S, Meyer M. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 2012;40:e3. doi: 10.1093/nar/gkr771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fu Q, et al. DNA analysis of an early modern human from Tianyuan Cave, China. Proc Natl Acad Sci USA. 2013;110:2223–2227. doi: 10.1073/pnas.1221359110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peltzer A, et al. EAGER: efficient ancient genome reconstruction. Genome Biol. 2016;17:60. doi: 10.1186/s13059-016-0918-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schubert M, Lindgreen S, Orlando L. AdapterRemoval v2: rapid adapter trimming, identification, and read merging. BMC Res Notes. 2016;9:88. doi: 10.1186/s13104-016-1900-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Skoglund P, et al. Separating endogenous ancient DNA from modern day contamination in a Siberian Neandertal. Proc Natl Acad Sci USA. 2014;111:2229–2234. doi: 10.1073/pnas.1318934111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fu Q, et al. The genetic history of Ice Age Europe. Nature. 2016;534:200–205. doi: 10.1038/nature17993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Korneliussen TS, Albrechtsen A, Nielsen R. ANGSD: Analysis of Next Generation Sequencing Data. BMC Bioinformatics. 2014;15:356. doi: 10.1186/s12859-014-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meyer M, et al. A mitochondrial genome sequence of a hominin from Sima de los Huesos. Nature. 2014;505:403–406. doi: 10.1038/nature12788. [DOI] [PubMed] [Google Scholar]
- 62.Renaud G, Slon V, Duggan AT, Kelso J. schmutzi: estimation of contamination and endogenous mitochondrial consensus calling for ancient DNA. Genome Biol. 2015;16:224. doi: 10.1186/s13059-015-0776-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 64.Moorjani P, et al. A genetic method for dating ancient genomes provides a direct estimate of human generation interval in the last 45,000 years. Proc Natl Acad Sci USA. 2016;113:5652–5657. doi: 10.1073/pnas.1514696113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461:489–494. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reich D, et al. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 2010;468:1053–1060. doi: 10.1038/nature09710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weissensteiner H, et al. HaploGrep 2: mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Res. 2016;44:W58–63. doi: 10.1093/nar/gkw233. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All newly reported ancient DNA data including nuclear DNA alignment files and mtDNA sequences are archived at the European Nucleotide Archive database (accession number PRJEB24810). Newly reported SNP genotyping and shotgun sequence data will be made available on request to H.C. (colleran@shh.mpg.de) and A.P (powell@shh.mpg.de), subject to a signed agreement to restrict usage to anonymized non-medical studies of population history, as outlined in the ethics and consent documentation.