Abstract
There are five peptidylarginine deiminase (PAD) isozymes designated PADs 1, 2, 3, 4 and 6, and many are expressed in female reproductive tissues. These enzymes post-translationally convert positively charged arginine amino acids into neutral citrulline residues. Targets for PAD catalyzed citrullination include arginine residues on histone tails which results in chromatin decondensation and changes in gene expression. Some of the first studies examining PADs found that they are localized to rodent uterine epithelial cells. Despite these findings, the function of PAD catalyzed citrullination in uterine epithelial cells is still unknown. To address this, we first examined PAD expression in uterine cross sections from pregnant ewes on gestation day 25 (d25). Immunohistochemistry revealed that the levels of PADs 2 and 4 are robust in luminal and glandular epithelia compared to PADs 1 and 3. Since PADs 2 and 4 have well characterized roles in histone citrullination, we next hypothesized that PADs citrullinate histones in these uterine cells. Examination of caruncle lysates from pregnant ewes on gestation d25 and an ovine luminal epithelial (OLE) cell line shows that histone H3 arginine residues 2, 8, 17 and 26 are citrullinated, but histone H4 arginine 3 is not. Using a pan-PAD inhibitor, we next attenuated histone citrullination in OLE cells which resulted in a significant decrease in the expression of insulin like growth factor binding protein 1 (IGFBP1) mRNA. Since IGFBP1 is important for migration and attachment of the trophectoderm to uterine endometrium, our results suggest that PAD catalyzed citrullination may be an important post-translational mechanism for establishment of pregnancy in ewes.
Introduction
There are five peptidylarginine deiminase (PAD) enzymes (PADs 1–4 and 6) with widespread, and often overlapping, expression in tissues and cells (Vossenaar et al. 2003). This calcium dependent enzyme family post-translationally converts arginine amino acids in proteins to the non-coded residue citrulline. One exception to this is PAD6, which does not possess catalytic activity and is restricted to fibrous cytoplasmic lattices in mouse oocytes (Wright et al. 2003). PAD activity, termed deimination or citrullination, results in the loss of the positively charged guanidinium group from arginine residues. This post-translational modification occurs on arginine residues in cytoplasmic proteins such as β-actin and β-tubulin and in multiple histone isotypes (Hagiwara et al. 2005, Jiang et al. 2013, Kholia et al. 2015). Although the functional consequences of this modification on cytoplasmic proteins remains largely unclear, histone citrullination results in changes in chromatin structure to alter gene expression (Wang et al. 2004). To date, only PADs 2 and 4 have been shown to localize to the nucleus and citrullinate arginine residues on histone tails (Wang et al. 2004, Cherrington et al. 2012). Some of the first studies investigating PAD enzymes discovered robust expression and catalytic activity in rodent uteri (Takahara et al. 1989, Terakawa et al. 1991). Despite this, the functional role of PAD catalyzed citrullination in the uterus is still unknown. To address this, we have investigated PAD catalyzed citrullination in uterine tissues from pregnant ewes on gestation day 25 (d25) and an ovine luminal epithelial (OLE) cell line.
PADs 1, 2 and 4 are expressed in rodent uterine tissue in luminal and glandular epithelial cells (Takahara et al. 1992, Horibata et al. 2012). In addition, a genomic study found that PAD mRNA levels are highest in mouse uterine tissue compared to all of the other 50 tissues examined (Barrett et al. 2009). Expression changes over the phases of the estrous cycle with highest levels of PADs 2 and 4 during estrus, while PAD1 is elevated in proestrus (Terakawa et al. 1991, Takahara et al. 1992). Our past work and that of others shows that ovariectomy of mice eliminates PAD expression in the uterine luminal and glandular epithelial cells, but that it is restored by exogenous estrogen treatment (Terakawa et al. 1991, Takahara et al. 1992, Rus’d et al. 1999, Horibata et al. 2012). In terms of uterine physiology, estrogen produced by the growing follicle is absolutely critical during the proliferative phase of the estrous cycle for regeneration and growth of the endometrium. Although their expression is clearly estrogen regulated, it is not known whether PADs play a role in endometrial growth during the proliferative phase of the estrous cycle.
PADs are also expressed in mouse uterine tissue during pregnancy (Arai et al. 1995). Arai et al. found that PAD activity sharply increases from days 8–10 of pregnancy in mice, but decreases at later stages (Arai et al. 1995). PADs are expressed in mouse decidual cells, which differentiate from the uterine stromal cells surrounding the site of embryo implantation; however, it is important to note that decidualization is not thought to occur in ovine stromal cells. Importantly, no PAD expression is detected in mouse embryo-derived trophoblast cells (Arai et al. 1995). Despite these initial findings, the function of PAD catalyzed citrullination in uterine tissues during pregnancy is unclear.
In the ewe, the placentome unit is composed of the fetal cotyledon and maternal caruncle. At approximately day 13 to 14 of pregnancy, the trophoblast begins to attach and forms a strong contact with maternal luminal epithelial cells. To achieve attachment, luminal and glandular epithelial cells initiate gene programs to produce numerous molecules collectively termed the histotroph, which is then secreted into the uterine lumen (Bazer 1975, Kane et al. 1997). The histotroph is required for conceptus survival and growth and to stimulate uterine receptivity for implantation/attachment (Roberts et al. 1987, Carson et al. 2000). Supporting this, the ewe uterine gland knockout model, which lacks uterine glandular epithelial cells, cannot maintain pregnancy (Gray et al. 2001a, Gray et al. 2001b, Gray et al. 2002). Many important genes expressed in ewe uterine luminal and glandular epithelial cells during early pregnancy have been identified (Gray et al. 2006, Spencer et al. 2008, Satterfield et al. 2009). For example, insulin like growth factor binding protein 1 (IGFBP1) is produced and secreted by uterine epithelial cells to stimulate migration and attachment of the trophectoderm to maternal tissue (Gleeson et al. 2001). To the best of our knowledge, little work has investigated the role of histone modifications in regulating gene expression of the histotroph in ewe uterine luminal epithelial cells.
Our studies show that PADs 2 and 4 are highly expressed, catalytically active, and citrullinate histone H3 arginine residues 2, 8, 17, and 26 in gestation d25 ewe uterine epithelia. To investigate gene expression in uterine cells, we used the OLE cell line which only expresses PADs 2 and 4 (Johnson et al. 1999). These cells display strong PAD2 and PAD4 staining in their nuclei and only citrullinate histone H3 arginine residues similar to patterns identified in gestation d25 ewe caruncle lysates. Following treatment of OLE cells with the pan-PAD inhibitor biphenyl-benzimidazole-Cl-amidine (BB-ClA), expression of IGFBP1 mRNA is significantly decreased. Overall, our work indicates that PAD catalyzed citrullination regulates gene expression in uterine luminal epithelial cells and this mechanism may be necessary for maintaining early pregnancy in the ewe.
Material and Methods
Materials
The rabbit anti-PAD2 antibody was purchased from ProteinTech (12110-1-AP, Rosemont, IL) and the rabbit anti-histone H4cit 3 antibody was purchased from EMD Milipore (07-596, Billerica, MA). The rabbit anti-β-actin (ab8227), histone H3 total (ab1791), H3cit 2,8,17 (ab5103), H3Cit 26 (ab19847), and PAD1 (ab24008) antibodies were purchased from Abcam (Cambridge, MA). Rabbit anti-PAD3 and PAD4 antibodies were purchased from Antibodies-online Inc. (ABIN347067 Atlanta, GA) and Sigma Aldrich (P4749, St. Louis, MO), respectively. The goat anti-rabbit HRP secondary antibody was purchased from Jackson ImmunoResearch (111-035-003, West Grove, PA). The PAD inhibitor, biphenyl-benzimidazole-Cl-amidine (BB-ClA) was synthesized by Dr. Paul R. Thompson (University of Massachusetts) as previously described (Knight et al. 2015).
Cell culture
OLE cells, a generous gift from Dr. Greg Johnson (Texas A&M), were maintained in high glucose DMEM containing 2 mM glutamine, 100 U penicillin/ml, 100 μg streptomycin/ml and 10% fetal bovine serum (FBS) (HyClone, Logan, UT). All cells were grown in 5% CO2 at 37 °C in a humidified environment.
Ewe uterine tissue samples
Rambouillet ewes (Ovis aries) were maintained with ad libitum access to food and water. Animal treatment and tissue collection are described in detail in Quinn et al. (Quinn et al. 2014). Euthanasia and tissue collection was performed in accordance with the guidelines outlined in the Report of the AVMA on Euthanasia. The New Mexico State University Animal Care and Use Committee reviewed and approved all experimental procedures using animals.
Immunohistochemistry (IHC) and immunofluorescence (IF)
IHC and IF experiments were carried out as previously described (Cherrington et al. 2010). Briefly, slides were deparaffinized in 3 × 5 minute washes in xylene followed by sequential 5 minute rehydrations in 100, 95, and 75% EtOH. Endogenous peroxidase activity was blocked by incubating slides in 0.5% hydrogen peroxide in cold methanol for 10 minutes. Next, antigen retrieval was conducted by submerging the slides in 0.01 M sodium citrate and boiling 2× for 12 minutes. After cooling, slides were washed in 1X PBS and then blocked in 10% normal goat serum and 1X casein (Vector Labs, Burlingame, CA) diluted in 1X PBS for 20 minutes at room temperature in a humidified chamber. After removing excess blocking solution, slides were incubated with anti-PAD antibodies diluted 1:100 in 1X PBS for 2 hours at 37 °C. For negative controls, slides were incubated with an equal mass of non-specific rabbit IgG. After washing three times in PBS, slides were incubated for 20 minutes at room temperature with a biotinylated secondary antibody diluted 1:200 in 1X PBS. Following three washes in PBS, IHC slides were incubated in DAB chromagen (Vector Labs) solutions according to the manufacturer’s protocol, washed and then counterstained with hematoxylin and coverslip mounted. Images were taken with a Zeiss Axio Vert.A1 microscope using the 10 and 40× objectives.
For IF, OLE cells were grown in MatTek 35 mm glass bottom dishes (Ashland, MA). Cells were fixed, permeabilized and then incubated in sequential steps with primary antibodies and fluorophore conjugated (488) secondary antibodies (A11008, Thermo Scientific, Waltham, MA). After washing three times in 1X PBS, cells were stained with DAPI. For each experiment, duplicate dishes were incubated with an equal mass of non-specific rabbit IgG as a negative control (1–1000, Vector labs). All samples were imaged on a Zeiss LSM 710 confocal microscope under a 40× objective.
PAD activity assay
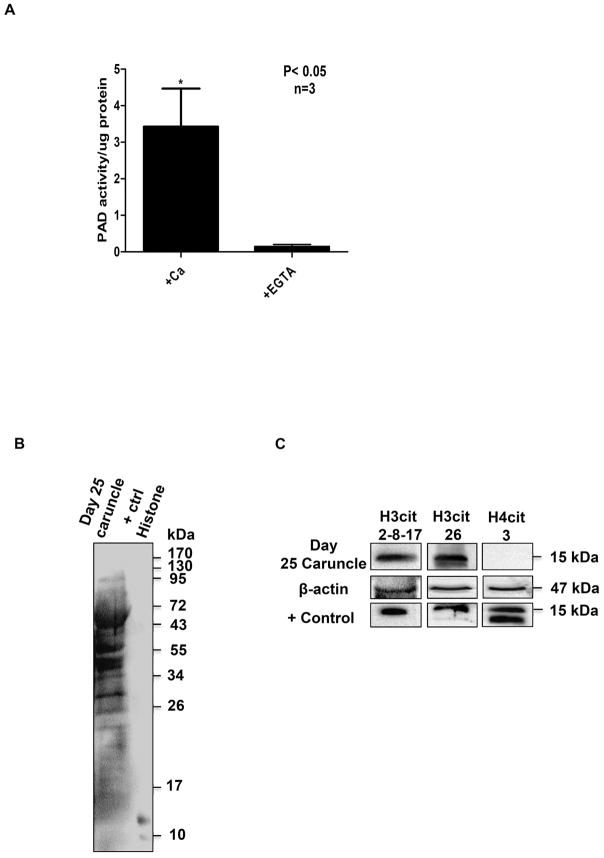
The Color Development Reagent (COLDER) assay was carried out as previously described (Knuckley et al. 2010). Briefly, caruncles were lysed in modified RIPA buffer (50 mM Tris-Cl pH 8, 150 mM NaCl, 1%NP-40, 0.25% Na-deoxycholate), sonicated, and then cleared by centrifugation. Lysates were incubated with 50 μg/ml of urease (Sigma Aldrich) to eliminate interference from urea and methylurea. 20 μgs from three independent d25 ewe caruncle lysates were then incubated with Nα-benzoyl-arginine ethyl-ester hydrochloride (BAEE) (Sigma-Aldrich) for 18 hours at 55 °C in the presence of 10 mM calcium chloride or 50 mM EGTA. Identical samples lacking BAEE were also incubated as a no-substrate control. Concentration was compared to a citrulline standard curve that was linear throughout the test range. Experiments were repeated at least three times and values are expressed as means + SEM. Means were separated using Student’s T-Test, and * indicates significantly different means (P<0.05).
Histone purification
For PAD inhibitor studies, OLE cells were treated with vehicle or 2 μM BB-ClA for 3 hours or for 6 hours which included a second spike after the first 3 hours. Histones were purified using an acid extraction method as previously described (Shechter et al. 2007). Briefly, cells were washed with 1X PBS and lysed with hypotonic lysis buffer (10 mM Tris pH 8, 1 mM KCl, 1.5 mM MgCl2, 0.5% NP-40, 1 mM DTT, 1X Protease Inhibitor, 1X PMSF, dH2O). The lysate was placed on ice for 30 minutes then nuclei were isolated by centrifugation (10,000xg for 10 minutes at 4 °C). The nuclei pellet was dissolved in 0.4 M H2SO4 and rotated overnight at 4 °C. Following centrifugation at 17,000xg for 10 minutes at 4 °C, the supernatant was collected, 100% TCA was added (20% of total volume) and incubated on ice for 20 minutes. Centrifugation at 17,000xg for 10 minutes at 4 °C was again conducted, the supernatant was then discarded, and the pellet was washed with 300 μl of cold acetone. The purified histone pellets were air dried, resuspended in 50 μl of nanopure H2O, and quantified using a Pierce 660 nm protein assay (Rockford, IL).
Western blots
Positive controls for PAD antibodies were generated by overexpressing human PAD 1–4 plasmids for 24 hours following a Mirus Bio (Madison, WI) TranIT-2020 transfection protocol. Positive controls for histone antibodies were generated by in vitro citrullination of bulk histones. OLE cells and snap-frozen d25 ewe caruncles were lysed with RIPA buffer containing 50 mM Tris, 150 mM NaCl, 0.1% SDS, 0.5% deoxycholate, 1% tritionX-100, 1 mM PMSF and 1X protease inhibitor (Thermo Scientific). Protein concentration of lysates and purified histones was measured by Pierce 660 nm protein assay prior to gel loading to ensure equal protein loading. 6X sample buffer consisting of 0.5 M Tris-HCl (pH 6.8), 60% glycerol, 30 mM DTT, 6% SDS was added into samples to yield a final concentration of 1X sample buffer and then boiled at 95 °C for 5 minutes. The samples were subjected to SDS-PAGE using 12 or 15% gels (acrylamide:bis-acrylamide ratio of 29:1) and subsequently transferred to Immobilin PVDF membranes (EMD Millipore). Membranes were blocked in 1X casein (Vector Labs) diluted in Tris buffered saline containing 0.1% Tween-20 (TBS-T) overnight at 4 °C. Primary antibodies were incubated overnight at 4 °C: Anti-PAD1 (1:1000), Anti-PAD2 (1:2000), Anti-PAD3 (1:800), or Anti-PAD4 (1:2000), H3cit 2,8,17 (1:1000), H3cit 26 (1:500), H4cit 3 (1:500), H3 total (1:5000). The following morning, membranes were washed in TBS-T, followed by a 2 hour incubation at room temperature with 1:10,000 anti-rabbit HRP secondary antibody. All blots were washed for 50 minutes (5×10 minutes) with TBS-T after secondary antibody incubation and then visualized with a BioRad Chemidoc XRS using SuperSignal West Pico and Femto chemiluminescence substrate (Pierce, Rockford, IL). To confirm equal protein loading, membranes were stripped and re-probed with anti-β-actin or anti-total histone H3. Quantitative densitometry analysis was conducted with BioRad Image Lab software. Anti-Modified Citrulline (AMC) western blots were performed according to the manufacture’s protocol (EMD Millipore). Experiments were repeated at least independent times and values are expressed as the mean ± SEM. Means were separated using separated using Newman-Keuls ANOVA and * indicates significantly different means (P<0.05).
qPCR
RNA from OLE cells was purified according to the Omega Bio-Tek Total RNA Kit protocol (Omega Bio-Tek, Inc., Norcross, GA). 1 μg of resulting RNA was reverse transcribed using iScript Reverse Transcription Supermix for RT-qPCR (BIO-RAD, Hercules, CA). Complementary DNA was subject to real time PCR analysis with SYBR Green (BIO-RAD) using intron spanning primers: HSD11B1; FWD 5′-TAGGTTCTCTCTGTGTGTCCCA-3′, REV 5′-TCCTCGAAGCATCTCTGGTCT-3′; IGFBP1 FWD 5′-CAGCAAACAGTGTGAGACTTCG-3′, REV 5′-TCCCACTCCAAGGGTAGACA-3′; GAPDH FWD 5′-CGTTCTCTGCCTTGACTGTG-3′, REV 5′-TGACCCCTTCATTGACCTTC-3′. Data was analyzed using the ΔΔ ct method in which ct values of target genes are adjusted to corresponding ct value of reference gene (GAPDH) (Livak & Schmittgen 2001, Coleson et al. 2015). The experiment was repeated 3 independent times and values are expressed as the mean ± SEM. Means were separated using Student’s T-Test, and *** indicates significantly different means (P<0.0001).
Statistical analysis
All experiments were independently repeated at least three times and resulting values are expressed as the mean ± SEM. Statistical analysis was done with GraphPad Prism 6.0. Means were separated using Newman-Keuls ANOVA or Student’s T-Test and * indicates significantly different means (* P<0.05 and *** P<0.0001).
Results
PADs 2 and 4 are expressed in luminal and glandular epithelial cells in gestation day 25 ewe uterine cross sections
Since PADs are expressed in mouse uterine luminal and glandular epithelial cells, we first examined if PADs are also expressed in ewe uterine cells on d25 of gestation. Paraffin embedded uterine tissue was cut into 5 μM sections and examined by IHC. Tissue sections were probed with an equal mass of non-specific IgG as a control or primary antibodies against PADs 1–4. Imaging revealed that PAD2 and PAD4 expression localizes to luminal and glandular epithelial cells with sparse staining in adjacent stromal tissue (Figure 1, panels B, I and D, J). In contrast, PAD1 and PAD3 display low levels of expression in luminal and glandular epithelial cells (Figure 1, panels A and C). This is the first work to show that PADs are expressed in pregnant ewe uterine luminal and glandular epithelial cells and display a cellular localization pattern similar to that found in mouse uteri.
Figure 1. PADs 2 and 4 are expressed in luminal and glandular epithelial cells in gestation day 25 ewe uterine cross sections.
Uterine tissue from pregnant ewes on d25 of gestation was harvested, fixed in 4% paraformaldehyde, embedded in paraffin and sectioned. 5 μm uterine tissue sections were subject to a standard IHC protocol using rabbit anti-PAD1, PAD2, PAD3, PAD4 antibodies or an equal amount of non-specific rabbit IgG as a control. Images were taken with a Zeiss Axio Vert.A1 microscope using the 10 and 40× objectives, and DAB staining represents PAD expression. Scale bars for 10× images are 100 μm, while scale bars in 40× images are 20 μm.
PADs are catalytically active in gestation day 25 ewe caruncle lysates and citrullinate multiple proteins including histone H3 arginine residues
Given that PADs are expressed in ewe uterine luminal and glandular epithelia, we hypothesized that the enzymes would be catalytically active and citrullinate proteins. To test this, we first examined PAD activity in caruncle lysates (n=3) from ewes on d25 of gestation using the COLDER assay as previously described (Li et al. 2016). Since PAD activity is highly calcium dependent, 20 μg of caruncle lysate was incubated in the presence of 10 mM calcium chloride or 50 mM EGTA. Our results show that PADs are catalytically active and that chelation of calcium significantly decreases activity (P<0.05) (Figure 2A). We next tested if citrullinated proteins are present in caruncle lysates. To do this, caruncle lysate was subjected to an anti-modified citrulline (AMC) western blot which included in vitro citrullinated histones as a positive control (Li et al. 2016). Multiple citrullinated protein bands were detected (Figure 2B); however, we achieved poor resolution of smaller sized bands. Therefore, we next used western blots to examine if characterized antibodies could detect citrullinated arginine residues on histone tails in caruncle lysates. Membranes were probed with anti-H3cit 2, 8, 17, H3cit 26, and H4cit 3 antibodies and β-actin as a loading control. Western blots included in vitro citrullinated bulk histones as a positive control. Our results show that histone H3 arginine residues 2, 8, 17, and 26 are citrullinated, however histone H4 arginine 3 is not (Figure 2C). Taken together, our results suggest that PAD catalyzed citrullination occurs in uterine luminal and glandular epithelial cells of ewes during pregnancy and one target is arginine residues on histone H3 tails.
Figure 2. PADs are catalytically active in gestation day 25 ewe caruncle lysates and citrullinate multiple proteins including histone H3 arginine residues.
(A) 20 μgs of caruncle lysate (n=3) was incubated with 10 mM calcium chloride or 50 mM EGTA then subject to the COLDER assay. Values represent the mean ± SEM. Means were separated using Student’s T-Test with * designating significant differences with treatment (P<0.05). (B) Caruncle lysate was examined by western blot following the AMC protocol. The positive control is in vitro citrullinated bulk histones. (C) Caruncle lysates were examined by western blot and membranes were probed with rabbit anti-histone H3cit 2, 8 and 17, anti-histone H3cit 26, and anti-histone H4cit 3 antibodies and an anti-β-actin antibody as a loading control. The positive control is in vitro citrullinated bulk histones.
PADs 2 and 4 are expressed in the nuclei of the OLE cell line
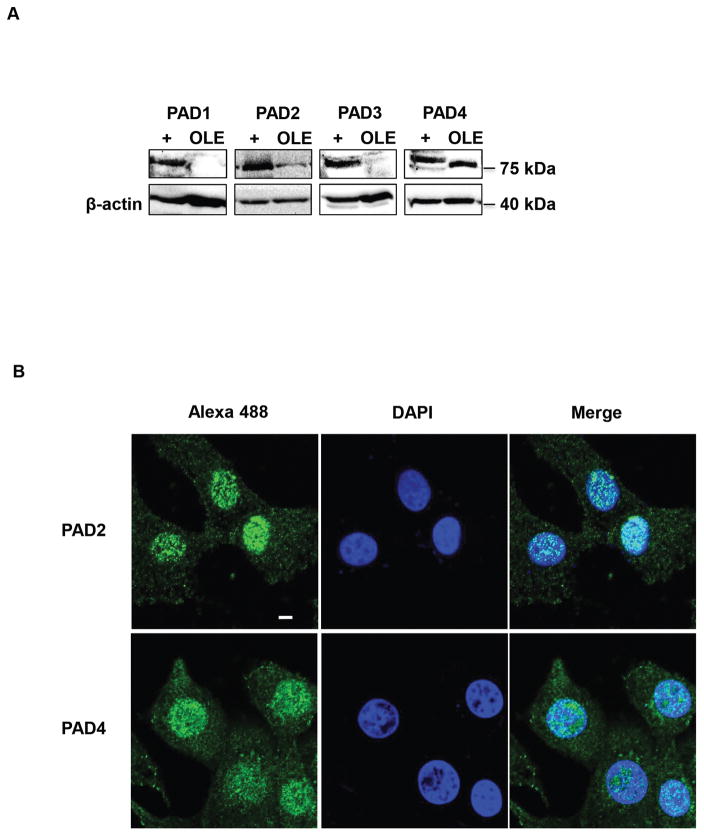
We next wanted to test if genes are regulated by PAD catalyzed histone citrullination in ewe uterine epithelial cells. To do so, we first needed to validate that OLE cells, which were initially isolated and characterized by Johnson et al., accurately recapitulate PAD expression patterns compared to gestation d25 ewe uterine tissue (Johnson et al. 1999). OLE cell lysates were examined by western blot in parallel with lysates in which human PAD 1–4 plasmids were overexpressed to generate positive controls (+) for PAD antibodies. Membranes were probed with anti-PAD 1–4 antibodies or β-actin as a loading control. Our result highlights that PADs 2 and 4 are the predominant isozymes expressed in OLE cells (Figure 3A).
Figure 3. PADs 2 and 4 are expressed in the nuclei of the OLE cell line.
(A) Equal amounts of OLE lysates were examined by western blot. Membranes were probed with rabbit anti-PAD1, PAD2, PAD3, PAD4 antibodies or anti-β-tubulin for a loading control. Positive controls (+) were generated by overexpression of PAD1–4 plasmids. (B) OLE cells were grown on glass bottom dishes then fixed, permeabilized and subjected to an IF protocol using anti-PAD2 and anti-PAD4 antibodies (green) and stained with DAPI (blue). Cells were imaged using a Zeiss LSM 710 confocal microscope using a 40× objective and scale bar is 5 μm.
We next used IF confocal microscopy to examine the subcellular localization of PADs 2 and 4 in OLE cells. OLE cells were fixed, probed with anti-PAD2 and PAD4 antibodies and then stained with DAPI. Our imaging results revealed that OLE cells have strong PAD2 and PAD4 staining within the nucleus (Figure 3B). Thus, the OLE cells recapitulate PAD expression patterns detected in gestation d25 ewe uterine tissue and may serve as a suitable model to examine PAD regulated gene expression.
Histone H3 arginine residues 2, 8, 17 and 26 are citrullinated in OLE cells
Since PADs 2 and 4 are expressed in the nuclei of OLE cells, we next tested if OLE cells display a similar citrullinated histone profile as that identified in gestation d25 caruncle lysates. To test this, equal concentrations of purified OLE histones were examined by western blot, which included in vitro citrullinated bulk histones as a positive control. Membranes were probed with anti-H3cit 2, 8, 17, H3cit 26, and H4cit 3 antibodies or total H3 as a loading control. Similar to gestation d25 caruncle lysates, histone arginine residues 2, 8, 17, and 26 are citrullinated, however H4 arginine 3 is not (Figure 4). These results indicate that citrullination of histone H3 arginine residues occurs in OLE cells.
Figure 4. Histone H3 arginine residues 2, 8, 17 and 26 are citrullinated in OLE cells.
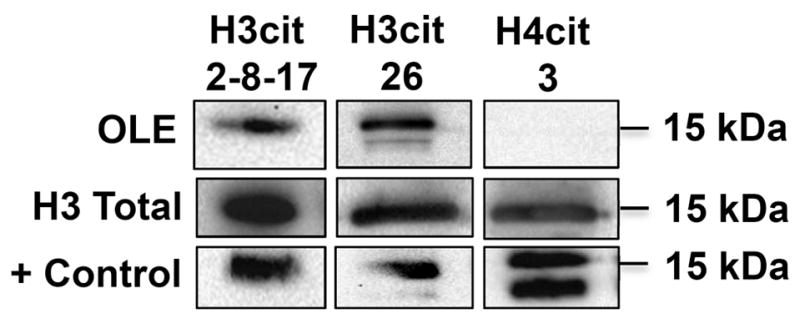
OLE histones were purified by acid extraction, quantified and equal amounts examined by western blot. Membranes were probed with rabbit anti-histone H3cit 2, 8 and 17, anti-histone H3cit 26, and anti-histone H4cit 3 antibodies and an anti-histone H3 total antibody as a loading control. The positive control is in vitro citrullinated bulk histones.
Treatment of OLE cells with the pan-PAD inhibitor BB-ClA decreases histone citrullination
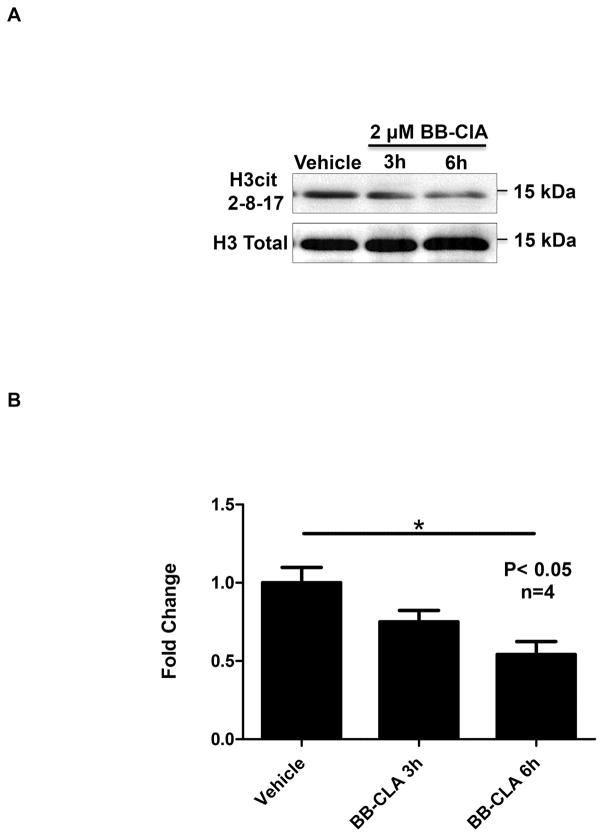
We next tested if treatment with BB-ClA decreases histone citrullination in OLE cells. OLE cells were treated with vehicle (DMSO) or 2 μM BB-ClA for 3 and 6 hours at which point histones were isolated and quantified. Equal concentrations of histones were examined by western blot and membranes were probed with anti-H3cit 2, 8, 17 antibody and total H3 as a loading control. The representative western blot illustrates that 6 hours of treatment with 2 μM BB-ClA reduces histone citrullination compared to vehicle treated controls (Figure 5A). Quantification of blots (n=4) revealed that BB-ClA significantly decreases histone H3 citrullination by approximately 50% following 6 hours of treatment compared to vehicle treated controls (P<0.05) (Figure 5B). Thus, BB-ClA significantly inhibits PAD catalyzed citrullination of histones in OLE cells.
Figure 5. Treatment of OLE cells with the pan-PAD inhibitor BB-ClA decreases histone citrullination.
(A) OLE cells were treated with vehicle (DMSO) or 2 μM BB-ClA for 3 and 6 hours. Post-treatment, OLE histones were purified by acid extraction, quantified and equal amounts examined by western blot. Membranes were probed with a rabbit anti-histone H3cit 2, 8 and 17 antibody, then stripped and reprobed with an anti-histone H3 total antibody as a loading control. (B) Quantitative analysis of the western blots (n=4) was conducted using BioRad ImageLab software and normalized to total histone H3 levels. Means were separated using Newman-Keuls ANOVA and * indicates significant differences (P< 0.05), while error bars represent the SEM.
PAD inhibition decreases expression of IGFBP1 mRNA in OLE cells
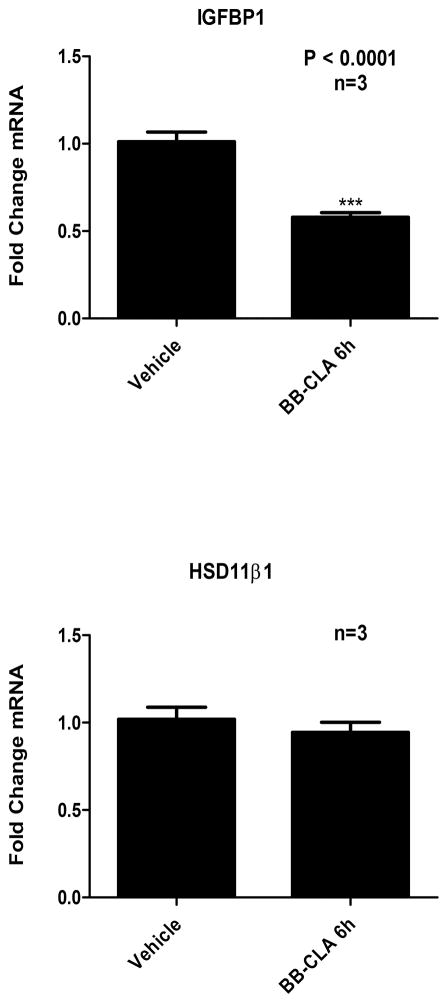
In order to determine if PAD catalyzed citrullination regulates gene expression in OLE cells, we first identified two genes that are important during early pregnancy in the ewe. We examined expression of IGFBP1 and hydroxysteroid (11-B) dehydrogenase 1 (HSD11B1) based on their characterized expression patterns in uterine luminal epithelia (Gray et al. 2006, Spencer et al. 2008, Satterfield et al. 2009). OLE cells were treated with vehicle (DMSO) or 2 μM BB-ClA for 6 hours, RNA was then purified from cells, reverse transcribed, and resulting cDNA examined by qPCR with primers specific for IGFBP1, HSD11B1 and GAPDH as the reference control gene. Our results show that inhibition of PAD catalyzed citrullination in OLE cells results in a significant decrease in IGFBP1 mRNA expression (P<0.0001), while having no effect on HSB11B1 (Figure 6). This result suggests that PAD catalyzed citrullination normally stimulates IGFBP1 expression in uterine luminal epithelial cells.
Figure 6. PAD inhibition decreases expression of IGFBP1 mRNA in OLE cells.
OLE cells were treated with vehicle (DMSO) or 2 μM BB-ClA for 3 and 6 hours. Total RNA was purified from OLE cells, reverse transcribed and resulting cDNA was examined by qPCR with intron spanning primers for IGFBP1, HSD11B1 or GAPDH as the reference gene control. Data was analyzed using the ΔΔct method where IGFBP1 and HSD11B1 ct values were normalized to GAPDH and are represented as fold change in mRNA expression. Data are expressed as means ± SEM. Means were separated using Student’s T-Test and *** indicating significant differences (P< 0.0001).
Discussion
The majority of pregnancy loss in humans and livestock occurs during early pregnancy (Macklon et al. 2002, Diskin et al. 2006, Dixon et al. 2007, Kwak-Kim et al. 2010). As such, significant effort has been expended to understand the relevant mechanisms to prevent such loss. One area under investigation is how the histotroph, produced by uterine luminal and glandular epithelial cells, regulates conceptus-uterine interactions. Underlying histotroph production is the activation of specific gene programs that encode the complex mixture of proteins, cytokines, growth factors and hormones required for implantation/attachment. Our work here has investigated the function of PAD catalyzed citrullination in ewe uterine epithelial cells and identified a possible role in expression of a histotroph protein.
In order to address PAD function in uterine tissue, we first examined expression in uterine cross sections from pregnant ewes on d25 of gestation and found that PADs 2 and 4 are highly expressed, while the expression of PADs 1 and 3 is low. This finding is supported by our previous work showing strong PAD2 and PAD4 expression in mouse uterine luminal and glandular epithelial cells (Horibata et al. 2012). At issue is what stimulates PAD expression in uterine luminal and glandular epithelial cells. In estrogen receptor α knockout (αERKO) and KI/KO (ER binding mutant) mice, uteri show decreased levels of PAD expression compared to wild type mice indicating that ERα is important for expression (Hewitt et al. 2003). However, low ERα mRNA and protein in the luminal and shallow glandular epithelia, combined with low serum estrogen on d25 of gestation in ewes indicates that this mechanism does not likely regulate PAD expression (Spencer & Bazer 1995). In contrast, serum progesterone levels increase during the peri-implantation period, but exogenous progesterone treatment of ovariectomized mice does not alter PAD expression over vehicle treated controls (Takahara et al. 1992). Thus, auto- and paracrine signals from maternal and embryonic tissues most likely regulate PAD expression during this period, and studies are currently underway to address this interesting question.
Multiple proteins are citrullinated in gestation d25 caruncle lysates. The identity and function of these citrullinated proteins is unknown, but it is an active area of research in our lab. Our studies did determine that histone H3 arginine residues 2, 8, 17, and 26 are citrullinated in OLE cells and gestation d25 caruncle lysates, but that histone H4 arginine 3 is not. Collectively, these results suggest that citrullination of specific arginine residues may govern the expression of gene programs in uterine luminal epithelial cells. Our previous studies in mammary epithelial cells also support that PAD2 and PAD4 localize to the nucleus and citrullinate specific histone tail arginine residues to regulate gene expression (Cherrington et al. 2010, Zhang et al. 2011, Cherrington et al. 2012). Once in the nucleus, PADs hydrolyze the positive guanidinium group of arginine residues on histone H2A, H3 and H4 tails to alter the charge association between DNA and histone octomers to decondense chromatin and subsequently alter gene expression (Hagiwara et al. 2002, Wang et al. 2004). Functionally, PADs antagonize histone arginine methylation by coactivator associated arginine methyltransferase (CARM1) and protein arginine methyltransferase (PRMT1) (Wang et al. 2004). In addition to PADs, multiple isoforms of histone lysine demethylase are expressed in caruncle tissue from gestation day 90 ewes (Cleys et al. 2015). Interestingly, both epigenetic enzymes function as demethylases, which may be an important mechanism to regulate gene programs during pregnancy in ewe caruncle tissues.
The OLE cell line was isolated from ewe uterine luminal epithelial cells on day 5 of the estrous cyclic (Johnson et al. 1999). It is important to note that endometrial cells from both pregnant and cyclic ewes, cattle and humans express similar genes important for receptivity and implantation (Kao et al. 2002, Brooks et al. 2014). In OLE cells, histone citrullination is attenuated by BB-ClA, which blocks PAD activity by covalently binding to the enzyme active site (Knight et al. 2015). BB-CLA significantly decreases PAD activity both in vitro and in vivo (Horibata et al. 2015, Knight et al. 2015). We next identified two potential target genes, IGFBP1 and HSD11B1, expressed in uterine luminal epithelial cells based on microarrays data sets; both genes have characterized roles during early pregnancy in ewes (Johnson et al. 2001, Spencer et al. 2004, Spencer et al. 2008, Satterfield et al. 2009). In OLE cells, the inhibition of PAD catalyzed citrullination did not alter expression HSD11B1; however, IGFBP1 mRNA levels were significantly decreased. Given this decrease, it is likely that PAD catalyzed citrullination normally stimulates expression of IGFBP1. IGFBP1 is secreted by uterine luminal and glandular epithelial cells to facilitate migration and attachment of the trophectoderm (Gleeson et al. 2001, Brooks et al. 2014). During the peri-implantation period, luminal epithelial cells secrete increasing amounts of IGFBP1, which temporally corresponds with the transformation of the blastocyst into a filamentous conceptus (Spencer et al. 2004, Spencer et al. 2007). From a functional standpoint, IGFBP1 contains a tripeptide Arg-Gly-Asp (RGD) motif that binds the integrin heterodimer α5β1 to promote cell attachment. Mutation of the motif results in decreased migration of human trophoblast cells (Irwin & Giudice 1998, Gleeson et al. 2001). In addition to the ewe, IGFBP1 is implicated as a regulator of implantation and placental development in other species (Giudice & Saleh 1995, Robinson et al. 2000). Our work suggests that PAD catalyzed citrullination facilitates the expression of IGFBP1, and therefore may represent a novel mechanism contributing to trophoblast attachment in the ewe. It is also likely that PAD catalyzed citrullination regulates the expression of additional genes involved in this process. Our follow up studies are designed to identify the full cohort of genes regulated by PAD catalyzed citrullination in uterine luminal epithelial cells.
Our work here further advances our knowledge of PAD function in uterine epithelial cells and begins to address a role for theses enzymes during early pregnancy. We propose that PAD catalyzed citrullination may be an important mechanism utilized by uterine luminal epithelial cells to regulate histotroph gene expression during early pregnancy. If true, then modulating PAD catalyzed citrullination could be a novel approach to regulate conceptus attachment to uterine tissue. Thus, this line of research may represent a new approach to investigate early pregnancy loss which occurs in species ranging from humans to livestock.
Acknowledgments
Funding: This project was supported by grants from the National Center for Research Resources (P20RR016474) and the National Institute of General Medical Sciences (P20GM103432) from the National Institutes of Health (B.D.C) and by National Institute of General Medical Sciences grants GM079357 and GM110394 (P.R.T.). The content is solely the responsibility of the author and does not necessarily represent the official views of the National Institutes of Health. This project was also supported by a grant from the New Mexico Agriculture Experiment Station (R.A.A).
Footnotes
Declaration of interest: The authors have no known conflict of interest and nothing to disclose except that P.R.T is a paid consultant to Bristol Myers Squibb.
References
- Arai T, Kusubata M, Kohsaka T, Shiraiwa M, Sugawara K, Takahara H. Mouse uterus peptidylarginine deiminase is expressed in decidual cells during pregnancy. J Cell Biochem. 1995;58:269–278. doi: 10.1002/jcb.240580302. [DOI] [PubMed] [Google Scholar]
- Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Muertter RN, Edgar R. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazer FW. Uterine protein secretions: Relationship to development of the conceptus. J Anim Sci. 1975;41:1376–1382. doi: 10.2527/jas1975.4151376x. [DOI] [PubMed] [Google Scholar]
- Brooks K, Burns G, Spencer TE. Conceptus elongation in ruminants: roles of progesterone, prostaglandin, interferon tau and cortisol. J Anim Sci Biotechnol. 2014;5:53. doi: 10.1186/2049-1891-5-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson DD, Bagchi I, Dey SK, Enders AC, Fazleabas AT, Lessey BA, Yoshinaga K. Embryo implantation. Dev Biol. 2000;223:217–237. doi: 10.1006/dbio.2000.9767. [DOI] [PubMed] [Google Scholar]
- Cherrington BD, Morency E, Struble AM, Coonrod SA, Wakshlag JJ. Potential role for peptidylarginine deiminase 2 (PAD2) in citrullination of canine mammary epithelial cell histones. PLoS One. 2010;5:e11768. doi: 10.1371/journal.pone.0011768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherrington BD, Zhang X, McElwee JL, Morency E, Anguish LJ, Coonrod SA. Potential role for PAD2 in gene regulation in breast cancer cells. PLoS One. 2012;7:e41242. doi: 10.1371/journal.pone.0041242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleys ER, Halleran JL, Enriquez VA, da Silveira JC, West RC, Winger QA, Anthony RV, Bruemmer JE, Clay CM, Bouma GJ. Androgen receptor and histone lysine demethylases in ovine placenta. PLoS One. 2015;10:e0117472. doi: 10.1371/journal.pone.0117472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleson MP, Sanchez NS, Ashley AK, Ross TT, Ashley RL. Human chorionic gonadotropin increases serum progesterone, number of corpora lutea and angiogenic factors in pregnant sheep. Reproduction. 2015;150:43–52. doi: 10.1530/REP-14-0632. [DOI] [PubMed] [Google Scholar]
- Diskin MG, Murphy JJ, Sreenan JM. Embryo survival in dairy cows managed under pastoral conditions. Anim Reprod Sci. 2006;96:297–311. doi: 10.1016/j.anireprosci.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Dixon AB, Knights M, Winkler JL, Marsh DJ, Pate JL, Wilson ME, Dailey RA, Seidel G, Inskeep EK. Patterns of late embryonic and fetal mortality and association with several factors in sheep. J Anim Sci. 2007;85:1274–1284. doi: 10.2527/jas.2006-129. [DOI] [PubMed] [Google Scholar]
- Giudice LC, Saleh W. Growth factors in reproduction. Trends Endocrinol Metab. 1995;6:60–69. doi: 10.1016/1043-2760(94)00205-i. [DOI] [PubMed] [Google Scholar]
- Gleeson LM, Chakraborty C, McKinnon T, Lala PK. Insulin-like growth factor-binding protein 1 stimulates human trophoblast migration by signaling through alpha 5 beta 1 integrin via mitogen-activated protein Kinase pathway. J Clin Endocrinol Metab. 2001;86:2484–2493. doi: 10.1210/jcem.86.6.7532. [DOI] [PubMed] [Google Scholar]
- Gray CA, Abbey CA, Beremand PD, Choi Y, Farmer JL, Adelson DL, Thomas TL, Bazer FW, Spencer TE. Identification of endometrial genes regulated by early pregnancy, progesterone, and interferon tau in the ovine uterus. Biol Reprod. 2006;74:383–394. doi: 10.1095/biolreprod.105.046656. [DOI] [PubMed] [Google Scholar]
- Gray CA, Bazer FW, Spencer TE. Effects of neonatal progestin exposure on female reproductive tract structure and function in the adult ewe. Biol Reprod. 2001a;64:797–804. doi: 10.1095/biolreprod64.3.797. [DOI] [PubMed] [Google Scholar]
- Gray CA, Burghardt RC, Johnson GA, Bazer FW, Spencer TE. Evidence that absence of endometrial gland secretions in uterine gland knockout ewes compromises conceptus survival and elongation. Reproduction. 2002;124:289–300. [PubMed] [Google Scholar]
- Gray CA, Taylor KM, Ramsey WS, Hill JR, Bazer FW, Bartol FF, Spencer TE. Endometrial glands are required for preimplantation conceptus elongation and survival. Biol Reprod. 2001b;64:1608–1613. doi: 10.1095/biolreprod64.6.1608. [DOI] [PubMed] [Google Scholar]
- Hagiwara T, Hidaka Y, Yamada M. Deimination of histone H2A and H4 at arginine 3 in HL-60 granulocytes. Biochemistry. 2005;44:5827–5834. doi: 10.1021/bi047505c. [DOI] [PubMed] [Google Scholar]
- Hagiwara T, Nakashima K, Hirano H, Senshu T, Yamada M. Deimination of arginine residues in nucleophosmin/B23 and histones in HL-60 granulocytes. Biochem Biophys Res Commun. 2002;290:979–983. doi: 10.1006/bbrc.2001.6303. [DOI] [PubMed] [Google Scholar]
- Hewitt SC, Deroo BJ, Hansen K, Collins J, Grissom S, Afshari CA, Korach KS. Estrogen receptor-dependent genomic responses in the uterus mirror the biphasic physiological response to estrogen. Mol Endocrinol. 2003;17:2070–2083. doi: 10.1210/me.2003-0146. [DOI] [PubMed] [Google Scholar]
- Horibata S, Coonrod SA, Cherrington BD. Role for peptidylarginine deiminase enzymes in disease and female reproduction. J Reprod Dev. 2012;58:274–282. doi: 10.1262/jrd.2011-040. [DOI] [PubMed] [Google Scholar]
- Horibata S, Vo TV, Subramanian V, Thompson PR, Coonrod SA. Utilization of the Soft Agar Colony Formation Assay to Identify Inhibitors of Tumorigenicity in Breast Cancer Cells. J Vis Exp. 2015:e52727. doi: 10.3791/52727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin JC, Giudice LC. Insulin-like growth factor binding protein-1 binds to placental cytotrophoblast alpha5beta1 integrin and inhibits cytotrophoblast invasion into decidualized endometrial stromal cultures. Growth Horm IGF Res. 1998;8:21–31. doi: 10.1016/s1096-6374(98)80318-3. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Cui Y, Wang L, Zhao Y, Yan S, Chang X. Investigating citrullinated proteins in tumour cell lines. World J Surg Oncol. 2013;11:260. doi: 10.1186/1477-7819-11-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GA, Bazer FW, Jaeger LA, Ka H, Garlow JE, Pfarrer C, Spencer TE, Burghardt RC. Muc-1, integrin, and osteopontin expression during the implantation cascade in sheep. Biol Reprod. 2001;65:820–828. doi: 10.1095/biolreprod65.3.820. [DOI] [PubMed] [Google Scholar]
- Johnson GA, Burghardt RC, Newton GR, Bazer FW, Spencer TE. Development and characterization of immortalized ovine endometrial cell lines. Biol Reprod. 1999;61:1324–1330. doi: 10.1095/biolreprod61.5.1324. [DOI] [PubMed] [Google Scholar]
- Kane MT, Morgan PM, Coonan C. Peptide growth factors and preimplantation development. Hum Reprod Update. 1997;3:137–157. doi: 10.1093/humupd/3.2.137. [DOI] [PubMed] [Google Scholar]
- Kao LC, Tulac S, Lobo S, Imani B, Yang JP, Germeyer A, Osteen K, Taylor RN, Lessey BA, Giudice LC. Global gene profiling in human endometrium during the window of implantation. Endocrinology. 2002;143:2119–2138. doi: 10.1210/endo.143.6.8885. [DOI] [PubMed] [Google Scholar]
- Kholia S, Jorfi S, Thompson PR, Causey CP, Nicholas AP, Inal JM, Lange S. A novel role for peptidylarginine deiminases in microvesicle release reveals therapeutic potential of PAD inhibition in sensitizing prostate cancer cells to chemotherapy. J Extracell Vesicles. 2015;4:26192. doi: 10.3402/jev.v4.26192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight JS, Subramanian V, O’Dell AA, Yalavarthi S, Zhao W, Smith CK, Hodgin JB, Thompson PR, Kaplan MJ. Peptidylarginine deiminase inhibition disrupts NET formation and protects against kidney, skin and vascular disease in lupus-prone MRL/lpr mice. Ann Rheum Dis. 2015;74:2199–2206. doi: 10.1136/annrheumdis-2014-205365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuckley B, Causey CP, Jones JE, Bhatia M, Dreyton CJ, Osborne TC, Takahara H, Thompson PR. Substrate specificity and kinetic studies of PADs 1, 3, and 4 identify potent and selective inhibitors of protein arginine deiminase 3. Biochemistry. 2010;49:4852–4863. doi: 10.1021/bi100363t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak-Kim J, Park JC, Ahn HK, Kim JW, Gilman-Sachs A. Immunological modes of pregnancy loss. Am J Reprod Immunol. 2010;63:611–623. doi: 10.1111/j.1600-0897.2010.00847.x. [DOI] [PubMed] [Google Scholar]
- Li G, Hayward IN, Jenkins BR, Rothfuss HM, Young CH, Nevalainen MT, Muth A, Thompson PR, Navratil AM, Cherrington BD. Peptidylarginine Deiminase 3 (PAD3) Is Upregulated by Prolactin Stimulation of CID-9 Cells and Expressed in the Lactating Mouse Mammary Gland. PLoS One. 2016;11:e0147503. doi: 10.1371/journal.pone.0147503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Macklon NS, Geraedts JP, Fauser BC. Conception to ongoing pregnancy: the ‘black box’ of early pregnancy loss. Hum Reprod Update. 2002;8:333–343. doi: 10.1093/humupd/8.4.333. [DOI] [PubMed] [Google Scholar]
- Quinn KE, Ashley AK, Reynolds LP, Grazul-Bilska AT, Ashley RL. Activation of the CXCL12/CXCR4 signaling axis may drive vascularization of the ovine placenta. Domest Anim Endocrinol. 2014;47:11–21. doi: 10.1016/j.domaniend.2013.12.004. [DOI] [PubMed] [Google Scholar]
- Roberts RM, Murray MK, Burke MG, Ketcham CM, Bazer FW. Hormonal control and function of secretory proteins. Adv Exp Med Biol. 1987;230:137–150. doi: 10.1007/978-1-4684-1297-0_8. [DOI] [PubMed] [Google Scholar]
- Robinson RS, Mann GE, Gadd TS, Lamming GE, Wathes DC. The expression of the IGF system in the bovine uterus throughout the oestrous cycle and early pregnancy. J Endocrinol. 2000;165:231–243. doi: 10.1677/joe.0.1650231. [DOI] [PubMed] [Google Scholar]
- Rus’d AA, Ikejiri Y, Ono H, Yonekawa T, Shiraiwa M, Kawada A, Takahara H. Molecular cloning of cDNAs of mouse peptidylarginine deiminase type I, type III and type IV, and the expression pattern of type I in mouse. Eur J Biochem. 1999;259:660–669. doi: 10.1046/j.1432-1327.1999.00083.x. [DOI] [PubMed] [Google Scholar]
- Satterfield MC, Song G, Kochan KJ, Riggs PK, Simmons RM, Elsik CG, Adelson DL, Bazer FW, Zhou H, Spencer TE. Discovery of candidate genes and pathways in the endometrium regulating ovine blastocyst growth and conceptus elongation. Physiol Genomics. 2009;39:85–99. doi: 10.1152/physiolgenomics.00001.2009. [DOI] [PubMed] [Google Scholar]
- Shechter D, Dormann HL, Allis CD, Hake SB. Extraction, purification and analysis of histones. Nat Protoc. 2007;2:1445–1457. doi: 10.1038/nprot.2007.202. [DOI] [PubMed] [Google Scholar]
- Spencer TE, Bazer FW. Temporal and spatial alterations in uterine estrogen receptor and progesterone receptor gene expression during the estrous cycle and early pregnancy in the ewe. Biol Reprod. 1995;53:1527–1543. doi: 10.1095/biolreprod53.6.1527. [DOI] [PubMed] [Google Scholar]
- Spencer TE, Johnson GA, Bazer FW, Burghardt RC. Implantation mechanisms: insights from the sheep. Reproduction. 2004;128:657–668. doi: 10.1530/rep.1.00398. [DOI] [PubMed] [Google Scholar]
- Spencer TE, Johnson GA, Bazer FW, Burghardt RC. Fetal-maternal interactions during the establishment of pregnancy in ruminants. Soc Reprod Fertil Suppl. 2007;64:379–396. doi: 10.5661/rdr-vi-379. [DOI] [PubMed] [Google Scholar]
- Spencer TE, Sandra O, Wolf E. Genes involved in conceptus-endometrial interactions in ruminants: insights from reductionism and thoughts on holistic approaches. Reproduction. 2008;135:165–179. doi: 10.1530/REP-07-0327. [DOI] [PubMed] [Google Scholar]
- Takahara H, Kusubata M, Tsuchida M, Kohsaka T, Tagami S, Sugawara K. Expression of peptidylarginine deiminase in the uterine epithelial cells of mouse is dependent on estrogen. J Biol Chem. 1992;267:520–525. [PubMed] [Google Scholar]
- Takahara H, Tsuchida M, Kusubata M, Akutsu K, Tagami S, Sugawara K. Peptidylarginine deiminase of the mouse. Distribution, properties, and immunocytochemical localization. J Biol Chem. 1989;264:13361–13368. [PubMed] [Google Scholar]
- Terakawa H, Takahara H, Sugawara K. Three types of mouse peptidylarginine deiminase: characterization and tissue distribution. J Biochem. 1991;110:661–666. doi: 10.1093/oxfordjournals.jbchem.a123636. [DOI] [PubMed] [Google Scholar]
- Vossenaar ER, Zendman AJ, van Venrooij WJ, Pruijn GJ. PAD, a growing family of citrullinating enzymes: genes, features and involvement in disease. Bioessays. 2003;25:1106–1118. doi: 10.1002/bies.10357. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wysocka J, Sayegh J, Lee YH, Perlin JR, Leonelli L, Sonbuchner LS, McDonald CH, Cook RG, Dou Y, Roeder RG, Clarke S, Stallcup MR, Allis CD, Coonrod SA. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 2004;306:279–283. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- Wright PW, Bolling LC, Calvert ME, Sarmento OF, Berkeley EV, Shea MC, Hao Z, Jayes FC, Bush LA, Shetty J, Shore AN, Reddi PP, Tung KS, Samy E, Allietta MM, Sherman NE, Herr JC, Coonrod SA. ePAD, an oocyte and early embryo-abundant peptidylarginine deiminase-like protein that localizes to egg cytoplasmic sheets. Dev Biol. 2003;256:73–88. doi: 10.1016/s0012-1606(02)00126-4. [DOI] [PubMed] [Google Scholar]
- Zhang X, Gamble MJ, Stadler S, Cherrington BD, Causey CP, Thompson PR, Roberson MS, Kraus WL, Coonrod SA. Genome-wide analysis reveals PADI4 cooperates with Elk-1 to activate c-Fos expression in breast cancer cells. PLoS Genet. 2011;7:e1002112. doi: 10.1371/journal.pgen.1002112. [DOI] [PMC free article] [PubMed] [Google Scholar]