Abstract
Purpose
Application of whole exome and whole genome sequencing is likely to increase in clinical practice, public health contexts, and research. We examined how parental preferences for learning information from genome-scale testing is influenced by the characteristics of non-medically actionable genetic disorders in children, and assessed if preferences differed by gender and between African-American and White respondents.
Methods
We conducted a web-based discrete choice experiment with 1 289 parents of young children. Participants completed “choice tasks” using pairs of profiles describing sequencing results for hypothetical genetic disorders, selected the profile they believed would be more important to know, and answered questions that measured their level of distress.
Results
The likelihood that the disorder would develop given a true-positive test result was most important to parents. Parents showed greater interest in learning sequencing results for disease profiles with more severe manifestations. This was associated with greater distress. Differences by gender and race reflected small differences in magnitude, not direction.
Conclusion
Parents’ preferred learning results about genetic disorders with more severe manifestations, even when this knowledge was associated with increased distress. These results may help clinicians support parental decision making by revealing which types of sequencing results parents are interested in learning.
Keywords: genomic sequencing, discrete choice experiment, child’s genetic information, decision making, parental preferences, pediatrics
INTRODUCTION
Next-generation sequencing has decreased the cost and time for genome-scale testing and has increased the potential for sequencing a child’s genome. As both the predictive validity and diagnostic validity of genome-scale testing technologies have increased, discussion about their application—for example, newborn screening—has increased in pediatrics1 and public health contexts.2 These approaches produce information about a broad range of conditions potentially relevant to the child’s and family’s health. Consequently, genome-scale testing may result in a corresponding increase in the amount of secondary genetic information potentially available to parents and clinicians, requiring stakeholders to make difficult decisions about what types of information should be disclosed.
Research suggests that a majority of parents are interested in the hypothetical possibility of obtaining genomic sequencing results for their newborns3–5 or young children5 but preferences for research results decline when characterized on dimensions related to actionability, severity, age of onset or likelihood.6 What remains unclear from previous research is if and how preferences for this information vary when jointly characterized along these dimensions. Understanding how parents consider this information could help clinicians support parents or could be used to inform educational and decision support strategies when genome-scale testing is used in clinical practice or research.7
To address this gap, we elicited parental preferences for learning health-related information about a young child through genome-scale testing. Specifically, we examined parents’ interest in receiving their child’s test results related to non-medically actionable conditions. Information about the health conditions that could be revealed by the results was characterized along multiple dimensions, such as age of onset, level of disability the condition could cause, and the likelihood the condition would develop given a true-positive test result. The choice of dimensions was informed by models that describe systems for categorizing incidental findings8 and by factors relevant to child and family health, such as disability or quality of life. A guiding assumption of our approach is that the value parents place on learning genome-scale test results is influenced by multiple characteristics of the health conditions for which a child may be found to be at increased risk. We also examined if subgroup differences exist based on gender or race, as these could signal the importance of communicating differently with parental subgroups to support decision making.9
We conducted a discrete choice experiment (DCE) to quantify how parental preferences for learning a child’s genome-scale test results are affected by differences in health condition characteristics. DCE is a type of conjoint analysis that assumes that individuals value “products” (in this case, test results) based on their features. A recent review article found 179 health-related DCEs published between 2009 and 2012.10 This method is being used more frequently to study complex health decision making, and has been applied to study adult preferences to learn incidental genomic information.11,12
MATERIALS AND METHODS
The study protocol received Institutional Review Board (IRB) review and approval from the RTI International Committee for the Protection of Human Subjects. Study design and analyses were conducted following current guidance for conducting DCEs in health contexts.13–16
Study Sample
Respondents were eligible to participate in the study if they lived in the United States, and were English-speaking adults aged 18 to 40 who self-identified as either White or Black, and who had at least one child aged 5 years old or younger in the home. Qualtrics—an online data collection company based in Provo, Utah—hosted the survey and recruited the sample from their proprietary online subject panel. The recruitment procedures were not intended to draw a probability-based sample from the full population of adults meeting the eligibility criteria.
Because sample size calculations can present a challenge in choice experiments, minimum sample size depends on multiple criteria, including the question format, the complexity of the choice task, the desired precision of the results, and the need to conduct subgroup analyses.17 Most published choice experiments have a sample size between 100 and 300 respondents.18 The sample size for this survey was set at 1 200, with the goal of recruiting a sample split between women and men and Black and White respondents.
Survey Instrument
The primary goal was to elicit parental preferences for health-related information about a child that could be learned through genome-scale testing. Specifically, we examined parents’ interest in learning their child’s test results that would indicate a significant risk for non-medically actionable genetic health conditions, defined in the survey as conditions leading to mental and/or physical disabilities for which effective preventive treatments are not currently available, although there may be interventions to improve quality of life over the course of the condition or to relieve symptoms.
Based on a literature review19 and input from clinicians, genetic counselors, and geneticists at the University of North Carolina at Chapel Hill (UNC), we developed our DCE using an initial set of attributes and attribute levels. These attributes and levels were then refined using input from 10 cognitive interviews conducted with parents of children under the age of 5 years. The final survey allowed us to measure the relative importance of seven attributes: (1) penetrance, (2) typical age of onset of the health condition, (3) level of mental disability that the health condition may cause, (4) level of physical disability that the health condition may cause, (5) rate of progression (the speed with which the health condition may worsen), (6) availability of options to improve quality of life, and (7) impact on lifespan (Table 1). Each choice question comprised two generically labeled profiles—Profile A and Profile B—and participants chose the profile that would be more important for them to know (see Figure 1 for an example choice question).
Table 1.
List of attributes and attribute levels used in constructing the discrete choice experiment
| Attribute | Attribute Level |
|---|---|
| Chance that the condition will develop | 90% |
| 75% | |
| 50% | |
| 10% | |
|
| |
| Age of onset | Less than 1 year old |
| 1 to 5 years old | |
| 6 to 12 years old | |
| 13 to 18 years old | |
|
| |
| Speed that the condition may get worse | Rapid |
| Moderate | |
| Slow | |
| Stable | |
|
| |
| Options to improve quality of life | Limited options available |
| Effective options available | |
|
| |
| Level of mental disability | Severe |
| Mild to moderate | |
| None | |
|
| |
| Level of physical disability | Severe |
| Mild to moderate | |
| None | |
|
| |
| Shortened | |
| Normal | |
Figure 1.
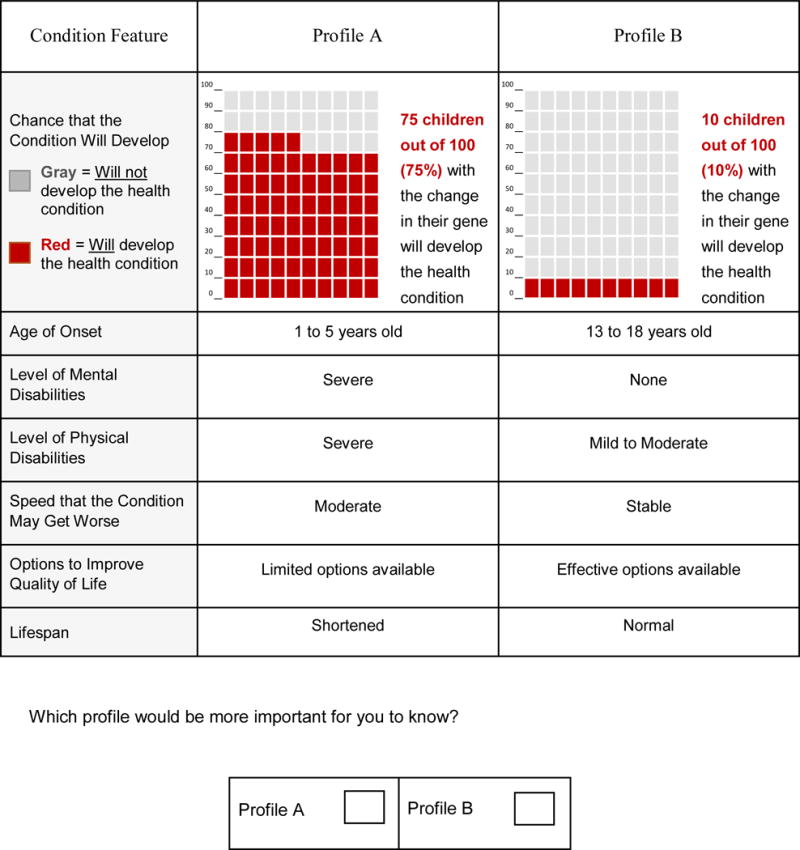
Example of a choice question from the discrete choice experiment
Because the full number of all possible pairwise combinations of the attribute levels is large, an experimental design program was used to create a smaller set of profiles and choice questions. The simplified design provides enough information to accommodate both main-effects and interaction-effects models; this approach has been shown to optimize the design when it is unclear if interaction effects exist between attributes in the model.20 In this study, a variation of the Fedorov21 algorithm was used to construct an orthogonal fractional factorial experimental design with 1 147 treatment profiles. Respondents were randomly assigned to one of 99 test blocks, each composed of eight distinct unordered profile pairs. Consequently, each respondent completed a series of eight choice questions. This design was statistically efficient in maximizing the amount of information gathered from respondents.
The final survey was designed for participants who may have limited genetic knowledge. Respondents were first asked to read background information defining several basic genetic concepts used throughout the survey. The survey then described the attributes and levels that were included in the DCE. Using Likert-type scales ranging from 1 (least distressing) to 7 (most distressing), respondents were asked to rate how distressing it would be for them to learn information for each level of the seven attributes. For the attribute describing the chance that the condition will develop, the chance (or risk) was presented both numerically and graphically. The probability levels were presented graphically using a 100 square grid, in which each square represented one person with a gene identifying a genetic disorder. Squares presented in color represented people for whom the condition will develop. Research suggests that pictograms such as risk grids are an effective method of communicating risk.21
Respondents reviewed an example risk grid and completed a comprehension question. Respondents who answered incorrectly were presented the correct answer and an explanation before proceeding with the survey, whereas respondents who answered correctly were presented with the correct answer to reinforce their understanding.
The attribute levels were presented in a scale array with one level per row (Figure 1). The study started with two practice trials involving two and four attributes, respectively. The survey also collected basic demographic information and respondents’ personal experience with genetic disorders and testing, their mistrust in the healthcare system,22 objective numeracy,23 and intolerance of uncertainty.24 We included the last three of these measures as potential moderators in exploratory analyses, but did not retain them in the final models because we found no evidence that they influenced preferences. The median time respondents took to complete the survey was 15 minutes.
Statistical Analysis
We analyzed the data using a conditional logistic regression model,25 using Stata 13.0.26 We ran a main-effect random parameters logit model27 for comparison, and the results in terms of significance and ranking of attribute levels were identical to the conditional logit. The dependent variable was coded “1” if respondents selected a test-result profile and “0” if they did not for each of the test-result profiles in the eight choice questions. The independent variables corresponded to the attribute levels for the test-result profile. The attribute levels were coded as categorical variables using effect coding. With effects coding, zero indicates the mean effect across all attribute levels, rather than the omitted level, as in dummy coding.28 This procedure produces a parameter estimate for all level categories, where the omitted-category parameter is the negative sum of the included-category parameters. Conditional logit regression uses choice information to estimate the relative contribution of different attribute levels on participant preferences. In a choice set comprising two alternative test-result profiles, respondents are assumed to choose the option they view as providing greater utility. In turn, the choices respondents make are a function of differences in the attributes of alternative test-result profiles. When aggregated over many choices, we are able to measure respondents’ relative preferences for the various attribute levels.
The regression coefficients from a conditional logit model are statistical estimates of the average relative contribution across the sample of each attribute level to the likelihood of choosing one test-result profile over another, with larger coefficients indicating greater relative preference for that level. We also calculated the relative importance of each attribute over the range of levels by subtracting the coefficient associated with the least preferred level of each attribute from the most preferred level, dividing by the sum of ranges over all attributes and multiplying by 100. Changes in attributes displaying a wider range from the most to least preferred level are more important in predicting parents’ choices and have a greater impact on determining the kinds of test results parents think would be important to know. We estimated a main-effects model of preferences for the attribute levels of the genome-scale test results for the full sample. Interaction terms were included to test for subgroup interactions testing attribute preferences by the gender and race of participants. In these models, gender and race variables were dummy coded so that women and White participants served as the reference category.
We conducted a descriptive analysis of the distress items using SPSS 21.0.29 We used one way repeated measures ANOVAs with a Greenhouse-Geisser correction to determine if the mean level of distress varied across levels within an attribute. We also conducted post-hoc tests with the Bonferroni correction for pairwise comparisons of levels. We conducted paired t tests for the two attributes that had only two levels.
RESULTS
The survey was conducted online from August 24, 2014, through September 8, 2014. Qualtrics screened a total of 3032 respondents, 2 021 of whom met the study eligibility criteria. Of these, 671 did not complete the survey or were excluded after screening because their relevant subgroup quota had already been met. To ensure that only respondents who read and answered survey materials in the expected time allotted were included in the analyses, we removed an additional 55 respondents who completed the survey in less than 6 minutes. Finally, six respondents were excluded from the analyses because they completed fewer than eight discrete choice tasks.
The final sample included 1 289 adult parents with young children living in the United States, for a minimum cooperation rate of 62.0%. By design, approximately half the respondents were women (n=643, 49.9%) and half were Black (n=638, 49.5%). The majority of respondents were married (n=762, 59.1%), non-Hispanic (n=1 132, 87.8%), and had completed at least some college (n=984, 76.4%). The median age was 30 years (M=29.8, standard deviation [SD]=5.6). About half the respondents (n=648, 50.3%) had two children under the age of 18 years. Among the sample, 61% (n=783) had never personally had a genetic test, nor did they know of a family member or someone close who had a genetic test. The demographic characteristics of the study sample are shown in Table 2. When compared to 2010 Census population estimates filtered by age and stratified by race, respondents in our sample had attained a higher level of education on average, were more likely to be married, were more likely to be enrolled in Medicare or Medicaid, and less likely to have privately-purchased health insurance. White respondents were also more likely than the White population at large to have employer provided insurance and had lower average household income. Black respondents in our sample had a higher average household income than the Black population at large.
Table 2.
Demographic characteristics of the study sample
| Variable | N | % | ||
|---|---|---|---|---|
| Gender | ||||
| Male | 646 | 50.1 | ||
| Female | 643 | 49.9 | ||
|
| ||||
| Race | ||||
| White | 651 | 50.5 | ||
| Black | 638 | 49.5 | ||
|
| ||||
| Ethnicity | ||||
| Not Hispanic or Latino | 1,132 | 87.8 | ||
| Hispanic or Latino | 136 | 10.6 | ||
| Refused | 21 | 1.6 | ||
|
| ||||
| Marital Status | ||||
| Married | 762 | 59.1 | ||
| Widowed | 3 | 0.2 | ||
| Divorced or Separated | 46 | 3.6 | ||
| Single | 444 | 34.5 | ||
| Other | 34 | 2.6 | ||
|
| ||||
| Education | ||||
| High school or less | 305 | 23.7 | ||
| Some college but no degree | 269 | 20.9 | ||
| 2-year college degree or technical school | 229 | 17.8 | ||
| 4-year college degree | 370 | 28.7 | ||
| Graduate or professional degree | 109 | 8.5 | ||
| Refused | 7 | 0.5 | ||
|
| ||||
| Household income | ||||
| Less than $20,000 | 291 | 22.6 | ||
| $20,000–$49,999 | 274 | 21.3 | ||
| $50,000–$69,999 | 236 | 18.3 | ||
| $70,000–$99,999 | 257 | 19.9 | ||
| $100,000 or more | 231 | 17.9 | ||
|
| ||||
| Age | ||||
| M | 29.8 | |||
| SD | 5.6 | |||
| Mdn | 30 | |||
| Min | 18 | |||
| Max | 40 | |||
Note. N=1 289. Percentages may not sum to 100 due to rounding error.
Discrete Choice Experiment Analysis
The main-effects model is presented in Table 3. The model fit statistics provide evidence that health-condition characteristics, as defined by the experiment’s attribute levels, influence parental choices about the kinds of test results that are important to know, χ2(15)=1407.80, P<.001, Pseudo R2=.10. The relative importance percentages indicate how much difference each attribute as a whole across the range of levels contributed to parental decisions about learning their child’s results. Because respondent characteristics are not taken into account, estimates can be interpreted as the average contribution of attribute levels on relative preferences for the sample as a whole. When deciding what kinds of genetic test results are most important to know, parents showed preference for conditions with a high degree of penetrance (38.5%), those that begin earlier in life (12.9%), those that progress rapidly (9.8%), those that have severe mental (11.8%) and/or physical symptoms (12.4%), and those that are characterized by a shortened lifespan (13.6%). However, knowing whether there were limited options versus effective options to improve quality of life did not have a significant effect on the choice of test results (B=0.02; standard error (SE)=0.01; 95% confidence interval (CI): −0.01–0.04], z=1.07, P=.284). Except for options to increase quality of life, all attributes significantly contributed to respondents’ choices.
Table 3.
Conditional logistic regression analysis results for each attribute and level predicting parent interest in learning child’s sequencing information: Main-effects model (N=1 289)
| Attribute | Level | Coef. | SE | 95% CI | p | Relative Importance (%) |
|---|---|---|---|---|---|---|
| Likelihood | 90% | 0.52 | 0.03 | 0.47–0.57 | < 0.001 | 38.5 |
| 75% | 0.21 | 0.02 | 0.16–0.26 | < 0.001 | ||
| 50% | −0.08 | 0.02 | -0.13–-0.03 | 0.001 | ||
| 10% | −0.65 | 0.03 | −0.71–−0.60 | < 0.001 | ||
|
| ||||||
| Age of onset | Less than 1 year | 0.15 | 0.03 | 0.10–0.21 | < 0.001 | 12.9 |
| 1–5 years | 0.14 | 0.03 | 0.09–0.19 | < 0.001 | ||
| 6–12 years | −0.06 | 0.02 | −0.11–0.01 | 0.018 | ||
| 13–18 years | −0.24 | 0.03 | −0.29–−0.19 | < 0.001 | ||
|
| ||||||
| Rate of progression | Rapid | 0.18 | 0.02 | 0.13–0.22 | < 0.001 | 9.8 |
| Moderate | 0.04 | 0.03 | −0.01–0.09 | 0.081 | ||
| Slow | −0.12 | 0.03 | −0.17–−0.07 | < 0.001 | ||
| Stable | −0.10 | 0.03 | −0.15–−0.05 | < 0.001 | ||
|
| ||||||
| Quality of life | Limited options | 0.02 | 0.01 | −0.01–0.04 | 0.284 | 1.0 |
| Effective options | −0.02 | 0.01 | −0.04–0.01 | 0.283 | ||
|
| ||||||
| Level of mental disability | Severe | 0.20 | 0.02 | 0.16–0.25 | < 0.001 | 11.8 |
| Mild to moderate | −0.03 | 0.02 | −0.07–0.01 | 0.100 | ||
| None | −0.17 | 0.02 | −0.21–−0.13 | < 0.001 | ||
|
| ||||||
| Level of physical disability | Severe | 0.19 | 0.02 | 0.15–0.23 | < 0.001 | 12.4 |
| Mild to moderate | −0.01 | 0.02 | −0.05–0.03 | 0.529 | ||
| None | −0.17 | 0.02 | −0.21–−0.13 | < 0.001 | ||
|
| ||||||
| Lifespan | Shortened | 0.21 | 0.01 | 0.18–0.24 | < 0.001 | 13.6 |
| Normal | −0.21 | 0.01 | −0.24–−0.18 | < 0.001 | ||
Note. Number of observations=20 624; χ2(15)=1407.80; P<.001; Pseudo R2=.10.
Differences in relative importance by each attribute’s levels are shown in Figure 2. All attribute level preference weights were significantly different from each other, with a few exceptions. For instance, on average, age of onset from less than 1 year (B=0.15; SE=0.03; 95% CI: 0.10–0.21) to 1 to 5 years (B=0.14; SE=0.03, 95% CI: 0.09–0.19) did not affect which profile was chosen. Also, preference rates among three of four rate-of-progression levels were not statistically distinct. Profiles with a stable rate of progression (B=−0.10; SE=0.03; 95% CI: −0.15–−0.05) were just as likely to be selected as important to know as those with a slow rate of progression (B=−0.12; SE=0.03; 95% CI: −0.17–−0.21]), which in turn were not significantly different from profiles with a moderate rate of progression (B=0.04; SE=0.03; 95% CI: −0.01– 0.09).
Figure 2. Plot showing preference weights (line graphs) and mean distress ratings (bar charts) by attribute level.
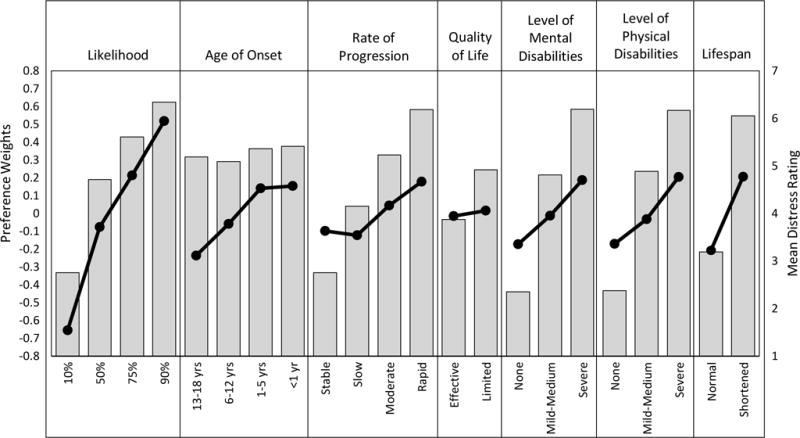
Mean distress ratings within an attribute are all significantly different at P<0.001, except for the difference between conditions with an age of onset at 6 to12 years versus 13 to18 years, t(1249)=-3.08, P=0.013, and a nonsignificant pairwise comparison between conditions with an age of onset less than 1 year versus 1 to 5 years, t(1249)=1.56, P=0.707.
Adding the parent gender interaction terms to the main-effects model significantly improved model fit, Δχ2 (15)=37.30, P=.001. Although the interaction analysis revealed differences between mothers’ and fathers’ preferences for select attribute levels, these were all differences of magnitude and not of direction. Consistent with the main-effects model for the full sample, mothers and fathers both showed greater interest in knowing information about health conditions with more severe or pressing implications. Fathers were less interested than mothers in conditions that had only a 10% chance of happening given a true-positive genetic test result (B=−0.12; SE=0.05; 95% CI: −0.22–−0.02; z=−2.25; P=.025), were likely to decrease a child’s lifespan (B=−0.09; SE=0.03; 95% CI: −0.14–−0.04; z=−3.14, P=.002), or were characterized by lasting and severe physical consequences (B=−0.11; SE=0.04; 95% CI: −0.18–−0.04; z=−2.59, P=.010). Fathers also showed greater relative preference for genetic test results pertaining to conditions that were not associated with any physical disability (B=0.09; SE=0.04; 95% CI: 0.01–0.17; z=2.15, P=.031). No other statistical differences by gender were observed.
We also tested for differences in attribute-level relative preferences between White and Black respondents; adding these interaction terms to the main-effects model significantly improved model fit, Δχ2(15)=38.57, P=.001. As with the observed moderation effects by gender, differences by race were reflected in the relative weights of preferences for specific attribute levels but did not alter the direction of these effects. The interaction estimates representing differences in coefficients by race showed that Black respondents were more likely than White respondents to show interest in learning about conditions that had a 10% chance of developing given a true-positive genetic test result (B=0.14; SE=0.05; 95% CI: 0.03–0.24, z=2.26; P=.010), were unlikely to impact a child’s lifespan (B=0.09; SE=0.03; 95% CI: 0.03–0.15; z=3.00, P=.003), or did not pose a threat of mental disability (B=0.15; SE=0.04; 95% CI: 0.06–0.23; z=3.44; P=.001). Black respondents also showed less relative preference than White respondents for test results about conditions associated with severe mental disability (B=−0.12; SE=0.04; 95% CI: 0.01–0.17, z=−2.72, P=.007). Overall, however, Black respondents placed greater value on knowing test results for conditions characterized by severe mental disability than on results that had no threat of mental disability. The interaction analysis did not reveal any other statistically significant differences by race.
Distress Questions
Respondents rated how distressing they found each level of the seven attributes. The mean distress ratings are grouped by attribute and presented in Figure 2. Notably, the pattern of distress ratings mirrors respondent preferences found in our conditional logistic regression analysis. Within any given attribute, respondents rated attribute levels corresponding to more severe manifestations as more distressing. There were statistically significant differences in mean distress ratings across levels of all seven attributes, with F tests ranging from F(1.48, 1852.66)=18.72, P<0.001 for age of onset to F(1.38, 1731.99)=2136.80, P<0.001 for level of mental disability. All but one pairwise difference between levels within attributes were statistically significant (Ps<.001). Specifically, parents did not find the prospect of a condition with an age of onset under 1 year any more or less distressing than an age of onset of 1 to 5 years, t(1249)=1.57, P=.707.
DISCUSSION
Parental interest in genomic sequencing information is high,5 at least when hypothetical scenarios are proposed. This study revealed that for childhood-onset non-medically actionable genetic disorders leading to mental and/or physical disabilities for which effective preventive treatments are not currently available, parents are relatively more interested in knowing about highly penetrant conditions. Further, the levels of each dimension of information parents most wanted to know were the most severe manifestations, such as greater penetrance, earlier age of onset, greater level of mental and physical disability, rapid rate of progression, and greater impact on lifespan. These were also rated as more distressing. This may be because the information, although perceived as valuable, was also recognized as being likely to cause distress. Only options to enhance quality of life were not significant and did not match the overall pattern of results.
Based on previous research that shows Blacks have greater distrust of the health system.30 we might have expected differences in parental interest in certain attributes associated with non-medically actionable genetic conditions. In general, we did not find meaningful differences between Black and White parents on most of the attributes studied. We did find some statistically significant differences along some attributes, such as desire to know information that was unlikely to impact a child’s lifespan, but the overall direction and magnitude of the findings across groups were quite similar. Past studies demonstrating mistrust focused on the personal receipt of healthcare31 or on being involved in health research.32 It is possible that in the context of receiving information about one’s child, these differences are diminished. Another possibility is that differences in mistrust in the health care system by race among participants in this study were less pronounced than those observed in prior research. Although we measured mistrust in the health care system for this study, disentangling the complex associations among mistrust, race and preferences is beyond the scope of this paper. Future studies should continue to examine racial and ethnic differences in preferences and responses to disclosure of genomic sequencing information, given that recent research shows that Blacks are less likely to view genetic testing and research as favorably as Whites.33
We found relatively few meaningful differences between mothers and fathers. Once again, statistically significant differences were revealed, but these differences were quite small. Previous research studying parental preferences for their child’s genomic sequencing has not typically reported differences by gender.34 Given limited research to date examining parental preferences in this context, future research should nonetheless continue to examine group differences by gender to extend these findings. Continued research is warranted because parental preferences may vary. Some of these preferences could require additional counseling, support or interventions to achieve informed consent. This examination could also inform decision support strategies if future differences emerge, and it could be used to develop support resources for family adaptation as technological advances in screening become more pervasive.35
Consistent results showing parental preferences to learn about conditions with more severe manifestations, which are also more distressing, may reflect the perceived value of knowing such information when making hypothetical decisions. However, it is important to recognize that in actual experience, learning more severe or distressing information could disrupt family functioning. Previous research shows that parents value the personal utility of genomic information in healthcare36 and research contexts.37 However, the long-term individual, familial, or societal effects of learning genetic information about non-medically actionable conditions are not known.
The strengths of our study include an experimental design, use of a large sample balanced across gender and two racial categories, and the examination of both the full sample results and subgroup differences for a variety of dimensions that could characterize genetic disorders in childhood that are not medically actionable. Our findings can facilitate an evidence-based approach to informed consent, interpretation and analysis, and disclosure of results, because we were able to obtain parental input using a structured framework that could guide decisions about which genomic variants should be interpreted and reported back to clinicians and families based on parental preferences. The results suggest that parents find utility in learning information that is not medically actionable and that could be distressing.
Despite these strengths, there are some limitations. Our approach focused on examining parental preferences for this one category of childhood-onset genetic disorders. Future research should examine other categories of information, such as those that are medically actionable in childhood or adulthood or non-medically actionable in adulthood, to understand the full spectrum of information that could be made available through genome-scale testing. The potential bias of using an online convenience sample cannot be quantified, although research using online panels has shown that this approach has comparable validity and reliability to more traditional survey methods.38,39 Additionally, the hypothetical nature of the choice experiment may not accurately reflect decisions made by parents when presented with such decisions in real-life situations. We did not include a “no test” option for parents who were not in the market for test results, and it is possible that these parents might have had more trouble answering the choice questions, contributing to higher variance in the responses.
Despite these potential limitations, this study contributes to the growing body of research illuminating parental preferences for genomic sequencing information for their children and the return of incidental or optional findings whether in a clinical or public health context. As genome-scale testing becomes more widely available, understanding what type of information should be disclosed to parents and why parents value certain types of information will help both clinicians and parents during clinical and research encounters.
Acknowledgments
Funding for this research was provided by Eunice Kennedy Shriver National Institute of Child Health and Human Development/National Human Genome Research Institute (NICHD/NHGRI) grant 1 U19 HD077632-01 to Drs. Cynthia Powell and Jonathan Berg at the University of North Carolina at Chapel Hill. The findings and conclusions are those of the authors and do not necessarily reflect the views of NICHD/NHGRI. The funder had no role in the design of the study, collection and analysis of the data, and the decision to publish the findings.
Footnotes
Disclosure/Conflict of Interest Notification
Megan Lewis – Declares no conflict of interest.
Alex Stine – Declares no conflict of interest.
Ryan Paquin – Declares no conflict of interest.
Carol Mansfield – Declares no conflict of interest
Dallas Wood – Declares no conflict of interest
Christine Rini – Declares no conflict of interest.
Myra Roche – Declares no conflict of interest.
Cynthia Powell – Declares no conflict of interest.
Jonathan Berg – Declares no conflict of interest.
Donald Bailey currently receives funding from The John Merck Fund and Ovid Therapeutics.
Authorship
Megan A. Lewis – Contributed to the conception, design, interpretation, and drafted the article.
Alex Stine – Contributed to the design, data acquisition, analysis and drafts of the article.
Ryan Paquin – Contributed to the analysis, interpretation and drafts of the article.
Carol Mansfield – Contributed to the design, analysis, interpretation and drafts of the article.
Dallas Wood – Contributed to the analysis, interpretation, and drafts of the article.
Christine Rini – Contributed to the design, interpretation and revisions of the article.
Myra Roche – Contributed to the design, interpretation and revisions of the article.
Cynthia Powell – Contributed to the design, interpretation and revisions of the article.
Jonathan Berg – Contributed to the design, interpretation and revisions of the article.
Donald Bailey – Contributed to the design, interpretation and revisions of the article.
References
- 1.Thiffault I, Lantos J. The Challenge of Analyzing the Results of Next-Generation Sequencing in Children. Pediatrics. 2016;137(Suppl 1):S3–7. doi: 10.1542/peds.2015-3731C. [DOI] [PubMed] [Google Scholar]
- 2.Howard HC, Knoppers BM, Cornel MC, Wright Clayton E, Senecal K, Borry P. Whole-genome sequencing in newborn screening? A statement on the continued importance of targeted approaches in newborn screening programmes. Eur J Hum Genet. 2015;23(12):1593–1600. doi: 10.1038/ejhg.2014.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waisbren SE, Back DK, Liu C, et al. Parents are interested in newborn genomic testing during the early postpartum period. Genet Med. 2015;17(6):501–504. doi: 10.1038/gim.2014.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waisbren SE, Weipert CM, Walsh RC, Petty CR, Green RC. Psychosocial factors influencing parental interest in genomic sequencing of newborns. Pediatrics. 2016;137(Suppl 1):S30–35. doi: 10.1542/peds.2015-3731G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dodson DS, Goldenberg AJ, Davis MM, Singer DC, Tarini BA. Parent and public interest in whole-genome sequencing. Public Health Genomics. 2015;18(3):151–159. doi: 10.1159/000375115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ziniel SI, Savage SK, Huntington N, et al. Parents’ preferences for return of results in pediatric genomic research. Public Health Genomics. 2014;17(2):105–114. doi: 10.1159/000358539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis MA, Paquin RS, Roche MI, et al. Supporting parental decisions about genomic sequencing for newborn screening: the NC NEXUS Decision Aid. Pediatrics. 2016;137(Suppl 1):S16–S23. doi: 10.1542/peds.2015-3731E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berg JS, Foreman AK, O’Daniel JM, et al. A semiquantitative metric for evaluating clinical actionability of incidental or secondary findings from genome-scale sequencing. Genet Med. 2015 doi: 10.1038/gim.2015.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kreuter MW, Wray RJ. Tailored and targeted health communication: strategies for enhancing information relevance. Am J Health Behav. 2003;27(Suppl 3):S227–232. doi: 10.5993/ajhb.27.1.s3.6. [DOI] [PubMed] [Google Scholar]
- 10.Clark MD, Determann D, Petrou S, Moro D, de Bekker-Grob EW. Discrete choice experiments in health economics: a review of the literature. Pharmacoeconomics. 2014;32(9):883–902. doi: 10.1007/s40273-014-0170-x. [DOI] [PubMed] [Google Scholar]
- 11.Regier DA, Peacock SJ, Pataky R, et al. Societal preferences for the return of incidental findings from clinical genomic sequencing: a discrete-choice experiment. CMAJ. 2015;187(6):E190–197. doi: 10.1503/cmaj.140697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bennette CS, Trinidad SB, Fullerton SM, et al. Return of incidental findings in genomic medicine: measuring what patients value [mdash] development of an instrument to measure preferences for information from next-generation testing (IMPRINT) Genet Med. 2013;15(11):873–881. doi: 10.1038/gim.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bridges JF, Hauber AB, Marshall D, et al. Conjoint analysis applications in health–a checklist: a report of the ISPOR Good Research Practices for Conjoint Analysis Task Force. Value Health. 2011;14(4):403–413. doi: 10.1016/j.jval.2010.11.013. [DOI] [PubMed] [Google Scholar]
- 14.Carson RT, Louviere JJ, Anderson DA, et al. Experimental analysis of choice. Marketing Letters. 1994;5:351–368. [Google Scholar]
- 15.Reed Johnson F, Lancsar E, Marshall D, et al. Constructing experimental designs for discrete-choice experiments: report of the ISPOR Conjoint Analysis Experimental Design Good Research Practices Task Force. Value Health. 2013;16(1):3–13. doi: 10.1016/j.jval.2012.08.2223. [DOI] [PubMed] [Google Scholar]
- 16.Ryan M, Watson V, Gerard K. Practical issues in conducting a discrete choice experiment. In: Ryan M, Gerard K, Amaya-Amaya M, editors. Using discrete choice experiments to value health and health care. Dordrecht, The Netherlands: Springer; 2008. pp. 73–97. [Google Scholar]
- 17.Louviere JJ, Hensher DA, Swait JD. Stated choice methods: analysis and applications. New York: Cambridge University Press; 2000. [Google Scholar]
- 18.Marshall D, Bridges JF, Hauber B, et al. Conjoint Analysis Applications in Health - How are Studies being Designed and Reported?: An Update on Current Practice in the Published Literature between 2005 and 2008. Patient. 2010;3(4):249–256. doi: 10.2165/11539650-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 19.Whitehead NS, Brown DS, Layton CM. Developing a conjoint analysis survey of parental attitudes regarding voluntary newborn screening. MR-0014-1003. Research Triangle Park, NC: RTI Press; 2010. [Google Scholar]
- 20.Yu J, Goos P, Vandebroek M. Model-robust design of conjoint choice experiments. Communications in Statistics-Simulation and Computation. 2008;37(8):1603–1621. doi: 10.1080/03610910802244638. [DOI] [Google Scholar]
- 21.Fagerlin A, Zikmund-Fisher BJ, Ubel PA. Helping patients decide: ten steps to better risk communication. J Natl Cancer Inst. 2011;103(19):1436–1443. doi: 10.1093/jnci/djr318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shea JA, Micco E, Dean LT, McMurphy S, Schwartz JS, Armstrong K. Development of a revised Health Care System Distrust scale. J Gen Intern Med. 2008;23(6):727–732. doi: 10.1007/s11606-008-0575-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weller JA, Dieckmann NF, Tusler M, Mertz CK, Burns WJ, Peters E. Development and testing of an abbreviated numeracy scale: A Rasch analysis approach. Journal of Behavioral Decision Making. 2013;26(2):198–212. doi: 10.1002/bdm.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacoby RJ, Fabricant LE, Leonard RC, Riemann BC, Abramowitz JS. Just to be certain: Confirming the factor structure of the intolerance of uncertainty scale in patients with obsessive-compulsive disorder. J Anxiety Disord. 2013;27(5):535–542. doi: 10.1016/j.janxdis.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 25.McFadden D. Conditional logit analysis of qualitative choice behavior. In: Zarembka P, editor. Frontiers in Econometrics. New York, NY: Academic Press; 1974. pp. 105–142. [Google Scholar]
- 26.StataCorp Release 13.0 [computer program] College Station, TX: StataCorp LP; 2013. [Google Scholar]
- 27.Train K. Discrete choice methods with simulation. Cambridge, UK: Cambridge University Press; 2009. [Google Scholar]
- 28.Hensher D, Rose J, Greene WH. Applied choice analysis: A primer. Cambridge, UK: Cambridge University Press; 2005. [Google Scholar]
- 29.IBM SPSS Statistics for Windows, Version 21.0 [computer program] Armonk, NY: IBM Corp; 2012. [Google Scholar]
- 30.LaVeist TA, Nickerson KJ, Bowie JV. Attitudes about racism, medical mistrust, and satisfaction with care among African American and white cardiac patients. Med Care Res Rev. 2000;57(Suppl 1):146–161. doi: 10.1177/1077558700057001S07. [DOI] [PubMed] [Google Scholar]
- 31.Lillie-Blanton M, Brodie M, Rowland D, Altman D, McIntosh M. Race, ethnicity, and the health care system: public perceptions and experiences. Med Care Res Rev. 2000;57(Suppl 1):218–235. doi: 10.1177/1077558700057001S10. [DOI] [PubMed] [Google Scholar]
- 32.Corbie-Smith G, Thomas SB, St George DM. Distrust, race, and research. Arch Intern Med. 2002;162(21):2458–2463. doi: 10.1001/archinte.162.21.2458. [DOI] [PubMed] [Google Scholar]
- 33.Dye T, Li D, Demment M, et al. Sociocultural Variation in Attitudes Toward use of Genetic Information and Participation in Genetic Research by Race in the United States: Implications for Precision Medicine. J Am Med Inform Assoc. 2016 doi: 10.1093/jamia/ocv214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goldenberg AJ, Dodson DS, Davis MM, Tarini BA. Parents’ interest in whole-genome sequencing of newborns. Genet Med. 2014;16(1):78–84. doi: 10.1038/gim.2013.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bailey DB, Jr, Armstrong FD, Kemper AR, Skinner D, Warren SF. Supporting family adaptation to presymptomatic and “untreatable” conditions in an era of expanded newborn screening. J Pediatr Psychol. 2009;34(6):648–661. doi: 10.1093/jpepsy/jsn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Townsend A, Adam S, Birch PH, Lohn Z, Rousseau F, Friedman JM. “I want to know what’s in Pandora’s Box”: comparing stakeholder perspectives on incidental findings in clinical whole genomic sequencing. Am J Med Genet A. 2012;158a(10):2519–2525. doi: 10.1002/ajmg.a.35554. [DOI] [PubMed] [Google Scholar]
- 37.Harris ED, Ziniel SI, Amatruda JG, et al. The beliefs, motivations, and expectations of parents who have enrolled their children in a genetic biorepository. Genet Med. 2012;14(3):330–337. doi: 10.1038/gim.2011.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Evans JR, Mathur A. The value of online surveys. Internet Research-Electronic Networking Applications and Policy. 2005;15(2):195–219. doi: 10.1108/10662204510590360. [DOI] [Google Scholar]
- 39.Eysenbach G, Wyatt J. Using the Internet for Surveys and Health Research. J Med Internet Res. 2002;4(2):e13. doi: 10.2196/jmir.4.2.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]


