ABSTRACT
During ontogeny, hematopoietic stem and progenitor cells arise from hemogenic endothelium through an endothelial-to-hematopoietic transition that is strictly dependent on the transcription factor RUNX1. Although it is well established that RUNX1 is essential for the onset of hematopoiesis, little is known about the role of RUNX1 dosage specifically in hemogenic endothelium and during the endothelial-to-hematopoietic transition. Here, we used the mouse embryonic stem cell differentiation system to determine if and how RUNX1 dosage affects hemogenic endothelium differentiation. The use of inducible Runx1 expression combined with alterations in the expression of the RUNX1 co-factor CBFβ allowed us to evaluate a wide range of RUNX1 levels. We demonstrate that low RUNX1 levels are sufficient and necessary to initiate an effective endothelial-to-hematopoietic transition. Subsequently, RUNX1 is also required to complete the endothelial-to-hematopoietic transition and to generate functional hematopoietic precursors. In contrast, elevated levels of RUNX1 are able to drive an accelerated endothelial-to-hematopoietic transition, but the resulting cells are unable to generate mature hematopoietic cells. Together, our results suggest that RUNX1 dosage plays a pivotal role in hemogenic endothelium maturation and the establishment of the hematopoietic system.
KEY WORDS: RUNX1, Hemogenic endothelium, Dosage, EHT, CBFβ, SOX7
Summary: At the onset of hematopoiesis, regulation of RUNX1 dosage is crucial for the successful maturation of hemogenic endothelium and for both the initiation and completion of the endothelial-to-hematopoietic transition.
INTRODUCTION
During development, the first definitive blood cells arise from a specialized endothelium (hemogenic endothelium or HE), via a process termed endothelial-to-hematopoietic transition (EHT). EHT has been elegantly visualized both in vivo and in vitro using multiple vertebrate model systems (Bertrand et al., 2010; Boisset et al., 2010; Eilken et al., 2009; Jaffredo et al., 1998; Kissa and Herbomel, 2010; Lam et al., 2010; Lancrin et al., 2009). The transcription factor RUNX1 is crucial for EHT and the emergence of definitive blood cells from HE (Chen et al., 2009; Kissa and Herbomel, 2010; Lacaud et al., 2002; Lancrin et al., 2009; North et al., 1999). Within the context of the definitive adult blood system, alterations in RUNX1 dosage or activity have been associated with several blood-related disorders with both reduction (thrombocytopenia, myelodysplastic syndrome) and gain (Down syndrome hematopoietic disorders) of functional Runx1 alleles leading to abnormalities (Banno et al., 2016; De Vita et al., 2010; Rio-Machin et al., 2012; Song et al., 1999). RUNX1 dosage also plays a crucial role in the maintenance of leukemias harboring core-binding factor-related translocations (Ben-Ami et al., 2013; Goyama et al., 2013; Ptasinska et al., 2014; Yanagida et al., 2005). RUNX1 dosage has also been extensively studied in ontogeny, with several studies clearly establishing that Runx1 haploinsufficiency or mutations result in a decrease in generation of hematopoietic stem and/or progenitor cells both in vitro and in vivo (Cai et al., 2000; Lacaud et al., 2002, 2004; Matheny et al., 2007; Wang et al., 1996a). However, little is known about the precise role of RUNX1 dosage in HE and during EHT at the onset of hematopoiesis.
Runx1 transcription is controlled by two alternative Runx1 promoters that generate transcripts coding for the two main RUNX1 isoforms (Miyoshi et al., 1995). The P1, or distal, promoter controls the expression of the distal RUNX1 isoform RUNX1C, and the P2, or proximal, promoter controls the proximal isoform RUNX1B. On a protein level the two isoforms are mostly identical and only differ in their N-terminal region (Fujita et al., 2001; Miyoshi et al., 1995). The dual promoter structure and the difference in N-terminal amino acid sequence are conserved across all RUNX genes and also across different mammalian species (Levanon and Groner, 2004). Although clear biochemical differences between the two isoforms remain relatively poorly defined (Bonifer et al., 2017; Nieke et al., 2017), specific expression patterns for each isoform in adult hematopoiesis and different requirements in megakaryocytic and lymphoid lineage commitment have been demonstrated (Brady et al., 2013; Challen and Goodell, 2010; Draper et al., 2017, 2016; Telfer and Rothenberg, 2001).
Runx1 P2 promoter activity starts early during hematopoietic development and is detected in HE, in which it is the sole active Runx1 promoter in mice (Bee et al., 2009; Sroczynska et al., 2009a) indicating that the RUNX1B isoform is responsible for the initiation of EHT. Experiments in mice have demonstrated that lowering the levels of RUNX1B by creating Runx1 heterozygote knockouts or by attenuating P2 proximal promoter activity does not dramatically affect the onset of hematopoiesis as all these animals develop to term (Bee et al., 2010; North et al., 1999; Pozner et al., 2007; Wang et al., 1996a). However, there are some indications that the RUNX1 levels change as the cells differentiate from hemangioblasts (HBs) via HE to the first CD41 (ITGA2B)+ hematopoietic progenitors (HPs). One line of evidence was provided by Swiers et al. who analyzed single cells derived from +23Runx1 enhancer-reporter transgenic mice (23GFP) (Swiers et al., 2013). In this study, Runx1 mRNA expression was found to be lower in embryo-derived 23GFP+ HE cells compared with CD41+ HPs (Swiers et al., 2013). In contrast to P2, the Runx1 P1 promoter is activated later in development during EHT in committed CD41+ HPs (Bee et al., 2009; Sroczynska et al., 2009a). In the adult hematopoietic system, P1 is the dominant active promoter (Bee et al., 2009; Draper et al., 2016).
Several transcription factors have been shown to regulate RUNX1 protein activity. CBFβ is a crucial RUNX1 co-factor that heterodimerizes with RUNX1, enhances its DNA-binding affinity and protects it from degradation (Huang et al., 2001; Nagata et al., 1999; Tang et al., 2000). Cbfb knockout is embryonic lethal and shows an almost identical phenotype to that of Runx1 knockout embryos (Niki et al., 1997; Okuda et al., 1996; Sasaki et al., 1996; Wang et al., 1996a,b). RUNX1 protein is barely detectable in these Cbfb−/− embryos, suggesting that this phenotype is mainly caused by a reduction in RUNX1 levels (Huang et al., 2001; Yokomizo et al., 2008). More recently, SOXF transcription factor family members SOX7 and SOX17 have emerged as novel important regulators of RUNX1 activity. These transcription factors have established roles in vasculogenesis and angiogenesis, suggesting that they are potentially able to interact with RUNX1 in the context of HE development (Lilly et al., 2016b). Indeed, both SOX7 and SOX17 are co-expressed with RUNX1 during a narrow temporal window of hematopoietic development that encompasses HE (Clarke et al., 2013; Costa et al., 2012; Gandillet et al., 2009; Lizama et al., 2015). Furthermore, both factors have been proposed to be negative regulators of RUNX1 and overexpression of either transcription factor can block hematopoietic development (Clarke et al., 2013; Costa et al., 2012; Gandillet et al., 2009; Lilly et al., 2016a; Lizama et al., 2015).
Here, we sought to define the role and importance of RUNX1B dosage in HE and EHT. To circumvent the limitations of in vivo gene dosage experiments, we took advantage of the in vitro mouse embryonic stem cell (mESC) differentiation system. Mouse ESCs have been shown to recapitulate key events of early embryonic yolk sac hematopoiesis, including HE formation and differentiation (Choi et al., 1998; Eilken et al., 2009; Frame et al., 2016; Lancrin et al., 2010; Sroczynska et al., 2009b). To evaluate the effects of different RUNX1B expression levels on HE and EHT, we utilized an mESC line in which Runx1b transcription is under control of a doxycycline inducible tet-on system (Lancrin et al., 2009). Analyses of this line in the presence of either a wild-type or a disrupted Cbfb locus allowed us to evaluate the effects of a wide range of RUNX1B levels on blood cell development. Overall, our results indicate that RUNX1B is required for both the initiation and the completion of the EHT. Initially low levels of RUNX1B in HE are crucial for the maturation of the HE and the initiation of EHT. Circumventing HE maturation by increasing RUNX1 levels results in abortive differentiation, highlighting the need for a phase of low level of RUNX1 expression in the HE to initiate EHT successfully.
RESULTS
Runx1b transcripts are low and Runx1c transcripts are absent prior to EHT
Upon mESC differentiation as embryoid bodies (EBs), the differential activity of the two Runx1 promoters results in sequential expression of Runx1b and Runx1c (Sroczynska et al., 2009a). Proximal Runx1b transcripts are detected first during the early stage of the EB culture whereas distal Runx1c transcripts are only detected at later stages. To determine the transcript levels of Runx1b and Runx1c at key developmental stages of blood formation, we queried our recently published genome-scale resource encompassing enriched mesoderm (Mes), HB, HE and HP cell populations (Goode et al., 2016; Lancrin et al., 2010). Analysis of the RNA-seq datasets revealed that Runx1 has a biphasic expression pattern across the different populations (Fig. 1A,B). Relatively low expression of Runx1 in HBs and HE is followed by a 4.6-fold increase in HPs (Fig. 1A,B). Furthermore, only the Runx1b isoform, generated from the P2 promoter, was detected in HB and HE cells whereas both Runx1b and Runx1c were present in HPs (Fig. 1B). The biphasic pattern of expression and the specific expression of the Runx1b isoform were confirmed by qPCR (Fig. 1C). These data indicate that the initiation of EHT is associated with relatively low levels of Runx1b transcripts.
Fig. 1.
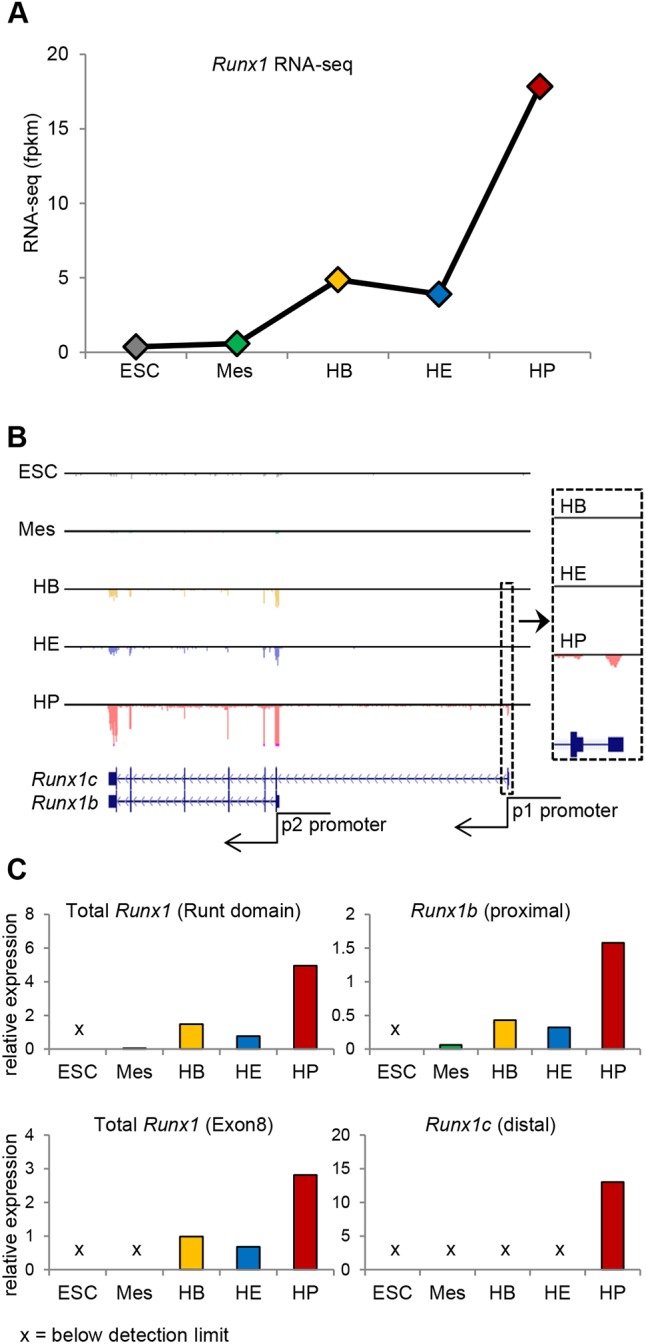
Biphasic Runx1 expression during hematopoietic differentiation of mESCs. (A) Runx1 RNA expression determined by RNA-seq across five stages of differentiation. Fpkm, fragments/kilobase of transcript/million mapped reads. (B) Mouse Runx1 locus depicting Runx1b (P2-promoter driven) and Runx1c (P1 promoter driven) with mapped RNA-seq data. (C) qPCR across five stages of differentiation. Runt domain and exon 8 primers detect both Runx1b and Runx1c transcripts. Mean of n=5 or n=6 technical replicates is shown. Cross indicates below detection limit.
High Runx1 induction in iRunx1ko induces accelerated EHT
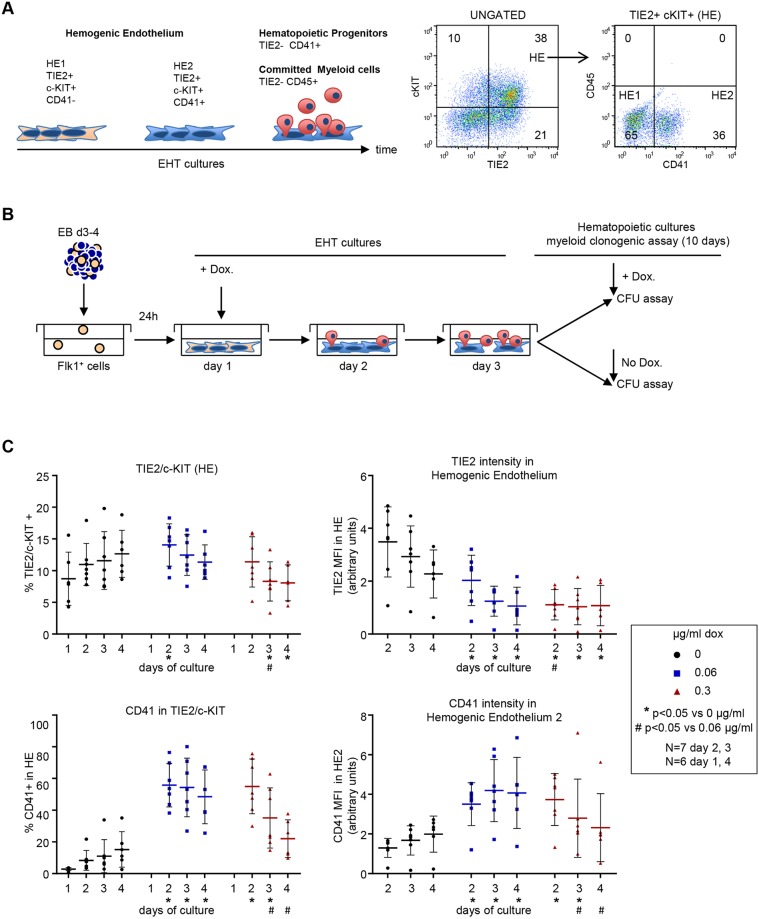
HE generated from mESC differentiation is defined as a cell population that is positive for the endothelial markers TIE2 (TEK) or CDH5 and the stem cell growth factor receptor c-KIT (Fig. 2A, Fig. S1) (Eilken et al., 2009; Frame et al., 2016; Lancrin et al., 2009). Initially, the HE population is negative for the expression of the HP marker CD41 (termed hemogenic endothelium 1 or HE1). HE1 cells proceed to a HE2 stage by gaining cell surface expression of CD41, while retaining TIE2 and c-KIT. Subsequently, TIE2 expression is lost and HE2 cells become HP cells (TIE2−/CD41+). Most HP cells commit to myeloid hematopoiesis and acquire CD45 (PTPRC) expression. Because primitive erythropoiesis in the yolk sac does not depend on either RUNX1 or CBFβ (Niki et al., 1997; Okuda et al., 1996; Sasaki et al., 1996; Wang et al., 1996a,b), we focused our analyses on the generation of myeloid hematopoietic cells.
Fig. 2.
High Runx1 induction in iRunx1ko induces accelerated EHT. (A) Left: schematic of hemogenic endothelium differentiation in mESC-derived EHT cultures. HE1 (TIE2+/c-KIT+/CD41−) cells transition via the HE2 stage (TIE2+/c-KIT+/CD41+) into TIE2−/CD41+ hematopoietic progenitors and TIE2−/CD45+ committed myeloid cells. Right: flow cytometry of HE1 and HE2 populations in day 2 wild-type mESC-derived EHT cultures. (B) Schematic of hematopoietic differentiation experiments. FLK1+ cells, isolated from mESCs differentiated as EBs, were induced with doxycycline 1 day after re-plating in EHT-culture conditions. Day 2 and 3 EHT cultures were subjected to hematopoietic CFU assays with or without additional doxycycline. (C) Flow cytometry data of a 4-day time course depicting the transition through HE1 and HE2 in iRunx1ko cells after induction by low (0.06 µg/ml) and high (0.3 µg/ml) levels of doxycycline. Top left: percentage HE cells. Top right: median fluorescence intensity of TIE2+ cells in HE. Bottom left: percentage of HE2 cells within HE. Bottom right: median fluorescence intensity of CD41+ cells within HE2. Individual biological experiments and mean±s.d. are shown. Two-way ANOVA. N=biological replicates.
To investigate the effects of RUNX1 dosage on the dynamics of EHT and the formation of myeloid hematopoietic cells, we made use of a Runx1−/− mESC line containing an exogenous Runx1b cDNA under the control of the doxycycline inducible tet-on system (iRunx1Runx1−/−, referred to as iRunx1ko hereafter) (Lancrin et al., 2009). Previous experiments have demonstrated that both myeloid and erythroid development can be rescued by doxycycline treatment of this line (Fig. S2A,B; Lancrin et al., 2009). Doxycycline titration experiments demonstrated that induction with 0.06 µg/ml and 0.3 µg/ml resulted in, respectively, low and high levels of Runx1b induction in iRunx1ko (Fig. S2C,D). Doxycycline titration in mESC lines in which GFP was under the control of the tet-on system demonstrated that a concentration of 0.3 µg/ml induced GFP in more than 90% of the cells (Fig. S2C). In contrast, although induction with 0.06 µg/ml still resulted in GFP expression in the majority of the cells (>70%), the mean fluorescence intensity was reduced by at least 50% when compared with the 0.3 µg/ml induction (Fig. S2C,D).
To study the effect of different levels of RUNX1B on EHT and subsequent myeloid blood cell formation, we differentiated mESCs using an EHT-culture system (Fig. 2B). One day after re-plating hemangioblast-enriched cells [FLK1 (KDR)+], Runx1 transcription was induced with doxycycline. Some of these cells were re-plated into myeloid hematopoietic colony forming unit assays (CFU assay) after 2 or 3 days of EHT culture. In the EHT cultures, a clear difference (6.3-fold increase in relative density) in RUNX1B protein expression could be detected between 0.06 and 0.3 µg/ml doxycycline induction (Fig. S3A, lanes 1, 4, 7).
We first analyzed the progression of HE cells during the first 4 days of EHT culture by flow cytometry. When Runx1b is not induced in the iRunx1ko line, the differentiation process is blocked at the HE1 stage, as indicated by the very low frequency of HE2 (TIE2+/c-KIT+/CD41+) cells within the total HE (TIE2+/c-KIT+) population (Fig. 2C, left). Both low and high doxycycline induction released the iRunx1ko cultures from their HE1 arrest and resulted in the generation of HE2 cells (Fig. 2C). Both doxycycline concentrations induced a reduction in the expression of the surface marker TIE2 within the HE population (Fig. 2C, top right), further indicating that the doxycycline-induced samples were undergoing EHT. There was, however, a striking difference between the two treatments with regards to the persistence of the HE in general and of the HE2 population in particular. The high doxycycline dose resulted in an initial peak of HE2 (day 2) followed by a rapid decline. In line with this, we also observed a reduction in the cell surface levels of TIE2 (within HE) and CD41 (within HE2) over time (Fig. 2C, right). The cells induced with low doxycycline appeared to have a more gradual EHT with the HE2 cells persisting longer and the surface expression of CD41 in the HE2 remaining high throughout the 4 day culture period (Fig. 2C).
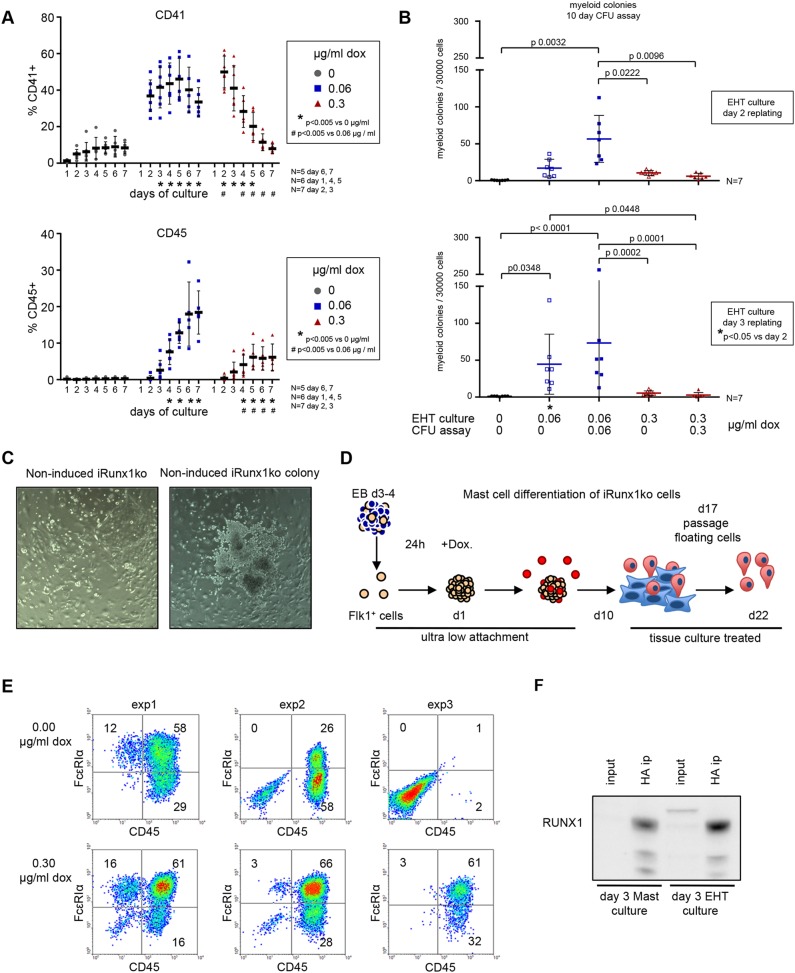
High Runx1 induction in iRunx1ko hampers myeloid hematopoietic potential
To determine whether the seemingly rapid transition of iRunx1ko through EHT initiated by the high doxycycline induction resulted in an accelerated and/or an increased myeloid hematopoietic potential, we followed CD41 (marking HE2 and HP cells) and CD45 (marking committed myeloid cells) expression over 7 days of EHT culture (Fig. 3A). Initially, at day 2, we observed a higher percentage of CD41+ cells in the high doxycycline-induced samples compared with the low doxycycline samples. However, over time the percentage of CD41+ cells in the high doxycycline-induced group dropped below that of the low doxycycline-induced group (day 4 onwards) and eventually it was reduced to the amounts seen in the non-induced group (day 6-7). In contrast, in the low doxycycline-induced group the percentage of CD41+ cells remained relatively stable during the whole experiment. The rapid rise and fall in the CD41+ cells in the high doxycycline-induced group did not coincide with a faster conversion into CD45+ hematopoietic cells nor did it result in an increase in the percentage of CD45+ cells compared with the low doxycycline-induced group (Fig. 3A). In fact, the low doxycycline-induced group was more efficient at generating CD45+ cells at any time point during the culture (Fig. 3A). We also performed myeloid CFU assays on cells from day 2 and day 3 EHT cultures (Fig. 3B). Only low doxycycline efficiently generated myeloid colonies whereas high doxycycline-induced cells were severely impaired in their ability to generate myeloid colonies (Fig. 3B).
Fig. 3.
High Runx1 induction in iRunx1ko hampers hematopoietic potential. (A) Flow cytometry time course depicting the percentage of CD41+ (top) and CD45+ (bottom) cells in iRunx1ko upon doxycycline induction. Individual biological experiments and mean±s.d. are shown. Two-way ANOVA. N=biological replicates. (B) Myeloid hematopoietic colony-formation assay with iRunx1ko re-plated after 2 (top) or 3 (bottom) days of EHT culture. Individual biological experiments and mean±s.d. are shown. Two-way ANOVA. N=biological replicates. (C) iRunx1ko day 3 EHT-cultures cultured for 5 days in liquid hematopoietic mix. Right-hand panel shows a rare colony of budding cells in non-induced iRunx1ko cells. (D) Schematic of mast cell differentiation. FLK1+ cells were re-plated in mast cell media under ultra-low-attachment conditions. One day after re-plating, the cells were induced with doxycycline. Mast assays were analyzed after 22 days of culture. (E) Flow cytometry analysis of three independent iRunx1ko mast assays with or without doxycycline. In two out of three experiments, CD45+ cells could be generated in the absence of doxycycline induction. (F) RUNX1 western blot on day 3 non-induced iRunx1ko mast and EHT culture HA-tag immunoprecipitations. Input represents 5% of the total lysates. HA antibody raised in mouse, RUNX1 antibody raised in rabbit.
Together, these results suggest that EHT cultures expressing high RUNX1B levels rapidly transit through HE and EHT. However, the resulting cells are not able to generate myeloid colonies efficiently. In contrast, lower levels of RUNX1B resulted in a more gradual EHT and the efficient generation of myeloid progenitors.
Leaky expression of RUNX1B in iRunx1ko drives low frequency hematopoiesis
Cells from non-induced day 3 iRunx1ko EHT cultures did not generate hematopoietic colonies in CFU assays (Fig. 3B). However, when these cells were cultured in liquid hematopoiesis-supporting media for 5 days, we observed occasional patches of cells resembling the round hematopoietic cells generated during EHT (Fig. 3C). These patches were very rare, possibly derived from the low frequency CD41+ cells observed in the non-induced EHT cultures (Fig. 3A). To assess this low hematopoietic potential further, we re-aggregated FLK1+ iRunx1ko cells under culture conditions favoring the generation of mast cells (Fig. 3D). Although this mast assay is not quantitative, the highly proliferative nature of the mast cells combined with an extended 3 weeks of culture makes it a very sensitive detection system for a low frequency of hematopoietic potential. In two out of three independent differentiations, non-induced iRunx1ko cells were able to produce phenotypic mast cells characterized by expression of CD45 and the mature mast cell marker FcεRIα (Fig. 3E). Upon doxycycline induction, mast cell generation was more efficient and was observed in all experiments (Fig. 3E). We verified that generation of hematopoietic CD45+ cells in this assay is dependent on the presence of intact Runx1 by performing lentiviral rescue experiments on Runx1−/− mESCs (Fig. S4). Mast cell production could only be rescued by lentiviral transduction with a wild-type Runx1b cDNA and not by mutated or truncated Runx1b cDNAs. Immunoprecipitation experiments (utilizing the HA tag in the iRunx1ko Runx1b cDNA and performed with large amounts of cells) confirmed the presence of low levels of RUNX1 in non-induced iRunx1ko mast and EHT cultures (Fig. 3F). These data indicate that under appropriate culture conditions the leaky activity of the non-induced tet-on system is capable of initiating EHT, although at very low frequency, suggesting that low levels of RUNX1B could be sufficient to initiate this process.
Cbfb−/− have reduced RUNX1 protein levels and cannot initiate EHT
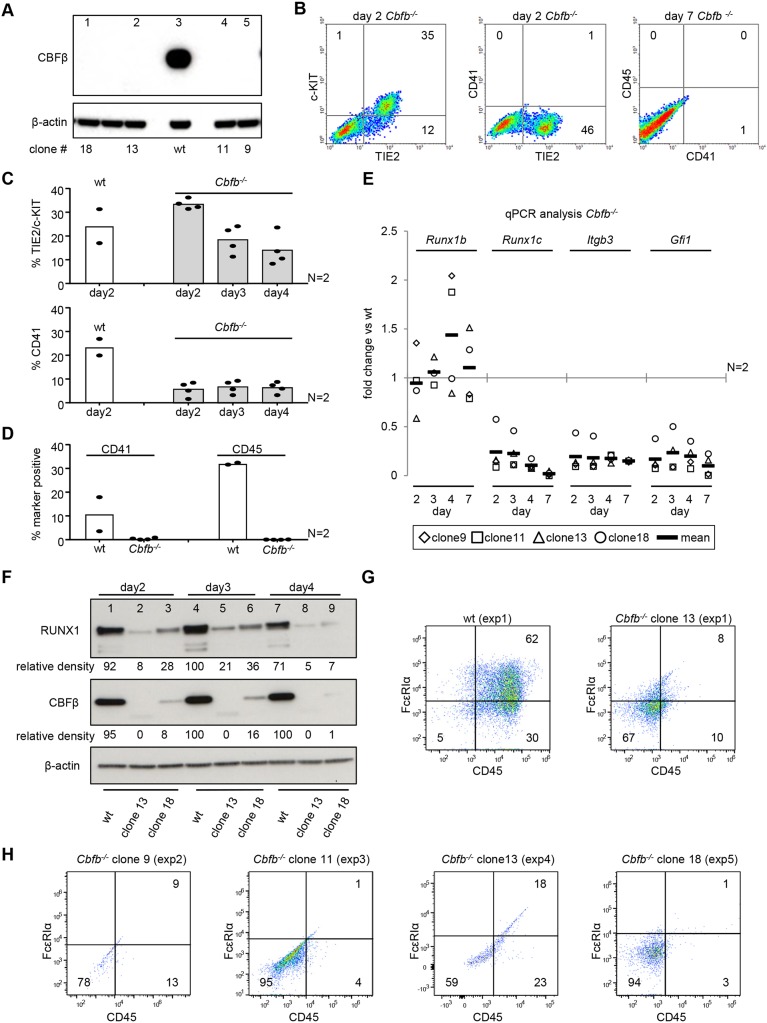
The results of the mast assays prompted us to investigate the role of RUNX1B concentrations lower than those generated with 0.06 µg/ml doxycycline. However, lowering the concentration of doxycycline below 0.06 µg/ml rendered a large proportion of the cells unresponsive (Fig. S2C). To investigate a lower range of RUNX1B levels and activity we decided to target the RUNX1 binding partner CBFβ. Heterodimerization of RUNX1 with CBFβ has been shown to enhance RUNX1 DNA-binding capacity and to protect it from degradation (Huang et al., 2001; Nagata et al., 1999; Ogawa et al., 1993; Tang et al., 2000). To confirm that CBFβ interacts with RUNX1 in our EHT cultures and could potentially enhance its DNA-binding capacity, we performed Duolink proximity ligation assays for RUNX1 and CBFβ in HE1, HE2 and HP cells (Fig. S5A,B). Interactions between endogenous RUNX1 and CBFβ, detected as individual puncta, were abundant in the nuclei of all populations analyzed. This suggests that the enhancement of RUNX1 DNA-binding capacity that has been shown to result from heterodimerization with CBFβ (Nagata et al., 1999; Ogawa et al., 1993; Tang et al., 2000) could also occur within our culture system. We therefore proceeded to disrupt Cbfb using CRISPR/Cas9 targeting exon 4, which is part of the RUNX1-binding domain of CBFβ (Kagoshima et al., 1996; Ogawa et al., 1993). The resulting Cbfb knockout cells are not only expected to contain less RUNX1 protein but the remaining RUNX1 protein should be less efficient at activating RUNX1 target genes.
Following CRISPR/Cas9 targeting of Cbfb in wild-type mESCs, knockout clones were identified by the absence of detectable CBFβ protein on western blot (Fig. 4A). To analyze the effect of Cbfb deletion on EHT and hematopoietic development, four different Cbfb−/− mESC clones were differentiated once across two biological experiments. Flow cytometry analysis of the EHT cultures indicated that, similarly to the non-induced iRunx1ko cells, the Cbfb−/− lines fail to proceed beyond the HE1 stage (Fig. 4B,C). The Cbfb−/− lines also failed to generate CD45+ mature hematopoietic cells after 7 days of EHT culture (Fig. 4B,D). This block in hematopoiesis was observed despite the fact that Runx1b transcript levels in these Cbfb−/− lines were similar to those observed in the wild-type parent line (Fig. 4E). By day 4, two out of four clones displayed a two-fold increase in Runx1b RNA (suggestive of transcriptional compensation or an accumulation of HE1 cells) whereas the other clones had similar levels of Runx1b RNA as the parent line. Additional qPCR further confirmed a defect in the ability of Cbfb−/− cells to undergo EHT as only minimal expression of the RUNX1B early target genes Gfi1 and Itgb3 were detected as well as minimal expression of Runx1c transcripts, which are indicative of HP formation (Fig. 4E).
Fig. 4.
Cbfb−/− have reduced RUNX1 protein levels and cannot initiate EHT. (A) Western blot for CBFβ on CRISPR/Cas9-generated Cbfb−/− mESC clones. Lane 3 shows a wild-type (wt) mESC positive control. (B) Representative flow cytometry plots for Cbfb−/− day 2 and day 7 EHT cultures. (C-E) Four Cbfb−/− clones were differentiated across two independent experiments. Individual clones and mean are shown. (C,D) Flow cytometry data for four Cbfb−/− clones analyzed on day 2-4 (C) and day 7 (D) of EHT culture. (E) Time-course qPCR analysis of Cbfb−/− EHT cultures. Data are normalized to the wild-type line. (F) Time-course western blot analysis of EHT cultures for RUNX1, CBFβ and β-actin on two Cbfb−/− clones (lanes 2, 5, 8, and 3, 6, 9) and wild type (lanes 1, 4, 7). β-actin-normalized relative densities for RUNX1 and CBFβ are shown. (G,H) Flow cytometry analysis of four Cbfb−/− clones across five independent mast cell differentiations.
As shown for two Cbfb−/− clones (13 and 18), RUNX1 protein levels were reduced at all time points analyzed (Fig. 4F). In clone 18, a residual level of CBFβ was observed suggesting that some of the CRISPR/Cas9-generated indels did not completely abrogate CBFβ production. The reduction of RUNX1 protein in the Cbfb−/− lines suggests that in the EHT cultures RUNX1 is no longer protected by CBFβ from proteasomal degradation, as has been described in other culture systems (Huang et al., 2001). This is further supported by our finding that the reduction in RUNX1 could, at least partially, be rescued by treating the Cbfb−/− EHT cultures with the proteasome inhibitor MG132 (Fig. S5C). To confirm that the Cbfb−/− cells had lost their ability to initiate EHT, we performed mast assays to assess for any residual hematopoietic activity. The four Cbfb−/− clones were differentiated across five biological experiments (Fig. 4G,H) and none of them was able to generate mast cells.
Altogether, these data demonstrate that, in the absence of CBFβ, endogenous RUNX1 protein levels and activity drop below the level required to initiate EHT.
Knockout of Cbfb in iRunx1ko eliminates hematopoietic potential induced by leaky RUNX1B expression
Having established that the RUNX1 levels in the Cbfb−/− lines failed to induce EHT, we next used the same CRISPR/Cas9 approach to disrupt the Cbfb locus in the iRunx1ko line in order to generate iRunx1koCbfb−/− lines. Clones were selected by screening for the absence of detectable CBFβ protein by western blot (Fig. S6A). Induction of Runx1b with either 0.06 or 0.3 µg/ml doxycycline resulted in similar Runx1b transcript levels in both the parental iRunx1ko line and the iRunx1koCbfb−/− lines (Fig. S6B). However, as expected, RUNX1B protein levels were markedly lower in the iRunx1koCbfb−/− lines with relative amounts reduced by more than 38% (0.3 μg/ml doxycycline) and more than 81% (0.06 μg/ml doxycycline) (Fig. S3A). Next, we tested the hematopoietic potential of the iRunx1koCbfb−/− lines by performing mast assays (Fig. S6C). The four iRunx1koCbfb−/− clones were each differentiated once across four biological experiments. None of the clones was able to generate mast cells, demonstrating that the absence of CBFβ eliminated the low hematopoietic potential observed in the non-induced iRunx1ko cells (Fig. 3E). These results demonstrate that in iRunx1koCbfb−/− lower levels of RUNX1B can be achieved than in the parent iRunx1ko line.
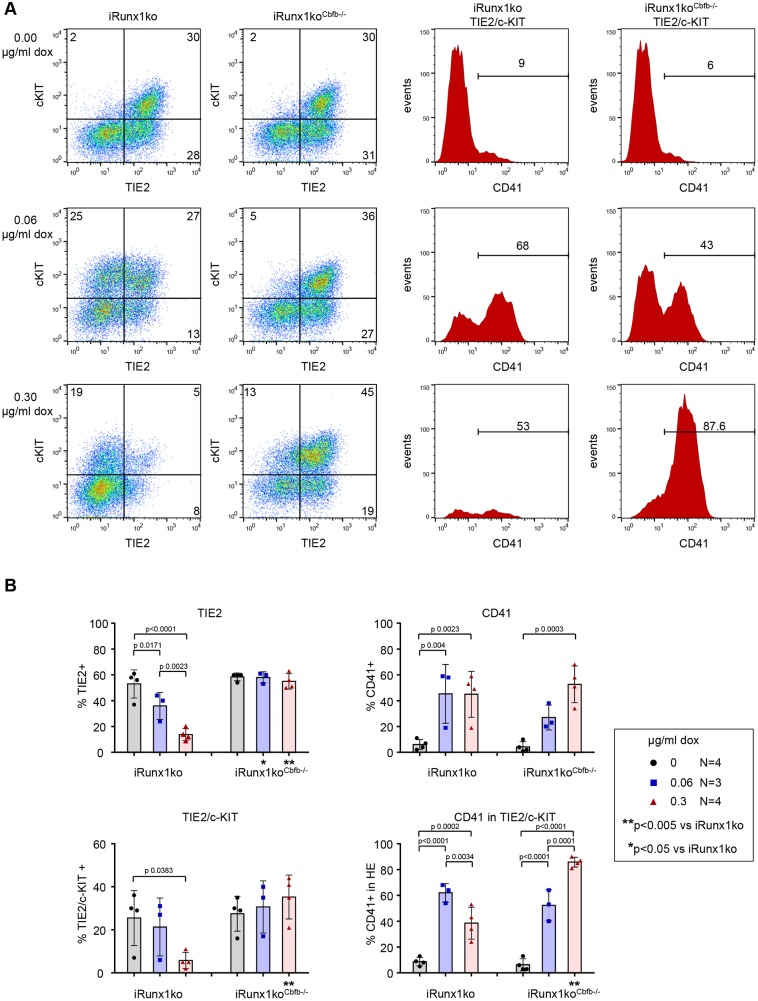
Low-level Runx1b induction is sufficient to initiate EHT in iRunx1koCbfb−/−
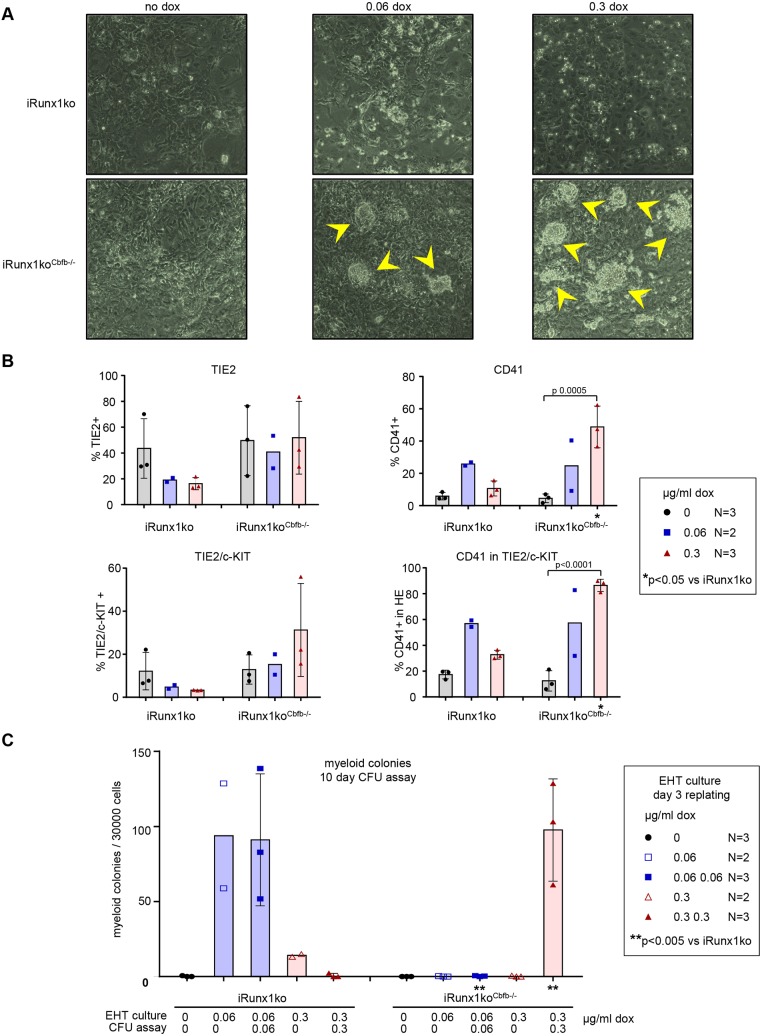
We next evaluated the ability of the iRunx1koCbfb−/− cells to progress through EHT as described above (Fig. 2B). Flow cytometry analysis of day 2 (Fig. S7) and 3 (Fig. 5) EHT cultures showed that upon Runx1b induction with either concentration of doxycycline the iRunx1koCbfb−/− lines were able to progress to HE2. However, further differentiation was clearly perturbed compared with the parent iRunx1ko line (Fig. 5, Fig. S7). Upon high doxycycline induction, the percentage of CD41+ cells was comparable to that of the parent iRunx1ko line but the iRunx1koCbfb−/− cells did not downregulate TIE2 expression as efficiently as the parent line. This was reflected by the fact that the HE population remained high in the induced iRunx1koCbfb−/− lines. On day 3, nearly all of the HE population in the high doxycycline-induced iRunx1Cbfb−/− consisted of HE2 cells whereas in the identically induced parent iRunx1ko only very few HE cells remained (Fig. 5). Consistent with this, we observed, after extended culture, multiple HE core-like structures in doxycycline-induced iRunx1koCbfb−/− cultures (Fig. 6A). This was particularly apparent in the high doxycycline-induced iRunx1koCbfb−/− cultures and was not observed in non-induced iRunx1koCbfb−/− cultures or in the original parent line (Fig. 6A). Flow cytometry revealed that the late high doxycycline-induced iRunx1koCbfb−/− cultures contained high frequencies of TIE2 and CD41 cells, similar to those observed in early day 2 and 3 doxycycline-induced iRunx1koCbfb−/− EHT cultures (Figs 5 and 6, Figs S7 and S9). These late cultures still retained an appreciable HE population that consisted of mainly HE2 cells (Fig. 6B, Fig. S9).
Fig. 5.
Low RUNX1 levels can induce EHT-like changes. (A,B) Representative flow cytometry plots (A) and flow cytometry data (B) of day 3 iRunx1ko and iRunx1koCbfb−/− EHT cultures. (A) Left: HE (TIE2+/c-KIT+). Right: TIE2+/c-KIT+ gated histograms depicting the percentage CD41+ cells within HE. Horizontal bars indicate the CD41+ population. (B) Individual biological experiments and mean±s.d. are shown. Two-way ANOVA. N=biological replicates.
Fig. 6.
Persistence of HE core-like structures in doxycycline-induced iRunx1koCbfb−/− cultures. (A) Representative pictures of day 5 iRunx1ko (top) and iRunx1koCbfb−/− (bottom) EHT cultures. HE core-like structures (arrowheads) can be observed in doxycycline-induced iRunx1koCbfb−/−. (B) Flow cytometry analysis of day 5 iRunx1ko and iRunx1koCbfb−/− EHT cultures with or without doxycycline. Individual biological experiments and mean±s.d. (only for N>2) are shown. Two-way ANOVA (only for N>2). (C) Myeloid hematopoietic colony-formation assay with iRunx1ko and iRunx1koCbfb−/− re-plated after 3 days of EHT culture. Individual biological experiments and mean±s.d. (only for N>2) are shown. Two-way ANOVA (only for N>2). N=biological replicates.
Quantitative PCR analysis confirmed the downregulation of the endothelial program in doxycycline-induced iRunx1ko EHT cultures, as indicated by reduced levels of Cdh5, Sox7 and Sox17 transcripts (Fig. S8A), which are known to be downregulated during EHT and subsequent HP formation (Clarke et al., 2013; Costa et al., 2012; Lilly et al., 2016b; Lizama et al., 2015). In contrast, the iRunx1koCbfb−/− cultures showed only a modest reduction in Sox7 and Sox17 transcripts with high doxycycline and Cdh5 did not decline at all (Fig. S8A). We also determined the levels of early hematopoietic target genes, such as Itgb3 (CD61) and Gfi1, which are both early RUNX1 targets and mark the early stages of EHT (Lancrin et al., 2012; Lie-A-Ling et al., 2014; Thambyrajah et al., 2016), and Gfi1b and AI467606 (SAIL), which mark the next progenitor stage (Ferreras et al., 2011; Kim et al., 2015; Thambyrajah et al., 2016). These early hematopoietic target genes were activated to a similar level by both doses of doxycycline in iRunx1ko cultures (Fig. S8B). In contrast, in the iRunx1koCbfb−/− lines Itgb3, Gfi1 and AI467606 transcripts were significantly induced only by high doxycycline induction (Fig. S8B). Altogether, these results suggest that the lower levels of RUNX1 protein and activity achieved in the induced iRunx1koCbfb−/− lines (compared with the iRunx1ko parent line) are sufficient to progress to the HE2 stage but that higher doses/activity of RUNX1 are required for the generation of hematopoietic progenitors.
Myeloid hematopoiesis in iRunx1koCbfb−/− requires continuous high-level Runx1b induction
To investigate further the RUNX1B levels required to generate hematopoietic cells in the iRunx1koCbfb−/− clones, we performed myeloid CFU assays following EHT culture (Fig. 2B). These experiments revealed that a single dose of doxycycline at day 1 of the EHT culture did not allow the iRunx1koCbfb−/− cells to generate hematopoietic colonies (Fig. 6C). This is consistent with our findings that these cells are blocked at the HE2 stage (Figs 4 and 5). This result was further corroborated by the observation that a single dose of doxycycline applied at day 1 of iRunx1koCbfb−/− mast assays also failed to produce hematopoietic cells (Fig. S6C). However, with an additional high dose of doxycycline at the start of the myeloid hematopoietic CFU assay we found that iRunx1koCbfb−/− cells generated myeloid hematopoietic colonies (Fig. 6C). This is in contrast to the parent iRunx1ko line, in which a single dose of low doxycycline at day 1 was sufficient for colony formation whereas high doxycycline abolished most colony-formation capability (Fig. 6C).
Altogether, these results show that the reduced levels of RUNX1B protein and activity obtained upon a single dose of doxycycline induction of the iRunx1koCbfb−/− lines is sufficient to initiate EHT. However, the cells arrest at the HE2 stage and are not able to complete EHT. This process can only be completed, and hematopoiesis rescued, by treating the HE2-arrested cells with an additional high dose of doxycycline. This indicates that a critical level of RUNX1B activity is required not only to initiate but also to complete EHT and suggests that this critical level might be different for initiation and completion of EHT. These data also establish that enforcing the expression of RUNX1B rescues the block of hematopoietic development observed in the absence of CBFβ.
The CBFβ competitor SOX7 can bind RUNX1 in mESC-derived HE
Our findings indicate that different levels and activities of RUNX1 are crucial at different stages of EHT. This is likely to be achieved by a combination of regulation of the transcription of Runx1b as well as modulation of RUNX1 activity by interactions with other proteins and post-translational modifications. In this context, it is interesting that the SOX7 transcription factor has been recently shown to regulate hematopoietic RUNX1 activity at a protein level by directly competing with CBFβ (Lilly et al., 2016a). To determine whether such a mechanism could also play a role in the control of RUNX1 activity in our mESC-derived EHT-culture system, we queried previously generated RNA-seq datasets (Goode et al., 2016) and performed qPCR. We observed that Sox7 is preferentially expressed in the HE population (Fig. S10A). In addition, Duolink proximity ligation assays demonstrated that interactions between endogenous RUNX1 and SOX7 were abundant in the nuclei of mESC-derived HE cells (Fig. S10B). These findings suggest that SOX7 might control hematopoietic RUNX1 activity in mESC-derived HE by counteracting CBFβ-mediated enhancement of RUNX1 DNA binding and/or protection from degradation. To investigate the mechanism of RUNX1 regulation by SOX7 further, we introduced an additional doxycycline-inducible Sox7 expression cassette (tet-on system) into two iRunx1koCbfb−/− clones (iRunx1ko_Sox7Cbfb−/−). In iRunx1ko_Sox7Cbfb−/− mESCs, both Runx1b and Sox7 could be induced by the addition of doxycycline (Fig. S10C,D). Both iRunx1koCbfb−/− and iRunx1ko_Sox7Cbfb−/− mESCs produced similar amounts of Runx1b transcripts upon doxycycline induction (Fig. S10C,D). However, RUNX1B protein levels were consistently higher in the induced iRunx1ko_Sox7Cbfb−/− mESCs (Fig. S10C,D). This suggests that in the HE populations SOX7 might modulate RUNX1 activity by protecting it from degradation and sequestering it from its activator, CBFβ. Such a mechanism could allow for rapid and precise control of RUNX1 activity within the HE population.
DISCUSSION
RUNX1 is a master regulator of hematopoiesis, which is indispensable for the establishment of the hematopoietic system during embryogenesis (Chen et al., 2009; Lacaud et al., 2002; North et al., 1999; Okuda et al., 1996). Although RUNX1 is not essential for survival once the adult blood system has been established, its absence or the reduction of its level still results in perturbations in the specification and differentiation of multiple blood lineages (Chen et al., 2009; Growney et al., 2005; Ichikawa et al., 2004). Similarly, during ontogeny Runx1 haploinsufficiency or mutations result in decreased numbers of hematopoietic stem and/or progenitor cells (Cai et al., 2000; Lacaud et al., 2002, 2004; Matheny et al., 2007; Wang et al., 1996a). However, the specific RUNX1 dosage requirements in HE and during different stages of EHT remain largely unknown.
By combining transcriptional control with the modulation of RUNX1 stability and activity by deleting Cbfb, we demonstrate that RUNX1 is required for both the initiation and completion of EHT. Furthermore, at the onset of hematopoiesis RUNX1 dosage must be carefully controlled in HE. A high dose of RUNX1B rapidly induced a post-EHT phenotype but the resulting cells had only minor hematopoietic potential. In contrast, a lower dose of RUNX1B allowed for a more gradual EHT induction. On a Cbfb wild-type background, a temporally restricted low dose of RUNX1B at the initiation of the EHT is sufficient to proceed through EHT fully and produce hematopoietic colonies. However, when RUNX1 stability and activity was perturbed (by disrupting the Cbfb locus), continuous high-level Runx1b induction was required to rescue hematopoiesis. Interestingly, when Runx1b was temporarily induced in a Cbfb−/− background, using either low or high Runx1b induction, the cells were able to progress from the HE1 stage to the HE2 stage. This suggests that although both the initiation of EHT and the subsequent generation of hematopoietic precursors require RUNX1 activity the required threshold of RUNX1 activity might be different for EHT initiation and completion.
A recent study suggests that in zebrafish Runx1 can drive the generation of hematopoietic stem cells (HSCs) in the absence of Cbfβ (Bresciani et al., 2014), implying that Runx1 can initiate EHT independently of Cbfβ. In contrast, in mice Cbfb knockouts display a similar phenotype as Runx1 knockouts (Niki et al., 1997; Okuda et al., 1996; Sasaki et al., 1996; Wang et al., 1996a,b). However, it is unclear whether in Cbfb−/− mice EHT changes can be initiated by residual RUNX1 activity. Our in vitro results suggest that this is probably not the case. During the differentiation of Cbfb−/− mESCs, we did not detect robust expression of the early HE RUNX1 target genes Itgb3 and Gfi1. Furthermore, the cells did not activate appreciable Runx1c transcription and did not generate CD41+ HP or CD45+ hematopoietic cells. This difference with the phenotype observed in zebrafish could be due to different requirements for RUNX1 activity between the two species. Indeed, it has been reported that in zebrafish adult HSCs can be formed without wild-type Runx1 (Sood et al., 2010).
Using sustained high-level induction of Runx1b, we were able to rescue myeloid hematopoiesis in Cbfb−/− cells. This indicates that if RUNX1 levels are sufficiently high, it can function independently of CBFβ. However, a previous study by Yokomizo et al. performed in Cbfb−/− mice demonstrated, in contrast, that Runx1 overexpression was not able to restore hematopoiesis (Yokomizo et al., 2008). These contrasting findings could be due to the use, by Yokomizo et al., of a GATA-1 gene regulatory cassette (GATA-1-HRD) to overexpress Runx1. The most robust activity of this cassette is restricted to erythroid progenitors (Yoon et al., 2008). In addition, in our hands Runx1 overexpression only rescued hematopoiesis in Cbfb−/− cells if overexpression was maintained throughout the EHT and subsequent differentiation steps. Furthermore, we found that temporal pre-EHT Runx1 expression can initiate EHT, or at least promote similar early changes, but is not sufficient to complete the process in the absence of CBFβ. Thus, the absence of hematopoietic rescue when using the GATA-1-HRD cassette to express Runx1 in the context of a Cbfb−/− background might be explained by insufficient levels of Runx1 transcription in cell types relevant to the onset of hematopoiesis. Our data, in concordance with previous reports, show that under normal physiological conditions CBFβ is essential for hematopoiesis and reveals that, in the mESC system, CBFβ is required at the very onset of EHT.
The RUNX1B dosage experiments also highlight the need for relatively moderate levels of RUNX1B activity in order to initiate a productive EHT. This suggests that RUNX1B dosage or activity must be tightly controlled in pre-hematopoietic progenitor populations, in particular in the HE. Quantification of Runx1 transcripts suggests that transcriptional control and/or mRNA turnover plays a role in determining RUNX1 dosage at the early stages of differentiation (this study; Swiers et al., 2013). However, given the transient nature of the HE and the dynamic nature of EHT, dependence on transcriptional control alone is unlikely. Recently, the SOX7 transcription factor has been shown to regulate hematopoietic RUNX1 activity at the protein level by directly competing with CBFβ (Lilly et al., 2016a). At the onset of hematopoiesis, Runx1 and Sox7 are co-expressed in the HE population (this study; Costa et al., 2012; Lilly et al., 2016a) and here we further demonstrate that endogenous SOX7 and RUNX1 can interact specifically in mESC-derived HE. We further demonstrate that SOX7 can increase RUNX1 levels independently of transcription in the absence of CBFβ. Interestingly, SOX7 overexpression in pre-EHT populations has been shown to cause accumulation of cells in HE2 both in mESC-derived cells and in mouse yolk sac explant cultures (Costa et al., 2012; Lilly et al., 2016a). This phenotype is similar to the HE2 block we observed when we induced Runx1b expression on a Cbfb−/− background. It would be therefore interesting to determine whether some of the early RUNX1 target genes in HE are in part controlled by the RUNX1-SOX7 complexes. However, although the binding sites of RUNX1B in mouse ESC-derived HE have been mapped (Lie-A-Ling et al., 2014), this has not yet been achieved for SOX7. However, our finding underscores the intriguing possibility that interaction of RUNX1 with proteins other than CBFβ plays a role in regulating or re-directing RUNX1 activity in HE and EHT. Further studies will be required to elucidate whether such mechanisms are in place and to what extent the RUNX1-SOX7 complex can participate.
The data presented here suggests a model in which RUNX1 is crucial for the initiation and completion of EHT and the regulation of RUNX1B dosage in HE is crucial for the efficient progression of EHT and blood formation (Fig. S11). In early HE (HE1), RUNX1B protein is present, but its activity might be minimal. This facilitates HE maturation and the initiation and progression of EHT. Circumventing this HE maturation phase, by increasing RUNX1B levels too early, results in abortive hematopoiesis. Once EHT is initiated RUNX1 activity is still required in order to produce mature hematopoietic cells. This process might be facilitated by modulating RUNX1 activity, potentially by changing RUNX1 binding partners, and also later by increasing Runx1 transcription and switching from the Runx1b to the Runx1c isoform.
In summary, this study reveals that sustained RUNX1 activity is required from the initiation to the completion of the EHT and indicates that RUNX1 transcription and activity need to be strictly controlled during this process.
MATERIALS AND METHODS
Mouse ESC lines and culture
The following ESC lines were used: Ainv18 (wild-type ESC), Runx1−/−, doxycycline-inducible line iRunxRunx1−/− (iRunx1ko), Sox7-GFP (Gandillet et al., 2009; Kyba et al., 2002; Lancrin et al., 2009). All lines were authenticated and regularly checked for mycoplasma contamination. ESCs were maintained and differentiated as previously described (Sroczynska et al., 2009b). Briefly, hemangioblast-enriched cell populations isolated from day 3.5 EBs by magnetic-activated cell sorting for FLK1 (Miltenyi Biotec) (Table S1) were cultured in liquid hemangioblast colony-forming media (EHT cultures). Hematopoietic assays and hematopoietic colony-forming assays were performed by transferring EHT cultures to liquid or semisolid hematopoietic mix. For mast-assays, hemangioblast-enriched cells were aggregated in EB medium with IL3, KIT ligand (both 0.5% conditioned media) and IL6 (5 ng/ml) in ultra-low attachment plates (CoStar) as previously described (Fehling et al., 2003). After 10 days, cells were transferred to tissue-culture-treated dishes and cultured for 7 days. Non-attached cells were passaged and cultured for 5 days. All tissue culture images were taken on a DMI 3000B with a DFC310 FX camera (Leica).
CRISPR-Cas9 Cbfb knockout
Cbfb CRISPR knockout lines were generated with D10A mutant Cas9 nickase (Cas9n) (Addgene plasmid #48140, deposited by Feng Zhang), as previously described (Ran et al., 2013). Primers used to generate guides, targeting exon 4 of mouse Cbfb, are provided in Table S2.
Quantitative reverse transcription PCR
RNA was extracted using the RNeasy Micro Kit (Qiagen). For the confirmation of RNA-seq data, RNA was reverse transcribed with random hexamer oligos and SuperScriptIII First Strand Synthesis (Invitrogen). To maximize the limited quantity of sample, cDNA was run on a 96.96 Array (Fluidigm) on the BioMark HD platform according to the manufacturer’s protocol. For all other samples, cDNA was prepared with Omniscript RT (Qiagen) and random hexamer oligos. Real-time PCR was performed using the TaqMan assay (Applied Biosystems) and Universal Probe Library (Roche) and run on an ABI 7900. Primer sequences are listed in Table S2.
Flow cytometry
Antibodies are listed in Table S1. Cells dissociated by trypsinisation were analyzed by flow cytometry (Table S3; BD Biosciences/ACEA Biosciences).
Western blot/immunoprecipitation
Total proteins were extracted with the Nuclear Extract Kit (Active Motif) according to the manufacturer’s protocol. Lysates were subjected to SDS-PAGE and transferred to PVDF membranes. Immunoprecipitations were performed with EZview Red Anti-HA Affinity Gel (Sigma-Aldrich) according to the manufacturer’s protocol. Antibodies are listed in Table S1.
Duolink proximity ligation assay
The Duolink assay was performed and analyzed as described previously (Lilly et al., 2016a). Antibodies are listed in Table S1. Images were taken using a Zeiss Axiovert 200M microscope at 40× or 100× magnification and analyzed using ImageJ software.
Runx1b lentiviral vectors
Lentiviral vectors with murine Runx1b cDNA were produced as described previously (Dull et al., 1998; Seppen et al., 2000; Zufferey et al., 1998). Mutations were introduced with QuikChangeII Site-Directed Mutagenesis Kit (Agilent Technologies). Transduction of FLK1+ cells was performed as previously described (Gandillet et al., 2009). For details, see supplementary Materials and Methods.
Inducible Sox7 PiggyBAC transposon
Inducible Sox7 transposon-containing mESCs were established by PEI co-transfection with PiggyBAC transposase (Bartman et al., 2015). Lines were established by magnetic-activated cell sorting for thNGF (Table S1). For details, see supplementary Materials and Methods.
Statistics
Two-way ANOVA was used to assess differences (GraphPad Prism 7).
Supplementary Material
Acknowledgements
We thank Dr Pentao Liu (Wellcome Trust Sanger Institute) for providing Piggy BAC transposase reagents and Dr Julia Draper for critically reviewing the manuscript. We thank the staff at the Flow Cytometry, Advanced Imaging and Molecular Biology Core facilities of CRUK Manchester Institute for technical support.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: M.L., E.M., A.J.L., C.L., V.K., G.L.; Methodology: M.L., E.M., A.J.L., M.C., R.P., C.L., V.K., G.L.; Writing - original draft: M.L.; Writing - review & editing: M.L., V.K., G.L.; Supervision: V.K., G.L.; Funding acquisition: G.L.
Funding
Research in the authors' laboratory is supported by the Medical Research Council (MR/P000673/1 to V.K.), the Biotechnology and Biological Sciences Research Council (BB/I001794/1 to G.L. and V.K.), Bloodwise (12037), the European Union's Horizon 2020 (GA6586250) and Cancer Research UK (C5759/A20971). Deposited in PMC for immediate release.
Data availability
The RNAseq data used in this study have been previously deposited by Lie-A-Ling et al. (2014) and Goode et al. (2016), and are available in GEO under accession numbers GSE69101 and GSE55335.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.149419.supplemental
References
- Banno K., Omori S., Hirata K., Nawa N., Nakagawa N., Nishimura K., Ohtaka M., Nakanishi M., Sakuma T., Yamamoto T. et al. (2016). Systematic cellular disease models reveal synergistic interaction of trisomy 21 and GATA1 mutations in hematopoietic abnormalities. Cell Rep. 15, 1228-1241. 10.1016/j.celrep.2016.04.031 [DOI] [PubMed] [Google Scholar]
- Bartman C. M., Egelston J., Ren X., Das R. and Phiel C. J. (2015). A simple and efficient method for transfecting mouse embryonic stem cells using polyethylenimine. Exp. Cell Res. 330, 178-185. 10.1016/j.yexcr.2014.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bee T., Liddiard K., Swiers G., Bickley S. R. B., Vink C. S., Jarratt A., Hughes J. R., Medvinsky A. and de Bruijn M. F. T. R. (2009). Alternative Runx1 promoter usage in mouse developmental hematopoiesis. Blood Cells Mol. Dis. 43, 35-42. 10.1016/j.bcmd.2009.03.011 [DOI] [PubMed] [Google Scholar]
- Bee T., Swiers G., Muroi S., Pozner A., Nottingham W., Santos A. C., Li P. S., Taniuchi I. and de Bruijn M. F. T. R. (2010). Nonredundant roles for Runx1 alternative promoters reflect their activity at discrete stages of developmental hematopoiesis. Blood 115, 3042-3050. 10.1182/blood-2009-08-238626 [DOI] [PubMed] [Google Scholar]
- Ben-Ami O., Friedman D., Leshkowitz D., Goldenberg D., Orlovsky K., Pencovich N., Lotem J., Tanay A. and Groner Y. (2013). Addiction of t(8;21) and inv(16) acute myeloid leukemia to native RUNX1. Cell Rep. 4, 1131-1143. 10.1016/j.celrep.2013.08.020 [DOI] [PubMed] [Google Scholar]
- Bertrand J. Y., Chi N. C., Santoso B., Teng S., Stainier D. Y. R. and Traver D. (2010). Haematopoietic stem cells derive directly from aortic endothelium during development. Nature 464, 108-111. 10.1038/nature08738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisset J.-C., van Cappellen W., Andrieu-Soler C., Galjart N., Dzierzak E. and Robin C. (2010). In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature 464, 116-120. 10.1038/nature08764 [DOI] [PubMed] [Google Scholar]
- Bonifer C., Levantini E., Kouskoff V. and Lacaud G. (2017). Runx1 structure and function in blood cell development. Adv. Exp. Med. Biol. 962, 65-81. 10.1007/978-981-10-3233-2_5 [DOI] [PubMed] [Google Scholar]
- Brady G., Elgueta Karstegl C. and Farrell P. J. (2013). Novel function of the unique N-terminal region of RUNX1c in B cell growth regulation. Nucleic Acids Res. 41, 1555-1568. 10.1093/nar/gks1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresciani E., Carrington B., Wincovitch S., Jones M., Gore A. V., Weinstein B. M., Sood R. and Liu P. P. (2014). CBFbeta and RUNX1 are required at 2 different steps during the development of hematopoietic stem cells in zebrafish. Blood 124, 70-78. 10.1182/blood-2013-10-531988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z., de Bruijn M., Ma X., Dortland B., Luteijn T., Downing J. R. and Dzierzak E. (2000). Haploinsufficiency of AML1 affects the temporal and spatial generation of hematopoietic stem cells in the mouse embryo. Immunity 13, 423-431. 10.1016/S1074-7613(00)00042-X [DOI] [PubMed] [Google Scholar]
- Challen G. A. and Goodell M. A. (2010). Runx1 isoforms show differential expression patterns during hematopoietic development but have similar functional effects in adult hematopoietic stem cells. Exp. Hematol. 38, 403-416. 10.1016/j.exphem.2010.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. J., Yokomizo T., Zeigler B. M., Dzierzak E. and Speck N. A. (2009). Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature 457, 887-891. 10.1038/nature07619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K., Kennedy M., Kazarov A., Papadimitriou J. C. and Keller G. (1998). A common precursor for hematopoietic and endothelial cells. Development 125, 725-732. [DOI] [PubMed] [Google Scholar]
- Clarke R. L., Yzaguirre A. D., Yashiro-Ohtani Y., Bondue A., Blanpain C., Pear W. S., Speck N. A. and Keller G. (2013). The expression of Sox17 identifies and regulates haemogenic endothelium. Nat. Cell Biol. 15, 502-510. 10.1038/ncb2724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa G., Mazan A., Gandillet A., Pearson S., Lacaud G. and Kouskoff V. (2012). SOX7 regulates the expression of VE-cadherin in the haemogenic endothelium at the onset of haematopoietic development. Development 139, 1587-1598. 10.1242/dev.071282 [DOI] [PubMed] [Google Scholar]
- De Vita S., Canzonetta C., Mulligan C., Delom F., Groet J., Baldo C., Vanes L., Dagna-Bricarelli F., Hoischen A., Veltman J. et al. (2010). Trisomic dose of several chromosome 21 genes perturbs haematopoietic stem and progenitor cell differentiation in Down's syndrome. Oncogene 29, 6102-6114. 10.1038/onc.2010.351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper J. E., Sroczynska P., Tsoulaki O., Leong H. S., Fadlullah M. Z. H., Miller C., Kouskoff V. and Lacaud G. (2016). RUNX1B expression is highly heterogeneous and distinguishes megakaryocytic and erythroid lineage fate in adult mouse hematopoiesis. PLoS Genet. 12, e1005814 10.1371/journal.pgen.1005814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper J. E., Sroczynska P., Leong H. S., Fadlullah M. Z. H., Miller C., Kouskoff V. and Lacaud G. (2017). Mouse RUNX1C regulates premegakaryocytic/erythroid output and maintains survival of megakaryocyte progenitors. Blood 130, 271-284. 10.1182/blood-2016-06-723635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dull T., Zufferey R., Kelly M., Mandel R. J., Nguyen M., Trono D. and Naldini L. (1998). A third-generation lentivirus vector with a conditional packaging system. J. Virol. 72, 8463-8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilken H. M., Nishikawa S.-I. and Schroeder T. (2009). Continuous single-cell imaging of blood generation from haemogenic endothelium. Nature 457, 896-900. 10.1038/nature07760 [DOI] [PubMed] [Google Scholar]
- Fehling H. J., Lacaud G., Kubo A., Kennedy M., Robertson S., Keller G. and Kouskoff V. (2003). Tracking mesoderm induction and its specification to the hemangioblast during embryonic stem cell differentiation. Development 130, 4217-4227. 10.1242/dev.00589 [DOI] [PubMed] [Google Scholar]
- Ferreras C., Lancrin C., Lie-A-Ling M., Kouskoff V. and Lacaud G. (2011). Identification and characterization of a novel transcriptional target of RUNX1/AML1 at the onset of hematopoietic development. Blood 118, 594-597. 10.1182/blood-2010-06-294124 [DOI] [PubMed] [Google Scholar]
- Frame J. M., Fegan K. H., Conway S. J., McGrath K. E. and Palis J. (2016). Definitive hematopoiesis in the Yolk Sac emerges from Wnt-responsive hemogenic endothelium independently of circulation and arterial identity. Stem Cells 34, 431-444. 10.1002/stem.2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Nishimura M., Taniwaki M., Abe T. and Okuda T. (2001). Identification of an alternatively spliced form of the mouse AML1/RUNX1 gene transcript AML1c and its expression in early hematopoietic development. Biochem. Biophys. Res. Commun. 281, 1248-1255. 10.1006/bbrc.2001.4513 [DOI] [PubMed] [Google Scholar]
- Gandillet A., Serrano A. G., Pearson S., Lie-A-Ling M., Lacaud G. and Kouskoff V. (2009). Sox7-sustained expression alters the balance between proliferation and differentiation of hematopoietic progenitors at the onset of blood specification. Blood 114, 4813-4822. 10.1182/blood-2009-06-226290 [DOI] [PubMed] [Google Scholar]
- Gilham D. E., Lie-A-Ling M., Taylor N. and Hawkins R. E. (2010). Cytokine stimulation and the choice of promoter are critical factors for the efficient transduction of mouse T cells with HIV-1 vectors. J. Gene Med. 12, 129-136. [DOI] [PubMed] [Google Scholar]
- Goode D. K., Obier N., Vijayabaskar M. S., Lie-A-Ling M., Lilly A. J., Hannah R., Lichtinger M., Batta K., Florkowska M., Patel R. et al. (2016). Dynamic gene regulatory networks drive hematopoietic specification and differentiation. Dev. Cell 36, 572-587. 10.1016/j.devcel.2016.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyama S., Schibler J., Cunningham L., Zhang Y., Rao Y., Nishimoto N., Nakagawa M., Olsson A., Wunderlich M., Link K. A. et al. (2013). Transcription factor RUNX1 promotes survival of acute myeloid leukemia cells. J. Clin. Invest. 123, 3876-3888. 10.1172/JCI68557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Growney J. D., Shigematsu H., Li Z., Lee B. H., Adelsperger J., Rowan R., Curley D. P., Kutok J. L., Akashi K., Williams I. R. et al. (2005). Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood 106, 494-504. 10.1182/blood-2004-08-3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G., Shigesada K., Ito K., Wee H.-J., Yokomizo T. and Ito Y. (2001). Dimerization with PEBP2beta protects RUNX1/AML1 from ubiquitin-proteasome-mediated degradation. EMBO J. 20, 723-733. 10.1093/emboj/20.4.723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa M., Asai T., Saito T., Yamamoto G., Seo S., Yamazaki I., Yamagata T., Mitani K., Chiba S., Hirai H., et al. (2004). AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat. Med. 10, 299-304. 10.1038/nm997 [DOI] [PubMed] [Google Scholar]
- Jaffredo T., Gautier R., Eichmann A. and Dieterlen-Lievre F. (1998). Intraaortic hemopoietic cells are derived from endothelial cells during ontogeny. Development 125, 4575-4583. [DOI] [PubMed] [Google Scholar]
- Kagoshima H., Akamatsu Y., Ito Y. and Shigesada K. (1996). Functional dissection of the alpha and beta subunits of transcription factor PEBP2 and the redox susceptibility of its DNA binding activity. J. Biol. Chem. 271, 33074-33082. 10.1074/jbc.271.51.33074 [DOI] [PubMed] [Google Scholar]
- Kim S. Y., Theunissen J.-W., Balibalos J., Liao-Chan S., Babcock M. C., Wong T., Cairns B., Gonzalez D., van der Horst E. H., Perez M. et al. (2015). A novel antibody-drug conjugate targeting SAIL for the treatment of hematologic malignancies. Blood Cancer J. 5, e316 10.1038/bcj.2015.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissa K. and Herbomel P. (2010). Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 464, 112-115. 10.1038/nature08761 [DOI] [PubMed] [Google Scholar]
- Kyba M., Perlingeiro R. C. R. and Daley G. Q. (2002). HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell 109, 29-37. 10.1016/S0092-8674(02)00680-3 [DOI] [PubMed] [Google Scholar]
- Lacaud G., Gore L., Kennedy M., Kouskoff V., Kingsley P., Hogan C., Carlsson L., Speck N., Palis J. and Keller G. (2002). Runx1 is essential for hematopoietic commitment at the hemangioblast stage of development in vitro. Blood 100, 458-466. 10.1182/blood-2001-12-0321 [DOI] [PubMed] [Google Scholar]
- Lacaud G., Kouskoff V., Trumble A., Schwantz S. and Keller G. (2004). Haploinsufficiency of Runx1 results in the acceleration of mesodermal development and hemangioblast specification upon in vitro differentiation of ES cells. Blood 103, 886-889. 10.1182/blood-2003-06-2149 [DOI] [PubMed] [Google Scholar]
- Lam E. Y. N., Hall C. J., Crosier P. S., Crosier K. E. and Flores M. V. (2010). Live imaging of Runx1 expression in the dorsal aorta tracks the emergence of blood progenitors from endothelial cells. Blood 116, 909-914. 10.1182/blood-2010-01-264382 [DOI] [PubMed] [Google Scholar]
- Lancrin C., Sroczynska P., Stephenson C., Allen T., Kouskoff V. and Lacaud G. (2009). The haemangioblast generates haematopoietic cells through a haemogenic endothelium stage. Nature 457, 892-895. 10.1038/nature07679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancrin C., Sroczynska P., Serrano A. G., Gandillet A., Ferreras C., Kouskoff V. and Lacaud G. (2010). Blood cell generation from the hemangioblast. J. Mol. Med. (Berl.) 88, 167-172. 10.1007/s00109-009-0554-0 [DOI] [PubMed] [Google Scholar]
- Lancrin C., Mazan M., Stefanska M., Patel R., Lichtinger M., Costa G., Vargel O., Wilson N. K., Moroy T., Bonifer C. et al. (2012). GFI1 and GFI1B control the loss of endothelial identity of hemogenic endothelium during hematopoietic commitment. Blood 120, 314-322. 10.1182/blood-2011-10-386094 [DOI] [PubMed] [Google Scholar]
- Levanon D. and Groner Y. (2004). Structure and regulated expression of mammalian RUNX genes. Oncogene 23, 4211-4219. 10.1038/sj.onc.1207670 [DOI] [PubMed] [Google Scholar]
- Lie-A-Ling M., Marinopoulou E., Li Y., Patel R., Stefanska M., Bonifer C., Miller C., Kouskoff V. and Lacaud G. (2014). RUNX1 positively regulates a cell adhesion and migration program in murine hemogenic endothelium prior to blood emergence. Blood 124, e11-e20. 10.1182/blood-2014-04-572958 [DOI] [PubMed] [Google Scholar]
- Lilly A. J., Costa G., Largeot A., Fadlullah M. Z., Lie-A-Ling M., Lacaud G. and Kouskoff V. (2016a). Interplay between SOX7 and RUNX1 regulates hemogenic endothelial fate in the yolk sac. Development. 143, 4341.-. 10.1242/dev.140970 [DOI] [PubMed] [Google Scholar]
- Lilly A. J., Lacaud G. and Kouskoff V. (2016b). SOXF transcription factors in cardiovascular development. Semin. Cell Dev. Biol. 63, 50.-. 10.1016/j.semcdb.2016.07.021 [DOI] [PubMed] [Google Scholar]
- Lizama C. O., Hawkins J. S., Schmitt C. E., Bos F. L., Zape J. P., Cautivo K. M., Borges Pinto H., Rhyner A. M., Yu H., Donohoe M. E. et al. (2015). Repression of arterial genes in hemogenic endothelium is sufficient for haematopoietic fate acquisition. Nat. Commun. 6, 7739 10.1038/ncomms8739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheny C. J., Speck M. E., Cushing P. R., Zhou Y., Corpora T., Regan M., Newman M., Roudaia L., Speck C. L., Gu T.-L. et al. (2007). Disease mutations in RUNX1 and RUNX2 create nonfunctional, dominant-negative, or hypomorphic alleles. EMBO J. 26, 1163-1175. 10.1038/sj.emboj.7601568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavilio F., Ferrari G., Rossini S., Nobili N., Bonini C., Casorati G., Traversari C. and Bordignon C. (1994). Peripheral blood lymphocytes as target cells of retroviral vector-mediated gene transfer. Blood 83, 1988-1997. [PubMed] [Google Scholar]
- Miyoshi H., Ohira M., Shimizu K., Mitani K., Hirai H., Imai T., Yokoyama K., Soceda E. and Ohkl M. (1995). Alternative splicing and genomic structure of the AML1 gene involved in acute myeloid leukemia. Nucleic Acids Res. 23, 2762-2769. 10.1093/nar/23.14.2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata T., Gupta V., Sorce D., Kim W.-Y., Sali A., Chait B. T., Shigesada K., Ito Y. and Werner M. H. (1999). Immunoglobulin motif DNA recognition and heterodimerization of the PEBP2/CBF Runt domain. Nat. Struct. Biol. 6, 615-619. 10.1038/10658 [DOI] [PubMed] [Google Scholar]
- Nieke S., Yasmin N., Kakugawa K., Yokomizo T., Muroi S. and Taniuchi I. (2017). Unique N-terminal sequences in two Runx1 isoforms are dispensable for Runx1 function. BMC Dev. Biol. 17, 81 10.1186/s12861-017-0156-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki M., Okada H., Takano H., Kuno J., Tani K., Hibino H., Asano S., Ito Y., Satake M. and Noda T. (1997). Hematopoiesis in the fetal liver is impaired by targeted mutagenesis of a gene encoding a non-DNA binding subunit of the transcription factor, polyomavirus enhancer binding protein 2/core binding factor. Proc. Natl. Acad. Sci. USA 94, 5697-5702. 10.1073/pnas.94.11.5697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- North T., Gu T. L., Stacy T., Wang Q., Howard L., Binder M., Marin-Padilla M. and Speck N. A. (1999). Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development 126, 2563-2575. [DOI] [PubMed] [Google Scholar]
- Ogawa E., Inuzuka M., Maruyama M., Satake M., Naito-Fujimoto M., Ito Y. and Shigesada K. (1993). Molecular cloning and characterization of PEBP2β, the heterodimeric partner of a novel Drosophila runt-related DNA binding protein PEBP2α. Virology 194, 314-331. 10.1006/viro.1993.1262 [DOI] [PubMed] [Google Scholar]
- Okuda T., van Deursen J., Hiebert S. W., Grosveld G. and Downing J. R. (1996). AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84, 321-330. 10.1016/S0092-8674(00)80986-1 [DOI] [PubMed] [Google Scholar]
- Pozner A., Lotem J., Xiao C., Goldenberg D., Brenner O., Negreanu V., Levanon D. and Groner Y. (2007). Developmentally regulated promoter-switch transcriptionally controls Runx1 function during embryonic hematopoiesis. BMC Dev. Biol. 7, 84 10.1186/1471-213X-7-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptasinska A., Assi S. A., Martinez-Soria N., Imperato M. R., Piper J., Cauchy P., Pickin A., James S. R., Hoogenkamp M., Williamson D. et al. (2014). Identification of a dynamic core transcriptional network in t(8;21) AML that regulates differentiation block and self-renewal. Cell Rep. 8, 1974-1988. 10.1016/j.celrep.2014.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A. and Zhang F. (2013). Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281-2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio-Machin A., Menezes J., Maiques-Diaz A., Agirre X., Ferreira B. I., Acquadro F., Rodriguez-Perales S., Juaristi K. A., Alvarez S. and Cigudosa J. C. (2012). Abrogation of RUNX1 gene expression in de novo myelodysplastic syndrome with t(4;21)(q21;q22). Haematologica 97, 534-537. 10.3324/haematol.2011.050567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K., Yagi H., Bronson R. T., Tominaga K., Matsunashi T., Deguchi K., Tani Y., Kishimoto T. and Komori T. (1996). Absence of fetal liver hematopoiesis in mice deficient in transcriptional coactivator core binding factor beta. Proc. Natl. Acad. Sci. USA 93, 12359-12363. 10.1073/pnas.93.22.12359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppen J., Barry S. C., Klinkspoor J. H., Katen L. J., Lee S. P., Garcia J. V. and Osborne W. R. A. (2000). Apical gene transfer into quiescent human and canine polarized intestinal epithelial cells by lentivirus vectors. J. Virol. 74, 7642-7645. 10.1128/JVI.74.16.7642-7645.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W.-J., Sullivan M. G., Legare R. D., Hutchings S., Tan X., Kufrin D., Ratajczak J., Resende I. C., Haworth C., Hock R. et al. (1999). Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat. Genet. 23, 166-175. 10.1038/13793 [DOI] [PubMed] [Google Scholar]
- Sood R., English M. A., Belele C. L., Jin H., Bishop K., Haskins R., McKinney M. C., Chahal J., Weinstein B. M., Wen Z. et al. (2010). Development of multilineage adult hematopoiesis in the zebrafish with a runx1 truncation mutation. Blood 115, 2806-2809. 10.1182/blood-2009-08-236729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sroczynska P., Lancrin C., Kouskoff V. and Lacaud G. (2009a). The differential activities of Runx1 promoters define milestones during embryonic hematopoiesis. Blood 114, 5279-5289. 10.1182/blood-2009-05-222307 [DOI] [PubMed] [Google Scholar]
- Sroczynska P., Lancrin C., Pearson S., Kouskoff V. and Lacaud G. (2009b). In vitro differentiation of embryonic stem cells as a model of early hematopoietic development. Methods Mol. Biol. 538, 317-334. 10.1007/978-1-59745-418-6_16 [DOI] [PubMed] [Google Scholar]
- Swiers G., Baumann C., O'Rourke J., Giannoulatou E., Taylor S., Joshi A., Moignard V., Pina C., Bee T., Kokkaliaris K. D. et al. (2013). Early dynamic fate changes in haemogenic endothelium characterized at the single-cell level. Nat. Commun. 4, 2924 10.1038/ncomms3924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y.-Y., Crute B. E., Kelley J. J., Huang X., Yan J., Shi J., Hartman K. L., Laue T. M., Speck N. A. and Bushweller J. H. (2000). Biophysical characterization of interactions between the core binding factor alpha and beta subunits and DNA. FEBS Lett. 470, 167-172. 10.1016/S0014-5793(00)01312-0 [DOI] [PubMed] [Google Scholar]
- Telfer J. C. and Rothenberg E. V. (2001). Expression and function of a stem cell promoter for the murine CBFalpha2 gene: distinct roles and regulation in natural killer and T cell development. Dev. Biol. 229, 363-382. 10.1006/dbio.2000.9991 [DOI] [PubMed] [Google Scholar]
- Thambyrajah R., Mazan M., Patel R., Moignard V., Stefanska M., Marinopoulou E., Li Y., Lancrin C., Clapes T., Möröy T. et al. (2016). GFI1 proteins orchestrate the emergence of haematopoietic stem cells through recruitment of LSD1. Nat. Cell Biol. 18, 21-32. 10.1038/ncb3276 [DOI] [PubMed] [Google Scholar]
- Wang Q., Stacy T., Binder M., Marin-Padilla M., Sharpe A. H. and Speck N. A. (1996a). Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl. Acad. Sci. USA 93, 3444-3449. 10.1073/pnas.93.8.3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Stacy T., Miller J. D., Lewis A. F., Gu T.-L., Huang X., Bushweller J. H., Bories J.-C., Alt F. W., Ryan G. et al. (1996b). The CBFbeta subunit is essential for CBFalpha2 (AML1) function in vivo. Cell 87, 697-708. 10.1016/S0092-8674(00)81389-6 [DOI] [PubMed] [Google Scholar]
- Yanagida M., Osato M., Yamashita N., Liqun H., Jacob B., Wu F., Cao X., Nakamura T., Yokomizo T., Takahashi S. et al. (2005). Increased dosage of Runx1/AML1 acts as a positive modulator of myeloid leukemogenesis in BXH2 mice. Oncogene 24, 4477-4485. 10.1038/sj.onc.1208675 [DOI] [PubMed] [Google Scholar]
- Yokomizo T., Yanagida M., Huang G., Osato M., Honda C., Ema M., Takahashi S., Yamamoto M. and Ito Y. (2008). Genetic evidence of PEBP2β-independent activation of Runx1 in the murine embryo. Int. J. Hematol. 88, 134-138. 10.1007/s12185-008-0121-4 [DOI] [PubMed] [Google Scholar]
- Yoon D., Kim B. and Prchal J. T. (2008). Cre recombinase expression controlled by the hematopoietic regulatory domain of Gata-1 is erythroid-specific. Blood Cells Mol. Dis. 40, 381-387. 10.1016/j.bcmd.2007.10.008 [DOI] [PubMed] [Google Scholar]
- Zufferey R., Dull T., Mandel R. J., Bukovsky A., Quiroz D., Naldini L. and Trono D. (1998). Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J. Virol. 72, 9873-9880. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.