ABSTRACT
The entire lung epithelium arises from SRY box 9 (SOX9)-expressing progenitors that form the respiratory tree and differentiate into airway and alveolar cells. Despite progress in understanding their initial specification within the embryonic foregut, how these progenitors are subsequently maintained is less clear. Using inducible, progenitor-specific genetic mosaic mouse models, we showed that β-catenin (CTNNB1) maintains lung progenitors by promoting a hierarchical lung progenitor gene signature, suppressing gastrointestinal (GI) genes, and regulating NK2 homeobox 1 (NKX2.1) and SRY box 2 (SOX2) in a developmental stage-dependent manner. At the early, but not later, stage post-lung specification, CTNNB1 cell-autonomously maintained normal NKX2.1 expression levels and suppressed ectopic SOX2 expression. Genetic epistasis analyses revealed that CTNNB1 is required for fibroblast growth factor (Fgf)/Kirsten rat sarcoma viral oncogene homolog (Kras)-mediated promotion of the progenitors. In silico screening of Eurexpress and translating ribosome affinity purification (TRAP)-RNAseq identified a progenitor gene signature, a subset of which depends on CTNNB1. Wnt signaling also maintained NKX2.1 expression and suppressed GI genes in cultured human lung progenitors derived from embryonic stem cells.
KEY WORDS: Lung development, Epithelial progenitors, Beta-catenin, Mouse
Summary: CTNNB1-mediated Wnt signaling maintains lung fate and suppresses gastrointestinal fate of mouse and human SOX9 progenitors, maintaining NKX2.1 and inhibiting SOX2 in a developmental stage-specific manner.
INTRODUCTION
The epithelium in a mature lung consists of proximal air-conducting airways and distal gas exchange alveoli. Despite such spatial and functional distinction, both the airway and alveolar epithelia arise from common embryonic epithelial progenitors that express SRY box 9 (SOX9) and inhibitor of DNA binding 2 (ID2) in mice (Rawlins et al., 2009; Alanis et al., 2014; Yang and Chen, 2014; Volckaert and De Langhe, 2015). These SOX9-expressing progenitors (hereafter ‘SOX9 progenitors’) constitute the lung buds as early as their specification and emergence from the lateral side of the embryonic foregut, around embryonic day (E) 9.5, and before they extend and branch away from the foregut (Yang and Chen, 2014). The SOX9 progenitors undergo branching morphogenesis to build the respiratory tree until at least E18 and leave behind daughter cells that differentiate into airway and alveolar cells at early and late stages of lung development, respectively (Rawlins et al., 2009; Alanis et al., 2014). Thus, formation of a functional lung epithelium depends on the precise control of the morphogenesis and differentiation of these SOX9 progenitors, calling for a better understanding of the associated molecular mechanisms.
Although mouse genetics experiments have shown that CTNNB1-mediated canonical Wnt signaling is required for the foregut specification of NK2 homeobox 1 (NKX2.1)-expressing lung cells, which include SOX9 progenitors (Goss et al., 2009; Harris-Johnson et al., 2009), how these progenitors are subsequently maintained is less understood (Warburton et al., 2005; Cardoso and Lu, 2006; Morrisey and Hogan, 2010; Volckaert and De Langhe, 2015). Epithelial deletion of fibroblast growth factor receptor 2 (Fgfr2) leads to a complete loss of SOX9 progenitors and no branch formation beyond the two primary lung buds (De Moerlooze et al., 2000; Abler et al., 2009; Chang et al., 2013). Overexpression of an Fgf ligand-encoding gene, Fgf10, or epithelial expression of a hyperactive mutant of Kras, a small GTPase downstream of receptor tyrosine kinases, including FGFR2, leads to excessive SOX9 progenitors and overgrown branches that interfere with normal branching (Chang et al., 2013; Volckaert et al., 2013). Besides Fgf signaling, although multiple other signaling pathways are required for lung branching and patterning (Morrisey and Hogan, 2010), their specific role in the SOX9 progenitors is unclear, in part because prior studies have not specifically targeted and analyzed these cells. For example, epithelial Ctnnb1 deletion using a Sftpc-rtTA driver led to a higher proportion of proximal airways versus distal alveoli (Mucenski et al., 2003; Shu et al., 2005), which might result from inefficient expansion of the SOX9 progenitors and/or their excessive differentiation into SOX2-expressing cells. Moreover, the relationship between CTNNB1-mediated Wnt signaling and Fgf signaling in SOX9 progenitors is still unclear (Shu et al., 2005; Wang et al., 2012; Volckaert et al., 2013). In addition, it is unknown to what extent the molecular program of the SOX9 progenitors depends on CTNNB1.
SOX9 is not only a progenitor marker, but is also required for normal progenitor branching (Chang et al., 2013; Rockich et al., 2013). However, the epithelial Sox9 mutant lung still branches and expresses many genes that have the same expression pattern as SOX9 (Chang et al., 2013), suggesting the presence of additional upstream regulators of the progenitor program. In the current study, after screening multiple signaling pathways, we focused on the CTNNB1-mediated canonical Wnt signaling. Using a genetic model that allowed inducible, progenitor-specific deletion of Ctnnb1, we analyzed the SOX9 progenitors at different developmental stages and within a few days of Cre recombination to minimize secondary phenotypes. Using a combination of 3D imaging, genetic mosaic and epistasis analyses, as well as genome-wide database mining and expression profiling, we demonstrate developmental stage-dependent roles of CTNNB1, including its unexpected role in maintaining lung fate and suppressing GI fate after lung specification; identify a 119-gene signature for the SOX9 progenitors; and construct an initial progenitor regulatory network. We further show that the role of Wnt signaling in maintaining NKX2.1 expression and suppressing GI genes is conserved in human lung progenitors generated from embryonic stem cells (ESCs).
RESULTS
Deletion of Ctnnb1 at E11 leads to loss of SOX9, derepression of GI genes, decreased NKX2.1 and ectopic SOX2
Given that SOX9 is a marker and regulator of lung epithelial progenitors (Chang et al., 2013; Rockich et al., 2013), we reasoned that identification of cell-autonomous regulators of SOX9 expression should provide insights into progenitor biology. For this, we revisited several published signaling pathways involved in lung development (Eblaghie et al., 2006; Xing et al., 2010) by generating pan-epithelial mutants using ShhCre (Harris et al., 2006) and examining branch morphology and SOX9 expression. We found that pan-epithelial deletion of Bmp4 or Bmpr1a had no detectable phenotype (Fig. S1A) (Alanis et al., 2014). Although the pan-epithelial Tgfbr1 mutant was defective in branching, SOX9 expression was still present in branch tips, suggesting the disruption of other progenitor genes (Fig. S1B). Given that sonic hedgehog (Shh) is considered to signal toward the mesenchyme (Morrisey and Hogan, 2010), these data led us to focus in this study on the CTNNB1-mediated Wnt signaling and Fgf signaling.
To bypass the requirement of Ctnnb1 in lung specification from the foregut (Goss et al., 2009; Harris-Johnson et al., 2009), we induced recombination specifically in the progenitors using Sox9CreER at E11, after the left and right lung buds had extended away from the foregut (Yang and Chen, 2014). We also used a limited dose of tamoxifen to induce mosaic deletion of Ctnnb1 to assess its cell-autonomous function and to minimize secondary effects from gross disruption of mesenchymal signals and tissue morphology. Given that recombination at the Ctnnb1 and Rosa loci does not match with a low dose of tamoxifen, we identified mutant cells by CTNNB1 immunostaining instead of using a Rosa reporter.
We performed a detailed time-course analysis to correlate Ctnnb1 deletion with SOX9 expression (Fig. 1). At 2 days post-tamoxifen injection, whereas CTNNB1 was present throughout the epithelium and mesenchyme in the control lung, the mutant lung had epithelial patches that had lost CTNNB1 expression (Fig. 1, middle). Loss of CTNNB1 correlated precisely with loss of SOX9, with sharp boundaries between control and mutant cells, indicating a cell-autonomous regulation of SOX9 by CTNNB1. Loss of SOX9 was likely to be an immediate consequence of Ctnnb1 deletion because, as early as 1 day after tamoxifen injection, targeted progenitors had lost SOX9 expression (Fig. 1, top). This occurred despite only a small decrease in the level of total CTNNB1 protein, which was only apparent in merged images of CTNNB1 and E-cadherin (ECAD) staining. This suggested that regulation of SOX9 by CTNNB1 depended on a labile pool of CTNNB1 protein, possibly the nuclear pool that mediated the canonical Wnt signaling but was undetectable by our immunostaining protocol because of its low abundance. We also observed that mutant cells formed clusters of ∼5-10 cells on single sections (Fig. 1). Given the observed low frequency of recombination at E12 and the isolated distribution of mutant clusters at E13 and E14, we reasoned that these mutant clusters were derived from a few or even a single recombined cell, suggesting that Ctnnb1 mutant progenitors were still able to proliferate, which was supported by their expression of a proliferation marker, Ki67 (Fig. S2A).
Fig. 1.

Ctnnb1 deletion at E11 leads to cell-autonomous loss of SOX9. Confocal images of immunostained lung sections from E12 (top), E13 (middle) and E14 (bottom) Ctnnb1CKO/−; Sox9CreER/+ mutant and littermate control embryos. Cre recombination was induced at E11 with a low dosage (0.5 mg) of tamoxifen (Tam) to generate a genetic mosaic. The epithelium is marked by ECAD and also identifiable by its compact cell arrangement. Boxed regions are enlarged to show sharp boundaries between SOX9-expressing control and non-SOX9-expressing Ctnnb1 mutant (open arrowheads) cells. Square brackets indicate additional examples of mosaic Ctnnb1 deletion and cell-autonomous loss of SOX9. At E12, loss of CTNNB1 is minimal and only obvious in the merged images with ECAD, which appears purple instead of white. At E14, Ctnnb1 mutant cells are often found in branch stalk regions where SOX9 expression is downregulated. Asterisk indicates tissue loss because of sectioning artifact. Scale bars: 10 µm. At least three sections for each lung and at least three lungs for each genotype were examined, with consistent results.
Given the loss of SOX9 and many other progenitor markers, as described below, we sought a positive marker for the Ctnnb1 mutant cells. We chose the lung identity marker NKX2.1 because the timing of our tamoxifen injection was selected to bypass lung specification when NKX2.1 expression is initially established. Surprisingly, Ctnnb1 mutant cells had a minimal, but detectable, level of NKX2.1; this decrease in NKX2.1 expression occurred in a cell-autonomous manner, as best demonstrated among juxtaposed control and mutant cells (Fig. 2A). Furthermore, Ctnnb1 mutant cells ectopically expressed SOX2, a marker of the anterior GI tract and the airways, again in a cell-autonomous manner (Fig. 2A). The concurrent decreased NKX2.1 expression and ectopic SOX2 expression were substantiated by examining consecutive sections (Fig. S2B). Remarkably, at least a subset of the Ctnnb1 mutant cells expressed liver and gastric genes, such as Alb, Apoa1 and Gkn2 (Fig. 2C), candidate genes selected from our subsequent RNAseq analysis.
Fig. 2.
Ctnnb1 deletion at E11 leads to cell-autonomous downregulation of NKX2.1 and ectopic expression of SOX2 and GI genes. (A) Confocal images of immunostained lung sections from E13 Ctnnb1CKO/−; Sox9CreER/+ mutant and littermate control embryos with Cre recombination induced at E11 with 1 mg of tamoxifen (Tam). The epithelium is marked by ECAD and also identifiable by its compact cell arrangement. Boxed regions are enlarged to show the sharp boundaries between control and Ctnnb1 mutant (open arrowheads) cells and the associated cell-autonomous changes in NKX2.1 and SOX2. Square brackets indicate additional such examples. At least three sections for each lung and at least three lungs for each genotype were examined, with consistent results. (B) OPT projection images of whole-mount immunostained lungs from E15 Ctnnb1CKO/−; Sox9CreER/+ mutant and littermate control embryos with Cre recombination induced at E11 with 1 mg Tam. Boxed regions are enlarged to show the matched branch lineages (RCd.L1.V1 and RCd.L2.V1, dashed circles) where the SOX2-negative branching rosettes in the control are replaced by SOX2-expressing stumps (compare ECAD staining) with reduced NKX2.1 (open arrowheads). Such stumps do not express SOX9 (right panels; branch lineages unknown without SOX2 staining). Asterisk indicates speckle because of staining artifact. The esophagus (eso) separates normally from the trachea. Given the limited cellular resolution of OPT, only branch tips mostly made of Ctnnb1 mutant cells are discernible. At least two lungs for each genotype were examined, with consistent results. (C) Sections of whole-mount in situ hybridization of E13 Ctnnb1CKO/−; Sox9CreER/+ mutant and littermate control lungs with Cre recombination induced by 3.5 mg Tam at E11. Arrowheads indicate depression of GI genes, including Alb, Apoa1 and Gkn2. At least three sections for each lung and at least two lungs for each genotype were examined, with consistent results. (D) Confocal images of immunostained lung sections from E17 Ctnnb1CKO/−; Sox9CreER/+ mutant and littermate control embryos with Cre recombination induced at E11 with 2 mg Tam. The epithelium is marked by ECAD and also identifiable by its compact cell arrangement. Ctnnb1 mutant cells (open arrowheads) are excluded from SOX9-expressing branch tips and limited to the proximal airways. At least three sections for each lung and at least three lungs for each genotype were examined, with consistent results. Scale bars: 10 µm in A,C,D; 250 µm in B.
We also observed that, although induced by the branch tip driver Sox9CreER, Ctnnb1 mutant cells did not occupy the very distal regions, with most branch tips dominated by SOX9-expressing control cells over time, so much so that mutant cells were excluded from tips and limited to the airways by E17 (Fig. 1, bottom; Fig. 2B,D). A notable exception was observed for lateral branches in late-forming domains (Metzger et al., 2008), where branching rosettes were replaced by SOX2-expressing stumps that were devoid of SOX9 progenitors and had a minimal level of NKX2.1 (Fig. 2B), which also confirmed the concurrent changes in SOX9, SOX2 and NKX2.1. Collectively, the decreased NKX2.1 and ectopic SOX2 and GI gene expression manifested as a milder version of the lung specification defects described in Wnt/Ctnnb1 mutants (Goss et al., 2009; Harris-Johnson et al., 2009). This suggests that CTNNB1 is still required after lung specification to maintain the full respiratory fate and to promote SOX9 progenitors.
CTNNB1 is required for Fgf/Kras-dependent promotion of SOX9 progenitors
To address how such CTNNB1-dependent promotion of SOX9 progenitors is related to previous reports of Fgf/Kras-dependent promotion of these cells (Chang et al., 2013; Volckaert et al., 2013), we performed three sets of genetic epistasis analyses of SOX9 progenitors. First, as an expected positive control, we deleted the Fgf receptor, Fgfr2, and expressed the hyperactive KrasG12D allele throughout the lung epithelium using ShhCre (Harris et al., 2006) (Fig. S3). As published previously (Chang et al., 2013), the Fgfr2 single mutant had only left and right main bronchi comprising SOX2-expressing cells with no SOX9 progenitors, whereas the Kras single mutant had overgrown branches made of excessive SOX9 progenitors and ectopic branches off the trachea. Consistent with the signaling cascade downstream of receptor tyrosine kinase receptors that included FGFR2, the Fgfr2 and Kras double mutant resembled the Kras single mutant and had overgrown SOX9-expressing branches and ectopic tracheal branches. This led us to conclude that Kras functions downstream of, and bypasses the requirement for, Fgfr2 in promoting the SOX9 progenitors.
We next expressed the hyperactive KrasG12D allele and deleted Ctnnb1 throughout the lung epithelium using ShhCre (Fig. S4). Consistent with previous reports (Goss et al., 2009; Harris-Johnson et al., 2009), the Ctnnb1 single mutant failed to form NKX2.1-expressing respiratory cells, including SOX9 progenitors, and failed to separate the tracheal and esophageal tubes. The same phenotype was recapitulated in the Ctnnb1 and Kras double mutant and, thus, was dominant over the Kras-induced overgrowth phenotype, supporting the notion that the Kras-dependent promotion of SOX9 progenitors requires their specification by CTNNB1.
Last, in a key experiment to test their epistatic relationship after lung specification and specifically in the progenitors, we used the Sox9CreER driver to activate Kras and delete Ctnnb1 (Fig. 3). This Ctnnb1 and Kras double mutant resembled the Ctnnb1 single mutant with only a few remaining SOX9 progenitors, which were likely escapees from Cre recombination, and an expansion of SOX2 expression to the very distal region. This indicated that Ctnnb1 is required for Fgf/Kras-dependent promotion of SOX9 progenitors after lung specification, a genetic relationship the biochemical basis of which requires future study but possibly results from the requirement of CTNNB1 to maintain the full respiratory fate and SOX9 progenitor state (Fig. 2).
Fig. 3.

CTNNB1 is required for Fgf/Kras-dependent promotion of SOX9 progenitors. OPT projection images of whole-mount immunostained E14 lungs of indicated mutant and littermate control embryos with Cre recombination induced at E11 with 3.5 mg of tamoxifen (Tam). Control and overgrown Kras mutant branch tips express SOX9 (filled arrowheads), whereas both the Ctnnb1 single mutant and the Ctnnb1/Kras double mutant have thwarted SOX2-expressing branch tips (open arrowheads) with a few patches of SOX9 cells that have escaped recombination. The Ctnnb1/Kras double mutant is larger than the Ctnnb1 single mutant, possibly because of faster kinetics of Kras activation than of Ctnnb1 inactivation. Scale bar: 250 µm. At least three lungs for each genotype were examined, with consistent results.
To test whether CTNNB1 activation was sufficient to promote SOX9 progenitors, we activated a stabilized version of CTNNB1 in E11 SOX9 progenitors, which would contribute to most of the lung epithelium by E15 (Rawlins et al., 2009; Alanis et al., 2014). Different from previous mouse models using Spc drivers that were possibly active before E11 (Hashimoto et al., 2012; Ustiyan et al., 2016), we did not observe any changes in branch morphology or expression of SOX9 and SOX2, even though the cartilage rings, which were also targeted by Sox9CreER, were defective in the same mutant lung, and stabilization of CTNNB1 before lung specification using ShhCre completely blocked branching (Fig. S5). This, together with the epistasis experiments above, suggested that CTNNB1-mediated Wnt signaling is permissive for, but not directly downstream of, Fgf/Kras signaling.
Deletion of Ctnnb1 at E13 leads to loss of SOX9 and derepression of GI genes with no change in NKX2.1 and SOX2
The complete loss of NKX2.1 when Ctnnb1 was deleted before lung specification (∼E9) and the partial loss of NKX2.1 when Ctnnb1 was deleted shortly after lung specification (E11) suggest that the respiratory fate of these cells is gradually specified. To test this, we deleted Ctnnb1 at a later stage (E13) specifically in the SOX9 progenitors (Fig. 4A). Similar to the E11 deletion model (Fig. 1), SOX9 expression was lost in a cell-autonomous manner. Although variable in size, patches of phosphorylated ERK (pERK)-positive cells were still present even in branches composed entirely of Ctnnb1 mutant cells (Fig. 4A), suggesting that at least an immediate event downstream of Fgf/Kras signaling was independent of CTNNB1. Unlike the E11 deletion model, there was no decrease in NKX2.1 expression or ectopic SOX2 expression in the very distal region. The apparent smaller distance between the SOX2 domain and the distal edge resulted from the thwarted expansion and branching of SOX9 progenitors in the Ctnnb1 mutant. The same requirement of CTNNB1 for SOX9 expression, but not SOX2 or NKX2.1 expression, was also observed when Ctnnb1 was deleted from E14 and E15 progenitors (Fig. S6A). Therefore, after E13, CTNNB1 continues to promote SOX9 progenitors without affecting NKX2.1 or SOX2 expression. However, the respiratory fate was not completely fixed at E13 because, even though NKX2.1 expression was unaffected, the Ctnnb1 mutant cells still expressed liver and gastric genes, including Alb, Apoa1 and Gkn2 (Fig. 4B). Such cell fate plasticity decreased over time because, when recombination was induced at E15, fewer progenitors ectopically expressed the GI genes (Fig. S6B).
Fig. 4.
Ctnnb1 deletion at E13 leads to cell-autonomous loss of SOX9 with ectopic GI gene expression but no change in NKX2.1 and SOX2 expression. (A) Confocal images of immunostained lung sections from E15 Ctnnb1 mutant and littermate control embryos with Cre recombination induced at E13 with 2 mg of tamoxifen (Tam). The epithelium is marked by ECAD and also identifiable by its compact cell arrangement. Ctnnb1 mutant cells (open arrowheads) lose SOX9 expression without a reduction in NKX2.1 expression or ectopic SOX2 expression. Patches of pERK staining are unaffected in Ctnnb1 mutant branch tips. SOX2 expression in the proximal region of the control lung is expected. At least three sections for each lung and at least three lungs for each genotype were examined, with consistent results. (B) Sections of whole-mount in situ hybridization of E15 Ctnnb1CKO/−; Sox9CreER/+ mutant and littermate control lungs with Cre recombination induced by 3.5 mg Tam at E13. Arrowheads indicate depression of GI genes, including Alb, Apoa1 and Gkn2. At least three sections for each lung and at least two lungs for each genotype were examined, with consistent results. Scale bars: 10 µm in A,B.
Genome-level database mining identifies a 119-gene signature for the SOX9 progenitors
We previously found that SOX9 regulates some progenitor genes (e.g. Clu and Mia), but not others (e.g. Bmp4 and Spry2) (Chang et al., 2013), suggesting the existence of a hierarchical progenitor gene network with SOX9 at a lower level and, thus, controlling a small subset of progenitor genes. Furthermore, the suppression of the Kras overgrown phenotype was more dramatic following Ctnnb1 deletion (Fig. 3) than following Sox9 deletion (Chang et al., 2013). These data led us to hypothesize that CTNNB1 functions further upstream in the progenitor gene hierarchy and regulates additional progenitor genes that are independent of SOX9.
As a first step to construct such a hierarchical gene network, we sought to systematically identify progenitor genes by searching a public database, Eurexpress, which contains in situ hybridization data for nearly 20,000 genes on sagittal sections of E15 mouse embryos (Diez-Roux et al., 2011). We developed a computer script to download images from Eurexpress for any genes on any sections of interest and to assign each gene to a predefined expression pattern (Fig. S7). Although assignment of expression pattern was manual, this process was facilitated by our script to such an extent that 200 genes could be analyzed in an hour. Notably, a small modification of the script allowed expression pattern analysis of any organs, such as the brain (Yackle et al., 2017). Using this script, we identified genes expressed in distinct patterns for the epithelium, mesenchyme and vasculature (Table S1). In particular, we identified 119 genes with the SOX9-like expression pattern and a high signal:noise ratio in Eurexpress images (Table S2). This group was considered a progenitor signature because it included known progenitor genes (e.g. Bmp4, Spry2 and Id2), as well as new ones (e.g. Col18a1 and Mapk6), some of which we confirmed by whole-mount in situ hybridization (Fig. S8).
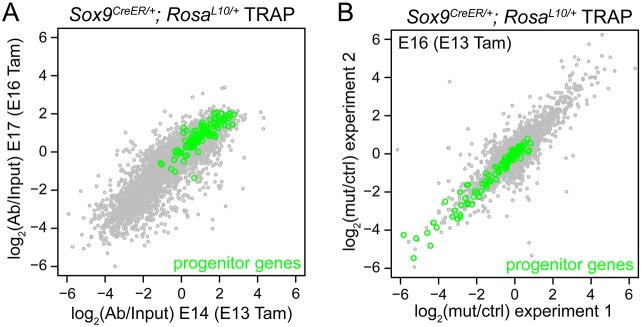
To further validate our progenitor gene list, we adapted the translating ribosome affinity purification (TRAP) method, where a Cre-dependent tagged ribosomal protein is expressed in cell types of interest allowing affinity purification of the associated translating mRNAs, which are considered representative of the total mRNAs (Heiman et al., 2014; Liu et al., 2014). Compared with cell sorting, TRAP has the advantage of purifying the mRNAs in cold tissue homogenate and, thus, minimizes artifacts associated with cell dissociation at a higher temperature. A sufficient amount of mRNAs could be obtained from a single embryonic lung as early as 24 h after recombination by Sox9CreER (Fig. S9). Compared with the total lung homogenate (input), the purified ribosome-bound mRNAs from the Sox9CreER lineage showed an obvious reduction in the expression of mesenchymal and proximal epithelial genes, such as Eln and Sox2, respectively (Fig. S9A). Importantly, RNAseq comparison of the input versus purified mRNAs revealed some of the 119 progenitor genes described above to be among the most enriched genes (GSEA P=3.0×10−45), supporting the general validity of both our Eurexpress screen and progenitor TRAP method (Fig. 5A, Table S3). We note that the fold changes in enriched progenitor genes, including Sox9, were smaller than those of the depleted genes, presumably because of the abundance of progenitor cells in embryonic lungs.
Table 1.
Eurexpress screen summary

Fig. 5.
Sox9CreER TRAP-RNAseq reveals enrichment of the progenitor gene signature and its downregulation in the Ctnnb1 mutant. (A) Scatterplot of log2-fold change in fragments per kilobase of transcript per million mapped reads (FPKM) values comparing GFP antibody-purified (Ab) versus input mRNAs from E14 (x-axis) and E17 (y-axis) RosaL10/+; Sox9CreER/+ lungs with Cre recombination induced by 2 mg of tamoxifen (Tam) 1 day before. Progenitor genes from the Eurexpress screen (Table 1) are highlighted in green and some are among the most enriched genes (P=3.0×10−45, gene set enrichment analysis). (B) Scatterplot of log2-fold change in FPKM values comparing GFP antibody-purified mRNAs from mutant (Ctnnb1CKO/−; RosaL10/+; Sox9CreER/+) versus littermate control (Ctnnb1CKO/+; RosaL10/+; Sox9CreER/+) lungs. Cre recombination is induced by 3.5 mg Tam at E13 and mRNAs are purified at E16. Progenitor genes from the Eurexpress screen (Table 1) are highlighted in green, a subset of which are among the most downregulated genes (P=4.9×10−22, gene set enrichment analysis). In A,B, the tight distribution along the diagonal indicates good reproducibility between biological replicates. Only genes with an average FPKM value greater than 1 are plotted.
CTNNB1 is required for a subset of progenitor genes as revealed by TRAP-RNAseq and confirmed by in situ hybridization
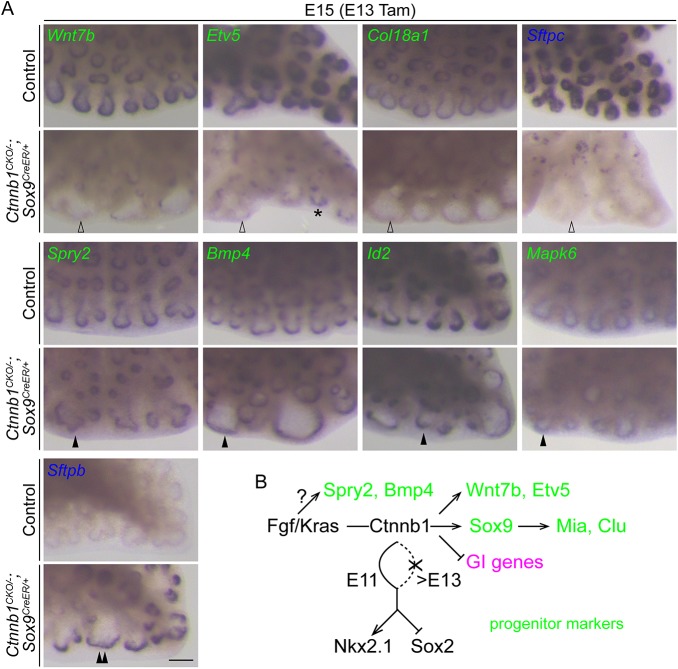
As reasoned above, we hypothesized that these progenitor genes organize into a hierarchical regulatory network that defines the SOX9 progenitors and collectively ensures their full expansion and branching capability. To test this hypothesis and to begin to construct the hierarchical progenitor gene network, we examined changes in gene expression after Ctnnb1 deletion using TRAP-RNAseq, which again profiles cell-specific translating mRNAs as a proxy of the total mRNAs without cell dissociation (Heiman et al., 2014; Liu et al., 2014). Given that Ctnnb1 deletion at E11 led to the downregulation of NKX2.1, and genetic deletion of Nkx2.1 led to the cell-autonomous loss of SOX9 (Fig. 2, Fig. S10), we induced recombination at E13, a time when NKX2.1 expression was no longer affected (Fig. 4A), to separate the requirement of CTNNB1 for progenitor genes from that for NKX2.1 expression, as well as to obtain sufficient mRNAs for RNAseq. To filter out secondary changes in nonepithelial cells, we used our progenitor TRAP method and administered a high dose of tamoxifen to recombine both the Ctnnb1CKO and RosaL10 alleles in most progenitors (Fig. S9B). We purified mRNAs at E16 to allow enough time for mRNA degradation and dilution, albeit with the trade-off of additionally targeting SOX2-expressing progeny of the SOX9 progenitors. Nevertheless, RNAseq comparisons of control versus Ctnnb1 mutant SOX9-lineage cells revealed that some of our 119 progenitor genes were among the most downregulated in the mutant (GSEA P=4.9×10−22; Fig. 5B, Table S4), some of which were confirmed by whole-mount in situ hybridization (Fig. 6A) and included genes, such as Wnt7b and Etv5, that were not affected in the Sox9 mutant (Chang et al., 2013). However, a subset of these progenitor genes (e.g. Spry2 and Bmp4) were still unaffected and, thus, independent of CTNNB1 (Fig. 6A), supporting the notion that these progenitor genes are organized in a hierarchical manner and additional regulators function upstream of CTNNB1.
Fig. 6.
CTNNB1 is required for a subset of progenitor genes. (A) Whole-mount in situ hybridization of E15 Ctnnb1CKO/−; Sox9CreER/+ mutant and littermate control lungs with Cre recombination induced by 3.5 mg of tamoxifen (Tam) at E13. A subset of progenitor genes (green; top row) depend on Ctnnb1 (open arrowheads), whereas others (green, middle row) are independent on Ctnnb1 (filled arrowheads). Asterisk indicates small epithelial patches with normal gene expression from cells that have escaped recombination. A marker of differentiated alveolar type 2 cells (Sftpb; blue; bottom row) is not detected in the control lung at this developmental stage, but is derepressed in the Ctnnb1 mutant progenitors (double arrowhead), whereas another differentiation marker (Sftpc; blue; top row) is expressed at a detectable level in the control progenitors but lost in the Ctnnb1 mutant progenitors. Scale bar: 100 µm. At least two lungs for each genotype were examined, with consistent results. (B) An initial hierarchical gene network of the SOX9 progenitors. CTNNB1 mediates Fgf/Kras-dependent promotion of the SOX9 progenitors and regulates a subset of progenitor genes (green). CTNNB1 is required to maintain normal NKX2.1 expression and suppress ectopic SOX2 expression at the early (E11), but not later (after E13), stage post-lung specification. CTNNB1 also suppresses GI genes, including Alb, Apoa1 and Gkn2 (magenta).
Interestingly, although CTNNB1 was upstream of SOX9 (Figs 1 and 4) and SOX9 suppressed the precocious expression of alveolar differentiation genes, including Sftpc, Sftpb and Lamp3 (Chang et al., 2013), only Sftpb was upregulated in the Ctnnb1 mutant, whereas Sftpc and Lamp3 were downregulated (Fig. 6, Fig. S11). This apparent paradox raised the possibility that CTNNB1 also promoted alveolar differentiation, a function that was in addition to, and dominant over, its SOX9-mediated suppression of Sftpc and Lamp3 expression. Consistent with this possibility, Ctnnb1 mutant cells lost LAMP3 expression and were defective in forming lamella bodies, a hallmark of mature alveolar type 2 cells (Fig. S11), reminiscent of a recent report deleting Ctnnb1 in postnatal alveolar type 2 cells (Frank et al., 2016).
Wnt signaling is required to maintain NKX2.1 expression and suppress GI genes in lung progenitors generated from human ESCs
Finally, to test whether the requirement of CTNNB1 and the presumed Wnt signaling in maintaining NKX2.1 expression in the mouse lung can be extended to human cells, we used an established culture system that generates NKX2.1-expressing lung progenitors from human ESC line RUES2 at an efficiency of more than 95% (Huang et al., 2014, 2015). We withdrew a Wnt agonist CHIR99021 (Bennett et al., 2002; Huang et al., 2014; Naujok et al., 2014; McCauley et al., 2017) on day 12 of our culture protocol, when the lung progenitors are already specified because most of the cells expressed NKX2.1 and its expression level did not further increase at day 15, as measured by RNAseq (Fig. 7A, Table S5). We observed a marked loss of NKX2.1 at day 15 in the absence of CHIR99021 (Fig. 7A), suggesting that Wnt signaling is also required to maintain NKX2.1 expression in specified human ESC-derived lung progenitors. RNAseq analysis confirmed the downregulation of NKX2.1 and revealed the upregulation of multiple GI genes; however, these GI genes were not the same ones upregulated in the Ctnnb1 mutant mouse lung, possibly because of differences between species or culture versus in vivo conditions (Fig. 7B, Table S5). Interestingly, the human orthologs of the mouse progenitor gene signature had a low expression level and contained a comparable number of upregulated and downregulated genes, possibly because of the lack of clear branching structures and bona fide branching tip/stalk genes in cultured human lung progenitors (Fig. 7B,C, Table S6). Further inhibition of endogenous WNT production by IWP2 led to comparable changes in NKX2.1, GI genes and SOX9-like progenitor genes (Fig. S12, Table S5).
Fig. 7.
Wnt signaling is required to maintain NKX2.1 expression and suppresses GI genes in human ESC-derived lung cells. (A) (Top) Schematics of the culture protocol and samples analyzed (orange). See Materials and Methods section for details. (Bottom) Immunostained differentiated REUS2 cells at day (d) 12 (first column) and cultured for 3 additional days with (middle column) or without (last column) the Wnt agonist CHIR99021 (CHIR). The number and percentage of NKX2.1-positive cells are shown. Despite robust NKX2.1 expression at d12, Wnt agonist withdrawal causes a significant decrease in NKX2.1 positive cells: P<10−15 with versus without CHIR (χ2-squared test). Scale bar: 50 µm. At least three regions for each condition and an independent differentiated culture were examined, with consistent results. (B) Scatterplot of log2-fold change in RNAseq FPKM values comparing d15 without CHIR (noCHIR) versus d12 control (ctrl; x-axis) or d15 control (y-axis). Gene names are approximately aligned horizontally to the corresponding color-coded data points to avoid interference with the scatterplot. Blue indicates GI genes according to annotations by the Human Protein Atlas (www.proteinatlas.org). Green indicates the human orthologs of the mouse progenitor genes, which are not consistently downregulated in comparison to the Ctnnb1 mutant mouse lung (Fig. 5B). Only genes with an average FPKM value greater than 1 are plotted. (C) Scatterplot of average log2-fold change in FPKM values of mouse versus human progenitor genes, showing that genes downregulated in the Ctnnb1 mutant mouse lung are not consistently downregulated in the noCHIR human cells. The mouse data are the average of the two data sets in Fig. 5B and the human data are the average of the two data sets in Fig. 7B (see Table S6 for numbers). Only genes with an average FPKM value greater than 1 are plotted.
DISCUSSION
Our data support a central role of CTNNB1 in maintaining lung epithelial progenitors after their foregut specification: it promotes a hierarchical progenitor gene network, suppresses the dormant GI fate, and regulates NKX2.1 and SOX2 expression in a developmental stage-dependent manner (Fig. 6B). Notably, Wnt signaling similarly maintains the lung progenitor fate and suppresses the GI fate in specified ESC-derived human lung progenitors. Our study establishes an experimental path to incorporate other signaling pathways into our initial progenitor network, including epithelial-mesenchymal crosstalk, and to further explore its relevance in human lung development and regeneration (Morrisey and Hogan, 2010; Ornitz and Yin, 2012; Herriges et al., 2015).
In the classical Waddington's landscape of cell fate determination (Waddington, 1942), emphasis is usually placed on bifurcation nodes and the associated signaling events, whereas the internode downhill roll is often taken for granted as the result of ‘gravity’. In the lung, demonstration of CTNNB1/Wnt signaling as the key event in specifying epithelial progenitors from the embryonic foregut (Goss et al., 2009; Harris-Johnson et al., 2009) has focused current research on the morphogenesis and differentiation of these progenitors, whereas their lung fate is generally considered fixed (Morrisey and Hogan, 2010). However, our data show unexpectedly that CTNNB1 continues to be required to maintain the lung fate and suppress the GI fate of cells: the ‘momentum’ past the bifurcation node in the Waddington's landscape. Interestingly, CTNNB1 is no longer required after E13 for NKX2.1 expression, which is presumably sustained by alternative mechanisms. Future analyses of the epigenetic state of Nkx2.1 should shed light on its gradual independence from CTNNB1. More broadly, a detailed time-course analysis of the genomics and epigenomics of the progenitors will better define their normal specification and maintenance, as well as their aberrant lung-GI hybrid fate.
Besides the lung versus GI fate choice, maintenance of lung progenitors also requires precise control of the entire 119-progenitor gene program. Indeed, a larger subset of these progenitor genes is downregulated in the Ctnnb1 mutant (this study) than in the Sox9 mutant (Chang et al., 2013), consistent with the different severity in the branching defect between these mutants. It is conceivable that, depending on where in the progenitor program hierarchy a given gene acts, the resulting mutant will exhibit a branching defect accordingly; this is now a testable idea, given the availability of the described progenitor gene signature.
What are the molecular events underlying the CTNNB1-dependent regulation of SOX9 progenitors and its epistasis relationship with Fgf/Kras signaling? A previous mouse model of whole-lung overexpression of Fgf10 and Dkk1, a Wnt signaling inhibitor, suggests that FGF10 directly activates CTNNB1 via AKT-mediated phosphorylation (Volckaert et al., 2013). By contrast, another study proposed that Forkhead box M1 (FOXM1), as a target of Kras, inhibits canonical Wnt signaling (Wang et al., 2012). However, we show that CTNNB1 stabilization is insufficient to expand SOX9 progenitors or disrupt branching morphogenesis (Fig. S5). Thus, it is possible that CTNNB1-mediated Wnt signaling constitutes a permissive factor on the promoter/enhancer of progenitor genes, such as Sox9. Chromatin immunoprecipitation experiments should reveal the mechanisms underlying our genetic and gene expression data, but are technically challenging because they require purification of the limited number of the lung progenitors, given the ubiquitous expression of CTNNB1 and its likely distinct function in different cell types.
The mature lung epithelium consists of proximal airways and distal alveoli; the embryonic lung epithelium consists of the proximal SOX2-expressing future airways and the distal SOX9 progenitors that give rise to both the airways and alveoli. The proportion of the proximal and distal compartments in either the mature or embryonic lung is frequently disrupted in lung malformation mutants. Although this resembles the classical proximal-distal patterning in the Drosophila embryos that is determined by morphogen gradients (St Johnston and Nusslein-Volhard, 1992), the proximal-distal proportion in the developing lung differs in that it is not a one-time decision made after branching is complete. Instead, as branching proceeds, the distal SOX9 progenitors continuously leave behind daughter cells that turn off SOX9 and turn on SOX2, such that the proximal-distal boundary continuously shifts distally (Yang and Chen, 2014). According to this model, alteration of the proximal-distal proportion can result from the less efficient expansion of SOX9 progenitors without affecting their differentiation into SOX2-expressing daughter cells, a scenario different from the proximal-distal patterning in the Drosophila embryos, where the expansion of one compartment occurs at the expense of the other compartment. Indeed, SOX9 progenitor expansion is defective in both the Ctnnb1 mutant (this study) and, to a lesser extent, in the Sox9 mutant (Chang et al., 2013), whereas their SOX2-expressing daughter cells are still restricted to the proximal region. Perhaps more direct evidence is that, in the Ctnnb1 mosaic deletion model, the Ctnnb1 mutant cells in the distal region lose SOX9 expression without expressing SOX2 (Fig. 4) and are defective progenitors that still express CTNNB1-independent progenitor genes (Fig. 6). The notable exception is the early (E11) Ctnnb1 mosaic deletion model, in which loss of SOX9 precisely correlates with ectopic expression of SOX2 in the distal Ctnnb1 mutant cells (Fig. 2, Fig. S2). However, this is associated with the downregulation of NKX2.1 and enhancement of the GI fate (Figs 2 and 4), for which SOX2 can also be a marker. Such temporal difference should be taken into account when examining Spc-rtTA-induced mutants that receive doxycycline at early developmental stages. Similar mosaic analyses of other lung mutants with an abnormal proximal-distal proportion are expected to pinpoint the underlying changes at the level of individual cells.
Given the central importance of NKX2.1 expression in differentiating lung cells from human ESCs and induced pluripotent cells, elucidating the signals and epigenetic marks underlying NKX2.1 expression should facilitate protocol optimization (Huang et al., 2014; Jacob et al., 2017; McCauley et al., 2017). Indeed, we show that, similar to their mouse counterparts, human ESC-derived lung progenitors also depend on CTNNB1-mediated Wnt signaling to maintain NKX2.1 expression and suppress GI genes (Fig. 7), an example of conservation and parallels between in vivo mouse lung development and in vitro human lung cell generation. However, the 119-progenitor gene signature is not consistently affected by loss of Wnt signaling in cultured human lung progenitors. Although such discrepancy between mice and humans might be attributed to species difference (Nikolic et al., 2017), it is also conceivable that, in vivo, the progenitor gene network is mostly used to promote progenitor branching, a structural requirement that is bypassed in culture. If true, progenitor expansion in culture is uncoupled from branching and might be largely driven by the proliferative nature of stem cells, reminiscent of the unstructured expansion of naïve ESCs in culture. As a result, cultured lung cells skip branching morphogenesis and transition directly from NKX2.1 cell specification to alveolar/airway differentiation, a process analogous to the in vivo development of the frog lung (Chang et al., 2013). A thorough comparison of in vivo mouse lungs and in vitro human lung cells, especially the role of Wnt signaling, is necessary to minimize potential GI differentiation from Wnt signaling manipulation (Jacob et al., 2017; McCauley et al., 2017).
MATERIALS AND METHODS
Mice
The following mouse (Mus musculus) strains were used: Bmpr1aCKO (Mishina et al., 2002), Ctnnb1CKO (Brault et al., 2001), Ctnnb1Ex3 (Harada et al., 1999), Fgfr2CKO (Yu et al., 2003), KrasG12D (Jackson et al., 2001), Nkx2.1CKO (Kusakabe et al., 2006), RosaL10 (Liu et al., 2014), ShhCre (Harfe et al., 2004), Sox9CreER (Soeda et al., 2010) and Tgfbr1CKO (Larsson et al., 2001). Mice carrying the Ctnnb1CKO conditional allele were mated to female HprtCre/+ mice (Tang et al., 2002) to generate mice carrying the germ line Ctnnb1 null allele (Ctnnb1−). The day of observance of a vaginal plug was designated as E1. To induce Cre recombination, tamoxifen (T5648, Sigma) dissolved in corn oil (C8267, Sigma) was injected intraperitoneally. The tamoxifen dosage varied depending on the developmental stage and the desired level of recombination and is specified in the figure legends. In general, 0.5-2 mg tamoxifen was used to induce mosaic recombination and 3-3.5 mg tamoxifen for near-complete recombination. All replicates were control-mutant lungs from different animals. Outliers were excluded only if there were technical errors, such as failed immunostaining. The number of control-mutant pairs and sections analyzed is stated in the figure legends. Unless specified in the text, littermate controls included heterozygotes mice regardless of Cre or homozygotes without Cre. Investigators were not blind to the genotypes. Control and mutant samples were processed in the same tube or block to minimize experimental variations. No power analysis was used to determine the sample size. All animal experiments were approved by the institutional animal care and use committee at Texas A&M Health Science Center Institute of Biosciences and Technology.
Antibodies
The following antibodies were used for immunostaining: rabbit anti-ATP-binding cassette, sub-family A, member 3 (ABCA3, 1:1000, WRAB70565, Seven Hills Bioreagents), rabbit anti-β-catenin (CTNNB1, 1:1000, sc-7199, Santa Cruz Biotechnology), goat anti-β-catenin (CTNNB1, 1:500, AF1329, R&D Systems), chicken anti-green fluorescent protein (GFP, 1:1000, AB13970, Abcam), rat anti-E-cadherin (ECAD, 1:2000, 131900, Invitrogen), Alexa488-conjugated rat anti-E-cadherin (ECAD, 1:250, 53-3249-80, eBioscience), eFluor570-conjugated rat anti-Mki67 (KI67, 1:500, 41-5698-82, eBioscience), rat anti-lysosomal-associated membrane protein 3 (LAMP3, 1:1000, DDX0192, Imgenex), rabbit anti-phospho-ERK1/2 (pERK, 1:250, 4370, Cell Signaling Technology), rabbit anti-NK2 homeobox 1 (NKX2.1, 1:1000, sc-13040, Santa Cruz Biotechnology), rabbit anti-NK2 homeobox 1 (NKX2.1, 1:1000, WRAB-1231, Seven Hills Bioreagents), rabbit anti-SRY-box-containing gene 9 (SOX9, 1:2000, AB5535, Millipore), goat anti-SRY-box-containing gene 9 (SOX9, 1:1000, AF3075, R&D Systems) and goat anti-SRY-box-containing gene 2 (SOX2, 1:250, sc-17320, Santa Cruz Biotechnology).
Section and whole-mount immunostaining
Immunostaining was performed following published protocols with minor modifications (Chang et al., 2013; Alanis et al., 2014). Dissected embryo torsos were opened along the sternum and fixed in phosphate-buffered saline (PBS, pH 7.4) with 0.5% paraformaldehyde (PFA; P6148, Sigma) for 3 h on a rocker at room temperature. For section immunostaining, dissected lungs were cryoprotected in 20% (wt/vol) sucrose in PBS overnight at 4°C and embedded in optimal cutting temperature compound (OCT; 4583, Tissue-Tek). OCT sections at 10 µm thickness were blocked in PBS with 0.3% Triton X-100 and 5% normal donkey serum (017-000-121, Jackson ImmunoResearch) and then incubated with primary antibodies diluted in PBS with 0.3% Triton X-100 in a humidified chamber at 4°C overnight. The sections were washed with PBS in a coplin jar for 1 h and incubated with secondary antibodies (1:1000, Jackson ImmunoResearch or Invitrogen) and 4′,6-diamidino-2-phenylindole (DAPI, 0.5 µg/ml final concentration) diluted in PBS with 0.3% Triton X-100 at room temperature for 2 h. After another 1 h wash with PBS, the sections were mounted with Aquamount (18606, Polysciences).
For whole-mount immunostaining, dissected lungs were blocked in 2 ml Eppendorf tubes containing PBS with 0.3% Triton X-100 and 5% normal donkey serum and then incubated with primary antibodies diluted in PBS with 0.3% Triton X-100 on a rocker at 4°C overnight. The following day, tissue was washed in PBS with 1% Triton X-100 and 1% Tween-20 for 3-6 h and incubated with secondary antibodies diluted in PBS with 0.3% Triton X-100 on a rocker at 4°C overnight. On the third day, tissue was washed as before and fixed with 2% PFA in PBS for ∼2-3 h. To minimize experimental variation, littermate control and mutant lungs were embedded in the same OCT block or whole-mount stained in the tube. Images were captured on an optical projection tomography (OPT) microscope (Bioptonics) as published previously (Chang et al., 2013; Alanis et al., 2014) or confocal microscopes (FV1000, Olympus; A1plus, Nikon).
Whole-mount in situ hybridization
This was performed essentially as previously published (Chang et al., 2013; Alanis et al., 2014). Briefly, PFA-fixed embryo torsos were dissected in PFA, lungs were rinsed once with diethylpyrocarbonate-treated PBS (DEPC-PBS), dehydrated quickly through a methanol/DEPC-PBS gradient and stored in methanol at −20°C. Samples were rehydrated in a methanol/DEPC-PBS gradient at room temperature and permeabilized in DEPC-PBS with 10 µg/ml protease K for 15 min for E15 lungs or 20 µg/ml protease K for 20 min for E17 lungs. Samples were then fixed in PBS with 4% PFA and 0.25% glutaraldehyde for 20 min and then washed with DEPC-PBS twice and a rinse solution (500 µl 4×SSC, 50% formamide, 0.1% Tween-20) once. Samples were blocked in prehybridization solution (50% formamide, 4×SSC, 1×Denhardt's solution, 250 µg/µl salmon sperm DNA, 250 µg/µl yeast tRNA, 50 µg/ml heparin, 0.1% Tween-20) at 62°C for 1 h in a slide oven. Subsequently, 1 µg/ml riboprobes diluted in 1 ml of hybridization solution (prehybridization solution with 10% >500 kDa dextran sulfate) was preheated at 70°C for 5 min, chilled on ice and then added to samples followed by incubation at 62°C in a slide oven overnight. The next day, samples were washed in 50% formamide with 4×SSC and 0.1% Tween-20 at 65°C twice for 1 h, incubated in 100 µg/ml RNase A diluted in 100 mM Tris (pH 7.5) with 500 mM sodium chloride and 0.1% Tween-20 at 37°C for 1 h, washed in 50% formamide with 2×SSC and 0.1% Tween-20 at 65°C three times for 2 h and then washed again overnight in a water bath. The following day, samples were washed in TBST (100 mM pH 7.5 Tris, 150 mM sodium chloride, 0.1% Tween-20) twice for 10 min, followed by blocking with TBST+5% normal sheep serum for 1 h. An anti-digoxigenin alkaline phosphatase Fab fragment (1:2000, Roche, 11093274910) was then added to TBST for 2 h followed by TBST washes four times for 2 h and an overnight wash in TBST with 0.5 mg/ml levamisole (L9756, Sigma) at 4°C. On the final day, samples were washed with TBST for 1 h followed by detection of alkaline phosphatase activity with Nitroblue Tetrazolium and 5-bromo-4-chloro-3′-indolylphosphate (Roche, 11681451001) in the alkaline phosphatase reaction buffer (100 mM pH 9.5 Tris, 100 mM sodium chloride, 5 mM magnesium chloride). Following the alkaline phosphatase reaction, samples were fixed in 4% PFA in PBS overnight, cleared via a 25%-50%-75% glycerol gradient and mounted on a depression well slide for imaging. Digoxigenin-labeled riboprobes (Roche, 11277073910) were transcribed with a T7 RNA polymerase (18033-019, Invitrogen) using cloned cDNAs or PCR products as templates. To minimize experimental variation, littermate control and mutant lungs were processed in the same tube for each riboprobe throughout the in situ hybridization experiment. Images were taken on a stereomicroscope (M80, Leica) or an upright microscope (BX60, Olympus). When indicated, 75% glycerol-cleared lungs were embedded in OCT, sectioned at 10 µm, mounted with Aquamount after drying, and imaged on an upright microscope (BX60, Olympus).
Eurexpress database screen
A custom Perl script was developed to generate a user interface that downloaded user-specified images from Eurexpress and recorded users' annotations of expression patterns. Given that in situ signals would vary depending on probe strength and colorimetric reaction time, the gene expression levels (0-3) were defined as qualitative assessment of the relative strength of the in situ signal to the nonspecific background, such that genes ubiquitously expressed were assigned a low expression level (0 or 1), whereas genes with clear cell type specificity were assigned a high expression level (2 or 3). Only genes with the highest cell type specificity (level 3) were reported. The progenitor genes were combined in groups C (distal tip+stalk) and D (distal tip) of the original expression patterns because of difficulty in assessing branch tips and stalks on sections. Some genes were expressed in more than one tissue compartment. The Perl script is available upon request.
TRAP-RNAseq
Embryonic lung TRAP was performed as published previously, with minor modifications (Heiman et al., 2014; Liu et al., 2014). Sox9CreER lineage-labeled embryos were screened under a fluorescence stereomicroscope (M205C, Leica) and GFP-positive lungs were quickly dissected to remove extrapulmonary tissues and tails, which were saved for genotyping. In separate tubes, each lung was homogenized in 500 µl polysome extraction buffer [20 mM pH 7.4 HEPES, 150 mM potassium chloride, 10 mM magnesium chloride, EDTA-free protease inhibitor (Roche 5892791001), 1% NP-40, 0.5 mM DTT, 1 mg/ml heparin (Sigma, H3393), 80 U/ml RNasin (Promega N2615), 100 µg/ml cycloheximide (freshly dissolved in methanol as 100 mg/ml, Sigma, C7698)]. The lung homogenate was centrifuged at 4°C for 10 min at 2000 rpm (350 g) and the supernatant centrifuged for another 10 min at 13,000 rpm (14,500 g). The resulting supernatant was precleared with 10 µl protein G Dynabeads (Invitrogen, 10004D) with rotation at 4°C for 30 min. While preparing this lung homogenate, 0.5 µg GFP antibodies (rabbit anti-GFP from Invitrogen, G10362; or mouse anti-GFP from Antibody & Bioresource Core Facility at Memorial Sloan Kettering Cancer Center, HTZ-19C8) were allowed to bind to 5 µl protein G Dynabeads in binding buffer (20 mM pH 7.4 HEPES, 150 mM potassium chloride, 10 mM magnesium chloride, 1% NP-40, 0.5 mM DTT, 100 µg/ml cycloheximide) with rotation at 4°C for 1 h and subsequently washed with binding buffer four times in 15 min. Precleared lung homogenate was incubated with GFP antibodies prebound to protein G Dynabeads with rotation at 4°C for 1 h and subsequently washed with wash buffer (20 mM pH 7.4 HEPES, 350 mM potassium chloride, 10 mM magnesium chloride, 1% NP-40, 0.5 mM DTT, 100 µg/ml cycloheximide) six times in 30 min. During the final wash, Dynabeads were transferred to a new tube and 1 ml Trizol reagents (Invitrogen 15596018) were added to extract the RNAs using an RNeasy Micro kit (Qiagen, 74004). Throughout the immunoprecipitation procedure, 50 µl precleared lung homogenate was kept on ice and its RNA extracted to assess RNA integrity and serve as input control.
RT-PCR was performed using a SuperScript IV First-Strand Synthesis System (Invitrogen, 18091050). Samples from both the rabbit (Ab1) and mouse (Ab2) anti-GFP antibodies were used in the RT-PCR experiments, whereas only the rabbit anti-GFP antibody samples were used for RNAseq because of higher RNA yield compared with the mouse antibody samples. An RNAseq library was prepared using ∼100-200 ng total RNA and an mRNA isolation kit (New England BioLabs, E7490) and a NEBNext Ultra RNA library prep kit (New England BioLabs, E7530S) with a final double (0.65×–1×bead volume) size selection step using a SPRIselect reagent kit (Beckman Coulter, B23318). The libraries were bar-coded (New England BioLabs, E7335S) and sequenced on an Illumina HiSeq 4000 or NextSeq 500 at the UT MD Anderson Sequencing and Microarray Facility to obtain 20 million-30 million 76-nucleotide pair-end reads for each sample. Reads were aligned to the mm10 Mus musculus genome assembly and transcriptome using a bowtie2, cufflinks and cuffdiff pipeline (Trapnell et al., 2010, 2013; Song et al., 2014) at the MD Anderson High Powered Computing core facility. Gene set enrichment analysis (GSEA) was performed using the GAGE package in an R environment (Luo et al., 2009). Genes with an average FPKM value greater than 1 were plotted. The Mouse Genome Informatics batch converter (http://www.informatics.jax.org/batch) was used to obtain the official gene names for annotated progenitor genes from Eurexpress and duplicated genes removed before plotting. Three progenitor gene names were matched manually: Cnmd (official) for Lect1 (Eurexpress), Epb41l3 for Epb4.1l3 and Stk26 for 2610018G03Rik.
Transmission electron microscopy
Dissected embryonic lungs were fixed with a solution containing 3% glutaraldehyde plus 2% paraformaldehyde in 0.1 M cacodylate buffer, pH 7.3, for 1 h. After fixation, the samples were washed and treated with 0.1% Millipore-filtered cacodylate buffered tannic acid, post-fixed with 1% buffered osmium tetroxide for 30 min, and stained en bloc with 1% Millipore-filtered uranyl acetate. The samples were dehydrated in increasing concentrations of ethanol, infiltrated, and embedded in LX-112 medium. The samples were polymerized in a 60°C oven for 2 days. Ultrathin sections were cut in a Leica Ultracut microtome, stained with uranyl acetate and lead citrate in a Leica EM Stainer, and examined in a JEM 1010 transmission electron microscope (JEOL) at an accelerating voltage of 80 kV. Digital images were obtained using AMT Imaging System (Advanced Microscopy Techniques).
Directed differentiation of human lung progenitors from hESC
Human lung progenitors were derived from Rockefeller University Embryonic Stem Cell Line 2 [RUES2, NIH Registration number 0013, a gift from Dr Ali Brivanlou (Rockefeller University, NY, USA), tested for contamination] as previously described with small modifications (Huang et al., 2014, 2015). Briefly, RUES2 were maintained on irradiated mouse embryonic fibroblasts (Global Stem, GSC-6001G) before starting a differentiation experiment. Definitive endoderm (DE) induction started on day 1 and took 3 days. DE induction medium contained 10 μM Y-27632 dihydrochloride, 0.5 ng/ml human BMP4, 2.5 ng/ml human bFGF (FGF2) and 100 ng/ml human Activin A. DE induction was performed in six-well Ultra-Low Attachment plates to form embryoid bodies (EBs). On day 4, EBs were dissociated with trypsin into single cells and plated onto 24-well fibronectin-coated tissue culture plates for anterior foregut endoderm (AFE) induction and subsequent lung progenitor induction. For AFE induction on days 4-6, cells were cultured in the presence of 2 μM dorsomorphin dihydrochloride (BMP signaling antagonist; abbreviated as DSM) and 10 μM SB431542 (TGFβ signaling antagonist; abbreviated as SB) for 24 h, and then switched to 24 h of 10 μM SB431542 and 1 μM IWP2 (Wnt production inhibitor) treatment. Day 6-15 lung progenitor induction and expansion medium contained 3 μM CHIR99021 (WNT signaling agonist), 10 ng/ml human FGF10, 10 ng/ml human FGF7 (or KGF), 10 ng/ml human BMP4 and 50 nM all-trans retinoic acid (ATRA). All the cytokines were from R&D Systems and all the small molecules were from Tocris (R&D Systems). To examine the role of the CTNNB1-mediated Wnt signaling pathway in maintaining NKX2.1-expressing lung progenitors, at day 12 of the differentiation, some cultures were switched to a medium lacking the WNT signaling agonist (i.e. no CHIR99021) or additionally containing 1 μM IWP2 (no CHIR99021 and with IWP2), but in the presence of the other lung induction morphogens (FGF7, FGF10, BMP4 and ATRA), and carried in such medium for 3 days before immunofluorescence analysis of NKX2.1 expression at day 15 (see schematics in Fig. 7A). As reported previously, around day 15 of the differentiation, the cultured cells became confluent and required replating (Huang et al., 2014, 2015). In the current study, differentiated RUES2 cells analyzed on day 15 were replated on day 10.
For NKX2.1 immunostaining, cells were fixed with 4% paraformaldehyde, washed with PBS, permeabilized with 0.25% Triton and 2% normal donkey serum (Jackson ImmunoResearch), and blocked in 2% normal donkey serum. The cell cultures were stained with primary antibody NKX2.1 in 2% normal donkey serum in PBS at 4°C overnight. The cultures were then incubated with donkey anti-rabbit whole IgG-Alexa Fluor 488 (Jackson ImmunoResearch) at 1:500 dilutions in 2% normal donkey serum at room temperature for 2 h, washed twice and incubated with DAPI for 5 min at room temperature. The stained cultures were preserved in VECTASHIELD Mounting Media (Vector Laboratories) in the dark at 4°C. Stained samples were imaged using the Leica DMi8 Automated Inverted Microscope System. All images were acquired with HC PL FL L 20×/0.40 CORR PH1 objective. The images were exported as JPG files, processed (contrast and brightness adjustments) and quantified with ImageJ (NIH). A total number of ∼600-1500 cells were scored for each of the control and experimental groups, and the percentage of NKX2.1 nuclei over DAPI were reported. Statistical significance was determined using the χ2 test.
Supplementary Material
Acknowledgements
We thank Drs Myriam Heiman (MIT) and Herman A. Dierick (Baylor College of Medicine) for advice on TRAP, and the Antibody & Bioresource Core Facility at Memorial Sloan Kettering Cancer Center for the mouse anti-GFP antibody. The Eurexpress screen was initiated when J.C. was a postdoctoral fellow in Dr Mark Krasnow's lab at Stanford University and was completed in his own lab at MD Anderson Cancer Center. The University of Texas MD Anderson Cancer Center DNA Analysis Facility and High Resolution Electron Microscopy Facility are supported by the Cancer Center Support Grant (CA #16672). We thank Kenneth Dunner Jr for help with transmission electron microscopy.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: E.J.O., S.X.L.H., J.C.; Methodology: E.J.O., D.R.L., K.N.G.-M., E.A.S., R.R.-C., E.A., S.E.H., S.X.L.H., J.C.; Software: E.J.O.; Validation: E.A., J.C.; Formal analysis: E.J.O., D.R.L., K.N.G.-M., E.A.S., R.R.-C., E.A., S.E.H., S.X.L.H., J.C.; Investigation: E.J.O., D.R.L., K.N.G.-M., E.A.S., R.R.-C., S.E.H., N.R.F.-C., S.X.L.H., J.C.; Resources: H.A., S.K., S.X.L.H.; Writing - original draft: J.C.; Writing - review & editing: E.J.O., S.M.H., S.K., S.X.L.H., J.C.; Supervision: S.M.H., S.X.L.H., J.C.; Project administration: J.C.; Funding acquisition: J.C.
Funding
This work was supported by the University of Texas System Rising STARS Award, the University Cancer Foundation via the Institutional Research Grant Program at the University of Texas MD Anderson Cancer Center, the University of Texas MD Anderson Cancer Center Start-up Fund, and National Institutes of Health grant R01HL130129 (J.C.). Deposited in PMC for release after 12 months.
Data availability
RNAseq data have been deposited at Gene Expression Omnibus under accession numbers GSE101582 and GSE108252.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.160788.supplemental
References
- Abler L. L., Mansour S. L. and Sun X. (2009). Conditional gene inactivation reveals roles for Fgf10 and Fgfr2 in establishing a normal pattern of epithelial branching in the mouse lung. Dev. Dyn. 238, 1999-2013. 10.1002/dvdy.22032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alanis D. M., Chang D. R., Akiyama H., Krasnow M. A. and Chen J. (2014). Two nested developmental waves demarcate a compartment boundary in the mouse lung. Nat. Commun. 5, 3923 10.1038/ncomms4923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett C. N., Ross S. E., Longo K. A., Bajnok L., Hemati N., Johnson K. W., Harrison S. D. and MacDougald O. A. (2002). Regulation of Wnt signaling during adipogenesis. J. Biol. Chem. 277, 30998-31004. 10.1074/jbc.M204527200 [DOI] [PubMed] [Google Scholar]
- Brault V., Moore R., Kutsch S., Ishibashi M., Rowitch D. H., McMahon A. P., Sommer L., Boussadia O. and Kemler R. (2001). Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development 128, 1253-1264. [DOI] [PubMed] [Google Scholar]
- Cardoso W. V. and Lu J. (2006). Regulation of early lung morphogenesis: questions, facts and controversies. Development 133, 1611-1624. 10.1242/dev.02310 [DOI] [PubMed] [Google Scholar]
- Chang D. R., Martinez Alanis D., Miller R. K., Ji H., Akiyama H., McCrea P. D. and Chen J. (2013). Lung epithelial branching program antagonizes alveolar differentiation. Proc. Natl. Acad. Sci. USA 110, 18042-18051. 10.1073/pnas.1311760110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Moerlooze L., Spencer-Dene B., Revest J. M., Hajihosseini M., Rosewell I. and Dickson C. (2000). An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development 127, 483-492. [DOI] [PubMed] [Google Scholar]
- Diez-Roux G., Banfi S., Sultan M., Geffers L., Anand S., Rozado D., Magen A., Canidio E., Pagani M., Peluso I. et al. (2011). A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 9, e1000582 10.1371/journal.pbio.1000582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eblaghie M. C., Reedy M., Oliver T., Mishina Y. and Hogan B. L. M. (2006). Evidence that autocrine signaling through Bmpr1a regulates the proliferation, survival and morphogenetic behavior of distal lung epithelial cells. Dev. Biol. 291, 67-82. 10.1016/j.ydbio.2005.12.006 [DOI] [PubMed] [Google Scholar]
- Frank D. B., Peng T., Zepp J. A., Snitow M., Vincent T. L., Penkala I. J., Cui Z., Herriges M. J., Morley M. P., Zhou S. et al. (2016). Emergence of a wave of Wnt Signaling that regulates lung alveologenesis by controlling epithelial self-renewal and differentiation. Cell Rep. 17, 2312-2325. 10.1016/j.celrep.2016.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss A. M., Tian Y., Tsukiyama T., Cohen E. D., Zhou D., Lu M. M., Yamaguchi T. P. and Morrisey E. E. (2009). Wnt2/2b and beta-catenin signaling are necessary and sufficient to specify lung progenitors in the foregut. Dev. Cell 17, 290-298. 10.1016/j.devcel.2009.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada N., Tamai Y., Ishikawa T., Sauer B., Takaku K., Oshima M. and Taketo M. M. (1999). Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. EMBO J. 18, 5931-5942. 10.1093/emboj/18.21.5931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe B. D., Scherz P. J., Nissim S., Tian H., McMahon A. P. and Tabin C. J. (2004). Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell 118, 517-528. 10.1016/j.cell.2004.07.024 [DOI] [PubMed] [Google Scholar]
- Harris K. S., Zhang Z., McManus M. T., Harfe B. D. and Sun X. (2006). Dicer function is essential for lung epithelium morphogenesis. Proc. Natl. Acad. Sci. USA 103, 2208-2213. 10.1073/pnas.0510839103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris-Johnson K. S., Domyan E. T., Vezina C. M. and Sun X. (2009). beta-Catenin promotes respiratory progenitor identity in mouse foregut. Proc. Natl. Acad. Sci. USA 106, 16287-16292. 10.1073/pnas.0902274106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto S., Chen H., Que J., Brockway B. L., Drake J. A., Snyder J. C., Randell S. H. and Stripp B. R. (2012). beta-Catenin-SOX2 signaling regulates the fate of developing airway epithelium. J. Cell Sci. 125, 932-942. 10.1242/jcs.092734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M., Kulicke R., Fenster R. J., Greengard P. and Heintz N. (2014). Cell type-specific mRNA purification by translating ribosome affinity purification (TRAP). Nat. Protoc. 9, 1282-1291. 10.1038/nprot.2014.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herriges J. C., Verheyden J. M., Zhang Z., Sui P., Zhang Y., Anderson M. J., Swing D. A., Zhang Y., Lewandoski M. and Sun X. (2015). FGF-regulated ETV transcription factors control FGF-SHH feedback loop in lung branching. Dev. Cell 35, 322-332. 10.1016/j.devcel.2015.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S. X. L., Islam M. N., O'Neill J., Hu Z., Yang Y.-G., Chen Y.-W., Mumau M., Green M. D., Vunjak-Novakovic G., Bhattacharya J. et al. (2014). Efficient generation of lung and airway epithelial cells from human pluripotent stem cells. Nat. Biotechnol. 32, 84-91. 10.1038/nbt.2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S. X., Green M. D., de Carvalho A. T., Mumau M., Chen Y.-W., D'Souza S. L. and Snoeck H.-W. (2015). The in vitro generation of lung and airway progenitor cells from human pluripotent stem cells. Nat. Protoc. 10, 413-425. 10.1038/nprot.2015.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson E. L., Willis N., Mercer K., Bronson R. T., Crowley D., Montoya R., Jacks T. and Tuveson D. A. (2001). Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 15, 3243-3248. 10.1101/gad.943001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob A., Morley M., Hawkins F., McCauley K. B., Jean J. C., Heins H., Na C. L., Weaver T. E., Vedaie M., Hurley K. et al. (2017). Differentiation of human pluripotent stem cells into functional lung alveolar epithelial cells. Cell Stem Cell 21, 472-488.e10. 10.1016/j.stem.2017.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusakabe T., Kawaguchi A., Hoshi N., Kawaguchi R., Hoshi S. and Kimura S. (2006). Thyroid-specific enhancer-binding protein/NKX2.1 is required for the maintenance of ordered architecture and function of the differentiated thyroid. Mol. Endocrinol. 20, 1796-1809. 10.1210/me.2005-0327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson J., Goumans M. J., Sjostrand L. J., van Rooijen M. A., Ward D., Leveen P., Xu X., ten Dijke P., Mummery C. L. and Karlsson S. (2001). Abnormal angiogenesis but intact hematopoietic potential in TGF-beta type I receptor-deficient mice. EMBO J. 20, 1663-1673. 10.1093/emboj/20.7.1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Krautzberger A. M., Sui S. H., Hofmann O. M., Chen Y., Baetscher M., Grgic I., Kumar S., Humphreys B. D., Hide W. A. et al. (2014). Cell-specific translational profiling in acute kidney injury. J. Clin. Invest. 124, 1242-1254. 10.1172/JCI72126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W., Friedman M. S., Shedden K., Hankenson K. D. and Woolf P. J. (2009). GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics 10, 161 10.1186/1471-2105-10-161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauley K. B., Hawkins F., Serra M., Thomas D. C., Jacob A. and Kotton D. N. (2017). Efficient derivation of functional human airway epithelium from pluripotent stem cells via temporal regulation of Wnt signaling. Cell Stem Cell 20, 844-857.e6. 10.1016/j.stem.2017.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger R. J., Klein O. D., Martin G. R. and Krasnow M. A. (2008). The branching programme of mouse lung development. Nature 453, 745-750. 10.1038/nature07005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina Y., Hanks M. C., Miura S., Tallquist M. D. and Behringer R. R. (2002). Generation of Bmpr/Alk3 conditional knockout mice. Genesis 32, 69-72. 10.1002/gene.10038 [DOI] [PubMed] [Google Scholar]
- Morrisey E. E. and Hogan B. L. M. (2010). Preparing for the first breath: genetic and cellular mechanisms in lung development. Dev. Cell 18, 8-23. 10.1016/j.devcel.2009.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucenski M. L., Wert S. E., Nation J. M., Loudy D. E., Huelsken J., Birchmeier W., Morrisey E. E. and Whitsett J. A. (2003). beta-Catenin is required for specification of proximal/distal cell fate during lung morphogenesis. J. Biol. Chem. 278, 40231-40238. 10.1074/jbc.M305892200 [DOI] [PubMed] [Google Scholar]
- Naujok O., Lentes J., Diekmann U., Davenport C. and Lenzen S. (2014). Cytotoxicity and activation of the Wnt/beta-catenin pathway in mouse embryonic stem cells treated with four GSK3 inhibitors. BMC Res. Notes 7, 273 10.1186/1756-0500-7-273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolic M. Z., Caritg O., Jeng Q., Johnson J. A., Sun D., Howell K. J., Brady J. L., Laresgoiti U., Allen G., Butler R. et al. (2017). Human embryonic lung epithelial tips are multipotent progenitors that can be expanded in vitro as long-term self-renewing organoids. eLife 6, e26575 10.7554/eLife.26575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz D. M. and Yin Y. (2012). Signaling networks regulating development of the lower respiratory tract. Cold Spring Harb. Perspect Biol. 4, a008318 10.1101/cshperspect.a008318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins E. L., Clark C. P., Xue Y. and Hogan B. L. M. (2009). The Id2+ distal tip lung epithelium contains individual multipotent embryonic progenitor cells. Development 136, 3741-3745. 10.1242/dev.037317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockich B. E., Hrycaj S. M., Shih H. P., Nagy M. S., Ferguson M. A. H., Kopp J. L., Sander M., Wellik D. M. and Spence J. R. (2013). Sox9 plays multiple roles in the lung epithelium during branching morphogenesis. Proc. Natl. Acad. Sci. USA 110, E4456-E4464. 10.1073/pnas.1311847110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu W., Guttentag S., Wang Z., Andl T., Ballard P., Lu M. M., Piccolo S., Birchmeier W., Whitsett J. A., Millar S. E. et al. (2005). Wnt/beta-catenin signaling acts upstream of N-myc, BMP4, and FGF signaling to regulate proximal-distal patterning in the lung. Dev. Biol. 283, 226-239. 10.1016/j.ydbio.2005.04.014 [DOI] [PubMed] [Google Scholar]
- Soeda T., Deng J. M., de Crombrugghe B., Behringer R. R., Nakamura T. and Akiyama H. (2010). Sox9-expressing precursors are the cellular origin of the cruciate ligament of the knee joint and the limb tendons. Genesis 48, 635-644. 10.1002/dvg.20667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Florea L. and Langmead B. (2014). Lighter: fast and memory-efficient sequencing error correction without counting. Genome Biol. 15, 509 10.1186/s13059-014-0509-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D. and Nusslein-Volhard C. (1992). The origin of pattern and polarity in the Drosophila embryo. Cell 68, 201-219. 10.1016/0092-8674(92)90466-P [DOI] [PubMed] [Google Scholar]
- Tang S.-H. E., Silva F. J., Tsark W. M. K. and Mann J. R. (2002). A Cre/loxP-deleter transgenic line in mouse strain 129S1/SvImJ. Genesis 32, 199-202. 10.1002/gene.10030 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., van Baren M. J., Salzberg S. L., Wold B. J. and Pachter L. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511-515. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Hendrickson D. G., Sauvageau M., Goff L., Rinn J. L. and Pachter L. (2013). Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31, 46-53. 10.1038/nbt.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustiyan V., Zhang Y., Perl A. T., Whitsett J. A., Kalin T. V. and Kalinichenko V. V. (2016). beta-Catenin and Kras/Foxm1 signaling pathway are critical to restrict Sox9 in basal cells during pulmonary branching morphogenesis. Dev. Dyn. 245, 590-604. 10.1002/dvdy.24393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volckaert T. and De Langhe S. P. (2015). Wnt and FGF mediated epithelial-mesenchymal crosstalk during lung development. Dev. Dyn. 244, 342-366. 10.1002/dvdy.24234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volckaert T., Campbell A., Dill E., Li C., Minoo P. and De Langhe S. (2013). Localized Fgf10 expression is not required for lung branching morphogenesis but prevents differentiation of epithelial progenitors. Development 140, 3731-3742. 10.1242/dev.096560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington C. H. (1942). Canalization of development and the inheritance of acquired characteristics. Nature 150, 563-565. 10.1038/150563a0 [DOI] [Google Scholar]
- Wang I.-C., Snyder J., Zhang Y., Lander J., Nakafuku Y., Lin J., Chen G., Kalin T. V., Whitsett J. A. and Kalinichenko V. V. (2012). Foxm1 mediates cross talk between Kras/mitogen-activated protein kinase and canonical Wnt pathways during development of respiratory epithelium. Mol. Cell. Biol. 32, 3838-3850. 10.1128/MCB.00355-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton D., Bellusci S., De Langhe S., Del Moral P.-M., Fleury V., Mailleux A., Tefft D., Unbekandt M., Wang K. and Shi W. (2005). Molecular mechanisms of early lung specification and branching morphogenesis. Pediatr. Res. 57, 26R-37R. 10.1203/01.PDR.0000159570.01327.ED [DOI] [PubMed] [Google Scholar]
- Xing Y., Li C., Li A., Sridurongrit S., Tiozzo C., Bellusci S., Borok Z., Kaartinen V. and Minoo P. (2010). Signaling via Alk5 controls the ontogeny of lung Clara cells. Development 137, 825-833. 10.1242/dev.040535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yackle K., Schwarz L. A., Kam K., Sorokin J. M., Huguenard J. R., Feldman J. L., Luo L. and Krasnow M. A. (2017). Breathing control center neurons that promote arousal in mice. Science 355, 1411-1415. 10.1126/science.aai7984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. and Chen J. (2014). Developmental programs of lung epithelial progenitors: a balanced progenitor model. Wiley Interdiscip. Rev. Dev. Biol. 3, 331-347. 10.1002/wdev.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K., Xu J., Liu Z., Sosic D., Shao J., Olson E. N., Towler D. A. and Ornitz D. M. (2003). Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development 130, 3063-3074. 10.1242/dev.00491 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.