ABSTRACT
Under stress conditions, the coactivator Multiprotein bridging factor 1 (Mbf1) translocates from the cytoplasm into the nucleus to induce stress-response genes. However, its role in the cytoplasm, where it is mainly located, has remained elusive. Here, we show that Drosophila Mbf1 associates with E(z) mRNA and protects it from degradation by the exoribonuclease Pacman (Pcm), thereby ensuring Polycomb silencing. In genetic studies, loss of mbf1 function enhanced a Polycomb phenotype in Polycomb group mutants, and was accompanied by a significant reduction in E(z) mRNA expression. Furthermore, a pcm mutation suppressed the Polycomb phenotype and restored the expression level of E(z) mRNA, while pcm overexpression exhibited the Polycomb phenotype in the mbf1 mutant but not in the wild-type background. In vitro, Mbf1 protected E(z) RNA from Pcm activity. Our results suggest that Mbf1 buffers fluctuations in Pcm activity to maintain an E(z) mRNA expression level sufficient for Polycomb silencing.
KEY WORDS: Mbf1, Enhancer of zeste, E(z), Polycomb silencing, Pcm, Exoribonuclease, Drosophila melanogaster
Highlighted Article: In addition to its role as a nuclear coactivator, a cytoplasmic mRNA-stabilizing function of Multiprotein bridging factor 1 may contribute to various types of stress defense, metabolic processes and neurogenesis in Drosophila.
INTRODUCTION
Polycomb silencing is essential for the developmental regulation of gene expression (Grossniklaus and Paro, 2014; Comet et al., 2016; Kundu et al., 2017). The silencing needs to be robust to tightly repress the expression of developmental genes in undifferentiated cells, such as stem cells, but should also be flexible for rapid release upon differentiation. However, this paradoxical aspect of Polycomb silencing is not well understood.
Mbf1 was originally identified as an evolutionarily conserved coactivator that connects a transcriptional activator with the TATA element-binding protein (Li et al., 1994; Takemaru et al., 1997, 1998). Usually, Mbf1 is present in the cytoplasm; however, under stress conditions, Mbf1 translocates into the nucleus to induce stress-response genes (Kabe et al., 1999; Jindra et al., 2004; Ballabio et al., 2004). Previous studies have revealed roles for the coactivator in axon guidance (Liu et al., 2003), oxidative stress response (Jindra et al., 2004; Arce et al., 2010), heat-shock response (Suzuki et al., 2008), defense against microbial infection (Suzuki et al., 2005; Kim et al., 2007), and resistance to drugs such as tamoxifen (Mendes-Pereira et al., 2012). However, the cytoplasmic role of Mbf1 has remained elusive, except for mRNA or ribosomal binding (Baltz et al., 2012; Klass et al., 2013; Kwon et al., 2013; Blombach et al., 2014).
Pacman (Pcm/Xrn1) is an evolutionarily conserved 5′-3′ exoribonuclease that degrades decapped mRNA (Till et al., 1998; Jones et al., 2012). Genetic studies have demonstrated that Drosophila pcm is involved in epithelial closure, male fertility, apoptosis and growth control (Grima et al., 2008; Lim et al., 2009; Jones et al., 2012, 2016; Waldron et al., 2015). Null mutants of pcm are lethal during early pupal stages, suggesting the enzyme plays an essential role in development (Waldron et al., 2015; Jones et al., 2016).
Using a genetic approach in Drosophila, we show that cytoplasmic Mbf1 ensures Polycomb silencing by protecting E(z) mRNA from degradation by Pcm. Our results thus demonstrate an unexpected component of the regulatory mechanism underlying Polycomb silencing. This mechanism might also allow flexibility in Polycomb silencing, as Mbf1 protein expression declines upon differentiation.
RESULTS AND DISCUSSION
Mbf1 enhances Polycomb silencing by protecting E(z) mRNA in the cytoplasm
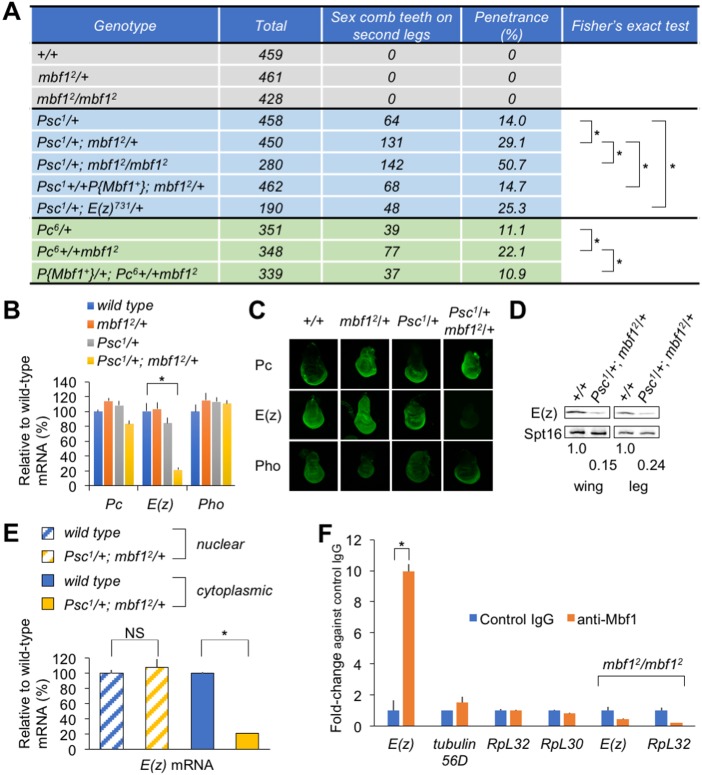
To address the cytoplasmic role of Mbf1, we searched for novel genes that interact with mbf1. Surprisingly, the mbf1 mutation enhanced a classical Polycomb phenotype of Psc and Pc mutants, namely the appearance of an ectopic sex comb tooth or teeth on the male mid-leg (Fig. 1A). Although mbf12/+ or mbf12/mbf12 flies never exhibited the Polycomb phenotype, penetrance of the phenotype in Psc1/+ increased significantly in Psc1/+; mbf12/+, and further increased in Psc1/+; mbf12/mbf12. The penetrance was restored to the Psc1/+ level by expressing wild-type Mbf1 protein from a transgene. Similar effects of the mbf12 allele were observed with the Pc6 mutation.
Fig. 1.
Compromised E(z) expression in the mbf1 mutant in a Polycomb group mutant background. (A) Genetic interactions between mbf1 and Polycomb group mutants. The P-element vector P{mbf1+} expresses wild-type Mbf1 from a transgene. *P<0.01 (Fisher's exact test). (B) RT-qPCR analysis of the indicated Polycomb group mRNAs in whole extracts from third instar male larvae. Data are mean±s.d., relative to the wild-type mRNA level; *P<0.01 (Student's t-test). (C) Immunofluorescence analyses of indicated Polycomb group proteins in wing discs of third instar larvae. (D) Western blot analyses of E(z) in wing or leg discs. Numbers indicate relative E(z) levels normalized to those of Spt16. (E) RT-qPCR analysis of E(z) mRNA in the nuclear or cytoplasmic fraction of wing discs. NS, not significant; *P<0.01 (Student's t-test). (F) Mbf1 binds to E(z) mRNA. RIP samples from wild-type or mbf12 embryonic extracts were analyzed by RT-qPCR. Data are mean±s.d. of fold-change versus control IgG; *P<0.01 (Student's t-test).
To gain insight into the mechanism underlying the genetic interaction between Psc and mbf1, we examined the expression of the representative Polycomb group genes Pc, E(z) and pho. Results of reverse transcription-quantitative PCR (RT-qPCR) analyses demonstrated a prominent reduction in the expression level of E(z) mRNA in Psc1/+; mbf12/+ larvae, whereas Pc and pho mRNA levels remained unchanged (Fig. 1B). Immunostaining of wing discs demonstrated that E(z) protein expression was severely compromised in Psc1/+; mbf12/+ compared with that in wild type, mbf12/+ or Psc1/+ (Fig. 1C). By contrast, the expression of Pc and Pho proteins was not significantly affected. Western blot analyses confirmed the marked decrease in the E(z) protein level in both wing and leg discs from Psc1/+; mbf12/+ (Fig. 1D). Consistently, Psc1/+; E(z)731/+ exhibited the extra sex comb phenotype, which was comparable to Psc1/+; mbf12/+ (Fig. 1A).
It is unlikely that Mbf1 affects E(z) transcription because no significant difference was detected in the E(z) mRNA level between wild-type and mbf12/mbf12 larvae (Fig. S1A). Consistently, we were unable to detect any significant difference in the expression of E(z) in the wing disc upon knockdown or overexpression of Mbf1 using a posterior compartment-specific Gal4 driver (Fig. S1B). When cytoplasmic and nuclear RNA fractions from wing discs were analyzed by RT-qPCR, the nuclear E(z) mRNA level was similar between wild type and Psc1/+; mbf12/+. However, the cytoplasmic E(z) mRNA level in Psc1/+; mbf12/+ decreased to ∼20% of the wild-type level (Fig. 1E). Collectively, these results suggest that mbf1 regulates the E(z) mRNA level post-transcriptionally in the cytoplasm.
Considering that Mbf1 binds to mRNA (Baltz et al., 2012; Klass et al., 2013; Kwon et al., 2013), we hypothesized that cytoplasmic Mbf1 might bind to E(z) mRNA to protect it from degradation, and thereby regulates the E(z) mRNA level. Results of RNA-immunoprecipitation (RIP) experiments revealed a preferential binding of Mbf1 to E(z) mRNA. We found a ∼10-fold enrichment of E(z) mRNA in the anti-Mbf1 antibody pull-down fraction from cytoplasmic extracts of embryos (Fig. 1F). The pull-down was clearly selective, as enrichment of abundant mRNAs, such as RpL32 and RpL30, was not observed. By contrast, E(z) mRNA was barely detectable in the anti-Mbf1 antibody pull-down fraction from embryonic extracts of the mbf1 mutant, used as a negative control. This is not due to absence of E(z) mRNA in the mbf1 mutant (Fig. S1A).
Pcm counteracts Polycomb silencing
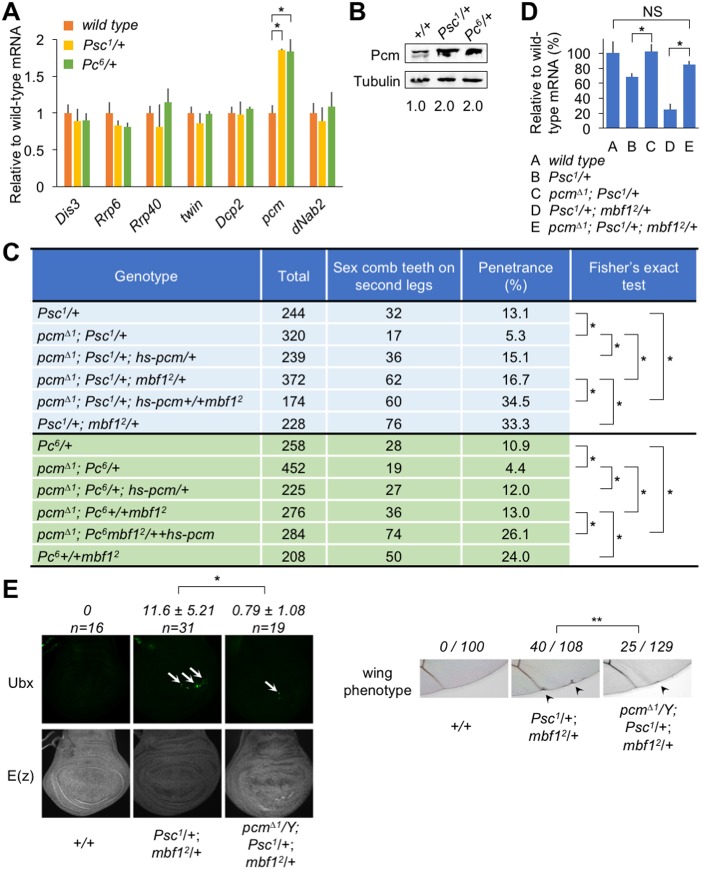
Following the observed preferential binding of Mbf1 to E(z) mRNA, we focused on the Polycomb phenotype and reduced E(z) mRNA expression level, which were not caused by the mbf1 mutation alone. Enhancement of the Polycomb phenotype and the reduction of E(z) mRNA were only detected in the double mbf1 and Polycomb group gene mutant. To explain the synergistic effect of mbf1 and Polycomb group mutations, we posited that a component of the mRNA degradation pathway was only activated in the Polycomb group mutant background. Therefore, we sought to identify the component of the pathway that was activated in the Psc or Pc mutants. Among the mRNAs tested, only pcm mRNA, which encodes the 5′-exoribonuclease, was upregulated in Psc1/+ and Pc6/+ larvae (Fig. 2A). Neither the decapping enzyme (Dcp2), components of the exosome [Dis3, Prp6 (CG6841) and Prp40 (CG3542)] (Siwaszek et al., 2014), nor components in the 3′-deadenylation-mediated pathway (twin and Nab2) (Morris et al., 2005; Pak et al., 2011) appeared to be activated. Western blot analyses revealed a 2-fold increase in the Pcm protein level in wing discs from Psc1/+ or Pc6/+ larvae compared with that from wild type (Fig. 2B). These results led us to further investigate the effects of the pcm mutation on Polycomb silencing and E(z) mRNA expression.
Fig. 2.
Functional relationship among mbf1, E(z) and pcm. (A) pcm is downregulated by Psc and Pc. Expression of the indicated genes in third instar male larvae was analyzed by RT-qPCR in the wild-type or Polycomb group mutant background. pcm does not appear to be a direct target of Polycomb silencing (Zeng et al., 2012). Data are mean±s.d. relative to the wild-type mRNA level; *P<0.01 (Student's t-test). (B) Western blot analysis of Pcm in wing discs from the indicated lines. Numbers indicate relative Pcm levels normalized to Tubulin levels. (C) pcm mutation suppresses the extra sex comb phenotype. *P<0.01 (Fisher's exact test). (D) pcm mutation restores the E(z) mRNA level in Psc1/+ or Psc1/+; mbf12/+. The E(z) mRNA levels in third instar male larvae of the indicated lines were analyzed by RT-qPCR. Data are mean±s.d. relative to the wild-type mRNA level. NS, not significant; *P<0.01 (Student's t-test). (E) (Top) Misexpression of Ubx in the wing disc of Psc1/+; mbf12/+ and its suppression by pcmΔ1. Arrows indicate Ubx-positive spots. (Bottom) Immunostaining of E(z) protein in the wing discs shown above. (Right) Adult wing defect (arrowheads) in Psc1/+; mbf12/+ and its suppression by pcmΔ1. The number of wings with the defect among the total number of wings examined is indicated. **P<0.05 (Fisher's exact test).
Strikingly, the pcmΔ1 mutation resulted in significant suppression of the Polycomb phenotype in Psc1/+ and Psc1/+; mbf12/+ (Fig. 2C). This suppression was rescued by expressing the wild-type Pcm protein from a transgene. Similar results were obtained using the Pc6 mutant (Fig. 2C) and another pcm allele, pcm5 (Fig. S2). Consistent with this result, the pcmΔ1 mutation restored the E(z) mRNA levels in Psc1/+ and Psc1/+; mbf12/+ to near wild-type levels (Fig. 2D).
In addition to the extra sex comb phenotype, Psc1/+; mbf12/+ exhibited misexpression of Ubx in wing discs (Fig. 2E, top). The signals appeared as spots consisting of clusters of Ubx-positive cells. The pcmΔ1 mutation decreased the number of spots per wing disc. The misexpression occurred predominantly around the dorsoventral border in the posterior compartment. Consistently, we observed adult wing defects along the posterior wing margin, which was also suppressed by pcmΔ1 (Fig. 2E, right).
Mbf1 protects E(z) mRNA from Pcm activity
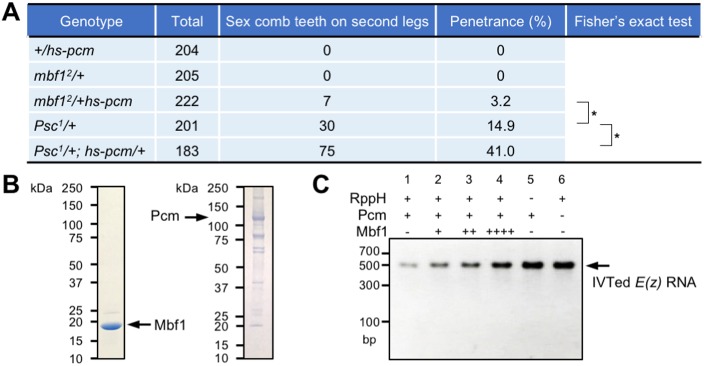
Importantly, we detected the extra sex comb phenotype under mild overexpression of pcm in mbf12/hs-pcm double heterozygotes at 25°C, even in the wild-type Polycomb group background (Fig. 3A). hs-pcm/+ exhibited an ∼2.5-fold overexpression of Pcm at 25°C (Fig. S3A). Nevertheless, hs-pcm heterozygotes in the wild-type mbf1 background did not show any Polycomb phenotype. These results suggest that Mbf1 stabilizes Polycomb silencing against fluctuations in the Pcm protein level in vivo. We also observed enhancement of the Polycomb phenotype in Psc1/+; hs-pcm/+ compared with that in Psc1/+ (Fig. 3A).
Fig. 3.
Mbf1 protein directly counteracts 5′-3′ exoribonuclease activity in vivo and in vitro. (A) mbf12/hs-pcm double heterozygotes exhibit the Polycomb phenotype in the wild-type Polycomb group background. *P<0.01. (B) Recombinant Drosophila Mbf1 and Pcm preparations were resolved by 5-20% SDS-PAGE and the gel stained with Coomassie Brilliant Blue. (C) Recombinant Drosophila Mbf1 inhibits Pcm activity in vitro. In vitro-transcribed E(z) RNA [IVTed E(z) RNA] was used as substrate for Pcm. Reactions included the indicated components, and purified RNAs were resolved on a 1.5% agarose gel. dsDNA marker size is indicated (bp). Amounts of Mbf1 added: +, 2.5 µg; ++, 5 µg; ++++, 10 µg.
Biochemical analyses using purified recombinant Mbf1 and Pcm proteins (Fig. 3B) revealed that Mbf1 protects E(z) RNA from degradation by Pcm. RNA protection assays were performed in which in vitro-synthesized E(z) RNA was treated with the RNA pyrophosphatase RppH to convert the 5′-triphosphoryl end into the 5′-monophosphoryl form, which is a Pcm substrate. The RNA was digested with Pcm in the presence or absence of Mbf1. Mbf1 inhibited the digestion of E(z) RNA (Fig. 3C, lanes 2-4 versus lane 1). In the absence of RppH, RNA degradation was barely detectable (Fig. 3C, lane 5), suggesting that the digestion was due to 5′-exoribonuclease activity. Gel filtration of a mixture of Pcm and Mbf1 resulted in the elution of each protein in a clearly separated peak (Fig. S3B). Furthermore, Mbf1 did not co-immunoprecipitate with Pcm and vice versa (Fig. S3C). These results suggest that Mbf1 does not inhibit Pcm activity through protein-protein interactions. Collectively, we conclude that Mbf1 protects E(z) mRNA from degradation by Pcm both in vivo and in vitro.
Model and implications of Mbf1 binding to mRNA
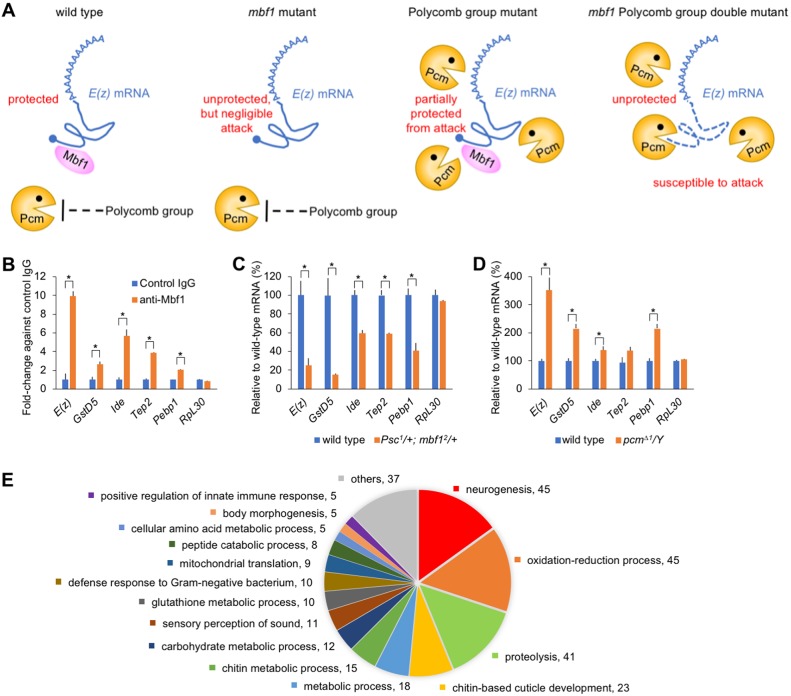
We propose that cytoplasmic Mbf1 ensures Polycomb silencing by protecting E(z) mRNA from the activity of Pcm (Fig. 4A). In the mbf1 mutant, E(z) mRNA is free from Mbf1 protein, but pcm expression is downregulated by Polycomb group genes. In the Polycomb group mutant, Pcm expression is upregulated, but E(z) mRNA is partly protected by Mbf1. In the mbf1 Polycomb group double mutant, E(z) mRNA is free from Mbf1 protein and is subject to Pcm attack. Whereas Mbf1 is highly expressed in undifferentiated cells, such as those of embryos, larval testis, ovary, imaginal discs and neuroblasts, its expression is reduced in differentiated tissues (Fig. S4A; see also Jindra et al., 2004), similar to the situation in the mbf1 mutant. This would facilitate the rapid release of developmental genes from Polycomb silencing upon differentiation. Interestingly, expression of mammalian Mbf1 [also termed endothelial differentiation-related factor 1 (Edf1)] (Dragoni et al., 1998) and Ezh2 (Ezhkova et al., 2009) declines immediately after the onset of differentiation.
Fig. 4.
Conceivable functions of cytoplasmic Mbf1 protein via binding to mRNAs. (A) Model for Mbf1-ensured Polycomb silencing. In wild-type and mbf1 mutant lines, Pcm is not upregulated. Therefore, the steady-state level of E(z) mRNA is well balanced irrespective of Mbf1 expression. In Polycomb group mutants, Pcm expression is upregulated so that E(z) mRNA could become susceptible to Pcm attack. However, Mbf1 protects E(z) mRNA to ensure robustness of Polycomb silencing. In mbf1 and Polycomb group double mutants, loss of Mbf1 allows extensive degradation of E(z) mRNA by derepressed Pcm, thereby affecting Polycomb silencing. (B) The enrichment of four representative mRNAs (GstD5, Ide, Tep2 and Pebp1) identified in the RIP-seq results was confirmed by RIP RT-qPCR analysis. Results for E(z) and RpL30 mRNAs from Fig. 1F are included for comparison. Data are mean±s.d. of fold-change versus control IgG; *P<0.01. (C,D) RT-qPCR analysis of the indicated mRNAs in whole extracts of third instar male larvae from Psc1/+; mbf12/+ (C) or pcmΔ1/Y (D). Data are mean±s.d. relative to the wild-type mRNA level; *P<0.01. All P-values obtained using Student's t-test. (E) Gene ontology analysis of the RNA-seq results is consistent with the known Mbf1 functions. The number of genes in each term is indicated.
A recent study demonstrated that Pcm prevents apoptosis in imaginal discs and downregulates specific transcripts such as hid and reaper (Waldron et al., 2015). However, suppression of apoptosis did not rescue the lethality of a pcm null mutation at the early pupal stage. Therefore, there might be other targets of Pcm that are essential for early pupal development. The present study indicates that E(z) mRNA could be one such target.
The mRNA-binding activity of Mbf1 was selective, but might not be strictly specific to E(z) mRNA. Although Polycomb silencing is central to the developmental regulation of gene expression, there could be other mRNAs that bind to Mbf1 in a similar manner, thereby modulating another biological function. Therefore, we conducted RIP-seq analysis to identify Mbf1-bound mRNAs. To ensure robustness of our RIP-seq data, we compared our results independently with two publically available datasets (Fig. S5A) and identified 804 commonly enriched mRNAs (Table S1). Among these, the enrichment of four representative mRNAs (GstD5, Ide, Tep2 and Pebp1) was confirmed by RIP RT-qPCR analyses (Fig. 4B). Interestingly, the expression levels of these four mRNAs decreased in Psc1/+; mbf12/+ and increased in pcmΔ1/Y compared with those in wild type (Fig. 4C,D), suggesting that the model (Fig. 4A) can be applied to a wider range of mRNAs than just E(z). However, dependency on the Mbf1/Pcm antagonism appears to differ among the mRNAs.
Gene ontology and pathway analyses of the 804 genes revealed some interesting properties of the Mbf1-associated mRNAs (Fig. 4E, Fig. S5B, Tables S2 and S3). The gene ontology terms ‘glutathione metabolic process’, ‘oxidation-reduction process’ and ‘neurogenesis’, which includes E(z), are consistent with the fact that we previously found defects in oxidative stress defense and axon guidance in the mbf1 mutant (Liu et al., 2003; Jindra et al., 2004). Also of interest are the groups ‘positive regulation of innate immune response’ and ‘defense response to Gram-negative bacterium’, as Arabidopsis MBF1 is involved in host defense against microbial infection (Suzuki et al., 2005; Kim et al., 2007). Moreover, pathway analysis of the enriched genes implicated Mbf1 in ‘drug metabolism’, as previously suggested for tamoxifen resistance (Mendes-Pereira et al., 2012). This raises an intriguing possibility that Mbf1 contributes to various types of stress defense, metabolic processes and neurogenesis as both a nuclear coactivator and as a cytoplasmic mRNA-stabilizing protein. Although mbf1 null mutants are viable under laboratory conditions, evolutionary conservation of mbf1 suggests that it has essential role(s) under real-world stress conditions.
MATERIALS AND METHODS
Fly lines
yw; mbf12 and yw; P{Mbf1+}; mbf12 have been described (Jindra et al., 2004). pcmΔ1/FM7 (Lim et al., 2009) was a gift from Dr T. Kai (Osaka University, Japan). pcm5 and P{CaSpeR hs-pcm} (Grima et al., 2008) (designated hs-pcm here) were from Dr S. F. Newbury (University of Sussex, Brighton, UK). w*; E(z)731 FRT2A/TM6C was from Dr J. Müller (Max Planck Institute of Biochemistry, Munich, Germany). yw: UAS-GFP; hh-Gal4 and yw; UAS-mbf1 were from Dr Q.-X. Liu (Shandong Agricultural University, China). Psc1/CyO and Pc6/TM3 were obtained from The Kyoto Stock Center. yw; UAS-mbf1RNAi was from The Bloomington Stock Center. Pc6/TM6B w+ GFP, Psc1/CyO GFP, yw; mbf12 Pc6/TM6B w+ GFP, pcmΔ1; mbf12, pcmΔ1; hs-pcm, pcm5; hs-pcm and yw; Psc1/CyO; mbf12/TM6B lines were established through appropriate crosses. The expression level of pcm in wild type is extremely low and approximately half that of leaky expression from the hsp70 promoter at 25°C in hs-pcm/+. Therefore, the pcm mutation can be rescued in pcmΔ1; hs-pcm/+ without any heat shock.
Polycomb phenotype
Females of yw, yw; mbf12, yw; P{Mbf1+}; mbf12, Psc1/CyO, yw; Psc1/CyO; mbf12/TM6B, pcmΔ1, pcmΔ1; mbf12, pcmΔ1; hs-pcm, pcm5, pcm5; hs-pcm or hs-pcm were crossed with males of Psc1/CyO GFP, Pc6/TM6B w+ GFP, yw; mbf12 Pc6/TM6B w+ GFP, yw; mbf12, w*; E(z)731 FRT2A/TM6C or hs-pcm. After rearing at 25°C, male progeny of desired genotype were used for inspection of the Polycomb phenotype. As the Polycomb phenotype is significantly affected by rearing conditions, such as ingredients of the fly diet, the penetrance should be compared within the same experiment. Statistical analysis was performed by Fisher's exact test.
Immunostaining
Rabbit polyclonal antiserum was raised against bacterially expressed polypeptides carrying the C-terminal region of Pc, the N-terminal region of E(z) or the N-terminal region of Pho. Immunoblot data using these antibodies are shown in Fig. S6. Immunostaining of imaginal discs was carried out as described previously (Liu et al., 2003). Antibodies were used at the following dilutions: anti-Pc (1:1000), anti-E(z) (1:1000), anti-Pho (1:1000), anti-Ubx (Developmental Studies Hybridoma Bank; 1:1000), anti-Mbf1 (Jindra et al., 2004; 1:500), goat anti-rabbit IgG or anti-mouse IgG Alexa488 (Molecular Probes, A72731 and A32723; 1:2000) and anti-rabbit IgG-Cy5 (Jackson ImmunoResearch, 111-225-144; 1:500). Images were acquired with an LSM510 META confocal microscope (Zeiss).
RIP
Approximately 0.8 ml packed volume of Drosophila embryos (0-22 h after egg laying) from yw or yw; mbf12 was homogenized with 2 ml lysis buffer comprising 100 mM Na phosphate (pH 7.1), 10 mM NaCl, 3 mM MgCl2, 0.5% (v/v) NP-40, 0.5 mM DTT, 1× Protease Inhibitor Cocktail (Sigma-Aldrich), 0.5 mM PMSF and 0.5 units/ml porcine liver RNase inhibitor (Takara) in a Dounce homogenizer. The homogenate was centrifuged at 20,000 g for 10 min and the supernatant collected as the cytoplasmic fraction. After addition of 5 M NaCl to a final concentration of 360 mM, 1 ml of the cytoplasmic fraction was mixed with anti-Mbf1 antibody (Abcam, ab 174651)-loaded or rabbit IgG-loaded Dynabeads protein A (Life Technologies) at 4°C for 2 h. The beads were washed with PBS containing 0.1% (v/v) Tween 20 and the immunoprecipitated materials were dissolved in 0.2 ml 6 M guanidine-HCl in 0.4 M Tris-acetate/1 mM EDTA (pH 8.0). RNA was purified using a Direct-zol RNA MiniPrep Kit (Zymo Research) with a DNase I treatment step and quantitated by RT-qPCR (see below). Data are presented as fold-change compared with the IgG control experiment. Each mean±s.d. was calculated from qPCRs performed in triplicate. Statistical analysis was performed by Student's t-test.
The previous antibody against Drosophila Mbf1 (Jindra et al., 2004) did not precipitate any RNA. As Mbf1 binds to mRNA through its N-terminal region (Klass et al., 2013), the antibody might mask the RNA-binding region. Therefore, we used antibody ab174651 (Abcam), which was raised against a peptide carrying the C-terminal region (amino acids 98-148) of human MBF1 (EDF1) (Fig. S7A). It was able to immunoprecipitate Drosophila Mbf1 from the cytoplasmic fraction of embryos (Fig. S7B).
RT-qPCR
Total RNA was prepared from ten heads of third instar male larvae, or a cytoplasmic or nuclear fraction from 60 wing discs of the desired genotype. GFP signals were used to exclude larvae with the GFP balancers. cDNAs were prepared from RNA samples of two biological replicates. qPCR was performed using a Roche LightCycler 2.0 as described previously (Nakayama et al., 2012) on each cDNA in three to five technical replicates. Primer sequences are listed in Table S4. Data were normalized by the βTub56D mRNA level and then presented as relative to the wild-type mRNA level. As the fluctuation between biological replicates did not differ substantially from that among technical replicates, the data (mean±s.d.) from a representative biological replicate are shown in figures. Statistical analysis was performed by Student's t-test.
Western blot
Western blotting was performed as described previously (Nakayama et al., 2012) on samples containing 60 wing or leg discs. Antibodies were used at the following dilutions: anti-E(z) (1:4000), anti-Pc (1:4000), anti-Pho (1:4000), anti-Pcm (gift of S. F. Newbury; 1:2000), anti-Spt16 (Dre4) (Nakayama et al., 2012; 1:4000), anti-Mbf1 (1:5000), anti-FLAG M2 (Sigma, F3165; 1:2000), anti-tubulin (Developmental Studies Hybridoma Bank, a gift of K. Saito, National Institute of Genetics, Japan; 1:5000), anti-rabbit and anti-mouse IgG-HRP (GE Healthcare, NA9340 and NA9310; 1:5000) and anti-mouse IgG-HRP.
RNA protection assay
E(z) RNA was in vitro transcribed from T7 vector template (a derivative of Drosophila Genomics Resource Center clone LD30505) using the MEGAscript Kit (Ambion). A reaction comprised 0.5 µg E(z) RNA, 2.5 units recombinant E. coli RNA 5′-pyrophosphohydrolase (RppH) (NEB), 0.5 µg baculovirus-expressed recombinant Drosophila Pcm protein, 5 units RNase inhibitor (Roche), 10 µg BSA and 0.1% Triton X-100 in 10 µl 1× NEB2 buffer (New England Biolabs), with the titrating amount of bacterially expressed Drosophila Mbf1 protein (2.5-10 µg). RNA was purified using a DNA Clean&Concentrator Kit (Zymo Research). An aliquot of the purified RNA was subject to agarose gel electrophoresis and stained with ethidium bromide.
Purification of bacterially expressed recombinant proteins
Drosophila mbf1 or mouse Mbf1 cDNA was cloned into pET28a (EMD Biosciences) or pQE80L (Qiagen) vector, respectively. To prepare antigen for expression, Drosophila cDNA encoding the C-terminal region (amino acids 190-390) of Pc, the N-terminal region (amino acids 2-219) of E(z) or the N-terminal region (amino acids 2-302) of Pho was cloned into pQE80L. E. coli BL21(DE3) was transformed with each vector, and expression of recombinant proteins was induced by adding IPTG to 0.5 mM. Recombinant proteins were purified using Ni-NTA agarose (Qiagen) according to the supplier's protocol.
The recombinant Mbf1 protein was further purified by passing through a 30 kDa cut-off spin-concentrator and then concentrated on a 10 kDa cut-off spin-concentrator (both Amicon Ultra, Millipore) while exchanging the buffer to 50 mM Na phosphate (pH 7.0), 0.2 mM EDTA.
Purification of baculovirus-expressed recombinant Pcm protein
Recombinant Drosophila Pcm protein (residues 1-1141) with the C-terminal FLAG tag was expressed as previously described (Jinek et al., 2011) using the Bac-to-Bac system (Invitrogen), and was purified using Ni-NTA agarose (Qiagen). The protein was concentrated on a 30 kDa cut-off spin-concentrator (Amicon Ultra, Millipore) while exchanging the buffer to 20 mM Tris-HCl (pH 8.3), 420 mM NaCl, 0.1% Triton X-100, 0.2 mM EDTA, 20% glycerol.
Gel filtration analysis of Pcm and Mbf1 proteins
A Sephacryl S-200 HR (GE Healthcare) column (5 mm×185 mm) was prepared and equilibrated with 10 mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.2 mM EDTA, 1 mM DTT. Input material consisted of 50 µg each of Pcm and Mbf1 in 30 µl. Fractions were collected of 50 µl after void fractions.
Co-immunoprecipitation
Anti-FLAG M2 (Sigma), anti-Mbf1 or rabbit normal IgG were each conjugated to 10 µl Dynabeads Protein G (Dynal) in TBST (Tris-buffered saline pH 8.0 containing 0.1% Tween 20) and 5% skimmed milk overnight at 4°C, and the beads then washed extensively with TBST. Pcm (5 µg) and Mbf1 (5 µg) proteins were mixed in 200 µl TBST, and added to each bead preparation. The mixtures were rotated for 3 h at room temperature. After washing the beads with TBST five times, bound proteins were eluted in 100 mM Tris-HCl (pH 6.8) and 4% SDS. An aliquot of each eluate was subject to western blot.
RNA-seq and data analysis
Immunoprecipitated RNA was subjected to a library generation protocol using the SENSE mRNA-Seq Library Prep Kit (Lexogen). The library was sequenced using an Illumina HiSeq 2500. Reads were mapped on the custom dm6 transcriptome with 3′-untranslated regions using TopHat v2.1.1 (Trapnell et al., 2009). Transcript abundance was quantified as fragments per kilobase of transcript per million fragments mapped (FPKM) values using Cufflinks v2.1.1 (Trapnell et al., 2010), and analyzed using Cuffdiff v2.1.1 (Trapnell et al., 2013). Enrichment scores were calculated in log10 transformation of the modified FPKM value ratio between immunoprecipitated sample and the means of two independent publically available transcriptome datasets using embryonic poly(A)+ RNAs [modENCODE datasets (http://data.modencode.org/?Organism=D.%20melanogaster) (IDs: 2019-2023) and GSE57517], in which all the FPKM values were modified by addition of 1 to minimize dispersion effect. Genes demonstrating log10 of modified FPKM value >1 were extracted from each dataset, and common genes between the datasets (804 genes; Fig. S5A, Table S1) were subjected to gene ontology and pathway analyses using DAVID (https://david.ncifcrf.gov).
Supplementary Material
Acknowledgements
We thank T. Kai, S. F. Newbury, J. Müller and K. Saito for gifts of fly lines and antibodies; Q.-X. Liu for gift of fly lines and support in use of the confocal microscope; Y. Tsukada for gift of baculovirus backbone vector; H. Ihara for gift of Sf21 cells; A. Vaquero for critical review of the manuscript; and H. Sasaki, H. Toh and Y. Kabayama for support of HiSeq run.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: K.N., H.M., S.H.; Investigation: K.N., X.-F.W., H.S., S.H.; Resources: X.-F.W.; Writing - original draft: K.N.; Writing - review & editing: S.H.; Supervision: S.H.; Project administration: K.N., S.H.; Funding acquisition: K.N.
Funding
This work was supported by the Japan Science and Technology Agency PRESTO program and Japan Society for the Promotion of Science (JSPS) KAKENHI grant number JP16K07451 to K.N., and was partly performed in the Cooperative Research Project Program of the Medical Institute of Bioregulation, Kyushu University. X.-F.W. is a postdoctoral fellow of JSPS. Deposited in PMC for immediate release.
Data availability
RNA-seq data are available at DNA Data Bank of Japan (DDBJ) under accession number DRA005292.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.162461.supplemental
References
- Arce D. P., Godoy A. V., Tsuda K., Yamazaki K., Valle E. M., Iglesias M. J., Di Mauro M. F. and Casalongué C. A. (2010). The analysis of an Arabidopsis triple knock-down mutant reveals functions for MBF1 genes under oxidative stress conditions. J. Plant Physiol. 167, 194-200. 10.1016/j.jplph.2009.09.003 [DOI] [PubMed] [Google Scholar]
- Ballabio E., Mariotti M., De Benedictis L. and Maier J. A. M. (2004). The dual role of endothelial differentiation-related factor-1 in the cytosol and nucleus: modulation by protein kinase A. Cell. Mol. Life Sci. 61, 1069-1074. 10.1007/s00018-004-4016-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltz A. G., Munschauer M., Schwanhäusser B., Vasile A., Murakawa Y., Schueler M., Youngs N., Penfold-Brown D., Drew K., Milek M. et al. (2012). The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell 46, 674-690. 10.1016/j.molcel.2012.05.021 [DOI] [PubMed] [Google Scholar]
- Blombach F., Launay H., Snijders A. P. L., Zorraquino V., Wu H., de Koning B., Brouns S. J. J., Ettema T. J. G., Camilloni C., Cavalli A. et al. (2014). Archaeal MBF1 binds to 30S and 70S ribosomes via its helix-turn-helix domain. Biochem. J. 462, 373-384. 10.1042/BJ20131474 [DOI] [PubMed] [Google Scholar]
- Comet I., Riising E. M., Leblanc B. and Helin K. (2016). Maintaining cell identity: PRC2-mediated regulation of transcription and cancer. Nat. Rev. Cancer 16, 803-810. 10.1038/nrc.2016.83 [DOI] [PubMed] [Google Scholar]
- Dragoni I., Mariotti M., Consalez G. G., Soria M. R. and Maier J. A. M. (1998). EDF-1, a novel gene product down-regulated in human endothelial cell differentiation. J. Biol. Chem. 273, 31119-31124. 10.1074/jbc.273.47.31119 [DOI] [PubMed] [Google Scholar]
- Ezhkova E., PasollI H. A., Parker J. S., Stokes N., Su I.-H., Hannon G., Tarakhovsky A. and Fuchs E. (2009). Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell 136, 1122-1135. 10.1016/j.cell.2008.12.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grima D. P., Sullivan M., Zabolotskaya M. V., Browne C., Seago J., Wan K. C., Okada Y. and Newbury S. F. (2008). The 5′-3′ exoribonuclease pacman is required for epithelial sheet sealing in Drosophila and genetically interacts with the phosphatase puckered. Biol. Cell 100, 687-701. 10.1042/BC20080049 [DOI] [PubMed] [Google Scholar]
- Grossniklaus U. and Paro R. (2014). Transcriptional silencing by Polycomb-group proteins. Cold Spring Harb. Perspect. Biol. 6, a019331 10.1101/cshperspect.a019331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jindra M., Gaziova I., Uhlirova M., Okabe M., Hiromi Y. and Hirose S. (2004). Coactivator MBF1 preserves the redox-dependent AP-1 activity during oxidative stress in Drosophila. EMBO J. 23, 3538-3547. 10.1038/sj.emboj.7600356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Coyle S. M. and Doudna J. A. (2011). Coupled 5′ nucleotide recognition and processivity in Xrn1-mediated mRNA decay. Mol. Cell 41, 600-608. 10.1016/j.molcel.2011.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. I., Zabolotskaya M. V. and Newbury S. F. (2012). The 5′→3′ exoribonuclease XRN1/Pacman and its functions in cellular processes and development. Wiley Interdiscip. Rev. RNA 3, 455-468. 10.1002/wrna.1109 [DOI] [PubMed] [Google Scholar]
- Jones C. I., Pashler A. L., Towler B. P., Robinson S. R. and Newbury S. F. (2016). RNA-seq reveals post-transcriptional regulation of Drosophila insulin-like peptide dilp8 and the neuropeptide-like precursor Nplp2 by the exoribonuclease Packman/Xrn1. Nucl. Acids Res. 44, 267-280. 10.1093/nar/gkv1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabe Y., Goto M., Shima D., Imai T., Wada T., Morohashi K., Shirakawa M., Hirose S. and Handa H. (1999). The role of human MBF1 as a transcriptional coactivator. J. Biol. Chem. 274, 34196-34202. 10.1074/jbc.274.48.34196 [DOI] [PubMed] [Google Scholar]
- Kim M.-J., Lim G.-H., Kim E.-S., Ko C.-B., Yang K.-Y., Jeong J.-A., Lee M.-C. and Kim C.-S. (2007). Abiotic and biotic stress tolerance in Arabidopsis overexpressing the multiprotein bridging factor 1a (MBF1a) transcriptional coactivator gene. Biochem. Biophys. Res. Commun. 354, 440-446. 10.1016/j.bbrc.2006.12.212 [DOI] [PubMed] [Google Scholar]
- Klass D. M., Scheibe M., Butter F., Hogan G. J., Mann M. and Brown P. O. (2013). Quantitative proteomic analysis reveals concurrent RNA-protein interactions and identifies new RNA-binding proteins in Saccharomyces cerevisiae. Genome Res. 23, 1028-1038. 10.1101/gr.153031.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu S., Ji F., Sunwoo H., Jain G., Lee J. T., Sadreyev R. I., Dekker J. and Kongston R. E. (2017). Polycomb repressive complex 1 generates discrete compacted domains that change during differentiation. Mol. Cell 65, 432-446. 10.1016/j.molcel.2017.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon S. C., Yi H., Eichelbaum K., Föhr S., Fischer B., You K. T., Castello A., Krijgsveld J., Hentze M. W. and Kim V. N. (2013). The RNA-binding protein repertoire of embryonic stem cells. Nat. Struct. Mol. Biol. 20, 1122-1130. 10.1038/nsmb.2638 [DOI] [PubMed] [Google Scholar]
- Li F. Q., Ueda H. and Hirose S. (1994). Mediators of activation of fushi tarazu gene transcription by BmFTZ-F1. Mol. Cell. Biol. 14, 3013-3021. 10.1128/MCB.14.5.3013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim A. K., Tao L. and Kai T. (2009). piRNAs mediate posttranscriptional retroelement silencing and localization to pi-bodies in the Drosophila germline. J. Cell Biol. 186, 333-342. 10.1083/jcb.200904063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q.-X., Jindra M., Ueda H., Hiromi Y. and Hirose S. (2003). Drosophila MBF1 is a co-activator for Tracheae Defective and contributes to the formation of tracheal and nervous systems. Development 130, 719-728. 10.1242/dev.00297 [DOI] [PubMed] [Google Scholar]
- Mendes-Pereira A. M., Sims D., Dexter T., Fenwick K., Assiotis I., Kozarewa I., Mitsopoulos C., Hakas J., Zvelebil M., Lord C. J. et al. (2012). Genome-wide functional screen identifies a compendium of genes affecting sensitivity to tamoxifen. Proc. Natl. Acad. Sci. USA 109, 2730-2735. 10.1073/pnas.1018872108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J. Z., Hong A., Lilly M. A. and Lehmann R. (2005). twin, a CCR4 homolog, regulates cyclin poly(A) tail length to permit Drosophila oogenesis. Development 132, 1165-1174. 10.1242/dev.01672 [DOI] [PubMed] [Google Scholar]
- Nakayama T., Shimojima T. and Hirose S. (2012). The PBAP remodeling complex is required for histone H3.3 replacement at chromatin boundaries and for boundary functions. Development 139, 4582-4590. 10.1242/dev.083246 [DOI] [PubMed] [Google Scholar]
- Pak C., Garshasbi M., Kahrizi K., Gross C., Apponi L. H., Noto J. J., Kelly S. M., Leung S. W., Tzschach A., Behjati F. et al. (2011). Mutation of the conserved polyadenosine RNA binding protein, ZC3H14/dNab2, impairs neural function in Drosophila and humans. Proc. Natl. Acad. Sci. USA 108, 12390-12395. 10.1073/pnas.1107103108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siwaszek A., Ukleja M. and Dziembowski A. (2014). Proteins involved in the degradation of cytoplasmic mRNA in the major eukaryotic model systems. RNA Biol. 11, 1122-1136. 10.4161/rna.34406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Rizhsky L., Liang H., Shuman J., Shulaev V. and Mittler R. (2005). Enhanced tolerance to environmental stress in transgenic plants expressing the transcriptional coactivator multiprotein bridging factor 1c. Plant Physiol. 139, 1313-1322. 10.1104/pp.105.070110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Bajad S., Shuman J., Shulaev V. and Mittler R. (2008). The transcriptional co-activator MBF1c is a key regulator of thermotolerance in Arabidopsis thaliana. J. Biol. Chem. 283, 9269-9275. 10.1074/jbc.M709187200 [DOI] [PubMed] [Google Scholar]
- Takemaru K., Li F.-Q., Ueda H. and Hirose S. (1997). Multiprotein bridging factor 1 (MBF1) is an evolutionarily conserved transcriptional coactivator that connects a regulatory factor and TATA element-binding protein. Proc. Natl. Acad. Sci. USA 94, 7251-7256. 10.1073/pnas.94.14.7251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemaru K., Harashima S., Ueda H. and Hirose S. (1998). Yeast coactivator MBF1 mediates GCN4-dependent transcriptional activation. Mol. Cell. Biol. 18, 4971-4976. 10.1128/MCB.18.9.4971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till D. D., Linz B., Seago J. E., Elgar S. J., Marujo P. E., Elias M. L., Arraiano C. M., McClellan J. A., McCarthy J. E. G. and Newbury S. F. (1998). Identification and developmental expression of a 5'-3′ exoribonuclease from Drosophila melanogaster. Mech. Dev. 79, 51-55. 10.1016/S0925-4773(98)00173-7 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L. and Salzberg S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105-1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., van Baren M. J., Salzberg S. L., Wold B. J. and Pachter L. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511-515. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Hendrickson D. G., Sauvageau M., Goff L., Rinn J. L. and Pachter L. (2013). Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31, 46-53. 10.1038/nbt.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron J. A., Jones C. I., Towler B. P., Pashler A. L., Grima D. P., Hebbes S., Crossman S. H., Zabolotskaya M. V. and Newbury S. F. (2015). Xrn1/Packman affects apoptosis and regulates expression of hid and reaper. Biol. Open 4, 649-660. 10.1242/bio.201410199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J., Kirk B. D., Gou Y., Wang Q. and Ma J. (2012). Genome-wide Polycomb target gene prediction in Drosophila melanogaster. Nucleic Acids Res. 40, 5848-5863. 10.1093/nar/gks209 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.