ABSTRACT
Oxygen concentrations vary between tissues of multicellular organisms and change under certain physiological or pathological conditions. Multiple methods have been developed for measuring oxygenation of biological samples in vitro and in vivo. However, most require complex equipment, are laborious and have significant limitations. Here we report that oxygen concentration determines the choice between two maturation pathways of DsRed FT (Timer). At high oxygen levels, this DsRed derivate matures predominantly into a red fluorescent isoform. By contrast, a green fluorescent isoform is favored by low oxygen levels. Ratiometric analysis of green and red fluorescence after a pulse of Timer expression in Drosophila larvae provides a record of the history of tissue oxygenation during a subsequent chase period, for the whole animal with single-cell precision. Tissue spreads revealed fine differences in oxygen exposure among different cells of the same organ. We expect that the simplicity and robustness of our approach will greatly impact hypoxia research, especially in small animal models.
KEY WORDS: Oxygen, Reporter, Fluorescence, Hypoxia
Summary: DsRed FT forms the basis of a simple and robust method for direct visualization of oxygenation exposure that is applicable to whole animals with single-cell precision.
INTRODUCTION
Oxidative phosphorylation, the most efficient pathway for ATP generation in cells, depends on the availability of molecular oxygen. Therefore, sophisticated oxygen transport systems, such as lungs combined with blood circulation in tetrapods or trachea in insects, have evolved in complex multicellular organisms. Local differences in tissue oxygenation are of crucial importance during development (Eliasson and Jönsson, 2010; Gustafsson et al., 2005; Mohyeldin et al., 2010) and adult life, as well as during cancer development (Forster et al., 2017; Noman et al., 2015). The importance of oxygen gradients has inspired the development of a variety of oxygen-responsive chemical probes and imaging techniques (see e.g. Carreau et al., 2011; Shao and Ashkenazi, 2015).
Over 20 oxygen-responsive plasma membrane-permeable molecules have been reported during the last decade. Reversible and ratiometric nanoprobes allow accurate and time-resolved measurements (Dmitriev and Papkovsky, 2015; Fercher et al., 2010; Lee et al., 2009; O'Riordan et al., 2007). However, these approaches suffer from complexity and require carefully controlled cell permeation, causing issues such as toxicity for example (reviewed by Dmitriev and Papkovsky, 2015). Dye-based techniques are not sufficiently advanced for targeting reporter molecules to specific organelles. Moreover, many of these methods also demand sophisticated equipment (e.g. lifetime phosphorescence microscopes), display modest spatial resolution and are time consuming (Spencer et al., 2014; Yoshihara et al., 2015). Limited tissue transparency is yet another problem encountered with available methods of oxygen imaging. The accessible depth is currently limited by the z-resolution of two-photon microscopy (Spencer et al., 2014), while physical dissection of the experimental animal or other complex multicellular object might lead to a contamination with atmospheric gases and distort the analysis.
Another family of oxygen sensors comprises genetically encoded fluorescent protein (FP) variants. Widely used biosensors are based on hypoxia inducible factor 1α (HIF1-α)-derived promoter sequences and/or degrons fused to FPs. Although these biosensors are more convenient, they are indirect reporters. They measure a complex biochemical response to oxygen concentration rather than its concentration directly (Danhier et al., 2015; Ivan et al., 2001; Misra et al., 2017; Santhakumar et al., 2012).
However, FPs can also be used to sense oxygen levels more directly. All FPs are synthesized as non-fluorescent precursors. The fluorophore develops during a maturation process that involves cyclization of a defined region of the protein backbone into a heterocycle, taking minutes to hours (Chudakov et al., 2010). While fluorophore buildup and fluorescence in FPs are usually independent of physiological variations in the intracellular milieu, two factors are known to be important: temperature and the concentration of molecular oxygen. Temperature affects the speed of the maturation process, which is known to include one or several oxygen-dependent oxidation reactions in the case of GFP/RFP maturation (Verkhusha et al., 2004). The understanding of oxygen effects is incomplete. It is clear that purified GFP/RFP-type FPs are unable to mature in deoxygenated buffers (Heim et al., 1994; Inouye and Tsuji, 1994) as well as in severely anoxic cells in vivo (e.g. Kaida and Miura, 2012). Molecular oxygen is believed to affect fluorophore oxidation, but the exact parameters of this reaction have not been analyzed in detail, presumably in part owing to experimental complications of oxygen control and the short maturation times of commonly used FPs [e.g. 14 min for EGFP (Iizuka et al., 2011), 15-30 min for mCherry (Merzlyak et al., 2007; Shaner et al., 2004)].
In contrast to GFP-like FPs, flavin mononucleotide-binding fluorescent proteins (FbFPs) do not require molecular oxygen and are able to develop fluorescence under anoxic conditions (Drepper et al., 2007). A FRET-based intracellular oxygen sensor FluBO has been created in E. coli by fusing FbFP and YFP (Potzkei et al., 2012). YPF is unable to mature under oxygen-deficient conditions, leading to scorable changes in the FbFP/YFP fluorescence ratio. However, the irreversibility and high speed of maturation limit the use of this biosensor to cells with a very rapid division rate, where the mature probe is constantly diluted by cell growth, resulting in low levels of mature FP. Furthermore, although the ability of FluBO to serve as a reporter at nearly anoxic conditions was well demonstrated, its functionality in conditions close to physiological range remains to be determined. Therefore, it is questionable whether it can be used in eukaryotic cells or model animals. Recently, another oxygen-independent FP, UnaG (Kumagai et al., 2013), was used to improve HIF1-α-mediated hypoxia response sensing in cultured cells as well as in a mouse model (Erapaneedi et al., 2016). UnaG fused to the oxygen-sensitive protein mOrange was placed under the control of HIF1-α-responsive elements. Exposure to hypoxia resulted in solely UnaG (green) fluorescence, since the mOrange fluorophore was unable to mature. Dual (green and red) labeling marked reoxygenized cells that pass the period of HIF1-α activation. Unfortunately, the authors did not investigate the utility of the UnaG-mOrange fusion for direct oxygen sensing outside the context of the HIF1-α pathway.
Here we demonstrate that the oxygen concentration present during maturation of the two-color DsRed FT protein in vitro and in Drosophila larvae determines the resulting red-to-green fluorescence ratio. The differences between hypoxic and normoxic cells could be observed directly with a very simple experimental setup (e.g. a fluorescence stereomicroscope). DsRed FT readouts are very robust and stable upon fixation. Thus, DsRed FT can be used for recording the history of oxygen exposure in model animals with single-cell resolution. For the first time, to our knowledge, we report a whole-animal map of oxygen exposure history, acquired with single-cell precision.
RESULTS
Identifying a fluorophore pair with different oxygen requirements
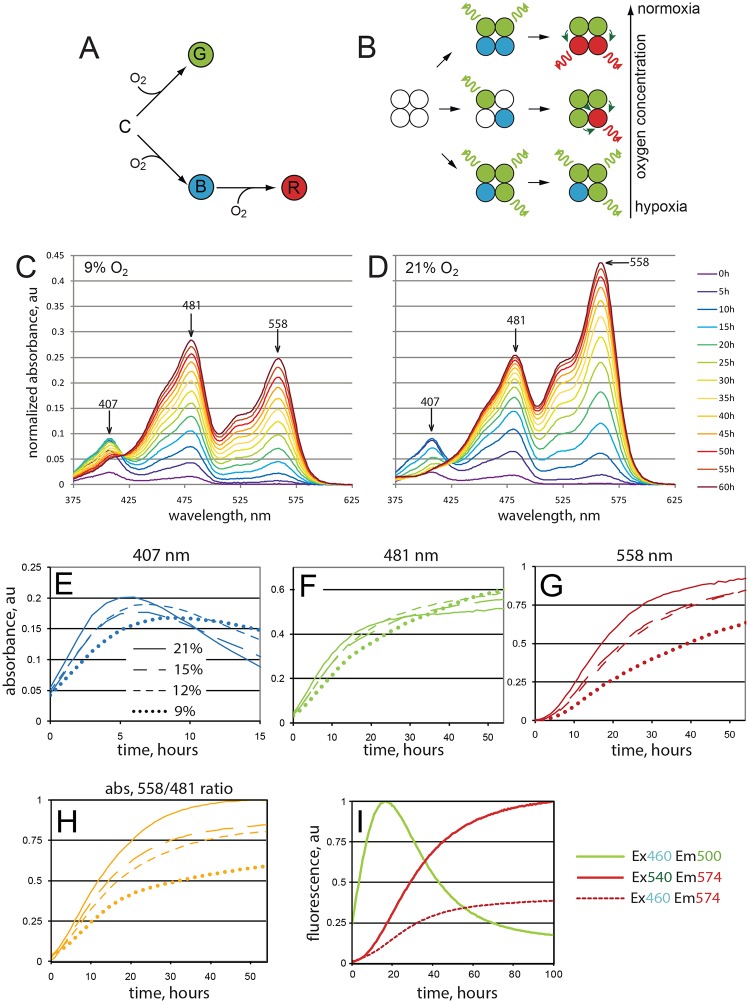
To investigate the effects of oxygen concentration on the maturation of different fluorophore species, we have chosen DsRed FT (also designated DsRed-E5NA), a non-aggregating form of DsRed that can be used as a fluorescent timer protein (Terskikh et al., 2000; Yanushevich et al., 2002). This DsRed derivate is able to mature competitively into either (1) a green fluorescent form or (2) a blue fluorescent intermediate form that slowly transforms further into (3) a red fluorescent form (Fig. 1A). Maturation of the green form occurs more rapidly than the blue to red transformation (Subach and Verkhusha, 2012; Verkhusha et al., 2004). Therefore, after a pulse of DsRed FT expression, blue fluorescence is detected initially, followed by green and eventually red fluorescence (Verkhusha et al., 2004). The green and red fluorescent species are terminal forms that do not mature further. Nevertheless, green fluorescence disappears eventually in parallel with an increase in red fluorescence. This green to red change results from robust quenching of the green fluorescence by Förster resonance energy transfer (FRET). FRET from the green to the red fluorophore occurs with very high efficiency within the tetrameric DsRed FT complexes (Fig. 1B,I) (Verkhusha et al., 2004). While this feature contributes to the time-dependent change from green to red fluorescence, it precludes the simple translation of the fluorescence ratio into precise ratios of the different fluorophore isoforms. In contrast to fluorescence, absorbance is not significantly affected by tetramerization. Therefore, in our in vitro studies we analyzed mainly absorbance as a measure of the different DsRed FT species composition.
Fig. 1.
Fluorescence color changes during maturation of DsRed FT. (A) DsRed FT maturation can occur via distinct oxygen-dependent pathways that generate fluorophores emitting the indicated colors. (B) Efficient FRET from green to red fluorophores (bent arrows) in tetramers quenches the emission (wavy arrows) of green but not red fluorophores. Quenching further enhances the differential oxygen dependence of the two maturation pathways, resulting in a very high oxygen sensitivity of the red-to-green fluorescence ratio. (C,D) Oxygen concentration affects in vitro DsRed FT maturation into spectroscopically distinct forms. Newly synthesized DsRed-FT was purified and exposed to 9% (C) or 21% (D) oxygen. Absorbance spectra were determined at the indicated time points. (E-G) Maturation dynamics of distinct isoforms. Whereas the green isoform (F) is relatively insensitive to oxygen concentration, the blue (E) and red (G) isoforms are strongly dependent on oxygen. (H) Ratio of red/green isoforms depends on oxygen concentration. (I) Fluorescence of the green isoform declines with an increase of red fluorescence. Excitation and emission values used are shown. Note the increase of red (Em574 nm) fluorescence excited by blue (Ex460 nm) light (dashed line). This signal largely corresponds to the intratetrameric FRET. The experiment was performed at normoxic conditions. A representative experiment out of three replicates is shown.
At atmospheric oxygen concentration (21% O2), full maturation of DsRed FT protein takes more than 8 h at 37°C. At room temperature (25°C), maturation was even slower, providing good temporal resolution for the analysis of fluorophore transformations. To analyze the effects of oxygen concentration, we placed purified immature DsRed FT samples in gas-tight cuvettes and saturated it with oxygen-depleted (9, 12 or 15% O2) or normal (21% O2) air. Our analysis revealed that the maturation dynamics of the different spectral forms over time strongly depends on oxygen concentration (Fig. 1C-H). Whereas the green form appeared to mature with a comparable speed irrespective of oxygen concentration (Fig. 1F), the red form experienced a significant maturation delay under hypoxic conditions (Fig. 1G). Therefore, the red fluorophore maturation pathway is significantly less efficient in hypoxia than that resulting in the green fluorophore species. The blue-to-red oxidation step appears to be the most oxygen-sensitive stage of the maturation process.
nlsTimer is an in vivo sensor that responds to cell oxygenation in Drosophila
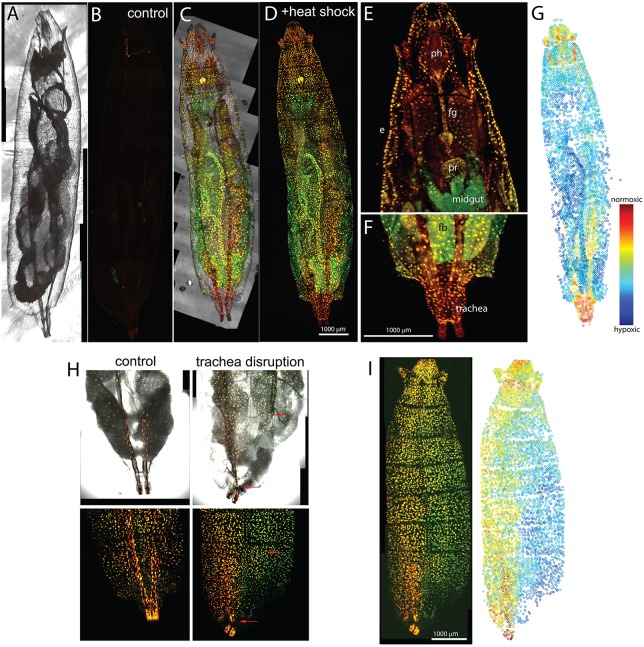
The differential oxygen dependency of the two maturation pathways observed in vitro suggested that DsRed FT protein might allow analyses of differential oxygenation in tissues and organisms. To evaluate this possibility we performed experiments with Drosophila melanogaster. Fruit flies develop successfully in persistent hypoxic conditions, tolerating as little as 7.5% O2 (Harrison and Haddad, 2011). Moreover, temporally regulated pulse-chase expression under heat shock promoter control is well established in D. melanogaster. To extend the versatility for future applications, a transgenic line encoding a nuclear localization signal fused to DsRed FT under the control of the UAS promoter (Brand and Perrimon, 1993), UASt-nlsTimer, was established and used in combination with an hs-GAL4 driver insertion. Progeny possessing both transgenes (hs>nlsTimer) were collected and aged to the second larval instar (50-72 h after egg deposition) before the application of a heat shock (1 h at 37°C). After the heat shock, larvae were aged for three additional days at 25°C and subjected to rapid freeze fixation and imaging (Fig. 2C-G). As expected, strong nuclear fluorescent signals were observed throughout the heat-treated hs>nlsTimer larvae. Control experiments with larvae that were not exposed to a heat shock (Fig. 2A,B), as well as with heat-treated larvae carrying only UASt-nlsTimer but not hs-GAL4, demonstrated that the vast majority of the observed nuclear signals reflected heat-induced hs-GAL4-mediated UASt-nlsTimer expression. Heat shock-independent UASt-nlsTimer expression was detected only at low levels in single cells within salivary glands, eye discs, brain and other tissues (Fig. 2B, Fig. 4D-G). The high signal-to-background ratio in the heat-treated hs>nlsTimer larvae allowed whole-animal confocal imaging and fully automated recognition of fluorescent nuclei in z-stacks, followed by quantification and accurate numeric determination of the ratio between the red and green fluorescent signals (r/g ratio) in these nuclei (Fig. 2G). This r/g ratio clearly varied regionally, and the spatial distribution of nuclei with particularly high or low r/g ratios appeared to be in line with expectations concerning tissue oxygenation. For example, the most reddish nuclei were located in the posterior spiracles, i.e. in the posterior ends of the tracheal trunks where ambient air diffuses into the tracheal system (Fig. 2F).
Fig. 2.
Variable red/green ratio after nlsTimer expression in vivo. Drosophila larvae with an hs-GAL4 and a UASt-nlsTimer transgene were subjected to heat shock (C-G), or not (A,B), and imaged after 3 days of recovery at 25°C. Maximum intensity projections (A-D) or single focal planes of the anterior (E) and posterior (F) region of the larvae shown in C and D displayed at higher magnification. Different tissues display distinct red/green ratios. e, epidermis; ph, pharynx; fg, foregut; pr, proventriculus; fb, fat body. (G) Heat map of the red/green ratio in the larva shown in D. Each circle corresponds to one nucleus. See also Movie 1. n=5. (H,I) Disruption of tracheal trunk leads to lateral asymmetry in the color of nlsTimer. Larvae were prepared as in C-G. Tracheal trunks were collapsed with forceps immediately after the heat shock. (H) Larval abdomen. Dorsal trunk breakages are marked with red arrows. (I) Whole larva view. Note the dramatic decrease in red/green ratio in the corresponding damaged half of the larva. Contrast is set differently from C-G to cover the full dynamic range. Representative data from three consistent experiments are shown. At least three animals were used in each experiment.
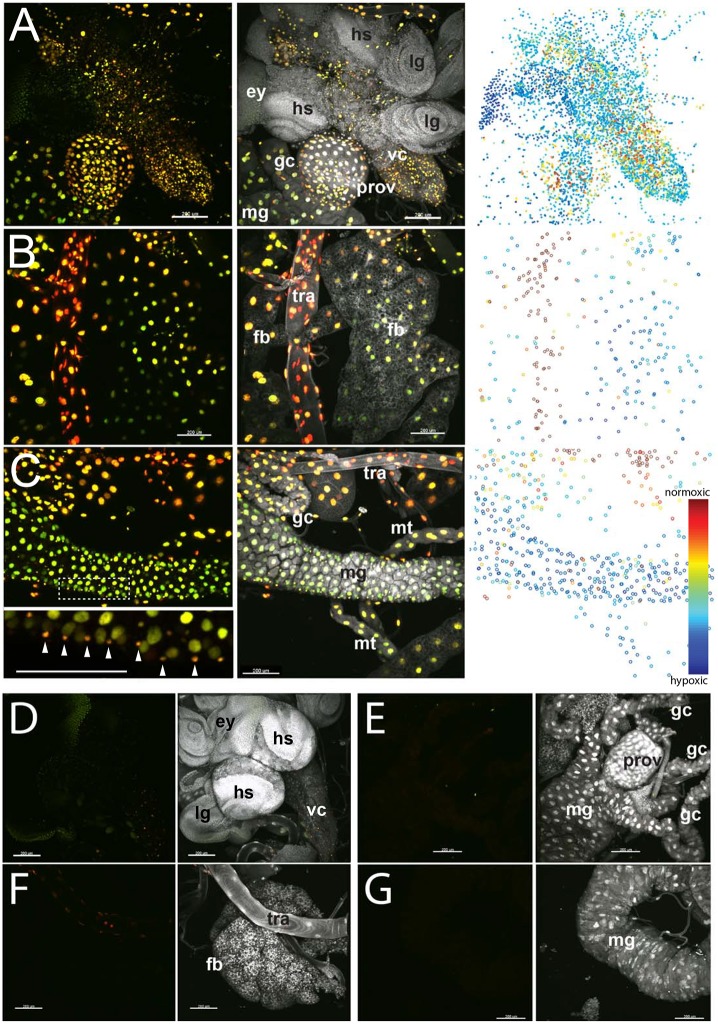
Fig. 4.
Variable nlsTimer red/green ratios in fixed organ preparations. (A-C) Organs from hs>nlsTimer larvae after heat shock and normoxic recovery were dissected, fixed and labeled with a DNA stain (DAPI, white). Variation in the red/green ratio was observed between and also within different tissues. (A) Central nervous system with ventral nerve cord (vc) and brain lobes (hs), proventriculus (prov), gastric caeca (gc) and midgut (mg). Eye (ey) and leg (lg) imaginal discs, as well as the outer layers of the brain lobes, display low levels of reporter expression, presumably as a result of nlsTimer protein dilution by extensive growth and cell proliferation during recovery. (B) Fat body (fb) cells adjacent to trachea (tra) have a higher red/green ratio than those further away. (C) Midgut region with Malpighian tubules (mt) and additional tissues (see above). The inset below shows the same panel with the color dynamic range adjusted to display the difference between enterocyte and visceral muscle nuclei (arrowheads). (D-G) Control organs from hs>nlsTimer larvae that were not exposed to a heat shock. Imaging was performed with the same settings as in panels A-C. Representative data from three consistent experiments are shown. Scale bars: 200 μm.
To demonstrate that r/g ratios were indeed reflecting oxygenation, we selectively destroyed one of the tracheal trunks in 3-day-old larvae, just after the heat shock performed as described above. After 3 days of incubation at 25°C, the larvae displayed a clear lateral asymmetry in r/g ratios (Fig. 2H,I), with increased red fluorescence in the intact domain, indicating that nlsTimer maturation responds to trachea-mediated oxygenation.
Color of nlsTimer fluorescence correlates with the level of external oxygen supply
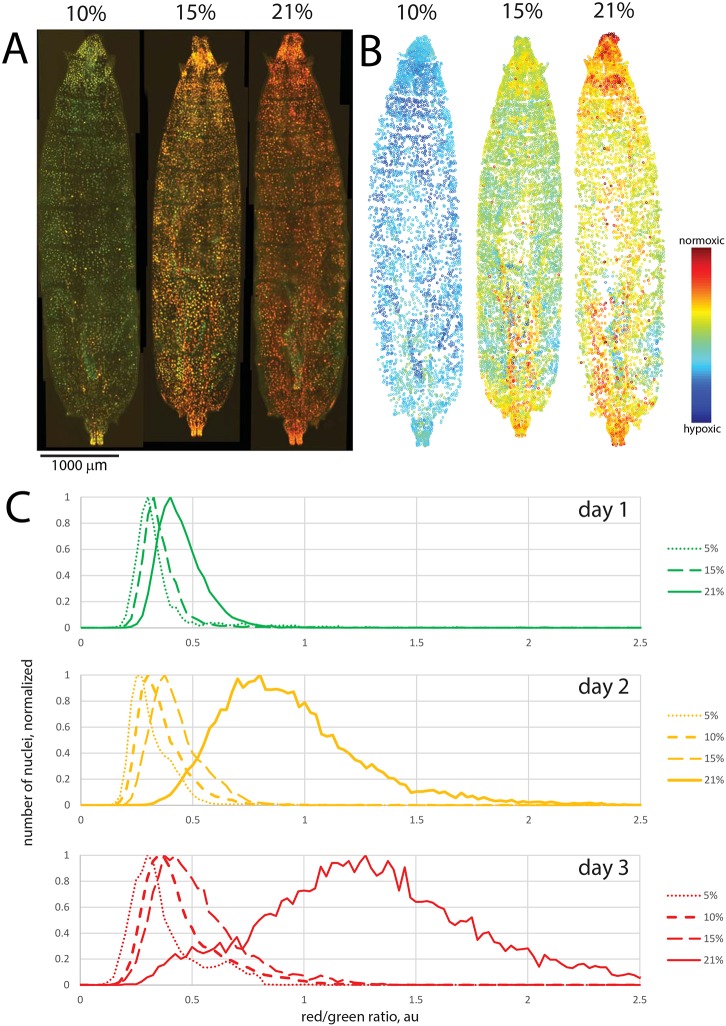
To confirm that oxygen concentration is indeed a major determinant of the r/g ratio resulting after nlsTimer expression, we incubated hs>nlsTimer larvae after heat shock at different oxygen tensions (5%, 10%, 15% and 21%). Thereafter, aliquots of larvae were collected at daily intervals and analyzed by ratiometric imaging. These analyses revealed a dependence of nlsTimer fluorescence on oxygen concentration, as expected based on the in vitro experiments. The r/g ratio correlated well with oxygen concentration (Fig. 3A-C). We note that the vast majority of nuclei in larvae raised in normoxic conditions showed an r/g ratio above the average values from animals exposed to 5% hypoxia (Fig. 3C). Therefore, the dynamic range of the nlsTimer reporter appears to cover the physiological range of oxygen concentrations present in cells of larvae developing under normoxic conditions.
Fig. 3.
Oxygen concentration determines red/green ratio after nlsTimer expression in vivo. (A-C) hs>nlsTimer larvae were heat treated as in Fig. 2, followed by recovery in different oxygen concentrations (5%, 10%, 15%, 21%, as indicated). (A,B) Red and green fluorescence (A) and a heat map of the red/green ratio (B) after 3 days of maturation. Note that contrast is set differently from Fig. 2 to cover the full dynamic range. (C) Dependence of red/green ratio on recovery time and oxygen concentration. The histograms were produced from images similar to those shown in A. Curves are based on data points from three or more animals and 1500 or more individual nuclei from each animal.
Parental DsRed is considered one of the most pH-tolerant FPs (Baird et al., 2000). DsRed FT variant maturation itself has been shown to tolerate a wide range of ionic strength, pH variations within the physiological range (7.0-8.0), variation in protein concentration and the presence of EDTA (Terskikh et al., 2000). Although we have not excluded these factors experimentally, we consider it unlikely that these parameters vary persistently in vivo to an extent that could produce a major effect on the readout.
The color of mitochondria-targeted DsRed1-E5 protein has been shown to change upon incubation of cells in conditions promoting the generation of reactive oxygen species (ROS) (Laker et al., 2014). To exclude direct ROS effects on our readouts we performed several experiments. First, we exposed maturing purified DsRed FT protein to H2O2. We found that in a wide range of concentrations (up to 1 mM, which is much higher than is physiologically relevant), H2O2 has no effect on DsRed FT maturation (Fig. S1A), as has been described for its parental protein DsRed (Gross et al., 2000). Also, feeding larvae with the ROS-inducing compounds paraquat and rotenone did not result in significant changes in r/g ratios (Fig. S1B).
Color differences of nlsTimer fluorescent isoforms are retained upon fixation
To show that the variations in r/g ratio resulting after a nlsTimer expression pulse and chase period are not destroyed by standard fixation, we opened and fixed hs>nlsTimer larvae treated as described above. Selected organs were dissected and mounted on microscope slides. In fixed samples, the r/g ratio varied between different organs (Fig. 4) with a slight red shift when compared with unfixed samples (Fig. S2). Highly tracheated organs were more red-shifted than less tracheated organs. The most oxygenized organ appeared to be the trachea and the most hypoxic structures the posterior regions of the fat body. Interestingly, differences in the r/g ratio were also apparent within a given tissue. Regions of the fat body adjacent to trachea showed elevated levels of red fluorescence (Fig. 4B). Within the central nervous system, reddish cells were more numerous in the ventral nerve cord than in the brain lobes (Fig. 4A). Another interesting example of fine variation was observed in the midgut. Comparison of fluorescence in the large nuclei of enterocytes with that in the small nuclei of circular muscles around the gut revealed an elevated level of red fluorescence in the latter (Fig. 4C, inset), indicating a higher oxygen supply in these cells.
These results demonstrate the capacity of our technique for identification of cell-to-cell variation and also indicate that fixation does not interfere with the use of nlsTimer as a reporter of the cellular history of oxygen exposure.
Approaches shown above for larvae (Figs 2 and 4) can be also applied to adult Drosophila animals (Fig. S3A) and organs (Fig. S3B).
DISCUSSION
Oxygen concentrations within the human body vary from tissue to tissue and are known to be severely disturbed under pathological conditions such as diabetes, coronary heart disease or stroke. Furthermore, hypoxia affects malignant growth and is a predictive marker of poor clinical cancer outcome. Some strategies of cancer therapy target hypoxic regions of tumors by drugs with a cytotoxicity that is alleviated by reversible O2-dependent oxidation (for reviews, see Carreau et al., 2011; Harris, 2002; Vaupel and Mayer, 2007; Wilson and Hay, 2011). Thus, accurate measurement of oxygen concentrations in patients or in animal models is important for medical research.
In this study, we found that the maturation of distinct fluorophores of DsRed FT is differentially sensitive to oxygen concentration. The DsRed FT-based probe (nlsTimer) that we expressed in vivo provides a clear example that fluorophore maturation can be strongly influenced by physiological variations in oxygenation. We demonstrate that the history of oxygen exposure of individual cells within a multicellular organism can be recorded with nlsTimer. This allowed the generation of a whole-animal map of intracellular oxygen supply. We also show that the dynamic range of nlsTimer allows differentiation within the biologically relevant range close to normoxia. This physiological range is assessed relatively poorly by other genetically encoded sensors. The simple experimental setup, direct observation, stable and very robust output signals make nlsTimer a useful tool for high-throughput analyses, such us genetic screenings and oxygen exposure-mediated fluorescence-activated cell sorting (FACS). Targeting of DsRed FT to other organelles than the nucleus might also provide information about compartment-specific oxygen supply.
The slow and irreversible maturation of nlsTimer prevents the tracking of rapid changes in oxygen concentration, and in this sense nlsTimer differs from sensors such as the GCaMP reporter family for Ca2+ ions (Chen et al., 2013) or Hyper for peroxide (Belousov et al., 2006). However, current methods to assess intracellular oxygen concentrations with high temporal resolution suffer from slow dye penetration into cultured cells (Fercher et al., 2011) or are functional only at very low oxygen concentrations (Elowitz et al., 1997; Kaida and Miura, 2012; Potzkei et al., 2012; Takahashi and Sato, 2011) and are hardly applicable in animals. Moreover, if created, a hypothetical cell-permeable or genetically encoded, highly dynamic oxygen reporter with a broad dynamic range might encounter problems when used in poorly transparent or large animal models. Low optical transparency of a tissue of interest and the risk of gas contamination upon intervention will likely make some parts of the animal inaccessible for imaging. Although Drosophila larvae are relatively transparent, allowing direct imaging through the body wall, even more detailed results can be obtained with dissected and fixed organ samples in the case of our nlsTimer approach. This opens up opportunities for combining it with immunostaining or other labeling techniques. Furthermore, tissue dissociation followed by acquisition of relatively hypoxic or relatively normoxic cell populations should be feasible with the help of FACS. The limitation of the probe is its slow maturation speed that requires 3 days of chase at 25°C for the best resolution (Fig. 3C). It should also be stated that the multiplicity of maturation reactions of DsRed FT protein might result in non-additive readouts if the cell of interest moves from a normoxic to a hypoxic environment or vice versa during the prolonged time of the chase. Oxidation of the blue form, which seems to be a limiting oxygen-dependent stage (Fig. 1C,D), might occur within relatively short periods of time. Therefore, the probe might tend to report the maximum, and not the average, oxygen concentration experienced by the cell (not shown).
We expect our assay to work well in other small poikilothermic animal models (e.g. C. elegans, D. rerio) where pulse-chase expression experiments are well established. Combining the two-color oxygen reporter with a destabilization sequence might also be helpful in model organisms, such as mice, where pulsed protein expression is not an established routine. A future direction of research is to engineer more rapidly maturing dsRed-FT derivatives that will increase temporal resolution.
MATERIALS AND METHODS
DsRedFT-NA isolation and measurements
DsRedFT-NA in pQE-30 (Yanushevich et al., 2002) was transformed into E. coli XL-1 Blue. A single colony in PBS was spread onto five to seven Petri dishes and grown at 30°C for 20 h. Under these conditions of expression, most DsRedFT-NA protein is still immature at the time of collection and purification. Bacteria were washed from the dishes with PBS and lysed by sonication. Soluble FP was isolated using the metal affinity sorbent TALON (Clontech) according to the manufacturer's recommendations.
Protein samples in sealable quartz cuvettes (117F-QS, Hellma Analytics) were saturated with air containing 9, 12, 15 or 21% O2 using bubbling with an Everest Summit II generator (Hypoxico) for 10 min. The tightly closed cuvettes were placed into a Varian Carry 100 multichannel spectrophotometer and measured in parallel for 3 days at room temperature (see Fig. 1). In the experiment shown in Fig. S1A, protein samples were supplemented with hydrogen peroxide at the concentrations indicated.
Generation of transgenic flies
To construct pUASt-nlsDsRedFT-NA-attB, the DsRedFT-NA-encoding fragment was amplified by PCR and introduced into the pUASt-attB vector cut with BglII/XbaI. The nls sequence preceded by an initiation codon in an optimal sequence context was introduced as a double-stranded oligonucleotide after annealing a forward oligo (5′-TATAAGATCTCAACATGCCAAAGAAGAAACGTAAAGTTGCCTCCTCCGAGAACGTC-3′) and a reverse oligo (5′-TATATCTAGATTACAGGAACAGGTGGTGGCGG-3′). pUASt-nlsDsRedFT-NA-attB was integrated into the M {3xP3-RFP.attP} ZH-86Fb landing site as described (Bischof et al., 2007). The 3xP3-dsRed marker was removed by crossing in a Cre recombinase-expressing transgene followed by outcrossing of the cleaned UASt-nlsDsRedFT-NA insertion. The resulting homozygous flies were used for further experiments.
Fly stocks
In addition to the UASt-nlsDsRedFT-NA line we used a stock with a GAL4 transgene under control of the Hsp70 promoter and two copies of the sevenless enhancer (Ruberte et al., 1995). Another heat shock GAL4 driver, as used in Fig. S3B, was chosen for lower background expression in adult brain and was a gift from Dr Maksim Erokhin (Institute of Gene Biology, RAS). Animals with one copy each of hs-GAL4 and UASt-nlsTimer (hs>UASt-nlsTimer) were used in all experiments.
Animal collection and sample preparation
Embryos were allowed to develop at 25°C. 50-72 h after egg deposition larvae collections were subject to a heat shock (1 h at 37°C) followed by incubation at 25°C for 1-3 days. To perform the incubation at lower O2 concentrations, collection plates or tubes with larvae were placed into hermetic chambers filled with pre-cast O2/N2 mixtures of 5%, 10% and 15% oxygen tensions.
Feeding larvae with ROS-inducing drugs was performed by adding rotenone (0.4 mM, Matrix Scientific) or paraquat (10 mM, Sigma) to the fly food. Larvae were placed into drug-containing food immediately after the heat shock and incubated for 3 days at 25°C.
For whole larvae preparations, the animals were mounted on a drop of 90% glycerol/PBS, covered and flattened with a coverslip, incubated at −20°C for 20 min to prevent movements and imaged. To reveal intestinal organs, which are masked by light scattering resulting from the presence of a large fat body, early third instar larvae with small fat bodies were selected for Fig. 2. To prepare fixed tissue samples, larvae were opened in cold PBS, inverted and fixed in 4% paraformaldehyde in PBS for 20 min. After three washes with PBS the organs were dissected and mounted in 90% glycerol/PBS. Adult brains were dissected in PBS and fixed as described above. For comparison of native and fixed tissue, organs were placed on poly-L-lysine (Sigma) coated slides, imaged, fixed as above and re-imaged.
Microscopy and image analysis
Acquisition of image stacks was performed with a Leica TCS SP2 confocal microscope equipped with a 10× objective, with an Olympus FV1000 confocal microscope equipped with a 10× or 60× objective, and with a Nikon FN-1 microscope equipped with a 10× or 20× lens. z-spacing was 1 µm. Optical slices were processed individually in CellProfiler (Kamentsky et al., 2011) for object recognition and quantification of red and green intensities. Single-slice data were then imported into MATLAB (MathWorks) software and the r/g ratios plotted as histograms or scatter plots. The 3D animations shown in Movie 1 were generated using Imaris software (Bitplane).
Image analysis
All confocal images were acquired on a Carl Zeiss 700 microscope and analyzed using ImageJ.
Supplementary Material
Acknowledgements
We thank Dr Maksim Erokhin (Institute of Gene Biology, RAS) for hs-GAL4 fly stock and Prof. Ma Denke (UCSF) for access to the hypoxia chamber.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: P.V.L., K.A.L., C.F.L.; Methodology: P.V.L.; Software: P.V.L.; Investigation: P.V.L., K.A.L., T.M., B.H., A.S.M.; Writing - original draft: P.V.L.; Writing - review & editing: P.V.L., K.A.L., C.F.L.; Visualization: P.V.L.; Supervision: P.V.L., C.F.L.; Funding acquisition: P.V.L., K.A.L., C.F.L.
Funding
This work was supported by grants from the Swiss National Science Foundation (Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung) to C.F.L. (grant number 31003A_152667) and National Institutes of Health grants R01 AI36178, R01 AI40085 and P01 AI091575. Imaging has been partially performed with IGB RAS equipment supported by the Ministry of Education and Science of the Russian Federation (grant number 16.552.11.7067). In vitro protein characterization was supported by the Russian Science Foundation (project 14-25-00129). Experiments were partially carried out using equipment provided by the IBCH core facility (CKP IBCH, supported by the Ministry of Education and Science of the Russian Federation, grant RFMEFI62117X0018). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.156257.supplemental
References
- Baird G. S., Zacharias D. A. and Tsien R. Y. (2000). Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proc. Natl. Acad. Sci. USA 97, 11984-11989. 10.1073/pnas.97.22.11984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belousov V. V., Fradkov A. F., Lukyanov K. A., Staroverov D. B., Shakhbazov K. S., Terskikh A. V. and Lukyanov S. (2006). Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat. Methods 3, 281-286. 10.1038/nmeth866 [DOI] [PubMed] [Google Scholar]
- Bischof J., Maeda R. K., Hediger M., Karch F. and Basler K. (2007). An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA 104, 3312-3317. 10.1073/pnas.0611511104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A. H. and Perrimon N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401-415. [DOI] [PubMed] [Google Scholar]
- Carreau A., El Hafny-Rahbi B., Matejuk A., Grillon C. and Kieda C. (2011). Why is the partial oxygen pressure of human tissues a crucial parameter? Small molecules and hypoxia. J. Cell. Mol. Med. 15, 1239-1253. 10.1111/j.1582-4934.2011.01258.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T.-W., Wardill T. J., Sun Y., Pulver S. R., Renninger S. L., Baohan A., Schreiter E. R., Kerr R. A., Orger M. B., Jayaraman V. et al. (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295-300. 10.1038/nature12354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chudakov D. M., Matz M. V., Lukyanov S. and Lukyanov K. A. (2010). Fluorescent proteins and their applications in imaging living cells and tissues. Physiol. Rev. 90, 1103-1163. 10.1152/physrev.00038.2009 [DOI] [PubMed] [Google Scholar]
- Danhier P., Krishnamachary B., Bharti S., Kakkad S., Mironchik Y. and Bhujwalla Z. M. (2015). Combining optical reporter proteins with different half-lives to detect temporal evolution of hypoxia and reoxygenation in tumors. Neoplasia 17, 871-881. 10.1016/j.neo.2015.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitriev R. I. and Papkovsky D. B. (2015). Intracellular probes for imaging oxygen concentration: how good are they? Methods Appl. Fluorescence 3, 034001 10.1088/2050-6120/3/3/034001 [DOI] [PubMed] [Google Scholar]
- Drepper T., Eggert T., Circolone F., Heck A., Krauß U., Guterl J.-K., Wendorff M., Losi A., Gärtner W. and Jaeger K.-E. (2007). Reporter proteins for in vivo fluorescence without oxygen. Nat. Biotechnol. 25, 443-445. 10.1038/nbt1293 [DOI] [PubMed] [Google Scholar]
- Eliasson P. and Jönsson J.-I. (2010). The hematopoietic stem cell niche: low in oxygen but a nice place to be. J. Cell. Physiol. 222, 17-22. 10.1002/jcp.21908 [DOI] [PubMed] [Google Scholar]
- Elowitz M. B., Surette M. G., Wolf P.-E., Stock J. and Leibler S. (1997). Photoactivation turns green fluorescent protein red. Curr. Biol. 7, 809-812. 10.1016/S0960-9822(06)00342-3 [DOI] [PubMed] [Google Scholar]
- Erapaneedi R., Belousov V. V., Schäfers M. and Kiefer F. (2016). A novel family of fluorescent hypoxia sensors reveal strong heterogeneity in tumor hypoxia at the cellular level. EMBO J. 35, 102-113. 10.15252/embj.201592775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fercher A., O'Riordan T. C., Zhdanov A. V., Dmitriev R. I. and Papkovsky D. B. (2010). Imaging of cellular oxygen and analysis of metabolic responses of mammalian cells. Methods Mol. Biol. 591, 257-273. 10.1007/978-1-60761-404-3_16 [DOI] [PubMed] [Google Scholar]
- Fercher A., Borisov S. M., Zhdanov A. V., Klimant I. and Papkovsky D. B. (2011). Intracellular O2 sensing probe based on cell-penetrating phosphorescent nanoparticles. ACS Nano 5, 5499-5508. 10.1021/nn200807g [DOI] [PubMed] [Google Scholar]
- Forster J. C., Harriss-Phillips W. M., Douglass M. J. J. and Bezak E. (2017). A review of the development of tumor vasculature and its effects on the tumor microenvironment. Hypoxia 5, 21-32. 10.2147/HP.S133231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross L. A., Baird G. S., Hoffman R. C., Baldridge K. K. and Tsien R. Y. (2000). The structure of the chromophore within DsRed, a red fluorescent protein from coral. Proc. Natl. Acad. Sci. USA 97, 11990-11995. 10.1073/pnas.97.22.11990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson M. V., Zheng X., Pereira T., Gradin K., Jin S., Lundkvist J., Ruas J. L., Poellinger L., Lendahl U. and Bondesson M. (2005). Hypoxia requires notch signaling to maintain the undifferentiated cell state. Dev. Cell 9, 617-628. 10.1016/j.devcel.2005.09.010 [DOI] [PubMed] [Google Scholar]
- Harris A. L. (2002). Hypoxia--a key regulatory factor in tumour growth. Nat. Rev. Cancer 2, 38-47. 10.1038/nrc704 [DOI] [PubMed] [Google Scholar]
- Harrison J. F. and Haddad G. G. (2011). Effects of oxygen on growth and size: synthesis of molecular, organismal, and evolutionary studies with Drosophila melanogaster. Annu. Rev. Physiol. 73, 95-113. 10.1146/annurev-physiol-012110-142155 [DOI] [PubMed] [Google Scholar]
- Heim R., Prasher D. C. and Tsien R. Y. (1994). Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc. Natl. Acad. Sci. USA 91, 12501-12504. 10.1073/pnas.91.26.12501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka R., Yamagishi-Shirasaki M. and Funatsu T. (2011). Kinetic study of de novo chromophore maturation of fluorescent proteins. Anal. Biochem. 414, 173-178. 10.1016/j.ab.2011.03.036 [DOI] [PubMed] [Google Scholar]
- Inouye S. and Tsuji F. I. (1994). Evidence for redox forms of the Aequorea green fluorescent protein. FEBS Lett. 351, 211-214. 10.1016/0014-5793(94)00859-0 [DOI] [PubMed] [Google Scholar]
- Ivan M., Kondo K., Yang H., Kim W., Valiando J., Ohh M., Salic A., Asara J. M., Lane W. S. and Kaelin W. G. Jr. (2001). HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292, 464-468. 10.1126/science.1059817 [DOI] [PubMed] [Google Scholar]
- Kaida A. and Miura M. (2012). Visualizing the effect of hypoxia on fluorescence kinetics in living HeLa cells using the fluorescent ubiquitination-based cell cycle indicator (Fucci). Exp. Cell Res. 318, 288-297. 10.1016/j.yexcr.2011.10.016 [DOI] [PubMed] [Google Scholar]
- Kamentsky L., Jones T. R., Fraser A., Bray M.-A., Logan D. J., Madden K. L., Ljosa V., Rueden C., Eliceiri K. W. and Carpenter A. E. (2011). Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics 27, 1179-1180. 10.1093/bioinformatics/btr095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A., Ando R., Miyatake H., Greimel P., Kobayashi T., Hirabayashi Y., Shimogori T. and Miyawaki A. (2013). A bilirubin-inducible fluorescent protein from eel muscle. Cell 153, 1602-1611. 10.1016/j.cell.2013.05.038 [DOI] [PubMed] [Google Scholar]
- Laker R. C., Xu P., Ryall K. A., Sujkowski A., Kenwood B. M., Chain K. H., Zhang M., Royal M. A., Hoehn K. L., Driscoll M. et al. (2014). A novel mitotimer reporter gene for mitochondrial content, structure, stress, and damage in vivo. J. Biol. Chem. 289, 12005-12015. 10.1074/jbc.M113.530527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.-E. K., Smith R. and Kopelman R. (2009). Nanoparticle PEBBLE sensors in live cells and in vivo. Annu. Rev. Anal. Chem. 2, 57-76. 10.1146/annurev.anchem.1.031207.112823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merzlyak E. M., Goedhart J., Shcherbo D., Bulina M. E., Shcheglov A. S., Fradkov A. F., Gaintzeva A., Lukyanov K. A., Lukyanov S., Gadella T. W. J. et al. (2007). Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat. Methods 4, 555-557. 10.1038/nmeth1062 [DOI] [PubMed] [Google Scholar]
- Misra T., Baccino-Calace M., Meyenhofer F., Rodriguez-Crespo D., Akarsu H., Armenta-Calderón R., Gorr T. A., Frei C., Cantera R., Egger B. et al. (2017). A genetically encoded biosensor for visualising hypoxia responses in vivo. Biol. Open 6, 296-304. 10.1242/bio.018226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohyeldin A., Garzón-Muvdi T. and Quiñones-Hinojosa A. (2010). Oxygen in stem cell biology: a critical component of the stem cell niche. Cell Stem Cell 7, 150-161. 10.1016/j.stem.2010.07.007 [DOI] [PubMed] [Google Scholar]
- Noman M. Z., Hasmim M., Messai Y., Terry S., Kieda C., Janji B. and Chouaib S. (2015). Hypoxia: a key player in antitumor immune response. A review in the theme: cellular responses to hypoxia. Am. J. Physiol. Cell Physiol. 309, C569-C579. 10.1152/ajpcell.00207.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Riordan T. C., Zhdanov A. V., Ponomarev G. V. and Papkovsky D. B. (2007). Analysis of intracellular oxygen and metabolic responses of mammalian cells by time-resolved fluorometry. Anal. Chem. 79, 9414-9419. 10.1021/ac701770b [DOI] [PubMed] [Google Scholar]
- Potzkei J., Kunze M., Drepper T., Gensch T., Jaeger K.-E. and Buechs J. (2012). Real-time determination of intracellular oxygen in bacteria using a genetically encoded FRET-based biosensor. BMC Biol. 10, 28 10.1186/1741-7007-10-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruberte E., Marty T., Nellen D., Affolter M. and Basler K. (1995). An absolute requirement for both the type II and type I receptors, punt and thick veins, for dpp signaling in vivo. Cell 80, 889-897. 10.1016/0092-8674(95)90292-9 [DOI] [PubMed] [Google Scholar]
- Santhakumar K., Judson E. C., Elks P. M., McKee S., Elworthy S., van Rooijen E., Walmsley S. S., Renshaw S. A., Cross S. S. and van Eeden F. J. M. (2012). A zebrafish model to study and therapeutically manipulate hypoxia signaling in tumorigenesis. Cancer Res. 72, 4017-4027. 10.1158/0008-5472.CAN-11-3148 [DOI] [PubMed] [Google Scholar]
- Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N. G., Palmer A. E. and Tsien R. Y. (2004). Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22, 1567-1572. 10.1038/nbt1037 [DOI] [PubMed] [Google Scholar]
- Shao Q. and Ashkenazi S. (2015). Photoacoustic lifetime imaging for direct in vivo tissue oxygen monitoring. J. Biomed. Opt. 20, 036004 10.1117/1.JBO.20.3.036004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer J. A., Ferraro F., Roussakis E., Klein A., Wu J., Runnels J. M., Zaher W., Mortensen L. J., Alt C., Turcotte R. et al. (2014). Direct measurement of local oxygen concentration in the bone marrow of live animals. Nature 508, 269-273. 10.1038/nature13034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subach F. V. and Verkhusha V. V. (2012). Chromophore transformations in red fluorescent proteins. Chem. Rev. 112, 4308-4327. 10.1021/cr2001965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi E. and Sato M. (2011). Impact of intracellular diffusion of oxygen in hypoxic sensing. Adv. Exp. Med. Biol. 701, 301-306. 10.1007/978-1-4419-7756-4_40 [DOI] [PubMed] [Google Scholar]
- Terskikh A., Fradkov A., Ermakova G., Zaraisky A., Tan P., Kajava A. V., Zhao X., Lukyanov S., Matz M., Kim S. et al. (2000). “Fluorescent timer”: protein that changes color with time. Science 290, 1585-1588. 10.1126/science.290.5496.1585 [DOI] [PubMed] [Google Scholar]
- Vaupel P. and Mayer A. (2007). Hypoxia in cancer: significance and impact on clinical outcome. Cancer Metastasis Rev. 26, 225-239. 10.1007/s10555-007-9055-1 [DOI] [PubMed] [Google Scholar]
- Verkhusha V. V., Chudakov D. M., Gurskaya N. G., Lukyanov S. and Lukyanov K. A. (2004). Common pathway for the red chromophore formation in fluorescent proteins and chromoproteins. Chem. Biol. 11, 845-854. 10.1016/j.chembiol.2004.04.007 [DOI] [PubMed] [Google Scholar]
- Wilson W. R. and Hay M. P. (2011). Targeting hypoxia in cancer therapy. Nat. Rev. Cancer 11, 393-410. 10.1038/nrc3064 [DOI] [PubMed] [Google Scholar]
- Yanushevich Y. G., Staroverov D. B., Savitsky A. P., Fradkov A. F., Gurskaya N. G., Bulina M. E., Lukyanov K. A. and Lukyanov S. A. (2002). A strategy for the generation of non-aggregating mutants of Anthozoa fluorescent proteins. FEBS Lett. 511, 11-14. 10.1016/S0014-5793(01)03263-X [DOI] [PubMed] [Google Scholar]
- Yoshihara T., Hosaka M., Terata M., Ichikawa K., Murayama S., Tanaka A., Mori M., Itabashi H., Takeuchi T. and Tobita S. (2015). Intracellular and in vivo oxygen sensing using phosphorescent Ir(III) complexes with a modified acetylacetonato ligand. Anal. Chem. 87, 2710-2717. 10.1021/ac5040067 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.