ABSTRACT
Genetic studies in Drosophila have been instrumental in characterizing the Hippo pathway, which converges on the co-activator Yorkie to regulate target gene transcription. A routinely used strategy to interrogate upstream regulators of Yorkie involves the examination of selected Hippo target genes upon loss or gain of function of a suspected pathway regulator. A caveat with this strategy is that aberrant expression of a given Hippo target per se does not distinguish whether it is caused by changes in Yorkie or Yorkie-independent inputs converging on the same target gene. Building on previous findings that the DNA-binding transcription factor Scalloped mediates both Yorkie overexpression and loss-of-function phenotypes yet is itself dispensable for normal eye development, we describe a simple strategy to distinguish these possibilities by analyzing double-mutant clones of scalloped and a suspected Yorkie regulator. We provide proof of principle that this strategy can be used effectively to validate canonical Yorkie regulators and to exclude proteins that impact target expression independent of Yorkie. The described methodology and reagents should facilitate efforts to assess the expanding repertoire of proteins implicated in regulation of Yorkie activity.
KEY WORDS: Hippo signaling, Sd, Yki, Bantam, Expanded, Interommatidial cells
Summary: Analysis of double-mutant clones of sd and putative Yki regulators provides a simple and efficient strategy to validate upstream regulators of Yki activity in Hippo signaling.
INTRODUCTION
The Hippo signaling pathway is an evolutionarily conserved mechanism that regulates diverse physiological processes such as organ size control, cell fate determination, tissue regeneration and stem cell renewal (Harvey and Tapon, 2007; Johnson and Halder, 2014; Pan, 2010). This pathway comprises a core kinase cascade involving Hippo (Hpo; Mst1/2, also known as Stk4/3 in mammals), Salvador (Sav; Sav1 in mammals), Warts (Wts; Lats1/2 in mammals) and Mob as tumor suppressor (Mats; Mob1A/B in mammals) that converges on the transcriptional co-activator Yorkie (Yki; YAP/TAZ in mammals). Phosphorylation of Yki/YAP/TAZ excludes it from the nucleus, where it normally functions as a co-activator for the transcription of growth-promoting genes. Consistent with the requirement of Hippo signaling for normal tissue homeostasis, YAP is a bona fide oncogene and is activated/overexpressed in a wide range of human cancers (Pan, 2010).
The TEF/TEAD family transcription factors, Sd in Drosophila and TEAD1/2/3/4 in mammals, are the primary DNA-binding partners for the Yki/YAP/TAZ co-activators. Not only do they bind to Hippo target genes, such as Diap1 in Drosophila and Ctgf in mammals (Wu et al., 2008; Zhang et al., 2008; Zhao et al., 2008), the TEF/TEAD transcription factors have been identified as Yki/YAP-binding proteins in multiple unbiased protein-protein interaction screens in both Drosophila and mammals (Giot et al., 2003; Vassilev et al., 2001; Wu et al., 2008). The physiological importance of TEF/TEAD-Yki/YAP interactions is further supported by the discovery of a disease-causing point mutation in human TEAD1 (TEAD1Y421H underlying Sveinsson's chorioretinal atrophy) (Kitagawa, 2007) and the unbiased recovery of a missense mutant allele in Drosophila Yki (YkiP88L) that specifically disrupts this interaction (Wu et al., 2008), as well as structural studies of TEAD-YAP co-crystals that independently pinpoint these residues in the protein-binding interface (Chen et al., 2010; Li et al., 2010; Tian et al., 2010). Accentuating the physiological importance of this interaction, there is great interest in developing small molecule inhibitors of TEAD-YAP interactions as potential therapeutics against the YAP oncogene in human cancers (Liu-Chittenden et al., 2012).
Given its crucial role in normal development and tumorigenesis, there has been much interest in understanding the regulation of Yki/YAP/TAZ activity in Hippo signaling. In contrast to the relatively simple molecular organization of the core kinase cascade leading from Hpo/Mst to phosphorylation of Yki/YAP/TAZ, studies in Drosophila and mammalian cells have reported a complex array of upstream inputs converging on Yki/YAP/TAZ, such as cell polarity, adhesion, mechanical forces and secreted ligands (Boggiano and Fehon, 2012; Enderle and McNeill, 2013; Yu and Guan, 2013). A challenge for the field is to understand how these diverse upstream inputs intersect the Hippo pathway at a molecular level, and to define the exact physiological contexts in which these inputs impinge on Hippo signaling in vivo. Indeed, among the ever-expanding list of proteins implicated in regulating Yki/YAP/TAZ activity, few have been genetically validated in vivo. Thus, there is a need for the development of simple and robust assays for validating these upstream regulators in vivo.
As Yki/YAP/TAZ represents the ultimate convergence of Hippo signaling, characterization of upstream regulators of Hippo signaling often involves examining the subcellular localization of Yki/YAP/TAZ, and, more sensitively/reliably in vivo, the expression of selected target genes such as Diap1 in Drosophila or Ctgf in mammals. However, it is important to bear in mind that changes in the expression of a given Hippo target gene per se do not necessarily indicate changes in Yki/YAP/TAZ activity, because any Hippo target gene is likely to be regulated by a myriad of transcriptional regulators in parallel with Yki/YAP/TAZ. For example, Diap1, one of the most commonly analyzed Hippo target genes in Drosophila, is also regulated by parallel inputs, such as the JAK-STAT signaling component Stat92E (Betz et al., 2008). Similarly, Ctgf, one of the best-characterized Hippo targets in mammals, is regulated by other pathways such as TGFβ/Smad, Ras/MEK/ERK and JNK (Leask et al., 2003). Thus, it is important to distinguish whether any observed changes in the expression of a Hippo target gene are actually due to modulation of Yki/YAP/TAZ activity. In Drosophila, this can be interrogated through genetic epistasis analysis in vivo by combining loss of function of a tumor suppressor (or gain of function of an oncogene) that is suspected to converge on Yki with loss of function of yki. In such analysis, one would place a suspected gene upstream of yki if the resulting double mutants display identical phenotypes to that induced by loss of function of yki alone. On the other hand, if a gene impacts growth or Hippo target gene expression independently of changes in Yki activity, the double mutants should present an intermediate phenotype. Indeed, epistasis analysis of this nature was used to support the suggestion that yki is genetically epistatic to the core Hippo pathway components hpo, sav or wts, by analyzing double-mutant clones of each tumor suppressor gene with yki (Huang et al., 2005). A potential complication with this approach is the prominent requirement of yki for cell viability and basal-level Hippo target gene expression (Huang et al., 2005), which could make it difficult to distinguish whether a double-mutant combination displays an intermediate phenotype or yki mutant phenotype. This point is especially relevant given that many of the reported upstream regulators of Yki display relatively subtle mutant phenotypes. Another limitation is that epistasis analysis cannot be conducted between loss of function of yki and a suspected regulator of Yki that functions in the same direction as yki, as classical epistasis test requires combining two mutations with opposite phenotypes. Lastly, from a technical standpoint, generating double-mutant clones is time-consuming because it requires complicated genetic crosses and a yki rescue construct on the same FRT chromosome as the tumor suppressor gene being tested (Huang et al., 2005). Thus, a definitive, efficient and generally applicable strategy for epistasis analysis would greatly facilitate the genetic characterization of the expanding list of proteins that are thought to converge on the regulation of Yki activity.
RESULTS AND DISCUSSION
As an alternative to yki, we explored the possibility of using loss-of-function mutations in Sd, the DNA-binding partner of Yki, in genetic epistasis analysis. As expected of a critical Yki partner in Hippo signaling, loss of sd not only fully rescues tissue overgrowth and elevated target gene transcription induced by Yki overexpression (Wu et al., 2008), it also fully rescues tissue undergrowth and decreased Hippo target gene transcription in yki loss-of-function mutations (Koontz et al., 2013). However, unlike yki, which is required for normal growth and Hippo target gene expression in Drosophila, sd is genetically dispensable for normal growth and basal-level expression of Hippo target genes in most imaginal discs as a result of its default repressor activity (Koontz et al., 2013). These unique properties make sd a better choice than yki for genetic epistasis analysis, as the former is not complicated by genetic requirement in normal tissue growth or Hippo target gene expression. Thus, it provides a more robust assay to distinguish whether changes in the expression of a Hippo target gene are due to modulation of Yki activity or Yki-independent inputs into the same target gene; only the former is expected to be rescued by loss of sd. Furthermore, because loss of sd rescues both gain- and loss-of-yki phenotypes, it provides a more generally applicable assay than yki-based epistasis, irrespective of whether a gene of interest functions in the same or opposite direction as yki.
To facilitate double-mutant analysis with sd, we developed a clonal marking strategy based on double FRT chromosomes carrying a different fluorescent protein (GFP or RFP) on each FRT chromosome, together with an eye-specific FLP source. We have previously used this strategy to show that the defects in growth and Diap1 expression in yki mutant clones were completely rescued in sd; yki double-mutant clones (Koontz et al., 2013). Here, we extend this strategy to the analysis of negative regulators of Yki (potential tumor suppressors), loss-of-function mutations of which presumably lead to gain-of-Yki activity. As a proof-of-concept, we first tested whether the elevated expression of Diap1 in mutants of the core kinase cassette of the Hippo pathway, hpo, sav and wts, was rescued by loss of sd. As illustrated schematically in Fig. 1 using sd; hpo double-mutant analysis as an example, sd mutant clones and hpo mutant clones can be generated independently and labeled with different markers: loss of GFP for sd clones and loss of RFP for hpo clones. In the merged channel, one should unambiguously distinguish four different genotypes in the same eye disc based on fluorescent markers: (1) double heterozygous genetic background (essentially a wild-type control): marked as GFP and RFP positive (appearing yellow); (2) sd single mutant clones: marked as GFP negative and RFP positive (appearing red); (3) hpo single mutant clones: marked as RFP negative and GFP positive (appearing green); (4) sd; hpo double-mutant clones: marked as both GFP and RFP negative (appearing black). This marking strategy therefore allows one to reliably compare the expression of any Hippo pathway target gene (such as Diap1) in hpo single mutant, sd; hpo double mutant, and wild-type cells right next to each other in the same eye disc.
Fig. 1.
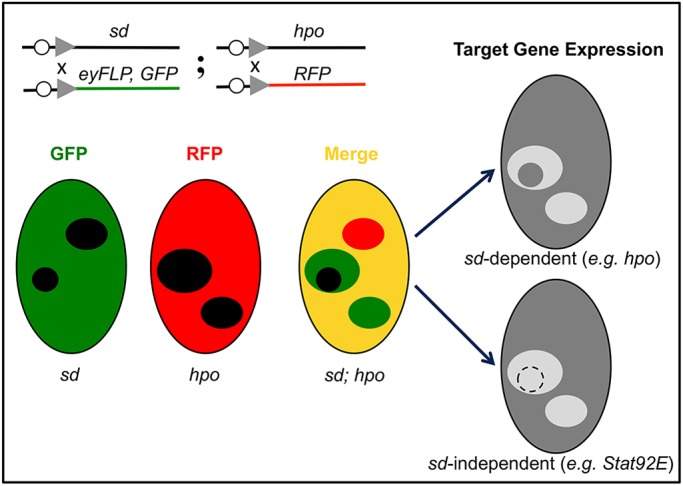
Schematic of sd-based genetic epistasis test for validating Hippo pathway components and regulators, using sd; hpo double-mutant clones as an example. Double-mutant clones are generated in flies containing double FRT chromosomes carrying sd and hpo mutations in trans to double FRT chromosomes carrying GFP and RFP markers, together with an eye-specific flippase (FLP) source. This allows unambiguous marking of the double heterozygous background (yellow), sd single mutant clones (red), hpo single mutant clones (green), and sd; hpo double-mutant clones (black) in the same eye disc. By determining whether aberrant expression of a Hippo target gene (either upregulation or downregulation) in a given mutant is genetically sd dependent (such as hpo) or sd independent (such as Stat92E), one can infer whether the mutation impacts target gene expression through Yki or Yki-independent inputs converging on the same Hippo target gene.
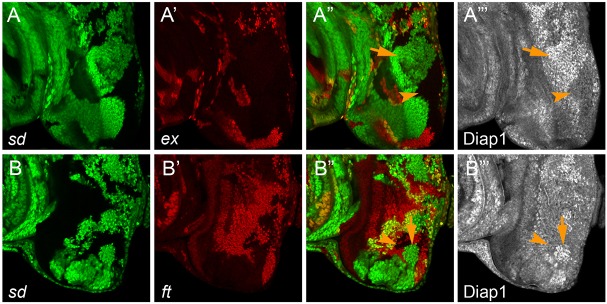
Consistent with its established role in inhibiting Yki activity, loss of hpo caused a robust increase in Diap1 expression in the eye disc (Fig. 2A-A‴). When sd was simultaneously removed, the elevated Diap1 expression was completely rescued in sd; hpo double-mutant clones, as the double-mutant clones showed a similar level of Diap1 expression as the neighboring wild-type cells (Fig. 2A-A‴). Similarly, loss of sd completely rescued the elevated Diap1 expression in sav and wts mutant clones, as evidenced by the similar level of Diap1 protein and Diap1-lacZ in sd; sav or sd; wts double-mutant clones compared with the neighboring wild-type cells (Fig. 2B-C‴, Fig. S1A-B‴). We also applied this strategy to two reported upstream regulators of the Hippo pathway, ex and ft. Consistent with these proteins being bona fide upstream regulators of Yki, loss of sd completely rescued the elevated Diap1 expression in ex and ft mutant clones (Fig. 3). Besides Diap1 and Diap1-lacZ, we also tested ex-lacZ, another widely used reporter of Yki activity. Similarly, loss of sd completely rescued the elevated ex-lacZ expression in hpo mutant clones (Fig. 2D-D‴).
Fig. 2.
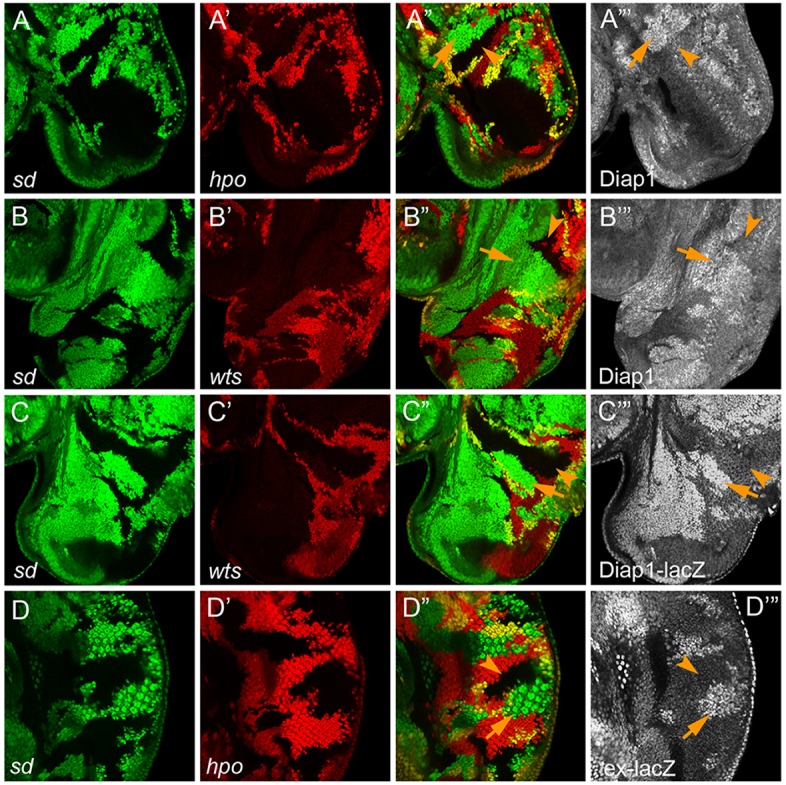
Validation of Hippo pathway core components by sd-based double-mutant analysis. In all panels, mutant clones of sd and the tumor suppressor gene being tested were marked by loss of GFP and RFP, respectively. (A-B‴) Eye discs containing mutant clones of the indicated genotypes were stained for Diap1 protein. Note the increased Diap1 staining in hpo or wts single mutant clones (arrows, green areas in the merged channel), but not the corresponding double-mutant clones with sd (arrowheads, black areas in the merged channel). (C-C‴) An eye disc containing mutant clones of sd (GFP negative) and wts (RFP negative) was stained for Diap1-lacZ. Note the increased Diap1-lacZ staining in wts single mutant clones (arrows, green areas in the merged channel), but not sd; wts double-mutant clones (arrowheads, black areas in the merged channel). (D-D‴) An eye disc containing mutant clones of sd (GFP negative) and hpo (RFP negative) was stained for ex-lacZ. Note the increased ex-lacZ staining in hpo single mutant clones (arrows, green areas in the merged channel), but not sd; hpo double-mutant clones (arrowheads, black areas in the merged channel).
Fig. 3.
Validation of Hippo pathway upstream regulators by sd-based double-mutant analysis. (A-B‴) In all panels, mutant clones of sd and the tumor suppressor gene being tested were marked by loss of GFP and RFP, respectively. Eye discs containing mutant clones of the indicated genotypes were stained for Diap1 protein. Note the increased Diap1 staining in ex or ft single mutant clones (arrows, green areas in the merged channel), but not the corresponding double-mutant clones with sd (arrowheads, black areas in the merged channel).
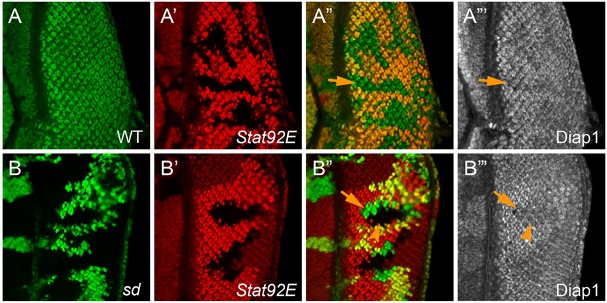
After demonstrating the efficacy of our double-mutant strategy in validating known Yki regulators, we wished to test whether this strategy can be used to exclude genes that influence Diap1 expression independently of changes in Yki activity. For this purpose, we tested the JAK-STAT signaling component Stat92E. Stat92E mutant clones have been reported to show a modest cell-autonomous decrease in Diap1 expression in the wing discs (Betz et al., 2008; Recasens-Alvarez et al., 2017). After confirming that Stat92E mutant clones in the eye discs showed a similar decrease in Diap1 expression (Fig. 4A-A‴), we used our double-mutant strategy to analyze sd; Stat92E double-mutant clones. In contrast to the complete rescue of decreased Diap1 expression in yki mutant clones by simultaneous loss of sd (Koontz et al., 2013), loss of sd did not rescue the decreased Diap1 expression in Stat92E mutant clones (Fig. 4B-B‴). Mechanistically, such findings are consistent with the distinct location of the STAT-responsive element (within the promoter region) and Hippo/Yki-responsive element (within the first intron) in the Diap1 genomic locus (Betz et al., 2008; Wu et al., 2008; Zhang et al., 2008). Taken together, these results demonstrate that the sd-based epistasis test can be used not only to validate known Yki regulators but also to exclude proteins that impact Hippo target gene expression independently of Yki activity. To facilitate the sd-based double-mutant analysis, we have developed a set of fly stocks that can be used to test mutations on every autosomal arm, allowing one to obtain definitive epistasis information in two generations (see Materials and Methods for details).
Fig. 4.
Loss of sd did not rescue the decreased Diap1 expression in Stat92E mutant clones. (A-A‴) An eye disc containing Stat92E mutant clones (RFP negative) was stained for Diap1 protein. Note the modest decrease of Diap1 protein level in Stat92E mutant clones (arrows). (B) An eye disc containing sd; Stat92E mutant clones was stained for Diap1. Mutant clones of sd and Stat92E were marked by loss of GFP and RFP, respectively. Note the decreased Diap1 protein level in both Stat92E single mutant clones (arrows, green areas in the merged channel) and sd; Stat92E double-mutant clones (arrowheads, black areas in the merged channel).
In principle, one could apply the sd-based double-mutant analysis to the expression of any Yki target genes besides Diap1 and ex, as long as target gene expression can be followed in a different detection channel as the GFP and RFP markers in confocal microscopy. We tested this possibility by examining the expression of the miRNA bantam (Nolo et al., 2006; Thompson and Cohen, 2006). bantam presents a particularly interesting Yki target gene as previous studies suggested that, unlike Diap1, transcriptional regulation of bantam by Yki is mediated by other DNA-binding proteins, including Mad, Tsh and Hth (Oh and Irvine, 2011; Peng et al., 2009). We note, however, that these non-Sd transcription factors acting as mediators of Yki's regulation of bantam expression cannot be readily reconciled with the observation that loss of sd completely rescues the growth defects of yki mutant clones (Koontz et al., 2013), as the latter would support Sd as a physiological partner of Yki in the expression of all growth-relevant genes, including bantam. It is also hard to imagine how Yki could switch between different DNA-binding partners to regulate different target genes as proposed by these studies (Oh and Irvine, 2011; Peng et al., 2009). Thus, the most parsimonious model is that, like the other Yki targets, any changes in bantam expression resulting from loss or gain of Yki activity should be dependent on sd.
To test this model, we applied the double-mutant labeling strategy described above to examine whether aberrant bantam expression resulting from defective Hippo signaling is dependent on sd function. As expected of a Yki target, bantam expression was strongly upregulated in loss-of-function mutant clones of hpo or sav, as revealed by a bantam-lacZ reporter inserted into the endogenous locus (Herranz et al., 2012) (Fig. 5A-A‴, Fig. S1C-C‴). Strikingly, when sd was simultaneously removed from hpo or sav mutant clones, the elevated bantam expression was completely rescued, as sd; hpo or sd; sav double-mutant clones showed a similar level of bantam expression as the neighboring wild-type cells (Fig. 5A-A‴, Fig. S1C-C‴). Conversely, loss of yki resulted in decreased bantam expression (Fig. 5C-C‴), and the decreased bantam expression in yki mutant clones was completely rescued in sd; yki double-mutant clones (Fig. 5D-D‴). As an additional, independent test for the genetic requirement of sd in Yki-mediated bantam expression, we used the MARCM (mosaic analysis with a repressible cell marker) technique to generate sd mutant clones with Yki overexpression. As expected, Yki overexpression induced strong upregulation of bantam expression (Fig. 5E-E″), and such upregulation was completely rescued by simultaneous loss of sd (Fig. 5F-F″). We applied the same double-mutant labeling strategy to examine the regulation of brC12-lacZ, a Yki-dependent bantam reporter that was reported to be regulated in a Mad-dependent but Sd-independent manner (Oh and Irvine, 2011). Contrary to the previous report, we found that when sd was simultaneously removed from hpo mutant clones, the elevated brC12-lacZ expression was completely rescued to a similar level to the neighboring wild-type cells (Fig. 5B-B‴). We conclude that, like Diap1 and ex, aberrant bantam expression resulting from both gain and loss of Yki activity is completely dependent on Sd function. We further infer from these results that Sd functions as a default repressor for the transcription of bantam, similar to its role in Diap1 and ex transcription (Koontz et al., 2013). Taken together, these findings support the central role of Sd in Yki-mediated transcriptional regulation. We emphasize that this conclusion is not intended to imply that Yki target genes cannot be regulated by transcription factors other than Sd. Like Diap1, which contains distinct STAT- and Yki-responsive enhancers (Betz et al., 2008; Wu et al., 2008; Zhang et al., 2008), any Yki target gene is likely to be regulated by several DNA-binding transcription factors that bind to distinct enhancers, and these enhancer-binding transcription factors can still interact with each other on chromatin to influence transcriptional output. The essence of our conclusion is that any changes of Yki target genes upon loss or gain of Yki activity cannot be realized without Sd binding to these target loci.
Fig. 5.
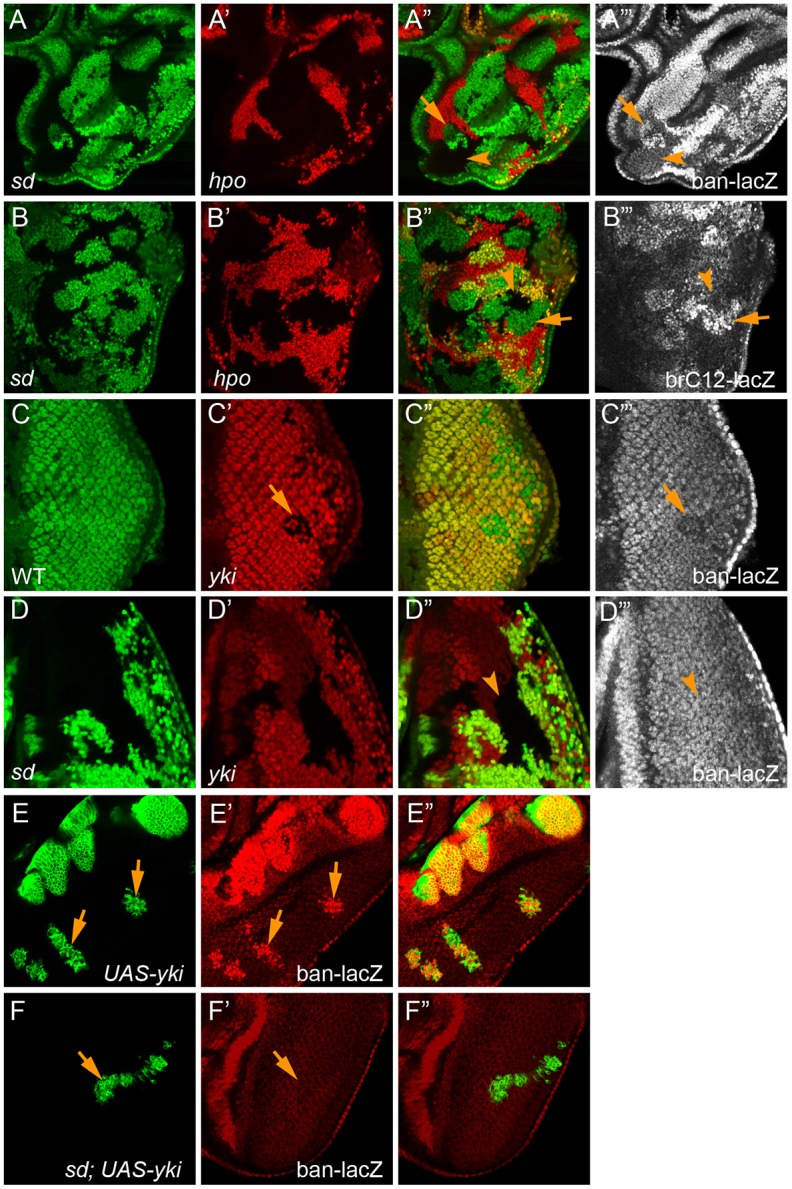
Loss of sd completely rescues aberrant bantam expression resulting from defective Hippo signaling. (A-A‴) An eye disc containing mutant clones of sd (GFP negative) and hpo (RFP negative) was stained for bantam-lacZ. Note the increased bantam-lacZ expression in hpo single mutant clones (arrows, green areas in the merged channel), but not sd; hpo double-mutant clones (arrowheads, black areas in the merged channel). (B-B‴) An eye disc containing mutant clones of sd (GFP negative) and hpo (RFP negative) was stained for brC12-lacZ. Note the increased brC12-lacZ expression in hpo single mutant clones (arrows, green areas in the merged channel), but not sd; hpo double-mutant clones (arrowheads, black areas in the merged channel). (C-C‴) An eye disc containing yki mutant clones (RFP negative) was stained for bantam-lacZ. Note the modest decrease of bantam-lacZ staining in the yki mutant clones (arrow). (D-D‴) An eye disc containing sd; yki double-mutant clones was stained for bantam-lacZ. Note the similar bantam-lacZ level in the double-mutant clone (arrowheads) as in the neighboring wild-type cells. (E-E″) An eye disc containing yki-overexpressing clones (GFP positive) and stained for bantam-lacZ. Note the dramatic increase of bantam-lacZ staining in the Yki-overexpressing clones (arrows). (F-F″) An eye disc containing sd mutant clones with yki overexpression (GFP positive), showing similar bantam-lacZ staining in the clones (arrows) as the neighboring wild-type cells.
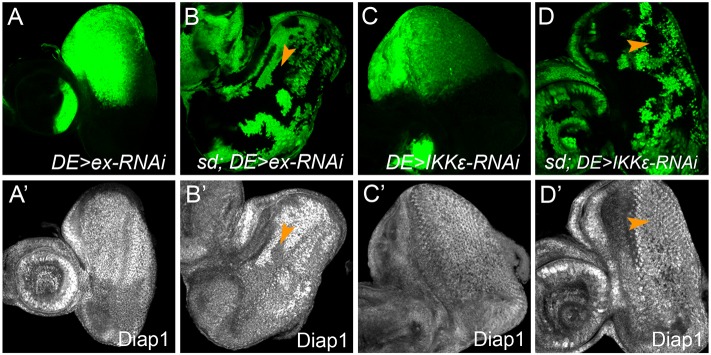
Although loss-of-function mutations are preferred in epistasis analysis, there might be situations in which mutant alleles are unavailable for a suspected Yki regulator, or when mutant alleles are unsuitable for clonal analysis for reasons such as cell lethality, protein endurance and genetic redundancy. Indeed, many studies of Hippo signaling have relied on the use of the UAS-Gal4 system to knockdown or overexpress a gene of interest, often in a specific region of the wing or eye imaginal disc. In order to extend sd-based epistasis analysis to such genes, we devised a way to combine UAS-Gal4-mediated transgene expression with sd mutant clones in the eye. For this purpose, we took advantage of a Gal4 driver (DE-Gal4) that is specifically expressed in the dorsal half of the eye discs (Morrison and Halder, 2010). The expectation is that if aberrant Diap1 expression (or any other Hippo targets) upon perturbation of a given gene is truly due to abnormal Yki activity, such aberration should be sd dependent. We first tested Ex, a known regulator of Yki activity, as a proof of concept. As expected, knockdown of ex by DE-Gal4 caused increased Diap1 expression specifically in the dorsal half of the eye disc (Fig. 6A,A′), and the upregulation of Diap1 expression was completely rescued by loss of sd (Fig. 6B,B′). As a negative control, we tested the Drosophila IKK-related kinase DmIKKɛ (IKKε). RNAi knockdown of DmIKKɛ by the en-Gal4 driver was previously reported to result in a mild upregulation of Diap1 protein level in the posterior compartment of the wing discs (Kuranaga et al., 2006). We confirmed that knockdown of DmIKKɛ in the eye discs resulted in a similar increase of Diap1 level (Fig. 6C,C′). In contrast to the knockdown of ex, however, DmIKKɛ RNAi-induced upregulation of Diap1 expression was not rescued by loss of sd (Fig. 6D,D′). Indeed, whereas Hippo signaling controls Diap1 transcription, DmIKKɛ is known to impact Diap1 expression post-transcriptionally by regulating Diap1 protein stability (Kuranaga et al., 2006). Together with our analysis of Stat92E, these results demonstrate the efficacy of sd-based epistasis analysis in identifying proteins that impinge on Hippo target expression independently of Yki activity.
Fig. 6.
Application of sd-based epistasis analysis to genetic background created by UAS-Gal4-mediated transgene expression. The eye disc is oriented with dorsal side up in all panels. (A,A′) An eye disc expressing UAS-exRNAi and UAS-GFP under the control of DE-Gal4 was stained for Diap1 protein. Note the enhanced Diap1 staining in the dorsal half of the eye disc. (B,B′) An eye disc containing sd mutant clones (GFP negative) and expressing UAS-exRNAi under the control of DE-Gal4 was stained for Diap1 protein. The increased Diap1 staining in the dorsal half of the eye disc was specifically reduced in sd mutant clones (arrowheads) to a level comparable to that in the ventral half of the eye. (C,C′) An eye disc expressing UAS-DmIKKɛRNAi and UAS-GFP under the control of DE-Gal4 was stained for Diap1 protein. Note the enhanced Diap1 staining in the dorsal half of the eye disc. (D,D′) An eye disc containing sd mutant clones (GFP negative) and expressing UAS-DmIKKɛRNAi under the control of DE-Gal4 was stained for Diap1 protein. The increased Diap1 staining in the dorsal half of the eye disc was not affected in sd mutant clones (arrowheads).
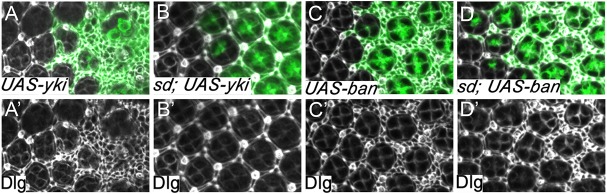
Besides aberrant target gene expression, another routinely assayed phenotype suggestive of elevated Yki activity is increased interommatidial cell number in pupal retina. As with our concerns about target gene expression, it is crucial to distinguish whether an observed alteration in interommatidial cell number is due to changes in Yki activity or other modalities impacting interommatidial cell number. We reasoned that sd-based epistasis analysis can be similarly applied in this context, as any alteration in interommatidial cell number caused by changes in Yki activity should be completely rescued by loss of sd. We tested this idea by comparing two different genetic backgrounds resulting in increased interommatidial cell number in pupal retina: overexpression of Yki or its downstream target bantam. Our prediction is that only the former can be rescued by loss of sd. Indeed, Yki overexpression induced a massive increase in interommatidial cells [average >40 extra cells per cluster (ECPC), n=11; Fig. 7A,A′], and this increase was completely rescued by simultaneous loss of sd (0 ECPC, n=16; Fig. 7B,B′). In contrast, even though bantam overexpression caused a milder increase of interommatidial cells (average 16.4 ECPC, n=15; Fig. 7C,C′) compared with Yki overexpression, the bantam-induced increase in interommatidial cell number was not rescued at all by loss of sd (average 16.1 ECPC, n=13; Fig. 7D,D′). Thus, besides Hippo target genes, sd-based epistasis analysis can be applied as a specific and definitive way to determine the causality of the interommatidial cell phenotype with respect to changes in Yki activity.
Fig. 7.
Application of sd-based epistasis analysis to determine the causality of interommatidial cell phenotype. (A,A′) A mid-pupal retina containing yki-overexpressing clones (GFP positive) and stained for Discs large (Dlg). Note the dramatic increase of interommatidial cells (average >40 ECPC, n=11) in yki-overexpressing clones. (B,B′) A mid-pupal retina containing sd mutant clones with yki overexpression (GFP positive) and stained for Dlg. Note the similar number of interommatidial cells (0 ECPC, n=16) in the clones as the neighboring wild-type cells. (C,C′) A mid-pupal retina containing bantam-overexpressing clones (GFP positive) and stained for Dlg. Note the increase of interommatidial cells (average 16.4 ECPC, n=15) in bantam-overexpressing clones. (D,D′) A mid-pupal retina containing sd mutant clones with bantam overexpression (GFP positive) and stained for Dlg, showing an average of 16.1 ECPC (n=13).
Conclusions
Genetic epistasis analyses are instrumental in dissecting developmental pathways and determining the relationships between genes of interest (Huang and Sternberg, 2006). We have presented a simple genetic epistasis test, as well as the necessary fly reagents, to validate proteins implicated in regulating Yki activity, taking advantage of the unique genetic property of sd as an essential mediator of both loss- and gain-of-Yki phenotypes. A key feature of this strategy is that it examines the ability of loss of sd, which by itself does not affect normal growth and basal-level Hippo target gene expression in the eye, to revert/rescue the mutant phenotype of another gene potentially linked to the regulation of Yki activity, irrespective of whether the gene acts positively or negatively on yki. This method provides a specific, sensitive and versatile assay to ascertain whether aberrant expression of a given Hippo target gene is due to changes in Yki activity, as we have shown for hpo, sav, wts, ex and ft, or Yki-independent inputs converging on the same target gene, as we have shown for Stat92E and DmIKKɛ. Such sd-based epistasis analysis is not limited to target gene expression, and can be extended to any Yki-dependent phenotypes, as we have shown for the interommatidial cell phenotype. We suggest that sd-based epistasis analysis should be broadly applied to assess the expanding repertoire of proteins and inputs that have been suggested to converge on the regulation of Yki activity.
MATERIALS AND METHODS
Drosophila genetics
The bantam-lacZ reporter P{lacW}banL1170a (stock ID 10154 from Bloomington Drosophila Stock Center) is a lacZ-containing P-element enhancer trap line inserted in the promoter of bantam (Herranz et al., 2012; Dent et al., 2015). The UAS-DmIKKɛ RNAi line was also obtained from Bloomington Drosophila Stock Center (stock ID 35266). UAS-ex RNAi flies were obtained from the Vienna Drosophila Resource Center (transformant ID 22994). The following flies have been described previously: Diap1-lacZ reporter thj5c8 (Wu et al., 2003), brC12-lacZ (Oh and Irvine, 2011), UAS-yki and ykiB5 (Huang et al., 2005), UAS-ban (Brennecke et al., 2003), sd47M (Srivastava et al., 2004), hpo42-47 (Wu et al., 2003), sav3 (Tapon et al., 2002), wtsX1 (Xu et al., 1995), ft8 (Bryant et al., 1988), exe1 (Hamaratoglu et al., 2006), Stat92EP1681 (Hou et al., 1996).
For MARCM, all clones were induced 68-72 h after egg deposition and heat-shocked at 38°C for 30 min. Double-mutant clones in the eye imaginal discs were generated using flies containing double FRT chromosomes with GFP and RFP markers together with an eye-specific FLP source as described previously (Koontz et al., 2013).
The following genotypes were used:
yki overexpression clones: tub-Gal80 FRT19A/FRT19A; UAS-GFP hs-FLP/UAS-yki, bantam-lacZ
sd clones overexpressing yki: tub-Gal80 FRT19A/sd47M FRT19A; UAS-GFP hs-FLP/UAS-yki, bantam-lacZ
sd clones overexpressing bantam: tub-Gal80 FRT19A/sd47M FRT19A; UAS-GFP hs-FLP/UAS-ban
sd; hpo double-mutant clones: ey-FLP, Ubi-GFP FRT19A/sd47M FRT19A; FRT42D hpo42-47/FRT42D Ubi-RFP
sd; sav double-mutant clones: ey-FLP, Ubi-GFP FRT19A/sd47M FRT19A; FRT82B sav3/FRT82B Ubi-RFP
sd; wts double-mutant clones: ey-FLP, Ubi-GFP FRT19A/sd47M FRT19A; FRT82B wtsx1/FRT82B Ubi-RFP
ex RNAi clones: UAS-GFP/+; DE-Gal4/UAS-exRNAi
sd clones with ex RNAi: ey-FLP, Ubi-GFP FRT19A/sd47M FRT19A; DE-Gal4/UAS-exRNAi
DmIKKɛ RNAi clones: UAS-GFP/+; DE-Gal4/UAS-IKKɛRNAi
sd clones with DmIKKɛ RNAi: ey-FLP, Ubi-GFP FRT19A/sd47M FRT19A; DE-Gal4/UAS-IKKɛRNAi
sd clones with yki overexpression: ey-FLP, Ubi-GFP FRT19A/sd47M FRT19A; DE-Gal4/UAS-yki
Stat92E mutant eyes: ey-FLP; FRT82B Stat92EP1681/FRT82B Ubi-GFP
sd; Stat92E double-mutant clones: ey-FLP, Ubi-GFP FRT19A/sd47M FRT19A; FRT82B Stat92EP1681/FRT82B Ubi-RFP
yki mutant eyes: ey-FLP, Ubi-GFP FRT19A/+; FRT42D ykiB5/FRT42D Ubi-RFP
sd; yki double-mutant clones: ey-FLP, Ubi-GFP FRT19A/sd47M FRT19A; FRT42D ykiB5/FRT42D Ubi-RFP
sd; ft double-mutant clones: ey-FLP, Ubi-GFP FRT19A/sd47M FRT19A; ft8FRT40A/Ubi-RFP FRT40A
sd; ex double-mutant clones: ey-FLP, Ubi-GFP FRT19A/sd47M FRT19A; exe1FRT40A/Ubi-RFP FRT40A.
Tool flies developed for sd-based genetic epistasis test: ey-FLP, Ubi-GFP FRT19A; Adv/T(2;3)SM6-TM6B, sd47M FRT19A/FM6; FRT40A Ubi-RFP/CyO (for candidate genes on 2L), sd47M FRT19A/FM6; FRT42D Ubi-RFP/CyO (for candidate genes on 2R), sd47M FRT19A/FM6; Ubi-RFP FRT80B/TM6B (for candidate genes on 3L), sd47M FRT19A/FM6; Ubi-RFP FRT80B/TM6B (for candidate genes on 3R), sd47M FRT19A/FM6; DE-Gal4/TM6B.
To generate sd; hpo double-mutant clones, male FRT42D hpo/CyO flies were first crossed to ey-FLP, Ubi-GFP FRT19A; Adv/T(2;3)SM6-TM6B virgin females. The resulting F1 male progeny of the genotype ey-FLP, Ubi-GFP FRT19A/Y; FRT42D hpo/T(2;3)SM6-TM6B were crossed to sd47M FRT19A/FM6; FRT42D Ubi-RFP/Cyo virgin females. Female third instar larva without the T(2;3)SM6-TM6B balancer from the cross are predicted to be ey-FLP, Ubi-GFP FRT19A/sd47M FRT19A; FRT42D hpo42-47/FRT42D Ubi-RFP, the genotype that allows the generation of sd; hpo double-mutant clones. A similar mating scheme can be used to examine double mutants of sd and candidate genes on other chromosome arms, by replacing the sd47M FRT19A/FM6; FRT42D Ubi-RFP/Cyo flies with a fly stock for a candidate gene's chromosome arm (see the list of tool flies above).
To generate sd mutant clones in the genetic background of UAS-Gal4-mediated transgene expression (ex RNAi as an example) in the eye, male UAS-exRNAi flies were first crossed to ey-FLP, Ubi-GFP FRT19A; Adv/T(2;3)SM6-TM6B virgin females. The resulting F1 male progeny of the genotype ey-FLP, Ubi-GFP FRT19A/Y; UAS-exRNAi/T(2;3)SM6-TM6B were crossed to sd47M FRT19A/FM6; DE-Gal4/TM6B virgin females. Female third instar larva without the T(2;3)SM6-TM6B balancer from the cross are predicted to be ey-FLP, Ubi-GFP FRT19A/sd47M FRT19A; DE-Gal4/UAS-exRNAi, the genotype that allows the generation of sd mutant clones in the presence of ex RNAi.
Image analysis
All confocal images were acquired on a Carl Zeiss 700 microscope and analyzed using ImageJ.
Supplementary Material
Acknowledgements
We thank the Confocal Microscopy Core and the Molecular Biology Core funded by KSU-CVM.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: J.Y., D.P.; Methodology: J.Y.; Validation: J.Y.; Formal analysis: J.Y., D.P.; Investigation: J.Y., D.P.; Resources: J.Y.; Data curation: J.Y.; Writing - original draft: J.Y.; Writing - review & editing: J.Y., D.P.; Supervision: D.P.; Project administration: J.Y., D.P.; Funding acquisition: D.P.
Funding
This study was supported in part by grants from the National Institutes of Health (EY015708). D.P. is an investigator of the Howard Hughes Medical Institute. Deposited in PMC for release after 6 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.157545.supplemental
References
- Betz A., Ryoo H. D., Steller H. and Darnell J. E. Jr. (2008). STAT92E is a positive regulator of Drosophila inhibitor of apoptosis 1 (DIAP/1) and protects against radiation-induced apoptosis. Proc. Natl. Acad. Sci. USA 105, 13805-13810. 10.1073/pnas.0806291105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggiano J. C. and Fehon R. G. (2012). Growth control by committee: intercellular junctions, cell polarity, and the cytoskeleton regulate Hippo signaling. Dev. Cell 22, 695-702. 10.1016/j.devcel.2012.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J., Hipfner D. R., Stark A., Russell R. B. and Cohen S. M. (2003). bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113, 25-36. 10.1016/S0092-8674(03)00231-9 [DOI] [PubMed] [Google Scholar]
- Bryant P. J., Huettner B., Held L. I. Jr, Ryerse J. and Szidonya J. (1988). Mutations at the fat locus interfere with cell proliferation control and epithelial morphogenesis in Drosophila. Dev. Biol. 129, 541-554. 10.1016/0012-1606(88)90399-5 [DOI] [PubMed] [Google Scholar]
- Chen L., Chan S. W., Zhang X., Walsh M., Lim C. J., Hong W. and Song H. (2010). Structural basis of YAP recognition by TEAD4 in the hippo pathway. Genes Dev. 24, 290-300. 10.1101/gad.1865310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent L. G., Poon C. L. C., Zhang X., Degoutin J. L., Tipping M., Veraksa A. and Harvey K. F. (2015). The GTPase regulatory proteins Pix and Git control tissue growth via the Hippo pathway. Curr. Biol. 25, 124-130. 10.1016/j.cub.2014.11.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enderle L. and McNeill H. (2013). Hippo gains weight: added insights and complexity to pathway control. Sci. Signal. 6, re7 10.1126/scisignal.2004208 [DOI] [PubMed] [Google Scholar]
- Giot L., Bader J. S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y. L., Ooi C. E., Godwin B., Vitols E. et al. (2003). A protein interaction map of Drosophila melanogaster. Science 302, 1727-1736. 10.1126/science.1090289 [DOI] [PubMed] [Google Scholar]
- Hamaratoglu F., Willecke M., Kango-Singh M., Nolo R., Hyun E., Tao C., Jafar-Nejad H. and Halder G. (2006). The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat. Cell Biol. 8, 27-36. 10.1038/ncb1339 [DOI] [PubMed] [Google Scholar]
- Harvey K. and Tapon N. (2007). The Salvador-Warts-Hippo pathway - an emerging tumour-suppressor network. Nat. Rev. Cancer 7, 182-191. 10.1038/nrc2070 [DOI] [PubMed] [Google Scholar]
- Herranz H., Hong X. and Cohen S. M. (2012). Mutual repression by bantam miRNA and Capicua links the EGFR/MAPK and Hippo pathways in growth control. Curr. Biol. 22, 651-657. 10.1016/j.cub.2012.02.050 [DOI] [PubMed] [Google Scholar]
- Hou X. S., Melnick M. B. and Perrimon N. (1996). Marelle acts downstream of the Drosophila HOP/JAK kinase and encodes a protein similar to the mammalian STATs. Cell 84, 411-419. 10.1016/S0092-8674(00)81286-6 [DOI] [PubMed] [Google Scholar]
- Huang L. S. and Sternberg P. W. (2006). Genetic dissection of developmental pathways. WormBook, 1-19. 10.1895/wormbook.1.88.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Wu S., Barrera J., Matthews K. and Pan D. (2005). The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 122, 421-434. 10.1016/j.cell.2005.06.007 [DOI] [PubMed] [Google Scholar]
- Johnson R. and Halder G. (2014). The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat. Rev. Drug Discov. 13, 63-79. 10.1038/nrd4161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M. (2007). A Sveinsson's chorioretinal atrophy-associated missense mutation in mouse Tead1 affects its interaction with the co-factors YAP and TAZ. Biochem. Biophys. Res. Commun. 361, 1022-1026. 10.1016/j.bbrc.2007.07.129 [DOI] [PubMed] [Google Scholar]
- Koontz L. M., Liu-Chittenden Y., Yin F., Zheng Y., Yu J., Huang B., Chen Q., Wu S. and Pan D. (2013). The Hippo effector Yorkie controls normal tissue growth by antagonizing scalloped-mediated default repression. Dev. Cell 25, 388-401. 10.1016/j.devcel.2013.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuranaga E., Kanuka H., Tonoki A., Takemoto K., Tomioka T., Kobayashi M., Hayashi S. and Miura M. (2006). Drosophila IKK-related kinase regulates nonapoptotic function of caspases via degradation of IAPs. Cell 126, 583-596. 10.1016/j.cell.2006.05.048 [DOI] [PubMed] [Google Scholar]
- Leask A., Holmes A., Black C. M. and Abraham D. J. (2003). Connective tissue growth factor gene regulation: requirements for its induction by transforming growth factor-beta 2 in fibroblasts. J. Biol. Chem. 278, 13008-13015. 10.1074/jbc.M210366200 [DOI] [PubMed] [Google Scholar]
- Li Z., Zhao B., Wang P., Chen F., Dong Z., Yang H., Guan K.-L. and Xu Y. (2010). Structural insights into the YAP and TEAD complex. Genes Dev. 24, 235-240. 10.1101/gad.1865810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Chittenden Y., Huang B., Shim J. S., Chen Q., Lee S.-J., Anders R. A., Liu J. O. and Pan D. (2012). Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 26, 1300-1305. 10.1101/gad.192856.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison C. M. and Halder G. (2010). Characterization of a dorsal-eye Gal4 line in Drosophila. Genesis 48, 3-7. 10.1002/dvg.20608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolo R., Morrison C. M., Tao C., Zhang X. and Halder G. (2006). The bantam microRNA is a target of the hippo tumor-suppressor pathway. Curr. Biol. 16, 1895-1904. 10.1016/j.cub.2006.08.057 [DOI] [PubMed] [Google Scholar]
- Oh H. and Irvine K. D. (2011). Cooperative regulation of growth by Yorkie and Mad through bantam. Dev. Cell 20, 109-122. 10.1016/j.devcel.2010.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D. (2010). The hippo signaling pathway in development and cancer. Dev. Cell 19, 491-505. 10.1016/j.devcel.2010.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H. W., Slattery M. and Mann R. S. (2009). Transcription factor choice in the Hippo signaling pathway: homothorax and yorkie regulation of the microRNA bantam in the progenitor domain of the Drosophila eye imaginal disc. Genes Dev. 23, 2307-2319. 10.1101/gad.1820009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recasens-Alvarez C., Ferreira A. and Milán M. (2017). JAK/STAT controls organ size and fate specification by regulating morphogen production and signalling. Nat. Commun. 8, 13815 10.1038/ncomms13815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A., Simmonds A. J., Garg A., Fossheim L., Campbell S. D. and Bell J. B. (2004). Molecular and functional analysis of scalloped recessive lethal alleles in Drosophila melanogaster. Genetics 166, 1833-1843. 10.1534/genetics.166.4.1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapon N., Harvey K. F., Bell D. W., Wahrer D. C. R., Schiripo T. A., Haber D. A. and Hariharan I. K. (2002). salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell 110, 467-478. 10.1016/S0092-8674(02)00824-3 [DOI] [PubMed] [Google Scholar]
- Thompson B. J. and Cohen S. M. (2006). The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell 126, 767-774. 10.1016/j.cell.2006.07.013 [DOI] [PubMed] [Google Scholar]
- Tian W., Yu J., Tomchick D. R., Pan D. and Luo X. (2010). Structural and functional analysis of the YAP-binding domain of human TEAD2. Proc. Natl. Acad. Sci. USA 107, 7293-7298. 10.1073/pnas.1000293107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev A., Kaneko K. J., Shu H., Zhao Y. and DePamphilis M. L. (2001). TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 15, 1229-1241. 10.1101/gad.888601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Huang J., Dong J. and Pan D. (2003). hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell 114, 445-456. 10.1016/S0092-8674(03)00549-X [DOI] [PubMed] [Google Scholar]
- Wu S., Liu Y., Zheng Y., Dong J. and Pan D. (2008). The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev. Cell 14, 388-398. 10.1016/j.devcel.2008.01.007 [DOI] [PubMed] [Google Scholar]
- Xu T., Wang W., Zhang S., Stewart R. A. and Yu W. (1995). Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development 121, 1053-1063. [DOI] [PubMed] [Google Scholar]
- Yu F.-X. and Guan K.-L. (2013). The Hippo pathway: regulators and regulations. Genes Dev. 27, 355-371. 10.1101/gad.210773.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Ren F., Zhang Q., Chen Y., Wang B. and Jiang J. (2008). The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev. Cell 14, 377-387. 10.1016/j.devcel.2008.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Ye X., Yu J., Li L., Li W., Li S., Yu J., Lin J. D., Wang C.-Y., Chinnaiyan A. M. et al. (2008). TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 22, 1962-1971. 10.1101/gad.1664408 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.