2.1 Introduction
Biological membranes provide stable barriers to the external environment. Lipid bilayers maintain the integrity of both cells and enveloped viruses. The infectious program of viruses necessitates shedding of the viral envelope to release the genome-containing nucleocapsid. The merging or fusion of two distinct membranes is a key step at several junctures of the herpesvirus replication cycle. The membrane fusion reaction is driven by host cell-triggered refolding of a viral fusion protein, either acting alone or, in the case of herpesviruses, in concert with additional viral proteins. Herpesviral entry, assembly, and spread all require fusion events (Fig. 2.1). The execution and regulation of these processes require distinct yet often overlapping sets of viral proteins and host cell factors. Each of the distinct fusion processes described here likely has variations, in part due to redundant functions harbored by HSV-1 and differences in cell types. Glycoprotein B (gB) and the heterodimer of gH/gL are highly conserved among the Herpesviridae and constitute the core fusion machinery. gB is thought to be the central fusion protein, with gH/gL having a less defined but essential role. In addition, there are several examples of herpesvirus subfamily-specific proteins that are required for fusion. These proteins such as gD of the alphaherpesviruses often have critical receptor-binding activities.
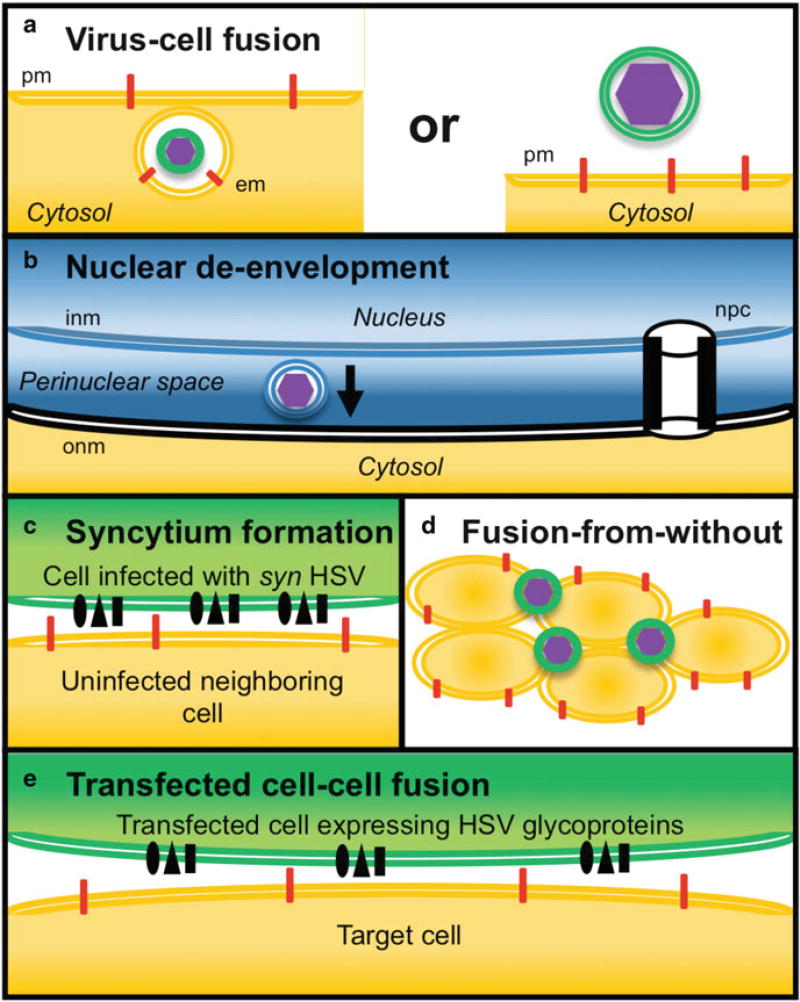
Fig. 2.1.
Types of HSV-1 membrane fusion. (a) Virus-cell fusion during entry. HSV entry proceeds by either an endocytosis mechanism (left) or by direct penetration at the plasma membrane. There is fusion of the viral membrane (green) with either the cell endosomal membrane (em; orange) or the host plasma membrane (pm; orange), respectively, (b) Fusion of primary enveloped HSV-1 with the outer nuclear membrane. The primary envelope of HSV is derived from the inner nuclear membrane (inm) of the infected cell. The membrane of primary enveloped virions (blue) fuses with the outer nuclear membrane (onm; black). Shown spanning the inm and onm is a nuclear pore complex (npc), which is too narrow to allow passage of HSV particles, (c) Syncytium formation. The surface of a cell infected with a syncytial strain of HSV-1 (green) fuses directly with a neighboring uninfected cell (orange). (d) Fusion from without. The envelope of an FFWO strain of HSV triggers cell fusion in the absence of de novo protein synthesis. (e) Transfected cell-cell fusion. A cell transiently expressing HSV glycoproteins (green) fuses with a permissive target cell (orange)
2.2 Types of Fusion Mediated by HSV Glycoproteins
2.2.1 Virus-Cell Fusion During Entry
To initiate entry and infection, enveloped viruses must fuse with a host cell membrane to release their genetic material into the cytosol. Most enveloped animal viruses fuse with an intracellular membrane following endocytosis of viral particles from the cell surface (Barrow et al. 2013). Some viruses fuse directly with the plasma membrane. A single herpesvirus species can enter cells via endocytic or non-endocytic (direct fusion) pathways depending on the target cell type (Frampton et al. 2007; Miller and Hutt-Fletcher 1992; Nicola et al. 2003, 2005; Raghu et al. 2009; Van de Walle et al. 2008). HSV is proposed to enter human mucosal epithelial cells via endocytosis followed by low-pH fusion with an endosomal membrane and peripheral neurons by fusion with the cell surface (Lycke et al. 1988; Nicola 2016; Nicola et al. 2003, 2005).
Vero cells are a model cell line that support fusion of HSV with the plasma membrane. Entry into Vero cells, and by extension virus-cell fusion, requires viral envelope glycoproteins gB and gD and the gH/gL heterodimer. Virus mutants devoid of these glycoproteins fail to penetrate the Vero cell surface (Cai et al. 1987; Forrester et al. 1992; Ligas and Johnson 1988; Roop et al. 1993). Furthermore, antibodies specific for gB, gD, or gH can neutralize entry into and infection of Vero cells (Cohen et al. 1972; Fuller and Spear 1987; Gompels and Minson 1986; Highlander et al. 1987, 1988; Navarro et al. 1992; Nicola et al. 1998; Peng et al. 1998). Viruses lacking gB, gD, or gH/gL also fail to enter CHO-nectin-1 cells, a model cell type that supports entry by endocytosis, suggesting that this set of four glycoproteins is required for fusion following endocytosis (Nicola and Straus 2004).
HSV-1 null mutants that lack gC, gE, gG, gl, gJ, gM, gN, UL45, or Us9 are competent for entry via either endocytic or non-endocytic pathways, suggesting they are dispensable for virus-cell fusion (Baines and Roizman 1991; Balan et al. 1994; Dingwell et al. 1994; Dollery et al. 2010b; Komala Sari et al. 2013; Longnecker et al. 1987; Longnecker and Roizman 1987; Nicola and Straus 2004; Polcicova et al. 2005; Ruyechan et al. 1979; Striebinger et al. 2016; Visalli and Brandt 1991; Weber et al. 1987). The HSV-1 polytopic membrane glycoprotein K (gK) is nonessential for viral entry into Vero cells but is reportedly important for entry in a cell-specific manner (Chowdhury et al. 2013; David et al. 2012; Hutchinson and Johnson 1995).
The molecular mechanism of HSV-cell fusion is also based on studies of viral entry. Initial attachment of HSV to the cell surface occurs via HSV binding to glycosaminoglycans (GAGs), principally heparan sulfate proteoglycans (HSPGs) (see review by Azab—Chap. 1). HSV attachment to GAGs is not an absolute requirement for virus-cell fusion, as HSPG-negative cells still support HSV entry (Shukla and Spear 2001). Attachment is mediated by HSV gC and to a lesser extent gB. HSV-1 lacking gC or bearing a gB that is defective in HSPG-binding remains competent for entry (Laquerre et al. 1998). Nonetheless the HSV-heparan sulfate interaction enhances the probability of subsequent events in entry leading to fusion and may be critical in vivo.
Fusion of entering HSV with the host cell target membrane is governed by a cascade of interactions between gB, gD, and gH/gL. Glycoprotein D binding to one of its cognate receptors is critical. Host cell receptors for HSV gD include the calcium-dependent cell-cell adhesion molecules, nectin-1 and nectin-2, the tumor necrosis factor receptor-related molecule herpesvirus entry mediator (HVEM), and 3-O-sulfonated heparan sulfate (Geraghty et al. 1998; Montgomery et al. 1996; Shukla et al. 1999; Warner et al. 1998). Expression of a gD receptor in cells that are resistant to entry such as CHO or B78 cells (Montgomery et al. 1996) alleviates the block to entry by restoring the receptor-binding step critical for virus-cell fusion. Electron microscopic analysis of HSV-1 added to CHO cells reveals accumulation of damaged enveloped virions in large vesicular compartments that have failed to fuse and are likely destined for degradation (Nicola and Straus 2004). HSV can engage nectin-1 at the surface of CHO-nectin-1 cells (unpublished data). It is not clear whether this interaction ultimately leads to fusion with an endocytic compartment or whether productive interaction of virion gD with a gD receptor can occur exclusively in endosomes and lead to fusion.
gD is a 369-amino acid type I membrane glycoprotein with a short cytoplasmic tail. Orthologs of gD are only found in alphaherpesviruses. The HSV-1 gD ectodomain is comprised of an immunoglobulin-like core flanked by N- and C-terminal extensions (Carfi et al. 2001). Nectin-1 and HVEM bind to the same face of gD but at distinct sites (Carfi et al. 2001; Connolly et al. 2002, 2003, 2005; Di Giovine et al. 2011; Krummenacher et al. 1998, 2005; Lazear et al. 2008; Manoj et al. 2004; Nicola et al. 1998; Yoon and Spear 2004; Yoon et al. 2003). The current model of fusion initiation posits that binding of gD to either receptor results in the movement of the C-terminal extension, revealing receptor contact sites on the core. The extension contains residues 260–285, the profusion domain of gD (Cocchi et al. 2004; Gallagher et al. 2013). The receptor-triggered, pH-independent conformational change in gD is thought to initiate the membrane fusion cascade. Regions of gD important for viral entry have been determined by assessing the ability of mutant gDs to complement the infectivity of a gD-null virus. Mutations that affect receptor binding adversely affect entry. gD mutants that bind receptors yet fail to function in entry have separated gD’s receptor-binding activity and its additional role(s) in virus-cell fusion (Eisenberg et al. 2012; Spear et al. 2006).
HSV-1 gB is an 898-amino acid glycoprotein with an extended rodlike ectodomain (Heldwein et al. 2006). gB is highly conserved among herpesviruses. Herpesvirus gB is a class III fusion protein, along with vesicular stomatitis virus G and baculovirus gp64 (Weissenhom et al. 2007). The latter two mediate membrane fusion on their own (Blissard and Wenz 1992; Florkiewicz and Rose 1984). HSV gB is likely the central fusion protein but is nonfunctional on its own, requiring assistance from both gD and gH/gL. Mapping of virus-neutralizing antibodies and complementation analysis of gB insertion mutants revealed that gB contains four functional regions critical for viral entry into Vero cells (Bender et al. 2007; Connolly and Longnecker 2012; Lin and Spear 2007). HSV-1 gB contains internal hydrophobic fusion loops, two per monomer. Mutagenesis of specific loop residues yields gB that is nonfunctional for entry, likely because gB is rendered incapable of productive interaction with the host cell membrane (Hannah et al. 2007; Lin and Spear 2007). Mutations and truncations in the cytoplasmic tail domain of gB affect HSV-1 infectivity of Vero cells (Beitia Ortiz de Zarate et al. 2004; Bzik et al. 1984; Gage et al. 1993; Ruel et al. 2006).
Several gB-binding host cell proteins have been proposed as receptors for HSV-1 entry, including paired immunoglobulin-like type 2 receptor alpha, non-muscle myosin IIA and IIB, and myelin-associated glycoprotein (Arii et al. 2010, 2015; Satoh et al. 2008; Suenaga et al. 2010). For each of these, a gD-binding receptor is also needed for entry to occur.
Lysosomotropic agents that block the normally low pH of endosomes block HSV entry into a subset of cell types including human epithelial cells (Nicola et al. 2003, 2005). We have proposed that intracellular low pH serves as a host cell trigger for fusion during HSV entry into a subset of cells (Nicola 2016). The triggered refolding of fusion proteins drives the merging of viral and host membranes. Endosomal low pH is the most common inducer of conformational changes that mediate fusion. Following exposure to mildly acidic pH, the pre-fusion form of gB in virions undergoes conformational alterations, including changes in the antigenic structure of the fusion domain (Cairns et al. 2011; Dollery et al. 2010a, 2011; Siekavizza-Robles et al. 2010). Consistent with other class III fusion proteins, most of the pH-triggered changes are reversible. Notably, an irreversible, low-pH-induced change in the gB fusion domain was recently identified (Weed et al. 2017).
The entry of several strains of HSV-1 and HSV-2 into CHO-nectin-1 cells occurs via a well-characterized low-pH, endocytic pathway (Nicola 2016). However, when nectin-2 is expressed in CHO cells, HSV-1 strains ANG path and ANG are directed to a pH-independent, non-endocytic pathway (Delboy et al. 2006; Roller et al. 2008). When PILRalpha is expressed in CHO cells, wild-type HSV-1 enters in a pH-independent, non-endocytic manner (Arii et al. 2009). The same receptor may direct HSV entry to diverse pathways, depending on the cell in which it is expressed, indicating the involvement of additional host cell factors that remain to be identified. For example, nectin-1 mediates low-pH entry into CHO cells (Nicola et al. 2003), whereas nectin-1 expressed in the J1.1–2 (Gianni et al. 2004) or B78 (Milne et al. 2005) cell lines initiates entry that is pH independent.
HSV-1 gH is 838 amino acids in length (Gompels and Minson 1986). It is a type I membrane glycoprotein with a single pass transmembrane domain and a short cytoplasmic tail of 14 amino acids. HSV-1 gL contains 224 amino acids, lacks a transmembrane domain, and is non-covalently bound near the N-terminus of gH (Chowdary et al. 2010; Hutchinson et al. 1992). The 1:1 gH/gL heterodimer is absolutely required for HSV fusion dining entry, yet its specific role is not well understood. HSV-1 gH/gL interacts with integrins (Gianni et al. 2013; Parry et al. 2005). Binding to alpha-V-beta-6 or alpha-V-beta-8 integrins leads to the release of gL (Gianni et al. 2015). Mutations in the gH transmembrane region, in the cytoplasmic tail, or in the membrane-proximal H3 domain of the gH ectodomain impair HSV-1 infectivity of Vero cells (Galdiero et al. 1997; Harman et al. 2002).
2.2.2 Fusion of Primary Enveloped HSV-1 with the Outer Nuclear Membrane
Herpesviruses have a complex assembly and egress strategy. Progeny nucleocapsids are assembled in the nucleus and bud through the inner nuclear membrane to acquire a primary envelope. In the perinuclear space, primary enveloped virions fuse with the outer nuclear membrane (ONM) and deliver capsids and tegument to the cytosol. This fusion process termed de-envelopment is unique to herpesviruses. HSV acquires its mature envelope at Golgi-derived cytoplasmic membranes followed by exocytosis of progeny infectious virions. A distinct feature of fusion during de-envelopment is that the effector membrane (the primary virion envelope) and the target membrane (the ONM of the infected cell) both contain viral proteins. The compositions of primary and mature virions are distinct, which may result in different fusion mechanisms. For example, UL31p and UL34p are components of perinuclear HSV particles but are not detected in mature, extracellular virions (Fuchs et al. 2002; Loret et al. 2008; Reynolds et al. 2002; Padula et al. 2009).
Perinuclear virions deleted for gB, gD, or gH exhibit little to no defects in de-envelopment, suggesting a fusion mechanism very different from that occurring during viral entry. Since gB-null virus is competent for ONM fusion, this suggests the possibility that HSV-1 contains a fusogen other than gB. Alternately, cellular fusion factors may play a role. Viruses lacking both gB and gH are defective in ONM fusion, suggesting that gB and gH play redundant roles in de-envelopment fusion (Farnsworth et al. 2007; Johnson et al. 2011). However, considerable numbers of mature virions are produced, suggesting there are alternate mechanisms of de-envelopment independent of gB and gH/gL (Johnson and Baines 2011; Klupp et al. 2008). Phosphorylation of the gB cytoplasmic tail by the viral Us3 kinase is important for de-envelopment (Wisner et al. 2009).
Additional viral proteins are proposed to positively (VP16 and UL51p) or negatively (UL20p and gK) regulate ONM fusion (Baines et al. 1991; Hutchinson and Johnson 1995; Mossman et al. 2000; Nozawa et al. 2005). Two host cell molecules, CD98 heavy chain and beta-1 integrin, promote ONM fusion (Hirohata et al. 2015). Host cell p32 regulates de-envelopment in an HSV-1 UL47p-dependent manner (Liu et al. 2014). The complete viral and cellular requirements and mechanistic details of ONM fusion are still being elucidated. The complexity of de-envelopment fusion rivals or exceeds that of the other herpesviral fusion events.
2.2.3 HSV Syncytium Formation
Based on the physical phenotype of infected cells in culture, HSV strains can be divided into those that form plaques, clusters of rounded, infected cells often with clearings in the center, or those that form multinucleated giant cells or syncytia. Syncytial and non-syncytial (plaque-forming) strains have been referred to as syn and syn+, respectively. Syncytium formation is caused by fusion of an infected cell with neighboring uninfected cells, resulting in the clustering of nuclei that share the same cytoplasm. This cell-to-cell fusion has also been referred to as fusion-from-within. Microscopic visualization of tissue from the lesion of an HSV-infected individual typically reveals syncytia. However, patient isolates usually form plaques in culture, not syncytia, for reasons that are not understood. Plaque formation likely involves limited fusion of infected cells with neighboring uninfected cells. The gE/gl complex facilitates cell-to-cell spread (Dingwell et al. 1994), but the fusion mechanism associated with spread of non-syncytial strains is not well characterized.
The syncytial phenotype of HSV in culture results from defined mutations in one or more viral genes. Syncytium formation can in turn be modulated by additional viral proteins. Syncytium-forming mutants arise readily in culture. Truncations or single amino acid mutations in the gB cytoplasmic tail can cause syncytia (Bzik et al. 1984; Cai et al. 1988a, b; Engel et al. 1993; Gage et al. 1993). In the presence of wild-type gB, gD, gE, gH/gL, and gM, viruses with specific mutations in gK, UL20, or UL24 form syncytia in culture (Baines et al. 1991; Bzik et al. 1984; Debroy et al. 1985; Jacobson et al. 1989; Ruyechan et al. 1979; Sanders et al. 1982; Tognon et al. 1991). Most mutations in gK are located in its N-terminal, extracellular/luminal domain (Dolter et al. 1994). Deletion of the gB, gD, gE, gH/gL, gl, gM, UL11, UL16, or UL21 genes from a syncytial virus abolishes or reduces syncytium formation (Balan et al. 1994; Cai et al. 1987, 1988a; Davis-Poynter et al. 1994; Han et al. 2012; Ligas and Johnson 1988), suggesting that these envelope proteins play required or key roles in fusion during syncytium formation. A gE deletion mutant that retains its full syncytial phenotype has also been reported (Neidhardt et al. 1987). Antibodies to gB, gD, gE, gH, or gL inhibit cell fusion by syncytial strains (Chatterjee et al. 1989; Gompels and Minson 1986; Minson et al. 1986; Navarro et al. 1992, Noble et al. 1983; Novotny et al. 1996; Sanchez-Pescador et al. 1993), underscoring the importance of these glycoproteins. gB, gD, and gH/gL must be in same membrane (in cis) to mediate syncytium formation (Davis-Poynter et al. 1994). Mutations in HSV-1 UL20 (Foster et al. 2004), UL45 (Haanes et al. 1994), or the gH cytoplasmic tail (Browne et al. 1996; Wilson et al. 1994) negatively regulate syncytium formation. Overexpression of gN causes syncytium formation in wild-type HSV-infected cells (El Kasmi and Lippe 2015). The gC gene is often deleted in syncytial mutants of HSV-1 for reasons that are not clear (DeLuca et al. 1982; Heine et al. 1974; Zezulak and Spear 1984).
The mechanism of syncytium formation is poorly understood. Changes in cytoplasmic tail domains of HSV envelope proteins may affect the structure and function of ectodomains or they may affect an interaction of the tail with the membrane. The cytoplasmic tails may interact with unidentified host cell components that are important for syncytium formation. Syncytial strains frequently cause fusion in some cell types but not others (Roizman 1962), consistent with a role for cellular factors. The host cell gD-binding receptors nectin-1, nectin-2, or HVEM are required to mediate syncytium formation in CHO cells, provided that the proper form of gD is present (Terry-Allison et al. 1998, 2001). Heparan sulfate appears to be less important for syncytium formation than for viral entry (Shieh and Spear 1994; Terry-Allison et al. 2001).
2.2.4 Fusion from Without
Fusion from without (FFWO) is cell fusion triggered by contact of virions with the target cell surface at high multiplicity in the absence of HSV protein synthesis (Fig. 2.2) (Falke et al. 1985). A subset of syncytial strains of HSV-1 has FFWO activity. A V553A mutation in the gB ectodomain and the A855V syncytial mutation in the cytoplasmic tail of gB are both required for FFWO (Saharkhiz-Langroodi and Holland 1997). The ectodomain mutation V553A maps to domain III of the gB structure (Heldwein et al. 2006). This mutation has been described as a rate-of-entry determinant (Bzik et al. 1984). Transfer of FFWO gB to a non-FFWO HSV-1 strain is sufficient to bestow FFWO activity (Saharkhiz-Langroodi and Holland 1997). FFWO is cell-type dependent and temperature dependent and occurs at an optimal pH of 7.8–8.5 (Falke et al. 1985). Low-pH pretreatment inactivates the infectivity of virions with FFWO activity to a similar extent as wild-type HSV-1 (Siekavizza-Robles et al. 2010).
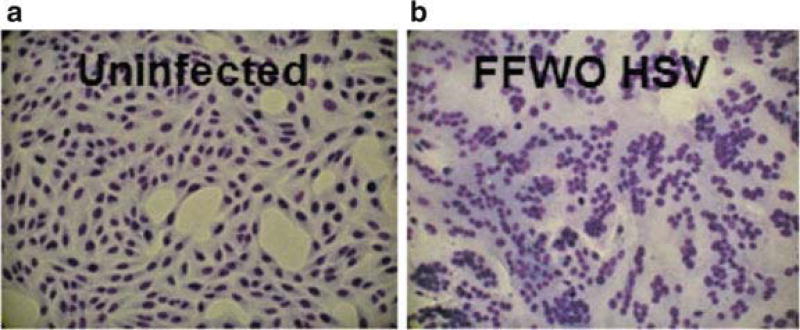
Fig. 2.2.
Fusion from without induced by HSV. (a) Uninfected Vero cells. (b) HSV-1 strain ANG path added to Vero cells for 3 h in the presence of cycloheximide
The cell fusion triggered by FFWO strains is likely due to a virus-cell fusion event. A single viral particle that simultaneously binds to two adjacent cells may result in a bipartite fusion event. This is consistent with the high MOI requirement for FFWO.
Alternately, FFWO may be mediated by fusion of input virions with cells followed by fusion of the cells with each other. FFWO is a useful surrogate for virus-cell fusion during entry as both processes share several criteria (Delboy et al. 2008). The effector and target membranes for FFWO and plasma membrane entry are the same. FFWO like viral entry depends on the presence of an appropriate cognate gD receptor in the target membrane. Nectin-1, nectin-2, or HVEM can each mediate FFWO, provided that the effector virus bears a form of gD that can interact with the given receptor (Delboy et al. 2006; Roller et al. 2008). Further, the efficiency of gD receptor usage for an FFWO strain correlates with the efficiency of entry mediated by the same receptor. Virus-neutralizing antibodies block FFWO (Falke et al. 1985). Monoclonal antibodies to gB and gD that block HSV-1 FFWO also neutralize viral entry (Roller et al. 2008). FFWO requires host cell cholesterol, and the cholesterol precursor desmosterol can also function in this capacity (Wudiri et al. 2014).
Clinical isolates with fusion-from-without activity have not been reported. FFWO gB may be considered hyperfusogenic. However, FFWO gB does not alone promote pH-independent fusion with the plasma membrane of cells that support endocytic low-pH entry of wild-type HSV-1. The presence of FFWO gB in HSV does not alter the pH dependence of entry. In other words, in cell types that require low pH for wild-type HSV entry, FFWO gB does not promote pH-independent fusion with the cell surface (Roller et al. 2008). FFWO gB has reduced reactivity with MAbs DL16 and H126 and is thus antigenically distinct. The antigenic changes in FFWO gB are similar to those induced in wild-type gB by mildly acidic pH, suggesting that changes in gB antigenic conformation correlate with fusion activity (Dollery et al. 2010a; Roller et al. 2008).
2.2.5 Transfected Cell Fusion
The most detailed models of HSV-1 membrane fusion are based on a virus-free experimental system in which effector cells transiently transfected with viral glycoproteins are mixed with target cells (Fig. 2.3). This reductionist approach has yielded critical information about the fusion capabilities of HSV glycoproteins and their interactions. Results from transfected cell assays, hereafter referred to as cell-cell fusion, ultimately require confirmation in experiments that measure fusion events relevant to the HSV replication cycle. This is complicated by the broad landscape of viral and cellular factors at play during HSV-1 entry and infection.
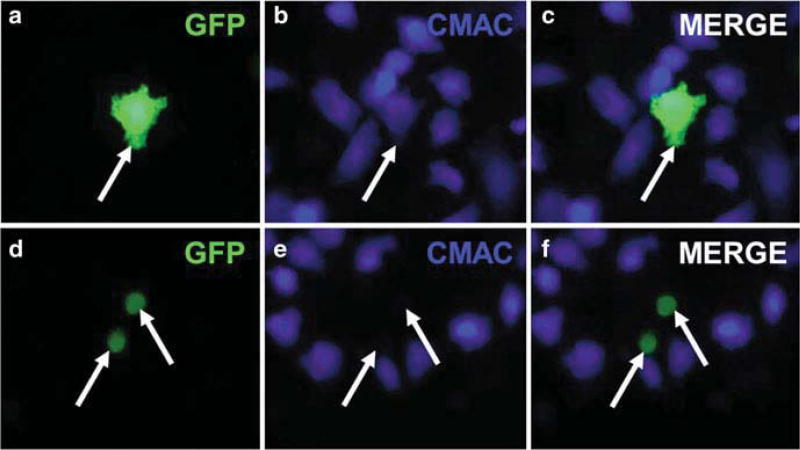
Fig. 2.3.
Transfected cell-cell fusion. Effector CHO-K1 cells were transfected with plasmids for HSV-1 gB, gD, gH, gL, and GFP (a–c) or gD, gH, gL, and GFP (d–f). Target CHO-nectin-1 cells were labeled with CMAC CellTracker Blue. Effector and target cells were mixed for 6 h. Arrow in (a), (b), and (c) indicates at least one target and one effector cell that have fused. Arrows in (d), (e), and (f) indicate effector cells that have not fused with a target cell
Four HSV envelope proteins, gB, gD, and gH/gL, are necessary and sufficient for cell-cell fusion (Muggeridge 2000; Turner et al. 1998). A gD receptor but not cell surface heparan sulfate is required in the target cell (Browne et al. 2001; Pertel et al. 2001). αvβ6 integrin or αvβ8 integrin enhances nectin-1-mediated cell-cell fusion (Gianni et al. 2013). The roles of additional host factors such as gB receptors and gH receptors remain to be elucidated. Soluble or lipid-anchored forms of gD can trigger cell-cell fusion provided that native gB and gH/gL are present (Atanasiu et al. 2007; Cocchi et al. 2004; Jones and Geraghty 2004). Similarly, soluble forms of gH/gL in the context of authentic gB and gD can also mediate low levels of fusion. However, gB must be membrane anchored in order for cell-cell fusion to occur, consistent with the notion that it is the core fusogen (Atanasiu et al. 2010a).
Mutations in the transmembrane region of gH or its cytoplasmic tail result in undetectable or reduced cell-cell fusion (Harman et al. 2002; Jackson et al. 2010; Rogalin and Heldwein 2015). However a soluble, membrane-truncated form of gH/gL can trigger fusion, provided that full-length gD and gB are in the membrane. Mutation of the gB fusion loops at hydrophobic residues 174, 179, or 261 ablates cell-cell fusion activity (Hannah et al. 2007; Lin and Spear 2007). Single amino acid mutations in gB domain V reduce cell-cell fusion (Connolly and Longnecker 2012).
Transient cell-cell fusion can be regulated by additional HSV-1 envelope proteins. Expression of alphaherpesviral gM or the gM/gN complex reduces fusion mediated by gB, gD, and gH/gL. gM/gN reduces surface expression of gD and gH/gL, which at least partly explains the reduction in cell-cell fusion (Crump et al. 2004; Klupp et al. 2000; Koyano et al. 2003). Transfection of gM or gM/gN also downregulates surface expression of glycoproteins of unrelated viruses, of some cellular proteins, and also inhibits cell-cell fusion of unrelated viruses. Expression of gK reduces fusion mediated by gB, gD, and gH/gL, and co-expression of gK and UL20p decreases surface expression of the fusion glycoproteins (Avitabile et al. 2003, 2004). The gM/gN complex and gK and UL20 are dispensable for HSV-1 entry, yet play regulatory roles in cell-cell fusion and syncytium formation. In this regard, the cell-cell fusion experimental system may be more aligned with syncytium formation than with fusion during viral entry.
Many details of the HSV-1 fusion mechanism are derived from cell-cell fusion experiments. Transfected cell experiments coupled with bimolecular complementation (BiMC) have been particularly informative, detecting interactions between gB, gD, and gH/gL (Atanasiu et al. 2007, 2010a, b, 2013; Avitabile et al. 2007, 2009). For BiMC, split fluorescent proteins are fused to HSV-1 glycoproteins. When the two glycoproteins are in close enough proximity, the two halves come together to produce fluorescence that is detected by microscopy. Interactions revealed by BiMC must be interpreted cautiously, however, as non-specific interactions between HSV-1 glycoproteins and unrelated paramyxovirus glycoproteins were detected using this approach (Connolly et al. 2009). The affinity between the N- and C-terminal halves of the fluorescent protein may drive non-specific interactions.
A prevailing model of fusion is as follows: receptor-activated gD interacts with gH, which in turn interacts with gB, culminating in membrane fusion (Atanasiu et al. 2010a). Deletion of the N-terminal 28 residues of gH permits cell-cell fusion in the presence of gB and gL, but notably in the absence of gD, supporting a model whereby the fusion activation signal passes from gD to gH to gB (Atanasiu et al. 2013). However, interactions between gD and gB have also been detected (Atanasiu et al. 2010b; Avitabile et al. 2007), consistent with the notion that there are variations on the cell-cell fusion mechanism.
2.3 Summary and Future Directions
Successful HSV infection in cell culture requires several membrane fusion events. Each process involves a distinct pair of membranes (Fig. 2.1). The viral and host requirements, execution, and regulation of each fusion type are likely unique. Thus, there is no unified HSV-1 fusion mechanism. With the possible exception of nuclear de-envelopment, however, HSV-1 fusion events likely require gB, gH/gL, and gD as the core fusion machinery. A given HSV-1 fusion process has variations, which poses additional challenges. Defining the HSV and cell factors that are both necessary and sufficient for a given fusion reaction is fundamental to our understanding. The recent work to identify the functional interactions between and among these factors must also be continued and expanded. Elucidating the conformational changes in gB and other proteins that drive the fusion reaction is key. The incomplete understanding of the mechanism of herpesviral fusion has been a roadblock to developing therapeutic inhibitors of fusion and entry. Once we learn how herpesviruses mediate fusion, we can devise strategies to prevent it.
References
- Arii J, Uema M, Morimoto T, Sagara H, Akashi H, Ono E, Arase H, Kawaguchi Y. Entry of herpes simplex virus 1 and other alphaherpesviruses via the paired immunoglobulin-like type 2 receptor alpha. J Virol. 2009;83:4520–4527. doi: 10.1128/JVI.02601-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arii J, Goto H, Suenaga T, Oyama M, Kozuka-Hata H, Imai T, Minowa A, Akashi H, Arase H, Kawaoka Y, Kawaguchi Y. Non-muscle myosin IIA is a functional entry receptor for herpes simplex virus-1. Nature. 2010;467:859–862. doi: 10.1038/nature09420. [DOI] [PubMed] [Google Scholar]
- Arii J, Hirohata Y, Kato A, Kawaguchi Y. Nonmuscle myosin heavy chain IIb mediates herpes simplex virus 1 entry. J Virol. 2015;89:1879–1888. doi: 10.1128/JVI.03079-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanasiu D, Whitbeck JC, Cairns TM, Reilly B, Cohen GH, Eisenberg RJ. Bimolecular complementation reveals that glycoproteins gB and gH/gL of herpes simplex virus interact with each other during cell fusion. Proc Natl Acad Sci USA. 2007;104:18718–18723. doi: 10.1073/pnas.0707452104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanasiu D, Saw WT, Cohen GH, Eisenberg RJ. Cascade of events governing cell-cell fusion induced by herpes simplex virus glycoproteins gD, gH/gL, and gB. J Virol. 2010a;84:12292–12299. doi: 10.1128/JVI.01700-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanasiu D, Whitbeck JC, de Leon MP, Lou H, Hannah BP, Cohen GH, Eisenberg RJ. Bimolecular complementation defines functional regions of herpes simplex virus gB that are involved with gH/gL as a necessary step leading to cell fusion. J Virol. 2010b;84:3825–3834. doi: 10.1128/JVI.02687-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanasiu D, Cairns TM, Whitbeck JC, Saw WT, Rao S, Eisenberg RJ, Cohen GH. Regulation of herpes simplex virus gB-induced cell-cell fusion by mutant forms of gH/gL in the absence of gD and cellular receptors. MBio. 2013;4(2):e00046–e00013. doi: 10.1128/mBio.00046-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avitabile E, Lombardi G, Campadelli-Fiume G. Herpes simplex virus glycoprotein K, but not its syncytial allele, inhibits cell-cell fusion mediated by the four fusogenic glycoproteins, gD, gB, gH, and gL. J Virol. 2003;77:6836–6844. doi: 10.1128/JVI.77.12.6836-6844.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avitabile E, Lombardi G, Gianni T, Capri M, Campadelli-Fiume G. Coexpression of UL20p and gK inhibits cell-cell fusion mediated by herpes simplex virus glycoproteins gD, gH-gL, and wild-type gB or an endocytosis-defective gB mutant and downmodulates their cell surface expression. J Virol. 2004;78:8015–8025. doi: 10.1128/JVI.78.15.8015-8025.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avitabile E, Forghieri C, Campadelli-Fiume G. Complexes between herpes simplex virus glycoproteins gD, gB, and gH detected in cells by complementation of split enhanced green fluorescent protein. J Virol. 2007;81:11532–11537. doi: 10.1128/JVI.01343-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avitabile E, Forghieri C, Campadelli-Fiume G. Cross talk among the glycoproteins involved in herpes simplex virus entry and fusion: the interaction between gB and gH/gL does not necessarily require gD. J Virol. 2009;83:10752–10760. doi: 10.1128/JVI.01287-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines JD, Roizman B. The open reading frames UL3, UL4, UL10, and UL16 are dispensable for the replication of herpes simplex virus 1 in cell culture. J Virol. 1991;65:938–944. doi: 10.1128/jvi.65.2.938-944.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines JD, Ward PL, Campadelli-Fiume G, Roizman B. The UL20 gene of herpes simplex virus 1 encodes a function necessary for viral egress. J Virol. 1991;65:6414–6424. doi: 10.1128/jvi.65.12.6414-6424.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balan P, Davis-Poynter N, Bell S, Atkinson H, Browne H, Minson T. An analysis of the in vitro and in vivo phenotypes of mutants of herpes simplex virus type 1 lacking glycoproteins gG, gE, gl or the putative gJ. J Gen Virol. 1994;75(Pt 6):1245–1258. doi: 10.1099/0022-1317-75-6-1245. [DOI] [PubMed] [Google Scholar]
- Barrow E, Nicola AV, Liu J. Multiscale perspectives of virus entry via endocytosis. Virol J. 2013;10:177. doi: 10.1186/1743-422X-10-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beitia Ortiz de Zarate I, Kaelin K, Rozenberg F. Effects of mutations in the cytoplasmic domain of herpes simplex virus type 1 glycoprotein B on intracellular transport and infectivity. J Virol. 2004;78:1540–1551. doi: 10.1128/JVI.78.3.1540-1551.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender FC, Samanta M, Heldwein EE, de Leon MP, Bilman E, Lou H, Whitbeck JC, Eisenberg RJ, Cohen GH. Antigenic and mutational analyses of herpes simplex virus glycoprotein B reveal four functional regions. J Virol. 2007;81:3827–3841. doi: 10.1128/JVI.02710-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blissard GW, Wenz JR. Baculovirus gp64 envelope glycoprotein is sufficient to mediate pH-dependent membrane fusion. J Virol. 1992;66:6829–6835. doi: 10.1128/jvi.66.11.6829-6835.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne HM, Bruun BC, Minson AC. Characterization of herpes simplex virus type 1 recombinants with mutations in the cytoplasmic tail of glycoprotein H. J Gen Virol. 1996;77(Pt 10):2569–2573. doi: 10.1099/0022-1317-77-10-2569. [DOI] [PubMed] [Google Scholar]
- Browne H, Bruun B, Minson T. Plasma membrane requirements for cell fusion induced by herpes simplex virus type 1 glycoproteins gB, gD, gH and gL. J Gen Virol. 2001;82:1419–1422. doi: 10.1099/0022-1317-82-6-1419. [DOI] [PubMed] [Google Scholar]
- Bzik DJ, Fox BA, DeLuca NA, Person S. Nucleotide sequence of a region of the herpes simplex virus type 1 gB glycoprotein gene: mutations affecting rate of virus entry and cell fusion. Virology. 1984;137:185–190. doi: 10.1016/0042-6822(84)90022-9. [DOI] [PubMed] [Google Scholar]
- Cai WZ, Person S, Warner SC, Zhou JH, DeLuca NA. Linker-insertion nonsense and restriction-site deletion mutations of the gB glycoprotein gene of herpes simplex virus type 1. J Virol. 1987;61:714–721. doi: 10.1128/jvi.61.3.714-721.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai WH, Gu B, Person S. Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. J Virol. 1988a;62:2596–2604. doi: 10.1128/jvi.62.8.2596-2604.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai WZ, Person S, DebRoy C, Gu BH. Functional regions and structural features of the gB glycoprotein of herpes simplex virus type 1. An analysis of linker insertion mutants. J Mol Biol. 1988b;201:575–588. doi: 10.1016/0022-2836(88)90639-0. [DOI] [PubMed] [Google Scholar]
- Cairns TM, Whitbeck JC, Lou H, Heldwein EE, Chowdary TK, Eisenberg RJ, Cohen GH. Capturing the herpes simplex virus core fusion complex (gB-gH/gL) in an acidic environment. J Virol. 2011;85:6175–6184. doi: 10.1128/JVI.00119-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carfi A, Willis SH, Whitbeck JC, Krummenacher C, Cohen GH, Eisenberg RJ, Wiley DC. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol Cell. 2001;8:169–179. doi: 10.1016/s1097-2765(01)00298-2. [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Koga J, Whitley RJ. A role for herpes simplex virus type 1 glycoprotein E in induction of cell fusion. J Gen Virol. 1989;70(Pt 8):2157–2162. doi: 10.1099/0022-1317-70-8-2157. [DOI] [PubMed] [Google Scholar]
- Chowdary TK, Cairns TM, Atanasiu D, Cohen GH, Eisenberg RJ, Heldwein EE. Crystal structure of the conserved herpesvirus fusion regulator complex gH-gL. Nat Struct Mol Biol. 2010;17:882–888. doi: 10.1038/nsmb.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury S, Chouljenko VN, Naderi M, Kousoulas KG. The amino terminus of herpes simplex virus 1 glycoprotein K is required for virion entry via the paired immunoglobulin-like type-2 receptor alpha. J Virol. 2013;87:3305–3313. doi: 10.1128/JVI.02982-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocchi F, Fusco D, Menotti L, Gianni T, Eisenberg RJ, Cohen GH, Campadelli-Fiume G. The soluble ectodomain of herpes simplex virus gD contains a membrane-proximal pro-fusion domain and suffices to mediate virus entry. Proc Natl Acad Sci USA. 2004;101:7445–7450. doi: 10.1073/pnas.0401883101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen GH, Ponce de Leon M, Nichols C. Isolation of a herpes simplex virus-specific antigenic fraction which stimulates the production of neutralizing antibody. J Virol. 1972;10:1021–1030. doi: 10.1128/jvi.10.5.1021-1030.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly SA, Longnecker R. Residues within the C-terminal arm of the herpes simplex virus 1 glycoprotein B ectodomain contribute to its refolding during the fusion step of virus entry. J Virol. 2012;86:6386–6393. doi: 10.1128/JVI.00104-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly SA, Landsburg DJ, Carfi A, Wiley DC, Eisenberg RJ, Cohen GH. Structure-based analysis of the herpes simplex virus glycoprotein D binding site present on herpesvirus entry mediator HveA (HVEM) J Virol. 2002;76:10894–10904. doi: 10.1128/JVI.76.21.10894-10904.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly SA, Landsburg DJ, Carfi A, Wiley DC, Cohen GH, Eisenberg RJ. Structure-based mutagenesis of herpes simplex virus glycoprotein D defines three critical regions at the gD-HveA/HVEM binding interface. J Virol. 2003;77:8127–8140. doi: 10.1128/JVI.77.14.8127-8140.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly SA, Landsburg DJ, Carfi A, Whitbeck JC, Zuo Y, Wiley DC, Cohen GH, Eisenberg RJ. Potential nectin-1 binding site on herpes simplex virus glycoprotein D. J Virol. 2005;79:1282–1295. doi: 10.1128/JVI.79.2.1282-1295.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly SA, Leser GP, Jardetzky TS, Lamb RA. Bimolecular complementation of paramyxovirus fusion and hemagglutinin-neuraminidase proteins enhances fusion: implications for the mechanism of fusion triggering. J Virol. 2009;83:10857–10868. doi: 10.1128/JVI.01191-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump CM, Bruun B, Bell S, Pomeranz LE, Minson T, Browne HM. Alphaherpesvirus glycoprotein M causes the relocalization of plasma membrane proteins. J Gen Virol. 2004;85:3517–3527. doi: 10.1099/vir.0.80361-0. [DOI] [PubMed] [Google Scholar]
- David AT, Saied A, Charles A, Subramanian R, Chouljenko VN, Kousoulas KG. A herpes simplex virus 1 (McKrae) mutant lacking the glycoprotein K gene is unable to infect via neuronal axons and egress from neuronal cell bodies. MBio. 2012;3:e00144–12. doi: 10.1128/mBio.00144-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis-Poynter N, Bell S, Minson T, Browne H. Analysis of the contributions of herpes simplex virus type 1 membrane proteins to the induction of cell-cell fusion. J Virol. 1994;68:7586–7590. doi: 10.1128/jvi.68.11.7586-7590.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debroy C, Pederson N, Person S. Nucleotide sequence of a herpes simplex virus type 1 gene that causes cell fusion. Virology. 1985;145:36–48. doi: 10.1016/0042-6822(85)90199-0. [DOI] [PubMed] [Google Scholar]
- Delboy MG, Patterson JL, Hollander AM, Nicola AV. Nectin-2-mediated entry of a syncytial strain of herpes simplex virus via pH-independent fusion with the plasma membrane of Chinese hamster ovary cells. Virol J. 2006;3:105. doi: 10.1186/1743-422X-3-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delboy MG, Roller DG, Nicola AV. Cellular proteasome activity facilitates herpes simplex virus entry at a postpenetration step. J Virol. 2008;82:3381–3390. doi: 10.1128/JVI.02296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca N, Bzik DJ, Bond VC, Person S, Snipes W. Nucleotide sequences of herpes simplex virus type 1 (HSV-1) affecting virus entry, cell fusion, and production of glycoprotein gb (VP7) Virology. 1982;122:411–423. doi: 10.1016/0042-6822(82)90240-9. [DOI] [PubMed] [Google Scholar]
- Di Giovine P, Settembre EC, Bhargava AK, Luftig MA, Lou H, Cohen GH, Eisenberg RJ, Krummenacher C, Carfi A. Structure of herpes simplex virus glycoprotein D bound to the human receptor nectin-1. PLoS Pathog. 2011;7:e1002277. doi: 10.1371/journal.ppat.1002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwell KS, Brunetti CR, Hendricks RL, Tang Q, Tang M, Rainbow AJ, Johnson DC. Herpes simplex virus glycoproteins E and I facilitate cell-to-cell spread in vivo and across junctions of cultured cells. J Virol. 1994;68:834–845. doi: 10.1128/jvi.68.2.834-845.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollery SJ, Delboy MG, Nicola AV. Low pH-induced conformational change in herpes simplex virus glycoprotein B. J Virol. 2010a;84:3759–3766. doi: 10.1128/JVI.02573-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollery SJ, Lane KD, Delboy MG, Roller DG, Nicola AV. Role of the UL45 protein in herpes simplex virus entry via low pH-dependent endocytosis and its relationship to the conformation and function of glycoprotein B. Virus Res. 2010b;149:115–118. doi: 10.1016/j.virusres.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollery SJ, Wright CC, Johnson DC, Nicola AV. Low-pH-dependent changes in the conformation and oligomeric state of the prefusion form of herpes simplex virus glycoprotein B are separable from fusion activity. J Virol. 2011;85:9964–9973. doi: 10.1128/JVI.05291-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolter KE, Ramaswamy R, Holland TC. Syncytial mutations in the herpes simplex virus type 1 gK (UL53) gene occur in two distinct domains. J Virol. 1994;68:8277–8281. doi: 10.1128/jvi.68.12.8277-8281.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg RJ, Atanasiu D, Cairns TM, Gallagher JR, Krummenacher C, Cohen GH. Herpes virus fusion and entry: a story with many characters. Virus. 2012;4:800–832. doi: 10.3390/v4050800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Kasmi I, Lippe R. Herpes simplex virus 1 gN partners with gM to modulate the viral fusion machinery. J Virol. 2015;89:2313–2323. doi: 10.1128/JVI.03041-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel JP, Boyer EP, Goodman JL. Two novel single amino acid syncytial mutations in the carboxy terminus of glycoprotein B of herpes simplex virus type 1 confer a unique pathogenic phenotype. Virology. 1993;192:112–120. doi: 10.1006/viro.1993.1013. [DOI] [PubMed] [Google Scholar]
- Falke D, Knoblich A, Muller S. Fusion from without induced by herpes simplex virus type 1. Intervirology. 1985;24:211–219. doi: 10.1159/000149645. [DOI] [PubMed] [Google Scholar]
- Farnsworth A, Wisner TW, Webb M, Roller R, Cohen G, Eisenberg R, Johnson DC. Herpes simplex virus glycoproteins gB and gH function in fusion between the virion envelope and the outer nuclear membrane. Proc Natl Acad Sci USA. 2007;104:10187–10192. doi: 10.1073/pnas.0703790104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florkiewicz RZ, Rose JK. A cell line expressing vesicular stomatitis virus glycoprotein fuses at low pH. Science. 1984;225:721–723. doi: 10.1126/science.6087454. [DOI] [PubMed] [Google Scholar]
- Forrester A, Farrell H, Wilkinson G, Kaye J, Davis-Poynter N, Minson T. Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J Virol. 1992;66:341–348. doi: 10.1128/jvi.66.1.341-348.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster TP, Melancon JM, Baines JD, Kousoulas KG. The herpes simplex virus type 1 UL20 protein modulates membrane fusion events dining cytoplasmic virion morphogenesis and virus-induced cell fusion. J Virol. 2004;78:5347–5357. doi: 10.1128/JVI.78.10.5347-5357.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frampton AR, Jr, Stolz DB, Uchida H, Goins WF, Cohen JB, Glorioso JC. Equine herpesvirus 1 enters cells by two different pathways, and infection requires the activation of the cellular kinase ROCK1. J Virol. 2007;81:10879–10889. doi: 10.1128/JVI.00504-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs W, Klupp BG, Granzow H, Osterrieder N, Mettenleiter TC. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J Virol. 2002;76:364–378. doi: 10.1128/JVI.76.1.364-378.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller AO, Spear PG. Anti-glycoprotein D antibodies that permit adsorption but block infection by herpes simplex virus 1 prevent virion-cell fusion at the cell surface. Proc Natl Acad Sci USA. 1987;84:5454–5458. doi: 10.1073/pnas.84.15.5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage PJ, Levine M, Glorioso JC. Syncytium-inducing mutations localize to two discrete regions within the cytoplasmic domain of herpes simplex virus type 1 glycoprotein B. J Virol. 1993;67:2191–2201. doi: 10.1128/jvi.67.4.2191-2201.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galdiero M, Whiteley A, Bruun B, Bell S, Minson T, Browne H. Site-directed and linker insertion mutagenesis of herpes simplex virus type 1 glycoprotein H. J Virol. 1997;71:2163–2170. doi: 10.1128/jvi.71.3.2163-2170.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher JR, Saw WT, Atanasiu D, Lou H, Eisenberg RJ, Cohen GH. Displacement of the C terminus of herpes simplex virus gD is sufficient to expose the fusion-activating interfaces on gD. J Virol. 2013;87:12656–12666. doi: 10.1128/JVI.01727-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraghty RJ, Krummenacher C, Cohen GH, Eisenberg RJ, Spear PG. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science. 1998;280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- Gianni T, Campadelli-Fiume G, Menotti L. Entry of herpes simplex virus mediated by chimeric forms of nectin1 retargeted to endosomes or to lipid rafts occurs through acidic endosomes. J Virol. 2004;78:12268–12276. doi: 10.1128/JVI.78.22.12268-12276.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianni T, Salvioli S, Chesnokova LS, Hutt-Fletcher LM, Campadelli-Fiume G. αvβ6- and αvβ8-integrins serve as interchangeable receptors for HSV gH/gL to promote endocytosis and activation of membrane fusion. PLoS Pathog. 2013;9:e1003806. doi: 10.1371/journal.ppat.1003806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianni T, Massaro R, Campadelli-Fiume G. Dissociation of HSV gL from gH by alphavbeta6- or alphavbeta8-integrin promotes gH activation and virus entry. Proc Natl Acad Sci USA. 2015;112:E3901–E3910. doi: 10.1073/pnas.1506846112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompels U, Minson A. The properties and sequence of glycoprotein H of herpes simplex virus type 1. Virology. 1986;153:230–247. doi: 10.1016/0042-6822(86)90026-7. [DOI] [PubMed] [Google Scholar]
- Haanes EJ, Nelson CM, Soule CL, Goodman JL. The UL45 gene product is required for herpes simplex virus type 1 glycoprotein B-induced fusion. J Virol. 1994;68:5825–5834. doi: 10.1128/jvi.68.9.5825-5834.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Chadha P, Starkey JL, Wills JW. Function of glycoprotein E of herpes simplex virus requires coordinated assembly of three tegument proteins on its cytoplasmic tail. Proc Natl Acad Sci USA. 2012;109:19798–19803. doi: 10.1073/pnas.1212900109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah BP, Heldwein EE, Bender FC, Cohen GH, Eisenberg RJ. Mutational evidence of internal fusion loops in herpes simplex virus glycoprotein B. J Virol. 2007;81:4858–4865. doi: 10.1128/JVI.02755-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harman A, Browne H, Minson T. The transmembrane domain and cytoplasmic tail of herpes simplex virus type 1 glycoprotein H play a role in membrane fusion. J Virol. 2002;76:10708–10716. doi: 10.1128/JVI.76.21.10708-10716.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heine JW, Honess RW, Cassai E, Roizman B. Proteins specified by herpes simplex virus. XII. The virion polypeptides of type 1 strains. J Virol. 1974;14:640–651. doi: 10.1128/jvi.14.3.640-651.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldwein EE, Lou H, Bender FC, Cohen GH, Eisenberg RJ, Harrison SC. Crystal structure of glycoprotein B from herpes simplex virus 1. Science. 2006;313:217–220. doi: 10.1126/science.1126548. [DOI] [PubMed] [Google Scholar]
- Highlander SL, Sutherland SL, Gage PJ, Johnson DC, Levine M, Glorioso JC. Neutralizing monoclonal antibodies specific for herpes simplex virus glycoprotein D inhibit virus penetration. J Virol. 1987;61:3356–3364. doi: 10.1128/jvi.61.11.3356-3364.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Highlander SL, Cai WH, Person S, Levine M, Glorioso JC. Monoclonal antibodies define a domain on herpes simplex virus glycoprotein B involved in virus penetration. J Virol. 1988;62:1881–1888. doi: 10.1128/jvi.62.6.1881-1888.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirohata Y, Arii J, Liu Z, Shindo K, Oyama M, Kozuka-Hata H, Sagara H, Kato A, Kawaguchi Y. Herpes simplex virus 1 recruits CD98 heavy chain and beta1 integral to the nuclear membrane for viral de-envelopment. J Virol. 2015;89:7799–7812. doi: 10.1128/JVI.00741-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson L, Browne H, Wargent V, Davis-Poynter N, Primorac S, Goldsmith K, Minson AC, Johnson DC. A novel herpes simplex virus glycoprotein, gL, forms a complex with glycoprotein H (gH) and affects normal folding and surface expression of gH. J Virol. 1992;66:2240–2250. doi: 10.1128/jvi.66.4.2240-2250.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson L, Johnson DC. Herpes simplex virus glycoprotein K promotes egress of virus particles. J Virol. 1995;69:5401–5413. doi: 10.1128/jvi.69.9.5401-5413.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JO, Lin E, Spear PG, Longnecker R. Insertion mutations in herpes simplex virus 1 glycoprotein H reduce cell surface expression, slow the rate of cell fusion, or abrogate functions in cell fusion and viral entry. J Virol. 2010;84:2038–2046. doi: 10.1128/JVI.02215-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson JG, Martin SL, Coen DM. A conserved open reading frame that overlaps the herpes simplex virus thymidine kinase gene is important for viral growth in cell culture. J Virol. 1989;63:1839–1843. doi: 10.1128/jvi.63.4.1839-1843.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DC, Baines JD. Herpesviruses remodel host membranes for virus egress. Nat Rev Microbiol. 2011;9:382–394. doi: 10.1038/nrmicro2559. [DOI] [PubMed] [Google Scholar]
- Johnson DC, Wisner TW, Wright CC. Herpes simplex virus glycoproteins gB and gD function in a redundant fashion to promote secondary envelopment. J Virol. 2011;85:4910–4926. doi: 10.1128/JVI.00011-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones NA, Geraghty RJ. Fusion activity of lipid-anchored envelope glycoproteins of herpes simplex virus type 1. Virology. 2004;324:213–228. doi: 10.1016/j.virol.2004.03.024. [DOI] [PubMed] [Google Scholar]
- Klupp BG, Nixdorf R, Mettenleiter TC. Pseudorabies virus glycoprotein M inhibits membrane fusion. J Virol. 2000;74:6760–6768. doi: 10.1128/jvi.74.15.6760-6768.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klupp B, Altenschmidt J, Granzow H, Fuchs W, Mettenleiter TC. Glycoproteins required for entry are not necessary for egress of pseudorabies virus. J Virol. 2008;82:6299–6309. doi: 10.1128/JVI.00386-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komala Sari T, Pritchard SM, Cunha CW, Wudiri GA, Laws EI, Aguilar HC, Taus NS, Nicola AV. Contributions of herpes simplex virus type 1 envelope proteins to entry by endocytosis. J Virol. 2013;87:13922–13926. doi: 10.1128/JVI.02500-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyano S, Mar EC, Stamey FR, Inoue N. Glycoproteins M and N of human herpesvirus 8 form a complex and inhibit cell fusion. J Gen Virol. 2003;84:1485–1491. doi: 10.1099/vir.0.18941-0. [DOI] [PubMed] [Google Scholar]
- Krummenacher C, Nicola AV, Whitbeck JC, Lou H, Hou W, Lambris JD, Geraghty RJ, Spear PG, Cohen GH, Eisenberg RJ. Herpes simplex virus glycoprotein D can bind to poliovirus receptor-related protein 1 or herpesvirus entry mediator, two structurally unrelated mediators of virus entry. J Virol. 1998;72:7064–7074. doi: 10.1128/jvi.72.9.7064-7074.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krummenacher C, Supekar VM, Whitbeck JC, Lazear E, Connolly SA, Eisenberg RJ, Cohen GH, Wiley DC, Carfi A. Structure of unliganded HSV gD reveals a mechanism for receptor-mediated activation of virus entry. EMBO J. 2005;24:4144–4153. doi: 10.1038/sj.emboj.7600875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laquerre S, Argnani R, Anderson DB, Zucchini S, Manservigi R, Glorioso JC. Heparan sulfate proteoglycan binding by herpes simplex virus type 1 glycoproteins B and C, which differ in their contributions to virus attachment, penetration, and cell-to-cell spread. J Virol. 1998;72:6119–6130. doi: 10.1128/jvi.72.7.6119-6130.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear E, Carfi A, Whitbeck JC, Cairns TM, Krummenacher C, Cohen GH, Eisenberg RJ. Engineered disulfide bonds in herpes simplex virus type 1 gD separate receptor binding from fusion initiation and viral entry. J Virol. 2008;82:700–709. doi: 10.1128/JVI.02192-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligas MW, Johnson DC. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by beta-galactosidase sequences binds to but is unable to penetrate into cells. J Virol. 1988;62:1486–1494. doi: 10.1128/jvi.62.5.1486-1494.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin E, Spear PG. Random linker-insertion mutagenesis to identify functional domains of herpes simplex virus type 1 glycoprotein B. Proc Natl Acad Sci USA. 2007;104:13140–13145. doi: 10.1073/pnas.0705926104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Kato A, Shindo K, Noda T, Sagara H, Kawaoka Y, Arii J, Kawaguchi Y. Herpes simplex virus 1 UL47 interacts with viral nuclear egress factors UL31, UL34, and Us3 and regulates viral nuclear egress. J Virol. 2014;88:4657–4667. doi: 10.1128/JVI.00137-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longnecker R, Roizman B. Clustering of genes dispensable for growth in culture in the S component of the HSV-1 genome. Science. 1987;236:573–576. doi: 10.1126/science.3033823. [DOI] [PubMed] [Google Scholar]
- Longnecker R, Chatterjee S, Whitley RJ, Roizman B. Identification of a herpes simplex virus 1 glycoprotein gene within a gene cluster dispensable for growth in cell culture. Proc Natl Acad Sci USA. 1987;84:4303–4307. doi: 10.1073/pnas.84.12.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loret S, Guay G, Lippe R. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J Virol. 2008;82:8605–8618. doi: 10.1128/JVI.00904-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lycke E, Hamark B, Johansson M, Krotochwil A, Lycke J, Svennerholm B. Herpes simplex virus infection of the human sensory neuron. An electron microscopy study. Arch Virol. 1988;101:87–104. doi: 10.1007/BF01314654. [DOI] [PubMed] [Google Scholar]
- Manoj S, Jogger CR, Myscofski D, Yoon M, Spear PG. Mutations in herpes simplex virus glycoprotein D that prevent cell entry via nectins and alter cell tropism. Proc Natl Acad Sci USA. 2004;101:12414–12421. doi: 10.1073/pnas.0404211101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller N, Hutt-Fletcher LM. Epstein-Barr virus enters B cells and epithelial cells by different routes. J Virol. 1992;66:3409–3414. doi: 10.1128/jvi.66.6.3409-3414.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne RS, Nicola AV, Whitbeck JC, Eisenberg RJ, Cohen GH. Glycoprotein D receptor-dependent, low-pH-independent endocytic entry of herpes simplex virus type 1. J Virol. 2005;79:6655–6663. doi: 10.1128/JVI.79.11.6655-6663.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minson AC, Hodgman TC, Digard P, Hancock DC, Bell SE, Buckmaster EA. An analysis of the biological properties of monoclonal antibodies against glycoprotein D of herpes simplex virus and identification of amino acid substitutions that confer resistance to neutralization. J Gen Virol. 1986;67:1001–1013. doi: 10.1099/0022-1317-67-6-1001. [DOI] [PubMed] [Google Scholar]
- Montgomery RI, Warner MS, Lum BJ, Spear PG. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- Mossman KL, Sherburne R, Lavery C, Duncan J, Smiley JR. Evidence that herpes simplex virus VP16 is required for viral egress downstream of the initial envelopment event. J Virol. 2000;74:6287–6299. doi: 10.1128/jvi.74.14.6287-6299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muggeridge MI. Characterization of cell-cell fusion mediated by herpes simplex virus 2 glycoproteins gB, gD, gH and gL in transfected cells. J Gen Virol. 2000;81:2017–2027. doi: 10.1099/0022-1317-81-8-2017. [DOI] [PubMed] [Google Scholar]
- Navarro D, Paz P, Pereira L. Domains of herpes simplex virus I glycoprotein B that function in virus penetration, cell-to-cell spread, and cell fusion. Virology. 1992;186:99–112. doi: 10.1016/0042-6822(92)90064-v. [DOI] [PubMed] [Google Scholar]
- Neidhardt H, Schroder CH, Kaemer HC. Herpes simplex virus type 1 glycoprotein E is not indispensable for viral infectivity. J Virol. 1987;61:600–603. doi: 10.1128/jvi.61.2.600-603.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicola AV. Herpesvirus entry into host cells mediated by endosomal low pH. Traffic. 2016;17:965–975. doi: 10.1111/tra.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicola AV, Straus SE. Cellular and viral requirements for rapid endocytic entry of herpes simplex virus. J Virol. 2004;78:7508–7517. doi: 10.1128/JVI.78.14.7508-7517.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicola AV, Ponce de Leon M, Xu R, Hou W, Whitbeck JC, Krummenacher C, Montgomery RI, Spear PG, Eisenberg RJ, Cohen GH. Monoclonal antibodies to distinct sites on herpes simplex virus (HSV) glycoprotein D block HSV binding to HVEM. J Virol. 1998;72:3595–3601. doi: 10.1128/jvi.72.5.3595-3601.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicola AV, McEvoy AM, Straus SE. Roles for endocytosis and low pH in herpes simplex virus entry into HeLa and Chinese hamster ovary cells. J Virol. 2003;77:5324–5332. doi: 10.1128/JVI.77.9.5324-5332.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicola AV, Hou J, Major EO, Straus SE. Herpes simplex virus type 1 enters human epidermal keratinocytes, but not neurons, via a pH-dependent endocytic pathway. J Virol. 2005;79:7609–7616. doi: 10.1128/JVI.79.12.7609-7616.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble AG, Lee GT, Sprague R, Parish ML, Spear PG. Anti-gD monoclonal antibodies inhibit cell fusion induced by herpes simplex virus type 1. Virology. 1983;129:218–224. doi: 10.1016/0042-6822(83)90409-9. [DOI] [PubMed] [Google Scholar]
- Novotny MJ, Parish ML, Spear PG. Variability of herpes simplex virus 1 gL and anti-gL antibodies that inhibit cell fusion but not viral infectivity. Virology. 1996;221:1–13. doi: 10.1006/viro.1996.0347. [DOI] [PubMed] [Google Scholar]
- Nozawa N, Kawaguchi Y, Tanaka M, Kato A, Kato A, Kimura H, Nishiyama Y. Herpes simplex virus type 1 UL51 protein is involved in maturation and egress of virus particles. J Virol. 2005;79:6947–6956. doi: 10.1128/JVI.79.11.6947-6956.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padula ME, Sydnor ML, Wilson DW. Isolation and preliminary characterization of herpes simplex virus 1 primary enveloped virions from the perinuclear space. J Virol. 2009;83:4757–4765. doi: 10.1128/JVI.01927-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry C, Bell S, Minson T, Browne H. Herpes simplex virus type 1 glycoprotein H binds to alphavbeta3 integrins. J Gen Virol. 2005;86:7–10. doi: 10.1099/vir.0.80567-0. [DOI] [PubMed] [Google Scholar]
- Peng T, Ponce-de-Leon M, Jiang H, Dubin G, Lubinski JM, Eisenberg RJ, Cohen GH. The gH-gL complex of herpes simplex virus (HSV) stimulates neutralizing antibody and protects mice against HSV type 1 challenge. J Virol. 1998;72:65–72. doi: 10.1128/jvi.72.1.65-72.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertel PE, Fridberg A, Parish ML, Spear PG. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH-gL requires a gD receptor but not necessarily heparan sulfate. Virology. 2001;279:313–324. doi: 10.1006/viro.2000.0713. [DOI] [PubMed] [Google Scholar]
- Polcicova K, Biswas PS, Banerjee K, Wisner TW, Rouse BT, Johnson DC. Herpes keratitis in the absence of anterograde transport of virus from sensory ganglia to the cornea. Proc Natl Acad Sci USA. 2005;102:11462–11467. doi: 10.1073/pnas.0503230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghu H, Sharma-Walia N, Veettil MV, Sadagopan S, Chandran B. Kaposi’s sarcoma-associated herpesvirus utilizes an actin polymerization-dependent macropinocytic pathway to enter human dermal microvascular endothelial and human umbilical vein endothelial cells. J Virol. 2009;83:4895–4911. doi: 10.1128/JVI.02498-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds AE, Wills EG, Roller RJ, Ryckman BJ, Baines JD. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J Virol. 2002;76:8939–8952. doi: 10.1128/JVI.76.17.8939-8952.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogalin HB, Heldwein EE. Interplay between the herpes simplex virus 1 gB cytodomain and the gH cytotail during cell-cell fusion. J Virol. 2015;89:12262–12272. doi: 10.1128/JVI.02391-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roizman B. Polykaryocytosis. Cold Spring Harb Symp Quant Biol. 1962;27:327–342. doi: 10.1101/sqb.1962.027.001.031. [DOI] [PubMed] [Google Scholar]
- Roller DG, Dollery SJ, Doyle JL, Nicola AV. Structure-function analysis of herpes simplex virus glycoprotein B with fusion-from-without activity. Virology. 2008;382:207–216. doi: 10.1016/j.virol.2008.09.015. [DOI] [PubMed] [Google Scholar]
- Roop C, Hutchinson L, Johnson DC. A mutant herpes simplex virus type 1 unable to express glycoprotein L cannot enter cells, and its particles lack glycoprotein H. J Virol. 1993;67:2285–2297. doi: 10.1128/jvi.67.4.2285-2297.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruel N, Zago A, Spear PG. Alanine substitution of conserved residues in the cytoplasmic tail of herpes simplex virus gB can enhance or abolish cell fusion activity and viral entry. Virology. 2006;346:229–237. doi: 10.1016/j.virol.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Ruyechan WT, Morse LS, Knipe DM, Roizman B. Molecular genetics of herpes simplex virus. II. Mapping of the major viral glycoproteins and of the genetic loci specifying the social behavior of infected cells. J Virol. 1979;29:677–697. doi: 10.1128/jvi.29.2.677-697.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saharkhiz-Langroodi A, Holland TC. Identification of the fusion-from-without determinants of herpes simplex virus type 1 glycoprotein B. Virology. 1997;227:153–159. doi: 10.1006/viro.1996.8327. [DOI] [PubMed] [Google Scholar]
- Sanchez-Pescador L, Pereira L, Charlebois ED, Kohl S. Antibodies to epitopes of herpes simplex virus type 1 glycoprotein B (gB) in human sera: analysis of functional gB epitopes defined by inhibition of murine monoclonal antibodies. J Infect Dis. 1993;168:844–853. doi: 10.1093/infdis/168.4.844. [DOI] [PubMed] [Google Scholar]
- Sanders PG, Wilkie NM, Davison AJ. Thymidine kinase deletion mutants of herpes simplex virus type 1. J Gen Virol. 1982;63:277–295. doi: 10.1099/0022-1317-63-2-277. [DOI] [PubMed] [Google Scholar]
- Satoh T, Arii J, Suenaga T, Wang J, Kogure A, Uehori J, Arase N, Shiratori I, Tanaka S, Kawaguchi Y, Spear PG, Lanier LL, Arase H. PILRalpha is a herpes simplex virus-1 entry coreceptor that associates with glycoprotein B. Cell. 2008;132:935–944. doi: 10.1016/j.cell.2008.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh MT, Spear PG. Herpesvirus-induced cell fusion that is dependent on cell surface heparan sulfate or soluble heparin. J Virol. 1994;68:1224–1228. doi: 10.1128/jvi.68.2.1224-1228.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla D, Spear PG. Herpesviruses and heparan sulfate: an intimate relationship in aid of viral entry. J Clin Invest. 2001;108:503–510. doi: 10.1172/JCI13799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla D, Liu J, Blaiklock P, Shworak NW, Bai X, Esko JD, Cohen GH, Eisenberg RJ, Rosenberg RD, Spear PG. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell. 1999;99:13–22. doi: 10.1016/s0092-8674(00)80058-6. [DOI] [PubMed] [Google Scholar]
- Siekavizza-Robles CR, Dollery SJ, Nicola AV. Reversible conformational change in herpes simplex virus glycoprotein B with fusion-from-without activity is triggered by mildly acidic pH. Virol J. 2010;7:352. doi: 10.1186/1743-422X-7-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear PG, Manoj S, Yoon M, Jogger CR, Zago A, Myscofski D. Different receptors binding to distinct interfaces on herpes simplex virus gD can trigger events leading to cell fusion and viral entry. Virology. 2006;344:17–24. doi: 10.1016/j.virol.2005.09.016. [DOI] [PubMed] [Google Scholar]
- Striebinger H, Funk C, Raschbichler V, Bailer SM. Subcellular trafficking and functional relationship of the HSV-1 glycoproteins N and M. Virus. 2016;8:83. doi: 10.3390/v8030083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suenaga T, Satoh T, Somboonthum P, Kawaguchi Y, Mori Y, Arase H. Myelin-associated glycoprotein mediates membrane fusion and entry of neurotropic herpesviruses. Proc Natl Acad Sci USA. 2010;107:866–871. doi: 10.1073/pnas.0913351107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry-Allison T, Montgomery RI, Whitbeck JC, Xu R, Cohen GH, Eisenberg RJ, Spear PG. HveA (herpesvirus entry mediator A), a coreceptor for herpes simplex virus entry, also participates in virus-induced cell fusion. J Virol. 1998;72:5802–5810. doi: 10.1128/jvi.72.7.5802-5810.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry-Allison T, Montgomery RI, Warner MS, Geraghty RJ, Spear PG. Contributions of gD receptors and glycosaminoglycan sulfation to cell fusion mediated by herpes simplex virus 1. Virus Res. 2001;74:39–45. [Google Scholar]
- Tognon M, Guandalini R, Romanelli MG, Manservigi R, Trevisani B. Phenotypic and genotypic characterization of locus syn 5 in herpes simplex virus 1. Virus Res. 1991;18:135–150. doi: 10.1016/0168-1702(91)90014-m. [DOI] [PubMed] [Google Scholar]
- Turner A, Bruun B, Minson T, Browne H. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J Virol. 1998;72:873–875. doi: 10.1128/jvi.72.1.873-875.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Walle GR, Peters ST, Vander Ven BC, O’Callaghan DJ, Osterrieder N. Equine herpesvirus 1 entry via endocytosis is facilitated by alphaV integrins and an RSD motif in glycoprotein D. J Virol. 2008;82:11859–11868. doi: 10.1128/JVI.00868-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visalli RJ, Brandt CR. The HSV-1 UL45 gene is not required for growth in Vero cells. Virology. 1991;185:419–423. doi: 10.1016/0042-6822(91)90790-i. [DOI] [PubMed] [Google Scholar]
- Warner MS, Geraghty RJ, Martinez WM, Montgomery RI, Whitbeck JC, Xu R, Eisenberg RJ, Cohen GH, Spear PG. A cell surface protein with herpesvirus entry activity (HveB) confers susceptibility to infection by mutants of herpes simplex virus type 1, herpes simplex virus type 2, and pseudorabies virus. Virology. 1998;246:179–189. doi: 10.1006/viro.1998.9218. [DOI] [PubMed] [Google Scholar]
- Weber PC, Levine M, Glorioso JC. Rapid identification of nonessential genes of herpes simplex virus type 1 by Tn5 mutagenesis. Science. 1987;236:576–579. doi: 10.1126/science.3033824. [DOI] [PubMed] [Google Scholar]
- Weed DJ, Pritchard SM, Gonzalez F, Aguilar HC, Nicola AV. Mildly acidic pH triggers an irreversible conformational change in the fusion domain of herpes simplex virus 1 glycoprotein B and inactivation of viral entry. J Virol. 2017;91 doi: 10.1128/JVI.02123-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenhom W, Hinz A, Gaudin Y. Virus membrane fusion. FEBS Lett. 2007;581:2150–2155. doi: 10.1016/j.febslet.2007.01.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DW, Davis-Poynter N, Minson AC. Mutations in the cytoplasmic tail of herpes simplex virus glycoprotein H suppress cell fusion by a syncytial strain. J Virol. 1994;68:6985–6993. doi: 10.1128/jvi.68.11.6985-6993.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisner TW, Wright CC, Kato A, Kawaguchi Y, Mou F, Baines JD, Roller RJ, Johnson DC. Herpesvirus gB-induced fusion between the virion envelope and outer nuclear membrane during virus egress is regulated by the viral US3 kinase. J Virol. 2009;83:3115–3126. doi: 10.1128/JVI.01462-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wudiri GA, Pritchard SM, Li H, Liu J, Aguilar HC, Gilk SD, Nicola AV. Molecular requirement for sterols in herpes simplex virus entry and infectivity. J Virol. 2014;88:13918–13922. doi: 10.1128/JVI.01615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon M, Spear PG. Random mutagenesis of the gene encoding a viral ligand for multiple cell entry receptors to obtain viral mutants altered for receptor usage. Proc Natl Acad Sci USA. 2004;101:17252–17257. doi: 10.1073/pnas.0407892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon M, Zago A, Shukla D, Spear PG. Mutations in the N termini of herpes simplex virus type 1 and 2 gDs alter functional interactions with the entry/fusion receptors HVEM, nectin-2, and 3-O-sulfated heparan sulfate but not with nectin-1. J Virol. 2003;77:9221–9231. doi: 10.1128/JVI.77.17.9221-9231.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zezulak KM, Spear PG. Mapping of the structural gene for the herpes simplex virus type 2 counterpart of herpes simplex virus type 1 glycoprotein C and identification of a type 2 mutant which does not express this glycoprotein. J Virol. 1984;49:741–747. doi: 10.1128/jvi.49.3.741-747.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]