Abstract
Translation is a pivotal step in the regulation of gene expression as well as one of the most energy consuming processes in the cell. Dysregulation of translation caused by the aberrant function of upstream signaling pathways and/or perturbations in the expression or function of components of the translation machinery is frequent in cancer. In this review, we discuss emerging findings that highlight hitherto unappreciated aspects of signaling to the translation apparatus with the particular focus on emerging connections between protein synthesis, autophagy and energy homeostasis in cancer.
Introduction
Dysregulation of mRNA translation is common in cancer. Oncogenes (e.g. MYC, RAS, PI3KCA) and tumor suppressors (PTEN, LKB1, TSC1/2, p53) impinge on the translation apparatus [1]. Changes in expression and/or mutations of the components of translational machinery (e.g. eIFs, ribosomal proteins) are also frequent in neoplasia [1]. Translation is an essential step in the regulation of gene expression. Although the contribution of the translatome to the composition of the proteome is debated, at least under certain conditions translation plays a major role in regulation of gene expression. For instance, various stressors (e.g. ER-stress) impede the ternary complex (TC) recycling [2]. Limited availability of TC (composed of the eIF2, GTP and initiator tRNA) results in reprograming of the translatome, whereby decrease in global protein synthesis is accompanied by translational activation of some uORF-containing mRNAs (e.g. ATF4) [2]. Cancer cells are exposed to various stressors (e.g. hypoxia, nutrient deprivation) as the neoplastic growth outstrips vascular supply, ergo it is reasonable to postulate that translation is important for shaping malignant proteomes. Translation is also one of the most energy costly processes [3]. Signaling nodes (e.g. mTOR and AMPK) are linked to both regulation of translation and energy homeostasis (Figure 1). Herein, we focus on recent findings highlighting the role of signaling pathways in the orchestration of protein synthesis and energy balance.
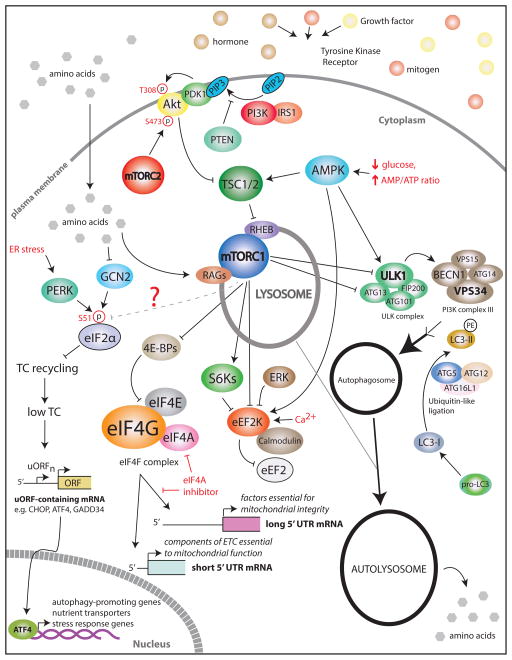
Figure 1. Schematic presentation of the orchestration of protein synthesis, energy metabolism and autophagy.
mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2) are two functionally and structurally distinct complexes. In response to growth factors (e.g. IGFs) and hormones (e.g. insulin), mTORC1 is activated via the PI3K/AKT/TSC/RHEB pathway, whereas amino acids activate mTORC1 via RAG GTPases (reviewed in [4]). mTORC1 phosphorylates and inactivates translational inhibitor 4E-BPs, which then allows eIF4F complex assembly. Increased levels of eIF4F lead to reprogramming of the translatome which in part leads to selective increase in translation of long 5′UTR mRNAs that encode factors that protect mitochondrial integrity, and short 5′UTR mRNAs which encode components of the electron transport chain (ETC) complex. When energy resources are limiting, AMPK is activated to reduce anabolic processes, while stimulating catabolic ones (reviewed in [5]. This is in part achieved by inhibition of mTORC1. eEF2K phosphorylates eEF2 which interferes with its ribosomal association and reduces elongation rates. This suppresses protein synthesis and reduces energy consumption. mTORC1 and ERK inactivate eEF2K thereby increasing protein synthesis, whereas AMPK activates eEF2K and reduces translation rates. GCN2 and PERK are major eIF2α kinases that sense nutritional (e.g. limited glucose, amino acids etc.) and endoplasmic reticulum (ER) stress, respectively. eIF2α phosphorylation coincides with reduction in ternary complex (TC) levels. This leads to the suppression of global translation while translationally activating some of the upstream open reading frame (uORF) containing mRNAs including CHOP, ATF4, GADD34. ATF4 induces upregulation of amino acid transporters and aminoacyl-tRNA synthetases as well as a number of autophagy protein-encoding genes, including p62, ATG16L, LC3B, ATG12, ATG3, and BECN1. AMPK is a major positive regulator of the autophagy protein ULK1, while mTORC1 is a major negative regulator of autophagy. The ULK1 complex functions to initiate autophagosome formation along with the PI3K complex. This increased autophagic flux allows for cytoplasmic components to be recycled during acute nutrient starvation to feed back into the synthesis of vital proteins. Collectively, mTOR, AMPK, and eIF2α kinases coordination show multifaceted signaling nodes of nutrient sensing, translation, and autophagy. Of note, this representation of the pathways is simplified to highlight the mechanisms of coordination of mRNA translation, metabolism and autophagy. Detailed representation of mTOR, AMPK pathways and autophagy can be found in [4,5,64].
The mTOR/4E-BP/eIF4F axis coordinates translation and cancer energetics
Cancer cells must adjust their protein synthesis output to adapt to fluctuations in nutrient and oxygen availability. Initiation is thought to be the most regulated phase of protein synthesis [2]. One of the rate-limiting steps of initiation is the eIF4F complex assembly. eIF4F consists of the 5′mRNA cap-binding subunit eIF4E, the scaffold eIF4G and the DEAD-box helicase eIF4A [2]. eIF4F recruits mRNA to the ribosome and its levels are largely determined by mTOR, a serine/threonine kinase which integrates a number of stimuli (e.g. nutrients, growth factors, hormones) to adjust growth rates to cellular energy status [4]. In mammals, there are two functionally and structurally distinct mTOR complexes, mTORC1 and mTORC2 [4]. mTORC1 is a major stimulator of protein synthesis and other anabolic processes [4]. In addition to mTOR, AMPK plays a central role in maintenance of energy balance [5]. AMPK was thought to be activated when the AMP/ATP ratio is increased, but recently it was shown that AMPK may be activated by glucose withdrawal even before AMP/ATP ratios raise [6]. AMPK reduces anabolic processes while bolstering catabolism, in part by inhibiting mTORC1 [5].
mTORC1 phosphorylates translational inhibitors 4E-BPs (4E-BP1-3 in mammals) that block eIF4G:eIF4E binding [1]. 4E-BP phosphorylation dissociates them from eIF4E, which facilitates eIF4E:eIF4G interaction and eIF4F assembly. mTORC1 activity is frequently upregulated in cancer, which in addition to common overexpression of eIF4E results in elevated eIF4F levels in neoplasia [1]. High eIF4F levels are linked to resistance to both chemotherapy and targeted therapies, and predict poor patient outcome [1]. Hyperactivation of mTORC1 in cancer not only increases global protein synthesis, but also results in dramatic translational reprograming [1]. In part, mTORC1 stimulates translation of nuclear-encoded mRNAs which are translated into proteins with mitochondrial functions (e.g. TFAM, ETC complex components) [7,8]. Increase in energy consumption upon mTORC1 activation is thus compensated by enhanced translation of mRNAs that encode proteins that impact on mitochondrial number, function and dynamics. This is mediated by 4E-BPs [7–9]. These findings demonstrate that the mTORC1/4E-BP axis plays a major role in maintaining energy balance by coordinating mitochondrial ATP production, and protein synthesis rates (Figure 1).
Regulation of synthesis of nuclear-encoded mitochondrial proteins
Allosteric mTOR inhibitors (rapamycin and rapalogs) are employed for a variety of oncological indications, whereas second-generation mTOR inhibitors that target its active site are undergoing clinical trials [10]. Third-generation bivalent mTOR inhibitors that target both allosteric and active site have also been developed [11]. In most cell lines, however, mTOR inhibitors exert anti-proliferative, but not cytotoxic effects. This may be explained by emerging data that show that although mTOR inhibition results in decreased energy production, it concomitantly leads to reduced energy consumption, thereby resulting in a state of metabolic dormancy [8]. Accordingly, anti-neoplastic effects of anti-diabetic biguanides (e.g. metformin), which induce stress by decreasing mitochondrial ATP production, are potentiated in LKB1-deficient cells as they fail to induce AMPK, inhibit mTOR, and reduce energy consumption, thus resulting in energy crisis and cell death [12,13]. mTOR inhibition leads to simultaneous suppression of translation of mRNAs that encode proteins with mitochondrial function (i.e. subunits of ETC) and those that protect mitochondrial integrity (i.e. BCL-2 family members) [14]. This results in reduction in mitochondrial respiration and number, which is compensated by reduction in protein synthesis, and stimulation of autophagy, a vital cytoplasmic recycling process[14].
A large proportion of the transcripts which encode ETC subunits are characterized by short 5′UTRs (<30 nucleotides), whereas most transcripts encoding pro-survival factors harbor long 5′UTRs (>150 nucleotides). Transcripts with long, but not short 5′UTRs exhibit enhanced sensitivity to eIF4A inhibition [14]. eIF4A inhibitors induce cell death through translational suppression of BCL-2 family members and survivin, without reducing translation of ETC component-encoding mRNAs, or mTORC1 activity [14,15]. This induces mitochondrial depolarization and Bax/Bak-mediated apoptosis [14]. Hence, while mTOR inhibitors concurrently suppress translation of mRNAs that encode proteins involved in mitochondrial functions and protection of mitochondrial integrity, resulting in metabolic dormancy and cytostatic effects, eIF4A inhibitors induce apoptosis at least in part by selective inhibition of synthesis of proteins that maintain mitochondrial integrity without reducing mTOR activity, which results in energy crisis and cell death (Figure 1).
TISU element and coordination of transcriptional and translational mechanisms
The precise mechanism of how metabolic stress impacts on the translatome remains to be determined. A subset of short 5′UTR mRNAs contains a Translation Initiator of Short 5′UTR (TISU) element (C/GAAC/GATGGCGGC) which also serves as YY1 transcription regulatory element [16]. TISU mRNAs are actively translated under glucose deprivation despite reduction in global protein synthesis [17,18]. mTOR inhibitor rapamycin suppresses translation of TISU mRNAs [18]. Mechanistically, eIF4G:eIF1 interaction is thought to stimulate eIF4F release upon 48S complex formation on TISU mRNAs [18], followed by eIF1A-directed interaction between RPS3 and TISU and subsequent RPS3/RPS10e exchange upon 80S assembly [19]. Simultaneous monitoring of the effects of glucose starvation on transcription start site selection and translation suggested that whereas 5′UTR length plays a role in regulation of mRNA translation in unstressed cells, this appears to be mostly mediated by the nature of cap-proximal nucleotides in glucose-deprived cells [20]. Intriguingly, glucose starvation appeared to induce alternative promoter selection in genes encoding translation factors including eIF4A1 and PABP. This appears to stimulate their translation under stress and results in eIF4A1 and PABP proteoforms with altered function [20]. Although the generality and physiological significance of these findings remains to be established, these data suggest the presence of complex orchestration of transcriptional and translational reprograming during adaptation to energy stress that is likely to play a major, yet underexplored role in neoplasia.
eIF2 and nutrient sensing
Cells in solid tumors are exposed to limited nutrients (e.g. glucose, amino acids) and in this context eIF2α phosphorylation is thought to act as a pro-survival mechanism [21,22]. eIF2α phosphorylation limits TC recycling and is stimulated via four kinases: GCN2, PERK, PKR, and HRI [2]. GCN2 is a major eIF2α kinase which senses nutritional stress [2]. In yeast, GCN2 is activated by binding directly to uncharged tRNAs via a protein domain related to histidyl-tRNA synthetase, which induces autophosphorylation and derepression of its kinase domain [23]. eIF2α phosphorylation inhibits GEF activity of eIF2B, thereby limiting TC levels [2]. This suppresses global translation while inducing synthesis of proteins encoded by uORF mRNAs, including the transcription factor ATF4 [2]. ATF4 upregulates expression of amino acid transporters and aminoacyl-tRNA synthetases [24]. Prostate cancer cells appear to depend on increased expression of multiple amino acid transporters, including LAT family (L-type amino acid transporters) members which are involved in uptake of amino acids with neutral side chains, including L-leucine [25]. LAT1 is required for growth of androgen-insensitive PC3 cells [26] and ATF4 stimulates its expression. ATF4 also stimulates expression of genes encoding autophagic factors (see below) to increase amino acid availability. Accordingly, ATF4 has been suggested as a potential target in various malignancies [27]. Importantly, a number of studies have suggested a cross-talk between mTOR, eIF2α phosphorylation [28–30] and/or ATF4 [31,32]. This positions eIF2 and ATF4 as central nodes of regulatory networks which orchestrate protein synthesis, autophagy and energy metabolism to maintain energy balance of cancer cells (Figure 1).
Emerging roles of elongation in cancer energetics
Cross-talk between the translation machinery and energetics also occurs at the elongation step, which is the most energy consuming translation phase. Each elongation cycle consumes two GTPs, one during delivery of aminoacyl (aa)-tRNA to the ribosomal A-site by eEF1A, and another during translocation of the ribosome which is mediated by eEF2 [33]. In addition, elongation consumes aa-tRNAs, whereby aminoacylation of tRNAs requires hydrolysis of ATP to AMP, which is equivalent of 2 ATPs [33].
eEF2K, a member of the atypical α-kinase family, phosphorylates eEF2 (T56 in humans) which interferes with its ribosomal association and reduces elongation rates[34]. Under physiological conditions eEF2K is regulated by insulin and Ca2+. In muscle, reduction in ATP levels releases Ca2+, which stimulates eEF2K association with calmodulin, eEF2K activation and eEF2 phosphorylation [35]. This shuts down protein synthesis, thereby reducing energy consumption and allowing for ATP replenishment.
mTORC1 phosphorylates and inactivates eEF2K (S366 in humans) via S6K, which leads to increased elongation rates [36]. S366 is also phosphorylated by ERK/p90RSK pathway [36]. In addition, mTORC1 has been shown to directly phosphorylate eEF2K (S78 in humans), which interferes with calmodulin binding and also leads to inactivation of eEF2K [36]. AMPK phosphorylates eEF2K (S398 in humans), thereby leading to an overall decrease in protein synthesis [37]. eEF2K activity is also stimulated in hypoxia in an HIF-independent manner that appears to involve reduction in hydroxylation of Pro98 (in humans). Hydroxylation of Pro98 likely compromises calmodulin:eEF2K association[38].
Since mTORC1 and ERK are upregulated in neoplasia, it is plausible that increased protein synthesis rates in cancer are at least in part caused by eEF2K inactivation. Considering that elevated protein synthesis rates parallel autonomous growth observed in neoplasia, it is thus expected that inactivation of eEF2K endows cancer cells with proliferative advantage. Indeed, in a model of APC-loss-driven intestinal carcinogenesis, eEF2K inactivation by the mTORC1/S6K axis reprograms elongation to increase cyclin D3 levels and support aberrant WNT signaling [39]. Ablation of eEF2K alleviated the effects of mTORC1 inactivation on proliferation of enterocytes [39], which suggests that eEF2K exhibits tumor suppressive properties. In contrast, eEF2K is thought to prevent energy crisis and cancer cell death under conditions when energy resources are limiting. For instance, it has been demonstrated that eEF2K may exhibit tumor protective effects in cell culture and xenograft models under the conditions of nutrient starvation [40,41]. This seemingly contradictory role of eEF2K in cancer is reminiscent of AMPK function in tumor initiation and progression. AMPK appears to impede tumor formation, but when the tumors reach the size whereby the nutrients and oxygen become limiting, AMPK takes a cytoprotective role [42]. By analogy, it is plausible that eEF2K is involved in suppressing tumor formation (e.g. in the APC loss-driven colorectal cancer model), while promoting tumor survival via reducing energy consumption by the translational machinery when the tumors reach certain mass and the nutrients become scarce. Since eEF2K is not an essential gene [43], there is a heightened interest to delineate the precise role of eEF2K in neoplasia to facilitate employment of eEF2K inhibitors in the clinic.
Cross-talk between autophagy and protein synthesis
While mTORC1 stimulates protein synthesis, it is a major negative regulator of autophagy (Figure 1). Nutrient deprivation, which causes mTORC1 inhibition, induces autophagy, thereby supplying the free amino acids needed for the synthesis of crucial proteins [4]. Indeed, as shown in yeast, autophagy maintains protein synthesis when amino acids are limiting [44]. ULK1 is a serine/threonine kinase that resides in an initiatory complex baring its name and regulates other autophagy factors such as AMBRA, ATG9, and BECN1, thereby stimulating autophagosome formation [45–47]. mTORC1 inhibits ULK1 function by phosphorylating it at S758 and S638 (in human), whereas phosphorylation by AMPK on Ser317 and Ser556 (in human) results in ULK1 activation [48,49]. mTORC1 also phosphorylates and inactivates other autophagy proteins including AMBRA and ATG13 [50,51].
Autophagy proteins also feedback to regulate mTORC1, and therefore indirectly affect protein synthesis. For example, the autophagy receptor protein SQSTM1/p62, which brings ubiquitinated substrates to the autophagosome, has been reported to interact with mTOR, raptor and RAG GTPases, and is important for amino acid sensing [52]. Once the autophagosome is formed, it must fuse with lysosomes to degrade its contents which results in release of free amino acids [53]. This implies that autophagy can self-regulate via its later stages through modulation of mTORC1 and stimulation of protein synthesis, once sufficient cytoplasmic contents are recycled.
Changes in eIF levels and/or activity have also been demonstrated to impact on autophagy. For instance, increased eIF4G1 levels are paralleled by translational activation of mRNAs implicated in cell survival that consequently prevent autophagy and apoptosis [54]. Moreover, the eIF2α/ATF4 axis upregulates transcription of many autophagy genes, including p62, ATG16L, LC3B, ATG12, ATG3, and BECN1 [55]. Cross-regulation of mTOR and eIF2α phosphorylation also impacts on autophagy. mTORC1 inhibition results in eIF2α phosphorylation via PP6C phosphatase dependent activation of GCN2. Depletion of PP6C attenuates autophagy in response to mTORC1 inhibition, whereas PP6C mutations present in melanoma increase autophagy [29].
Post-transcriptional regulation of autophagy mRNAs has also been reported. HuD binds to the 3′-UTR of ATG5 mRNA and increases its translation [56]. In Drosophila, deadenylation by Orb-regulated CCR4 represses translation of ATG12 mRNA, which consequently inhibits autophagy [57]. Decapping of autophagy protein-encoding mRNAs was reported to inhibit their synthesis when nutrients are not limiting [58]. Others have however reported the opposite effects of mRNA capping on autophagy rates in other experimental systems [59].
Therefore, autophagy generates amino acids for protein synthesis, which consequently consumes them. Both cellular processes are both controlled by the same signaling hubs (mTORC1, AMPK, and GCN2) to maintain energy balance (Figure 1).
Concluding remarks and future challenges
Recent data indicate that mTOR, AMPK and eIF2α kinases coordinate translation with cellular energetics and autophagy, which allows simultaneous modulation of the proteome and maintenance of energy balance. As understanding of the molecular underpinnings of the crosstalk between translation and energy metabolism is expanding, new regulatory nodes are starting to emerge. For example, eIF6, which binds to 60S ribosome and interferes with subunit joining, stimulates translation of transcriptional factors which increase lipogenesis and glycolysis in a mTOR-independent manner [60]. Similarly, eIF3, a large multiprotein complex that plays a role in recruitment of the mRNA to the ribosome, increases translation of mRNAs encoding mitochondrial components [61]. Changes in mRNA methylation also impact on the translatome [62]. Cancer-specific metabolic perturbations, including increase in D-2HG in tumors harboring IDH1/2 mutations (e.g. glioma and leukemia) are expected to disrupt mRNA methylation patterns and alter translation by inhibiting mRNA demethylases (e.g. ALKBH5) [63]. Collectively, findings outlined in this review show complex cross-regulation of nutrient sensing, metabolism, translation and autophagy via signaling pathways. Future research is required to dissect the molecular mechanisms which orchestrate these processes in homeostasis and when dysregulated result in cancer.
Table 1.
List of Abbreviations
| Definition | |
|---|---|
| 4E-BP | Eukaryotic initiation factor 4E-binding protein |
| AKT | Protein Kinase B |
| ALKBH5 | Alpha-Ketoglutarate-Dependent Dioxygenase AlkB Homolog 5 |
| AMBRA | Activating Molecule In BECN1-Regulated Autophagy Protein 1 |
| AMPK | Adenosine Monophosphate-activated Protein Kinase |
| APC | Adenomatous polyposis coli |
| ATF4 | Activating Transcription Factor 4 |
| ATG | Autophagy Related |
| ATG16L1 | Autophagy Related 16 Like 1 |
| BCL-2 | B-Cell CLL/Lymphoma 2 |
| BECN1 | Beclin1 |
| CCR4 | C-C motif Chemokine Receptor 4 |
| D-2HG | D-2-hydroxyglutarate |
| eEF2 | Eukaryotic Elongation factor 2 |
| eEF2K | Eukaryotic Elongation factor 2 kinase |
| eIF | Eukaryotic Initiation Factor |
| ER | Endoplasmic Reticulum |
| ERK | Extracellular Signal–regulated kinase |
| ETC | Electron Transport Chain |
| GCN2 | General Control Nonderepressible 2 |
| GDP | Guanosine diphosphate |
| GEF | Guanine nucleotide exchange factor |
| HIF | Hypoxia-inducible factor |
| HRI | Heme-regulated Eukaryotic Initiation Factor 2α kinase |
| HuD | Hu Antigen D |
| IDH1/2 | Isocitrate Dehydrogenase (NADP+) 1/2 |
| LAT | L-type Amino Acid Transporter |
| LAT1 | L-type Amino Acid Transporter 1 |
| LC3B | Microtubule Associated Protein 1 Light Chain 3 Beta |
| LKB1/STK11 | Serine/Threonine Kinase 11 |
| mRNA | Messenger RNA |
| mTOR | Mammalian Target of Rapamycin |
| mTORC1 | Mammalian Target of Rapamycin complex 1 |
| mTORC2 | Mammalian Target of Rapamycin complex 2 |
| P90RSK | 90 KDa Ribosomal Protein S6 Kinase |
| PABP | Poly-A binding protein |
| PC-3 | Prostate cancer cells |
| PERK | protein kinase R (PKR)-like endoplasmic reticulum kinase |
| PI3KCA | Phosphatidylinositol 3-kinase |
| PKR | Protein Kinase R |
| PP6C | Protein Phosphatase 6 Catalytic Subunit |
| Pro98 | Proline residue 98 |
| PTEN | Phosphatase and Tensin Homolog |
| RPS10e | Ribosomal protein S10e |
| RPS3 | Ribosomal protein S3 |
| S6K | Ribosomal Protein S6 Kinase |
| SQSTM1/p62 | Sequestome 1/p62 |
| TC | Ternary complex |
| TFAM | Transcription Factor A, Mitochondrial |
| TFEB | Transcription Factor EB |
| TISU | Translation Initiator of Short 5′Untranslated Region |
| tRNA | Transfer RNA |
| TSC1/2 | Tuberous sclerosis 1/2 |
| ULK1 | Unc-51 Like Autophagy Activating Kinase 1 |
| uORF | Upstream Open Reading Frame |
| UTR | Untranslated region |
Highlights.
Protein synthesis is one of the most energy consuming processes in the cell
Nutrient availability modulates growth signaling and protein synthesis rates
Autophagy is a survival mechanism providing metabolic substrates during stress
Nutrient sensing, protein synthesis and autophagy are coordinated via mTORC1
Acknowledgments
Due to the brevity of the format we apologize to the colleagues whose work we did not reference. We thank all the members of our labs for their hard work. Many thanks to Laura Hulea and Oro Uchenunu for proof-reading the manuscript. LF is supported by the Department of Health and Human Services acting through the Victorian Cancer Agency (MCRF16007) and Cancer Australia grant (CA 1084546). This work was made possible through Independent Research Institutes Infrastructure Support Scheme Grant (361646) from the Australian National Health and Medical Research Council and a Victorian State Government Operational Infrastructure Support Grant to The Walter & Eliza Hall Institute of Medical Research (L.M.L.). I.T. is a Junior 2 Research Scholars of the Fonds de Recherche du Québec – Santé (FRQ-S) and the work in his lab is supported in part by grants from Canadian Institutes of Health Research (PJT-148603), National Institutes of Health (CA202021-01-A1) and Terry Fox New Frontiers Program in Cancer (TFRI 242115).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Bhat M, Robichaud N, Hulea L, Sonenberg N, Pelletier J, Topisirovic I. Targeting the translation machinery in cancer. Nat Rev Drug Discov. 2015;14:261–278. doi: 10.1038/nrd4505. [DOI] [PubMed] [Google Scholar]
- 2.Hinnebusch AG. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem. 2014;83:779–812. doi: 10.1146/annurev-biochem-060713-035802. [DOI] [PubMed] [Google Scholar]
- 3.Buttgereit F, Brand MD. A hierarchy of ATP-consuming processes in mammalian cells. Biochem J. 1995;312(Pt 1):163–167. doi: 10.1042/bj3120163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saxton RA, Sabatini DM. mTOR Signaling in Growth, Metabolism, and Disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13:251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang CS, Hawley SA, Zong Y, Li M, Wang Z, Gray A, Ma T, Cui J, Feng JW, Zhu M, et al. Fructose-1,6-bisphosphate and aldolase mediate glucose sensing by AMPK. Nature. 2017;548:112–116. doi: 10.1038/nature23275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morita M, Gravel SP, Chenard V, Sikstrom K, Zheng L, Alain T, Gandin V, Avizonis D, Arguello M, Zakaria C, et al. mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab. 2013;18:698–711. doi: 10.1016/j.cmet.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 8.Morita M, Gravel SP, Hulea L, Larsson O, Pollak M, St-Pierre J, Topisirovic I. mTOR coordinates protein synthesis, mitochondrial activity and proliferation. Cell Cycle. 2015;14:473–480. doi: 10.4161/15384101.2014.991572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •9.Morita M, Prudent J, Basu K, Goyon V, Katsumura S, Hulea L, Pearl D, Siddiqui N, Strack S, McGuirk S, et al. mTOR Controls Mitochondrial Dynamics and Cell Survival via MTFP1. Mol Cell. 2017;67:922–935. e925. doi: 10.1016/j.molcel.2017.08.013. This article shows that the translation of MTFP1 is tightly regulated by mTORC1 and that uncoupling this regulation leads to cell death by accumulation of fragmented dysfunctional mitochondria. [DOI] [PubMed] [Google Scholar]
- 10.Liko D, Hall MN. mTOR in health and in sickness. J Mol Med (Berl) 2015;93:1061–1073. doi: 10.1007/s00109-015-1326-7. [DOI] [PubMed] [Google Scholar]
- ••11.Rodrik-Outmezguine VS, Okaniwa M, Yao Z, Novotny CJ, McWhirter C, Banaji A, Won H, Wong W, Berger M, de Stanchina E, et al. Overcoming mTOR resistance mutations with a new-generation mTOR inhibitor. Nature. 2016;534:272–276. doi: 10.1038/nature17963. In this article, the authors have chemically joined rapamycin with TOR active site inhibitors as a way to bypass resistance to either inhibitors alone. They termed their new inhibitor series, rapalink. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shackelford DB, Abt E, Gerken L, Vasquez DS, Seki A, Leblanc M, Wei L, Fishbein MC, Czernin J, Mischel PS, et al. LKB1 inactivation dictates therapeutic response of non-small cell lung cancer to the metabolism drug phenformin. Cancer Cell. 2013;23:143–158. doi: 10.1016/j.ccr.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Algire C, Amrein L, Bazile M, David S, Zakikhani M, Pollak M. Diet and tumor LKB1 expression interact to determine sensitivity to anti-neoplastic effects of metformin in vivo. Oncogene. 2011;30:1174–1182. doi: 10.1038/onc.2010.483. [DOI] [PubMed] [Google Scholar]
- •14.Gandin V, Masvidal L, Hulea L, Gravel SP, Cargnello M, McLaughlan S, Cai Y, Balanathan P, Morita M, Rajakumar A, et al. nanoCAGE reveals 5′ UTR features that define specific modes of translation of functionally related MTOR-sensitive mRNAs. Genome Res. 2016;26:636–648. doi: 10.1101/gr.197566.115. Using transcription start site profiling applied to mRNA fractionated by density gradients (polysome profiling) this article revealed distinct subsets of mTOR sensitive mRNAs with varying dependence on eIF4A and that do not contain 5′TOP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindqvist LM, Vikström I, Chambers JM, McArthur K, Ann Anderson M, Henley KJ, Happo L, Cluse L, Johnstone RW, Roberts AW, et al. Translation inhibitors induce cell death by multiple mechanisms and Mcl-1 reduction is only a minor contributor. Cell Death Dis. 2012;3:e409. doi: 10.1038/cddis.2012.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elfakess R, Dikstein R. A translation initiation element specific to mRNAs with very short 5′UTR that also regulates transcription. PLoS One. 2008;3:e3094. doi: 10.1371/journal.pone.0003094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elfakess R, Sinvani H, Haimov O, Svitkin Y, Sonenberg N, Dikstein R. Unique translation initiation of mRNAs-containing TISU element. Nucleic Acids Res. 2011;39:7598–7609. doi: 10.1093/nar/gkr484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••18.Sinvani H, Haimov O, Svitkin Y, Sonenberg N, Tamarkin-Ben-Harush A, Viollet B, Dikstein R. Translational tolerance of mitochondrial genes to metabolic energy stress involves TISU and eIF1-eIF4GI cooperation in start codon selection. Cell Metab. 2015;21:479–492. doi: 10.1016/j.cmet.2015.02.010. This article showed that eIF4F needs to be displaced from the 5′mRNA Cap by a eIF1/eiF4G1 complex for maximal translation of TISU-containing mRNAs under stress conditions. [DOI] [PubMed] [Google Scholar]
- 19.Haimov O, Sinvani H, Martin F, Ulitsky I, Emmanuel R, Tamarkin-Ben-Harush A, Vardy A, Dikstein R. Efficient and Accurate Translation Initiation Directed by TISU Involves RPS3 and RPS10e Binding and Differential Eukaryotic Initiation Factor 1A Regulation. Mol Cell Biol. 2017:37. doi: 10.1128/MCB.00150-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamarkin-Ben-Harush A, Vasseur JJ, Debart F, Ulitsky I, Dikstein R. Cap-proximal nucleotides via differential eIF4E binding and alternative promoter usage mediate translational response to energy stress. Elife. 2017:6. doi: 10.7554/eLife.21907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muaddi H, Majumder M, Peidis P, Papadakis AI, Holcik M, Scheuner D, Kaufman RJ, Hatzoglou M, Koromilas AE. Phosphorylation of eIF2alpha at serine 51 is an important determinant of cell survival and adaptation to glucose deficiency. Mol Biol Cell. 2010;21:3220–3231. doi: 10.1091/mbc.E10-01-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye J, Kumanova M, Hart LS, Sloane K, Zhang H, De Panis DN, Bobrovnikova-Marjon E, Diehl JA, Ron D, Koumenis C. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J. 2010;29:2082–2096. doi: 10.1038/emboj.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong J, Qiu H, Garcia-Barrio M, Anderson J, Hinnebusch AG. Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol Cell. 2000;6:269–279. doi: 10.1016/s1097-2765(00)00028-9. [DOI] [PubMed] [Google Scholar]
- 24.Krokowski D, Han J, Saikia M, Majumder M, Yuan CL, Guan BJ, Bevilacqua E, Bussolati O, Broer S, Arvan P, et al. A self-defeating anabolic program leads to beta-cell apoptosis in endoplasmic reticulum stress-induced diabetes via regulation of amino acid flux. J Biol Chem. 2013;288:17202–17213. doi: 10.1074/jbc.M113.466920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Q, Tiffen J, Bailey CG, Lehman ML, Ritchie W, Fazli L, Metierre C, Feng YJ, Li E, Gleave M, et al. Targeting amino acid transport in metastatic castration-resistant prostate cancer: effects on cell cycle, cell growth, and tumor development. J Natl Cancer Inst. 2013;105:1463–1473. doi: 10.1093/jnci/djt241. [DOI] [PubMed] [Google Scholar]
- 26.Wang Q, Bailey CG, Ng C, Tiffen J, Thoeng A, Minhas V, Lehman ML, Hendy SC, Buchanan G, Nelson CC, et al. Androgen receptor and nutrient signaling pathways coordinate the demand for increased amino acid transport during prostate cancer progression. Cancer Res. 2011;71:7525–7536. doi: 10.1158/0008-5472.CAN-11-1821. [DOI] [PubMed] [Google Scholar]
- 27.Singleton DC, Harris AL. Targeting the ATF4 pathway in cancer therapy. Expert Opin Ther Targets. 2012;16:1189–1202. doi: 10.1517/14728222.2012.728207. [DOI] [PubMed] [Google Scholar]
- 28.Gandin V, Masvidal L, Cargnello M, Gyenis L, McLaughlan S, Cai Y, Tenkerian C, Morita M, Balanathan P, Jean-Jean O, et al. mTORC1 and CK2 coordinate ternary and eIF4F complex assembly. Nat Commun. 2016;7:11127. doi: 10.1038/ncomms11127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •29.Wengrod J, Wang D, Weiss S, Zhong H, Osman I, Gardner LB. Phosphorylation of eIF2alpha triggered by mTORC1 inhibition and PP6C activation is required for autophagy and is aberrant in PP6C-mutated melanoma. Sci Signal. 2015;8:ra27. doi: 10.1126/scisignal.aaa0899. Wengrod and colleagues showed that the well characterized increase in autophagy observed following mTORC1 inhibition is in fact dependent of the phosphatase activity of PP6C. Thereby identifying a key regulatory phosphatase linking mTORC1 and GCN2 activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tenkerian C, Krishnamoorthy J, Mounir Z, Kazimierczak U, Khoutorsky A, Staschke KA, Kristof AS, Wang S, Hatzoglou M, Koromilas AE. mTORC2 Balances AKT Activation and eIF2alpha Serine 51 Phosphorylation to Promote Survival under Stress. Mol Cancer Res. 2015;13:1377–1388. doi: 10.1158/1541-7786.MCR-15-0184-T. [DOI] [PubMed] [Google Scholar]
- 31.Park Y, Reyna-Neyra A, Philippe L, Thoreen CC. mTORC1 Balances Cellular Amino Acid Supply with Demand for Protein Synthesis through Post-transcriptional Control of ATF4. Cell Rep. 2017;19:1083–1090. doi: 10.1016/j.celrep.2017.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ben-Sahra I, Hoxhaj G, Ricoult SJH, Asara JM, Manning BD. mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science. 2016;351:728–733. doi: 10.1126/science.aad0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hershey JW, Sonenberg N, Mathews MB. Principles of translational control: an overview. Cold Spring Harb Perspect Biol. 2012:4. doi: 10.1101/cshperspect.a011528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Proud CG. Regulation and roles of elongation factor 2 kinase. Biochem Soc Trans. 2015;43:328–332. doi: 10.1042/BST20140323. [DOI] [PubMed] [Google Scholar]
- 35.Rose AJ, Alsted TJ, Jensen TE, Kobbero JB, Maarbjerg SJ, Jensen J, Richter EA. A Ca(2+)-calmodulin-eEF2K-eEF2 signalling cascade, but not AMPK, contributes to the suppression of skeletal muscle protein synthesis during contractions. J Physiol. 2009;587:1547–1563. doi: 10.1113/jphysiol.2008.167528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, Li W, Williams M, Terada N, Alessi DR, Proud CG. Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO J. 2001;20:4370–4379. doi: 10.1093/emboj/20.16.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Browne GJ, Finn SG, Proud CG. Stimulation of the AMP-activated protein kinase leads to activation of eukaryotic elongation factor 2 kinase and to its phosphorylation at a novel site, serine 398. J Biol Chem. 2004;279:12220–12231. doi: 10.1074/jbc.M309773200. [DOI] [PubMed] [Google Scholar]
- 38.Moore CE, Mikolajek H, Regufe da Mota S, Wang X, Kenney JW, Werner JM, Proud CG. Elongation Factor 2 Kinase Is Regulated by Proline Hydroxylation and Protects Cells during Hypoxia. Mol Cell Biol. 2015;35:1788–1804. doi: 10.1128/MCB.01457-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •39.Faller WJ, Jackson TJ, Knight JR, Ridgway RA, Jamieson T, Karim SA, Jones C, Radulescu S, Huels DJ, Myant KB, et al. mTORC1-mediated translational elongation limits intestinal tumour initiation and growth. Nature. 2015;517:497–500. doi: 10.1038/nature13896. This article demonstrates that a reduction in eEF2 kinase activity following mTORC1 inhibition using rapamycin causes a decrease in elongation rate which is responsible for cell cycle arrest in APC deficient cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moore CE, Wang X, Xie J, Pickford J, Barron J, Regufe da Mota S, Versele M, Proud CG. Elongation factor 2 kinase promotes cell survival by inhibiting protein synthesis without inducing autophagy. Cell Signal. 2016;28:284–293. doi: 10.1016/j.cellsig.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •41.Leprivier G, Remke M, Rotblat B, Dubuc A, Mateo AR, Kool M, Agnihotri S, El-Naggar A, Yu B, Somasekharan SP, et al. The eEF2 kinase confers resistance to nutrient deprivation by blocking translation elongation. Cell. 2013;153:1064–1079. doi: 10.1016/j.cell.2013.04.055. This article shows that tumor cells require eEF2K activity in order to decrease translation elongation rate in response to nutrient deprivation as a survival mechanism. Cells lacking eEF2K are impaired in their ability to adapt to low nutrients condition. Suppression of eEF2K in tumor cells lead to resensitization to caloric restriction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Faubert B, Vincent EE, Poffenberger MC, Jones RG. The AMP-activated protein kinase (AMPK) and cancer: many faces of a metabolic regulator. Cancer Lett. 2015;356:165–170. doi: 10.1016/j.canlet.2014.01.018. [DOI] [PubMed] [Google Scholar]
- 43.Ryazanov AG. Elongation factor-2 kinase and its newly discovered relatives. FEBS Lett. 2002;514:26–29. doi: 10.1016/s0014-5793(02)02299-8. [DOI] [PubMed] [Google Scholar]
- 44.Onodera J, Ohsumi Y. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J Biol Chem. 2005;280:31582–31586. doi: 10.1074/jbc.M506736200. [DOI] [PubMed] [Google Scholar]
- 45.Di Bartolomeo S, Corazzari M, Nazio F, Oliverio S, Lisi G, Antonioli M, Pagliarini V, Matteoni S, Fuoco C, Giunta L, et al. The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J Cell Biol. 2010;191:155–168. doi: 10.1083/jcb.201002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Papinski D, Schuschnig M, Reiter W, Wilhelm L, Barnes CA, Maiolica A, Hansmann I, Pfaffenwimmer T, Kijanska M, Stoffel I, et al. Early steps in autophagy depend on direct phosphorylation of Atg9 by the Atg1 kinase. Mol Cell. 2014;53:471–483. doi: 10.1016/j.molcel.2013.12.011. The authors conducted a screen to identify new substrates of the Atg1 kinase and identified Atg9 as a target. Phosphorylated Atg9 recruits Atg8 and Atg18 to induce the early steps of autophagosome formation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •47.Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15:741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •48.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim C, Kim W, Lee H, Ji E, Choe YJ, Martindale JL, Akamatsu W, Okano H, Kim HS, Nam SW, et al. The RNA-binding protein HuD regulates autophagosome formation in pancreatic β cells by promoting autophagy-related gene 5 expression. J Biol Chem. 2014;289:112–121. doi: 10.1074/jbc.M113.474700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •51.Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M, Gretzmeier C, Dengjel J, Piacentini M, Fimia GM, et al. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol. 2013;15:406–416. doi: 10.1038/ncb2708. [DOI] [PubMed] [Google Scholar]
- 52.Duran A, Amanchy R, Linares JF, Joshi J, Abu-Baker S, Porollo A, Hansen M, Moscat J, Diaz-Meco MT. p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol Cell. 2011;44:134–146. doi: 10.1016/j.molcel.2011.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 54.Badura M, Braunstein S, Zavadil J, Schneider RJ. DNA damage and eIF4G1 in breast cancer cells reprogram translation for survival and DNA repair mRNAs. Proc Natl Acad Sci U S A. 2012;109:18767–18772. doi: 10.1073/pnas.1203853109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •55.B’chir W, Maurin AC, Carraro V, Averous J, Jousse C, Muranishi Y, Parry L, Stepien G, Fafournoux P, Bruhat A. The eIF2α/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 2013;41:7683–7699. doi: 10.1093/nar/gkt563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••56.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rojas-Ríos P, Chartier A, Pierson S, Séverac D, Dantec C, Busseau I, Simonelig M. Translational Control of Autophagy by Orb in the Drosophila Germline. Dev Cell. 2015;35:622–631. doi: 10.1016/j.devcel.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 58.Hu G, McQuiston T, Bernard A, Park YD, Qiu J, Vural A, Zhang N, Waterman SR, Blewett NH, Myers TG, et al. A conserved mechanism of TOR-dependent RCK-mediated mRNA degradation regulates autophagy. Nat Cell Biol. 2015;17:930–942. doi: 10.1038/ncb3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chu C, Shatkin AJ. Apoptosis and autophagy induction in mammalian cells by small interfering RNA knockdown of mRNA capping enzymes. Mol Cell Biol. 2008;28:5829–5836. doi: 10.1128/MCB.00021-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •60.Brina D, Miluzio A, Ricciardi S, Clarke K, Davidsen PK, Viero G, Tebaldi T, Offenhauser N, Rozman J, Rathkolb B, et al. eIF6 coordinates insulin sensitivity and lipid metabolism by coupling translation to transcription. Nat Commun. 2015;6:8261. doi: 10.1038/ncomms9261. In this article, Brina and colleagues show that eIF6, which binds to the 60S ribosome and interferes with subunit joining, stimulates translation of transcriptional factors which increase lipogenesis and glycolysis in an mTOR-independent manner. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shah M, Su D, Scheliga JS, Pluskal T, Boronat S, Motamedchaboki K, Campos AR, Qi F, Hidalgo E, Yanagida M, et al. A Transcript-Specific eIF3 Complex Mediates Global Translational Control of Energy Metabolism. Cell Rep. 2016;16:1891–1902. doi: 10.1016/j.celrep.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat Rev Genet. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- 63.LMG, Boulay K, Topisirovic I, Huot ME, Mallette FA. Oncogenic Activities of IDH1/2 Mutations: From Epigenetics to Cellular Signaling. Trends Cell Biol. 2017;27:738–752. doi: 10.1016/j.tcb.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 64.Lindqvist LM, Simon AK, Baehrecke EH. Current questions and possible controversies in autophagy. Cell Death Discov. 2015:1. doi: 10.1038/cddiscovery.2015.36. [DOI] [PMC free article] [PubMed] [Google Scholar]