Abstract
Chimeric RNAs have been believed to be solely produced by gene fusions resulting from chromosomal rearrangement, thus unique features of cancer. Detected chimeric RNAs have also been viewed as surrogates for the presence of gene fusions. However, more and more research has demonstrated that chimeric RNAs in general are not a hallmark of cancer, but rather widely present in non-cancerous cells and tissues. At the same time, they may be produced by other mechanisms other than chromosomal rearrangement. The field of non-canonical chimeric RNAs is still in its infancy, with many challenges ahead, including the lack of a unified terminology. However, we believe that these non-canonical chimeric RNAs will have significant impacts in cancer detection and treatment.
Introduction
The term “chimeric RNAs” is not foreign to the field of cancer research. Starting from the discovery of BCR-ABL fusion resulting from “Philadelphia Chromosome”[1], chimeric RNAs are known to be products of gene fusions and considered ideal biomarkers for cancer. With modern technologies, they are being uncovered at an unprecedented rate. Several large databases including Mitelman [2], ChimerDB [3], ChiTaRs [4], FusionDB [5], dbCRID [6], TICdb [7], ConjoinG [8], FusionCancer [9] and HYBRIDdb [10], have collected thousands or more chimeric RNAs (Table 1). However, as next generation sequencing becoming popular and additional chimeric RNAs being discovered, increasing evidence is mounting toward a realization that goes against two traditional dogmas: chimeric RNAs are strictly the result of gene fusions, and that chimeric RNAs are unique to cancer.
Table 1.
Different databases hosting chimeric RNAs/gene fusions.
| Database | Tumor | Non-tumor | Total entry | Web link |
|---|---|---|---|---|
| Mitleman | Yes | No | 10861 | https://ulib.iupui.edu/databases/mitelman-database-chromosome-aberrations-and-gene-fusions-cancer |
| ChimerDB3.0 | Yes | No | 33316 | http://203.255.191.229:8080/chimerdbv31/mindex.cdb |
| ChiTaRs-3.1 | Yes | No | 42000 | http://chitars.md.biu.ac.il/ |
| dbCRID | Yes | Yes | 2643 | http://dbcrid.biolead.org |
| TICdb | Yes | No | 28171 | http://www.unav.es/genetica/TICdb/ |
| ConjoinGene | Yes | Yes | 800 | https://metasystems.riken.jp/conjoing/index |
| FusionCancer | Yes | No | 11839 | http://donglab.ecnu.edu.cn/databases/FusionCancer/index.html |
| HYBRIDdb | Yes | Yes | 46492 | http://www.primate.or.kr/hybriddb/ |
| FusionDB | Web link no longer active |
We now know that chimeric RNAs can be made at the RNA level by intergenic splicing, and that they are not specific to cancer. Despite these new and exciting discoveries, the field of non-traditional chimeric RNA is still in its infancy. In addition, the discoveries of these new chimeric RNAs bring confusion to some current nomenclatures, and raise alarms for some traditional practices. In this communication, we try to clarify some of the terminology. We also reviewed the current knowledge about the mechanisms of chimeric RNA formation, and their categorizations, as well as discussed their implications in cancer diagnosis and treatment (Fig. 1).
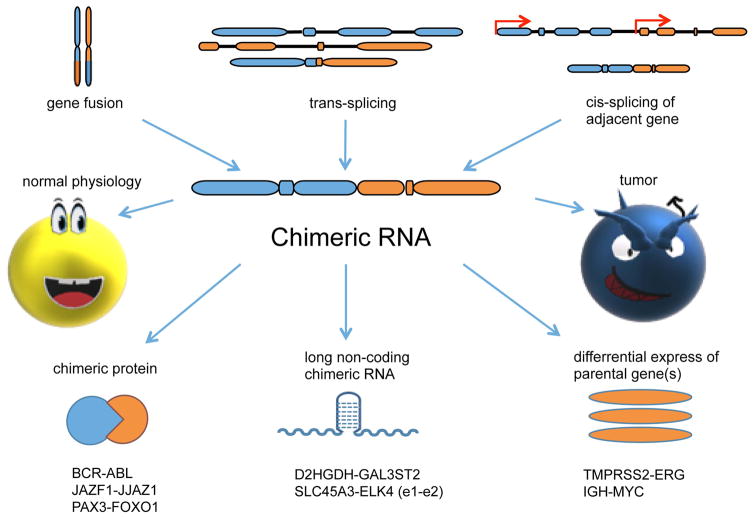
Figure 1. Chimeric RNA centric view.
Chimeric RNAs can be generated by gene fusion, trans-splicing, and cis-splicing between adjacent genes (cis-SAGe). They can function as chimeric proteins, long non-coding chimeric RNAs (lnccRNA), or affect parental gene expression. Traditionally, they have been thought to be specific to cancer. However, increasing numbers are being found in normal physiology.
Terminology
Firstly, there is a need to clarify some terminology. Although tens of thousands of chimeric RNAs are deposited into various databases, the way in which they are defined is unclear, partially due to the ill-defined “gene” and unclear mechanism of generation for the chimeric RNAs [11**]. Various names have been used, including: transcription mediated fusions [12], gene fusions [13], conjoined genes [8,14*], complex genes [15], co-transcribed genes [16], spanning genes [17], hybrid genes [10], and tandem chimerism [18]. We prefer the definition of “gene” as a nucleotide sequence in a DNA molecule that acts as a functional unit for the production of a protein, a structural RNA, or a catalytic or regulatory RNA molecule [19]. We then define “chimeric RNA” as a fusion transcript composed of exons, or fragments of exons from different genes.
We propose to differentiate “chimeric RNA” from the traditional term, “gene fusion”. Even though in the past, labs including our own had been using the two terms interchangeably, the term “gene fusion” tends to leave the impression of a fusion event happening at the DNA (gene) level. “Chimeric RNA” on the other hand can be a general term to describe fusion transcripts regardless of their mechanism of formation. In this regard, we consider the terms “chimeric RNA”, “chimeric transcript”, “fusion RNA”, and “fusion transcript” to be the same. Additionally, when it comes to referencing chimeric RNAs that are composed of exons from two adjacent genes transcribing in the same direction, many terms have also been used, including: transcription induced chimeras [12], tandem RNA chimeras [18,20], conjoined genes [8], and read-through fusions [21**,22]. However, these terms do not differentiate cis-splicing (precursor RNA transcribing through gene boundary and splicing together exons of neighboring genes) from trans-splicing events. In fact, trans-splicing may have a higher incidence rate with genes in close proximity to each other. To distinguish between the two, we prefer to use the term “cis-splicing of adjacent genes (cis-SAGe)” [23] or “gene read-through” for the first mechanism. Since “gene read-through” was originally used to describe protein translation processes that skip a stop codon, we propose to use the term cis-SAGe to avoid confusion. Recently Yuan et al., proposed to restrict the term “chimeric RNA” to those that are formed by separate RNA transcripts [11**]. By their definition, fusion RNAs from gene fusion or cis-SAGe should not be called “chimeric RNAs”. Even though this restriction holds some merit, it is not practical, and will cause more confusion, as it requires additional mechanistic insights, that are currently missing for the vast majority of fusion RNAs. Our definition is simple, as it relies only on the genome annotation, and requires no additional knowledge regarding how the fusion RNAs are formed.
Generating Mechanism
Given the above definition, chimeric RNAs are known to be produced hitherto by three mechanisms (Fig. 2) [24]. The classic and well-studied mechanism is through gene fusions. Chromosomal rearrangements that include insertions, deletions, and translocations can result in cytogenetically distinct gene loci juxtaposed together. In the past, due to technical limitations, most gene fusions were discovered in haematological and childhood malignant tumors, including BCR-ABL resulting from t(9;22) in chronic myelogenous leukemia (CML) [1,25] and in acute lymphoblastic leukemia (ALL) [26] or acute myelogenous leukemia (AML) [27] ;TEL-AML1 (ALL with t(12 ; 21) [28], AML1-ETO (M2 AML with t(8 ; 21)) [29]; PAX3-FOXO1 (t(2;13) in alveolar rhabdomyosarcoma [30]. With modern high throughput technologies, recurrent gene fusions are uncovered in common epithelial cancers, such as TMPRSS2-ERG in prostate cancers [31], and BCAM-AKT2 in serous ovarian cancer [32**].
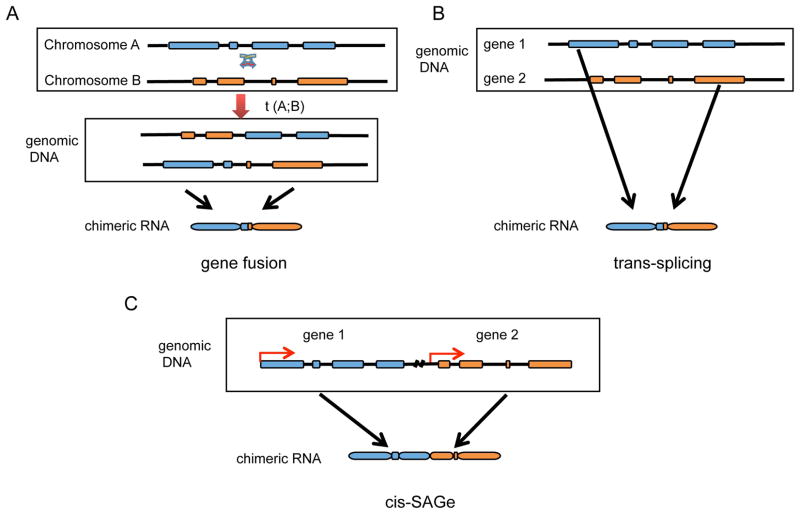
Figure 2. Three known generating mechanisms for chimeric RNAs.
Exons are depicted as blocks, and introns are indicated by lines. (A) Chromosomal rearrangement including translocation, deletion, and inversion. Shown here is a case of translocation. Gene fragments from different genomic loci are juxtaposed together. (B) RNA trans-splicing. Two separate pre-mRNA transcripts are spliced together. (C) cis-splicing between adjacent genes. The transcription machinery reads through two neighboring genes, and the exons from the two genes are spliced together.
The other two mechanisms sometimes are grouped together as “intergenic splicing”. One is trans-splicing, in which exons from two separate RNA transcripts are spliced together. Trans-splicing is well documented in lower eukaryotes [33,34], while it was first observed in vitro using mammalian cell extracts [35–38], and then in vivo in higher eukaryotes, including humans [39–46**,47*]. The molecular mechanism of trans-splicing in vertebrates is still elusive. It likely involves multiple factors, including transcriptional and splicing machinery, in conjunction with some sequence specificity, and three-dimensional proximity (detail discussion see [48**]).
Another intergenic splicing mechanism is cis-SAGe, which involves same strand neighbor genes. Traditionally, such chimeric RNAs containing exons of neighboring genes have been considered rare in mammalian cells, but recent studies incorporating systematic in silico analysis and paired-end RNA-Seq have identified many potential cis-SAGe chimeric RNAs [49,50]. In a recent study involving the analysis of both prostate cancer and non-cancerous samples, over 300 chimeric RNA events were observed, of which 30%, were characterized as cis-SAGe chimeras [21]. Another study classified 76% of their candidates as deriving from adjacent genes [51*]. Intuitively, cis-SAGe requires an active transcription from the 5′ parental gene, a readthrough transcript across gene boundaries, and alternative splicing between exons of the two genes. Factors involved in any of the three steps are likely to contribute to the regulation of cis-SAGe. Take SLC45A3-ELK4 e1e2 form for example. It is a recurrent cis-SAGe fusion in prostate cancer. Insulator binding factor CTCF negatively correlates with the expression of the fusion RNA, and silencing CTCF resulted induction of the fusion [52*]. In addition, CTCF silencing combined with RNA-Seq indeed revealed additional cis-SAGe fusions [21]. However, it is not the only factor. Even though CTCF binding to the insulators can regulate the fusion expression in different culture conditions for the same cell line [52*], among different cell lines the expression of SLC45A3-ELK4 correlates with the expression of the parental gene SLC45A3. In addition, forced RNA polII pausing by flavopiridol can induce the fusion RNA expression (unpublished). Recently, Vilborg et al. reported upregulation of ‘downstream of gene’-containing transcripts (DoG) under osmotic stress [53**]. It is thus likely that these same stresses may also enhance the level of cis-SAGe fusions.
Yuan et al. discussed three options for categorizing cis-SAGe fusions: a variant of upstream gene, dubbed as gene A; or a variant of downstream gene, dubbed gene B; or a totally new gene, gene C [11**]. Among the three, they favor the last option. We disagree with such classification, and propose to still dub them as A-B. Many of the parental genes involved in cis-SAGe are genuine genes that have well annotated TSS, and polyadenylation and termination sites. Their expression patterns are not identical, and they may have distinctive functions. In the case of SLC45A3-ELK4, SLC45A3 encodes a solute carrier, which is expressed on cell membrane. It is almost exclusively expressed in prostate tissue. ELK4 encodes an ETS family transcription factor, which is a nuclear protein. It is expressed in many tissues and cell types. Grouping the fusion into either of the parental gene variants, or calling it a new gene is not accurate, and will cause unnecessary confusion.
Categories
Chimeric RNAs could be classified into different categories by various criteria (Fig. 3). They can be grouped according to their generating mechanism as outlined above: gene fusion products, trans-splicing and cis-SAGe [48]. However, chimeric RNAs can also be sorted by other criteria. For instance, chimeras could be classified according to the chromosomal locations of their parental genes: parental genes located on different chromosomes (INTERCHR), neighboring genes transcribing the same strand (INTRACHR-SS-0GAP), and parental genes on the different strands of the same chromosome or with gaps in between (INTRACHR-OTHER) [21**]. Chimeras can be divided by the junction position relative to the exons of the parental genes, which include: both sides being known exon ends (E/E), one side being exon end, the other not (E/M or M/E); or both sides falling into the middle of exons (M/M) [46**]. Chimeras can also be grouped by their protein coding potentials: those in which the chimera’s coding sequence is in-frame with both parental genes (in-frame), those in which the reading frame of the downstream parental gene is different from the upstream parental genes (frame-shift), and those whose junction site falls outside of the open reading frame of either parental genes, or one or both parental genes are non-coding [46]. One note of caution is that the protein coding potential is only a prediction, which may not mirror how individual chimeric RNAs truly function. One example is the SLC45A3-ELK4 e1e2 fusion. Even though it encodes the ELK4 protein, it functions as a long non-coding chimeric RNA (lnccRNA) [54**]. All the above mentioned and even more categorizations may be used according to the topics researchers are investigating.
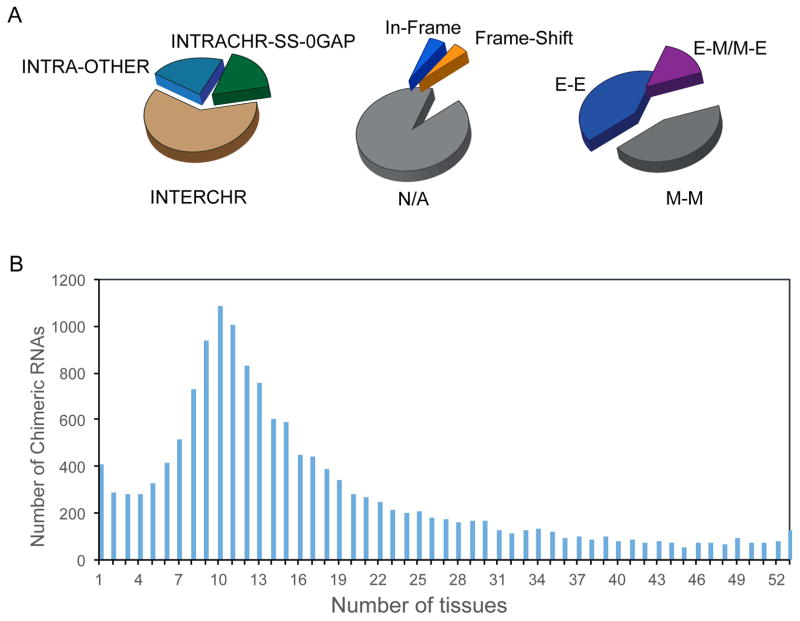
Figure 3. Example of the landscape of chimeric RNAs.
The data is from our analysis of over 7000 RNA-Seq samples covering 53 different tissues. (A) Chimeric RNAs can be grouped by different criteria. They can be grouped according to the location of parental genes, the fusion junction site relative to exons, and protein coding reading frames. (B) Chimeric RNAs may have different tissue distributions.
Chimeric RNAs in Cancer Diagnosis and Treatment
Gene fusions and their fusion products (RNA and protein) have had major impacts in cancer diagnosis and treatment. The oncogenic fusion BCR-ABL1 resulted from the t(9,22) translocation loci, named the ‘ Philadelphia chromosome’, was the target of an effective cancer drug known as, Gleevec (Imantinib) [25,55]. Recent advances in high-throughput sequencing and computational capabilities, which provide single nuclear-base resolution and long single strand coverage, have enabled the systematic discovery of novel chimeric RNAs in solid cancers [21**,49,56**,57–59]. EML4-ALK was detected in non-small cell lung cancer, which was treated with ALK inhibitors to improve prognosis [60,61]. In prostate cancer, fusions with promoters of certain inducible, or highly expressed genes were identified and characterized. The most well-know fusion is TMPRSS2-ERG, harbored by the majority of prostate cancers. In this fusion, the androgen-regulated TMPRSS2 promoter fuses to the coding region of ERG [62]. This fusion drives a unique transcriptional program, inducing DNA damage, invasion and metastasis [63*,64–67]. Wang et al., recently identified a series of peptides that interact specifically with the DNA binding domain of the ERG part of this fusion [63*]. This peptide inhibited ERG-mediated transcription, with reduced cell invasion, proliferation and tumor growth.
These textbook successes have led to great enthusiasm for using chimeric RNAs as biomarkers and drug targets, and at the same time three assumptions: 1, chimeric RNAs are solely produced by gene fusions at chromosomal level; 2, detecting chimeric RNA is equivalent of detecting gene fusions, and; 3, chimeric RNAs are unique to cancer cells.
However, increasing amounts of research has found that chimeric RNAs are also present in non-cancerous tissues and cells [44,45,68,69]. Some of them may also play important roles in normal physiology [46,70*]. In a recent survey of 117 RNA-Seq datasets covering 30 different non-cancerous tissues and cells, 291 recurrent chimeric RNAs were found [46]. Our ongoing research with over 7,000 RNA-Seq datasets of 44 tissues have also yield a large number of chimeric RNAs, of which many were identified in more than ten samples (unpublished). Even though the vast majority have not been experimentally validated, let alone functionally studied, their presence has already posed a challenge to the traditional view that fusion RNAs are cancer-specific features. In the field of cancer researchers, one common practice is still associate fusion RNAs discovered by RNA-Seq in certain cancer cell lines and tissues being equivalent to gene fusions, as well as to immediately assign them the title of cancer biomarkers. This practice has resulted in an explosion in the number of entries deposit into the Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer (https://cgap.nci.nih.gov/Chromosomes/Mitelman) [2]. As of August 4th of 2017, 10,841 “ gene fusions” are included. However, many were not validated to be product of gene fusion or being cancer specific. We would likely to raise the alarm against this practice. Chimeric RNAs are not a phenomenon unique to cancer, and they are not all the results of gene fusions. Therefore, extensive validations involving sufficient numbers of non-cancer control samples need to be conducted before assigning a chimeric RNA being a cancer marker [70*]. Similarly, more evidence is needed in order to claim that a chimeric RNA is a product of gene fusion.
On the other hand, chimeric RNAs generated by non-traditional gene fusions add another layer of transcriptional regulation, which can go awry in cancer. Therefore, they represent a new repertoire for cancer biomarkers and/or therapeutic targets. For instance, the CYCLIN D1-TROP2 chimeric RNA was found in multiple types of cancer, with no involvement of chromosomal rearrangement [71]. YPEL5-PPP1CB, was found in 28% of CLL samples with no evidence for a genomic fusion between YPEL5 and PPP1CB [72]. The SLC45A3-ELK4 e1e2 form is an example of cis-SAGe fusion in the absence of corresponding fusions at the DNA level. Its level correlates with prostate cancer development, and yet the parental genes do not share that correlation [23,58]. In addition, Wen et al. identified seven fusions in acute myeloid leukemia with normal karyotyping [73]. These examples, and others, support that the abnormality of intergenically spliced chimeric RNAs may play an underappreciated role in cancer biology. Whole exome or even whole genome sequencing will miss the discovery of this pool of potential biomarkers and/or drug targets that are produced abnormally at the RNA level.
Conclusions and Future Perspectives
We aim to clarify some discrepancies in nomenclature within the field of chimeric RNAs. Gene fusions and their corresponding chimeric RNAs have had major impacts on cancer diagnosis and treatment. We predicted that the non-traditional chimeric RNAs produced by trans-splicing, cis-SAGe, and possibly other unknown mechanisms will contribute to the discovery of new biomarkers and potential drugs. At the same time, we want to sound the alarm for some of the misconceptions and incorrect practices.
In the future, new sequencing technologies, especially full-length sequencing, and development of new software will facilitate the discovery of additional chimeric RNAs. One of the bottlenecks is the lack of a high throughput approach to study the functions of chimeric RNAs. Mechanisms of formation for the non-traditional chimeric RNAs, as well as their relationships with gene fusions are also burning questions that are begging for answers.
Acknowledgments
We thank Loryn Facemire for her help in English editing. This work is supported by NCI grant CA190713, a Research Scholar Grant [126405-RSG-14-065-01-RMC] from the American Cancer Society, and St. Baldrick’s V Scholarship.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Recommended Reading
- 1.Nowell PC. The minute chromosome (Phl) in chronic granulocytic leukemia. Blut. 1962;8:65–66. doi: 10.1007/BF01630378. [DOI] [PubMed] [Google Scholar]
- 2.Mitelman F. Recurrent chromosome aberrations in cancer. Mutat Res. 2000;462:247–253. doi: 10.1016/s1383-5742(00)00006-5. [DOI] [PubMed] [Google Scholar]
- 3.Lee M, Lee K, Yu N, Jang I, Choi I, Kim P, Jang YE, Kim B, Kim S, Lee B, et al. ChimerDB 3.0: an enhanced database for fusion genes from cancer transcriptome and literature data mining. Nucleic Acids Res. 2017;45:D784–D789. doi: 10.1093/nar/gkw1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gorohovski A, Tagore S, Palande V, Malka A, Raviv-Shay D, Frenkel-Morgenstern M. ChiTaRS-3.1-the enhanced chimeric transcripts and RNA-seq database matched with protein-protein interactions. Nucleic Acids Res. 2017;45:D790–D795. doi: 10.1093/nar/gkw1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suhre K, Claverie JM. FusionDB: a database for in-depth analysis of prokaryotic gene fusion events. Nucleic Acids Res. 2004;32:D273–276. doi: 10.1093/nar/gkh053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kong F, Zhu J, Wu J, Peng J, Wang Y, Wang Q, Fu S, Yuan LL, Li T. dbCRID: a database of chromosomal rearrangements in human diseases. Nucleic Acids Res. 2011;39:D895–900. doi: 10.1093/nar/gkq1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Novo FJ, de Mendibil IO, Vizmanos JL. TICdb: a collection of gene-mapped translocation breakpoints in cancer. BMC Genomics. 2007;8:33. doi: 10.1186/1471-2164-8-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prakash T, Sharma VK, Adati N, Ozawa R, Kumar N, Nishida Y, Fujikake T, Takeda T, Taylor TD. Expression of conjoined genes: another mechanism for gene regulation in eukaryotes. PLoS One. 2010;5:e13284. doi: 10.1371/journal.pone.0013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Wu N, Liu J, Wu Z, Dong D. FusionCancer: a database of cancer fusion genes derived from RNA-seq data. Diagn Pathol. 2015;10:131. doi: 10.1186/s13000-015-0310-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim DS, Huh JW, Kim HS. HYBRIDdb: a database of hybrid genes in the human genome. BMC Genomics. 2007;8:128. doi: 10.1186/1471-2164-8-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11**.Yuan C, Han Y, Zellmer L, Yang W, Guan Z, Yu W, Huang H, Liao DJ. It Is Imperative to Establish a Pellucid Definition of Chimeric RNA and to Clear Up a Lot of Confusion in the Relevant Research. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18040714. In this assay, the authros proposed to only name the fusion events at RNA level to be chimeric RNAs. They also discussed flaws, technical constraints and understudied tasks in the field of chimeric RNAs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akiva P, Toporik A, Edelheit S, Peretz Y, Diber A, Shemesh R, Novik A, Sorek R. Transcription-mediated gene fusion in the human genome. Genome Res. 2006;16:30–36. doi: 10.1101/gr.4137606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 14*.Funnell T, Tasaki S, Oloumi A, Araki S, Kong E, Yap D, Nakayama Y, Hughes CS, Cheng SG, Tozaki H, et al. CLK-dependent exon recognition and conjoined gene formation revealed with a novel small molecule inhibitor. Nat Commun. 2017;8:7. doi: 10.1038/s41467-016-0008-7. In this study, the authors reported that inhibiting CDC-like kinase increased conjoined genes (cis-SAGe candidates). Consistently, siRNA targeting CLK2 also increases conjoined gene formation. Collectively, their results reveal an unexpected role for CDC-like kinase in conjoined gene formation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roginski RS, Mohan Raj BK, Birditt B, Rowen L. The human GRINL1A gene defines a complex transcription unit, an unusual form of gene organization in eukaryotes. Genomics. 2004;84:265–276. doi: 10.1016/j.ygeno.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 16.Lauer B, Russwurm R, Schwarz W, Kalmanczhelyi A, Bruntner C, Rosemeier A, Bormann C. Molecular characterization of co-transcribed genes from Streptomyces tendae Tu901 involved in the biosynthesis of the peptidyl moiety and assembly of the peptidyl nucleoside antibiotic nikkomycin. Mol Gen Genet. 2001;264:662–673. doi: 10.1007/s004380000352. [DOI] [PubMed] [Google Scholar]
- 17.Petit MA, Yoshida K, Fujita Y, Ehrlich SD. The 409 bp tandem repeat spanning genes yxaK and yxaL is absent from the Bacillus subtilis chromosome. Microbiology. 2000;146( Pt 9):2091–2092. doi: 10.1099/00221287-146-9-2091. [DOI] [PubMed] [Google Scholar]
- 18.Parra G, Reymond A, Dabbouseh N, Dermitzakis ET, Castelo R, Thomson TM, Antonarakis SE, Guigo R. Tandem chimerism as a means to increase protein complexity in the human genome. Genome Res. 2006;16:37–44. doi: 10.1101/gr.4145906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bruce Alberts AJ, Lewis Julian, Morgan David, Raff Martin, Roberts Keith, Walter Peter. Molecular Biology of THE CEL. 6. 2014. [Google Scholar]
- 20.Greger L, Su J, Rung J, Ferreira PG, Geuvadis c, Lappalainen T, Dermitzakis ET, Brazma A. Tandem RNA chimeras contribute to transcriptome diversity in human population and are associated with intronic genetic variants. PLoS One. 2014;9:e104567. doi: 10.1371/journal.pone.0104567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21**.Qin F, Song Z, Babiceanu M, Song Y, Facemire L, Singh R, Adli M, Li H. Discovery of CTCF-sensitive Cis-spliced fusion RNAs between adjacent genes in human prostate cells. PLoS Genet. 2015;11:e1005001. doi: 10.1371/journal.pgen.1005001. This is the first report combining CTCF silencing and RNA-sequencing to identify genome wide cis-SAGe fusions. By studying the features associated with these fusions, a set of criteria were developed. With those criteria, 20% of randomly selected neighboring genes were shown to produce cis-SAGe fusion. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Varley KE, Gertz J, Roberts BS, Davis NS, Bowling KM, Kirby MK, Nesmith AS, Oliver PG, Grizzle WE, Forero A, et al. Recurrent read-through fusion transcripts in breast cancer. Breast Cancer Res Treat. 2014;146:287–297. doi: 10.1007/s10549-014-3019-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Gong M, Yuan H, Park HG, Frierson HF, Li H. Chimeric transcript generated by cis-splicing of adjacent genes regulates prostate cancer cell proliferation. Cancer Discov. 2012;2:598–607. doi: 10.1158/2159-8290.CD-12-0042. [DOI] [PubMed] [Google Scholar]
- 24.Jividen K, Li H. Chimeric RNAs generated by intergenic splicing in normal and cancer cells. Genes Chromosomes Cancer. 2014;53:963–971. doi: 10.1002/gcc.22207. [DOI] [PubMed] [Google Scholar]
- 25.Rowley JD. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 26.Lucas GS, Ardern JC. BCR-ABL rearrangements in acute lymphoblastic leukaemia. Lancet. 1991;337:1548. doi: 10.1016/0140-6736(91)93241-z. [DOI] [PubMed] [Google Scholar]
- 27.Cuenco GM, Ren R. Cooperation of BCR-ABL and AML1/MDS1/EVI1 in blocking myeloid differentiation and rapid induction of an acute myelogenous leukemia. Oncogene. 2001;20:8236–8248. doi: 10.1038/sj.onc.1205095. [DOI] [PubMed] [Google Scholar]
- 28.Romana SP, Mauchauffe M, Le Coniat M, Chumakov I, Le Paslier D, Berger R, Bernard OA. The t(12;21) of acute lymphoblastic leukemia results in a tel-AML1 gene fusion. Blood. 1995;85:3662–3670. [PubMed] [Google Scholar]
- 29.Nucifora G, Birn DJ, Erickson P, Gao J, LeBeau MM, Drabkin HA, Rowley JD. Detection of DNA rearrangements in the AML1 and ETO loci and of an AML1/ETO fusion mRNA in patients with t(8;21) acute myeloid leukemia. Blood. 1993;81:883–888. [PubMed] [Google Scholar]
- 30.Barr FG, Galili N, Holick J, Biegel JA, Rovera G, Emanuel BS. Rearrangement of the PAX3 paired box gene in the paediatric solid tumour alveolar rhabdomyosarcoma. Nat Genet. 1993;3:113–117. doi: 10.1038/ng0293-113. [DOI] [PubMed] [Google Scholar]
- 31.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao XH, Tchinda J, Kuefer R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 32**.Kannan K, Coarfa C, Chao PW, Luo L, Wang Y, Brinegar AE, Hawkins SM, Milosavljevic A, Matzuk MM, Yen L. Recurrent BCAM-AKT2 fusion gene leads to a constitutively activated AKT2 fusion kinase in high-grade serous ovarian carcinoma. Proc Natl Acad Sci U S A. 2015;112:E1272–1277. doi: 10.1073/pnas.1501735112. This is the first report demonstrating a recurrent cancer-specific gene fusion between BCAM and AKT2 in high-grade serous ovarian cancer (HGSC). The resulting AKT2 fusion kinase escapes the regulation from external stimuli. Such BCAM-AKT2 fusion gene generated via chromosomal translocation using the CRISPR/Cas9 system leads to focus formation in both OVCAR8 and HEK-293T cell lines. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sutton RE, Boothroyd JC. Evidence for trans splicing in trypanosomes. Cell. 1986;47:527–535. doi: 10.1016/0092-8674(86)90617-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang H, Hou Y, Miranda L, Campbell DA, Sturm NR, Gaasterland T, Lin S. Spliced leader RNA trans-splicing in dinoflagellates. Proc Natl Acad Sci U S A. 2007;104:4618–4623. doi: 10.1073/pnas.0700258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boothroyd JC, Cross GA. Transcripts coding for variant surface glycoproteins of Trypanosoma brucei have a short, identical exon at their 5′ end. Gene. 1982;20:281–289. doi: 10.1016/0378-1119(82)90046-4. [DOI] [PubMed] [Google Scholar]
- 36.Van der Ploeg LH, Liu AY, Michels PA, De Lange T, Borst P, Majumder HK, Weber H, Veeneman GH, Van Boom J. RNA splicing is required to make the messenger RNA for a variant surface antigen in trypanosomes. Nucleic Acids Res. 1982;10:3591–3604. doi: 10.1093/nar/10.12.3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Konarska MM, Padgett RA, Sharp PA. Trans splicing of mRNA precursors in vitro. Cell. 1985;42:165–171. doi: 10.1016/s0092-8674(85)80112-4. [DOI] [PubMed] [Google Scholar]
- 38.Solnick D. Trans splicing of mRNA precursors. Cell. 1985;42:157–164. doi: 10.1016/s0092-8674(85)80111-2. [DOI] [PubMed] [Google Scholar]
- 39.Sullivan PM, Petrusz P, Szpirer C, Joseph DR. Alternative processing of androgen-binding protein RNA transcripts in fetal rat liver. Identification of a transcript formed by trans splicing. J Biol Chem. 1991;266:143–154. [PubMed] [Google Scholar]
- 40.Bruzik JP, Maniatis T. Spliced leader RNAs from lower eukaryotes are trans-spliced in mammalian cells. Nature. 1992;360:692–695. doi: 10.1038/360692a0. [DOI] [PubMed] [Google Scholar]
- 41.Yu C, Chen J, Lin S, Liu J, Chang CC, Chang TY. Human acyl-CoA:cholesterol acyltransferase-1 is a homotetrameric enzyme in intact cells and in vitro. J Biol Chem. 1999;274:36139–36145. doi: 10.1074/jbc.274.51.36139. [DOI] [PubMed] [Google Scholar]
- 42.Zaphiropoulos PG. RNA molecules containing exons originating from different members of the cytochrome P450 2C gene subfamily (CYP2C) in human epidermis and liver. Nucleic Acids Res. 1999;27:2585–2590. doi: 10.1093/nar/27.13.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dorn R, Reuter G, Loewendorf A. Transgene analysis proves mRNA trans-splicing at the complex mod(mdg4) locus in Drosophila. Proc Natl Acad Sci U S A. 2001;98:9724–9729. doi: 10.1073/pnas.151268698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, Wang J, Mor G, Sklar J. A neoplastic gene fusion mimics trans-splicing of RNAs in normal human cells. Science. 2008;321:1357–1361. doi: 10.1126/science.1156725. [DOI] [PubMed] [Google Scholar]
- 45.Yuan H, Qin F, Movassagh M, Park H, Golden W, Xie Z, Zhang P, Sklar J, Li H. A chimeric RNA characteristic of rhabdomyosarcoma in normal myogenesis process. Cancer Discov. 2013;3:1394–1403. doi: 10.1158/2159-8290.CD-13-0186. [DOI] [PubMed] [Google Scholar]
- 46**.Babiceanu M, Qin F, Xie Z, Jia Y, Lopez K, Janus N, Facemire L, Kumar S, Pang Y, Qi Y, et al. Recurrent chimeric fusion RNAs in non-cancer tissues and cells. Nucleic Acids Res. 2016;44:2859–2872. doi: 10.1093/nar/gkw032. This is a genome wide study demonstrating a large number of chimeric RNAs in non-cancer cells and tissues. A subset was validated at RNA and protein level. Some recurrent ones were shown to play functional roles. The implications of these non-pathological fusions in cancer and in evolution were also explored. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47*.Xie Z, Babiceanu M, Kumar S, Jia Y, Qin F, Barr FG, Li H. Fusion transcriptome profiling provides insights into alveolar rhabdomyosarcoma. Proc Natl Acad Sci U S A. 2016;113:13126–13131. doi: 10.1073/pnas.1612734113. This is the first report to demonstrate that fusion RNA profiles can be used to investigate connections between biological samples. They used this approach to provide insights into the cell of origin for a mysterious tumor, alveolar rhabdomyosarcoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48**.Chwalenia K, Facemire L, Li H. Chimeric RNAs in cancer and normal physiology. Wiley Interdiscip Rev RNA. 2017 doi: 10.1002/wrna.1427. In this review, the authors went through the histories of chimeric RNAs, summarized current knowledge about the generating mechanisms of chimeric RNAs, and discussed some challenges. [DOI] [PubMed] [Google Scholar]
- 49.Kannan K, Wang L, Wang J, Ittmann MM, Li W, Yen L. Recurrent chimeric RNAs enriched in human prostate cancer identified by deep sequencing. Proc Natl Acad Sci U S A. 2011;108:9172–9177. doi: 10.1073/pnas.1100489108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nacu S, Yuan W, Kan Z, Bhatt D, Rivers CS, Stinson J, Peters BA, Modrusan Z, Jung K, Seshagiri S, et al. Deep RNA sequencing analysis of readthrough gene fusions in human prostate adenocarcinoma and reference samples. BMC Med Genomics. 2011;4:11. doi: 10.1186/1755-8794-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51*.Lai J, An J, Seim I, Walpole C, Hoffman A, Moya L, Srinivasan S, Perry-Keene JL, Wang C, et al. Australian Prostate Cancer B. Fusion transcript loci share many genomic features with non-fusion loci. BMC Genomics. 2015;16:1021. doi: 10.1186/s12864-015-2235-4. In this report, the authors identified 185 fusion transcripts from RNA-seq datasets. The majority (76 %) of these fusion transcripts were ‘read-through chimeras’ derived from adjacent genes in the genome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52*.Qin F, Song Y, Zhang Y, Facemire L, Frierson H, Li H. Role of CTCF in Regulating SLC45A3-ELK4 Chimeric RNA. PLoS One. 2016;11:e0150382. doi: 10.1371/journal.pone.0150382. This report demonstrated that CTCF is not the only factor influencing cis-SAGe fusion RNA, SLC45A3-ELK4. The authors concluded that CTCF and its bindings to the insulators are not the primary reasons for differential SLC45A3-ELK4 expression in different cell lines, or clinical cases. However, they are the likely mechanism for the same cells to respond to different environmental cues, in order to regulate the expression of SLC45A3-ELK4 chimeric RNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53**.Vilborg A, Passarelli MC, Yario TA, Tycowski KT, Steitz JA. Widespread Inducible Transcription Downstream of Human Genes. Mol Cell. 2015;59:449–461. doi: 10.1016/j.molcel.2015.06.016. This is the first report demonstrating a widespread phenomenon of transcripts downstream of polyadenylation and termination sites (DoGs). DoGs are inducible by osmotic stress through an IP3 receptor signaling-dependent pathway. The authors reported DoGs in several human cell lines and provided evidence for thousands of DoGs genome wide. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54**.Qin F, Zhang Y, Liu J, Li H. SLC45A3-ELK4 functions as a long non-coding chimeric RNA. Cancer Lett. 2017;404:53–61. doi: 10.1016/j.canlet.2017.07.007. This is the first report of a long non-coding chimeric RNA, SLC45A3-ELK4 e1e2 form. Even though the fusion RNA encodes the same protein as ELK4, the fusion RNA functions as a long non-coding RNA (lnccRNA) [DOI] [PubMed] [Google Scholar]
- 55.Druker BJ. Current treatment approaches for chronic myelogenous leukemia. Cancer J. 2001;7(Suppl 1):S14–18. [PubMed] [Google Scholar]
- 56**.Kumar-Sinha C, Kalyana-Sundaram S, Chinnaiyan AM. Landscape of gene fusions in epithelial cancers: seq and ye shall find. Genome Med. 2015;7:129. doi: 10.1186/s13073-015-0252-1. In this manuscript, the authors reviewed the landscape of gene fusions across solid cancers, focusing on the more recent discoveries made through sequencing. They reviewed common features of “driver” fusions (those that contribute to tumorigenesis), the major functional classes of fusions that have been described, and their clinical, diagnostic and/or therapeutic implications. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stransky N, Cerami E, Schalm S, Kim JL, Lengauer C. The landscape of kinase fusions in cancer. Nat Commun. 2014;5:4846. doi: 10.1038/ncomms5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rickman DS, Pflueger D, Moss B, VanDoren VE, Chen CX, de la Taille A, Kuefer R, Tewari AK, Setlur SR, Demichelis F, et al. SLC45A3-ELK4 is a novel and frequent erythroblast transformation-specific fusion transcript in prostate cancer. Cancer Res. 2009;69:2734–2738. doi: 10.1158/0008-5472.CAN-08-4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maher CA, Kumar-Sinha C, Cao X, Kalyana-Sundaram S, Han B, Jing X, Sam L, Barrette T, Palanisamy N, Chinnaiyan AM. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458:97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 61.Shaw AT, Yeap BY, Solomon BJ, Riely GJ, Gainor J, Engelman JA, Shapiro GI, Costa DB, Ou SH, Butaney M, et al. Effect of crizotinib on overall survival in patients with advanced non-small-cell lung cancer harbouring ALK gene rearrangement: a retrospective analysis. Lancet Oncol. 2011;12:1004–1012. doi: 10.1016/S1470-2045(11)70232-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 63*.Wang X, Qiao Y, Asangani IA, Ateeq B, Poliakov A, Cieslik M, Pitchiaya S, Chakravarthi B, Cao X, Jing X, et al. Development of Peptidomimetic Inhibitors of the ERG Gene Fusion Product in Prostate Cancer. Cancer Cell. 2017;31:844–847. doi: 10.1016/j.ccell.2017.05.001. The authors identified a series of peptides that interact specifically with the DNA binding domain of ERG (EIPs). The EIPs attenuated ERG-mediated transcription, chromatin recruitment, protein-protein interactions, cell invasion and proliferation, and tumor growth. [DOI] [PubMed] [Google Scholar]
- 64.Brenner JC, Ateeq B, Li Y, Yocum AK, Cao Q, Asangani IA, Patel S, Wang X, Liang H, Yu J, et al. Mechanistic rationale for inhibition of poly(ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer Cell. 2011;19:664–678. doi: 10.1016/j.ccr.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang J, Cai Y, Yu W, Ren C, Spencer DM, Ittmann M. Pleiotropic biological activities of alternatively spliced TMPRSS2/ERG fusion gene transcripts. Cancer Res. 2008;68:8516–8524. doi: 10.1158/0008-5472.CAN-08-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tomlins SA, Laxman B, Varambally S, Cao X, Yu J, Helgeson BE, Cao Q, Prensner JR, Rubin MA, Shah RB, et al. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia. 2008;10:177–188. doi: 10.1593/neo.07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sun C, Dobi A, Mohamed A, Li H, Thangapazham RL, Furusato B, Shaheduzzaman S, Tan SH, Vaidyanathan G, Whitman E, et al. TMPRSS2-ERG fusion, a common genomic alteration in prostate cancer activates C-MYC and abrogates prostate epithelial differentiation. Oncogene. 2008;27:5348–5353. doi: 10.1038/onc.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chase A, Ernst T, Fiebig A, Collins A, Grand F, Erben P, Reiter A, Schreiber S, Cross NC. TFG, a target of chromosome translocations in lymphoma and soft tissue tumors, fuses to GPR128 in healthy individuals. Haematologica. 2010;95:20–26. doi: 10.3324/haematol.2009.011536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu CS, Yu CY, Chuang CY, Hsiao M, Kao CF, Kuo HC, Chuang TJ. Integrative transcriptome sequencing identifies trans-splicing events with important roles in human embryonic stem cell pluripotency. Genome Res. 2014;24:25–36. doi: 10.1101/gr.159483.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70*.Tang Y, Qin F, Liu A, Li H. Recurrent fusion RNA DUS4L-BCAP29 in non-cancer human tissues and cells. Oncotarget. 2017;8:31415–31423. doi: 10.18632/oncotarget.16329. The authors challenged the previous claim that DUS4L-BCAP29 is a cancer specific fusion and promotes tumorigenesis. They demonstrated that DUS4L-BCAP29 fusion transcript exists in a variety of normal tissues. Overexpression of DUS4L-BCAP29 promotes cell growth and motility, even in non-cancer cells. Furthermore, it is shown to be a product of cis-SAGe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guerra E, Trerotola M, Dell’ Arciprete R, Bonasera V, Palombo B, El-Sewedy T, Ciccimarra T, Crescenzi C, Lorenzini F, Rossi C, et al. A bicistronic CYCLIN D1-TROP2 mRNA chimera demonstrates a novel oncogenic mechanism in human cancer. Cancer Res. 2008;68:8113–8121. doi: 10.1158/0008-5472.CAN-07-6135. [DOI] [PubMed] [Google Scholar]
- 72.Velusamy T, Palanisamy N, Kalyana-Sundaram S, Sahasrabuddhe AA, Maher CA, Robinson DR, Bahler DW, Cornell TT, Wilson TE, Lim MS, et al. Recurrent reciprocal RNA chimera involving YPEL5 and PPP1CB in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2013;110:3035–3040. doi: 10.1073/pnas.1214326110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wen H, Li Y, Malek SN, Kim YC, Xu J, Chen P, Xiao F, Huang X, Zhou X, Xuan Z, et al. New fusion transcripts identified in normal karyotype acute myeloid leukemia. PLoS One. 2012;7:e51203. doi: 10.1371/journal.pone.0051203. [DOI] [PMC free article] [PubMed] [Google Scholar]