Abstract
Background
Relatively high serum carotenoids are associated with reduced risks of chronic diseases, but inter-individual variability in serum carotenoid concentrations is modestly explained by diet. The bacterial community in the colon could contribute to the bio-accessibility of carotenoids by completing digestion of plant cells walls and by modulating intestinal permeability.
Objective
To evaluate whether colonic bacterial composition is associated with serum and colon carotenoid concentrations.
Design
The study was a randomized dietary intervention trial in healthy individuals who were at increased risk of colon cancer. Colon mucosal biopsies were obtained before and after six months of intervention without prior preparation of the bowels.
Participants/Setting
Participants were recruited from Ann Arbor, MI and nearby areas July 2007 to November 2010. Biopsy data was available from 88 participants at baseline and 82 participants after six months.
Intervention
Study participants were randomized to counseling for either a Mediterranean or a Healthy Eating diet for six months.
Results
At baseline, bacterial communities in biopsies from study participants in the highest versus the lowest tertile of total serum carotenoids differed by several parameters. Linear discriminant analysis effect size (LEfSe) identified 11 operational taxonomic units that were significantly associated with higher serum carotenoids. In linear regression analyses, three of these accounted for an additional 12% of the variance in serum total carotenoid concentrations after including body mass index, smoking, and dietary intakes in the model. These factors together explained 36% of the inter-individual variance in serum total carotenoid concentrations. The bacterial community in the colonic mucosa, however, was resistant to change after dietary intervention with either a Mediterranean or Healthy Eating diet, each of which doubled fruit and vegetable intakes.
Conclusions
The colonic mucosal bacterial community was associated with serum carotenoid concentrations at baseline but was not appreciably changed by dietary intervention.
Introduction
One of the beneficial aspects of a Mediterranean diet is its high content of fruits and vegetables. Relatively high dietary carotenoid intakes are linked with decreased risks of several cancer types 1. Although the data linking colon cancer risk with dietary intakes of carotenoids is equivocal, high serum carotenoid concentrations were significantly associated with lower rates of colon polyp recurrence and of colon cancer risk when repeated measures of β-carotene concentrations were analyzed 2, 3. A caveat is that in those studies dietary fat was concomitantly being reduced. In an intervention trial, the Polyp Prevention Trial, individuals who had the best compliance with the low-fat, high-dietary fiber, high-fruit and -vegetable eating pattern had lower polyp recurrence rates 4.
Inter-individual variation in serum carotenoids is generally large and stem from many factors 5. In the Healthy Eating Study from which colon biopsies were obtained for the present study, the coefficient of variation for total serum carotenoids was about 65% 6. Carotenoids are obtained from fruits and vegetables, but the correlation coefficients between serum carotenoids and dietary intakes are typically not greater than 0.5 7, 8. Behavioral and metabolic factors therefore might be important in governing serum concentrations of carotenoids. Many polymorphisms have been identified that either affect carotenoid uptake (eg. lipoproteins that transport carotenoids) or metabolism, including the β-carotene oxygenases 9. Demographic factors associated with relatively low serum carotenoid concentrations include male sex, body mass index (BMI), smoking, and possibly alcohol intake, although the latter does not display consistent associations with serum carotenoid concentrations across population groups and/or types of carotenoids 10–12. In a middle-aged French population, age, diet, alcohol intake, serum cholesterol, BMI and smoking status explained 15.2% of the variance of serum β-carotene in men and 13.9% in women 12. In another study of European middle-aged adults, gender, BMI, smoking, age, education, alcohol consumption, season, population center and supplement use explained 15–25% of the variance in individual serum carotenoids 11. Concurrent intakes of food components other than carotenoids is also important: dietary fiber can inhibit carotenoid absorption by interfering with micelle formation and micelle interactions with enterocytes, and a small amount of dietary fat is needed for complete absorption 13–15.
Carotenoid bioavailability from foods also varies. In a review of studies, the bioavailability of β-carotene from vegetables compared with purified β-carotene ranged between 3% and 24%, depending on the vegetable and method of preparation 13. Breakdown of plant cells walls appears important, and the bioavailability of carotenoids is improved by heat-treatment of foods 16, 17. It is also possible that undigested plant cells trap carotenoids and other phytochemicals which are then released upon microbial digestion in the colon. Several studies show that intestinal bacteria have a role in maximizing bio-accessibility of phytochemicals by degrading plant cell walls 13. In vitro, colonic fermentation increases release of carotenoids from the food matrix 18. Polyphenols in whole plant foods, as opposed to polyphenols in juices, are estimated to be more bioaccessible in the colon than in the small intestine 19, which also may be the case for carotenoids.
Thus far, the vast majority of human gut microbiome studies have been based on fecal bacteria, but we postulate that mucosal bacteria play a more direct role in mediating availability and absorption of dietary carotenoids. Certain bacteria, such as Akkermansia, are known to reside in the mucin of the mucosal surface but their physiological roles are not yet well-defined and this is the subject of ongoing research 20.
In the present study, colonic biopsies were available from individuals with an elevated risk of colon cancer who were enrolled in a dietary intervention trial 21. The trial randomized participants to receive dietary counseling for either a Healthy Eating or a Mediterranean diet for 6 months. Blood, colon biopsies and dietary data was obtained before and after intervention. The primary goal of the intervention was to reduce pro-inflammatory eicosanoids in the colon. The present study evaluated whether the relative abundance of the bacterial populations adhering to the mucosal surface are associated with inter-individual variation in carotenoid concentrations in serum. In addition, changes in the relative abundance of specific bacterial taxa were evaluated after intervention with either a Healthy Eating or Mediterranean diet for 6 months.
Methods
Participants and samples
Details of the Healthy Eating Study have been published previously 6, 21. The study was approved by the University of Michigan Institutional Review Board (HUM00007622) and registered at clinicaltrials.org (registration number NCT00475722). Briefly, a total 120 individuals at increased risk of colon cancer were enrolled from Ann Arbor, MI and surrounding areas from July 2007 to November 2010. Study participants provided signed, informed consent, and were randomized to a Healthy Eating or Mediterranean diet. A total of 93 participants completed six months of study. Increased risk was defined as a family history of colon cancer in a first-degree relative or two second-degree relatives, or a personal history of an adenoma or colon cancer. The primary goal of the study was to evaluate changes in colonic eicosanoid concentrations. Dietary intakes of carotenoids, from analysis of food records, roughly doubled in both study arms 6.
Fasting blood samples were obtained in ethylenediaminetetraacetic acid (EDTA) tubes, and plasma was stored at −80°C before analysis as described 6, 21. C-reactive protein, lipopolysaccharide (LPS) binding protein, cholesterol, high density lipoprotein (HDL), low density lipoprotein (LDL) and triglycerides were measured using commercial kits as previously published 22. The homeostasis model of assessment for insulin resistance (HOMA2-IR) was calculated from fasting C-peptide and glucose using an online calculator from the University of Oxford (The HOMA Calculator version 2.2 ) 23. Eight colon biopsies were obtained without prior preparation of the bowels by flexible sigmoidoscopy from each participant at each time point. The biopsies were all collected in the colon 20 to 25 cm from the anal sphincter. Biopsies were flash frozen in liquid nitrogen exactly 5 seconds after harvesting and were frozen at −70 ºC until analysis. After biopsies were used for the primary study endpoints, one colon biopsy was available for microbiome analysis from 94 participants at baseline and from 85 participants after dietary intervention (179 biopsies of the 212 biopsies originally collected for the study).
Dietary intervention
Subjects were randomized to receive counseling with a registered dietitian for either a Healthy Eating or a Mediterranean diet for six months. The Healthy Eating arm had goals for consuming at least five ½ cup servings/day of fruit and vegetables (including at least one serving that is a dark green or dark orange fruit or vegetables), at least three servings/day from whole grains, and less than 10% of calories from saturated fat. The Mediterranean arm goals were to maintain 30% of calories from fat while achieving a polyunsaturated: saturated: monounsaturated fatty acids (PUFA: SFA: MUFA) ratio of 1:2:5. Other goals were to consume foods high in omega 3 fatty acids at least twice a week, at least three servings/day from whole grains, at least 7–9 ½ cup servings of fruits and vegetables per day, depending on energy intake, and to include both culinary herbs and allium vegetables daily. The counseling was done mainly by telephone. Details of the interventions, including adherence, have been published previously and indicated that the main difference between arms after six month was in fat intakes while fruit, vegetable and fiber intakes all increased by similar amounts 6, 21.
Bacterial 16S rRNA Gene Sequencing
Bacteria adhering to the biopsies were identified by isolating DNA and sequencing the V4 region of the bacterial-specific 16S rRNA gene. DNA was isolated from biopsies with a PowerMag Microbiome RNA/DNA Isolation Kit (Mo Bio Laboratories, Inc.) using an epMotion 5075 liquid handling system. The hypervariable V4 region of the 16srRNA gene was amplified from each sample using primers described previously: forward sequence GTGCCAGCMGCCGCGGTAA and reverse sequence GGACTACHVGGGTWTCTAAT 24, 25. Quality control samples included a water blank, a mock bacterial community and a rinse of the biopsy instrument that had been opened in the biopsy procedure room. These control samples indicated no problems with assay performance. Polymerase chain reaction (PCR) products were visualized using an E-Gel 96 with SYBR Safe DNA Gel Stain, 2% (Life technologies cat# G7208-02).
Libraries were prepared using a dual-indexing strategy according to Illumina’s protocol for Preparing Libraries for Sequencing on the MiSeq (part# 15039740 Rev. D) for 2nM or 4nM libraries. Libraries were normalized using Life technologies SequalPrep Normalization Plate Kit (cat # A10510-01) following the manufactures protocol for sequential elution. The concentration of PCR products in the pooled samples was determined using Kapa Biosystems Library Quantification kit for Illumina platforms (KapaBiosystems KK4824). The sizes of the amplicons in the library were determined using an Agilent Bioanalyzer High Sensitivity DNA analysis kit (cat# 5067-4626). The final library consisted of equal molar amounts from each of the plates, and it was normalized to the pooled plate at the lowest concentration. If the library concentration was below 1nM, modified hybridization buffers were used for denaturation 26. The final load concentration was 4pM with a 10% PhiX spike to add diversity. Sequencing reagents were prepared according to a published protocol 25, 27; custom read 1, read 2 and index primers were added to the reagent cartridge.
Sequencing was done on the Illumina MiSeq platform (San Diego, CA), using a MiSeq Reagent Kit V2 500 cycles (Cat# MS-102-2003), according to the manufacturer’s instructions with published modifications that are available in a web-based standard operating procedure for generating libraries 25, 27. The Accuprime High Fidelity Taq (Life Cat # 12346094) was used instead of Accuprime Pfx supermix. The bacterial 16S sequence data can be found at the Sequence Read Archive maintained by the National Center for Biotechnology Information at the National Institutes of Health (SRP115434: Mediterranean Diet Study, release date August 15, 2018).
Bacterial sequence processing and analysis
The analysis of bacteria present in the biopsies was based on the procedures developed by the Schloss laboratory 25, 27. The 16S rRNA gene sequence data was processed and analyzed using the software package “mothur” version 1.35.0 and the MiSeq standard operating procedure using MiSeq Control Software version 2.5.0 released September 15, 2014 (http://www.mothur.org/wiki/MiSeq_SOP) 25, 27. FASTQ files were generated for paired end reads. Reads were aligned when making contigs. Pre-clustering was used and a uchime chimera filter was run. The SILVA version 119 reference alignment was used, and quality filtering was done with the screen.seqs command, filtered by length and by alignment to the V4 region of the SILVA data 28. Sequences were binned into operational taxonomic units (OTUs) based on 97% sequence similarity using the average neighbor cluster method. A distance matrix was built with the dist.seq command. An OTU is defined as a group of sequences clustered together by similarity. Sequence counts per OTU for each sample were calculated (average 21,136 sequences/sample, SD 15,582) and no OTU filtering was applied. Subsampling was done to 1047 sequences per sample which eliminated 9 samples. Subsampling to 3067 sequences per sample yielded similar results but eliminated 18 samples and therefore was not used. Use of at least 1000 sequences per sample is adequate for the taxonomic, relative abundance and diversity analyses that were done for this study, and as reviewed by Jovel et al. 29. Genus level taxonomic classifications were made using a modified version of the Ribosomal Database Project (RDP) training set 9 within mothur 30, 31. For graphical presentation of changes in bacterial taxa, only the 30 most abundant taxa were utilized.
Calculation of diversity indices and principal coordinates analysis
Since the number of OTUs identified is large, methods were used to describe the nature of the bacterial communities in the biopsies as a whole. The Shannon diversity index (H), that accounts for both richness and evenness of the species present, and the inverse Simpson index, another diversity metric, were calculated as previously described 32. The community distance index (θYC) was calculated using the method of Yue and Clayton that accounts for the proportions of both shared and non-shared species, producing a “Nonparametric Maximum Likelihood Estimator” index, with values between 0 and 1 indicating increasing community diversity 33. The mean inter-individual θYC was calculated for samples both at baseline and at 6 months after averaging the θYC for all pairs of values at each time point for each sample. The θYC values for each pair of samples before and after dietary intervention (intra-individual θYC) was also generated and analyzed separately. The principal coordinates analysis (PCoA) of θYC values were calculated at baseline in mothur using OTU relative abundance. Results were plotted using the pca3d package with the R program 34, 35.
Statistical analyses
The basic analysis approach was to evaluate associations of bacterial communities with carotenoids at baseline and to evaluate changes in bacterial communities as a result of dietary intervention. Linear discriminant analysis effect size (LEfSe) methods of Segata et al. were used to identify bacterial OTUs that differ across tertiles of total serum carotenoids at baseline 36. Subsequent analyses were performed with SPSS software, version 22 37. Kruskal-Wallis non-parametric tests were used to identify pairs of values that differ across carotenoid tertiles. Significant trends across tertiles were identified by the Jonckheere-Terpstra test for ordered alternatives. Continuous variables of participant characteristics with a normal distribution or a distribution that could be normalized using log transformation were compared across carotenoid tertiles using ANOVA after log transformation if appropriate. Categorical variables were compared across tertiles using Chi-square analyses (Tables 1–2). Spearman correlations of variables associated with metabolic health and bacterial variables were performed with correction of p-values for false discovery rates using the method of Benjamini-Hochberg 38. Paired t-tests were used to evaluate changes in bacterial diversity and taxa in each diet arm over time.
Table 1.
Demographic characteristics of 88 study subjects at study baseline by serum total carotenoid tertile. The tertiles represent low, medium and high serum carotenoids, and the mean and range for each tertile is shown in the column headings. Data shown is mean (standard deviation) or number (%). The mean and range of serum total carotenoids for each tertile is shown in the table column headings.
| Characteristic | Tertile 1 N=29 487 (110–720) μg/ml |
Tertile 2 N=29 859 (730–1038) μg/ml |
Tertile 3 N=30 1627 (1060–4760) μg/ml |
|---|---|---|---|
| Dietary carotenoids (mg/1000 kcal/day) | 4.8 (2.6) | 5.7 (3.3) | 6.4 (2.3)c |
| Colon Carotenoids (pg/μg protein) | 19 (26) | 16 (12) | 18 (15) |
| Age (Years) | 54 (10) | 51 (11) | 54 (11) |
| Femalea | 20 (69%) | 20 (69%) | 25 (83%) |
| Smokera | 4 (14%) | 1 (3%) | 2 (7%) |
| Aspirin Usera | 6 (21%) | 5 (17%) | 5 (17%) |
| Marrieda | 18 (62%) | 21 (72%) | 21 (70%) |
| HOMA2-IRb | 2.1 (1.1) | 1.5 (0.9) c | 1.2 (0.4) c |
| C-Reactive Protein (mg/L) | 3.8 (4.7) | 1.9 (1.8) | 1.5 (1.4) c |
| LPS binding proteinb | 20 (7) | 15 (8) | 15 (7) c |
| BMI (kg/m2)b | 28.8 (3.7) | 25.9 (4.5) c | 25.7 (2.9) c |
| Total Cholesterol (mg/dL) | 197 (47) | 207 (38) | 214 (33) |
| Triglycerides (mg/dL) | 152 (74) | 105 (44) c | 99 (57) c |
| HDL (mg/dL)b | 57 (15) | 67 (15) c | 73 (17) c |
For categorical variables, Chi-square tests were used and no significant differences between tertiles were found. These subjects were 88% white and there was no significant difference in race by tertile. Subjects consuming aspirin regularly for cardiovascular disease prevention (defined as 81 mg/day or 325 mg every other day) were classified as aspirin users.
Abbreviations used in the table are homeostatic model of assessment for insulin resistance (HOMA2-IR, from the HOMA Calculator version 2.2, University of Oxford) body mass index (BMI) and high density lipoprotein (HDL).
These values are significantly different than tertile 1 from ANOVA. Log transformation was used before conducting ANOVA for the HOMA2-IR, C-reactive protein, lipopolysaccharide (LPS) binding protein, dietary carotenoids, serum carotenoids, triglycerides and HDL. Serum low density lipoprotein, interleukins (IL1β, IL6, IL10, IL13), interferon gamma (IFNγ) and tumor necrosis factor alpha (TNFα) displayed no significant differences across serum carotenoid tertiles, not shown.
Table 2.
Characteristics of colonic mucosal microbiota at baseline among 88 study subjects by serum carotenoid tertile. Data shown are the median and inter-quartile range of the summed relative abundance for each phylum, family or genus using abundance data subsampled to 1047 sequences per sample. Major phyla, families and genera shown were generated from summing appropriate taxonomic-based operational taxonomic units (OTUs). The 11 OTUs identified to differ by serum carotenoid tertile from linear discriminant analysis effect size (LEfSE) analyses are also shown, given using the taxonomic classification for each OTU (phylum, family, genus).
| Characteristic | Tertile 1 | Tertile 2 | Tertile 3 |
|---|---|---|---|
| Indices | |||
| Shannon Diversity Index | 3.43 (0.53) | 3.68 (0.94) | 3.60 (1.03) |
| Inverse Simpson Index | 19.9 (10.5) | 19.4 (24.7) | 18.1 (19.1) |
| θYC Distances a,b | 0.69 (0.11) | 0.81 (0.16) d | 0.84 (0.15) d |
| Major Phylac | |||
| Actinobacteria | 18 (31) | 18 (24) | 14 (27) |
| Bacteroidetes | 247 (157) | 230 (206) | 280 (192) |
| Firmicutes a,b | 725 (117) | 634 (280) | 594 (276) d |
| Proteobacteria a,b | 12 (17) | 31 (78) d | 23 (33) d |
| Verrucomicrobia | 2 (14) | 3 (20) | 3 (33) |
| Major Familiesc | |||
| Firmicutes, Lachnospiraceae a,b | 450 (216) | 341 (243) d | 287 (177) d |
| Firmicutes, Ruminoccaceae | 156 (111) | 128 (129) | 119 (110) |
| Generac | |||
| Bacteroidetes, Bacteroidaceae, Bacteroides | 203 (151) | 96 (166) | 137 (179) |
| Bacteroidetes, Prevotellaceae, Prevotella a,b | 0 (4) | 1 (34) | 2 (37) d |
| Firmicutes, Lachnospiraceae, Blautia a,b | 121 (72) | 49 (91) d | 64 (76) d |
| Firmicutes, Lachnospiraceae, Roseburia a,b | 70 (66) | 31 (38) d | 13 (42) d |
| Verrucomicrobia, Verrucomicrobiaceae, Akkermansia | 2 (14) | 3 (20) | 3 (32) |
| OTUs identified from LEfSe analysis to differ by carotenoid tertile, with taxonomic classification of each OTU | |||
| OTU24: Actinobacteria, Coriobacteriaceae, Collinsella | 6 (24) | 6 (17) | 2 (8) |
| OTU1: Bacteroidetes, Bacteroidaceae, Bacteroides | 92 (90) | 57 (74) | 54 (100) |
| OTU102: Bacteroidetes, Porphyromonadaceae, Odoribacter b | 1 (4) | 0 (1) | 0 (1) |
| OTU3: Firmicutes, Lachnospiraceae, Blautia a,b | 55 (62) | 18 (39) d | 26 (34) d |
| OTU4: Firmicutes, Lachnospiraceae, Roseburia a,b | 65 (60) | 22 (37) d | 13 (41) d |
| OTU75: Firmicutes unclassified | 0 (0) | 2 (5) | 0 (3) |
| ‘OTU20: Firmicutes, Lachnospiraceae, Lachnospiracea incertae sedis a,b | 11 (10) | 9 (21) | 7 (10) d |
| OTU12: Firmicutes, Lachnospiraceae unclassified a,b | 25 (39) | 11 (23) | 6 (17) d |
| OTU45: Firmicutes, Lachnospiraceae unclassified a,b | 3 (10) | 1 (3) | 1 (3) d |
| OTU118: Firmicutes, Lachnospiraceae unclassified | 1 (0) | 1 (2) | 0 (2) |
| OTU63: Proteobacteria, Sutterellaceae, Parasutterella a | 2 (7) | 0 (2) d | 1 (7) e |
Significant difference in relative bacterial abundance across tertiles of serum carotenoids by the Kruskal-Wallis test, after adjustment for false discovery rates. The θYC distances were calculated by the method of Yue and Clayton as given in the Methods section.
Significant trend across tertiles by the Jonckheere-Terpstra test for ordered alternatives.
OTUs in the subsampled data were summed based on their taxonomic classification to generate phyla, families and/or genera shown. For genera, the number of OTUs summed was as follows: Prevotella 19 OTUs, Bacteroides 23 OTUs, Akkermansia 1 OTU, Blautia 11 OTUs, Roseburia 2 OTUs.
Significantly different than tertile 1 by the Kruskal-Wallis test with pairwise comparisons, p<0.05.
Significantly different than tertile 2 by the Kruskal-Wallis test with pairwise comparisons, p<0.05.
To evaluate the contribution of differences in bacterial abundance to inter-individual variation in serum carotenoids at baseline relative to demographic factors that are known to affect carotenoids, linear regression models were used. The bacterial OTUs identified in LEfSe to differ by carotenoid tertile at baseline were included in linear regression analyses shown in Figure 2. In these models, serum carotenoid concentrations were transformed as needed to satisfy normality assumptions: this was log for serum and colon lycopene and natural log for all other carotenoids. Variables entered stepwise were those published to affect serum carotenoid concentrations (BMI in kg/m2, current smoking yes/no, serum cholesterol in mg/ml, dietary intake in μg carotenoid/1000 kcal) and each of the 11 OTUs identified in the LEfSe analysis which had the following taxonomic classifications: Bacteroides, Roseburia, Blautia, three OTUs of Lachnospiraceae unclassified, Collinsella, Lachnospiraceae incertae sedis, Parasuttrella, Odoribacter and Firmicutes unclassified.
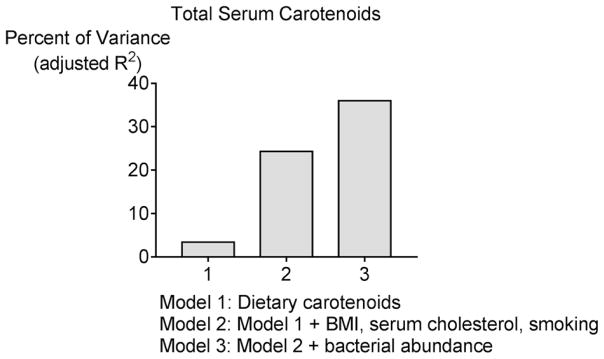
Figure 2.
Graphical representation of the regression models for baseline serum carotenoids. Models were constructed including dietary intakes of respective carotenoids per 1000 kcal, variables known to affect carotenoid concentrations (body mass index denoted as “BMI”, smoking, gender, serum total cholesterol) and the relative abundance of 11 bacterial operational taxonomic units (OTUs) that were identified to be associated with serum carotenoids from linear discriminant analysis effect size (LEfSe) analysis. OTUs are constructed based on sequence similarity and represent bacteria at the genus level. Only the variables that remained significant after stepwise selection are shown, and the final model explained 36% of the variance in total serum carotenoids. For bacterial relative abundance, the retained OTUs were OTU1 (Bacteroides), OTU118 (an unclassified species of Lachnospiraceae) and OTU4 (Roseburia). The adjusted R2 was significant across models (R2 = 0.034 for model 1, R2 = 0.243 for model 2 and R2 = 0.360 for model 3, p<0.05 for the F change in each case).
Results
Bacterial Sequencing and Participant Characteristics
The sequencing of colonic mucosal bacteria was successful, yielding at least 1047 reads in 170 of 179 biopsies after subsampling, representing a 95% success rate. This included 156 successfully sequenced biopsies from 76 participants with paired baseline and six month samples, 12 additional biopsies from participants at the baseline visit and six additional biopsies from the six-month visit for which a paired baseline biopsy was not available. The 94 study participants for whom a successfully sequenced biopsy was available at one or both time points were 75% female, 89% white, mean age 53 years (SD 11 years) and mean body mass index (BMI) of 27 kg/m2 (SD 4 kg/m2). Most were college graduates (79%) and a small proportion were current smokers (9%) or taking non-steroidal anti-inflammatory drugs (19%). None of these participant characteristics were significantly different (p<0.05) versus that of the 120 participants recruited for the study, and these demographic characteristics were published previously 39. Table 1 shows select characteristics of the 88 participants who provided baseline, subsampled, data by tertile of baseline serum total carotenoids.
Characteristics of Mucosal Bacteria by Serum Carotenoid Tertile
The baseline data was categorized by tertile of total serum carotenoid concentrations to facilitate analysis of potential associations of carotenoids with bacterial communities: low (110–720 μg/ml), medium (730–1038 μg/ml) and upper (60–4760 μg/ml) tertiles. Insulin resistance, C-reactive protein, lipopolysaccharide (LPS) binding protein, BMI and triglycerides were lower in the highest tertile while HDL was higher, versus the lowest tertile, as shown in Table 1. Several bacterial taxa differed across serum carotenoid tertiles at baseline, as shown in Table 2. Higher bacterial variation, indicated by a larger bacterial community θYC distance, was found in tertiles 2 and 3. Principal Covariate Analysis (PCoA) of colonic mucosal bacterial community θYC distances across tertiles of serum total carotenoids at baseline indicated clustering of θYC distances in the lowest carotenoid tertile with more variability of θYC distances in the two upper tertiles.
At the phylum level, there was a significantly lower abundance of Firmicutes and higher abundance of Proteobacteria with higher serum carotenoids (p<0.027 after adjusting for false discovery rates), with no significant differences in the other major phyla. The relative abundance of OTUs in the Lachnospiraceae family, and the OTUs classified in the genera Blautia and Roseburia were lower with higher serum carotenoids but the relative abundance of the OTUs in the Prevotella genus was higher (Table 2). LEfSe analysis of bacterial communities identified 11 individual OTUs that differed by tertile of total serum carotenoids (p<0.05). Using the Kruskal-Wallis test, six of these remained significant (p<0.027). In addition, significant trends across tertiles were identified for Blautia, Prevotella, Roseburia and six of the OTUs from LEfSe analysis using the Jonckheere-Terpstra test for ordered alternatives with p<0.05 (Table 2).
Associations of Demographic Factors with Relative Bacterial Abundance at Baseline
The relationships of participant characteristics associated with metabolic health and the identified bacterial taxa were explored using Spearman correlations at baseline (Supplemental Table 1). The strongest correlations with BMI were positive correlations with abundance of the Firmicutes phylum and Roseburia genera and three OTUs in the Lachnospiraceae family (R2>0.3 in each case). Correlations with triglycerides, CRP, HOMA2-IR and lipopolysaccharide (LPS) binding protein were similar in magnitude and direction as the correlations with BMI. The correlations of the relative abundance of these taxa was opposite in direction with HDL and serum carotenoids versus that with the aforementioned measures that are indicative of adverse metabolic health (Supplemental Table 1). LDL cholesterol, however, did not display any significant correlations with bacterial taxa (using p>0.015 to control for false discovery rates).
The correlations of serum and colon carotenoids with the relative abundance of bacterial taxa are shown in Supplemental Table 2 as a heat map. In serum, correlations of bacterial variables with α-carotene were weakest. In general, most of the correlations with individual carotenoids and bacterial taxa were in the same direction, and the pattern of correlations with xanthophylls tended to differ somewhat from that with the other carotenoids (Supplemental Table 2A). The correlations of the relative abundance of bacterial taxa with concentrations of carotenoids in colonic biopsies (expressed as in pg per mg protein) were much weaker versus that that with serum carotenoids, as shown in Supplemental Table 2B.
Bacterial Predictors of Serum Carotenoids
Regression analyses were performed to determine if the abundance of specific bacterial populations is associated with inter-individual variability in serum carotenoid concentrations after accounting for demographic factors previously identified to be associated with carotenoids. Models were constructed using stepwise selection of factors known to be associated serum carotenoids: BMI, smoking, serum total cholesterol, gender, age, dietary intakes of total carotenoids, dietary fat and dietary soluble fiber 10–13, 40. Bacterial OTUs for inclusion in the linear regression models were selected based on the LEfSe results of OTUs that differed significantly across carotenoid tertiles, p<0.05, as shown in Table 2. Adding season of blood collection or carotenoid supplement use (which was in 14% of participants) did not improve the models. The significant predictors of total serum carotenoids were dietary intakes of total carotenoids per 1000 kcal, BMI, serum cholesterol, smoking and the relative abundance of OTUs that were classified as Bacteroides, Roseburia and a genus of the Lachnospiraceae family (Figure 2). These variables together accounted for 36% of the variance in serum total carotenoid concentrations at baseline (p<0.001 overall model). For colon carotenoids, similar analyses of demographic factors and the same bacterial populations explained a much smaller proportion of the inter-individual variance (<15%, not shown), which is consistent with the relatively weak correlations of bacterial taxa with colon carotenoids shown in Supplemental Table 2.
Changes in Bacterial Populations with Dietary Intervention
Finally, we evaluated if dietary intervention to increase carotenoid intakes would alter bacterial populations in the colonic mucosa. We reported previously that carotenoid intakes increased in both arms by a similar amount, about 2-fold versus baseline 21. Total carotenoid intakes increased from a mean of 5.3 to 11.4 mg/1000 kcal/day in the Healthy Eating arm and from 6.0 to 11.2 mg/1000 kcal/day in the Mediterranean arm in the participants from whom biopsies were analyzed by 16s rRNA sequencing. A unique feature of the Healthy Eating arm was a decrease in total fat intake while in the Mediterranean arm, total fat intake remained constant while monounsaturated fat intake increased 21. The relative abundance of 30 of the most abundant genera before and after six months of dietary intervention are shown in Supplemental Figure 1. Paired t-tests for pre/post-intervention differences in the bacterial taxa listed in Table 2 revealed no significant differences after adjustment for false discovery rates (not shown). The inverse Simpson and Shannon indices also showed no significant changes after intervention. There was, however, a decrease in the community θYC distance (indicating increased similarity) in both diet arms, and this was significant for the Healthy Eating arm after adjustment for false discovery raters with p=0.000004 (Table 3). This is consistent with participants pursuing the same dietary goals as a result of the counseling provided. Similar results were obtained when analyzing the log of θYC inter-individual distance using mixed models (which allows for analysis of all the data) with BMI, serum choelsterol and smoking as covariates (not shown). The intra-individual θYC distance also was calculated for each participant from baseline to 6 months. The intra-individual θYC distance was smaller than the inter-individual θYC shown in Table 3 and did not differ by diet group assignment (mean intra-individual θYC of 0.55, SD 0.32 for the Healthy Eating group and mean of 0.59, SD 0.28, for the Mediterranean group).
Table 3.
Bacterial diversity indices in the colonic mucosal biopsies before and after dietary intervention. Data shown is mean and standard deviation (SD) for diversity measures in biopsies from subjects from whom paired baseline and 6 month samples were available.
| Bacterial Diversity Index | Healthy Eating Arm, n=39 | Mediterranean Arm, n=37 | ||
|---|---|---|---|---|
|
| ||||
| Baseline | 6 Months | Baseline | 6 Months | |
| Shannon Diversity, H | 3.35 (0.78) | 3.62 (0.40) | 3.46 (0.54) | 3.52 (0.36) |
| Inverse Simpson | 19.6 (11.1) | 23.3 (10.1) | 19.7 (10.1) | 20.5 (8.8) |
| Inter-individual Distances θYC | 0.79 (0.10) | 0.72 (0.10)a | 0.78 (0.10) | 0.74 (0.10) |
Paired t-tests were conducted on the indices shown and on all taxa and operational taxonomic units (OTUs) shown in Table 2. After correction for false discovery rates (FDR), the only pair that was statistically different pre/post intervention was for the θYC distances shown in the Healthy Eating arm (p<0.001).
Discussion
There is much interest in deciphering the role of the intestinal microbiota in various aspects of human health and disease, and this area of research is now rapidly growing. Interactions of the microbiota and intestinal epithelium are important for maintaining homeostasis of the intestinal epithelium and host immune or inflammatory status that in turn affects multiple health risks 41. In the present study, six months of dietary intervention did not appreciably alter the colonic mucosal bacteria (Table 3 and Results). This is consistent with other studies showing that changes in dietary patterns have a minimal effect on the nature of the gut microbiota and that long-term dietary patterns may be more important for determining the gut enterotype 42–45. Larger changes in the gut bacteria have been reported with prebiotics and bariatric surgery 46–48. One study reported that initial intestinal bacterial richness was associated with smaller changes in bacteria after fiber supplementation 49, but other studies found no significant bacterial changes after dietary supplementation with fiber containing foods 47, 50, 51. In rodent models, irreversible changes in the gut microbiota after a low fiber diet occurred only after several generations 52. This is consistent with a weight loss study in humans that found initial changes in the gut microbiota followed by return to the baseline composition after one year 53.
We therefore focused additional data analyses on the relationships between bacterial communities in the colonic mucosa with carotenoid concentrations in serum and colon at study baseline. There were stronger associations of bacterial abundance with carotenoid concentrations in the serum versus that in the colon (Supplemental Table 2). We previously reported that diet more directly affects serum versus colon carotenoids, indicating that host metabolic factors are relatively more important in governing tissue versus serum concentrations 6. We therefore explored the associations of relative bacterial abundance with serum carotenoids in more depth.
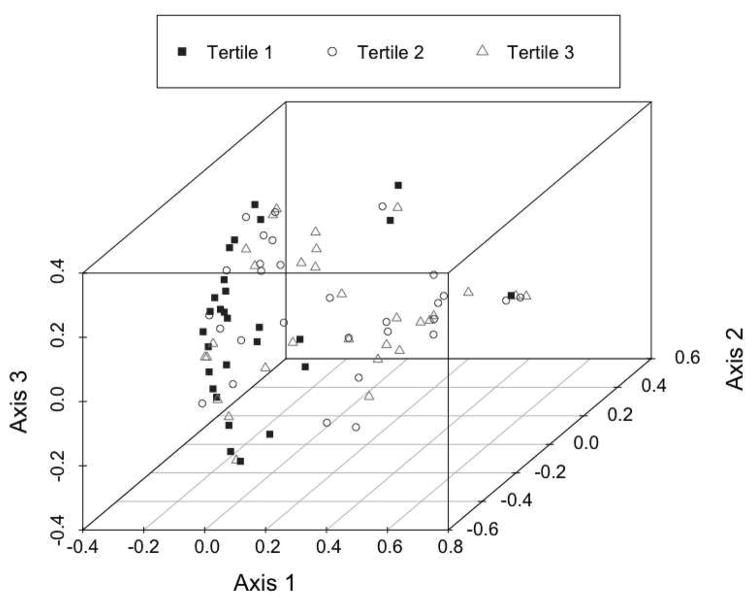
Bacterial communities in biopsies from individuals in the highest tertile of carotenoids did tend to cluster with each other in the PCoA analysis (Figure 1). In evaluating differences in bacterial abundance across tertiles of total serum carotenoids at baseline, several differences emerged as shown in Table 2. The most notable difference was a lower abundance in Firmicutes taxa, particularly in the Lachnospiraceae family, in the highest carotenoid tertile. A high Firmicutes to Bacteroidetes ratio has been associated with a Western diet and with a high fat diet 54, 55. Although studies do vary, positive associations of a high Firmicutes to Bacteroidetes abundance ratio also have been observed with obesity 45, 56–59, and increased intestinal Lachnospiraceae abundance has been associated with development of diabetes 60, 61. Consistent with these observations, our data also showed that individuals in the highest tertile of carotenoids had lower prevalence of obesity and insulin resistance (Table 1). Several studies have suggested that the gut microbiota with relatively high abundance of Prevotella and a low abundance of Bacteroides is characteristic of a diet high in plant foods 62. In our study Prevotella abundance was higher in the highest versus the lowest carotenoid tertile, but the decrease in Bacteroides was not significant (Table 2).
Figure 1.
Principal Covariate Analysis (PCoA) of colonic mucosal bacterial community θYC distances and tertile of serum total carotenoids at baseline. PCoA is a multidimensional scaling metric that is constructed using the θYC distance matrix of the samples. The analysis produces a set of uncorrelated (orthogonal) axes that summarize the variability in the data. Axis 1 contributed to 18.9% of the variance, axis 2 contributed 11.5% and axis contributed 7.8%. Objects ordinated closer to one another are more similar than those ordinated further away. Samples from study participants in the lowest tertile of serum carotenoids (filled black squares) are shown to cluster in a different pattern than samples from participants in the 2nd and 3rd tertiles (open circles and triangles, respectively).
In linear regression analyses shown in Figure 2, three colonic mucosal OTUs were retained as statistically significant predictors of serum carotenoids; one OTU was classified as a genus of Bacteroides, another was a genus of Roseburia (family Lachnospiraceae), and the third was an unclassified OTU in the family Lachnospiraceae. The relative abundance of each was lower in the highest tertile of serum carotenoids (Table 2), making it difficult to discern a functional role for these observations. There is in vitro evidence that certain natural products from plant-based foods have anti-bactericidal activity 63, 64. Decreased abundance of select bacterial populations would allow other types of bacteria to expand 65, and if these changes are distributed among many species they would be difficult to detect.
Despite this significant association of colonic mucosal bacterial abundance with serum carotenoids at baseline, dietary intervention to increase serum carotenoids with either a Mediterranean or Healthy Eating approach did not result in large changes in the relative abundance of any bacterial taxa (Supplemental Figure 1). The only notable change was a decrease in the θYC distance after intervention, indicating greater community similarity, likely since all participants were given dietary advice with the same goals (Table 3). The decrease in the θYC distance after dietary intervention to increase carotenoids therefore contrasted with the result at baseline showing a relatively larger θYC distance in the highest carotenoid tertile. In a small cross-sectional study, lower abundances of Firmicutes and Lachnospiraceae, and higher abundances of Bacteroidetes, Prevotellacea and Prevotella, were associated with a higher Mediterranean diet score 66. Our dietary intervention results suggest that such associations of bacterial abundance with diet are more likely due to other characteristics of the participants who consume specific types of diets versus that of the diet per se. Alternatively, these associations may depend on how the Mediterranean diet is defined.
Our study differs from published studies in which microbiota typically were quantified in excreted stool of rodents or humans, or in the ceca of rodents. The bacterial species present in the mucin layer lining the luminal colonic epithelium differ from those in the lumen and stool 67, 68. The mucin layer provides protection to the colonic epithelium, but it is also a niche for bacterial growth 69, 70. Bacteria localized in the mucin vs. that in the lumen layer might have greater impact on the biology of the colonic epithelium 69, 71, 72. Alternatively, we speculate that the bacteria adhering to the mucin layer might alter the absorptive abilities of the epithelial layer by degrading the mucin to increase permeability of the intestinal barrier. Several species of normal human gut bacteria have the ability to degrade mucin glycoproteins 73. A study of 24 healthy humans showed that the luminal and mucosal bacterial populations differ markedly, and large differences have been found in animal studies as well 74–76. This makes it difficult to compare our results with that of other studies that largely evaluated fecal bacteria.
There are several limitations to this study. Genetic polymorphisms in the metabolism of carotenoids do contribute somewhat to serum carotenoid concentrations, and this was not measured 77–79. The dietary intervention in this study was only 6 months long 21. With longer periods of intervention, changes in bacterial populations may become more evident. The data in this study were analyzed up to the genus level. Therefore, this might not be sufficient to uncover all important bacterial contributors to carotenoid accessibility. Methods to describe similarities and differences among complex bacterial populations are still evolving. In addition, we only analyzed bacteria in the colonic mucosal biopsies. Bacteria localized in the mucin may be relatively resistant to dietary change versus the bacteria in the lumen. Our results are, however, consistent with data from other studies that stool enterotypes (based on the relative abundance of Prevotella and Bacteroides) are resistant to change with diet, although transient changes can be observed in the first 1–4 days 42, 43, 80.
Conclusions
Changes in the intestinal microbiota were not evident after six months of dietary intervention. The relative abundance of specific OTUs was, however, significantly associated with serum carotenoid concentrations at baseline, indicating that long-term dietary exposures might be relatively more important in governing the bacterial community in the colonic mucosa. Further research is therefore warranted to better understand the role of the intestinal microbiota in modulating carotenoid accessibility and absorption.
Supplementary Material
Acknowledgments
We thank all the individuals who volunteered for the Healthy Eating Study. We thank Dr. Vincent Young for helpful discussions in planning the study and Ms. Judy Opp in the Microbiome Core for supervising the laboratory analysis of the samples. We acknowledge support from the University of Michigan Medical School Host Microbiome Initiative and NIH grants RO1 CA120381, Cancer Center Support Grant P30 CA046592, and the Rose and Lawrence C. Page, Sr. Family Charitable Foundation (to Dr. D. Kim Turgeon). Rena Chan was supported by the Cancer Biology Training Program grant T32 CA009676. The research used core resources supported by a Clinical Translational Science Award, NIH grant UL1RR024986 (the Michigan Clinical Research Unit), by the Michigan Diabetes Research Center, NIH grant 5P60 DK20572 (Chemistry Laboratory), the Michigan Nutrition and Obesity Research Center, and NIH grant P30 DK089503.
Abbreviations
- ANOVA
analysis of variance
- BMI
body mass index
- HDL
high density lipoprotein
- HOMA2-IR
homeostasis model of assessment for insulin resistance
- LDL
low density lipoprotein
- LEfSe
linear discriminant analysis effect size
- OTU
operational taxonomic unit
- PCoA
principal covariate analysis
- SD
standard deviation
Footnotes
None of the authors have conflicts of interest with the work reported in the following submission:
“Colonic Mucosal Bacteria Contribute to Inter-Individual Variability in Serum Carotenoid Concentrations”
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.World Cancer Research Fund, American Institute for Cancer Research. Food, Nutrition, Physical Activity, and the Prevention of Cancer: a Global Perspective. Washington, DC: American Institute for Cancer Research; 2007. [Google Scholar]
- 2.Steck-Scott S, Forman MR, Sowell A, et al. Carotenoids, vitamin A and risk of adenomatous polyp recurrence in the polyp prevention trial. Int J Cancer. 2004;112:295–305. doi: 10.1002/ijc.20364. [DOI] [PubMed] [Google Scholar]
- 3.Kabat GC, Kim MY, Sarto GE, Shikany JM, Rohan TE. Repeated measurements of serum carotenoid, retinol and tocopherol levels in relation to colorectal cancer risk in the Women’s Health Initiative. Eur J Clin Nutr. 2012;66:549–554. doi: 10.1038/ejcn.2011.207. [DOI] [PubMed] [Google Scholar]
- 4.Sansbury LB, Wanke K, Albert PS, et al. The effect of strict adherence to a high-fiber, high-fruit and -vegetable, and low-fat eating pattern on adenoma recurrence. Am J Epidemiol. 2009;170:576–584. doi: 10.1093/aje/kwp169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forman MR, Borkowf CB, Cantwell MM, et al. Components of variation in serum carotenoid concentrations: the Polyp Prevention Trial. Eur J Clin Nutr. 2009;63:763–770. doi: 10.1038/ejcn.2008.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sen A, Ren J, Ruffin MT, et al. Relationships between serum and colon concentrations of carotenoids and fatty acids in randomized dietary intervention trial. Cancer Prev Res (Phila) 2013;6:558–565. doi: 10.1158/1940-6207.CAPR-13-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al-Delaimy WK, Ferrari P, Slimani N, et al. Plasma carotenoids as biomarkers of intake of fruits and vegetables: individual-level correlations in the European Prospective Investigation into Cancer and Nutrition (EPIC) Eur J Clin Nutr. 2005;59:1387–1396. doi: 10.1038/sj.ejcn.1602252. [DOI] [PubMed] [Google Scholar]
- 8.Tucker KL, Chen H, Vogel S, Wilson PW, Schaefer EJ, Lammi-Keefe CJ. Carotenoid intakes, assessed by dietary questionnaire, are associated with plasma carotenoid concentrations in an elderly population. J Nutr. 1999;129:438–445. doi: 10.1093/jn/129.2.438. [DOI] [PubMed] [Google Scholar]
- 9.Bohn T, Desmarchelier C, Dragsted LO, et al. Host-related factors explaining interindividual variability of carotenoid bioavailability and tissue concentrations in humans. Mol Nutr Food Res. 2017 doi: 10.1002/mnfr.201600685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chai W, Conroy SM, Maskarinec G, Franke AA, Pagano IS, Cooney RV. Associations between obesity and serum lipid-soluble micronutrients among premenopausal women. Nutr Res. 2010;30:227–232. doi: 10.1016/j.nutres.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woodside JV, Young IS, Gilchrist SE, et al. Factors associated with serum/plasma concentrations of vitamins A, C, E and carotenoids in older people throughout Europe: the EUREYE study. Eur J Nutr. 2013;52:1493–1501. doi: 10.1007/s00394-012-0456-8. [DOI] [PubMed] [Google Scholar]
- 12.Galan P, Viteri FE, Bertrais S, et al. Serum concentrations of beta-carotene, vitamins C and E, zinc and selenium are influenced by sex, age, diet, smoking status, alcohol consumption and corpulence in a general French adult population. Eur J Clin Nutr. 2005;59:1181–1190. doi: 10.1038/sj.ejcn.1602230. [DOI] [PubMed] [Google Scholar]
- 13.Palafox-Carlos H, Ayala-Zavala JF, Gonzalez-Aguilar GA. The role of dietary fiber in the bioaccessibility and bioavailability of fruit and vegetable antioxidants. J Food Sci. 2011;76:R6–R15. doi: 10.1111/j.1750-3841.2010.01957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Het Hof KH, West CE, Weststrate JA, Hautvast JG. Dietary factors that affect the bioavailability of carotenoids. J Nutr. 2000;130:503–506. doi: 10.1093/jn/130.3.503. [DOI] [PubMed] [Google Scholar]
- 15.Riedl J, Linseisen J, Hoffmann J, Wolfram G. Some dietary fibers reduce the absorption of carotenoids in women. J Nutr. 1999;129:2170–2176. doi: 10.1093/jn/129.12.2170. [DOI] [PubMed] [Google Scholar]
- 16.Stahl W, Sies H. Uptake of lycopene and its geometrical isomers is greater from heat-processed than from unprocessed tomato juice in humans. J Nutr. 1992;122:2161–2166. doi: 10.1093/jn/122.11.2161. [DOI] [PubMed] [Google Scholar]
- 17.Rock CL, Lovalvo JL, Emenhiser C, Ruffin MT, Flatt SW, Schwartz SJ. Bioavailability of beta-carotene is lower in raw than in processed carrots and spinach in women. J Nutr. 1998;128:913–916. doi: 10.1093/jn/128.5.913. [DOI] [PubMed] [Google Scholar]
- 18.Goni I, Serrano J, Saura-Calixto F. Bioaccessibility of beta-carotene, lutein, and lycopene from fruits and vegetables. J Agric Food Chem. 2006;54:5382–5387. doi: 10.1021/jf0609835. [DOI] [PubMed] [Google Scholar]
- 19.Saura-Calixto F, Serrano J, Goni I. Intake and bioaccessibility of total polyphenols in a whole diet. Food Chemistry. 2007;101:492–501. [Google Scholar]
- 20.Derrien M, Belzer C, de Vos WM. Akkermansia muciniphila and its role in regulating host functions. Microb Pathog. 2016 doi: 10.1016/j.micpath.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 21.Sidahmed E, Cornellier ML, Ren J, et al. Development of exchange lists for Mediterranean and Healthy Eating Diets: implementation in an intervention trial. J Hum Nutr Diet. 2014;27:413–425. doi: 10.1111/jhn.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Umoh FI, Kato I, Ren J, et al. Markers of systemic exposures to products of intestinal bacteria in a dietary intervention study. Eur J Nutr. 2016;55:793–798. doi: 10.1007/s00394-015-0900-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Menendez JA, Lupu R, Colomer R. Exogenous supplementation with omega-3 polyunsaturated fatty acid docosahexaenoic acid (DHA; 22:6n-3) synergistically enhances taxane cytotoxicity and downregulates Her-2/neu (c-erbB-2) oncogene expression in human breast cancer cells. Eur J Cancer Prev. 2005;14:263–270. doi: 10.1097/00008469-200506000-00011. [DOI] [PubMed] [Google Scholar]
- 24.Koenigsknecht MJ, Theriot CM, Bergin IL, Schumacher CA, Schloss PD, Young VB. Dynamics and establishment of Clostridium difficile infection in the murine gastrointestinal tract. Infect Immun. 2015;83:934–941. doi: 10.1128/IAI.02768-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quail MA, Kozarewa I, Smith F, et al. A large genome center’s improvements to the Illumina sequencing system. Nat Methods. 2008;5:1005–1010. doi: 10.1038/nmeth.1270. supplementary protocol 1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schloss PD, Westcott SL, Ryabin T, et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schloss PD. A High-Throughput DNA Sequence Aligner for Microbial Ecology Studies. PLoS ONE. 2009;4:e8230. doi: 10.1371/journal.pone.0008230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jovel J, Patterson J, Wang W, et al. Characterization of the Gut Microbiome Using 16S or Shotgun Metagenomics. Front Microbiol. 2016;7:459. doi: 10.3389/fmicb.2016.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cole JR, Wang Q, Fish JA, et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Research (Online) 2014;42:D633–642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Begon M, Harper JL, Townsend CR. Ecology: Individuals, Populations, and Communities. 3. Cambridge, MA: Blackwell Science Ltd; 1996. [Google Scholar]
- 33.Yue JC, Clayton MK. A Similarity Measure Based on Species Proportions. Communications in Statistics - Theory and Methods. 2005;34:2123–2131. [Google Scholar]
- 34.Weiner J. Three Dimensional PCA Plots. 2015. p. pca3d. [Google Scholar]
- 35.Team RC. R: A Language and Environment for Statistical Computing. Vol. 2016. Vienna, Austria: R Foundation for Statistical Computing; 2014. [Google Scholar]
- 36.Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.IBM SPSS statistics for Windows, Version 22.0. Armonk, NY: IBM Corp; 2013. [Google Scholar]
- 38.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society B. 1995;57:289–300. [Google Scholar]
- 39.Djuric Z, Ruffin MTt, Rapai ME, et al. A Mediterranean dietary intervention in persons at high risk of colon cancer: recruitment and retention to an intensive study requiring biopsies. Contemp Clin Trials. 2012;33:881–888. doi: 10.1016/j.cct.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rock CL, Swendseid ME. Plasma beta-carotene response in humans after meals supplemented with dietary pectin. Am J Clin Nutr. 1992;55:96–99. doi: 10.1093/ajcn/55.1.96. [DOI] [PubMed] [Google Scholar]
- 41.Okumura R, Takeda K. Roles of intestinal epithelial cells in the maintenance of gut homeostasis. Exp Mol Med. 2017;49:e338. doi: 10.1038/emm.2017.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roager HM, Licht TR, Poulsen SK, Larsen TM, Bahl MI. Microbial enterotypes, inferred by the prevotella-to-bacteroides ratio, remained stable during a 6-month randomized controlled diet intervention with the new nordic diet. Appl Environ Microbiol. 2014;80:1142–1149. doi: 10.1128/AEM.03549-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haro C, Montes-Borrego M, Rangel-Zuniga OA, et al. Two Healthy Diets Modulate Gut Microbial Community Improving Insulin Sensitivity in a Human Obese Population. J Clin Endocrinol Metab. 2016;101:233–242. doi: 10.1210/jc.2015-3351. [DOI] [PubMed] [Google Scholar]
- 45.Remely M, Tesar I, Hippe B, Gnauer S, Rust P, Haslberger AG. Gut microbiota composition correlates with changes in body fat content due to weight loss. Benef Microbes. 2015;6:431–439. doi: 10.3920/BM2014.0104. [DOI] [PubMed] [Google Scholar]
- 46.Ordiz MI, May TD, Mihindukulasuriya K, et al. The effect of dietary resistant starch type 2 on the microbiota and markers of gut inflammation in rural Malawi children. Microbiome. 2015;3:37. doi: 10.1186/s40168-015-0102-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brahe LK, Le Chatelier E, Prifti E, et al. Dietary modulation of the gut microbiota--a randomised controlled trial in obese postmenopausal women. Br J Nutr. 2015;114:406–417. doi: 10.1017/S0007114515001786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seganfredo FB, Blume CA, Moehlecke M, et al. Weight-loss interventions and gut microbiota changes in overweight and obese patients: a systematic review. Obes Rev. 2017 doi: 10.1111/obr.12541. [DOI] [PubMed] [Google Scholar]
- 49.Tap J, Furet JP, Bensaada M, et al. Gut microbiota richness promotes its stability upon increased dietary fibre intake in healthy adults. Environ Microbiol. 2015;17:4954–4964. doi: 10.1111/1462-2920.13006. [DOI] [PubMed] [Google Scholar]
- 50.Pedersen C, Gallagher E, Horton F, et al. Host-microbiome interactions in human type 2 diabetes following prebiotic fibre (galacto-oligosaccharide) intake. Br J Nutr. 2016;116:1869–1877. doi: 10.1017/S0007114516004086. [DOI] [PubMed] [Google Scholar]
- 51.Eid N, Osmanova H, Natchez C, et al. Impact of palm date consumption on microbiota growth and large intestinal health: a randomised, controlled, cross-over, human intervention study. Br J Nutr. 2015;114:1226–1236. doi: 10.1017/S0007114515002780. [DOI] [PubMed] [Google Scholar]
- 52.Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, Sonnenburg JL. Diet-induced extinctions in the gut microbiota compound over generations. Nature. 2016;529:212–215. doi: 10.1038/nature16504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Louis S, Tappu RM, Damms-Machado A, Huson DH, Bischoff SC. Characterization of the Gut Microbial Community of Obese Patients Following a Weight-Loss Intervention Using Whole Metagenome Shotgun Sequencing. PLoS One. 2016;11:e0149564. doi: 10.1371/journal.pone.0149564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Musso G, Gambino R, Cassader M. Obesity, diabetes, and gut microbiota: the hygiene hypothesis expanded? Diabetes Care. 2010;33:2277–2284. doi: 10.2337/dc10-0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zeng H, Ishaq SL, Zhao FQ, Wright AD. Colonic inflammation accompanies an increase of beta-catenin signaling and Lachnospiraceae/Streptococcaceae bacteria in the hind gut of high-fat diet-fed mice. J Nutr Biochem. 2016;35:30–36. doi: 10.1016/j.jnutbio.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 56.Mathur R, Barlow GM. Obesity and the microbiome. Expert Rev Gastroenterol Hepatol. 2015;9:1087–1099. doi: 10.1586/17474124.2015.1051029. [DOI] [PubMed] [Google Scholar]
- 57.Kasai C, Sugimoto K, Moritani I, et al. Comparison of the gut microbiota composition between obese and non-obese individuals in a Japanese population, as analyzed by terminal restriction fragment length polymorphism and next-generation sequencing. BMC Gastroenterol. 2015;15:100. doi: 10.1186/s12876-015-0330-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sweeney TE, Morton JM. The human gut microbiome: a review of the effect of obesity and surgically induced weight loss. JAMA Surg. 2013;148:563–569. doi: 10.1001/jamasurg.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Verdam FJ, Fuentes S, de Jonge C, et al. Human intestinal microbiota composition is associated with local and systemic inflammation in obesity. Obesity (Silver Spring) 2013;21:E607–615. doi: 10.1002/oby.20466. [DOI] [PubMed] [Google Scholar]
- 60.Qin J, Li Y, Cai Z, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 61.Kameyama K, Itoh K. Intestinal colonization by a Lachnospiraceae bacterium contributes to the development of diabetes in obese mice. Microbes Environ. 2014;29:427–430. doi: 10.1264/jsme2.ME14054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Greiner AK, Papineni RV, Umar S. Chemoprevention in gastrointestinal physiology and disease. Natural products and microbiome. Am J Physiol Gastrointest Liver Physiol. 2014;307:G1–15. doi: 10.1152/ajpgi.00044.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Okeke MI, Okoli AS, Eze EN, Ekwume GC, Okosa EU, Iroegbu CU. Antibacterial activity of Citrus limonum fruit juice extract. Pak J Pharm Sci. 2015;28:1567–1571. [PubMed] [Google Scholar]
- 64.Borges A, Abreu AC, Ferreira C, Saavedra MJ, Simoes LC, Simoes M. Antibacterial activity and mode of action of selected glucosinolate hydrolysis products against bacterial pathogens. J Food Sci Technol. 2015;52:4737–4748. doi: 10.1007/s13197-014-1533-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Petersen C, Round JL. Defining dysbiosis and its influence on host immunity and disease. Cell Microbiol. 2014;16:1024–1033. doi: 10.1111/cmi.12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gutierrez-Diaz I, Fernandez-Navarro T, Sanchez B, Margolles A, Gonzalez S. Mediterranean diet and faecal microbiota: a transversal study. Food Funct. 2016;7:2347–2356. doi: 10.1039/c6fo00105j. [DOI] [PubMed] [Google Scholar]
- 67.Gosiewski T, Strus M, Fyderek K, et al. Horizontal Distribution Of The Fecal Microbiota In Adolescents With Inflammatory Bowel Disease. J Pediatr Gastroenterol Nutr. Jul 27;2011 doi: 10.1097/MPG.0b013e31822d53e5. in press 2011. [DOI] [PubMed] [Google Scholar]
- 68.Lavelle A, Lennon G, O’Sullivan O, et al. Spatial variation of the colonic microbiota in patients with ulcerative colitis and control volunteers. Gut. 2015;64:1553–1561. doi: 10.1136/gutjnl-2014-307873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol. 2004;54:1469–1476. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- 70.De Lisle RC, Roach E, Jansson K. Effects of laxative and N-acetylcysteine on mucus accumulation, bacterial load, transit, and inflammation in the cystic fibrosis mouse small intestine. Am J Physiol Gastrointest Liver Physiol. 2007;293:G577–584. doi: 10.1152/ajpgi.00195.2007. [DOI] [PubMed] [Google Scholar]
- 71.Derrien M, Collado MC, Ben-Amor K, Salminen S, de Vos WM. The Mucin degrader Akkermansia muciniphila is an abundant resident of the human intestinal tract. Appl Environ Microbiol. 2008;74:1646–1648. doi: 10.1128/AEM.01226-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van Passel MW, Kant R, Zoetendal EG, et al. The genome of Akkermansia muciniphila, a dedicated intestinal mucin degrader, and its use in exploring intestinal metagenomes. PLoS One. 2011;6:e16876. doi: 10.1371/journal.pone.0016876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pudlo NA, Urs K, Kumar SS, German JB, Mills DA, Martens EC. Symbiotic Human Gut Bacteria with Variable Metabolic Priorities for Host Mucosal Glycans. MBio. 2015;6:e01282–01215. doi: 10.1128/mBio.01282-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ringel Y, Maharshak N, Ringel-Kulka T, Wolber EA, Sartor RB, Carroll IM. High throughput sequencing reveals distinct microbial populations within the mucosal and luminal niches in healthy individuals. Gut Microbes. 2015;6:173–181. doi: 10.1080/19490976.2015.1044711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mao B, Li D, Ai C, Zhao J, Zhang H, Chen W. Lactulose Differently Modulates the Composition of Luminal and Mucosal Microbiota in C57BL/6J Mice. J Agric Food Chem. 2016;64:6240–6247. doi: 10.1021/acs.jafc.6b02305. [DOI] [PubMed] [Google Scholar]
- 76.Yasuda K, Oh K, Ren B, et al. Biogeography of the intestinal mucosal and lumenal microbiome in the rhesus macaque. Cell Host Microbe. 2015;17:385–391. doi: 10.1016/j.chom.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moran NE, Erdman JW, Jr, Clinton SK. Complex interactions between dietary and genetic factors impact lycopene metabolism and distribution. Arch Biochem Biophys. 2013;539:171–180. doi: 10.1016/j.abb.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tan HL, Moran NE, Cichon MJ, et al. beta-Carotene-9′,10′-oxygenase status modulates the impact of dietary tomato and lycopene on hepatic nuclear receptor-, stress-, and metabolism-related gene expression in mice. J Nutr. 2014;144:431–439. doi: 10.3945/jn.113.186676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yonova-Doing E, Hysi PG, Venturini C, et al. Candidate gene study of macular response to supplemental lutein and zeaxanthin. Exp Eye Res. 2013;115:172–177. doi: 10.1016/j.exer.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.