Abstract
Protein design is a useful strategy to interrogate the protein structure-function relationship. We demonstrate using a highly modular 3-stranded Coiled Coil (TRI-peptide system) that a functional type 2 copper center exhibiting copper nitrite reductase (NiR) activity exhibits the highest homogeneous catalytic efficiency under aqueous conditions for the reduction of nitrite to NO and H2O. Modification of the amino acids in the second coordination sphere of the copper center increases the nitrite reductase activity up to 75-fold compared to previously reported systems. We find also that steric bulk can be used to enforce a three-coordinate CuI in a site, which tends toward two-coordination with decreased steric bulk. This study demonstrates the importance of the second coordination sphere environment both for controlling metal center ligation and enhancing the catalytic efficiency of metalloenzymes and their analogues.
Keywords: de novo design, copper nitrite reductase, TRI peptide, second coordination sphere, sterics
Graphical Abstract
A significant increase in nitrite reductase activity is achieved by modification of sterics in the second coordination sphere of a type 2 copper center. We demonstrate that sterics can be harnessed to control metal coordination and reactivity in a TRI peptide scaffold.
De novo protein design is a powerful tool to assess the concept that biological function is a direct consequence of structure[1–3] and to probe evolutionary conversion of one activity to another. One example is the insertion of functional carbonic anhydrase and nitrite reductase active sites into alpha-helical 3-stranded-coiled-coil (3SCC) scaffolds. These constructs provide protein folds that are completely unrelated to that of the native enzymes while retaining the core features of the catalytic metal binding site.[4–6] Furthermore, studies have revealed how proteins containing identical primary sequences and 3-dimensional structures can be easily converted from hydrolytic to redox enzymes by the simple expedient of exchanging ZnII by CuII. These advances have utilized the TRI family of peptides which are designed with a heptad repeat strategy to form 3SCCs.[7] Hydrophobic amino acids that make up the 3SCC interior can be substituted to create transition metal binding sites while maintaining the overall fold of the structure. This strategy has successfully produced both a Zn-binding carbonic anhydrase mimic with activity that approaches that of the native system, as well as a Cu-binding nitrite reductase (CuNiR) mimic.[8] Other successful non-heme redox active de novo constructs include the Due Ferri system[9, 10] and 4Fe-4S clusters.[11, 12]
Copper proteins play important roles in many biological processes.[13, 14] Type 2 copper (T2Cu) centers are mononuclear CuII(His)n(X)4-n metal binding sites that serve various functions in native proteins.[14] CuNiR, which carries out the dissimilatory reduction of NO2− to NO, contains a T2Cu catalytic center [CuI(His)3(OH2)]. We have previously described a structural and functional model for the CuNiR T2Cu center using a TRI peptide derivative capable of catalyzing nitrite reduction at a rate of 5 turnovers over the course of 3.7 hours.[6, 15] Despite exhibiting similar reactivity, this activity is seven orders of magnitude below that of comparable natural CuNiRs. This may be due to the difficulty of incorporating non-heme redox centers into de novo scaffolds, in which one must consider the preferred coordination of both Cu oxidation states in the catalytic cycle. In this first generation model we have shown CuI to exist in a three-coordinate state, the same coordination number as the native, ascorbate-reduced T2Cu center in CuNiR.[16] Based on EPR spectroscopy, the oxidized form of this designed protein likely binds to CuII with a higher coordination number (five coordinate) than is seen in the native form of oxidized CuNiR (four coordinate). The de novo construct also possessed a higher reduction potential than native CuNiR, suggesting significant deviation from native CuNiR redox behavior. To address how these factors influence reactivity, we carried out stepwise modifications of the helical interface to create a series of peptides with reduction potentials spanning ~200mV and NiR reaction rates that varied by a factor of four.[15] Exploiting remote interactions, we were able to successfully alter copper-binding affinities, reduction potentials, and protonation equilibria.
We now turn our attention to the interior of the coiled coils to examine the impact of changes in the second coordination sphere. Our initial aim was to evaluate how steric constraints of the hydrophobic side chains around the metal center (either above or below) influence NiR activity. Previous studies comparing the coordination number of CdII bound in TRI systems revealed that while a mixture of CdS3 and CdS3O species were formed in Cd(TRIL16C)3−, only CdS3O was obtained in Cd(TRIL12AL16C)3− or Cd(TRL16DC)3−.[17, 18] Furthermore, the introduction of greater steric encumbrance in the Cd(TRIL12DLL16C)3− construct resulted in exclusive CdS3 while providing greater proximal space to the metal binding site in Cd(GRCSL16CL19DL)3− led to the formation of CdS3O2.[19] These studies demonstrated that modification of steric bulk around the metal-binding site could control solvent/substrate access to the metal. The first generation CuNiR model peptide, TRIW-H, contains a Leu residue at the d-sites[20] above and below the Cu-binding histidines in the 23rd position, in a fashion analogous to TRIL16C. Therefore, we substituted the Leu residues at the 19th and 26th positions in three different ways. First, we incorporated residues (Ile and D-Leu) that would be more sterically crowding around the metal center. Second, we replaced Leu with a less sterically demanding Ala residue that might enhance substrate access or allow greater conformational flexibility of the active site. Crystal structures of similar constructs (Hg(GRAND-CSL12AL16C)3− and Zn(GRAND-CSL12AL16C)3−) show that the pocket created by substituting a Leu3 layer for Ala3 residues allows up to 4 molecules of water within the 3SCC interior near the metal binding site.[21, 22] Third, we incorporated a potentially hydrogen bonding ligand (Asp) that might assist in catalysis (Figure 1). These modifications were predicted to impact the primary coordination sphere of the copper center and in turn modify NiR reactivity.
Figure 1.
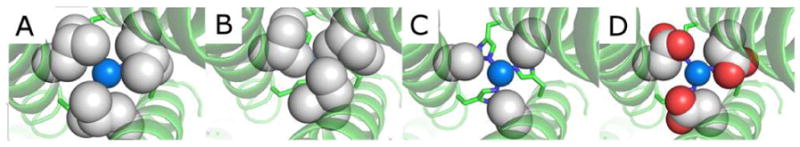
PyMol models of TRIW-H (A) L19I, (B) L19, (C) L19A, and (D) L19D showing the change of steric packing above the copper center made based on the crystal structure of ZnII-NHgII-S(CSL9PenL23H)3+ (PDB code: 3PBJ) or ZnII(H2O)(GRANDCSL12AL16C)3− (5KB2). Peptide strands are represented by helices and His23 side chains are represented by sticks. Residues on the 19th position and CuI are shown in spheres to illustrate the space-filling above the copper center.
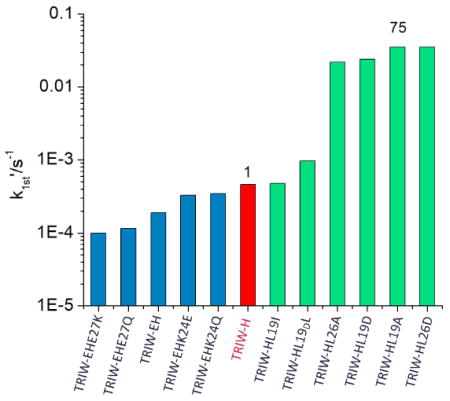
The NiR activities (pseudo-first order rate constants) of these peptides were measured following previously reported procedures.[6] L19I and L19DL variants showed no notable rate enhancement compared to the parent TRIW-H. Variants of TRIW-H with substitution of L19 or L26 with Ala had a marked increase in NiR rates compared to the parent from 4.6 × 10−4 s−1 for TRIW-H to 3.5 × 10−2 s−1 for L19A or 2.2 × 10−2 s−1 for L26A. Variants with L19 or L26 mutated to Asp had similarly enhanced rates from 4.6 × 10−4 s−1 for TRIW-H to 2.4 × 10−2 s−1 for L19D or 3.5 × 10−2 s−1 for L26D. Our study shows no evidence of additional catalytic enhancement from the use of protonated side chains compared to Ala. Yamaguchi and coworkers have previously reported a copper nitrite reductase model system [CuMe2bpa(H2O)(ClO4)]+ (Me2bpa: bis(6-methyl 2-pyrideymethyl)amine) which, when attached to an electrode surface, has a catalytic efficiency of 57 s−1M−1.[23] However, based on the difference in the turnover number at pH 5.5 provided in their report, the catalytic efficiency, under homogeneous aqueous conditions, is estimated to be reduced to 2.2 × 10−2 s−1M−1. Therefore, our TRIW-H L19A system has, thus far, the highest reported catalytic efficiency of an NiR model under homogeneous aqueous conditions at 1.0(0.3) s−1M−1 at pH 5.8 as determined by Michaelis-Menton kinetics (see Supporting Information).
Under catalytic conditions, the oxidized CuII(3SCC) is rapidly reduced by ascorbate to CuI(3SCC), followed by the slow re-oxidation of CuI into CuII coupled to nitrite reduction.[6] Therefore, changes in the catalytic rate induced by secondary coordination sphere modifications could be the result of alterations in the nitrite interactions with the CuI ‘resting’ state. For this reason, we examined the structure of the CuI center in the second-sphere-TRIW-H variants and compared them to the parent TRIW-H by X-ray Absorption Spectroscopy (XAS).
Previously, we have shown that CuI-bound TRIW-H was best described as a three coordinate CuI with 1.93 Å Cu-N bond lengths at both pH 7.4 and pH 5.8.[6]. The XANES region of CuI contains an edge feature corresponding to the 1s → 4p transition, whose relative intensity serves as a useful diagnostic of the coordination of CuI. As the coordination number of CuI increases, the degree of 4s-4p mixing increases as well, resulting in a greater degree of parity forbidden 1s → 4s character and a decrease in intensity for this edge feature.[24] Intensity ratios near 1:1 of this edge feature relative to the edge jump are indicative of 2-coordinate CuI, while a decrease to around 1:2 is typical of 3-coordinate, and less than 1:2 of 4-coordinate.[25] Figure 3 demonstrates that the L19A and L19D constructs have a significantly more intense edge feature compared to the parent construct TRIW-H, or L19I and L19DL, consistent with a 2-coordinate CuI. While this decrease in coordination is not seen for L26A and L26D, all four constructs with enhanced NiR activity show an increase in the energy of the edge feature to 8983.6±0.2 eV compared to 8982.8±0.2 eV observed for TRIW-H. Exploring the EXAFS region (SI), we find two different categories of constructs based upon the Cu-N distance as well as number of imidazoles used in the model, which follow the categories defined by the XANES analysis. TRIW-H, L19I, L26A, and L26D are all best fit by a model utilizing three imidazoles at 1.91 – 1.93 Å with Debye-Waller values between 9.3 and 10.2 × 10−3 Å2. Meanwhile, L19A and L19D make up the second category, fitting best with two imidazoles at 1.86 – 1.88 Å with Debye-Waller values of 8.0 or 7.8 × 10−3 Å2, respectively (Table 2). The fitted Debye-Waller factors are somewhat larger than usual, suggesting that there is some disorder in these sites.
Figure 3.
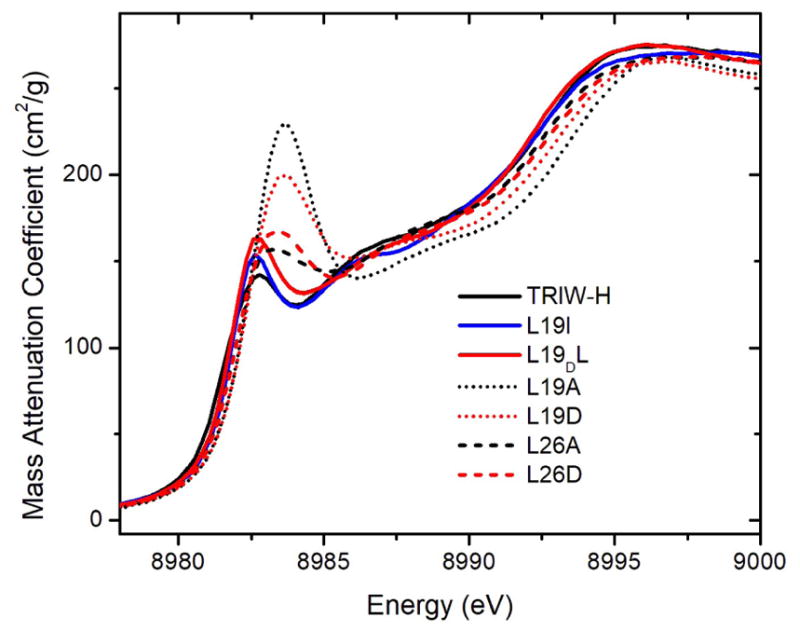
(A) XANES of CuI bound constructs reported in this work, measured at pH 5.8.
Table 2.
. EXAFS fitting parameters and first order NiR kinetics of interior mutation constructs reported in this work compared to TRIW-H[6]
| Construct | Model | Cu-N R (Å) | Cu-N σ2 × 10−3 (Å2) | Rate (s−1) |
|---|---|---|---|---|
| TRIW-H | 3 Cu-His | 1.93 | ~9 | 4.6×10−4 |
| L19DL | 3 Cu-His | 1.92 | 9.8 | 9.7×10−4 |
| L19A | 2 Cu-His | 1.86 | 8.0 | 3.5×10−2 |
| L19D | 2 Cu-His | 1.88 | 7.8 | 2.4×10−2 |
| L26A | 3 Cu-His | 1.91 | 10.1 | 2.2×10−2 |
| L26D | 3 Cu-His | 1.91 | 8.6 | 3.4×10−2 |
These structural observations indicate that the steric bulk in the second coordination sphere above the His3 layer prevents CuI from adopting a linear geometry, thereby forcing the binding of an ‘extra’ imidazole to CuI. Once this steric bulk is removed, as in the case of L19D and L19A, CuI readily adopts its preferred linear geometry, leaving an unbound imidazole. In previous studies with HgII, we demonstrated that providing three cysteine sulfurs that were predisposed for metal binding led to a higher than normal metal coordination environment [from HgII(SR)2 to HgII(SR)3].[7, 26, 27] We have also explored how increased steric bulk in the layer above a CdII and a three Cys plane can decrease the coordination number of the cation [from CdII(SR)3(H2O) to CdII(SR)3].[17, 28, 29] The present system provides the first example in which enhancement of steric bulk in the second coordination sphere leads to an increase in the coordination number of the bound metal ion (from CuI(His)2 to CuI(His)3).
At this point, it is worth assessing why the L19A, L19D, L26A, or L26D peptides have such an increased NiR rate compared to all previously prepared 3SCC NiR mimics. As discussed above, increased substrate access is one possibility, though our observation that L19I, with greater steric bulk, does not exhibit a decreased rate relative to TRIW-H leaves this uncertain. It would be tempting to conclude that the enhanced NiR rates of the L19A and L19D constructs are a consequence of their two-coordinate nature, but in this case, one would expect that neither L26A or L26D would show enhanced NiR reaction rates. One striking commonality between all the constructs presented with enhanced NiR activity, is the ~1 eV shift observed in the XANES edge feature. This shift is greater in the two coordinate constructs but is observed in all four enhanced activity constructs. Previous work has shown that both coordination number and geometry can affect the energy of the 1s→4p transition, where an increase in 4s-4p mixing results both in a decrease in intensity and shift to lower energy, which may already account for the differences seen in Figure 3.[25] Another possibility is that small changes in the 3SCC bundle may affect the net electron density on the bound Cu(I) which leads to an energy shift of the edge. This last explanation is particularly intriguing as it provides a plausible explanation as to how constructs with 3 or 2 coordinate Cu(I) could have similarly enhanced NiR rates. The change in the net charge of the bound Cu would then lead to similarly increased rates of nitrite reduction amongst all four constructs.
In summary, steric bulk above the histidine layer has been used to enforce a non-preferred metal coordination geometry in TRIW-H, L19I and L19DL, providing a novel strategy for the incorporation of desired metal coordination in metallopeptides. The substitution of Leu to Ala or Asp at the 19th or 26th positions of TRIW-H leads to a 45 - 75-fold increase in NiR activity with a catalytic efficiency of 1.0(0.3) s−1M−1 at pH 5.8 for L19A. The cause for this increase in NiR activity is still under investigation, but XANES gives evidence that this enhancement is likely significant for the CuI species, with all increased rate constructs showing a similar 1 eV shift in their edge transition energies. This shift may be caused by geometric or electrostatic changes, and this will require further investigation. It should be noted that L19A, L19D, L26A, and L26D could differ structurally in the Cu(I) state in the absence of substrate, while all constructs may ligate three His once nitrite binds as we have previously shown with CO binding to these constructs.[30, 31] Future designs will attempt to distinguish between these two possible effects and will provide guidance for NiR models with even higher catalytic efficiency.
Experimental Section
Supplementary Material
Figure 2.
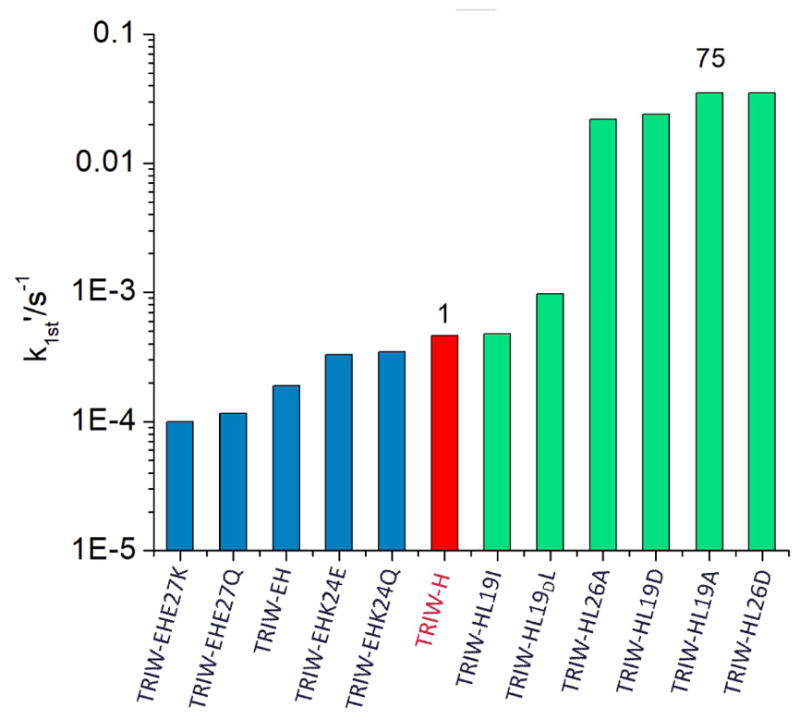
Pseudo-first order rate constants at pH 5.8 for the original construct TRIW-H (red), previously reported exterior mutations (blue), and currently reported interior mutations (green)[15]
Table 1.
TRI family peptide sequences used in this study
| Peptides[a] | 1 | 2 | 9 | 16 19 | 23 | 30 |
|---|---|---|---|---|---|---|
| abcdefg | abcdefg | Abcdefg | abcdefg | |||
| TRIW-H | G | WKALEEK | LKALEEK | LKALEEK | HKALEEK | G |
|
| ||||||
| L19I | G | WKALEEK | LKALEEK | LKAIEEK | HKALEEK | G |
|
| ||||||
| L19DL | G | WKALEEK | LKALEEK | LKADLEEK | HKALEEK | G |
|
| ||||||
| L19A | G | WKALEEK | LKALEEK | LKAAEEK | HKALEEK | G |
|
| ||||||
| L26A | G | WKALEEK | LKALEEK | LKALEEK | HKAAEEK | G |
|
| ||||||
| L19D | G | WKALEEK | LKALEEK | LKADEEK | HKALEEK | G |
|
| ||||||
| L26D | G | WKALEEK | LKALEEK | LKALEEK | HKADEEK | G |
The C- and N-termini are amidated and acetylated, respectively.
Acknowledgments
V.L.P thanks the National Institutes of Health for financial support of this research (ES012236). The authors thank Dr. Leela Ruckthong and Dr. Tyler Pinter for the helpful comments and discussion.
References
- 1.Yu F, Cangelosi VM, Zastrow ML, Tegoni M, Plegaria JS, Tebo AG, Mocny CS, Ruckthong L, Qayyum H, Pecoraro VL. Chemical Reviews. 2014;114:3495–3578. doi: 10.1021/cr400458x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mocny CS, Pecoraro VL. Accounts of Chemical Research. 2015;48:2388–2396. doi: 10.1021/acs.accounts.5b00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeGrado WF, Summa CM, Pavone Vincenzo, Nastri F, Lombardi A. Annual Review of Biochemistry. 1999;68:779–819. doi: 10.1146/annurev.biochem.68.1.779. [DOI] [PubMed] [Google Scholar]
- 4.Zastrow ML, Peacock AFA, Stuckey JA, Pecoraro VL. Nat Chem. 2012;4:118–123. doi: 10.1038/nchem.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zastrow ML, Pecoraro VL. Journal of the American Chemical Society. 2013;135:5895–5903. doi: 10.1021/ja401537t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tegoni M, Yu F, Bersellini M, Penner-Hahn JE, Pecoraro VL. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:21234–21239. doi: 10.1073/pnas.1212893110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dieckmann GR, McRorie DK, Lear JD, Sharp KA, DeGrado WF, Pecoraro VL. Journal of Molecular Biology. 1998;280:897–912. doi: 10.1006/jmbi.1998.1891. [DOI] [PubMed] [Google Scholar]
- 8.Wijma HJ, Jeuken LJC, Verbeet MP, Armstrong FA, Canters GW. Journal of Biological Chemistry. 2006;281:16340–16346. doi: 10.1074/jbc.M601610200. [DOI] [PubMed] [Google Scholar]
- 9.Bell CB, Calhoun JR, Bobyr E, Wei P-p, Hedman B, Hodgson KO, DeGrado WF, Solomon EI. Biochemistry. 2009;48:59–73. doi: 10.1021/bi8016087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reig AJ, Pires MM, Snyder RA, Wu Y, Jo H, Kulp DW, Butch SE, Calhoun JR, Szyperski TG, Solomon EI, DeGrado WF. Nat Chem. 2012;4:900–906. doi: 10.1038/nchem.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roy A, Sarrou I, Vaughn MD, Astashkin AV, Ghirlanda G. Biochemistry. 2013;52:7586–7594. doi: 10.1021/bi401199s. [DOI] [PubMed] [Google Scholar]
- 12.Roy A, Sommer DJ, Schmitz RA, Brown CL, Gust D, Astashkin A, Ghirlanda G. Journal of the American Chemical Society. 2014;136:17343–17349. doi: 10.1021/ja510621e. [DOI] [PubMed] [Google Scholar]
- 13.Kaim W, Rall J. Angewandte Chemie International Edition in English. 1996;35:43–60. [Google Scholar]
- 14.MacPherson IS, Murphy MEP. Cellular and Molecular Life Sciences. 2007;64:2887–2899. doi: 10.1007/s00018-007-7310-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu F, Penner-Hahn JE, Pecoraro VL. Journal of the American Chemical Society. 2013;135:18096–18107. doi: 10.1021/ja406648n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy MEP, Turley S, Adman ET. Journal of Biological Chemistry. 1997;272:28455–28460. doi: 10.1074/jbc.272.45.28455. [DOI] [PubMed] [Google Scholar]
- 17.Peacock AFA, Iranzo O, Pecoraro VL. Dalton Transactions. 2009:2271–2280. doi: 10.1039/b818306f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruckthong L, Peacock AFA, Pascoe CE, Hemmingsen L, Stuckey JA, Pecoraro VL. Chemistry – A European Journal. 2017;23:8232–8243. doi: 10.1002/chem.201700660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruckthong L, Deb A, Hemmingsen L, Penner-Hahn JE, Pecoraro VL. J Biol Inorg Chem. 2017 doi: 10.1007/s00775-017-1515-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghosh D, Pecoraro VL. Inorganic Chemistry. 2004;43:7902–7915. doi: 10.1021/ic048939z. [DOI] [PubMed] [Google Scholar]
- 21.Ruckthong L, Zastrow ML, Stuckey JA, Pecoraro VL. Journal of the American Chemical Society. 2016;138:11979–11988. doi: 10.1021/jacs.6b07165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruckthong L. University of Michigan; Ann Arbor, MI: 2016. [Google Scholar]
- 23.Isoda N, Yokoyama H, Nojiri M, Suzuki S, Yamaguchi K. Bioelectrochemistry. 2010;77:82–88. doi: 10.1016/j.bioelechem.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 24.Blackburn NJ, Strange RW, Reedijk J, Volbeda A, Farooq A, Karlin KD, Zubieta J. Inorganic Chemistry. 1989;28:1349–1357. [Google Scholar]
- 25.Kau LS, Spira-Solomon DJ, Penner-Hahn JE, Hodgson KO, Solomon EI. Journal of the American Chemical Society. 1987;109:6433–6442. [Google Scholar]
- 26.Dieckmann GR, McRorie DK, Tierney DL, Utschig LM, Singer CP, O’Halloran TV, Penner-Hahn JE, DeGrado WF, Pecoraro VL. Journal of the American Chemical Society. 1997;119:6195–6196. [Google Scholar]
- 27.Farrer BT, Harris NP, Balchus KE, Pecoraro VL. Biochemistry. 2001;40:14696–14705. doi: 10.1021/bi015649a. [DOI] [PubMed] [Google Scholar]
- 28.Iranzo O, Cabello C, Pecoraro VL. Angewandte Chemie (International ed in English) 2007;46:6688–6691. doi: 10.1002/anie.200701729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peacock AFA, Hemmingsen L, Pecoraro VL. Proceedings of the National Academy of Sciences. 2008;105:16566–16571. doi: 10.1073/pnas.0806792105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu F. PhD thesis. University of Michigan; Ann Arbor, MI: 2014. [Google Scholar]
- 31.Ross MR, White AM, Yu F, King JT, Pecoraro VL, Kubarych KJ. Journal of the American Chemical Society. 2015;137:10164–10176. doi: 10.1021/jacs.5b02840. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


