Abstract
Nonsense-mediated mRNA decay (NMD) is a conserved mRNA surveillance pathway that cells use to ensure the quality of transcripts and to fine-tune transcript abundance. The role of NMD in cancer development is complex. In some cases, tumors have exploited NMD to downregulate gene expression by apparently selecting for mutations causing destruction of key tumor-suppressor mRNAs. In other cases, tumors adjust NMD activity to adapt to their microenvironment. Understanding how particular tumors exploit NMD for their benefit may augment the development of new therapeutic interventions.
Introduction: Nonsense-mediated mRNA Decay (NMD)
The fidelity of genetic information as it is passed from DNA to mRNA to protein is important for the survival of cells and organisms. mRNA is unique in this flow of information since a single gene often gives rise to multiple mRNA species through alternative pre-mRNA splicing or alternative 3′-end formation, and each mRNA species may encode a unique protein. Cells therefore tightly control the quality and quantity of mRNAs using various surveillance pathways. Among these is nonsense-mediated mRNA decay (NMD), which inspects mRNAs for premature termination codons (PTCs) introduced through DNA mutations or pre-mRNA processing defects [1].
The most well-studied form of NMD detects PTCs located >50–55-nucleotides upstream of an exon-exon junction—a situation that is usually aberrant since nearly all normal termination codons lie within the final exon. The splicing history of an mRNA is recorded through deposition of an exon-junction complex (EJC) of proteins ~20–24-nucleotides upstream of exon-exon junctions [2]. NMD is a cytoplasmic and translation-dependent process. As the ribosome plows through a transcript during translation, it strips off any EJCs until it reaches a PTC or a normal termination codon, leaving any remaining downstream EJCs, which would generally be present only in the case of PTCs. A series of protein complexes, composed of NMD factors that include up-frameshift proteins (UPF1, UPF2, and UPF3X) and suppressors with morphological effects on genitalia proteins (SMG1, SMG5, SMG6, SMG7, SMG8, and SMG9), are bound to mRNAs via EJCs either before or after a translation termination event and lead to mRNA degradation should an EJC lie downstream of a termination codon. This prevents the potentially negative consequences of dominant effects from truncated proteins [3]. UPF3X, also called UPF3B, decorates the EJC and anchors UPF2. When an EJC lies downstream of a termination event, the ATP-dependent RNA helicase UPF1 becomes part of the translation termination complex. UPF1 interacts with EJC-bound UPF2, undergoing a conformational change that activates its RNA-helicase activity [4]*. EJC-bound UPF1 is phosphorylated by the protein kinase SMG1 committing the mRNA to degradation [5], since phosphorylated UPF1 recruits SMG6 endonuclease and/or the heterodimer SMG5 and SMG7, which further recruits decapping and deadenylating enzymes, promoting mRNA destruction. A closely related paralog of UPF3X, UPF3 (also called UPF3A), inhibits NMD and is regulated by UPF3X expression, possibly allowing for fine-tuning of NMD activity [6].
Endogenous mRNAs that are not the consequence of mutations or aberrant pre-mRNA processing can be degraded by the same set of proteins. Upstream open reading frames (uORFs), unusually long 3′-untranslated regions (3′UTRs), regulated alternative splicing that introduces a PTC, normal termination codons situated >50–55-nucleotides upstream of an exon-exon junction as a result of regulated alternative 3′-end formation, and the recognition of a UGA selenocysteine codon within some selenoprotein-encoding mRNAs as a termination codon, can target a normal transcript for NMD [1], allowing the cell to utilize the NMD apparatus to fine-tune gene expression. Tumors have found ways to exploit both quality-control and fine-tuning aspects of NMD, using the quality-control function to selectively eliminate the expression of genes through PTC acquisition to better promote unconstrained growth, and also adjusting NMD activity to affect the abundance of endogenous transcripts that allow successful adaptation to the growth environment [7].
Nonsense mutations in cancer
Relative to oncogenes, which often contain missense mutations, tumor-suppressor genes are characterized more by NMD-inducing nonsense mutations [8]. Examples include genes encoding E-cadherin in stomach cancers [9], BRCA1 in breast and ovarian cancers [10], BRCA2 in ovarian cancers [11], p53 in breast cancers [12], Rb in lymphoma [13], and WT1 in kidney cancers [14]. In these cases, the cell uses NMD for its protective function, eliminating transcripts that might produce truncated proteins with dominant-negative properties, as has been shown for WT1 [14], leading to tumorigenesis. The downside of this strategy is that, with the gene product from the PTC-bearing allele destroyed, the cell is now one step closer to achieving biallelic tumor-suppressor gene inactivation. Additionally, in some cases, the NMD of an mRNA deriving from a PTC-bearing allele might eliminate production of a protein that does not function in a dominant-negative manner, but is partially functional and could help prevent disease, as has been shown for CFTR mRNA in cystic fibrosis [15].
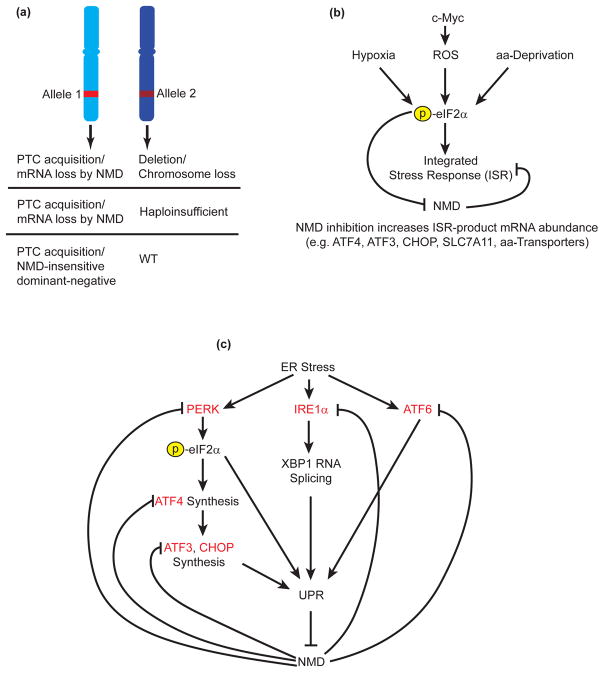
By interrogating genomes and their matching transcriptomic datasets from human cancer patients, PTCs were found to be enriched in regions of known tumor-suppressor genes predicted to trigger NMD. Tumors leveraged PTCs to ablate tumor-suppressor gene function in several ways: by combining a PTC-bearing allele with a heterozygous deletion, by selecting for a PTC in haploinsufficient tumor-suppressor genes, or by selecting for PTCs in NMD-insensitive regions that likely generate dominant-negative proteins [16] (Fig. 1A). Another study comparing somatic mutations in The Cancer Genome Atlas (TCGA) with their matching transcriptomic datasets discovered that particular cancers types, including stomach adenocarcinoma, kidney cancer and colon cancer, had higher numbers of NMD-causing mutations relative to other cancer types with similar mutation frequencies [17]. Tumor-suppressor genes were frequently inactivated by NMD in combination with deletion of the remaining wild-type allele. In tumors with hypermutations, non-tumor-suppressor genes with NMD-causing mutations showed a more modest decrease in expression (i.e. likely possessed hypofunction), and these mutated genes clustered in particular pathways such as DNA repair, chromatin remodeling, and RNA binding. Possibly, hypofunction in these pathways facilitated by NMD passively allows the establishment and survival of hypermutated tumor cells [17].
Fig. 1.
The roles that NMD plays in cancer. (a) Tumor-suppressor gene function can be inactivated by NMD. PTC introduction that leads to the loss of tumor-suppressor mRNA via NMD can be combined with either a deletion or chromosomal loss of the wild-type (WT) version, or the remaining wild-type version may be haploinsufficient. Alternatively, PTC acquisition in a region that fails to trigger NMD may lead to production of a dominant-negative allele that interferes with wild-type function. (b) NMD is involved in adaptation to the tumor microenvironment. Stresses in the microenvironment such as hypoxia, production of reactive oxygen species (ROS), and amino acid (aa)-deprivation, lead to phosphorylation of eIF2α and the initiation of the integrated stress response (ISR) in an attempt to adapt to these insults. One outcome of eIF2α phosphorylation is the inhibition of NMD by a poorly understood mechanism. Among the endogenous NMD targets upregulated are components of the ISR. (c) Mutual regulation by the unfolded-protein response (UPR) and NMD. Endoplasmic reticulum (ER) stress causes activation of three ER sensors—PERK, IRE1α, and ATF6. PERK activation leads to phosphorylation of eIF2α and a decrease in global protein synthesis as well as production of ATF4, which upregulates ATF3 and CHOP. These gene products are necessary for the initiation of the UPR, where the cell attempts to increase its protein-folding capacity. IRE1α activation leads to cytoplasmic splicing of XBP1 RNA and activation of the UPR. ATF6 cleavage likewise leads to activation of the UPR. UPR activation attenuates NMD, upregulating a suite of endogenous NMD targets that include UPR components (red). This mutual regulation controls the timing and magnitude of the UPR.
Mutations in the NMD apparatus
Mutations in the NMD machinery have been described in certain cancers. Pancreatic adenosquamous carcinomas (ASC) are more aggressive and metastatic than adenocarcinomas. Somatic UPF1 mutations that disrupt UPF1 pre-mRNA splicing, and thus function, were frequently detected in ASC pancreatic tumors but not in adjacent normal tissue [18]. Somatic UPF1 mutations that cause exon skipping in lung inflammatory myofibroblastic tumors (IMTs), a type of tumor characterized by infiltration of immune cells, reduced NMD activity, allowing upregulated expression of NF-kappa-B-inducing kinase (NIK; encoded by the MAP3K14 gene), an NMD target [19]. NIK stimulates the NF-kB pathway, generating chemokines that recruit the immune cells typifying these tumors [19]. Reduced UPF1 expression, attributed to UPF1 promoter methylation, was also noted in hepatocellular carcinoma (HCC) tumor tissue relative to adjacent normal tissue and was correlated with poorer prognosis than patients with normal UPF1 expression [20]. Similarly, UPF1 expression is lower in lung adenocarcinoma (ADC) relative to adjacent normal tissue. This promotes characteristics of the epithelial-to-mesenchymal transition (EMT) since decreased NMD activity allows for upregulation of TGF-β signaling components, which are vital to the EMT [21]. Because of the many distinct NMD targets that could affect cell physiology in diverse ways, further experiments need to be done to prove that the inactivating mutations in UPF1 observed in these cases drive progression of the cancer phenotype rather than merely represent a molecular remnant of a barrier that cancer cells have overcome by selecting for advantageous mutations in other genes.
Inactivating mutations in UPF1 [22], UPF2 [23], SMG1 [24], and SMG6 [25] are not compatible with life in mice. In the case of UPF1, blastocysts undergo cell death via apoptosis after five days in culture, suggesting that NMD plays a key role in development. Lethality in the absence of UPF1 extends to zebrafish, flies, and plants. In Drosophila, this is largely due to NMD-mediated control of a single transcript encoding Growth Arrest and DNA Damage 45 (GADD45) [26]. GADD45 is a signaling protein that promotes apoptosis by upregulating the Mitogen-activated protein kinase (MAPK) signaling pathway. Amazingly, this same regulation of apoptosis occurs in mammalian cells, where depletion of GADD45β (also encoded by an NMD target) partially rescues apoptosis caused by UPF1 depletion [26]. Thus, an intact NMD pathway serves a protective function, preventing cells from undergoing premature apoptosis. Yet another pro-apoptotic NMD target is GAS5 (growth arrest-specific 5) RNA, which is classified as a noncoding RNA despite being translationally active [27–29]. When overexpressed, GAS5 RNA causes cell-cycle arrest and cell death by binding to and antagonizing the glucocorticoid receptor. How are these results, which suggest that experimental disruptions of NMD promote cell death, reconciled with the seemingly contradictory fact that aggressive tumors have a reduced capacity for NMD? The observed inactivation of NMD in some tumors may indicate an unsuccessful attempt at apoptosis, which the remaining cells comprising the tumor were able to avoid in other ways. Varying degrees of NMD down-modulation, as well as the tissue-specific expression of key NMD targets mediating either a pro-growth or pro-apoptotic effect, may also offer an explanation. In fact, the activity of NMD can be modulated to achieve distinct outcomes in cellular responses to stress.
Stress, NMD, and tumorigenesis
The efficiency of cellular NMD can be regulated to achieve a desired cellular outcome. Tumor cells encounter various stresses in the microenvironments where they develop and must adapt to survive. Hypoxia inhibits NMD, stabilizing a suite of NMD-target transcripts [30]. Among those transcripts stabilized are mRNAs encoding components of the integrated stress response (ISR), a pathway used to reestablish homeostasis after acute stress. Stresses, in this case hypoxia, lead to eIF2α phosphorylation, which attenuates general protein synthesis and, paradoxically, promotes translation of the transcription factor ATF4. ATF4 activates the production of other transcription factors, namely CHOP and ATF3, which initiate a transcriptional program aimed at maintaining homeostasis. ATF4, ATF3, and CHOP all derive from direct NMD targets, and thus suppression of NMD during hypoxia serves to augment the ISR. The ability of eIF2α to be phosphorylated is key to NMD suppression, but the detailed molecular mechanism behind how this attenuates NMD is unclear (Fig. 1B).
In situations where the MYC oncogene is constitutively active, reactive oxygen species (ROS) are generated. Here too, eIF2α is phosphorylated, and NMD is suppressed [31]. One of the NMD-target transcripts stabilized encodes the cystine/glutamate exchanger SLC7A11, which is a component of the xCT amino-acid transporter system, the rate-limiting step for the import of cystine into cells for glutathione production. Glutathione is a ROS scavenger, explaining why UPF1-knockdown cells are more tolerant of oxidative stress [32]. Amino-acid deprivation, another stress encountered in the tumor microenvironment, dampens NMD and upregulates a suite of amino-acid transporters in an attempt to maintain homeostasis [33].
In addition to its role in the ISR and blunting NMD activity, eIF2α phosphorylation is required for the induction of autophagy during amino-acid deprivation [34]. Autophagy is a process where cytoplasmic macromolecules and organelles are sequestered in double-membrane structures termed autophagosomes, which merge with the lysosomal pathway to recycle proteins into their constituent amino acids. Inhibiting NMD is one way that cells activate autophagy to restore amino-acid homeostasis [35], possibly clearing mutated or misfolded proteins that may result from the attenuation of NMD. This occurs at least partially due to ATF4 transcript stabilization resulting from NMD downregulation. Autophagy has previously been reported to be important for tumor-cell survival and tumorigenesis [36].
Since NMD attenuation promotes the ISR to all of the above cellular stresses encountered during tumor formation, and given that the ISR is required for tumor growth [37], does the inhibition of NMD promote tumor growth? This is the case because in cells where NMD cannot be inhibited due to UPF1 overexpression, tumor formation in nude mice is dramatically reduced, and cells from these tumors form fewer and smaller colonies in soft agar [38]. Inhibiting NMD not only could promote tumor-cell survival but also could allow for the generation of dominant-negative mutated tumor-suppressor proteins (see above) that downregulate the activity of the tumor-suppressor protein produced by the non-mutated allele, further driving tumorigenesis [39]. To illustrate, in the case of pancreatic ASC tumors where UPF1 was mutated and NMD function was attenuated, a dominant-negative PTC-bearing p53 tumor-suppressor allele was expressed [18], and this could also be the case in situations where NMD activity is inhibited in response to stresses.
Many of the stresses in the tumor microenvironment described above—e.g. hypoxia, amino-acid deprivation, lack of other nutrients, insufficient vasculature, decreased pH—can activate the unfolded-protein response (UPR), which is centered around the endoplasmic reticulum (ER) [40]. Briefly, three sensors in the ER—PERK, IRE1, and ATF6—discern when the protein-folding load is mismatched relative to protein-folding capacity [1]. PERK phosphorylates eIF2α in an attempt to decrease protein production, also activating ATF4 synthesis. IRE1 activates its own nuclease activity, unconventionally splicing a single substrate, XBP1 RNA, which encodes a protein that coordinates the synthesis of chaperone proteins to assist in folding. Additionally, ATF6 is cleaved, liberating a transcriptional activator that likewise assists in chaperone production. Here again, many of the components involved in the UPR, including ATF4, ATF3, and CHOP [30,33], as well as ATF6, FSD1L, HERP, IRE1a, PERK, PRDG1, TNRC1, and TRAF2 [41], derive from NMD targets, indicating that the blunting of NMD seen in the tumor microenvironment assists in promoting the UPR. The UPR is tightly controlled: innocuous low-level stresses should not trigger the UPR, while transient or moderate stresses should lead to activation followed by termination. Depletion of UPF3X in tissue-culture cells or mice lowers the threshold for UPR activation and also prolongs the UPR response, indicating that NMD normally suppresses the UPR [41]. Suppression of the UPR by NMD is driven largely through control of IRE1α mRNA levels, which has a long 3′UTR [41]. ER stressors that trigger the UPR can themselves inhibit NMD [30,41,42], likely because PERK is able to phosphorylate eIF2α. This symbiotic regulation—NMD inhibits the UPR, and the UPR inhibits NMD—creates several desirable characteristics: (i) it allows the cell to achieve a rapid, switch-like response where insults below a certain threshold will not trigger the UPR (because NMD suppresses it); (ii) when appropriate stresses are encountered, the UPR is activated in full (because the UPR attenuates NMD allowing full production of its effectors); and (iii) when stresses cease, the UPR fully shuts down (because NMD presumably resumes) (Fig. 1C).
If the stresses encountered exceed the threshold that the cell can tolerate even with all of its adaptive mechanisms in place, it must terminate itself through programmed cell death, i.e. apoptosis. Here too, NMD plays a role. When cells are treated with apoptosis-inducing small molecules, some of which are clinically used chemotherapeutics, NMD activity is inhibited [43,44]. Caspases, the proteases responsible for dismantling the cell during apoptosis, become active, indicating that the cell cannot adequately adapt to the insult, and cleave UPF1 [43,44] and UPF2 [43]. In the case of UPF1, a dominant-interfering fragment is generated that antagonizes NMD. These events upregulate the production of gene products that derive from endogenous NMD targets and promote apoptosis such as GADD45α/β and GAS5 transcripts (discussed in “Mutations in the NMD apparatus”). Thus, much like the UPR, NMD normally inhibits apoptosis. However, when apoptosis is triggered, NMD is attenuated, augmenting the rate and robustness of cell death.
Conclusion
The role that NMD plays in cancer is complex. Disabling mutations in UPF1 are observed in some tumors. A priori, one could think that this allows cells to better respond to stresses encountered in the tumor microenvironment by increasing expression of gene products needed for the ISR. How is this reconciled with the fact that depletion of UPF1 and reduced NMD activity causes cell-cycle arrest [45] and promotes cell death [26]? The answer likely depends on the tissue from which the tumor originates, the evolutionary history of the tumor, and other compensatory mutations present in the tumor, reinforcing the notion that cancers should be viewed as a collection of unique diseases. Although difficult to develop, NMD-activating small molecules may be of clinical utility. More realistic may be oligonucleotide therapies that re-engage NMD in order to degrade PTC-containing but NMD-insensitive transcripts that create dominant-negative truncated proteins such as for BRCA1 [46,47], p53 [48], and WT1 [14,49]. This would only work in situations where the patient has a remaining wild-type allele. In other situations where the NMD pathway is intact and tumor-suppressor genes have acquired PTCs promoting tumorigenesis, read-through therapies may be of utility. These drugs can promote the insertion of an amino acid at the PTC and thus some production of a possibly partially functional protein [50]. Adding further complexity is the finding that NMD inhibition causes the production of neoantigens that aid an immune system attack on tumors [51], indicating that, in some situations, small molecules that inhibit NMD may be of therapeutic value.
It is not surprising that a pathway that controls the stability of the products of ~30% of all genes (at least in mouse liver [52]) is itself regulated and exploited to tune cellular responses. Undoubtedly the list of pathways and diseases that NMD affects will grow in the future.
Acknowledgments
Work on NMD in the Maquat laboratory is supported by NIH R01 GM59614.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Popp MW, Maquat LE. Organizing principles of mammalian nonsense-mediated mRNA decay. Annu Rev Genet. 2013;47:139–165. doi: 10.1146/annurev-genet-111212-133424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Le Hir H, Moore MJ, Maquat LE. Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon-exon junctions. Genes Dev. 2000;14:1098–1108. [PMC free article] [PubMed] [Google Scholar]
- 3.Lykke-Andersen S, Jensen TH. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat Rev Mol Cell Biol. 2015;16:665–677. doi: 10.1038/nrm4063. [DOI] [PubMed] [Google Scholar]
- 4*.Kurosaki T, Maquat LE. Nonsense-mediated mRNA decay in humans at a glance. J Cell Sci. 2016;129:461–467. doi: 10.1242/jcs.181008. This reference gives a more comprehensive overview of the molecular details that are known about how NMD is initiated and how the NMD machinery degrades mRNAs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kurosaki T, Li W, Hoque M, Popp MW, Ermolenko DN, Tian B, Maquat LE. A post-translational regulatory switch on UPF1 controls targeted mRNA degradation. Genes Dev. 2014;28:1900–1916. doi: 10.1101/gad.245506.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan WK, Bhalla AD, Le Hir H, Nguyen LS, Huang L, Gecz J, Wilkinson MF. A UPF3-mediated regulatory switch that maintains RNA surveillance. Nat Struct Mol Biol. 2009;16:747–753. doi: 10.1038/nsmb.1612. [DOI] [PubMed] [Google Scholar]
- 7.Goetz AE, Wilkinson M. Stress and the nonsense-mediated RNA decay pathway. Cell Mol Life Sci. 2017 doi: 10.1007/s00018-017-2537-6. [DOI] [PMC free article] [PubMed]
- 8.Mort M, Ivanov D, Cooper DN, Chuzhanova NA. A meta-analysis of nonsense mutations causing human genetic disease. Hum Mutat. 2008;29:1037–1047. doi: 10.1002/humu.20763. [DOI] [PubMed] [Google Scholar]
- 9.Karam R, Carvalho J, Bruno I, Graziadio C, Senz J, Huntsman D, Carneiro F, Seruca R, Wilkinson MF, Oliveira C. The NMD mRNA surveillance pathway downregulates aberrant E-cadherin transcripts in gastric cancer cells and in CDH1 mutation carriers. Oncogene. 2008;27:4255–4260. doi: 10.1038/onc.2008.62. [DOI] [PubMed] [Google Scholar]
- 10.Perrin-Vidoz L, Sinilnikova OM, Stoppa-Lyonnet D, Lenoir GM, Mazoyer S. The nonsense-mediated mRNA decay pathway triggers degradation of most BRCA1 mRNAs bearing premature termination codons. Hum Mol Genet. 2002;11:2805–2814. doi: 10.1093/hmg/11.23.2805. [DOI] [PubMed] [Google Scholar]
- 11.Ware MD, DeSilva D, Sinilnikova OM, Stoppa-Lyonnet D, Tavtigian SV, Mazoyer S. Does nonsense-mediated mRNA decay explain the ovarian cancer cluster region of the BRCA2 gene? Oncogene. 2006;25:323–328. doi: 10.1038/sj.onc.1209033. [DOI] [PubMed] [Google Scholar]
- 12.Anczukow O, Ware MD, Buisson M, Zetoune AB, Stoppa-Lyonnet D, Sinilnikova OM, Mazoyer S. Does the nonsense-mediated mRNA decay mechanism prevent the synthesis of truncated BRCA1, CHK2, and p53 proteins? Hum Mutat. 2008;29:65–73. doi: 10.1002/humu.20590. [DOI] [PubMed] [Google Scholar]
- 13.Pinyol M, Bea S, Pla L, Ribrag V, Bosq J, Rosenwald A, Campo E, Jares P. Inactivation of RB1 in mantle-cell lymphoma detected by nonsense-mediated mRNA decay pathway inhibition and microarray analysis. Blood. 2007;109:5422–5429. doi: 10.1182/blood-2006-11-057208. [DOI] [PubMed] [Google Scholar]
- 14.Reddy JC, Morris JC, Wang J, English MA, Haber DA, Shi Y, Licht JD. WT1-mediated transcriptional activation is inhibited by dominant negative mutant proteins. J Biol Chem. 1995;270:10878–10884. doi: 10.1074/jbc.270.18.10878. [DOI] [PubMed] [Google Scholar]
- 15.Wilschanski M, Yahav Y, Yaacov Y, Blau H, Bentur L, Rivlin J, Aviram M, Bdolah-Abram T, Bebok Z, Shushi L, et al. Gentamicin-induced correction of CFTR function in patients with cystic fibrosis and CFTR stop mutations. N Engl J Med. 2003;349:1433–1441. doi: 10.1056/NEJMoa022170. [DOI] [PubMed] [Google Scholar]
- 16**.Lindeboom RG, Supek F, Lehner B. The rules and impact of nonsense-mediated mRNA decay in human cancers. Nat Genet. 2016;48:1112–1118. doi: 10.1038/ng.3664. The authors interrogate matching exome and transcriptome data from human tumors to reveal how tumors select for PTC acquisition in key genes, especially tumor-supressor genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17*.Hu Z, Yau C, Ahmed AA. A pan-cancer genome-wide analysis reveals tumour dependencies by induction of nonsense-mediated decay. Nat Commun. 2017;8:15943. doi: 10.1038/ncomms15943. The authors use data from The Cancer Genome Atlas to identify cancer-specific gene signatures characterized by NMD causing mutations in tumor-supressor genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu C, Karam R, Zhou Y, Su F, Ji Y, Li G, Xu G, Lu L, Wang C, Song M, et al. The UPF1 RNA surveillance gene is commonly mutated in pancreatic adenosquamous carcinoma. Nat Med. 2014;20:596–598. doi: 10.1038/nm.3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu J, Plank TD, Su F, Shi X, Liu C, Ji Y, Li S, Huynh A, Shi C, Zhu B, et al. The nonsense-mediated RNA decay pathway is disrupted in inflammatory myofibroblastic tumors. J Clin Invest. 2016;126:3058–3062. doi: 10.1172/JCI86508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang L, Li C, Guo T, Wang H, Ma W, Yuan Y, Liu Q, Ye Q, Liu Z. The human RNA surveillance factor UPF1 regulates tumorigenesis by targeting Smad7 in hepatocellular carcinoma. J Exp Clin Cancer Res. 2016;35:8. doi: 10.1186/s13046-016-0286-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao L, Qi L, Zhang L, Song W, Yu Y, Xu C, Li L, Guo Y, Yang L, Liu C, et al. Human nonsense-mediated RNA decay regulates EMT by targeting the TGF-ss signaling pathway in lung adenocarcinoma. Cancer Lett. 2017 doi: 10.1016/j.canlet.2017.06.021. [DOI] [PubMed] [Google Scholar]
- 22.Medghalchi SM, Frischmeyer PA, Mendell JT, Kelly AG, Lawler AM, Dietz HC. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum Mol Genet. 2001;10:99–105. doi: 10.1093/hmg/10.2.99. [DOI] [PubMed] [Google Scholar]
- 23.Weischenfeldt J, Damgaard I, Bryder D, Theilgaard-Monch K, Thoren LA, Nielsen FC, Jacobsen SE, Nerlov C, Porse BT. NMD is essential for hematopoietic stem and progenitor cells and for eliminating byproducts of programmed DNA rearrangements. Genes Dev. 2008;22:1381–1396. doi: 10.1101/gad.468808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McIlwain DR, Pan Q, Reilly PT, Elia AJ, McCracken S, Wakeham AC, Itie-Youten A, Blencowe BJ, Mak TW. Smg1 is required for embryogenesis and regulates diverse genes via alternative splicing coupled to nonsense-mediated mRNA decay. Proc Natl Acad Sci U S A. 2010;107:12186–12191. doi: 10.1073/pnas.1007336107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li T, Shi Y, Wang P, Guachalla LM, Sun B, Joerss T, Chen YS, Groth M, Krueger A, Platzer M, et al. Smg6/Est1 licenses embryonic stem cell differentiation via nonsense-mediated mRNA decay. EMBO J. 2015;34:1630–1647. doi: 10.15252/embj.201489947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26**.Nelson JO, Moore KA, Chapin A, Hollien J, Metzstein MM. Degradation of Gadd45 mRNA by nonsense-mediated decay is essential for viability. Elife. 2016:5. doi: 10.7554/eLife.12876. This study shows that NMD degrades the pro-apoptotic Gadd45 transcript, and that degradation is critical for cells to avoid undergoing apoptosis. This mechanism is conserved in human cells and likely helps to explain why NMD has been found to be essential for embryonic viability. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith CM, Steitz JA. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5′-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol Cell Biol. 1998;18:6897–6909. doi: 10.1128/mcb.18.12.6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tani H, Torimura M, Akimitsu N. The RNA degradation pathway regulates the function of GAS5 a non-coding RNA in mammalian cells. PLoS One. 2013;8:e55684. doi: 10.1371/journal.pone.0055684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mourtada-Maarabouni M, Williams GT. Growth arrest on inhibition of nonsense-mediated decay is mediated by noncoding RNA GAS5. Biomed Res Int. 2013;2013:358015. doi: 10.1155/2013/358015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gardner LB. Hypoxic inhibition of nonsense-mediated RNA decay regulates gene expression and the integrated stress response. Mol Cell Biol. 2008;28:3729–3741. doi: 10.1128/MCB.02284-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang D, Wengrod J, Gardner LB. Overexpression of the c-myc oncogene inhibits nonsense-mediated RNA decay in B lymphocytes. J Biol Chem. 2011;286:40038–40043. doi: 10.1074/jbc.M111.266361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin L, Gardner LB. Stress-induced inhibition of nonsense-mediated RNA decay regulates intracellular cystine transport and intracellular glutathione through regulation of the cystine/glutamate exchanger SLC7A11. Oncogene. 2015;34:4211–4218. doi: 10.1038/onc.2014.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- 34.Talloczy Z, Jiang W, Virgin HWt, Leib DA, Scheuner D, Kaufman RJ, Eskelinen EL, Levine B. Regulation of starvation- and virus-induced autophagy by the eIF2alpha kinase signaling pathway. Proc Natl Acad Sci U S A. 2002;99:190–195. doi: 10.1073/pnas.012485299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wengrod J, Martin L, Wang D, Frischmeyer-Guerrerio P, Dietz HC, Gardner LB. Inhibition of nonsense-mediated RNA decay activates autophagy. Mol Cell Biol. 2013;33:2128–2135. doi: 10.1128/MCB.00174-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gelinas C, Fan Y, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye J, Kumanova M, Hart LS, Sloane K, Zhang H, De Panis DN, Bobrovnikova-Marjon E, Diehl JA, Ron D, Koumenis C. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J. 2010;29:2082–2096. doi: 10.1038/emboj.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang D, Zavadil J, Martin L, Parisi F, Friedman E, Levy D, Harding H, Ron D, Gardner LB. Inhibition of nonsense-mediated RNA decay by the tumor microenvironment promotes tumorigenesis. Mol Cell Biol. 2011;31:3670–3680. doi: 10.1128/MCB.05704-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gardner LB. Nonsense-mediated RNA decay regulation by cellular stress: implications for tumorigenesis. Mol Cancer Res. 2010;8:295–308. doi: 10.1158/1541-7786.MCR-09-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giampietri C, Petrungaro S, Conti S, Facchiano A, Filippini A, Ziparo E. Cancer Microenvironment and Endoplasmic Reticulum Stress Response. Mediators Inflamm. 2015;2015:417281. doi: 10.1155/2015/417281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41**.Karam R, Lou CH, Kroeger H, Huang L, Lin JH, Wilkinson MF. The unfolded protein response is shaped by the NMD pathway. EMBO Rep. 2015;16:599–609. doi: 10.15252/embr.201439696. This study demonstrates that there is mechanistic interplay between the UPR and NMD. It shows that NMD affects the magnitude and duration of the UPR since many of the critical components of the UPR derive from transcripts that are endogenous NMD targets. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Usuki F, Fujimura M, Yamashita A. Endoplasmic reticulum stress preconditioning attenuates methylmercury-induced cellular damage by inducing favorable stress responses. Sci Rep. 2013;3:2346. doi: 10.1038/srep02346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jia J, Furlan A, Gonzalez-Hilarion S, Leroy C, Gruenert DC, Tulasne D, Lejeune F. Caspases shutdown nonsense-mediated mRNA decay during apoptosis. Cell Death Differ. 2015;22:1754–1763. doi: 10.1038/cdd.2015.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Popp MW, Maquat LE. Attenuation of nonsense-mediated mRNA decay facilitates the response to chemotherapeutics. Nat Commun. 2015;6:6632. doi: 10.1038/ncomms7632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Azzalin CM, Lingner J. The human RNA surveillance factor UPF1 is required for S phase progression and genome stability. Curr Biol. 2006;16:433–439. doi: 10.1016/j.cub.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 46.Fan S, Yuan R, Ma YX, Meng Q, Goldberg ID, Rosen EM. Mutant BRCA1 genes antagonize phenotype of wild-type BRCA1. Oncogene. 2001;20:8215–8235. doi: 10.1038/sj.onc.1205033. [DOI] [PubMed] [Google Scholar]
- 47.Sylvain V, Lafarge S, Bignon YJ. Dominant-negative activity of a Brca1 truncation mutant: effects on proliferation, tumorigenicity in vivo, and chemosensitivity in a mouse ovarian cancer cell line. Int J Oncol. 2002;20:845–853. [PubMed] [Google Scholar]
- 48.Cardinali M, Kratochvil FJ, Ensley JF, Robbins KC, Yeudall WA. Functional characterization in vivo of mutant p53 molecules derived from squamous cell carcinomas of the head and neck. Mol Carcinog. 1997;18:78–88. doi: 10.1002/(sici)1098-2744(199702)18:2<78::aid-mc3>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 49.Englert C, Vidal M, Maheswaran S, Ge Y, Ezzell RM, Isselbacher KJ, Haber DA. Truncated WT1 mutants alter the subnuclear localization of the wild-type protein. Proc Natl Acad Sci U S A. 1995;92:11960–11964. doi: 10.1073/pnas.92.26.11960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Welch EM, Barton ER, Zhuo J, Tomizawa Y, Friesen WJ, Trifillis P, Paushkin S, Patel M, Trotta CR, Hwang S, et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007;447:87–91. doi: 10.1038/nature05756. [DOI] [PubMed] [Google Scholar]
- 51.Pastor F, Kolonias D, Giangrande PH, Gilboa E. Induction of tumour immunity by targeted inhibition of nonsense-mediated mRNA decay. Nature. 2010;465:227–230. doi: 10.1038/nature08999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weischenfeldt J, Waage J, Tian G, Zhao J, Damgaard I, Jakobsen JS, Kristiansen K, Krogh A, Wang J, Porse BT. Mammalian tissues defective in nonsense-mediated mRNA decay display highly aberrant splicing patterns. Genome Biol. 2012;13:R35. doi: 10.1186/gb-2012-13-5-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]