Abstract
Here we review the evolving story of the coronavirus endoribonuclease (EndoU). Coronavirus EndoU is encoded within the sequence of nonstructural protein (nsp) 15, which was initially identified as a component of the viral replication complex. Biochemical and structural studies revealed the enzymatic nature of nsp15/EndoU, which was postulated to be essential for the unique replication cycle of viruses in the order Nidovirales. However, the role of nsp15 in coronavirus replication was enigmatic as EndoU-deficient coronaviruses were viable and replicated to near wild-type virus levels in fibroblast cells. A breakthrough in our understanding of the role of EndoU was revealed in recent studies, which showed that EndoU mediates the evasion of viral double-stranded RNA recognition by host sensors in macrophages. This new discovery of nsp15/EndoU function leads to new opportunities for investigating how a viral EndoU contributes to pathogenesis and exploiting this enzyme for therapeutics and vaccine design against pathogenic coronaviruses.
Keywords: Coronavirus, Nsp15, Endoribonuclease, Double-stranded RNA, Interferon, Host recognition, Antiviral defense
1. History of coronavirus endoribonuclease
We provide a brief outline of the major research findings related to coronavirus (CoV) endoribonucleases (EndoU) in Table 1. In the text below, we describe the experimental approaches that led to these findings and compare the activity of CoV EndoU with reports of other viral and host ribonucleases.
Table 1.
A review of CoV nsp15 research.
| Research topic | Virus | Findings | References |
|---|---|---|---|
| Proteolytic processing of pp1ab and characterizing its cleaved products | HCoV-229E | Identifies a 35 kDa (MHV) and a 41 kDa (229E) proteolytic product of pp1ab. Antibodies characterize p35 and p41 localization to the perinuclear regions. | Heusipp et al., JGV (1997) |
| Shi et al., Virology (1999) | |||
| MHV | |||
| Bioinformatic analysis of emerging viruses | Nidoviruses | Predicts that nsp15 encodes an endoribonuclease as a nidovirus-wide genetic marker. | Snijder et al., JMB (2003) |
| Biochemical purification and enzymatic analysis | SARS-CoV | Demonstrates the endoribonuclease activity of nsp15 in vitro. | Ivanov et al., PNAS (2004) |
| Bhardwaj et al., J.Virol (2004) | |||
| HCoV-229E | |||
| MHV | Cao et al., Intervirology (2008) | ||
| IBV | |||
| Turkey CoV | |||
| SARS-CoV | Characterizes nsp15 oligomerization, RNA binding, cleavage preference, and restriction factors. | Guarino et al., JMB (2005) | |
| Bhardwaj et al., JMB (2006) | |||
| Kang et al., J.Virol (2007) | |||
| MHV | |||
| Structural studies of nsp15 | SARS-CoV | Structure studies of nsp15 reveal a unique fold of EndoU. | Bhardwaj et al., JMB (2006) |
| Ricagno et al., PNAS (2006) | |||
| Xu et al., J.Virol (2006) | |||
| MHV | Joseph et al., J.Virol (2007) | ||
| Bhardwaj et al., JBC (2008) | |||
| Evaluating the role of nsp15 in virus replication | MHV | Mutations of the nsp15 active site residues resulted in minimal reduction of viral RNA synthesis and viral titers in fibroblast cell lines. | Kang et al., J.Virol (2007) |
| SARS-CoV | |||
| HCoV-229E | Ulferts and Ziebuhr, RNA Biol (2011) | ||
| Activities of nsp15 detected by overexpression studies | SARS-CoV | Overexpression of nsp15 inhibits the IFN response and MAVS-mediated apoptosis; epitope tagged-nsp15 exhibits diffuse pattern of localization. | Frieman et al., J.Virol (2009) |
| Lei et al., PLoS One (2009) | |||
| Cao and Zhang, Virus Res (2012) | |||
| MHV | |||
| TGEV | |||
| Identifying inhibitors and interacting partners | SARS-CoV | Small molecule inhibitors of Rnase A inhibit EndoU activity and viral replication. | Ortiz-Alcantara et al., Virus Adaptation and Treatment (2012) |
| MHV | |||
| SARS-CoV | Nsp15 interacts with pRb and affects pRb function. | Bhardwaj et al., J.Virol (2012) | |
| MHV | |||
| Evading host sensors | MHV | Nsp15 acts as an IFN antagonist and mediates the evasion of dsRNA sensing; nsp15 mutant viruses exhibit severe replication defects in macrophages. MHV nsp15 mutants are highly attenuated in mice. | Kindler et al., PLoS Pathog (2017) |
| HCoV-229E | Deng et al., PNAS (2017) |
1.1. Identifying nsp15 as a component of the CoV replicase polyprotein
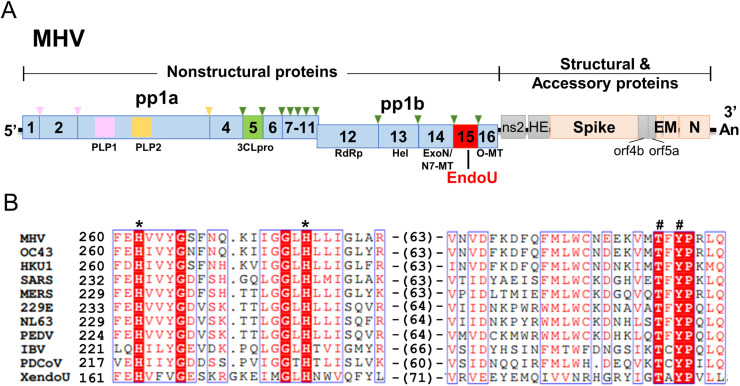
Initial studies focused on identifying the products of the CoV replicase polyprotein, pp1ab (depicted in Fig. 1A). Heusipp et al. used a murine monoclonal antibody (10G11) that recognizes amino acid residues 6158–6164 of human CoV 229E pp1ab (Heusipp et al., 1997). They identified a 41-kDa polypeptide in virus-infected cells and found that this polypeptide was a product of the viral 3C-like protease-mediated cleavage of pp1ab. This protein localizes to the perinuclear region, as detected by immunofluorescence assay, similar to other pp1ab-derived polypeptides (Heusipp et al., 1997). Shi and coworkers obtained similar findings while studying mouse hepatitis virus (MHV) (Shi et al., 1999). They generated a rabbit antiserum against amino acids 6679–6821 of MHV pp1ab and found that this antiserum detected a 35-kDa product in infected cells. They also found that this protein co-localized with de novo synthesized viral RNA, and therefore postulated that this viral protein associated with the viral RNA replication/transcription machinery (Shi et al., 1999). Later, the corresponding polypeptides that these antibodies recognized were defined as the 15th cleavage product of pp1ab (called nsp15), counting from the amino-terminus to the carboxyl terminus of pp1ab (Ziebuhr et al., 2000, Snijder et al., 2003).
Fig. 1.
Coronavirus nsp15 is an endoribonuclease. (A) Schematic diagram of MHV-A59 genome. Triangles indicate cleavage sites recognized by three viral proteases: papain-like proteases PLP1 (pink) and PLP2 (yellow), and 3C-like protease (3CLpro, green). RdRp, RNA-dependent RNA polymerase; Hel, helicase; ExoN, exoribonuclease; N7-MT, guanosine-N7-methyltransferase; EndoU, endoribonuclease; O-MT, O-methyltransferase; HE, Hemagglutinin-Esterase; E, envelope; M, matrix; N, nucleocapsid. (B) Alignment of the core domains of CoV EndoU and XendoU of X. laevis. Putative residues involved in catalysis (*) or substrate specificity (#). Abbreviations: MHV, mouse hepatitis virus; OC43, human CoV OC43; HKU1, human CoV HKU1; SARS, severe acute respiratory syndrome CoV; 229E, human CoV 229E; NL63, human CoV NL63; PEDV, porcine epidemic diarrhea virus; IBV, infectious bronchitis virus; PDCoV, porcine delta CoV; XendU, endoribonuclease of X. laevis.
1.2. Bioinformatic analysis of nidovirus replicase polyproteins
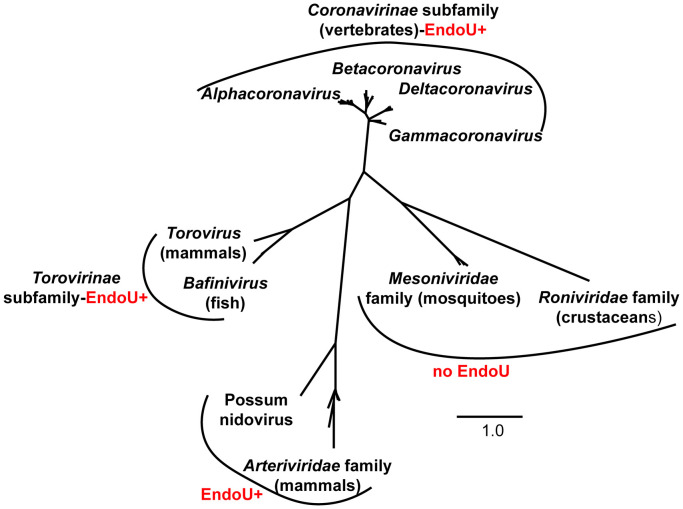
After the outbreak of Severe Acute Respiratory Syndrome (SARS) in 2002–2003, and once a CoV had been confirmed as the etiological agent of SARS, researchers intensively scrutinized CoV genomic sequences to better understand this novel human pathogen. By comparative genomic characterization of CoV replicases, Snijder et al. reported that the C-terminus of nsp15 has high sequence similarity to the Xenopus laevis poly(U)-specific endoribonuclease and therefore predicted that nsp15 possesses EndoU activity (Snijder et al., 2003). Based on the available sequence information of viruses from the order Nidovirales at that time, the EndoU was considered a nidovirus-specific marker (called NendoU) (Fig. 1B) (Snijder et al., 2003). The members of the family Arteriviridae, including equine arteritis virus (EAV) and porcine respiratory and reproductive syndrome virus (PRRSV), also have EndoU domains within nsp11. However, it was later discovered that the presence of the EndoU domain is not universal in all nidoviruses. Nam Dinh virus, the first insect nidovirus belonging to the family Mesoniviridae, and roniviruses that infect invertebrates, do not encode an EndoU domain (Nga et al., 2011, Lauber et al., 2012). These findings suggest that the EndoU domain may only serve as a signature for vertebrate nidoviruses, including CoVs and arteriviruses ( Fig. 2).
Fig. 2.
Virus-encoded endoribonuclease is a genetic signature of nidoviruses that infect vertebrates. A phylogenic tree of 32 representative nidoviruses was generated based on a conserved region of RdRp (Sequnces and Genbank Assession numbers are available upon request). Multiple sequence alignment and phylogeny analyses were conducted with the programs MUSCLE and PhyML, respectively (available at http://www.phylogeny.fr/). The phylogenic tree was generated using Dendroscope software version 3 with default parameters.
1.3. Biochemical purification and enzymatic analysis of EndoU
Previous reviews have comprehensively summarized the biochemical and structural features of nidovirus ribonucleases (Ulferts and Ziebuhr, 2011, Snijder et al., 2016). Here, we highlight the key experiments with respect to nidovirus EndoU and provide recent updates. Studies performed by Ivanov et al. (2004) and Bhardwaj et al. (2004) first demonstrated the EndoU activity of SARS-CoV nsp15 in vitro ( Ivanov et al., 2004; Bhardwaj et al., 2004). The wild-type (WT) nsp15 and its mutants with alanine (Ala) substitutions of the putative catalytic residues were expressed in E. coli. The recombinant nsp15-WT, but not the mutants, could efficiently cleave single-stranded (ss) and double-stranded (ds) RNAs. In contrast, neither ssDNA nor dsDNA molecules could be processed by nsp15, demonstrating its predicted ribonuclease—as opposed to deoxyribonuclease—activity. Blocking either the 5′ or the 3′ terminus of the RNA substrates by covalent modifications with fluorescein or puromycin, respectively, had no effect on the RNA cleavage (Bhardwaj et al., 2004), confirming that nsp15 is an endo- rather than an exoribonuclease. Similar EndoU activity was detected in other CoV nsp15s, including human CoV 229E, MHV, infectious bronchitis virus (IBV), and turkey CoV (Ivanov et al., 2004, Bhardwaj et al., 2004, Cao et al., 2008), and in the nsp11s of arteriviruses EAV and PRRSV (Nedialkova et al., 2009).
Hexamerization of nsp15 is critical for its EndoU activity. Guarino and coworkers found that CoV nsp15 could be present in solution as either monomers or hexamers in a protein concentration-dependent manner. The hexamer is the fully active form of EndoU that binds RNA and executes optimal EndoU activity (Guarino et al., 2005). The residues in the N-terminal domain of nsp15 are critical for hexamer formation (Guarino et al., 2005). In addition, nsp15 requires divalent metal ions as a co-factor for RNA cleavage and prefers Mn2+ over Mg2+ or other divalent cations (Ivanov et al., 2004, Bhardwaj et al., 2004). Addition of Mn2+ significantly affects the protein conformation, enhances RNA binding, and increases EndoU activity (Ivanov et al., 2004; Bhardwaj et al., 2004). In contrast to CoV nsp15, arterivirus nsp11 forms dimers in solution and does not require divalent cations as a cofactor for activity in vitro (Nedialkova et al., 2009, Shi et al., 2016). Instead, a concentration of Mn2+ that greatly stimulated the activity of CoV nsp15 inhibited the EndoU activity of EAV and PRRSV nsp11s.
CoV EndoU was revealed biochemically to hydrolyze the 3′ end of pyrimidines, with a preference for uridylates, and release products with 2′, 3′-cyclic phosphate and 5′-hydroxyl ends (Ivanov et al., 2004, Bhardwaj et al., 2004). This finding seems to implicate a broad range of targets; however, the EndoU activity of CoV nsp15 can be affected by secondary structure and modification of the RNA substrate. Bhardwaj et al. found that nsp15 preferentially cleaved unpaired uridylates in hairpin-structured RNAs and that the neighboring nucleotides of targeted sites also influenced hydrolysis (Bhardwaj et al., 2006). On the other hand, Ivanov et al. found that 2′-O-ribose methyl groups present on the substrate RNA blocked EndoU-mediated cleavage (Ivanov et al., 2004). These data suggest that multiple factors might limit the range of EndoU targets. This is reasonable because the EndoU activity of nsp15 is likely to be tightly regulated during infection in cells to avoid unwanted cleavage on viral and/or cellular targets. For example, CoV nsp16 is a 2′-O-ribose methyltransferase, whose function could theoretically block the EndoU-mediated cleavage of viral RNAs. Similar to CoV nsp15, arterivirus nsp11 also prefers 3′ of uridylates for cleavage and yields products with 2′, 3′-cyclic phosphate ends (Nedialkova et al., 2009). Further studies are needed to address whether the EndoU activity of arterivirus nsp11 is restricted by RNA modifications or secondary structures.
1.4. Structural studies reveal unique features of CoV EndoU
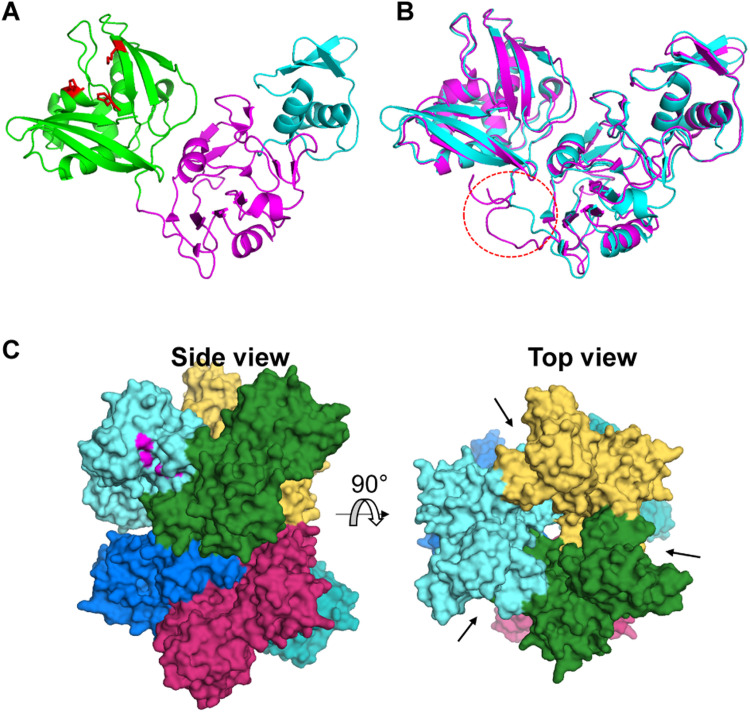
Guarino and coworkers visualized single particles of SARS-CoV nsp15 using electron microscopy (Guarino et al., 2005) and the same group later reported an 8.3 Å structure of nsp15 by cryoelectron microscopy (Bhardwaj et al., 2006). They reported that the nsp15 hexamer comprises a dimer of trimers and proposed that the RNA substrate binds to the inter-trimer interface. X-ray crystal structures of SARS-CoV and MHV nsp15s were first solved by Ricagno et al. and Xu et al., respectively (Ricagno et al., 2006; Xu et al., 2006). These high-resolution structures define CoV nsp15 as a separate EndoU family with unique folds that differ from cellular endoribonucleases ( Fig. 3A). The monomer of SARS-CoV nsp15 consists of three domains: a small N-terminal domain (NTD) (residues 1–62, cyan), a middle domain (residues 63–191, magenta), and a large C-terminal domain (CTD) (residues192–345, green). The EndoU is located in the CTD. The monomeric structure of MHV nsp15 can be superimposed onto that of SARS-CoV nsp15, except for a flexible loop structure in the middle domain of MHV nsp15 (Fig. 3B). This flexible loop is encoded by a viral RNA packaging signal sequence, which is present in MHV nsp15 but not in SARS-CoV nsp15 (Kuo and Masters, 2013). These structural studies demonstrated that the presence of the flexible loop did not alter the overall folding of MHV nsp15 relative to SARS-CoV nsp15. In addition, through these and other structural studies (Ricagno et al., 2006, Xu et al., 2006, Joseph et al., 2007, Bhardwaj et al., 2008), the nsp15 hexamer was again confirmed to be a dimer of trimers. As shown in Fig. 3C, three monomers form a trimer and two trimers interact head-to-head to form a hexamer. The NTDs line up in the center of the hexamer, while the CTDs face outward. This architecture allows the nsp15 hexamer to possess six active sites. The extensive contact between subunits through the NTD and middle domain is critical for hexamerization. The crystal structure of arterivirus nsp11 was also recently reported (Shi et al., 2016, Zhang et al., 2017). These structural studies revealed that the monomer of nsp11 contains only two domains: NTD and CTD (no middle domain). The monomeric structures of CoV nsp15 and arterivirus nsp11 could not be superimposed except in the CTD (catalytic domain). Distinct from the hexameric structure of CoV nsp15, nsp11 monomers assembled into an asymmetric dimer (Shi et al., 2016).
Fig. 3.
Structural features of nsp15. (A) The three domains of the SARS-CoV nsp15 monomer (Protein Data Bank code: 2H85): N-terminal domain (cyan), middle domain (magenta), and C-terminal domain (green). Catalytic residues are shown in red. (B) Structural comparison of SARS-CoV nsp15 (cyan) and MHV nsp15 (magenta, 2GTH). A flexible loop encoded by the packaging signal sequence of MHV nsp15 is circled. (C) Hexamer of SARS-CoV nsp15 (2RHB). The catalytic residues of one monomer are highlighted in magenta (left); the catalytic pockets of a hexamer are indicated by arrows (right).
1.5. EndoU activity can be inhibited by small molecules
Since the crystallographic studies of CoV nsp15 revealed that the active site of EndoU is structurally similar to that of RNase A, researchers evaluated small molecule inhibitors of RNase A for their ability to inhibit nsp15 activity. Ortiz-Alcantara et al. tested several commercially available RNase A inhibitors (Benzopurpurin B, Congo red, and others) and reported that their 50% inhibitory concentration (IC50) values for inhibiting the EndoU activity of SARS-CoV nsp15 ranged from 0.2 μM to 40 μM. Benzopurpurin B was shown to have a broad-spectrum activity and could inhibit nsp15 orthologs from MHV and IBV with IC50 values of 0.4 μM and 0.2 μM, respectively (Ortiz-Alcantara et al., 2010). These RNase A inhibitors had variable effects on CoV replication in cell cultures. In plaque formation assays, treatment with Congo red resulted in 27-fold and 4-fold reductions in MHV and SARS-CoV titers, respectively, while Benzopurpurin B led to marginal inhibition of both CoVs (Ortiz-Alcantara et al., 2010). Although the impact of these RNase A inhibitors on CoV replication requires more comprehensive investigation, these early results suggest that the similarity between nsp15/EndoU and RNase A may provide a basis for exploiting small molecule inhibitors to modulate viral EndoU activity.
1.6. Evaluating the role of nsp15 in coronavirus replication
Due to its unique enzymatic activity and co-localization with the replicating viral RNA, nsp15/EndoU was initially thought to play an important role in virus replication. However, MHV encoding catalytic-defective EndoU exhibited only a subtle defect in RNA synthesis and a slight reduction in viral titers (~ 1 log) compared to WT virus when evaluated in fibroblasts (Kang et al., 2007). Similar results were obtained for SARS-CoV and HCoV-229E nsp15 mutants generated using either reverse genetics or replicon systems (Ulferts and Ziebuhr, 2011). These data indicate that the EndoU activity of nsp15 is not essential for CoV replication, as was initially proposed (Snijder et al., 2003, Ivanov et al., 2004, Bhardwaj et al., 2004). Intriguingly, nsp15 may indirectly affect virus replication through other mechanisms. Ivanov et al. demonstrated that when the conserved aspartic acid (Asp)-298 of HCoV-229E nsp15 was replaced with an Ala, its EndoU activity was eliminated, viral RNA synthesis was completely abolished, and no viable virus was recovered (Ivanov et al., 2004). Similar phenotypes were observed for an MHV nsp15 mutant with an Ala substitution of Asp-324 (equivalent to Asp-298 of HCoV-229E) (Kang et al., 2007). It is unclear how the Asp residues affect viral RNA synthesis. Kang et al. predicted that Asp-324 is critical for an ionic-bond network and observed that the Ala substitution resulted in an insoluble protein when MHV nsp15 was expressed in E. coli ( Kang et al., 2007 ). These data suggest that the Ala substitution may prevent the nsp15 protein from folding correctly. Since the proteins adjacent to nsp15 are critical replicative components, any protein-folding issue with nsp15 may have an effect on the proteolytic processing of the neighboring components, thereby leading to a nonviable phenotype. Overall, current evidence indicates that the EndoU activity of CoV nsp15 is dispensable for viral RNA synthesis and virus replication in cell culture. Further work is required to address any non-EndoU-mediated role of nsp15 protein in virus replication.
Mutagenesis of arterivirus nsp11 revealed pleiotropic effects of EndoU on the viral life cycle (Posthuma et al., 2006, Sun et al., 2016). Similar to CoV nsp15, mutations in the Asp-3014 and Asp-3038 residues (corresponding to MHV nsp15 Asp-324 and Asp-351, respectively) in EAV nsp11 resulted in a nonviable phenotype. Compared with the mild effect of mutating the catalytic histidine (His) residues (His262Ala and His277Ala) of MHV nsp15, EAV infectious clones with catalytic residue mutations (His2963Ala/Gln and His2978Ala/Gln) exhibited smaller plaque sizes, reduced RNA synthesis, and dramatic titer reductions up to 5 log units. Other substitutions at conserved, non-catalytic residues of EAV nsp11 resulted in an intermediate phenotype: intermediate plaque sizes and ~1–2 log reduction in titers. Similar results were obtained with the PRRSV nsp11 mutant viruses (Sun et al., 2016). These data indicate that arterivirus nsp11 may be involved in viral RNA synthesis. However, similar to the Asp324Ala mutation of MHV nsp15, both Asp3014Ala and Asp3038Ala mutations in EAV nsp11 rendered the protein insoluble, again raising the question of whether or not these mutations influence viral RNA synthesis through interfering with proteolytic processing of the ORF1b polyprotein. The absence of an EndoU domain in insect nidoviruses and invertebrate roniviruses further indicates that EndoU activity is not required for the unique RNA synthesis strategy of nidoviruses (Nga et al., 2011, Lauber et al., 2012).
The aforementioned studies highlight the extensive efforts of the field to investigate the characteristics of nsp15/EndoU and its role in virus life cycle. The EndoU activity of CoV nsp15 was found to play a non-essential role in viral RNA synthesis and replication in immortalized fibroblast cells. Recent work with different cell types and in vivo experiments revealed a novel function of nsp15/EndoU in virus replication and pathogenesis and provided a new direction of study with respect to this “old” protein.
1.7. Ectopic expression of nsp15 reveals antagonism of reporters
CoV nsp15 was first suggested to possess interferon (IFN) antagonism capabilities through ectopic expression experiments. Frieman et al. used an alphavirus replication-defective vector (VRP) to screen SARS-CoV proteins that suppress VRP-induced IFN responses. One of the identified IFN antagonists of SARS-CoV was nsp15 (Frieman et al., 2009). Later, arterivirus nsp11 was also identified as an IFN antagonist and its EndoU activity was found to mediate inhibition of IFN-beta induction (Beura et al., 2010, Shi et al., 2011). Several other studies also reported that CoV nsp15 and arterivirus nsp11 inhibit cellular innate responses in ectopic expression experiments (Lei et al., 2009, Wang et al., 2015). These data seem to indicate that these two proteins function as IFN antagonists; however, the EndoU activity of overexpressed nsp15/nsp11 may unexpectedly affect the activities of the reporters used in these assays. We found that transfected MHV nsp15 reduced the signal of both IFN-reporter firefly luciferase and the internal control renilla luciferase (Hackbart M, Deng X, and Baker S, unpublished data). A similar result was obtained with the overexpressed PRRSV nsp11 (Shi et al., 2016). These observations imply that nsp15/nsp11 may execute non-specific cleavage when ectopically expressed. Since CoV nsp15 is part of the viral replicase/transcriptase complex (RTC), it is reasonable to predict that its EndoU activity is tightly regulated during viral infection to avoid unwanted cleavage. In line with this prediction, we and others reported a specific, punctate, perinuclear localization of CoV nsp15 during viral infection (Athmer, 2017, Deng, 2017, Heusipp et al., 1997, Shi et al., 1999), while ectopically expressed nsp15 was distributed throughout the cytoplasm (Cao and Zhang, 2012). Hence, we advise caution when interpreting the results of overexpression studies, as the nature of the EndoU activity was only revealed after studying nsp15/nsp11 in the context of viral infection.
1.8. EndoU is important for evading host sensors of dsRNA
It was first discovered that the EndoU activity of nsp15 mediates the evasion of host recognition of viral dsRNA by infecting primary macrophages with EndoU-deficient CoVs (Deng, 2017, Kindler et al., 2017). Infection with MHV EndoU-deficient mutants stimulated mouse bone-marrow derived macrophages (BMDMs) to produce a remarkably high level of type I IFNs during the early phase of infection compared to WT infection. This IFN response is MDA5-dependent, as both the IFN mRNA and protein levels were not elevated in MDA5-deficient BMDMs. Moreover, the replication of EndoU-deficient CoVs was severely impaired in primary macrophages. Interestingly, the IFN-induced antiviral response is not the only player responsible for this replication defect, as the titers of the EndoU-deficient CoVs were not completely restored in BMDMs that lack critical genes (e.g. MDA5, MAVS, and IRF3/5/7) involved in the IFN response (Deng, 2017, Kindler et al., 2017). These data suggest that other antiviral pathways may also contribute to the observed replication defect. In support of this, it was found that the infection of EndoU-deficient CoVs also activate the PKR and the OAS-RNase L pathways (Deng, 2017, Kindler et al., 2017), which both execute potent antiviral functions, discussed further below. Indeed, the replication of EndoU-deficient CoVs could be partially restored in PKR/RNase L-double knockout cells (Kindler et al., 2017). Replication was only fully restored in type I IFN receptor-knockout macrophages, as these cells not only have a defect in IFN signaling but also express very low basal levels of PKR and OAS relative to WT cells (Deng, 2017, Birdwell et al., 2016). Taken together, these studies using live viruses in primary cells effectively illustrated the IFN antagonistic properties of CoV EndoU.
As mentioned above, infection with EndoU-deficient CoVs also activates the PKR and OAS-RNase L pathways in macrophages (Deng, 2017, Kindler et al., 2017). PKR is a dsRNA-activated protein kinase and serves as a dsRNA sensor. Activated PKR phosphorylates eukaryotic initiation factor 2α (eIF2α), resulting in inhibition of host and viral mRNA translation. Thus, PKR-mediated translation shutoff plays an important role in the host antiviral defense (Barber, 2005). The EndoU-deficient CoV-infected macrophages exhibited increased levels of phosphorylated eIF2α and decreased levels of translation (Deng, 2017, Kindler et al., 2017), indicating that PKR was activated during infection. Another piece of evidence of PKR activation was that EndoU-deficient CoVs induced rapid apoptotic cell death in infected-macrophages (Deng et al., 2017). It has been shown that PKR-mediated translation shutoff leads to apoptosis in macrophages (Hsu et al., 2004). When the EndoU-deficient CoV-infected macrophages were treated with a PKR-specific inhibitor (C16), the level of apoptosis was significantly reduced (Deng et al., 2017). This result further supports the hypothesis that EndoU-deficient CoVs activate PKR. Interestingly, loss of PKR expression or inhibition of its activity only partially restored the replication of EndoU-deficient CoVs in macrophages (Kindler et al., 2017). Moreover, treatment with the PKR inhibitor did not affect IFN induction or RNase L-mediated ribosomal RNA degradation in the EndoU-deficient CoV infected-macrophages (Deng et al., unpublished data). These results imply that the infection of EndoU-deficient CoV activates multiple host dsRNA sensors independently, including MDA5, PKR, and OAS.
OAS is a protein family of 2′, 5′-oligoadenylate (2–5A) synthetases. Upon activation, OAS can synthesize 2–5A, which binds to and activates RNase L. RNase L is a host ribonuclease that executes global degradation of host and viral RNAs. Thus, OAS and RNase L constitute a potent host antiviral system. Macrophages infected by the EndoU-deficient CoVs exhibited an early, RNase L-mediated degradation of ribosomal RNA, demonstrating that the OAS-RNase L system was activated (Deng, 2017, Kindler et al., 2017). Lack of MDA5 expression or treatment with the PKR inhibitor did not affect virus-induced RNA degradation (Deng, 2017, Kindler et al., 2017), suggesting that the nsp15-mediated blockage of OAS-RNase L activation is independent of the MDA5-IFN and PKR pathways. Loss of RNase L expression does not restore the replication of EndoU-deficient CoVs in macrophages. Taken together, these results suggest again that multiple antiviral pathways, including MDA5-IFN, PKR, and the OAS-RNase L system, were activated during infection with EndoU-deficient CoVs. The antiviral defense executed by these pathways contribute together to the replication defect of EndoU-deficient CoVs in macrophages. Interestingly, some CoVs encode a 2′, 5′-phosphodiesterase (PDE) (e.g. MERS-CoV orf4b and MHV ns2) (Thornbrough et al., 2016, Zhao et al., 2012). This CoV PDE also prevents RNase L-mediated RNA degradation through digesting the 2–5A produced by OAS. Thus, the presence of two antagonists of the OAS-RNase L system in some CoV genomes represents a functional redundancy or tissue-specific roles for these antagonists. In fact, not all CoVs encode a PDE, and it has been reported that loss of the PDE activity mitigated MHV pathogenicity in the mouse liver but not in the brain, suggesting a liver-specific effect of CoV PDE activity in vivo (Roth-Cross et al., 2009; Zhao et al., 2011).
1.9. Lack of phenotype of EndoU-deficient CoVs in fibroblast cell lines
Several EndoU-deficient CoVs have been evaluated in fibroblast cell lines and no marked phenotypes were obtained (mild reduction of viral RNA synthesis and replication) (discussed above). We speculate that the activation of an IFN response and apoptosis in macrophages is due to high levels of basal expression of host sensors, such as MDA5, PKR, and OAS (Deng, 2017, Birdwell et al., 2016). Indeed, when the expression of OAS was induced by pre-treatment with IFN, the immortalized fibroblast cells also exhibited RNA degradation upon infection with EndoU-deficient CoV (Kindler et al., 2017). In line with this, without the stimulation of IFN, constitutively expressed OAS and PKR in the MDA5-deficient macrophages is capable of sensing the EndoU-deficient CoV infection and implementing antiviral processes (Deng et al., 2017). Although the production of IFN is dispensable for the activation of the OAS-RNase L system and PKR, type I IFN receptors or a direct signal is required for maintaining the high basal expression of these IFN-inducible genes (Deng, 2017, Birdwell et al., 2016). This is biologically relevant because macrophages and other myeloid cells are quick responders to early virus infection, responding even before IFN is highly induced. It is unclear whether other cell types, such as epithelial cells, behave similar to macrophages, but at least mouse embryonic fibroblasts have been shown to display the nsp15/EndoU-mediated effects (Kindler et al., 2017).
1.10. MHV EndoU-deficient mutants are highly attenuated in mice
Due to robust activation of antiviral responses, MHV EndoU-deficient mutants also exhibited a marked attenuation in vivo relative to MHV-WT. Depending on the inoculation route, MHV infection can result in hepatitis or encephalitis in C57/BL6 mice. Strikingly, regardless of which infection route (intraperitoneal or intracranial injection) was used, we found that MHV EndoU-deficient mutants were highly attenuated (Deng et al., 2017). When mice were inoculated intraperitoneally using a high dose of the mutant virus, there was no detectable viral titer in the liver or spleen at day 3 post-infection and only a minimal detection of viral mRNA in mesenteric lymph nodes at day 1 post-infection. When a sensitive encephalitis model was used, mice infected with MHV EndoU-deficient mutants exhibited only a transient loss of body weight and recovered completely. This significant attenuation is attributed to the loss of EndoU-mediated evasion of host antiviral defenses. We found that EndoU-deficient mutants maintained WT-level virulence in type I IFN receptor knockout (IFNAR-/-) mice (Deng et al., 2017). Similar results obtained by Kindler et al. showed that the mutant virus was only detected in the organs from IFNAR-/- mice but not wild-type or other gene-deficient (MDA5-/-, TLR7-/-, and MDA5-/-/TLR7-/-) mice at day 2 post-infection (Kindler et al., 2017). Interestingly, even though MHV EndoU-deficient mutants were highly attenuated and exhibited limited replication in vivo, the pre-infected mice were protected from subsequent lethal challenges of wild-type MHV in both disease models (Deng et al., 2017). These results demonstrate that nsp15 plays an important role in virus pathogenesis and illustrate the potential use of EndoU-deficient CoVs as vaccine candidates.
1.11. How does nsp15 mediate the evasion of host sensors?
As an EndoU, it is plausible that nsp15 may degrade viral dsRNA to prevent host recognition. Previous studies of pestivirus envelope glycoprotein (Erns) and Lassa virus nucleoprotein linked viral ribonuclease activity to type I IFN antagonism through degrading viral dsRNA (Python et al., 2013, Qi et al., 2010, Hastie et al., 2011), although no direct evidence for this linkage has been obtained. For CoV nsp15, Kindler et al. detected an increased level of dsRNA in EndoU mutant-infected cells by flow cytometry using a dsRNA-specific monoclonal antibody (mAb) (Kindler et al., 2017). This mAb recognizes dsRNA molecules longer than 40 bp in length, regardless of the sequence, meaning that a long dsRNA could potentially bind multiple mAb molecules. To saturate the mAb-dsRNA binding, we tested serial concentrations of antibody but did not detect any significant change of dsRNA levels by either flow cytometry or immunofluorescence analysis (Deng et al., 2017). The discrepancy between these two studies may be ascribed to the experimental settings in respective experiments. However, it is also possible that the methods used in these studies may not be sensitive enough to detect changes in dsRNA levels if the WT-nsp15 produces dsRNA cleavage products that are >40 bp, such that the uncleaved and cleaved dsRNAs are indistinguishable for binding by the anti-dsRNA antibody. Importantly, it has been demonstrated that the EndoU activity of CoV nsp15 is influenced by secondary structures and modifications of dsRNA (Ivanov et al., 2004, Bhardwaj et al., 2006). Because these factors may limit the number of cleavage events and/or the targets of EndoU, nsp15-mediated cleavage may produce few overall cleavage products that are <40 bp. Consequently, nsp15-mediated cleavage may not reduce the level of total dsRNA in the cell, but rather may hydrolyze long dsRNAs into shorter cleavage products that are sufficiently short to evade recognition by host sensors (e.g. MDA5) but not by the anti-dsRNA antibody.
It has been documented that CoV dsRNAs form cytoplasmic aggregated foci during replicating in cells (Becares et al., 2016, Hagemeijer et al., 2012). These dsRNA foci co-localize with the viral RTCs early during infection. Interestingly, we noted that during the early stages of infection, dsRNA foci were not co-localized with the RTCs in the EndoU-deficient CoV-infected cells. This decrease in co-localization relative to WT infection resulted in a dispersed pattern of dsRNA foci, such that some dsRNAs did not appear to associate with the RTCs. It is not known whether these “free” dsRNAs trigger host sensors, but this dispersed distribution of dsRNA does notably coincide with early activation of the IFN response and other dsRNA sensors (e.g. PKR and OAS). Overall, additional studies with new methods are needed to characterize the intracellular localizations and the fates of dsRNA in CoV-infected cells.
2. Significance and perspectives
The detailed strategy used by CoV EndoU to evade host recognition remains enigmatic. More studies are needed to address several key questions: (i) What is the natural target of EndoU? (ii) How does the EndoU activity of nsp15 alter the fate of dsRNA? (iii) How is this EndoU activity regulated to avoid unwanted cleavage events? (iv) Do any interaction partners (viral or cellular) of nsp15 participate in regulating its EndoU activity (Athmer, 2017, Bhardwaj et al., 2012)? Answers to these questions will be essential for understanding the mechanism of the EndoU-mediated evasion of host dsRNA sensors. Additionally, it is possible that EndoU serves as a conserved antagonist in vertebrate nidoviruses. A similar phenotype has been observed in human blood-derived macrophages infected with the HCoV-229E nsp15 mutant virus (Kindler et al., 2017), which is a representative alphacoronavirus. Whether arterivirus nsp11 also functions as an IFN antagonist during infection is still unknown. One obstacle to studying arterivirus nsp11 mutants in cell culture is that mutations of catalytic residues have been reported to severely affect viral replication even in IFN-deficient cells (Posthuma et al., 2006, Sun et al., 2016). Nonetheless, given the striking roles of CoV EndoU in macrophages and in vivo, it will be important to understand the underlying mechanism of EndoU-mediated evasion of host antiviral defenses in order to advance an ultimate goal of exploiting this enzyme as a target for therapeutics and vaccine design against existing and emerging pathogenic CoVs.
Acknowledgements
We thank Robert C. Mettelman and Aaron Volk for assistance with editing the manuscript. Our studies are supported by National Institutes of Health R01 AI085089 (to SCB).
References
- Athmer J. In situ tagged nsp15 reveals interactions with coronavirus replication/transcription complex-associated proteins. MBio. 2017;8(1) doi: 10.1128/mBio.02320-16. (e02320-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber G. The dsRNA-dependent protein kinase, PKR and cell death. Cell Death Differ. 2005;12:563–570. doi: 10.1038/sj.cdd.4401643. [DOI] [PubMed] [Google Scholar]
- Becares M. Mutagenesis of coronavirus nsp14 reveals its potential role in modulation of the innate immune response. J. Virol. 2016;90(11):5399–5414. doi: 10.1128/JVI.03259-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beura L.K. Porcine reproductive and respiratory syndrome virus nonstructural protein 1beta modulates host innate immune response by antagonizing IRF3 activation. J. Virol. 2010;84(3):1574–1584. doi: 10.1128/JVI.01326-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj K. Structural and functional analyses of the severe acute respiratory syndrome coronavirus endoribonuclease Nsp15. J. Biol. Chem. 2008;283(6):3655–3664. doi: 10.1074/jbc.M708375200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj K., Guarino L., Kao C.C. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J. Virol. 2004;78(22):12218–12224. doi: 10.1128/JVI.78.22.12218-12224.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj K., Sun J., Holzenburg A., Guarino L.A., Kao C.C. RNA recognition and cleavage by the SARS coronavirus endoribonuclease. J. Mol. Biol. 2006;361(2):243–256. doi: 10.1016/j.jmb.2006.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj K., Liu P., Leibowitz J.L., Kao C.C. The coronavirus endoribonuclease Nsp15 interacts with retinoblastoma tumor suppressor protein. J. Virol. 2012;86(8):4294–4304. doi: 10.1128/JVI.07012-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birdwell L.D. Activation of RNase L by murine coronavirus in myeloid cells is dependent on basal Oas gene expression and independent of virus-induced interferon. J. Virol. 2016;90(6):3160–3172. doi: 10.1128/JVI.03036-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Zhang X. Comparative in vivo analysis of the nsp15 endoribonuclease of murine, porcine and severe acute respiratory syndrome coronaviruses. Virus Res. 2012;167(2):247–258. doi: 10.1016/j.virusres.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Wu C.-C., Lin T.L. Turkey coronavirus non-structure protein NSP15 – An endoribonuclease. Intervirology. 2008;51(5):342–351. doi: 10.1159/000175837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc. Natl. Acad. Sci. USA. 2017;114(21):E4251–E4260. doi: 10.1073/pnas.1618310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M., Ratia K., Johnston R.E., Mesecar A.D., Baric R.S. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-kappaB signaling. J. Virol. 2009;83(13):6689–6705. doi: 10.1128/JVI.02220-08. (13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarino L.A. Mutational analysis of the SARS virus Nsp15 endoribonuclease: identification of residues affecting hexamer formation. J. Mol. Biol. 2005;353(5):1106–1117. doi: 10.1016/j.jmb.2005.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeijer M.C., Vonk A.M., Monastyrska I., Rottier P.J.M., de Haan C.A.M. Visualizing coronavirus RNA synthesis in time by using click chemistry. J. Virol. 2012;86(10):5808–5816. doi: 10.1128/JVI.07207-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie K.M. Crystal structure of the Lassa virus nucleoprotein-RNA complex reveals a gating mechanism for RNA binding. Proc Natl. Acad. Sci. 2011;108(48):19365–19370. doi: 10.1073/pnas.1108515108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heusipp G. Identification and subcellular localization of a 41 kDa, polyprotein 1ab processing product in human coronavirus 229E-infected cells. J. Gen. Virol. 1997;78(11):2789–2794. doi: 10.1099/0022-1317-78-11-2789. [DOI] [PubMed] [Google Scholar]
- Hsu L.-C. The protein kinase PKR is required for macrophage apoptosis after activation of Toll-like receptor 4. Nature. 2004;428(6980):341–345. doi: 10.1038/nature02405. [DOI] [PubMed] [Google Scholar]
- Ivanov K.A. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc Natl. Acad. Sci. USA. 2004;101(34):12694–12699. doi: 10.1073/pnas.0403127101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph J.S. Crystal structure of a monomeric form of severe acute respiratory syndrome coronavirus endonuclease nsp15 suggests a role for hexamerization as an allosteric switch. J. Virol. 2007;81(12):6700–6708. doi: 10.1128/JVI.02817-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H. Biochemical and genetic analyses of murine hepatitis virus Nsp15 endoribonuclease. J. Virol. 2007;81(24):13587–13597. doi: 10.1128/JVI.00547-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler E. Early endonuclease-mediated evasion of RNA sensing ensures efficient coronavirus replication. PLoS Pathog. 2017;13(2):e1006195. doi: 10.1371/journal.ppat.1006195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L., Masters P.S. Functional analysis of the murine coronavirus genomic RNA packaging signal. J. Virol. 2013;87(9):5182–5192. doi: 10.1128/JVI.00100-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber C. Mesoniviridae: a proposed new family in the order Nidovirales formed by a single species of mosquito-borne viruses. Arch. Virol. 2012;157(8):1623–1628. doi: 10.1007/s00705-012-1295-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y. MAVS-mediated apoptosis and its inhibition by viral proteins. PLoS One. 2009;4(5):e5466. doi: 10.1371/journal.pone.0005466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedialkova D.D. Biochemical characterization of arterivirus nonstructural protein 11 reveals the nidovirus-wide conservation of a replicative endoribonuclease. J. Virol. 2009;83(11):5671–5682. doi: 10.1128/JVI.00261-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nga P.T. Discovery of the first insect nidovirus, a missing evolutionary link in the emergence of the largest RNA Virus genomes. PLoS Pathog. 2011;7(9):e1002215. doi: 10.1371/journal.ppat.1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Alcantara J. Small molecule inhibitors of the SARS-CoV Nsp15 endoribonuclease. Virus Adapt. Treat. 2010;2(1):125–133. [Google Scholar]
- Posthuma C.C. Site-directed mutagenesis of the Nidovirus replicative endoribonuclease NendoU exerts pleiotropic effects on the arterivirus life cycle. J. Virol. 2006;80(4):1653–1661. doi: 10.1128/JVI.80.4.1653-1661.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Python S., Gerber M., Suter R., Ruggli N., Summerfield A. Efficient sensing of infected cells in absence of virus particles by blasmacytoid dendritic cells is blocked by the viral ribonuclease Erns. PLoS Pathog. 2013;9(6):e1003412. doi: 10.1371/journal.ppat.1003412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X. Cap binding and immune evasion revealed by Lassa nucleoprotein structure. Nature. 2010;468(7325):779–783. doi: 10.1038/nature09605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricagno S. Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proc. Natl. Acad. Sci. 2006;103(32):11892–11897. doi: 10.1073/pnas.0601708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth-Cross J.K. Organ-specific attenuation of murine hepatitis virus strain A59 by replacement of catalytic residues in the putative viral cyclic phosphodiesterase ns2. J. Virol. 2009;83(8):3743–3753. doi: 10.1128/JVI.02203-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S.T. Colocalization and membrane association of murine hepatitis virus gene 1 products and De novo-synthesized viral RNA in infected cells. J. Virol. 1999;73(7):5957–5969. doi: 10.1128/jvi.73.7.5957-5969.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X. Endoribonuclease activities of porcine reproductive and respiratory syndrome virus nsp11 was essential for nsp11 to inhibit IFN-β induction. Mol. Immunol. 2011;48(12–13):1568–1572. doi: 10.1016/j.molimm.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. A dimerization-dependent mechanism drives the endoribonuclease function of porcine reproductive and respiratory syndrome virus nsp11. J. Virol. 2016;90(9):4579–4592. doi: 10.1128/JVI.03065-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331(5):991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Decroly E., Ziebuhr J. The nonstructural proteins directing coronavirus RNA synthesis and processing. Adv. Virus Res. 2016:59–126. doi: 10.1016/bs.aivir.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. Nonstructural protein 11 of porcine reproductive and respiratory syndrome virus suppresses both MAVS and RIG-I expression as one of the mechanisms to antagonize Type I interferon production. PLoS One. 2016;11(12):e0168314. doi: 10.1371/journal.pone.0168314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornbrough J.M. Middle East respiratory syndrome coronavirus NS4b protein inhibits host RNase L activation. MBio. 2016;7(2) doi: 10.1128/mBio.00258-16. (e00258-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulferts R., Ziebuhr J. Nidovirus ribonucleases: structures and functions in viral replication. RNA Biol. 2011;8(2):295–304. doi: 10.4161/rna.8.2.15196. [DOI] [PubMed] [Google Scholar]
- Wang D. The nonstructural protein 11 of porcine reproductive and respiratory syndrome virus inhibits NF-κB signaling by means of its deubiquitinating activity. Mol. Immunol. 2015;68(2):357–366. doi: 10.1016/j.molimm.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X. New antiviral target revealed by the hexameric structure of mouse hepatitis virus nonstructural protein nsp15. J. Virol. 2006;80(16):7909–7917. doi: 10.1128/JVI.00525-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M. Structural biology of the arterivirus nsp11 endoribonucleases. J. Virol. 2017;91(1):e01309–e01316. doi: 10.1128/JVI.01309-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L. Antagonism of the interferon-induced OAS-RNase L pathway by murine coronavirus ns2 protein is required for virus replication and liver pathology. Cell Host Microbe. 2012;11(6):607–616. doi: 10.1016/j.chom.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Rose K.M., Elliott R., Van Rooijen N., Weiss S.R. Cell-type-specific type I interferon antagonism influences organ tropism of murine coronavirus. J. Virol. 2011;85(19):10058–10068. doi: 10.1128/JVI.05075-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J., Snijder E.J., Gorbalenya A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000;81(4):853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]