Abstract
Combining peroxidase activity based haem-staining (TMBZ/SDS-PAGE) with mass spectrometry analyses (Nano LC-MS/MS) of protein extracts from wild type and appropriate mutants, we provide evidence that the polychlorinated biphenyl (PCB) degrader Pseudomonas pseudoalcaligenes KF707 primarily expresses a caa3-type cytochrome c oxidase (caa3-Cox) using cytochrome (cyt) c4 as an electron donor in cells grown with biphenyl versus glucose as the sole carbon source. Homology modeling of KF707 caa3-Cox using the three-dimensional structure of that from Thermus thermophiles highlights multiple similarities and differences between the proton channels in subunit I of the aa3- and caa3-Cox of Paracoccus and Thermus spp., respectively. To our knowledge, this is the first report demonstrating the presence of a caa3-Cox using cyt c4 as an electron donor in a Pseudomonas species.
Keywords: caa3-type cytochrome c oxidase, growth with biphenyl, Nano LC-MS/MS, aerobic respiratory chain, Pseudomonas pseudoalcaligenes
1. Introduction
Cytochrome (cyt) c oxidases (or cyt c: O2 reductases) belong to the haem copper oxidase (HCO) superfamily [1]. HCO members are classified into three main families, type A, B, and C, based on the sequence signatures and conservation of their proton transport pathways (named D and K). Among the type A HCO, the mitochondrial cyt c oxidases (Cox) and their closely related counterparts from bacteria such as Paracoccus denitrificans and Rhodobacter sphaeroides, are referred to as aa3-Cox [2]. These enzymes contain a low-spin a-type haem and a haem a3-CuB binuclear center at their catalytic subunit (SU) I, as well as a haem binuclear CuA center at their SU II. Electrons are received initially from cyt c by the CuA center, and conveyed via haem a to the haem a3-CuB center where O2 is converted to H2O [1]. According to the amino acid motifs found at the hydrophobic end of the D-proton pathway, the type A HCOs can be further divided into two subclasses: type A1 contains a highly conserved glutamate residue in the sequence motif – XGHPEV – whereas in type A2 the gating glutamate is replaced by tyrosine - serine in the motif – YSHPXV [3]. The type B HCOs are found in bacteria and archaea, and contain a low-spin b-type haem and a high-spin haem a (ba3-Cox). The type C HCOs are quite different from both type A and B Cox with respect to their amino acid and subunit compositions, and contain three c-type haems and two b-type haems (cbb3-Cox) [4].
The first crystal structure of a type A2 HCO was described from the thermophilic Gram-negative bacterium Thermus thermophilus [5]. This HCO consists of two SUs, referred to as SU I/III and SU IIc that contain all of the metal centers characteristics of aa3-Cox [6]. In this enzyme, the SU I/III is a fusion of the classical SU I and SU III whereas SU IIc is a fusion between a canonical SU II and a cyt c domain. Due to this feature, the HCO of T. thermophilus is also called caa3-Cox. This type of HCO is present in the thermophilic Gram negative Rhodothermus marinus and the Gram-positive Bacillus subtilis species [3,7]. The occurrence of a cyt c domain fused with a canonical SU II seems to reflect the need for a better control of the respiratory electron transfer under harsh environmental conditions (e.g., high temperatures), or in the absence of a periplasmic space in Gram positive species [1,8]. Similar to aa3-Cox, the proton pumping stoichiometry of caa3-Cox (0.8–1 H+/e−) is higher than ba3- (0.5–0.75 H+/e−) and cbb3- (0.2–0.5 H+/e−) Cox, also present in thermophilic and mesophilic bacteria [3,9,10,44].
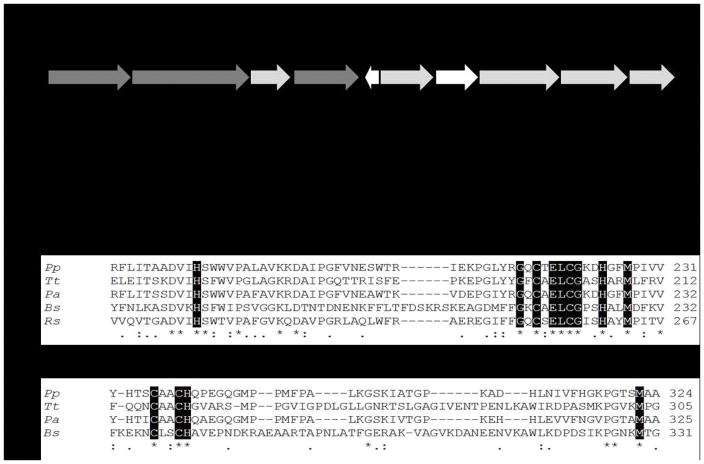
Recently, genome analyses of the polychlorinated biphenyl (PCBs) degrader Pseudomonas pseudoalcaligenes KF707 (hereafter called KF707) suggested the presence of a gene cluster (BAU71738-71737-71740) encoding an aa3-Cox composed of three subunits (SU I, SU IIc and SU III) [11]. The predicted SU IIc (42 kDa) of KF707 was annotated as a membrane-anchored cupredoxin domain containing one electron-accepting homo-binuclear copper center CuA, with a carboxyl terminal fusion to a cyt c domain [5,11]. The occurrence of a c-type haem in the KF707 aa3-Cox was also suggested by sequence alignments which showed, at the C-terminal region, the typical amino acid residues (CXXCH) that coordinate the c-type haem in B. subtilis and T. thermophilus caa3-Cox [5,8] (Figure 1). These haem binding residues are not found in the aa3-Cox from R. sphaeroides and P. denitrificans. Notably, the SU IIc amino acid sequence of KF707 shows 81% of identity with CoxB (SU II) of the opportunistic pathogen Pseudomonas aeruginosa PAO1 [11,12]. In this latter species, the presence of an orthodox aa3-Cox was documented by Arai and co-workers [13,14,15], although this group referred recently to the product of P. aeruginosa PAO1 cox genes [16] as caa3-Cox despite any supporting data.
Figure 1.
A. Organization of the Caa3-Cox gene cluster in KF707: dark gray, functional genes (coxIIc-I-III); light gray, genes for Caa3-Cox biogenesis and white, genes probably not directly related to Caa3-Cox. B. Alignment analysis for subunit II of the caa3-type Cox from KF707 (Pp - BAU71737), T. thermophilus (Tt – AFH37994), P. aeruginosa (Pa – ONM68414) and B. subtilis (Bs – OBA08775), and aa3-type Cox from R. sphaeroides (Rs – ASN35548). The first alignment focuses on the cupredoxin domain and the copper center CuA (N-terminal portion), and the second one shows the cytochrome c domain (C-terminal portion), absent in the aa3-type Cox from R. sphaeroides. The dark boxes are the amino acids that are directly involved in the CuA and haem c binding, asterisks and dots represent the fully and partially conserved amino acid residues, respectively. Analyses were performed using the ClustalW software [20].
Our studies aimed at understanding the metabolic abilities of KF707 in using biphenyl as a sole carbon source [17,18], indicated that membranes from KF707 cells grown with different carbon sources (e.g., glucose or biphenyl) contained several membrane-bound c-type haem proteins. In the present study, using tetramethylbenzidine (TMBZ) staining of SDS-PAGE and tandem mass spectrometry (MS) analyses, we defined the identities of these hemoproteins, and demonstrated that the protein band of 37 kDa corresponded to the c-type cyt domain containing SU IIc of KF707 caa3-Cox. As expected, this 37 kDa band was absent in a KF707 mutant lacking cox1-2 gene clusters (KFΔcox1-2, Caa3- and Ccaa3-minus). We also showed that this SU IIc is highly expressed in membranes from cells grown in the presence of biphenyl, while it is poorly expressed in those grown in the presence of glucose. This finding is consistent with our recent study related to the promoter activity of the cox1 (caa3-Cox) gene cluster [11]. To further support our findings, a homology based structural model of KF707 caa3-Cox was built using the X-ray structure of that from T. thermophilus (PDB code 2YEV) [5]. Finally, the TMBZ/SDS-PAGE, mass spectrometry and ascorbate/TMPD oxidase activity measurements using membranes from a KF707 triple mutant (KFΔc4/cco1-2), which lacks the di-haem cyt c4 (BAU71765) and the cbb3-Cox-1 and -Cox-2, grown in biphenyl showed that cyt c4 is the electron donor to caa3-Cox. To the best of our knowledge, this work represents the first report that unequivocally demonstrates the presence in a Pseudomonas species of a caa3-Cox that uses cyt c4 as an electron donor.
2. Material and Methods
2.1. Bacterial strains and construction of terminal oxidase deleted mutants
The bacterial strains used are listed in Table 1, and Pseudomonas pseudoalcaligenes KF707 was used as a wild type strain (W.T.). Nucleotide and amino acid sequences were from the complete genome sequence of KF707 available in GenBank (accession number AP014862). Sequence similarity searches were performed using the BLAST software (http://www.ncbi.nlm.nih.gov/blast/blast.cgi) [19] and the conserved domain database (http://www.ncbi.nlm.nih.gov/structure/cdd/), and multiple sequence alignments were performed with ClustalW software [20]. Single and multiple Cox-deletion mutants of KF707, Gene SOEing PCR [21], pG19II conjugative plasmid [22] and double crossover recombination were used as described previously [11]. Deletions of the cyt c4 (BAU71765) or cyt c5 (BAU71764) genes were constructed using appropriate primers listed in Table S3.
Table 1.
List of the Pseudomonas pseudoalcaligenes KF707 strains used in this study.
| Bacterial strains | Relevant Genotype |
|---|---|
| Wild type (W.T.) | Ampr |
| KFΔcco1-2 | Cbb31 and Cbb32 minus - Deletion of ccoN1O1Q1P1 and ccoN2O2Q2P2, Ampr |
| KFΔcox1-2 | Caa3 and Ccaa3 minus - Deletion of coxI-IIc-III and coxMNOP, Ampr |
| KFΔcox1-2/cco1-2 | Caa3, Ccaa3, Cbb31 and Cbb32 minus - Deletion of coxI-IIc-III, coxMNOP, coN1O1Q1P1 and ccoN2O2Q2P2, Ampr |
| KFΔCIO | CIO minus - Deletion of cioABC, Ampr |
| KFΔcox1-2/CIO | Cbb31, Cbb32 and CIO minus - Deletion of coxI-IIc-III, coxMNOP and cioABC, Ampr |
| KFΔc4 | cyt c4 minus – Deletion of c4 (BAU71765), Ampr |
| KFΔc5 | cyt c4 minus – Deletion of c4 (BAU71764), Ampr |
| KFΔc4/c5 | cyt c4 and cyt c5 minus – Deletion of c4 (BAU71764) and c5 (BAU71764), Ampr |
| KFΔc4/cco1-2 | cyt c4, Cbb31 and Cbb32 minus – Deletion of c4 (BAU71765), coN1O1Q1P1 and ccoN2O2Q2P2, Ampr |
2.2. Cell membrane fragments preparation, protein concentration and respiratory activities determinations
For preparation of cell membranes, KF707 W.T. and mutant strains were grown aerobically at 30°C and 130 rpm in 3 L Erlenmeyer flasks containing 1 L Mineral Salt Medium (MSM), pH 7.2 [18], supplemented with 6 mM glucose or biphenyl as the sole carbon source. Cells were harvested at stationary phase (OD600 nm 0.7–0.9), after 16 or 30 hours growth for glucose or biphenyl, respectively. Membrane fragments were prepared as previously described [23]. Protein concentrations were determined using the bicinchoninic (BCA) acid assay according to the supplier’s recommendations (Sigma Inc.; procedure TPRO-562). Respiratory activities in membrane fragments from appropriate strains were determined as reported previously [11].
SDS-PAGE and haem staining
The c-type cyt detection was performed using membrane fragments from KF707 W.T. and mutant strains grown in MSM medium with glucose or biphenyl. 150–200 μg of protein samples were denatured for 5 minutes at 37°C and separated by 15% SDS-PAGE [24]. The c-type cyts were revealed via their intrinsic peroxidase activity by using 3,3′,5,5′-tetramethylbenzidine (TMBZ) and H2O2 [25]. ImageJ served to quantify the intensity of the bands.
2.3. NADI staining
Cox phenotypes of colonies from KF707 W.T. and deletion mutants were determined using NADI staining, which visualizes their Cox activities. This staining is based on the oxidation of α-naphthol to indophenol blue via the artificial electron donor N,N,dimethyl-p-phenylenediamine (DMPD), coupled to Cox activity. NADI−negative indicates the absence, and NADI-positive (formation of blue staining) the presence of Cox activity in colonies [11].
2.4. Nano LC-MS/MS analysis and database searches
SDS-PAGE bands were excised, and after reduction (dithiothreitol, Sigma) and alkylation (iodoacetamide, Bio-Rad), subjected to in-gel trypsin digestion (Promega, Sequencing Grade Modified Trypsin) overnight at 37°C. Peptides eluted from the gel samples were dried and desalted using ZipTips (Milipore U-C18 P10), lyophilized and stored at −80 °C. They were resuspended in 10 uL 5% acetonitrile/0.1% formic acid prior to mass spectrometry, using Q-Exactive Quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific, San Jose, CA) coupled to an Easy-nLCTM 1000 nano liquid chromatography system (Thermo Fisher Scientific, San Jose, CA). Samples were loaded in buffer A (0.1% formic acid) onto a 20 cm long fused silica capillary column (75 μm ID), packed with reversed-phase Repro-Sil Pur C18-AQ 3 μm resin (Dr. Maisch GmbH, Germany). Peptides were eluted using a 45 min linear gradient from 4%–40% buffer B (100% acetonitrile, 0.1% formic acid), followed by a 7 min gradient from 40%–80% buffer B and 8 min wash with 80% B (constant flow rate 300 nL/min). The Q-Exactive was operated in data-dependent acquisition (DDA) mode with dynamic exclusion enabled (repeat count: 1, exclusion duration: 20 sec). Full MS survey scans (mass range 300–1600 m/z) at high resolution (70,000 at 200 m/z) were followed by MS/MS fragmentation of the top 15 most intense ions with higher energy collisional dissociation (HCD) at a normalized collision energy of 22 (resolution 17,500 at 200 m/z). Dual lock mass calibration was enabled with 371.101233 and 445.120024 m/z background ions. MS spectra were searched against the KF707 protein database (UniProtKB) using Proteome Discoverer 1.4 Software (Thermo) with Sequest-HT search engine. Search parameters were set to full trypsin digestion, with a maximum of 3 missed cleavages and 3 modifications per peptide. Oxidation of methionine (+16 Da) and carbamido-methylation of cysteine (+57 Da) were selected as dynamic modifications. Precursor and fragment ion tolerances were set to 10 ppm and 0.6 Da, respectively. False discovery rates by target-decoy search (FDR) were set to 0.01 (high confidence) with Xcorr of >2 for m/z=2, >2.5 for m/z=3, and > 2.6 for m/z=4 values.
2.5. Membrane protein modeling
Template searches for each of the three subunits (SU I, IIc, and III) of KF707 caa3-Cox (Pp-caa3) were performed using the HHsearch method implemented in the HHpred server [26]. The server performs up to eight iterative PSI-BLAST [27] searches through filtered versions of the non-redundant (nr) database from NCBI. Using the final target alignment, a hidden Markov model (HMM) [28,29] profile is calculated. Homologous templates are identified by searching through a database containing HMMs for a representative subset of PDB sequences. HHsearch ranks the database matches based on the probability of the match to be homologous to the target sequence to distinguish homologous from non-homologous matches. Excluding PDB structures that were not solved by X-ray crystallography, HHpred identified the caa3-Cox from T. thermophilus (Tt-caa3). Tt-caa3 is composed of two proteins homologous to Pp-caa3 SU I/III and SU IIc sequences. SU I/III is a fusion of the canonical SU I to SU III, connected with a linker of ca. 70 residues. The pronounced sequence identity/similarity of Pp-caa3 SU I, IIc and III with respect to the corresponding sequences of Tt-caa3 (45/62%, 30/45% and 31/42%, respectively) allows the generation of a reliable structural model. The target and template sequences were realigned using the Promals3D server [30] (see Supplementary Information for the alignments). The sequences of aa3-Cox subunit II and III from P. denitrificans (Pd-aa3) were included to align two large insertions not present in the Tt-caa3 sequences. The obtained alignment was then used to calculate 100 structures using the available crystal structures of Tt-caa3 (2YEV) [5] and Pd-aa3 (3HB3 [31] and 1QLE [32]) to model Pp-caa3 subunit IIc and III, respectively, using the Modeller 9.18 software [33]. The two haems a, the haem c, the binuclear and mononuclear copper centers (CuA and CuB, respectively) and the Mg(II) ion, together with the water molecules in direct contact with the metal ions and the lipid molecules found in the Tt-caa3 structure, were included in the modeling procedure. The best model was selected using the DOPE potential function built into Modeller [34]. A loop optimization routine was used to refine the regions that showed higher than average energy as calculated using the DOPE potential function. The stereo-chemical quality of the model structure was established using ProCheck [35] to confirm the reliability of the model structure. The obtained molecular model and its molecular surface were displayed using UCSF Chimera [36] and APBS [37] softwares. The position of the protein model in the membrane was predicted using the PPM web server [38], and the conservation of the surface residues was evaluated using the ConSurf 2016 server [39].
3. Results
3.1. Cyt c profile and Nano LC-MS/MS analysis of KF707 W.T. and appropriate mutants
Recently, we reported KF707 mutants carrying deletions of single or multiple terminal oxidases [11], which were crucial in understanding the unusual metabolic abilities of a polychlorinated biphenyls degrader such as KF707 [17]. Membranes from KF707 cells, grown with glucose or biphenyl as a carbon source, expressed several membrane-bound c-type cyts, corresponding to caa3- and cbb3-Cox. Genetic and functional analyses identified five terminal oxidases in KF707: two c(c)aa3-Cox (Caa3 and Ccaa3), two cbb3-Cox (Cbb31 and Cbb32) and one bd-type cyanide-insensitive quinol oxidase (CIO). While the expression of both cbb3-Cox was prevalent in glucose grown cells, the promoter activity of the caa3-Cox genes increased considerably when biphenyl was used as a single carbon source, whereas the promoter activity of cco2 gene corresponding to Cbb32 was repressed [11].
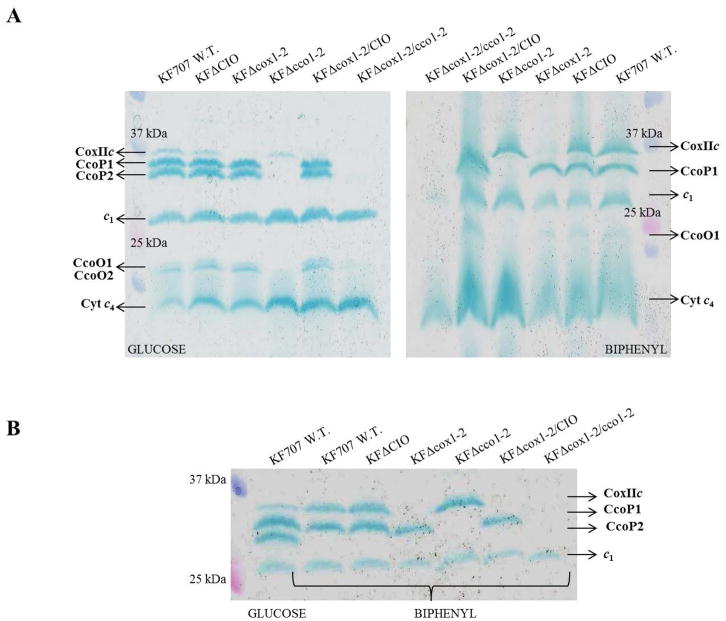
Presently, no biochemical or structural data are available to evidence the presence of any caa3-Cox in the Pseudomonas species [1,40]. In order to fill this knowledge-gap, the cyt c contents of membranes isolated from KF707 W.T. and the Cox-deletion mutants were examined using TMBZ/SDS-PAGE [24,25]. Proteins were extracted from membrane fragments of cells harvested at their stationary phase of growth (OD600nm 0.8–1.0) in MSM glucose or biphenyl (6 mM each). In membranes from glucose grown cells at least six TMBZ stained bands were found (Figure 2A, left), while those grown in biphenyl exhibited five such bands (Figure 2A, right). The lowest MW (~17 kDa) band, present in membranes of all strains grown on either carbon source (Figure 2A and B) was identified by MS analysis to be the cyt c4 (see Section 3.3 and Table 2). Similarly, the two bands of 20–22 kDa were assigned to the CcoO1 and CcoO2 subunits of Cbb31 and Cbb32, respectively (Table S1), whereas in cells grown with biphenyl only the band related to CcoO1 was detected. In both glucose and biphenyl grown cells, a band attributable to cyt c1 subunit (29 kDa) (Table S1) of the bc1 complex of KF707 was also present [11]. Of the haem c protein bands with MW between 34–36 kDa, two in glucose (Figure 2A, left) and one in biphenyl (Figure 2A, right) were identified as the CcoP subunits of Cbb31 (CcoP1, 35.5 kDa) and Cbb32 (CcoP2, 34 kDa), respectively (Table S1). As expected, both bands were undetectable in membranes from the KFΔcco1-2 mutant, lacking the Cbb31 and Cbb32. Moreover, while the CcoP1 band was present in both glucose and biphenyl grown cells, the CcoP2 band was absent in biphenyl grown cells (Figure 2A, right). These findings were in line with previous data showing that the Cbb32 genes were not expressed in the stationary phase of growth when biphenyl was the sole carbon source [11].
Figure 2.
Profiles of the c-type cytochromes detected in membranes of KF707 W.T. and mutant strains grown with glucose or biphenyl as the carbon source until stationary phase (OD600 nm 0.8–1.0). Membrane fragments were analyzed by SDS-PAGE and the c-type cytochromes were revealed via their peroxidase activities using TMBZ staining.
A – Membranes of cells grown in glucose (left) or biphenyl (right) are shown. Lane 1 and lane 7 on the left and the right gels contain the Precision Plus Protein Standards Ladder (BIO-RAD) molecular weight [MW] standards; the remaining lanes correspond to KF707 W.T., KFΔCIO, KFΔcox1-2, KFΔcco1-2, KFΔcox1-2/CIO and KFΔcox1-2/cco1-2, as indicated.
B – Bands of proteins containing cytochrome c domains with MW between 25 and 37 kDa, are shown in detail. KF707 W.T. cells grown with glucose was compared with other KF707 strains grown with biphenyl. Lane 1 contains Precision Plus Protein Standards Ladder (BIO-RAD) MW standards, lane 2 contains KF707 W.T. grown with glucose, Lane 3 to 8 correspond to KF707 W.T., KFΔCIO, KFΔcox1-2, KFΔcco1-2, KFΔcox1-2/CIO and KFΔcox1-2/cco1-2, respectively Mutant phenotypes are listed in Table 1. All experiments were repeated at least three times.
Table 2.
Nano LC-MS/MS analysis results of the 17 and the 37 kDa bands extracted from TMBZ gels of KF707 W.T. and mutant strains grown in MSM medium with glucose or biphenyl as the sole carbon source. The amino acids coverage, significant peptides and their modifications are listed.
|
17 kDa Glucose or Biphenyl |
Accession | Description |
|
| ||
| L8MT74 |
Cytochrome c4
OS=Pseudomonas pseudoalcaligenes KF707 = NBRC 110670 GN=ppKF707_770 PE=3 SV=1 - [L8Mt74_PSEPS] – NCBI Cyt c4 BAU71765 – Coverage 31,4 % |
|
|
| ||
| Sequence | Modifications | |
|
| ||
| DIEAVSSYIQGLH | ||
| AAVcGAcHGPDGNSAAPNFPK | C4-C7(Carbamidomethyl) | |
| TVLEmTGLLTNmSDQDmADLAAYFASQK | M5-M12-M17(Oxidation) | |
|
| ||
|
37 kDa Glucose |
Accession | Description |
|
| ||
| L8MR96 |
Cytochrome c oxidase subunit 2 OS=Pseudomonas pseudoalcaligenes KF707 = NBRC 110670 GN=ppKF707_5503 PE=3 SV=1 - [L8MR96_PSEPS] – NCBI CoxII BAU71737 – Coverage 32,4 % |
|
|
| ||
| Sequence | Modifications | |
|
| ||
| APKDEHYLLEVDQPLVVPVGTK | ||
| ELTDKEWTLDELVAR | ||
| DEHYLLEVDQPLVVPVGTK | ||
| DHGFmPIVVEAK | M5(Oxidation) | |
| YLGQDVEFFSNLATPSEQIHNK | ||
| ADHLNIVFHGKPGTSmAAFGK | M16(Oxidation) | |
| DAIPGFVNESWTR | ||
| NAWGNNTGDmVTPK | M10(Oxidation) | |
|
| ||
| Accession | Description | |
|
37 kDa Biphenyl |
|
|
| L8MR96 |
Cytochrome c oxidase subunit 2 OS=Pseudomonas pseudoalcaligenes KF707 = NBRC 110670 GN=ppKF707_5503 PE=3 SV=1 - [L8MR96_PSEPS] – NCBI CoxII BAU71737 – Coverage 38,7 % |
|
|
| ||
| Sequence | Modifications | |
|
| ||
| YLGQDVEFFSNLATPSEQIHNK | ||
| ADHLNIVFHGKPGTSmAAFGK | M16(Oxidation) | |
| DEHYLLEVDQPLVVPVGTK | ||
| ELTDKEWTLDELVAR | ||
| ADHLNIVFHGKPGTSMAAFGK | ||
| APKDEHYLLEVDQPLVVPVGTK | ||
| KDAIPGFVNESWTR | ||
| LKELTDKEWTLDELVAR | ||
| GQcTELcGKDHGFmPIVVEAK | C3-C7(Carbamidomethyl); M14(Oxidation) | |
| DHGFMPIVVEAK | ||
| DHGFmPIVVEAK | M5(Oxidation) | |
| NAWGNNTGDmVTPK | M10(Oxidation) | |
| EVLALKQAESQ | ||
| EWTLDELVAR | ||
| GQcTELcGK | C3-C7(Carbamidomethyl) | |
Notably, the ~ 37 kDa protein band seen in membranes from both glucose and biphenyl grown cells, was not present in mutants lacking the caa3-Cox (KFΔcox1-2, KFΔcox1-2/CIO, KFΔcox1-2/cco1-2), and was roughly seven times more abundant in cells grown on biphenyl (Figure 2B) than those on glucose (Materials and Methods). Similarly, this 37 kDa was roughly eight time less abundant in glucose grown cells (Figure 2A and B) as compared with the 34 or the 35.5 kDa bands (CcoP2 or CcoP1 subunits), and about two-three times less than CcoP1 present in biphenyl grown cells (Figure 2A and B). These experiments were repeated four times, and the intensities of the bands evaluated by ImageJ (6.8 ± 0.7 biphenyl versus glucose).
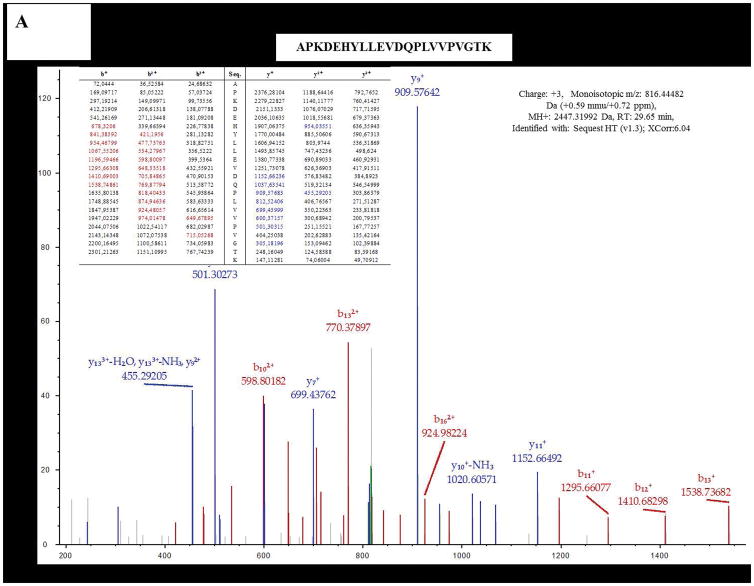
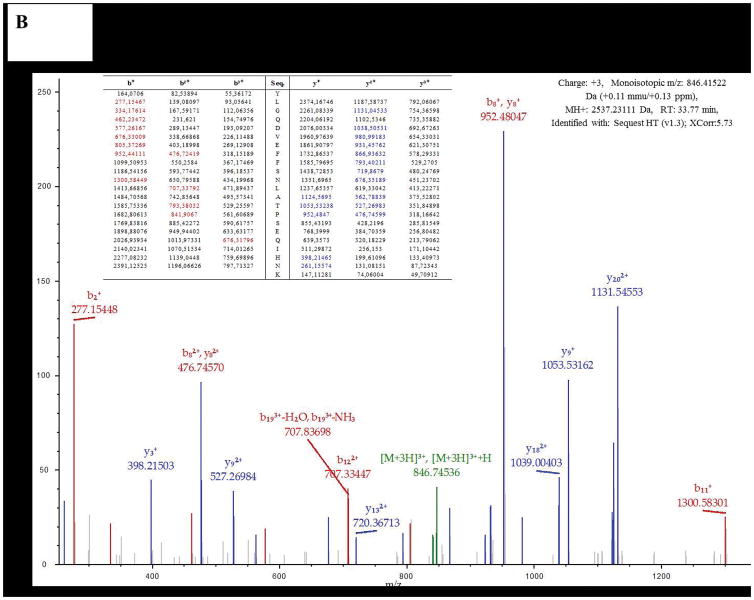
The 37 kDa band was excised from gels run with glucose or biphenyl grown cells membranes, and subjected to MS analysis, which identified over one hundred co-migrating proteins. CoxII (Uniprot code L8MR96 – 42 kDa) was present among these proteins and contained the canonical haem binding CXXCH motif of a c-type cyt, as previously seen with the CoxB subunit of T. thermophilus Caa3 [5]. In both cases, MS data identified several distinct peptides (8 for glucose and 15 for biphenyl grown cells) of the CoxII subunit, with an overall coverage of over 30% (Table 2, Figure 3A and B, Figure S1). These results strongly support the earlier conclusion made on the role of caa3-Cox in biphenyl grown cells of KF707 based on the expression of its gene cluster [11].
Figure 3.
Examples of two of Nano LC-MS/MS fragmentation spectra obtained with the 37 kDa band from cells grown with glucose (A) or biphenyl (B). These bands were identified as the subunit SU IIc of KF707 caa3-Cox.
3.2. Protein modeling of the caa3-Cox
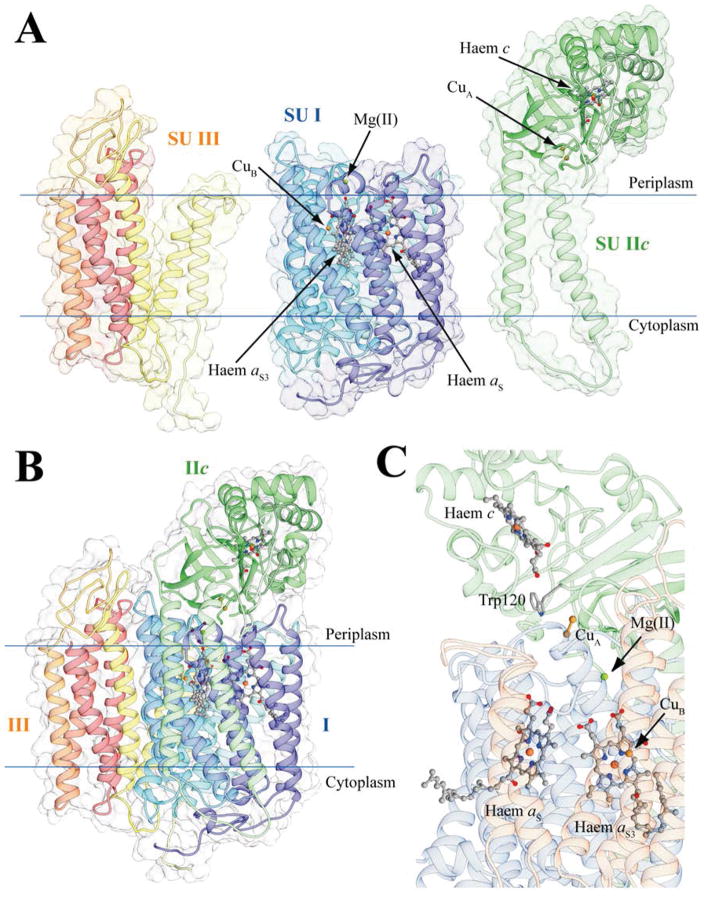
A model structure of KF707 caa3-Cox from (Pp-caa3), based on the X-ray structure of that from T. thermophilus (Tt-caa3, 2YEV) was generated [5]. Pp-caa3 consists of three subunits (I, IIc, and III) (Figure 4A). Its SU I is composed of 12 transmembrane α-helices connected to each other by unstructured linkers, and contains two a-type haem groups located in the hydrophobic core, and one mononuclear copper center (named CuB) (Figure 4A, B and C). SU IIc is a fusion protein between a canonical SU II and a cyt c domain, similar to that from T. thermophilus [41]. It is composed of two transmembrane α-helices in direct contact with SU I and a periplasmic domain consisting of a 7-strands antiparallel β-sheet fused to a typical cyt c protein. Hence, SU IIc contains a haem c group with the Fe-ion bound to its His282 and Met332, and a homobinuclear copper center (CuA) bound to its His181, Cys216, Glu218, Cys220, His224, and Met227 residues (Figure 4A and B). The SU III is composed of six transmembrane α-helices and is in direct contact with SU I.
Figure 4.
Ribbon representation of the caa3-type Cox homology model of KF707; SU I, SU IIc and SU III are shown as single entities (A) or together (B) and with their cofactors (C). The subunits are colored light blue, yellow and light green in the in the proximity of the N-terminus, and dark blue, dark red and dark green at the C-terminus of SU I, IIc and III, respectively. In this KF707 caa3-Cox model, Trp120 of Pp-caa3 is highlighted since it substitutes for the Tt-caa3 Phe126, which has similar electron transfer properties, while the copper binding Cys224 (Pp-caa3 numbering) is not indicated as it is conserved in both sequences. The haems are shown as ball and stick with the iron and copper metal centers as orange and ochre spheres, respectively. The Mg(II) ion in SU I is shown as a green sphere. Residues involved in electron transport are reported as sticks, and colored accordingly to atom type.
The protein surface of Pp-caa3 shows the typical charge distribution of a membrane protein (Figure S2), with a large hydrophobic region corresponding to the transmembrane portion of SU I, IIc and III, twined with electrostatically charged surfaces exposed to the aqueous environment of the periplasm and the cytoplasm. The protein surface of the membrane-extern portion of Pp-caa3 SU IIc analyzed by ConSurf 2016 [39] revealed three conserved residues, Ala280, Gln283 and Phe293, on a flat region located on the His282 side of the haem c group. Of these residues, only Ala280 is conserved in Tt-caa3, suggesting possibly different ways of interactions between the SU IIc and its soluble electron donor in T. thermophilus and KF707 species [11,42] (see Discussion).
3.3. A di-haem cyt c4 is the electron donor to KF707 caa3-Cox
The KF707 genome contains three putative cyt c4 (BAU71765, BAU75530 and BAU75507) and two cyt c5 (BAU77240 and BAU71764) genes (11). Of these, BAU71765 and BAU71764, corresponding to the cyt c4 and cyt c5, respectively, are clustered together. The c-type cyt profiles obtained by TMBZ SDS-PAGE gels from membranes of KF707 W.T. and mutant strains grown in glucose or biphenyl, show only one cyt c protein band of 17 kDa (Figure 2 and Table 2). This protein was identified by MS analysis as the di-haem cyt c4 encoded by the BAU71765 gene (Table 2). In order to confirm this identification and clarify the role of cyt c4 (BAU71765) or cyt c5 (BAU71764) as possible electron donor(s) to KF707 caa3-Cox, three different deletion mutants (KFΔc4, KFΔc5 and KFΔc4/c5) were constructed (Table 1 and Table S3). The NADI test, which indicates the Cox activity, was mostly negative for KFΔc4 and KFΔc4/c5 mutants (Figure S4), but was positive for KFΔc5 cells like the W.T. strain, indicating that only the absence of cyt c4, but not that of c5, affected KF707 Cox activity. Based on these and other findings (e.g., growth rates of mutants, not shown), the function of cyt c5 (BAU71764) in the KF707 respiratory chain was not pursued further.
The TMBZ SDS-PAGE analyses of W.T. and KFΔc4 membranes prepared from cells grown on glucose or biphenyl (Figure 5) indicated that the 17 kDa protein band corresponding to cyt c4 was absent in KFΔc4 mutants, while all of the remaining c-type cyts were present. Apparently, the lack of cyt c4 did not affect the regulation of KF707 terminal oxidases, a finding which was also confirmed using appropriate translational lacZ fusion constructs (not shown). In order to unequivocally establish the role of cyt c4 as an electron donor to KF707 caa3-Cox, the triple mutant KFΔc4/cco1-2 was constructed (Table 1). The rate of oxygen consumption initiated by addition of NADH (or ascorbate/TMPD) as electron donors was determined (Table 3). The data showed that in both KFΔc4 and KFΔc4/cco1-2 membranes from cells grown either with glucose- (Glu) or biphenyl- (Bph), NADH respiration was largely insensitive to 50 μM cyanide (CN−). This chemical is known to fully inhibit both caa3- and cbb31-2 Cox (11). Thus, in the absence of cyt c4, most of the NADH/oxygen consumption is sustained via the CIO (cyanide insensitive oxidase) activity. This latter activity was ~ 2–3 folds higher than the equivalent activity found in W.T. membranes, but similar to that seen with KFΔcco1-2 membranes. Most importantly, the ascorbate/TMPD initiated oxygen consumption activity in KFΔc4 membranes was 70% and 80% less than that in KF707 W.T membranes from cells grown on glucose or biphenyl, respectively, supporting the crucial role of cyt c4 in mediating TMPD oxidation via the terminal oxidases. Further, considering that the caa3-Cox was greatly expressed in biphenyl grown cells (Figure 4), the decrease of the ascorbate/TMPD activity seen by comparing membranes from biphenyl grown KFΔc4/cco1-2 with those from similarly grown KFΔcco1-2, unequivocally demonstrated that cyt c4 acts as a physiological electron donor to KF707 caa3-Cox.
Figure 5.
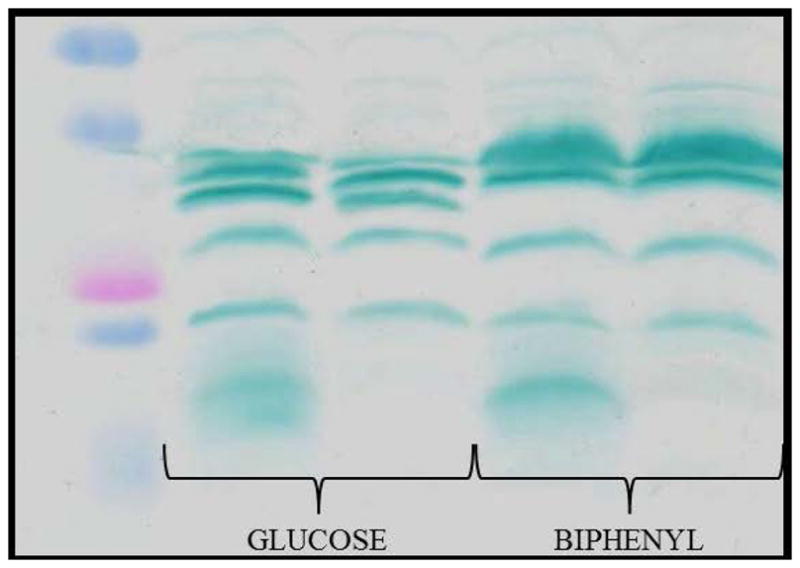
Profiles of the c-type cytochromes detected in membranes of KF707 W.T. and KFΔc4 mutant strains grown with glucose or biphenyl as the carbon source until stationary phase (OD600 nm 0.8–1.0). Membrane fragments were analyzed by SDS-PAGE, and the c-type cytochromes were revealed via their peroxidase activities using TMBZ staining, as in Figure 2.
Table 3.
Respiratory activities in KF707 membranes from W.T. and appropriate mutant cells grown until the stationary phase (OD600 nm 0.7–0.9) in glucose (Glu) or in biphenyl (Bph) as the sole carbon source. See the text for further details.
| Substrate | ||||
|---|---|---|---|---|
|
| ||||
| Carbon source/Strain | NADH | NADH/CN− | Asc-TMPD | Asc-TMPD/CN− |
| Glu/KF707 W.T. | 158 ± 9,0 | 31 ± 5,0 | 150 ± 15 | 10 ± 2,0 |
| Glu/KFΔc4 | 67 ± 2,0 | 55 ± 4,0 | 52 ± 2,0 | 4,5 ± 1,0 |
| Glu/KFΔcco1-2 | 160 ± 8,0 | 114 ± 0,1 | 38 ± 1,0 | 8,0 ± 1,0 |
| Glu/KFΔc4/cco1-2 | 85 ± 6,0 | 80 ± 5,0 | 10 ± 1,0 | 8,0 ± 1,0 |
|
| ||||
| Bph/KF707 W.T. | 110 ± 3,0 | 10 ± 0,5 | 161 ± 12 | 8,0 ± 2,0 |
| Bph/KFΔc4 | 34 ± 2,0 | 31 ± 2,0 | 32 ± 2,0 | 3,0 ± 1,0 |
| Bph/KFΔcco1-2 | 97 ± 8,0 | 27 ± 1,5 | 115 ± 11 | 5,0 ± 1,5 |
| Bph/KFΔc4/cco1-2 | 28 ± 1,5 | 27 ± 1,5 | 15 ± 1,0 | 5,3 ± 1,0 |
Non standard abbreviations used: Asc, sodium ascorbate (5 mM); TMPD, N,N,N′,N′-tetramethyl-p-phenylenediamine (50 μM); CN−, potassium cyanide (50 μM). Mutant strains: KFΔc4, lacks cytochrome c4; KFΔcco1-2, lacks both cbb31- and cbb32-Cox; KFΔc4/Δcco1-2, lacks cytochrome c4 and the cbb3-1- and cbb32-Cox. Oxygen consumption rates are expressed as nmoles of O2 consumed min−1 mg of protein−1, are the mean values of at least two to three assays.
4. Discussion
Most bacteria possess multiple respiratory terminal oxidases that differ in their proton pumping efficiency and oxygen affinity, enabling energy transduction under various O2 tensions and with different types and amounts of carbon sources available to cells, as shown for R. sphaeroides, P. aeruginosa PAO1 and Shewanella oneidensis MR-1 [13,15,40,43–47]. In Pseudomonas sp. B4, which is a PCB degrader closely related to KF707, a shift from growth in glucose to a medium with biphenyl was shown to induce a stress response highlighted by accumulation of inorganic polyphosphate and production of the general stress protein GroEL [44]. Although our present knowledge on KF707 cell stress response is quite limited [18,45], the use of biphenyl as a carbon source is likely to generate a stress response, as suggested by prolonged growth phase initial lag (several times longer than that seen in glucose) [11]. In this respect, the RelA/SpoT Homologue (RSH) proteins are known to play key roles in regulating the cellular response to nutrient stress signals [46]. Indeed, the KF707 RelA/SpoT double mutants (KFΔrelA/spot) show a longer initial lag phase as compared with the W.T. cells when grown with aromatic carbon sources like biphenyl or benzoate. Moreover, in these mutants the expression of the terminal oxidases does not respond anymore to the carbon source, unlike the W.T. strain, suggesting that RelA/SpoT proteins may be involved in this regulation as a stress response (unpublished data).
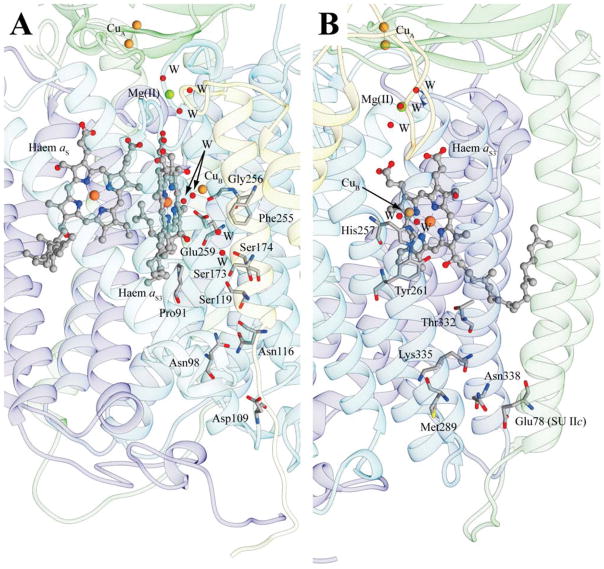
The expression levels of the terminal oxidases affect the respiratory energy transduction and the membrane electron transport rates, which in turn reflect the capacity of soluble redox carriers to connect the terminal oxidases with other membrane-integral redox complexes [8]. As these connections might be rate-limiting, bacterial species have adopted strategies to overcome these restrictions by forming respiratory super-complexes, or fusing a c-type haem electron carrier to a terminal oxidase subunit [8]. In the thermophiles T. thermophilus and R. marinus and the spore-forming B. subtilis the subunit II of an aa3-Cox has an extra domain carrying a c-type haem [1,48,49]. In S. oneidensis MR-1, a C-terminal extension of the aa3-Cox subunit II has two putative c-type haem binding sequences [40]. Interestingly, this fusion oxidase (named Ccaa3) is expressed under nutrient-starved conditions, similar to the aa3-Cox of PAO1 [15,47]. Analogous c-type haem domains containing aa3-Cox were also reported in the anaerobe Desulfovibrio vulgaris [50] and in the anoxygenic phototrophs Rubrivivax gelatinosus and R. sphaeroides [51,52]. Thus, the aa3-Cox with additional c-type haem containing subunits are frequently encountered among the bacterial genera and habitats [8]. The finding that KF707 also contains a caa3-Cox expressed in cells grown on biphenyl [11] now extends the occurrence of this type of metabolically regulated Cox enzymes to the Pseudomonas species. Here, using TMBZ/SDS-PAGE and Nano LC-MS/MS analyses, we demonstrated for the first time that the 37 kDa SU IIc of KF707 Caa3-Cox indeed contains a c-type haem. Moreover, this SU IIc is more abundant in membranes from cells grown on biphenyl than those on glucose, in agreement with the previously determined promoter activities of caa3-Cox, using cox1-lacZ fusion constructs [11]. In light of these findings, the amino acid sequence and predicted structure of KF707 caa3-Cox (Pp-caa3) were compared with those from T. thermophilus (Tt-caa3) (Figure S3). A large part of the haem c pocket of SU IIc, especially on the His282 side, is conserved between the two species. In contrast, the strand-loop-strand motif forming the bottom part of this haem c pocket in Tt-caa3 (residues 273-284 in T. thermophilus numbering) is not present in Pp-caa3, where it is replaced by a large insertion (residues 129-153 in Pp-caa3, numbering Tt-caa3). As this insertion is missing in Tt-caa3, this portion of the model built here relied on the structure of P. denitrificans aa3-Cox (Pd-aa3), and suggested that the haem c pocket of Pp-caa3 might be a combination of that from Tt-caa3 and Pd-aa3 together.
Comparing the homology model of Pp-caa3 with the structure of Tt-caa3 we note that the electron transfer pathway seen between the SU IIc haem c and its homo-binuclear CuA center is highly conserved in Pp-caa3, except for a few conservative substitutions. This observation suggests that the electron transfer pathways from CuA to aa3 haems in both enzymes are similar (Figure 4C). Likewise, the two proton channels (D- and K-pathways) located in SU I of the aa3-Cox [1], required for chemical reduction of O2 in the haem-Cu binuclear center, are conserved between the Tt-caa3 and Pp-caa3, with most of the residues constituting these pathways being conserved, or conservatively substituted (Figure 6A and B). A major difference is observed at the end of the D-pathway, near haem aS3, where Tt-caa3 Gln84, Ser249 and Thr252 were substituted with Pro91, Gly256, and Glu259 in Pp-caa3. Apparently, the so-called YS gateway seen in the Tt-caa3 structure [5] may not be present in of Pp-caa3. Whether these differences constitute the basis for our inability to obtain a viable KF707 mutant containing only the caa3-Cox, remains unknown [11]. Somewhat similarly, a P. aeruginosa PAO1 mutant that contains only the aa3-Cox was also unable to grow aerobically [14], unless it contained a suppressor mutation in the two-component regulatory system RoxSR [16]. Work is in progress to see whether a similar situation also occurs in KF707.
Figure 6.
Proton D- and K-pathways (A and B, respectively) in KF707 caa3-Cox model structure. The D-pathway in the Tt-caa3 structure begins near to Asp103 and leads through a solvent-filled cavity to Tyr248 in the vicinity of SU I haem aS3. In the mode, Tt-caa3 Tyr248 corresponds to Pp-caa3 Phe255. The K-pathway originates at Tt-caa3 with Glu84 of SU IIc and continues up to the binuclear copper center CuB via the cross-linked SU I/III His250-Tyr254, by means of Lys328. These residues are fully conserved in the Pp-caa3 model structure shown here (SU IIc Glu78 and SU I His257-Tyr261 and Lys335). The protein backbone is reported as ribbons colored as in Figure 3. The haem groups are in ball and stick, with the iron and copper metal centers as orange and ocher spheres, respectively. The Mg(II) ion in SU I is shown as a green, while the water molecules (W) are depicted as red spheres. The residues mentioned in the text are in sticks colored according to the atom type.
Structural comparisons of Tt-caa3 and Pp-caa3 raise intriguing issues, such as the membrane-external surface of Pp-caa3 SU IIc, which has three residues (Ala280, Gln283 and Phe293) of which only one (Ala280) is conserved in Tt-caa3. A possibility is that this difference might reflect the presence of different electron donors to the SU IIc of T. thermophilus versus that of KF707 caa3-Cox. In the case of KF707, this work showed for the first time in a Pseudomonas species that the 17 kDa di-haem c4 (BAU71765) functions as the main electron donor to the caa3-Cox (Table 3). In the case of T. thermophilus caa3-Cox, although the multi-domain di-haem cyt c550 acts as an electron donor [53], this subject remains a matter of controversy [42].
Supplementary Material
Acknowledgments
DZ thanks the University of Bologna for supporting this work (RFO grants 2013-16). FS was supported by a PhD fellowship and a Marco Polo fellowship provided by the University of Bologna. The contributions of NS and FD were supported partly by grants from the Division of Chemical Sciences, Geosciences and Biosciences, Office of Basic Energy Sciences of Department of Energy [DOE DE-FG02-91ER20052] and partly by the National Institute of Health [NIH GM 38237] to FD.
Footnotes
Author contribution
FS conceived, designed, and performed a significant part of the experiments, assisted in the design of the study and co-wrote the manuscript. FM performed part of the experiments and co-wrote the manuscript. NS assisted in design and performance of a part of the experiments, FD assisted in design of the study and co-wrote the manuscript. DZ conceived, directed, supervised, and co-wrote the manuscript.
References
- 1.Lyons JA, Hilbers F, Caffrey M. Cytochrome complexes: evolution, structures, energy transduction and signalling. In: Cramer WA, Kallas T, editors. Chapter. Vol. 16. Spinger; USA: 2016. pp. 307–329. [Google Scholar]
- 2.Sousa FL, Alves RJ, Ribeiro MA, Pereira-Leal JB, Teixeira M, Pereira MM. The superfamily of heme–copper oxygen reductases: Types and evolutionary considerations. Biochim Biophys Acta. 2012;1817:629–637. doi: 10.1016/j.bbabio.2011.09.020. [DOI] [PubMed] [Google Scholar]
- 3.Pereira MM, Santana M, Soares CM, Mendes J, Carita JN, Fernandes AS, Saraste M, Teixeira M. A novel scenario for the evolution of haem–copper oxygen reductases. Biochim Biophys Acta. 1999;1505:185–208. doi: 10.1016/s0005-2728(01)00169-4. [DOI] [PubMed] [Google Scholar]
- 4.Pereira MM, Sousa FL, Verissimo AF, Teixeira M. Looking for the minimum common denominator in haem–copper oxygen reductases: Towards a unified catalytic mechanism. Biochim Biophys Acta. 2008;1777:929–934. doi: 10.1016/j.bbabio.2008.05.441. [DOI] [PubMed] [Google Scholar]
- 5.Lyons JA, Aragäo D, Slattery O, Pisiliakov AV, Soulimane T, Caffrey M. Structural insights into electron transfer in caa3-type cytochrome oxidase. Nature. 2012;487:514–518. doi: 10.1038/nature11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferguson-Miller S, Babcock GT. Heme/Copper terminal oxidases. Chem Rev. 1996;96:2889–2908. doi: 10.1021/cr950051s. [DOI] [PubMed] [Google Scholar]
- 7.Bengtsson J, Tjalsma H, Rivolta C, Hederstedt L. Subunit II of Bacillus subtilis cytochrome c oxidase is a lipoprotein. J Bacteriol. 1999;181:685–688. doi: 10.1128/jb.181.2.685-688.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sone N, Hagerhall C, Sakamoto J. Respiration in Archaea and Bacteria – Diversity of prokaryotic respiratory systems. In: Zannoni D, editor. Chapter. Vol. 2. Springer; Netherlands: 2004. pp. 35–62. [Google Scholar]
- 9.Kannt A, Soulimane T, Buse G, Becker A, Bamberg E, Michel H. Electrical current generation and proton pumping catalyzed by the ba3-type cytochrome c oxidase from Thermus thermophilus. FEBS Lett. 1998;434:17–22. doi: 10.1016/s0014-5793(98)00942-9. [DOI] [PubMed] [Google Scholar]
- 10.Toledo-Cuevas M, Barquera B, Gennis RB, Wikstrom M, Garcia-Horsman JA. The cbb3-type cytochrome c oxidase from Rhodobacter sphaeroides, a proton-pumping heme-copper oxidase. Biochim Biophys Acta. 1998;1365:421–434. doi: 10.1016/s0005-2728(98)00095-4. [DOI] [PubMed] [Google Scholar]
- 11.Sandri F, Fedi S, Cappelletti M, Calabrese FM, Turner RJ, Zannoni D. Biphenyl modulates the expression and function of respiratory oxidases in the polychlorinated-biphenyls degrader Pseudomonas pseudoalcaligenes KF707. Front Microbiol. 2017;8:1223. doi: 10.3389/fmicb.2017.01223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 13.Arai H. Regulation and function of versatile aerobic and anaerobic respiratory metabolism in Pseudomonas aeruginosa. Front Microbiol. 2011;2:103. doi: 10.3389/fmicb.2011.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arai H, Kawakami T, Osamura T, Hirai T, Sakai Y, Ishii M. Enzymatic characterization and in vivo function of five terminal oxidases in Pseudomonas aeruginosa. J Bacteriol. 2014;196:4206–4215. doi: 10.1128/JB.02176-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawakami T, Kuroki M, Ishii M, Igarashi Y, Arai H. Differential expression of multiple terminal oxidases for aerobic respiration in Pseudomonas aeruginosa. Environ Microbiol. 2010;12:1399–1412. doi: 10.1111/j.1462-2920.2009.02109.x. [DOI] [PubMed] [Google Scholar]
- 16.Osamura T, Kawakami T, Kido R, Ishii M, Arai H. Specific expression and function of the A-type cytochrome oxidase under starvation conditions in Pseudomonas aeruginosa. PLOS One. 2017;12:e0177957. doi: 10.1371/journal.pone.0177957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fedi S, Carnevali M, Fava F, Andracchio A, Zappoli S, Zannoni D. Polychlorinated biphenyl degradation activities and hybridization analysis of fifteen aerobic strains isolated from PCB-contaminated site. Res Microbiol. 2001;152:583–592. doi: 10.1016/s0923-2508(01)01233-5. [DOI] [PubMed] [Google Scholar]
- 18.Tremaroli V, Vacchi-Suzzi C, Fedi S, Ceri H, Zannoni D, Turner RJ. Tolerance of Pseudomonas pseudoalcaligenes KF707 to metals, polychlobiphenyls and chlorobenzoates: effects on chemotaxis-, biofilm- and planktonic-grown cells. FEMS Microbiol Ecol. 2010;74:291–301. doi: 10.1111/j.1574-6941.2010.00965.x. [DOI] [PubMed] [Google Scholar]
- 19.Altschul FS, Gish W, Miller W, Myers EW, Lipman DJ. Basic logical alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 20.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Izumi K, Aramaki M, Kimura T, Naito Y, Udaka T, Uchikawa M, et al. Identification of a prosencephalic-specific enhancer of SALL1: comparative genomic approach using the chick embryo. Pediatr Res. 2007;61:660–665. doi: 10.1203/pdr.0b013e318053423a. [DOI] [PubMed] [Google Scholar]
- 22.Maseda H, Sawada I, Saito K, Uchiyama H, Nakae T, Nomura N. Enhancement of the mexAB-oprM efflux pump expression by a quorum-sensing auto inducer and its cancellation but a regulator, MexT, of the mexEF-oprN efflux pump operon in Pseudomonas aeruginosa. Antimicrob Agents Chemiother. 2004;48:1320–1328. doi: 10.1128/AAC.48.4.1320-1328.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Di Tomaso G, Fedi S, Carnevali M, Manegatti M, Taddei C, Zannoni D. The membrane-bound respiratory chain of Pseudomonas pseudoalcaligenes KF707 cells grown in the presence or absence of potassium tellurite. Microbiol. 2002;148:1699–1708. doi: 10.1099/00221287-148-6-1699. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Thomas PE, Ryan D, Levin DW. An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. (1976) Anal Biochem. 1976;75:168–176. doi: 10.1016/0003-2697(76)90067-1. [DOI] [PubMed] [Google Scholar]
- 26.Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:244–248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krogh A, Brown M, Mian IS, Sjölander K, Haussler D. Hidden Markow models in computational biology: applications to protein modeling. J Mol Biol. 1994;235:1501–1531. doi: 10.1006/jmbi.1994.1104. [DOI] [PubMed] [Google Scholar]
- 29.Jo T, Cheng J. Improving protein fold recognition by random forest. Bioinformatics. 2014:15. doi: 10.1186/1471-2105-15-S11-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pei J, Kim BH, Grishin NV. PROMALS 3D: a tool for multiple protein sequence and structure alignment. Nucleic Acids Res. 2008;36:2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koepke J, Olkhova E, Angerer H, Müller H, Peng G, Michel H. High resolution crystal structure of Paracoccus denitrificans cytochrome c oxidase: new insights into the active site and the proton transfer pathways. Biochim Biophys Acta. 2009;1787:635–645. doi: 10.1016/j.bbabio.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 32.Harrenga A, Michel H. The cytochrome c oxidase from Paracoccus denitrificans does not change the metal center ligation upon reduction. J Biol Chem. 1999;272:33296–33299. doi: 10.1074/jbc.274.47.33296. [DOI] [PubMed] [Google Scholar]
- 33.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 34.Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Science. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the sterochemical quality of protein structures. J App Cryst. 1993;26:283–291. [Google Scholar]
- 36.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Mang EC, Ferrin TE. UCSF Chimera – a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 37.Baker NA, Sept D, Joseph S, Holst MJ, Mac Cammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. PNAS. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lomize MA, Pogozheva ID, Joo H, Mosberg HI, Lomize AL. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 2012;40:D370–D376. doi: 10.1093/nar/gkr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, Ben-Tal N. ConSurf2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44:W344–W350. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le Laz S, Kpebe A, Bauzan M, Lignon S, Rousset M, Brugna M. A biochemical approach to study the role of the terminal oxidases in aerobic respiration of Shewanella oneidensis MR-1. PLOS One. 2014;9:e86343. doi: 10.1371/journal.pone.0086343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mather MW, Springer P, Hensel S, Buse G, Fee JA. Cytochrome oxidase genes from Thermus thermophilus. Nucleotide sequence of the fused gene and analysis of the deduced primary structures for subunits I and III of cytochrome caa3. J Biol Chem. 1993;268:5395–5408. [PubMed] [Google Scholar]
- 42.Noor MR, Soulimane T. Bioenergetics at extreme temperature: Thermus thermophilus ba3- and caa3-type cytochrome c oxidases. Biochim Biophys Acta. 2012;1817:638–649. doi: 10.1016/j.bbabio.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 43.Poole RK, Cook GM. Redundancy of aerobic respiratory chains in bacteria? Routes, reasons and regulation. Adv Microbiol Physiol. 2000;43:165–224. doi: 10.1016/s0065-2911(00)43005-5. [DOI] [PubMed] [Google Scholar]
- 44.Chàvez FP, Lünsdorf H, Jerez CA. Growth of polychlorinated-biphnely-degrading bacteria in the presence of biphenyl and chlorobiphenyls generates oxidative stress and massive accumulation of inorganic polyphosphate. Appl And Eviron Microbiol. 2004;70:3064–3072. doi: 10.1128/AEM.70.5.3064-3072.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morris RL, Schmidt TM. Shallow breathing: bacterial life at low O2. Nat Rev Microbiol. 2013;11:205–212. doi: 10.1038/nrmicro2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol. 2015;13:298–309. doi: 10.1038/nrmicro3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Le Laz S, Kpebe A, Bauzan M, Lignon S, Rousset M, Brugna M. Expression of terminal oxidases under nutrient-starved conditions in Shewanella oneidensis: detection of A-type cytochrome c oxidase. Sci Rep. 2016;6:19726. doi: 10.1038/srep19726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mather MW, Springer P, Fee JA. Cytochrome oxidase gene from Thermus thermophilus. Nucleotide sequence and analysis of the deduced primary structure of subunit IIc of cyt caa3. J Biol Chem. 1991;266:5025–5035. [PubMed] [Google Scholar]
- 49.Lauraeus M, Haltia T, Saraste M, Wikstrom M. Bacillus subtilis expresses two kinds of haem-A-containing terminal oxidases. Eur J Biochem. 1991;197:699–705. doi: 10.1111/j.1432-1033.1991.tb15961.x. [DOI] [PubMed] [Google Scholar]
- 50.Lobo SA, Almeida CC, Carita JN, Teixeira M, Saraiva LM. The haem-copper oxygen reductase of Desulfuvibrio vulgaris contains a dihaem cytochrome c in subunit II. Biochim Biophys Acta. 2008;1777:1528–1534. doi: 10.1016/j.bbabio.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 51.Khalfaoui-Hassani B, Stenou AS, Liotenberg S, Reiss-Husson F, Astier C, Ouchane S. Adaptation to oxygen: role of terminal oxidases in photosynthesis initiation in the purple photosynthetic bacterium, Rubrivivax Gelatinosus. J Biol Chem. 2010;285:19891–19899. doi: 10.1074/jbc.M109.086066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zannoni D, Schoepp-Cothenet B, Hosler J. In: The Purple Phototrophic Bacteria. Hunter CN, Daldal F, Thurnaure MC, Baetty JT, editors. Chapter 27. Springer; Netherlands: 2009. pp. 537–561. [Google Scholar]
- 53.Robin S, Arese M, Forte E, Sarti P, Kolaj-Robin O, Giuffrè A. Functional dissection of the multi-domain di-heme cytochrome c550 from Thermus thermophilus. PLOS One. 2013;8:e55129. doi: 10.1371/journal.pone.0055129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.