Abstract
A metabolic mechanism for oxalic acid biosynthesis in the wood-rotting basidiomycete Fomitopsis palustris has been proposed on the basis of biochemical analyses of glucose metabolism. There was a strong correlation between glucose consumption and oxalate production. Oxalic acid was found to accumulate in the culture fluid in about 80% of the theoretical yield or about 5-fold, on the basis of the fungal biomass harvested. The results clearly indicate that glucose was not completely oxidized to CO2 by the tricarboxylic acid (TCA) cycle but converted mainly to oxalate. The determination of the 12 enzymes concerned has revealed the occurrence of the unprecedented metabolic coupling of the TCA and glyoxylate cycles that support oxalate biosynthesis. In this metabolic system, isocitrate lyase (EC 4.1.3.1), together with oxaloacetase (EC 3.7.1.1), was found to play a pivotal role in yielding oxalate from oxaloacetate via the acetate-recycling routes. Moreover, malate dehydrogenase (EC 1.1.1.37), with an extraordinarily high activity among the enzymes tested, was shown to play an important role in generating NADH by oxidation of malate to oxaloacetate. Thus, it is proposed that the wood-rotting basidiomycete acquires biochemical energy by oxidizing glucose to oxalate.
Metabolism of oxalic acid, which occurs in a wide variety of higher plants, cultures of molds, and even in human urine as an end product, has long been investigated from a medicinal viewpoint (1). Oxalic acid has three chemical natures: it is as proton and electron source and a strong metal chelator, despite its simple chemical formula of (COOH)2. Due to its unique multiple chemical natures, it has been receiving much attention for its various ecological qualities, such as: (i) bioremediation of a wide variety of organic pollutants (2) with lignin biodegradation systems (3–7); (ii) inactivation of copper-containing wood preservatives by wood-rotting fungi (8, 9); (iii) detoxification of aluminum toxicity in Al-resistant buckwheat (10); (iv) crop damage caused by oxalic acid-producing phytopathogens (11, 12); (v) the biofertilizer effect of ectomycorrhizal fungi (13, 14); and (vi) being an electron source for nitrogen fixation in symbiotic rhizobia in a legume plant (15). In addition, oxalic acid is known as the general physiological trait that most brown-rot basidiomycetes, including Fomitopsis palustris, accumulate oxalic acid at greater concentrations in culture fluid, whereas white-rot ones do not because they metabolize and/or decompose oxalic acid by various mechanisms (16–19). Nevertheless, the white-rots were observed to accumulate Ca-oxalate during wood decay processes (20).
However, no systematic biochemical analysis of glucose metabolism for oxalate biosynthesis has been conducted in earlier investigations. Therefore, a physiological role of oxalate production in wood-rotting basidiomycetes has not yet been clarified. Previously, we have reported the occurrence of the two oxalate-producing enzymes, glyoxylate (GLOX) oxidase and oxaloacetase (OXA), in the wood-rotting fungus F. palustris (21, 22). We have purified and characterized a flavohemoprotein glyoxylate dehydrogenase (GLOXDH) that catalyzes dehydrogenation of glyoxylate to oxalate in the presence of cytochrome c (23). Quite recently, we have also found that the glyoxylate-producing enzyme, isocitrate lyase (ICL), and the glyoxylate-using enzyme, malate synthase (MS), the GLOX cycle key enzymes, commonly occur in white- and brown-rot basidiomycetes, although they were grown on the glucose medium. Thus, it has been hypothesized that oxalate biosynthesis may link with the TCA and GLOX cycles (24).
In this context, it is of importance to elucidate a physiologically intrinsic role of the oxalic acid biosynthesis as a metabolic model for a wide variety of oxalate-producing organisms. In this paper, biochemical analyses of glucose metabolism were conducted in relation to oxalate production and activities of all of the enzymes involved in the metabolic system. The results have revealed the occurrence of a metabolic coupling of the TCA and the GLOX cycles in coordination with the biosynthesis of oxalic acid in the brown-rot fungus F. palustris. Moreover, an acetate-recycling system has been discovered. Also, ICL (EC 4.1.3.1), which is shared by these two cycles, has been revealed to be a pivotal enzyme involved in a new biochemical device for producing oxalic acid. The oxalate biosynthesis in this fungus was characterized as the oxalate fermentation, through which biochemical energy is generated for the growth of the fungus. Here we report an unprecedented metabolic system that explains an important physiological role of the oxalate biosynthesis in the wood-rotting basidiomycete F. palustris.
Materials and Methods
Chemicals.
All chemical and biochemical reagents were of reagent grade. NADP, acetyl CoA, dl-isocitric acid, and 2-oxoglutaric acid were obtained from Nacalai Tesque (Kyoto). CoA and β-NAD+ were from Oriental Yeast (Tokyo), thiamine pyrophosphate was from Wako Pure Chemicals (Osaka), and glyoxylic acid and cytochrome c were purchased from Sigma. The protein assay kit was from Bio-Rad. Oxalic acid, acetic acid, and glucose assay kits were purchased from Boehringer Mannheim.
Organism and Growth Conditions.
One of the copper-tolerant brown-rot basidiomycetes, F. palustris (formerly called Tyromyces palustris) (Berkeley et Curtis) Murill, which is a Japanese industrial fungus for wood-preservative efficacy tests, was used as a model fungus. The fungus was cultivated in modified Kirk's medium (200 ml) containing 2% (wt/vol) glucose and 24 mM ammonium tartrate as the carbon and nitrogen sources, respectively (25). Alternatively, the fungus was grown on peptone-containing medium with 2% glucose (24) to compare enzyme activity between the two different culture media.
Preparation of Cell-Free Extracts and Enzyme Assays.
Cell-free extracts were prepared from the fungal mycelia in the same way as previously reported (24). All 12 enzyme activities in the fungal extracts were determined spectrophotometrically by using a double-beam spectrophotometer (Hitachi model U-3000, Hitachi, Tokyo) equipped with a temperature controller and external recorder. Each of the assayed enzymes (1–12) listed in Table 1 catalyzed the reaction step with the same number (1–12) as shown in Fig. 5. ICL and MS activities were determined by the methods of Dixon and Kornberg (26). ICL activity was assayed by measurement of the increase in absorbance at 324 nm because of the formation of phenylhydrazone derivative of glyoxylate produced from isocitrate. MS activity was determined on the basis of the consumption of acetyl-CoA with the decrease in absorbance at 232 nm. Malate dehydrogenase (MDH), 2-oxoglutarate dehydrogenase (ODH) and isocitrate dehydrogenase (IDH) activity was determined by measuring the increase in absorbance at 340 nm due to the reduction of NAD or NADP, on the basis of the reported methods (27, 28). Aconitase activity was determined by measurement of the increase in absorbance at 240 nm (29). Citrate synthase activity was assayed by measurement of the decrease in absorbance at 232 nm due to consumption of acetyl-CoA. Acetyl-CoA synthase activity was determined by the modified coupling assay method, as described by Londesborough et al. (30). Succinate dehydrogenase activity was determined on the basis of a modified Moore and Ewaze method (28). Fumarase activity was determined by measuring the conversion of malate to fumarate (31). GLOXDH activity was determined by measurement of the increase in absorbance at 550 nm due to the reduction of cytochrome c (23). OXA activity was determined by measurement of the decrease in absorbance at 255 nm due to the hydrolysis of oxaloacetate (22).
Table 1.
Activities of the enzymes involved in a metabolic system for oxalate biosynthesis
| Number* | Enzymes assayed | Specific activity† |
|---|---|---|
| 1. | Isocitrate lyase (ICL) | 671 |
| 2. | Isocitrate dehydrogenase (IDH) | 88 |
| 3. | 2-Oxoglutarate dehydrogenase (ODH) | 0 |
| 4. | Succinate dehydrogenase | 33 |
| 5. | Fumarase | 261 |
| 6. | Malate dehydrogenase (MDH) | 5,840 |
| 7. | Citrate synthase | 1,475 |
| 8. | Aconitase | 691 |
| 9. | Malate synthase (MS) | 568 |
| 10. | Oxaloacetase (OXA) | 300 |
| 11. | Acetyl-CoA synthase | 17‡ |
| 12. | Glyoxylate dehydrogenase (GLOXDH) | 65 |
The enzymes, listed in numerical order, correspond to enzymes that catalyze the reaction numbered in the metabolic system in Fig. 5.
Enzymatic activities (nmol min−1⋅mg−1 protein) were determined for fungus grown for 4 days on ammonium tartrate-containing medium.
The enzyme was assayed for the 2-day-old mycelia of the fungus.
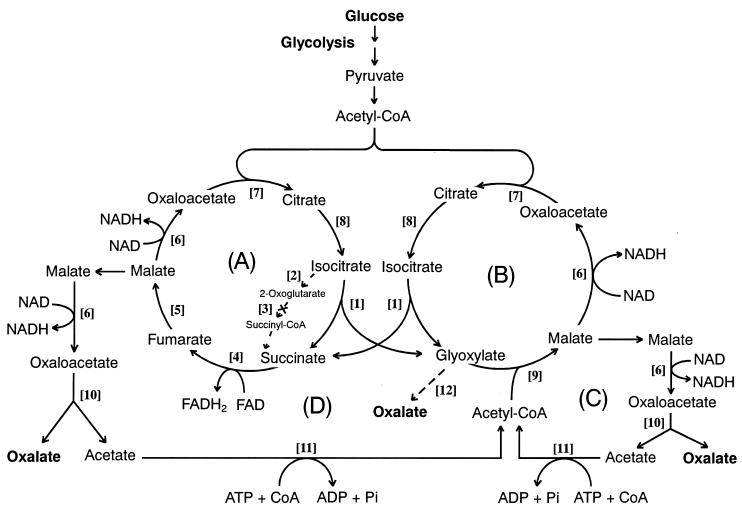
Figure 5.
A proposed metabolic mechanism for the oxalate biosynthesis in the wood-rotting basidiomycete F. palustris. (A) the TCA cycle; (B) GLOX cycle; (C and D) acetate-recycling routes. For the enzyme catalyzing each step reaction ([1]–[12]), see Table 1.
Enzyme activities were expressed in terms of total (μmol min−1⋅culture−1) and specific activity (μmol min−1⋅mg−1 protein), unless otherwise stated. One unit of enzyme activity is defined as the amount of enzyme that catalyzes the formation of 1 μmol product per minute or the consumption of 1 μmol substrate per minute under the conditions described.
Determination of Proteins, Glucose, Oxalate, Acetate, and Other Organic Acids.
Protein concentrations were determined by the Bio-Rad method (32) with BSA as the standard. Glucose, oxalic acid, and acetic acid content in the culture fluid was quantitatively determined enzymatically by using the glucose, oxalic acid, and acetic acid assay kits, respectively, following the methods provided by Boehringer. Other organic acids, including oxalic acid, were analyzed by GC-MS (Shimazu GC-MS QP-5050A) after derivatization of the acids with N-(t-butyldimethylsilyl)N-methyltrifluoroacetoamide, according to the reported method (33).
Results and Discussion
Metabolic Conversion of Glucose to Oxalic Acid in F. palustris.
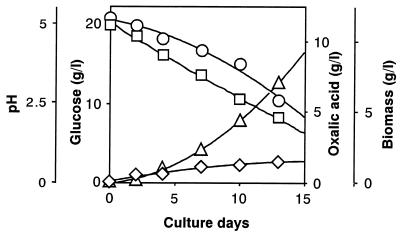
First, focusing on the prominent oxalic acid-producing fungus (F. palustris) known as the copper-tolerant brown-rot fungus, we determined the amount of glucose consumed, the amount of oxalic acid, and the fungal biomass produced during the cultivation period, showing a strong correlation between glucose consumption and oxalic acid production (Fig. 1). Fig. 1 also shows that the amounts of oxalic acid produced correlate with decrease in pH (from 5.0 to 2.0) in the culture fluid. At the end of cultivation (day 13), the amounts of glucose consumed, oxalic acid produced, and the fungal biomass obtained were found to be 10.5, 7.0, and 1.5 g, respectively. Thus, yields of oxalate and biomass produced were 67 and 14%, respectively, on the basis of total glucose consumption and the rest, 19%, was for CO2 released and other metabolites, if any. This finding suggests that glucose was not completely oxidized to CO2 by the TCA cycle. Correcting total glucose consumption with the weight of the biomass shows that about 9 g of glucose was consumed, mostly for oxalate production, because fungal biomass is assumed to be composed of mainly carbohydrates, which are roughly equivalent to the amount of glucose. Thus, the corrected oxalate yield was estimated to be 80%. As a rule, the following equation (Eq. 1) could be derived for the overall conversion of glucose to oxalic acid in this fungus. The equation expresses that 1 mol glucose (180 g) is converted to 2 mol oxalic acid (180 g).
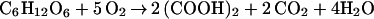 |
1 |
So, it is intriguing to inquire why the conversion of glucose to oxalate by this fungus is so high, what a kind of biochemical device is involved in producing this organic acid, and for what. To answer these questions, enzymatic analyses of biochemical systems involved in the metabolic conversion of glucose to oxalic acid were conducted.
Figure 1.
Relationship between glucose consumption and oxalate and fungal biomass production accompanied by pH reduction during the cultivation of F. palustris. Circles, glucose; squares, pH; triangles, oxalic acid; diamonds, biomass.
Enzymatic Analyses.
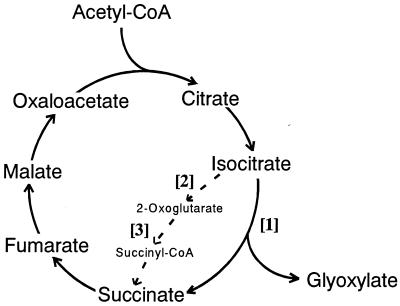
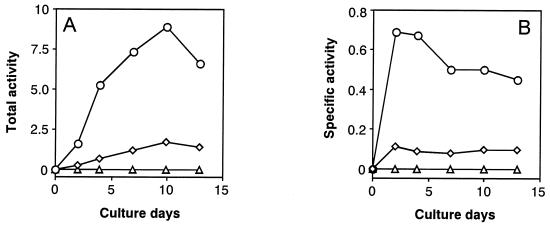
First, we attempted to compare activities of the two branch-point enzymes, i.e., NADP-specific isocitrate dehydrogenase (IDH) [2] as the TCA cycle key enzyme and ICL [1] as the GLOX cycle key enzyme, as shown in Fig. 2, because it has not been clarified yet which of these two enzymes is more major in yielding succinate from isocitrate as a common precursor in wood-rotting fungi. Fig. 3 A and B show the changes in total and specific activities, respectively, of the two branch-point enzymes (ICL and IDH) and ODH [3]. Interestingly, the results clearly indicate that ICL activity is much greater than IDH activity (the ratio of ICL to IDH is 8:1) throughout the cultivation period, which contrasts sharply with the reported findings that the former [1] was not or little detected, whereas the latter [2] was predominant when microorganisms were grown on glucose (34–37). Furthermore, ODH [3], as another key enzyme of the TCA cycle, was not detected, and NAD-specific IDH also was not found (data not shown). Thus, it has been shown that this fungus has the modified TCA cycle with ICL [1] as a major enzyme and does not contain the Krebs TCA cycle, which oxidizes 1 mol acetyl-CoA to 2 mol CO2 (dotted line in Fig. 2); a biochemical role of ICL in the TCA cycle in this fungus is to cleave isocitrate, yielding succinate and glyoxylate (Fig. 2). These findings are in good accordance with Eq. 1. Smaller amounts of oxoglutarate formed after decarboxylation of isocitrate by IDH [2] may be bypassed to succinate via glutamate and γ-aminobutyrate route as a minor pathway, as found in other bacteria (38, 39) and fungi (28, 40, 41).
Figure 2.
Occurrence of the modified TCA cycle with ICL in the wood-rotting fungus, F. palustris. Normal routes in the Krebs TCA cycle are indicated with dotted line. [1], ICL; [2], IDH; [3], ODH.
Figure 3.
Change in total and specific activities of ICL, IDH, and ODH during cultivation of F. palustris. The total (A) and the specific (B) activities are defined in the text. Circles, ICL; diamonds, IDH; triangles, ODH.
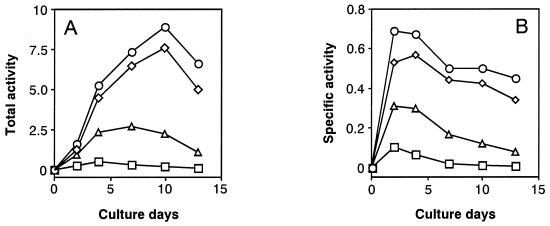
Second, we also measured activities of MS as another key enzyme of the GLOX cycle and the two oxalate-producing enzymes, OXA and GLOXDH, in comparison with ICL activity. Fig. 4 A and B show changes in total and specific activities, respectively, of these four enzymes from the same cultures during the cultivation period. The results show that activities of both ICL and MS increased more markedly at the earlier stage of cultivation than the two oxalate-producing enzymes (OXA and GLOXDH) and were more or less maintained at a higher level, with a gradual decrease in activity thereafter. This means that the activity of both GLOX key enzymes was not repressed by glucose but was strongly expressed as constitutive enzymes, in sharp contrast with the general behavior of the GLOX enzymes (34). However, the activity of OXA was greater than that of GLOXDH, which decreased more rapidly than the former after day 4 of the cultivation period, indicating that OXA was a major enzyme in producing oxalate. It is noteworthy that the changes in activity of OXA are in good accordance with those of the GLOX key enzymes. The result obtained is consistent with the previous finding that the activities of all four enzymes increased in parallel during cultivation (24). Thus, we infer that both TCA and GLOX cycles cooperatively couple with the oxalate-producing enzyme system, as proposed in Fig. 5.
Figure 4.
Change in total and specific activities of the GLOX key enzymes (ICL and MS) and the oxalate-producing enzymes (OXA and GLOXDH) during the cultivation of F. palustris. The total (A) and the specific (B) activities are defined in the text. Circles, ICL; diamonds, MS; triangles, OXA; squares, GLOXDH.
Then, in this experiment, we were motivated to detect all 12 enzymes involved in the metabolic system in this fungus. Table 1 shows that all of the enzymes except ODH were active to a varying degree, and that the greatest activity was found for MDH: 5,840 nmol min−1⋅mg−1 protein. The reason for the high activity of MDH may be that the enzyme is intrinsically required for catalyzing the reaction step [6] at the four different reaction sites in the metabolic system in Fig. 5. By changing the nitrogen nutrient from ammonium tartrate to peptone, similar results were obtained; MDH with 2,639 nmol min−1⋅mg−1 protein was recorded. Importantly, ICL and OXA predominate over IDH and GLOXDH, respectively, regardless of the culture media. Furthermore, acetyl-CoA synthase [11] was detected in a significant amount. This indicates that acetate liberated from oxaloacetate hydrolyzed by OXA was recycled by recycling routes C and D involving acetyl-CoA synthase [11] (Fig. 5). In fact, free accumulating acetate was not detected at all by rigorous enzymatic analysis. Moreover, no other free organic acid could be detected by GC-MS analysis. Thus, we conclude that acetate and other intermediary organic acids were not leaked out of the metabolic system into the culture fluid but were metabolized to oxalate with high efficiency as the result of the coordination of the acetate-recycling system (recycling routes C and D) with the TCA (A) and GLOX (B) cycles (Fig. 5).
Biochemical Role of a Metabolic System for Biosynthesis of Oxalic Acid.
On the basis of physiological and enzymatic analyses of the metabolism of glucose, a metabolic system for oxalate biosynthesis in F. palustris was discovered, as shown in Fig. 5. The metabolic system consists of two cycles, TCA and GLOX, which cooperatively couple with each other and also coordinate with the acetate-recycling routes (C and D) to bring the liberated acetate back into the GLOX cycle via acetyl-CoA. Because the activity of GLOXDH is much lower than that of OXA, glyoxylate may not be oxidized effectively to oxalate (step 12) but may be condensed with acetyl-CoA to form malate by MS (step 9). Thus, most oxalate is inferred to be produced from oxaloacetate and to be hydrolyzed by OXA involved in both recycling routes (C and D).
In principle, the metabolic system in Fig. 5 indicates that 2 mol acetyl-CoA, derived from 1 mol glucose through the glycolysis shunt, enter Cycles A and B first and finally exit as oxalate of the end product from recycling routes C and D. A biochemical role of the metabolic system is to primarily oxidize acetyl-CoA to yield oxalate, which accumulates in the culture fluid, because this fungus does not contain oxalate-decomposing enzyme systems such as oxalate oxidase (42), oxalate decarboxylase (43, 44), and the ligninolytic systems occurring in white-rot fungi (3–7).
It is noteworthy that, among all of the enzymes determined, MDH had extraordinarily high activity, which shows that MDH plays a major role in generating energy (NADH, equivalent to 3 ATP) by oxidation of malate to oxaloacetate, which is the direct precursor of oxalic acid. Thus, evidently, oxalate production is coupled with energy production.
Subsequently, it has been established that oxaloacetate cannot be shuttled through the membrane system of mitochondria and glyoxysomes, and not oxaloacetate but malate may flow out from Cycle A or B to be oxidized to form oxaloacetate (Fig. 5). To maintain the level of intermediates in both cycles, the TCA and GLOX cycles are replenished with succinate and glyoxylate, respectively. Consequently, the proposed mechanism indicates that not only is the GLOX cycle anaplerotic to the TCA cycle but also vice versa, to keep both cycles coupling. From the literature surveyed, the metabolic system found in this investigation is the first example of metabolic coupling or interdependency of the TCA and GLOX cycles, which has never been reported from any other organism. However, Li et al. (45) have discovered a different type of oxalate-producing system involving acetyl-CoA from the plant pathogen Burkholderia glumae. Moreover, ICL, which is shared by the two coupling cycles, is focused as a crucial enzyme for the oxalate biosynthesis in this fungus. Quite recently, we have reported that itaconate, as a specific inhibitor of ICL, potently inhibited the oxalate biosynthesis and consequently the growth of the fungus, confirming the pivotal role of the enzyme for the fungal growth (24), which supports the reaction mechanism proposed in Fig. 5.
Taking into account the biochemical energy generated and consumed in the metabolic system in Fig. 5, overall oxidation of 2 mol acetyl-CoA, yielding 2 mol oxalate, results in the generation of a net amount of 4 NADHs and 2 ATPs or production of 14 ATPs, on the assumption that no other metabolic pathway oxidizes acetyl-CoA. Because white-rot fungi not only biosynthesize oxalate from wood carbohydrates but also produce it from lignin degradation products, such as the side-chain cleavage (46) and aromatic ring cleavage products (47), and further metabolize it, they can get more energy from oxalate via formate, which is a source of NADH generated by formate dehydrogenase. In fact, formate dehydrogenase has been reported for the first time to occur in the white-rot fungi, including Coriolus versicolor (48). Because of their advantage in energy acquisition over brown-rot fungi, which cannot decompose lignin to yield oxalate nor metabolize it, white-rot fungi with a much greater number of species have probably succeeded in occupying higher positions in nature through biochemical evolution.
Taken together, the results of this investigation provide unprecedented evidence for the occurrence of metabolic linkage of oxalate biosynthesis with both TCA and GLOX cycles, which are coupled with each other to supply oxaloacetate of the oxalate precursor in the wood-rotting fungi. The highly ordered metabolic coupling system may serve as a metabolic machinery for the oxalate biosynthesis accompanied by energy production. Consequently, it has been proposed as a new concept that the wood-rotting basidiomycete F. palustris acquires biochemical energy for growth by oxidizing glucose to oxalate, which may be a general feature of both brown- and white-rot fungi during the wood decay process. However, further research remains to elucidate biochemical mechanisms controlled by the regulatory key enzymes involved in the metabolic system.
Acknowledgments
This work was supported by a Grant-in-Aid (13556024) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, which also has provided a scholarship for E.M., a student from Indonesia, through the Japan Society for the Promotion of Science Core University Program.
Abbreviations
- GLOX
glyoxylate cycle
- GLOXDH
glyoxylate dehydrogenase
- OXA
oxaloacetase
- ICL
isocitrate lyase
- MS
malate synthase
- IDH
isocitrate dehydrogenase
- MDH
malate dehydrogenase
References
- 1.Hodgkinson A. Oxalic Acid in Biology and Medicine. London: Academic; 1977. pp. 1–324. [Google Scholar]
- 2.Barr D P, Aust S D. Environ Sci Technol. 1994;28:79A–87A. doi: 10.1021/es00051a724. [DOI] [PubMed] [Google Scholar]
- 3.Akamatsu Y, Ma D B, Higuchi T, Shimada M. FEBS Lett. 1990;269:261–263. doi: 10.1016/0014-5793(90)81169-o. [DOI] [PubMed] [Google Scholar]
- 4.Ma D B, Hattori T, Akamatsu Y, Adachi M, Shimada M. Biosci Biotechnol Biochem. 1992;56:1378–1381. [Google Scholar]
- 5.Shimada M, Akamatsu Y, Tokimatsu T, Mii K, Hattori T. J Biotechnol. 1997;53:103–113. [Google Scholar]
- 6.Popp J L, Kalyanaraman B, Kirk T K. Biochemistry. 1990;29:10475–10480. doi: 10.1021/bi00498a008. [DOI] [PubMed] [Google Scholar]
- 7.Kuan I C, Tien M. Proc Natl Acad Sci USA. 1993;90:1242–1246. doi: 10.1073/pnas.90.4.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsunoda K, Nagashima K, Takahashi M. Mater Org. 1997;31:31–41. [Google Scholar]
- 9.Murphy R J, Levy J F. Trans Br Mycol Soc. 1983;81:165–168. [Google Scholar]
- 10.Ma J F, Zheng S J, Matsumoto H. Nature (London) 1997;390:569–570. [Google Scholar]
- 11.Maxwell D P, Bateman D F. Phytopathology. 1968;58:1635–1642. [Google Scholar]
- 12.Margo P, Marciano P, Lenna P D. FEMS Microbiol Lett. 1984;24:9–12. [Google Scholar]
- 13.Malajczuk N, Cromack K., Jr New Phytol. 1982;92:527–531. [Google Scholar]
- 14.Lapeyrie F, Chilaver G A, Bhem C A. New Phytol. 1987;106:139–146. [Google Scholar]
- 15.Trinchant J C, Guérin V, Rigaud J. Plant Physiol. 1994;105:555–561. doi: 10.1104/pp.105.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takao S. Appl Microbiol. 1965;13:732–737. doi: 10.1128/am.13.5.732-737.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dutton M V, Evans C S, Akey P T, Wood D A. Appl Microbiol Biotechnol. 1993;39:5–10. [Google Scholar]
- 18.Green F, Clausen C A. Holzforschung. 1999;53:563–568. [Google Scholar]
- 19.Akamatsu Y, Takahashi M, Shimada M. Mater Org. 1993;28:251–264. [Google Scholar]
- 20.Dutton M V, Evans C S. Can J Microbiol. 1996;42:881–895. [Google Scholar]
- 21.Akamatsu Y, Shimada M. Phytochemistry. 1994;37:649–653. [Google Scholar]
- 22.Akamatsu Y, Ohta A, Takahashi M, Shimada M. J Jpn Wood Res Soc. 1991;37:575–577. [Google Scholar]
- 23.Tokimatsu T, Nagai Y, Hattori T, Shimada M. FEBS Lett. 1998;437:117–121. doi: 10.1016/s0014-5793(98)01104-1. [DOI] [PubMed] [Google Scholar]
- 24.Munir E, Yoon J J, Tokimatsu T, Hattori T, Shimada M. J Wood Sci. 2001;47:368–373. doi: 10.1073/pnas.191389598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirk T K, Schultz E, Connors W J, Lorenz L F, Zeikus J G. Arch Microbiol. 1978;117:277–285. [Google Scholar]
- 26.Dixon G H, Kornberg H L. Biochem J. 1959;72:3P. [Google Scholar]
- 27.Labrou N E, Clonis Y D. Arch Biochem Biophys. 1997;337:103–114. doi: 10.1006/abbi.1996.9748. [DOI] [PubMed] [Google Scholar]
- 28.Moore D, Ewaze J O. J Gen Microbiol. 1976;97:313–322. [Google Scholar]
- 29.Kennedy M C, Emptage M H, Dreyer J-L, Beinert H. J Biol Chem. 1983;258:11098–11105. [PubMed] [Google Scholar]
- 30.Londesborough J C, Yuan S L, Webster L T., Jr Biochem J. 1973;133:23–36. doi: 10.1042/bj1330023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cooper T G, Beevers H. J Biol Chem. 1969;244:3507–3513. [PubMed] [Google Scholar]
- 32.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 33.Hattori T, Akitsu N, Seo G-S, Ohta A, Shimada M. Ann Report Interdiscipl Res Inst Environ Sci. 2000;19:59–65. [Google Scholar]
- 34.Kornberg H L. Biochem J. 1966;99:1–11. doi: 10.1042/bj0990001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O‘Connel B T, Paznokas J L. J Bacteriol. 1980;143:416–421. doi: 10.1128/jb.143.1.416-421.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rua J, Arriaga D D, Busto F, Soler J. J Bacteriol. 1989;171:6391–6393. doi: 10.1128/jb.171.11.6391-6393.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt G, Stahmann K P, Sahm H. Microbiology. 1996;142:411–417. doi: 10.1099/13500872-142-2-411. [DOI] [PubMed] [Google Scholar]
- 38.McDermott T R, Griffith S M, Vance C P, Graham P H. FEMS Microbiol Rev. 1989;63:327–340. [Google Scholar]
- 39.Green L S, Emerich D W. J Bacteriol. 1997;179:194–201. doi: 10.1128/jb.179.1.194-201.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moore D. Exp Mycol. 1984;8:283–297. [Google Scholar]
- 41.Kumar S, Punekar N S. Mycol Res. 1997;101:403–409. [Google Scholar]
- 42.Anguilar C, Urzúa U, Koenig C, Vicuña R. Arch Biochem Biophys. 1999;366:275–282. doi: 10.1006/abbi.1999.1216. [DOI] [PubMed] [Google Scholar]
- 43.Dutton M V, Kathiara M, Gallager I M, Evans C S. FEMS Microbiol Lett. 1994;116:321–325. [Google Scholar]
- 44.Mehta A, Datta A. J Biol Chem. 1991;266:23548–23553. [PubMed] [Google Scholar]
- 45.Li H-Q, Matsuda I, Fujise Y, Ichiyama A. J Biochem. 1999;126:243–253. doi: 10.1093/oxfordjournals.jbchem.a022429. [DOI] [PubMed] [Google Scholar]
- 46.Kersten P J, Kirk T K. J Bacteriol. 1987;169:2195–2201. doi: 10.1128/jb.169.5.2195-2201.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Umezawa T, Higuchi T. FEBS Lett. 1987;218:255–260. [Google Scholar]
- 48.Mii K, Hattori T, Shimada M. Wood Res. 1996;83:23–26. [Google Scholar]