Abstract
Introduction
Phelan-McDermid syndrome (PMS) is caused by SHANK3 haploinsufficiency. Its wide phenotypic variation is attributed partly to the type and size of 22q13 genomic lesion (deletion, unbalanced translocation, ring chromosome), partly to additional undefined factors. We investigated a child with severe global neurodevelopmental delay (NDD) compatible with her distal 22q13 deletion, complicated by bilateral perisylvian polymicrogyria (BPP) and urticarial rashes, unreported in PMS.
Methods
Following the cytogenetic and array-comparative genomic hybridization (CGH) detection of a r(22) with SHANK3 deletion and two upstream duplications, whole-genome sequencing (WGS) in blood and whole-exome sequencing (WES) in blood and saliva were performed to highlight potential chromothripsis/chromoanagenesis events and any possible BPP-associated variants, even in low-level mosaicism.
Results
WGS confirmed the deletion and highlighted inversion and displaced order of eight fragments, three of them duplicated. The microhomology-mediated insertion of partial Alu-elements at one breakpoint junction disrupted the topological associating domain joining NFAM1 to the transcriptional coregulator TCF20. WES failed to detect BPP-associated variants.
Conclusions
Although we were unable to highlight the molecular basis of BPP, our data suggest that SHANK3 haploinsufficiency and TCF20 misregulation, both associated with intellectual disability, contributed to the patient’s NDD, while NFAM1 interruption likely caused her skin rashes, as previously reported. We provide the first example of chromoanasynthesis in a constitutional ring chromosome and reinforce the growing evidence that chromosomal rearrangements may be more complex than estimated by conventional diagnostic approaches and affect the phenotype by global alteration of the topological chromatin organisation rather than simply by deletion or duplication of dosage-sensitive genes.
Keywords: polymicrogyria, SHANK3, NFAM1, TCF20, topologically associating domains
Introduction
The Phelan-McDermid syndrome (PMS; MIM#606232) is a neurodevelopmental disorder caused by the deletion or interruption of SHANK3 gene at chromosome 22q13,1–5 leading to haploinsufficiency of SHANK3 protein, which is a key structural component of the glutamatergic postsynaptic density.6 Patients with PMS mainly exhibit neurological and behavioural symptoms including hypotonia, intellectual disability (ID), impaired language development (delayed or absent speech), behavioural features consistent with disorders of the autistic spectrum and seizures.
The penetrance and expressivity of these symptoms may be variable, with different degrees of severity at different ages3 7–9 as well as congenital malformations,4 5 even among patients with the uniformly-sized recurrent 22q13.3 deletion2 or interstitial deletions affecting only SHANK3. 3 10
It is still uncertain whether the phenotypic variability is caused by variants regarding the genes and expression controlling elements present on the corresponding region of the intact chromosome 22, by a genome rearrangement more complex than estimated or even by a cryptic mosaic condition of the 22q rearrangement. The latter mechanism may particularly apply to the PMS individuals in whom the distal 22q deletion lies in a ring chromosome. Indeed, rings are intrinsically unstable, resulting in the continuous production of cells having duplicated rings, broken portions of the original ring or even missing the entire chromosome.
In this study, we investigated a patient with a de novo ring chromosome 22, present in all the 50 metaphases analysed, who showed a complex cortical brain abnormality and unusual dermatological features, in addition to the core phenotype of PMS. Since array-CGH had revealed a distal 22q deletion and two non-contiguous duplications, we performed whole-genome sequencing (WGS) to investigate a possible event of chromothripsis/chromoanagenesis. WGS confirmed this hypothesis, highlighting eight breakpoints along the chromosome 22 long arm and disordered assembly of the resulting fragments, with disruption of topological associating domains (TAD) and breakage of four genes.
Methods
Clinical report
A 3-year-old female child came to our attention after a ring chromosome 22 was detected by cytogenetics when she was 8 months old. The patient was the first child of healthy unrelated parents with uneventful family history. She was born by caesarean section to a 37-year-old mother and 32-year-old father at 37 weeks of gestational age because of umbilical cord wrapped around her neck. Birth weight was 2000 g (10th–25th centile), length 44.5 cm (25th centile), cranial circumference 31 cm (25th centile); APGAR scores were 8/9 at 1’/5’, respectively. At birth, she was initially transferred to the intensive care unit and then to the neonatal pathology unit for 28 days. Hypoglycaemia and thrombocytopaenia were solved in 4 and 12 days, respectively, while her feeding difficulties, characterised by poor sucking, required gavage. In the first months of life, generalised hypotonia and delayed psychomotor development were observed. She acquired head control at 12 months, sat at the age of 13 months, was standing at 20 months and walked with support at the age of 24 months. She uttered her first words at 12 months but her parents refer no subsequent progression of language. Hypersomnia was present since the age of 2 years.
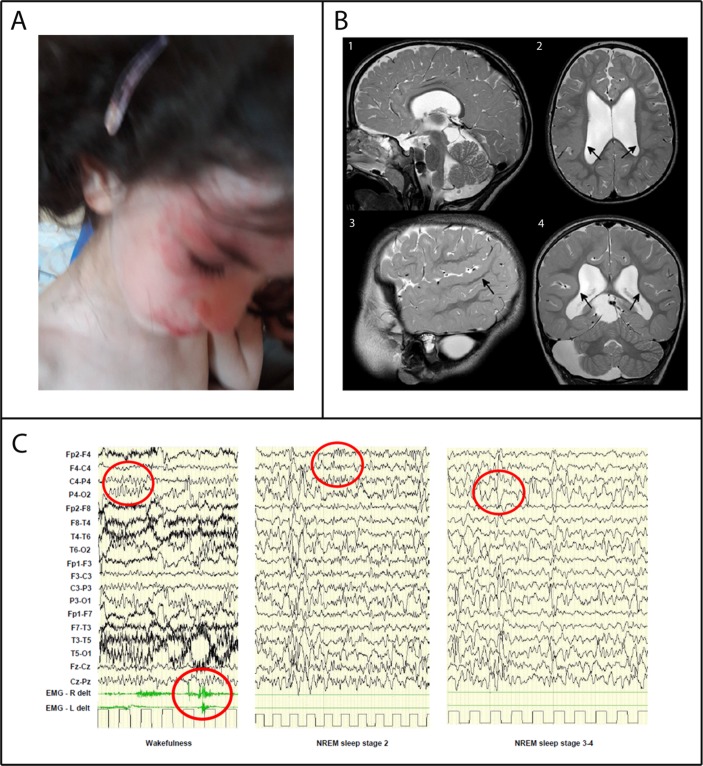
At the age of 22/12 years (26 months), she began to manifest atopic dermatitis characterised by recurrent pruritic urticarial rushes on the face and the trunk in addition to persistent limb pilaris keratosis. Food-specific IgE antibody tests resulted negative and her family history of atopy was unremarkable. At last evaluation, at the age of 3 years, her weight was 12.25 kg (25th centile), height 95.5 cm (75th centile), cranial circumference 50.50 cm (75th centile). She showed ptosis of her left eye and long face but no major dysmorphic features. Griffiths scale scores revealed severe ID (IQ: 24, mental age equivalent 8 months) and language was absent. Poor eye contact and stereotypic hand movements characterised her neurological picture. Diffuse mild muscular hypotrophy and weakness were present, with trunk hypotonia, unsteadiness of orthostatic position and ataxic gait with support. Bilateral coxa valga, hyporeflexia in the lower limbs, together with bilateral Babinski sign, were present. Orofacial hypotonia, with drooling and bruxism, and severe swallowing disorder were observed. She suffered episodes of gastro-oesophageal reflux. Kidney ultrasound, urinalysis and function blood tests were normal. Atopic dermatitis was still present (figure 1A). The brain 3T MRI showed complex brain anomalies including bilateral perisylvian polymicrogyria (BPP) (figure 1B). The white matter volume was diffusely reduced. The third and lateral ventricles were markedly enlarged, probably because of a partial stenosis of the aqueduct caused by a large cyst compressing the lamina quadrigemina. The corpus callosum was thinned. An arachnoid cyst with mass effect compressed the right cerebellar hemisphere. EEG recording showed slowing of background activity during wakefulness, while the physiologic features (K-complex and spindles) during sleep were rare with irregular morphologies. Slow waves, of greater amplitude than the background rhythm, were present in the posterior areas, bilaterally, without side predominance. On electromyography (EMG) derivation, isolated myoclonic jerks were recorded without EEG correlates (figure 1C). She never presented episodes of epileptic febrile or non-febrile seizures and no antiepileptic drugs were administered so far.
Figure 1.
Patient’s clinical findings: (A) Lateral view of the patient showing urticarial rashes on her face. (B) Brain MRI at the age of 3 years showed bilateral symmetric areas of polymicrogyric cortex in the posterior part of the Sylvian fissures, extending to the parietal lobes. In these areas, the cortex appears thickened and characterised by small gyri (arrows in 2, 3, 4). Corpus callosum is thinned. An arachnoid cyst is present in the right part of the posterior fossa (4) and posteriorly to the lamina quadrigemina (1). (C) EEG-polygraphic recording at the age of 3 years. The red circles indicate background activity during wakefulness, myoclonic jerks on EMG derivation and K-complex and spindles and slow waves during sleep stages 2 and 3–4.
Molecular studies
Genomic DNA was extracted from proband’s, parents’ and maternal grandparents’ blood and saliva with standard protocols.
Array-CGH investigations
Array-CGH analysis was performed on proband’s DNA from blood using an Agilent Human Genome CGH Microarray Kit 4×180 k with an overall median probe space of 13 Kb. All nucleotide positions refer to the Human Genome, February 2009 Assembly (hg19). Data analysis was performed using Agilent Cytogenomics V.2.5.8.1. High-resolution G-banding metaphase chromosomes and interphase nuclei were obtained from PHA-stimulated blood lymphocytes cultured for 72 hours, according to standard protocols. Dual and three-colour FISH experiments with prokaryotic artificial chromosome (PAC) probes, selected according to the UCSC Human Genome Browser (GRCh37/hg19) and obtained from the human library RPCI-4 and 3, were performed as previously described.11 The pan-telomeric peptide nucleic acid (PNA) probe (PNA FISH kit/Cy3, Dako Denmark A/S) which recognises the consensus sequence (TTAGGG)n of human telomeres was hybridised according to the manufacturer’s instructions.
Whole-genome sequencing (WGS) and Sanger confirmation of breakpoint junctions
Sequencing libraries to perform WGS were prepared with Illumina’s TruSeq DNA PCR Free kit (San Diego, California, USA) according to manufacturer’s protocol. We obtained an average coverage of 42X in the father’s sample and 37X in the proband’s sample. Reads from the fastq files were mapped to the human reference genome GRCh37/hg19 using Isaac Genome Alignment Software (V.iSAAC-03.16.06.06).12 Coverage graphs were performed plotting the average coverage of 1000 bp windows sliding over the whole chromosome length, in order to identify large deletions and duplications in each chromosome. Structural variants were called using Lumpy (V.0.2.12) and Manta (V.0.29.6).13 14 Then these structural variants were visualised and manually checked in Integrative Genomics Viewer (IGV) genome browser (V.2.3.72) in order to reconstruct chromosomes showing chromothripsis events. Sanger sequencing was performed in order to verify the structural variations predicted by WGS findings. Primer3 software was used to design primers for sequencing de novo breakpoint junctions of structural variations. PCR was performed by using Taq Polymerase (Invitrogen) with elongation times of 1 min. Sequencing reads were analysed manually to characterise the exact breakpoints.
Whole-exome sequencing (WES)
To test for any possible constitutional or mosaic mutation that could explain the polymicrogyria, proband’s DNA from both blood and saliva was analysed by OneSeq protocol, at an average coverage of 194.3x and 134.7x, respectively, on an Illumina HiSeq2500, as previously described.15 After reads alignment, variant calling, annotation and filtering for possible artefacts was done as previously reported.16
The sequences of polymicrogyria candidate genes (online supplementary table 1 reported in Parrini et al 17) were carefully examined together with PI4KA.18 Sanger sequencing was performed to validate the only variant possibly associated with polymicrogyria, SRPX2 at Xq22.1, by using standard methods, extending the analysis to the three-generation family members and performing X-inactivation investigation.
jmedgenet-2017-105125supp001.docx (4.7MB, docx)
Topologically associating domains (TADs) analysis
Search for TADs was performed by using the web-based 3D Genome Browser.19
Results
Cytogenetics investigations
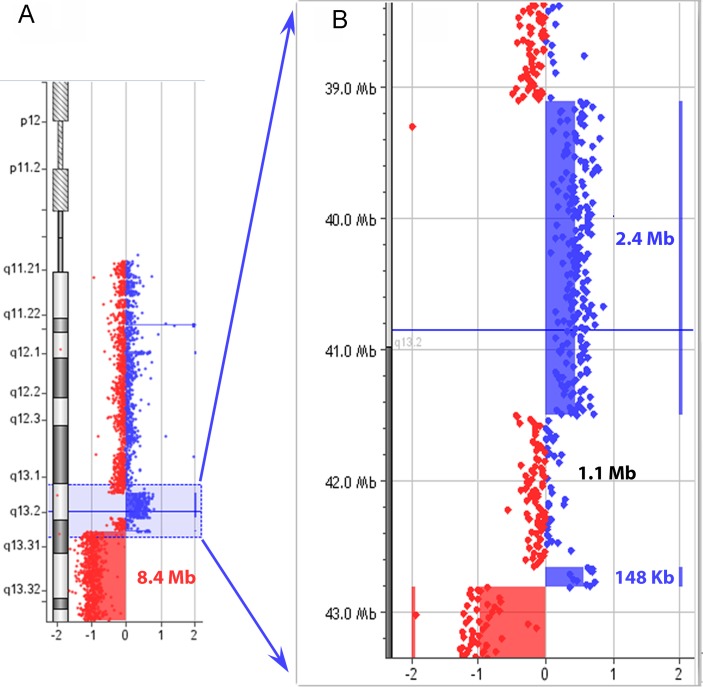
The patient’s karyotype was 46,XX,r(22)(::p11.2>q13::)dn with no evidence of ring instability in the 50 metaphases analysed. Array-CGH showed several copy number changes involving the ring chromosome 22, potentially representing a complex chromosome rearrangement. The 22q13 band showed a multiple rearrangement pattern: a duplication (dup) followed by a normal copy region (nlm), a second duplication and a terminal deletion (dup-del) (figure 2). This rearranged region (dup-nlm-dup-del) of 21 Mb spanned at least four breakpoints, with a copy-number loss of approximately 8.4 Mb and copy-number gain of approximately 2.5 Mb. Parental origin analysis showed that both duplication and deletion involved the paternal chromosome 22 (online supplementary table 1).
Figure 2.
Array CGH results: (A) Rearrangement pattern at 22q13: the profile of chromosome 22 shows a terminal deletion of 8.4 Mb at 22q13.2q13.3 (chr23: 42 817 697–51 219 009 bp) and two duplications at 22q13.1q13.2 (shaded in blue). (B) Enlargement of 22q13.1q13.2 showing the normal copy region of 1.1 Mb between two duplicated portions of 2.4 Mb (chr22: 39 106 714–41 497 969 bp) and 148 Kb (ch22: 42 663 298–42 810 963 bp).
Genome analysis
Whole-genome sequencing (WGS)
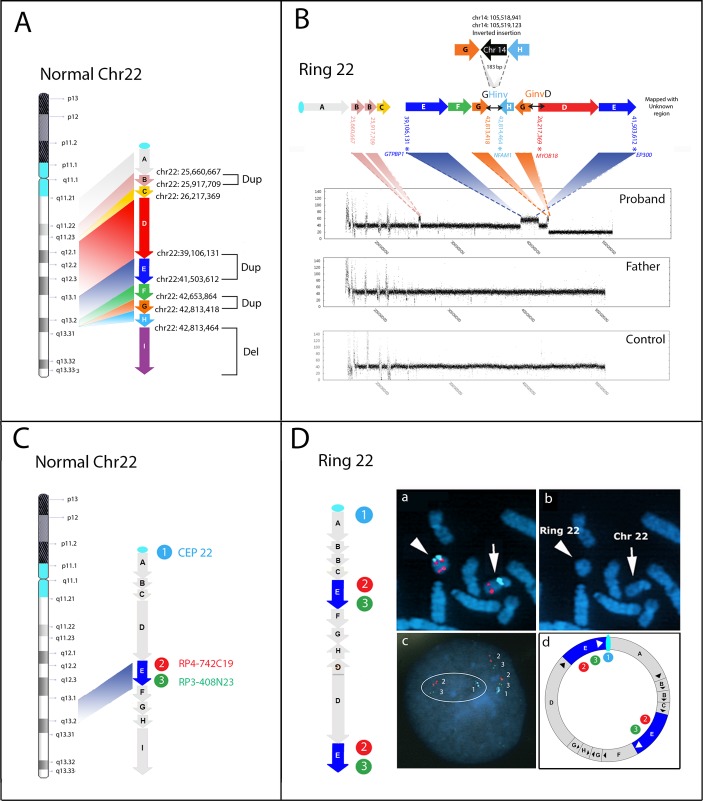
WGS analysis, performed on blood samples from proband and father (average coverage 37X and 42X, respectively), revealed eight breakpoints in the proband, producing nine fragments from A to I at the q arm of chromosome 22 (figure 3A). According to array-CGH analysis (figure 2) and visual inspection of chromosome 22 by IGV (Integrative Genome Viewer), fragment B (22q11.23–q12.1; size 257 Kb), fragment E (22q13.1–q13.2; size 2,4 Mb) and fragment G (22q13.2; size 160 Kb) were found to be duplicated, while fragment I (22q13.2q13.33; size 8.4 Mb) was deleted. In addition to the copy number variations detected by array-CGH (fragments E, G, I), inversions and displaced order of fragments were highlighted, indicating a chromoanagenesis event. In more detail, fragment H (1 kb) appeared to be inverted and fused with fragment G (termed as GHinv fusion joint), showing a 1 kb length inversion event between the distal duplicated and deleted segments (figure 3B, online supplementary figure 1). Additionally, a copy of fragment G (160 kb) was inverted and fused with the start site of fragment D (termed as GinvD fusion joint), showing an inverted insertion event within chromosome 22 (figure 3B). Sanger sequencing validated both GHinv and GinvD rearrangements (online supplementary figure 2 and table 2) and revealed an unexpected 183 bp inverted insertion between fragments G and H of a chromosome 14 sequence (chr14:105 518 942–105 519 123) (figure 3B), consisting of two portions of an AluSq sequence, as denoted by the UCSC RepeatMasker track (online supplementary figure 3). The breakpoint characteristics of the GHinv junction involved a microhomology of 2 bp (5’-GA) in fragment G-chr14 fusion junction and a microhomology of 2 bp (5’-TC) with the insertion of a single bp (5’-G) in chr14-Fragment H fusion junction (online supplementary figure 4). The breakpoint junction of H-I (chr22:42814464) falls within another subfamily of Alu element (AluSx3 sequence, as denoted by the UCSC RepeatMasker track, online supplementary figure 5). The GinvD rearrangement had a blunt junction without microhomology (figure 3B; online supplementary figure 4). Sanger investigations could not confirm the other predicted breakpoint-fusions due to the presence of repeated sequences. Dual-colour and three-colour FISH experiments with PACs probes (figure 4C), either in metaphases or interphase nuclei, proved that one copy of fragment E was an insertion-duplication, located close to the centromere of the r(22) (figure 4D). This might explain why we failed to define the fusion point between the end of fragment E (chr22: 41 503 612) and the repeated pericentromeric sequences (online supplementary figure 7). The FISH data fit well with the proposed model predicting a fusion between fragment E and the centromeric region of chromosome 22, thus forming a closed ring structure (figure 4D). We also verified the absence of interstitial pan-telomeric sequences with a PNA probe (online supplementary figure 8). Since the 22q deletion and duplications was of paternal origin, the father’s DNA was investigated by WGS for the presence of any genomic rearrangement predisposing to the chromosome 22 abnormality in the proband. We detected two tandem duplications, located at Chr22: 23 015 179–23 080 761 and Chr22: 25 684 091–25 980 630, reported in DGV as well in our in-house WGS database (5 out of 21 unrelated cases and 1 out of 11, respectively).
Figure 3.
(A,B) Reconstruction of ring 22 by WGS. (A) Coloured arrows indicate the orientation of the nine fragments relative to the normal chromosome 22 and are shown from proximal (fragment A) to distal (fragment I) with their breakpoints. Duplicated and deleted fragments are indicated by brackets. (B) Reconstruction of the ring 22; GHinv and GinvD fusion points, confirmed by Sanger sequencing, are indicated by horizontal two-way black arrows. Upper: schematic representation of the insertion of the 183 bp Alu-sequence from chromosome 14 in the junction of the GHinv rearrangement. Bottom: chromosome 22 WGS coverage plot from proband, father and control DNAs showing that the three duplications (shaded in pink for fragment B, blue for fragment E and orange for fragment G) and the distal deletion (fragment I), are present in the proband only, whereas the proximal copy number variants are common to all. (C,D) FISH analysis providing positional information on fragment E. Schematic visualisation of the probes’ location on the normal chromosome 22 (C) and the ring chromosome 22 (D) (ring (22)), according to WGS data. The light blue, red and green circles indicate the location of the differentially labelled probes. Fish results (D): (1) Metaphase dual-colour FISH analysis: RP4-742C19 (spectrum red, Genbank Accession: AL031846.2, at 22q13.1), located within fragment E (indicated by the blue arrow), showed two distinct hybridisation signals on the ring chromosome 22 (arrowhead), one of them positioned close to the centromere targeted by probe CEP22 (spectrum aqua, SureFish, Agilent); (2) DAPI-staining of the partial metaphase showing the normal chromosome 22 (arrow) and ring (22) (arrowhead). (3) interphase three-colour FISH (RP4-742C19, spectrum red, RP3-408N23, spectrum green, Genbank Accession: Z98048, at 22q13.1 and 22q13.2, respectively and CEP22, spectrum aqua, Sure Fish Agilent) shows the direct orientation of the two fragments E on the ring chromosome (aqua-green-red-green-red). (4) schematic representation of FISH results on the ring chromosome 22 model. WGS, whole-genome sequencing.
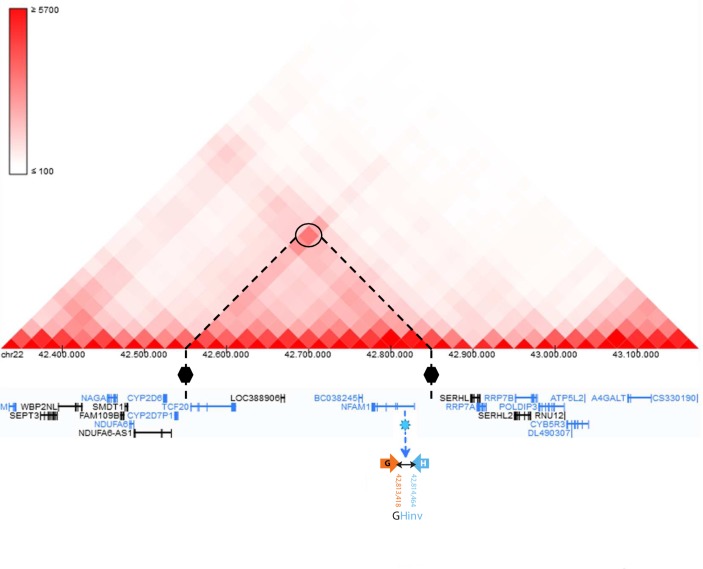
Figure 4.
View of the TAD region (chr22:42 550 000–42 850 000), disrupted by the GHinv breakpoint located within intron 1 of NFAM1. Inferred TAD boundaries in respect of the GHinv breakpoint (shown with a blue light star) are represented by black hexagons. The TAD connects NFAM1 to TCF20. Genes in black are on the forward strand (+), while those in blue are on the reverse strand (–). TAD, topologically associating domain.
Analysis of genes and TADs at breakpoint regions
Four of eight breakpoints detected by WGS disrupted four genes (figure 3B): EP300 (MIM 602700) has a role in the neurodevelopmental disorder Rubinstein-Taybi syndrome 2 (RSTS2, MIM#613684); NFAM1 (MIM 608740) is involved in immune system functioning; MYO18B (MIM 607295) is associated with Klippel-Feil syndrome 4 (KSF4, MIM#616549); GTPBP1 (MIM 602245) encodes a protein belonging to the family of GTP-binding proteins. TAD analysis showed that the GHinv breakpoint within NFAM1 (figure 2B) lies in a TAD that joins NFAM1 to TCF20 (figure 4).
Whole-exome (WES)
WES analysis was conducted on blood and saliva samples from the proband and her parents in order to exclude single nucleotide variants that could explain the BPP, a sign never before reported in PMS. In fact, the coverage obtained by WGS (30–40×) could have been insufficient to highlight any possible mosaic variant known to be responsible for polymicrogyria.20 WES analysis revealed a heterozygous variant in the exon 8 of SRPX2 at Xq22.1, NM_014467.2:c.893G>A (NP_055282.1:p.Arg298His), in both proband and mother as well as in the maternal grandmother (online supplementary figure 9). This substitution is reported in several databases (dbSNP, ExAC, ESP, COSMIC) and has a frequency in the population of about 0.01%. Moreover, it affects a highly conserved amino-acid residue of the Sushi/SCR/CCP domain (InterPro#IPR000436) but with contrasting predictions of pathogenicity, according to various in silico tools (eg, SIFT, Mutation Taster, Align GVGD). Since the BPP could also be caused by somatic mosaic variants in PIK3CA, we tried to exclude this possibility by analysing proband’s blood and saliva DNA by OneSeq, under experimental conditions allowing an average coverage higher than 130x. The analysis gave normal results. Similarly, careful analysis, including manual IGV inspection, of the exonic sequence of PI4KA 18 and SNAP29, 17 21 both on chromosome 22 and reported in association with cortex disorders, did not reveal any variants.
Discussion
We describe a complex genomic rearrangement associated with a constitutional ring chromosome 22 detected in a 3-year-old patient with severe neurodevelopmental disorder. Haploinsufficiency for SHANK3, fitting with the presence of the r(22), led to her initial framing within the PMS syndrome. However, the later MRI detection of BPP and the occurrence of recurrent episodes of scattered atopic dermatitis indicated a more complex molecular basis for her condition. Among the brain-imaging studies in patients with PMS, the most frequent abnormalities described are thinning of the corpus callosum (36%), white matter changes (39%), ventricular dilatation (32%) and arachnoid cysts (14%).22 Other findings showed that some patients with PMS have severe posterior cerebellar vermis hypoplasia and enlarged cisterna magna.23 Indeed, our patient’s complex brain malformations included many common brain abnormalities frequently observed in PMS individuals (thin corpus callosum, reduced volume of white matter, enlarged ventricles and arachnoid cysts). However, with the limitation that not all subjects with PMS have undergone MRI analysis, polymicrogyric cortex has never been reported in patients with 22q distal deletions,5 23 24 suggesting that mechanisms other than simple haploinsufficiency for genes located at distal 22q are responsible for it. By WGS and confirmatory WES data (the latter in both blood and saliva, at a coverage >100 x), we excluded pathogenic variants in genes associated with polymicrogyria and cortex abnormalities having a frequency <1%, and in theory, also mosaic condition, as most commonly reported for PIK3R2.17 20 Indeed, the c.893G>A (p.Arg298His) variant in the in SRPX2 gene on chromosome X, coding for a protein involved in synaptogenesis of the cerebral cortex, was the only one taken into consideration. A variant in exon 4 of SRPX2 was previously described in a male patient with BPP, his unaffected mother and two maternal aunts with mild ID and no MRI abnormality.25 Similarly, our variant was present in the unaffected mother and grandmother and X-inactivation studies revealed that the active X carried the variant in the proband but not in her mother (online supplementary figure 9). Amino-acid residue Arg298 is targeted by other missense or synonymous variations (namely p.Arg298Ser, p.Arg298Cys and p.Arg298Arg), but p.Arg298His is the most frequently observed, with 11 hemizygous subjects reported in the gnomAD browser. Therefore, based on currently available information, this variant does not seem to have a clear pathogenic role.
None of the genes disrupted by the breakpoints of the ring, namely EP300, MYO18B, NFAM1 and GTBP1, was associated with cortex abnormalities, although some of them may contribute to severity of the patient’s phenotype.
Regarding NFAM1, encoding a protein regulating B-cell development, the 183 bp insertion at GHinv breakpoint within intron 1, might theoretically impair its two transcript variants. The TAD region, disrupted by the GHinv breakpoint, joins NFAM1 to TCF20 (figure 4). De novo variants in TCF20 (OMIM 603107), encoding a widely expressed transcriptional coregulator structurally and functionally related to RAI1 (OMIM 607642), have been implicated in the aetiology of autism spectrum disorders, ID/developmental delay, schizophrenia and postnatal overgrowth.26–29 Thus, misregulation of TCF20 as a consequence of TAD perturbation may likely contribute to the neurodevelopmental features observed in our patient. Two patients with haploinsufficiency of NFAM1 resulting from interstitial 22q13.2 deletion have been reported with atopic dermatitis and urticarial rash,30 31 as observed in our patient (figure 1A), but on closer examination one of them30 has a terminal deletion that does not include NFAM1. With the limitation that RNA was not available for NFAM1 expression analysis in our patient, we cannot exclude that other genes may contribute to the urticarial rashes.
EP300 frameshift mutations were reported in Rubinstein-Taybi syndrome 2, with some phenotypic overlap with our patient (speech, gross motor delay, feeding/swallowing issues). The interruption of MYO18B does not seem to interfere with the phenotype of the patient because it is associated with an autosomal recessive condition, namely the Klippel-Feil syndrome 4, while the function of GTBP1 is still unknown. Altogether, these data explain only part of the patient’s clinical picture, including the dermatological manifestation. However, it seems likely that disruption of the TAD, either as a consequence of the duplication/deletion regions or the ring conformation itself,32 played an equally important role in her clinical manifestations
Chromoanagesis and ring chromosomes
We describe the first case of a constitutional ring chromosome derived by a chromoanagenesis event, previously reported only in cancer.33 In line with the predominant occurrence of chromoanagenesis constitutional events on paternally derived chromosomes,34 our r(22) was of paternal origin and we excluded through WGS that any bona fide predisposing rearrangement was present on paternal chromosomes 22. According to the mechanisms associated with chromothripsis and chromoanasynthesis, both belonging to the novel category of chromoanagenesis events,34 35 we have to assume that the rearrangement occurred during either paternal gametogenesis or early embryogenesis by isolation of a chromosome 22 within a micronucleus or, alternatively, as a consequence of the formation of a chromatin bridge interconnecting two daughter cells.36 In the first case, chromosome mis-segregation might have led to the trapping of chromosome 22 within the micronucleus, as well described in chromothripsis, while in the second the occurrence of a dicentric chromosome 22, pter>q13::q13-pter, possibly secondary to telomeric crisis, would have promoted chromothripsis events, as previously proposed in cancer genomes.37 38 The sequences at the breakpoints of our chromosome 22 are consistent with repair-based mechanisms, although further complexity was added at the GHinv fusion point by a 183 bp insertion from chromosome 14, containing two partial Alu elements, in contrast to the full length 300 bp Alu sequence.39 The sequence at the junction of AluSq insertion, did not resemble L1 endonuclease consensus site (5′-YYYY/RR-3′ where Y=pyrimidine, R=purine) nor its additional variations,40 41 making unlikely any L1 endonuclease role in the rearrangement. According to all these features at GHinv junction, we might predict an endonuclease-independent non-classical Alu insertion42 with a potential role in DSB repair. An Alu-Alu mediated insertion may also be an option since a second Alu was detected at H-I breakpoint (online supplementary figure 5) as well as in other chromothripsis/chromoanasynthesis rearrangements.43 44
Ring chromosomes are unstable pieces of circular chromatin that, as a consequence of sister chromatid exchanges, may lead, at least in vitro, to a mosaic condition with different cells carrying the entire duplicated or deleted ring chromosome or part of it.45 This in vivo instability should result in massive cell death, in turn causing the so-called ring syndrome that, independently from the chromosome involved, would cause extreme growth failure without major malformations, with only a few or no minor anomalies and mild to moderate ID.46 Although the existence of this syndrome remains highly hypothetical, a compensatory uniparental disomy mechanism has been demonstrated in induced pluripotent stem cells (iPSCs) obtained from patient fibroblasts containing deleted ring chromosomes, with loss of cells containing the abnormal chromosome and duplication of the wild type homologue.47 Whatever the meaning of this cell-autonomous correction of ring chromosomes in vivo, it seems clear that circular chromatin structures are potentially related to decompensation of cellular homeostasis via massive gene deregulation. In our study, where no ring chromosome instability was highlighted at least in blood, we were unable to detect the molecular basis for the patient’s BPP, although we can hypothesise that it may be linked to insufficient production during embryogenesis of proteins encoded by some chromosome 22 genes with a role in the formation of the brain cortex.
Although rearrangements more complex than the classical model, which considers the circular conformation a consequence of the deletion of either chromosome end, have been reported,48 it is likely that other ring chromosomes may be generated by a mechanism such as the one we describe here. This hypothesis might also explain the genotype–phenotype discrepancies reported for other ring chromosomes, particularly those of chromosomes 14 and 20, in which the severe phenotype cannot be explained based on the deleted region.49 50
Conclusion
We demonstrate for the first time the occurrence of a constitutional chromoanagenesis event resulting in the formation a ring chromosome, with both copy number losses and gains, leading to a PMS phenotype associated with BPP and atopic dermatitis with urticarial rashes. The outcome of concurrent deletions, duplications, gene interruptions and TAD is a dramatically complex phenotype. This study reinforces the growing evidence that chromosomal rearrangements (1) may be not as simple as they appear by conventional diagnostic approaches and (2) affect the phenotype by global alteration of the topological chromatin organisation rather than simply by the deletion or duplication of dosage-sensitive genes.
Web sources
DGV http://dgv.tcag.ca/dgv/app/home
dbSNP https://www.ncbi.nlm.nih.gov/projects/SNP/
ExAC http://exac.broadinstitute.org/
ESP http://evs.gs.washington.edu/EVS/
COSMIC http://cancer.sanger.ac.uk/cosmic
3D Genome Browser (http://3dgenome.org)
gnomAD browser (http://gnomad.broadinstitute.org/)
Acknowledgments
We would like to gratefully acknowledge the family participating in this study.
Footnotes
Contributors: NK, MD, LX, MR performed WGS analysis. EE performed TAD and WES analysis. FA interpreted Brain MRI analysis. CZ assessed EEG finding. CM and MGD conducted clinical neurological evaluations and managed the patient. SiB and RG performed segregation analysis of the SRPX2 variant and X-inactivation analysis. MCB interpreted cytogenetics, array-CGH and FISH investigations. SB provided technical assistance to cytogenetic analysis. MCB and OZ designed the study. MCB, OZ, NK and EE contributed significantly to this study, wrote the article and critically discussed the final editing of the manuscript.
Funding: MCB was supported by grants of the Italian Ministry of Health (Ricerca Corrente 2017). OZ was supported by Telethon Italy Grant GGP13060.
Competing interests: None declared.
Patient consent: Obtained.
Ethics approval: The study has E. Medea Scientific Institute Research Ethics Committee approval (Prot. no. 007/15-CE).
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1. Bonaglia MC, Giorda R, Borgatti R, Felisari G, Gagliardi C, Selicorni A, Zuffardi O. Disruption of the ProSAP2 gene in a t(12;22)(q24.1;q13.3) is associated with the 22q13.3 deletion syndrome. Am J Hum Genet 2001;69:261–8. 10.1086/321293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bonaglia MC, Giorda R, Mani E, Aceti G, Anderlid BM, Baroncini A, Pramparo T, Zuffardi O. Identification of a recurrent breakpoint within the SHANK3 gene in the 22q13.3 deletion syndrome. J Med Genet 2006;43:822–8. 10.1136/jmg.2005.038604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bonaglia MC, Giorda R, Beri S, De Agostini C, Novara F, Fichera M, Grillo L, Galesi O, Vetro A, Ciccone R, Bonati MT, Giglio S, Guerrini R, Osimani S, Marelli S, Zucca C, Grasso R, Borgatti R, Mani E, Motta C, Molteni M, Romano C, Greco D, Reitano S, Baroncini A, Lapi E, Cecconi A, Arrigo G, Patricelli MG, Pantaleoni C, D’Arrigo S, Riva D, Sciacca F, Dalla Bernardina B, Zoccante L, Darra F, Termine C, Maserati E, Bigoni S, Priolo E, Bottani A, Gimelli S, Bena F, Brusco A, di Gregorio E, Bagnasco I, Giussani U, Nitsch L, Politi P, Martinez-Frias ML, Martínez-Fernández ML, Martínez Guardia N, Bremer A, Anderlid BM, Zuffardi O. Molecular mechanisms generating and stabilizing terminal 22q13 deletions in 44 subjects with Phelan/McDermid syndrome. PLoS Genet 2011;7:e1002173 10.1371/journal.pgen.1002173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sarasua SM, Boccuto L, Sharp JL, Dwivedi A, Chen CF, Rollins JD, Rogers RC, Phelan K, DuPont BR. Clinical and genomic evaluation of 201 patients with Phelan-McDermid syndrome. Hum Genet 2014;133:847–59. 10.1007/s00439-014-1423-7 [DOI] [PubMed] [Google Scholar]
- 5. Soorya L, Kolevzon A, Zweifach J, Lim T, Dobry Y, Schwartz L, Frank Y, Wang AT, Cai G, Parkhomenko E, Halpern D, Grodberg D, Angarita B, Willner JP, Yang A, Canitano R, Chaplin W, Betancur C, Buxbaum JD. Prospective investigation of autism and genotype-phenotype correlations in 22q13 deletion syndrome and SHANK3 deficiency. Mol Autism 2013;4:18 18-2392-4-18. 10.1186/2040-2392-4-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kreienkamp HJ. Scaffolding proteins at the postsynaptic density: shank as the architectural framework. Handb Exp Pharmacol 2008;186:365–80. 10.1007/978-3-540-72843-6_15 [DOI] [PubMed] [Google Scholar]
- 7. Denayer A, Van Esch H, de Ravel T, Frijns JP, Van Buggenhout G, Vogels A, Devriendt K, Geutjens J, Thiry P, Swillen A. Neuropsychopathology in 7 patients with the 22q13 deletion syndrome: presence of bipolar disorder and progressive loss of skills. Mol Syndromol 2012;3:14–20. 10.1159/000339119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Reierson G, Bernstein J, Froehlich-Santino W, Urban A, Purmann C, Berquist S, Jordan J, O’Hara R, Hallmayer J. Characterizing regression in Phelan McDermid Syndrome (22q13 deletion syndrome). J Psychiatr Res 2017;91:139–44. 10.1016/j.jpsychires.2017.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Egger JI, Zwanenburg RJ, van Ravenswaaij-Arts CM, Kleefstra T, Verhoeven WM. Neuropsychological phenotype and psychopathology in seven adult patients with Phelan-McDermid syndrome: implications for treatment strategy. Genes Brain Behav 2016;15:395–404. 10.1111/gbb.12285 [DOI] [PubMed] [Google Scholar]
- 10. Delahaye A, Toutain A, Aboura A, Dupont C, Tabet AC, Benzacken B, Elion J, Verloes A, Pipiras E, Drunat S. Chromosome 22q13.3 deletion syndrome with a de novo interstitial 22q13.3 cryptic deletion disrupting SHANK3. Eur J Med Genet 2009;52:328–32. 10.1016/j.ejmg.2009.05.004 [DOI] [PubMed] [Google Scholar]
- 11. Bonaglia MC, Giorda R, Massagli A, Galluzzi R, Ciccone R, Zuffardi O. A familial inverted duplication/deletion of 2p25.1-25.3 provides new clues on the genesis of inverted duplications. Eur J Hum Genet 2009;17:179–86. 10.1038/ejhg.2008.160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Raczy C, Petrovski R, Saunders CT, Chorny I, Kruglyak S, Margulies EH, Chuang HY, Källberg M, Kumar SA, Liao A, Little KM, Strömberg MP, Tanner SW. Isaac: ultra-fast whole-genome secondary analysis on Illumina sequencing platforms. Bioinformatics 2013;29:2041–3. 10.1093/bioinformatics/btt314 [DOI] [PubMed] [Google Scholar]
- 13. Layer RM, Chiang C, Quinlan AR, Hall IM. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol 2014;15:R84 R84-2014-15-6-r84 10.1186/gb-2014-15-6-r84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen X, Schulz-Trieglaff O, Shaw R, Barnes B, Schlesinger F, Källberg M, Cox AJ, Kruglyak S, Saunders CT. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 2016;32:1220–2. 10.1093/bioinformatics/btv710 [DOI] [PubMed] [Google Scholar]
- 15. Vetro A, Goidin D, Lesende I, Limongelli I, Ranzani GN, Novara F, Bonaglia MC, Rinaldi B, Franchi F, Manolakos E, Lonardo F, Scarano F, Scarano G, Costantino L, Tedeschi S, Giglio S, Zuffardi O. Diagnostic application of a capture based NGS test for the concurrent detection of variants in sequence and copy number as well as LOH. Clin Genet 2017. 10.1111/cge.13060 [DOI] [PubMed] [Google Scholar]
- 16. Errichiello E, Mustafa N, Vetro A, Notarangelo LD, de Jonge H, Rinaldi B, Vergani D, Giglio SR, Morbini P, Zuffardi O. SMARCA4 inactivating mutations cause concomitant Coffin-Siris syndrome, microphthalmia and small-cell carcinoma of the ovary hypercalcaemic type. J Pathol 2017;243:9–15. 10.1002/path.4926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Parrini E, Conti V, Dobyns WB, Guerrini R. Genetic basis of brain malformations. Mol Syndromol 2016;7:220–33. 10.1159/000448639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pagnamenta AT, Howard MF, Wisniewski E, Popitsch N, Knight SJ, Keays DA, Quaghebeur G, Cox H, Cox P, Balla T, Taylor JC, Kini U. Germline recessive mutations in PI4KA are associated with perisylvian polymicrogyria, cerebellar hypoplasia and arthrogryposis. Hum Mol Genet 2015;24:3732–41. 10.1093/hmg/ddv117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yardımcı GG, Noble WS. Software tools for visualizing Hi-C data. Genome Biol 2017;18:26 26-017-1161-y 10.1186/s13059-017-1161-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mirzaa GM, Conti V, Timms AE, Smyser CD, Ahmed S, Carter M, Barnett S, Hufnagel RB, Goldstein A, Narumi-Kishimoto Y, Olds C, Collins S, Johnston K, Deleuze JF, Nitschké P, Friend K, Harris C, Goetsch A, Martin B, Boyle EA, Parrini E, Mei D, Tattini L, Slavotinek A, Blair E, Barnett C, Shendure J, Chelly J, Dobyns WB, Guerrini R. Characterisation of mutations of the phosphoinositide-3-kinase regulatory subunit, PIK3R2, in perisylvian polymicrogyria: a next-generation sequencing study. Lancet Neurol 2015;14:1182–95. 10.1016/S1474-4422(15)00278-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Diggle CP, Martinez-Garay I, Molnar Z, Brinkworth MH, White E, Fowler E, Hughes R, Hayward BE, Carr IM, Watson CM, Crinnion L, Asipu A, Woodman B, Coletta PL, Markham AF, Dear TN, Bonthron DT, Peckham M, Morrison EE, Sheridan E. A tubulin alpha 8 mouse knockout model indicates a likely role in spermatogenesis but not in brain development. PLoS One 2017;12:e0174264 10.1371/journal.pone.0174264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kolevzon A, Angarita B, Bush L, Wang AT, Frank Y, Yang A, Rapaport R, Saland J, Srivastava S, Farrell C, Edelmann LJ, Buxbaum JD. Phelan-McDermid syndrome: a review of the literature and practice parameters for medical assessment and monitoring. J Neurodev Disord 2014;6:39 10.1186/1866-1955-6-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Aldinger KA, Kogan J, Kimonis V, Fernandez B, Horn D, Klopocki E, Chung B, Toutain A, Weksberg R, Millen KJ, Barkovich AJ, Dobyns WB. Cerebellar and posterior fossa malformations in patients with autism-associated chromosome 22q13 terminal deletion. Am J Med Genet A 2013;161A:131–6. 10.1002/ajmg.a.35700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Philippe A, Boddaert N, Vaivre-Douret L, Robel L, Danon-Boileau L, Malan V, de Blois MC, Heron D, Colleaux L, Golse B, Zilbovicius M, Munnich A. Neurobehavioral profile and brain imaging study of the 22q13.3 deletion syndrome in childhood. Pediatrics 2008;122:e376–e382. 10.1542/peds.2007-2584 [DOI] [PubMed] [Google Scholar]
- 25. Roll P, Rudolf G, Pereira S, Royer B, Scheffer IE, Massacrier A, Valenti MP, Roeckel-Trevisiol N, Jamali S, Beclin C, Seegmuller C, Metz-Lutz MN, Lemainque A, Delepine M, Caloustian C, de Saint Martin A, Bruneau N, Depétris D, Mattéi MG, Flori E, Robaglia-Schlupp A, Lévy N, Neubauer BA, Ravid R, Marescaux C, Berkovic SF, Hirsch E, Lathrop M, Cau P, Szepetowski P. SRPX2 mutations in disorders of language cortex and cognition. Hum Mol Genet 2006;15:1195–207. 10.1093/hmg/ddl035 [DOI] [PubMed] [Google Scholar]
- 26. Babbs C, Lloyd D, Pagnamenta AT, Twigg SR, Green J, McGowan SJ, Mirza G, Naples R, Sharma VP, Volpi EV, Buckle VJ, Wall SA, Knight SJ, Parr JR, Wilkie AO; International Molecular Genetic Study of Autism Consortium (IMGSAC). De novo and rare inherited mutations implicate the transcriptional coregulator TCF20/SPBP in autism spectrum disorder. J Med Genet 2014;51:737–47. 10.1136/jmedgenet-2014-102582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schäfgen J, Cremer K, Becker J, Wieland T, Zink AM, Kim S, Windheuser IC, Kreiß M, Aretz S, Strom TM, Wieczorek D, Engels H. De novo nonsense and frameshift variants of TCF20 in individuals with intellectual disability and postnatal overgrowth. Eur J Hum Genet 2016;24:1739–45. 10.1038/ejhg.2016.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lelieveld SH, Reijnders MR, Pfundt R, Yntema HG, Kamsteeg EJ, de Vries P, de Vries BB, Willemsen MH, Kleefstra T, Löhner K, Vreeburg M, Stevens SJ, van der Burgt I, Bongers EM, Stegmann AP, Rump P, Rinne T, Nelen MR, Veltman JA, Vissers LE, Brunner HG, Gilissen C. Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat Neurosci 2016;19:1194–6. 10.1038/nn.4352 [DOI] [PubMed] [Google Scholar]
- 29. Smeland OB, Frei O, Kauppi K, Hill WD, Li W, Wang Y, Krull F, Bettella F, Eriksen JA, Witoelar A, Davies G, Fan CC, Thompson WK, Lam M, Lencz T, Chen CH, Ueland T, Jonsson EG, Djurovic S, Deary IJ, Dale AM, Andreassen OA. NeuroCHARGE (Cohorts for Heart and Aging Research in Genomic Epidemiology) Cognitive Working Group. Identification of genetic loci jointly influencing schizophrenia risk and the cognitive traits of verbal-numerical reasoning, reaction time, and general cognitive function. JAMA Psychiatry 2017;74:1065–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen CP, Lin SP, Chern SR, Tsai FJ, Wu PC, Lee CC, Chen YT, Chen WL, Wang W. A de novo 7.9 Mb deletion in 22q13.2→qter in a boy with autistic features, epilepsy, developmental delay, atopic dermatitis and abnormal immunological findings. Eur J Med Genet 2010;53:329–32. 10.1016/j.ejmg.2010.06.004 [DOI] [PubMed] [Google Scholar]
- 31. Simenson K, Õiglane-Shlik E, Teek R, Kuuse K, Õunap K. A patient with the classic features of Phelan-McDermid syndrome and a high immunoglobulin E level caused by a cryptic interstitial 0.72-Mb deletion in the 22q13.2 region. Am J Med Genet A 2014;164A:806–9. 10.1002/ajmg.a.36358 [DOI] [PubMed] [Google Scholar]
- 32. Timpson NJ, Greenwood CMT, Soranzo N, Lawson DJ, Richards JB. Genetic architecture: the shape of the genetic contribution to human traits and disease. Nat Rev Genet 2017. 10.1038/nrg.2017.101 [DOI] [PubMed] [Google Scholar]
- 33. Ortega V, Chaubey A, Mendiola C, Ehman W, Vadlamudi K, Dupont B, Velagaleti G. Complex Chromosomal Rearrangements in B-Cell Lymphoma: Evidence of Chromoanagenesis? A Case Report. Neoplasia 2016;18:223–8. 10.1016/j.neo.2016.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Collins RL, Brand H, Redin CE, Hanscom C, Antolik C, Stone MR, Glessner JT, Mason T, Pregno G, Dorrani N, Mandrile G, Giachino D, Perrin D, Walsh C, Cipicchio M, Costello M, Stortchevoi A, An JY, Currall BB, Seabra CM, Ragavendran A, Margolin L, Martinez-Agosto JA, Lucente D, Levy B, Sanders SJ, Wapner RJ, Quintero-Rivera F, Kloosterman W, Talkowski ME. Defining the diverse spectrum of inversions, complex structural variation, and chromothripsis in the morbid human genome. Genome Biol 2017;18:36 10.1186/s13059-017-1158-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fukami M, Shima H, Suzuki E, Ogata T, Matsubara K, Kamimaki T. Catastrophic cellular events leading to complex chromosomal rearrangements in the germline. Clin Genet 2017;91:653–60. 10.1111/cge.12928 [DOI] [PubMed] [Google Scholar]
- 36. Ly P, Cleveland DW. Rebuilding Chromosomes After Catastrophe: Emerging Mechanisms of Chromothripsis. Trends Cell Biol 2017;27:917–30. 10.1016/j.tcb.2017.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Maciejowski J, Li Y, Bosco N, Campbell PJ, de Lange T. Chromothripsis and Kataegis Induced by Telomere Crisis. Cell 2015;163:1641–54. 10.1016/j.cell.2015.11.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ernst A, Jones DT, Maass KK, Rode A, Deeg KI, Jebaraj BM, Korshunov A, Hovestadt V, Tainsky MA, Pajtler KW, Bender S, Brabetz S, Gröbner S, Kool M, Devens F, Edelmann J, Zhang C, Castelo-Branco P, Tabori U, Malkin D, Rippe K, Stilgenbauer S, Pfister SM, Zapatka M, Lichter P. Telomere dysfunction and chromothripsis. Int J Cancer 2016;138:2905–14. 10.1002/ijc.30033 [DOI] [PubMed] [Google Scholar]
- 39. Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet 2002;3:370–9. 10.1038/nrg798 [DOI] [PubMed] [Google Scholar]
- 40. Feng Q, Moran JV, Kazazian HH, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 1996;87:905–16. 10.1016/S0092-8674(00)81997-2 [DOI] [PubMed] [Google Scholar]
- 41. Hancks DC, Kazazian HH. Roles for retrotransposon insertions in human disease. Mob DNA 2016;7:9 10.1186/s13100-016-0065-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Srikanta D, Sen SK, Huang CT, Conlin EM, Rhodes RM, Batzer MA. An alternative pathway for Alu retrotransposition suggests a role in DNA double-strand break repair. Genomics 2009;93:205–12. 10.1016/j.ygeno.2008.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gu S, Yuan B, Campbell IM, Beck CR, Carvalho CM, Nagamani SC, Erez A, Patel A, Bacino CA, Shaw CA, Stankiewicz P, Cheung SW, Bi W, Lupski JR. Alu-mediated diverse and complex pathogenic copy-number variants within human chromosome 17 at p13.3. Hum Mol Genet 2015;24:4061–77. 10.1093/hmg/ddv146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nazaryan-Petersen L, Bertelsen B, Bak M, Jønson L, Tommerup N, Hancks DC, Tümer Z. Germline Chromothripsis Driven by L1-Mediated Retrotransposition and Alu/Alu Homologous Recombination. Hum Mutat 2016;37:385–95. 10.1002/humu.22953 [DOI] [PubMed] [Google Scholar]
- 45. Gisselsson D. Huret J-L, Ring chromosomes: vicious circles at the end and beginning of life: INIST-CNRS, 2002. (Editor-in-Chief) http://hdl.handle.net/2042/37826. [Google Scholar]
- 46. Kosztolányi G. The genetics and clinical characteristics of constitutional ring chromosomes. J Assoc Genet Technol 2009;35:44–8. [PubMed] [Google Scholar]
- 47. Bershteyn M, Hayashi Y, Desachy G, Hsiao EC, Sami S, Tsang KM, Weiss LA, Kriegstein AR, Yamanaka S, Wynshaw-Boris A. Cell-autonomous correction of ring chromosomes in human induced pluripotent stem cells. Nature 2014;507:99–103. 10.1038/nature12923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rossi E, Riegel M, Messa J, Gimelli S, Maraschio P, Ciccone R, Stroppi M, Riva P, Perrotta CS, Mattina T, Memo L, Baumer A, Kucinskas V, Castellan C, Schinzel A, Zuffardi O. Duplications in addition to terminal deletions are present in a proportion of ring chromosomes: clues to the mechanisms of formation. J Med Genet 2008;45:147–54. 10.1136/jmg.2007.054007 [DOI] [PubMed] [Google Scholar]
- 49. Rinaldi B, Vaisfeld A, Amarri S, Baldo C, Gobbi G, Magini P, Melli E, Neri G, Novara F, Pippucci T, Rizzi R, Soresina A, Zampini L, Zuffardi O, Crimi M. Guideline recommendations for diagnosis and clinical management of Ring14 syndrome-first report of an ad hoc task force. Orphanet J Rare Dis 2017;12:69 10.1186/s13023-017-0606-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vignoli A, Bisulli F, Darra F, Mastrangelo M, Barba C, Giordano L, Turner K, Zambrelli E, Chiesa V, Bova S, Fiocchi I, Peron A, Naldi I, Milito G, Licchetta L, Tinuper P, Guerrini R, Dalla Bernardina B, Canevini MP. Epilepsy in ring chromosome 20 syndrome. Epilepsy Res 2016;128:83–93. 10.1016/j.eplepsyres.2016.10.004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jmedgenet-2017-105125supp001.docx (4.7MB, docx)