ABSTRACT
The Salmonella type three secretion system (T3SS), encoded in the Salmonella pathogenicity island 1 (SPI1) locus, mediates the invasion of the host intestinal epithelium. SPI1 expression is dependent upon three AraC-like regulators: HilD, HilC, and RtsA. These regulators act in a complex feed-forward loop to activate each other and hilA, which encodes the activator of the T3SS structural genes. HilD has been shown to be the major integration point of most signals known to activate the expression of the SPI1 T3SS, acting as a switch to control induction of the system. HilE is a negative regulator that acts upon HilD. Here we provide genetic and biochemical data showing that HilE specifically binds to HilD but not to HilC or RtsA. This protein-protein interaction blocks the ability of HilD to bind DNA as shown by both an in vivo reporter system and an in vitro gel shift assay. HilE does not affect HilD dimerization, nor does it control the stability of the HilD protein. We also investigated the role of HilE during the infection of mice using competition assays. Although deletion of hilE does not confer a phenotype, the hilE mutation does suppress the invasion defect conferred by loss of FliZ, which acts as a positive signal controlling HilD protein activity. Together, these data suggest that HilE functions to restrict low-level HilD activity, preventing premature activation of SPI1 until positive inputs reach a threshold required to fully induce the system.
IMPORTANCE Salmonella is a leading cause of gastrointestinal and systemic disease throughout the world. The SPI1 T3SS is required for Salmonella to induce inflammatory diarrhea and to gain access to underlying tissue. A complex regulatory network controls expression of SPI1 in response to numerous physiological inputs. Most of these signals impinge primarily on HilD translation or activity. The system is triggered when HilD activity crosses a threshold that allows efficient activation of its own promoter. This threshold is set by HilE, which binds to HilD to prevent the inevitable minor fluctuations in HilD activity from inappropriately activating the system. The circuit also serves as a paradigm for systems that must integrate numerous environmental parameters to control regulatory output.
KEYWORDS: Salmonella, SPI1, HilD, HilE
INTRODUCTION
Salmonella enterica serovar Typhimurium is a Gram-negative pathogen that, when ingested, is capable of causing a range of diseases from gastroenteritis to potentially lethal systemic infections in a wide range of hosts, including humans. It is a leading cause of bacterial foodborne illness in the United States, with nontyphoid species of Salmonella estimated to cause over 1 million infections per year (1). Salmonella is defined and differentiated from other Enterobacteriaceae by the presence of Salmonella pathogenicity island 1 (SPI1). This 40-kb locus carries all of the genes required for a functional type III secretion system (T3SS) and the regulators HilD, HilC, and HilA (2). The additional regulators RtsA (STM14_5188) and HilE (STM14_4514) are encoded elsewhere on the chromosome (3–5). The T3SS forms a needle-like complex that injects bacterial effector proteins into host epithelial cells. These effectors induce a number of physiological responses, including inflammatory diarrhea and bacterial engulfment that can lead to a systemic infection (6).
Expression of the SPI1 T3SS is controlled by a complex regulatory network (Fig. 1A). HilA functions to activate all of the genes required to synthesize a functional T3SS (7–10). Transcriptional activation of hilA is dependent upon three AraC-like proteins, HilD, HilC, and RtsA, which bind to the same sites upstream of activated promoters (11, 12). Each of the three regulators activates, in addition to hilA, its own promoter as well as those of the other two regulators, thus forming a complex feed-forward loop (12–15).
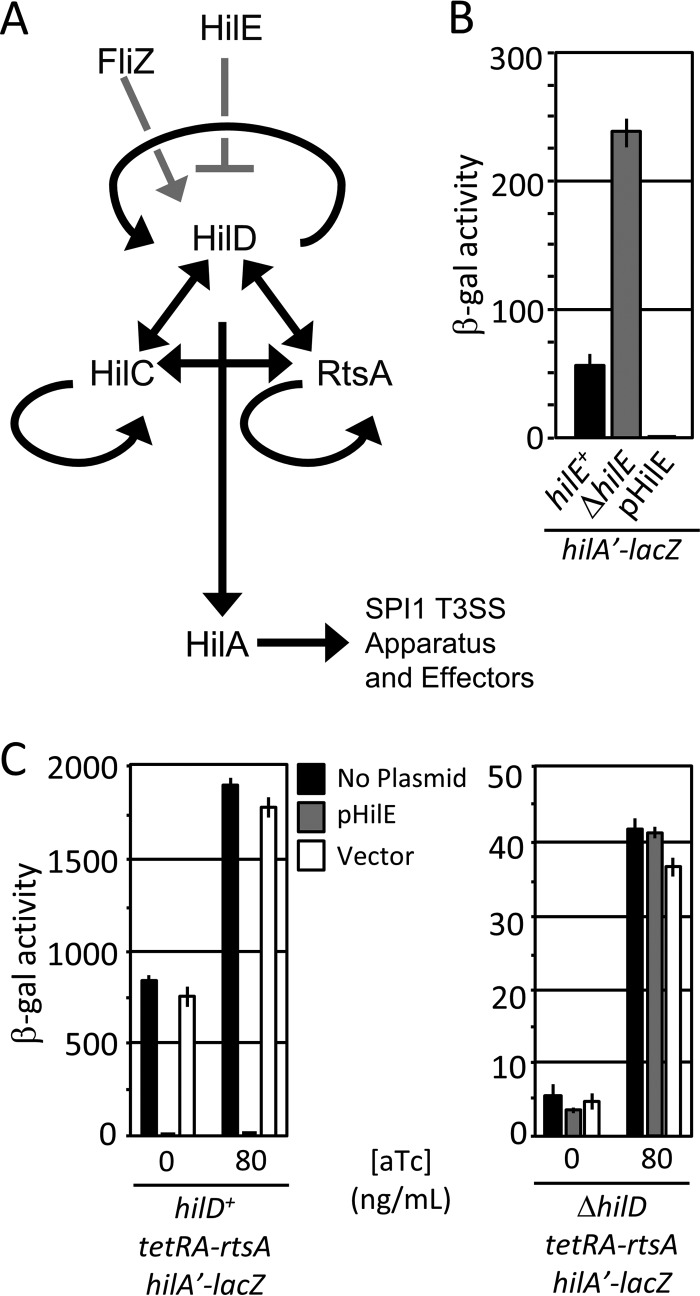
FIG 1.
(A) A simplified model for SPI1-T3SS regulation. Black lines indicate transcriptional regulation. Gray lines indicate posttranslational regulation. For clarity, the genes encoding SPI1 regulators are not shown. (B) HilE is a negative regulator of SPI1. Strains with hilE deleted or overexpressing hilE were grown under SPI1-inducing conditions and analyzed via β-galactosidase assay. β-Galactosidase activity units are defined as (micromoles of ONP formed per minute × 106)/(OD600 × milliliters of cell suspension) and reported as mean ± one standard deviation (n = 6, 3 sets of duplicate assays on 3 separate days). The strains used were JS749, JS749 with pHilE, and JS2103. (C) HilE acts via HilD. The rtsA gene was under the control of a tetA promoter, hilD was left unaltered or deleted, and HilE was overexpressed as indicated. Strains were grown overnight under SPI1-inducing conditions and subcultured 1:100 with the indicated concentration of anhydrotetracycline (aTc) (n = 4, 3 sets of duplicate assays on 2 separate days; error bars represent one standard deviation). The strains used were JS953 and JS2104 with pHilE or pWKS30.
Our data indicate that the majority of regulatory inputs into the SPI1 system are integrated at the level of HilD (16). The overall effect is to control the ability of HilD to activate its own promoter, the critical step in initiating expression of the SPI1 T3SS (17). Primary examples of factors that influence HilD activity are HilE, a major negative regulator of SPI1 T3SS expression, and the flagellar gene product FliZ, which acts indirectly upon HilD to increase activity (10, 16, 18–20). HilE and FliZ regulate the activity of HilD simultaneously and independently (20). Our immediate goal was to understand how HilE regulates HilD activity at the molecular level to further our understanding of overall integration of signals into the SPI1 regulatory circuit.
Fahlen et al. identified HilE as a negative regulator of hilA transcription, and Baxter et al. provided bacterial two-hybrid data indicating that HilE directly interacts with HilD protein (5, 21). We have also provided genetic data demonstrating that HilE operates at the level of the HilD protein (16, 20). HilE controls an ectopically expressed HilD but does not affect expression of hilD-lacZ transcriptional or translational fusions in the absence of HilD (20). In this study, we provide further genetic and biochemical data showing that HilE directly binds to HilD and blocks its ability to bind DNA but does not affect the dimerization or stability of HilD. Consistent with our previous genetic data, HilE is specific for HilD and does not bind or affect HilC or RtsA. In vivo data suggest that during infection, FliZ acts to overcome the negative regulation of HilE to switch on SPI1 T3SS expression in the intestine.
RESULTS
HilE acts specifically upon HilD.
Previous data suggest that HilE acts directly at the level of the HilD protein (5, 16, 20, 21). We sought to further understand the molecular mechanism by which HilE controls HilD protein activity. Figure 1B shows the effect of HilE on hilA expression. In the absence of HilE, hilA transcription was increased ∼4-fold, whereas overproduction of HilE decreased hilA transcription by ∼50-fold, consistent with previous data (20, 21). Our previous results showed that HilE does not regulate HilC or RtsA activity in the absence of HilD (16). To confirm this finding, we constructed a strain in which rtsA is placed under the control of the tetRA cassette. Otherwise isogenic variations of this strain included the deletion of hilD and/or overexpression of HilE, which should maximize any effect. Upon induction with anhydrotetracycline under SPI1-inducing conditions, we observed the effect of HilE only in the HilD-producing strain (Fig. 1C). Because these strains express both RtsA and HilC, we can conclude that HilE acts only through HilD, consistent with our previous experiments examining the effects of a hilE deletion (16).
HilE binds directly and specifically with HilD.
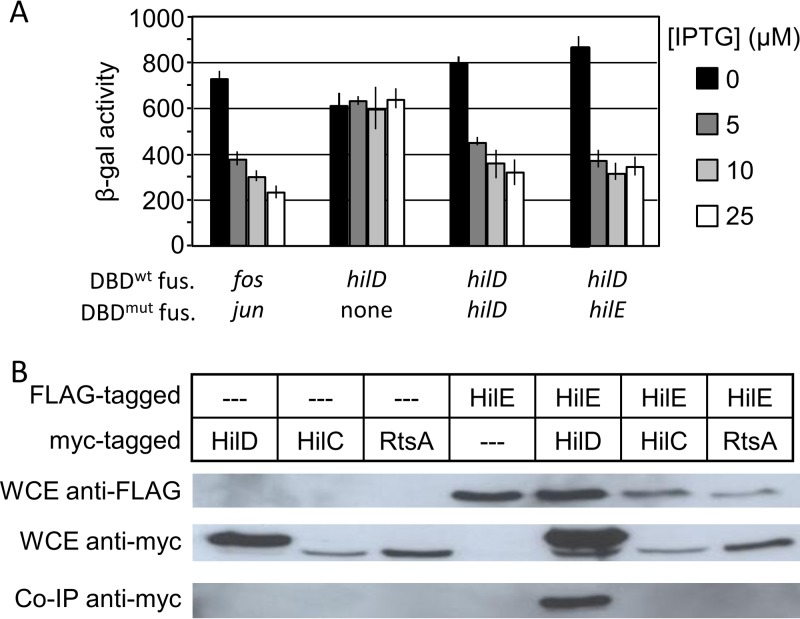
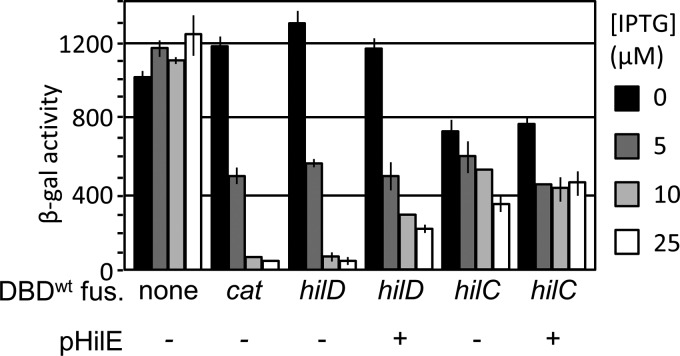
To characterize the interaction between HilE and HilD, we first confirmed the results of Baxter et al. (21) using a bacterial two-hybrid system. In this system, IPTG (isopropyl-β-d-thiogalactopyranoside) induces the expression of a LexA DNA binding domain (LexADBD) fused to the C terminus of a protein of interest. If the protein of interest dimerizes, the LexADBD binds to its cognate sulA promoter, repressing the expression of a transcriptionally linked lacZ gene. Alternatively, a mutant LexADBD and a half-mutant sulA promoter can be used to test for heterodimerization of two proteins (21–23). We created hilD-lexADBD and hilE-lexADBD constructs. The data in Fig. 2A suggest that first, HilD forms homodimers, and second, HilE and HilD indeed interact via protein-protein interaction.
FIG 2.
HilE specifically interacts with HilD. (A) A bacterial two-hybrid system with the indicated genes fused to either a wild-type or mutant lexA DNA binding domain were grown overnight in LSLB medium at 37°C and then subcultured 1:100 and allowed to incubate at 37°C for 18 h in LSLB medium containing the indicated concentration of IPTG. The genes fos and jun were employed as a positive control (n = 6, 3 sets of duplicate assays on 3 separate days; error bars represent one standard deviation). The strains used were SU202 with either pMS604 and pDP804, pSR659::hilD, pSR659::hilD and pSR658::hilE, or pSR659::hilD and pSR658::hilD. (B) Strains containing FLAG-tagged HilE and Myc-tagged HilD, HilC, or RtsA were grown overnight and lysed with a French pressure cell. Whole-cell extract (WCE) was then analyzed via Western blotting using either anti-FLAG or anti-Myc antibodies. Extracts were immunoprecipitated using anti-FLAG magnetic beads and subsequently analyzed via Western blotting using anti-Myc. The strains used were 14028 with pLS118, pLS119, or pCE81 and JS2105 with pLS118, pLS119, or pCE81.
To verify these data and to determine if HilE is capable of interacting with either HilC or RtsA, we performed a coimmunoprecipitation assay using strains containing FLAG-tagged HilE and Myc-tagged HilD, HilC, or RtsA constructs. HilE was immunoprecipitated using anti-FLAG antibodies, and the coprecipitation of HilD, HilC, or RtsA was monitored via Western analysis. The data in Fig. 2B indicate that HilE specifically binds HilD but not HilC or RtsA in vivo. Both the two-hybrid and coimmunoprecipitation data are consistent with our genetic model (Fig. 1A) and previous data indicating that HilE controls HilD protein activity but does not affect either HilC or RtsA (16).
HilE specifically disrupts HilD DNA binding activity.
One can envision several ways in which HilE could mechanistically regulate the activity of HilD. (i) HilE may simply interact with HilD in such a way that it blocks DNA binding. (ii) Given the results above and our unpublished data indicating that HilD forms homodimers in vivo, it is possible that HilE disrupts HilD dimerization and thereby indirectly blocks HilD DNA binding and activation of the hilA promoter. (iii) HilD has been reported to be degraded by Lon protease (24, 25). HilE could bind HilD and deliver it to Lon or another protease. (iv) HilE could regulate HilD function after HilD has bound to DNA. However, HilD functions partially by displacing H-NS (11), making the last model more difficult to imagine.
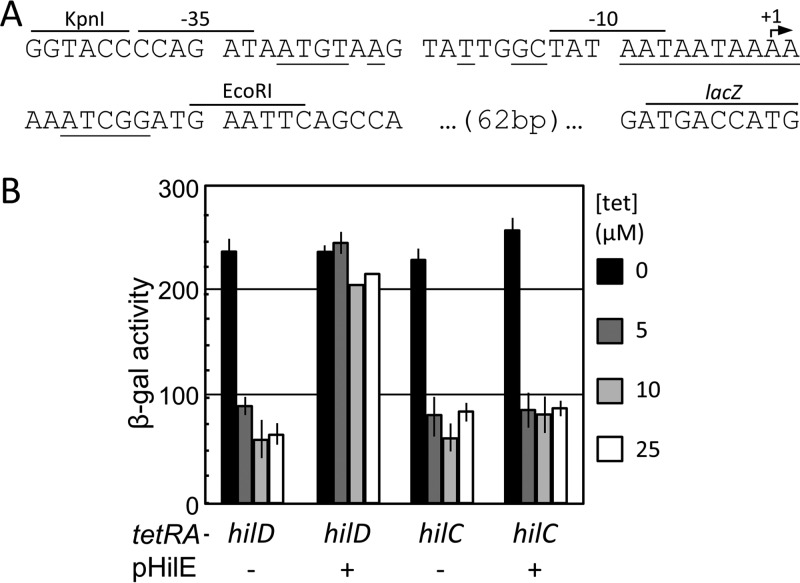
We first examined the ability of HilE to block HilD DNA binding. To provide a direct in vivo assay of HilD DNA binding, we created an artificial promoter that includes an overlapping HilD/HilC binding site and which drives the expression of lacZ, analogous to the method used by Luo and Farrand (26). In this construct, HilD or HilC should act as a simple repressor, occluding the binding of RNA polymerase. Thus, HilD and HilC DNA binding could be analyzed independent of displacement of H-NS or activation of RNA polymerase. We placed HilD or HilC under tetRA control in strains containing the synthetic promoter construct and in which the chromosomal hilD, hilC, rtsA, and hilE loci were deleted. These strains were grown in low-salt LB (LSLB) medium with various concentrations of tetracycline. When induced with tetracycline, there was marked a decrease in β-galactosidase activity in both the HilD- and HilC-producing strains (Fig. 3). These data suggest that both HilD and HilC act as simple repressors when bound to this synthetic promoter.
FIG 3.
HilE selectively blocks HilD DNA binding. (A) Sequence of the pJG1 HilD/HilC DNA binding reporter plasmid. Bases important for HilD/HilC binding are indicated by underlining (11). The recognition sequences for the restriction enzymes used in the creation of pJG1, the −35 and −10 promoter sequences, the predicted start site of transcription, and the start of the lacZ open reading frame are indicated. (B) Strains contained a HilD/HilC-repressible lac fusion and hilD or hilC under the control of a tetA promoter. Where indicated, HilE was overexpressed. Strains were incubated overnight at 37°C in LSLB medium with various concentrations of tetracycline (n = 6, 3 sets of duplicate assays on 3 separate days; error bars represent one standard deviation). The strains used were JS2106 and JS2107 with or without pHilE.
To determine if HilE prevents HilD from DNA binding, we introduced a plasmid encoding HilE into the above-mentioned strains. As shown in Fig. 3, HilE blocked the reduction in β-galactosidase activity in the HilD-producing strain but had no effect in the HilC-producing background. We interpret these data to indicate that HilE prevents HilD from binding DNA but does not affect HilC. This is consistent with our model (Fig. 1A) and with previous genetic data (14, 16, 20).
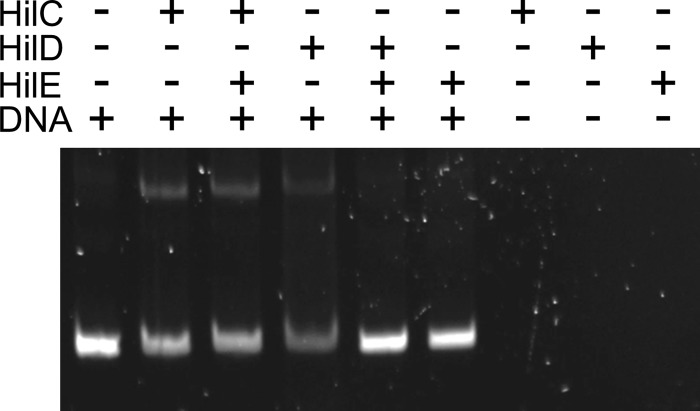
We next sought to biochemically verify these results. For these experiments, we used small ubiquitin-related modifier (SUMO)-tagged HilD and HilC. These SUMO-tagged constructs are functional in vivo and in vitro (see Fig. S1 in the supplemental material) and are more soluble than untagged proteins. We also purified HilE as a SUMO-tagged protein but then proteolytically removed the SUMO tag. We mixed either SUMO-tagged HilD or HilC with a fragment of the hilA promoter region and tested whether the addition of HilE would disrupt any shift of the DNA. As shown in Fig. 4, both HilD and HilC shifted a fragment of the hilA promoter, consistent with previous findings (11). Addition of HilE blocked DNA binding by HilD but had no effect on the HilC-mediated gel shift. Combined with our findings from Fig. 3, we interpret these data to indicate that HilE prevents HilD from binding DNA but does not affect HilC.
FIG 4.
HilE selectively blocks HilD DNA binding. The samples contained 0.7 μM purified HilD or HilC, 0.56 μM DNA, and, where indicated, 2.25 μM HilE. Samples were electrophoresed on a 10% continuous polyacrylamide gel and subsequently stained with SYBR Safe.
HilE does not affect HilD dimerization.
HilE could block HilD DNA binding indirectly by preventing HilD dimerization. To test whether HilD dimerization was affected, we employed the bacterial two-hybrid system (22, 23). The data in Fig. 5 suggest that both HilD and HilC homodimerize, as evidenced by an IPTG-dependent decrease in β-galactosidase activity. The addition of HilE to this system did not markedly alter the ability of the HilD- or HilC-LexADBD proteins to repress lacZ expression. Although not conclusive, these data suggested that HilE does not disrupt HilD or HilC homodimerization.
FIG 5.
HilE does not disrupt HilD dimerization. A bacterial monohybrid system with the indicated genes fused to a wild-type lexA DNA binding domain were grown under SPI1-inducing conditions with the indicated concentrations of IPTG. Where indicated, hilE was overexpressed from pWKS30 (n = 6, 3 sets of duplicate assays on 3 separate days; error bars represent one standard deviation). The strains used were SU101 with pSR658, pDD506, pSR658::hilD, or pSR658::hilC with and without pHilE.
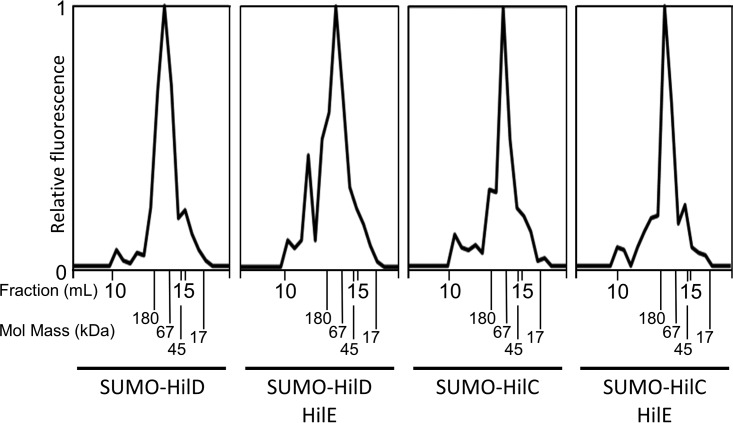
To further test whether HilE affects HilD dimerization, we fluorescently labeled either SUMO-tagged HilD or HilC and mixed the labeled protein in a 5% molar ratio with unlabeled SUMO-tagged HilD or HilC. These samples, with or without the addition of purified HilE, were separated via size exclusion chromatography over a Superose 12 HR 10/30 column, and the fluorescence of each fraction was measured. The use of fluorescently labeled protein allowed us to specifically monitor the elution of HilD or HilC but not HilE, which is unlabeled. The molecular masses of monomers and dimers of SUMO-HilD should be 47.7 kDa and 95.4 kDa, respectively; SUMO-HilC monomers and dimers should be 46.4 kDa and 92.7 kDa, respectively. In Fig. 6, we observe that both HilD and HilC eluted from the column consistent with the predicted mass of SUMO-tagged dimers. Structural prediction suggests that HilE should be a hexamer with a total molecular mass of 100 kDa. The addition of HilE in the HilD sample led to formation of a fluorescent species that eluted with an apparent molecular mass that was significantly greater than 180 kDa. The addition of HilE to the SUMO-tagged HilC sample had no effect on elution. This is consistent with a predicted protein complex composed of 2 HilD and 12 HilE monomers (295 kDa). The alternative model is that a hexamer of HilE would be bound to a HilD monomer, giving a predicted mass of 148 kDa. Combined, these data show that HilE binds to HilD without disrupting the HilD dimer. Further studies are required to determine the actual stoichiometry of the complex.
FIG 6.
HilE does not disrupt HilD dimerization. Purified SUMO-HilD or SUMO-HilC was fluorescently labeled and mixed in a 1:20 molar ratio with unlabeled SUMO-HilD or SUMO-HilC. Unlabeled HilE was mixed in a 12:1 molar ratio with either HilD or HilC where indicated. Proteins were separated by size exclusion chromatography, and the fluorescence of each fraction was assayed. The elution fraction and molecular mass of protein standards are labeled.
HilD remains stable in the presence of HilE.
We next determined whether HilE regulates HilD activity via modulating its stability. To avoid the complication of HilD autoactivation, we constructed strains in which the expression of a HilD-3×FLAG is under the control of tetRA. Furthermore, these strains contained a hilA′-lacZ construct, allowing us to measure HilD activity. We constructed hilE+, ΔhilE, and pHilE variations of these strains. It has been reported that HilD, HilC, and RtsA are degraded by Lon protease (24, 25). We therefore created otherwise isogenic Δlon Δwca variations of the above-mentioned strains. (The Δwca blocks capsule production, a complicating phenotype of Δlon strains.) All strains were grown overnight and subcultured for 2.5 h with tetracycline at a concentration that leads to activation of hilA to approximately wild-type levels. After incubation, an aliquot was withdrawn to determine hilA promoter activity via β-galactosidase assay. The remaining cells were immediately boiled in sodium dodecyl sulfate (SDS) loading buffer. The proteins in each aliquot were then resolved via SDS-PAGE and visualized using anti-FLAG antibody. As shown in Fig. 7, we observed HilE-dependent hilA promoter activity consistent with our genetic model and with previous findings (20). The effect of deleting hilE is dampened in these backgrounds, likely due to the lack of HilD autoregulation. Importantly, overproduction of HilE completely blocked hilA expression. However, the steady-state levels of HilD-3×FLAG were only slightly different in the various backgrounds. Although HilD activity was slightly increased in the Δlon background, overproduction of HilE blocks this activity without having any significant effect on the steady-state levels of the protein. These results suggest that HilE binds and directly blocks HilD function and that degradation of HilD is not a significant mechanism of regulation.
FIG 7.

HilE regulates HilD activity independently of stability. The hilD-3XFLAG construct is expressed from a tetA promoter. Strains with the indicated genotypes were grown to mid-log phase and induced with tetracycline. Western blotting and β-galactosidase assay were performed using aliquots from the same sample. Densitometry of the Western blot was performed using ImageJ. HilD levels and β-galactosidase activity were normalized to those of the hilE+ strain. The strains used were JS2108, JS2109, JS2110, JS2111, JS2112, and JS2113.
A hilE mutation partially suppresses the invasion defect conferred by loss of FliZ.
HilE is a major negative regulator of HilD and therefore of SPI1 T3SS expression. We therefore wished to determine whether HilE-mediated regulation is important during intestinal invasion. To test this, we performed oral and intraperitoneal (i.p.) competition assays, infecting BALB/c mice with a 1:1 mixture of ΔhilE and wild-type strains and, after 2.5 days, determining the ratio of the two strains in the small intestine and the spleen. As shown in Table 1, the ΔhilE strain conferred no significant phenotype in either oral or i.p. competition assays. This is perhaps not surprising, given that the negative regulatory effects of HilE must be normally overcome during intestinal infection.
TABLE 1.
Role of HilE during infection
| Genotype of straina: |
Infection route | Inoculum (CFU) | No. of mice | Spleen |
Small intestine |
|||
|---|---|---|---|---|---|---|---|---|
| A | B | Median CIb | Pc | Median CI | P | |||
| ΔhilE | WT | i.p. | 103 | 5 | 1.41 | NS | 0.79 | NS |
| Oral | 107 | 5 | 0.66 | NS | 0.93 | NS | ||
| 108 | 15 | 2.08 | NS | 1.41 | NS | |||
| ΔhilE ΔfliZ | ΔfliZ | i.p. | 103 | 9 | 0.92 | NS | 0.85 | NS |
| Oral | 107 | 9 | 3.35 | 0.01 | 1.88 | 0.04 | ||
| 108 | 5 | 4.97 | 0.01 | 4.21 | 0.06 | |||
The strains used were JS135, JS996, JS2115, and JS2116.
The competitive index (CI) was calculated as described in Materials and Methods.
The Student t test was used to compare the CIs to those for the inocula. NS, not significant.
It has been previously shown that FliZ significantly affects SPI1 T3SS regulation both in vivo and in vitro, with a ΔfliZ strain conferring an ∼12.5-fold SPI1-dependent invasion phenotype (20). HilE and FliZ act both independently and simultaneously to regulate HilD (16, 20). We therefore asked whether a ΔhilE mutation would suppress a ΔfliZ phenotype. We performed both oral and i.p. competition assays using a 1:1 mixture of ΔhilE ΔfliZ and ΔfliZ strains. After 3 days, we measured the ratio of the strains in the spleen and small intestine. As shown in Table 1, the ΔhilE ΔfliZ strain was 5-fold more invasive than the ΔfliZ strain. In all cases, no phenotype was observed after i.p. infection, indicating that the differences in infectivity between the strains were due to differences in invasiveness alone and not survivability within the animal. These data suggest that the deletion of hilE is capable of partially rescuing the invasion deficit conferred by ΔfliZ.
DISCUSSION
Salmonella enterica has evolved a complex regulatory network to control the expression of the SPI1 T3SS, which is central to the ability of Salmonella to cause gastroenteritis and invade the intestinal epithelium. We have developed a model for the SPI1 regulatory network and have begun to understand the mechanisms by which a large number of signals impinge upon this regulatory system (14, 16). In this work, we focused on HilE, a major negative regulator of HilD with a central role in SPI1 regulation.
HilE was previously identified as a negative regulator of the SPI1 T3SS, and two-hybrid data suggested that HilE binds directly to HilD (5, 21). We previously provided genetic evidence that the HilE regulatory effect is mediated through HilD (16, 20). Here we provide genetic and biochemical data showing that HilE directly binds to HilD and blocks its ability to bind DNA. These studies included the use of a synthetic promoter in which a HilD/HilC binding site is integrated into the −10 and −35 sequences, resulting in a promoter that is repressible by HilD and HilC, allowing us to study HilD and HilC DNA binding in vivo, independent of any interactions with H-NS or RNA polymerase. HilE does not block dimerization, nor does it significantly affect the stability of HilD. This regulation by direct protein-protein interaction explains the phenotype conferred by both hilE null and HilE overproduction, which completely blocks HilD activity.
Accumulated data suggest that HilD acts as the integration point for many of the environmental signals that control SPI1 expression. HilC and RtsA act as amplifiers of these signals. Although the actual mechanism is known in only a few cases, we have shown that regulation is primarily through either translation of the hilD message or control of HilD protein activity (14, 16, 19, 27). HilE falls into the latter class. Saini et al. demonstrated that the interaction of HilD with the hilD promoter is the trigger for inducing the expression of SPI1 through a stepwise increase of hilD promoter activity and autoactivation (17). Thus, the system is switched on when HilD activity reaches some threshold sufficient to efficiently activate its own expression. It is the amount of HilE that sets this threshold, by blocking the activity of low levels of HilD and thus preventing the inevitable minor fluctuations in HilD activity (noise) from inappropriately activating the system.
The N-terminal domains of HilD, HilC, and RtsA share approximately 10% identity, whereas the C-terminal DNA binding domains share 46% identity. The three proteins bind to the same DNA sequences to activate transcription of the hilD, hilC, rtsA, and hilA genes (11, 12, 15). Despite this homology, HilE functions solely through HilD. HilE does not interact with HilC or RtsA, nor does it have any regulatory effect in the absence of HilD. Our unpublished data suggest that HilD, HilC, and RtsA form both homodimers and heterodimers. HilE does not apparently affect dimerization. We propose that HilE binds independently to the individual HilD proteins in both homodimers and heterodimers containing HilD. This would allow HilE to efficiently block induction. Otherwise, heterodimers containing HilD would be capable of inappropriately activating expression of the SPI1 system.
How the level of HilE and, thus, the threshold of activation is controlled is only partially understood. Expression of hilE is apparently activated primarily by FimZ, but this regulation is modest under normal conditions. Loss of FimZ results in less than a 2-fold increase in hilA expression and a slight delay in the timing of SPI1 turning off in an in vitro experiment (17, 28). Only when FimZ is artificially overexpressed is there a dramatic effect on SPI1 expression (17, 28). Mlc is a transcriptional repressor considered a global regulator of carbohydrate metabolism (29–31). Lim et al. showed that loss of Mlc leads to a 2-fold increase in hilE mRNA levels and a 4-fold HilE-dependent decrease in invasion (32). They provide evidence that Mlc acts directly in the hilE promoter region, but the effects of various carbon sources on SPI1 expression and the physiological relevance are not clear. The transcriptional regulator LeuO is normally repressed by H-NS under laboratory conditions. Ectopic expression of LeuO in Salmonella transcriptionally induces hilE and leads to decreased expression of all of the SPI1 regulators. However, importantly, production of LeuO decreases hilD transcription in the absence of HilD, decreases hilC and rtsA transcription in the absence of both HilD and HilE, and leads to an invasion defect in the absence of HilE (33). Thus, although there is an apparent effect on hilE transcription, the predominant regulatory effects on SPI1 are HilE independent and seem to be global, analogous to the effects of H-NS (16). The PhoPQ two-component system strongly represses SPI1 gene expression (16). Baxter and Jones recently reported that PhoP activates hilE transcription in a manner that is somehow dependent on FimZ (34). However, our data show that PhoPQ can repress the system independently of HilD and HilE (reference 6 and unpublished data); thus, this transcriptional induction of hilE is apparently superfluous. Baxter and Jones also report that deletion of pstS, which induces the PhoBR two-component system, leads to transcriptional induction of fimZ and hilE and decreased expression of hilA (34). Understanding the overall impact of PhoBR in the SPI1 system will require further investigation.
Wang et al. provided evidence that the small RNA InvS (STnc470) negatively affects FimZ levels by an undefined mechanism (35). Deletion of invS leads to increased levels of FimZ, which should lead to increased expression of hilE (not tested). However, deletion of invS did not affect transcription of HilD-dependent invF, as would be expected if the phenotype were mediated through HilE. Rather, secretion, but not production, of effector proteins was reduced in an invS deletion strain, reportedly via control of translation of prgH, encoding a component of secretion machinery. The small RNA IsrM is reported to directly block translation of hilE by base pairing at the initiation codon (36). Deletion of isrM decreased SPI1 gene expression, although transcriptional effects were never quantified. However, the isrM deletion mutant is significantly attenuated in an animal, apparently during systemic stages of the disease, in which SPI1 is neither expressed nor required. These data suggest that IsrM has pleiotropic effects, and further studies are required to determine the relative role of hilE translation in these phenotypes. Together, these data suggest that although there is some regulation of hilE transcription and translation, this regulation is not dramatic under normal conditions and/or the identified regulators have effects on the system that are independent of HilE. We believe that a low-level regulation is consistent with the primary role of HilE in setting the threshold for activation rather than being a primary mechanism to repress the system in response to particular environmental signals.
Competition assays between ΔhilE and wild-type strains indicated no significant phenotype as a result of the loss of HilE (Table 1). This observation is consistent with previously published data indicating that the deletion of hilE confers no invasion phenotype in HEp-2 cells (28). It has been previously demonstrated that the deletion of fliZ, encoding a strong positive regulator of HilD, causes a defect in the ability of Salmonella to invade the host intestinal epithelium (20). However, deletion of hilE in a ΔfliZ background led to a 3- to 5-fold increase of the competitive index (CI) over a ΔfliZ background. These data together suggest that the negative regulatory effects that HilE has upon HilD are overcome by the point at which Salmonella reaches the small intestine and the site of invasion, consistent with the overall role of HilE in setting the threshold of activation that is overcome at the appropriate time and place during infection.
It has been suggested that Lon protease regulates SPI1 expression by the degradation of SPI1 regulatory components (24, 25), and HilD stability has been invoked numerous times to help explain regulatory effects (37–41). We previously demonstrated, however, that loss of Lon causes increased SPI1 expression by stabilizing FliZ and not the SPI1 regulatory components per se (20). We did note a change in the steady-state levels of HilD in response to HilE. The half-life of HilD-FLAG has been reported to be from 40 to 155 min by various investigators (20, 37, 38, 40, 41). Under our conditions, the HilD half-life changed from ∼75 min in a wild-type background to ∼40 min when HilE was overproduced (data not shown). However, the effect of HilE upon HilD activity does not correlate with the relative levels of HilD; overexpression of HilE resulted in a 50-fold decrease in hilA expression, while there was only 10% less HilD protein at the time that activity was measured (Fig. 7). This shows that the primary effect of HilE is to directly regulate HilD DNA binding activity and that any effect on HilD stability is a secondary effect. We propose that the changes in HilD half-life conferred by changing HilE levels are a result of HilD being slightly more unstable when free in solution than its DNA-bound form. This likely contributes to the effects seen for other regulatory inputs. Proteins that are truly regulated by modulating stability, such as RpoS, RpoH, and SulA, have half-lives on the order of 1 to 2 min under the unstable condition and are stabilized to >30 min in the induced state (42). In those cases where the stability of HilD has been invoked in regulation, the measured half-life under the “unstable” condition remained >15 min. It is difficult to imagine that these purported 2- to 3-fold changes in half-life in what is a relatively stable protein constitute the primary mechanism of regulation. It seems more likely that these various factors have some more direct effect on HilD activity.
Overall, our data suggest that the role of HilE is to simply bind to HilD, disrupting its ability to bind to DNA, thereby putting HilD into an inactive or low-activity state. The ability of HilE to maintain HilD in such a state is then overcome by the positive regulatory effects, including FliZ, at the time when Salmonella reaches the site of invasion in the small intestine. At this point, HilD is able to autoactivate its own promoter, as well as those of HilC and RtsA, leading to robust and rapid induction of the SPI1 T3SS.
MATERIALS AND METHODS
Construction of strains and plasmids.
All Salmonella strains are isogenic derivatives of Salmonella enterica serovar Typhimurium strain 14028 (American Type Culture Collection) and are listed in Table S1 in the supplemental material. Gene deletions and concomitant insertion of an antibiotic resistance cassette were constructed using λ Red-mediated recombination as previously described (43, 44). All constructs were verified by PCR and moved into a clean background via P22HTint105 phage transduction (45). In some cases, the antibiotic resistance cassette was removed by FLP-mediated recombination via the introduction of pCP20 (46).
Media, reagents, and enzymatic assays.
Strains were routinely grown using high-salt lysogeny broth (HSLB) (0.5% yeast extract, 1% tryptone, 1% NaCl) medium (47) at 37°C, except for strains containing the temperature-sensitive plasmid pKD46, pλInt, or pCP20, which were grown at 30°C. Where indicated, strains grown under Salmonella pathogenicity island 1 (SPI1)-inducing conditions were cultured overnight in 3 ml low-salt LB (LSLB) medium (0.5% NaCl) in 13-mm culture tubes at 37°C on a rotary drum and then subcultured 1:100 and grown in 3 ml HSLB medium in standing cultures (low oxygen) in 13-mm tubes at 37°C for 12 to 16 h. For non-SPI1-inducing conditions, cells were grown in LSLB medium at 37°C in a rotary drum for 12 to 16 h. Antibiotics were used at the following concentrations: ampicillin, 25 or 50 μg/ml; chloramphenicol, 20 μg/ml; kanamycin, 50 μg/ml; apramycin, 50 μg/ml; and tetracycline, 13 μg/ml. Antibodies were purchased from Sigma (monoclonal anti-FLAG M2 and monoclonal anti-c-Myc) or R&D Systems (rabbit anti-mouse IgG conjugated to horseradish peroxidase). Enzymes were purchased from Invitrogen, New England BioLabs, or Stratagene and used according to the manufacturers' recommendations. Primers were obtained from IDT.
β-Galactosidase assays were performed using a 96-well microtiter plate assay as previously described (48). Unless otherwise indicated, cells were grown under SPI1-inducing conditions for these assays. The indicated amount of anhydrotetracycline (aTc) or isopropyl-β-d-thiogalactopyranoside (IPTG) was added for specific gene induction. β-Galactosidase activity units are defined as (micromoles of ortho-nitrophenol [ONP] formed per minute × 106)/(optical density at 600 nm [OD600] × milliliters of cell suspension) and are reported as mean ± one standard deviation.
Coimmunoprecipitation.
We constructed strains containing Myc-tagged HilD, HilC, or RtsA on a plasmid and a FLAG (SPA)-tagged HilE inserted into the chromosome. These strains were grown overnight and subsequently harvested by centrifugation and then lysed in a French pressure cell. The protein concentration was determined by bicinchoninic acid (BCA) assay (Pierce). Anti-Myc magnetic beads (Pierce/Thermo Fisher) were washed twice in Tris-buffered saline (TBS) with Tween (TBST) (25 mM Tris, 150 mM NaCl, 0.05% Tween 20) after being collected in a magnetic stand. Whole-cell extracts (WCE) containing 100 μg of protein were mixed with a 4-fold excess of TBST and magnetic beads and then allowed to incubate for 30 min while mixing via rotation. The beads were then collected using a magnetic stand and washed twice in 5× TBST. Bound protein was then eluted from the magnetic beads using SDS-PAGE sample buffer (50 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 1% β-mercaptoethanol, 12.5 mM EDTA, 0.02% bromphenol blue) and heating the samples to 95°C for 15 min before analysis via Western blotting. The samples were separated on a 10% discontinuous SDS-polyacrylamide gel and transferred to Hybond ECL membranes (Amersham). The membranes were blocked three times for 10 min each using 5% (wt/vol) nonfat dry milk in Tris-buffered saline (TBS) (5% milk, 50 mM Tris, 150 mM NaCl, pH 8.0). The blots were then exposed to anti-FLAG M2 or anti-c-Myc antibody diluted 1:1,000 in TBS–5% milk overnight at 4°C. The blots were washed three times for 10 min each time in TBS–5% milk and then exposed to rabbit polyclonal anti-mouse IgG conjugated to horseradish peroxidase (R&D Systems) diluted 1:1,000 in TBS–5% milk for 1 h. The blots were then washed in Tris-buffered saline (TBS) (50 mM Tris, 150 mM NaCl, pH 8.0) three times for 5 min each repetition. The membranes were then exposed to Amersham ECL Western blot detection reagents (GE Healthcare) for 1 min and exposed to film (Kodak).
HilD/HilC DNA binding reporter.
To build a construct in which HilD or HilC acts as a simple transcriptional repressor, we synthesized four pairs of complementary oligonucleotides. Each pair consisted of a consensus promoter into which a HilD/HilC binding sequence had been integrated at various positions relative to the −10 and −35 promoter sequences. These fragments were cloned into pDX1 upstream of the promoterless lacZ gene between the unique KpnI and EcoRI sites (10). After passage through the Salmonella strain JS198 (r− m+), the plasmids were transformed and stably integrated in single copy at attλ by Int-mediated recombination (49), thus creating single-copy transcriptional fusions of the synthetic promoter and lacZ. The resulting constructs were verified to have a single integrated copy of the plasmid by PCR. These lacZ fusions were then moved via P22 transduction into ΔhilE ΔrtsA ΔSPI1 strains containing either tetracycline-inducible hilD or hilC. The resulting colonies were then screened by β-galactosidase assay to determine the extent to which HilD or HilC was able to repress expression from the synthetic promoter. The strain with the greatest repression upon tetracycline induction was selected for further studies.
To further increase the responsiveness of the strain to the induction of HilD or HilC, we mutagenized the −35 region of the synthetic promoter via PCR mutagenesis and reconstructed the fusion strains as described above. The resulting mutants were screened on medium containing tetracycline and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). Colonies demonstrating a robust reduction of β-galactosidase activity on X-Gal were selected and analyzed further. The colony with the most robust decrease of β-galactosidase activity upon induction with tetracycline was then selected, and the DNA sequence was determined. Final isogenic strains were then grown overnight in HSLB medium with 0, 0.78, 1.56, or 3.13 mg/ml tetracycline and analyzed via β-galactosidase assay.
Protein purification and gel filtration.
Translational fusions of hilD, hilC, or hilE to the small ubiquitin-related modifier (SUMO) were constructed using the pET-SUMO plasmid (Invitrogen; the SUMO-HilD construct was a gift from Marc Erhardt). These constructs were then transformed into Escherichia coli BL21(DE3) (50). These expression strains were grown overnight and subsequently subcultured 1:100 and incubated for 4 h at 37°C. At this point, 1 mM IPTG was added for induction, and the cells were incubated overnight at room temperature. The cells were then pelleted by centrifugation at 7,000 × g for 10 min and resuspended in NPI buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole) with a protease inhibitor cocktail (Roche). The cells were lysed with a French press and the lysate clarified by centrifugation at 22,000 × g for 1 h. The proteins were isolated by passing the clarified cell lysate over Ni-nitrilotriacetic acid (Ni-NTA)–agarose columns (Qiagen), washed with NPI buffer containing 90 mM imidazole, and eluted from the columns using NPI buffer with 250 mM imidazole. The proteins were concentrated by the use of centrifugal concentrators (Amicon). The protein was further purified via size exclusion chromatography over a Superose 12 HR 10/30 column and reconcentrated via centrifugal concentrators (Amicon) before use. The protein concentration was determined by the BCA assay (Pierce).
HilE was cleaved from the SUMO tag using SUMO protease (Invitrogen) and the protease and cleaved SUMO tag removed from the solution by passage over an Ni-NTA column (Qiagen). The HilE was subsequently reconcentrated with a centrifugal concentrator and further purified by size exclusion chromatography. The protein elutes as a series of overlapping peaks, consistent with structural predictions of multimerization to a hexamer (see Fig. S2 in the supplemental material). The Superose 12 HR 10/30 column used for both purification and size exclusion chromatography of the HilD-HilE complex has a resolution 1 to 300 kDa and an exclusion limit of 2,000 kDa.
Electromobility shift assay.
HilD, HilC, and HilE were purified as described above. The hilA promoter region is a known target of both HilD and HilC binding (11). Part of the hilA promoter (corresponding to base pairs 3033211 to 3033421, inclusive; NCBI genome accession number CP001363.1) was amplified via PCR and purified using a DNA spin column (Qiagen). Purified HilD or HilC was mixed with DNA in protein binding buffer (20 mM HEPES, 20 mM KCl, 1% glycerol, 50 μM EDTA) to establish a solution of 1 μM HilD or HilC and 15 nM DNA. HilE was added as indicated to a final concentration of 3 μM. A continuous 10% polyacrylamide gel was prerun for 30 min before loading the samples. The gel was then stained with SYBR Safe DNA gel stain (Invitrogen) and the image obtained on a transilluminator (Bio-Rad).
Bacterial mono- and two-hybrid methods.
Translational fusions of hilD, hilC, or hilE to the LexA DNA binding domain were constructed in pSR659 and pSR659 (23). These constructs were then transformed into either the SU101 (homodimer) or SU202 (heterodimer) E. coli strain containing a LexA-repressible sulA promoter that drives the expression of lacZ. These strains were grown overnight in 3 ml HSLB medium at 37°C and then subcultured 1:100 and grown for 18 h at 37°C in 3 ml HSLB medium containing 0, 5, 10, or 25 μM IPTG. A plasmid producing only the LexA DNA binding domain (pSR658) or a Cat-LexADBD fusion served as a negative or positive control for the bacterial monohybrid system (SU101), respectively (22). Plasmids pSR658 and pSR659 encoding only the DNA binding domains or encoding fos and jun were used as negative and positive controls for the two-hybrid (SU202) system, respectively. The strains were then analyzed via β-galactosidase assay as described above.
Stability assay methods.
To measure HilD protein stability, we cultured a tetRA-hilD-3×FLAG attλ::pDX1::hilA′-lacZ+ strain in 3 ml LSLB medium in 13-mm culture tubes overnight at 37°C on a rotary drum. The cells were subcultured 1:100 in 10 ml HSLB medium containing 0.8 μg/ml tetracycline in 18-mm culture tubes and grown in a rotary drum for 2.5 h at 37°C. After 2.5 h, the OD600 of each sample was measured and the volume adjusted with HSLB medium to equalize the concentration of cells. One milliliter of each sample was then withdrawn for β-galactosidase assays as described above. Additional one-milliliter aliquots were removed from each culture and immediately centrifuged for 1 min at 14,000 × g. The pellets were resuspended in 120 μl sodium dodecyl sulfate (SDS) loading buffer (50 mM Tris-HCl [pH 6.8], 2% SDS, 10% glycerol, 1% β-mercaptoethanol, 12.5 mM EDTA, 0.02% bromphenol blue) and boiled for 10 min before being analyzed via Western blotting as described above using anti-FLAG antibodies. Densitometry of the resulting blots was performed using NIH ImageJ, and protein levels were determined by measuring the area under the curve for the resulting protein bands.
Virulence assays.
We inoculated 6- to 8-week-old BALB/c mice (Envigo) via oral gavage or intraperitoneal (i.p.) injection with 0.2 ml of bacterial cell suspension. In all the competition assays, the inoculum consisted of a 1:1 mixture of two bacterial strains. The actual CFU and relative percentage of each strain were determined by direct plating of the inocula. Cells were cultured overnight in HSLB medium in 13-mm tubes at 37°C in a rotary drum. For oral infections, the cells were washed in sterile 0.1 M phosphate-buffered saline (PBS) (pH 8.0) and suspended at 5 × 107 or 5 × 108 cells/ml. For i.p. infections, the cells were washed in sterile saline and diluted to 5 × 103 cells/ml in 0.15 M NaCl. Food was withdrawn 4 h prior to oral infection. Food and water were provided ad libitum after infection. The mice were sacrificed via CO2 asphyxiation 2.5 days after infection, and the small intestines and spleens were harvested. The organs were homogenized, and aliquots of serial dilutions were plated to appropriate selective medium to determine the number of CFU per organ. The ratio of the two strains was determined by replica plating to appropriate antibiotic-containing medium. The competitive index (CI) was calculated as (CFU of strain A recovered/CFU of strain B recovered)/(CFU of strain A inoculated/CFU of strain B inoculated). Each mutant strain was reconstructed at least once to verify that the observed phenotypes were a result of the designated mutation. The Student t test was used to determine whether the output ratio was significantly different from the input ratio. All animal work was reviewed and approved by the University of Illinois Institutional Animal Care and Use Committee and performed under protocols 10050 and 13030.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by NIH grant GM120182 to J.M.S.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We thank Marc Erhardt for the pET-SUMO::hilD plasmid and the University of Illinois Roy J. Carver Biotechnology Center Protein Sciences facility for gel filtration analysis.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00750-17.
REFERENCES
- 1.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis 17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lostroh CP, Lee CA. 2001. The Salmonella pathogenicity island-1 type III secretion system. Microbes Infect 3:1281–1291. doi: 10.1016/S1286-4579(01)01488-5. [DOI] [PubMed] [Google Scholar]
- 3.Ellermeier CD, Slauch JM. 2003. RtsA and RtsB coordinately regulate expression of the invasion and flagellar genes in Salmonella enterica serovar Typhimurium. J Bacteriol 185:5096–5108. doi: 10.1128/JB.185.17.5096-5108.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hansen-Wester I, Hensel M. 2002. Genome-based identification of chromosomal regions specific for Salmonella spp. Infect Immun 70:2351–2360. doi: 10.1128/IAI.70.5.2351-2360.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fahlen TF, Mathur N, Jones BD. 2000. Identification and characterization of mutants with increased expression of hilA, the invasion gene transcriptional activator of Salmonella typhimurium. FEMS Immunol Med Microbiol 28:25–35. doi: 10.1111/j.1574-695X.2000.tb01453.x. [DOI] [PubMed] [Google Scholar]
- 6.Moest TP, Méresse S. 2013. Salmonella T3SSs: Successful mission of the secret(ion) agents. Curr Opin Microbiol 16:38–44. doi: 10.1016/j.mib.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 7.Lee CA, Jones BD, Falkow S. 1992. Identification of a Salmonella typhimurium invasion locus by selection for hyperinvasive mutants. Proc Natl Acad Sci U S A 89:1847–1851. doi: 10.1073/pnas.89.5.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bajaj V, Hwang C, Lee CA. 1995. HilA is a novel OmpR/ToxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol Microbiol 18:715–727. doi: 10.1111/j.1365-2958.1995.mmi_18040715.x. [DOI] [PubMed] [Google Scholar]
- 9.Ahmer BM, van Reeuwijk J, Watson PR, Wallis TS, Heffron F. 1999. Salmonella SirA is a global regulator of genes mediating enteropathogenesis. Mol Microbiol 31:971–982. doi: 10.1046/j.1365-2958.1999.01244.x. [DOI] [PubMed] [Google Scholar]
- 10.Lin D, Rao CV, Slauch JM. 2008. The Salmonella SPI1 type three secretion system responds to periplasmic disulfide bond status via the flagellar apparatus and the RcsCDB system. J Bacteriol 190:87–97. doi: 10.1128/JB.01323-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olekhnovich IN, Kadner RJ. 2002. DNA-binding activities of the HilC and HilD virulence regulatory proteins of Salmonella enterica serovar Typhimurium. J Bacteriol 184:4148–4160. doi: 10.1128/JB.184.15.4148-4160.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olekhnovich IN, Kadner RJ. 2006. Crucial roles of both flanking sequences in silencing of the hilA promoter in Salmonella enterica. J Mol Biol 357:373–386. doi: 10.1016/j.jmb.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 13.Lucas RL, Lee CA. 2001. Roles of HilC and HilD in regulation of hilA expression in Salmonella enterica serovar Typhimurium. J Bacteriol 183:2733–2745. doi: 10.1128/JB.183.9.2733-2745.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellermeier CD, Ellermeier JR, Slauch JM. 2005. HilD, HilC and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator hilA in Salmonella enterica serovar Typhimurium. Mol Microbiol 57:691–705. doi: 10.1111/j.1365-2958.2005.04737.x. [DOI] [PubMed] [Google Scholar]
- 15.Olekhnovich IN, Kadner RJ. 2007. Role of nucleoid-associated proteins Hha and H-NS in expression of Salmonella enterica activators HilD, HilC, and RtsA required for cell invasion. J Bacteriol 189:6882–6890. doi: 10.1128/JB.00905-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Golubeva YA, Sadik AY, Ellermeier JR, Slauch JM. 2012. Integrating global regulatory input into the Salmonella pathogenicity Island 1 type III secretion system. Genetics 190:79–90. doi: 10.1534/genetics.111.132779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saini S, Ellermeier JR, Slauch JM, Rao CV. 2010. The role of coupled positive feedback in the expression of the SPI1 type three secretion system in Salmonella. PLoS Pathog 6:1–16. doi: 10.1371/journal.ppat.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iyoda S, Kamidoi T, Hirose K, Kutsukake K, Watanabe H. 2001. A flagellar gene fliZ regulates the expression of invasion genes and virulence phenotype in Salmonella enterica serovar Typhimurium. Microb Pathog 30:81–90. doi: 10.1006/mpat.2000.0409. [DOI] [PubMed] [Google Scholar]
- 19.Ellermeier JR, Slauch JM. 2008. Fur regulates expression of the Salmonella pathogenicity island 1 type III secretion system through HilD. J Bacteriol 190:476–486. doi: 10.1128/JB.00926-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cott Chubiz JE, Golubeva YA, Lin D, Miller LD, Slauch JM. 2010. FliZ regulates expression of the Salmonella pathogenicity island 1 invasion locus by controlling HilD protein activity in Salmonella enterica serovar Typhimurium. J Bacteriol 192:6261–6270. doi: 10.1128/JB.00635-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baxter MA, Fahlen TF, Wilson RL, Jones BD. 2003. HilE interacts with HilD and negatively regulates hilA transcription and expression of the Salmonella enterica serovar Typhimurium invasive phenotype. Infect Immun 71:1295–1305. doi: 10.1128/IAI.71.3.1295-1305.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daines DA, Granger-Schnarr M, Dimitrova M, Silver RP. 2002. Use of LexA-based system to identify protein-protein interactions in vivo. Methods Enzymol 358:153–161. doi: 10.1016/S0076-6879(02)58087-3. [DOI] [PubMed] [Google Scholar]
- 23.Daines DA, Silver RP. 2000. Evidence for multimerization of Neu proteins involved in polysialic acid synthesis in Escherichia coli K1 using improved LexA-based vectors. J Bacteriol 182:5267–5270. doi: 10.1128/JB.182.18.5267-5270.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boddicker JD, Jones BD. 2004. Lon protease activity causes down-regulation of Salmonella pathogenicity island 1 invasion gene expression after infection of epithelial cells. Infect Immun 72:2002–2013. doi: 10.1128/IAI.72.4.2002-2013.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takaya A, Kubota Y, Isogai E, Yamamoto T. 2005. Degradation of the HilC and HilD regulator proteins by ATP-dependent Lon protease leads to downregulation of Salmonella pathogenicity island 1 gene expression. Mol Microbiol 55:839–852. doi: 10.1111/j.1365-2958.2004.04425.x. [DOI] [PubMed] [Google Scholar]
- 26.Luo ZQ, Farrand SK. 1999. Signal-dependent DNA binding and functional domains of the quorum-sensing activator TraR as identified by repressor activity. Proc Natl Acad Sci U S A 96:9009–9014. doi: 10.1073/pnas.96.16.9009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Golubeva YA, Ellermeier JR, Cott Chubiz JE, Slauch JM. 2016. Intestinal long-chain fatty acids act as a direct signal to modulate expression of the Salmonella pathogenicity island 1 type III secretion system. mBio 7:e02170-15. doi: 10.1128/mBio.02170-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baxter MA, Jones BD. 2005. The fimYZ genes regulate Salmonella enterica serovar Typhimurium invasion in addition to type 1 fimbrial expression and bacterial motility. Infect Immun 73:1377–1385. doi: 10.1128/IAI.73.3.1377-1385.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee S, Boos W, Bouche JP, Plumbridge J. 2000. Signal transduction between a membrane-bound transporter, PtsG, and a soluble transcription factor, Mlc, of Escherichia coli. EMBO J 19:5353–5361. doi: 10.1093/emboj/19.20.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanaka Y, Kimata K, Aiba H. 2000. A novel regulatory role of glucose transporter of Escherichia coli: membrane sequestration of a global repressor Mlc. EMBO J 19:5344–5352. doi: 10.1093/emboj/19.20.5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nam TW, Cho SH, Shin D, Kim JH, Jeong JY, Lee JH, Roe JH, Peterkofsky A, Kang SO, Ryu S, Seok YJ. 2001. The Escherichia coli glucose transporter enzyme IICB Glc recruits the global repressor Mlc. EMBO J 20:491–498. doi: 10.1093/emboj/20.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim S, Yun J, Yoon H, Park C, Kim B, Jeon B, Kim D, Ryu S. 2007. Mlc regulation of Salmonella pathogenicity island I gene expression via hilE repression. Nucleic Acids Res 35:1822–1832. doi: 10.1093/nar/gkm060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dillon SC, Espinosa E, Hokamp K, Ussery DW, Casadesús J, Dorman CJ. 2012. LeuO is a global regulator of gene expression in Salmonella enterica serovar Typhimurium. Mol Microbiol 85:1072–1089. doi: 10.1111/j.1365-2958.2012.08162.x. [DOI] [PubMed] [Google Scholar]
- 34.Baxter MA, Jones BD. 2015. Two-component regulators control hilA expression by controlling fimZ and hilE expression within Salmonella enterica serovar Typhimurium. Infect Immun 83:978–985. doi: 10.1128/IAI.02506-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L, Cai X, Wu S, Bomjan R, Nakayasu ES, Händler K, Hinton JCD, Zhou D. 2017. InvS coordinates expression of prgH and fimZ and is required for invasion of epithelial cells by Salmonella enterica serovar Typhimurium. J Bacteriol 199:e00824-16. doi: 10.1128/JB.00824-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gong H, Vu G-P, Bai Y, Chan E, Wu R, Yang E, Liu F, Lu S. 2011. A Salmonella small non-coding RNA facilitates bacterial invasion and intracellular replication by modulating the expression of virulence factors. PLoS Pathog 7:e1002120. doi: 10.1371/journal.ppat.1002120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hung C-C, Garner CD, Slauch JM, Dwyer ZW, Lawhon SD, Frye JG, Mcclelland M, Ahmer BMM, Altier C. 2013. The intestinal fatty acid propionate inhibits Salmonella invasion through the post-translational control of HilD. Mol Microbiol 87:1045–1060. doi: 10.1111/mmi.12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De la Cruz MA, Pérez-Morales D, Palacios IJ, Fernández-Mora M, Calva E, Bustamante VH. 2015. The two-component system CpxR/A represses the expression of Salmonella virulence genes by affecting the stability of the transcriptional regulator HilD. Front Microbiol 6:807. doi: 10.3389/fmicb.2015.00807. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Hung C-C, Eade CR, Altier C. 2016. The protein acyltransferase Pat post-transcriptionally controls HilD to repress Salmonella invasion. Mol Microbiol 102:121–136. doi: 10.1111/mmi.13451. [DOI] [PubMed] [Google Scholar]
- 40.Eade CR, Hung C-C, Bullard B, Gonzalez-Escobedo G, Gunn JS, Altier C. 2016. Bile acids function synergistically to repress invasion gene expression in Salmonella by destabilizing the invasion regulator HilD. Infect Immun 84:2198–2208. doi: 10.1128/IAI.00177-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sang Y, Ren J, Qin R, Liu S, Cui Z, Cheng S, Liu X, Lu J, Tao J, Yao YF. 2017. Acetylation regulates protein stability and DNA-binding ability of HilD, thus modulating Salmonella Typhimurium virulence. J Infect Dis 216:1018–1026. doi: 10.1093/infdis/jix102. [DOI] [PubMed] [Google Scholar]
- 42.Gottesman S. 1996. Proteases and their targets in Escherichia coli. Annu Rev Genet 30:465–506. doi: 10.1146/annurev.genet.30.1.465. [DOI] [PubMed] [Google Scholar]
- 43.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ellermeier CD, Janakiraman A, Slauch JM. 2002. Construction of targeted single copy lac fusions using λ Red and FLP-mediated site-specific recombination in bacteria. Gene 290:153–161. doi: 10.1016/S0378-1119(02)00551-6. [DOI] [PubMed] [Google Scholar]
- 45.Maloy SR, Stewart VJ, Taylor RK. 1996. Genetic analysis of pathogenic bacteria: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 46.Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14. doi: 10.1016/0378-1119(95)00193-A. [DOI] [PubMed] [Google Scholar]
- 47.Silhavy TJ, Berman ML, Enquist LW. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 48.Slauch JM, Silhavy TJ. 1991. Genetic fusions as experimental tools. Methods Enzymol 204:213–248. doi: 10.1016/0076-6879(91)04011-C. [DOI] [PubMed] [Google Scholar]
- 49.Haldimann A, Wanner BL. 2001. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J Bacteriol 183:6384–6393. doi: 10.1128/JB.183.21.6384-6393.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Studier FW, Moffatt BA. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol 189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.