ABSTRACT
The Pseudomonas fluorescens genome encodes more than 50 proteins predicted to be involved in c-di-GMP signaling. Here, we demonstrated that, tested across 188 nutrients, these enzymes and effectors appeared capable of impacting biofilm formation. Transcriptional analysis of network members across ∼50 nutrient conditions indicates that altered gene expression can explain a subset of but not all biofilm formation responses to the nutrients. Additional organization of the network is likely achieved through physical interaction, as determined via probing ∼2,000 interactions by bacterial two-hybrid assays. Our analysis revealed a multimodal regulatory strategy using combinations of ligand-mediated signals, protein-protein interaction, and/or transcriptional regulation to fine-tune c-di-GMP-mediated responses. These results create a profile of a large c-di-GMP network that is used to make important cellular decisions, opening the door to future model building and the ability to engineer this complex circuitry in other bacteria.
IMPORTANCE Cyclic diguanylate (c-di-GMP) is a key signaling molecule regulating bacterial biofilm formation, and many microbes have up to dozens of proteins that make, break, or bind this dinucleotide. A major open issue in the field is how signaling specificity is conferred in the unpartitioned space of a bacterial cell. Here, we took a systems approach, using mutational analysis, transcriptional studies, and bacterial two-hybrid analysis to interrogate this network. We found that a majority of enzymes are capable of impacting biofilm formation in a context-dependent manner, and we revealed examples of two or more modes of regulation (i.e., transcriptional control with protein-protein interaction) being utilized to generate an observable impact on biofilm formation.
KEYWORDS: c-di-GMP, biofilm, network, P. fluorescens, Pseudomonas fluorecens
INTRODUCTION
Cyclic diguanylate (c-di-GMP) is a second messenger used across a range of bacterial species to control important life style decisions. c-di-GMP is produced by enzymes called diguanylate cyclases (DGCs) with the canonical GGDEF domain and is in turn degraded by phosphodiesterases (PDEs) with the canonical EAL domain. This second messenger governs cellular processes by binding to and activating effector proteins that govern such behaviors as biofilm formation, motility, virulence activation, macromolecular synthesis, cell division, and other processes (1–4).
Many bacteria have large c-di-GMP signaling networks, with dozens of DGCs, PDEs, and possible effectors. Several models have been developed to help explain how large c-di-GMP signaling networks may effectively communicate with specific effector proteins at the right time or location in the cell. One model relies on global levels of c-di-GMP signaling to different effectors based on their binding affinity. Pultz et al. demonstrated that two Salmonella enterica serovar Typhimurium effector proteins with PilZ domains bound c-di-GMP with greater than 40-fold differences in affinity (5). This finding was generalized to Pseudomonas aeruginosa, where greater than 140-fold differences were found among its eight PilZ domain-containing proteins (5, 6). Consistent with this “affinity model,” modulating global pools of c-di-GMP has been shown to alter expression and activity of c-di-GMP-metabolizing enzymes, as well as outputs of the network such as exopolysaccharide production and control of flagellar synthesis (7–11). A second model put forth makes use of DGCs, PDEs, and effectors that physically interact with one another, allowing local signaling. Evidence for physical interaction among c-di-GMP enzymes has been found in Yersinia pestis (12), and examples of physical interaction being required for signaling include a DGC-PDE interaction in Escherichia coli and a DGC-effector interaction in Pseudomonas fluorescens (13, 14). Transcriptional control of the genes coding for c-di-GMP-related enzymes, including rapA in P. fluorescens and several genes encoding DGCs and PDEs in Vibrio cholerae, provides a third mechanism of signaling specificity (15–18). Finally, ligand binding may mediate the activity state of various DGCs and PDEs, as is the case in E. coli for the oxygen-sensing DosC-DosP DGC-PDE pair or the inactivating effect of zinc on DgcZ (19, 20). Both transcriptional regulation and ligand-mediated activation could conceivable fit in either a global or local signaling model. Thus, there does not appear to be any single mechanism used by bacteria to ensure a specific output from this complex network.
In order to understand how large c-di-GMP networks are organized and utilized, we focused on P. fluorescens Pf01. The mechanism of biofilm formation in P. fluorescens is one of the best understood of such signaling systems, from input to output (21, 22). The effector protein LapD binds cytoplasmic c-di-GMP, which in turns leads to an accumulation of the large adhesion LapA on the cell surface via an inside-out signaling mechanism (23, 24). P. fluorescens contains a large c-di-GMP network, with 21 GGDEF domain-containing proteins, 5 EAL domain-containing proteins, and 17 dual-domain proteins containing both a GGDEF domain and an EAL domain (Fig. 1). These dual-domain proteins may function as DGCs or PDEs or both or as effectors that can bind c-di-GMP (23, 25–27). P. fluorescens also contains 6 proteins with PilZ domains, a domain previously characterized as a c-di-GMP binding effector (28, 29), as well as a FleQ homolog (8). In a previous study, a mutant library of P. fluorescens was assayed for the impact of a majority of these genes on biofilm formation using a single medium containing glycerol and tryptone. Four DGCs and five putative PDEs appeared to impact biofilm formation under this minimal medium (MM) condition, leaving most enzymes without an apparent function (30). This conundrum of “extra enzymes” is not unique to P. fluorescens. Similar results have been obtained in V. cholerae, P. aeruginosa, and E. coli (31–33). This observation leads to two important questions. First, can these other c-di-GMP-related enzymes provide fine control over biofilm formation under nonlaboratory conditions? And second, in what ways can such a large network based on a diffusible molecule be organized to provide order to the signaling process?
FIG 1.
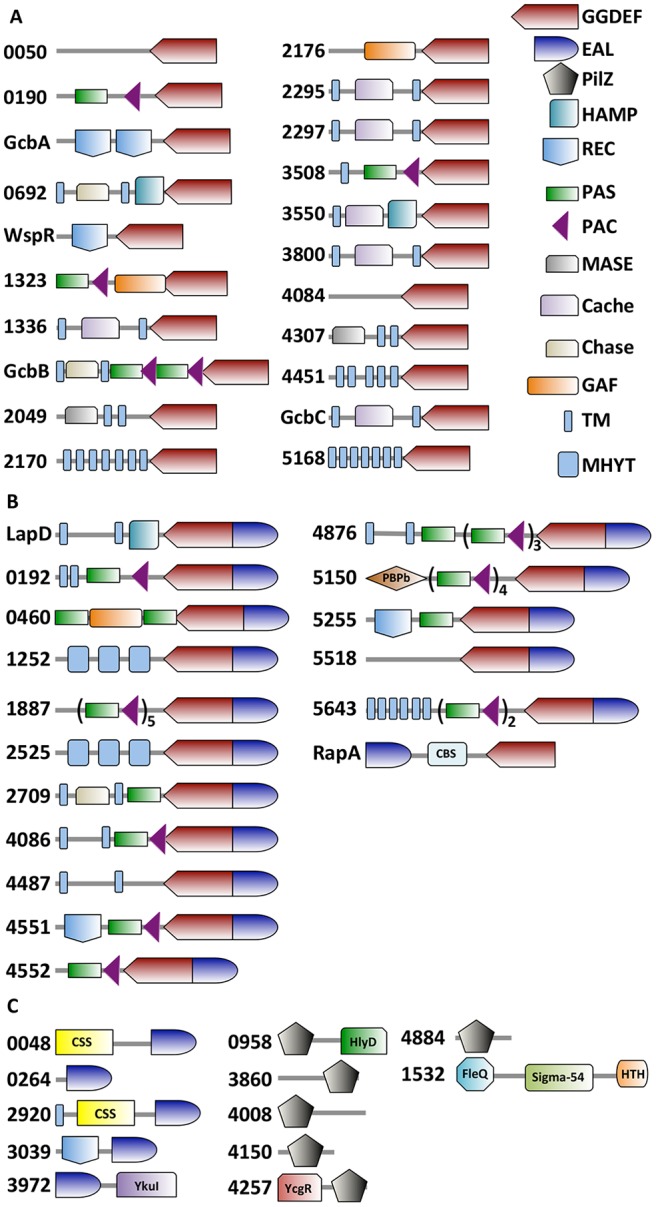
Domain-level depiction of c-di-GMP-related proteins in P. fluorescens. All proteins in P. fluorescens with an EAL, GGDEF, PilZ, or other known c-di-GMP-binding domain are depicted. The illustrated domains include those predicted by Pfam and SMART. In cases where these two databases predicted different domain architectures over the same amino acid span, the Pfam prediction was used, and Pfam domain-naming conventions are used in all cases. If a gene or protein name has been assigned or reported, it is also shown here. All recurrent domains are listed in the key at the right of panel A, with the remainder of domains labeled on each protein. Models are not drawn to scale, although large amino acid stretches without a domain depicted represent space in the protein where no known domains are predicted. (A) GGDEF domain-containing proteins. (B) Dual-domain-containing proteins. (C) EAL and PilZ domain-containing proteins and the predicted FleQ homolog of P. aeruginosa. Note that the predicted function has been demonstrated biochemically for only a small number of these proteins.
Here, we attempted to resolve those questions using P. fluorescens Pf01 as a model system for a large c-di-GMP signaling network. We found that, tested under a broad spectrum of conditions, a large majority of the c-di-GMP-related enzymes and effectors of P. fluorescens can impact biofilm formation. Further, we found that transcriptional regulation, nontranscriptional responses to putative ligands, and protein-protein interactions are common features among network members. Furthermore, these modes of regulation can be combined, for example, with some pairs of proteins interacting while also responding to common extracellular cues. These findings provide a road map for approaching c-di-GMP signaling in bacteria of clinical and environmental relevance, with particular emphasis on understanding how multimodal regulatory strategies modulate biofilm formation in the context of a complex network.
RESULTS
A majority of proteins with predicted roles as c-di-GMP metabolizing enzymes or effectors participate in biofilm formation.
A previous study performed by our group involved construction of a mutant library of a majority of DGCs and PDEs in P. fluorescens and testing of those mutants for biofilm formation on our standard laboratory minimal medium with glycerol and tryptone (30). Four putative DGCs—GcbA, GcbB, GcbC, and WspR—were found to positively contribute to biofilm formation in this medium. Mutations in the genes coding for five dual-domain proteins—Pfl01_0192, Pfl01_1887, Pfl01_2709, Pfl01_4086, and Pfl01_4876—were found to increase biofilm formation under these conditions, indicating that they likely act as PDEs. Given the large array of remaining GGDEF and EAL domain-containing proteins that demonstrated no altered phenotype under this single growth condition, we hypothesized that some number of these c-di-GMP metabolizing enzymes and/or effectors may also contribute to biofilm formation under conditions not previously tested. Further, most predicted c-di-GMP-related enzymes in P. fluorescens are fused to a variety of putative ligand-binding domains, including CACHE, CHASE, and GAF domains (Fig. 1). It therefore seemed possible that the c-di-GMP metabolizing enzymes could be activated by growing P. fluorescens under different conditions. To this end, we tested the wild-type (WT) strain and 50 strains each carrying a mutation in an individual predicted c-di-GMP-related gene (CDG gene) (see Table S1 in the supplemental material) using Biolog plates (Biolog, Inc.) to test 188 different nutrients (Table S2) for their impact on biofilm formation. While we refer to the compounds in each of the wells of the Biolog plates as “nutrients” because they are all organic compounds, these compounds could conceivably serve as carbon sources or input signals or both or neither. The term “nutrients” is used here as convenient shorthand to generally describe the chemical compounds in the Biolog plates.
The minimal medium typically used to grow P. fluorescens biofilms for our studies, referred to as K10T-1 (K10), is a high-phosphate medium containing tryptone and glycerol as carbon and energy sources (30). For the purposes of testing biofilm formation in the Biolog plates, we used a similar medium lacking the tryptone with its diverse array of carbon/energy sources. Because the nutrient sources tested here may not be metabolized by P. fluorescens and yet may still impact the activity of the c-di-GMP network, glycerol was retained in this medium to permit growth. We refer to this glycerol-containing medium as “base minimal medium” (BMM) (see Materials and Methods for details).
In-frame deletions of genes encoding the predicted DGCs, PDEs, and effectors as well as of the six genes predicted to encode PilZ domain-containing proteins and a FleQ homolog were constructed. The remainder of tested strains originated from a previously reported (30) single-crossover mutant library (Table S1). Each strain was grown overnight in LB with aeration, normalized to an optical density at 600 nm (OD600) of ∼0.716, and added to the BMM with each nutrient found in the Biolog plates before allowing biofilm formation to commence. The nutrients that were toxic to P. fluorescens, as indicated by lack of growth of the wild-type strain (not shown), were eliminated from the analysis. Analyzing the remaining nutrients, it was clear that different nutrients have differential effects on biofilm formation.
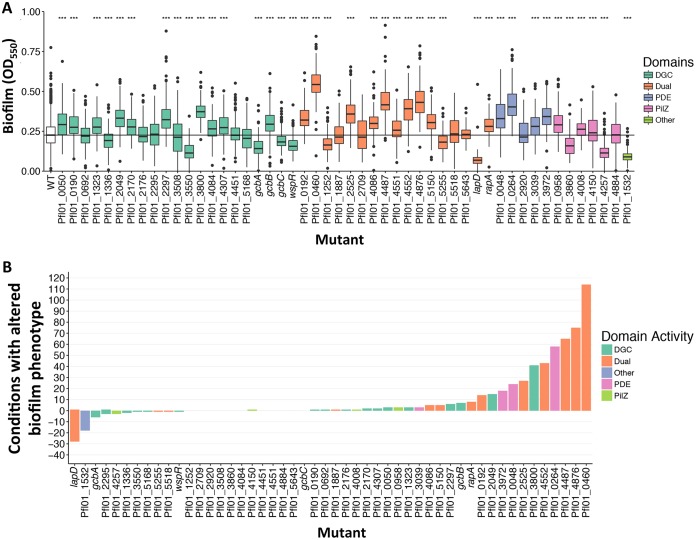
Among the 50 mutants tested, 44 demonstrated a significant difference (P < 0.001) from the wild-type results in comparisons of the median levels of ability to promote biofilm formation across all nutrients tested (Fig. 2A). Among strains lacking the GGDEF-containing proteins, 6 showed a significant reduction in biofilm formation across all conditions such as might canonically be expected for this class of mutants. Surprisingly, 10 of these strains carrying mutations in GGDEF-encoding genes showed a significant increase in biofilm formation across all conditions, suggesting that some putative DGCs may be able to negatively impact biofilm formation, in contrast to the activity canonically associated with their enzymatic activity. Another 5 mutants associated with GGDEF-encoding genes showed no significant impact. Results from strains lacking EAL-containing proteins were more straightforward, with 4 such mutants showing elevated levels of biofilm formation and a single strain that did not significantly differ from the wild type.
FIG 2.
Strains with mutations in genes coding for predicted c-di-GMP signaling proteins show a defect in biofilm formation compared to the WT strain. (A) Data from biofilm formation assays conducted using all 188 tested nutrients for each gene indicated are displayed as a box plot. Each box is color-coded based on the domain associated with the indicated gene (see legend on right side). The boxes indicate the range between the first and third quartiles (i.e., the 25th and 75th percentiles). The horizontal line inside the box is the median. The size of the box represents the interquartile range (IQR), which is used to determine the size of the whiskers. The whiskers indicate spread of data outside the box at up to 1.5 times the IQR from the edge of the box (i.e., 1.5 times the size of the box below the 25th percentile or above the 75th percentile). Data further than 1.5 times the IQR from the box are designated outliers and are plotted individually. Mutants were compared to the wild type using a Wilcoxon signed-rank test, and P values were adjusted using Bonferroni correction. ***, P < 0.001. (B) Bar graph of each of the mutants versus the number of nutrient conditions, where the mutant shows an altered biofilm phenotype, defined as a value that is 2 standard deviations higher or lower than the WT value using the test described in Materials and Methods. The height of the bar above and below the origin indicates the number of conditions under which each mutant had a significantly enhanced level of biofilm formation and a significantly reduced level of biofilm formation, respectively. The bars are color-coded to match the domain of the protein encoded by the mutated gene with the legend shown on the right side of the panel. Please note that the number of conditions with reduced biofilm formation is smaller given the relatively narrow dynamic range of decreasing biofilm formation versus the larger possible dynamic range of increasing biofilm formation.
Analysis of strains lacking the dual-domain proteins revealed a range of results. Among the 17 mutant strains, 10 demonstrated elevated levels of biofilm formation, 3 showed a decrease, and 4 showed no significant impact across all conditions (Fig. 2A). While prediction of catalytic activity from sequence alone can be unreliable, it is noteworthy that the bulk of mutants lacking dual-domain proteins showed elevated biofilm formation (Fig. 2A). Four of the five strains carrying mutations in genes predicted to encode phosphodiesterases showed increased biofilm formation across all conditions, with the strain carrying a mutation in Pfl01_2920 showing no significant differences in biofilm formation across all conditions. Given that P. fluorescens harbors 21 GGDEF-containing proteins while having only five EAL domain-containing proteins, it is perhaps not surprising that most of the dual-domain proteins participate in c-di-GMP turnover.
Of the putative effectors, the PilZ domain-containing mutants of Pfl01_0958, Pfl01_4008, and Pfl01_4150 showed significant increases across all nutrients tested, the mutant of Pfl01_4884 showed no significant difference across all nutrients tested, and the Pfl01_3860 and Pfl01_4257 mutants showed a significant decrease across all nutrients compared to the wild type. When their encoding genes were mutated, two of the effectors, the FleQ homolog Pfl01_1532 and LapD, both showed significant loss of biofilm formation across a majority of the conditions tested, demonstrating their general relevance to promoting biofilm formation.
Interestingly, 11 mutants showed no aggregate change in biofilm formation across all conditions. These included GGDEF domain-containing Pfl01_0692, Pfl01_2176, Pfl01_2295, Pfl01_4451, and Pfl01_5168; EAL domain-containing Pfl01_2920; dual-domain-containing Pfl01_1887, Pfl01_2709, Pfl01_5518, and Pfl01_5643; and PilZ domain-containing Pfl01_4884. However, while there may have been no global effect detected, there were individual nutrients that appeared to impact biofilm formation for these mutants. For example, the Pfl01_1887 mutant showed no significant difference from the wild-type strain in this overall comparison, despite being previously identified as a critical contributor to biofilm formation in this organism in minimal K10 medium, where its mutation results in elevated biofilm formation (30). Taken together, our data suggest that while most genes encoding components of the c-di-GMP network, when mutated, show an altered biofilm phenotype under conditions that included at least some small set of nutrients, other genes appear to play a much broader role in biofilm formation across many environments.
Differential impact of nutrients as a function of the presence of genes in the c-di-GMP network.
To display the relative levels of impact of the different nutrients tested here on biofilm formation by strains carrying mutations in the network, we plotted the number of conditions (among the 188 tested) with an altered biofilm phenotype (decreased or increased) as a function of each mutant (Fig. 2B). We considered the mutant different from the WT if the value for that mutant showed a change of ≥2 standard deviations (SDs) from the WT value under that specific nutrient condition. These data show that some mutants displayed an altered biofilm phenotype under a large number of the conditions tested. For example, the lapD mutant showed significantly reduced biofilm formation under ∼25 of the conditions tested, confirming its broad importance in biofilm formation. In contrast, other mutants showed changes in biofilm formation in few of the conditions tested, with the strain carrying a mutation in Pfl01_5643 showing an altered biofilm phenotype in only 1 of the 188 nutrients tested. Interestingly, the mutants with increased biofilm formation were not uniformly disrupting genes that are predicted to code for PDEs and thus would be predicted to result in increased c-di-GMP levels and enhanced biofilm formation. Indeed the classes of proteins represented on the right side of Fig. 2B, indicating increased biofilm formation, also include putative DGCs. These data speak to the differential results seen with respect to the impact of mutating different components of the network across the environments that this microbe may encounter.
c-di-GMP-related genes are transcriptionally regulated in response to nutrients.
Given that different DGCs, PDEs, and effectors appeared to play roles in biofilm formation depending on the conditions tested, we hypothesized that transcriptional regulation may be responsible, at least in part, for determining which enzymes are present to contribute to c-di-GMP signaling. While very few transcriptional studies of genes coding for DGCs/PDEs/effectors have previously been conducted, there is some precedence for transcriptional regulation. For example, transcription of the rapA gene, which codes for a dual-domain protein of P. fluorescens shown to have PDE activity, has been previously demonstrated to be upregulated when cells are grown in a low-phosphate medium (15).
Our goal was to capture roles for transcriptional regulation that may be responsible for the altered biofilm phenotypes observed in the Biolog plates. To this end, we ranked carbon sources based on their ability to promote biofilm across all tested strains and selected 8 low-biofilm-promoting nutrients (“low”), 6 medium-biofilm-promoting nutrients (“medium”), 13 high-biofilm-promoting carbon sources (“high”), and 18 very-high-biofilm-promoting carbon sources (“very high”) (Table S3). To assess transcription of all the genes encoding proteins with GGDEF, EAL, dual, and PilZ domains, cells were grown on the BMM with the selected nutrients at a concentration of 2 mM, unless otherwise indicated (see Table S3), for 6 h before RNA was extracted. Expression analysis was conducted using a Nanostring nCounter system, which directly measures transcript abundance without an amplification step, and raw data (Table S4A) were expressed as numbers of reads per 1,000 counts (Table S4B; see also Materials and Methods). This normalization strategy has been successfully employed previously in P. aeruginosa (34), and we have adopted it here for the purposes of comparing transcripts across many conditions.
To assess the impact of nutrients on expression of the genes in the c-di-GMP network, we plotted the ratios of the number of transcript reads produced by each of the genes from each of the 46 nutrients tested to the number of reads obtained in the BMM (i.e., ratio = number of reads on nutrient X/number of reads on base medium). Figure 3A shows the individual data points, with significant changes in expression indicated by the blue dots (see Materials and Methods for statistical analyses and Fig. S1A in the supplemental material for a figure with a legend showing the nutrients). Figure 3A and Fig. S1A show that for most genes and for most nutrients, the change in gene expression is <2-fold. However, there are several instances where specific nutrients were associated with gene expression that had increased or decreased 2-fold to 8-fold. We found that the gene showing the highest fold change encoded RapA, a verified PDE. The expression level of rapA was previously shown to be strongly upregulated under low-phosphate conditions (15), thus serving to validate our transcriptional analysis. These data indicate that the degree of transcriptional response to any particular nutrient, when such a response was detected, was quite modest (∼2-fold), although small transcriptional changes with respect to enzyme-encoding genes can result in more-robust changes in intracellular c-di-GMP levels. The idea that the modest regulation of several of these genes could govern cellular functions is discussed in the next section.
FIG 3.
Analysis of the expression of genes in the network. (A) Log2 expression ratio for each gene in the network across the 50 nutrients tested. The ratio was calculated by dividing the number of counts per 1,000 normalized mRNA transcripts for cells grown on BMM plus the indicated nutrient by the number of mRNA transcripts for cells grown on BMM. A positive value indicates an increase in expression in response to the nutrient, while a negative value indicates a decrease in expression. All ratios were transformed to their log2 values and then plotted. Each red dot indicates a condition under which expression was not significantly different from the expression on BMM alone, while blue dots indicate conditions under which expression was significantly different from that seen with BMM (determined as described in Materials and Methods). A version of this plot with the individual dots assigned to nutrients is shown in Fig. S1A. (B) Plot relating the number of nutrient conditions under which transcription of the indicated gene was changed and a biofilm defect in a strain carrying a mutation in that same gene was observed in that same nutrient. Data are shown for all the genes in the network. (C) Conditions under which a gene with both a significant change in transcription and the mutation of that gene had a significant impact on biofilm formation are plotted and color-coded by nutrient.
A few nutrients drive the largest changes in expression of genes in the network.
In a large network of many related c-di-GMP proteins, one may consider two general methods of gene regulation. The first is to have one or a small number of c-di-GMP-related genes dramatically up- or downregulated under a given condition, such as the response of the rapA gene under low-phosphate conditions. An alternative strategy would be to finely regulate a larger number of genes in the network at the same time to produce a desired c-di-GMP output. To test whether cells might use this second strategy, we examined which nutrients cause the greatest fold increase and decrease in each gene compared to BMM. If groups of genes were not regulated together under the same conditions, we would expect that the nutrients causing the largest changes compared to the results seen with base medium would be different for each gene. In contrast, if genes were regulated together, we would expect the same nutrient(s) to be responsible for the largest fold changes in expression of several genes across the network.
We found that six nutrients—m-tartaric acid, glycogen, d-ribose, d-galactose, l-proline, and phosphate (corresponding to the low-phosphate condition)—are responsible for the highest recorded expression of 26 genes in the network (Table S5, bolded nutrient designations). Intriguingly, this same group of nutrients is responsible for the lowest relative levels of expression of an overlapping group of 29 genes. Fold change compared to the BMM conditions among these genes varied greatly, from near the baseline value to up to 5-fold change. Furthermore, m-tartaric acid and glycogen were found to be among the highest- and lowest-biofilm-promoting nutrients that we tested (Table S3). Notably, those two nutrients were responsible for a majority of the peak high and low transcriptional values, impacting 18 and 10 genes, respectively (Table S5). Overall, 12% of the tested nutrients appeared to create the largest transcriptional changes in half of the genes in the network, suggesting that, at least for some nutrients that do impact transcription, they may do so across many genes simultaneously.
Growth on a surface minimally impacts expression of genes in the c-di-GMP network.
Previous studies of pseudomonads showed that growth on a surface increases c-di-GMP levels compared to growth as planktonic cells (35–37). Thus, we compared the levels of expression of all genes in the c-di-GMP network for a selected set of nutrients in liquid-medium-grown bacteria to the levels seen with bacteria grown on the same medium solidified with 1.5% agar. Specifically, we investigated gene expression on K10T-1, K10T-1 with low phosphate, and BMM, as well as on BMM supplemented with citrate, pyruvate, and l-methionine (Table S6A). The data were plotted as a ratio of the solid-grown cells to the liquid-grown cells on a per-nutrient basis and were analyzed by a Wilcoxon signed-rank test with multiple-comparison correction (Fig. S1B). Only three genes were found to have significantly different levels of expression on a surface compared to liquid across all conditions, correcting for multiple comparisons after a Bonferroni correction (Fig. S1B, marked with an asterisk). The gcbA gene, which has previously been described as being important in early surface attachment (38), was significantly more highly expressed when cells were grown in liquid, whereas genes Pfl01_0050 and Pfl01_1336 were significantly more highly expressed when cells were grown on a solid. The gcbA and Pfl01_1336 genes were both differentially regulated by ∼2-fold to 4-fold, while Pfl01_0050 exhibited a less than 2-fold change. Based on the modest dynamic range that we observed for most genes in the c-di-GMP network under conditions of exposure to different nutrients (Fig. 3A), it may be possible that these modest differences in expression would still be sufficient to impact the role of these genes in cells grown on a surface.
Modulation of gene expression in the network by c-di-GMP.
In P. aeruginosa, increased levels of c-di-GMP positively regulate the expression of exopolysaccharide production and downregulate expression of flagellar genes (8, 10, 39). Furthermore, previous reports have shown that c-di-GMP can regulate transcriptional levels of genes through riboswitches, although there have been no reports of such structures in P. fluorescens (7, 40). Thus, we assessed the impact of modulating c-di-GMP levels on expression of genes in the network by selecting two strains of P. fluorescens that produce low and high levels of c-di-GMP, respectively. The previously reported Δ4DGC mutant lacks the GcbA, GcbB, GcbC, and WspR DGCs, fails to make a biofilm in K10 medium, and produces 2-fold-lower levels of c-di-GMP than the wild-type strain (41). In contrast, the GcbC R366E mutant contains a point mutation in the autoinhibitory site of this DGC, producing over 10-fold-higher levels of c-di-GMP than the Δ4DGC strain (41). Three biological replicates of each strain were grown on BMM and analyzed via Nanostring (Table S6B).
Most genes showed changes of <2-fold (Table S6B), indicating that this large c-di-GMP network does not broadly rely on a core global level of intracellular c-di-GMP to regulate the transcription of other members of the network, although there is a small set of genes whose expression is significant impacted by c-di-GMP levels (Table S6B, in bold). Gene Pfl01_4451 (the homolog of the P. aeruginosa sadC gene) was upregulated 2.4-fold in the high-c-di-GMP condition. This result was significant (P < 0.01 [t test with Benjamini-Hochberg correction comparing the low- and high-c-di-GMP-producing strains]). Additionally, Pfl01_0190, Pfl01_5255, and Pfl01_5518 also showed significant and greater than 2-fold differences in expression between the low- and high-c-di-GMP-producing strains tested here, indicating that transcription of only a small portion of the network is sensitive to large swings in intracellular c-di-GMP concentrations.
Broad trends observed in the transcriptional network of P. fluorescens are also seen in P. aeruginosa.
Our analysis of the expression of c-di-GMP-related genes (CDG genes) in P. fluorescens showed that, with the exception of rapA under low-phosphate conditions, those genes did not show large variations across the conditions tested. To address whether a similar pattern of expression of CDG genes might be found in another, related organism, we analyzed the large number of microarray experiments that have been conducted in P. aeruginosa. Using the tools developed by Tan et al. (42), we assembled a compendium of all published microarrays in P. aeruginosa. This compendium includes data for 5,549 genes in 1,185 experiments, including 78 different growth media.
First, to assess whether the broad characteristics of expression of CDG genes in P. fluorescens Pf01 would also be seen in P. aeruginosa, we compared the overall levels of expression of CDG genes in the two organisms. To do this analysis, we normalized the expression of genes between experiments in the compendium by converting all expression data to values between 0 and 1 according to how they compared to the highest and lowest reported values for each experiment, with the highest value becoming 1 and the lowest value becoming 0. We compared summary statistics corresponding to the normalized data to our counts determined per 1,000 normalized Nanostring data (see Table S4B). We calculated the median expression for CDG genes in the two data sets, as well as the median for non-CDG genes, which we defined as genes coding for proteins without a GGDEF, EAL, or PilZ domain or the HD-GYP domain (HD-GYP domain-containing proteins are predicted to have c-di-GMP phosphodiesterase activity). We also included the Pho regulon in the P. aeruginosa PAO1 compendium for a comparison with a group of genes that are under the control of robust transcriptional regulation. The pattern of expression observed for P. fluorescens Pf01, where most CDG genes showed relatively low levels of expression whereas PilZ-domain-encoding genes were more highly expressed (Fig. S2A), was also observed for P. aeruginosa PAO1 (Fig. S2B), although the difference in the magnitude of expression of the PilZ-encoding genes was lower in P. aeruginosa PAO1.
Given that the methods used to assess gene expression differed between P. aeruginosa PAO1 (microarrays) and P. fluorescens (Nanostring), we sought to use a common metric to compare the variations in gene expression in the two microbes. We calculated the coefficient of variance for each data set by dividing the standard deviation by the mean corresponding to the expression of each gene across the conditions tested. The levels of variability in the expression of these genes across conditions were similar for the two organisms as judged by this metric, with both organisms exhibiting variability in the CDG genes that was lower than the median variability exhibited by non-CDG genes (Fig. S2C and D). In contrast, the variability of genes in the Pho regulon was higher than that of most non-CDG genes (Fig. S2D). Together, these data suggest that large changes in transcription across multiple growth conditions are not likely to represent a major factor controlling the contribution of genes involved in the c-di-GMP network in either P. fluorescens or P. aeruginosa.
To investigate potential transcriptional coregulation common to both organisms, we applied a simple pairwise Spearman correlation analysis of all genes in the compendium and Nanostring data sets. In P. fluorescens, we found that several genes seem to correlate strongly and significantly with one another (Fig. S2E) but that fewer genes were significantly and strongly correlated with one another than was the case with P. aeruginosa (Fig. S2F). These correlation analyses suggest that whereas the magnitude of transcriptional changes is not high, groups of CDG genes may be under the control of related transcriptional regulation in both organisms.
Evidence for nutrient-mediated, nontranscriptional control of proteins in the network.
We next explored how much of the response of the network to the nutrients tested can be explained by transcriptional regulation. That is, among the 188 nutrients tested, how often does the defect in a given mutant grown on “nutrient X” correlate with a change in the transcription of that gene under conditions of growth on “nutrient X”? To address this question, we assessed what portions of the conditions tested in both Biolog and Nanostring were associated with changes in biofilm formation or in gene expression or with both or neither of these phenotypes (Fig. 3B).
In most cases, there was no link between a change in biofilm formation in a particular mutant and a change in expression of a c-di-GMP-related gene under that condition (Fig. 3B). However, we did observe instances of altered biofilm phenotypes that were associated with differences in gene expression (Fig. 3B), suggesting that gene expression may explain the impact of these conditions. This was true for diverse genes, including the GGDEF domain-encoding Pfl01_2297 gene, the EAL domain-encoding Pfl01_0048 gene, and the dual-domain-encoding Pfl01_0460 gene. Data from the nutrients corresponding to both biofilm impact and gene expression differences are also displayed in Fig. 3C. Interestingly, m-tartaric acid impacted both the level of biofilm formation and the level of expression seen with 13 different genes. m-Tartaric acid also induced one of the highest overall levels of biofilm tested, suggesting that m-tartaric acid may be an important signal or nutrient for P. fluorescens, and exerted at least some degree of its impact on biofilm formation via transcriptional control. As noted above, in a majority of circumstances, when biofilm formation occurred, it was attributable to the nutrient added in the absence of any detectable transcriptional change (Fig. 3B). Given the large number of putative sensory domains present in many of the c-di-GMP-related proteins in P. fluorescens Pf0-1 (Fig. 1), we suggest that nutrients may be impacting the c-di-GMP network by controlling enzyme activity.
Physical interaction is common for GGDEF-containing and dual-domain proteins.
While patterns of biofilm formation and transcriptional regulation emerged from the differing nutrient conditions tested, these findings did not appear to be sufficient to explain how this larger network might be ordered to prevent cross talk among its many enzymes and effectors. Furthermore, physical interaction as a mechanism of signaling specificity has precedence in a number of bacterial c-di-GMP systems, including E. coli, P. aeruginosa, and Xanthomonas axonopodis (13, 43, 44). In P. fluorescens, physical interaction between DGC GcbC and the LapD effector has previously been shown to be required for specificity of GcbC signaling in biofilm formation (14). However, it is largely unknown how common physical interaction is among enzymes and effectors in the network, whether a given protein may have a single interaction partner or many interaction partners, or if a given type of c-di-GMP protein is more or less likely to form physical interactions than others.
To address these issues, each c-di-GMP-related gene in P. fluorescens was cloned into both “bait” and “prey” vectors of a bacterial two-hybrid (B2H) system to assess the capacity of each protein for physical interaction. Each member of every protein type—GGDEF, EAL, dual domain, and PilZ—was tested against all members of the other types. Additionally, all dual-domain proteins were also tested against all other dual-domain proteins as it is possible that such proteins act as DGCs or as PDEs or both or as effectors. Summaries of this analysis, which represented close to 2,000 bacterial two-hybrid assays, are presented in Fig. 4 and in Fig. S3 and Table S7A.
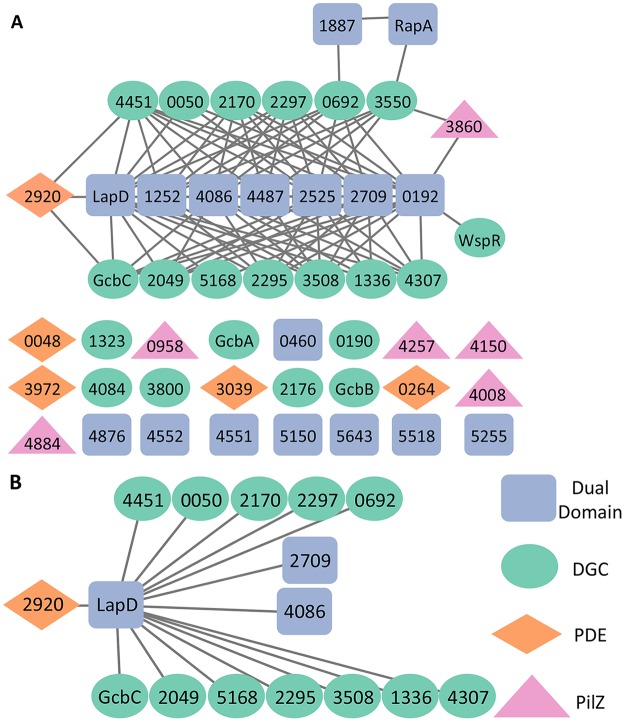
FIG 4.
Physical interaction map of c-di-GMP-related proteins in P. fluorescens. (A) BTH101 E. coli cells were grown on LB medium supplemented with X-Gal for 20 h, and colonies were assessed for blue coloration. Protein pairs of each domain type were tested for physical interaction in the bacterial two-hybrid system against each member of every other domain type, with dual-domain proteins also being tested against each other. Each gene was tested on both the pUT18 and pKNT25 vectors; a positive result was recorded if either the T18-fused protein or the T25-fused protein produced blue colonies with another member of the network. Previous studies demonstrated a one-way interaction that was confirmed by pulldown studies (14); thus, these one-way interactions were included in the data. Proteins that failed to interact are shown at the bottom of the panel. FleQ was not tested in this analysis. The interaction map was generated by Cytoscape v 3.5.1. (B) LapD serves as a node of interaction among several proteins shown here. The legend is shown at bottom right. GGDEF-containing proteins are represented by green, EAL-containing proteins are indicated by orange, dual-domain proteins are indicated by blue, and PilZ-containing proteins are indicated by pink.
Several dual-domain proteins showed a relatively high capacity for interaction with other network members (Fig. 4A). In particular, LapD, Pfl01_0192, Pfl01_1252, Pfl01_2525, Pfl01_2709, Pfl01_4086, and Pfl01_4487 interacted with 7 to 18 partners each. A subset of 15 proteins interacted with LapD (Fig. 4B) (Table S7B), and 4 additional dual-domain proteins interacted with more than 10 partners (Table S7A and Fig. S3). In contrast, RapA and Pfl01_1887 are not a part of these larger hubs, interacting with only two partners and one partner, respectively (Fig. 4A and Table S7A).
Putative DGCs showed a highly variable propensity to interact (Table S7A). A total of 14 putative DGCs appeared to participate in physical interaction, having between one and eight interaction partners. Interestingly, the four DGCs previously found to be necessary for biofilm formation under K10T-1 medium conditions had different interaction profiles. GcbA and GcbB failed to interact with other proteins in this assay, while, by contrast, GcbC interacted with several dual-domain proteins (including LapD) and with an EAL domain-containing protein. WspR has a single interaction partner—Pfl01_0192. The remaining seven putative DGCs did not engage in detectable physical interactions under our assay conditions.
Finally, little evidence was found to indicate that EAL and PilZ domain-containing proteins interacted with other members of the network (Fig. 4A). Among the five EAL domain-containing proteins, only Pfl01_2920 was found to interact with two GGDEF domain-containing proteins and LapD. Likewise, a single PilZ domain-containing protein, Pfl01_3860, interacted with GGDEF domain-containing protein Pfl01_3550 and dual-domain protein Pfl01_0192. The Pfl01_3860 PilZ domain-containing protein is part of a small node made up of a GGDEF domain and two dual-domain proteins (Fig. 4A). Together, these data indicate that a number of the proteins in the network have the potential to interact and thereby potentially form signaling complexes.
Relationship between interacting proteins and shared biofilm phenotypes.
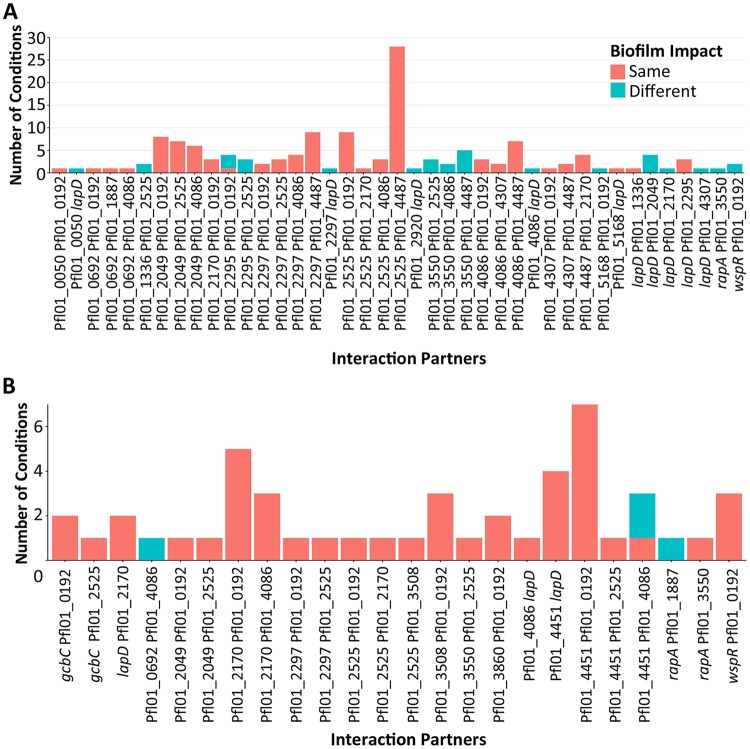
We next tried to determine how much of a role physical interaction might play in contributing to signaling specificity in the network. To perform this analysis, we assessed the conditions under which both members of an interacting pair of proteins also displayed a significantly altered biofilm phenotype when mutated. We scored these biofilm phenotypes based on whether they were in the same direction (i.e., whether both enhanced biofilm formation or both reduced biofilm formation) and calculated the number of the total biofilm phenotypes that each member of the interacting pairs shared (Fig. 5A). The results of the analysis showed that most interaction partners impacted biofilm under only one or two of the same conditions; however, some pairs did demonstrate a large overlap in the nutrient conditions, resulting in altered biofilm phenotypes for the respective mutants. For example, interaction pairs such as wspR plus Pfl01_0192 and Pfl01_0692 plus Pfl01_1887, which impact biofilm formation under only a few sets of conditions, overlapped under most of those conditions. Furthermore, there were several instances where interacting pairs, when mutated, showed opposite biofilm phenotypes under a particular set of nutrient conditions. Such a finding is likely physiologically relevant given that the biofilm formation is impacted under common growth conditions. It is also not surprising that the overlapping conditions for interaction partners were generally consistent in being in the same direction, as mutants generally only impacted biofilm formation in one direction. These data suggest that there may be numerous examples of both antagonistic and cooperative interactions in this network, often involving the same proteins and possibly modulated via physical interaction.
FIG 5.
Assessing the protein-protein interactions in the network. (A) Plot of interacting pairs of proteins in the network versus the number of nutrient conditions under which each pair shared a biofilm phenotype. Orange indicates the number of conditions under which the two interactors affected biofilm formation in the same way, while blue indicates the number of conditions under which interactors affected biofilm formation in opposite directions. The number of possible growth conditions interrogated here was 188. We refer the reader to Fig. 2A to determine the number of conditions under which a mutation in a particular c-di-GMP-related gene resulted in an altered biofilm phenotype for the particular gene. (B) The stacked plot shows the relationships between interacting pairs of proteins in the network versus a change in expression of both genes in the interacting pair in the same nutrient. Orange indicates the number of conditions under which both interactors showed a change in transcription in the same direction, while blue indicates the number of conditions under which interactors showed a change in transcription in the opposite direction. The number of possible growth conditions interrogated for the expression analysis is 46. We refer the reader to Fig. 2A to determine the number of conditions under which a mutation in a particular c-di-GMP-related gene results in an altered biofilm phenotype for the particular gene. Note the differences between panels A and B in the y-axis scales.
Intersection between B2H and transcriptional control.
We next examined possible overlaps of proteins that interacted with one another and were coordinately expressed. We scored the number of conditions under which genes of an interacting pair changed in expression under the same condition. A number of interacting pairs showed coordinated transcriptional changes under one to seven sets of conditions (Fig. 5B) (Table S7B). Interestingly, some genes that showed expression significantly different from that seen with BMM under only one condition or under a small number of conditions (see Table S4B) did so under the same nutrient conditions as genes encoding their interaction partner. For example, for the Pfl01_0192-Pfl01_4451 interacting pair, Pfl01_0192 shares all seven sets of conditions that produce differential expression from the BMM with the GGDEF domain-containing Pfl01_4451. Together, these results suggest that, in some cases, a combination of transcriptional control and physical interactions may be utilized to modulate signaling specificity.
DISCUSSION
In this study, we investigated the c-di-GMP network of P. fluorescens. We selected this model organism because it has one of the best-understood c-di-GMP circuits, from input to output, with the control of localization of the cell surface biofilm adhesion LapA as a key output (21, 22). LapA appears to contribute to or be required for biofilm formation under each condition that we have examined to date, including the base minimal medium used in this study and the same medium supplemented with the 188 nutrients investigated here (data not shown). Our analysis led to one simple, central conclusion: no one mode of regulation outlined above (nutrient input, transcriptional regulation, or protein-protein interaction) adequately describes the function of the network. Instead, these modes of regulation can function in combination to contribute to tuning of the network. We propose that this multimodal strategy of controlling c-di-GMP-mediated outputs ultimately provides fidelity and specificity to the network.
We found that a majority of c-di-GMP-related genes in P. fluorescens impacted biofilm formation when mutated and tested under a large array of conditions, indicating that context is key for understanding the function of these large signaling networks. Further, this context appears to shape the organization of the network in several ways. We found that transcriptional regulation in P. fluorescens rarely resulted in greater than 5-fold changes in transcription across all tested conditions. It was also rare for any given nutrient and any single gene for a transcriptional change to be solely responsible for an observed biofilm defect. Despite this observation, we did find that a small number of nutrients were responsible for the largest (albeit still modest) change in expression for many genes. For example, we identified several groups of genes whose expression changed under the same nutrient conditions (i.e., m-tartaric acid, d-ribose, glycogen), possibly suggesting coregulation. Given this finding, we speculate that some genes in the c-di-GMP network may be regulated as “suites” and that coordination of 2-fold-to-5-fold changes to a single nutrient across 8 to 12 genes may be an effective method of governing the network by controlling which proteins are present under that given environmental condition. The idea that sets of c-di-GMP enzymes may be transcriptionally regulated together as suites has some precedence in Vibrio cholerae, where a group of 14 enzymes have been shown to be transcriptionally regulated in response to quorum sensing (45). In the case of P. fluorescens, why would these nutrients in particular regulate suites of genes? Interestingly, m-tartaric acid is produced and metabolized by fluorescent pseudomonads (46) and plays a critical role in solubilizing inorganic phosphate (47); phosphate is a key regulator of biofilm formation by this microbe (48). The physiological relevance of d-ribose and glycogen is less clear. Finally, given that many of the members of the c-di-GMP network, including those encoded by these suites of genes, also have putative ligand binding domains, it is quite possible that more than one mechanism regulates the function and/or specificity of these network members.
Physical interaction was also found to play a role in the network and to potentially be a common means of conferring signaling specificity. In comparison to our transcriptional results, we found many discrete examples where two genes that, when mutated, shared an altered biofilm phenotype on the same nutrient source also physically interacted with one another. We interpret the frequency of interacting pairs sharing biofilm phenotypes to indicate that physical interaction occurs among network members that can exert their influence under similar environmental conditions. Such a combination of common nutrient inputs among interacting proteins represents one of the strongest findings supporting the concept of multimodal regulation. In the case of those interaction pairs which showed little or no overlap in the biofilm phenotypes of their mutants, it possible that we have not yet identified the correct conditions under which the interaction pair is relevant. Alternatively, some of these enzymes and effectors might interact but might be alternatively activated in differing environments.
The potential for physical interaction, especially among the putative DGCs and the LapD receptor, is potentially informative regarding signaling specificity. We reported previously that GcbC and LapD interact (14), and a recent report by Cooley et al. (49) proposed that LapD forms a dimer-of-dimers “basket,” with space for a DGC to nestle within the basket—it is possible, for example, that all DGCs that interact with LapD have the potential to form this signaling complex. Furthermore, given that many DGCs have associated ligand-binding domains and show relatively few examples of ligand-mediated transcriptional control, we propose that interaction of putative DGCs with the LapD receptor, coupled with ligand-mediated control of DGC activity, could be a general mechanism of signaling specificity. Finally, the propensity of members of the network to interact indicates the possibility that “local” signaling is a common aspect of the network, and any models describing how the network functions cannot ignore this feature (50). This possibility is especially relevant to the several dual-domain proteins that appeared to serve as interaction hubs—a observation that has also recently been made in E. coli, where several DGCs and PDEs serve as highly interactive “supermodules” whereas other supporting enzymes are far more selective with respect to their interaction partners (51). Interestingly, these P. fluorescens hub proteins, which show interaction with 10 or more other proteins (see Table S7A in the supplemental material), displayed no particular pattern in regard to their biofilm-forming capacity: strains carrying mutations in Pfl_0192, Pfl_2525, and Pfl_4086 all showed modest increases in biofilm formation across all nutrients tested, biofilm formation of the lapD mutant was reduced compared to the WT, and Pfl_2709 showed no significant change across the conditions tested (Fig. 1A). Thus, it is possible that such networks impact both promotion and reduction of biofilms or perhaps modulate other aspects of this microbe's biology.
We also identified a number of putative DGCs that worked “against type,” whereby their absence actually resulted in an increase in biofilm formation under a number of conditions at various frequencies. Previous work has shown that the DGC GcbA in P. aeruginosa is partly responsible for modulating a PDE linked to dispersal of biofilm (52), providing precedence for the conjecture that DGCs do not necessarily have to promote biofilm formation. However, we note here that two GGDEF domain-containing proteins (i.e., Pfl01_2049 and Pfl01_2297) that interact with LapD demonstrated the analogous phenotype under some conditions, indicating the possibility that some putative DGCs exert their impact by binding and blocking effector proteins. Additionally, strains carrying mutations in some GGDEF domain-containing proteins were found to either enhance or reduce biofilm formation depending on the nutrient tested, suggesting that the mechanism of their impact on biofilm formation associated with some of these enzymes may be different from what was reported for the P. aeruginosa GcbA enzyme. It is not inconceivable that physical interaction may play a role in this phenomenon as well, with DGCs blocking effectors until the correct nutrient/ligand activates the DGC, whereupon catalytic activity is initiated or structural rearrangements cause the enzyme to change its interaction profile.
While we have provided insight into the manner in which a large c-di-GMP network is organized and regulated, we note that there are also several hurdles to interpreting the results presented here. One alternative explanation for the high percentage of c-di-GMP-related proteins impacting biofilm formation is that they are in fact responsible for different cellular processes that have an indirect impact on biofilm formation. It is also likely that the nutrients tested here do not represent the complete collection of environmental cues that these enzymes and effectors respond to in terms of catalytic activation and transcriptional regulation. Indeed, the largest transcriptional change observed was for rapA, the PDE upregulated under a low-phosphate condition, suggesting that inputs other than organic compounds can impact the network. Further, we note that several transcripts measured in our Nanostring experiments showed very low expression under most conditions. Still, we find it compelling that a small subset of nutrients produced both the highest and lowest transcriptional changes for a small number of genes, indicating that those genes may be regulated as suites in addition to being regulated individually by highly specific input signals. Finally, we note that the bacterial two-hybrid system presents only evidence of which proteins interact with each other (or do not) in a heterologous host under one growth condition (i.e., LB). That is, it is conceivable that the ligand-bound state of a DGC or PDE not only may activate/deactivate enzyme activity but also may cause structural changes making the protein competent or incompetent for interaction. Additionally, there may be further interactions that we were not able to observe in this system, either because the protein is unstable/unable to function outside its native host or, alternatively, because the interaction was not observable under our assay conditions. Finally, we cannot rule out the possibility that some of the B2H findings were the result of false-positive or -negative results.
Taken together, our results show that this large c-di-GMP network is a dynamic system capable of responding in specific ways to a variety of inputs, with individual members able to take on different functions under differing circumstances. Importantly, we identified numerous instances where combined modes of regulation were observed (nutrient input/protein-protein interactions, transcription control/protein-protein interactions, nutrient input/transcriptional control). Such multimodal regulation would allow the integration of multiple inputs to fine-tune the output(s) of c-di-GMP-regulated processes, likely enhancing the specificity of the response by this network. Future studies will be required to examine the detailed mechanisms of ligand recognition, physical interaction among member proteins, and transcriptional regulation of genes in the network.
MATERIALS AND METHODS
Strains and media.
P. fluorescens Pf0-1 E. coli S17, and E. coli BTH101 (53, 54) were used throughout this study. P. fluorescens and E. coli BTH101 were grown at 30°C on 1.5% agar LB plates unless otherwise indicated. E. coli S17 was grown at 37°C. Media used in this study included K10T-1, as reported previously (54), and base minimal medium (BMM). BMM is composed of 50 mM Tris HCl (pH 7.4), 7.56 mM (NH4)2SO4, 0.15% glycerol, 1 mM K2HPO4, and 0.6 mM MgSO4. P. fluorescens strains harboring the pMQ72 expression plasmid were grown overnight with 10 μg/ml gentamicin. Expression was induced during experimentation with 0.2% arabinose. In-frame deletions were constructed as previously reported (55). Ligation cloning of plasmids was conducted using standard laboratory techniques. All strains and plasmids used in this study are listed in Table S1 in the supplemental material.
Biofilm assay.
P. fluorescens strains were struck out on LB plates overnight. In the case of mutants coming from a previously generated mutant library, plates were supplemented with 30 μg/ml tetracycline (30). Single colonies were picked the following day to grow overnight in liquid LB medium and were supplemented with 15 μg/ml tetracycline when appropriate (see Table S1). Cell densities were measured and normalized to an OD600 of 0.716. Cells were mixed 1.5:100 with the BMM. A 125-μl volume of the inoculated BMM was pipetted up and down in the Biolog PM1 and PM2A nutrient plates; 100 μl of this mixture was then transferred from the Biolog plates to a standard 96-well polystyrene plate (Costar) to perform the biofilm assays. Plates were covered and placed in a humidified microchamber at 30°C for 6 h, at which time the liquid in the wells was discarded and the wells were stained with 125 μl of 0.1% crystal violet (CV) at room temperature for ∼20 min and then rinsed 2 or 3 times with water. Wells were allowed to dry overnight and were destained the following day using 150 μl of a solution of water, methanol, and acetic acid (45:45:10 ratio by volume) for 20 min at room temperature. A 125-μl volume of the solubilized CV solution was transferred to a flat-bottom 96-well plate, and the OD was recorded at 550 nm.
Analysis of biofilm data.
For all analyses of experiments assaying biofilm formation using nutrients from the Biolog plates, OD550 values were adjusted in the following ways. First, a baseline staining was established for each condition by growing a strain lacking the genes gcbA, gcbB, gcbC, and wspR (referred to as the Δ4DGC mutant) and subtracting the value determined under each condition from the corresponding values determined under each condition in all other experiments. Second, the variability of WT values for each of the conditions was assessed, and conditions under which the WT exhibited a coefficient of variance of greater than 0.35 were excluded from further analysis to reduce errors due to noisy conditions. Third, the value corresponding to one batch of data (batch 11) (Table S2) was higher than those for all others, so that batch was normalized by multiplying all data corresponding to that day by the ratio between the WT median determined on that day and the median WT value across all other days.
Mutant values were compared to the mean WT value for each condition, and the difference between these values was expressed in terms of the number of standard deviations of WT data under that condition. The portion of values in a normal distribution that would be expected to be this many or more standard deviations from the mean was expressed as a P value. All P values were adjusted using a Benjamini-Hochberg adjustment.
An example of a calculation of the P value is as follows. The WT strain in carbon source X has a mean value of 0.3 and a standard deviation value of 0.1. Mutant A has a value of 0.496. The difference between WT mean value and the value for mutant A is as follows: 0.496 − 0.3 = 0.196. The number of WT standard deviations by which this value differs from the WT mean is as follows: 0.196/0.1 = 1.96. The portion of the normally distributed population that one would expect to differ from the mean by 1.96 SDs or more equals 0.05. Therefore, the unadjusted P value for mutant A under condition X is 0.05.
Transcriptional expression.
A total of 11 replicates of 5 μl each from overnight LB liquid cultures of the wild-type and mutant strains were spotted onto 1.5% agar plates containing either K10T-1 medium or the BMM alone or containing a 2 mM concentration of the desired nutrient sources. Exceptions were made for nutrients that have previously been published as having altered biofilm phenotypes, including arginine (0.4%), citric acid (0.4%), and l-arabinose (0.2%), or where exact measurements were infeasible (dextrin; soluble fraction, ∼0.2%). Cells were grown at 30°C for 6 h, scraped from the plate and resuspended in Tris-EDTA (TE) buffer, and processed using an RNAeasy extraction kit (Qiagen). A 75-ng volume of each RNA sample was added to a Nanostring nCounter kit, and the protocol provided by the manufacturer (Nanostring) was followed without modification.
Transcriptional analysis.
Raw reads from the Nanostring system were normalized as a fraction of the total number of reads measured for targeted genes, expressed as reads per 1,000 transcripts as described previously (34). A K10T-1 sample and a base medium sample were processed on each cartridge, yielding six biological replicates. The six replicates of the BMM were used to test the significance of the results of determinations of the differences between BMM and BMM plus other compounds. This analysis was conducted in the same way as the biofilm data analysis described above. Briefly, the value corresponding to each reading in the BMM plus a compound was compared to the mean value in base medium alone. The standard deviation of the value corresponding to that gene in the base medium was then used to estimate the portion of values one would expect to find to be as different from the mean as or more different from the mean than the value being tested. This percentage was taken to be the P value. All P values were adjusted using the Benjamini-Hochberg method. Of the 62 genes analyzed, 48 showed a less than 2-fold variation from the lowest count to highest count for six replicate experiments grown on K10T-1, and another 12 showed variation levels of between 2-fold and 3-fold. Two genes showed a greater than 3-fold change; those genes were Pfl01_1252 and gcbC, and both of those genes were found to have low counts across all experiments.
Bacterial two-hybrid studies and analysis.
BTH101 cells were electroporated with 50 ng of the pUT18 and pKNT25 plasmids containing the construct to be tested. Cells were recovered for 1 h in 1 ml LB at 37°C. A 50-μl volume was then spread on selective agar containing 50 μg/ml carbenicillin, 50 μg/ml kanamycin, 40 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), and 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). Plates were incubated at 30°C for 20 h, and colonies were examined for blue coloration. A leucine zipper protein served as a positive control, and the empty plasmids acted as the negative control. Each gene of interest was fused into both plasmids, and a positive result was recorded in the event that either orientation produced blue colonies at the 20-h time point.
Supplementary Material
ACKNOWLEDGMENTS
We thank Colleen Harty and T. Jarrod Smith for their contributions in developing use of the Biolog plates, James Rudd for consulting on hierarchical clustering methods in R, and Tom Hampton for advice on the statistical analysis of the data.
This work was supported by NIGMS R01GM123609-06 (G.A.O.) and grant T32-GM08704 (K.M.D.).
Footnotes
For a commentary on this article, see https://doi.org/10.1128/JB.00030-18.
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00703-17.
REFERENCES
- 1.Kulasakara H, Lee V, Brencic A, Liberati N, Urbach J, Miyata S, Lee DG, Neely AN, Hyodo M, Hayakawa Y, Ausubel FM, Lory S. 2006. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc Natl Acad Sci U S A 103:2839–2844. doi: 10.1073/pnas.0511090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tischler AD, Camilli A. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol Microbiol 53:857–869. doi: 10.1111/j.1365-2958.2004.04155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paul K, Nieto V, Carlquist WC, Blair DF, Harshey RM. 2010. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol Cell 38:128–139. doi: 10.1016/j.molcel.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abel S, Bucher T, Nicollier M, Hug I, Kaever V, Abel Zur Wiesch P, Jenal U. 2013. Bi-modal distribution of the second messenger c-di-GMP controls cell fate and asymmetry during the Caulobacter cell cycle. PLoS Genet 9:e1003744. doi: 10.1371/journal.pgen.1003744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pultz IS, Christen M, Kulasekara HD, Kennard A, Kulasekara B, Miller SI. 2012. The response threshold of Salmonella PilZ domain proteins is determined by their binding affinities for c-di-GMP. Mol Microbiol 86:1424–1440. doi: 10.1111/mmi.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christen M, Kulasekara HD, Christen B, Kulasekara BR, Hoffman LR, Miller SI. 2010. Asymmetrical distribution of the second messenger c-di-GMP upon bacterial cell division. Science 328:1295–1297. doi: 10.1126/science.1188658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. 2008. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science 321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hickman JW, Harwood CS. 2008. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol 69:376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pérez-Mendoza D, Coulthurst SJ, Sanjuán J, Salmond GP. 2011. N-Acetylglucosamine-dependent biofilm formation in Pectobacterium atrosepticum is cryptic and activated by elevated c-di-GMP levels. Microbiology 157(Pt 12):3340–3348. doi: 10.1099/mic.0.050450-0. [DOI] [PubMed] [Google Scholar]
- 10.Hickman JW, Tifrea DF, Harwood CS. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci U S A 102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krasteva PV, Fong JC, Shikuma NJ, Beyhan S, Navarro MV, Yildiz FH, Sondermann H. 2010. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science 327:866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bobrov AG, Kirillina O, Forman S, Mack D, Perry RD. 2008. Insights into Yersinia pestis biofilm development: topology and co-interaction of Hms inner membrane proteins involved in exopolysaccharide production. Environ Microbiol 10:1419–1432. doi: 10.1111/j.1462-2920.2007.01554.x. [DOI] [PubMed] [Google Scholar]
- 13.Lindenberg S, Klauck G, Pesavento C, Klauck E, Hengge R. 2013. The EAL domain protein YciR acts as a trigger enzyme in a c-di-GMP signalling cascade in E. coli biofilm control. EMBO J 32:2001–2014. doi: 10.1038/emboj.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dahlstrom KM, Giglio KM, Collins AJ, Sondermann H, O'Toole GA. 2015. Contribution of physical interactions to signaling specificity between a diguanylate cyclase and its effector. mBio 6:e01978-15. doi: 10.1128/mBio.01978-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monds RD, Newell PD, Gross RH, O'Toole GA. 2007. Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens P biofilm formation by controlling secretion of the adhesin LapA. Mol Microbiol 63:656–679. doi: 10.1111/j.1365-2958.2006.05539.x. [DOI] [PubMed] [Google Scholar]
- 16.Lee SH, Angelichio MJ, Mekalanos JJ, Camilli A. 1998. Nucleotide sequence and spatiotemporal expression of the Vibrio cholerae vieSAB genes during infection. J Bacteriol 180:2298–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim B, Beyhan S, Meir J, Yildiz FH. 2006. Cyclic-diGMP signal transduction systems in Vibrio cholerae: modulation of rugosity and biofilm formation. Mol Microbiol 60:331–348. doi: 10.1111/j.1365-2958.2006.05106.x. [DOI] [PubMed] [Google Scholar]
- 18.Yildiz FH, Liu XS, Heydorn A, Schoolnik GK. 2004. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol Microbiol 53:497–515. doi: 10.1111/j.1365-2958.2004.04154.x. [DOI] [PubMed] [Google Scholar]
- 19.Tuckerman JR, Gonzalez G, Sousa EHS, Wan XH, Saito JA, Alam M, Gilles-Gonzalez MA. 2009. An oxygen-sensing diguanylate cyclase and phosphodiesterase couple for c-di-GMP control. Biochemistry 48:9764–9774. doi: 10.1021/bi901409g. [DOI] [PubMed] [Google Scholar]
- 20.Zähringer F, Lacanna E, Jenal U, Schirmer T, Boehm A. 2013. Structure and signaling mechanism of a zinc-sensory diguanylate cyclase. Structure 21:1149–1157. doi: 10.1016/j.str.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 21.Dahlstrom KM, O'Toole GA. 2017. A symphony of cyclases: specificity in diguanylate cyclase signaling. Annu Rev Microbiol 71:179–195. doi: 10.1146/annurev-micro-090816-093325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyd CD, O'Toole GA. 2012. Second messenger regulation of biofilm formation: breakthroughs in understanding c-di-GMP effector systems. Annu Rev Cell Dev Biol 28:439–462. doi: 10.1146/annurev-cellbio-101011-155705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newell PD, Monds RD, O'Toole GA. 2009. LapD is a bis-(3′,5′)-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0-1. Proc Natl Acad Sci U S A 106:3461–3466. doi: 10.1073/pnas.0808933106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newell PD, Boyd CD, Sondermann H, O'Toole GA. 2011. A c-di-GMP effector system controls cell adhesion by inside-out signaling and surface protein cleavage. PLoS Biol 9:e1000587. doi: 10.1371/journal.pbio.1000587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu N, Pak T, Boon EM. 2010. Characterization of a diguanylate cyclase from Shewanella woodyi with cyclase and phosphodiesterase activities. Mol Biosyst 6:1561–1564. doi: 10.1039/c002246b. [DOI] [PubMed] [Google Scholar]
- 26.Navarro MV, De N, Bae N, Wang Q, Sondermann H. 2009. Structural analysis of the GGDEF-EAL domain-containing c-di-GMP receptor FimX. Structure 17:1104–1116. doi: 10.1016/j.str.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Navarro MV, Newell PD, Krasteva PV, Chatterjee D, Madden DR, O'Toole GA, Sondermann H. 2011. Structural basis for c-di-GMP-mediated inside-out signaling controlling periplasmic proteolysis. PLoS Biol 9:e1000588. doi: 10.1371/journal.pbio.1000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryjenkov DA, Simm R, Romling U, Gomelsky M. 2006. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J Biol Chem 281:30310–30314. doi: 10.1074/jbc.C600179200. [DOI] [PubMed] [Google Scholar]
- 29.Pratt JT, Tamayo R, Tischler AD, Camilli A. 2007. PilZ domain proteins bind cyclic diguanylate and regulate diverse processes in Vibrio cholerae. J Biol Chem 282:12860–12870. doi: 10.1074/jbc.M611593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newell PD, Yoshioka S, Hvorecny KL, Monds RD, O'Toole GA. 2011. Systematic analysis of diguanylate cyclases that promote biofilm formation by Pseudomonas fluorescens P. J Bacteriol 193:4685–4698. doi: 10.1128/JB.05483-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Townsley L, Yildiz FH. 2015. Temperature affects c-di-GMP signalling and biofilm formation in Vibrio cholerae. Environ Microbiol 17:4290–4305. doi: 10.1111/1462-2920.12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ha DG, Richman ME, O'Toole GA. 2014. Deletion mutant library for investigation of functional outputs of cyclic diguanylate metabolism in Pseudomonas aeruginosa PA14. Appl Environ Microbiol 80:3384–3393. doi: 10.1128/AEM.00299-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spurbeck RR, Tarrien RJ, Mobley HL. 2012. Enzymatically active and inactive phosphodiesterases and diguanylate cyclases are involved in regulation of motility or sessility in Escherichia coli CFT073. mBio 3:e00307-12. doi: 10.1128/mBio.00307-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hollomon JM, Grahl N, Willger SD, Koeppen K, Hogan DA. 2016. Global role of cyclic AMP signaling in pH-dependent responses in Candida albicans. mSphere 1:e00283-16. doi: 10.1128/mSphere.00283-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo Y, Zhao K, Baker AE, Kuchma SL, Coggan KA, Wolfgang MC, Wong GC, O'Toole GA. 2015. A hierarchical cascade of second messengers regulates Pseudomonas aeruginosa surface behaviors. mBio 6:e02456-14. doi: 10.1128/mBio.02456-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Güvener ZT, Harwood CS. 2007. Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol Microbiol 66:1459–1473. doi: 10.1111/j.1365-2958.2007.06008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuchma SL, Griffin EF, O'Toole GA. 2012. Minor pilins of the type IV pilus system participate in the negative regulation of swarming motility. J Bacteriol 194:5388–5403. doi: 10.1128/JB.00899-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petrova OE, Cherny KE, Sauer K. 2014. The Pseudomonas aeruginosa diguanylate cyclase GcbA, a homolog of P. fluorescens GcbA, promotes initial attachment to surfaces, but not biofilm formation, via regulation of motility. J Bacteriol 196:2827–2841. doi: 10.1128/JB.01628-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baraquet C, Harwood CS. 2013. Cyclic diguanosine monophosphate represses bacterial flagella synthesis by interacting with the Walker A motif of the enhancer-binding protein FleQ. Proc Natl Acad Sci U S A 110:18478–18483. doi: 10.1073/pnas.1318972110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee ER, Baker JL, Weinberg Z, Sudarsan N, Breaker RR. 2010. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science 329:845–848. doi: 10.1126/science.1190713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dahlstrom KM, Giglio KM, Sondermann H, O'Toole GA. 2016. The inhibitory site of a diguanylate cyclase is a necessary element for interaction and signaling with an effector protein. J Bacteriol 198:1595–1603. doi: 10.1128/JB.00090-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan J, Doing G, Lewis KA, Price CE, Chen KM, Cady KC, Perchuk B, Laub MT, Hogan DA, Greene CS. 2017. Unsupervised extraction of stable expression signatures from public compendia with an ensemble of neural networks. Cell Syst 5:63–71 e66. doi: 10.1016/j.cels.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baker AE, Diepold A, Kuchma SL, Scott JE, Ha DG, Orazi G, Armitage JP, O'Toole GA. 2016. PilZ domain protein flgz mediates cyclic di-GMP-dependent swarming motility control in Pseudomonas aeruginosa. J Bacteriol 198:1837–1846. doi: 10.1128/JB.00196-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andrade MO, Alegria MC, Guzzo CR, Docena C, Rosa MC, Ramos CH, Farah CS. 2006. The HD-GYP domain of RpfG mediates a direct linkage between the Rpf quorum-sensing pathway and a subset of diguanylate cyclase proteins in the phytopathogen Xanthomonas axonopodis pv citri. Mol Microbiol 62:537–551. doi: 10.1111/j.1365-2958.2006.05386.x. [DOI] [PubMed] [Google Scholar]
- 45.Waters CM, Lu W, Rabinowitz JD, Bassler BL. 2008. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J Bacteriol 190:2527–2536. doi: 10.1128/JB.01756-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Allen RH, Jakoby WB. 1969. Tartaric acid metabolism. IX. Synthesis with tartrate epoxidase. J Biol Chem 244:2078–2084. [PubMed] [Google Scholar]
- 47.Hurlbert RE, Jakoby WB. 1965. Tartaric acid metabolism. I. Subunits of L(+)-tartaric acid dehydrase. J Biol Chem 240:2772–2777. [PubMed] [Google Scholar]
- 48.Park KH, Lee CY, Son HJ. 2009. Mechanism of insoluble phosphate solubilization by Pseudomonas fluorescens RAF15 isolated from ginseng rhizosphere and its plant growth-promoting activities. Lett Appl Microbiol 49:222–228. doi: 10.1111/j.1472-765X.2009.02642.x. [DOI] [PubMed] [Google Scholar]
- 49.Cooley RB, O'Donnell JP, Sondermann H. 2016. Coincidence detection and bi-directional transmembrane signaling control a bacterial second messenger receptor. Elife 5:e21848. doi: 10.7554/eLife.21848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yan J, Deforet M, Boyle KE, Rahman R, Liang R, Okegbe C, Dietrich LEP, Qiu W, Xavier JB. 2017. Bow-tie signaling in c-di-GMP: machine learning in a simple biochemical network. PLoS Comput Biol 13:e1005677. doi: 10.1371/journal.pcbi.1005677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sarenko O, Klauck G, Wilke FM, Pfiffer V, Richter AM, Herbst S, Kaever V, Hengge R. 2017. More than enzymes that make or break cyclic di-GMP-local signaling in the interactome of GGDEF/EAL domain proteins of Escherichia coli. mBio 8:e01639-17. doi: 10.1128/mBio.01639-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petrova OE, Cherny KE, Sauer K. 2015. The diguanylate cyclase GcbA facilitates Pseudomonas aeruginosa biofilm dispersion by activating BdlA. J Bacteriol 197:174–187. doi: 10.1128/JB.02244-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simon R, Priefer U, Puhler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Biotechnology (NY) 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 54.Monds RD, Newell PD, Schwartzman JA, O'Toole GA. 2006. Conservation of the Pho regulon in Pseudomonas fluorescens P. Appl Environ Microbiol 72:1910–1924. doi: 10.1128/AEM.72.3.1910-1924.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shanks RM, Caiazza NC, Hinsa SM, Toutain CM, O'Toole GA. 2006. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl Environ Microbiol 72:5027–5036. doi: 10.1128/AEM.00682-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.