ABSTRACT
Streptococcus pyogenes (group A Streptococcus [GAS]) causes a wide range of human infections. The pathogenesis of GAS infections is dependent on the temporal expression of numerous secreted and surface-associated virulence factors that interact with host proteins. Streptococcal pyrogenic exotoxin B (SpeB) is one of the most extensively studied toxins produced by GAS, and the coordinate growth phase-dependent regulation of speB expression is linked to disease severity phenotypes. Here, we identified the endopeptidase PepO as a novel growth phase-dependent regulator of SpeB in the invasive GAS M1 serotype strain 5448. By using transcriptomics followed by quantitative reverse transcriptase PCR and Western blot analyses, we demonstrate through targeted mutagenesis that PepO influences growth phase-dependent induction of speB gene expression. Compared to wild-type and complemented mutant strains, we demonstrate that the 5448ΔpepO mutant strain is more susceptible to killing by human neutrophils and is attenuated in virulence in a murine model of invasive GAS infection. Our results expand the complex regulatory network that is operating in GAS to control SpeB production and suggest that PepO is a virulence requirement during GAS M1T1 strain 5448 infections.
IMPORTANCE Despite the continuing susceptibility of S. pyogenes to penicillin, this bacterial pathogen remains a leading infectious cause of global morbidity and mortality. A particular subclone of the M1 serotype (M1T1) has persisted globally for decades as the most frequently isolated serotype from patients with invasive and noninvasive diseases in Western countries. One of the key GAS pathogenicity factors is the potent broad-spectrum cysteine protease SpeB. Although there has been extensive research interest on the regulatory mechanisms that control speB gene expression, its genetic regulation is not fully understood. Here, we identify the endopeptidase PepO as a new regulator of speB gene expression in the globally disseminated M1T1 clone and as being essential for virulence.
KEYWORDS: endopeptidase, PepO, SpeB, Streptococcus pyogenes, pathogenesis, virulence regulation
INTRODUCTION
Streptococcus pyogenes, the beta-hemolytic group A streptococcus (GAS), is a strictly human bacterial pathogen with a global distribution that is often associated with mild infections of the respiratory tract (pharyngitis) and the skin (impetigo). Less commonly, scarlet fever may develop following pharyngeal infection, predominantly afflicting children. GAS has the capacity to penetrate deeper tissues, causing much more severe potentially life-threatening infections, such as septicemia, necrotizing fasciitis, and streptococcal toxic shock syndrome. GAS may also cause autoimmune sequelae, such as rheumatic fever and acute glomerulonephritis, upon repeated infection. GAS is a leading infectious cause of human mortality, with an estimated global burden of over half a million deaths annually (1). Hence, the worldwide resurgence of scarlet fever outbreaks and invasive GAS strains in recent years raises major public health concerns (2–8).
While many different emm types and subtypes of GAS are capable of causing serious infections, the globally disseminated M1T1 clone has become one of the most prevalent isolates and is associated with a disproportionate number of invasive disease cases (5, 9). This association appears to be linked to the propensity of the M1T1 clone to switch to an invasive disease phenotype through the acquisition of mutations in a two-component regulatory system, covRS, during the course of an infection (10). One key factor that may promote the transition from localized to invasive infections is the abrogation of extracellular cysteine protease SpeB expression as a result of covRS mutations (10–13).
SpeB is one of the most abundant secreted proteins of GAS and a major virulence factor that contributes to adhesion and colonization of the host epithelium, and it has the ability to degrade a wide array of host proteins (14, 15). SpeB has also been shown to degrade numerous secreted GAS virulence factors implicated in processes that include bacterial invasion, immune evasion, and several toxins and superantigens (13). The diverse activities of SpeB are echoed by its extraordinarily tight regulation, which involves both transcriptional and posttranscriptional mechanisms that are still not fully elucidated (for details, see reference 16). SpeB is cotranscribed with its cognate inhibitor protein Spi (17) and translated into a 43-kDa preproprotein, followed by removal of a signal peptide generating the 40-kDa zymogen form (18, 19). Under reducing conditions that can be replicated in vitro, the zymogen form undergoes autocatalytic processing to the 28-kDa mature enzymatically active form of SpeB. While SpeB is an important virulence factor of GAS, its role during the course of streptococcal infection remains incompletely understood (15).
One of the regulators that directly controls the transcription of speB is the regulator of proteinase B (RopB), also known as Rgg, which is located adjacent to the speB gene in the GAS genome (16, 20, 21). RopB belongs to the RRNPP family of transcriptional regulators and binds to the speB promoter to facilitate transcription initiation in a growth phase-dependent manner (21–24). Like other members of the RRNPP family, RopB was recently shown to sense and respond to a cognate secreted peptide pheromone signal to mediate speB gene regulation (24, 25). The open reading frame (ORF) encoding the SpeB-inducing peptide is located in the ropB-speB intergenic region and belongs to a new class of short leaderless intercellular peptide signals in bacteria (25).
Gram-positive bacteria, such as GAS, use small peptides as communication signals in response to environmental stimuli and population density, in a process called quorum sensing (QS), to coordinate gene expression (26, 27). To date, four Rgg paralogs have been identified in GAS: RopB (Rgg), Rgg2, Rgg3, and ComR (Rgg4) (28, 29). All four Rgg paralogs respond to signaling peptides and have been shown to regulate genes involved in virulence, biofilm formation, and competence (29).
A novel mechanism for regulating activity of RRNPP-type QS systems has recently been identified in GAS by Wilkening and colleagues (30). These authors discovered that proteolysis of short signaling peptides by the endopeptidase PepO, a member of the M13 family of metallopeptidases, abrogated the Rgg2/3 signaling pathway under stress conditions, suggesting a role in biofilm formation (30). This CovRS-dependent role of PepO in Rgg2/3 signaling, however, was not conserved among different GAS serotypes and thus might implicate additional roles for PepO, such as the recently reported role in evasion of complement-mediated bacteriolysis by direct binding to human complement factor C1q (31).
In the present study, we examined the role of PepO in the representative M1T1 GAS strain 5448. We conducted a microarray transcriptome comparison of wild-type 5448 and isogenic pepO mutant strains and demonstrate that PepO specifically inhibits speB gene expression during logarithmic-growth phase. This inhibitory role of PepO on speB gene expression was pronounced under peptide-rich and carbohydrate-poor growth conditions that are consistent with skin and soft tissue infections (20, 32). We further show that mutation of pepO rendered GAS strain 5448 more susceptible to human neutrophil clearance and resulted in reduced virulence in a murine model of invasive GAS disease.
RESULTS
Subcellular localization of endopeptidase PepO in GAS strain 5448.
GAS endopeptidase PepO is a 71-kDa (631-amino-acid) protein, which belongs to the ubiquitous neprilysin (M13) family of zinc-metalloenzymes that are potent regulators of peptide signaling (33). The pepO gene is highly conserved in sequenced GAS genomes, and the encoded protein shows 28% amino acid sequence identity with chain A of human neutral endopeptidase (PDB: 1DMT_A). Previous studies have detected PepO either solely in the cytoplasm (30) or both in the cytoplasm and as a secreted protein in the culture supernatant, depending on the GAS serotype (31).
To address the serotype-specific subcellular localization of PepO in GAS strain 5448, we prepared streptococcal cell lysates and collected culture supernatants of wild-type and pepO mutant strains grown in Todd-Hewitt broth supplemented with 1% yeast extract (THY) medium. The pepO mutant has been constructed by replacing the pepO coding region with the erythromycin resistance gene (erm), preserving the integrity of coding sequences and the regulatory elements of neighboring genes. The chromosomal context of the gene rules out any polar effect on neighboring ORFs (Fig. 1A). The presence of the PepO protein was detected using polyclonal rabbit anti-PepO antiserum. Immunoblot analysis revealed that PepO is already expressed at early growth stages, where it is predominantly localized in the cytoplasmic fraction (Fig. 1B). In addition, PepO was detectable in the culture supernatant after early logarithmic-growth phase (Fig. 1B). No classical sequence motifs have been found that target GAS PepO to the extracellular surroundings (30); therefore, the mechanism by which PepO is released into the supernatant is unknown.
FIG 1.
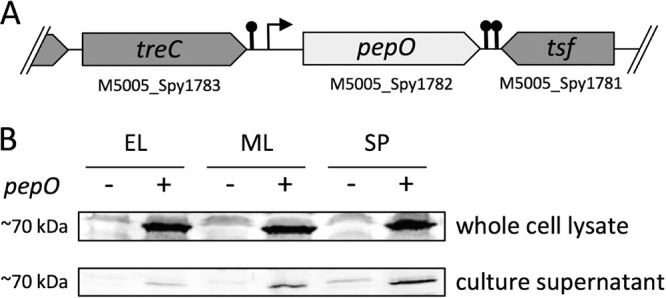
Subcellular localization of PepO. (A) Schematic of the pepO open reading frame in wild-type GAS strain 5448. Predicted promoters and Rho-independent transcriptional terminators are indicated. (B) Cytoplasmic fractions and culture supernatant of GAS strains 5448 (+) and 5448ΔpepO (−) were prepared at early logarithmic phase (EL; A600, ∼ 0.4), mid-logarithmic phase (ML; A600, ∼0.8), and stationary-growth phase (SP; A600, ∼1.2) in THY medium. All fractions were subjected to Western blot analysis with anti-PepO antiserum. Data shown are representative of at least three independent experiments.
Transcriptional comparison of GAS strains 5448 and 5448ΔpepO.
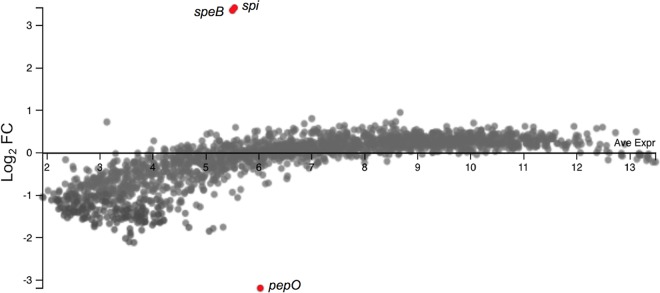
It was previously reported that PepO modulated Rgg2/3 QS signaling, suggesting a role in regulating the global transcriptional response (30). To investigate this in M1T1 GAS, we performed transcriptional profiling of wild-type 5448 and an isogenic pepO mutant strain. Each strain was grown to mid-logarithmic-growth phase (A600, ∼0.8), and the gene expression profiles of both strains were compared by microarrays (Fig. 2). Overall, 182 genes had a >2-fold change in expression. Two genes encoding the protease-inhibitor pair SpeB-Spi were identified as being the only genes that were upregulated in the pepO mutant strain (listed in Table 1), supporting the prior demonstration that these genes are cotranscribed (17). Both genes had a >5-fold change in expression, which was the threshold change required to be included in further analyses. However, we were unable to detect any difference in the expression of previously described target genes of the Rgg2/3 QS system (30). Despite an ∼10-fold increase in speB-spi mRNA abundance, none of the known regulators of speB gene expression were affected at the transcriptional level by the deletion of pepO. As expected, pepO was the most downregulated gene in the mutant strain. To validate the microarray results and confirm a regulatory role of PepO in speB expression, we performed quantitative reverse transcriptase PCR (qPCR). The qPCR results verified that speB expression was strongly upregulated in the pepO mutant strain by ∼20-fold (Fig. 3). The expression of the gene encoding RopB, the major positive regulator of SpeB, was unaffected by the loss of pepO (Fig. 3).
FIG 2.
Gene expression profiling of an M1T1 5448 pepO mutant strain. Microarray analysis (MA) plot comparison of gene expression in a pepO mutant relative to the wild-type strain based on microarray analyses. The fold change (FC) of each gene (represented as dot) is plotted against the average expression level. Genes with a >5-fold change in expression are highlighted in red. The plot was generated using the Degust Web server (http://degust.erc.monash.edu).
TABLE 1.
Genes found to be strongly differentially expressed in a 5448ΔpepO mutant versus wild-type strain, as identified by DNA microarray profiling
| Locus | Gene | Function | log2 FCa | P value | FDRb |
|---|---|---|---|---|---|
| M5005_Spy1734 | spi | Streptopain protease inhibitor | 3.42 | 1.81e−5 | 1.16e−2 |
| M5005_Spy1735 | speB | Streptococcal pyrogenic exotoxin B | 3.36 | 5.04e−6 | 4.87e−3 |
FC, fold change.
FDR, false-discovery rate.
FIG 3.
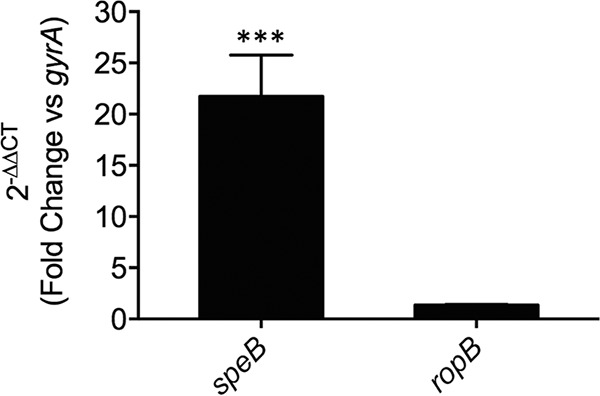
Confirmation of microarray data by quantitative reverse transcriptase PCR analysis of gene expression. Gene expression of the two genes speB and ropB in 5448ΔpepO relative to the wild-type strain grown in THY broth to mid-log phase (A600, ∼0.8). Relative gene expression was calculated using the 2−ΔΔCT method using gyrA as the reference gene. A 2−ΔΔCT value equal to 1 (ratio of a ΔΔCT value of 0) indicates no change in gene expression compared to the wild-type control. Data represent the mean 2−ΔΔCT values ± standard deviations from 3 independent biological replicates. A two-tailed unpaired Student t test was used for comparisons to the wild-type control (***, P < 0.001).
PepO reduces SpeB protein expression in a growth phase-dependent manner.
We next investigated whether increased speB mRNA levels in the pepO mutant resulted in increased SpeB production and whether this is a growth phase-related phenotype. To address this issue, we examined SpeB production in a growth medium (C medium) rich in peptides and poor in carbohydrates, which supports high-level expression of speB and was reported to reflect in vivo gene expression patterns observed during infection of mice (32). Western blot analysis of culture supernatants taken at different growth stages in C medium demonstrated that SpeB protein levels were significantly increased by 6-fold in the pepO mutant during early growth phase (one-way analysis of variance [ANOVA], P < 0.05, n = 4) and by 2-fold during mid-logarithmic-growth phase (one-way ANOVA, P < 0.01, n = 4) (Fig. 4), correlating with speB transcript abundance in THY medium. However, PepO-mediated repression of SpeB production was insignificant by the time stationary phase was reached. These results highlight a previously undescribed role for PepO to provide temporal control of speB expression under in vitro growth conditions reported to mimic the physiological conditions found during GAS infection.
FIG 4.
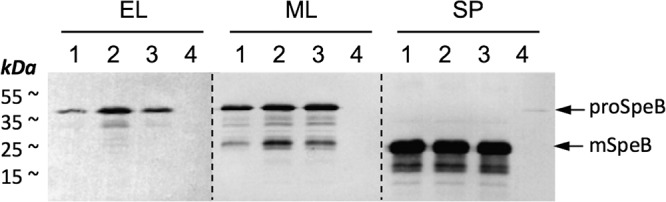
Inhibitory effect of PepO on SpeB production. Culture supernatants prepared from different growth phases in C medium were analyzed by Western blotting, as detailed in Materials and Methods, using the anti-SpeB antibody. Arrows indicate the 40-kDa zymogen form (proSpeB) and 28-kDa mature form of SpeB (mSpeB) Lane 1, 5448 wild type (WT); lane 2, 5448ΔpepO; lane 3, 5448ΔpepO::pepO; lane 4, 5448 ΔspeB. EL, early log phase (A600, ∼0.4); ML, mid-log phase (A600, ∼0.8); SP, stationary phase (A600, ∼1.2).
Contribution of the CovRS system to PepO-dependent speB gene regulation.
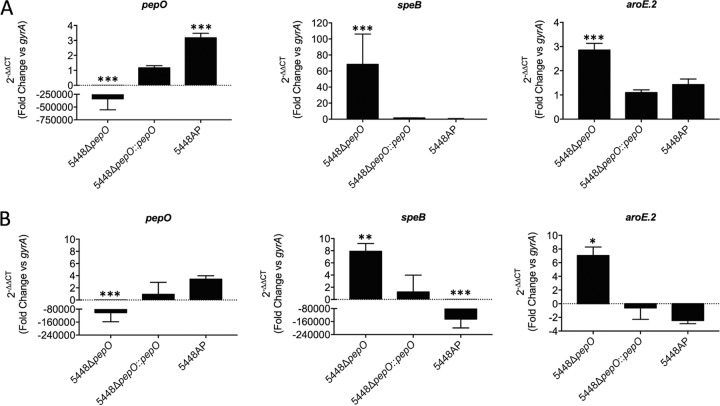
The CovRS two-component system temporally coordinates the transcriptional network of multiple GAS virulence-associated genes in response to environmental changes. The recent report that pepO is part of the CovRS regulatory circuit, being significantly repressed by CovR in GAS strain NZ131 (30), prompted us to investigate the influence of the CovRS system on pepO gene regulation in strain 5448. We therefore used an animal-passaged (AP) hypervirulent covRS mutant strain of 5448 to study relative gene expression patterns in C medium using qPCR (13). During the early logarithmic phase of growth, at the onset of speB expression, the levels of pepO mRNA were indeed increased by 3-fold in the 5448AP strain compared to the wild-type strain (Fig. 5A). However, despite significantly elevated levels of pepO mRNA, there was no detectable difference in speB expression compared to the wild-type strain at this time point. In contrast, the levels of the speB transcript were dramatically increased by ∼60-fold in the pepO mutant strain. Considering the pronounced phenotype of the pepO mutant in C medium, we also measured the expression patterns of aroE.2 (shikimate dehydrogenase gene) as a target gene of the Rgg2/3 QS circuit (28). Although the expression of the aroE.2 gene was increased by 3-fold in the 5448ΔpepO mutant, confirming the results of a previous study (30), this change in expression is relatively lower than the strong increase in speB expression. Again, there was no detectable difference in aroE.2 expression in the 5448AP strain regardless of increased pepO transcription. Gene expression patterns in the examined strains changed when cells entered mid-logarithmic-growth phase (Fig. 5B). Although still significantly increased compared to the wild-type strain and pepO gene-complemented strain, relative speB gene expression in the pepO mutant was approximately 10-fold lower than during early logarithmic growth, at levels similar to that observed for aroE.2 (Fig. 5B). Strain 5448AP showed a severe impairment in speB expression (>100,000-fold) at this growth phase, which confirms the results of previous studies (34, 35). Of note, the expression of the gene encoding RopB, the major positive regulator of SpeB, was again unaffected by the loss of pepO (data not shown). Taken together, our data show that genes of the speB operon are the primary target of PepO regulation in GAS M1T1 strain 5448, and they suggest that pepO is part of the CovRS regulon mainly during the early stages of growth, albeit that the expression of both speB and aroE.2 was unaffected in the AP mutant of GAS 5448. However, these genes could already be at baseline expression at this growth phase, which would provide an explanation as to why there was no significant differential expression.
FIG 5.
Quantitative transcript abundance analysis. Gene expression of indicated genes in 5448ΔpepO, 5448ΔpepO::pepO, and 5448AP mutants relative to the wild-type strain grown in C medium- to early log phase (A600, ∼0.4) (A) and mid-log phase (A600, ∼0.8) (B). Relative gene expression was calculated using the 2−ΔΔCT method using gyrA as the reference gene. A 2−ΔΔCT value equal to 1 (ratio of a ΔΔCT value of 0) indicates no change in gene expression compared to the wild-type control. Data represent the mean 2−ΔΔCT values ± standard deviations from 3 independent biological replicates. The two-way ANOVA test was used for all comparisons to the wild-type control for each strain (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
PepO plays a role in protecting GAS strain 5448 from human neutrophil-mediated killing.
SpeB is a potent broad-spectrum cysteine protease that has been shown to possess proteolytic activity against multiple secreted GAS virulence factors, including the Sda1 DNase and the pore-forming toxin streptolysin O (SLO) (13). It is established that both of these virulence factors render GAS more resistant to neutrophil-mediated killing. Sda1 attenuates the host antimicrobial defense by degrading DNA-based neutrophil extracellular traps (NETs), liberating entrapped GAS (11, 36, 37). SLO, on the other hand, was shown to induce neutrophil apoptosis and to rapidly impair the neutrophil oxidative burst during phagocytosis (38, 39). We hypothesized that the increased secretion of SpeB could consequently affect the susceptibility of the pepO mutant strain to neutrophil-mediated killing. To test this hypothesis, we investigated in vitro GAS survival after incubation with neutrophils purified from human blood. We observed that loss of pepO was significantly associated with reduced survival upon exposure to human neutrophils. Complementation of the 5448ΔpepO mutant with pepO restored survival to wild-type levels (Fig. 6A).
FIG 6.
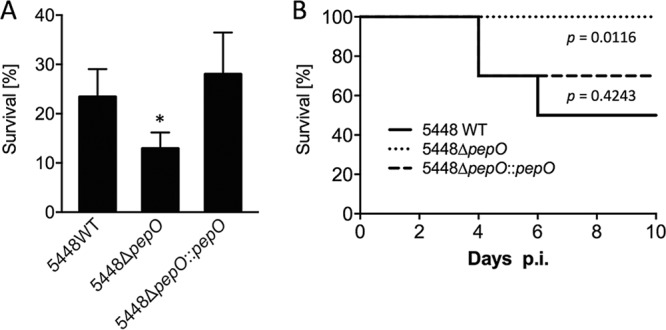
Contribution of PepO to GAS virulence. (A) Human neutrophil killing assay. Bacteria were incubated with neutrophils isolated from blood of healthy donors at an MOI of 10. After 30 min of incubation, bacteria were plated onto agar plates for enumeration. The survival rate is calculated as the number of CFU at a given time point divided by the initial CFU at the initiation of the experiment. Error bars represent the standard deviations of the results from three independent experiments. Statistical analyses were performed using the unpaired Student's t test; *, P < 0.05. (B) Survival curves following subcutaneous infection of humanized plasminogen transgenic mice with the indicated strains of GAS 5448. Mice (10 per group) were challenged with an injection of GAS at a dose of 1 × 108 CFU. Statistical analyses of the survival curves were performed using the log-rank (Mantel-Cox) test.
Loss of PepO attenuates virulence of GAS strain 5448 in a murine model of invasive disease.
To ascertain the contribution of PepO to GAS virulence in vivo, we examined the phenotype of the 5448ΔpepO mutant in a transgenic humanized plasminogen mouse model of invasive infection (40). Following subcutaneous infection, mouse survival was monitored daily over a period of 10 days. Fifty percent of the mice succumbed to infection between days 4 and 6 postinfection in the group infected with the 5448 wild-type strain. In contrast, all mice infected with the 5448ΔpepO mutant strain survived challenge. Complementation of the mutant strain with pepO restored virulence (Fig. 6B). These data demonstrate that the presence of an intact pepO gene is required for full virulence of this highly virulent GAS strain 5448 in a murine model of invasive disease.
DISCUSSION
The neprilysin (M13) family of peptidases is composed of evolutionary conserved zinc metalloproteases which play diverse roles in physiological processes (33). Members of this family are described in a wide range of organisms, including mammals and bacteria, and are recognized as important regulators involved in the processing and degradation of neuropeptides and peptide hormones (41, 42). In recent years, the endopeptidase PepO, a homolog of neprilysin/neutral endopeptidase, has gained increasing attention in the genus Streptococcus for its influence on multiple virulence-related phenotypes, having evolved remarkable functional regulatory diversity. Interestingly, homologues of the PepO endopeptidase occur only in a limited number of bacteria, most commonly in Gram-positive pathogenic bacteria (43).
PepO is a highly conserved enzyme that shares about 90% amino acid sequence similarity among streptococci (44). It has been shown to display endopeptidase activity involved in the degradation of small peptides, including the human opioid neuropeptide Met-enkephalin, a substrate for human neprilysin (30, 43, 45, 46). Besides its role in peptide signaling, studies in Streptococcus pneumoniae revealed multifaceted functions of PepO in host-pathogen interactions. Agarwal and colleagues reported growth phase-dependent secretion of PepO to the culture supernatants, with the capability to bind to both human plasminogen and fibronectin in a dose-dependent manner (47). A later report discovered a direct interaction of PepO with the complement C1q protein that was required for adhesion to host cells in vitro (48). Further, S. pneumoniae PepO was shown to be involved in the activation of the host immune response partially through Toll-like receptor 2 (TLR2)- and TLR4-mediated signaling pathways, both in vitro and in vivo (49–51). The importance of this endopeptidase in pathogenesis has also been reported in the important porcine and human pathogen Streptococcus suis, where secreted and immunogenic PepO has been suggested as a potential vaccine candidate (52). In addition, the mutation of pepO in Streptococcus mutans was associated with a strong induction of bacteriocin mutacin I expression, although the mechanism of action of PepO in this process remains to be determined (44). More recently, evidence has been obtained to show that PepO is a potent regulator of the Rgg2/3 quorum sensing circuit facilitating biofilm formation in GAS, albeit in a strain-dependent fashion (30). This study thereby provided the first direct experimental evidence demonstrating that bacterial cytoplasmic PepO functions as an enzymatic silencer of an RRNPP-type QS pathway. While the cytoplasmic localization of PepO appears to be conserved in GAS, clinical isolates showed varied abilities to release PepO into the extracellular milieu (31). Secreted PepO was reported to directly bind human complement factor C1q, thereby helping GAS to escape complement-mediated bacteriolysis, similar to that observed in S. pneumoniae (31, 48). Together, these findings suggest distinct localization-dependent functions of PepO in streptococci, a phenomenon referred to as moonlighting. Moonlighting proteins are highly conserved enzymes that can carry out diverse functions involved in bacterial virulence (53). Hypothetically, it seems possible that secreted PepO is directly involved in host-pathogen interactions, while intracellular PepO could actively affect peptide pheromone signaling and regulate virulence gene expression in Gram-positive bacteria.
With this work, we extend recent knowledge of PepO as an important virulence factor in GAS. By using DNA microarrays for whole-genome transcriptome profiling, we identified the extracellular SpeB cysteine protease as a primary regulatory target of PepO. Our findings bring to light a new player in the complex regulatory control of speB expression (16). SpeB is a major virulence factor of GAS, yet its role in streptococcal infection remains controversial (15). While SpeB is essential for the establishment for localized skin infection, speB expression is inversely correlated with the severity of infection, i.e., a significant proportion of GAS isolates from severe invasive infections exhibit a SpeB-negative phenotype (12, 54, 55).
The pepO mutant showed a reduction in virulence compared to the wild type. The increased susceptibility of the mutant to human neutrophil killing might be a consequence of reduced levels of Sda1 as a result of changes in extracellular levels of SpeB, which is known to degrade Sda1 (13). However, a direct interaction of PepO with the human complement system offers an alternative explanation for the reduced virulence phenotype of the mutant, as PepO has recently been shown to aid S. mutans evasion of complement-mediated opsonization (56). The deletion of pepO enhanced levels of C3b deposition and opsonophagocytosis of S. mutans by human neutrophils. Since PepO has already been shown to degrade human C1q in GAS (31), it therefore seems possible that pepO mutation renders GAS more prone to complement-dependent opsonophagocytosis by neutrophils. However, it remains to be determined whether the reduced virulence of the pepO mutant in the mouse model of invasive GAS disease is due to enhanced bacterial clearance by neutrophils or if dysregulation of speB caused the low virulence phenotype in mice.
The activation of speB transcription is dependent on the RRNPP family regulator RopB, located immediately adjacent to speB (21). While there is a strain-specific variation in the RopB regulon, the genes of the speB operon are likely the only core regulon of RopB (57–59). Like other members of the RRNPP family of transcriptional regulators, RopB responds to short signaling peptides to control gene expression. RopB has recently been shown to respond to a cell density-specific activation peptide signal to induce speB expression (24, 25). The conserved peptide is encoded within the ropB-speB intergenic region and is a requirement for the robust induction of speB expression (25). Furthermore, RopB was shown to bind to an N-terminal signal peptide of the virulence factor-related (Vfr) protein, inhibiting RopB-mediated speB transcription in response to changes in bacterial cell density (60), which adds a more fine-tuned level of regulation to the RopB signaling pathway in GAS.
The speB-specific and growth phase-dependent regulatory role of PepO led us to hypothesize that PepO may be involved in the degradation of a RopB-activating peptide signal. To test this, we performed culture supernatant-swapping experiments, as previously described (24), but our attempts to demonstrate an accumulation of a potential activating peptide in the pepO mutant were unsuccessful (data not shown). Alternatively, we hypothesized that PepO could be involved in the processing of the N-terminal peptide signal of Vfr, even though substrates for M13 peptidases are typically smaller peptides. However, the addition of different concentrations of the synthetic Vfr peptide (10 nM to 1 μM; LAVLFGVR, purity of >95%; Neo Scientific) to culture medium had no effect on increased speB transcript levels in the pepO mutant strain (data not shown). Despite these observations, degradation of a RopB-activating proteinaceous factor still seems a plausible explanation for temporal PepO-mediated regulation of speB expression.
We suggest that this regulatory role of PepO in virulence gene expression could occur in other bacteria. Of note, PepO has been shown to control mutacin I transcription in S. mutans through an as-yet-unknown mechanism (44). The authors report that mutations in pepO increased mutA gene expression by more than 100-fold. Interestingly, mutA is under the control of MutR, an Rgg family transcription regulator indispensable for mutacin I production, whose activity is further linked to biofilm formation in S. mutans (61, 62). These results therefore suggest a possible conserved role of PepO in Rgg QS pathways in Gram-positive pathogenic bacteria. Altogether, our results suggest a role for PepO in controlling growth phase-dependent expression of the GAS virulence factor SpeB.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The S. pyogenes M1T1 clinical isolate 5448 (5) and isogenic derivatives were grown statically at 37°C in liquid Todd-Hewitt broth supplemented with 1% yeast extract (THY) medium or, where indicated, in C medium adjusted to pH 7.5 with NaOH (20). Escherichia coli strains MC1061 and BL21(DE3) (Stratagene) were routinely used for cloning experiments and grown in Luria-Bertani (LB) medium. For plasmid selection and maintenance, antibiotics were added at the following final concentrations: ampicillin at 100 μg/ml for E. coli, and spectinomycin and erythromycin at 100 μg/ml for both GAS and E. coli. All bacterial strains and plasmids used are listed in Table 2.
TABLE 2.
Bacterial strains and plasmids used in the study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| MC1061 | Laboratory cloning strain | 69 |
| BL21(DE3) | Laboratory expression strain | Stratagene |
| Streptococcus pyogenes | ||
| 5448 | Invasive M1T1 strain | 13 |
| 5448ΔpepO | 5448 ΔpepO::spec mutant | This study |
| 5448ΔpepO::pepO | 5448 ΔpepO::pepO-complemented strain | This study |
| 5448AP | AP hyperinvasive covRS mutant strain of 5448 | 13 |
| Plasmids | Description | Reference |
| pHY304 | Temperature-sensitive shuttle plasmid::Emr | 64 |
| pHY304-pepOKO | pHY304 + pepO knockout construct inserted at the EcoRI and XhoI site | This study |
| pHY304-pepO | pHY304 + pepO complement construct inserted at the EcoRI and XhoI | This study |
| pUCSpec | pUC18-derived plasmid::Spr | 63 |
| pET151 | Directional TOPO expression plasmid::Apr | Invitrogen |
| pET151-pepO | pET151 + pepO expression construct | This study |
Emr, erythromycin resistance; Spr, spectinomycin resistance; Apr, ampicillin resistance.
DNA manipulation and genetic techniques.
The GAS pepO mutant strain 5448ΔpepO was constructed as follows. The 1-kb region upstream of pepO was amplified using primers PepO-5U-F (containing EcoRI restriction site) and PepO-5U-R (Table 3). The 1-kb downstream region was amplified using primers PepO-3D-F and PepO-3D-R (containing XhoI restriction site) (Table 3). The spectinomycin resistance cassette was amplified from pUCSpec (63) using Spec-F and Spec-R (Table 3). The three resulting PCR fragments (5′- and 3′-flanking regions with spectinomycin cassette) were joined together with primers PepO-5U-F and PepO-3D-R. The subsequent fragment was cloned via restriction ligation into the pHY304 shuttle vector (64) and transformed into E. coli MC1061. Electrotransformation and allelic replacement mutagenesis were undertaken using standard protocols (37, 65). For complementation, the entire wild-type region of pepO, including the 1-kb upstream and downstream sequences, was amplified using primers PepO-5U-F and PepO-3D-R. The resulting fragment was cloned via restriction ligation into pHY304 and transformed into E. coli MC1061. Electrotransformation and allelic replacement mutagenesis were undertaken as described above. Successful complementation was assessed by a loss of spectinomycin resistance. All strains were confirmed by DNA sequencing (Australian Equine Genome Research Centre, University of Queensland, Brisbane, Australia).
TABLE 3.
List of primers used in the study
| Primer by function | Sequence (5′–3′) |
|---|---|
| Deletion mutation and reverse complementation constructs | |
| PepO-5U-F | TAGAGAATTCCTACCAACGCTTGATTGCTTTGCG |
| PepO-5U-R | CCATAAAACTAAAGTCGTATCTCCTCGTATCTCCTTATATCATTTTAAATTTGTAGAATCTTTATC |
| PepO-3D-F | TTCATTCTAATTGGTAATCAGTTAGGTTAGGTAGTTGATAAACCTTCAAATGCCTGTGAT |
| PepO-3D-R | ATAGCTCGAGAAGCTCAATTATCAGACGATGTGATTACTG |
| Spec-F | CCTAACTGATTACCAATTAGAATGAATATTTCCCAAATATTAAATAATAAAAC |
| Spec-R | GGAGATACGACTTTAGTTTTATGGAAATGAAAGATCATATCATATATAATCTAG |
| Protein expression studies | |
| pET151PepO-F | CACCATGACAACTTATCAAGATGAT |
| pET151PepO-R | TTACCAAATAATCACACGGTCTTTCGG |
| qPCR studies | |
| gyrA-F | CGACTTGTCTGAACGCCAAA |
| gyrA-R | GTCAGCAATCAAGGCCAACA |
| pepO-F | TGCCTTTAAAGAGCGCACAG |
| pepO-R | CTACCCCACCTAGATCAGCG |
| speB-F | TGCTGACGGACGTAACTTCT |
| speB-R | CCACCAGTACCAAGAGCTGA |
| aroE.2-F | ACAGTTGTATACAGGGAGCCA |
| aroE.2-R | ATTTGTTGATCAGCCAATTCCTT |
| ropB-F | TGAACGGTGTTGTGTGTCTT |
| ropB-R | TCTGCACACACTTTTGCTCA |
Production of polyclonal antiserum to recombinant PepO.
The full-length pepO gene was amplified by PCR from the chromosomal DNA of GAS M1T1 isolate 5448 with primers pET151PepO-F and pET151PepO-R (Table 3). The amplified DNA was cloned in vector pET151 for protein expression. GAS PepO with an N-terminal 6×His tag was expressed from the pET151 vector in E. coli BL21(DE3) (Stratagene) and purified using a nickel-nitrilotriacetic acid column (GE Healthcare), according to the manufacturer's instructions. The 6×His tag was removed after on-column cleavage by recombinant 6×His-tagged tobacco etch virus (TEV) protease, followed by passage through a second nickel-nitrilotriacetic acid column (GE Healthcare). Antibodies were raised against purified recombinant PepO protein in a rabbit at the South Australian Health and Medical Research Institute (SAHMRI).
Immunoblotting.
Cultures of GAS strains were either grown in THY broth containing the cysteine protease inhibitor E64 (28 μM) or, where indicated, in C medium. GAS supernatants were precipitated with 15% trichloroacetic acid (TCA). Cellular extracts and protein pellets from TCA precipitates were resuspended in loading buffer normalized to the optical density. Samples were boiled for 10 min, subjected to SDS-PAGE, and then transferred to nitrocellulose membrane for detection by Western immunoblotting. Western blot analyses were performed using rabbit anti-SpeB (PBI222; Toxin Technology) and rabbit anti-PepO (SAHMRI), with horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (GenWay Biotech) as the second antibody for colorimetric detection with diaminobenzidine (DAB; Sigma).
Transcriptome analysis.
M1T1 GAS 5448 strains were grown in THY to mid-logarithmic-growth phase (A600, ∼0.8). Bacterial cells were collected by centrifugation, and cell pellets were frozen immediately on dry ice and stored at −80°C. Total RNA was extracted using an RNeasy minikit (Qiagen), according to the manufacturer's protocol. Labeled cDNA was generated from RNA samples by using the FairPlay III microarray labeling kit (Agilent Technologies, Stratagene). RNA from the 5448 wild type was labeled with Cy5 monoreactive dye (catalog no. Q15108; GE Healthcare), whereas 5448ΔpepO mutant RNA was labeled with Cy3 monoreactive dye (catalog no. Q13108; GE Healthcare), according to the manufacturer's instructions. Labeled cDNA was purified using the MinElute kit (Qiagen). The amount of incorporated label was determined by measuring the absorbance at 550 nm (Cy3) or 650 nm (Cy5).
Glass slide DNA microarrays used in this study contained 1,932 70-mer oligonucleotide probes of nonrepetitive open reading frames present in the sequenced genome of M1 strain SF370, as described previously by Dalton et al. (66). The slides were rehydrated, cross-linked, and blocked in a prehybridization solution (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% SDS, 0.1 mg/ml bovine serum albumin) according to the manufacturer's instructions (Corning).
For each set of microarray experiments, Cy5-labeled cDNA and Cy3-labeled cDNA (both 200 pmol) were pooled, concentrated, and resuspended in hybridization buffer solution (40% formamide, 5× SSC, 0.1% SDS, 0.1% dithiothreitol, 0.6 μg/μl salmon sperm DNA). The Cy3-Cy5 probe mixture was heated at 95°C for 10 min before being applied to the array. The probe was allowed to hybridize to the array at 42°C for at least 16 h. Following hybridization, the arrays were washed twice with 2× SSC–0.1% SDS (preheated to 55°C), twice with 0.1× SSC–0.1% SDS, and three times in 0.1× SSC.
The washed array slides were scanned using a GenePix 4000B scanner (Axon Instruments) and analyzed using the GenePix Pro 4.0 software (Axon Instruments). Data from each microarray had undergone strict threshold and normalization, as described previously (66). Statistical analysis was performed using the Degust Web server (http://degust.erc.monash.edu).
Quantitative gene expression studies.
GAS strains were harvested at the desired growth phase grown either in THY broth or C medium, where indicated. Bacterial cells were first mechanically disrupted using FastPrep lysing matrix B tubes (MP Biomedicals). Total RNA was extracted using the RNeasy minikit (Qiagen), according to the manufacturer's protocol. RNA samples were treated with Turbo DNase (Ambion) to eliminate contaminating genomic DNA. cDNA was subsequently synthesized using the SuperScript Vilo cDNA synthesis kit (Invitrogen). Quantitative reverse transcriptase PCR was performed with the QuantStudio 6 Flex system (Applied Biosystems) using the SYBR green PCR master mix (Applied Biosystems), according to the manufacturer's instructions. Relative gene expression was calculated using the threshold cycle (2−ΔΔCT) method with gyrA as the reference housekeeping gene. Gene-specific primer sequences are listed in Table 3. All reactions were performed in triplicate from three independently isolated RNA samples.
Neutrophil killing assay.
GAS survival was determined following exposure to human neutrophils ex vivo, as previously described (11). Neutrophils were isolated from the blood of healthy donors, and experiments were performed in triplicate using early logarithmic-phase (A600, ∼0.4) GAS at a multiplicity of infection (MOI) of 10:1 (GAS-to-neutrophil ratio).
Murine infection model.
Transgenic humanized plasminogen mice heterozygous for the human plasminogen transgene (AlbPLG1+/−) were infected with a dose of approximately 1 × 108 CFU of either the 5448, 5448ΔpepO, or 5448ΔpepO::pepO strain. Sex- and age-matched mice (n = 10 per group) were subcutaneously infected with freshly prepared GAS strains in 100 μl phosphate-buffered saline (PBS), and virulence was assessed as previously described (11).
Ethics approval.
All animal experiments were conducted according to the Australian code for the care and use of animals for scientific purposes (National Health and Medical Research Council, Australia [67]) and were approved by the University of Queensland animal ethics committee. Human blood donation for use in neutrophil killing assays was conducted in accordance with the National statement on ethical conduct in human research (68), in compliance with the regulations governing experimentation on humans, and was approved by the University of Queensland medical research ethics committee.
Statistical analyses.
Statistical analysis of bacterial killing by human neutrophils was performed using an unpaired two-tailed Student t test. Differences in relative gene expression were analyzed using a two-tailed unpaired Student t test and two-way analysis of variance (ANOVA), followed post hoc with Dunnett's t test. Murine survival curves were analyzed using the Mantel-Cox log rank test. Statistical analyses were performed using the GraphPad Prism 7 software package.
Accession number(s).
Microarray data have been deposited in the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo) (accession no. GSE108748).
ACKNOWLEDGMENT
This work was supported by the National Health and Medical Research Council (Australia).
REFERENCES
- 1.Carapetis JR, Steer AC, Mulholland EK, Weber M. 2005. The global burden of group A streptococcal diseases. Lancet Infect Dis 5:685–694. doi: 10.1016/S1473-3099(05)70267-X. [DOI] [PubMed] [Google Scholar]
- 2.Tse H, Bao JYJ, Davies MR, Maamary P, Tsoi H-W, Tong AHY, Ho TCC, Lin C-H, Gillen CM, Barnett TC, Chen JHK, Lee M, Yam W-C, Wong C-K, Ong C-lY, Chan Y-W, Wu C-W, Ng T, Lim WWL, Tsang THF, Tse CWS, Dougan G, Walker MJ, Lok S, Yuen K-Y. 2012. Molecular characterization of the 2011 Hong Kong scarlet fever outbreak. J Infect Dis 206:341–351. doi: 10.1093/infdis/jis362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ben Zakour NL, Davies MR, You Y, Chen JHK, Forde BM, Stanton-Cook M, Yang R, Cui Y, Barnett TC, Venturini C, Ong C-lY, Tse H, Dougan G, Zhang J, Yuen K-Y, Beatson SA, Walker MJ. 2015. Transfer of scarlet fever-associated elements into the group A Streptococcus M1T1 clone. Sci Rep 5:15877. doi: 10.1038/srep15877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davies MR, Holden MT, Coupland P, Chen JHK, Venturini C, Barnett TC, Zakour NLB, Tse H, Dougan G, Yuen K-Y, Walker MJ. 2015. Emergence of scarlet fever Streptococcus pyogenes emm12 clones in Hong Kong is associated with toxin acquisition and multidrug resistance. Nat Genet 47:84–87. doi: 10.1038/ng.3147. [DOI] [PubMed] [Google Scholar]
- 5.Aziz RK, Kotb M. 2008. Rise and persistence of global M1T1 clone of Streptococcus pyogenes. Emerg Infect Dis 14:1511–1517. doi: 10.3201/eid1410.071660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turner CE, Abbott J, Lamagni T, Holden MT, David S, Jones MD, Game L, Efstratiou A, Sriskandan S. 2015. Emergence of a new highly successful acapsular group A Streptococcus clade of genotype emm89 in the United Kingdom. mBio 6:e00622-. doi: 10.1128/mBio.00622-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu L, Olsen RJ, Nasser W, Beres SB, Vuopio J, Kristinsson KG, Gottfredsson M, Porter AR, DeLeo FR, Musser JM. 2015. A molecular trigger for intercontinental epidemics of group A Streptococcus. J Clin Invest 125:3545–3559. doi: 10.1172/JCI82478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walker MJ, Barnett TC, McArthur JD, Cole JN, Gillen CM, Henningham A, Sriprakash KS, Sanderson-Smith ML, Nizet V. 2014. Disease manifestations and pathogenic mechanisms of group A Streptococcus. Clin Microbiol Rev 27:264–301. doi: 10.1128/CMR.00101-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cleary PP, Kaplan EL, Handley JP, Wlazlo A, Kim MH, Hauser AR, Schlievert PM. 1992. Clonal basis for resurgence of serious Streptococcus pyogenes disease in the 1980s. Lancet 339:518–521. doi: 10.1016/0140-6736(92)90339-5. [DOI] [PubMed] [Google Scholar]
- 10.Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM. 2006. Genome-wide analysis of group A streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog 2:e5. doi: 10.1371/journal.ppat.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walker MJ, Hollands A, Sanderson-Smith ML, Cole JN, Kirk JK, Henningham A, McArthur JD, Dinkla K, Aziz RK, Kansal RG, Simpson AJ, Buchanan JT, Chhatwal GS, Kotb M, Nizet V. 2007. DNase Sda1 provides selection pressure for a switch to invasive group A streptococcal infection. Nat Med 13:981–985. doi: 10.1038/nm1612. [DOI] [PubMed] [Google Scholar]
- 12.Cole JN, Barnett TC, Nizet V, Walker MJ. 2011. Molecular insight into invasive group A streptococcal disease. Nat Rev Microbiol 9:724–736. doi: 10.1038/nrmicro2648. [DOI] [PubMed] [Google Scholar]
- 13.Aziz RK, Pabst MJ, Jeng A, Kansal R, Low DE, Nizet V, Kotb M. 2004. Invasive M1T1 group A Streptococcus undergoes a phase-shift in vivo to prevent proteolytic degradation of multiple virulence factors by SpeB. Mol Microbiol 51:123–134. doi: 10.1046/j.1365-2958.2003.03797.x. [DOI] [PubMed] [Google Scholar]
- 14.Brouwer S, Barnett TC, Rivera-Hernandez T, Rohde M, Walker MJ. 2016. Streptococcus pyogenes adhesion and colonization. FEBS Lett 590:3739–3757. doi: 10.1002/1873-3468.12254. [DOI] [PubMed] [Google Scholar]
- 15.Nelson DC, Garbe J, Collin M. 2011. Cysteine proteinase SpeB from Streptococcus pyogenes—a potent modifier of immunologically important host and bacterial proteins. Biol Chem 392:1077–1088. doi: 10.1515/BC.2011.208. [DOI] [PubMed] [Google Scholar]
- 16.Carroll RK, Musser JM. 2011. From transcription to activation: how group A Streptococcus, the flesh-eating pathogen, regulates SpeB cysteine protease production. Mol Microbiol 81:588–601. doi: 10.1111/j.1365-2958.2011.07709.x. [DOI] [PubMed] [Google Scholar]
- 17.Kagawa TF, O'Toole PW, Cooney JC. 2005. SpeB-Spi: a novel protease-inhibitor pair from Streptococcus pyogenes. Mol Microbiol 57:650–666. doi: 10.1111/j.1365-2958.2005.04708.x. [DOI] [PubMed] [Google Scholar]
- 18.Doran JD, Nomizu M, Takebe S, Menard R, Griffith D, Ziomek E. 1999. Autocatalytic processing of the streptococcal cysteine protease zymogen: processing mechanism and characterization of the autoproteolytic cleavage sites. Eur J Biochem 263:145–151. doi: 10.1046/j.1432-1327.1999.00473.x. [DOI] [PubMed] [Google Scholar]
- 19.Rosch JW, Caparon MG. 2005. The ExPortal: an organelle dedicated to the biogenesis of secreted proteins in Streptococcus pyogenes. Mol Microbiol 58:959–968. doi: 10.1111/j.1365-2958.2005.04887.x. [DOI] [PubMed] [Google Scholar]
- 20.Lyon WR, Gibson CM, Caparon MG. 1998. A role for trigger factor and an rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J 17:6263–6275. doi: 10.1093/emboj/17.21.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neely MN, Lyon WR, Runft DL, Caparon M. 2003. Role of RopB in growth phase expression of the SpeB cysteine protease of Streptococcus pyogenes. J Bacteriol 185:5166–5174. doi: 10.1128/JB.185.17.5166-5174.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chaussee MS, Ajdic D, Ferretti JJ. 1999. The rgg gene of Streptococcus pyogenes NZ131 positively influences extracellular SPE B production. Infect Immun 67:1715–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anbalagan S, Dmitriev A, McShan WM, Dunman PM, Chaussee MS. 2012. Growth phase-dependent modulation of Rgg binding specificity in Streptococcus pyogenes. J Bacteriol 194:3961–3971. doi: 10.1128/JB.06709-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makthal N, Gavagan M, Do H, Olsen RJ, Musser JM, Kumaraswami M. 2016. Structural and functional analysis of RopB: a major virulence regulator in Streptococcus pyogenes. Mol Microbiol 99:1119–1133. doi: 10.1111/mmi.13294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Do H, Makthal N, VanderWal AR, Rettel M, Savitski MM, Peschek N, Papenfort K, Olsen RJ, Musser JM, Kumaraswami M. 2017. Leaderless secreted peptide signaling molecule alters global gene expression and increases virulence of a human bacterial pathogen. Proc Natl Acad Sci U S A 40:E8498–E8507. doi: 10.1073/pnas.1705972114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cook LC, Federle MJ. 2014. Peptide pheromone signaling in Streptococcus and Enterococcus. FEMS Microbiol Rev 38:473–492. doi: 10.1111/1574-6976.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perez-Pascual D, Monnet V, Gardan R. 2016. Bacterial cell-cell communication in the host via RRNPP peptide-binding regulators. Front Microbiol 7:706. doi: 10.3389/fmicb.2016.00706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang JC, LaSarre B, Jimenez JC, Aggarwal C, Federle MJ. 2011. Two group A streptococcal peptide pheromones act through opposing Rgg regulators to control biofilm development. PLoS Pathog 7:e1002190. doi: 10.1371/journal.ppat.1002190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jimenez JC, Federle MJ. 2014. Quorum sensing in group A Streptococcus. Front Cell Infect Microbiol 4:127–127. doi: 10.3389/fcimb.2014.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilkening RV, Chang JC, Federle MJ. 2016. PepO, a CovRS-controlled endopeptidase, disrupts Streptococcus pyogenes quorum sensing. Mol Microbiol 99:71–87. doi: 10.1111/mmi.13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Honda-Ogawa M, Sumitomo T, Mori Y, Hamd DT, Ogawa T, Yamaguchi M, Nakata M, Kawabata S. 2017. Streptococcus pyogenes endopeptidase O contributes to evasion from complement-mediated bacteriolysis via binding to human complement factor C1q. J Biol Chem 292:4244–4254. doi: 10.1074/jbc.M116.749275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loughman JA, Caparon M. 2006. Regulation of SpeB in Streptococcus pyogenes by pH and NaCl: a model for in vivo gene expression. J Bacteriol 188:399–408. doi: 10.1128/JB.188.2.399-408.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bland ND, Pinney JW, Thomas JE, Turner AJ, Isaac RE. 2008. Bioinformatic analysis of the neprilysin (M13) family of peptidases reveals complex evolutionary and functional relationships. BMC Evol Biol 8:16. doi: 10.1186/1471-2148-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Treviño J, Perez N, Ramirez-Peña E, Liu Z, Shelburne SA III, Musser JM, Sumby P. 2009. CovS simultaneously activates and inhibits the CovR-mediated repression of distinct subsets of group A Streptococcus virulence factor-encoding genes. Infect Immun 77:3141–3149. doi: 10.1128/IAI.01560-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aziz RK, Kansal R, Aronow BJ, Taylor WL, Rowe SL, Kubal M, Chhatwal GS, Walker MJ, Kotb M. 2010. Microevolution of group A streptococci in vivo: capturing regulatory networks engaged in sociomicrobiology, niche adaptation, and hypervirulence. PLoS One 5:e9798. doi: 10.1371/journal.pone.0009798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sumby P, Barbian KD, Gardner DJ, Whitney AR, Welty DM, Long RD, Bailey JR, Parnell MJ, Hoe NP, Adams GG, Deleo FR, Musser JM. 2005. Extracellular deoxyribonuclease made by group A Streptococcus assists pathogenesis by enhancing evasion of the innate immune response. Proc Natl Acad Sci U S A 102:1679–1684. doi: 10.1073/pnas.0406641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buchanan JT, Simpson AJ, Aziz RK, Liu GY, Kristian SA, Kotb M, Feramisco J, Nizet V. 2006. DNase expression allows the pathogen group A Streptococcus to escape killing in neutrophil extracellular traps. Curr Biol 16:396–400. doi: 10.1016/j.cub.2005.12.039. [DOI] [PubMed] [Google Scholar]
- 38.Timmer AM, Timmer JC, Pence MA, Hsu LC, Ghochani M, Frey TG, Karin M, Salvesen GS, Nizet V. 2009. Streptolysin O promotes group A Streptococcus immune evasion by accelerated macrophage apoptosis. J Biol Chem 284:862–871. doi: 10.1074/jbc.M804632200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uchiyama S, Dohrmann S, Timmer AM, Dixit N, Ghochani M, Bhandari T, Timmer JC, Sprague K, Bubeck-Wardenburg J, Simon SI, Nizet V. 2015. Streptolysin O rapidly impairs neutrophil oxidative burst and antibacterial responses to group A Streptococcus. Front Immunol 6:581. doi: 10.3389/fimmu.2015.00581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cole JN, McArthur JD, McKay FC, Sanderson-Smith ML, Cork AJ, Ranson M, Rohde M, Itzek A, Sun H, Ginsburg D, Kotb M, Nizet V, Chhatwal GS, Walker MJ. 2006. Trigger for group A streptococcal M1T1 invasive disease. FASEB J 20:1745–1747. doi: 10.1096/fj.06-5804fje. [DOI] [PubMed] [Google Scholar]
- 41.Turner AJ, Isaac RE, Coates D. 2001. The neprilysin (NEP) family of zinc metalloendopeptidases: genomics and function. Bioessays 23:261–269. doi:. [DOI] [PubMed] [Google Scholar]
- 42.Sitnik JL, Francis C, Hens K, Huybrechts R, Wolfner MF, Callaerts P. 2014. Neprilysins: an evolutionarily conserved family of metalloproteases that play important roles in reproduction in Drosophila. Genetics 196:781–797. doi: 10.1534/genetics.113.160945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Froeliger EH, Oetjen J, Bond JP, Fives-Taylor P. 1999. Streptococcus parasanguis pepO encodes an endopeptidase with structure and activity similar to those of enzymes that modulate peptide receptor signaling in eukaryotic cells. Infect Immun 67:5206–5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nguyen T, Zhang Z, Huang IH, Wu C, Merritt J, Shi W, Qi F. 2009. Genes involved in the repression of mutacin I production in Streptococcus mutans. Microbiology 155:551–556. doi: 10.1099/mic.0.021303-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chavagnat F, Meyer J, Casey MG. 2000. Purification, characterisation, cloning and sequencing of the gene encoding oligopeptidase PepO from Streptococcus thermophilus A. FEMS Microbiol Lett 191:79–85. doi: 10.1111/j.1574-6968.2000.tb09322.x. [DOI] [PubMed] [Google Scholar]
- 46.Oetjen J, Fives-Taylor P, Froeliger E. 2001. Characterization of a streptococcal endopeptidase with homology to human endothelin-converting enzyme. Infect Immun 69:58–64. doi: 10.1128/IAI.69.1.58-64.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Agarwal V, Kuchipudi A, Fulde M, Riesbeck K, Bergmann S, Blom AM. 2013. Streptococcus pneumoniae endopeptidase O (PepO) is a multifunctional plasminogen- and fibronectin-binding protein, facilitating evasion of innate immunity and invasion of host cells. J Biol Chem 288:6849–6863. doi: 10.1074/jbc.M112.405530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Agarwal V, Sroka M, Fulde M, Bergmann S, Riesbeck K, Blom AM. 2014. Binding of Streptococcus pneumoniae endopeptidase O (PepO) to complement component C1q modulates the complement attack and promotes host cell adherence. J Biol Chem 289:15833–15844. doi: 10.1074/jbc.M113.530212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang H, Kang L, Yao H, He Y, Wang X, Xu W, Song Z, Yin Y, Zhang X. 2016. Streptococcus pneumoniae endopeptidase O (PepO) elicits a strong innate immune response in mice via TLR2 and TLR4 signaling pathways. Front Cell Infect Microbiol 6:23. doi: 10.3389/fcimb.2016.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yao H, Zhang H, Lan K, Wang H, Su Y, Li D, Song Z, Cui F, Yin Y, Zhang X. 2017. Purified Streptococcus pneumoniae endopeptidase O (PepO) enhances particle uptake by macrophages in a Toll-like receptor 2- and miR-155-dependent manner. Infect Immun 85:e01012-16. doi: 10.1128/IAI.01012-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zou J, Zhou L, Hu C, Jing P, Guo X, Liu S, Lei Y, Yang S, Deng J, Zhang H. 2017. IL-8 and IP-10 expression from human bronchial epithelial cells BEAS-2B are promoted by Streptococcus pneumoniae endopeptidase O (PepO). BMC Microbiol 17:187. doi: 10.1186/s12866-017-1081-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li J, Xia J, Tan C, Zhou Y, Wang Y, Zheng C, Chen H, Bei W. 2011. Evaluation of the immunogenicity and the protective efficacy of a novel identified immunogenic protein, SsPepO, of Streptococcus suis serotype 2. Vaccine 29:6514–6519. doi: 10.1016/j.vaccine.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 53.Henderson B, Martin A. 2011. Bacterial virulence in the moonlight: multitasking bacterial moonlighting proteins are virulence determinants in infectious disease. Infect Immun 79:3476–3491. doi: 10.1128/IAI.00179-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kansal RG, McGeer A, Low DE, Norrby-Teglund A, Kotb M. 2000. Inverse relation between disease severity and expression of the streptococcal cysteine protease, SpeB, among clonal M1T1 isolates recovered from invasive group A streptococcal infection cases. Infect Immun 68:6362–6369. doi: 10.1128/IAI.68.11.6362-6369.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Svensson MD, Scaramuzzino DA, Sjobring U, Olsen A, Frank C, Bessen DE. 2000. Role for a secreted cysteine proteinase in the establishment of host tissue tropism by group A streptococci. Mol Microbiol 38:242–253. doi: 10.1046/j.1365-2958.2000.02144.x. [DOI] [PubMed] [Google Scholar]
- 56.Alves LA, Harth-Chu EN, Palma TH, Stipp RN, Mariano FS, Hofling JF, Abranches J, Mattos-Graner RO. 2017. The two-component system VicRK regulates functions associated with Streptococcus mutans resistance to complement immunity. Mol Oral Microbiol 32:419–431. doi: 10.1111/omi.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dmitriev AV, McDowell EJ, Chaussee MS. 2008. Inter- and intraserotypic variation in the Streptococcus pyogenes Rgg regulon. FEMS Microbiol Lett 284:43–51. doi: 10.1111/j.1574-6968.2008.01171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anbalagan S, McShan WM, Dunman PM, Chaussee MS. 2011. Identification of Rgg binding sites in the Streptococcus pyogenes chromosome. J Bacteriol 193:4933–4942. doi: 10.1128/JB.00429-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Friães A, Pato C, Melo-Cristino J, Ramirez M. 2015. Consequences of the variability of the CovRS and RopB regulators among Streptococcus pyogenes causing human infections. Sci Rep 5:12057–12057. doi: 10.1038/srep12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shelburne SA, Olsen RJ, Makthal N, Brown NG, Sahasrabhojane P, Watkins EM, Palzkill T, Musser JM, Kumaraswami M. 2011. An amino-terminal signal peptide of Vfr protein negatively influences RopB-dependent SpeB expression and attenuates virulence in Streptococcus pyogenes. Mol Microbiol 82:1481–1495. doi: 10.1111/j.1365-2958.2011.07902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kreth J, Merritt J, Bordador C, Shi W, Qi F. 2004. Transcriptional analysis of mutacin I (mutA) gene expression in planktonic and biofilm cells of Streptococcus mutans using fluorescent protein and glucuronidase reporters. Oral Microbiol Immunol 19:252–256. doi: 10.1111/j.1399-302X.2004.00148.x. [DOI] [PubMed] [Google Scholar]
- 62.Merritt J, Qi F. 2012. The mutacins of Streptococcus mutans: regulation and ecology. Mol Oral Microbiol 27:57–69. doi: 10.1111/j.2041-1014.2011.00634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Husmann LK, Scott JR, Lindahl G, Stenberg L. 1995. Expression of the Arp protein, a member of the M protein family, is not sufficient to inhibit phagocytosis of Streptococcus pyogenes. Infect Immun 63:345–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chaffin DO, Beres SB, Yim HH, Rubens CE. 2000. The serotype of type Ia and III group B streptococci is determined by the polymerase gene within the polycistronic capsule operon. J Bacteriol 182:4466–4477. doi: 10.1128/JB.182.16.4466-4477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sanderson-Smith ML, Dinkla K, Cole JN, Cork AJ, Maamary PG, McArthur JD, Chhatwal GS, Walker MJ. 2008. M protein-mediated plasminogen binding is essential for the virulence of an invasive Streptococcus pyogenes isolate. FASEB J 22:2715–2722. doi: 10.1096/fj.07-105643. [DOI] [PubMed] [Google Scholar]
- 66.Dalton TL, Collins JT, Barnett TC, Scott JR. 2006. RscA, a member of the MDR1 family of transporters, is repressed by CovR and required for growth of Streptococcus pyogenes under heat stress. J Bacteriol 188:77–85. doi: 10.1128/JB.188.1.77-85.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.National Health and Medical Research Council. 2013. Australian code for the care and use of animals for scientific purposes, 8th ed National Health and Medical Research Council, Canberra, Australia: https://www.nhmrc.gov.au/guidelines-publications/ea28. [Google Scholar]
- 68.National Health and Medical Research Council. 2015. National statement on ethical conduct in human research. National Health and Medical Research Council, Canberra, Australia: https://www.nhmrc.gov.au/guidelines-publications/e72. [Google Scholar]
- 69.Wertman KF, Wyman AR, Botstein D. 1986. Host/vector interactions which affect the viability of recombinant phage lambda clones. Gene 49:253–262. doi: 10.1016/0378-1119(86)90286-6. [DOI] [PubMed] [Google Scholar]